How collective cell migration mediates tissue shape changes during the development of normal tissue or tumors is unclear. Fessenden et al. show that the formin Dia1 stabilizes adhesions of protruding MDCK cells during branching morphogenesis, permitting force generation against collagen and eventual tissue shape changes.
Abstract
Developing tissues change shape and tumors initiate spreading through collective cell motility. Conserved mechanisms by which tissues initiate motility into their surroundings are not known. We investigated cytoskeletal regulators during collective invasion by mouse tumor organoids and epithelial Madin–Darby canine kidney (MDCK) acini undergoing branching morphogenesis in collagen. Use of the broad-spectrum formin inhibitor SMIFH2 prevented the formation of migrating cell fronts in both cell types. Focusing on the role of the formin Dia1 in branching morphogenesis, we found that its depletion in MDCK cells does not alter planar cell motility either within the acinus or in two-dimensional scattering assays. However, Dia1 was required to stabilize protrusions extending into the collagen matrix. Live imaging of actin, myosin, and collagen in control acini revealed adhesions that deformed individual collagen fibrils and generated large traction forces, whereas Dia1-depleted acini exhibited unstable adhesions with minimal collagen deformation and lower force generation. This work identifies Dia1 as an essential regulator of tissue shape changes through its role in stabilizing focal adhesions.
Introduction
Tissue shape changes encompass multiple developmental and pathological processes. To form branched tubular networks, developing tissues such as mammalian vasculature or the Drosophila melanogaster trachea undergo extensive elongation and remodeling known as branching morphogenesis (Lubarsky and Krasnow, 2003; Lecaudey and Gilmour, 2006; Wang et al., 2017). In many cases, branching morphogenesis is initiated when growth factors stimulate a few individual cells within the developing tissue to extend protrusions that adhere to the surrounding ECM. These cells subsequently lead cohorts of their neighbors out of their initial site, migrating collectively through the ECM to form extensively branched tubules (O’Brien et al., 2002; Affolter et al., 2009). Malignant tissue can exhibit similar, if deregulated, shape changes during local invasion from the site of tumor formation (Friedl et al., 2012). Invasion by tumors is often accomplished by collective cell migration in a manner that frequently mimics development (Gray et al., 2010; Friedl and Alexander, 2011). In both developmental and pathological contexts, shape changes undertaken by tissues rely on the coordination of cell motility and cell adhesions to neighboring cells and the ECM.
An outstanding question is how tissues transition from compact structures dominated by cell–cell adhesions to invading cohorts of cells that interact extensively with their ECMs. A well-established framework describing the acquisition of invasive behaviors is the epithelial–mesenchymal transition (EMT; Thiery et al., 2009). EMT comprises a gene-regulatory program that simultaneously suppresses cells’ epithelial traits while activating mesenchymal traits, thereby stimulating invasion. However, EMT does not adequately describe tissue shape changes when epithelial traits such as cell–cell adhesion are maintained (Kowalski et al., 2003; Affolter et al., 2009; Shamir et al., 2014). In these cases, a partial or transient EMT has been proffered to account for invasive behaviors exhibited by intact tissues (O’Brien et al., 2002; Christiansen and Rajasekaran, 2006; Revenu and Gilmour, 2009; Friedl et al., 2012; Lambert et al., 2017). But this model leaves unclear how the partial loss or gain of epithelial or mesenchymal traits, respectively, can orchestrate collective cell invasion (O’Brien et al., 2004; Ewald et al., 2012). For example, cell movements within tissues are required in some cases to maintain epithelial homeostasis (Haigo and Bilder, 2011; Wang et al., 2013; Isabella and Horne-Badovinac, 2016), but in other cases, they are required to drive branching morphogenesis (Ewald et al., 2008; Wang et al., 2017). Thus, we lack precise mechanisms to describe how motility and adhesions to the ECM are shifted in individual cells to accomplish tissue shape changes.
Cell motility and adhesions rely on the actin cytoskeleton, which is organized in space and time into protrusive, contractile, and adhesive organelles (Lauffenburger and Horwitz, 1996). Protrusion of the cell’s leading edge is typically driven by Arp2/3–mediated lamellipodia (Pollard and Borisy, 2003; Gardel et al., 2010). Proximal to the lamellipodium and within a RhoA-dependent lamellum, actomyosin networks construct actin bundles and generate contractile forces. Coordinated with the actin cytoskeleton is the assembly and maturation of focal adhesions, which serve as sites of biochemical signaling and as mechanical linkages between the cell and its surroundings (Gardel et al., 2010; Geiger and Yamada, 2011). Focal adhesions assemble within the lamellipodia (Zaidel-Bar et al., 2003), but they undergo increases in size and changes in composition in a maturation process that relies on the Rho effectors myosin II (Riveline et al., 2001) and Dia1 (Chrzanowska-Wodnicka and Burridge, 1996; Oakes et al., 2012). Focal adhesion maturation has been extensively studied in cells on 2D planar surfaces and exerts context-dependent effects on matrix deposition, front–rear polarity, and migration speed (Hoffman et al., 2006; Oakes et al., 2012; Thievessen et al., 2013; Horton et al., 2016; Rahman et al., 2016). In fibrillar 3D environments, focal adhesion morphology is significantly altered, and the role of focal adhesion maturation is less well defined (Fraley et al., 2010; Harunaga and Yamada, 2011; Kubow et al., 2013; Doyle et al., 2015; Owen et al., 2017). Although branching morphogenesis and tumor invasion require canonical focal adhesion components (Jiang et al., 2001; Wei et al., 2009; Hunter and Zegers, 2010; Friedl et al., 2012), the mechanisms underlying focal adhesion regulation and the physiological consequences of their maturation are not known.
We used 3D culture and organotypic models to dissect the contributions of cytoskeletal organelles to tissue shape changes. A tractable in vitro model to study the regulation of tissue remodeling is provided by MDCK cells undergoing branching morphogenesis. MDCK cells cultured in 3D matrices form hollow acini that resemble simple cuboidal epithelial tissues (Bryant and Mostov, 2008). These undergo robust and well-described branching morphogenesis in response to hepatocyte growth factor (HGF; Yu et al., 2003). In this study, we show that the formin Dia1 is dispensable for growth and polarization of MDCK acini and, interestingly, that Dia1 depletion does not affect HGF-mediated planar cell motility in either cell-scattering assays or within acini. Rather, it is selectively required for cells to form stable adhesions that generated large traction forces and deformed collagen fibrils during the initiation of branching morphogenesis. Thus, Dia1 conditions tissue shape changes by controlling the stabilization of cell–collagen adhesions.
Results
Formin activity is required for invasion and branching morphogenesis
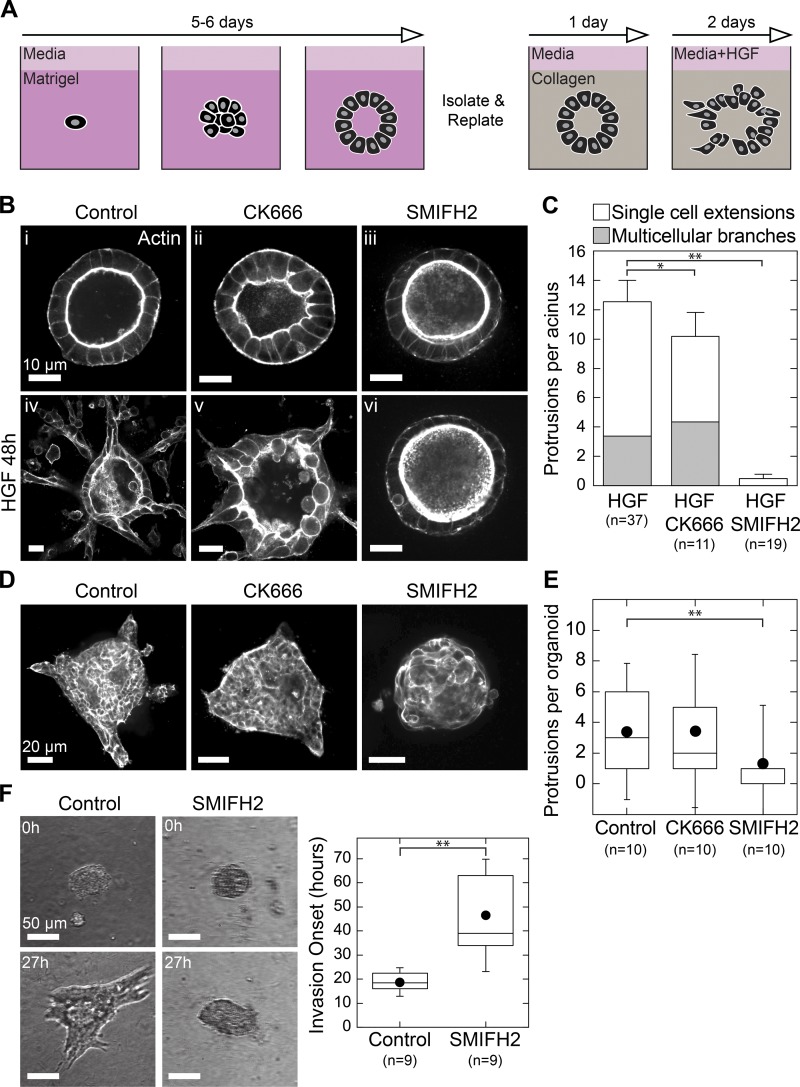
To explore the roles of Arp2/3 and formins in tissue shape changes, we used two cell models: MDCK acini undergoing branching morphogenesis and invasive motility by murine tumor explants. Single MDCK cells embedded in Matrigel grew into polarized acini with clear lumens over 4–5 d. Using a protocol modified from Rubashkin et al. (2014), we isolated acini and plated them intact into 2-mg/ml collagen gels. Once plated in collagen, acini remained quiescent for at least 1 wk or could be induced to undergo branching morphogenesis by the addition of 20 ng/ml HGF (Fig. 1 A; see also the Culture and manipulation of acini section of Materials and methods). After 48 h, branching morphogenesis resulted in multiple protrusive fronts extending into the collagen gel from each acinus. These protrusions started as extensions by single cells that then lengthened to form chains and tubules of cells as shown by phalloidin staining for actin (Fig. S1 A). The size and growth rate of protrusions we observed agreed with the stages of branching morphogenesis described previously (Yu et al., 2003; Zegers et al., 2003).
Figure 1.
Formin activity is required for invasion and branching morphogenesis. (A) Strategy for 3D culture and branching morphogenesis of MDCK acini. (B) i–iii: Equatorial confocal sections of MDCK acini showing phalloidin stain for actin. Acini were plated in collagen and treated with DMSO, 50 µM CK666, or 30 µM SMIFH2 for 48 h. iv–vi: Confocal sections showing actin in acini treated for 48 h with 20 ng/ml HGF with or without the indicated inhibitors. (C) Stacked plot showing protrusion type and number formed per acinus in each condition. Error bars indicate SEM. (D) Mouse mammary tumor organoids were harvested and digested, and resulting organoids were plated directly in collagen gels and treated with serum plus DMSO, 50 µM CK666, or 30 µM SMIFH2. Shown are equatorial confocal sections of phalloidin stain for actin after fixation at 24 h. (E) Box plot of protrusions per organoid, with number of organoids scored indicated below. (F) Organoids were plated in collagen, treated with serum alone or with 30 µM SMIFH2, and imaged in brightfield for 72 h by time-lapse microscopy (see also Video 1). Invasion onset was scored as the time of initial extension into collagen gel and was plotted in a box plot with the number of organoids scored indicated below. Box plots show the 25th and 75th percentiles and the median, circles indicate means, and whiskers mark 1.5 SDs. *, P < 0.01; **, P < 0.05 by a Student’s two-tailed t test assuming unequal variance.
To explore the role of Arp2/3 and formins in regulating branching morphogenesis, we treated acini with the Arp2/3 inhibitor CK666 (Nolen et al., 2009) or the pan-formin inhibitor SMIFH2 (Rizvi et al., 2009). Phalloidin staining in control acini before branching morphogenesis revealed that F-actin was localized exclusively to the cell membranes. The apical surface was marked by a dense band of actin because of the presence of microvilli as previously reported (Fig. 1 B, i; McAteer et al., 1987). Addition of 20 ng/ml HGF resulted in prototypical branching morphogenesis (Fig. 1 B, iv). Treatment with 50 µM CK666 in the absence of HGF caused individual cells to bulge within acini as indicated by convex curvature at the apical membrane (Fig. 1 B, ii). This suggested an acute polarity defect consistent with reported roles for Arp2/3 in forming polarized membrane domains (Martin-Belmonte et al., 2007). Stimulating acini with HGF in the presence of CK666 did not prevent acini from forming multiple protrusions into the collagen gel even as cell morphology remained perturbed (Fig. 1 B, v). These phenotypes were unchanged when we increased CK666 concentration to 100 µM (Fig. S1, C–E). In contrast with Arp2/3 inhibition, treating acini with 30 µM SMIFH2 did not appreciably alter acinar morphology relative to controls (Fig. 1 B, iii). When stimulated with HGF, however, SMIFH2-treated acini did not form any discernable protrusions into the collagen gel. Rather, these acini were indistinguishable from untreated controls (Fig. 1 B, vi).
