Ogungbenro et al. use reverse genetics in human hTERT-RPE1 cells and zebrafish to demonstrate novel roles for the centriolar BRCA2 interactor, centrobin, in controlling primary cilium formation through a C-terminal tubulin- and CP110-interacting region.
Abstract
The BRCA2 interactor, centrobin, is a centrosomal protein that has been implicated in centriole duplication and microtubule stability. We used genome editing to ablate CNTROB in hTERT-RPE1 cells and observed an increased frequency of monocentriolar and acentriolar cells. Using a novel monoclonal antibody, we found that centrobin primarily localizes to daughter centrioles but also associates with mother centrioles upon serum starvation. Strikingly, centrobin loss abrogated primary ciliation upon serum starvation. Ultrastructural analysis of centrobin nulls revealed defective axonemal extension after mother centriole docking. Ciliogenesis required a C-terminal portion of centrobin that interacts with CP110 and tubulin. We also depleted centrobin in zebrafish embryos to explore its roles in an entire organism. Centrobin-depleted embryos showed microcephaly, with curved and shorter bodies, along with marked defects in laterality control, morphological features that indicate ciliary dysfunction. Our data identify new roles for centrobin as a positive regulator of vertebrate ciliogenesis.
Introduction
Centrosomes are the major microtubule organizing center in animal somatic cells and contribute to cell division, cell movement, and cell polarity. Mature centrosomes consist of two centrioles, cylindrical assemblies of triplet microtubules arranged with a ninefold symmetry, within the pericentriolar material. The centrioles differ from one another: the older of the two carries distal and subdistal appendages and is termed the mother centriole, as distinct from the younger, daughter centriole (Vorobjev and Chentsov, 1982; Nigg and Stearns, 2011).
Primary cilia are membrane-bounded, antenna-like structures that transduce extracellular signals at the surface of most human cell types (Goetz and Anderson, 2010). Defects in primary cilium formation or activity cause a diverse range of human developmental disorders that particularly affect the kidney, eye, liver, brain, and skeleton, collectively termed the ciliopathies (Waters and Beales, 2011; Braun and Hildebrandt, 2017). The centrosome has an important function in primary ciliation (Sorokin, 1962; Seeley and Nachury, 2010; Ishikawa and Marshall, 2011): the basal body, the structure at the base of the cilium, is established during primary ciliogenesis by plasma membrane docking of the mother centriole (Anderson, 1972; Ishikawa et al., 2005; Tanos et al., 2013).
Centrobin (also known as NIP2 or LIP8) is a centrosome component first described as an in vitro interactor of the C-terminal region of the BRCA2 tumor suppressor (Zou et al., 2005; Jeffery et al., 2010). Centrobin was initially described as a component of daughter centrioles and is required for efficient centriole duplication, which is at least partly because of its interactions with tubulin (Zou et al., 2005; Jeong et al., 2007; Jeffery et al., 2010; Lee et al., 2010; Gudi et al., 2011). Work in Drosophila melanogaster has indicated centrobin as a negative regulator of ciliogenesis in specialized sensory neurons (Gottardo et al., 2015). Here we describe the function of centrobin in vertebrate ciliogenesis.
Results and discussion
Centrobin-deficient cells show centriole duplication and ciliogenesis defects
We generated monoclonal antibody 6D4F4 against amino acids 113–361 of the human protein (Fig. S1 A). 6D4F4 recognized a centrosomal protein slightly larger than 100 kD (Fig. S1 B). To confirm 6D4F4’s specificity, we used siRNA to deplete centrobin and lost the signal seen in immunoblot and immunofluorescence (IF) microscopy analyses (Fig. S1, B and C). This signal localized to the interphase microtubule organizing center and to the spindle poles, and predominantly to one of the two centrioles detected by CEP135 and CPAP staining, within the pericentriolar material revealed by pericentrin labeling (Fig. S1 D). Centrobin localized adjacently to the ninein signal, consistent with its being associated with the daughter centriole, as previously observed (Zou et al., 2005).
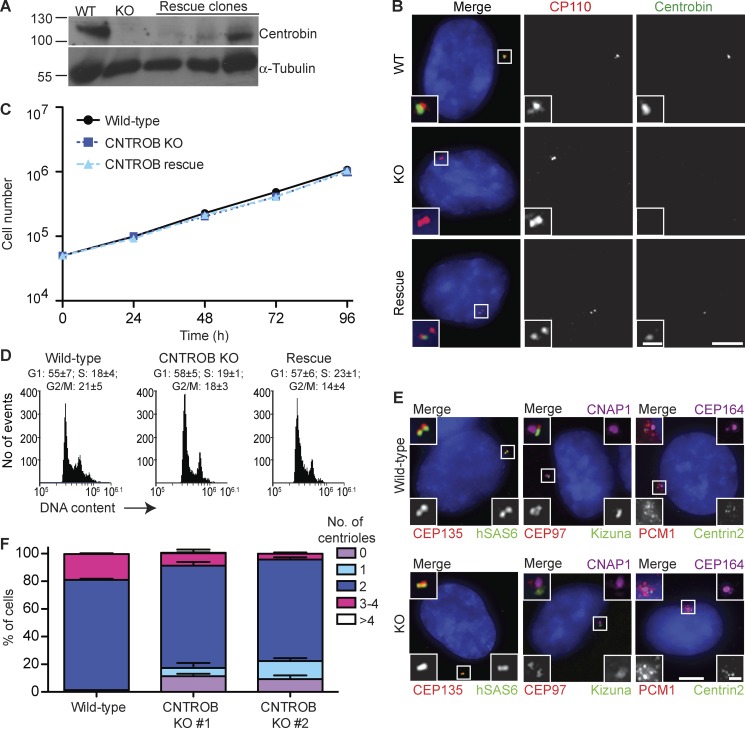
We then used CRISPR-Cas9 genome editing to disrupt exons 1 and 4 of CNTROB in the immortalized hTERT-RPE1 cell line. Immunoblot screening of candidates yielded four clones that lacked detectable centrobin. Genomic PCR and DNA sequencing was used to confirm that CNTROB disruption generated premature stop codons that we verified in RT-PCR experiments. The analysis presented in this paper is based predominantly on a clone in which targeting of exon 4 led to a 43-nucleotide deletion (KO1), but we also examined a second clone where exon 1 disruption caused deletion of seven bases (KO2) and found no difference in the phenotypes we observed (Fig. S2, A–C). Western blotting and IF microscopy confirmed the absence of centrobin, and stable expression of full-length centrobin was used to obtain rescue clones (Fig. 1, A and B). Proliferative analysis showed no significant impact on cell doubling times in the absence of centrobin (Fig. 1 C), with a similar cell cycle profile being observed in CNTROB nulls and in WT cells (Fig. 1 D). Although centriole proteins, centriolar appendage proteins, and the centriolar satellites localized normally in the absence of centrobin, we observed an increased number of acentriolar and monocentriolar centrobin null cells (Fig. 1, E and F), confirming the requirement for centrobin in centriole duplication (Zou et al., 2005). Although it is possible that a more significant impact on centriole duplication may have been selected against in the p53-positive hTERT-RPE1 cells, the centriole duplication phenotypes seen in our knockout cells were similar to those seen in the original study of siRNA-treated HeLa cells, which have defective p53 signaling (Zou et al., 2005).
Figure 1.
CNTROB null cells are viable but show a defect in centriole duplication. (A) CNTROB-edited clones and rescue candidates were identified by immunoblot for centrobin. (B) Confirmation of loss of centrobin protein expression by IF microscopy using antibodies to CP110 (red) and centrobin (green). Bars: 5 µm; (inset) 1 µm. (C) Growth curves show mean ± SEM of five independent experiments. No significant difference was observed between WT and centrobin-deficient cells at any time point. (D) Flow cytometry analysis of cell cycle profiles in asynchronous cells. Numbers indicate the mean percentage ± SEM of cells in each cell cycle phase (n = 3). (E) IF microscopy of the indicated PCM, centriole, and centriolar satellite makers in asynchronous WT and centrobin null cells. Bars: 5 µm; (inset) 1 µm. (F) The number of centrioles per cell were counted in 100 cells in three separate experiments. Centrioles were visualized by centrin2 and CEP135 staining. Bar graph shows mean + SEM.
