PRC2 is a known chromatin regulator. Here the authors show novel cytosolic components of PRC2 and that PRC2 inactivation results in attenuation of MAPK/Erk signaling and impaired T cell activation. Systemic PRC2 inhibition in vivo cures the autoimmune syndrome caused by regulatory T cell depletion.
Abstract
Differentiation and activation of T cells require the activity of numerous histone lysine methyltransferases (HMT) that control the transcriptional T cell output. One of the most potent regulators of T cell differentiation is the HMT Ezh2. Ezh2 is a key enzymatic component of polycomb repressive complex 2 (PRC2), which silences gene expression by histone H3 di/tri-methylation at lysine 27. Surprisingly, in many cell types, including T cells, Ezh2 is localized in both the nucleus and the cytosol. Here we show the presence of a nuclear-like PRC2 complex in T cell cytosol and demonstrate a role of cytosolic PRC2 in T cell antigen receptor (TCR)–mediated signaling. We show that short-term suppression of PRC2 precludes TCR-driven T cell activation in vitro. We also demonstrate that pharmacological inhibition of PRC2 in vivo greatly attenuates the severe T cell–driven autoimmunity caused by regulatory T cell depletion. Our data reveal cytoplasmic PRC2 is one of the most potent regulators of T cell activation and point toward the therapeutic potential of PRC2 inhibitors for the treatment of T cell–driven autoimmune diseases.
Introduction
Polycomb repressive complex 2 (PRC2) is a multiprotein complex that is best known for its contribution to transcriptional gene silencing (Margueron and Reinberg, 2011). This function of PRC2 is mediated by the lysine methyltransferases Ezh1 or Ezh2, which catalyze the di/tri-methylation of lysine 27 of histone H3 (H3K27me3; Cao and Zhang, 2004; Margueron and Reinberg, 2011). In T cells, the relative contribution of Ezh1 and Ezh2 to PRC2 function differs between resting and dividing cells. Ezh1 expression levels are very similar in resting and dividing T cells, whereas Ezh2 expression significantly increases after mitotic stimulation (Fig. 1, G and H). The gene regulatory function of PRC2 has been implicated in many aspects of T cell development, differentiation, and activation (Dobenecker et al., 2015; Yang et al., 2015). However, the interpretation of these findings is rather controversial because of the multiplicity of the histone H3–independent Ezh2 protein substrates (He et al., 2012; Lee et al., 2012; Kim et al., 2013b; Gunawan et al., 2015). One of the least understood aspects of the histone H3–independent PRC2 functions concerns Ezh2’s role in signaling (Su et al., 2005; Su and Tarakhovsky, 2006). Our earlier studies showed the presence of Ezh2 in the T cell cytosol, where it contributes to TCR-driven actin polymerization (Su et al., 2005). The signaling capacity of Ezh2 was further underscored by the identification of the membrane associated protein talin-1, which plays an important role in adhesion, as a cytosolic Ezh2 substrate in dendritic cells (Gunawan et al., 2015). Here we describe the composition of the cytoplasmic PRC2 (cPRC2) complex in T cells. We show that although the cytoplasmic and nuclear PRC2 share common subunits, cPRC2 is uniquely associated with key signaling proteins that control TCR signaling and T cell activation. Using short-term pharmacological PRC2 suppression, we show that cPRC2 is required for TCR-mediated activation of MAPK/Erk and expression of IL2 and IL2RA, which support T cell proliferation. We also show that pharmacological suppression of PRC2 in vivo leads to immunosuppression, characterized by greatly diminished T cell responses. We demonstrate that pharmacological PRC2 inhibition could be used for the treatment of severe autoimmune inflammation caused by excessive T cell activation.
Figure 1.
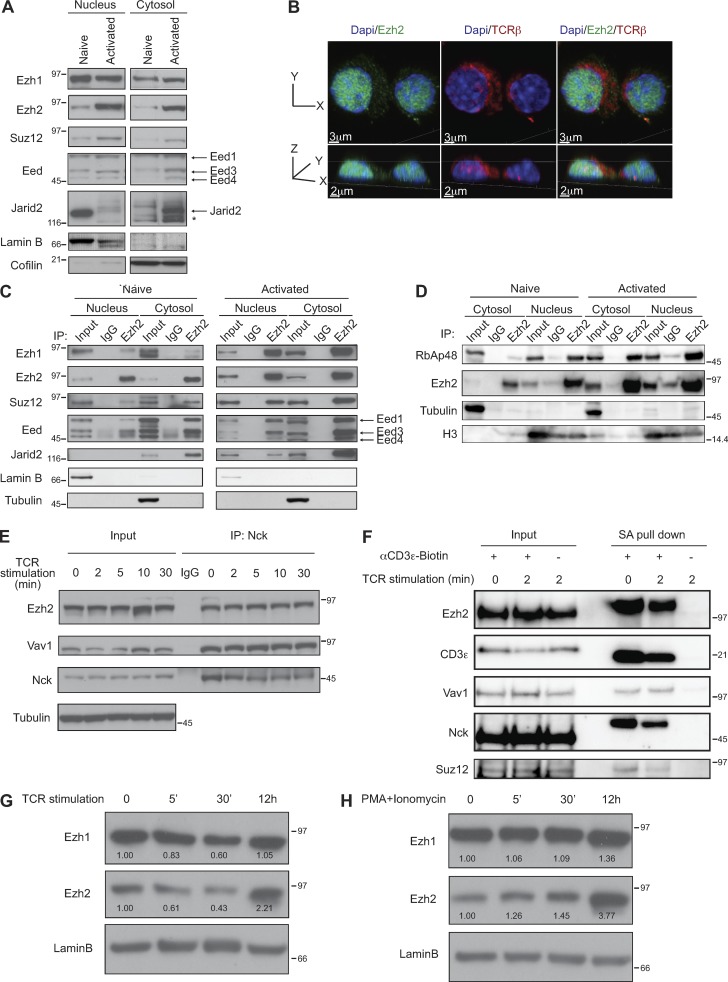
Composition of the cytoplasmic PRC2 complex. (A) Expression levels of the individual PRC2 components in T cell nuclei and cytosol in naive and TCR-activated splenic T cells were measured by Western blotting. Lamin B or cofilin were used as loading controls for the nuclear and cytoplasmic extracts, respectively. The asterisk indicates an unspecific band. Results from one of more than three independent experiments are shown. (B) Ezh2 is present in the cytosol of activated T cells. Cells were stained with fluorescently labeled antibodies against Ezh2 (green) and TCRβ (red), and chromatin was stained with DAPI (blue). Experiments were performed twice. (C and D) Ezh2 binds to the core PRC2 components in T cell cytosol. Ezh2 was immunoprecipitated from nuclear or cytoplasmic extracts derived from naive or activated T cells. Western blotting of the immunoprecipitates revealed the indicated Ezh2-associated proteins. Immunoprecipitation with IgG was used as control. Lamin B and tubulin or histone 3 (H3) were used as loading controls for the nuclear and cytoplasmic extracts, respectively. Results from one of more than three independent experiments are shown. (E) Nck1 is associated with Ezh2 and Vav1 in naive and activated CD4+ T cells. The cytosolic lysates were immunoprecipitated using an Nck-specific antibody followed by Western blot analysis of Nck, Vav1, and Ezh2. Tubulin was used as a loading control for the input. Results from one of more than three independent experiments are shown. (F) Ezh2 and Suz12 as well as Vav1 and Nck1 bind to CD3ε before and after TCR stimulation. CD3ε was immunoprecipitated with SA beads before and after TCR cross-linking for 2 min. Western blotting of the immunoprecipitates revealed the indicated associated proteins. Results from one of more than three independent experiments are shown. (G and H) Expression levels of Ezh1 and Ezh2 in whole cell lysate from naive and TCR- (G) or PMA/ionomycin– (H) activated CD4+T cells were measured by Western blotting. Ezh1 and Ezh2 levels were normalized to lamin B, and numbers indicate the relative abundance in comparison to naive cells. Results from one of two independent experiments are shown. Molecular mass is indicated in kilodaltons.