We scored the response of acini to HGF in these conditions by counting protrusive structures, whether by single cells or multicellular chains or branches invading into the collagen gel. This confirmed that SMIFH2 but not CK666 prevented protrusion formation in response to HGF (Fig. 1 C). We scored protrusion lengths under these conditions and found that branching morphogenesis is moderately impacted by Arp2/3 inhibition but is blocked completely upon formin inhibition (Fig. S1 B). Collectively, these data demonstrate that Arp2/3 signaling acutely impacts cell shape within acini and moderately affects invasion during branching morphogenesis. Meanwhile, formin activity is dispensable for acinar morphology but is required for HGF-mediated invasion into the surrounding collagen matrix.
To confirm the generality of these results, we performed invasion assays with primary tumor organoids from mice. In contrast with MDCK acini, tumors from the murine mammary tumor virus–polyoma middle-T (MMTV-PyMT) mouse strain comprise heterogeneous cell populations, do not form a basement membrane, and do not require a defined signal to invade into collagen gels (Lin et al., 2003; Cheung et al., 2013). Mice with advanced tumors were sacrificed, and tumor tissue was harvested and digested into multicellular organoids. Organoids were first cultured in Matrigel for 48 h in low-serum media and then replated intact into collagen gels. To stimulate invasion, media containing serum was added, and after 24 h, tumor organoids were fixed to visualize the actin cytoskeleton. Control organoids were uniformly invasive, extending multicellular fronts into the collagen matrix (Fig. 1 D). Inhibition of Arp2/3 did not affect the total number of fronts per organoid. However, SMIFH2-treated organoids did not form invasive fronts after 24 h (Fig. 1 E). To clarify the motility defects rendered by formin inhibition, we analyzed invasion in tumor organoids via time-lapse imaging (Video 1). Cells in control and SMIFH2-treated tumor organoids appeared to actively rearrange over 48 h. However, by 20 h after plating, control organoids had formed subcellular extensions from which collective invasion proceeded. Formin inhibition prevented organoids from generating such extensions until ∼40 h after plating (Fig. 1 F).
Collectively, these data implicate formin activity as a previously unappreciated determinant of both branching morphogenesis by MDCK acini and collective invasion by mouse tumor organoids. In MDCK acini, formin activity appears to be dispensable for cell shape and acinar homeostasis before HGF stimulation. However, in both acini and tumor organoids, formin inhibition blocked tissue shape changes by preventing the formation of invasive fronts into the surrounding collagen matrix. In this study, we examine tissue shape changes using MDCK branching morphogenesis, and we refer hereafter to invasive behaviors of MDCK cells within the context of branching morphogenesis.
Dia1 and FHOD1 are required for branching morphogenesis
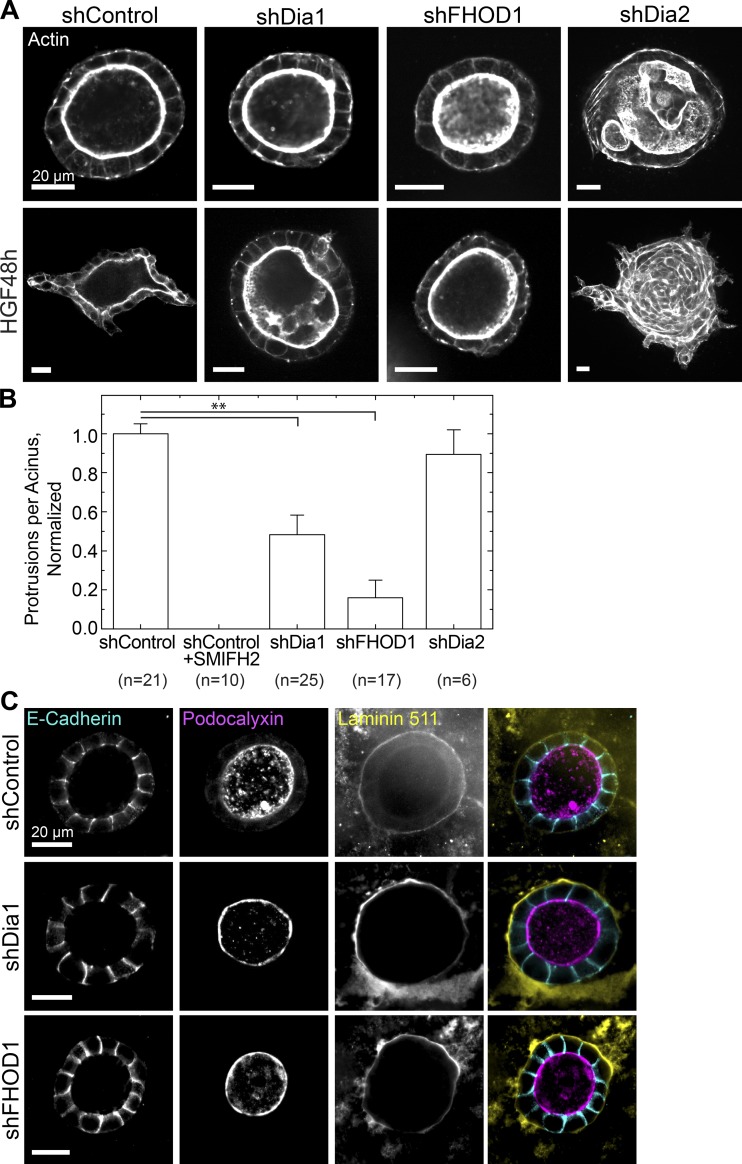
Because SMIFH2 is a pleiotropic inhibitor of formins (Rizvi et al., 2009), we sought to identify formin family members required for invasion. We initially chose to explore the roles of three formin family members: Diaphanous 1 (Dia1), diaphanous 2 (Dia2), and formin homology 2 domain–containing 1 (FHOD1). We and others have previously established roles for Dia1 in cell motility and focal adhesion maturation (Riveline et al., 2001; Oakes et al., 2012). Like Dia1, Dia2 is activated by Rho and participates in stress fiber formation and cell motility, but it is also implicated in cytokinetic ring formation (Gupton et al., 2007; Pollard, 2010; Pettee et al., 2014). FHOD1 is a noncanonical formin activated through the Rho effector Rho-associated protein kinase (ROCK) and participates in focal adhesion maturation (Takeya et al., 2008; Iskratsch et al., 2013). shRNA constructs targeting these genes reduced their expression to 28–50% of control levels (Fig. S2, A–C).
Stable cell lines expressing each of these shRNA constructs could proliferate in 2D culture and in Matrigel. After replating acini from each into collagen gels, we fixed and stained them to analyze acinar morphology and actin architecture. Although shDia1 and shFHOD1 cells could form acini with cleared lumens, shDia2 cells formed acini of aberrant shapes without clear lumens (Figs. 2 A and S2 D).
Figure 2.
Dia1 and FHOD1 are required for branching morphogenesis. (A) Equatorial confocal sections of actin in acini of each genotype before (top) and after stimulation with HGF for 48 h (bottom). (B) The number of protrusions per acinus relative to matched shControl acini; numbers of acini scored are indicated below. SMIFH2 treatment serves as a negative control. See also Fig. S2. **, P < 0.05 by a Student’s two-tailed t test assuming unequal variance. Error bars indicate SD. (C) Equatorial confocal sections of acini stained for E-cadherin to mark cell–cell junctions, podocalyxin to mark the apical surface, and the basement membrane component laminin 511 in acini generated from control (top), shDia1 (middle), and shFHOD1 (bottom) cells.
To test their capacity to undergo branching morphogenesis, we treated acini from each cell type with 20 ng/ml HGF. After 48 h, shDia1 and shFHOD1 acini formed significantly fewer invasive protrusions relative to shControl acini (Fig. 2 A, bottom row). We analyzed protrusions as above and compared the mean number of protrusions formed per acinus in each genotype (Fig. S2 E). Combining data from all acini showed a significant decrease in protrusions formed by shDia1 and shFHOD1 acini but not shDia2 acini relative to matched controls (Fig. 2 B). When protrusion lengths were measured across these conditions, we found that the protrusions formed by shDia1 and shFHOD1 acini were shorter on average than matched controls (Fig. S2 F).
Although they appeared morphologically similar to controls, knockdown of Dia1 and FHOD1 may still alter cell polarity within the acini. To test for polarity defects in shDia1 and shFHOD1 acini, we stained for E-cadherin, podocalyxin, and laminin 511 by immunofluorescence. Localization of these proteins to lateral, apical, and basal surfaces, respectively, resembled that in control acini (Fig. 2 C). These results suggest that as they develop and polarize, shDia1 and shFHOD1 acini could generate and orient an apical domain marked by podocalyxin and form adherens junctions marked by E-cadherin. After replating in collagen, these acini also deposited basement membrane laminins similar to their control counterparts.
We conclude from these data that depletion of Dia1 or FHOD1 does not impair the growth and polarization of acini. These formins are, however, required to initiate branching morphogenesis by establishing and elongating protrusions into the collagen gel. In contrast, Dia2 is required for normal acinar development but is dispensable for generating invasive protrusions in response to HGF. Dia1 and FHOD1 appear to play overlapping roles, perhaps because of their activation directly and indirectly, respectively, through Rho signaling. ROCK regulates myosin contractility and restricts rather than promotes protrusion formation during branching morphogenesis (Yu et al., 2003). In this study, we chose to focus on the role of Dia1 in this process.
Dia1 is dispensable for HGF-mediated planar motility in 2D and within acini
Branching morphogenesis requires that cells migrate into the surrounding collagen matrix (Yu et al., 2003). Previous work has shown the importance of Dia1 in the establishment of a polarized leading edge for motility on 2D substrates (Dachsel et al., 2013; Isogai et al., 2015). Thus, one possibility is that a cell migration defect underlies the role of Dia1 in branching morphogenesis.
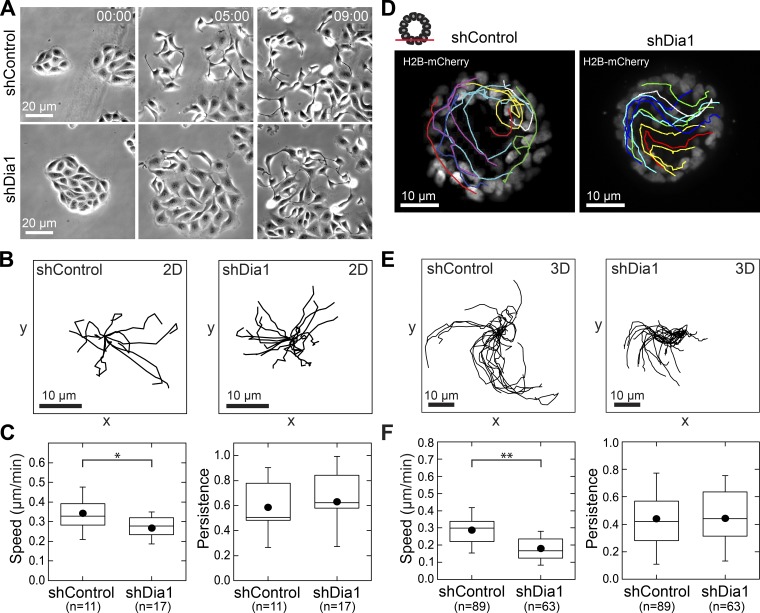
To test this hypothesis, we first used a cell-scattering assay in which HGF drives the dissociation of cell islands on glass coverslips into individual, highly motile cells (Stoker et al., 1987). We plated shControl and shDia1 MDCK cells on glass coverslips and serum starved them for 24 h before adding 20 ng/ml HGF to stimulate scattering. Time-lapse microscopy revealed that shDia1 cells could break cell–cell contacts and migrate as single cells similarly to shControl cells (Fig. 3 A and Video 2). We tracked individual cells (Fig. 3 B) and found only a modest reduction in the mean instantaneous speed of shDia1 cells and no change in their persistence (Fig. 3 C). Thus, Dia1 is dispensable for HGF-mediated motility in 2D.
Figure 3.
Dia1 is dispensable for HGF-mediated planar motility in 2D and within acini. (A) Transmitted light images at 0, 5, and 9 h of shControl and shDia1 cell island scattering after 20 ng/ml HGF addition at 0 h. Time is indicated in h:min. See also Video 2. (B) Rose plots of 10 cell trajectories from the cell islands scattering in A and Video 2 over 9 h. The initial location of each trajectory was positioned at 0, 0. (C) Box plots of mean instantaneous speed and persistence for shControl and shDia1 cells. (D) Acini of shControl- and shDia1-expressing H2B-mCherry. Acini were stimulated with 20 ng/ml HGF and imaged by time-lapse fluorescence microscopy for 12 h. Shown are stills with individual tracks overlaid. See also Video 3. (E) Rose plots from cell trajectories obtained from D and Video 3. (F) Box plots of mean instantaneous speed and persistence to characterize cell motility, with the number of cell trajectories obtained from at least five acini per condition indicated below each plot. Box plots show the 25th and 75th percentiles and the median, circles indicate means, and whiskers mark 1.5 SDs. *, P < 0.01; **, P < 0.05 by a Student’s two-tailed t test assuming unequal variance.
