Guan et al. identify genetic cooperativity between the transcription factor ZEB1 and the miR-200 family in memory CD8+ T cell development, which contrasts with that observed in the EMT. This study also shows that ZEB1 and its closely related homologue, ZEB2, play functionally distinct roles in CD8+ T cell differentiation.
Abstract
Long-term immunity depends partly on the establishment of memory CD8+ T cells. We identified a counterregulatory network between the homologous transcription factors ZEB1 and ZEB2 and the miR-200 microRNA family, which modulates effector CD8+ T cell fates. Unexpectedly, Zeb1 and Zeb2 had reciprocal expression patterns and were functionally uncoupled in CD8+ T cells. ZEB2 promoted terminal differentiation, whereas ZEB1 was critical for memory T cell survival and function. Interestingly, the transforming growth factor β (TGF-β) and miR-200 family members, which counterregulate the coordinated expression of Zeb1 and Zeb2 during the epithelial-to-mesenchymal transition, inversely regulated Zeb1 and Zeb2 expression in CD8+ T cells. TGF-β induced and sustained Zeb1 expression in maturing memory CD8+ T cells. Meanwhile, both TGF-β and miR-200 family members selectively inhibited Zeb2. Additionally, the miR-200 family was necessary for optimal memory CD8+ T cell formation. These data outline a previously unknown genetic pathway in CD8+ T cells that controls effector and memory cell fate decisions.
Introduction
Our immune system has two primary goals upon infection: (1) to rapidly fight off and eliminate the current invading pathogen, and (2) to generate long-term immunity, protecting us from future infection. This phenomenon, called immunological memory, is the basis for vaccination—one of the greatest achievements of modern medicine (Pulendran and Ahmed, 2011). However, not all vaccines (or infections) effectively induce protective and long-lasting memory, which accounts in part for the existing failure of effective prophylactic vaccines against many types of infections.
CD8+ T cells are a vital arm of adaptive immunity because they directly locate and kill virus-infected cells, limiting viral dissemination. Achieving present and future protection is accomplished within the CD8+ T cell population by the simultaneous generation of shorter-lived effector and longer-lived memory CD8+ T cells. Understanding how these cell fate decisions are regulated within CD8+ T cells is an important endeavor for developing better forms of vaccination and immunotherapy. Here, we describe a genetic network, previously not known to function in immune cells, that plays a critical role in establishing both the effector response and future immunity through the coordinated activities of two transcription factors (TFs), zinc-finger E-box–binding homeobox 1 (ZEB1) and ZEB2, and the miR-200 microRNA family.
Our understanding of the biological processes and molecular mechanisms regulating effector and memory CD8+ T cell development has extensively advanced over the past two decades. During many acute viral infections, naive CD8+ T cells expand into a heterogeneous population of effector cells, the majority of which become highly differentiated CTLs that we refer to as terminal effector (TE) cells and distinguish by high killer cell lectin-like receptor G1 (KLRG1) and fractalkine receptor (CX3CR1) and low IL-7 receptor α (IL-7R) expression (Kaech et al., 2003; Huster et al., 2004; Joshi et al., 2007; Gerlach et al., 2016). Most of these TE cells undergo apoptosis after viral clearance, but some persist long-term, mainly circulating in the blood (Joshi et al., 2007; Jameson and Masopust, 2009; Olson et al., 2013; Gerlach et al., 2016). A smaller fraction of effector cells, referred to as memory precursor (MP) cells, up-regulate IL-7R and seed multiple memory cell compartments, including central memory (TCM), effector memory (TEM), peripheral memory, and resident memory (TRM) T cells (Joshi et al., 2007; Kaech and Cui, 2012; Gerlach et al., 2016; Mackay and Kallies, 2017). Several TFs have been identified that regulate the ability of CD8+ T cells to adopt effector or memory CD8+ T cell fates, with many operating in a dynamic and graded manner, generating an intricate layered system of transcriptional states reflecting the integration of environmental signals individual cells experience over the course of an infection (Chang et al., 2007; Kaech and Cui, 2012; Kakaradov et al., 2017; Yu et al., 2017). For example, runt-related TF3 (RUNX3), IFN regulatory factor 4 (IRF4), T-box TF21 (Tbx21, also known as T-BET), eomesodermin (EOMES), inhibitor of DNA binding 2 (ID2), and B lymphocyte–induced maturation protein 1 (BLIMP-1) promote the early stages of cytotoxic effector CD8+ T cell differentiation (Intlekofer et al., 2005, 2008; Cannarile et al., 2006; Joshi et al., 2007; Cruz-Guilloty et al., 2009; Kallies et al., 2009; Kwon et al., 2009; Rutishauser et al., 2009; Banerjee et al., 2010; Yang et al., 2011; Knell et al., 2013; Xin et al., 2016). However, as the expression and/or activities of some of these pro–effector TFs is intensified and prolonged, they can promote the terminal differentiation of effector cells, namely through the transcriptional and epigenetic repression of pro–memory TFs, such as T cell factor 1 (TCF1; Tcf7), BTB domain and CNC homologue 2 (BACH2), Forkhead box protein O1 (FOXO1), B cell lymphoma 6 (BCL6), EOMES, and ID3, that govern circulating memory cell differentiation (Ichii et al., 2004; Intlekofer et al., 2005; Banerjee et al., 2010; Jeannet et al., 2010; Zhou et al., 2010; Ji et al., 2011; Yang et al., 2011; Kim et al., 2013; Tejera et al., 2013; Roychoudhuri et al., 2016; Gray et al., 2017; Kakaradov et al., 2017; Yu et al., 2017). Less is understood about the transcriptional networks governing TRM formation, but in contrast to circulating TEM and TCM cells, TRM cell development relies on up-regulation of BLIMP-1 and homologue of Blimp-1 in T cells (HOBIT) and down-regulation of Krüppel-like factor 2 (KLF2), T-BET, EOMES, and TCF1 (Mackay et al., 2016; Mackay and Kallies, 2017). There are still many open questions regarding how these TFs interact with one another to instruct or repress effector and memory fates, which has important implications on the generation of a diverse array of memory T cells. Furthermore, as memory T cells traffic to many different tissue compartments, what are the environmental signals and underlying genetic modules regulated by such signals that influence their development and maintenance?
Prior work investigating the graded action of T-BET in terminal differentiation of effector CD8+ T cells identified ZEB2 as a translator of high T-BET expression that switched on TE cell differentiation (Dominguez et al., 2015; Omilusik et al., 2015). Both ZEB2 and its highly conserved homologue, ZEB1, are well-known activators of the epithelial-to-mesenchymal transition (EMT), a process centrally involved in embryogenesis and tumorigenesis, particularly metastasis, in which they operate in a well-described negative-feedback pathway with the miR-200 microRNA family (Bracken et al., 2008; Brabletz and Brabletz, 2010; Brabletz et al., 2011; Gregory et al., 2011). Given our discovery of ZEB2 in CD8+ T cell differentiation, we sought to investigate whether ZEB1 and the miR-200 family also played a role in this process. Surprisingly, rather than cooperating with ZEB2, we found that ZEB1 and ZEB2 were expressed in a reciprocal manner at temporally distinct phases of the immune response. Although ZEB2 promoted TE cell differentiation and survival, ZEB1 was critical for normal maintenance of memory CD8+ T cells and protective immunity. Our data also demonstrated that well-described regulators of the EMT, miR-200 family members and TGF-β differentially regulated Zeb1 and Zeb2 expression in CD8+ T cells. In contrast to the EMT, where miR-200 family members repress both Zeb1 and Zeb2 to maintain epithelial states, in CD8+ T cells, miR-200 members selectively inhibited Zeb2, but not Zeb1, expression. Likewise, TGF-β induced Zeb1 yet repressed Zeb2 expression in CD8+ T cells. Ectopic expression of miR-200 family members in CD8+ T cells impaired TE and promoted memory cell development, whereas miR-200 deficiency resulted in the loss of memory CD8+ T cells. Altogether, our study revealed a novel gene regulatory network involving ZEB1, ZEB2, and the miR-200 family that governs effector and memory CD8+ T cells fate decisions during viral infection.
Results
Zeb1 and Zeb2 are reciprocally expressed in CD8+ T cells during lymphocytic choriomeningitis virus (LCMV) infection
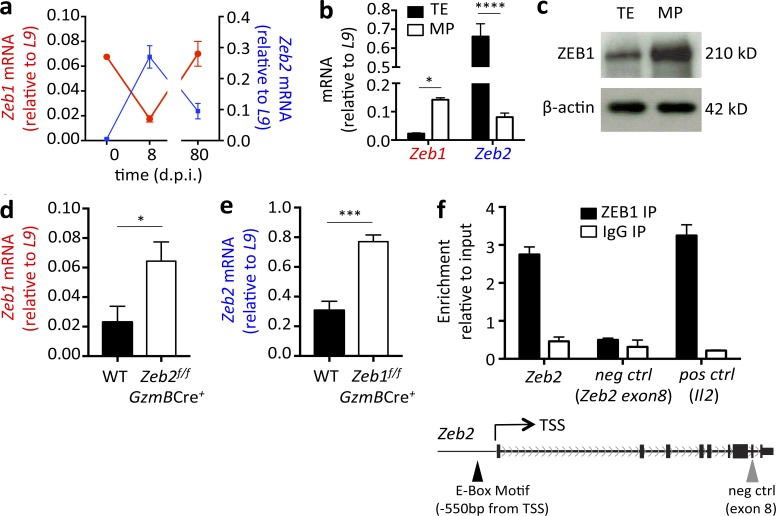
To investigate the role of ZEB1 in CD8+ T cell differentiation and determine whether it cooperated with ZEB2 in this process, we first examined Zeb1 and Zeb2 expression in CD8+ T cells during the course of viral infection. WT C57BL/6 (B6) mice were infected with LCMV-Arm, which led to an acute viral infection. Naive (CD44lo CD62Lhi) CD8+ T cells (day 0) were sorted from uninfected mice and MHC class I DbGP33-41 tetramer+ LCMV-specific effector and memory CD8+ T cells were isolated from 8 and 80 d postinfection (dpi), respectively. Zeb1 and Zeb2 mRNA expression was determined by quantitative RT-PCR, which revealed their reciprocal pattern of expression in naive, effector, and memory CTLs. Zeb1 mRNA was expressed in naive CD8+ T cells, reduced approximately fourfold in effector CD8+ T cells, and then up-regulated again in mature memory CD8+ T cells. Conversely, Zeb2 mRNA displayed the opposite expression pattern, as previously reported (Dominguez et al., 2015; Omilusik et al., 2015), and was below detection in naive CD8+ T cells, increased substantially in effector cells (8 dpi), and then decreased in memory CD8+ T cells (Fig. 1 a). Further subdividing of the LCMV-specific CD8+ T cells into more terminally differentiated TE (KLRG1hi IL-7Rlo) and less differentiated MP (KLRG1lo IL-7Rhi) cell subsets at 8 dpi showed an inverse expression pattern, with Zeb1 being more highly expressed in MP cells and Zeb2 being more dominant in TE cells (Fig. 1 b). ZEB1 expression in TE and MP subsets was examined using Western blot on protein lysates from LCMV-specific CD8+ T cells, and this confirmed MP cells contain more ZEB1 compared with the TE cells (Fig. 1 c). These data demonstrate that during an immune response, Zeb1 and Zeb2 are expressed in a reciprocal manner in CD8+ T cells, suggesting that rather than functioning cooperatively, as observed during the EMT, ZEB1 and ZEB2 may function in distinct processes in CD8+ T cells.
Figure 1.