Results and discussion
We found that Ezh1 and Ezh2 as well as other known components of the PRC2 complex such as Eed, Suz12, RbAp48, and Jarid2 are present in the cytosol of peripheral mouse T cells (Fig. 1, A–D). Immunofluorescence analysis showed that Ezh2 could be detected both in the nucleus and in the cytoplasm of activated T cells, where Ezh2 appears to be proximal to TCRβ (Fig. 1 B). Immunoprecipitation of Ezh2 from the cytoplasmic fraction of naive or activated T cells shows that the PRC2 components are assembled into a complex (cPRC2; Fig. 1, C and D). Previously we showed association of cytoplasmic Ezh2 with the protein Vav1 (Su et al., 2005). Vav1, which has GTP/GDP exchange activity, also functions as an adapter for numerous proteins (Katzav, 2007; Bustelo, 2014), and a recent study highlighted the functional significance of this Ezh2-Vav1 interaction (Gunawan et al., 2015). In our current studies we observed association of Ezh2 with both Vav1 and the Vav1-associated protein Nck (Fig. 1 E), which play major roles in multiple protein–protein interactions and facilitate assembly of signaling complexes downstream of the TCR (Davis, 2002; Barda-Saad et al., 2005). In support of the possible direct role of cPRC2 in TCR signaling, we found that cPRC2 also binds to CD3ε (Fig. 1 F). The interaction between CD3ε and Ezh2 was independent of TCR stimulation in ex vivo isolated peripheral T cells (Fig. 1 F) and could also be detected in thymocytes (not depicted). Of note, treatment with the PRC2 inhibitor UNC1999 had no impact on the CD3ε-Ezh2 association, showing that CD3e binding to cPRC2 does not require its methyltransferase activity (not depicted).
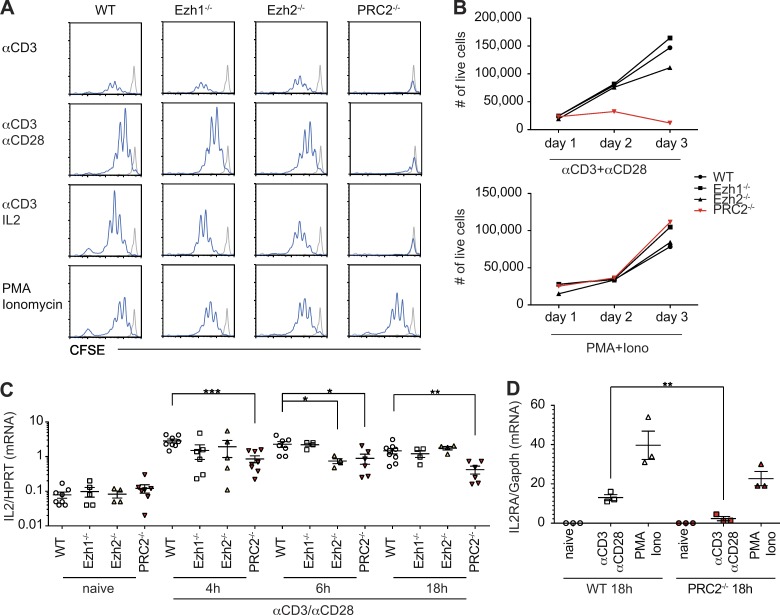
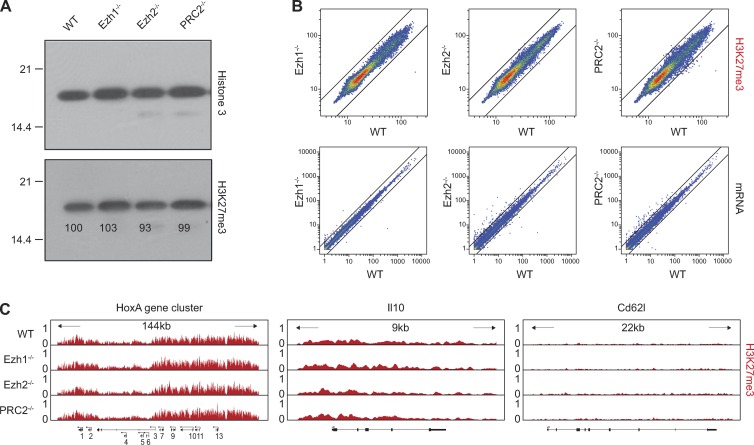
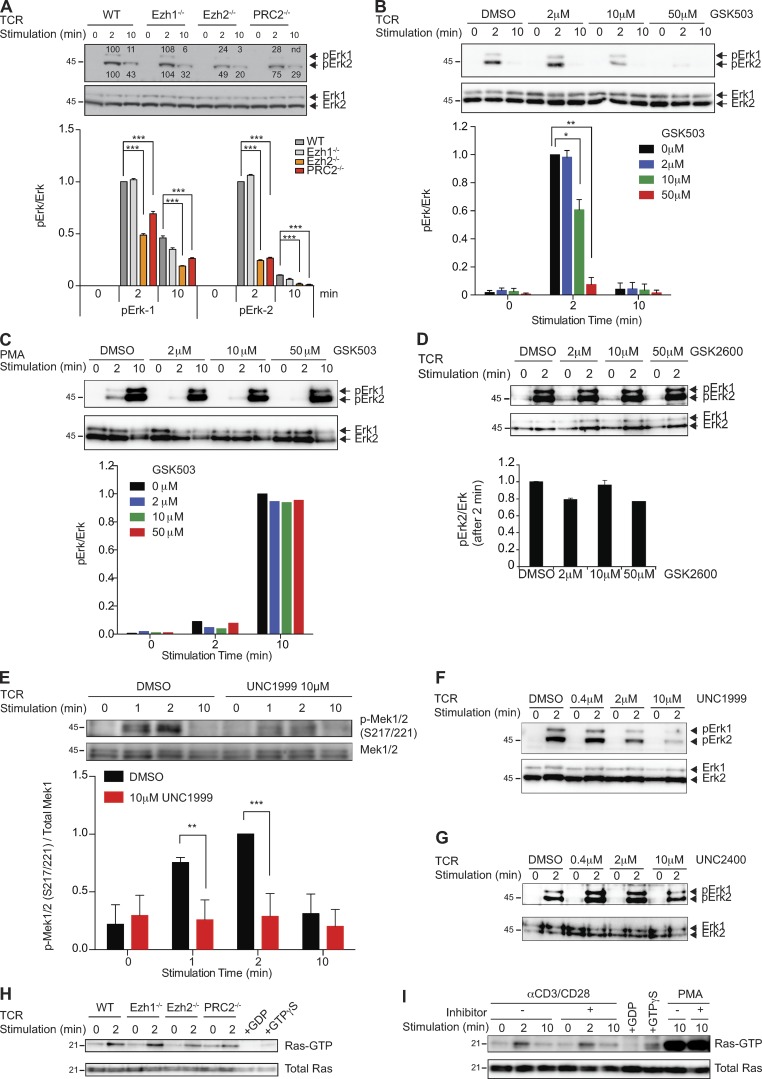
The association of PRC2 with the TCR points toward a role for PRC2 in TCR-mediated signaling. To test this hypothesis, we used genetic models that ablate Ezh1, Ezh2, or both enzymes in T cells. Ezh1-deficient mice are viable and healthy with a WT-like immune system, including T cells (Fig. S1). To generate PRC2-deficient T cells, we combined germline Ezh1 deficiency with a conditional inactivation of Ezh2 in CD4+CD8+ double-positive thymocytes that give rise to CD4+ or CD8+ peripheral T cells. Overall, peripheral PRC2-deficient T cells that lack both Ezh1 and Ezh2 (hereafter defined as PRC2-deficient) show no changes in the expression of characteristic T cell surface–expressed differentiation or activation markers and are present in secondary lymphoid organs in slightly reduced numbers compared with control mice (Fig. S1). Despite the overall WT-like phenotype, PRC2-deficient T cells fail to proliferate in response to TCR triggering in vitro (Fig. 2, A and B). This proliferative defect was associated with greatly reduced TCR-induced transcription of IL2 and IL2RA (Fig. 2, C and D). Additionally, treatment with exogenous IL2 could not rescue the proliferation defect of PRC2-deficient T cells (Fig. 2 A), suggesting that the reduced IL2RA expression may at least partially explain this defect. The failure of PRC2-deficient T cells to express IL2 and IL2RA is unlikely to reflect an altered transcriptional state of the mutant cells, as PRC2 deficiency impacts neither gene expression nor levels or pattern of H3K27me3 at PRC2 target genes in naive T cells (Fig. 3). However, deficiency in PRC2 is associated with diminished TCR-mediated activation of Erk, which plays a key role in IL2 and IL2RA activation (Whitehurst and Geppert, 1996; Fig. 4 A). The effect of PRC2 deficiency on Erk activation is rather selective as PRC2 deficiency has no obvious impact on TCR-induced calcium mobilization or the phosphorylation of numerous key components of the TCR signaling pathway, including PLCγ1, Zap70, and Vav1 (Fig. S2, A and B). We also found no evidence for PRC2 involvement in NF-κB or p38 activation (not depicted).