We next explored cell motility within acini. Cell rearrangements and rotation within 3D tissues have been reported in vitro (Pearson and Hunter, 2007) and can contribute to tissue morphogenesis (Ewald et al., 2008; Tanner et al., 2012) and ECM deposition (Wang et al., 2013; Isabella and Horne-Badovinac, 2016). We speculated that MDCK cells may exhibit similar in-plane motility before and perhaps during branching morphogenesis. To track cell motility within acini, we generated Dia1-knockdown cell lines and matched controls expressing the nuclear marker H2B-mCherry. Immediately after stimulating with HGF, we imaged acini from these cell lines via time-lapse confocal microscopy. Shortly after the start of imaging, shControl cells began moving within the acinus (Fig. 3 D and Video 3). This motility resulted in rotation of the entire acinus, with occasional cell rearrangements and rare single cells moving independently of their neighbors. The cell motility within shDia1 acini was virtually indistinguishable from that of control acini (Fig. 3 D). Analysis of individual cell tracks (Fig. 3 E) showed a slight decline in mean instantaneous speed of shDia1 cells from 0.3 µm/min to 0.2 µm/min, whereas mean persistence did not change relative to shControl cells (Fig. 3 F).
These data show that in-plane cell motility driven by HGF encompassing both scattering of small islands on 2D substrates and motility within acini does not require Dia1. This suggests that HGF-mediated motility within the plane of a tissue or on 2D substrata is largely independent of Dia1.
Dia1 is required to stabilize protrusions into the collagen matrix
We next explored whether Dia1 regulates protrusions extending from acini into the surrounding collagen matrix. Defects in branching morphogenesis rendered by Dia1 depletion could arise from reduced formation of protrusions. Alternatively, focal adhesion defects could impair protrusion stability because of reduced adhesion to collagen fibrils.
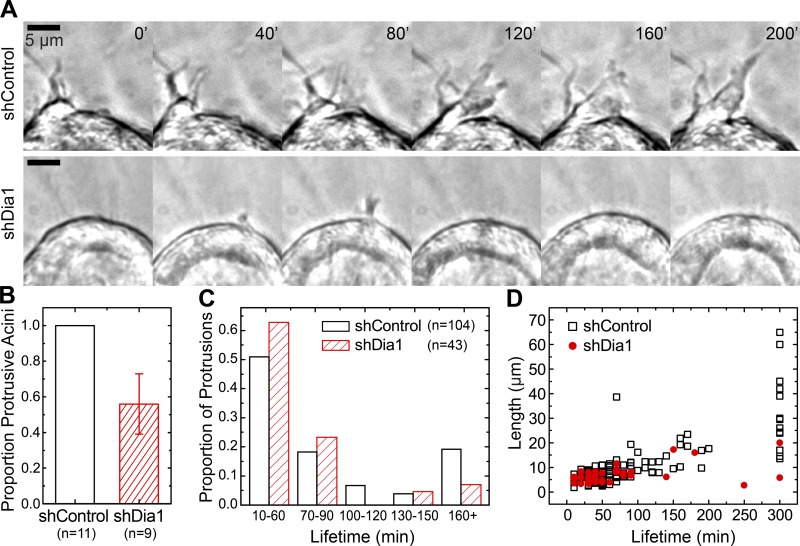
To adjudicate between these hypotheses, we turned to time-lapse imaging in transmitted light and analyzed protrusive activity at the onset of branching morphogenesis. After HGF stimulation, shControl acini extended narrow protrusions into the collagen gel over a period of tens of minutes (Fig. 4 A and Video 4). All shControl acini imaged formed protrusions over 12 h of imaging (Fig. 4 B). We analyzed protrusion lifetimes by measuring the duration over which protrusions remained before they underwent retraction. This analysis revealed that 60% of all protrusions retracted partially or completely within 1 h (Fig. 4 C). A smaller proportion was stable for ≤2 h, and 20% of protrusions were stable over several hours. These long-lived protrusions eventually mediated cell egress from control acini into the collagen matrix (Video 4). Comparing the lifetime of each protrusion to its maximum length confirmed that the subpopulation of most stable protrusions grew to the greatest lengths of >20 µm (Fig. 4 D).
Figure 4.
Dia1 is required to stabilize protrusions into the collagen matrix. (A) Time-lapse imaging in transmitted light during initiation of branching morphogenesis showing the formation and growth of protrusions in shControl and shDia1 acini over a period of 200 min. Images were obtained starting 3 h after addition of 20 ng/ml HGF. See also Video 4. (B) The proportion of acini from each cell type that formed any protrusions over 12 h of imaging with numbers of acini scored indicated below each plot. Error bars indicate SD. (C) Histogram summarizing the distribution of protrusion lifetimes from acini for each cell type from videos obtained 1 h after HGF addition. Numbers of protrusions scored are indicated in the legend. (D) Plot of maximum protrusion length reached as a function of lifetime.
In contrast with control acini, shDia1 acini exhibited impaired protrusion stability (Fig. 4 A and Video 4). Although 50% of shDia1 acini formed protrusions, the distribution of protrusion lifetimes was weighted toward short protrusions lasting <90 min (Fig. 4, B and C). The loss of stable protrusions lasting >2 h in shDia1 acini was matched by a reduction in the maximum length they reached (Fig. 4 D). Meanwhile, protrusions lasting <90 min were indistinguishable between shDia1 and shControl acini (Fig. S3 A). To test whether this failure of protrusion stability caused motility defects that were cell autonomous, we performed time-lapse imaging of single MDCK cells in collagen gels. This revealed a significant decrease in the motility of single shDia1 cells relative to their shControl counterparts after stimulation with HGF (Fig. S3, B and C; and Video 5).
These data show that under normal conditions, the early stages of branching morphogenesis are characterized by numerous small protrusions extending from the acinus into the surrounding collagen matrix. Although a majority of these protrusions are unstable and retract within 90 min, a minority of protrusions adhere stably and extend into the collagen matrix. The correlation observed between protrusion lifetime and maximum length indicates that protrusion growth depends acutely on cells’ ability to adhere to collagen. Dia1 knockdown did not abolish protrusions, nor did it alter lifetimes and sizes of the short-lived protrusions relative to controls. Rather, Dia1 was required for protrusions to stably adhere to collagen matrix and subsequently elongate. These data therefore strongly suggest that Dia1 conditions branching morphogenesis through its role in focal adhesion stability, which permits migration in 3D collagen networks.
Dia1 is required to adhere to and displace individual collagen fibrils
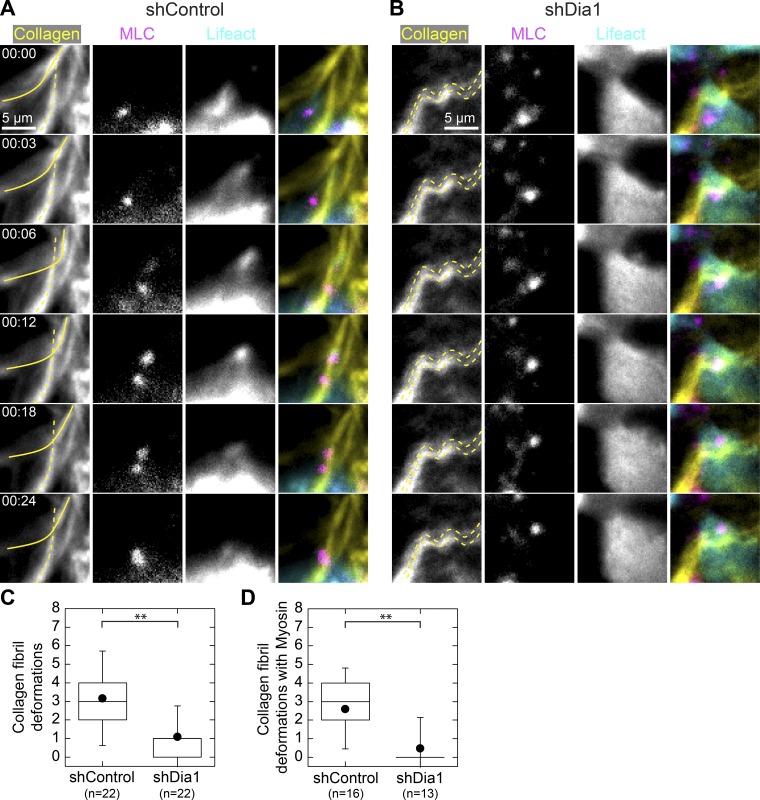
To directly assess cell interactions with collagen fibrils, we used fluorescence live-cell imaging to examine actin, myosin, and collagen fibrils during branching morphogenesis. We imaged shControl and shDia1 acini coexpressing GFP-LifeAct and mCherry-tagged myosin light chain (MLC) plated in collagen matrices labeled with Alexa Fluor 647. After incubating acini for 4 h in HGF, we captured fluorescence confocal images at the lower surface of acini juxtaposed with the collagen matrix. To better capture dynamic cell adhesions to collagen, we acquired images at 3-min intervals for 3 h. We noted passive responses of the collagen matrix as cells moved within acini. However, we also observed shControl cells deforming single-collagen fibrils at discrete points with displacements of ∼2 µm while leaving surrounding fibrils undisturbed (Fig. 5 A and Video 6). We captured deformations within each field of view and found a mean rate of one per hour per acinus (Fig. 5 C). Interestingly, we observed dense MLC puncta and increased actin intensity at sites of fibril deformation, which tracked the collagen fibril as it deformed over tens of minutes (Fig. 5 A and Video 6).
Figure 5.
Dia1 is required to adhere to and displace individual collagen fibrils. (A and B) Fluorescence images at the basal surface of GFP-LifeAct (cyan), mCherry-MLC (magenta), and Alexa Fluor 647–labeled collagen (yellow) obtained 4 h after addition of 20 ng/ml HGF in shControl acini (A) and shDia1 acini (B). The montage in A shows an shControl cell deforming a single collagen fibril, outlined in a solid yellow line, over a period of 24 min. An unaffected fibril is outlined with a dashed line. See also Video 6. (B) Example of an shDia1 cell showing collagen fibrils (dashed lines) that remain immobile as a cell moves over them. See also Video 7. (C) Box plot indicating the frequency of collagen fibril deformations with numbers of acini scored indicated below. (D) Collagen deformations at which MLC appeared or was recruited. Box plots show the 25th and 75th percentiles and the median, circles indicate means, and whiskers mark 1.5 SDs. **, P < 0.01 by a Student’s two-tailed t test assuming unequal variance.
We observed substantially fewer instances of acute collagen deformations in shDia1 acini, although nonspecific movements of the collagen matrix occurred as with controls (Fig. 5 B and Video 7). On average, we observed a threefold decrease in collagen fibril deformations by shDia1 acini relative to controls (Fig. 5 C). When we scored only those deformations with coincident MLC puncta, these were even more suppressed in shDia1 acini relative to controls (Fig. 5 D).
These data identify Dia1-dependent interactions between cells and the collagen matrix during the earliest stages of HGF-stimulated protrusion. Adhesions to collagen are marked by dense myosin puncta through which cells deform single collagen fibrils while leaving surrounding fibrils unaffected. The planar motility observed in shDia1 acini suggests that Dia1 is not necessary for weak adhesion to collagen fibrils that can support planar motility and rotation but is required specifically for adhesion to and contraction against collagen fibrils during the onset of branching morphogenesis.
Dia1 is required for cell spreading and force generation on collagen gels
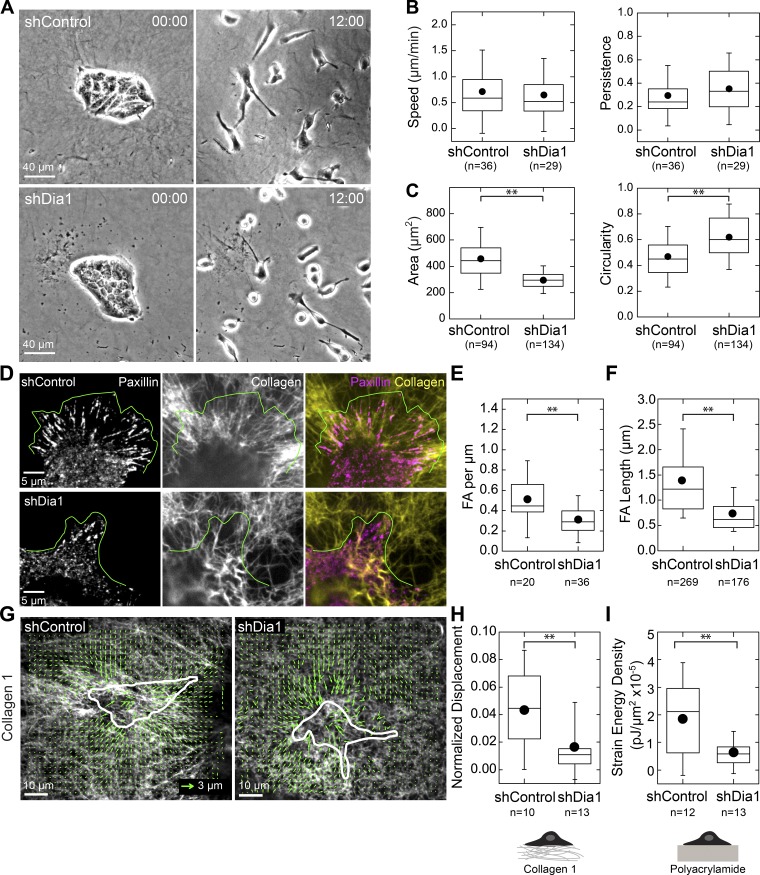
Next, we directly tested MDCK cells’ ability to form adhesions to and exert forces against collagen fibrils. First, we examined individual cells adhered to a collagen network identical to that used for branching morphogenesis assays. We sparsely plated MDCK cells on top of collagen gels prepared using the same protocol as above, and then we stimulated cell islands with 20 ng/ml HGF to induce scattering (Fig. 6 A). Tracking cell motility by time-lapse microscopy showed no difference in cell speed or persistence between scattering shControl and shDia1 cells (Fig. 6 B). We did, however, observe significantly reduced spread area and decreased elongation of scattering shDia1 cells relative to controls (Fig. 6 C).