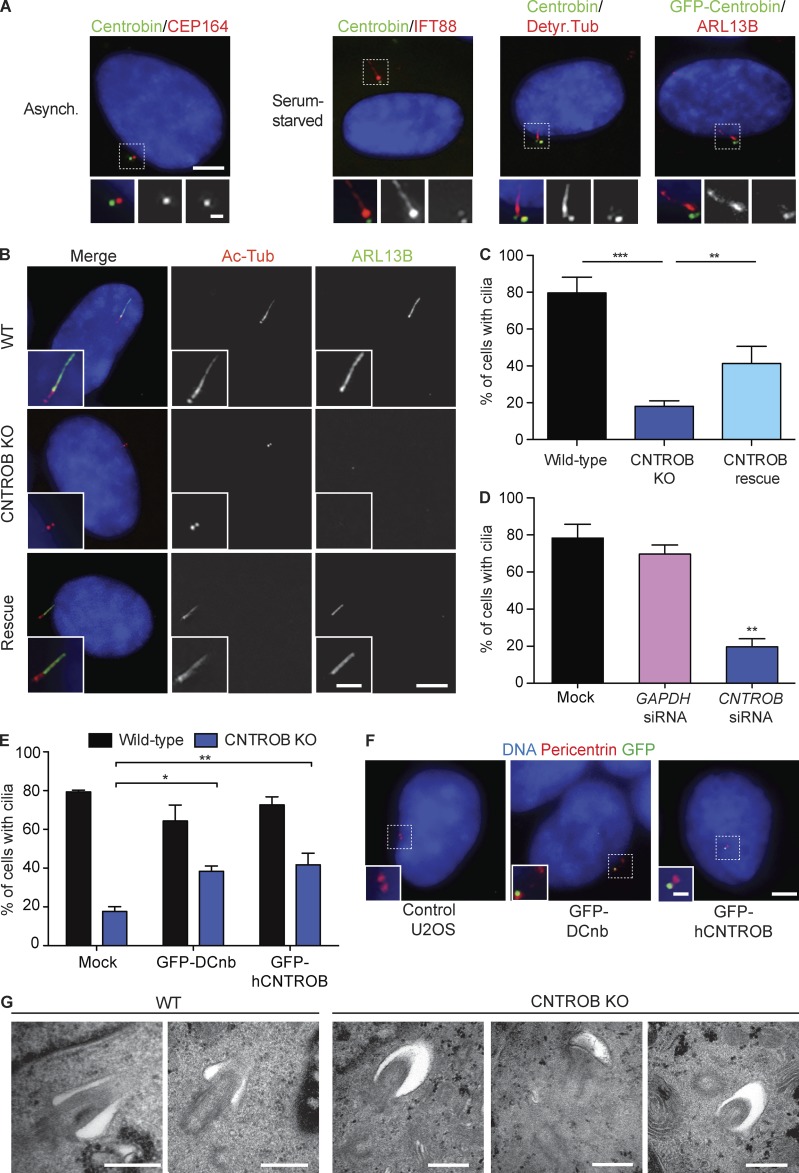
A negative role in ciliogenesis had been indicated for centrobin in Drosophila (Gottardo et al., 2015). Therefore, we examined the localization of centrobin after serum starvation, which induces ciliogenesis in RPE1 cells. Although centrobin was largely restricted to the daughter centriole in asynchronous cells, a small fraction was found at the basal body in ciliated cells (Fig. 2 A). We observed this association with both the endogenous centrobin protein and with GFP-tagged, overexpressed centrobin. Although centrobin has mainly been observed at daughter centrioles, it localized to both centrioles in cultured mouse hippocampal neurons (Jeong et al., 2007; Gudi et al., 2011; Shin et al., 2015). Strikingly, we found that centrobin-deficient cells had a marked defect in forming primary cilia, which could be rescued by reexpression of centrobin (Fig. 2, B and C). RNAi knockdown of centrobin in hTERT-RPE1 cells also reduced primary ciliation (Fig. 2 D). GFP-tagged Drosophila centrobin localized to centrosomes and overexpression of Drosophila centrobin in human CNTROB null cells restored ciliogenesis (Fig. 2, E and F). This suggests that there may be species- or tissue-specific determinants of centrobin function that differ between vertebrates and flies (Gottardo et al., 2015). For example, the roles of CP110 in controlling centriole overduplication differ between human cells and flies (Franz et al., 2013). Electron microscopy analysis revealed that 6 of 7 centrioles docked to the primary ciliary vesicle had failed to extend the axoneme from the ciliary bud, leaving the nascent cilia in the “mushroom” stage of cilium formation (Sorokin, 1962; Fig. 2 G).
Figure 2.
CNTROB null cells are defective in primary ciliogenesis. (A) Localization of centrobin (green) in asynchronous (Asynch.) or 48-h serum-starved cells. Detyr. Tub, detyrosinated tubulin. Bars: 5 µm; (inset) 1 µm. (B) IF microscopy of the cilium markers ARL13B (green) and acetylated tubulin (red) in cells after 48-h serum starvation. Bars: 5 µm; (inset) 2 µm. (C) Quantitation of the ciliation frequency after 48-h serum starvation showing mean + SEM of three independent experiments in which at least 100 cells were quantitated by acetylated tubulin staining. **, P < 0.01; ***, P < 0.001, in comparison to indicated samples by unpaired t test. (D) siRNA knockdown of CNTROB was used to ablate centrobin, with a GAPDH siRNA used as negative control (siGAPDH). Bar chart shows quantitation of the ciliation frequency after 24 h serum starvation. (E) Quantitation of the ciliation frequency after 48-h serum starvation of cells transiently transfected with GFP-tagged centrobin constructs. Bar charts show mean + SEM of three independent experiments in which at least 100 cells were quantitated by ARL13B staining. *, P < 0.05; **, P < 0.01, in comparison to controls by unpaired t test. (F) IF microscopy of U2OS cells transiently transfected with GFP-tagged human or Drosophila centrobin (green) with pericentrin (red) as a marker for centrosomes. Bars: 5 µm; (inset) 1 µm. (G) Transmission electron microscopy analysis of WT and centrobin null (knockout [KO]) cells after 48-h serum starvation. Bars, 500 nm.