Figure 2.
PRC2 is essential for TCR-mediated T cell proliferation and transcription of IL2 and IL2RA in vitro. (A) The histograms show the pattern of CFSE dilution between WT or mutant splenic CD4+ T cells triggered by indicated stimuli for 72 h. The histograms marked in gray and blue show amount of CFSE in nonstimulated cells or stimulated cells, respectively. (B) The amounts of viable T cells at different time points after stimulation with anti-CD3/anti-CD28 or PMA/ionomycin (Iono) are shown. (C and D) The expression levels of IL-2 (C) and IL-2RA (D) in TCR-triggered WT and PRC2-deficient splenic T cells were measured by qPCR at different time points after T cell stimulation. All data are representative of three independent experiments (n = 3–6) and significance was determined by the unpaired Student’s t test: *, P ≤ 0.05; **, P ≤ 0.01; and ***, P ≤ 0.001. Error bars show mean ± SEM.
Figure 3.
PRC2 deficiency in naive T cells does not affect H3K27me3 level and gene expression. (A) The levels of H3K7me3 in nuclear extracts of WT control and indicated mutant splenic CD4+ T cells were measured by Western blot analysis. Total histone H3 was used as a loading control. Results from one of more than three independent experiments are shown. Molecular mass is indicated in kilodaltons. (B) Top: Scatter plots show identical pattern of the genome-wide H3K27me3 association with individual genes in control and indicated mutant T cells. The pattern and levels of H3K27me3 were determined by ChIP sequencing analysis of the H3K27me3 associated chromatin. Bottom: Scatter plots show WT-like individual mRNA expression in mutant cells. (C) Representative examples of the H3K27me3 distribution at individual gene loci. The HoxA cluster represents known targets of PRC2, whereas Il10 and Cd62L correspond to silent or actively transcribed genes in naive T cells.
Figure 4.
PRC2 regulates TCR-mediated Mek-Erk1/2 phosphorylation. (A) Phosphorylation of Erk1/2 (pErk1/2) in naive and TCR-triggered WT and indicated mutant CD4+ T cells were measured and normalized to the total amount of Erk1/2. Cells were stimulated by a combination of anti-CD3 and anti-CD28 antibodies for the indicated time periods. The bar graph shows the quantification of these measurements. Results from one of three independent experiments are shown. (B and C) Phosphorylation of Erk1/2 (pErk1/2) in naive and TCR-triggered WT CD4+ T cells in the absence (DMSO) or presence of the indicated amounts of the Ezh2 inhibitor GSK503 was measured and normalized to the total amount of Erk1/2. Cells were stimulated by a combination of anti-CD3 and anti-CD28 antibodies (B) or PMA (C) for the indicated time periods. The bar graphs show the quantification of these measurements. Results from one of three independent experiments are shown. (D) Phosphorylation of Erk1/2 (pErk1/2) in naive and TCR-triggered WT CD4+ T cells in the absence (DMSO) or presence of the indicated amounts of the control compound GSK2792600 (GSK2600) was measured and normalized to the total amount of Erk1/2. Cells were stimulated by a combination of anti-CD3 and anti-CD28 antibodies for the indicated time periods. The bar graph shows the quantification of these measurements. Results from one of three independent experiments are shown. (E) WT CD4+ T cells were stimulated with anti-CD3 and anti-CD28 antibodies in the absence (DMSO) or presence of 10 µM UNC1999. The levels of pMek1/2 (S217/221) were measured and normalized to the total amount of Mek1. The bar graph shows the quantification of these measurements. Results from one of three independent experiments are shown. (F) Phosphorylation of Erk1/2 (pErk1/2) in naive and TCR-triggered WT CD4+ T cells in the absence (DMSO) or presence of the indicated amounts of the Ezh2 inhibitor UNC1999 was measured and normalized to the total amount of Erk1/2. Cells were stimulated by a combination of anti-CD3 and anti-CD28 antibodies for the indicated time periods. Results from one of three independent experiments are shown. (G) Phosphorylation of Erk1/2 (pErk1/2) in naive and TCR-triggered WT CD4+ T cells in the absence (DMSO) or presence of the indicated amounts of control compound UNC2400 was measured and normalized to the total amount of Erk1/2. Cells were stimulated by a combination of anti-CD3 and anti-CD28 antibodies for the indicated time periods. The experiments were performed independently three times, and significance, when calculated, was determined by the unpaired Student’s t test: *, P ≤ 0.05; **, P ≤ 0.01; and ***, P ≤ 0.001. (H) GTP-bound Ras in naive and TCR-stimulated WT and indicated mutant CD4+ T cells was detected by Raf1-RBD pulldown assay. (I) WT CD4+ T cells were treated with 10 µM UNC1999 or DMSO and stimulated with anti-CD3 and anti-CD28 antibodies. Ras-GTP pulldown assay was performed. Data from one representative of two independent experiments are shown for H and I. Molecular mass is indicated in kilodaltons. Error bars show mean ± SEM.
The signaling function of PRC2 was underscored by our experiments that showed very potent suppression of the TCR-mediated Erk activation by pharmacological PRC2 inhibition. We used two Ezh2 small molecule inhibitors, GSK503 (Béguelin et al., 2013) and UNC1999 (Konze et al., 2013). GSK503 suppresses both human EZH1 and EZH2 activity, although at different IC50s (633 nM and 8 nM, respectively; Béguelin et al., 2013). UNC1999 suppresses the activity of Ezh1 and Ezh2 at very similar concentrations (2 nM and 45 nM, respectively). We found that treatment of T cells with GSK503 at a concentration that has the potential to inhibit both Ezh1 and Ezh2, as judged by the intracellular accumulation of the inhibitor (Fig. S2, C and D), leads to a drastic reduction in TCR-mediated Erk phosphorylation (Fig. 4 B). The inhibitory effect of GSK503 on Erk activation in T cells could be observed after 1 h preincubation with the compound on ice. The selective effect of GSK503 on TCR-mediated Erk phosphorylation is exemplified by the overall unaltered pattern of TCR-mediated protein tyrosine phosphorylation in general and of PLCγ1 in particular in GSK503-treated naive and TCR-triggered T cells even at very high concentrations (Fig. S2, E and F). Importantly, the PRC2 inhibitor has no impact on Erk phosphorylation induced by PMA (Fig. 4 C), a phorbol ester derivative analogous to diacylglycerol that directly activates RasGRP and PKCs (Coudronniere et al., 2000; Dower et al., 2000). The specificity of PRC2-dependent Erk phosphorylation was further supported by the lack of an Erk phosphorylation defect in T cells treated with the inactive control compound GSK2792600 (Fig. 4 D). Finally, the Ezh2 and Ezh1 inhibitor UNC1999 (Konze et al., 2013) was also able to nearly abolish TCR-mediated Erk phosphorylation in TCR-triggered CD4+ T cells, whereas its control compound UNC2400 did not decrease the level of TCR-induced Erk phosphorylation (Fig. 4, F and G). In addition to TCR signaling in peripheral T cells, the dependence of TCR-mediated Erk phosphorylation on PRC2 methyltransferase activity was also observed in thymocytes (not depicted). The timing of the inhibitory effect of these pharmacological agents argues strongly in favor of a direct PRC2 involvement in Erk activation. Decreased TCR-mediated activation of Erk by PRC2 inhibitors suggests that cytoplasmic PRC2 could regulate signaling processes upstream of Erk. Indeed, we found that PRC2 inhibition causes reduced phosphorylation of Mek1/2 at S217/221 upon TCR stimulation (Fig. 4 E). Importantly, the PRC2 inhibitors affect neither the kinetics of Erk phosphorylation (Fig. S3 A) nor the phosphorylation of Mek1 at S298, which is mediated by PAK1 in response to TCR triggering (Rouquette-Jazdanian et al., 2012; Fig. S3 B). Mek1/2 phosphorylation at S217/221 is mediated by Raf kinase, which is controlled by Ras. Ras activity, as measured by Ras-GTP loading, is not affected by PRC2 inactivation or inhibition (Fig. 4, H and I). We also found no evidence for direct modification of Raf by cPRC2. Incubation of B-Raf or of C-Raf, purified from HeLa cells, with a minimal PRC2 complex did not result in Raf methylation in vitro (not depicted). Collectively, these data point to a novel mechanism of TCR-mediated Ras-Raf-Erk1/2 regulation where PRC2 controls the function of an unknown component downstream of Ras and upstream of Erk.