Figure 6.
Dia1 is required for cell spreading and force generation on collagen gels. (A) Transmitted light images at 0 and 9 h showing shControl and shDia1 cell island scattering after 20 ng/ml HGF addition at 0 h. (B) Box plots of instantaneous speed and persistence for shControl and shDia1 cells. (C) Box plots of cell area and circularity for shControl and shDia1 cells measured at 12 h after HGF addition. (D) Immunofluorescence stain for paxillin in cells plated on Alexa Fluor 647–labeled collagen gels 4 h after HGF addition. The green line indicates cell leading edges. (E) Box plot of focal adhesion (FA) number per micron leading edge. (F) Box plot of focal adhesion length. (G) Vector plot of Alexa Fluor 647–labeled collagen fibril displacements by single cells 12 h after HGF addition. Cell outline is indicated in white. (H) Box plot of the normalized displacement of collagen fibrils by single cells. (I) Box plot showing strain energy density of cells on PA gels measured by traction force microscopy (see also Fig. S3 D). Box plots show the 25th and 75th percentiles and the median, circles indicate means, and whiskers mark 1.5 SDs. **, P < 0.01 by a Student’s two-tailed t test assuming unequal variance.
To assess focal adhesions formed during scattering atop collagen gels, we performed immunofluorescence staining for paxillin. This staining confirmed that cells assemble focal adhesion plaques along fluorescently labeled collagen fibrils (Fig. 6 D). However, shDia1 cells formed ∼50% fewer focal adhesions per µm of leading edge (Fig. 6 E), and those focal adhesions formed were smaller compared with control cells (Fig. 6 F). These data reveal a role for Dia1 in cell spreading and adhesion assembly on collagen gels.
Finally, we sought to quantify the ability of cells to deform and generate forces on collagen matrices. Taking an approach similar to traction force microscopy, we collected images of the collagen gel underlying individual cells 12 h after HGF addition. We removed cells with detergent and acquired a second image of the relaxed collagen gel. With the resulting image pairs, we used particle intensity velocimetry (PIV) to derive strain fields that report the magnitude and distribution of collagen gel contractions (Fig. 6 G). When we normalized to the total area deformed under each cell, we observed a significant decrease in the normalized contraction driven by shDia1 cells relative to shControl cells (Fig. 6 H). To confirm these results, we also performed traditional traction force microscopy of cells plated on collagen-coated polyacrylamide (PA) substrates and found the strain energy density exerted by shDia1 cells was reduced relative to controls (Figs. 6 I and S3 D).
Collectively, these observations demonstrate that loss of Dia1 impairs focal adhesion assembly and elongation on fibrillar collagen networks. This adhesion defect correlates with reduced cell spreading and force generation on collagen fibrils. However, in agreement with our observations of planar motility (Fig. 3), Dia1 is dispensable for rapid cell migration in 2D. These results distinguish weak, Dia1-independent adhesions sufficient for planar motility from larger, Dia1-dependent adhesions through which cells deform collagen fibrils and initiate branching morphogenesis. We next examined whether Dia1 activity supports focal adhesion maturation by directing actomyosin organization at the leading edge of MDCK cells during branching morphogenesis.
Myosin colocalizes with Dia1-dependent focal adhesions
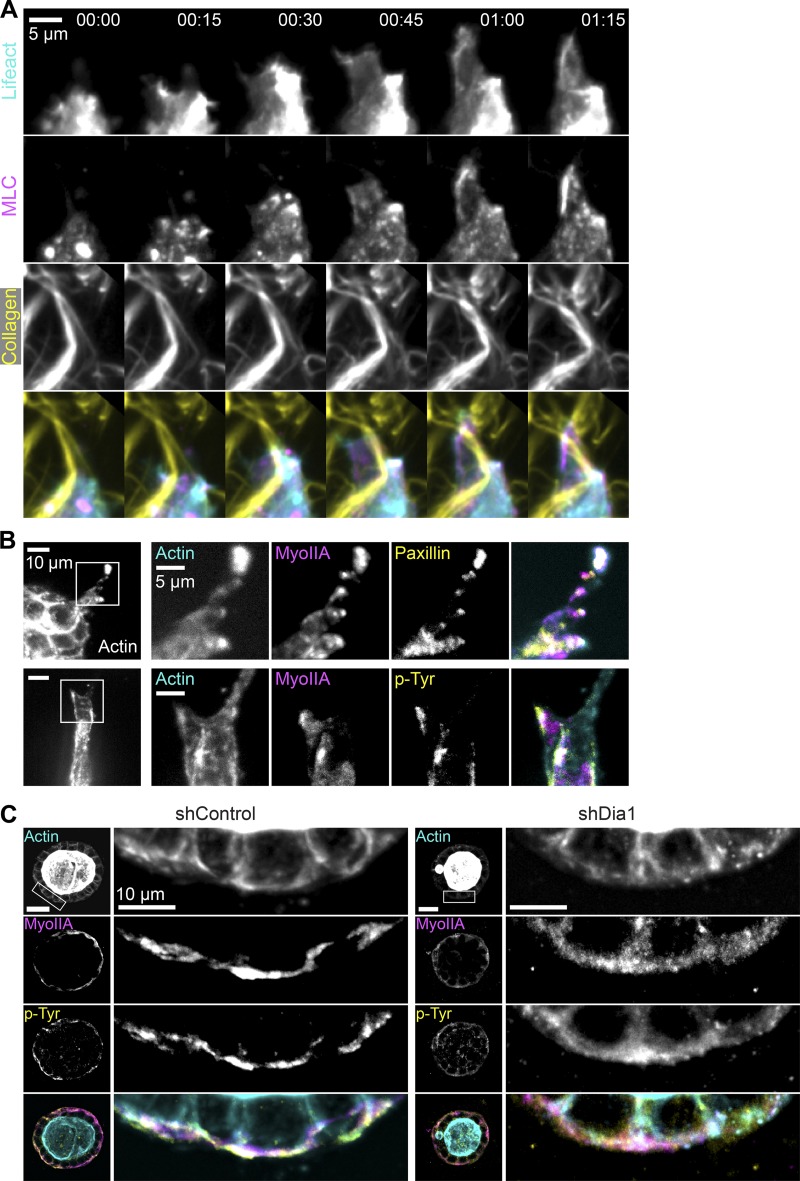
Our observation that myosin accumulates into discrete puncta at sites of adhesion to collagen fibrils (Fig. 5) suggests that myosin-rich puncta contain focal adhesions. To confirm this, we imaged actin, MLC, and collagen, as leader cells protruded away from acini into the surrounding collagen 24 h after stimulation with HGF (Video 8). Leader cells advanced via bursts of actin followed by the accumulation of MLC in puncta at the cell front (Fig. 7 A). MLC puncta at the cell front coincided with points of collagen fibril deformation.
Figure 7.
Myosin colocalizes with Dia1-dependent focal adhesions. (A) Time-lapse images of a leader cell protruding into the surrounding collagen matrix from an shControl acinus stimulated with HGF for 24 h. Images of GFP-LifeAct (cyan), mCherry-MLC (magenta), and Alexa Fluor 647–labeled collagen (yellow) are shown. Time is indicated in h:min. See also Video 8. (B) Immunofluorescence images of actin (cyan), myosin IIA (magenta), and phosphotyrosine (p-Tyr; top) or paxillin (bottom) shown in yellow in leader cells extending from shControl acini after stimulation with HGF for 48 h. (C) Immunofluorescence images of actin (cyan), myosin IIA (magenta), and phosphotyrosine acini 4 h after stimulation with HGF combined into maximum-intensity projections spanning 3 µm. Insets of the indicated boxed regions are shown at right.
To confirm these myosin-rich sites contained canonical focal adhesion proteins, we performed immunofluorescence staining for actin, myosin IIA, and either paxillin or phosphotyrosine. These images revealed that actin- and myosin-rich puncta within leader cells also contained paxillin and phosphotyrosine (Fig. 7 B). We found that around half (0.48 ± 0.16; n = 283) of all phosphotyrosine-positive puncta were also enriched in myosin IIA (Fig. S4 A). These observations confirm that focal adhesions along collagen fibrils retain a punctate appearance and that a subpopulation of them are rich in actin and myosin. Although the myosin localization we observed contrasts with canonical descriptions of myosin exclusion from focal adhesions on planar 2D surfaces (Vicente-Manzanares et al., 2009), our observations agree with studies identifying myosin recruitment at leading edge adhesions in 2D and 3D contexts (Wyckoff et al., 2006; Pasapera et al., 2015).
We next assessed the impact of Dia1 depletion on adhesion formation in acini, to confirm observations performed in single cells overlaid on collagen gels (Fig. 6). We analyzed acini after 4 h of HGF stimulation, when acinar rotation and collagen interactions can be observed. Acini were fixed and immunostained for actin, phosphotyrosine, and myosin IIA, and maximum-intensity projections at the equatorial plane were analyzed. Phosphotyrosine was enriched in dense puncta at the basal surface in shControl acini (Fig. 7 C, left). Meanwhile, phosphotyrosine was diffusely distributed at the basal surface of shDia1 acini (Fig. 7 C, right) and organized into fewer discrete puncta per acinus (Fig. S4 B). This defect was accompanied by a redistribution of phosphotyrosine away from the basal surface in shDia1 acini (Fig. S4 C), which was paralleled by myosin IIA and paxillin localization (Fig. S4 D). Together with data in Fig. 6, these results are consistent with prior research (Oakes et al., 2012) and demonstrate an important role for Dia1 in focal adhesion maturation both in acini and single cells on glass (Fig. S4, E and F) or collagen fibrils.
These data demonstrate that a subset of adhesions to collagen fibrils are marked by actin- and myosin-rich puncta. Myosin accumulation at the leading edge correlates with force generation against collagen fibrils. We find these adhesions do not form in the absence of Dia1, preventing stabilization of protrusive fronts into the collagen matrix. Dia1 has been previously shown to be essential for focal adhesion maturation on 2D surfaces (Oakes et al., 2012), and we suspect that Dia1 plays a similar role in this study. Clearly, because Dia1 is not required for planar cell motility, adhesions to collagen are capable of forming, consistent with prior observations (Oakes et al., 2012). Rather, we hypothesize that Dia1-mediated focal adhesion maturation is necessary to form stable adhesions that facilitate the egress of cells away from the acini into the surrounding collagen.
Dia1 regulates actomyosin organization and dynamics of the basal cortex
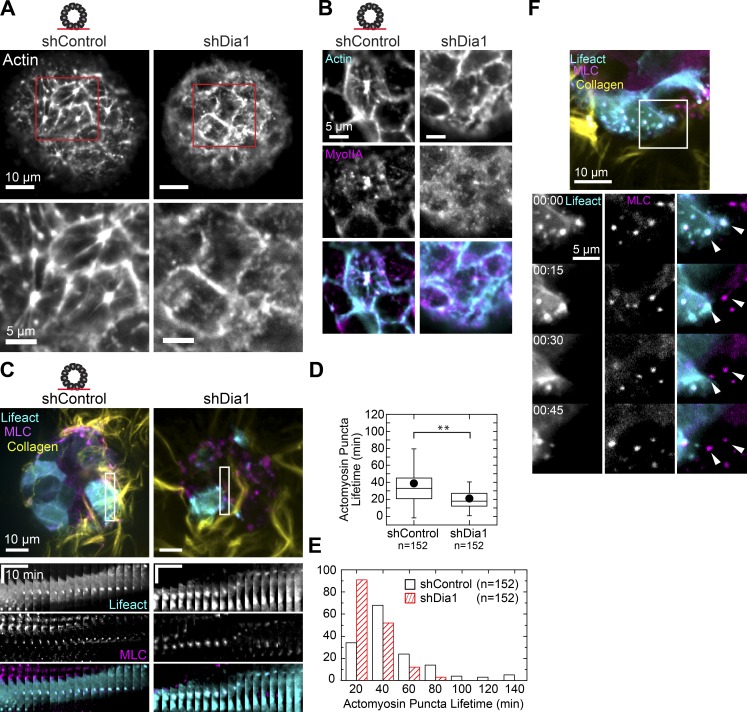
Prior studies have demonstrated that formin-dependent stress fibers act as templates for the compositional and morphological maturation of focal adhesions (Oakes et al., 2012; Iskratsch et al., 2013; Dolat et al., 2014). We hypothesized that Dia1 performs a similar role at the basal actin cortex of acini and may control focal adhesion maturation through its effects on actin architecture. Indeed, confocal sections of acini stained with phalloidin revealed striking differences in the organization of cortical actin between shControl and shDia1 acini. In the absence of HGF, cortical actin in shControl acini was organized into bundles often emanating from dense puncta in the middle of cells (Fig. 8 A, left). In shDia1 acini, the basal cortex was largely absent of such actin bundles and their associated puncta (Fig. 8 A, right). Instead, actin was diffusely spread across the cortex. These differences in basal actin organization remained after HGF stimulation and were matched by altered myosin localization (Fig. 8 B). This confirmed an important role for Dia1 in regulating the organization of the basal actomyosin cortex of acini.