Zeb1 and Zeb2 are reciprocally expressed in CD8+ T cells during LCMV infection. (a) Zeb1 and Zeb2 mRNA was measured in purified CD44lo CD62Lhi naive CD8+ T cells (day 0) or DbGP33-41+ LCMV-specific CD8+ T cells from 8 and 80 dpi using quantitative RT-PCR. LCMV-specific CD8+ T cells were purified based on DbGP33–41-tetramer staining. (b and c) Effector P14+ CD8+ T cells were isolated 8 dpi based on the expression of KLRG1hi IL-7Rlo (TE) and KLRG1lo IL-7Rhi (MP) and Zeb1 and Zeb2 mRNA was measured using quantitative RT-PCR (b) or ZEB1 protein expression was measured using Western blotting (c; β-actin was used as a loading control). (d) Zeb1 mRNA expression was compared between WT and Zeb2f/f GzmBCre+ P14+ CD8+ T cells at 8 dpi. (e) Zeb2 mRNA expression was compared between WT and Zeb1f/f GzmBCre+ P14+ CD8+ T cells 30 dpi. (f) ZEB1 ChIP-quantitative PCR was performed on purified CD44lo CD62Lhi naive CD8+ T cells using anti–ZEB1 (black bars) or IgG control (white bars) antibodies and primers to Zeb2 promoter (−550 bp), exon 8 and Il2 promoter. Data shown are representative of two (c) independent or cumulative of three (a, b, d, and e) independent experiments; n = 3–4 mice/group/experiment (c), n = 9–12 mice/group (a, b, d, and e). Data are expressed as mean ± SEM. *, P < 0.05; ***, P < 0.001; ****, P < 0.0001.
Given the reciprocal mRNA expression pattern of Zeb1 and Zeb2, we next asked whether they could directly antagonize each other’s expression in CD8+ T cells. To explore this possibility, Zeb1 mRNA was quantified in Zeb2-deficient LCMV-specific effector CD8+ T cells 8 dpi (Fig. 1 d), and Zeb2 mRNA was measured in Zeb1-deficient memory CD8+ T cells at 30 dpi (Fig. 1 e). Mice containing floxed alleles of Zeb2 (Zeb2flox/flox) have been previously described (Higashi et al., 2002; Dominguez et al., 2015; Omilusik et al., 2015). To generate Zeb1 loss-of-function T cells, we created mice containing two loxP sites flanking Zeb1 exon 6 using CRISPR/Cas9 technology, because exon 6 is the largest exon within Zeb1 and encodes the central homeodomain and part of both N- and C-terminal zinc-finger clusters (see Fig. S1 a for more details on generation of Zeb1flox/flox mice). Zeb1flox/flox and Zeb2flox/flox mice were crossed to mice expressing the Granzyme B-Cre (GzmB-Cre) transgene to inducibly delete Zeb1 or Zeb2, respectively, after CD8+ T cell activation during LCMV infection (we refer to these mice as Zeb1f/f GzmBCre+ and Zeb2f/f GzmBCre+ mice, and littermate controls lacking GzmB-Cre are referred to as WT). This analysis indicated that Zeb1 and Zeb2 mutually repress each other’s expression, because Zeb1 mRNA was significantly elevated in Zeb2f/f GzmBCre+ effector CD8+ T cells and, likewise, Zeb2 mRNA was increased in Zeb1f/f GzmBCre+ memory CD8+ T cells (Fig. 1, d and e).
Given their reciprocal expression and the fact that ZEB1 and ZEB2 have both been described as transcriptional repressors that bind to paired E-boxes, we hypothesized that perhaps they might directly repress each other’s expression. TF motif analysis indicated both Zeb1 and Zeb2 loci contain conserved E-box domains in their proximal promoter regions. Unfortunately, based on our experience, none of the commercial anti–mouse ZEB2 antibodies specifically recognize ZEB2, thereby preventing analysis of ZEB2 binding at the Zeb1 locus. However, to determine whether the repression by ZEB1 on Zeb2 transcription could be regulated via direct binding of ZEB1 to the Zeb2 locus, chromatin immunoprecipitation (ChIP) using a ZEB1-specific antibody followed by quantitative RT-PCR of two regions of the Zeb2 locus was performed. As a positive control, we also probed ZEB1 binding to a previously described target in the Il2 locus (Williams et al., 1991; Wang et al., 2009). The results of this analysis showed clear enrichment of ZEB1 binding at the Zeb2 promoter and the Il2 locus, but not an irrelevant region in exon 8 of Zeb2 (Fig. 1 f). This result demonstrated that ZEB1 can directly bind Zeb2, supporting the possibility ZEB1 may directly repress Zeb2 to coordinate their inverse expression patterns.
ZEB1 promotes the homeostasis of memory CD8+ T cells
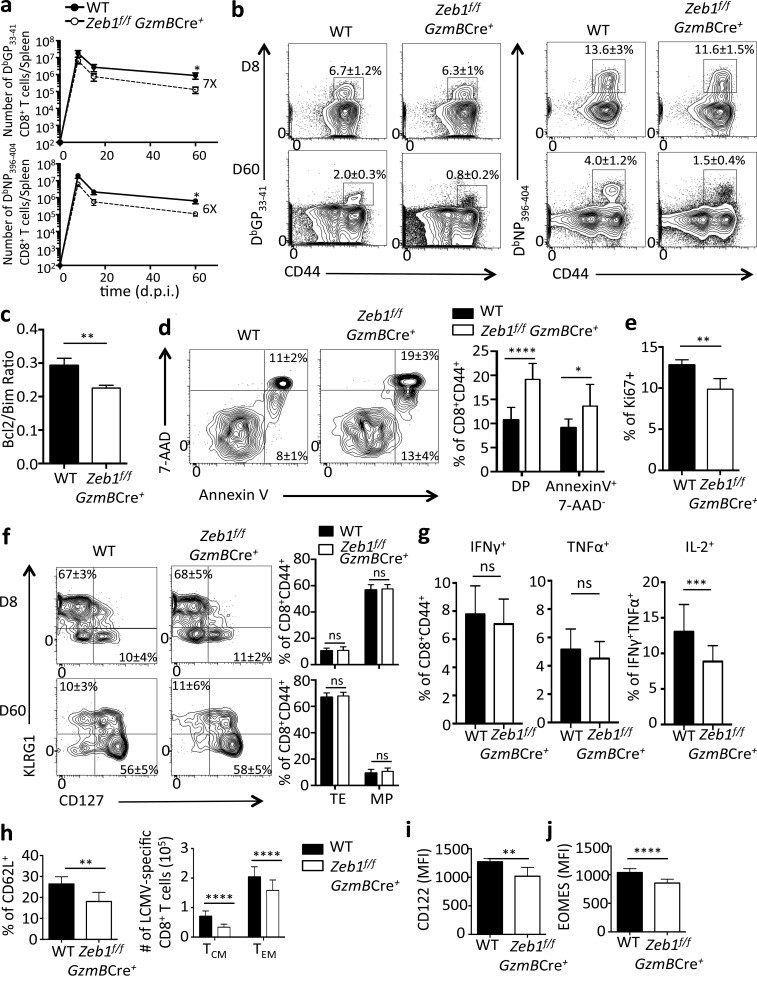
To determine the role of ZEB1 in CD8+ T cell differentiation during the course of viral infection and compare it with the role of ZEB2 previously described (Dominguez et al., 2015; Omilusik et al., 2015), Zeb1f/f GzmBCre− (WT) and Zeb1f/f GzmBCre+ mice were infected with LCMV-Arm, and splenic DbGP33–41 and DbNP396–404 tetramer+ CD8+ T cells were enumerated and characterized at 8, 15, and 60 dpi. Zeb1-deficient CD8+ T cells underwent clonal expansion upon LCMV challenge and displayed frequencies of tetramer-specific CTLs at 8 dpi similar to controls, but there was an approximately twofold reduction in cell numbers (Fig. 2, a and b). The decline of Zeb1-deficient CD8+ T cells became more apparent with time, demonstrating a profound defect in the establishment and/or maintenance of memory CD8+ T cells (Fig. 2, a and b). In general, there were approximately six- to sevenfold fewer Zeb1-deficient memory CD8+ T cells than WT cells, and this memory cell attrition was associated with a decreased ratio of BCL-2 to BIM within the Zeb1f/f GzmBCre+ memory CD8+ T cells (Fig. 2 c). Accordingly, Zeb1f/f GzmBCre+ memory CD8+ T cells contained a higher frequency of early- and late-stage apoptotic cells according to Annexin V and 7-AAD staining (Fig. 2 d). Homeostatic proliferation based on Ki67 staining revealed a modest but consistent decrease in the frequency of proliferating cells in Zeb1-deficient as compared with the WT memory CD8+ T cells (Fig. 2 e). Altogether, these data demonstrate that ZEB1 plays an important role in maintaining survival and homeostasis of memory CD8+ T cells, distinguishing it as a novel regulator of memory T cell development.
Figure 2.
ZEB1 promotes the survival of memory CD8+ T cells. (a) WT (filled circle) and Zeb1f/f GzmBCre+ (open circle) mice were infected with LCMV-Arm, and splenic DbGP33-41 and DbNP396-404 tetramer+ CD8+ T cells were quantified at 8, 15, and 60 dpi. (b) Representative FACS plots of DbGP33-41 and DbNP396-404 tetramer+ CD8+ T cells at 8 and 60 dpi in WT and Zeb1f/f GzmBCre+ mice (percentage of tetramer+ CD8+ T cells ± SEM is indicated). (c–e) Graphs are gated on DbGP33-41-tetramer+ CD8+ T cells from WT or Zeb1f/f GzmBCre+ mice at 30 dpi and show the ratio of BCL-2/BIM expression (c), percentage of Annexin V+ and 7-AAD+ cells (d), or percentage of Ki67+ cells (e) based on flow cytometry. (f) Expression of KLRG1 and IL-7R in DbGP33-41 tetramer+ CD8+ T cells at 8 and 60 dpi in WT and Zeb1f/f GzmBCre+ mice based on flow cytometry. (g–j) WT and Zeb1f/f GzmBCre+ CD8+ T cells from 30 dpi were analyzed for production of IFNγ, TNFα, or IL-2 as indicated using intracellular cytokine staining (g) or expression of CD62L (together with absolute number of TCM and TEM cells; h), CD122 (i), and EOMES (j). Data shown are representative of five (b and f) or two (d) independent or cumulative of three (a) or five (c, e, and g–j) independent experiments; n = 3–5 mice per group per experiment (b, d, and f), n = 8–14 mice/group (a), amd n = 15–25 mice/group (c, e, and g–j). Data are expressed as mean ± SEM. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
Surprisingly, despite the defect in long-term survival of Zeb1f/f GzmBCre+ memory cells and preferential expression of Zeb1 mRNA in IL-7Rhi MP cells, there was no discernible defect in the development of IL-7Rhi effector and memory cells in Zeb1fl/fl GzmBCre+ CD8+ T cells 8 dpi (Figs. 1 b and 2 f). That is, similar frequencies of MP- or TE-like subsets formed in both groups of mice based on KLRG1 and IL-7R expression (Fig. 2 f). However, fewer numbers of these phenotypically distinct subsets formed in the absence of Zeb1 (Fig. 2 a). These data suggest that unlike ZEB2, ZEB1 was dispensable for specialized aspects of CTL differentiation, but it played a more dominant role in the survival of all virus-specific CD8+ T cells during the effector to memory transition.
Although Zeb1-deficient memory CD8+ T cells produced similar amounts of IFNγ and TNFα compared with WT cells when restimulated with DbGP33-41 peptide, they produced less IL-2, indicating they were less polyfunctional (Fig. 2 g). Coinciding with the reduction in IL-2, a property of central memory CD8+ T cells (Sallusto et al., 1999; Wherry et al., 2003), we observed a modest reduction in other TCM properties in memory CD8+ T cells lacking ZEB1, including decreased l-selectin (CD62L), IL-15 receptor β (IL-15Rβ, CD122), and EOMES expression (Fig. 2, h–j). EOMES sustains memory CD8+ T cell homeostatic turnover by maintaining CD122 expression (Intlekofer et al., 2005; Banerjee et al., 2010), and this may explain the aforementioned decreased proliferation of Zeb1-deficient memory CD8+ T cells.
