Centriole duplication is tightly regulated throughout the cell cycle to ensure one duplication event per centriole. McLamarrah et al. show that a stepwise pattern of Ana2 phosphorylation by Plk4 facilitates proper centriole duplication.
Abstract
Polo-like kinase 4 (Plk4) initiates an early step in centriole assembly by phosphorylating Ana2/STIL, a structural component of the procentriole. Here, we show that Plk4 binding to the central coiled-coil (CC) of Ana2 is a conserved event involving Polo-box 3 and a previously unidentified putative CC located adjacent to the kinase domain. Ana2 is then phosphorylated along its length. Previous studies showed that Plk4 phosphorylates the C-terminal STil/ANa2 (STAN) domain of Ana2/STIL, triggering binding and recruitment of the cartwheel protein Sas6 to the procentriole assembly site. However, the physiological relevance of N-terminal phosphorylation was unknown. We found that Plk4 first phosphorylates the extreme N terminus of Ana2, which is critical for subsequent STAN domain modification. Phosphorylation of the central region then breaks the Plk4–Ana2 interaction. This phosphorylation pattern is important for centriole assembly and integrity because replacement of endogenous Ana2 with phospho-Ana2 mutants disrupts distinct steps in Ana2 function and inhibits centriole duplication.
Introduction
Centriole duplication begins with the formation of a procentriole, which assembles orthogonally from the proximal end of a parent centriole (Fu et al., 2015). Procentriole assembly involves the hierarchical recruitment of a conserved set of proteins, SPD-2/DSpd-2/Cep192, Plk4/ZYG-1, SAS-5/Ana2/STIL, Sas6, and Sas4/CPAP, to a single assembly site on the mother centriole (Avidor-Reiss and Gopalakrishnan, 2013). In addition, the activity of the regulatory kinase, Polo-like kinase 4 (Plk4), is essential for centriole assembly, and Plk4 overexpression induces not only amplification from preexisting centrioles but also de novo assembly (Bettencourt-Dias et al., 2005; Habedanck et al., 2005; Kleylein-Sohn et al., 2007; Peel et al., 2007; Rodrigues-Martins et al., 2007; Holland et al., 2010; Lopes et al., 2015). Characterizing Plk4 regulation of specific substrates is key to understanding centriole biogenesis.
Drosophila melanogaster anastral spindle 2 (Ana2; STIL in humans) is an essential core centriole protein that follows Plk4 to the procentriole assembly site and contains an N-terminal (NT) Sas4-binding site, a central coiled-coil (CC), and a C-terminal (CT) STil/ANa2 (STAN) domain (Fig. S1 A; Goshima et al., 2007; Stevens et al., 2010a; Tang et al., 2011; Cottee et al., 2013; Hatzopoulos et al., 2013). Ana2 tetramerizes through the CC and binds Sas6, a rod-shaped protein, through the STAN domain (Stevens et al., 2010a; Shimanovskaya et al., 2013; Dzhindzhev et al., 2014; Slevin et al., 2014; Cottee et al., 2015). Ana2 tetramers may bind and facilitate Sas6 assembly into rings on the mother centriole’s surface, ultimately creating the stack of Sas6 rings that form the cartwheel of the nascent procentriole (Stevens et al., 2010b; Guichard et al., 2012; Dzhindzhev et al., 2014; Cottee et al., 2015; Moyer et al., 2015; Rogala et al., 2015).
Plk4 extensively phosphorylates Ana2/STIL, especially the STAN domain, which promotes Sas6 binding (Dzhindzhev et al., 2014; Ohta et al., 2014; Kratz et al., 2015; Moyer et al., 2015), a critical event that is required for Sas6 recruitment to procentrioles. In addition, Plk4 phosphorylates several upstream residues in Ana2/STIL with unknown functional consequences (Dzhindzhev et al., 2014; Ohta et al., 2014; Kratz et al., 2015). Plk4 also binds the CC domain in STIL (Ohta et al., 2014; Kratz et al., 2015; Moyer et al., 2015), using Plk4’s Polo-box 3 (PB3) domain and an obscure second site located within its linker (L1) region (Arquint et al., 2015). Current models suggest that Ana2 is recruited to centrioles through its interaction with Plk4, as CC deletion mutants prevent Plk4 binding and centriole targeting (Ohta et al., 2014; Arquint et al., 2015; Cottee et al., 2015; Moyer et al., 2015). Our results here suggest that Ana2 binds Plk4 at two distinct sites. In turn, Plk4 phosphorylates clusters of Ana2 residues in an ordered pattern, and these modifications are critical for centriole assembly and integrity in cells.
Results
Ana2 interacts with two distinct regions of Plk4
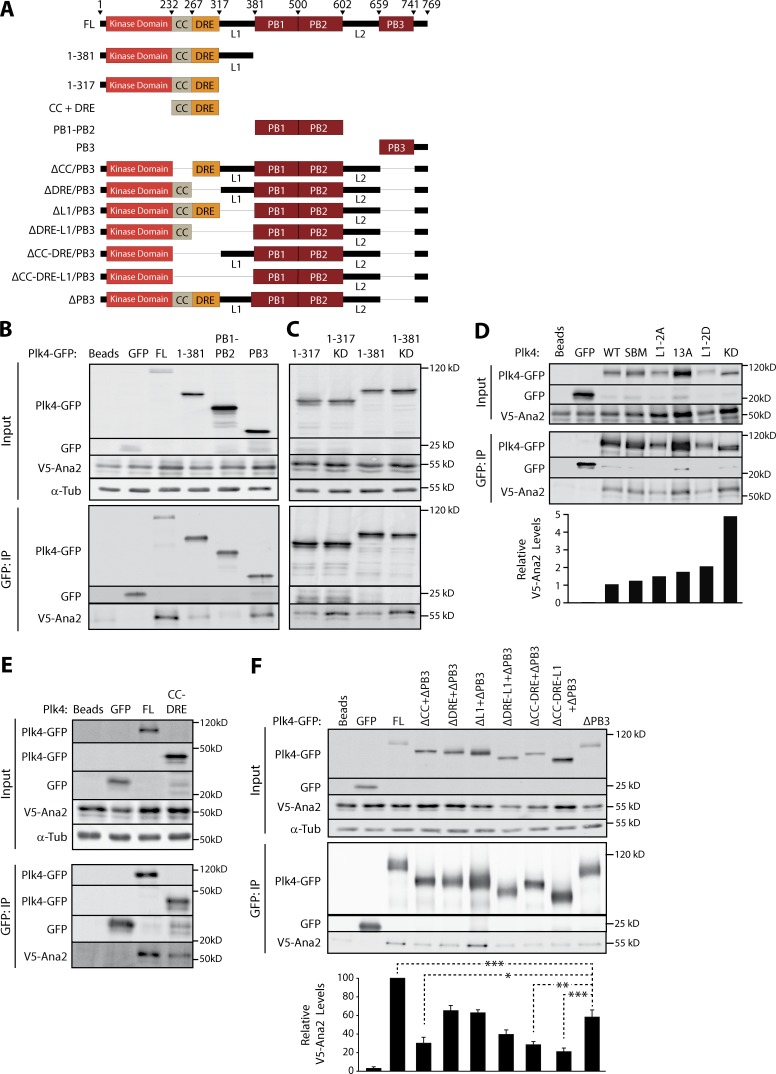
To test whether Plk4 and Ana2 binding occurs in Drosophila, we coexpressed transgenic V5-Ana2 and Plk4-GFP in S2 cells depleted of endogenous Ana2 (Fig. S1, B and C). Because Ana2 oligomerizes, we eliminated the influence of endogenous Ana2 on the binding assay by targeting its UTR with RNAi. Whereas full-length (FL) Ana2 coimmunoprecipitated with Plk4, Ana2 lacking the CC (ΔCC) did not (Fig. S1 D). Thus, the Plk4–Ana2 interaction is conserved in flies and requires the Ana2-CC.
Plk4 contains several functional domains (Klebba et al., 2015a): an NT kinase domain followed by the downstream regulatory element (DRE), and three Polo-boxes (PB1–3) interrupted by linkers L1 and L2 (Fig. 1 A). Previous efforts to map the STIL binding domain in human Plk4 have generated conflicting results and implicate multiple sites of interaction (Ohta et al., 2014; Arquint et al., 2015). We performed coimmunoprecipitation (coIP) experiments using V5-Ana2 with either FL or truncated Plk4-GFP proteins to map Ana2-binding sites (Fig. 1 A). Ana2 associates with Plk4-FL and PB3 but not PB1 or 2 (Fig. 1 B), in agreement with Arquint et al. (2015). Ana2 also weakly coimmunoprecipitates with Plk4 1–381, which lacks PBs. Yeast two-hybrid (Y2H) analysis confirmed these results: Ana2 interacts with 1–381 and PB3, but not PB1 or 2 (Fig. S1 E). Therefore, in contrast to a recent in vitro study indicating that Ana2 does not bind PB3 (Cottee et al., 2017), our results suggest that Ana2 associates with PB3 as well as an NT region restricted to 1–381. The possibility that Plk4 kinase activity regulates its interaction with STIL has been investigated but remains unresolved (Ohta et al., 2014; Moyer et al., 2015). We examined this in S2 cells by coexpressing Ana2 with either WT or kinase-dead (KD) Plk4 truncation fragments 1–317 and 1–381 and then evaluating their interactions by coIP. Consistently, Ana2 associated more with inactive Plk4-KD than WT (Fig. 1 C), suggesting that kinase activity suppresses this interaction in Drosophila. Furthermore, when combined with our finding that purified Plk4 1–317 binds GST-Ana2 using an in vitro pull-down assay (Fig. S1 F), these results also demonstrate that L1 is not necessary for Ana2 binding.
Figure 1.
Ana2 binds both the putative CC-DRE and PB3 domains in Plk4. (A) Plk4 constructs used in IP experiments. L1 and L2, linkers. (B) Ana2 associates with Plk4 NT (1–381) and CT (PB3). S2 cells were cotransfected with Plk4-GFP (FL or deletion constructs) and V5-Ana2. Anti-GFP IPs were performed from lysates of the transfected cells and probed for GFP, V5, and α-tubulin. (C) L1 is not necessary to associate with Ana2, and Ana2 interacts preferentially with KD Plk4. IPs performed as in B. (D) Ana2-Plk4 binding is greatest when the kinase is inactive (KD). FL Plk4 constructs evaluated were WT; SBM, active nondegradable mutant; L1-2A and 2PM, contain two Plk4-targeted L1 serines mutated to alanines (2A) or aspartates (2PM); and 13A, contains 13 Plk4-targeted DRE residues mutated to alanines. Each relative Ana2 level measured using the integrated intensity of a bound Ana2 band relative to its corresponding immunoprecipitated Plk4-GFP band. Graph shows relative Ana2 levels normalized to the WT result. (E) The Plk4 CC-DRE domain is sufficient to associate with Ana2. IPs performed as in B. (F) Ana2 associates with the putative CC and PB3 regions of Plk4. Graph depicts relative intensity of V5-Ana2 normalized to Plk4-GFP in the IP. Asterisks mark significant differences between treatments: *, P < 0.05; **, P < 0.01; ***, P < 0.001. Error bars, SEM; n = 3 independent experiments.