Figure 8.
Dia1 regulates actomyosin organization and dynamics of the basal cortex. (A) Actin stain of the basal surface of acini before HGF addition with regions of interest enlarged below. (B) Immunofluorescence of the basal surface of actin (cyan) and myosin IIA (magenta) in shControl and shDia1 acini stimulated with 20 ng/ml HGF for 4 h. (C) shControl and shDia1 acini expressing GFP-LifeAct (cyan) and mCherry-MLC (magenta) were stimulated for 4 h with HGF, and fluorescence images were taken every 3 min at the basal surface. Kymographs show representative MLC and actin puncta, time scale bar represents 10 min, and distance scale is 10 µm. See also Video 9. (D) Box plot of actomyosin puncta lifetimes per field of view observed over a 3-h window in n = 16 acini of each type. **, P < 0.01 by a Student’s two-tailed t test assuming unequal variance. (E) Histogram showing the distribution of actomyosin puncta lifetimes. (F) Fluorescence images of GFP-LifeAct– (cyan), mCherry-MLC– (magenta), and Alexa Fluor 647–labeled collagen (yellow) in an shControl acinus. Montage shows the movement of a cell with high expression of GFP-LifeAct followed by a low-expressing cell. As the first cell moves to the left, MLC and LifeAct puncta remain stationary. Once the cell moves past this region, the next cell forms puncta at the same points (arrowheads). See also Video 10. Box plot shows the 25th and 75th percentiles and the median, circles indicate means, and whiskers mark 1.5 SDs.
To explore changes in dynamics of the actomyosin cortex, we used time-lapse fluorescence microscopy to image the basal surface of GFP-LifeAct/MLC-mCherry acini (Fig. 8 C). We found MLC puncta in shControl acini remained immobile in cells moving within the plane of the acinus over tens of minutes (Fig. 8 C and Video 9). In contrast, MLC puncta in shDia1 were more dynamic, undergoing assembly and motion over similar time intervals (Fig. 8 C and Video 9). Counting only immobilized actomyosin puncta, we measured an ∼50% decrease in the mean puncta lifetimes between shControl and shDia1 acini. The distribution of actomyosin puncta lifetimes in shControl acini included many short-lived puncta and a few long-lived puncta. Meanwhile, the distribution of puncta in shDia1 acini were shifted toward short lifetimes with no long-lived puncta (Fig. 8, D and E). These distributions agree with those measured for cell protrusion lifetimes (Fig. 4), suggesting that similar cytoskeletal defects rendered by Dia1 depletion underlie both cell protrusions and subcellular actomyosin puncta. Overall, these data show that actomyosin puncta colocalized with focal adhesions are unstable in Dia1-depleted cells.
How do individual cells adhere to and migrate along collagen fibrils in the midst of ongoing planar motility within the acinus? During rotation of control acini, we observed isolated instances of multiple cells forming MLC puncta at the same location on collagen fibrils as they encountered it in sequence (Fig. 8 E and Video 10). Interestingly, a cell encountering the same region of collagen assembled puncta in the same location as the cell before it. This example illustrates that certain locations within the collagen matrix are primed for repeated adhesion assembly in subsequent cells during planar acinar motility.
Discussion
The suite of molecular mechanisms governing cell motility through 3D environments has enjoyed much attention as it forms the basis for profound tissue shape changes during development and cancer invasion (Gray et al., 2010). Detailed models exist to describe how cytoskeletal organelles control the motility of single cells (Petrie and Yamada, 2012), but how these organelles are regulated in space and time to affect tissue shape changes is less clear.
Data presented in this study suggest a previously unappreciated mechanism by which Dia1-mediated focal adhesion maturation regulates branching morphogenesis in a model epithelial tissue. We and others have previously reported that focal adhesion maturation for cells plated on planar substrates requires a stress fiber template through the activities of α-actinin, septins, and formins such as Dia1 (Gupton et al., 2007; Choi et al., 2008; Oakes et al., 2012; Dolat et al., 2014). We hypothesize a similar model applies for adhesion to collagen matrices. When an MDCK cell initially contacts a collagen fibril, nascent adhesion assembly occurs concomitantly with actin polymerization and myosin recruitment (Fig. 9 A). Our data indicate that this localized actomyosin activity requires Dia1 and promotes focal adhesion maturation marked by phosphotyrosine-rich proteins such as focal adhesion kinase (FAK) and paxillin (Mitra et al., 2005; Zaidel-Bar et al., 2007). Mature adhesions are resistant to disassembly as cells change shape and move within the acinus. On planar surfaces, focal adhesion maturation is not required for rapid cell migration (Oakes et al., 2012). This is consistent with our observation that motility within the plane of the acinus that underlies acinar rotation is independent of Dia1 (Fig. 9 B). Instead, loss of Dia1 selectively abrogates motility away from the acinus. We suspect that the stability of mature focal adhesions in concert with myosin contractility enables cells to pull themselves and their neighbors away from the acinus into the collagen matrix (Fig. 9 B).
Figure 9.
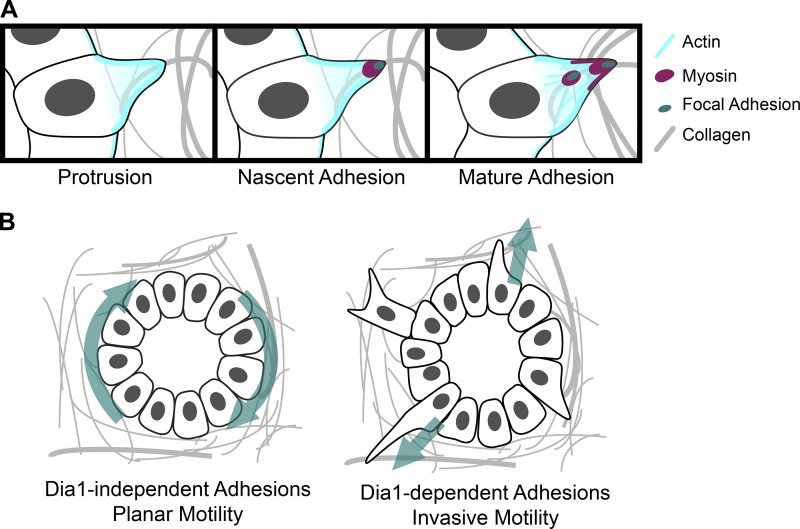
Maturation of focal adhesions through Dia1 during tissue shape changes. (A) Model for focal adhesion maturation during the onset of MDCK branching morphogenesis. Cells generate actin-rich protrusions from their basal surface into the collagen matrix. Protrusions are initially weakly adhered to collagen fibrils through nascent focal adhesions. Localized actin polymerization through Dia1 and myosin recruitment promote stabilization and maturation of focal adhesions. Mature adhesions resist turnover and allow cells to exert contractile forces against collagen fibrils to enable branching morphogenesis. (B) Dia1 as a mechanism by which tissues can regulate noninvasive and invasive motility. In the absence of Dia1 activity, cells can adhere sufficiently to the collagen matrix to mediate planar motility within the acinus and acinar rotation. Invasive motility into the collagen matrix requires that cells form mature adhesions dependent on Dia1.
This work provides evidence to expand the biological functions of focal adhesion maturation. Despite the exquisite detail with which focal adhesion assembly and maturation has been defined (Geiger and Yamada, 2011), the acute consequences of maturation defects on cell motility and tissue morphogenesis remain poorly characterized (Sieg et al., 2000; Hoffman et al., 2006; Fraley et al., 2010; Thievessen et al., 2013). Our observations suggest that tissues require focal adhesion maturation to carry out complex shape changes in 3D fibrillar environments. This model is supported by a study demonstrating that focal adhesion maturation through septins is similarly required for MDCK branching morphogenesis (Dolat et al., 2014). Regulation of focal adhesions may therefore act as a switch from planar motility to invasive motility in tissues. Of special interest for future studies is whether rotation of acini plays an instrumental role in tissue shape changes as has been reported in other systems (Wang et al., 2013; Cetera et al., 2014; Isabella and Horne-Badovinac, 2016). Overall, data presented in this study suggest an expanded repertoire for the biological functions of focal adhesions and their regulators and prompt testable predictions about the interplay among adhesion stability, cell motility, and tissue shape.
Our observation that formins are required for invasion by mouse tumor explants suggests that these findings could be generalized to tissue shape changes more broadly. We note, however, that both of the cell models reported in this study do not maintain an intact basement membrane during tissue shape changes. This contrasts with morphogenesis of tissues such as the mammary gland, for which focal adhesions may play a different role (Ewald et al., 2008; Wang et al., 2017). In addition, the role we identify for Dia1 does not exclude other effects that formins may exert on cell motility. Others have reported that both pharmacologic activation and inhibition of formins can abrogate cell invasion by interfering with directional polarity, suggesting that formins must be tightly regulated to achieve optimal motility in 3D (Arden et al., 2015).
Finally, the role we identify for Dia1 carries important implications for its primary regulator Rho. Rho activation is thought to impact cell motility primarily through ROCK and myosin contractility (Petrie and Yamada, 2012). Prior studies have shown that reducing myosin activity during branching morphogenesis results in increased branching, implicating Rho as a negative regulator of tissue shape changes (Yu et al., 2003; Fischer et al., 2009). Conversely, we show that Dia1 is required for branching morphogenesis through stabilizing focal adhesions to collagen fibrils. Collectively, these observations suggest competing effects of Rho signaling during tissue shape changes: one as a negative regulator via ROCK-mediated contractility and one as a positive regulator via Dia1-mediated focal adhesion maturation. In the absence of ROCK-mediated contractility, focal adhesion maturation is not needed to facilitate migration of cells away from the acinus because of the overall low levels of contractile tension within the tissue. In the presence of ROCK-mediated contractility, Dia1-mediated focal adhesion maturation is not required for weak adhesion to collagen fibrils and planar cell motility to support rotation. However, it is required to assemble adhesions that are sufficiently stable to withstand physiological contractile tension as cells migrate into the collagen and drive tissue shape change. Future studies may delineate these apparently opposing roles of Rho effectors during tissue morphogenesis and whether they indeed converge on focal adhesion stability and spatial regulation of contractility.
Materials and methods
Cell culture and reagents
MDCK type II/G cells (ATCC) were cultured in a humidified incubator with 5% CO2 using Dulbecco’s minimal essential medium (Corning) supplemented with 5% FBS (Corning), 2 mM l-glutamine, and penicillin-streptomycin (Corning). Selection media was supplemented with 5 µg/ml puromycin (Gibco). The MDCK cell line expressing GFP-LifeAct was a gift from T. Wittman (University of California, San Francisco, San Francisco, CA). The following antibodies were used: rat anti–E-cadherin (DECMA; Santa Cruz Biotechnologies, Inc.), mouse antipodocalyxin (a gift from the K. Mostov, University of California, San Francisco, San Francisco, CA), rabbit anti–laminin 511 (Sigma-Aldrich), rabbit anti–myosin IIA heavy chain (Covance), mouse antipaxillin (5H11; EMD Millipore), rabbit anti–FAK-pY397 (Thermo Fisher Scientific), Alexa Fluor 488 goat anti–mouse secondary antibody (Thermo Fisher Scientific), Alexa Fluor 568 goat anti–mouse secondary antibody (Thermo Fisher Scientific), Alexa Fluor 568 goat anti–rat secondary antibody (Thermo Fisher Scientific), rabbit anti-Dia1 (ProteinTech), rabbit anti-Dia2 (Cell Signaling Technology), rabbit anti-FHOD1 (Abcam), and rabbit anti-GAPDH (Sigma-Aldrich). For immunofluorescence, all primary antibodies were used at 1:200, and all secondary antibodies were used at 1:500. The formin inhibitor SMIFH2 (a gift from D. Kovar, University of Chicago, Chicago, IL) was dissolved in DMSO and used at 30 µM. The Arp2/3 inhibitor CK-666 (EMD Millipore) was dissolved in DMSO and used at 50 or 100 µM.