Centrobin microtubule binding and CP110 interaction regions are required for ciliogenesis
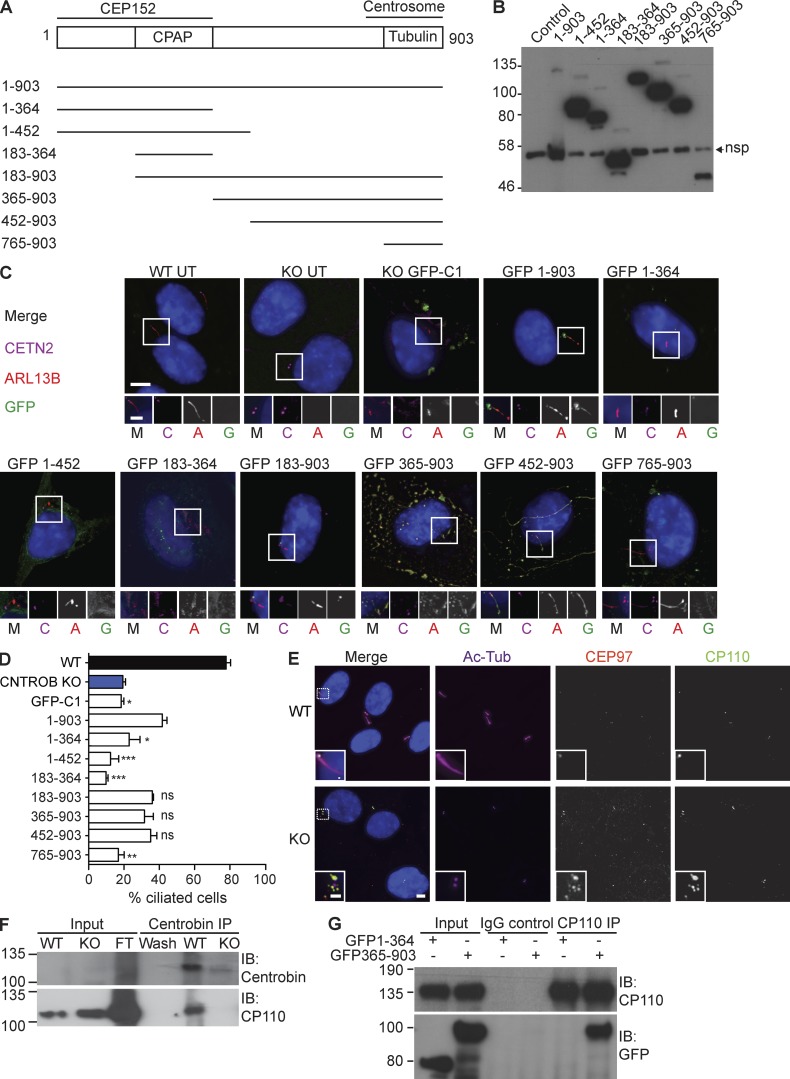
To explore how centrobin controls ciliogenesis, we dissected the protein on basis of its known interactions, as diagramed in Fig. 3 A. The tubulin-binding and centrosome-targeting region of centrobin has been mapped to amino acids 765–903 (Gudi et al., 2011) and the region required for interaction with the key centriole duplication and extension regulators, CEP152 and CPAP (CENPJ), within amino acids 1–364 (Gudi et al., 2014, 2015). Transiently expressed, GFP-tagged full-length centrobin localized to the basal body in CNTROB nulls and rescued ciliogenesis (Fig. 3, B–D). We also observed GFP-centrobin along the axoneme, a location that was only rarely seen with the endogenous protein. Strikingly, overexpression of centrobin in WT cells caused bulges along the axoneme (Fig. S2 D), suggestive of defects in cilium assembly or intraflagellar transport. Centrobin 1–452 was neither required nor sufficient for ciliogenesis, indicating that the N-terminal CEP152 and CPAP-binding region of centrobin is not necessary for ciliogenesis. Expression of centrobin 452–903 supported ciliogenesis, with the transgene product also localizing to microtubule bundles (Fig. 3, B–D), as was seen in HeLa cells transfected with centrobin 445–903 (Jeong et al., 2007). Centrobin 365–903 localized to cytoplasmic aggregates similar to those described in previous transfection experiments in HeLa cells (Gudi et al., 2014). These data indicate that the C-terminal 451 amino acids of centrobin are sufficient to allow ciliogenesis, although the centrosome-targeting region 765–903 alone was not. It remains to be determined whether centrosome localization of centrobin is required for ciliogenesis. Appropriate regulation of microtubule stability is an obvious candidate mechanism by which centrobin may contribute to ciliogenesis (Jeong et al., 2007; Gudi et al., 2011; Shin et al., 2015).
Figure 3.
The N-terminal CPAP-binding region of centrobin is dispensable for ciliogenesis. (A) Schematic of deletion analysis performed on centrobin, indicating regions required for CPAP, CEP152, or tubulin interactions or localization to the centrosome. (B) Immunoblot showing expression of each of the GFP-tagged centrobin fragments at 24 h after transfection into HCT116 cells. The control lane is from untransfected cells. The lane with full-length protein is overloaded relative to the others. Nsp, nonspecific band. (C) Fluorescence micrographs of CNTROB null cells 24 h after transfection with GFP-tagged expression constructs that were tested in B. In the merged images, GFP localization is shown in green, ARL13B in red, and centrin2 in magenta. The same order is used for all single channels in the blowups. Bars: 5 µm; (inset) 2 µm. (D) Quantitation of the frequency of primary cilia in CNTROB null cells transfected with the indicated constructs. Cells were serum starved for 24 h, starting 24 h after transfection. Scoring was based on ARL13B staining and bar chart shows mean + SEM from 100 transfected cells in each of three experiments. **, P < 0.01; ***, P < 0.001, compared with full-length rescue using one-way ANOVA and Tukey’s multiple comparison test. (E) IF microscopy of CP110 and CEP97 localization in WT and centrobin-null hTERT-RPE1 cells. Bars: 5 µm; (inset) 2 µm. (F) Coimmunoprecipitation (Co-IP) of endogenous CNTROB and CP110 from hTERT-RPE1 cell extract using anticentrobin monoclonal antibody 6D4F4 for the pulldown. CNTROB-null cells were used as the negative control. (G) Coimmunoprecipitation experiment using polyclonal anti-CP110 for pulldown 24 h after transfection of HCT116 WT cells with the indicated GFP constructs. IgG incubated with cell extracts was used as negative control. Size markers are in kilodaltons.
We next examined the relationship between centrobin and CP110, a key negative regulator of ciliogenesis. CP110 and its partner, CEP97, were aberrantly localized around the distal end of the centriole after serum starvation of centrobin-deficient cells (Fig. 3 E), rather than being removed, as is the case during normal ciliogenesis (Spektor et al., 2007; Tsang et al., 2008). Immunoprecipitation of full-length centrobin copurified CP110 (Fig. 3 F), whereas coimmunoprecipitation with CP110 was observed with centrobin 365–903 but not with centrobin 1–364 (Fig. 3 G). Notably, expression of full-length centrobin or the ciliation-permissive 452–903 fragment led to the absence of CP110 from the distal end of the centriole (Fig. S2 E). Together, these findings indicate that the C-terminal region of centrobin interacts with CP110 and suggest that the centrobin–CP110 interaction regulates CP110 removal during ciliogenesis, possibly by facilitating access of another regulatory protein to CP110. We tested whether CP110 ablation could restore ciliogenesis in CNTROB nulls, as in CETN2 nulls (Prosser and Morrison, 2015). CP110 knockdown in CNTROB nulls rescued ciliogenesis to an extent that was significant compared with mock-treated cells but not to GAPDH knockdown controls (Fig. S2, E–G). This indicates that both microtubule stabilization and CP110 regulation by the C-terminal region of centrobin are required for ciliogenesis. Thus, our model is that centrobin contributes to the removal of CP110 from the mother centriole, as distinct from inhibiting its recruitment, although we do not know the mechanism for this.
Centrobin deficiency causes ciliopathy defects in zebrafish
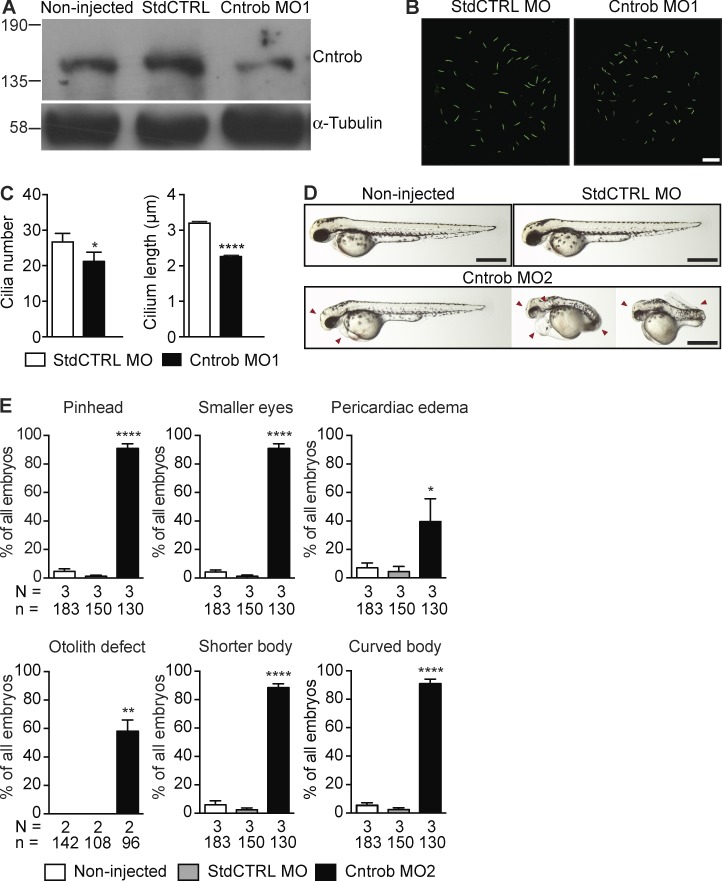
Next, to test whether centrobin is required for ciliary functions in another vertebrate system, we injected zebrafish embryos with two nonoverlapping antisense morpholinos (MOs) against the ATG and 5′-UTR of cntrob. As shown in Fig. 4 (A–C), Cntrob MO injection caused a decline in the levels of centrobin in embryos, along with significant declines in cilium number and length in the Kupffer’s vesicle (KV), an organelle that contains a ciliated epithelium critical for left–right asymmetry during development (Essner et al., 2005). Moreover, Cntrob MO-injected embryos showed morphological abnormalities impacted by ciliary defects, such as smaller heads and eyes, pericardiac edema, and defective otolith seeding, with shorter and aberrantly curved bodies (Fig. 4, D and E; and Fig. S3, A and B).