To more rigorously validate that these defects in memory CD8+ T cell development were caused by CD8+ T cell–autonomous functions of ZEB1, we cotransferred equal numbers of WT (Thy1.1/1.1) and Zeb1f/f GzmBCre+ (Thy1.1/1.2) P14+ CD8+ T cells into congenically mismatched (Thy1.2/1.2) B6 mice and assessed their response to LCMV infection. The P14+ TCR transgene is specific for the GP33–41 epitope in LCMV. Relative to the WT donor cells, similar phenotypes were observed in the P14+ Zeb1-deficient memory CD8+ T cells as described above, including reduced survival and homeostatic proliferation and impaired IL-2 production and formation of TCM cells (Fig. S2).
In summary, these data indicated that rather than functioning cooperatively, ZEB1 and ZEB2 have distinct roles in promoting memory and effector CD8+ T cell fates, respectively. ZEB1 is not involved in TE CD8+ T cell differentiation, unlike ZEB2, but rather is necessary for optimal memory cell survival, homeostasis, and TCM formation. These data indicate that ZEB1 and ZEB2 form a novel counterregulatory axis operating at distinct phases of the immune response to govern effector and memory CD8+ T cell differentiation.
ZEB1 is necessary for memory CD8+ T cell protective immunity
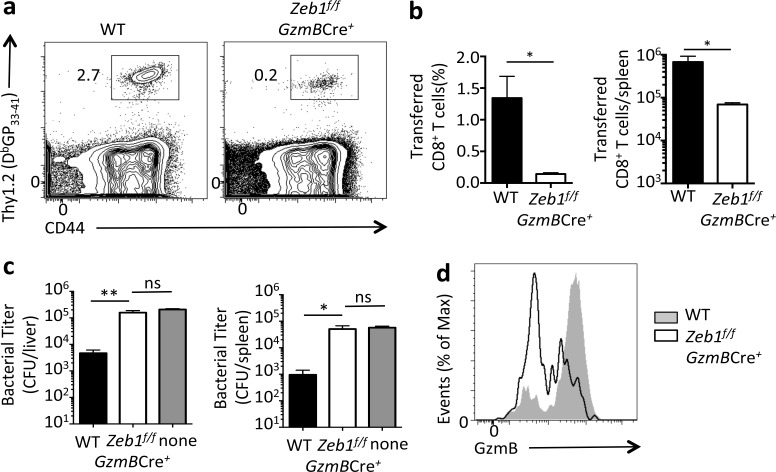
To assess whether the defect in Zeb1f/f GzmBCre+ memory CD8+ T cells extended beyond survival to affect memory function, we compared the protective qualities of the WT and Zeb1f/f GzmBCre+ memory CD8+ T cells during reinfection. WT and Zeb1f/f GzmBCre+ mice were infected with LCMV-Arm, and at 30 dpi, equal numbers of DbGP33–41 tetramer+ LCMV-specific CD8+ T cells were isolated from each cohort and transferred separately into congenically mismatched mice that were subsequently challenged the next day with a recombinant strain of Listeria monocytogenes expressing the GP33–41 epitope. Four days after the rechallenge, we compared the ability of transferred WT and Zeb1f/f GzmBCre+ cells to expand and mediate bacterial control upon secondary challenge. Although WT cells exhibited normal expansion and conducted robust bacterial clearance, Zeb1f/f GzmBCre+ memory CD8+ T cells displayed substantial impairment in their proliferative capacity (∼10-fold) and failed to reduce bacterial burden in the liver and spleen (Fig. 3, a–c). Additionally, secondary Zeb1f/f GzmBCre+ effector CD8+ T cells produced less Granzyme B compared with WT cells (Fig. 3 d). These data demonstrate that ZEB1 is important not only for the survival of memory CD8+ T cells but also for their protective recall responses.
Figure 3.
ZEB1 is necessary for memory CD8+ T cell protective immunity. 100,000 GP33–41–specific WT or Zeb1f/f GzmBCre+ memory CD8+ T cells (Thy1.2/1.2) were transferred into congenically mismatched (Thy1.1/1.1) naive B6 mice that were then infected with recombinant L. monocytogenes expressing the GP33-41 epitope (LM-33). (a and b) At day 4 after challenge, the frequency and numbers of recalled GP33–41–specific WT or Zeb1f/f GzmBCre+ CD8+ T cells were analyzed in the spleen. (c) LM-33 bacterial titers (CFUs) in the liver and spleen were determined at day 3 after challenge. (d) Histogram shows the expression of Granzyme B in GP33–41–specific WT or Zeb1f/f GzmBCre+ CD8+ T cells day 4 after LM-33 infection. Data shown are representative of two (a and d) or cumulative of two (b and c) independent experiments; n = 3–5 mice/group/experiment (a and d), n = 6–10 mice/group (b and c). Data are expressed as mean ± SEM. *, P < 0.05; **, P < 0.01.
Zeb1 and Zeb2 are inversely regulated by TGF-β
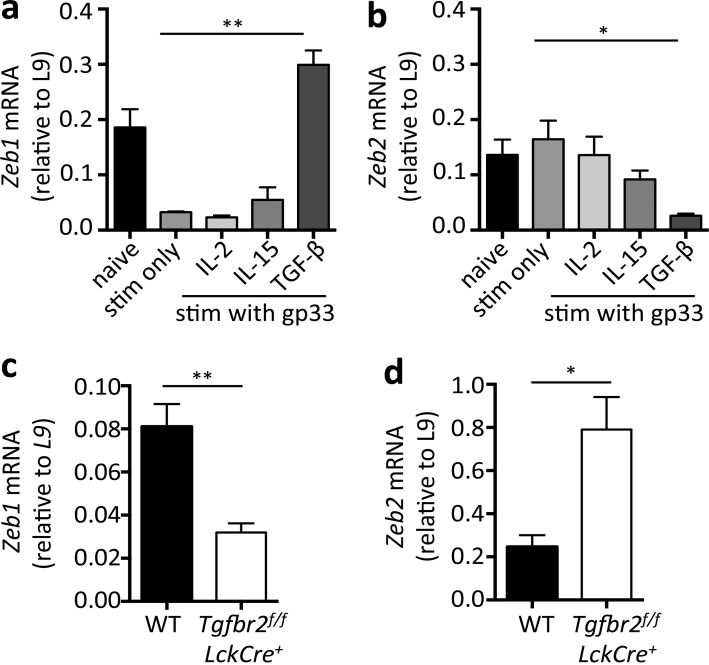
Given the reciprocal expression pattern of Zeb1 and Zeb2 in naive, effector, and memory CD8+ T cells, we hypothesized that they may be inversely regulated by environmental factors such as cytokines produced during or after viral infection. To examine this question, we tested the effects of several cytokines important for memory T cells, including TGF-β, IL-2, and IL-15, on Zeb1 and Zeb2 mRNA levels in in vitro activated P14+ CD8+ T cells. In addition to regulating the development of CD8+ TRM cells, CD4+ regulatory T cells, and Th17 cells, TGF-β is also a well-known inducer of ZEB1 and ZEB2 in mesenchymal cells during the EMT (Comijn et al., 2001; Eger et al., 2005; Pearce et al., 2009; Lee et al., 2011; Casey et al., 2012; Mackay et al., 2013). IL-2 and IL-15 were examined, because they regulate CTL differentiation and memory cell fitness and long-term survival (Becker et al., 2002; Tan et al., 2002; Williams et al., 2006; Kalia et al., 2010; Pipkin et al., 2010). These experiments showed that Zeb1 was down-regulated upon DbGP33–41 peptide stimulation and remained low with either IL-2 or IL-15 treatment. However, TGF-β treatment of the activated CD8+ T cells led to significant induction of Zeb1 mRNA (Fig. 4 a). In contrast, Zeb2 mRNA was lowly expressed in naive cells and not significantly affected by peptide stimulation alone or with IL-2 or IL-15 treatment. Furthermore, in contrast to the EMT, TGF-β further reduced Zeb2 mRNA levels in activated CD8+ T cells (Fig. 4 b). These opposing effects of TGF-β on Zeb1 and Zeb2 expression in activated CD8+ T cells provide greater insight for how TGF-β may fine-tune memory CD8+ T cell gene expression patterns and differentiation.
Figure 4.
Zeb1 and Zeb2 are inversely regulated by TGF-β. (a and b) Zeb1 and Zeb2 mRNA expression in naive P14+ CD8+ T cells or those stimulated (stim) with GP33–41 peptide for 3 d followed by 2-d culture alone or in the presence of the indicated cytokines (IL-2, IL-15, and TGF-β). (c and d) 50,000 WT or Tgfbr2f/f LckCre+ P14+ CD8+ T cells were transferred to naive B6 mice followed by LCMV-Arm infection. 45 dpi, the donor cells were purified using FACS and the amount of Zeb1 and Zeb2 mRNA was measured by quantitative RT-PCR. Data shown are cumulative of two (a–d) independent experiments; n = 8 mice (a and b), n = 6–10 mice/group (c and d). Data are expressed as mean ± SEM. *, P < 0.05; **, P < 0.01.
To investigate whether TGF-β inversely regulates Zeb1 and Zeb2 expression in a CD8+ T cell–intrinsic manner in vivo, we transferred small numbers of naive Tgfbr2f/f LckCre− (WT) and Tgfbr2f/f LckCre+ P14+ CD8+ T cells into B6 mice that were subsequently infected with LCMV-Arm. Memory CD8+ T cells were isolated from the two groups of mice 45 dpi, and the amounts of Zeb1 and Zeb2 mRNA was measured by quantitative RT-PCR. In agreement with the in vitro data, LCMV-specific memory CD8+ T cells lacking TGFβR2 failed to up-regulate Zeb1 and sustained abnormally high expression of Zeb2 (Fig. 4, c and d). Thus, in contrast to that observed in the EMT, where TGF-β induces both Zeb1 and Zeb2 expression, in developing memory CD8 T cells, TGF-β has differential effects and promotes Zeb1 while repressing Zeb2 mRNA expression.