DNA constructs and gene knockdown
Knockdown cell lines were generated using shRNA constructs against canine Dia1 (short hairpin 2, 5′-GATCCCCGCCACAGATGAGAGAGACATTCAAGAGATGTCTCTCTCATCTGTGGCTTTTTA-3′), FHOD1 (short hairpin 1, 5′-GATCCCCGAAGACGAGGACATACTGATTCAAGAGATCAGTATGTCCTCGTCTTCTTTTTA-3′), Dia2 (short hairpin 2, 5′-GATCCCCGCAACCTTACAGCAATGGATTCAAGAGATCCATTGCTGTAAGGTTGCTTTTTA-3′), or a nontargeting control (OligoEngine). These were annealed and cloned into the HindIIII and BglII sites of the pSuper retroviral vector (OligoEngine). Retrovirus was produced using the Phoenix cell line (from G. Nolan, Stanford University, Stanford, CA) using Fugene 6 transfection reagent (Roche) to transfect the retroviral vector and a VSV-G pseudotyping plasmid (a gift from M. Rosner, University of Chicago, Chicago, IL). Viral supernatant was collected, filtered, and incubated with target MDCK cells for 12 h in the presence of 8 µg/ml polybrene (EMD Millipore). After viral transfection and selection, knockdown was confirmed by Western blotting. For nuclear tracking, shControl and shDia1 cells were transfected using a pQCXIX retrovirus construct encoding H2B-mCherry (a gift from M. Burkard, University of Wisconsin, Madison, WI). Cells were purified using flow cytometry (University of Chicago Flow Cytometry Core). The lentiviral vector encoding MLC-mCherry was generated by cloning the MLC-mCherry sequence into pWPT lentiviral backbone (12255; Addgene) with the aid of SnapGene Software (GSL Biotech LLC). The virus was produced in 293T cells (a gift from G. Green, University of Chicago, Chicago, IL) using a pHR1-8.2-deltaR packaging plasmid and a VSV-G pseudotyping plasmid (gifts from M. Rosner). After viral transfection of GFP-LifeAct MDCK cells, positive cells were isolated by flow cytometry at the University of Chicago Flow Cytometry Core.
Culture and manipulation of acini
Matrigel (Corning) from a single lot containing 9.1 mg/ml protein was used for all 3D cell cultures. To generate acini, the lower surface of wells in a 24-well plate were coated with 30 µl Matrigel and allowed to gel at 37°C. MDCK cells were trypsinized and pipetted vigorously to break up cell clusters, and then 10,000 cells were added to 800 µl of a 1:1 solution of growth media and Matrigel, which was kept on ice to prevent gelation. The resulting cell suspension was immediately pipetted up and down to disperse cells and added to each Matrigel-coated well. The Matrigel cell suspension was immediately placed in a 37°C incubator and allowed to gel for 30 min before 800 µl growth media was gently added to each well. Media was changed every 3 d for 6–7 d until acini formed lumens. To isolate acini, a modified protocol by Rubashkin et al. (2014) was used to melt Matrigel by incubation at low temperatures. In brief, media was aspirated, and Matrigel was disrupted by pipetting up and down with 5 ml of warm PBS with Ca/Mg (Corning) per well. The PBS-Matrigel solution was pelleted at 500 g for 3 min and resuspended in 10 ml fresh PBS. Acini were incubated in a bucket of salted ice with rocking for 40 min. Acini were pelleted at 500 g, yielding a ∼50-µl slurry of residual Matrigel and acini, which were kept on ice to prevent gelation. Collagen gels were prepared 30 min before use and kept on ice. To prepare collagen gels, 1 M Hepes and 7.5% NaHCO3 were combined with media to achieve final ratios of 1:50 and 1:23.5, respectively. Rat tail collagen 1 (Corning) was gently added to a final concentration of 2 mg/ml. Collagen was fluorescently labeled using Alexa Fluor 647 succinimidyl ester (Thermo Fisher Scientific), dissolved at 10 µg/µl in DMSO, added to 3.4 mg/ml rat tail collagen at a ratio of 1:1,000 (vol/vol), and incubated at 4°C for at least 12 h. The resulting Alexa Fluor 647–labeled collagen was included in a ratio of 1:4 with unlabeled collagen. Acini were added to the collagen solution, mixed by pipetting, and plated in either four- or eight-well chambers (Ibidi) or in four-well Labtek chamber slides (Nunc) in volumes of 40 or 100 µl. All chambers were precoated with 10 or 30 µl of 2 mg/ml collagen solution for 10 min before plating acini. Collagen was allowed to gel for at least 30 min before growth media was gently added to the sides of wells.
Mouse tumor explants
One female day 70 mouse of the MMTV-PyMT (Jackson ImmunoResearch Laboratories, Inc.) was sacrificed, and the largest mammary tumor from each inguinal mammary gland was surgically excised. Each tumor was manually minced with a razor blade, and tumor tissue was digested by shaking for 30 min at 37°C in a conical tube containing 40 ml of DMEM/F12/50:50 (Corning) with 3 mg/ml collagenase A, 1 mg/ml hyaluronidase (Worthington), and 2 U/ml DNase 1 (Thermo Fisher Scientific). 5 ml of FBS was added to halt digestion, and tumor tissue suspension was pelleted at 500 g for 3 min and resuspended. Tumor organoids were placed in a 1:1 mixture of Matrigel and DMEM/F12/50:50 supplemented with 10 µg/ml human insulin (PromoCell), 5 µg/ml human transferrin, and penicillin-streptomycin (Corning). The bottom wells of a 24-well plate (Corning) were coated with Matrigel and allowed to gel before adding 800 µl of organoid-Matrigel suspension per well. Matrigel was allowed to gel for 30 min, and 800 µl media was gently added. Organoids were cultured in Matrigel for 24–48 h before replating into 2 mg/ml collagen 1 gels in four-well Ibidi chambers as described in the previous section with the ice incubation step reduced to 20 min. After replating in collagen, fresh media containing 10% FBS (Corning) with DMSO, 30 µM SMIFH2, or 50 µM CK666 was added, and chambers were placed on a heated microscope stage. Images were collected every 30 min for 72 h.
Western blot analysis
For Western blotting, cells were lysed in Laemmli buffer (4% sodium dodecyl sulfate, 20% glycerol, 120 mM Tris-Cl, pH 6.8, and 0.02% bromophenol blue). Lysates were separated by SDS-PAGE gel and electrotransferred to a nitrocellulose membrane. Blots were blocked in PBS with 5% nonfat dry milk and incubated with primary antibodies at 1:1,000 overnight at 4°C. Blots were incubated in secondary antibodies at 1:10,000 for 1 h at room temperature and developed with ECL Western blotting substrate (Thermo Fisher Scientific). Blots were scanned as film negatives on a photo scanner (Perfection v700; Epson) and analyzed using the gel analysis tool in ImageJ (National Institutes of Health). The intensity of the protein bands was normalized through comparison with the loading control bands.
Microscopy
All fluorescence images were acquired on an inverted microscope (Ti-E; Nikon) with a confocal scanhead (CSUX; Yokogawa Electric Corporation) with a laser-merge module containing 491-, 561-, and 642-nm laser lines (Spectral Applied Research), a stage controller (Prior), and a cooled charge-coupled device camera (HQ2; Roper Scientific). Images were acquired using either a 20× 0.75 NA Plan Fluor multiimmersion objective (Nikon) or a 40× 1.15 NA Plan Apochromat water immersion extra-long working distance objective (Nikon). Immunostained acini or organoids were imaged by acquiring z stacks with either 500-nm or 1-µm z spacing. All transmitted light images were acquired on an inverted microscope (Ti-E; Nikon), a stage controller (Prior), and a cooled charge-coupled device camera (HQ2; Roper Scientific). Images were acquired using a 20× 0.45 NA air extra-long working distance objective (Nikon). All hardware was controlled via MetaMorph acquisition software (Molecular Devices). Images of acini for Fig. S2 D were acquired on an Eclipse TS100 microscope (Nikon) using an iPhone 6S (Apple) and a SnapZoom adapter (SnapZoom).
Live-cell imaging
All live-cell imaging was performed with a stage incubator for temperature, humidity, and CO2 control (Chamlide TC and FC-5N; Quorum Technologies). The stage adapter, stage cover, and objective were maintained at 37°C, whereas humidified 5% CO2 air was maintained at 50°C at its source to prevent condensation within its tubing. Acini were transferred to collagen gels in four- or eight-well plastic chambers (Ibidi) at least 1 d before imaging. Imaging media was identical to growth media except that phenol red–free DMEM was used. Imaging media containing 20 ng/ml HGF was added to chambers immediately before transfer to the prewarmed microscope stage, where they equilibrated for ∼30 min before imaging. Image sequences were corrected for x/y drift using the Stackreg plugin in ImageJ and a custom MATLAB script (MathWorks). Fluorescence imaging of cells expressing H2B-mCherry was performed by collecting z stacks of 21 planes at 3-µm intervals every 10 min. For fluorescence imaging of GFP-LifeAct, mCherry-MLC, and Alexa Fluor 647–collagen, 20 ng/ml HGF was added to phenol red–free media 4 or 24 h before imaging. For each acinus, images were acquired at six planes separated by 3 µm every 3 min for 3 h. Only the lower surface of each acinus was imaged to capture collagen fibrils and cells simultaneously. Alexa Fluor 647–collagen images were bleach corrected in ImageJ.
Immunofluorescence
For immunofluorescence staining of acini, a 1.5-U/ml solution of clostridium collagenase (Sigma-Aldrich) in PBS was added to culture media before fixation, and acini were replaced in the incubator for 10 min. Acini were fixed in a solution of 4% paraformaldehyde with 0.1% Triton X-100 in PBS solution (Corning). Fixation solution was gently added to each well while simultaneously aspirating culture media and then was incubated with shaking for 20 min. Collagen autofluorescence was quenched with three rinses of 0.1 M glycine in PBS for 20 min each. Acini were permeabilized in 0.5% Triton X-100 for 10 min and blocked with 2.5% BSA and 0.1% Triton X-100 in PBS for 1 h. Acini were incubated with primary antibody at 1:200 in blocking solution overnight at 4°C and washed three times in 0.1% Triton X-100 for 20 min each, and then secondary antibody at 1:500 or Alexa Fluor 647 phalloidin at 1:1,000 (Thermo Fisher Scientific) was incubated in blocking solution for 1 h. After another three 20-min washes with 0.1% Triton X-100, chamber walls were removed, and 20 µl ProLong Gold (Thermo Fisher Scientific) was added per well. A clean coverslip had clear blobs of nail polish applied to its corners, was allowed to dry, and was placed over the chamber slide. Slides were allowed to dry, sealed with nail polish, and stored at 4°C. All incubations were performed on a rotary shaker. Immunofluorescence staining of cells in 2D culture was performed as above but without glycine rinses.
Branching morphogenesis assays
Acini plated in collagen gels were cultured for at least 1 d before inducing branching morphogenesis. In brief, growth media were removed and replaced with media containing 20 ng/ml human recombinant HGF (Sigma-Aldrich) with DMSO or inhibitors as indicated. Fresh HGF with inhibitors was added after 24 h. After 48 h, acini were fixed and stained with phalloidin.
Scattering assays
MDCK cells were plated sparsely in four-well plastic chambers (Ibidi) or on glass coverslips and allowed to form islands. Cells were serum starved by replacing growth media with phenol red–free media supplemented with 0.5% FBS and then cultured for 12 h. For time-lapse microscopy, chambers were placed on a heated microscope stage, and 20 ng/ml HGF was added to low-serum media to stimulate scattering. Cells were imaged in brightfield at 10-min intervals for 12 h. For immunofluorescence, cells were plated on glass coverslips and allowed to form islands, and then they were serum starved for 12 h, incubated in HGF for 6 h, and fixed for immunofluorescence staining.
For scattering assays on top of collagen gels, fluorescently labeled or unlabeled 2 mg/ml collagen gels were made as above, and 60–80 µl per well was added to either four-well plastic imaging chambers (Ibidi) or four-well chamber slides (Labtek). After gelation, single MDCK cells were sparsely plated and allowed to adhere overnight before serum starving for at least 12 h. After addition of 20 ng/ml HGF, cells were either fixed and stained by immunofluorescence or imaged for 12 h in transmitted light in imaging media as described in the Live-cell imaging section.
Image analysis
For branching morphogenesis assays, z stacks of each acinus were collected at 0.5- or 1-µm intervals, and the entire volume was scored for protrusions. These were scored as single- or multicellular extensions ≥5 µm in length emanating from acini into the collagen gel. Protrusion length was measured as a straight line from the basal surface of the cell or cells to the tip of the cellular extension. Protrusions extending in the z direction were measured by counting the confocal sections they spanned.
For nuclear tracking, image sequences for analysis were generated by combining lower z planes from each acinus into maximum projections. Cells were tracked manually in ImageJ (National Institutes of Health), and tracks were analyzed using custom MATLAB scripts. For cell tracking during scattering, cells were tracked manually using ImageJ, and tracks were analyzed using custom MATLAB scripts.