One possible explanation for an enhanced interaction of Ana2 with Plk4-KD is that Plk4 autophosphorylation prevents Ana2 binding. To test this, we mutated residues in the DRE and L1 regions known to be autophosphorylated (Cunha-Ferreira et al., 2013; Klebba et al., 2013, 2015a) and examined Ana2 association by immunoprecipitation (IP). Ana2 coimmunoprecipitated with both WT-Plk4 and a nondegradable Slimb-binding mutant (SBM; Fig. 1 D). Similar levels of Ana2 associated with Plk4 when 13 serines in the DRE were mutated to nonphosphorylatable alanines (13A), as well as when two serines of L1 were mutated to either nonphosphorylatable alanine or phosphomimetic (PM) aspartic acid (L1-2A/2D). Notably, Ana2 interacted with FL inactive Plk4-KD at approximately fivefold higher levels than WT, indicating that Plk4 kinase activity disrupts Ana2 association, and that this effect is not caused by autophosphorylation of the DRE or L1.
Using protein structure prediction software, we identified a conserved, previously uncharacterized CC within Plk4(1–317), immediately adjacent to the kinase domain (Fig. S1, G and H). To test the role of the putative Plk4-CC in Ana2 binding, we performed coIPs from S2 cells expressing Plk4 constructs lacking PB3 and one or more NT modules (Fig. 1 F). Ana2 association was decreased approximately twofold by deletion of PB3 and was significantly reduced a further approximately twofold by deletion of both the Plk4 putative CC and PB3 regions. Combined with the observation that Ana2 associates with a Plk4 fragment consisting of only CC-DRE (Fig. 1 E), this result pinpoints the putative CC of Plk4 as the other Ana2-binding domain besides PB3, although we cannot rule out additional Ana2-binding domains.
Plk4 phosphorylation of Ana2 residues upstream of the STAN domain is important for centriole duplication
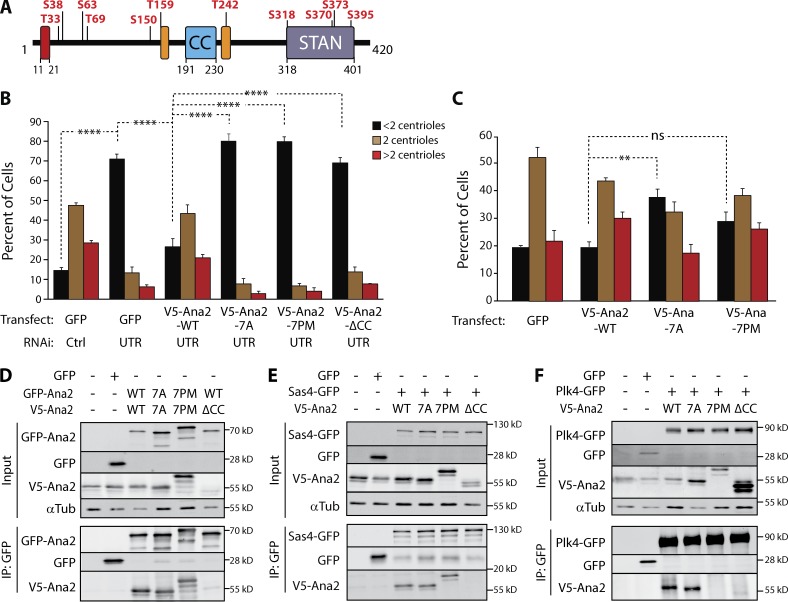
Previous studies show that Ana2/STIL is phosphorylated by Plk4 and that modification of the STAN domain promotes Sas6 binding (Dzhindzhev et al., 2014; Ohta et al., 2014; Kratz et al., 2015; Moyer et al., 2015). We used tandem mass spectrometry (MS/MS) to identify phosphorylated residues of Ana2 incubated with Plk4 in vitro. Plk4 phosphorylated Ana2 on 11 residues (Fig. 2 A and Table S1): four residues within the STAN domain and seven upstream residues that displayed varying conservation across phyla (Fig. S2 A). Plk4 is not a highly promiscuous kinase; e.g., it does not phosphorylate Sas6 (Fig. S2 B; Dzhindzhev et al., 2014). Notably, seven of the residues we mapped were previously shown to be either phosphorylated in cells coexpressing Plk4 or identified from samples mixed with Plk4 in vitro (Dzhindzhev et al., 2014). We also examined in vivo phosphorylation by isolating transgenic GFP-Ana2 from asynchronous S2 cells and performing MS/MS (Table S1). Although our peptide coverage lacked aa 1–65, we found that Ana2 is phosphorylated on numerous residues, including five residues also identified in vitro (Fig. S2 C).
Figure 2.
Ana2 is a Plk4 substrate; expression of phospho-Ana2 mutants inhibits centriole duplication. (A) Linear map of Ana2 depicting the Plk4 phosphorylation sites. Phospho-sites were identified by tandem MS of purified GST-Ana2 phosphorylated in vitro by purified Plk4 1–317-His6. Red box, Sas4-binding site (Cottee et al., 2013); orange boxes, LC8-binding sites (Slevin et al., 2014). (B) NT phosphorylation mutations in Ana2 disrupt centriole duplication. On days 0, 4, and 8, S2 cells were treated with control dsRNA or depleted of endogenous Ana2 by RNAi (UTR). On days 4 and 8, cells were transfected with the indicated V5-Ana2 or GFP construct and then induced to express with 0.1 mM CuSO4. Cells were immunostained for PLP and Asterless to mark centrioles, and the number of centrioles per cell was counted. n = 100 cells in each of three experiments. Asterisks indicate significant differences: **, P < 0.01; ****, P < 0.0001; ns, not significant. Error bars, SEM. (C) Expression of nonphosphorylatable Ana2-7A acts as a dominant/negative and inhibits centriole amplification. V5-Ana2 or GFP expression constructs were transfected on days 0 and 4 and induced with 0.5 mM CuSO4. Cells were fixed and immunostained on day 7. The mean percentages of cells containing the indicated number of centrioles are shown (n = 100 cells in each of three experiments). Error bars, SEM. Asterisks indicate significant difference: **, P < 0.01; ns, not significant. (D–F) The phosphorylation state of the Ana2 NT does not affect Ana2 binding to itself (D) or Sas4 (E). However, PM mutations in Ana2 (7PM) inhibit Plk4 binding, whereas Plk4 binding is not affected with nonphosphorylatable Ana2 (7A; F). In contrast, the CC of Ana2 is required for Ana2 self-association (D) and binding to Sas4 and Plk4 (E and F). S2 cells were depleted of endogenous Ana2 by RNAi for 7 d. On day 5, cells were cotransfected with the indicated constructs and the next day induced to express for 24 h. Anti-GFP IPs were then prepared from lysates, and Western blots of the inputs and IPs were probed for GFP, V5, and α-tubulin.
Because the physiological consequence of Ana2/STIL phosphorylation outside the STAN domain is unknown, we focused on the seven upstream phospho-residues (T33/S38/S63/T69/S150/T159/T242) that we mapped in vitro and examined their effects on centriole duplication. First, we generated nonphosphorylatable and PM Ana2 by mutating all seven residues to alanines (7A) or aspartate/glutamate (7PM) and transfected these into S2 cells depleted of endogenous Ana2. Centriole numbers were counted after immunostaining for centriole-markers PLP and Asterless (Asl; Fu and Glover, 2012; Mennella et al., 2012; Fig. 2 B). As expected, Ana2 depletion significantly decreased the percentage of cells with a normal number of centrioles (Fig. 2 B). In cells lacking endogenous Ana2, centriole numbers were rescued by expression of Ana2-WT, but, surprisingly, not by Ana2-7A, Ana2-7PM, or Ana2 lacking the CC domain (ΔCC), which is required for both centriole assembly and localization (Dzhindzhev et al., 2014; Ohta et al., 2014; Arquint et al., 2015; Cottee et al., 2015; Moyer et al., 2015).
The failure of both Ana2-7A and 7PM to rescue centriole duplication could be explained if the substitutions cause protein misfolding and therefore prevent functionality, or if the two Ana2 phospho mutants do fold properly but fail to support centriole duplication for unknown reasons. To evaluate these possibilities, we first purified recombinant Ana2-7A and 7PM and examined the structural stability of the mutant proteins versus the WT protein using circular dichroism (CD; Fig. S2, D and E). No significant difference was found between the CD spectra of WT and mutant proteins, suggesting that the 7A/7PM substitutions were not altering the secondary structure of Ana2. Next, we expressed Ana2 constructs in untreated cells, where they can hetero-oligomerize with endogenous Ana2, and measured centriole numbers. Whereas Ana2-WT and Ana2-7PM expression had no effect on centriole number, Ana2-7A suppressed centriole duplication (i.e., it significantly increased the percentage of cells with less than two centrioles; Fig. 2 C). Thus, unlike Ana2-7PM, Ana2-7A expression displays a dominant/negative effect on centriole duplication, suggesting that the Ana2-7A and 7PM mutants influence centriole assembly differently.
Ana2-7A interacts with Plk4 but fails to target to procentrioles, whereas Ana2-7PM fails to interact with Plk4 but is recruited to procentrioles
We next asked whether the phosphorylation state of the Ana2 NT alters Ana2’s interaction with known binding partners, including itself. Ana2 forms a tetramer through its CC, and point mutations that block oligomerization disrupt Ana2 function in vivo (Slevin et al., 2014; Cottee et al., 2015). We examined whether phospho-Ana2 mutants influence self-association using S2 cells depleted of endogenous Ana2. Ana2-ΔCC did not coimmunoprecipitate with Ana2-WT (Fig. 2 D), demonstrating that the CC is required for self-association, as it is for STIL (Arquint et al., 2015). However, both Ana2-7A and 7PM were able to coimmunoprecipitate with themselves, suggesting that the NT phosphorylation state of Ana2 does not influence its oligomerization.
Because Sas4 binds Ana2 (Cottee et al., 2013; Hatzopoulos et al., 2013), another possibility is that Ana2 phosphorylation could affect this interaction. Sas4 associated with Ana2 regardless of its phosphorylation state (Fig. 2 E), consistent with previous in vitro studies (Dzhindzhev et al., 2014). However, Ana2-ΔCC association with Sas4 was markedly diminished, a surprising result because the Sas4-binding site in Ana2 is well upstream of the Ana2-CC (Fig. S1 A). Thus, Ana2 oligomerization may be a prerequisite for Sas4 binding.
Last, we examined the interaction between Ana2 and Plk4 (Fig. 2 F). Both Ana2-WT and Ana2-7A coimmunoprecipitated with Plk4. Strikingly, however, Plk4 failed to immunoprecipitate with Ana2-7PM, similar to Ana2-ΔCC. In summary, our results suggest a complex relationship between Ana2 and Plk4: Ana2 binds Plk4, but when Ana2 is itself phosphorylated by Plk4, binding with Plk4 is inhibited. Thus far, our findings cannot explain how Ana2-7A acts as a dominant/negative of centriole duplication, but the inability of Ana2-7PM to bind Plk4 may explain why cells expressing this mutant cannot duplicate centrioles.
