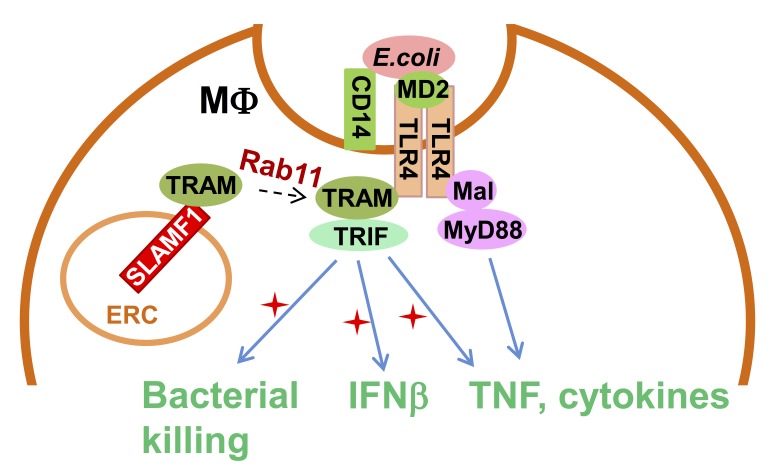
Yurchenko et al. discover that the Ig-like receptor molecule SLAMF1 enhances production of type I interferon induced by Gram-negative bacteria through modulation of MyD88-independent TLR4 signaling. This makes SLAMF1 a potential target for controlling inflammatory responses against Gram-negative bacteria.
Abstract
Signaling lymphocytic activation molecule family 1 (SLAMF1) is an Ig-like receptor and a costimulatory molecule that initiates signal transduction networks in a variety of immune cells. In this study, we report that SLAMF1 is required for Toll-like receptor 4 (TLR4)-mediated induction of interferon β (IFNβ) and for killing of Gram-negative bacteria by human macrophages. We found that SLAMF1 controls trafficking of the Toll receptor–associated molecule (TRAM) from the endocytic recycling compartment (ERC) to Escherichia coli phagosomes. In resting macrophages, SLAMF1 is localized to ERC, but upon addition of E. coli, it is trafficked together with TRAM from ERC to E. coli phagosomes in a Rab11-dependent manner. We found that endogenous SLAMF1 protein interacted with TRAM and defined key interaction domains as amino acids 68 to 95 of TRAM as well as 15 C-terminal amino acids of SLAMF1. Interestingly, the SLAMF1–TRAM interaction was observed for human but not mouse proteins. Overall, our observations suggest that SLAMF1 is a new target for modulation of TLR4–TRAM–TRIF inflammatory signaling in human cells.
Graphical Abstract
Introduction
Toll-like receptors (TLRs) are pivotal for the defense against multiple pathogens by recognizing pathogen-associated molecular patterns. TLR4 recognizes lipopolysaccharide (LPS) from Gram-negative bacteria in complex with the coreceptors myeloid differentiation factor 2 and CD14, and it recruits signaling adapters myeloid differentiation primary response gene 88 (MyD88) and MyD88 adapter–like (Mal). This results in an immediate activation of nuclear factor κB (NF-κB) and production of proinflammatory cytokines. TLR4 is also present on endosomes and phagosomes to which the signaling adapter Toll receptor–associated molecule (TRAM) is recruited (Husebye et al., 2006, 2010; Kagan et al., 2008). The mechanism controlling TRAM recruitment remains unclear but seems to be Rab11 dependent (Husebye et al., 2010; Klein et al., 2015).
TRAM is crucial for the subsequent recruitment of Toll/interleukin (IL)-1 receptor (TIR) domain–containing adapter-inducing IFN-β (TRIF) and other downstream molecules, leading to IFNβ secretion (Fitzgerald et al., 2003b; Oshiumi et al., 2003; Yamamoto et al., 2003; Husebye et al., 2010). The role of endogenous type I IFNs in host defense against bacterial infections could be either beneficial or detrimental. Type I IFNs make macrophages more sensitive to cell death–inducing stimuli that could favor bacterial replication and release (Trinchieri, 2010). At the same time, type I IFNs are required for the host resistance to group B streptococci, pneumococci, and Escherichia coli (Mancuso et al., 2007).
Assembly of the TLR4–TRAM–TRIF complex followed by the activation of TANK-binding kinase 1 (TBK1) results not only in the induction of type I IFNs but also is required for maintenance of the integrity of pathogen-containing vacuoles and restriction of bacterial proliferation in the cytosol (Radtke et al., 2007; Thurston et al., 2016). Moreover, TBK1 activates the Akt–mTOR–HIF1α signaling axis, which orchestrates metabolic reprogramming to aerobic glycolysis in immune cells (Krawczyk et al., 2010; Everts et al., 2014). Glycolysis provides ATP for driving phagocytosis, proinflammatory cytokine production, and NADPH for the NADPH oxidase 2 (NOX2) enzyme to generate reactive oxygen species (ROS; Kelly and O’Neill, 2015).
Signaling lymphocytic activation molecule family 1 (SLAMF1)/CD150 is a type I glycoprotein belonging to the SLAM subfamily of the CD2-like family of proteins (Sidorenko and Clark, 1993; Cocks et al., 1995). SLAMF1 acts as a coreceptor that can modulate signaling via the TNF family and antigen receptors (Mikhalap et al., 1999; Wang et al., 2004; Réthi et al., 2006; Makani et al., 2008). SLAMF1 is involved in the regulation of innate immune responses. Slamf1−/− bone marrow–derived macrophages (BMDMs) are deficient in bacterial killing as they produce less ROS in response to Escherichia coli. Mouse SLAMF1 positively regulates NOX2 activity by forming a complex with beclin-1–Vps34–Vps15–UVRAG (Berger et al., 2010; Ma et al., 2012). Thus, it was essential to explore the contribution of SLAMF1 to TLR4-mediated signaling in human cells. In this study, we show that in human macrophages, SLAMF1 acts as a critical regulator of TLR4-mediated signaling from the phagosome by interacting with TRAM adapters and class I Rab11 family interacting proteins (FIPs) and recruiting the adapter to the TLR4 signaling complex.