miR-200 family directly represses Zeb2 but not Zeb1 in CD8+ T cells
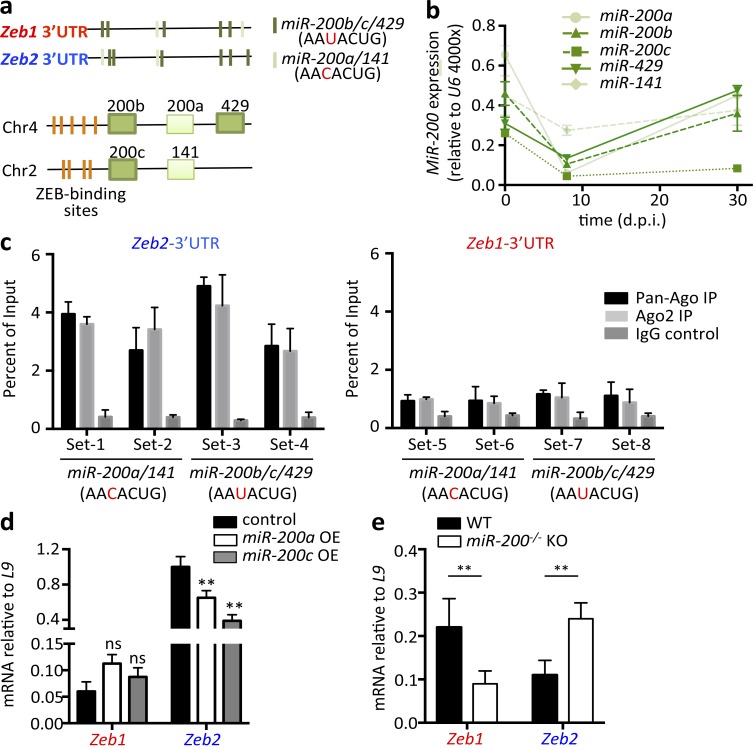
Our aforementioned findings identified a fundamental difference between the EMT and CD8+ T cell immune responses. In the former, Zeb1 and Zeb2 are coordinately expressed to promote the EMT, whereas in the latter, they are expressed in opposition to promote TE and memory CD8+ T cell development. This raises the question as to how Zeb1 and Zeb2 expression becomes uncoupled in CD8+ T cells. To explore this question, we turned our attention toward the miR-200 family members because they function in a negative-feedback pathway with Zeb1 and Zeb2 in the EMT. The miR-200 family members maintain epithelial identity by repressing expression of EMT drivers, principally Zeb1 and Zeb2, each of which contain several miR-200–binding targets in their 3′ untranslated regions (UTRs; Fig. 5 a; Mongroo and Rustgi, 2010). However, induction of the EMT by signals such as TGF-β induce ZEB1 and ZEB2, which in turn, directly bind to the highly conserved E-box–binding sequences in miR-200 gene promoters and transcriptionally repress miR-200 family expression (Brabletz et al., 2011; Fig. 5 a). The five members of miR-200 family are separated into two clusters on chromosomes 2 and 4 in Mus musculus (Fig. 5 a) and are distinguished by their conserved binding sequences (seed sequences). miR-200b, c, and 429 share the same seed sequence (AAUACUG), which differs by only one nucleic acid from the seed sequenced shared by miR-200a and 141 (AACACUG).
Figure 5.
miR-200 family directly represses Zeb2 but not Zeb1 mRNA in CD8+ T cells. (a) Schematic representation of Zeb1 and Zeb2 3′ UTRs (top) showing the miR-200 family seed sequences (dark and light green) and both clusters of miR-200 family genes (bottom) showing the corresponding conserved ZEB-binding sites (orange). (b) Naive (d0) and LCMV-specific effector (8 dpi) and memory (30 dpi) CD8+ T cells were sorted and measured for miR-200 family microRNA expression using quantitative RT-PCR. (c) LCMV-specific CD8+ T cells were sorted 12 dpi followed by UV cross-linking and immunoprecipitating (IP) with pan-Ago (black), Ago2 (light gray), or IgG control (dark gray) antibodies. Enrichment of specific miR-200a/141 or miR-200b/c/429 binding regions within Zeb2 (left) or Zeb1 (right) 3′ UTR over input was measured by site-specific primers using quantitative RT-PCR. (d) P14+ CD8+ T cells transduced with RVs designed to overexpress (OE) miR-200a (white bar) or miR-200c (gray bar) were sorted at 8 dpi, and Zeb1 and Zeb2 mRNA was measured by quantitative RT-PCR. (e) LCMV-specific CD8+ T cells were purified from WT (black bar) and miR-200−/− KO (white bar) mice at 30 dpi, and Zeb1 and Zeb2 mRNA was measured by quantitative RT-PCR. Data shown are cumulative of two (b, c, and e) or three (d) independent experiments; n = 6–10 mice/group (b, c, and e), n = 9–15 mice/group (d). Data are expressed as mean ± SEM. **, P < 0.01.
First, we examined the RNA expression patterns of the miR-200 family members in naive and LCMV-specific effector and memory CD8+ T cells using TaqMan microRNA quantitative RT-PCR. This revealed that all five members of miR-200 were expressed in naive CD8+ T cells, down-regulated in effector CTLs 8 dpi, and up-regulated again in memory CD8+ T cells (Fig. 5 b). Thus, the miR-200 members were expressed in a coordinated manner with Zeb1, but not Zeb2 (Fig. 1 a), suggesting that Zeb1 may escape miR-200 repression in CD8+ T cells, and vice versa. To better examine this point biochemically, we cross-linked LCMV-specific CD8+ T cells from 12 dpi under UV light, immunoprecipitated the RNA-induced silencing complex using anti-Argonaut (anti-AGO) antibodies, and measured the amount of Zeb1 or Zeb2 mRNA captured in the complex using quantitative RT-PCR with primers specifically designed to the predicted miR-200–binding sites within Zeb1 and Zeb2 3′ UTRs (Guo et al., 2014, 2015). We observed significantly greater enrichment of the Zeb2 3′ UTR in the RNA-induced silencing complex than Zeb1 3′ UTR (Fig. 5 c; note that Zeb1 and Zeb2 3′ UTRs both contain seed sequences of miR-200b/c/429 or miR-200a/14; Fig. 5 a). This finding fit nicely with the model proposed above in which Zeb2 mRNA is targeted by miR-200 repression in activated CD8+ T cells, whereas Zeb1 mRNA is not. This represents a novel mechanism uncoupling ZEB1 and ZEB2 expression that evolved in CD8+ T cells.
In addition to the binding assay, we also assessed how overexpression of miR-200 family members affects expression of Zeb1 and Zeb2 in CD8+ T cells. Consistent with the aforementioned data, retroviral overexpression of miR-200a or miR-200c (one representative member of each miR-200 seed sequence) selectively reduced Zeb2 mRNA levels, leaving Zeb1 mRNA unaffected or marginally increased in effector CD8+ T cells 8 dpi (Fig. 5 d). Conversely, memory CD8+ T cells lacking both miR-200 clusters (referred to as miR-200−/− and described in detail later) simultaneously displayed increased Zeb2 and decreased Zeb1 mRNA (Fig. 5 e). Altogether, these data illustrate the selective regulation of Zeb2 expression by miR-200 family members in CD8+ T cells, thus adding an additional layer of genetic control in the ZEB1 and ZEB2 counterregulatory network that guides effector and memory CD8+ T cell fate decisions.
miR-200 overexpression impairs TE and promotes MP cell development
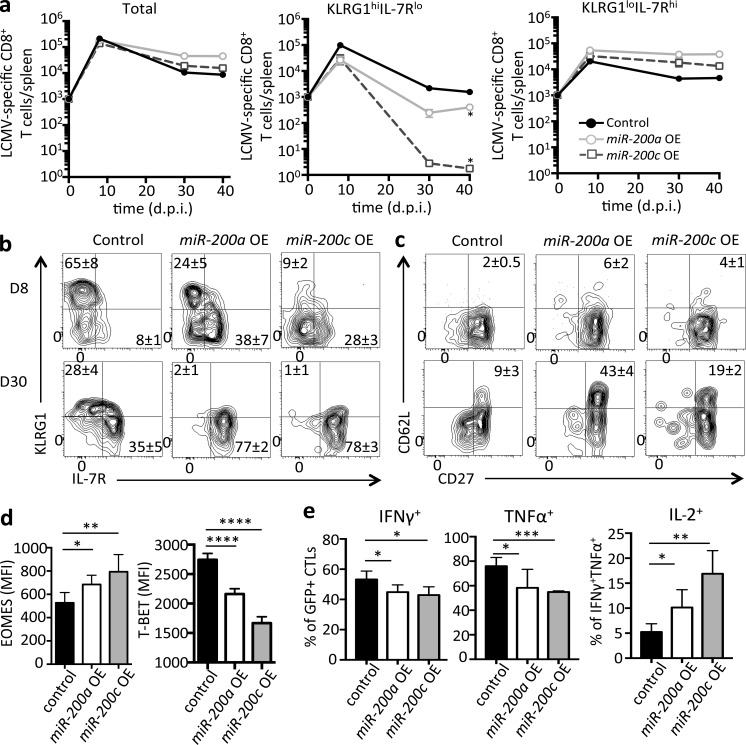
To determine whether miR-200 family members modulate Zeb2 expression and differentiation of virus-specific effector and memory CD8+ T cells, we individually overexpressed (OE) each miR-200 family member in P14+ CD8+ T cells using retroviruses (RVs) and transferred equal numbers of RV-transduced cells into naive B6 mice that were subsequently infected with LCMV-Arm. Control P14+ CD8+ T cells were transduced with empty RVs. Note, miR-200a and miR-200c were selected as representatives of each seed sequence and shown in Fig. 6, whereas the results of the remaining miR-200 family members are provided in Fig. S3. These experiments revealed that by 30 dpi, larger numbers (approximately three- to eightfold) of virus-specific CD8+ T cells were present from the individual overexpression of the miR-200 family members, indicating that these miR-200s enhanced memory CD8+ T cell formation (Fig. 6 a). At 8 and 30 dpi, the control and miR-200 OE P14+ CD8+ T cells were analyzed and in line with their preferential binding to Zeb2 mRNA, the overexpression of miR-200 family members largely phenocopied CTLs lacking Zeb2. That is, at 8 dpi, the frequency and number of KLRG1hi IL-7Rlo TE cells was greatly diminished, whereas the KLRG1lo IL-7Rhi MP subset was markedly increased with miR-200 overexpression compared with the control vector (Figs. 6 b and S3 a). Impressively, virtually the entire population of memory CD8+ T cells overexpressing miR-200 family members was IL-7Rhi and KLRG1lo and contained a substantially larger percentage of CD62L+ CD27+ IL-2+ TCM cells (Figs. 6c and S3 b). Moreover, pro-memory TFs such as EOMES and TCF1 were increased in miR-200 overexpressing CD8+ T cells, whereas the pro-TE TF T-BET was significantly down-regulated (Fig. 6 d and Fig. S3, c and d). Furthermore, at 8 dpi, we observed mildly reduced production of IFNγ and TNFα, but an increased proportion of IL-2–producing effector cells with miR-200 overexpression upon DbGP33-41 peptide stimulation (Fig. 6 e). These data indicated that the miR-200 family members are sufficient to inhibit the terminal differentiation of effector CD8+ T cells and promote formation of memory cells and their progenitors, which is likely due in part to the direct repression of Zeb2 by miR-200 family. That we observed a more profound phenotype with miR-200 OE compared with Zeb2 deficiency with regard to the loss of TE cells and gain of central memory properties suggests that miR-200 family members likely also target other pro-TE genes to achieve their biological function.
Figure 6.