In agreement with the reduced Erk activation and impaired IL2/IL2RA expression, treatment of ex vivo isolated peripheral T cells with GSK503 greatly reduces TCR-driven T cell proliferation, similar to TCR-triggered PRC2-deficient T cells (Fig. S3 C). It should be noted that neither PRC2 deficiency nor inhibition of PRC2 affect T cell proliferation induced by TCR-independent T cell activation with PMA in combination with ionomycin, underscoring the selective role of PRC2 in TCR-driven T cell activation.
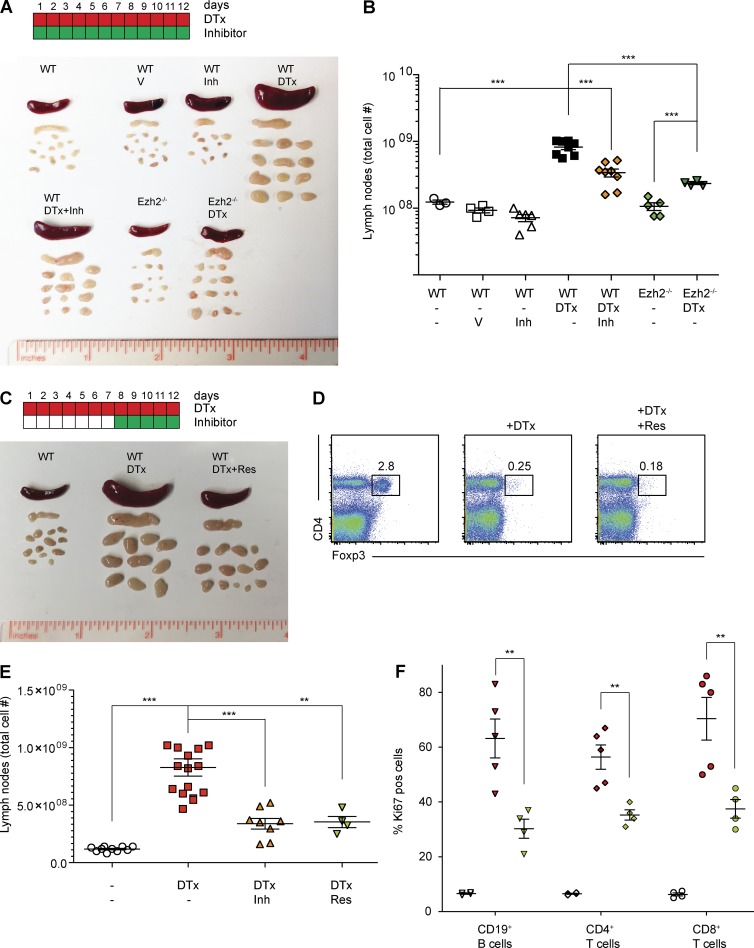
The failure of PRC2 inhibitor–treated T cells to respond to TCR stimulation suggests the possibility of pharmacological T cell immunosuppression by PRC2 inhibition in vivo. To address the immune-suppressive potential of PRC2 inhibition, we chose an experimental model that incorporates many of the clinically relevant aspects of aggressive T cell activation. It is well established that activation of T cells in vivo is governed by both cell intrinsic and extrinsic mechanisms, including immune suppressor regulatory T (T reg) cells (Josefowicz et al., 2012). Deficiency in T reg cells causes severe fatal T cell–driven autoimmunity in humans and mice (Bennett et al., 2001; Brunkow et al., 2001). In our experiments, we used inducible T reg deficiency in mice where Foxp3+ T reg cells, expressing a diphtheria toxin receptor transgene under the control of the Foxp3 locus (Foxp3DTR), could be rapidly and selectively eliminated by diphtheria toxin (DTx) administration in vivo (Kim et al., 2007). As reported, injection of 10 mg/kg DTx in Foxp3DTR mice results in rapid decline in T reg cell numbers followed by severe autoimmunity. The hallmarks of autoimmunity in the absence of T reg cells include splenomegaly, lymphadenopathy, and accumulation of activated lymphoid and myeloid cells in peripheral lymphoid organs within days (Fig. 5, A and B). Injection of the PRC2 inhibitor GSK503 at the dose of 150 mg/kg could not only prevent (Fig. 5, A and B) but, most important, cure (Fig. 5 C) the ongoing autoimmunity caused by T reg depletion. In each group, T reg cells were comparably and successfully depleted upon DTx treatment (Fig. 5 D). In the most illustrative case, administration of GSK503, to mice that already displayed signs of systemic autoimmunity including splenomegaly and lymphadenopathy, was able to reverse the disease (Fig. 5 C). The total numbers of cells in secondary lymphoid organs and inflammatory immune cells such as T cells, B cells, granulocytes, and macrophages were significantly decreased upon GSK503 treatment (Table 1), indicating that GSK503 treatment ameliorated T reg depletion–induced inflammation. Importantly, GSK503 had no effect on nonactivated T cells, but greatly reduced the frequency of dividing Ki67-positive cells in the peripheral lymphoid organs of GSK503-treated and T reg–depleted mice (Fig. 5 F). The negative effect of pharmacological inhibition of PRC2 on T cell activation in vivo most likely reflects the signaling function of cPRC2, as failure to proliferate in the absence of functional PRC2 would not give PRC2-deficient T cells the chance to undergo division-associated epigenetic changes that could be linked to the nuclear function of PRC2. On the other hand, the observed effects of PRC2 inhibition could be attributed to cell types other than T cells. Although this scenario is possible, we argue that the immune suppressive effect of PRC2 inhibition largely depends on suppression of T cell–mediated immunity. We found that selective genetic ablation of PRC2 in T cells alone precludes T cell activation caused by T reg depletion and the resulting autoimmune syndrome (Fig. 5, A and B). Therefore, it is likely that most of the therapeutic effects of the PRC2 inhibitor are related to the inhibitory effect on T cell activation and not other cell types. Our data also suggest that exclusively cytoplasmic inhibitors of PRC2 will allow for efficient and possibly nontoxic immunosuppression during autoimmunity caused by excessive T cell activation, including those that occur during cancer immunotherapy by checkpoint blockade inhibitors.
Figure 5.
Treatment of T cell autoimmunity by pharmacological Ezh2 inhibition reverses autoimmunity caused by T reg depletion. (A) The color scheme shows timing for the DTx-mediated T reg depletion (red) and treatment with the Ezh2 inhibitor (green). Each box represents a day of treatment. The photograph shows sizes of spleen and lymph nodes of Foxp3-DTR mice (wt), Foxp3-DTR mice treated with DTx (WT, DTx) or Foxp3-DTR mice that started to receive GSK503 on day 8 after the initiation of DTx treatment (WT, DTx+Res). (B) The frequency of Foxp3+ regulatory T cells in spleen of Foxp3-DTR mice (left), Foxp3-DTR mice treated with DTx (middle, +DTx) and Foxp3-DTR mice that started to receive GSK503 on day 8 after DTx treatment (right, +DTx +Res). (C) The total number of lymphoid cells in lymph nodes of Foxp3-DTR mice that receive indicated treatments are shown (untreated, white symbols; DTx injection, red symbols; DTx + GSK503 [Inh] treatment, orange symbols; and DTx + GSK503 [Res] treatment, green symbols. “Inh” indicates that the mice received GSK503 simultaneously with the DTx treatment whereas “Res” indicates that the mice started to receive GSK503 on day 8 after the initiation of DTx treatment). (D) The percentages of Ki67+ dividing CD19+ B and CD4+ or CD8+ T cells in lymph nodes of mice that receive indicated treatments are shown (untreated, white symbols; DTx injection, red symbols; DTx and GSK503 [Res] treatment, green symbols). Each symbol represents one mouse. Results from one of two representative experiments are shown (n = 4–15). Prevention of T cell autoimmunity by PRC2 inhibition or genetic deletion of Ezh2. (E) The color scheme shows timing for the DTx-mediated T reg depletion (red) and treatment with the Ezh2 inhibitor (green). Each box represents a day of treatment. The photograph shows sizes of spleen and lymph nodes derived from FoxP3-DTR mice (WT), FoxP3-DTR mice treated with vehicle (WT, V), FoxP3-DTR mice treated with PRC2 inhibitor GSK503 (WT, Inh), FoxP3-DTR mice treated with DTx (WT, DTx), FoxP3-DTR mice treated with DTx and with PRC2 inhibitor GSK503 (WT, DTx+Inh), Ezh2fl/fl; CD4-cre; FoxP3-DTR mice (Ezh2−/−), and Ezh2fl/fl; CD4-cre; FoxP3-DTR mice treated with DTx (Ezh2−/−, DTx). The total cell number in lymph nodes of mice that receive vehicle (V), DTx, or inhibitor (Inh) treatment are indicated. Each symbol represents a single mouse. All data are representative of two independent experiments (n = 3–8). (F) The percentage of Ki67+ dividing CD19+ B and CD4+ or CD8+ T cells in lymph nodes of mice that received indicated treatments is shown (white symbols, untreated; red symbols, DTx injection; green symbols, DTx and GSK503 (Res) treatment). Each symbol represents one mouse. Results from one of two representative experiments are shown, and significance was determined by unpaired Student’s t test: **, P ≤ 0.01; ***, P ≤ 0.001. Error bars show mean ± SEM.