Results
SLAMF1 is expressed in human macrophages and localized to the Rab11-positive endocytic recycling compartment (ERC)
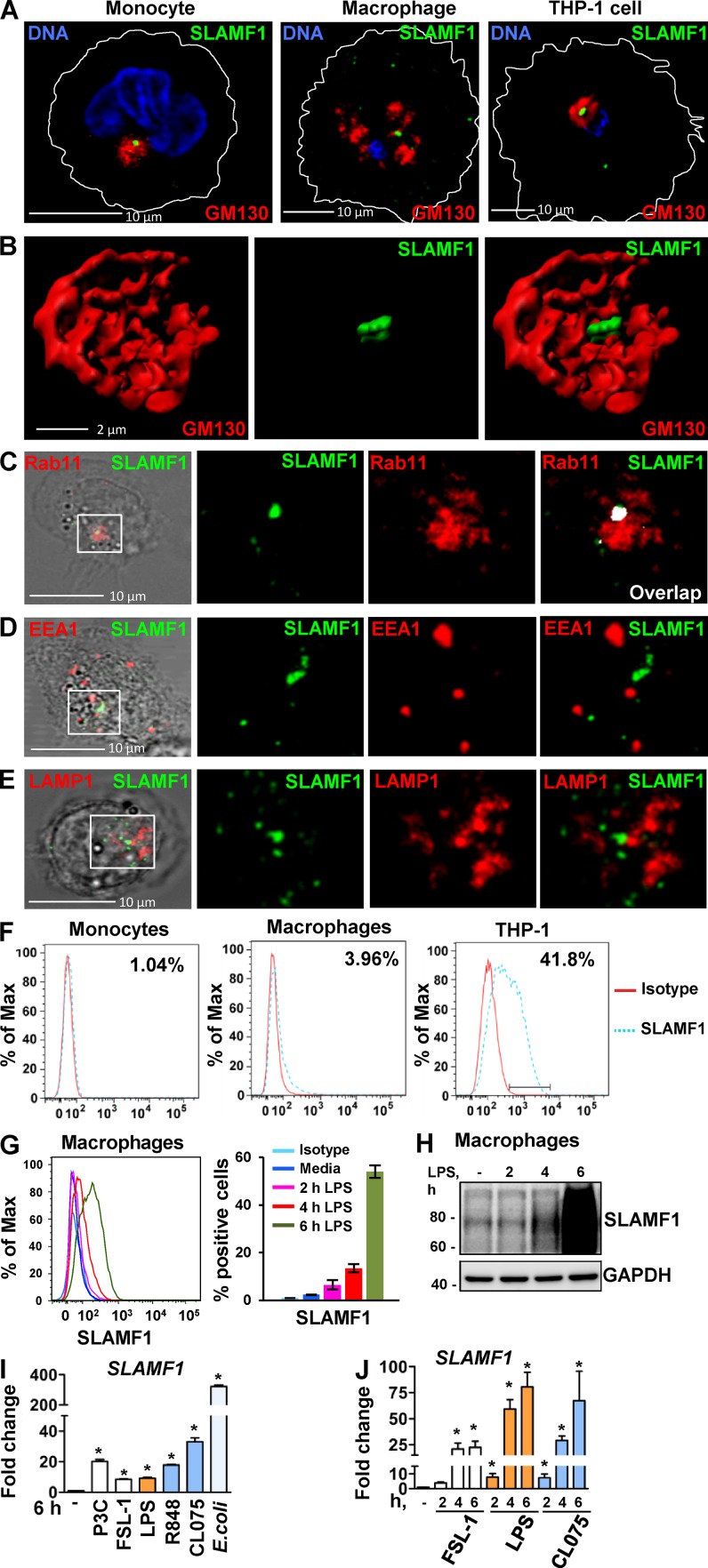
Previous studies have suggested that SLAMF1 is found in intracellular compartments of human primary dendritic cells and glioblastoma cells (Avota et al., 2011; Romanets-Korbut et al., 2015). Human peripheral blood monocytes do not express SLAMF1 on the plasma membrane (Farina et al., 2004; Romero et al., 2004). Therefore, we first analyzed the cellular distribution of SLAMF1 in human monocytes, macrophages, and THP-1 cells by confocal microscopy. In all the cell types examined, the major pool of SLAMF1 was located in a perinuclear area negative for the Golgi marker GM130 (Fig. 1, A and B). To further define SLAMF1 localization, monocytes were costained with markers for different types of endosomes: recycling (Rab11a; Fig. 1 C), early (EEA1), and late endosomes (LAMP1; Fig. 1, D and E). Rab11a also defines the ERC, a condensed perinuclear region containing tubular membrane structures that originate from the microtubule organizing center (Yamashiro et al., 1984). TRAM and TLR4 are also present in ERCs of human monocytes and macrophages (Husebye et al., 2010; Klein et al., 2015).
Figure 1.