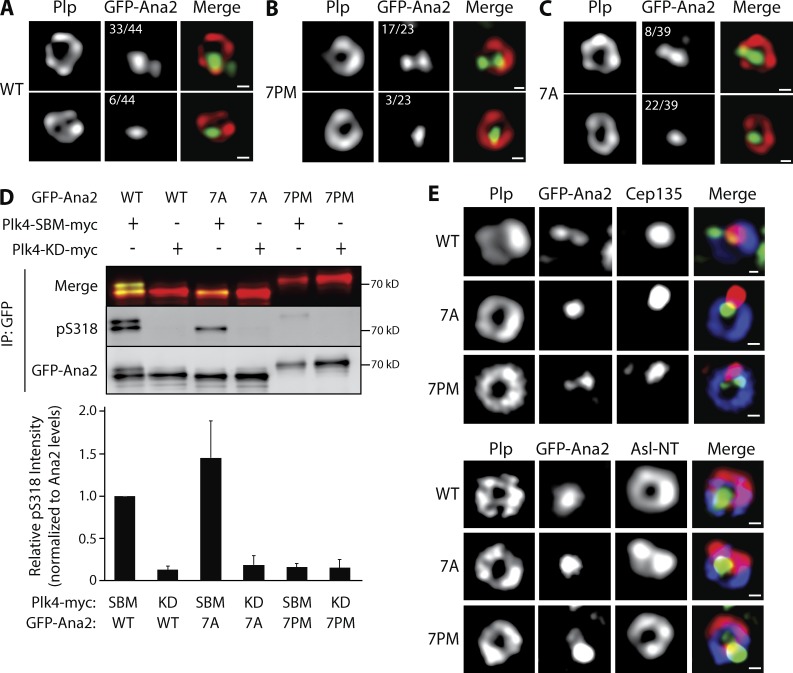
Ana2 phospho mutants can self-associate, bind Sas4, and in the case of Ana2-7A, bind Plk4, suggesting that NT substitutions do not cause protein misfolding in cells but instead interfere with specific functions of Ana2 that compromise centriole duplication. To further test this hypothesis, we used superresolution structured illumination microscopy (SR-SIM) to examine the centriolar localization of GFP-Ana2 (WT or mutant) in cells depleted of endogenous Ana2. In interphase cells, Ana2 staining typically appears as two spots that colocalize with a PLP-labeled mother centriole ring: a bright spot positioned near the center of the PLP ring and a second dimmer spot near the PLP periphery (Fig. 3, A–C, top; Dzhindzhev et al., 2014). The dim Ana2 spot is recruited after telophase disengagement of a centriole pair and marks the future site of procentriole assembly by recruiting Sas6 (Dzhindzhev et al., 2014). In cells whose endogenous Ana2 was replaced with transgenic Ana2-WT, 75% of interphase centrioles (n = 44) displayed the expected two-spot Ana2 pattern; only 14% contained a single Ana2 spot within the mother (Fig. 3 A). Previous studies have shown that the CC in Ana2/STIL is necessary for centriole localization, perhaps by directly binding Plk4 (Ohta et al., 2014; Arquint et al., 2015; Cottee et al., 2015; Moyer et al., 2015). Because Ana2-7PM fails to associate with Plk4 even though it has an intact CC, we predicted that Ana2-7PM would fail to localize to the procentriole site and instead appear only within the mother. Surprisingly, Ana2-7PM localized in a normal two-puncta centriole pattern (74%, n = 23; Fig. 3 B). This suggests that Ana2 targets centrioles by a means other than direct Plk4 binding. In contrast, Ana2-7A localized as a single spot within the majority of mother centrioles (56%, n = 39), and the expected two-spot Ana2 pattern was observed in only 21% of mothers (Fig. 3 C). Thus, in most cells, Ana2-7A recruitment to the procentriole site fails, providing a possible explanation for why Ana2-7A expression prevents centriole duplication.
Figure 3.
The phosphorylation state of the Ana2 NT affects Ana2 recruitment to procentrioles and STAN domain phosphorylation. (A–C) Ana2-7A recruitment to procentrioles is inhibited but that of Ana2-7PM is not. Interphase centrioles in transgenic Ana2-expressing cells were imaged using superresolution microscopy. S2 cell lines stably transfected with inducible GFP-Ana2 WT, 7PM, or 7A were depleted of endogenous Ana2 by RNAi (UTR) for 12 d and, during this period, were induced to express the indicated GFP-Ana2 (green) construct. Cells were immunostained with anti-PLP (red) to mark the surface of mature centrioles. DNA was visualized with Hoechst (not depicted). The number of centrioles that show either two (top) or one (bottom) Ana2 spots is indicated. Centrioles containing either no detectable Ana2 or >2 spots comprised 11% (WT), 13% (7PM), and 23% (7A) of the remaining centrioles. Bars, 200 nm. (D) Phosphorylation of S318 in the STAN domain is strongly reduced by Ana2-7PM expression. Cells were depleted of endogenous Ana2 by RNAi for 7 d. On day 5, cells were cotransfected with the indicated GFP-Ana2 and Plk4-myc constructs and the next day induced to express for 24 h. Anti-GFP IPs were then prepared from lysates, and blots of the IPs were probed for GFP and pS318. A merge of GFP-Ana2 and pS318 blots is shown (top). Graph shows relative pS318 intensities in the IPs. For each treatment, intensities of pS318 were measured, related to the corresponding GFP-Ana2 intensities, and normalized to control (lane 1). n = 3 experiments. Error bars, SEM. (E) Asterless (Asl) distribution in centrioles containing Ana2-7A is abnormal. Interphase centrioles in Ana2-expressing cells imaged using superresolution microscopy. Stable S2 cell lines were manipulated as in A. Cells were immunostained for Cep135 or Asl NT (red) and PLP (blue). Hoechst-stained DNA not depicted. Bars, 200 nm.
STAN domain phosphorylation by Plk4 is reduced in Ana2-7PM
Although Ana2-7PM localizes normally to interphase centrioles, duplication is inhibited when Ana2-7PM replaces endogenous Ana2 in cells. Possibly, 7PM’s inability to bind Plk4 reduces phosphorylation of the STAN domain and, consequently, procentrioles cannot assemble. To test this, we generated a phospho-specific antibody against S318 (Fig. S2 F), a conserved STAN residue phosphorylated by Plk4 and required for Sas6 binding (Dzhindzhev et al., 2014; Ohta et al., 2014). In cells depleted of endogenous Ana2, GFP-Ana2 was coexpressed with either KD Plk4 or a highly active nondegradable Plk4-SBM. Western blots of GFP-Ana2 purified by IP from cell lysates were probed with the anti-phospho-S318 (pS318) antibody. This antibody recognized Ana2-WT that was coexpressed with active Plk4-SBM-myc but not with inactive Plk4-KD-myc (Fig. 3 D, lanes 1 and 2). Likewise, pS318 was detected in Ana2-7A coexpressed with active (but not inactive) Plk4 (Fig. 3 D, lanes 3 and 4). In contrast, pS318 was almost entirely absent in Ana2-7PM, even when coexpressed with active Plk4 (Fig. 3 D, lanes 5 and 6). Therefore, the presence of phospho-mimicking residues within the NT of Ana2 prevents phosphorylation of the STAN domain (at least of S318) by Plk4 in cells. The reduction in STAN phosphorylation may account for the inability of Ana2-7PM to support centriole duplication. Collectively, our findings suggest that Ana2 binds Plk4 after being recruited to centrioles, and this interaction is required for STAN phosphorylation. Interestingly, the pS318 antibody reacts with a tight doublet of Ana2 WT but only a single, fast-migrating band of Ana2-7A, which suggests that the seven substituted residues in the mutant are responsible for the electrophoretic shift of Ana2-WT’s second slower-migrating band. Unfortunately, the pS318 antibody recognizes additional polypeptides in blots of whole-cell lysates and therefore was not useful for immunostaining studies.
Centrioles containing Ana2-7A display morphological abnormalities
Thus far, our results do not explain the disruption of Ana2-7A localization to the procentriole site because Ana2-7A retains several WT features, such as oligomerization, binding of Sas4 and Plk4, and phosphorylation of its STAN domain (specifically, S318). An untested possibility is that proper localization of other procentriole-targeted proteins is altered when Ana2-7A is expressed. To examine this, we raised antibodies against Cep135 and the Asl NT domain (Fig. S2 G) to evaluate the localization of these centriole proteins in cells. In centriole cross sections, Asl NT is positioned adjacent to PLP on the centriole surface, whereas Cep135 localizes as a single spot near the centriole center (Mennella et al., 2012; Fu et al., 2016). After depleting endogenous Ana2 and expressing the GFP-Ana2 constructs, we examined interphase centrioles using SR-SIM and observed normal localization patterns for both Cep135 and Asl in Ana2-WT– and Ana2-7PM–expressing cells (Fig. 3 E). Curiously, although Cep135 formed a normal single spot within centrioles in Ana2-7A cells, Asl staining was distorted and collapsed within the centriole as adjoining spots instead of assuming a normal ring shape (Fig. 3 E). Thus, Ala substitution of the NT phospho-sites of Ana2 causes the redistribution of at least two duplication-essential proteins, Ana2 and Asl, within centrioles.
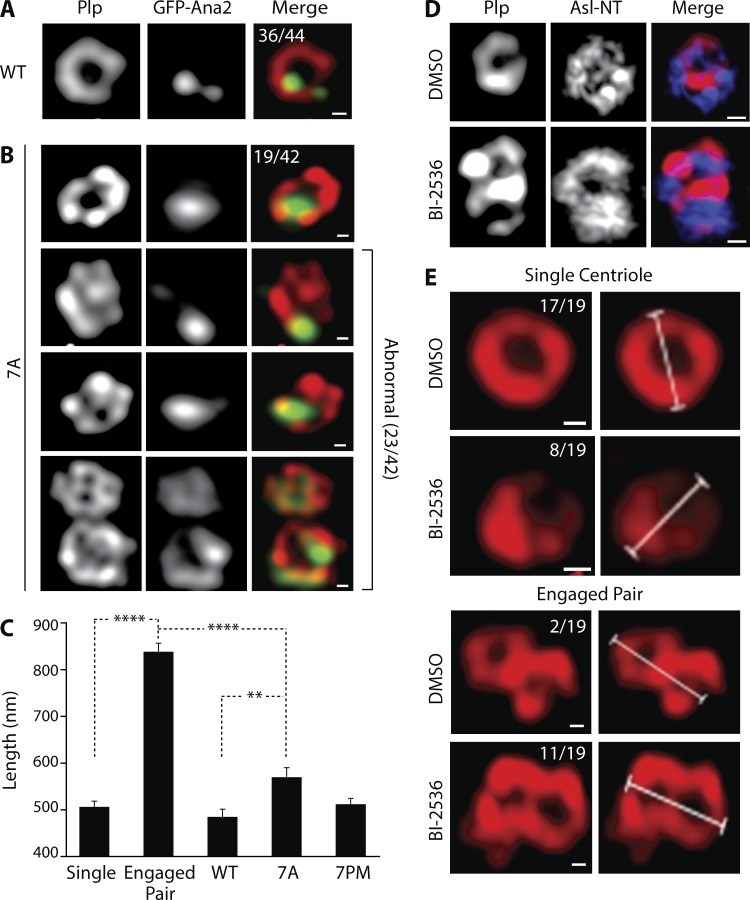
Next, we imaged PLP to examine centriole architecture in Ana2-7A–expressing cells. In this case, endogenous Ana2 was not depleted in order to preserve more centrioles for observation. Using SR-SIM, we imaged PLP-labeled centrioles in GFP-Ana2-WT– and Ana2 mutant–expressing interphase cells. We restricted our analysis to centrioles that were oriented orthogonally to the image plane (i.e., had a clear cross-sectional aspect) to more accurately measure the diameter (or major axis) of the PLP ring. In GFP-Ana2-WT–expressing cells, 82% of centrioles (36 of 44) displayed a normal PLP ring with quasi-ninefold symmetry and a mean diameter of 486 nm (Fig. 4, A and C). The mean diameter of the PLP ring in cells expressing GFP-Ana2-7PM was similar (515 nm; Fig. 4 C). Although normal PLP arrangements were observed in GFP-Ana2-7A–expressing cells (45%), the majority displayed an unusual spot or stripe of PLP in the centriole center (55%, 23 of 42; Fig. 4 B). In addition, the mean length of the major axis of PLP immunostaining (571 nm) was significantly greater than GFP-Ana2-WT–containing centrioles (Fig. 4 C). Despite these differences, centrioles containing GFP-Ana2-7A were functional in that they recruited γ-tubulin during mitosis (Fig. S2 H).
Figure 4.