Figure 4.
Centrobin loss causes ciliary and developmental defects in zebrafish embryos. MO1 was directed to the centrobin ATG and MO2 to the 5′ UTR. (A) Immunoblot analysis of efficacy of the centrobin knockdown in zebrafish embryos at 24 hpf. (B) Confocal stacks of cilia in KV in 8 ss zebrafish embryos injected with the standard control MO (StdCTRL MO) or MO targeting the translation start site of centrobin. Cilia were visualized with an antibody to acetylated tubulin (green). Bar, 10 µm. (C) Quantitation of ciliation frequency and length in KV after injection with the indicated MOs showing mean + SEM of four independent experiments in which 40 standard control MO and 33 Cntrob MO-treated KVs were quantitated by acetylated tubulin staining. In standard control embryos, 1067 cilia were measured, and in Cntrob MO embryos, 750 cilia. *, P < 0.01; ***, P < 0.0001; unpaired t test. (D) Live images show gross phenotypes of zebrafish embryos injected with control or Cntrob MOs at 24 hpf. Arrowheads indicate morphological abnormalities. Bars, 500 nm. (E) Quantitation of developmental phenotypes in centrobin-deficient embryos. Each phenotype was quantitated over three experiments in the indicated number of zebrafish embryos and graphs indicate means + SEM. *, P < 0.05; **, P < 0.01; ****, P < 0.0001, by one-way ANOVA.
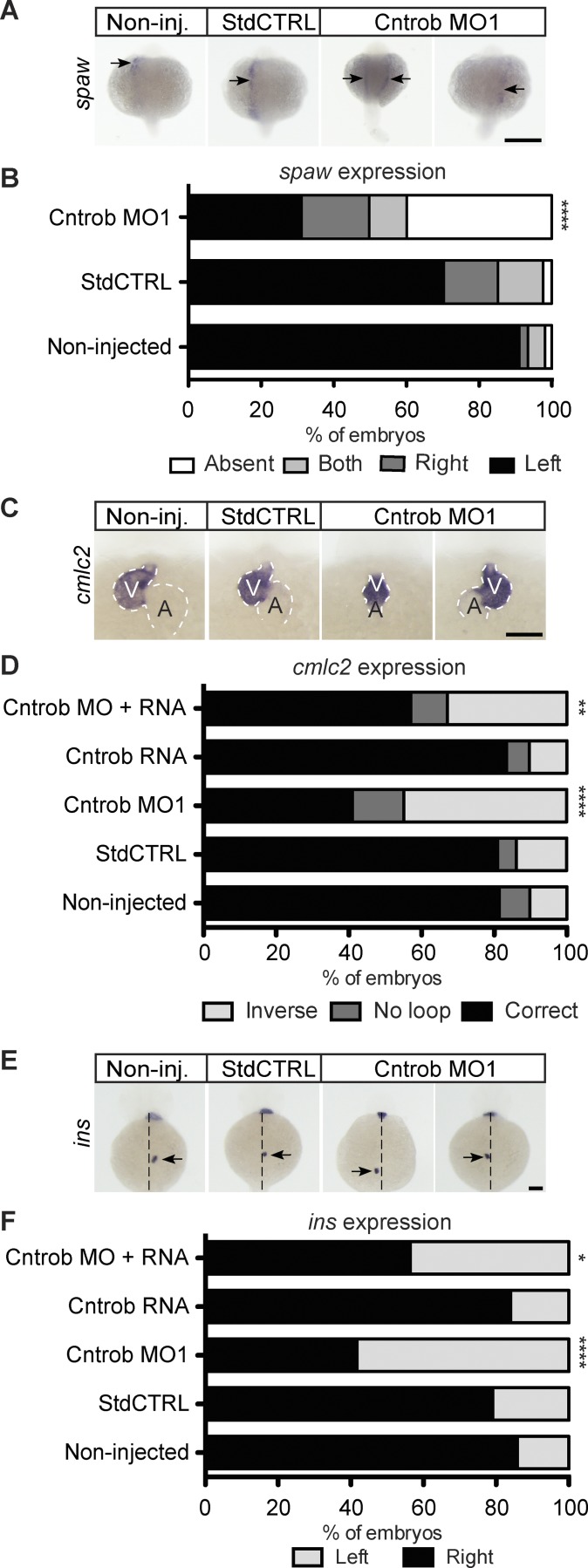
We also observed marked impacts on laterality, another feature observed in ciliopathies. Cntrob MO injection caused randomization of the localization of southpaw (spaw) expression, which is normally restricted to the left lateral plate mesoderm (Fig. 5, A and B). Zebrafish deficient in left–right asymmetry determination, however, possess mirror-imaged spaw expression right of the midline, or exhibit bilateral spaw expression. Consistently, analysis of heart development in Cntrob MO-treated embryos showed declines in the fraction with normal positioning of the ventricle to the left of the atrium, as determined by visualization of cardiac myosin light chain 2 (cmlc2; Fig. 5, C and D; and Fig. S3 C). Abdominal organ asymmetry was also disturbed, as the correct positioning of the endocrine pancreas was impaired during the development of centrobin-deficient embryos, as determined by insulin (ins) localization (Fig. 5, E and F; and Fig. S3 D). Importantly, normal heart and pancreas location was rescued by reexpression of cntrob in Cntrob MO-injected embryos, demonstrating the specificity of the MO phenotypes we describe here (Fig. 5, D and F). These phenotypes reveal defects in cilium formation and a range of developmental abnormalities that are consistent with defects in ciliation in the absence of centrobin. Together, our analyses of centrobin deficiency demonstrate that in vertebrates centrobin is required for ciliogenesis and cilium function.
Figure 5.
Centrobin loss causes laterality defects in zebrafish embryos. (A, C, and E) WMISH micrographs of showing localization of the messages of spaw for left lateral plate mesoderm, cmlc2 to indicate heart looping, and ins to label pancreata and assess abdominal situs development. Dashed line indicates midline. A, atrium; V, ventricle. Bars, 100 µm. (B) Quantitation of laterality of spaw expression (arrows) in WT embryos and embryos injected as indicated at 20–22 ss. Appropriate asymmetric expression is to the left. (D) Quantitation of appropriate heart looping in WT embryos and embryos injected as indicated at 48 hpf. (F) Quantitation of pancreas placement in WT embryos and embryos injected as indicated at 48 hpf. Correct placement is on the right side from the midline. *, P < 0.05; **, P < 0.01; ****, P < 0.0001. Significances were assessed using Fisher’s exact test.