SLAMF1 is enriched in the Rab11-positive ERCs in unstimulated macrophages, and SLAMF1 expression is induced by LPS and several other TLR ligands in primary human monocytes and macrophages. (A) Monocytes, macrophages, and differentiated THP-1 cells stained with antibodies against SLAMF1 (green) and GM130 (red) and imaged by confocal microscopy. (B) 3D model of cis-Golgi (GM130) and SLAMF1 in THP-1 cells. Z stacks from the GM130 and SLAMF1 channels were obtained using high-resolution confocal microscopy followed by 3D modeling in IMARIS software. (C) Macrophages stained for SLAMF1 and Rab11 (ERC marker). Representative image. Overlapping pixels for SLAMF1 and Rab11 are shown in the white overlap. tM1 = 0.683 ± 0.08 (mean with SD) for z stacks of ERCs as ROIs (30 ROIs analyzed per donor) where tM1 was the Manders’s colocalization coefficient with thresholds calculated in the Coloc 2 Fiji plugin with anti-SLAMF1 staining as first channel. (D) Macrophages costained for SLAMF1 and EEA1. (E) Macrophages costained for SLAMF1 and LAMP1. Colocalization accessed for z stacks for at least 30 cells for each experiment (four total) showing no colocalization for markers in both D and E. (F) Flow cytometry analysis of SLAMF1 surface expression by primary macrophages and differentiated THP-1 cells. Cells were costained for SLAMF1 and CD14 and gated for CD14-positive cells (primary cells) or stained for SLAMF1 (THP-1 cells). (G) Flow cytometry analysis of SLAMF1 surface expression by human macrophages stimulated by ultrapure K12 LPS (100 ng/ml) for 2, 4, and 6 h. (H) Western blot analysis of lysates from primary human macrophages stimulated by LPS for 2, 4, and 6 h. Graphs present mean values for three biological replicates with SD. Molecular weight is given in kilodaltons. (I and J) Quantification of SLAMF1 mRNA expression by qPCR in monocytes (I) and macrophages (J) stimulated by TLRs’ ligands FSL-1 (20 ng/ml), K12 LPS (100 ng/ml), and CL075 (1 μg/ml; both I and J) as well as R848 (1 μg/ml), Pam3Cys (P3C; 1 μg/ml), or K12 E. coli particles (20/cell; I only). Results are presented as means with SD. Statistical significance between groups was evaluated by a two-tailed t test. *, P < 0.01. Results are representative of at least four independent experiments/donors (A–H) or combined data for at least three donors (I and J).
A marked colocalization was found between SLAMF1 and Rab11 in ERCs of resting cells with a Manders’s colocalization coefficient of tM = 0.683 ± 0.08 (Fig. 1 C), whereas there was no colocalization with the other endosomal markers (Fig. 1, D and E). As determined by flow cytometry, only 1% of the monocytes and 4% of macrophages showed surface expression of SLAMF1, whereas 40% of the differentiated THP-1 cells were SLAMF1 positive (Fig. 1 F). LPS stimulation increased the surface expression of SLAMF1 in primary macrophages by >50% after 6 h of LPS stimulation, with an increase in the total SLAMF1 protein expression (Fig. 1, G and H; and Fig. S1 A). Moreover, various TLR ligands such as Pam3Cys (TLR1/2), FSL-1 (TLR2/6), R848 (TLR7 and -8), and CL075 (TLR8) increased SLAMF1 mRNA expression in monocytes and macrophages (Fig. 1, I and J), with E. coli being the most potent stimulator (Fig. 1 I). These results indicate that several TLRs control SLAMF1 expression in human cells.