Centrioles containing Ana2-7A display an abnormal architecture but this is not caused by a failure to disengage. (A and B) The morphology of the centriole outer surface protein, PLP, is abnormal in Ana2-7A–expressing cells. Interphase centrioles in inducible GFP-Ana2-WT (A) or GFP-Ana2-7A (B) stable cell lines were imaged using SR-SIM. Expression of GFP-Ana2 (green) was induced for 5 d. Cells were immunostained for PLP (red) and Hoechst-stained for DNA (not depicted). The number of centrioles with normal (A and B, row 1) or abnormal (B, rows 2–4) morphology is indicated. Bars, 200 nm. (C) Graph shows mean lengths of the major axis of centrioles in interphase cells. Measurements are shown for disengaged single centrioles in DMSO-treated cells (n = 17), engaged centriole pairs in BI-2536–treated cells (n = 9), and centrioles in stable GFP-Ana2–expressing cells (WT, n = 19; 7A, n = 13; 7PM, n = 34). Asterisks mark significant differences between treatments: **, P < 0.01; ****, P < 0.0001. Error bars, SEM. (D) Chemical inhibition of Polo blocks centriole disengagement in S2 cells. Cells were treated for 48 h with either DMSO or the BI-2536 Polo inhibitor, and interphase centrioles were imaged using SR-SIM. Cells were immunostained for PLP (red) and Asl NT (blue). Bars, 200 nm. (E) Examples of centriole measurements in control and Polo-inhibited interphase S2 cells. Cells were treated and prepared for SR-SIM as in D. The major axis lengths of single centrioles and engaged pairs was measured. The numbers of measured single centrioles (top) and engaged pairs (bottom) are indicated. Bars, 200 nm.
One potential explanation for the abnormal PLP pattern within centriole centers is that the centrioles we imaged were not single centrioles, but rather two centrioles in a tightly engaged configuration. In other words, Ana2-7A expression may block mother–daughter centriole disengagement; because disengagement of the mother–daughter pair is a prerequisite for duplication (Tsou and Stearns, 2006), then this would provide a mechanistic explanation for Ana2-7A’s inhibition of centriole duplication. To test this hypothesis, we generated engaged centriole pairs in S2 cells by using the drug BI2536 to chemically inhibit Polo kinase (Fig. 4 D), whose activity is required for centriole separation in spermatocytes (Riparbelli et al., 2014). After 48 h of drug treatment, most centrioles (58%, 11 of 19) in interphase cells remained engaged, compared with 11% (two of 19) in DMSO-control cells (Fig. 4 E). Not surprisingly, the mean length of the major axis of an engaged pair (840 nm) was significantly greater than a single centriole (506 nm; Fig. 4 C). The major axis length of an engaged pair was also significantly longer than centrioles in Ana2-7A–expressing cells (Fig. 4 C), suggesting that the PLP-labeled objects in 7A mutant–expressing cells are unlikely to be products of failed centriole disengagement. Instead, our findings support the conclusion that centrioles containing Ana2-7A are likely structurally abnormal and, to some extent, compromise both Ana2 recruitment to procentrioles and centriole duplication. If true, then a phosphorylated Ana2 NT is an important structural constituent of centrioles and necessary to recruit Ana2 to the procentriole.
Plk4 phosphorylates Ana2 in an ordered pattern
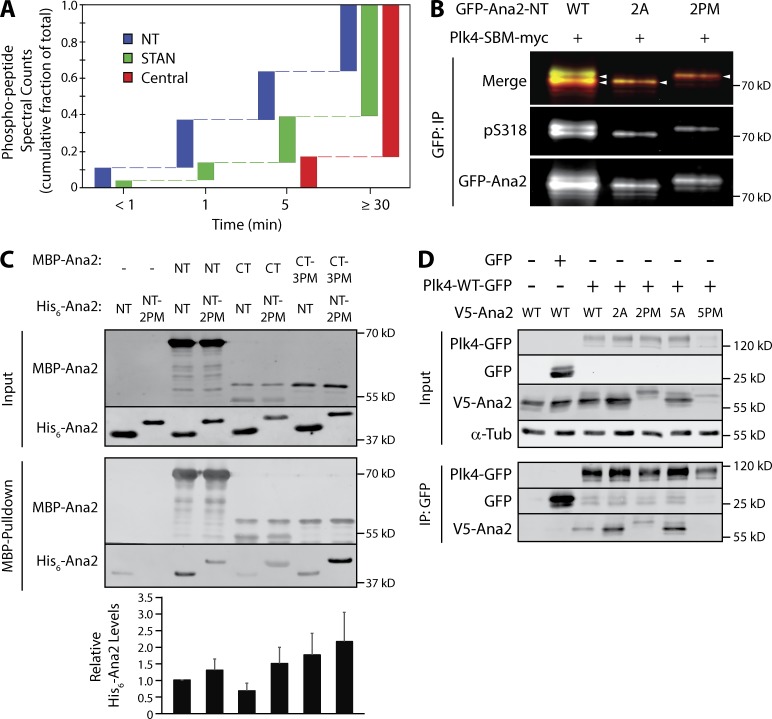
The fact that multiple domains of Ana2 are modified by Plk4 raises the intriguing possibility that the phosphorylations occur sequentially, perhaps causing Ana2’s multiple activities to switch states (or levels) in an ordered succession. For example, our findings with Ana2-7PM suggest that if Plk4 first phosphorylated residues upstream of the STAN domain, then it would release from Ana2 and fail to phosphorylate the STAN domain, which seems unlikely because STAN phosphorylation is required to recruit Sas6 (Dzhindzhev et al., 2014; Ohta et al., 2014; Kratz et al., 2015; Moyer et al., 2015). To test this, we sampled a mixture of purified Ana2, Plk4, and MgATP at different time points and then mapped phospho-residues in Ana2 by MS/MS (Fig. 5 A). Strikingly, we found that phosphorylation was rapid in our conditions; at the earliest time point (<1 min), Ana2 was phosphorylated on NT residues T33 and S38 as well as S318 within the STAN domain. At 1 and 5 min, Plk4 phosphorylated additional STAN domain residues. Phosphorylation of residues within the central region of Ana2 (S63/T69/S150/T159/T242) was first detected at 5 min and continued to accumulate until the final time point. Although the phosphorylation pattern of Ana2 may differ in cells, these in vitro findings suggest that Plk4 phosphorylates Ana2 residues in a specific order: (1) NT (T33/S38), (2) CT (STAN), and (3) central region.
Figure 5.
Plk4 phosphorylates Ana2 in an ordered pattern to regulate STAN domain activation and release Plk4. (A) Plk4 phosphorylates a specific pattern of Ana2 residues over time. Samples were taken at the indicated times from in vitro assays containing purified Plk4 1–317, GST-Ana2 and MgATP, and the phosphorylated residues of the sampled Ana2 were identified by MS/MS. Using data pooled from four analyses, the spectral counts of peptides containing phosphorylated residues within the NT (containing T33, S38), central (S63, T69, S150, T159, T242), or STAN (S318, S370, S373, S395) regions were examined. The fraction of the maximal spectral count number seen at each time point is plotted. (Maximum spectral counts were observed in the final time point in every assay.) Note that the fraction measured at the earliest time point is highest (0.11) for NT residues and lowest (0) for central region residues. (B) Phosphorylation of STAN domain residue S318 is reduced by Ana2-2A but not by the Ana2-2PM mutant. Cells were depleted of endogenous Ana2 by RNAi for 7 d. On day 5, cells were cotransfected with the indicated GFP-Ana2 construct and active Plk4-SBM-myc, and the next day induced to express for 24 h. Anti-GFP IPs were then prepared from lysates, and blots of the IPs were probed for pS318 and GFP. A merge of GFP-Ana2 and pS318 blots is shown. (C) Phosphorylation increases the interaction between Ana2 NT and CT. The indicated combinations of MBP- or His6-tagged Ana2 NT (aa 1–317) or CT (aa 318–420) proteins were mixed for 30 min, after which the MBP construct was recovered by pull-down with amylose resin. Constructs were either WT or mutated with PM substitutions: two residues (T33/S38) in NT (2PM), three residues (S318/S370/S373) in CT (3PM). Western blots of the washed resins were probed with anti-Ana2 antibody. Graph shows His6-Ana2 levels normalized to MBP-Ana2 NT. Error bars, SEM. (D) Mutations in Ana2 NT (2A or 2PM) and central domain 5A do not prevent binding to Plk4 and, in fact, display stronger association with phospho-null substitutions (2A and 5A). In contrast, mutation of the central residues to PM residues (5PM) eliminates Plk4 association. S2 cells were depleted of endogenous Ana2 by RNAi for 7 d; V5-Ana2 constructs were cotransfected with Plk4-GFP on day 5; cells were induced on day 6 to express the transgenes for 24 h. Anti-GFP IPs were then prepared from lysates, and Western blots of the inputs and IPs were probed for GFP, V5, and α-tubulin.
Whereas phosphorylation of the NT residues T33 and S38 promotes Ana2 folding and hyperphosphorylation, modification of Ana2 central residues disrupts Plk4 association
The identification of T33 and S38 as early phosphorylation targets of Plk4 prompted us to reexamine the effects of Ana2 mutants on centriole duplication. We generated Ana2 mutants with single nonphosphorylatable alanine substitutions and transfected these into cells depleted of endogenous Ana2. As before, Ana2 depletion caused significant centriole loss, which was rescued by replacement with Ana2-WT expression (Fig. S3 A). Interestingly, expression of only Ana2-S38A failed to rescue centriole numbers. Although S38 is near the Sas4-binding domain, both S38 mutants (S38A and S38D) bound Sas4 (when coimmunoprecipitated from transfected cell lysates; Fig. S3 B), and both also bound Plk4 (Fig. S3 C). Thus, expression of the S38A mutant inhibits centriole duplication, but this is likely not caused by a defect in either Sas4 or Plk4 binding, unlike Ana2-7PM.
To understand how phosphorylation of the NT contributes to Ana2 function, we next quantified STAN domain activation in phospho mutants (alanine [A] and PM) that cluster within the NT (T33/S38; 2A and 2PM). Endogenous Ana2-depleted cells were cotransfected with GFP-Ana2 (WT or mutant) and catalytically active Plk4-SBM. We then measured pS318 levels in GFP-Ana2 immunoprecipitates. As described previously, pS318 antibody reacted with Ana2-WT from cells coexpressing Plk4-SBM (Fig. 5 B, lane 1). Strikingly, detection of pS318 was confined to a single fast migrating band on SDS-PAGE, suggesting that expressed Ana2 cannot be hyperphosphorylated when T33/S38 are mutated to alanines (Fig. 5 B, lane 2). In contrast, expressed Ana2-2PM migrated entirely as a slower migrating band on SDS-PAGE, suggesting that Ana2 NT-2PM is completely hyperphosphorylated (Fig. 5 B, lane 3). CD analysis of purified Ana2-2A and 2PM proteins revealed no difference in their spectra compared with Ana2-WT (Fig. S2, D and E), suggesting that these mutations do not alter the structural integrity of Ana2. These findings suggest that modification of the NT is required for maximal Ana2 phosphorylation. Although pS318 is detected in the Ana2-2A mutant (similar to the 7A mutant), further phosphorylation of STAN domain residues may be absent in Ana2-2A, possibly explaining the failure of Ana2-S38A to rescue centriole duplication.