Analysis of individual collagen fibril deformations and MLC puncta lifetimes was performed on image sequences in which the lower basal surface of cells and the collagen matrix could be resolved in the same imaging plane. This surface ranged from 1,500–2,100 µm2, representing ∼0.19 of the total surface area for a typical acinus 50–60 µm in diameter. To quantify collagen fibril deformations, image sequences were analyzed for fibrils that were deformed by 1–2 µm, independently of their neighbors. MLC puncta lifetimes were measured for puncta that remained stable for at least 6 min.
Cell shape parameters during scattering on collagen gels were calculated from 12-h time points of imaging series. Cells not in contact with any neighbors were traced manually, and shape parameters were calculated using the Measure function in ImageJ.
Focal adhesion length and number per µm of leading edge in cells scattering on collagen gels were performed manually. Quantification of phosphotyrosine puncta was performed on maximum-intensity projections combining 3 µm at the acinar equator. The resulting images were background subtracted and binarized, and puncta were counted using the Analyze Particles feature of ImageJ set to a pixel range of 30–1,000 pixels squared. Maximum-intensity projections were also analyzed with line scans, each measuring 2 × 15 µm. Quantification of phospho-FAK was performed on background-subtracted binarized images using the Analyze Particles feature of ImageJ set to a pixel range of 30–1,000 pixels squared.
Collagen gel strain fields
To analyze cell-induced strain of collagen gels, cell islands plated on Alexa Fluor 647–labeled collagen gels were stimulated with HGF for 6 h. Z stacks spanning 15–20 µm of the collagen underlying individual migrating cells were captured before and after addition of 0.5% SDS to remove cells. Two or three planes from z stacks were combined in maximum projections to account for drift in z and slight variations in the topology of the gel. Total z height of the collagen gel was measured at each position to verify that proximity to the coverslip did not impact gel stiffness (shControl z height mean, 1,755 µm, and SD, 11 µm; shDia1 z height mean, 1,791 µm, and SD, 20 µm). Image pairs were aligned using the StackReg plugin in ImageJ. Collagen fibril displacement was analyzed using PIV (Mori and Chang, 2003) run in MATLAB with a grid spacing of 1.43 µm. Displacement vectors were interpolated and filtered using the Kirging interpolation method. Because estimating strain energy is not possible for nonlinearly elastic materials such as collagen 1 gels, total fibril displacement was instead measured beneath each cell. Vector magnitudes were summed and divided by the area over which the collagen was deformed (the apparent cell traction footprint). This dimensionless quantity corresponds with the strain of the collagen beneath each cell.
Traction force microscopy
Traction force microscopy was performed as previously described (Sabass et al., 2008; Oakes et al., 2012). In brief, PA gels with a Young’s modulus of 8.4 kPa were polymerized with 80-nm fluorescent microspheres (Invitrogen) on prepared glass coverslips and covalently conjugated with collagen 1 (BD) using sulphosanpah (Thermo Fisher Scientific). Cells were plated on gels and allowed to spread overnight. As cells readily formed pairs on PA gels, cell pairs were analyzed rather than single cells. Images of microspheres were captured before and after cell removal with a 0.5% SDS solution, and image pairs were registered and analyzed by PIV as in the previous section. Traction stresses were calculated from the displacement field by Fourier transform traction cytometry using zeroth-order regularization.
Statistical tests
To assess statistical significance, we used independent two-sample Student’s t tests of the mean to determine the significance with respect to WT or control. P-values are indicated by *, P < 0.01; **, P < 0.05.
Online supplemental material
Fig. S1 describes additional details of MDCK branching morphogenesis and effects of Arp2/3 inhibition. Fig. S2 includes Western blots of knockdown constructs and plots of branching morphogenesis for each knockdown cell line. Fig. S3 shows MDCK cell protrusion statistics to accompany Fig. 4 as well as traction force heatmaps to accompany Fig. 6 I. Fig. S4 furnishes quantification and additional immunofluorescence analysis of focal adhesions formed by control and shDia1 cells within acini and 2D. Video 1 shows mouse tumor explants invading into collagen gels. Video 2 shows MDCK cells scattering on coverslips. Video 3 shows cell nuclei movements during MDCK branching morphogenesis. Video 4 shows cell protrusion dynamics during MDCK branching morphogenesis. Video 5 shows motility of single MDCK cells in collagen gels. Videos 6 and 7 show collagen fibril deformations along with myosin and actin reporters during branching morphogenesis by shControl and shDia1 acini, respectively. Video 8 shows collagen fibril deformations and myosin and actin reporters at the leading edge of an advancing cell front. Video 9 shows movement of actomyosin puncta in individual shControl and shDia1 cells within acini during branching morphogenesis. Video 10 shows actomyosin puncta reassembly in adjacent shControl cells during branching morphogenesis.
Supplementary Material
Acknowledgments
The authors are grateful for many fruitful discussions with Patrick McCall, Kim Weirich, Bob Harmon, and Barbara Hissa. The authors also are indebted to Karl Matlin, Marsha Rosner, Valerie Weaver, and Keith Mostov for sharing reagents. Finally the authors thank Andrew Ewald and Thea Tlsty for providing critical guidance.
This work was supported by National Institute of General Medical Sciences grant GM104032 to M.L. Gardel.
The authors declare no competing financial interests.
Author contributions: T.B. Fessenden conceived the study, performed experiments and analysis, and wrote the manuscript. Y. Beckham, M. Perez-Neut, and G. Ramirez-San Juan performed experiments and analysis. A.H. Chourasia and K.F. MacCleod provided mice. P.W. Oakes performed analysis and reviewed and edited the manuscript. M.L. Gardel conceived the study, wrote, reviewed, and edited the manuscript, and provided funding.
References
- Affolter M., Zeller R., and Caussinus E.. 2009. Tissue remodelling through branching morphogenesis. Nat. Rev. Mol. Cell Biol. 10:831–842. 10.1038/nrm2797 [DOI] [PubMed] [Google Scholar]
- Arden J.D., Lavik K.I., Rubinic K.A., Chiaia N., Khuder S.A., Howard M.J., Nestor-Kalinoski A.L., Alberts A.S., and Eisenmann K.M.. 2015. Small-molecule agonists of mammalian Diaphanous-related (mDia) formins reveal an effective glioblastoma anti-invasion strategy. Mol. Biol. Cell. 26:3704–3718. 10.1091/mbc.E14-11-1502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant D.M., and Mostov K.E.. 2008. From cells to organs: building polarized tissue. Nat. Rev. Mol. Cell Biol. 9:887–901. 10.1038/nrm2523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cetera M., Ramirez-San Juan G.R., Oakes P.W., Lewellyn L., Fairchild M.J., Tanentzapf G., Gardel M.L., and Horne-Badovinac S.. 2014. Epithelial rotation promotes the global alignment of contractile actin bundles during Drosophila egg chamber elongation. Nat. Commun. 5:5511 10.1038/ncomms6511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung K.J., Gabrielson E., Werb Z., and Ewald A.J.. 2013. Collective invasion in breast cancer requires a conserved basal epithelial program. Cell. 155:1639–1651. 10.1016/j.cell.2013.11.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi C.K., Vicente-Manzanares M., Zareno J., Whitmore L.A., Mogilner A., and Horwitz A.R.. 2008. Actin and α-actinin orchestrate the assembly and maturation of nascent adhesions in a myosin II motor-independent manner. Nat. Cell Biol. 10:1039–1050. 10.1038/ncb1763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christiansen J.J., and Rajasekaran A.K.. 2006. Reassessing epithelial to mesenchymal transition as a prerequisite for carcinoma invasion and metastasis. Cancer Res. 66:8319–8326. 10.1158/0008-5472.CAN-06-0410 [DOI] [PubMed] [Google Scholar]
- Chrzanowska-Wodnicka M., and Burridge K.. 1996. Rho-stimulated contractility drives the formation of stress fibers and focal adhesions. J. Cell Biol. 133:1403–1415. 10.1083/jcb.133.6.1403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dachsel J.C., Ngok S.P., Lewis-Tuffin L.J., Kourtidis A., Geyer R., Johnston L., Feathers R., and Anastasiadis P.Z.. 2013. The Rho guanine nucleotide exchange factor Syx regulates the balance of dia and ROCK activities to promote polarized-cancer-cell migration. Mol. Cell. Biol. 33:4909–4918. 10.1128/MCB.00565-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolat L., Hunyara J.L., Bowen J.R., Karasmanis E.P., Elgawly M., Galkin V.E., and Spiliotis E.T.. 2014. Septins promote stress fiber-mediated maturation of focal adhesions and renal epithelial motility. J. Cell Biol. 207:225–235. 10.1083/jcb.201405050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle A.D., Carvajal N., Jin A., Matsumoto K., and Yamada K.M.. 2015. Local 3D matrix microenvironment regulates cell migration through spatiotemporal dynamics of contractility-dependent adhesions. Nat. Commun. 6:8720 10.1038/ncomms9720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewald A.J., Brenot A., Duong M., Chan B.S., and Werb Z.. 2008. Collective epithelial migration and cell rearrangements drive mammary branching morphogenesis. Dev. Cell. 14:570–581. 10.1016/j.devcel.2008.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewald A.J., Huebner R.J., Palsdottir H., Lee J.K., Perez M.J., Jorgens D.M., Tauscher A.N., Cheung K.J., Werb Z., and Auer M.. 2012. Mammary collective cell migration involves transient loss of epithelial features and individual cell migration within the epithelium. J. Cell Sci. 125:2638–2654. 10.1242/jcs.096875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer R.S., Gardel M., Ma X., Adelstein R.S., and Waterman C.M.. 2009. Local cortical tension by myosin II guides 3D endothelial cell branching. Curr. Biol. 19:260–265. 10.1016/j.cub.2008.12.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraley S.I., Feng Y., Krishnamurthy R., Kim D.-H., Celedon A., Longmore G.D., and Wirtz D.. 2010. A distinctive role for focal adhesion proteins in three-dimensional cell motility. Nat. Cell Biol. 12:598–604. 10.1038/ncb2062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedl P., and Alexander S.. 2011. Cancer invasion and the microenvironment: plasticity and reciprocity. Cell. 147:992–1009. 10.1016/j.cell.2011.11.016 [DOI] [PubMed] [Google Scholar]
- Friedl P., Locker J., Sahai E., and Segall J.E.. 2012. Classifying collective cancer cell invasion. Nat. Cell Biol. 14:777–783. 10.1038/ncb2548 [DOI] [PubMed] [Google Scholar]
- Gardel M.L., Schneider I.C., Aratyn-Schaus Y., and Waterman C.M.. 2010. Mechanical integration of actin and adhesion dynamics in cell migration. Annu. Rev. Cell Dev. Biol. 26:315–333. 10.1146/annurev.cellbio.011209.122036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiger B., and Yamada K.M.. 2011. Molecular architecture and function of matrix adhesions. Cold Spring Harb. Perspect. Biol. 3:a005033 10.1101/cshperspect.a005033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray R.S., Cheung K.J., and Ewald A.J.. 2010. Cellular mechanisms regulating epithelial morphogenesis and cancer invasion. Curr. Opin. Cell Biol. 22:640–650. 10.1016/j.ceb.2010.08.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupton S.L., Eisenmann K., Alberts A.S., and Waterman-Storer C.M.. 2007. mDia2 regulates actin and focal adhesion dynamics and organization in the lamella for efficient epithelial cell migration. J. Cell Sci. 120:3475–3487. 10.1242/jcs.006049 [DOI] [PubMed] [Google Scholar]