In summary, resting macrophages showed very low SLAMF1 surface level expression, and the major cellular pool of SLAMF1 was found to be in the ERC. THP-1 cells had more surface SLAMF1, but the major cellular pool was still located in the ERC. These observations suggest that ERC-located SLAMF1 may have a yet-undefined function in macrophages.
SLAMF1 is required for TLR4-mediated IFNβ production, but its expression is not regulated by the IFNα/β receptor (IFNAR)
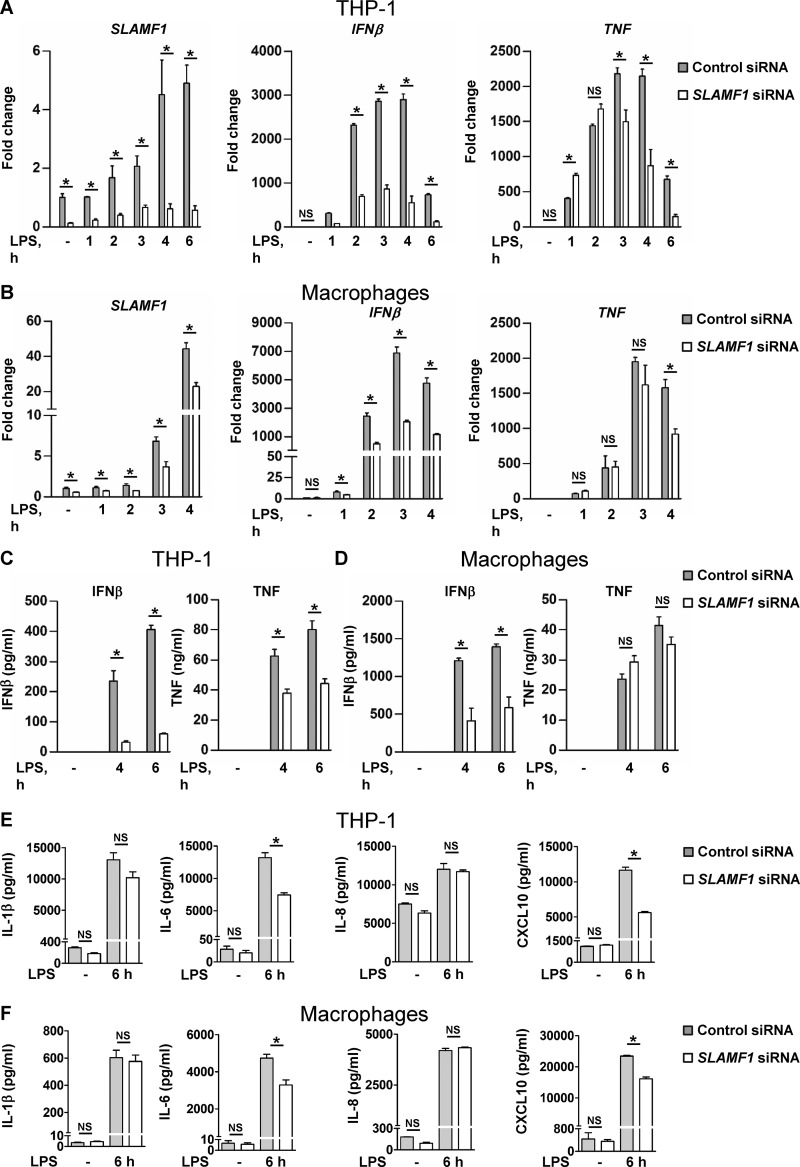
Next, we used siRNAs to target SLAMF1 in THP-1 cells (Fig. 2 A) and human macrophages (Fig. 2 B). We found that SLAMF1 silencing caused consistent reduction in LPS-mediated IFNβ mRNA levels (Fig. 2, A and B) and IFNβ secretion (Fig. 2, C and D). In contrast, TNF mRNA amounts were only reduced at late time points of LPS stimulation, and secretion was affected only in THP-1 cells (Fig. 2, A–D). SLAMF1 silencing impaired both IL-6 and CXCL10 secretion but did not affect the secretion of IL-1β and IL-8 in THP-1 cells and human macrophages (Fig. 2, E and F). The phosphorylation of STAT1 and the initiation of transcription of IFN-inducible genes like CXCL10 are readouts of IFNβ binding to the IFNAR (Toshchakov et al., 2002). IFNAR-dependent STAT1 phosphorylation (Y701) and CXCL10 mRNA expression in response to LPS were both significantly decreased in THP-1 cells pretreated by anti-IFNAR α/β chain 2 mAbs (Fig. S1, B and C). However, SLAMF1 mRNA expression was not altered by blocking IFNAR (Fig. S1 D). Thus, SLAMF1 mRNA expression was not driven by IFNβ-mediated signaling.
Figure 2.
Knockdown of SLAMF1 in macrophages results in strongly reduced TLR4-mediated IFNβ mRNA expression and protein secretion as well as some decrease of TNF, IL-6, and CXCL10 secretion. (A and B) Quantification of SLAMF1, IFNβ, and TNF mRNA expression by qPCR in THP-1 cells (A) and macrophages (B) treated by 100 ng/ml ultrapure K12 LPS. (C and D) IFNβ and TNF secretion levels by THP-1 cells (C) and macrophages (D) in response to LPS (4 and 6 h) assessed by ELISA. (E and F) Secretion levels of IL-1β, IL-6, IL-8, and CXCL-10 (6 h LPS) analyzed by multiplex assays. Data are presented as means with SD for combined data from three independent experiments (A, C, and E), for three biological replicates from one of six donors (B and D), or one of three donors (F). *, P < 0.01.
Upon stimulation with E. coli particles, SLAMF1-silenced THP-1 cells also showed a consistent reduction in IFNβ and TNF mRNA (Fig. S2 A). However, SLAMF1 silencing in macrophages had no effect on IFNβ or TNF mRNA expression in response to polyinosinic-polycytidylic acid (poly I:C) with or without transfection (RIG-I/MDA5 or TLR3) or to the TLR8 ligand CL075 (Fig. S2, B–D).
SLAMF1 regulates TLR4-mediated signaling upstream of TBK1 and IRF3
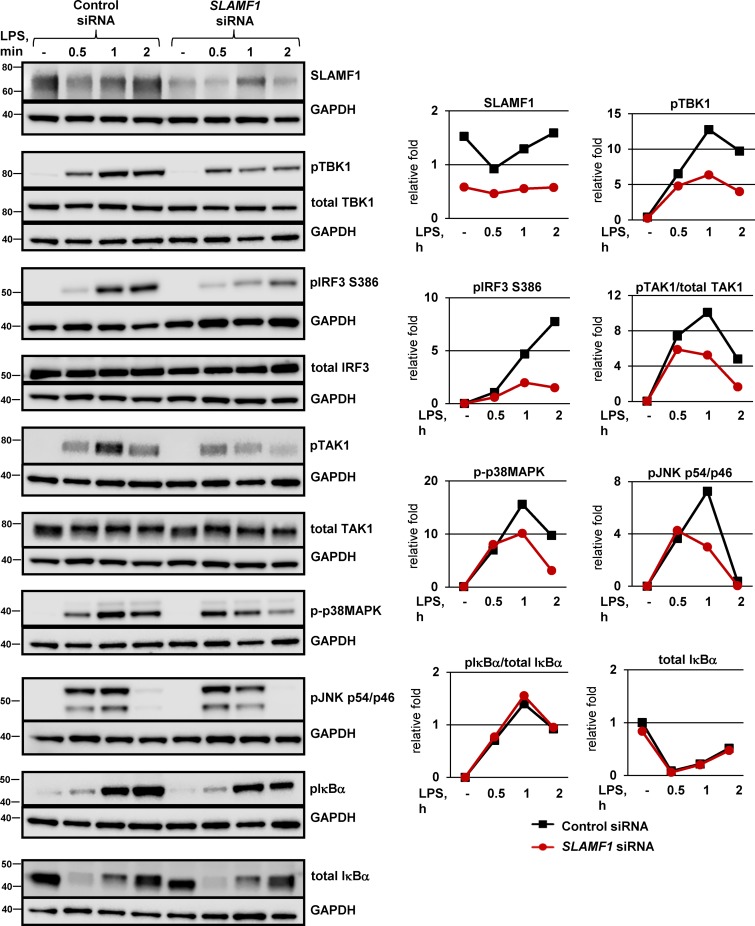
Phosphorylation of IFN regulatory factor 3 (IRF3) transcription factor is critical for the regulation of early IFNβ transcription in macrophages (Sakaguchi et al., 2003). TBK1 acts upstream of IRF3 and phosphorylates IRF3 by itself or together with inhibitor of NF-κB kinase subunit ε (IKKε; Fitzgerald et al., 2003a). Macrophages silenced for SLAMF1 showed decreased levels in both LPS-induced TBK1 and IRF3 phosphorylation (Fig. 3, top). This was also observed in SLAMF1-silenced THP-1 cells stimulated with LPS or E. coli particles (Fig. S3, A and B).