Overexpression of miR-200 family impairs TE and promotes memory cell formation. As in Figure 5, small numbers of P14+ CD8+ T cells were transduced with miR-200a (light gray open circle) or miR-200c (dark gray open square) RVs or empty vector control (black filled circle) RVs and transferred into B6 mice that were subsequently infected with LCMV-Arm. (a) Line plots show total numbers of donor RV-transduced P14+ CD8+ T cells (left) or subsets of KLRG1hi IL-7Rlo cells (middle) or KLRG1lo IL-7Rhi cells (right) at 8, 30 and 45 dpi. (b and c) Flow plots show expression of KLRG1 and IL-7R (b) CD62L and CD27 (c) in control, miR-200a and miR-200c OE P14+ CD8+ T cells at 8 and 30 dpi. (d and e) Bar graphs show amounts of EOMES and T-BET in donor P14+ CD8+ T cells at 30 dpi (d) or production of IFNγ, TNFα or IL-2 as indicated using intracellular cytokine staining (e; note that IL-2–producing cells were gated on IFNγ+ TNFα+ CD8+ T cells). See Fig. S3 for additional data on effects of miR-200b, miR-429, and miR-141 OE in LCMV-specific CD8+ T cells. Data shown are representative of five (b and c) or cumulative of two (a) or three (d and e) independent experiments; n = 3–5 mice/group/experiment (b and c), n = 6–8 mice/group (a), n = 8–10 (d and e). Data are expressed as mean ± SEM. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
miR-200 family deficiency results in loss of memory CD8+ T cells
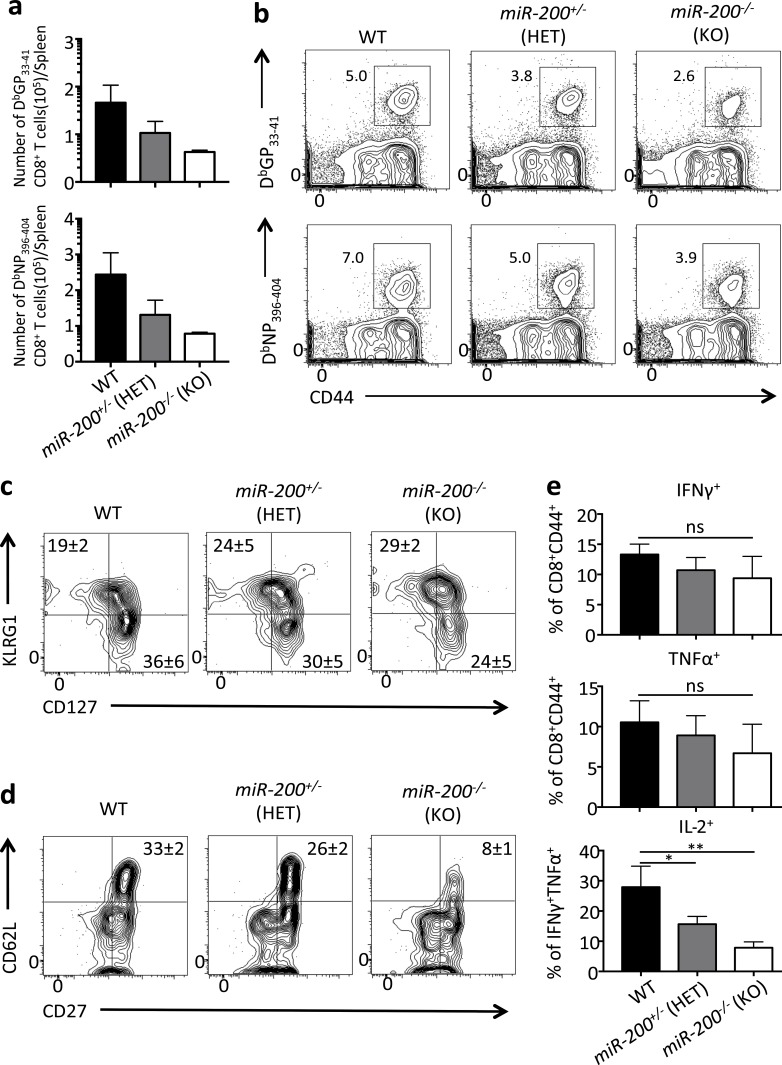
To further investigate the requirement of miR-200 for memory CD8+ T cell development, we generated a miR-200 double cluster KO mouse strain by crossing miR-200b/a/429 cluster germline KO mice (Hasuwa et al., 2013) with mice that contain flanking loxP sites around the miR-200c/141 cluster (miR-200c/141flox/flox; Cao et al., 2013) and the GzmB-Cre transgene to conditionally delete both clusters in activated CD8+ T cells. For simplicity, these mice will be referred to as miR-200−/−. We then infected WT mice (miR-200b/a/429+/+; miR-200c/141flox/flox; GzmB-Cre−) or those with heterozygous (miR-200b/a/42+/−; miR-200c/141+/flox; GzmB-Cre+) or homozygous (miR-200b/a/429−/−; miR-200c/141flox/flox; GzmB-Cre+) loss of both clusters with LCMV-Arm and analyzed effector and memory CD8+ T cell formation. We refer to these two groups of mice as miR-200+/− HET or miR-200−/− KO, respectively. This experiment revealed a dose-dependent requirement of both miR-200 clusters in the formation of virus-specific memory CD8+ T cells, because there was marked decrease in numbers and frequency of memory CD8+ T cell in miR200+/− HET mice and a further reduction in the miR200−/− KO mice (Fig. 7, a and b). Similar to the Zeb1-deficient memory CD8+ T cells, there was no gross difference in the expression of KLRG1 or IL-7R among WT, miR-200+/− HET, and miR-200−/− KO mice, suggesting that miR-200 is not required for phenotypic generation of these subsets of memory cells (Fig. 7 c); however, miR-200−/− mice exhibited a substantial decrease in the formation of CD62L+ CD27+ IL-2+ TCM cells (Fig. 7, d and e). In summary, these findings demonstrate that miR-200 members play a critical role in the development of long-lived memory CD8+ T cells, particularly TCM cells, and the loss of miR-200 family members in CD8+ T cells strongly phenocopies the loss of Zeb1. Collectively, these data indicate that the miR-200 family members temporally coordinate the reciprocal expression patterns of Zeb1 and Zeb2 during an immune response by selectively targeting Zeb2 in CD8+ T cells after effector cell development, allowing for Zeb1 re-expression and memory cell formation and persistence.
Figure 7.
miR-200 family deficiency results in the loss of memory CD8+ T cells. (a) WT (black bar), miR-200+/− (HET, gray bar), and miR-200−/− (KO, white bar) mice were infected with LCMV Arm and splenic DbGP33-41 and DbNP396-404 tetramer+ CD8+ T cells were quantitated at 45 dpi. (b) Representative contour plots of DbGP33-41 and DbNP396-404 tetramer+ CD8+ T cells at 45 dpi in WT, miR-200+/− HET, and miR-200−/− KO mice. (c and d) Representative flow cytometry data measuring KLRG1 and IL-7R expression (c) and CD62L and CD27 expression (d) in WT, miR-200+/− and miR-200−/− LCMV-specific CD8+ T cells 45 dpi. (e) WT (black bar), miR-200+/− (HET, gray bar), and miR-200−/− (KO, white bar) CD8+ T cells from 45 dpi were analyzed for IFNγ and TNFα (top two bar graphs) or IL-2 (bottom bar graph) expression using intracellular cytokine staining after a 5-h GP33–41 peptide stimulation. Note that IL-2 producing cells were gated on IFNγ+ TNFα+ CD8+ T cells. Data shown are representative of two (b–d) or cumulative of two (a and e) independent experiments; n = 2–3 mice/group/experiment (b–d), n = 5–6 mice/group (a and e). Data are expressed as mean ± SEM. *, P < 0.05; **, P < 0.01.
Discussion
During viral infection, our immune system seeks to achieve two principle outcomes: to eliminate the present invading pathogen and generate immunological memory. A deeper understanding of the molecular mechanisms regulating the formation of different types of effector and memory CD8+ T cells could propel the development of better vaccines and immunotherapies. This study identified a genetic circuit, previously unknown to function in the immune system, that regulates effector and memory CD8+ T cell fate decisions involving the TFs ZEB1 and ZEB2 and the miR-200 microRNA family, as well as the cytokine TGF-β—all critical regulators of the EMT. In particular, this is the first study of the functions of ZEB1 and miR-200 family members in CD8+ T cells and their important role in the establishment of a long-lived pool of memory CD8+ T cells and TCM differentiation.
The ZEB1, ZEB2, TGF-β–, and miR-200 network is best characterized in controlling epithelial versus mesenchymal cell fates as part of the EMT, wherein the miR-200 family promotes epithelial states and TGF-β induces both ZEB1 and ZEB2, which cooperatively drive mesenchymal states. However, in CD8+ T cells, we found that ZEB1 and ZEB2 work in opposition, possibly by inhibiting each other, in temporally distinct phases of an immune response to coordinate TE differentiation (ZEB2) and memory CD8+ T cell survival and TCM maturation (ZEB1). Moreover, our data show that in CD8+ T cells, Zeb1 and Zeb2 expression was inversely regulated by TGF-β and the miR-200 family members, outlining a novel mechanism for “splitting” the functions of ZEB1 and ZEB2 to generate alternative cell fates. These data present a working model wherein Zeb1 and miR-200 expression is repressed upon CD8+ T cell activation, allowing for the induction of Zeb2 by T-BET in cells exposed to increasing inflammation and stimulation of TE cell development. Although there is no direct binding data in CD8+ T cells, we postulate that ZEB2 operates to sustain repression of Zeb1 and miR-200 expression as TE cells form. Through TGF-β signaling, expression of Zeb1 and miR-200 is augmented in developing MP cells, which prevents Zeb2 expression and promotes the development of long-lived circulating memory CD8+ T cells, particularly TCM cells. Thus, this study identifies a new counterregulatory network in CD8+ T cells that temporally coordinates the formation of effector cells to fight present infection and memory cells to fight future infection.
There are other studies wherein ZEB1 and ZEB2 control alternative cell fate decisions in Xenopus laevis embryogenesis with ZEB1 guiding mesoderm fates and ZEB2 driving neural fates (Postigo, 2003; Postigo et al., 2003). In this context, it was found that ZEB1 and ZEB2 regulate TGF-β/BMP signaling in opposite ways wherein ZEB1 recruits transcriptional coactivators (p300 and P/CAF) and synergizes with Smad-mediated transcriptional activation, whereas ZEB2 binds to corepressors (CtBP) and suppresses downstream TGF-β signaling (Postigo, 2003; Postigo et al., 2003). Perhaps a similar scenario operates in CD8+ T cells where the presence of ZEB1 or ZEB2 alters the quality of TGF-β signaling in T cells. Our data showed that TGF-β enhanced Zeb1 while repressing Zeb2 expression in CD8+ T cells, but whether they modify downstream TGF-β signaling or TRM formation requires more exploration. Our data provide greater mechanistic insight into a recent study that demonstrated constitutive TGF-β signaling was required to maintain mature IL-7Rhi memory CD8+ T cells (Ma and Zhang, 2015); likely, this is due in part, to TGF-β–mediated maintenance of Zeb1 and miR-200 expression. Interestingly, despite increased Zeb2 mRNA in Zeb1-deficient CD8+ T cells, no overt changes in MP and TE cell differentiation was observed, indicating increased ZEB2 alone was not sufficient to repress MP cell fates. This result suggested that ZEB1 and ZEB2 do not simply compete for binding to the same gene loci with opposing effects on transcription, albeit there may be some loci for which this occurs (e.g., Il2). Rather, the data indicate that ZEB1 and ZEB2 largely drive different gene expression programs, with ZEB2 supporting TE differentiation during the naive→effector phase and ZEB1 supporting cell survival during the effector→memory transition. On this note, the expression patterns and loss-of-function phenotypes of Zeb1 and Zeb2 are highly overlapping with that of Id3 and Id2 in CD8+ T cells, respectively (Yang et al., 2011), and thus, it is tempting to speculate that functional cooperativity exists between ZEB1-ID3 and ZEB2-ID2 to control temporally and functionally distinct gene expression programs.