CNTROB as a candidate microcephaly or ciliopathy gene
Defects in centrosome regulation are implicated in primary microcephaly and primordial dwarfism, disorders that do not have ciliopathy features (Nigg and Raff, 2009; Klingseisen and Jackson, 2011). However, cilia assemble on the mother centriole, so there is clearly potential for overlap in the molecular pathology of these diseases. Mutations in PLK4, which encodes the key kinase that directs centriole duplication, were described in cases of microcephalic primordial dwarfism, and MO knockdown of plk4 in zebrafish led to ciliopathy phenotypes like those we describe for cntrob (Martin et al., 2014). Mutations in ATR give rise to Seckel syndrome, a microcephaly disorder (O’Driscoll et al., 2003). Recent work has demonstrated defects in ciliary signaling in ATR-SS cells and atr-depleted zebrafish embryos showed phenotypes indicative of ciliary dysfunction (Stiff et al., 2016). Although there is no published evidence of the clinical impact of CNTROB mutation to our knowledge, rat hypodactyly mutants, which have a truncating mutation in Cntrob, show skeletal abnormalities and male infertility caused by defective sperm flagellar axoneme assembly, a ciliopathy-related phenotype (Liska et al., 2009; Liška et al., 2013). The roles of centrobin in centriole duplication and its interactions with CPAP/CENPJ, a known microcephaly gene (Bond et al., 2005), link it to primary microcephaly. A human disease role for centrobin remains to be determined.
Material and methods
Cell culture and transfections
hTERT-RPE1 and U2OS cells were acquired from the American Type Culture Collection. hTERT-RPE1s were cultured in DMEM/F12 1:1 and U2OS in DMEM (Lonza). Matched HCT116 (colon carcinoma) TP53+/+ (40–16) and TP53−/−(379.2) clones were provided by B. Vogelstein (Johns Hopkins University, Baltimore, MD; Bunz et al., 1998) and cultured in DMEM. Media were supplemented with 10% vol/vol FBS (Sigma-Aldrich) and penicillin-streptomycin (100 U/ml and 100 µg/ml, respectively; Sigma-Aldrich). Cell lines were cultured in a humidified 5% CO2 atmosphere at 37°C, and mycoplasma testing was performed every 3 mo. Primary cilium formation was induced in hTERT-RPE1 by culturing cells in DMEM F-12 supplemented with 0.1% FBS and 1% penicillin-streptomycin for up to 48 h.
For transient transfections, we used Lipofectamine 2000 (Thermo Fisher Scientific) complexed with DNA at a 2:1 ratio in Opti-MEM (Gibco) for 20 min. For stable transfections, linearized plasmid was cotransfected with pLox-Bsr or pLox-Neo (Arakawa et al., 2001). Cells were incubated with the lipid–DNA complexes for 4–6 h, after which the medium was replaced and cells were allowed to recover for 24 h before trypsinization and serial dilution into media containing the necessary antibiotic (5 µg/ml Blasticidin S [Sigma-Aldrich] or 1 mg/ml G418 [Invivogen]) for 3 d. Single colonies were picked after 10–14 d and expanded using 3-mm Scienceware cloning discs (Sigma-Aldrich) before microscopy or immunoblot analysis.
Flow cytometry
Cells were fixed with ice-cold 70% ethanol at −20°C for at least 2 h or overnight. After fixation, cells were washed twice with 1× PBS, then resuspended in PBS containing 200 µg/ml RNase A and 40 µg/ml propidium iodide (Sigma-Aldrich) for 30 min. Cytometry was performed on an Accuri C6 Sampler (BD Biosciences).
Cloning
To clone human CNTROB cDNA isoform α (NM_053051), hTERT-RPE1 RNA was isolated using TRIzol (Invitrogen). Reverse transcription was performed using the High Capacity RNA to cDNA kit (Applied Biosystems) according to the manufacturer’s instructions, and PCR was performed using KOD Hot Start (EMD Millipore) with the primers 5′-AAAAAGAATTCTATGGCAACATCAGCTGAC-3′ and 5′-AAAAAGGTACCTCATCTCCAGACTCCC-3′. PCR products were cloned into pGEM-T Easy (Promega), sequenced, and subcloned into pEGFP-C1 (Clontech) using EcoRI and KpnI restriction sites. To generate pCNTROB-blasticidin, the EGFP coding sequence was removed using AgeI and XhoI, and a blasticidin resistance cassette was cloned into the neomycin resistance gene by blunt end ligation. CNTROB fragments were generated by PCR from pGEM-T Easy-hCNTROB and restriction cloned into pEGFP-C1. Primers used for fragment generation were as follows: hCNTROB-183-EcoR1F, 5′-TTTGGAATTCTTTTCAGGGGCTGAGAGATGCATTGG-3′; hCNTROB-365-EcoR1F, 5′-AAAAGAATTCACAAGAGCACCAGCTTAAGGAACACTACCAGG-3′; hCNTROB-452-EcoR1F, 5′-TTATGAATTCTGCTGTGCAGCTGGAGCAGCGGGTGAC-3′; hCNTROB-765-EcoR1F, 5′-TTTAGAATTCCGCCATGGCATCCAGTCTTTTCCGGGTCCC-3′; hCNTROB-364-Sal1R, 5′-TTTGTCGACCTACTGGGCCCAGGTCTGCCGTTCTTCTTCTAG-3′; and hCNTROB-452-Sal1R, 5′-AATGTCGACGCCAGCTCCGACTCCAGCTGGATCCGC-3′.
For CRISPR/Cas targeting of CNTROB in hTERT-RPE1 cells, we used the human exome Cas9 site catalog (Mali et al., 2013) to generate guide RNAs targeting CNTROB exons 1 and 4. We cloned the following annealed primers into pX330-U6-Chimeric_BB-CBh-hSpCas9 vector (Plasmid 43330; Addgene; Cong et al., 2013): exon 1, 5′-CACCGGTCTAACCCTTCATGAGGCG-3′ and 5′-AAACCGCCTCATGAAGGGTTAGACc-3′; and exon 4, 5′-CACCGGTTTGCATCTTACCTTGCGG-3′ and 5′-AAACCCGCAAGGTAAGATGCAAACc-3′.
To identify zebrafish centrobin EST sequence, we used Basic Local Alignment Search Tool (BLAST) comparison with the human CNTROB cDNA sequence. Rapid Amplification of cDNA ends (RACE) primers were designed within cDNA sequences that encode regions predicted to be highly conserved between mammals and fish. Next, we used a 5′/3′ RACE kit (Roche) to amplify centrobin sequences from zebrafish embryos at 8 h after fertilization (hpf) using the following primers: GSP1_ZF5, 5′-CAGCGGTGAGATCAGGAGAGGACAGAGCG-3′; GSP2_ZF5, 5′-CACTTTCCATCTCCTCTATGTGCTTCCTGC-3′; GSP3_ZF5, 5′-CCTCCACATGCACACGCAGATCAGACGCC-3′; and GSP4_ZF5, 5′-CGTCCGTGTGTCATGCGACTCTGTCTGG-3′. We then predicted a 2,610-bp full-length centrobin cDNA sequence and deposited this sequence in GenBank under accession no. MF461638. We then cloned the full-length cDNA in two fragments that we assembled into pGEM-T Easy after RT-PCR of 8-hpf embryo mRNA with the following primers: ZF CNTROB full F, 5′-ATGTCTGTGAGCCGAGCTGCTGCTGATGG-3′; ZF CNTROB full R, 5′-TCAGTGTTGTCCAGCTGTGGCTCCAGTG-3′; ZF CNTROB full R3, 5′-TCAGTACAGAAACTCCAACCTGCTCAGCCC-3′; and ZF7 CNTROB HindIII frag F, 5′-AGCAAAAGCTTAGCAGAGAAGCAGATAAACACAGAG-3′. A mutant form with a silent mutation to ensure MO resistance was cloned using the following primer: ZF7 CNTROB mut F, 5′-ATGTCAGTCAGTCGCGCTGCAGCAG-3′.
To generate pGFP-DCnb, reverse transcription was performed on Drosophila mRNA provided by C. Collins (National University of Ireland Galway, Galway, Ireland) as described above and the centrobin transcript cloned into pEGFP-C1 after PCR with primers we designed from centrobin transcript variant A (NM_139423): DCnb-Fwd, 5′-CCCTCAAGCTTCCATGAGTGATACCGATACGGACGAC-3′ and DCnb-Rev, 5′-GCCTTGGTACCTCAGCACTTCCAAGGTGGAGGCTTACT-3′.
Monoclonal antibody generation
A fragment of the human CNTROB cDNA encoding amino acids 113–361 was cloned into pGEX4T3 (GE Healthcare) and expressed in bacteria as a GST fusion protein. The centrobin fragment was purified from a glutathione column by thrombin cleavage and used for hybridoma preparation (Dundee Cell Products). The best-performing clone 6D4 was expanded and subcloned to give 6D4F4, which produces IgG1κ.