.
Table 1. Analysis of cell subsets in response to T reg elimination and PRC2 inhibition.
| Parameter | FoxP3DTR | FoxP3DTR | Fold change | FoxP3DTR | Fold change | FoxP3DTR | Fold change |
|---|---|---|---|---|---|---|---|
| +DTx | +DTx | +DTx | |||||
| +GSK503 | +Rescue | ||||||
| Lymph nodes | |||||||
| Total cell no. | 1.18E+08 | 8.28E+08 | 7.01 | 3.40E+08 | 0.41 | 3.54E+08 | 0.43 |
| ±SEM | 6.51E+06 | 7.53E+07 | *** | 4.62E+07 | *** | 4.80E+07 | ** |
| No. CD4+TCRb+ | 3.90E+07 | 2.50E+08 | 6.40 | 1.04E+08 | 0.42 | 9.90E+07 | 0.4 |
| ±SEM | 3.11E+06 | 2.53E+07 | *** | 1.85E+07 | *** | 1.20E+07 | * |
| No. CD8+TCRb+ | 3.72E+07 | 2.10E+08 | 5.66 | 8.94E+07 | 0.43 | 8.11E+07 | 0.39 |
| ±SEM | 2.49E+06 | 1.80E+07 | *** | 1.03E+07 | *** | 1.25E+07 | ** |
| No. CD19+B220+ | 2.66E+07 | 1.72E+08 | 6.48 | 4.97E+07 | 0.29 | 1.40E+08 | 0.81 |
| ±SEM | 5.30E+06 | 3.92E+07 | ** | 1.23E+07 | * | 2.29E+07 | NS |
| No. GR1hiCD11b+ | 5.92E+05 | 1.77E+07 | 29.9 | 3.84E+06 | 0.22 | 7.65E+06 | 0.43 |
| ±SEM | 2.14E+05 | 3.29E+06 | *** | 1.04E+06 | ** | 1.75E+06 | NS |
| No. F4/80+ CD11c− | 1.10E+06 | 1.11E+07 | 10.11 | 2.53E+06 | 0.23 | 6.32E+06 | 0.57 |
| ±SEM | 3.91E+05 | 1.90E+06 | *** | 9.77E+05 | ** | 2.75E+06 | NS |
| Spleen | |||||||
| Total cell no. | 1.22E+08 | 6.41E+08 | 5.26 | 2.04E+08 | 0.32 | 2.36E+08 | 0.37 |
| ±SEM | 1.34E+07 | 5.82E+07 | *** | 3.48E+07 | *** | 9.25E+07 | ** |
| No. CD4+TCRb+ | 1.90E+07 | 1.40E+08 | 7.36 | 3.62E+07 | 0.26 | 7.00E+07 | 0.5 |
| ±SEM | 2.67E+06 | 1.45E+07 | *** | 8.52E+06 | *** | 2.62E+07 | * |
| No. CD8+TCRb+ | 1.86E+07 | 1.13E+08 | 6.09 | 2.71E+07 | 0.24 | 4.94E+07 | 0.44 |
| ±SEM | 1.61E+06 | 1.45E+07 | *** | 6.61E+06 | *** | 2.23E+07 | NS |
| No. CD19+B220+ | 4.93E+07 | 1.09E+08 | 2.22 | 3.58E+07 | 0.33 | 6.94E+07 | 0.64 |
| ±SEM | 9.86E+06 | 1.65E+07 | * | 1.01E+07 | ** | 2.48E+07 | NS |
| No. GR1hiCD11b+ | 2.99E+06 | 5.95E+07 | 20.56 | 3.44E+07 | 0.58 | 2.48E+07 | 0.42 |
| ±SEM | 1.21E+06 | 1.19E+07 | *** | 5.72E+06 | NS | 1.14E+07 | NS |
| No. F4/80+CD11c− | 2.06E+06 | 5.57E+07 | 27.12 | 3.13E+07 | 0.56 | 2.19E+07 | 0.39 |
| ±SEM | 4.42E+05 | 1.14E+07 | ** | 1.24E+07 | NS | 9.71E+06 | NS |
Significance was determined by unpaired Student’s t test: *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001.
Materials and methods
Mice
Ezh2fl/fl mice were generated in our laboratory and have been described before (Su et al., 2003, 2005). They were crossed with Ezh1−/− mice, which were generated by D. O’Carroll (Thomas Jenuwein Lab, MPI-IE, Freiburg, Germany), to give rise to Ezh1−/−; Ezh2fl/fl; CD4-Cre mice on B6/129 background. Foxp3DTR knock-in mice (Kim et al., 2007), a gift from A. Rudensky (Memorial Sloan-Kettering Cancer Center, New York, NY), were bred with Ezh2fl/fl; CD4-Cre mice to generate Ezh2fl/fl; CD4-Cre; Foxp3DTR mice. To obtain a large number of cells for biochemical assays, C57BL/6 mice were used for harvesting lymph nodes and spleens. Mice were housed under specific pathogen-free conditions, and experimental protocols were approved by the Rockefeller University Institutional Animal Care and Use Committee.
Antibodies
For coimmunoprecipitations, Ezh2 (D2C9; Cell Signaling Technologies), Nck1/2 (C-19; Santa Cruz Biotechnology), and Vav1 (C-14; Santa Cruz Biotechnology) were used. For Western blot analysis, Ezh2 (Clone 11; BD Biosciences), Suz12 (P-15; Santa Cruz Biotechnology), Vav1 (05–219; Millipore), Nck (Clone 108; BD Biosciences), RbAp48 (11G10; Abcam), and Jarid2 (ab48137; Abcam) were used. The Eed and Ezh1 antibodies were provided by D. Reinberg (New York University, New York, NY). For measuring global levels of H3K27me3 and chromatin immunoprecipitation (ChIP)–sequencing experiments, H3K27me3 (07–449; Millipore) and H3K4me3 (17–614; Millipore) were used. For examining TCR-induced phosphorylation, PLCγ1 (05–163; Millipore), phospho-PLCγ1 (Y783, 2821; Cell Signaling Technologies), Vav1 (C-14, sc-132; Santa Cruz Biotechnology), phospho-Vav1 (Y174, sc-16408; Santa Cruz Biotechnology), Zap70 (29, 610239; BD Transduction Laboratories), phospho-Zap70 (Tyr319, 2701; Cell Signaling Technologies), JNK (C-17; Santa Cruz Biotechnology), phoshpo-JNK (T183/Y185, 9251; Cell Signaling Technologies), Akt (C67E7; Cell Signaling Technologies), phospho-Akt (Ser473, D9E; Cell Signaling Technologies), p38 (D13E1; Cell Signaling Technologies), phopho-p38 (D3F9; Cell Signaling Technologies), NF-κB p65 (L8F6; Cell Signaling Technologies), phospho-p65 (93H1; Cell Signaling Technologies), pan-phospho-tyrosine (4G10; Millipore), phospho-Mek1/2 (9121; Cell Signaling Technologies), phospho-Mek1 S298 (9128; Cell Signaling Technologies), Mek1 (61B12; Cell Signaling Technologies), and Mek1/2 (L38C12; Cell Signaling Technologies) were used.
CFSE labeling and stimulation of T cells
CD4+ T cells were isolated from spleen and lymph nodes by MACS purification. Purified CD4+ T cells were washed with PBS two times and labeled with CFSE at the final concentration of 2.5 µM for 10 min at 37°C. Labeling was stopped by complete medium, and cells were washed three times with PBS. Labeled cells were stimulated on 96-well plates coated with 10 µg/ml of anti-CD3 and 2.5 µg/ml anti-CD28 antibodies or with 5 ng/ml of PMA plus 0.3 µg/ml of ionomycin in a humidified 37°C, 5% CO2 incubator. Cells were harvested after overnight and 3 d after stimulation.