Recently, a superresolution microscopy study demonstrated that the NT and CT of Ana2 are positioned close to one another within the outer cartwheel of centrioles (Gartenmann et al., 2017), suggesting that Ana2 adopts a folded conformation. Therefore, we examined whether the NT and CT of Ana2 interact and, if so, whether this could be regulated by phosphorylation. We first examined in vitro binding of purified Ana2 NT (aa 1–317) and Ana2-CT (318–420; containing mostly STAN domain) using pull-downs of maltose-binding protein (MBP) and His6-tagged proteins. As expected, Ana2 NT interacted with itself, as both constructs contain the CC (Fig. 5 C); this interaction was not altered in Ana2 NT containing PM T33E/S38D (2PM) substitutions, suggesting that phosphorylation of these residues is not important for oligomerization through the CC. Notably, Ana2 NT and CT do not bind (Fig. 5 C), and we confirmed this inconsistent/weak interaction by Y2H analysis (Fig. S3 D). Next, we examined binding between Ana2 NT-2PM and Ana2 CT containing PM substitutions of its three most conserved phosphorylated STAN residues (S318/S370/S373). Surprisingly, mutation of the NT sites to PM residues did not interfere with the NT–CT interaction. In fact, when Ana2 pull-down bands were quantified (Fig. 5 C, graph), the opposite was found: PM residues in either the NT or CT construct increased the mean amount of interaction by two- to threefold. Interestingly, mutation to PM of both domains induces the highest interaction (∼3.5-fold), suggesting that phosphorylation of Ana2 increases the interaction between its NT and CT.
Next, we examined the effects of phosphorylation of the NT (2A or 2PM) and the Plk4-targeted residues of Ana2’s central region (S63/T69/S150/T159/T242; 5A or 5PM) on Plk4 association by coIP. When V5-Ana2 (WT or mutant) was cotransfected with Plk4-GFP into cells, nonphosphorylatable Ana2-2A and 5A mutants interacted strongly with Plk4 (Fig. 5 D, lanes 4 and 6), whereas WT and 2PM interacted weakly with Plk4 (Fig. 5 D, lanes 3 and 5). Strikingly, Ana2-5PM failed to interact with Plk4 (Fig. 5 D, lane 7), suggesting that phosphorylation of the central region causes Ana2 to dissociate from Plk4. Consistent with its inability to bind Plk4, expression of Ana2-5PM, but not Ana2-5A, failed to rescue centriole duplication in cells depleted of endogenous Ana2 (Fig. S3 E). Collectively, our data support the model that phosphorylation of NT residues T33/S38 and CT residues S318/S370/S373 promote Ana2 folding. Subsequently, phosphorylation of the five central residues causes Plk4 dissociation.
Discussion
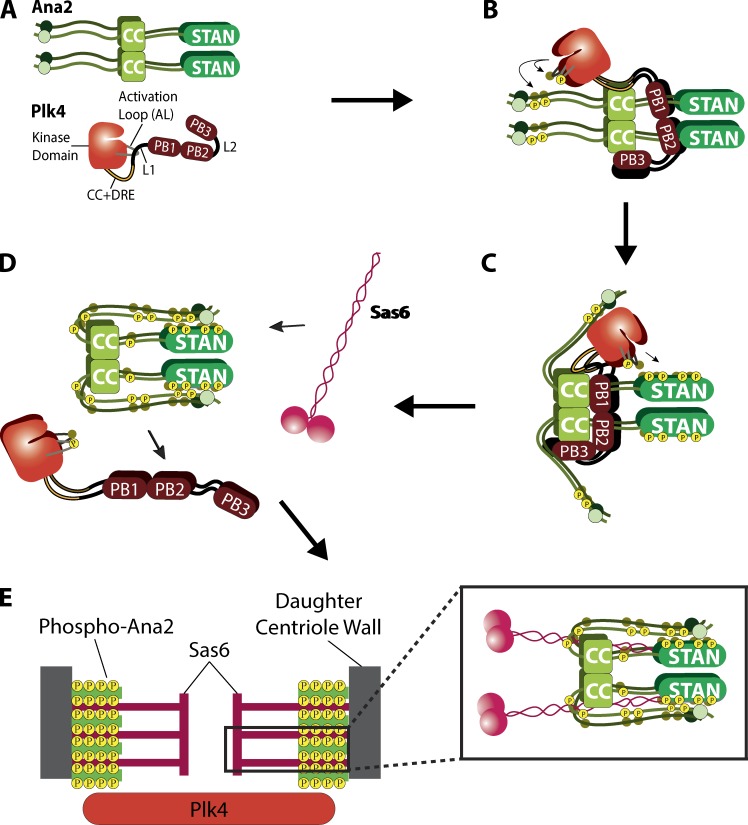
Our investigation of Plk4 and Ana2 reveals a complex interaction whereby Plk4 promotes centriole assembly through a series of Ana2-directed phosphorylation events (Fig. 6). The activities of Polo-like kinase family members are initially restrained by multiple mechanisms, including autoinhibition (Lowery et al., 2005). We demonstrated previously that Plk4 is no exception and uses L1 adjacent to the kinase domain to autoinhibit (Klebba et al., 2015a; Fig. 6 A), similar to Plk1 (Xu et al., 2013). We also showed that relief of autoinhibition requires PB3 as well as an unidentified extrinsic factor (Klebba et al., 2015a). STIL is likely the unidentified factor, because it binds PB3 and stimulates Plk4 activity in cells (Arquint et al., 2015; Moyer et al., 2015). Additionally, Arquint et al. (2015) identified a second STIL-binding site in Plk4, positioned somewhere between the kinase domain and PB1. Using a combination of IP, Y2H analysis, and in vitro protein binding methods, we show that Drosophila Plk4 also contains a second Ana2-binding site that maps to a conserved, leucine-rich segment (34 aa) located immediately downstream of the kinase domain and is predicted to form a CC. The crystal structure of the STIL-CC with the single α-helix within PB3 reveals a leucine zipper-type CC interaction (Arquint et al., 2015). We predict that a similar structure forms between Ana2-CC and the newly identified putative CC region of Plk4. Intriguingly, the corresponding region of human Plk4 possesses α-helical content (Wong et al., 2015), suggesting that this interaction site could be conserved between human Plk4 and STIL. However, we note that there are known differences in the quaternary structures of STIL and Ana2 with Plk4. For example, whereas the STIL-CC is monomeric when complexed with human PB3, Ana2-CC forms a tetramer (Cottee et al., 2015) and also has been shown to simultaneously bind eight dynein light chain (LC8) subunits (Slevin et al., 2014). Given the significance of this interaction, an important goal of future studies is to determine how the Ana2-CC associates with both PB3 and the putative CC in Plk4 and to determine the stoichiometry of this complex.
Figure 6.
Proposed model depicting the mechanism of Plk4 and Ana2 interdependent activation to achieve centriole assembly. (A) Plk4 and Ana2 exist initially in nonfunctional states. Nascent monomeric Plk4 is inactive, a state maintained by L1, which inhibits the phosphorylation of the activation loop needed for full enzymatic activity. Similarly, Ana2 is initially nonfunctional and exists in an open conformation, i.e., does not support centriole assembly until phosphorylated by Plk4. (B) Plk4 is active on centrioles before Ana2 recruitment. Centriole targeting of Ana2 occurs through an unidentified mechanism that requires Plk4 kinase activity and phosphorylation of Ana2 NT residues T33 and S38. Ana2 binding to Plk4 then initiates the next steps in procentriole assembly by relieving Plk4 autoinhibition to activate the kinase (Arquint et al., 2015; Klebba et al., 2015a; Moyer et al., 2015). (C) NT phosphorylation induces a conformational change of Ana2 that enhances Plk4 phosphorylation of the STAN domain, which is known to recruit Sas6 to the procentriole assembly site. (D) Ana2’s conformational change exposes five residues within the central region of Ana2 to Plk4, and their phosphorylation causes Ana2 to release from Plk4. (E) Ana2 tetramers incorporate into the growing procentriole to stabilize the vertical stacking of Sas6 cartwheels (as proposed by Cottee et al. [2015]). The hyperphosphorylated Ana2 NT may also promote additional stabilizing interactions with proteins positioned near centriolar microtubules. This is important to maintain structural integrity of the organelle and, if compromised, could interfere with procentriole assembly.
Although Ana2/STIL is the first described stimulator of Plk4 activity in cells, other unidentified activators probably exist. For example, work in Caenorhabditis elegans embryos has shown that Plk4/ZYG1 is recruited to centrioles before SAS-5 (the worm homologue of Ana2/STIL; Delattre et al., 2006; Pelletier et al., 2006). So what activates Plk4 before Ana2’s arrival? Drosophila Asterless (Asl) is an interesting candidate, because it targets Plk4 to centrioles and plays additional roles in stabilizing the kinase (Dzhindzhev et al., 2010; Klebba et al., 2015b). Identifying other Plk4 activators will be essential to determine how Ana2 targets centrioles because, intriguingly, Plk4 kinase activity is required for STIL’s centriolar recruitment (Moyer et al., 2015; Zitouni et al., 2016). Although it has been proposed that direct binding to Plk4 is responsible for STIL recruitment (Ohta et al., 2014; Moyer et al., 2015), we feel this is an open question. Previous work has shown that the Ana2-CC is required for its centriole localization and Plk4 binding (Ohta et al., 2014; Arquint et al., 2015; Moyer et al., 2015), and this observation forms the basis for the current model of Ana2’s centriole targeting. But we found that CC deletion is remarkably pleiotropic, because it also disrupts oligomerization and Sas4 binding. (Possibly, failure to oligomerize compromises most Ana2 interactions, including with Cdk1-cyclinB; Zitouni et al., 2016.) Our work demonstrates that Ana2-7PM fails to bind Plk4 but still successfully localizes to the procentriole. Therefore, although Plk4 kinase activity and Ana2 phosphorylation is required for Ana2 to target procentrioles, it is plausible that direct binding to Plk4 is not required for Ana2 localization (Fig. 6 B). Possibly other centriole surface proteins can, after phosphorylation by Plk4, function as high-affinity binding sites for Ana2/STIL. Additional work is needed to solve this problem.
Ana2 is extensively phosphorylated along its length by Plk4 (Dzhindzhev et al., 2014; Ohta et al., 2014; Kratz et al., 2015). Our data suggest that Plk4 likely initially targets NT residues T33/S38, which then promote STAN domain phosphorylation. However, our data are also compatible with a model in which residues of both NT and STAN regions are phosphorylated contemporaneously, with the end result being the same. We propose that Ana2 exists in an open conformation, where the NT and STAN domain are separated (Fig. 6 A). In this model, phosphorylation of T33/S38 induces Ana2 to fold, and the conformational change may be a functionally important event—it could, for example, facilitate Plk4’s further phosphorylation of key residues in the STAN domain that promote the subsequent recruitment of Sas6 to the procentriole assembly site (Fig. 6, C and D). This model predicts our observation that, when expressed in cells, the nonphosphorylatable Ana2 NT-2A mutant (T33A/S38A) does not display the mobility-shifted hyperphosphorylated band seen with Ana2 WT or 2PM expression (Fig. 5 B). Our interpretation of this result is that NT phosphorylation is a prerequisite for the complete modification of all the Plk4-targeted residues of Ana2. Further supporting this model is our finding that the Ana2 S38A single-point mutant completely fails to rescue centriole duplication in cells depleted of endogenous Ana2 (Fig. S3 A). If S38 phosphorylation is a prerequisite for the full course of Plk4-mediated phosphorylation, then failure of centriole duplication is expected for the S38A mutant. Similarly, the 7A mutant is also not hyperphosphorylated, likely because of the mutation of the two NT residues (Fig. 3 D). If Ana2 initially adopts an open conformation, its extensive phosphorylation could drive a conformational change that promotes Sas6 binding, thus enabling procentriole assembly.
Expression of phopsho-Ana2 mutants (7A or 7PM) had an unexpected effect; neither mutant rescued the loss of centrioles phenotype in endogenous Ana2-depleted cells. However, this phenotype is reached as a result of disrupting different Ana2-dependent steps in centriole assembly. Our results suggest that, after STAN domain modification, Plk4 targets several residues within the central region of Ana2, generating a hyperphosphorylated form of Ana2 that dissociates from Plk4. Extensive phosphorylation of the central region appears to inhibit Plk4 binding, as observed with the Ana2-5PM and -7PM mutants.