This study also identified a new manner by which ZEB1 and ZEB2 functions could be uncoupled through the selective actions of microRNAs. We observed that all five members of miR-200 family are expressed in naive and memory CD8+ T cells but down-regulated in effector CTLs. Ectopic expression of miR-200 family members markedly reduced TE subset formation and boosted memory CD8+ T cell maturation, phenotypes resembling a deficiency in Zeb2. Indeed, Zeb2 mRNA was diminished in CD8+ T cells overexpressing miR-200 family members, whereas Zeb1 was not. Although miR-200 family members can bind to both Zeb1 and Zeb2 3′ UTRs and repress their expression based on luciferase reporter assays in HEK 293T cells (Wang et al., 2013), in virus-specific CD8+ T cells, we detected a strong preference for miR-200 binding at the Zeb2 3′ UTR, but not the Zeb1 3′ UTR, using cross-linking immunoprecipitation assays. How Zeb1 mRNA escapes binding to miR-200 in CD8+ T cells is not clear, but several possibilities exist, the simplest being that CD8+ T cells express a Zeb1 isoform lacking the miR-200–binding sites in 3′ UTR; however, analysis of the Zeb1 transcripts in CD8+ T cells by RNA sequencing disproved this simple explanation (unpublished data). It is also possible that Zeb1 mRNA forms secondary structures in CD8+ T cells that masks the miR-200–binding sites or that other RNA-binding proteins such as HuR or microRNAs that promote mRNA stability and translation (Mukherjee et al., 2011) bind to the Zeb1 3′ UTR and prevent miR-200 recognition. Distinguishing between these mechanisms will be important to better understand the separation of ZEB1 and ZEB2 expression and function in T cells and possibly other immune cells in which they are expressed, such as CD4+ T cells, B cells, natural killer cells, dendritic cells (DCs), monocytes, and plasmacytoid DCs (Heng et al., 2008; van Helden et al., 2015). Interestingly, these data reveal that Zeb1 is predominantly expressed in CD103+ DCs from skin draining LNs, whereas Zeb2 is more highly expressed in spleen plasmacytoid DC populations (Heng et al., 2008). Thus, we postulate ZEB1 and ZEB2 may exhibit counterregulatory roles and modulate alternative cell fate decisions more broadly in the immune system.
Even though our data suggested that ZEB2 is a primary target of miR-200, clearly, miR-200 overexpression displayed a more profound phenotype compared with the Zeb2-deficient CD8+ T cells (Dominguez et al., 2015). In particular, Zeb2-deficient CD8+ T cells did not display enhanced survival of memory CD8+ T cells or diminished effector functions, whereas miR-200 overexpression affected both of these processes (Dominguez et al., 2015; Omilusik et al., 2015). These results suggest that, in addition to Zeb2, miR-200 family members regulate other genes involved in CD8+ T cell differentiation. Indeed, using microRNA-binding target prediction software, we found miR-200–binding sequences within the 3′ UTR of Prdm1 (Blimp-1), another critical pro–effector TF that suppresses memory CD8+ T cell formation and survival (Kallies et al., 2009; Rutishauser et al., 2009). Given the enhanced memory formation that resulted in the miR-200 OE, further characterization of their targets may have therapeutic implications.
Lastly, it is important to elucidate the connection between the EMT and T cell differentiation and why the ZEB1, ZEB2, TGF-β, and miR-200 network is used in both. That is, how do the cellular processes involved in the EMT relate to T cell function? One connection may be cellular motility and trafficking and induction of EMT-like processes are needed for T cell trafficking. However, we did not notice severe defects in LN egress or gross tissue infiltration of effector T cells during acute LCMV infection in either ZEB1- or ZEB2-deficient CD8+ T cells relative to their WT counterparts (Dominguez et al., 2015; Omilusik et al., 2015; unpublished data). Nonetheless, this does not rule out the possibility that this genetic network may fine-tune CD8+ T cell trafficking within tissues, for example between interstitial spaces versus epithelial linings versus the vasculature or in the skin between the dermis versus epidermis (i.e., tissue-resident vs. circulating memory T cells). In addition to TRM cells, it will be important to investigate how this network regulates the tissue surveillance of CX3CR1int peripheral memory T cells (Gerlach et al., 2016), particularly because ZEB2 is needed for CX3CR1 expression (Dominguez et al., 2015; Omilusik et al., 2015). In summary, this work identified a novel genetic circuit involving both transcriptional and posttranscriptional programs to guide T cell fate decision and provided new insights into the molecular regulation of T cell plasticity and heterogeneity, which could allow considerable improvement of vaccine and therapeutic development against infection and cancer.
Materials and methods
Mice
C57BL/6 (B6) mice were obtained from the National Cancer Institute. Zeb2flox/flox mice were originally generated by D. Hoylebroeck (University of Leuven, Leuven, Belgium; Higashi et al., 2002) and obtained from R. Aslopp (John A. Burns School of Medicine, University of Hawaii, Honolulu, HI). Granzyme B-Cre (GzB-Cre+) mice were provided by J. Jacobs (Emory University, Atlanta, GA) via R. Flavell’s laboratory (Yale University School of Medicine, New Haven, CT) and were crossed to Zeb2flox/flox mice for generation of GzB-cre+; Zeb2flox/flox (Zeb2-deficient) mice and GzB-cre+; Zeb2+/+ or GzB-cre−; Zeb2flox/flox (Zeb2-WT) mice. Zeb2f/f GzmBCre+ and Zeb2f/f GzmBCre− mice were further crossed to P14+ TCR transgenic mice so that P14+ Zeb2f/f GzmBCre+ and Zeb2f/f GzmBCre− mice could be obtained.
Zeb1flox/flox mice were generated using the CRIPSR/Cas9 technique in collaboration with R. Flavell’s laboratory under C57BL/6 (B6) mice background and were crossed to GzmBCre+ mice to generate Zeb1 conditional KO. P14+ Zeb1f/f GzmBCre+ mice were generated in the same way described above. To generate P14 chimeric mice, 10–50,000 P14+ CD8+ T cells were transferred into B6 mice by i.v. injection. Tgfbr2f/f LckCre mice were a gift from M. Bevan (University of Washington, Seattle, WA) and crossed to P14+ mice to generate P14+ Tgfbr2f/f LckCre mice. miR-200c/141f/f mice were purchased from The Jackson Laboratory and crossed to GzB-Cre+ mice to generate miR-200c/141−/− mice. miR-200b/a/429 KO mice were a gift from H. Hasuwa (Keio University, Keio, Japan). miR-200 family total KO mice were generated by crossing miR-200c/141−/− mice to miR-200b/a/429 KO mice. All animal experiments were done with approved Yale institutional animal care and use committee protocols.
Infections and treatments
For infections of mice, 2 × 105 PFU of the LCMV Armstrong strain were administered i.p. For recall experiments, mice were administered 2 × 104 CFU recombinant L. monocytogenes expressing the LCMV GP33–41 epitope.
Antibodies for surface and intracellular staining
Lymphocyte isolation, along with surface and intracellular staining, was performed as described previously (Joshi et al., 2007). For in vitro stimulation, splenocytes were stimulated with 100 ng/ml GP33–41 and NP396–404 peptides for 5 h in the presence of brefeldin A. Antibodies were purchased from eBioscience, BD Biosciences, or BioLegend and Cell Signal. Class I MHC tetramers were generated as described previously (Kaech et al., 2003). Flow cytometry data were acquired on BD LSRII with Diva software and analyzed with FlowJo software (Tree Star). Sorting was performed on a FACSAria (BD).
ChIP
ChIP experiments were performed with 10 million naive CD8+ T cells. The cells were cross-linked with 1% formaldehyde for 10 min and sonication to obtain an ∼200–500-bp DNA fragment. ChIP was performed with anti–ZEB1 antibody (Cell Signaling), and anti–mouse IgG was used as a negative control. Two independent experiments were performed. Immunoprecipitated DNA was analyzed by quantitative PCR.
Sybr-based quantitative PCR was performed with the following primers: Zeb1_Zeb2 forward, 5′-CCACATCTGGAAGTCAGCAA-3′; Zeb1_Zeb2 reverse, 5′-ACAAAACAGCAGAGCATTGG-3′; Zeb1_il2 forward, 5′-GGAGCTCCTGTAGGTCCATC-3′; Zeb1_il2 reverse, 5′-AAGCTCTACAGCGGAAGCAC-3′; Zeb2 exon 8 forward, 5′-CACCTAAGTGCTGCATTGGA-3′; and Zeb2 exon 8 reverse, 5′-TTAGTGGCAGCAGTCCCTTT-3′.
Cross-linking immunoprecipitation and quantitative RT-PCR
Cross-linking was performed using 5 million sorted LCMV-Specific CD8+ T cells 12 dpi according to a published protocol (Guo et al., 2014). RNA immunoprecipitation was performed with 10 µg pan-AGO antibody (MABE56; Millipore) and Ago2 antibody (015–22031; Wako Chemical), and quantitative RT-PCR was performed following a published protocol (Guo et al., 2015). RT reaction was performed using the Invitrogen Superscript III cDNA synthesis kit according to the manufacturer’s protocol. Sybr-based quantitative PCR was performed with the following primers: set-1 forward, 5′-GCAGTTCAGCCAAGACAGAG-3′: set-1 reverse, 5′-TGTAGTGATACATACGTAGAGTGCAA-3′; set-2 forward, 5′-CTGCAAGTGCCATCCTTGTA-3′; set-2 reverse, 5′-TGACCTAAAATTAAATGAATGCAAA-3′; set-3 forward, 5′-TTTAAAAGGTGCCCGCACTA-3′; set-3 reverse, 5′-TGCATCACTTCAAGTTCCTTCA-3′; set-4 forward, 5′-GGCAGCAGTTCCTTAGTTTACA-3′; set-4 reverse, 5′-GCCCAAATGATCAACGTCAT-3′; set-5 forward, 5′-GGCAGAATCAGTGTTCGTGA-3′; set-5 reverse, 5′-CAACAAACGAATCAACAACTGC-3′; set-6 forward, 5′-CAGTAGAGATGCAGTTGGTTCC-3′; set-6 reverse, 5′-AAAACTGGGGAAAGGGAGAA-3′; set-7 forward, 5′-AGGTTACAGGAGGCTGGATG-3′; set-7 reverse, 5′-TGCTCTGTGAAGGGAATTCTG-3′; set-8 forward, 5′-TTTGGTTCACAGCCGTTTTC-3′; and set-8 reverse, 5′-AAAAGTACGTGTCAGTAAGAAGGGTA-3′.
Retroviral transduction
Preparation of retroviral supernatants and transduction was performed as previously described (Dominguez et al., 2015). All five miR-200 family overexpression constructs were cloned by PCR-amplification of genomic DNA into the pMIRWAY-GFP vectors.
Immunoblot analysis
Protein lysates from 106 day 15 post–LCMV-infected mice were sorted on CD8+CD44+ KLRG1hi IL-7Rlo or KLRG1lo IL-7Rhi sorted effector CTLs were lysed and resolved by SDS-PAGE. ZEB1 (Cell Signaling Technology) and β-actin (Santa Cruz Biotechnology) were detected by immunoblotting.
In vitro cultures with cytokines
Naive P14+ CD8+ T cells were cultured for 72 h in the presence of 10 ng/ml GP33–41 peptide and 10 ng/ml IL-2 for 3 d followed by another 48 h culture in 10 ng/ml of IL-2, 20 ng/ml of IL-15, or 10ng/ml TGF-β (eBioscience or Peprotech). TGF-β was activated with citric acid as outlined in the product manual.
Gene expression by quantitative RT-PCR
For quantitative RT-PCR, RNA was isolated from 200,000–1,000,000 sorted cells using Qiagen RNeasy Mini kit. cDNA was synthesized using SSRTII (Life Technologies) and quantitative RT-PCR was performed on a Stratagene Mx3000P with iTaq Universal SYBR green super mix (Bio-Rad). Relative fold changes were calculated using Rpl9 (L9) expression.
The following primers were used in these studies: Zeb2 forward, 5′-GAGCAGGTAACCGCAAGTTC-3′; Zeb2 reverse, 5′-TGTTTCTCATTCGG-3′; Rpl9 forward, 5′-TGAAGAAATCTGTGGGTCG-3′; Rpl9 reverse, 5′-GCACTACGGACATAGGAACTC-3′; Zeb1 forward, 5′-CCGCCAACAAGCAGACTATT-3′; and Zeb1 reverse, 5′-GGCGTGGAGTCAGAGTCATT-3′.
For microRNA expression, sorted cells were lysed with TRIzol (Invitrogen) and RNA extraction was performed according to the manufacturer’s protocol. Taqman microRNA expression assays (for miR-200a, b, c, 429, and 141) were performed using Taqman-provided primers and following the manufacturer’s protocol.