Cell fractionation and Western blot
Purified CD4 T cells were lysed with 0.5% NP-40 containing TMSD lysis buffer (40 mM Tris-HCl, pH 7.9, 5 mM MgCl2, 250 mM sucrose, 1 mM DTT, 0.5% NP-40 [IGEPAL], protease cocktails [Sigma-Aldrich], and phosphatase cocktails [Millipore]) on ice for 15 min. The subcellular fractions were prepared by first centrifugation at 800g for 10 min, to remove nuclei. The supernatant was spun down a second time to remove residual cell and nuclear fragments. This supernatant was used as cytosolic fraction. The nuclei were washed twice with TMSD lysis buffer and then incubated with high-salt lysis buffer (50 mM Tris-HCl, pH 7.9, 420 mM NaCl, 20 mM NaF, 1 mM EDTA, 1 mM EGTA, 1.5 mM MgCl2, 10% glycerol, protease cocktails [Sigma-Aldrich], and phosphatase cocktails [Millipore]) for 10 min on ice. The nuclear extracts were sonicated for 10 cycles (30 s per cycle; Bioruptor Diagenode), followed by centrifugation at 13,000 rpm for 15 min, to obtain the final nuclear extract. Protein concentrations were determined by standard BCA assay. The extracts or whole cell lysates were separated by SDS-PAGE and transferred to a polyvinylidene difluoride membrane. The membrane was blocked, incubated with primary antibodies followed by horseradish peroxidase–conjugated secondary antibodies, and developed with ECL systems according to the manufacturer’s instructions.
Immunoprecipitation
Fractionated lysates or lysates obtained by using ODG lysis buffer (2% N-octyl-β-d-glucopyranoside, 50 mM Tris-HCl, pH 7.9, 150 mM NaCl, 20 mM NaF, 1 mM EDTA, 1 mM EGTA, 1.5 mM MgCl2, 10% glycerol, protease cocktails [Sigma-Aldrich], and phosphatase cocktails [Millipore]) were incubated with antibodies cross-linked to protein G dynabeads. The cross-linking of antibodies to beads was achieved by using BS3 cross-linker (Pierce). Immunoprecipitates were washed with lysis buffer and eluted by boiling in sample loading buffer. Coimmunoprecipitates were detected by Western blot analysis.
Erk phosphorylation assay
Purified CD4 T cells were incubated with 10 µg/ml of anti-CD3 antibody and 2.5 µg/ml of anti-CD28 antibody on ice for 20 min, followed by cross-linking with 20 µg/ml of anti–Armenian hamster IgG secondary antibody for 2 min or 10 min at 37°C. For PMA stimulation, cells were incubated with prewarmed media containing 5 ng/ml of PMA for 10 min at 37°C. For stimulation of thymocytes, thymocytes were incubated with 10 µg/ml of anti-CD3 biotinylated antibody and 10 µg/ml of anti-CD4 biotinylated antibody for 20 min on ice, followed by cross-linking with 20 µg/ml of streptavidin. Cells were washed with cold HBSS and then lysed with 0.5% of NP-40 lysis buffer (50 mM Tris-HCl, pH 7.9, 150 mM NaCl, 20 mM NaF, 1 mM EDTA, 1 mM EGTA, 1.5 mM MgCl2, 10% glycerol, 0.5% NP-40, and protease cocktails [Sigma-Aldrich]) containing phosphatase inhibitory cocktail (Millipore) for 15 min on ice. Lysates were obtained by centrifugation at 13,000 rpm for 15 min to remove insolubles. Phosphorylated Erk1/2 and total Erk were detected by Western blot analysis using rabbit anti–phospho-Erk1/2 (T202/Y204) antibody (D13.14.4E; Cell Signaling Technologies) and mouse anti-Erk antibody (L34F12; Cell Signaling Technologies), respectively.
Ras activation assay
Ras activation assay was performed using a Pan-Ras activation kit (Cell Biolabs) according to the manufacturer’s instructions. In brief, the cells were stimulated and lysed with the lysis buffer (25 mM Hepes, pH 7.5, 150 mM NaCl, 1% NP-40, 10 mM MgCl2, 1 mM EDTA, and 2% glycerol) supplemented with the protease/phosphatase inhibitor cocktails previously used. As control samples, cell lysates were incubated with GTPγS and GDP at 30°C for 30 min, followed by addition of MgCl2. Cell lysates were incubated with Raf1-RBD GST–conjugated agarose beads for 1 h at 4°C, followed by washing. Raf1-RBD–bound proteins were eluted by 2× sample loading buffer, and coprecipitated Ras protein was detected by Western blot analysis.
Differential salt wash
Coimmunoprecipitates were incubated with Tris-HCl, pH 7.9, 0.2 mM EDTA, 5% glycerol, 5 mM DTT, and different concentrations of NaCl (150 mM to 600 mM) for 30 min three times. Coimmunoprecipitates were further washed twice with 150 mM NaCl buffer and eluted by boiling in sample loading buffer.
Statistical analysis
Statistical analysis was performed in Prism (Graphpad Software) with the unpaired t test for total cell numbers and frequencies (*, P < 0.05; **, P < 0.01; and ***, P < 0.001).
Standard ChIP assays and ChIP-seq
ChIP was performed as previously described (Lee et al., 2006; Goldberg et al., 2010). In brief, 107 cells were cross-linked with 0.5% formaldehyde at room temperature for 10 min. Chromatin was sonicated to 300–500 bp in RIPA buffer with 0.3 M NaCl. 5–10 mg antibodies were preincubated with Dynabead Protein A/G (Invitrogen) for at least 8 h before incubating with sonicated chromatin overnight. After overnight incubation, beads were washed in modified RIPA wash buffer (100 mM LiCl) and 1× in TE. After overnight cross-link reversal at 65°C, RNase digestion, and proteinase K digestion, ChIP DNA and input DNA were purified using the Quiaquick PCR purification kit. For regular ChIP and for validation of ChIP-Seq, ChIP DNA was analyzed via qPCR using SYBR Green PCR Master Mix and the LightCycler 480. Primer sequences are available upon request.
For ChIP-Seq, 30 µl of ChIP DNA were used to generate blunt-ended DNA using reagents supplied with the Epicenter DNA EndRepair kit (Epicenter Biotechnologies) according to the manufacturer’s instructions. The end-repaired DNA was purified using the Quiaquick PCR purification kit. Using Klenow Fragment (3′ to 5′ exo-, NEB), “A” bases were added to the DNA. The DNA was purified using the MinElute kit. T4 DNA ligase (NEB) was used for ligation of Illumina/Solexa adapters to the DNA fragments. The adaptor-ligated DNA was purified with the MinElute kit. The DNA fragments were subjected to 18 cycles of PCR using the Illumina/Solexa primers 1.0 and 2.0 to generate the ChIP-Seq libraries. The ChIP-Seq libraries were purified with the MinElute kit.
Samples were sequenced on the Illumina Hiseq2000 platform for 50 cycles, and raw sequencing data were processed using the CASAVA_v1.8.2 software to generating fastq files. Sequencing reads were aligned to the mouse genome (mm9) using Bowtie v0.12.7 (Langmead et al., 2009). Reads were kept if they aligned with two errors or fewer and did not align to more than one location in the genome. A 25-bp density coverage map was created by extending each read for 100 bp to account for mean library fragment length and mapping the number of reads per 25 bp bin using igvtools (Thorvaldsdóttir et al., 2013). Values in each sample were normalized to fpkm values by calculating the fraction of mapped reads per bin in one million total reads.
For comparative analysis of promoter regions, the number of aligned reads in the area surrounding the transcriptional start site (±3 kb) of each gene was used.
Data availability
Sequencing data generated for this study are available through the GEO database: H3K27me3 ChIP-sequencing and RNA-Seq (accession no. GSE110598).
Preparation of libraries for RNA sequencing
2 µg total RNA were used per sample, ribosomal RNA was removed with the Ribo-Zero Magnetic kit (Epicentre), and libraries were prepared with the ScripSeq v2 RNA-Seq Library Prepapration kit (Epicentre) following the manufacturer’s instructions. In brief, the library preparation consisted of the following steps: fragmentation, cDNA synthesis, terminal tagging, and preamplification. Samples were sequenced in the same manner as the ChIP-seq samples but using 100 cycles instead of 50. Fastq reads were aligned to the mouse reference genome mm9 using Tophat (Kim et al., 2013a) to account for splicing and alternative promoter usage as well as insertions and deletions. Subsequently, the cufflinks RNA-seq analysis tool cuffdiff (Trapnell et al., 2013) was used to assess differential gene expression, alternative promoter usage, and splicing variation between experimental datasets. The resulting fpkm values were used for further data analysis and visualization.