Figure 3.
SLAMF1 silencing in macrophages impairs TLR4-mediated phosphorylation of TBK1, IRF3, and TAK1. Western blotting of lysate macrophages treated with a control nonsilencing oligonucleotide or SLAMF1-specific siRNA oligonucleotides and stimulated with 100 ng/ml LPS. The antibodies used are indicated in the figure. An antibody toward SLAMF1 was used to control for SLAMF1 silencing, and GAPDH was used as an equal loading control. Same GAPDH controls are presented for pTBK1, total TBK1, and phospho-p38MAPK, for total IRF3 and total TAK1, and for pTAK1 and pIκBα because they were probed on the same membranes. Western blots are representative of one of five donors. Molecular weight is given in kilodaltons. Graphs (right) show quantifications of protein levels relative to GAPDH levels obtained with Odyssey software.
Transcription of IFNβ is coordinately regulated by several transcription factor families such as IRFs, NF-κB, and ATF-2–c-Jun (Ford and Thanos, 2010). To explore events upstream of ATF-2–c-Jun activation, we analyzed the effect of SLAMF1 silencing on LPS-mediated activation of MAPKs. SLAMF1 silencing resulted in decreased phosphorylation of MAPK 7 (MAP3K)/TAK1 and downstream MAPKs (p38MAPK and JNK1/2; Figs. 3 and S3 A). Both p38MAPK and JNK1/2 positively regulate the transcriptional activity of AP1 (ATF-2–c-Jun; Chang and Karin, 2001), and it is therefore likely that the observed reduction in MAPK phosphorylation upon SLAMF1 silencing may contribute to decreased AP-1 activity.
The total level and phosphorylation of IκBα protein were not affected by LPS stimulation in the SLAMF1-depleted macrophages (Figs. 3 and S3 A, bottom), suggesting that SLAMF1 is not involved in the early NF-κB activation. This is consistent with our data showing that SLAMF1 silencing affected TNF levels only at late time points (Fig. 2, A–D).
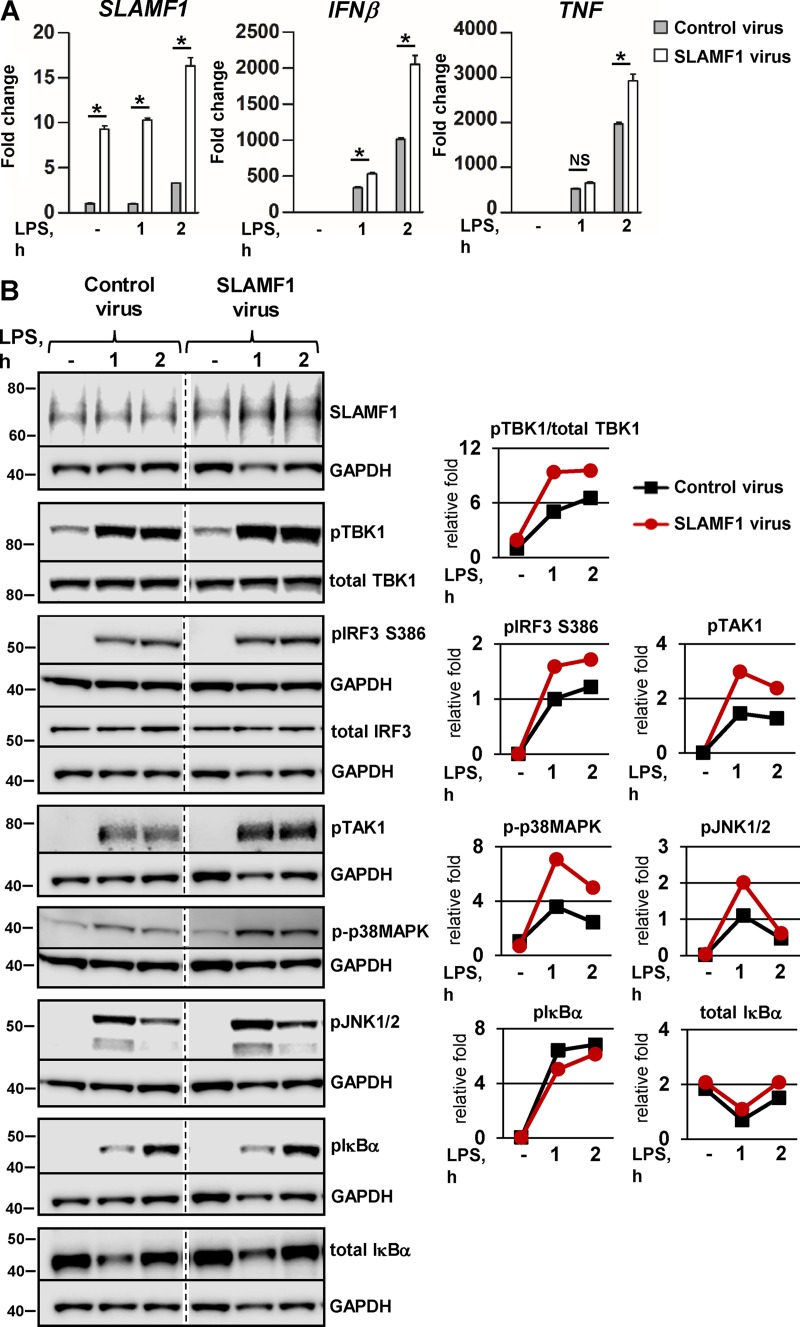
To further support the hypothesis that SLAMF1 regulates signaling from the endosome leading to IFNβ expression, we transduced primary macrophages with lentiviruses encoding SLAMF1. LPS-mediated IFNβ mRNA expression was significantly higher in SLAMF1-transduced cells, with only a modest effect on TNF mRNA expression (Fig. 4 A). Western blot analysis showed that the upregulation of IFNβ mRNA expression in SLAMF1-transduced cells was accompanied by higher amounts of phosphorylated TBK1, IRF3, and MAPK phosphorylation (Fig. 4 B). Thus, these results also suggest that SLAMF1 acts as a positive regulator of endosomal TLR4–TRAM–TRIF signaling.