Statistical analysis
Prism 6 (GraphPad Software) was used to calculate statistics for all bar graphs shown. For comparisons of two groups, a two-tailed t test was performed. For multiple-group comparisons, a one-way ANOVA with Tukey’s multiple comparisons test was used. For grouped multiple comparisons, a two-way ANOVA with Sidak’s multiple comparison test was used (*, P < 0.05; **, P < 0.01; ***, P < 0.001; and ****, P < 0.0001).
Online supplemental material
Fig. S1 shows the generation of Zeb1 conditional KO mice and validation of deletion. Fig. S2 shows that ZEB1 plays an intrinsic role in promoting memory CD8+ T cell survival. Fig. S3 shows that overexpression of miR-200 family promotes memory CD8+ T cell differentiation.
Supplementary Material
Acknowledgments
We thank the members of the Kaech, Craft, Flavell, and Lu laboratories for helpful comments and scientific advice in generation of Zeb1f/f mice, in particular X. Jiang, J. Stein, C. Hughes, and L. Borelli for technical support. We also thank H. Hasuwa for providing miR-200b/a/429 KO mice, M. Bevan for providing Tgfbr2f/f LckCre mice, and J. Cheng for the miR-200 overexpression constructs.
This work was supported by the National Institutes of Health (grants R01AI07469905, R01AI07469905S, and R37AI066232 to S.M. Kaech), National Institute of Allergy and Infectious Diseases (National Research Service Award, Ruth Kirschstein Predoctoral Fellowship F31AI084500-01 to C.X. Dominguez), Howard Hughes Medical Institute (to R.A. Amezquita and T. Guan), and Chinese Scholarship Council/Yale World Scholars Fellowship (to T. Guan).
The authors declare no competing financial interests.
Author contributions: T. Guan, C.X. Dominguez, and S.M. Kaech conceived and designed the experiments and analyzed data. T. Guan, C.X. Dominguez, B.J. Laidlaw, and J. Cheng performed experiments and analysis. R.A. Amezquita performed bioinformatics analysis. R.A. Flavell, J. Henao-Mejia, and A. Williams designed and performed experiments for generating Zeb1-floxed mice. J. Lu and J. Cheng assisted in miR-200 experiments. T. Guan, C.X. Dominguez, and S.M. Kaech wrote the manuscript.
References
- Banerjee A., Gordon S.M., Intlekofer A.M., Paley M.A., Mooney E.C., Lindsten T., Wherry E.J., and Reiner S.L.. 2010. Cutting edge: The transcription factor eomesodermin enables CD8+ T cells to compete for the memory cell niche. J. Immunol. 185:4988–4992. 10.4049/jimmunol.1002042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker T.C., Wherry E.J., Boone D., Murali-Krishna K., Antia R., Ma A., and Ahmed R.. 2002. Interleukin 15 is required for proliferative renewal of virus-specific memory CD8 T cells. J. Exp. Med. 195:1541–1548. 10.1084/jem.20020369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brabletz S., and Brabletz T.. 2010. The ZEB/miR-200 feedback loop--a motor of cellular plasticity in development and cancer? EMBO Rep. 11:670–677. 10.1038/embor.2010.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brabletz S., Bajdak K., Meidhof S., Burk U., Niedermann G., Firat E., Wellner U., Dimmler A., Faller G., Schubert J., and Brabletz T.. 2011. The ZEB1/miR-200 feedback loop controls Notch signalling in cancer cells. EMBO J. 30:770–782. 10.1038/emboj.2010.349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bracken C.P., Gregory P.A., Kolesnikoff N., Bert A.G., Wang J., Shannon M.F., and Goodall G.J.. 2008. A double-negative feedback loop between ZEB1-SIP1 and the microRNA-200 family regulates epithelial-mesenchymal transition. Cancer Res. 68:7846–7854. 10.1158/0008-5472.CAN-08-1942 [DOI] [PubMed] [Google Scholar]
- Cannarile M.A., Lind N.A., Rivera R., Sheridan A.D., Camfield K.A., Wu B.B., Cheung K.P., Ding Z., and Goldrath A.W.. 2006. Transcriptional regulator Id2 mediates CD8+ T cell immunity. Nat. Immunol. 7:1317–1325. 10.1038/ni1403 [DOI] [PubMed] [Google Scholar]
- Cao H., Jheon A., Li X., Sun Z., Wang J., Florez S., Zhang Z., McManus M.T., Klein O.D., and Amendt B.A.. 2013. The Pitx2:miR-200c/141:noggin pathway regulates Bmp signaling and ameloblast differentiation. Development. 140:3348–3359. 10.1242/dev.089193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casey K.A., Fraser K.A., Schenkel J.M., Moran A., Abt M.C., Beura L.K., Lucas P.J., Artis D., Wherry E.J., Hogquist K., et al. 2012. Antigen-independent differentiation and maintenance of effector-like resident memory T cells in tissues. J. Immunol. 188:4866–4875. 10.4049/jimmunol.1200402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang J.T., Palanivel V.R., Kinjyo I., Schambach F., Intlekofer A.M., Banerjee A., Longworth S.A., Vinup K.E., Mrass P., Oliaro J., et al. 2007. Asymmetric T lymphocyte division in the initiation of adaptive immune responses. Science. 315:1687–1691. 10.1126/science.1139393 [DOI] [PubMed] [Google Scholar]
- Comijn J., Berx G., Vermassen P., Verschueren K., van Grunsven L., Bruyneel E., Mareel M., Huylebroeck D., and van Roy F.. 2001. The two-handed E box binding zinc finger protein SIP1 downregulates E-cadherin and induces invasion. Mol. Cell. 7:1267–1278. 10.1016/S1097-2765(01)00260-X [DOI] [PubMed] [Google Scholar]
- Cruz-Guilloty F., Pipkin M.E., Djuretic I.M., Levanon D., Lotem J., Lichtenheld M.G., Groner Y., and Rao A.. 2009. Runx3 and T-box proteins cooperate to establish the transcriptional program of effector CTLs. J. Exp. Med. 206:51–59. 10.1084/jem.20081242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez C.X., Amezquita R.A., Guan T., Marshall H.D., Joshi N.S., Kleinstein S.H., and Kaech S.M.. 2015. The transcription factors ZEB2 and T-bet cooperate to program cytotoxic T cell terminal differentiation in response to LCMV viral infection. J. Exp. Med. 212:2041–2056. 10.1084/jem.20150186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eger A., Aigner K., Sonderegger S., Dampier B., Oehler S., Schreiber M., Berx G., Cano A., Beug H., and Foisner R.. 2005. DeltaEF1 is a transcriptional repressor of E-cadherin and regulates epithelial plasticity in breast cancer cells. Oncogene. 24:2375–2385. 10.1038/sj.onc.1208429 [DOI] [PubMed] [Google Scholar]
- Gerlach C., Moseman E.A., Loughhead S.M., Alvarez D., Zwijnenburg A.J., Waanders L., Garg R., de la Torre J.C., and von Andrian U.H.. 2016. The Chemokine Receptor CX3CR1 Defines Three Antigen-Experienced CD8 T Cell Subsets with Distinct Roles in Immune Surveillance and Homeostasis. Immunity. 45:1270–1284. 10.1016/j.immuni.2016.10.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray S.M., Amezquita R.A., Guan T., Kleinstein S.H., and Kaech S.M.. 2017. Polycomb Repressive Complex 2-Mediated Chromatin Repression Guides Effector CD8+ T Cell Terminal Differentiation and Loss of Multipotency. Immunity. 46:596–608. 10.1016/j.immuni.2017.03.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory P.A., Bracken C.P., Smith E., Bert A.G., Wright J.A., Roslan S., Morris M., Wyatt L., Farshid G., Lim Y.Y., et al. 2011. An autocrine TGF-beta/ZEB/miR-200 signaling network regulates establishment and maintenance of epithelial-mesenchymal transition. Mol. Biol. Cell. 22:1686–1698. 10.1091/mbc.E11-02-0103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y.E., Riley K.J., Iwasaki A., and Steitz J.A.. 2014. Alternative capture of noncoding RNAs or protein-coding genes by herpesviruses to alter host T cell function. Mol. Cell. 54:67–79. 10.1016/j.molcel.2014.03.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y., Liu J., Elfenbein S.J., Ma Y., Zhong M., Qiu C., Ding Y., and Lu J.. 2015. Characterization of the mammalian miRNA turnover landscape. Nucleic Acids Res. 43:2326–2341. 10.1093/nar/gkv057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasuwa H., Ueda J., Ikawa M., and Okabe M.. 2013. miR-200b and miR-429 function in mouse ovulation and are essential for female fertility. Science. 341:71–73. 10.1126/science.1237999 [DOI] [PubMed] [Google Scholar]
- Heng T.S.P., and Painter M.W.. Immunological Genome Project Consortium . 2008. The Immunological Genome Project: networks of gene expression in immune cells. Nat. Immunol. 9:1091–1094. 10.1038/ni1008-1091 [DOI] [PubMed] [Google Scholar]
- Higashi Y., Maruhashi M., Nelles L., Van de Putte T., Verschueren K., Miyoshi T., Yoshimoto A., Kondoh H., and Huylebroeck D.. 2002. Generation of the floxed allele of the SIP1 (Smad-interacting protein 1) gene for Cre-mediated conditional knockout in the mouse. Genesis. 32:82–84. 10.1002/gene.10048 [DOI] [PubMed] [Google Scholar]
- Huster K.M., Busch V., Schiemann M., Linkemann K., Kerksiek K.M., Wagner H., and Busch D.H.. 2004. Selective expression of IL-7 receptor on memory T cells identifies early CD40L-dependent generation of distinct CD8+ memory T cell subsets. Proc. Natl. Acad. Sci. USA. 101:5610–5615. 10.1073/pnas.0308054101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichii H., Sakamoto A., Kuroda Y., and Tokuhisa T.. 2004. Bcl6 acts as an amplifier for the generation and proliferative capacity of central memory CD8+ T cells. J. Immunol. 173:883–891. 10.4049/jimmunol.173.2.883 [DOI] [PubMed] [Google Scholar]
- Intlekofer A.M., Takemoto N., Wherry E.J., Longworth S.A., Northrup J.T., Palanivel V.R., Mullen A.C., Gasink C.R., Kaech S.M., Miller J.D., et al. 2005. Effector and memory CD8+ T cell fate coupled by T-bet and eomesodermin. Nat. Immunol. 6:1236–1244. 10.1038/ni1268 [DOI] [PubMed] [Google Scholar]
- Intlekofer A.M., Banerjee A., Takemoto N., Gordon S.M., Dejong C.S., Shin H., Hunter C.A., Wherry E.J., Lindsten T., and Reiner S.L.. 2008. Anomalous type 17 response to viral infection by CD8+ T cells lacking T-bet and eomesodermin. Science. 321:408–411. 10.1126/science.1159806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jameson S.C., and Masopust D.. 2009. Diversity in T cell memory: an embarrassment of riches. Immunity. 31:859–871. 10.1016/j.immuni.2009.11.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeannet G., Boudousquié C., Gardiol N., Kang J., Huelsken J., and Held W.. 2010. Essential role of the Wnt pathway effector Tcf-1 for the establishment of functional CD8 T cell memory. Proc. Natl. Acad. Sci. USA. 107:9777–9782. 10.1073/pnas.0914127107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji Y., Pos Z., Rao M., Klebanoff C.A., Yu Z., Sukumar M., Reger R.N., Palmer D.C., Borman Z.A., Muranski P., et al. 2011. Repression of the DNA-binding inhibitor Id3 by Blimp-1 limits the formation of memory CD8+ T cells. Nat. Immunol. 12:1230–1237. 10.1038/ni.2153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi N.S., Cui W., Chandele A., Lee H.K., Urso D.R., Hagman J., Gapin L., and Kaech S.M.. 2007. Inflammation directs memory precursor and short-lived effector CD8(+) T cell fates via the graded expression of T-bet transcription factor. Immunity. 27:281–295. 10.1016/j.immuni.2007.07.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaech S.M., and Cui W.. 2012. Transcriptional control of effector and memory CD8+ T cell differentiation. Nat. Rev. Immunol. 12:749–761. 10.1038/nri3307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaech S.M., Tan J.T., Wherry E.J., Konieczny B.T., Surh C.D., and Ahmed R.. 2003. Selective expression of the interleukin 7 receptor identifies effector CD8 T cells that give rise to long-lived memory cells. Nat. Immunol. 4:1191–1198. 10.1038/ni1009 [DOI] [PubMed] [Google Scholar]