Quantitative PCR
Total RNA was extracted using an RNeasy Mini kit (Qiagen) according to the manufacturer’s protocol. RNA was DNase-treated using an RNase free DNase set (Qiagen), and cDNA was synthesized using reagents supplied with a first-strand cDNA synthesis kit (Roche). Quantitative real-time PCR was performed using SYBR Green (Roche) on a Roche Lightcycler 480. Primer sequences are available upon request. Data are presented as mean ± SD.
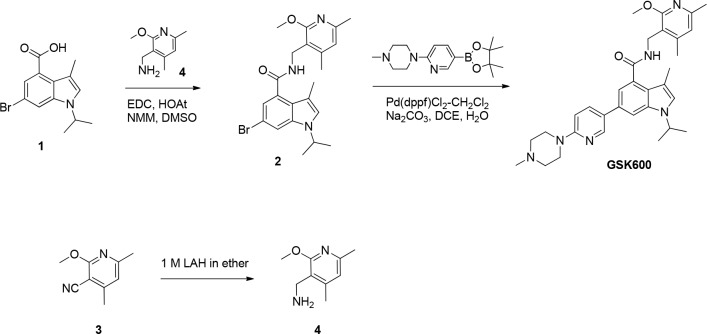
Preparation of GSK600
To a solution of 6-bromo-1-isopropyl-3-methyl-1H-indole-4-carboxylic acid (the synthesis of 6-bromo-1-isopropyl-3-methyl-1H-indole-4-carboxylic acid has been previously described; Béguelin et al., 2013; 1; 140 mg, 0.473 mmol) in dimethyl sulfoxide (10 ml) was added (2-methoxy-4,6-dimethylpyridin-3-yl) methanamine (98 mg, 0.591 mmol), 1-hydroxy-7-azabenzotriazole (129 mg, 0.945 mmol), EDC (181 mg, 0.945 mmol), and N-methylmorpholine (0.208 ml, 1.891 mmol). The reaction was maintained at room temperature for 16 h, at which time water (25 ml) was added. The mixture was stirred for 10 min and filtered. The solids were dissolved in dichloromethane and washed with water. They were dried over MgSO4 and concentrated to furnish 6-bromo-1-isopropyl-N-((2-methoxy-4,6-dimethylpyridin-3-yl)methyl)-3-methyl-1H-indole-4-carboxamide (2; 217 mg, 0.488 mmol) as a white solid. 1H NMR (400 MHz, DMSO-d6) d 1.38 (d, 6 H), 2.09 (s, 3 H), 2.32 (s, 3 H), 2.34 (s, 3 H), 3.82 (s, 3 H), 4.42 (d, J = 4.80 Hz, 2 H), 4.73 (quin, J = 6.63 Hz, 1 H), 6.68 (s, 1 H), 6.98 (d, J = 1.52 Hz, 1 H), 7.32 (s, 1 H), 7.76 (d, J = 1.77 Hz, 1 H), 8.43 (t, J = 4.93 Hz, 1 H). MS(ES) [M+H]+ 444.2, 446.2.
1-Isopropyl-N-((2-methoxy-4,6-dimethylpyridin-3-yl)methyl)-3-methyl-6-(6-(4-methylpiperazin-1-yl)pyridin-3-yl)-1H-indole-4-carboxamide (GSK600)
A mixture of 6-bromo-1-isopropyl-N-((2-methoxy-4,6-dimethylpyridin-3-yl)methyl)-3-methyl-1H-indole-4-carboxamide (200 mg, 0.450 mmol), 1-methyl-4-(5-(4,4,5,5-tetramethyl-1,3,2-dioxaborolan-2-yl)pyridin-2-yl)piperazine (164 mg, 0.540 mmol), sodium carbonate (0.675 ml, 1.350 mmol), 1,2-dimethoxyethane (3 ml), and water (1 ml) in a microwave vial was degassed for 10 min. The mixture was charged with 1,1'-bis(diphenylphosphino)ferrocene-palladium(II)dichloride dichloromethane complex (29.4 mg, 0.036 mmol) and heated in a microwave at 140°C for 10 min. The reaction was diluted with water and extracted with dichloromethane. The combined dichloromethane extracts were washed with water, dried (MgSO4), and concentrated. The residue was purified by reverse-phase HPLC (Gemini 5u C18 110A, AXIA, 50 × 30 mm 5 microncolumn; 40% acetonitrile/water/0.1% NH4OH to 90% acetonitrile/water/0.1% NH4OH) to give 1-isopropyl-N-((2-methoxy-4,6-dimethylpyridin-3-yl)methyl)-3-methyl-6-(6-(4-methylpiperazin-1-yl)pyridin-3-yl)-1H-indole-4-carboxamide (GSK600; 129 mg, 0.239 mmol) as an off-white foam. 1H NMR (400 MHz, CDCl3) d 1.50 (d, 6 H), 2.23 (s, 3 H), 2.36 - 2.44 (m, 6 H), 2.47 (s, 3 H), 2.62 (d, J = 5.31 Hz, 4 H), 3.65 (br. s., 4 H), 3.90 (s, 3 H), 4.57 - 4.83 (m, 3 H), 6.32 (t, J = 5.68 Hz, 1 H), 6.60 (s, 1 H), 6.74 (d, J = 8.84 Hz, 1 H), 7.03 (s, 1 H), 7.46 (d, J = 1.26 Hz, 1 H), 7.77 (dd, J = 8.84, 2.53 Hz, 1 H), 8.47 (d, J = 2.27 Hz, 1 H). MS(ES) [M+H]+ 541.4.
(2-Methoxy-4,6-dimethylpyridin-3-yl)methanamine
Lithium aluminum hydrate (2 M in tetrahydrofuran; 308 ml, 617 mmol) was added dropwise over 30 min to a cooled (ice/water bath) suspension of 2-methoxy-4,6-dimethylnicotinonitrile (3; 50 g, 308 mmol) in diethyl ether (1,000 ml). The reaction was stirred for 60 min, at which time the ice bath was removed. The reaction was stirred at ambient temperature for 20 h. The reaction was recooled (ice/water bath) and quenched dropwise with water (25 ml), followed by 3 M NaOH (25 ml) and more water (75 ml). The cooling bath was removed, the mixture was allowed to warm to room temperature, and Mg2SO4 (10 heaping scoops) was added. The mixture was stirred for 1 h, at which time it was filtered through Celite. The solids were washed with ether, and the mother liquors were concentrated to give a yellow oil, which solidified upon freezing. The residue was dried under vacuum (hivac) for 2 h to give (2-methoxy-4,6-dimethylpyridin-3-yl)methanamine (4; 50 g, 301 mmol). MS(ES) [M+H]+ 167.0.
Online supplemental material
Fig. S1 shows a detailed FACS analysis of WT and mutant mice with a focus on T cell development and peripheral populations. Fig. S2 shows unchanged activation of several signaling molecules downstream of the TCR and unchanged Ca2+-flux as well as mass spectrometry analysis of intracellular concentrations of GSK503 and normal total tyrosine phosphorylation and PLCγ1 activation after TCR cross-linking even in the presence of high amounts of GSK503. Fig. S3 shows unaltered kinetics of Erk activation, unaltered Mek phosphorylation at serine 298, and decreased proliferation after TCR cross-linking but not after stimulation with PMA and ionomycin, in the presence of PRC2 inhibitor.
Supplementary Material
Acknowledgments
We thank all members of the Tarakhovsky Lab for helpful feedback and discussion on this study.
This work was supported by National Institutes of Health grants R01GM112811 and R21 AI122093, by GlaxoSmithKline agreement 100026128, and by the Open Philanthropy Project/Good Ventures Foundation.
M.T. McCabe, R. Gregory, S.D. Knight, I. Rioja, A.K. Bassil, and R.K. Prinjha are employees of GlaxoSmithKline. The other authors declare no competing financial interests.
Author contributions: M.-W. Dobenecker and J.S. Park designed and performed the experiments, analyzed the data, and generated figures. J. Marcello analyzed sequencing data and generated figures. M.T. McCabe, R. Gregory, S.D. Knight, I. Rioja, A.K. Bassil, and R.K. Prinjha provided technical support and reagents necessary for this study. A. Tarakhovsky supervised this project. A. Tarakhovsky, M.-W. Dobenecker, and J.S. Park prepared the manuscript.