Figure 4.
Lentiviral transduction of SLAMF1 in macrophages results in the increase of IRF3 and TBK1 phosphorylation in response to LPS and upregulation of IFNβ and TNF expression. (A) Quantification of SLAMF1, IFNβ, and TNF mRNA expression by qPCR in macrophages transduced by Flag-tagged SLAMF1 coding or control virus and treated by LPS. The qPCR data are presented as means and SD for three biological replicates of one of three experiments. Significance was calculated by two-tailed t tests. *, P < 0.01. (B) Western blots showing LPS-induced phosphorylation of signaling molecules in cells transduced with the SLAMF1-expressing virus versus control virus. Dividing lines were added where the time point of 4 h was excised. The same GAPDH controls are presented for total IRF3 and total IκBα and for pIκBα and pTAK1 because they were probed on the same membranes. Molecular weight is given in kilodaltons. Graphs (right) show quantifications of protein levels relative to GAPDH levels obtained with Odyssey software.
SLAMF1 regulates TRAM recruitment to E. coli phagosomes in a Rab11-dependent manner
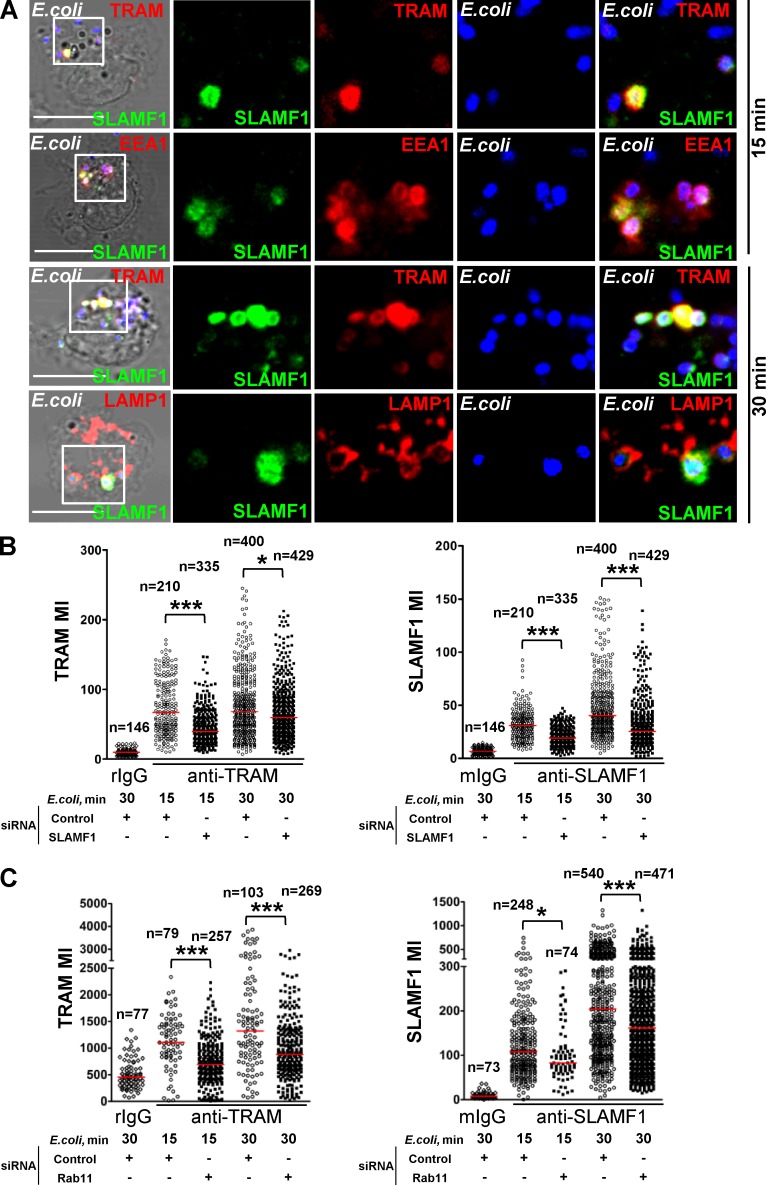
Both TLR4 and TRAM are rapidly recruited to E. coli phagosomes after phagocytosis and are required for induction of IFNβ (Husebye et al., 2010). Because SLAMF1 was needed for TRAM-TRIF signaling, we tested whether SLAMF1 was recruited to E. coli phagosomes containing TRAM. We found that TRAM and SLAMF1 were recruited to early (EEA1-positive) and late (LAMP1-positive) E. coli phagosomes (Fig. 5 A). This was consistent with data published for SLAMF1 in mouse macrophages, where SLAMF1 was found both on EEA1- and LAMP1-positive E. coli phagosomes (Berger et al., 2010). Moreover, we did not detect SLAMF1 on Staphylococcus aureus phagosomes (Fig. S3, A and B), similar to data reported for mouse macrophages (Berger et al., 2010).