- Kakaradov B., Arsenio J., Widjaja C.E., He Z., Aigner S., Metz P.J., Yu B., Wehrens E.J., Lopez J., Kim S.H., et al. 2017. Early transcriptional and epigenetic regulation of CD8+ T cell differentiation revealed by single-cell RNA sequencing. Nat. Immunol. 18:422–432. 10.1038/ni.3688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalia V., Sarkar S., Subramaniam S., Haining W.N., Smith K.A., and Ahmed R.. 2010. Prolonged interleukin-2Ralpha expression on virus-specific CD8+ T cells favors terminal-effector differentiation in vivo. Immunity. 32:91–103. 10.1016/j.immuni.2009.11.010 [DOI] [PubMed] [Google Scholar]
- Kallies A., Xin A., Belz G.T., and Nutt S.L.. 2009. Blimp-1 transcription factor is required for the differentiation of effector CD8(+) T cells and memory responses. Immunity. 31:283–295. 10.1016/j.immuni.2009.06.021 [DOI] [PubMed] [Google Scholar]
- Kim M.V., Ouyang W., Liao W., Zhang M.Q., and Li M.O.. 2013. The transcription factor Foxo1 controls central-memory CD8+ T cell responses to infection. Immunity. 39:286–297. 10.1016/j.immuni.2013.07.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knell J., Best J.A., Lind N.A., Yang E., D’Cruz L.M., and Goldrath A.W.. 2013. Id2 influences differentiation of killer cell lectin-like receptor G1(hi) short-lived CD8+ effector T cells. J. Immunol. 190:1501–1509. 10.4049/jimmunol.1200750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon H., Thierry-Mieg D., Thierry-Mieg J., Kim H.-P., Oh J., Tunyaplin C., Carotta S., Donovan C.E., Goldman M.L., Tailor P., et al. 2009. Analysis of interleukin-21-induced Prdm1 gene regulation reveals functional cooperation of STAT3 and IRF4 transcription factors. Immunity. 31:941–952. 10.1016/j.immuni.2009.10.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y.-T., Suarez-Ramirez J.E., Wu T., Redman J.M., Bouchard K., Hadley G.A., and Cauley L.S.. 2011. Environmental and antigen receptor-derived signals support sustained surveillance of the lungs by pathogen-specific cytotoxic T lymphocytes. J. Virol. 85:4085–4094. 10.1128/JVI.02493-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma C., and Zhang N.. 2015. Transforming growth factor-β signaling is constantly shaping memory T-cell population. Proc. Natl. Acad. Sci. USA. 112:11013–11017. 10.1073/pnas.1510119112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay L.K., and Kallies A.. 2017. Transcriptional Regulation of Tissue-Resident Lymphocytes. Trends Immunol. 38:94–103. 10.1016/j.it.2016.11.004 [DOI] [PubMed] [Google Scholar]
- Mackay L.K., Rahimpour A., Ma J.Z., Collins N., Stock A.T., Hafon M.-L., Vega-Ramos J., Lauzurica P., Mueller S.N., Stefanovic T., et al. 2013. The developmental pathway for CD103(+)CD8+ tissue-resident memory T cells of skin. Nat. Immunol. 14:1294–1301. 10.1038/ni.2744 [DOI] [PubMed] [Google Scholar]
- Mackay L.K., Minnich M., Kragten N.A., Liao Y., Nota B., Seillet C., Zaid A., Man K., Preston S., Freestone D., et al. 2016. Hobit and Blimp1 instruct a universal transcriptional program of tissue residency in lymphocytes. Science. 352:459–463. 10.1126/science.aad2035 [DOI] [PubMed] [Google Scholar]
- Mongroo P.S., and Rustgi A.K.. 2010. The role of the miR-200 family in epithelial-mesenchymal transition. Cancer Biol. Ther. 10:219–222. 10.4161/cbt.10.3.12548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee N., Corcoran D.L., Nusbaum J.D., Reid D.W., Georgiev S., Hafner M., Ascano M. Jr., Tuschl T., Ohler U., and Keene J.D.. 2011. Integrative regulatory mapping indicates that the RNA-binding protein HuR couples pre-mRNA processing and mRNA stability. Mol. Cell. 43:327–339. 10.1016/j.molcel.2011.06.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson J.A., McDonald-Hyman C., Jameson S.C., and Hamilton S.E.. 2013. Effector-like CD8+ T cells in the memory population mediate potent protective immunity. Immunity. 38:1250–1260. 10.1016/j.immuni.2013.05.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omilusik K.D., Best J.A., Yu B., Goossens S., Weidemann A., Nguyen J.V., Seuntjens E., Stryjewska A., Zweier C., Roychoudhuri R., et al. 2015. Transcriptional repressor ZEB2 promotes terminal differentiation of CD8+ effector and memory T cell populations during infection. J. Exp. Med. 212:2027–2039. 10.1084/jem.20150194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearce E.L., Walsh M.C., Cejas P.J., Harms G.M., Shen H., Wang L.-S., Jones R.G., and Choi Y.. 2009. Enhancing CD8 T-cell memory by modulating fatty acid metabolism. Nature. 460:103–107. 10.1038/nature08097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pipkin M.E., Sacks J.A., Cruz-Guilloty F., Lichtenheld M.G., Bevan M.J., and Rao A.. 2010. Interleukin-2 and inflammation induce distinct transcriptional programs that promote the differentiation of effector cytolytic T cells. Immunity. 32:79–90. 10.1016/j.immuni.2009.11.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postigo A.A. 2003. Opposing functions of ZEB proteins in the regulation of the TGFbeta/BMP signaling pathway. EMBO J. 22:2443–2452. 10.1093/emboj/cdg225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postigo A.A., Depp J.L., Taylor J.J., and Kroll K.L.. 2003. Regulation of Smad signaling through a differential recruitment of coactivators and corepressors by ZEB proteins. EMBO J. 22:2453–2462. 10.1093/emboj/cdg226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulendran B., and Ahmed R.. 2011. Immunological mechanisms of vaccination. Nat. Immunol. 12:509–517. 10.1038/ni.2039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roychoudhuri R., Clever D., Li P., Wakabayashi Y., Quinn K.M., Klebanoff C.A., Ji Y., Sukumar M., Eil R.L., Yu Z., et al. 2016. BACH2 regulates CD8(+) T cell differentiation by controlling access of AP-1 factors to enhancers. Nat. Immunol. 17:851–860. 10.1038/ni.3441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutishauser R.L., Martins G.A., Kalachikov S., Chandele A., Parish I.A., Meffre E., Jacob J., Calame K., and Kaech S.M.. 2009. Transcriptional repressor Blimp-1 promotes CD8(+) T cell terminal differentiation and represses the acquisition of central memory T cell properties. Immunity. 31:296–308. 10.1016/j.immuni.2009.05.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sallusto F., Lenig D., Förster R., Lipp M., and Lanzavecchia A.. 1999. Two subsets of memory T lymphocytes with distinct homing potentials and effector functions. Nature. 401:708–712. 10.1038/44385 [DOI] [PubMed] [Google Scholar]
- Tan J.T., Ernst B., Kieper W.C., LeRoy E., Sprent J., and Surh C.D.. 2002. Interleukin (IL)-15 and IL-7 jointly regulate homeostatic proliferation of memory phenotype CD8+ cells but are not required for memory phenotype CD4+ cells. J. Exp. Med. 195:1523–1532. 10.1084/jem.20020066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tejera M.M., Kim E.H., Sullivan J.A., Plisch E.H., and Suresh M.. 2013. FoxO1 controls effector-to-memory transition and maintenance of functional CD8 T cell memory. J. Immunol. 191:187–199. 10.4049/jimmunol.1300331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Helden M.J., Goossens S., Daussy C., Mathieu A.-L., Faure F., Marçais A., Vandamme N., Farla N., Mayol K., Viel S., et al. 2015. Terminal NK cell maturation is controlled by concerted actions of T-bet and Zeb2 and is essential for melanoma rejection. J. Exp. Med. 212:2015–2025. 10.1084/jem.20150809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G., Guo X., Hong W., Liu Q., Wei T., Lu C., Gao L., Ye D., Zhou Y., Chen J., et al. 2013. Critical regulation of miR-200/ZEB2 pathway in Oct4/Sox2-induced mesenchymal-to-epithelial transition and induced pluripotent stem cell generation. Proc. Natl. Acad. Sci. USA. 110:2858–2863. 10.1073/pnas.1212769110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Lee S., Teh C.E.Y., Bunting K., Ma L., and Shannon M.F.. 2009. The transcription repressor, ZEB1, cooperates with CtBP2 and HDAC1 to suppress IL-2 gene activation in T cells. Int. Immunol. 21:227–235. 10.1093/intimm/dxn143 [DOI] [PubMed] [Google Scholar]
- Wherry E.J., Teichgräber V., Becker T.C., Masopust D., Kaech S.M., Antia R., von Andrian U.H., and Ahmed R.. 2003. Lineage relationship and protective immunity of memory CD8 T cell subsets. Nat. Immunol. 4:225–234. 10.1038/ni889 [DOI] [PubMed] [Google Scholar]
- Williams M.A., Tyznik A.J., and Bevan M.J.. 2006. Interleukin-2 signals during priming are required for secondary expansion of CD8+ memory T cells. Nature. 441:890–893. 10.1038/nature04790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams T.M., Moolten D., Burlein J., Romano J., Bhaerman R., Godillot A., Mellon M., Rauscher F.J. III, and Kant J.A.. 1991. Identification of a zinc finger protein that inhibits IL-2 gene expression. Science. 254:1791–1794. 10.1126/science.1840704 [DOI] [PubMed] [Google Scholar]
- Xin A., Masson F., Liao Y., Preston S., Guan T., Gloury R., Olshansky M., Lin J.-X., Li P., Speed T.P., et al. 2016. A molecular threshold for effector CD8(+) T cell differentiation controlled by transcription factors Blimp-1 and T-bet. Nat. Immunol. 17:422–432. 10.1038/ni.3410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C.Y., Best J.A., Knell J., Yang E., Sheridan A.D., Jesionek A.K., Li H.S., Rivera R.R., Lind K.C., D’Cruz L.M., et al. 2011. The transcriptional regulators Id2 and Id3 control the formation of distinct memory CD8+ T cell subsets. Nat. Immunol. 12:1221–1229. 10.1038/ni.2158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu B., Zhang K., Milner J.J., Toma C., Chen R., Scott-Browne J.P., Pereira R.M., Crotty S., Chang J.T., Pipkin M.E., et al. 2017. Epigenetic landscapes reveal transcription factors that regulate CD8+ T cell differentiation. Nat. Immunol. 18:573–582. 10.1038/ni.3706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X., Yu S., Zhao D.-M., Harty J.T., Badovinac V.P., and Xue H.-H.. 2010. Differentiation and persistence of memory CD8(+) T cells depend on T cell factor 1. Immunity. 33:229–240. 10.1016/j.immuni.2010.08.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.