RNA-mediated interference
hTERT-RPE1 cells were plated to attain 30–40% confluency on the day of transfection. On the next day, 50 nmol of custom siRNA (5′-AAGGAUGGUUCUAAGCAUAUC-3′; Jeong et al., 2007) or Silencer Select siRNA oligonucleotides specific to GAPDH (s5573; Ambion) was complexed with Oligofectamine (Invitrogen) in Opti-MEM (Gibco) for 20 min before addition to cells. After 4-h incubation of cells with the Oligofectamine-siRNA complexes, medium supplemented with 30% FBS was added, and cells were incubated for 48 h. Where indicated, cells were transfected for 36 h before serum starvation for 24 h.
Zebrafish husbandry and manipulations
Zebrafish were housed in a fully automated water-circulating rack system (Tecniplast) and exposed to a 14-h light and 10-h dark cycle. Embryos from EK and AB WT strains were generated using natural matings and raised until the desired stage in an incubator set to 28.5°C. Microinjections of MOs or capped RNAs were done with the help of an Eppendorf Femtojet and a Narishige micromanipulator at the 1–2 cell stage. MOs were designed based on submitted sequences and synthesized by Gene Tools. Two nonoverlapping MOs against the ATG and 5′-UTR of zebrafish centrobin were used: MO1, 5′-GTCTGTGAGATGTCTGTGAGCCGAG-3′; and MO2, 5′-ATGAGAGTTTGTTTACCGTCCGAGT-3′. As control, the standard control MO was used. Capped RNAs for reconstitution experiments were in vitro transcribed from linearized plasmids using the mMessage mMachine SP6 kit (Ambion) according to the manufacturer’s instructions. All husbandry and experiments herein have been approved by local authorities and adhered to current European law.
IF microscopy
hTERT-RPE1 cells were grown on sterile coverslips and fixed in methanol containing 5 mM EGTA at −20 °C for 10 min or 4% paraformaldehyde for 10 min at room temperature followed by permeabilization with 0.15% Triton X-100 in 1× PBS for 2 min. To stain with antibodies against modified tubulins, cells were incubated on ice for 30 min to depolymerize microtubules. The cells were blocked in 1% BSA in 1× PBS before incubation in primary antibody for 1 h followed by 45 min incubation with fluorescently labeled secondary antibodies (Jackson Immunoresearch Laboratories Inc.). DNA was stained with Hoechst 33258 (Sigma-Aldrich), and slides were mounted in 80% vol/vol glycerol containing 3% N-propyl-gallate in 1× PBS. Cells were imaged using an IX81 microscope (Olympus) with a C4742-80-12AG camera (Hamamatsu) with a 100× oil objective, NA 1.35, using Volocity software (Perkin-Elmer). Images are presented as maximum intensity projections of z-stacks after deconvolution. Merges and individual channel images were exported as tagged image file formats (TIFFs) for publication and then cropped for publication using Photoshop CS6 (Adobe).
Primary antibodies used on human cells in this study were as follows: acetylated tubulin (1:2,000; T6793, clone 6-11B-1; Sigma-Aldrich); ARL13B (17711–1-AP; 1:2,000; ProteinTech); CP110 (1:2,000; 12780-1-AP; ProteinTech); IFT88 (13967–1-AP; 1:800; ProteinTech); detyrosinated tubulin (1:2,000; Ab48389; Abcam); pericentrin (Ab4448; 1:10,000; Abcam); Centrin2 (Poly6288; 1:1,000; Biolegend); Centrin (1:1,000; 20H5; Millipore); Centrin3 (1:1,000; 3E6; Abnova); CEP164 (Daly et al., 2016; 1F3G10; 1:100,000); α tubulin (1:2,000; B512, Sigma-Aldrich); CEP135 (Bird and Hyman, 2008; 1420 738; 1:1,000); CPAP (1:1,000; 11517-1-AP; ProteinTech); Ninein (Ab4447; 1:250; Abcam); SDCCAG8 (1:250; 13471-1-AP; ProteinTech); γ-tubulin (1:1,000; T3559; Sigma-Aldrich); centrobin (this study; 6D4F4; 1:10,000); CEP97 (1:1,000; 22050-1-AP; ProteinTech); hSAS6 (1:500; H00163786-B01P; Novus Biologicals); Kizuna (Oshimori et al., 2006; 1:2,000); C-NAP1 (Flanagan et al., 2017; 6F2C8; 1:2); PCM1 (Dammermann and Merdes, 2002; 817; 1:10,000); CEP164 (1:1,000; HPA037606, Sigma-Aldrich); and CEP97 (1:250; N-17; Santa Cruz Biotechnology).
For microscopy of zebrafish cilia, eight somite stage (ss) embryos were fixed overnight at 4°C using 4% PFA buffered in PBS and processed for cilia staining as previously described (Burkhalter et al., 2013). Cilia were labeled using anti-acetylated tubulin antibody (1:1,000; T6793; Sigma-Aldrich). An Alexa Fluor 488–labeled secondary antibody was used for detection (1:1;000; Molecular Probes). After staining, zebrafish embryos were manually deyolked using forceps, and the posterior part of the embryo was mounted in Vectashield (Vectorlabs) between two coverslips. Cilia were analyzed with a TCS SP5II confocal microscope (Leica). Cilia length was measured from confocal stacks using ImageJ.
Whole-mount in situ hybridization (WMISH)
Zebrafish embryos were fixed at the desired stages using 4% PFA buffered in PBS, dehydrated in a graded methanol series and stored at −20°C until further processing. For WMISH, embryos were gradually rehydrated and then processed according to standard protocols. WMISH probes against cmlc2, ins, and spaw have been described previously (Burkhalter et al., 2013). For detection of centrobin transcripts, a DIG-labeled in situ probe covering the whole coding sequence of centrobin was generated from the pGEM-T Easy plasmid after linearization with ApaI and using SP6 for in vitro transcription. Live embryos and those processed by WMISH were imaged on an M125 upright microscope (Leica) equipped with an IC80 HD camera (Lecia).
Transmission electron microscopy
Cells were harvested by trypsinization and pelleted at 250 g for 5 min. Cells were washed twice in 1× PBS, and then twice in 0.1 M sodium cacodylate buffer, pH 7.2 (Sigma-Aldrich), followed by overnight incubation at 4°C in primary fixative (2% glutaraldehyde and 2% paraformaldehyde [Electron Microscopy Sciences] in cacodylate buffer). The next day, cells were pelleted, washed, and fixed in secondary fixative (2% osmium tetroxide [Sigma-Aldrich] in cacodylate buffer) for a minimum of 2 h in the dark at room temperature until the cell pellet turned black. Cell pellets were washed three times in cacodylate buffer before dehydration through ethanol gradient (15 min each of 30, 60, and 90%), after which cells were further dehydrated three times for 30 min in 100% ethanol. To remove all of the alcohol, cell pellets were incubated in propylene oxide (Sigma-Aldrich) for 30 min followed by 4-h incubation in 50:50 propylene oxide/resin (TAAB) and overnight incubation in a 25:75 propylene oxide/resin mixture. The next day, samples were embedded in 100% low viscosity resin (TAAB). The blocks were sectioned on a microtome (Reichert-Jung Ultracut E; Leica) and stained with uranyl acetate and lead citrate before transmission electron microscopy (H-7000; Hitachi) with an ORCA-HRL camera (Hamamatsu). Images were processed using AMT version 6 (AMT Imaging).