Figure 5.
SLAMF1 regulates TRAM recruitment to E. coli phagosomes. (A) SLAMF1 costaining with TRAM, EEA1, or LAMP1 in primary macrophages coincubated with E. coli pHrodo particles for indicated time points. SLAMF1 (green), E. coli (blue), and TRAM, EEA1, or LAMP1 (red) are shown. The data shown are representative of one out of four donors. Bars, 10 μm. (B and C) TRAM and SLAMF1 MIs on E. coli phagosomes upon SLAMF1 silencing (B) or simultaneous Rab11a and Rab11b silencing (C) in primary human macrophages quantified from xyz images. The scatter plots are presented as median values of TRAM voxel intensity, and numbers of phagosomes are shown at the top. The nonparametric Mann-Whitney test was used to evaluate statistical significance. *, P < 0.01; ***, P ≤ 0.0001. Human macrophages were incubated with E. coli particles for indicated time points, fixed, and costained for SLAMF1 and TRAM, normal rabbit (rIgG), or mouse IgG (mIgG). The data shown are representative for one out of five (B) or four (C) donors.
We hypothesized that SLAMF1 could be involved in the transport of TRAM to E. coli phagosomes as this is a crucial step for TLR4-dependent IFNβ induction. Control and SLAMF1-silenced macrophages were pulsed with E. coli pHrodo particles for 15 min followed by 15 min chase in particle-free medium. The mean voxel intensities (MIs) for TRAM, SLAMF1, and pHrodo fluorescence on the phagosomes were calculated from z stacks obtained by confocal microscopy using 3D image analysis software (Fig. 5 B). We found that uptake of E. coli particles was not significantly affected by SLAMF1 silencing (Fig. S4 C). However, acidification of the E. coli phagosomes was significantly decreased upon SLAMF1 silencing (Fig. S4 D). Remarkably, we found that TRAM recruitment to E. coli phagosomes was markedly decreased upon SLAMF1 silencing (Fig. 5 B, left). As expected, SLAMF1-silenced cells showed decreased amounts of SLAMF1 on E. coli phagosomes (Fig. 5 B, right). Thus, SLAMF1 seems to positively regulate TRAM recruitment to E. coli phagosomes.
Transport of TRAM to phagosomes is known to be Rab11 dependent (Husebye et al., 2010). Moreover, SLAMF1 was located to the Rab11-positive compartment in resting cells (Fig. 1 C), positively regulated transport of TRAM to phagosomes (Fig. 5 B), and relocalized from the ERC in monocytes upon addition of E. coli or LPS (Fig. S4, E–G). Based on these observations, we tested whether SLAMF1 recruitment to phagosomes was Rab11 dependent. Two members of Rab11 subfamily, Rab11a and Rab11b, were simultaneously silenced in human macrophages. After silencing, macrophages were stimulated with E. coli for 15 and 30 min, and recruitment of TRAM and SLAMF1 to the phagosomes was quantified by evaluating MIs for TRAM and SLAMF1 staining (Fig. 5 C). Rab11 silencing significantly reduced the amounts of SLAMF1 and TRAM at the phagosomes (Fig. 5 C).
SLAMF1 interacts with the N-terminal part of the TRAM TIR domain
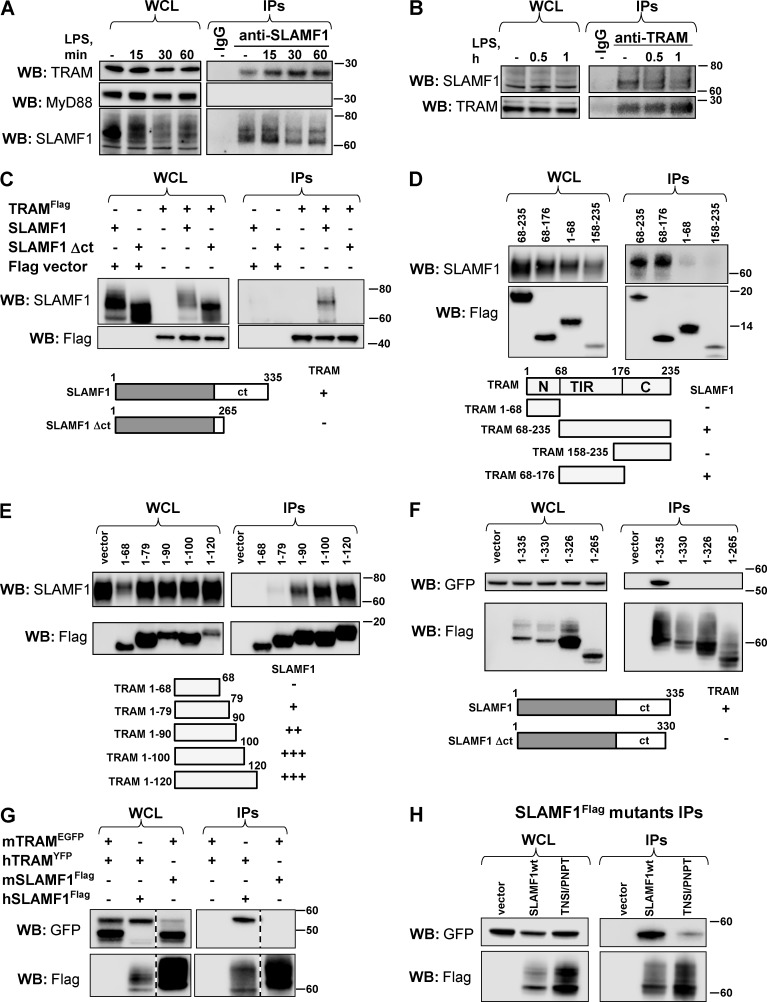
To investigate whether TRAM recruitment to E. coli phagosomes could be regulated by a physical interaction between SLAMF1 and TRAM, we performed endogenous immunoprecipitations (IPs) using anti-SLAMF1 and anti-TRAM antibodies. Endogenous SLAMF1 coprecipitated with TRAM in macrophages, and this interaction was enhanced upon LPS stimulation (Fig. 6 A). In contrast, the TIR-adapter MyD88 did not coprecipitate with SLAMF1 (Fig. 6 A, right), supporting the specificity of the SLAMF1–TRAM interaction. Endogenous TRAM also coprecipitated with SLAMF1 both before and after LPS treatment (Fig. 6 B). The bands detected by anti-TRAM antibody were specific as a similar band could be observed in coimmunoprecipitations (co-IPs) with TLR4 upon LPS stimulation (Fig. S5 A). Overall, we conclude that endogenous SLAMF1 interacts with TRAM in human macrophages and that the interaction is enhanced upon LPS stimulation.