Ours is not the first description of a mechanism that regulates the Plk4-STIL–Ana2 interaction. During mitosis, Cdk1-cyclinB binds the CC domain in STIL and prevents Plk4 association (Zitouni et al., 2016). This ensures that Plk4 activation and STAN phosphorylation are restricted to periods of the cell cycle when Cdk1 activity is low, such as during mitotic exit when Ana2 and Sas6 are first recruited to centrioles (Dzhindzhev et al., 2014). This same period is when the Ana2–Plk4 interaction that we describe probably occurs.
Last, we propose that hyperphosphorylated Ana2 releases Plk4 and incorporates into the nascent procentriole, where it makes contact with Sas6 and other centriole proteins (Fig. 6 E). Phosphorylation of Ana2/STIL may regulate its ability to organize Sas4/CPAP into the horizontal struts that connect the centriole spokes with microtubules (Hatzopoulos et al., 2013). Notably, STIL and Sas5 turnover is rapid between centrioles and cytoplasm (Delattre et al., 2004; Vulprecht et al., 2012) and may act as a transporter for centriolar components (Hatzopoulos et al., 2013)—yet another function that may be regulated by phosphorylation. Thus, we interpret centriole loss in cells expressing Ana2-7A to be a result of defective Ana2 recruitment to the procentriole. It will be instructive to determine the path of recruitment to the procentriole, whether Ana2 is first recruited to the lumen of the mother as demonstrated for Sas6 (Fong et al., 2014), or instead is directly recruited to the procentriole. Moreover, we recently reported that Ana2 interacts with multiple centriole proteins in a Y2H screen of centrosome components, including Asl, Cep135, and Ana1 (Galletta et al., 2016). Whether Plk4 activity alters these interactions remains to be determined. In any case, it is increasingly clear that Plk4 is a fundamentally important regulator of the interaction landscape of centriolar proteins that is needed for the seemingly complex molecular choreography that underlies centriole assembly.
Materials and methods
Cell culture and double-stranded RNAi
Drosophila S2 cell culture, in vitro dsRNA synthesis, and RNAi treatments were performed as previously described (Rogers and Rogers, 2008). Cells were cultured in Sf900II SFM media (Life Technologies). RNAi was performed in 6-well plates. For IP assays, cells (40–90% confluence) were treated with 10 µg dsRNA in 1 ml of medium and replenished with fresh medium/dsRNA every day for 7 d. For immunofluorescence microscopy, cells were transfected with 40 µg dsRNA on days 0, 4, and 8. An ∼550-bp control dsRNA was synthesized from DNA template amplified from a noncoding sequence of the pET28a vector (Clontech) using the primers 5′-ATCAGGCGCTCTTCCGC-3′ and 5′-GTTCGTGCACACAGCCC-3′. (All primers used for dsRNA synthesis begin with the T7 promoter sequence 5′-TAATACGACTCACTATAGGG-3′, followed by template-specific sequence). dsRNA targeting the Ana2 UTR was synthesized from EST LD22033 template by first deleting the Ana2 cDNA, and then joining 91 bp of 5′ UTR with 78 bp of 3′ UTR to create the following sequence: 5′-AGTTCCACCCCTAAGTCGATCGACTTCCAATTGGACAGATTCTCCCGCTCGAATTTAATTTAATCGGCAAATATAAACAAATACGCTCCAAAAGCATGTACAATGTTCGTTTTGTTATTTATGCATATGTCTATTTGCGATTTAAGTGGAAATATATTTCAATACACGG-3′. This template was amplified using the primers 5′-CAGATTCTCCCGCTCG-3′ and 5′-TTCCGTGTATTGAAATATATTTCC-3′. Immunoblotting confirmed that Ana2 UTR RNAi depleted endogenous Ana2 by ∼80–90% (Fig. S1 C).
Immunoblotting
S2 cell extracts were produced by lysing cells in cold PBS and 0.1% Triton X-100. Laemmli sample buffer was then added, and samples were boiled for 5 min. Samples of equal total protein were resolved by SDS-PAGE, blotted, probed with primary and secondary antibodies, and scanned on an Odyssey imager (Li-Cor Biosciences). Care was taken to avoid saturating the scans of blots. Antibodies used for Western blotting included rabbit anti-Ana2 (our laboratory), rat anti-Cep135 (our laboratory), rat anti–Asl A (aa 1–374; our laboratory), mouse anti–V5 monoclonal (Life Technologies), mouse anti–GFP monoclonal JL8 (Clontech), mouse anti-myc (Cell Signaling Technologies), mouse anti–α tubulin monoclonal DM1A (Sigma-Aldrich), mouse anti-GST (Cell Signaling Technologies), and mouse anti-FLAG monoclonal (Sigma-Aldrich) used at dilutions ranging from 1:1,000–30,000. IRDye 800CW secondary antibodies (Li-Cor Biosciences) were prepared according to the manufacturer’s instructions and used at 1:3,000 dilutions.
To generate the anti–phospho-specific pS318 Ana2 antibody, rat polyclonal antibodies were raised against the phosphor-peptide AKPNTEK{pSer}MVMNELAC. A nonphosphopeptide (AKPNTEKSMVMNELAC) was also synthesized (Thermo Fisher Scientific). Antibodies were affinity-purified from antisera using Affi-Gel 10/15 resin (Bio-Rad Laboratories) coupled to the phosphopeptide. The eluted material was then preabsorbed over an Affi-Gel column coupled to the non–phospho-specific peptide. The eluate was collected and concentrated using 10K Ultrafree 2 ml concentrators (Millipore). Antibodies were used at a 1:1,000 dilution.
Mass spectrometry
Samples for MS/MS analysis were first resolved by SDS-PAGE and Coomassie stained. Bands of interest were cut from gels and then processed for MS. Gel pieces containing Ana2 were reduced (10 µM dithiothreitol, 55°C, 1 h), alkylated (55 mM iodoacetamide at room temperature for 45 min), and trypsin digested (∼1 µg trypsin at 37°C for 12 h; for one experiment, a sequential chymotrypsin/trypsin digest was used) in-gel, and then extracted. Peptide samples were loaded onto a Zorbax C18 trap column (Agilent Technologies) to desalt the peptide mixture using an on-line Eksigent nano–liquid chromatography ultra-HPLC system. The peptides were then separated on a 10-cm Picofrit Biobasic C18 analytical column (New Objective). Peptides were eluted over a 90-min linear gradient of 5–35% acetonitrile/water containing 0.1% formic acid at a flow rate of 250 nl/min, ionized by electrospray ionization in positive mode, and analyzed on a LTQ Orbitrap Velos (Thermo Electron Corp.) mass spectrometer. All liquid chromatography-MS analyses were performed in “data-dependent” mode in which the top six most intense precursor ions detected in the MS1 precursor scan (m/z 300–2,000) were selected for fragmentation via collision-induced dissociation. Precursor ions were measured in the Orbitrap at a resolution of 60,000 (m/z 400), and all fragment ions were measured in the ion trap.
MS/MS data used for Fig. 2 A were accumulated from five experiments and generated by two facilities (Taplin Mass Spectrometry Facility [Harvard Medical School] and the NHLBI Proteomics Core Facility [National Institutes of Health]). Typically, coverage of Ana2 was ∼85%; coverage of residues 13–65 was often problematic. For Fig. 5 A, coverage ranged from ∼85 to 95%; residues 1–12 and 414–420 were never recovered. For Fig. S2 C, the best coverage was ∼80–85%; coverage of residues 13–65 was often problematic.
Immunofluorescence microscopy
S2 cells were fixed and processed as previously described (Rogers and Rogers, 2008) by spreading S2 cells on concanavalin A–coated glass-bottom dishes and fixing with ice-cold methanol. Primary antibodies were diluted to concentrations ranging from 1 to 20 µg/ml. They included rabbit anti–PLP (Rogers et al., 2009), guinea pig anti-Asl (Klebba et al., 2013), rat anti-Cep135, rat anti–Asl NT, and mouse anti-V5 (Life Technologies) antibodies. Goat secondary antibodies (conjugated with Cy2, Rhodamine red-X, or Cy5 [Jackson ImmunoResearch]) were used at 1:1,500. Hoechst 33342 (Life Technologies) was used at a final concentration of 3.2 µM. Cells were mounted in 0.1 M n-propyl galate, 90% (by volume) glycerol, and 10% PBS solution. Specimens were imaged using a DeltaVision Core deconvolution system (Applied Precision) equipped with an Olympus IX71 microscope, a 100× objective (NA 1.4), and a cooled CCD camera (CoolSNAP HQ2; Photometrics). Images were acquired with softWoRx v.1.2 software (Applied Science). Superresolution microscopy was performed using a Zeiss ELYRA S1 (SR-SIM) microscope equipped with an AXIO Observer Z1 inverted microscope stand with transmitted (HAL), UV (HBO), and solid-state (405/488/561-nm) laser illumination sources, a 100× objective (NA 1.4), and EM-CCD camera (Andor iXon). Images were acquired with ZEN 2011 software.
Constructs and transfection
FL cDNAs of Drosophila Ana2, Plk4, Sas4, and Sas6 were subcloned into a pMT vector containing in-frame coding sequences for EGFP, V5, or myc under control of the inducible metallothionein promoter. PCR-based site-directed mutagenesis with Phusion polymerase (Thermo Fisher Scientific) was used to generate the various Ana2 and Plk4 deletion and point mutants. Transient transfections of S2 cells were performed as described (Nye et al., 2014). In brief, ∼2–5 × 106 cells were pelleted by centrifugation, resuspended in 100 µl of transfection solution (5 mM KCl, 15 mM MgCl2, 120 mM sodium phosphate, and 50 mM d-mannitol, pH 7.2) containing 1–3 µg of purified plasmid, transferred to a cuvette (2-mm gap size), and then electroporated using a Nucleofector 2b (Lonza), program G-030. Transfected cells were diluted immediately with 0.5 ml SF-900 II medium and plated in a 6-well cell-culture plate. Typically, cells were allowed 24 h to recover before further manipulation. Expression of all constructs was induced by addition of 100 µM to 1 mM copper sulfate to the culture medium.
Y2H assay
Y2H experiments were performed using the Matchmaker Gold Y2H system (Clontech), with significant modifications. pDEST-GADT7 and pDEST-GBKT7, modified versions of Matchmaker vectors compatible with the Gateway cloning system (Life Technologies), were used (Rossignol et al., 2007). pDEST-GADT7 and pDEST-GBKT7 contain the 2 µ and pUC ori for growth in yeast and bacteria. Both contain the Gateway cassette and use the ADH1 promoter to drive expression in Saccharomyces cerevisiae. pDEST-GADT7 fuses the SV40 nuclear localization signal, the GAL4 activation domain, and the HA epitope tag to the NT of the protein encoded by DNA inserted into the Gateway cassette. pDEST-GBKT7 fuses the SV40 nuclear localization signal, the GAL4 DNA binding domain, and the c-Myc epitope tag to the NT of the protein encoded by DNA inserted into the Gateway cassette. pDest-pGBKT7 was modified by yeast-mediated recombination to confer resistance to ampicillin instead of kanamycin. pDEST-GADT7 and pDEST-GBKT7 plasmids containing fragments encoding the protein regions to be tested for interaction were transformed into Y187 and Y2HGold yeast strains, respectively, using standard techniques. Liquid cultures of yeast carrying these plasmids were grown at 30°C, with shaking, to OD600 ∼0.5 in SD –leu or SD –trp medium, as appropriate, to maintain plasmid selection. Interactions were tested by mating, mixing 20 µl each of a Y187 strain and a Y2HGold strain in 100 µl of 2× YPD media in a 96-well plate. Mating cultures were grown for 20–24 h at 30°C with shaking. Cells were pinned onto SD –leu –trp (DDO) plates to select for diploids carrying both plasmids, using a Multi-Blot Replicator (VP 407AH; V&P Scientific) and grown for 5 d at 30°C. These plates were then replica plated onto DDO, SD –ade –leu –trp –ura (QDO), SD –leu –trp + aureobasidin A (Clontech) + X-α-Gal (Clontech, Gold Biotechnology; DDOXA), and/or SD –ade –leu –trp –ura + aureobasidin A + X-α-Gal (QDOXA). Replica plates were grown for 5 d at 30°C. Interactions were scored based on growth and/or blue color, as appropriate.