Immunoblotting
Total cell extracts were prepared by lysing cells in lysis buffer (50 mM Tris HCl, pH 7.4, 150 mM NaCl, 5% glycerol, 1 mM EDTA, 0.5% sodium deoxycholate, 1% IGEPAL, protease inhibitor cocktail [Roche], and phosphatase inhibitors [Sigma-Aldrich]) for 20 min on ice. Samples were then centrifuged for 20 min at 18,000 g at 4°C, and supernatant was transferred to a fresh tube. 24-hpf zebrafish embryos were manually dechorioniated, deyolked (Burkhalter et al., 2013), and lysed in the same way. Protein concentration was determined by Bradford assay on a Nanodrop 2000c spectrophotometer (Thermo Fisher Scientific). For loading on SDS-PAGE gel, 20–80 µg of whole cell lysate was transferred into a fresh tube and 5× sample buffer containing 20% β-mercaptoethanol was added to the samples and boiled at 95°C for 5 min. After proteins were resolved on 6–10% SDS–polyacrylamide gels, they were transferred to a nitrocellulose membrane (GE Healthcare) using a semidry transfer unit (Hoeffer TE77) at 1 mA/cm2 for 2 h or a TransBlot wet transfer unit (BioRad) at 350 mA for 3 h. Blot detection was performed using ECL (GE Healthcare) after blocking and incubation in primary and secondary antibodies.
The primary antibodies used in this study were as follows: centrobin (6D4F4; 1:1,000), CP110 (1:2,000; 12780-1-AP; ProteinTech), α-tubulin (1:10,000; B512; Sigma-Aldrich), GFP (1:15,000; 66002-1-Ig; Proteintech), and GAPDH (1:10,000; 2118; Cell Signaling). HRP-labeled goat anti–mouse or anti–rabbit secondary antibodies were used at 1:10,000 (Jackson Immunoresearch Laboratories Inc.).
Coimmunoprecipitation
After trypsinization of HCT116 (WT) or hTERT-RPE1 cells, whole cell extracts were prepared by lysing cells in lysis buffer (50 mM Tris HCl, pH 7.4, 150 mM NaCl, 20% glycerol, 1 mM EDTA, 0.5% sodium deoxycholate, 1% IGEPAL, protease inhibitor cocktail [Roche], 1 mM sodium orthovanadate, 5 mM sodium fluoride, 1:10,000 dilution of benzonase nuclease [Sigma-Aldrich], and 1 mM PMSF) for 45 min at 4°C on a rotating wheel. Samples were then centrifuged for 20 min at 18,000 g at 4°C and supernatants transferred to a fresh tube. Protein concentration was determined by Bradford assay on a Thermo Fisher Scientific Nanodrop 2000c Spectrophotometer. Meanwhile, 20 µl of prewashed protein A/G beads (Santa Cruz Biotechnology) were incubated with 3–5 µg of primary antibody for 1 h at 4 °C with gentle agitation. The beads–antibody complex was washed twice with lysis buffer followed by incubation with 3–5 mg total cell extract for a further 2 h. After incubation, immunoprecipitates were spun down at 155 g for 3 min and the supernatant discarded. Beads were washed four times in lysis buffer, then boiled in 5× Laemmli buffer for 10 min and pelleted at 18,000 g before immunoblot analysis.
Quantification and statistical analysis
Data were analyzed using Prism v5.0 and v6.0 (GraphPad) with the statistical tests as indicated in the figure legends.
Online supplemental material
Fig. S1 shows generation and characterization of a monoclonal antibody for centrobin. Fig. S2 shows confirmation of CNTROB disruption by genome editing. Fig. S3 shows reproduction of the zebrafish phenotypes with a second MO.
Supplementary Material
Acknowledgments
We thank Caitríona Collins for Drosophila mRNA and Sandra Burczyk and Cornelia Donow for excellent fish care.
This work was funded by Science Foundation Ireland Principal Investigator award 10/IN.1/B2972 and European Commission grant SEC-2009-4.3-02, project 242361 “BOOSTER.” Y.A. Ogungbenro received a Government of Ireland Postgraduate Scholarship from the Irish Research Council (GOIPG/2013/318). We acknowledge the National Biophotonics and Imaging Platform Ireland and the National Centre for Biomedical Engineering Sciences Flow Cytometry core facility, which were supported by the Irish Government Program for Research in Third-Level Institutions. Zebrafish experiments were funded by Deutsche Forschungsgemeinschaft grant PH144/4-1.
The authors declare no competing financial interests.
Author contributions: Y.A. Ogungbenro and C.G. Morrison conceptualized the project. Methodology was by Y.A. Ogungbenro, T.C. Tena, D. Gaboriau, M. Philipp and C.G. Morrison. Investigation was by Y.A. Ogungbenro, T.C. Tena, D. Gaboriau, and P. Lalor. C.G. Morrison wrote the original draft. Y.A. Ogungbenro, T.C. Tena, D. Gaboriau, M. Philipp, and C.G. Morrison wrote, reviewed, and edited the paper. Funding acquisition was by M. Philipp, P. Dockery, and C.G. Morrison. M. Philipp, P. Dockery, and C.G. Morrison provided resources. M. Philipp, P. Dockery, and C.G. Morrison supervised the project.
References
- Anderson R.G. 1972. The three-dimensional structure of the basal body from the rhesus monkey oviduct. J. Cell Biol. 54:246–265. 10.1083/jcb.54.2.246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arakawa H., Lodygin D., and Buerstedde J.M.. 2001. Mutant loxP vectors for selectable marker recycle and conditional knock-outs. BMC Biotechnol. 1:7 10.1186/1472-6750-1-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird A.W., and Hyman A.A.. 2008. Building a spindle of the correct length in human cells requires the interaction between TPX2 and Aurora A. J. Cell Biol. 182:289–300. 10.1083/jcb.200802005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bond J., Roberts E., Springell K., Lizarraga S.B., Scott S., Higgins J., Hampshire D.J., Morrison E.E., Leal G.F., Silva E.O., et al. . 2005. A centrosomal mechanism involving CDK5RAP2 and CENPJ controls brain size. Nat. Genet. 37:353–355. 10.1038/ng1539 [DOI] [PubMed] [Google Scholar]
- Braun D.A., and Hildebrandt F.. 2017. Ciliopathies. Cold Spring Harb. Perspect. Biol. 9:a028191 10.1101/cshperspect.a028191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunz F., Dutriaux A., Lengauer C., Waldman T., Zhou S., Brown J.P., Sedivy J.M., Kinzler K.W., and Vogelstein B.. 1998. Requirement for p53 and p21 to sustain G2 arrest after DNA damage. Science. 282:1497–1501. 10.1126/science.282.5393.1497 [DOI] [PubMed] [Google Scholar]
- Burkhalter M.D., Fralish G.B., Premont R.T., Caron M.G., and Philipp M.. 2013. Grk5l controls heart development by limiting mTOR signaling during symmetry breaking. Cell Reports. 4:625–632. 10.1016/j.celrep.2013.07.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong L., Ran F.A., Cox D., Lin S., Barretto R., Habib N., Hsu P.D., Wu X., Jiang W., Marraffini L.A., and Zhang F.. 2013. Multiplex genome engineering using CRISPR/Cas systems. Science. 339:819–823. 10.1126/science.1231143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daly O.M., Gaboriau D., Karakaya K., King S., Dantas T.J., Lalor P., Dockery P., Kramer A., and Morrison C.G.. 2016. CEP164-null cells generated by genome editing show a ciliation defect with intact DNA repair capacity. J. Cell Sci. 129:1769–1774. 10.1242/jcs.186221 [DOI] [PubMed] [Google Scholar]
- Dammermann A., and Merdes A.. 2002. Assembly of centrosomal proteins and microtubule organization depends on PCM-1. J. Cell Biol. 159:255–266. 10.1083/jcb.200204023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essner J.J., Amack J.D., Nyholm M.K., Harris E.B., and Yost H.J.. 2005. Kupffer’s vesicle is a ciliated organ of asymmetry in the zebrafish embryo that initiates left-right development of the brain, heart and gut. Development. 132:1247–1260. 10.1242/dev.01663 [DOI] [PubMed] [Google Scholar]