- Haigo S.L., and Bilder D.. 2011. Global tissue revolutions in a morphogenetic movement controlling elongation. Science. 331:1071–1074. 10.1126/science.1199424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harunaga J.S., and Yamada K.M.. 2011. Cell-matrix adhesions in 3D. Matrix Biol. 30:363–368. 10.1016/j.matbio.2011.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman L.M., Jensen C.C., Kloeker S., Wang C.-L.A., Yoshigi M., and Beckerle M.C.. 2006. Genetic ablation of zyxin causes Mena/VASP mislocalization, increased motility, and deficits in actin remodeling. J. Cell Biol. 172:771–782. 10.1083/jcb.200512115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton E.R., Humphries J.D., Stutchbury B., Jacquemet G., Ballestrem C., Barry S.T., and Humphries M.J.. 2016. Modulation of FAK and Src adhesion signaling occurs independently of adhesion complex composition. J. Cell Biol. 212:349–364. 10.1083/jcb.201508080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter M.P., and Zegers M.M.. 2010. Pak1 regulates branching morphogenesis in 3D MDCK cell culture by a PIX and beta1-integrin-dependent mechanism. Am. J. Physiol. Cell Physiol. 299:C21–C32. 10.1152/ajpcell.00543.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isabella A.J., and Horne-Badovinac S.. 2016. Rab10-Mediated Secretion Synergizes with Tissue Movement to Build a Polarized Basement Membrane Architecture for Organ Morphogenesis. Dev. Cell. 38:47–60. 10.1016/j.devcel.2016.06.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iskratsch T., Yu C.-H., Mathur A., Liu S., Stévenin V., Dwyer J., Hone J., Ehler E., and Sheetz M.. 2013. FHOD1 is needed for directed forces and adhesion maturation during cell spreading and migration. Dev. Cell. 27:545–559. 10.1016/j.devcel.2013.11.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isogai T., van der Kammen R., Leyton-Puig D., Kedziora K.M., Jalink K., and Innocenti M.. 2015. Initiation of lamellipodia and ruffles involves cooperation between mDia1 and the Arp2/3 complex. J. Cell Sci. 128:3796–3810. 10.1242/jcs.176768 [DOI] [PubMed] [Google Scholar]
- Jiang S.-T., Chiu S.-J., Chen H.-C., Chuang W.-J., and Tang M.-J.. 2001. Role of α(3)β(1) integrin in tubulogenesis of Madin-Darby canine kidney cells. Kidney Int. 59:1770–1778. 10.1046/j.1523-1755.2001.0590051770.x [DOI] [PubMed] [Google Scholar]
- Kowalski P.J., Rubin M.A., and Kleer C.G.. 2003. E-cadherin expression in primary carcinomas of the breast and its distant metastases. Breast Cancer Res. 5:R217–R222. 10.1186/bcr651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubow K.E., Conrad S.K., and Horwitz A.R.. 2013. Matrix microarchitecture and myosin II determine adhesion in 3D matrices. Curr. Biol. 23:1607–1619. 10.1016/j.cub.2013.06.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert A.W., Pattabiraman D.R., and Weinberg R.A.. 2017. Emerging Biological Principles of Metastasis. Cell. 168:670–691. 10.1016/j.cell.2016.11.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauffenburger D.A., and Horwitz A.F.. 1996. Cell migration: a physically integrated molecular process. Cell. 84:359–369. 10.1016/S0092-8674(00)81280-5 [DOI] [PubMed] [Google Scholar]
- Lecaudey V., and Gilmour D.. 2006. Organizing moving groups during morphogenesis. Curr. Opin. Cell Biol. 18:102–107. 10.1016/j.ceb.2005.12.001 [DOI] [PubMed] [Google Scholar]
- Lin E.Y., Jones J.G., Li P., Zhu L., Whitney K.D., Muller W.J., and Pollard J.W.. 2003. Progression to malignancy in the polyoma middle T oncoprotein mouse breast cancer model provides a reliable model for human diseases. Am. J. Pathol. 163:2113–2126. 10.1016/S0002-9440(10)63568-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lubarsky B., and Krasnow M.A.. 2003. Tube morphogenesis: making and shaping biological tubes. Cell. 112:19–28. 10.1016/S0092-8674(02)01283-7 [DOI] [PubMed] [Google Scholar]
- Martin-Belmonte F., Gassama A., Datta A., Yu W., Rescher U., Gerke V., and Mostov K.. 2007. PTEN-mediated apical segregation of phosphoinositides controls epithelial morphogenesis through Cdc42. Cell. 128:383–397. 10.1016/j.cell.2006.11.051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAteer J.A., Evan A.P., and Gardner K.D.. 1987. Morphogenetic clonal growth of kidney epithelial cell line MDCK. Anat. Rec. 217:229–239. 10.1002/ar.1092170303 [DOI] [PubMed] [Google Scholar]
- Mitra S.K., Hanson D.A., and Schlaepfer D.D.. 2005. Focal adhesion kinase: in command and control of cell motility. Nat. Rev. Mol. Cell Biol. 6:56–68. 10.1038/nrm1549 [DOI] [PubMed] [Google Scholar]
- Mori N., and Chang K.-A.. 2003. Introduction to MPIV. http://www.oceanwave.jp/softwares/mpiv
- Nolen B.J., Tomasevic N., Russell A., Pierce D.W., Jia Z., McCormick C.D., Hartman J., Sakowicz R., and Pollard T.D.. 2009. Characterization of two classes of small molecule inhibitors of Arp2/3 complex. Nature. 460:1031–1034. 10.1038/nature08231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Brien L.E., Zegers M.M., and Mostov K.E.. 2002. Opinion: Building epithelial architecture: insights from three-dimensional culture models. Nat. Rev. Mol. Cell Biol. 3:531–537. 10.1038/nrm859 [DOI] [PubMed] [Google Scholar]
- O’Brien L.E., Tang K., Kats E.S., Schutz-Geschwender A., Lipschutz J.H., and Mostov K.E.. 2004. ERK and MMPs sequentially regulate distinct stages of epithelial tubule development. Dev. Cell. 7:21–32. 10.1016/j.devcel.2004.06.001 [DOI] [PubMed] [Google Scholar]
- Oakes P.W., Beckham Y., Stricker J., and Gardel M.L.. 2012. Tension is required but not sufficient for focal adhesion maturation without a stress fiber template. J. Cell Biol. 196:363–374. 10.1083/jcb.201107042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owen L.M., Adhikari A.S., Patel M., Grimmer P., Leijnse N., Kim M.C., Notbohm J., Franck C., and Dunn A.R.. 2017. A cytoskeletal clutch mediates cellular force transmission in a soft, three-dimensional extracellular matrix. Mol. Biol. Cell. 28:1959–1974. 10.1091/mbc.E17-02-0102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasapera A.M., Plotnikov S.V., Fischer R.S., Case L.B., Egelhoff T.T., and Waterman C.M.. 2015. Rac1-dependent phosphorylation and focal adhesion recruitment of myosin IIA regulates migration and mechanosensing. Curr. Biol. 25:175–186. 10.1016/j.cub.2014.11.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson G.W., and Hunter T.. 2007. Real-time imaging reveals that noninvasive mammary epithelial acini can contain motile cells. J. Cell Biol. 179:1555–1567. 10.1083/jcb.200706099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrie R.J., and Yamada K.M.. 2012. At the leading edge of three-dimensional cell migration. J. Cell Sci. 125:5917–5926. 10.1242/jcs.093732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettee K.M., Dvorak K.M., Nestor-Kalinoski A.L., and Eisenmann K.M.. 2014. An mDia2/ROCK signaling axis regulates invasive egress from epithelial ovarian cancer spheroids. PLoS One. 9:e90371 10.1371/journal.pone.0090371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard T.D. 2010. Mechanics of cytokinesis in eukaryotes. Curr. Opin. Cell Biol. 22:50–56. 10.1016/j.ceb.2009.11.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard T.D., and Borisy G.G.. 2003. Cellular motility driven by assembly and disassembly of actin filaments. Cell. 112:453–465. 10.1016/S0092-8674(03)00120-X [DOI] [PubMed] [Google Scholar]
- Rahman A., Carey S.P., Kraning-Rush C.M., Goldblatt Z.E., Bordeleau F., Lampi M.C., Lin D.Y., Garcia A.J., and Reinhart-King C.A.. 2016. Vinculin regulates directionality and cell polarity in two- and three-dimensional matrix and three-dimensional microtrack migration. Mol. Biol. Cell. 27:1431–1441. 10.1091/mbc.E15-06-0432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Revenu C., and Gilmour D.. 2009. EMT 2.0: shaping epithelia through collective migration. Curr. Opin. Genet. Dev. 19:338–342. 10.1016/j.gde.2009.04.007 [DOI] [PubMed] [Google Scholar]
- Riveline D., Zamir E., Balaban N.Q., Schwarz U.S., Ishizaki T., Narumiya S., Kam Z., Geiger B., and Bershadsky A.D.. 2001. Focal contacts as mechanosensors: externally applied local mechanical force induces growth of focal contacts by an mDia1-dependent and ROCK-independent mechanism. J. Cell Biol. 153:1175–1186. 10.1083/jcb.153.6.1175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizvi S.A., Neidt E.M., Cui J., Feiger Z., Skau C.T., Gardel M.L., Kozmin S.A., and Kovar D.R.. 2009. Identification and characterization of a small molecule inhibitor of formin-mediated actin assembly. Chem. Biol. 16:1158–1168. 10.1016/j.chembiol.2009.10.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubashkin M.G., Cassereau L., Bainer R., DuFort C.C., Yui Y., Ou G., Paszek M.J., Davidson M.W., Chen Y.-Y., and Weaver V.M.. 2014. Force engages vinculin and promotes tumor progression by enhancing PI3K activation of phosphatidylinositol (3,4,5)-triphosphate. Cancer Res. 74:4597–4611. 10.1158/0008-5472.CAN-13-3698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabass B., Gardel M.L., Waterman C.M., and Schwarz U.S.. 2008. High Resolution Traction Force Microscopy Based on Experimental and Computational Advances. Biophys. J. 94:207–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shamir E.R., Pappalardo E., Jorgens D.M., Coutinho K., Tsai W.-T., Aziz K., Auer M., Tran P.T., Bader J.S., and Ewald A.J.. 2014. Twist1-induced dissemination preserves epithelial identity and requires E-cadherin. J. Cell Biol. 204:839–856. 10.1083/jcb.201306088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sieg D.J., Hauck C.R., Ilic D., Klingbeil C.K., Schaefer E., Damsky C.H., and Schlaepfer D.D.. 2000. FAK integrates growth-factor and integrin signals to promote cell migration. Nat. Cell Biol. 2:249–256. 10.1038/35010517 [DOI] [PubMed] [Google Scholar]
- Stoker M., Gherardi E., Perryman M., and Gray J.. 1987. Scatter factor is a fibroblast-derived modulator of epithelial cell mobility. Nature. 327:239–242. 10.1038/327239a0 [DOI] [PubMed] [Google Scholar]
- Takeya R., Taniguchi K., Narumiya S., and Sumimoto H.. 2008. The mammalian formin FHOD1 is activated through phosphorylation by ROCK and mediates thrombin-induced stress fibre formation in endothelial cells. EMBO J. 27:618–628. 10.1038/emboj.2008.7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanner K., Mori H., Mroue R., Bruni-Cardoso A., and Bissell M.J.. 2012. Coherent angular motion in the establishment of multicellular architecture of glandular tissues. Proc. Natl. Acad. Sci. USA. 109:1973–1978. 10.1073/pnas.1119578109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiery J.P., Acloque H., Huang R.Y.J., and Nieto M.A.. 2009. Epithelial-mesenchymal transitions in development and disease. Cell. 139:871–890. 10.1016/j.cell.2009.11.007 [DOI] [PubMed] [Google Scholar]
- Thievessen I., Thompson P.M., Berlemont S., Plevock K.M., Plotnikov S.V., Zemljic-Harpf A., Ross R.S., Davidson M.W., Danuser G., Campbell S.L., and Waterman C.M.. 2013. Vinculin-actin interaction couples actin retrograde flow to focal adhesions, but is dispensable for focal adhesion growth. J. Cell Biol. 202:163–177. 10.1083/jcb.201303129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicente-Manzanares M., Ma X., Adelstein R.S., and Horwitz A.R.. 2009. Non-muscle myosin II takes centre stage in cell adhesion and migration. Nat. Rev. Mol. Cell Biol. 10:778–790. 10.1038/nrm2786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Lacoche S., Huang L., Xue B., and Muthuswamy S.K.. 2013. Rotational motion during three-dimensional morphogenesis of mammary epithelial acini relates to laminin matrix assembly. Proc. Natl. Acad. Sci. USA. 110:163–168. 10.1073/pnas.1201141110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Sekiguchi R., Daley W.P., and Yamada K.M.. 2017. Patterned cell and matrix dynamics in branching morphogenesis. J. Cell Biol. 216:559–570. 10.1083/jcb.201610048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei W.-C., Kopec A.K., and Tang M.-J.. 2009. Requirement of focal adhesion kinase in branching tubulogenesis. J. Biomed. Sci. 16:5 10.1186/1423-0127-16-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyckoff J.B., Pinner S.E., Gschmeissner S., Condeelis J.S., and Sahai E.. 2006. ROCK- and myosin-dependent matrix deformation enables protease-independent tumor-cell invasion in vivo. Curr. Biol. 16:1515–1523. 10.1016/j.cub.2006.05.065 [DOI] [PubMed] [Google Scholar]
- Yu W., O’Brien L.E., Wang F., Bourne H., Mostov K.E., and Zegers M.M.. 2003. Hepatocyte growth factor switches orientation of polarity and mode of movement during morphogenesis of multicellular epithelial structures. Mol. Biol. Cell. 14:748–763. 10.1091/mbc.E02-06-0350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaidel-Bar R., Ballestrem C., Kam Z., and Geiger B.. 2003. Early molecular events in the assembly of matrix adhesions at the leading edge of migrating cells. J. Cell Sci. 116:4605–4613. 10.1242/jcs.00792 [DOI] [PubMed] [Google Scholar]
- Zaidel-Bar R., Milo R., Kam Z., and Geiger B.. 2007. A paxillin tyrosine phosphorylation switch regulates the assembly and form of cell-matrix adhesions. J. Cell Sci. 120:137–148. 10.1242/jcs.03314 [DOI] [PubMed] [Google Scholar]
- Zegers M.M.P., O’Brien L.E., Yu W., Datta A., and Mostov K.E.. 2003. Epithelial polarity and tubulogenesis in vitro. Trends Cell Biol. 13:169–176. 10.1016/S0962-8924(03)00036-9 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.