In vitro kinase assays
Bacterially expressed constructs of Drosophila Plk4 (aa 1–317) C-terminally tagged with FLAG-His6 and FL Drosophila Ana2 N-terminally tagged with glutathione S-transferase (GST-Ana2) were purified on HisPur (Thermo Fisher Scientific), amylose (NEB), or glutathione resin (NEB), respectively, according to the manufacturers’ instructions. In vitro phosphorylation assays were performed by incubating 5–10 μM Plk4, 20 μM Ana2, and 100 μM ATP for 1–2 h at 22°C in RXN1 buffer (40 mM Na Hepes, pH 7.3, 150 mM NaCl, 5 mM MgCl2, 1 mM DTT, and 10% [by volume] glycerol). Phosphorylated residues within proteins were identified by MS/MS (Table S1) of purified bacterially expressed proteins phosphorylated in vitro (described above) in the presence of nonradioactive ATP.
In vitro binding assays
Bacterially expressed constructs of Drosophila Ana2 (aa 1–317 or 318–420) N-terminally tagged with His6 or MBP were purified on HisPur (Thermo Fisher Scientific) or amylose (NEB), respectively, according to the manufacturers’ instructions. MBP-Ana2 was not eluted from the resin for binding assays. Proteins were analyzed by SDS-PAGE, and equimolar amounts of MBP-Ana2 and His6-Ana2 were mixed together in buffer RXN1. Samples were incubated for 30 min at room temperature with shaking, followed by eight washes with buffer RXN1. Resin was resuspended in sample buffer and subsequently run on SDS-PAGE for analysis by Western blotting.
GFP immunoprecipitation assays
GFP-binding protein (GBP; Rothbauer et al., 2008) was fused to the Fc domain of human IgG (pIg-Tail; R&D Systems), tagged with His6 in pET28a (EMD Biosciences), expressed in Escherichia coli, and purified on HisPur resin (Thermo Fisher Scientific) according to the manufacturer’s instructions (Buster et al., 2013). Purified GBP was bound to magnetic Dyna Beads (Thermo Fisher Scientific) and cross-linked to the resin by incubating with 20 mM dimethyl pimelimidate dihydrochloride in PBS, pH 8.3, for 2 h at 22°C and then quenching the coupling reaction by incubating with 0.2 M ethanolamine, pH 8.3, for 1 h at 22°C. Antibody-coated beads were washed three times with PBS–Tween 20 (0.02%) then equilibrated in 1.0 ml cell lysis buffer (CLB; 50 mM Tris, pH 7.2, 125 mM NaCl, 2 mM DTT, 0.1% Triton X-100, 1× protease inhibitor cocktail [Roche], and 0.1 mM PMSF) or CLB containing phosphatase inhibitors for phospho-specific antibody detection (CLB+; CLB with 200 mM NaF, 150 mM β-glycerophosphate, and 1 mM Na3VO4). Transfected cells expressing recombinant proteins were lysed in CLB or CLB+, and the lysates were clarified by centrifugation at 16,100 g for 5 min at 4°C. 0.5–1% of the inputs were used for immunoblots. GBP-coated beads were rocked with lysate for 30 min at 4°C or 10 min at 22°C, washed four times with 1 ml CLB or CLB+, and boiled in Laemmli sample buffer.
CD
N-terminally GST-tagged Ana2 (WT or mutants) were bacterially expressed and then purified from clarified bacterial lysate using glutathione resin (Thermo Fisher Scientific) following the manufacturer’s instructions. The GST tag was removed by incubating the resin-bound protein with PreScission Protease (GE Healthcare) overnight at 4°C. Protease-cleaved protein was recovered in the column flow-through, and the material was then passed through new glutathione resin to remove any remaining GST-tagged protease and uncut GST-tagged Ana2. Samples were analyzed by SDS-PAGE. FL Ana2 WT, 2A, 2PM, 7A, and 7PM constructs were diluted to 0.10 mg/ml in CD buffer (10 mM sodium phosphate, pH 7.4, and 50 mM NaF). Spectra were acquired in a 1-mm–path length cuvette at 20°C from 260 to 185 nm with a step size of 0.5 nm every 1.25 s using a Chirascan-plus CD spectrometer (Applied Photophysics). A CD buffer spectrum was subtracted from each Ana2 spectrum, and the resulting curves were smoothed using Chirascan-plus software. Representative traces (Fig. S2 E) are shown from data collected on two independent experimental days.
Statistical analysis and curve fitting
Means of measurements were analyzed for significant differences by one-way ANOVA followed by Tukey’s posttest (to evaluate differences between treatment pairs) using Prism 7 software (GraphPad). Means are taken to be significantly different if P < 0.05. P-values shown for pairwise comparisons of Tukey’s posttest are adjusted for multiplicity. Asterisks indicate significant differences and ns indicates P ≥ 0.05 for the indicated pairwise comparison in figures. Error bars in all figures indicate SEM.
Online supplemental material
Fig. S1 shows the specificity of anti-Ana2 antibodies and that the Ana2-CC domain is necessary for Plk4 association. It also shows that the putative Plk4-CC and PB3 domains interact with Ana2 CT by Y2H analysis and that the putative Plk4-CC is conserved. Fig. S2 shows conservation of Ana2 NT phospho-residues and summarizes the in vivo Ana2 phosphorylation assay, that Plk4 does not phosphorylate Sas6 in vitro, and that Ana2 phospho mutants are not misfolded as determined by CD. It also shows that centrioles containing Ana2-7A recruit PCM during mitosis and demonstrates the specificity of anti-pS318 Ana2, Cep135, and Asl NT antibodies used in this study. Fig. S3 shows that Ana2 S38A and 5PM mutants fail to rescue centriole duplication, and that Ana2-S38A/PM does not prevent Sas4 or Plk4 binding. It also show that Ana2 NT and CT weakly interact via Y2H. Table S1 lists the Ana2 phospho-sites identified by MS/MS in this study.
Supplementary Material
Acknowledgments
We thank P. Krieg for editing the manuscript.
This work was supported by the Division of Intramural Research at the National Heart, Lung, and Blood Institute (1ZIAHL006104) to N.M. Rusan. G.C. Rogers is grateful for support from the National Cancer Institute (P30 CA23074), the National Institute of General Medical Sciences (R01GFM110166), the National Science Foundation (MCB1158151), and the Phoenix Friends.
The authors declare no competing financial interests.
Author contributions: T.A. McLamarrah, D.W. Buster, N.M. Rusan, and G.C. Rogers conceived the project. T.A. McLamarrah, D.W. Buster, and G.C. Rogers wrote the manuscript. T.A. McLamarrah, D.W. Buster, B.J. Galletta, C.J. Boese, J.M. Ryniawec, N.A. Hollingsworth, A.E. Byrnes, and C.W. Brownlee performed and analyzed the experiments. K.C. Slep, N.M. Rusan, and G.C. Rogers supervised the project.
References
- Arquint C., Gabryjonczyk A.M., Imseng S., Böhm R., Sauer E., Hiller S., Nigg E.A., and Maier T.. 2015. STIL binding to Polo-box 3 of PLK4 regulates centriole duplication. eLife. 4:e07888 10.7554/eLife.07888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avidor-Reiss T., and Gopalakrishnan J.. 2013. Building a centriole. Curr. Opin. Cell Biol. 25:72–77. 10.1016/j.ceb.2012.10.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bettencourt-Dias M., Rodrigues-Martins A., Carpenter L., Riparbelli M., Lehmann L., Gatt M.K., Carmo N., Balloux F., Callaini G., and Glover D.M.. 2005. SAK/PLK4 is required for centriole duplication and flagella development. Curr. Biol. 15:2199–2207. 10.1016/j.cub.2005.11.042 [DOI] [PubMed] [Google Scholar]
- Buster D.W., Daniel S.G., Nguyen H.Q., Windler S.L., Skwarek L.C., Peterson M., Roberts M., Meserve J.H., Hartl T., Klebba J.E., et al. 2013. SCFSlimb ubiquitin ligase suppresses condensin II-mediated nuclear reorganization by degrading Cap-H2. J. Cell Biol. 201:49–63. 10.1083/jcb.201207183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cottee M.A., Muschalik N., Wong Y.L., Johnson C.M., Johnson S., Andreeva A., Oegema K., Lea S.M., Raff J.W., and van Breugel M.. 2013. Crystal structures of the CPAP/STIL complex reveal its role in centriole assembly and human microcephaly. eLife. 2:e01071 10.7554/eLife.01071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cottee M.A., Muschalik N., Johnson S., Leveson J., Raff J.W., and Lea S.M.. 2015. The homo-oligomerisation of both Sas-6 and Ana2 is required for efficient centriole assembly in flies. eLife. 4:e07236 10.7554/eLife.07236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cottee M.A., Johnson S., Raff J.W., and Lea S.M.. 2017. A key centriole assembly interaction interface between human PLK4 and STIL appears to not be conserved in flies. Biol. Open. 6:381–389. 10.1242/bio.024661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunha-Ferreira I., Bento I., Pimenta-Marques A., Jana S.C., Lince-Faria M., Duarte P., Borrego-Pinto J., Gilberto S., Amado T., Brito D., et al. 2013. Regulation of autophosphorylation controls PLK4 self-destruction and centriole number. Curr. Biol. 23:2245–2254. 10.1016/j.cub.2013.09.037 [DOI] [PubMed] [Google Scholar]
- Delattre M., Leidel S., Wani K., Baumer K., Bamat J., Schnabel H., Feichtinger R., Schnabel R., and Gönczy P.. 2004. Centriolar SAS-5 is required for centrosome duplication in C. elegans. Nat. Cell Biol. 6:656–664. 10.1038/ncb1146 [DOI] [PubMed] [Google Scholar]
- Delattre M., Canard C., and Gönczy P.. 2006. Sequential protein recruitment in C. elegans centriole formation. Curr. Biol. 16:1844–1849. 10.1016/j.cub.2006.07.059 [DOI] [PubMed] [Google Scholar]
- Dzhindzhev N.S., Yu Q.D., Weiskopf K., Tzolovsky G., Cunha-Ferreira I., Riparbelli M., Rodrigues-Martins A., Bettencourt-Dias M., Callaini G., and Glover D.M.. 2010. Asterless is a scaffold for the onset of centriole assembly. Nature. 467:714–718. 10.1038/nature09445 [DOI] [PubMed] [Google Scholar]