References
- Barda-Saad M., Braiman A., Titerence R., Bunnell S.C., Barr V.A., and Samelson L.E.. 2005. Dynamic molecular interactions linking the T cell antigen receptor to the actin cytoskeleton. Nat. Immunol. 6:80–89. 10.1038/ni1143 [DOI] [PubMed] [Google Scholar]
- Béguelin W., Popovic R., Teater M., Jiang Y., Bunting K.L., Rosen M., Shen H., Yang S.N., Wang L., Ezponda T., et al. . 2013. EZH2 is required for germinal center formation and somatic EZH2 mutations promote lymphoid transformation. Cancer Cell. 23:677–692. 10.1016/j.ccr.2013.04.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett C.L., Christie J., Ramsdell F., Brunkow M.E., Ferguson P.J., Whitesell L., Kelly T.E., Saulsbury F.T., Chance P.F., and Ochs H.D.. 2001. The immune dysregulation, polyendocrinopathy, enteropathy, X-linked syndrome (IPEX) is caused by mutations of FOXP3. Nat. Genet. 27:20–21. 10.1038/83713 [DOI] [PubMed] [Google Scholar]
- Brunkow M.E., Jeffery E.W., Hjerrild K.A., Paeper B., Clark L.B., Yasayko S.A., Wilkinson J.E., Galas D., Ziegler S.F., and Ramsdell F.. 2001. Disruption of a new forkhead/winged-helix protein, scurfin, results in the fatal lymphoproliferative disorder of the scurfy mouse. Nat. Genet. 27:68–73. 10.1038/83784 [DOI] [PubMed] [Google Scholar]
- Bustelo X.R. 2014. Vav family exchange factors: an integrated regulatory and functional view. Small GTPases. 5:9 10.4161/21541248.2014.973757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao R., and Zhang Y.. 2004. The functions of E(Z)/EZH2-mediated methylation of lysine 27 in histone H3. Curr. Opin. Genet. Dev. 14:155–164. 10.1016/j.gde.2004.02.001 [DOI] [PubMed] [Google Scholar]
- Coudronniere N., Villalba M., Englund N., and Altman A.. 2000. NF-kappa B activation induced by T cell receptor/CD28 costimulation is mediated by protein kinase C-theta. Proc. Natl. Acad. Sci. USA. 97:3394–3399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis M.M. 2002. A new trigger for T cells. Cell. 110:285–287. 10.1016/S0092-8674(02)00865-6 [DOI] [PubMed] [Google Scholar]
- Dobenecker M.W., Kim J.K., Marcello J., Fang T.C., Prinjha R., Bosselut R., and Tarakhovsky A.. 2015. Coupling of T cell receptor specificity to natural killer T cell development by bivalent histone H3 methylation. J. Exp. Med. 212:297–306. 10.1084/jem.20141499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dower N.A., Stang S.L., Bottorff D.A., Ebinu J.O., Dickie P., Ostergaard H.L., and Stone J.C.. 2000. RasGRP is essential for mouse thymocyte differentiation and TCR signaling. Nat. Immunol. 1:317–321. 10.1038/79766 [DOI] [PubMed] [Google Scholar]
- Goldberg A.D., Banaszynski L.A., Noh K.M., Lewis P.W., Elsaesser S.J., Stadler S., Dewell S., Law M., Guo X., Li X., et al. . 2010. Distinct factors control histone variant H3.3 localization at specific genomic regions. Cell. 140:678–691. 10.1016/j.cell.2010.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunawan M., Venkatesan N., Loh J.T., Wong J.F., Berger H., Neo W.H., Li L.Y., La Win M.K., Yau Y.H., Guo T., et al. . 2015. The methyltransferase Ezh2 controls cell adhesion and migration through direct methylation of the extranuclear regulatory protein talin. Nat. Immunol. 16:505–516. 10.1038/ni.3125 [DOI] [PubMed] [Google Scholar]
- He A., Shen X., Ma Q., Cao J., von Gise A., Zhou P., Wang G., Marquez V.E., Orkin S.H., and Pu W.T.. 2012. PRC2 directly methylates GATA4 and represses its transcriptional activity. Genes Dev. 26:37–42. 10.1101/gad.173930.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Josefowicz S.Z., Lu L.F., and Rudensky A.Y.. 2012. Regulatory T cells: mechanisms of differentiation and function. Annu. Rev. Immunol. 30:531–564. 10.1146/annurev.immunol.25.022106.141623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katzav S. 2007. Flesh and blood: the story of Vav1, a gene that signals in hematopoietic cells but can be transforming in human malignancies. Cancer Lett. 255:241–254. 10.1016/j.canlet.2007.04.015 [DOI] [PubMed] [Google Scholar]
- Kim D., Pertea G., Trapnell C., Pimentel H., Kelley R., and Salzberg S.L.. 2013a TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14:R36 10.1186/gb-2013-14-4-r36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim E., Kim M., Woo D.H., Shin Y., Shin J., Chang N., Oh Y.T., Kim H., Rheey J., Nakano I., et al. . 2013b Phosphorylation of EZH2 activates STAT3 signaling via STAT3 methylation and promotes tumorigenicity of glioblastoma stem-like cells. Cancer Cell. 23:839–852. 10.1016/j.ccr.2013.04.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J.M., Rasmussen J.P., and Rudensky A.Y.. 2007. Regulatory T cells prevent catastrophic autoimmunity throughout the lifespan of mice. Nat. Immunol. 8:191–197. 10.1038/ni1428 [DOI] [PubMed] [Google Scholar]
- Konze K.D., Ma A., Li F., Barsyte-Lovejoy D., Parton T., Macnevin C.J., Liu F., Gao C., Huang X.P., Kuznetsova E., et al. . 2013. An orally bioavailable chemical probe of the Lysine Methyltransferases EZH2 and EZH1. ACS Chem. Biol. 8:1324–1334. 10.1021/cb400133j [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B., Trapnell C., Pop M., and Salzberg S.L.. 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10:R25 10.1186/gb-2009-10-3-r25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.M., Lee J.S., Kim H., Kim K., Park H., Kim J.Y., Lee S.H., Kim I.S., Kim J., Lee M., et al. . 2012. EZH2 generates a methyl degron that is recognized by the DCAF1/DDB1/CUL4 E3 ubiquitin ligase complex. Mol. Cell. 48:572–586. 10.1016/j.molcel.2012.09.004 [DOI] [PubMed] [Google Scholar]
- Lee T.I., Johnstone S.E., and Young R.A.. 2006. Chromatin immunoprecipitation and microarray-based analysis of protein location. Nat. Protoc. 1:729–748. 10.1038/nprot.2006.98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margueron R., and Reinberg D.. 2011. The Polycomb complex PRC2 and its mark in life. Nature. 469:343–349. 10.1038/nature09784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouquette-Jazdanian A.K., Sommers C.L., Kortum R.L., Morrison D.K., and Samelson L.E.. 2012. LAT-independent Erk activation via Bam32-PLC-γ1-Pak1 complexes: GTPase-independent Pak1 activation. Mol. Cell. 48:298–312. 10.1016/j.molcel.2012.08.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su I.H., and Tarakhovsky A.. 2006. Lysine methylation and ‘signaling memory’. Curr. Opin. Immunol. 18:152–157. 10.1016/j.coi.2006.01.012 [DOI] [PubMed] [Google Scholar]
- Su I.H., Basavaraj A., Krutchinsky A.N., Hobert O., Ullrich A., Chait B.T., and Tarakhovsky A.. 2003. Ezh2 controls B cell development through histone H3 methylation and Igh rearrangement. Nat. Immunol. 4:124–131. 10.1038/ni876 [DOI] [PubMed] [Google Scholar]
- Su I.H., Dobenecker M.W., Dickinson E., Oser M., Basavaraj A., Marqueron R., Viale A., Reinberg D., Wülfing C., and Tarakhovsky A.. 2005. Polycomb group protein ezh2 controls actin polymerization and cell signaling. Cell. 121:425–436. 10.1016/j.cell.2005.02.029 [DOI] [PubMed] [Google Scholar]
- Thorvaldsdóttir H., Robinson J.T., and Mesirov J.P.. 2013. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief. Bioinform. 14:178–192. 10.1093/bib/bbs017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C., Hendrickson D.G., Sauvageau M., Goff L., Rinn J.L., and Pachter L.. 2013. Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat. Biotechnol. 31:46–53. 10.1038/nbt.2450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitehurst C.E., and Geppert T.D.. 1996. MEK1 and the extracellular signal-regulated kinases are required for the stimulation of IL-2 gene transcription in T cells. J. Immunol. 156:1020–1029. [PubMed] [Google Scholar]
- Yang X.P., Jiang K., Hirahara K., Vahedi G., Afzali B., Sciume G., Bonelli M., Sun H.W., Jankovic D., Kanno Y., et al. . 2015. EZH2 is crucial for both differentiation of regulatory T cells and T effector cell expansion. Sci. Rep. 5:10643 10.1038/srep10643 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequencing data generated for this study are available through the GEO database: H3K27me3 ChIP-sequencing and RNA-Seq (accession no. GSE110598).