- Flanagan A.M., Stavenschi E., Basavaraju S., Gaboriau D., Hoey D.A., and Morrison C.G.. 2017. Centriole splitting caused by loss of the centrosomal linker protein C-NAP1 reduces centriolar satellite density and impedes centrosome amplification. Mol. Biol. Cell. 28:736–745. 10.1091/mbc.E16-05-0325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franz A., Roque H., Saurya S., Dobbelaere J., and Raff J.W.. 2013. CP110 exhibits novel regulatory activities during centriole assembly in Drosophila. J. Cell Biol. 203:785–799. 10.1083/jcb.201305109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goetz S.C., and Anderson K.V.. 2010. The primary cilium: a signalling centre during vertebrate development. Nat. Rev. Genet. 11:331–344. 10.1038/nrg2774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottardo M., Pollarolo G., Llamazares S., Reina J., Riparbelli M.G., Callaini G., and Gonzalez C.. 2015. Loss of Centrobin Enables Daughter Centrioles to Form Sensory Cilia in Drosophila. Curr. Biol. 25:2319–2324. 10.1016/j.cub.2015.07.038 [DOI] [PubMed] [Google Scholar]
- Gudi R., Zou C., Li J., and Gao Q.. 2011. Centrobin-tubulin interaction is required for centriole elongation and stability. J. Cell Biol. 193:711–725. 10.1083/jcb.201006135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gudi R., Zou C., Dhar J., Gao Q., and Vasu C.. 2014. Centrobin-centrosomal protein 4.1-associated protein (CPAP) interaction promotes CPAP localization to the centrioles during centriole duplication. J. Biol. Chem. 289:15166–15178. 10.1074/jbc.M113.531152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gudi R., Haycraft C.J., Bell P.D., Li Z., and Vasu C.. 2015. Centrobin-mediated regulation of the centrosomal protein 4.1-associated protein (CPAP) level limits centriole length during elongation stage. J. Biol. Chem. 290:6890–6902. 10.1074/jbc.M114.603423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa H., and Marshall W.F.. 2011. Ciliogenesis: building the cell’s antenna. Nat. Rev. Mol. Cell Biol. 12:222–234. 10.1038/nrm3085 [DOI] [PubMed] [Google Scholar]
- Ishikawa H., Kubo A., Tsukita S., and Tsukita S.. 2005. Odf2-deficient mother centrioles lack distal/subdistal appendages and the ability to generate primary cilia. Nat. Cell Biol. 7:517–524. 10.1038/ncb1251 [DOI] [PubMed] [Google Scholar]
- Jeffery J.M., Urquhart A.J., Subramaniam V.N., Parton R.G., and Khanna K.K.. 2010. Centrobin regulates the assembly of functional mitotic spindles. Oncogene. 29:2649–2658. 10.1038/onc.2010.37 [DOI] [PubMed] [Google Scholar]
- Jeong Y., Lee J., Kim K., Yoo J.C., and Rhee K.. 2007. Characterization of NIP2/centrobin, a novel substrate of Nek2, and its potential role in microtubule stabilization. J. Cell Sci. 120:2106–2116. 10.1242/jcs.03458 [DOI] [PubMed] [Google Scholar]
- Klingseisen A., and Jackson A.P.. 2011. Mechanisms and pathways of growth failure in primordial dwarfism. Genes Dev. 25:2011–2024. 10.1101/gad.169037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J., Jeong Y., Jeong S., and Rhee K.. 2010. Centrobin/NIP2 is a microtubule stabilizer whose activity is enhanced by PLK1 phosphorylation during mitosis. J. Biol. Chem. 285:25476–25484. 10.1074/jbc.M109.099127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liska F., Gosele C., Rivkin E., Tres L., Cardoso M.C., Domaing P., Krejcí E., Snajdr P., Lee-Kirsch M.A., de Rooij D.G., et al. . 2009. Rat hd mutation reveals an essential role of centrobin in spermatid head shaping and assembly of the head-tail coupling apparatus. Biol. Reprod. 81:1196–1205. 10.1095/biolreprod.109.078980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liška F., Gosele C., Popova E., Chylíková B., Křenová D., Křen V., Bader M., Tres L.L., Hubner N., and Kierszenbaum A.L.. 2013. Overexpression of full-length centrobin rescues limb malformation but not male fertility of the hypodactylous (hd) rats. PLoS One. 8:e60859 10.1371/journal.pone.0060859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali P., Yang L., Esvelt K.M., Aach J., Guell M., DiCarlo J.E., Norville J.E., and Church G.M.. 2013. RNA-guided human genome engineering via Cas9. Science. 339:823–826. 10.1126/science.1232033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin C.A., Ahmad I., Klingseisen A., Hussain M.S., Bicknell L.S., Leitch A., Nürnberg G., Toliat M.R., Murray J.E., Hunt D., et al. . 2014. Mutations in PLK4, encoding a master regulator of centriole biogenesis, cause microcephaly, growth failure and retinopathy. Nat. Genet. 46:1283–1292. 10.1038/ng.3122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nigg E.A., and Raff J.W.. 2009. Centrioles, centrosomes, and cilia in health and disease. Cell. 139:663–678. 10.1016/j.cell.2009.10.036 [DOI] [PubMed] [Google Scholar]
- Nigg E.A., and Stearns T.. 2011. The centrosome cycle: Centriole biogenesis, duplication and inherent asymmetries. Nat. Cell Biol. 13:1154–1160. 10.1038/ncb2345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Driscoll M., Ruiz-Perez V.L., Woods C.G., Jeggo P.A., and Goodship J.A.. 2003. A splicing mutation affecting expression of ataxia-telangiectasia and Rad3-related protein (ATR) results in Seckel syndrome. Nat. Genet. 33:497–501. 10.1038/ng1129 [DOI] [PubMed] [Google Scholar]
- Oshimori N., Ohsugi M., and Yamamoto T.. 2006. The Plk1 target Kizuna stabilizes mitotic centrosomes to ensure spindle bipolarity. Nat. Cell Biol. 8:1095–1101. 10.1038/ncb1474 [DOI] [PubMed] [Google Scholar]
- Prosser S.L., and Morrison C.G.. 2015. Centrin2 regulates CP110 removal in primary cilium formation. J. Cell Biol. 208:693–701. 10.1083/jcb.201411070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeley E.S., and Nachury M.V.. 2010. The perennial organelle: assembly and disassembly of the primary cilium. J. Cell Sci. 123:511–518. 10.1242/jcs.061093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin W., Yu N.K., Kaang B.K., and Rhee K.. 2015. The microtubule nucleation activity of centrobin in both the centrosome and cytoplasm. Cell Cycle. 14:1925–1931. 10.1080/15384101.2015.1041683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorokin S. 1962. Centrioles and the formation of rudimentary cilia by fibroblasts and smooth muscle cells. J. Cell Biol. 15:363–377. 10.1083/jcb.15.2.363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spektor A., Tsang W.Y., Khoo D., and Dynlacht B.D.. 2007. Cep97 and CP110 suppress a cilia assembly program. Cell. 130:678–690. 10.1016/j.cell.2007.06.027 [DOI] [PubMed] [Google Scholar]
- Stiff T., Casar Tena T., O’Driscoll M., Jeggo P.A., and Philipp M.. 2016. ATR promotes cilia signalling: links to developmental impacts. Hum. Mol. Genet. 25:1574–1587. 10.1093/hmg/ddw034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanos B.E., Yang H.J., Soni R., Wang W.J., Macaluso F.P., Asara J.M., and Tsou M.F.. 2013. Centriole distal appendages promote membrane docking, leading to cilia initiation. Genes Dev. 27:163–168. 10.1101/gad.207043.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsang W.Y., Bossard C., Khanna H., Peränen J., Swaroop A., Malhotra V., and Dynlacht B.D.. 2008. CP110 suppresses primary cilia formation through its interaction with CEP290, a protein deficient in human ciliary disease. Dev. Cell. 15:187–197. 10.1016/j.devcel.2008.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vorobjev I.A., and Chentsov Y.S.. 1982. Centrioles in the cell cycle. I. Epithelial cells. J. Cell Biol. 93:938–949. 10.1083/jcb.93.3.938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters A.M., and Beales P.L.. 2011. Ciliopathies: an expanding disease spectrum. Pediatr. Nephrol. 26:1039–1056. 10.1007/s00467-010-1731-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou C., Li J., Bai Y., Gunning W.T., Wazer D.E., Band V., and Gao Q.. 2005. Centrobin: A novel daughter centriole-associated protein that is required for centriole duplication. J. Cell Biol. 171:437–445. 10.1083/jcb.200506185 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.