- Dzhindzhev N.S., Tzolovsky G., Lipinszki Z., Schneider S., Lattao R., Fu J., Debski J., Dadlez M., and Glover D.M.. 2014. Plk4 phosphorylates Ana2 to trigger Sas6 recruitment and procentriole formation. Curr. Biol. 24:2526–2532. 10.1016/j.cub.2014.08.061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong C.S., Kim M., Yang T.T., Liao J.C., and Tsou M.F.. 2014. SAS-6 assembly templated by the lumen of cartwheel-less centrioles precedes centriole duplication. Dev. Cell. 30:238–245. 10.1016/j.devcel.2014.05.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu J., and Glover D.M.. 2012. Structured illumination of the interface between centriole and peri-centriolar material. Open Biol. 2:120104 10.1098/rsob.120104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu J., Hagan I.M., and Glover D.M.. 2015. The centrosome and its duplication cycle. Cold Spring Harb. Perspect. Biol. 7:a015800 10.1101/cshperspect.a015800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu J., Lipinszki Z., Rangone H., Min M., Mykura C., Chao-Chu J., Schneider S., Dzhindzhev N.S., Gottardo M., Riparbelli M.G., et al. 2016. Conserved molecular interactions in centriole-to-centrosome conversion. Nat. Cell Biol. 18:87–99. 10.1038/ncb3274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galletta B.J., Fagerstrom C.J., Schoborg T.A., McLamarrah T.A., Ryniawec J.M., Buster D.W., Slep K.C., Rogers G.C., and Rusan N.M.. 2016. A centrosome interactome provides insight into organelle assembly and reveals a non-duplication role for Plk4. Nat. Commun. 7:12476 10.1038/ncomms12476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gartenmann L., Wainman A., Qurashi M., Kaufmann R., Schubert S., Raff J.W., and Dobbie I.M.. 2017. A combined 3D-SIM/SMLM approach allows centriole proteins to be localized with a precision of ∼4-5 nm. Curr. Biol. 27:R1054–R1055. 10.1016/j.cub.2017.08.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goshima G., Wollman R., Goodwin S.S., Zhang N., Scholey J.M., Vale R.D., and Stuurman N.. 2007. Genes required for mitotic spindle assembly in Drosophila S2 cells. Science. 316:417–421. 10.1126/science.1141314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guichard P., Desfosses A., Maheshwari A., Hachet V., Dietrich C., Brune A., Ishikawa T., Sachse C., and Gönczy P.. 2012. Cartwheel architecture of Trichonympha basal body. Science. 337:553 10.1126/science.1222789 [DOI] [PubMed] [Google Scholar]
- Habedanck R., Stierhof Y.D., Wilkinson C.J., and Nigg E.A.. 2005. The Polo kinase Plk4 functions in centriole duplication. Nat. Cell Biol. 7:1140–1146. 10.1038/ncb1320 [DOI] [PubMed] [Google Scholar]
- Hatzopoulos G.N., Erat M.C., Cutts E., Rogala K.B., Slater L.M., Stansfeld P.J., and Vakonakis I.. 2013. Structural analysis of the G-box domain of the microcephaly protein CPAP suggests a role in centriole architecture. Structure. 21:2069–2077. 10.1016/j.str.2013.08.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland A.J., Lan W., Niessen S., Hoover H., and Cleveland D.W.. 2010. Polo-like kinase 4 kinase activity limits centrosome overduplication by autoregulating its own stability. J. Cell Biol. 188:191–198. 10.1083/jcb.200911102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klebba J.E., Buster D.W., Nguyen A.L., Swatkoski S., Gucek M., Rusan N.M., and Rogers G.C.. 2013. Polo-like kinase 4 autodestructs by generating its Slimb-binding phosphodegron. Curr. Biol. 23:2255–2261. 10.1016/j.cub.2013.09.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klebba J.E., Buster D.W., McLamarrah T.A., Rusan N.M., and Rogers G.C.. 2015a Autoinhibition and relief mechanism for Polo-like kinase 4. Proc. Natl. Acad. Sci. USA. 112:E657–E666. 10.1073/pnas.1417967112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klebba J.E., Galletta B.J., Nye J., Plevock K.M., Buster D.W., Hollingsworth N.A., Slep K.C., Rusan N.M., and Rogers G.C.. 2015b Two Polo-like kinase 4 binding domains in Asterless perform distinct roles in regulating kinase stability. J. Cell Biol. 208:401–414. 10.1083/jcb.201410105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleylein-Sohn J., Westendorf J., Le Clech M., Habedanck R., Stierhof Y.D., and Nigg E.A.. 2007. Plk4-induced centriole biogenesis in human cells. Dev. Cell. 13:190–202. 10.1016/j.devcel.2007.07.002 [DOI] [PubMed] [Google Scholar]
- Kratz A.S., Bärenz F., Richter K.T., and Hoffmann I.. 2015. Plk4-dependent phosphorylation of STIL is required for centriole duplication. Biol. Open. 4:370–377. 10.1242/bio.201411023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopes C.A., Jana S.C., Cunha-Ferreira I., Zitouni S., Bento I., Duarte P., Gilberto S., Freixo F., Guerrero A., Francia M., et al. 2015. PLK4 trans-autoactivation controls centriole biogenesis in space. Dev. Cell. 35:222–235. 10.1016/j.devcel.2015.09.020 [DOI] [PubMed] [Google Scholar]
- Lowery D.M., Lim D., and Yaffe M.B.. 2005. Structure and function of Polo-like kinases. Oncogene. 24:248–259. 10.1038/sj.onc.1208280 [DOI] [PubMed] [Google Scholar]
- Mennella V., Keszthelyi B., McDonald K.L., Chhun B., Kan F., Rogers G.C., Huang B., and Agard D.A.. 2012. Subdiffraction-resolution fluorescence microscopy reveals a domain of the centrosome critical for pericentriolar material organization. Nat. Cell Biol. 14:1159–1168. 10.1038/ncb2597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moyer T.C., Clutario K.M., Lambrus B.G., Daggubati V., and Holland A.J.. 2015. Binding of STIL to Plk4 activates kinase activity to promote centriole assembly. J. Cell Biol. 209:863–878. 10.1083/jcb.201502088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nye J., Buster D.W., and Rogers G.C.. 2014. The use of cultured Drosophila cells for studying the microtubule cytoskeleton. Methods Mol. Biol. 1136:81–101. 10.1007/978-1-4939-0329-0_6 [DOI] [PubMed] [Google Scholar]
- Ohta M., Ashikawa T., Nozaki Y., Kozuka-Hata H., Goto H., Inagaki M., Oyama M., and Kitagawa D.. 2014. Direct interaction of Plk4 with STIL ensures formation of a single procentriole per parental centriole. Nat. Commun. 5:5267 10.1038/ncomms6267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peel N., Stevens N.R., Basto R., and Raff J.W.. 2007. Overexpressing centriole-replication proteins in vivo induces centriole overduplication and de novo formation. Curr. Biol. 17:834–843. 10.1016/j.cub.2007.04.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelletier L., O’Toole E., Schwager A., Hyman A.A., and Müller-Reichert T.. 2006. Centriole assembly in Caenorhabditis elegans. Nature. 444:619–623. 10.1038/nature05318 [DOI] [PubMed] [Google Scholar]
- Riparbelli M.G., Gottardo M., Glover D.M., and Callaini G.. 2014. Inhibition of Polo kinase by BI2536 affects centriole separation during Drosophila male meiosis. Cell Cycle. 13:2064–2072. 10.4161/cc.29083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues-Martins A., Riparbelli M., Callaini G., Glover D.M., and Bettencourt-Dias M.. 2007. Revisiting the role of the mother centriole in centriole biogenesis. Science. 316:1046–1050. 10.1126/science.1142950 [DOI] [PubMed] [Google Scholar]
- Rogala K.B., Dynes N.J., Hatzopoulos G.N., Yan J., Pong S.K., Robinson C.V., Deane C.M., Gönczy P., and Vakonakis I.. 2015. The Caenorhabditis elegans protein SAS-5 forms large oligomeric assemblies critical for centriole formation. eLife. 4:e07410 10.7554/eLife.07410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers S.L., and Rogers G.C.. 2008. Culture of Drosophila S2 cells and their use for RNAi-mediated loss-of-function studies and immunofluorescence microscopy. Nat. Protoc. 3:606–611. 10.1038/nprot.2008.18 [DOI] [PubMed] [Google Scholar]
- Rogers G.C., Rusan N.M., Roberts D.M., Peifer M., and Rogers S.L.. 2009. The SCFSlimb ubiquitin ligase regulates Plk4/Sak levels to block centriole reduplication. J. Cell Biol. 184:225–239. 10.1083/jcb.200808049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossignol P., Collier S., Bush M., Shaw P., and Doonan J.H.. 2007. Arabidopsis POT1A interacts with TERT-V(I8), an N-terminal splicing variant of telomerase. J. Cell Sci. 120:3678–3687. 10.1242/jcs.004119 [DOI] [PubMed] [Google Scholar]
- Rothbauer U., Zolghadr K., Muyldermans S., Schepers A., Cardoso M.C., and Leonhardt H.. 2008. A versatile nanotrap for biochemical and functional studies with fluorescent fusion proteins. Mol. Cell. Proteomics. 7:282–289. 10.1074/mcp.M700342-MCP200 [DOI] [PubMed] [Google Scholar]
- Shimanovskaya E., Qiao R., Lesigang J., and Dong G.. 2013. The SAS-5 N-terminal domain is a tetramer, with implications for centriole assembly in C. elegans. Worm. 2:e25214 10.4161/worm.25214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slevin L.K., Romes E.M., Dandulakis M.G., and Slep K.C.. 2014. The mechanism of dynein light chain LC8-mediated oligomerization of the Ana2 centriole duplication factor. J. Biol. Chem. 289:20727–20739. 10.1074/jbc.M114.576041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens N.R., Dobbelaere J., Brunk K., Franz A., and Raff J.W.. 2010a Drosophila Ana2 is a conserved centriole duplication factor. J. Cell Biol. 188:313–323. 10.1083/jcb.200910016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens N.R., Roque H., and Raff J.W.. 2010b DSas-6 and Ana2 coassemble into tubules to promote centriole duplication and engagement. Dev. Cell. 19:913–919. 10.1016/j.devcel.2010.11.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang C.J., Lin S.Y., Hsu W.B., Lin Y.N., Wu C.T., Lin Y.C., Chang C.W., Wu K.S., and Tang T.K.. 2011. The human microcephaly protein STIL interacts with CPAP and is required for procentriole formation. EMBO J. 30:4790–4804. 10.1038/emboj.2011.378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsou M.F., and Stearns T.. 2006. Mechanism limiting centrosome duplication to once per cell cycle. Nature. 442:947–951. 10.1038/nature04985 [DOI] [PubMed] [Google Scholar]
- Vulprecht J., David A., Tibelius A., Castiel A., Konotop G., Liu F., Bestvater F., Raab M.S., Zentgraf H., Izraeli S., and Krämer A.. 2012. STIL is required for centriole duplication in human cells. J. Cell Sci. 125:1353–1362. 10.1242/jcs.104109 [DOI] [PubMed] [Google Scholar]
- Wong Y.L., Anzola J.V., Davis R.L., Yoon M., Motamedi A., Kroll A., Seo C.P., Hsia J.E., Kim S.K., Mitchell J.W., et al. 2015. Reversible centriole depletion with an inhibitor of Polo-like kinase 4. Science. 348:1155–1160. 10.1126/science.aaa5111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J., Shen C., Wang T., and Quan J.. 2013. Structural basis for the inhibition of Polo-like kinase 1. Nat. Struct. Mol. Biol. 20:1047–1053. 10.1038/nsmb.2623 [DOI] [PubMed] [Google Scholar]
- Zitouni S., Francia M.E., Leal F., Montenegro Gouveia S., Nabais C., Duarte P., Gilberto S., Brito D., Moyer T., Kandels-Lewis S., et al. 2016. CDK1 prevents unscheduled PLK4-STIL complex assembly in centriole biogenesis. Curr. Biol. 26:1127–1137. 10.1016/j.cub.2016.03.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.