Figure 6.
SLAMF1 interacts with TRAM protein. (A) Endogenous IPs using specific anti-SLAMF1 mAbs from macrophages stimulated by LPS. (B) Endogenous IPs using anti-TRAM polyclonal antibodies from macrophages stimulated by LPS. (C) TRAMFlag-precipitated SLAMF1 and SLAMF1ct was needed for interaction with TRAM. (D) Coprecipitation of TRAM deletion mutants: TIR domain (68–235), short TRAM TIR domain (68–176 aa), and N-terminal (1–68 aa) or C-terminal (158–235 aa) domains with SLAMF1 protein. (E) Coprecipitation of TRAM deletion mutants containing the N-terminal part of TRAM TIR domain with SLAMF1. (F) Coprecipitation of SLAMF1Flag deletion mutants with TRAMYFP. (G) Coprecipitation of human SLAMF1Flag with human TRAMYFP and of mouse SLAMF1Flag with mouse TRAMEGFP. Black dashed lines indicate that intervening lanes have been spliced out. (H) Human SLAMF1 cytoplasmic tail coprecipitation with TRAMYFP with or without amino acid substitutions at 321–324. Graphs under C–F summarize the IPs’ results. Indicated constructs were transfected to HEK293T cells, and anti-Flag agarose was used for the IPs. For endogenous IPs, specific SLAMF1 or TRAM antibodies were covalently coupled to beads. At least three independent experiments were carried out for anti-Flag IPs, and five independent experiments were carried out for the endogenous IPs, and one representative experiment is shown for each. Molecular weight is given in kilodaltons. WB, Western blot; WCL, whole-cell lysate.
Furthermore, HEK293T cells were transiently transfected with Flag-tagged TRAM (TRAMFlag) and full-length SLAMF1 or deletion mutant lacking the C terminus of SLAMF1 (SLAMF1Δct). We found that full-length SLAMF1 but not SLAMF1Δct coprecipitated with TRAMFlag, suggesting that the TRAM interaction site was located at the C terminus of SLAMF1 protein (Fig. 6 C). To map the TRAM region responsible for the interaction with SLAMF1, we generated several Flag-tagged TRAM deletion mutants. The mutants contained the N terminus (1–68), C terminus (158–225), TIR domain (68–176), or the TIR domain plus the C terminus (68–235) of TRAM (1–235; UniProtKB, Q86XR7). Of all these mutants, only TRAM 68–235 and TRAM 68–176 coprecipitated with SLAMF1 (Fig. 6 D).
To further define the subdomain of TRAM involved in the TRAM–SLAMF1 interaction, we made a series of TRAM deletion mutants, which contained the N-terminal part of TRAM with 10–20-aa increments. Although TRAM 1–68 mutant did not bind SLAMF1 (Fig. 6, D and E), a weak interaction was found with TRAM 1–79 that increased markedly for TRAM 1–90 and further for TRAM 1–100. Collectively, these results suggest that the SLAMF1 binding site in TRAM is located within the first 30–35 aa of the TRAM TIR domain (68–95 aa; Fig. 6 E).
TRAM interacts with the C-terminal part of SLAMF1, and the interaction occurs for human but not mouse proteins
We used a similar strategy to establish the TRAM-interacting subdomain of SLAMF1 by deleting amino acids from the C-terminal part of SLAMF1. Cells were cotransfected with Flag-tagged SLAMF1 WT or deletion mutants along with TRAMYFP (Fig. 6 F). A deletion mutant (1–330) lacking the last five C-terminal amino acids did not interact with TRAMYFP (Fig. 6 F), pinpointing the TRAM interaction site at the very C terminus of SLAMF1.
There are two tyrosine residues in the cytoplasmic tail of human SLAMF1 in the signaling motifs designated as immunoreceptor tyrosine–based switch motifs (Shlapatska et al., 2001). SLAMF1 tyrosine phosphorylation and interaction with other proteins via immunoreceptor tyrosine–based switch motifs could potentially alter SLAMF1–TRAM interaction. Point mutations Y281F, Y327F, and double mutation Y281/327F did not alter the interaction with TRAM (Fig. S5 B). Endogenous SLAMF1 was tyrosine phosphorylated in resting macrophages and subsequently dephosphorylated within the 45 min of LPS stimulation (Fig. S5, C and D). Furthermore, both nonphosphorylated and tyrosine-phosphorylated (pY) recombinant SLAMF1ct GST fusion proteins effectively pulled out TRAM from lysates of untreated or LPS-treated macrophages (Fig. S5 E). Thus, SLAMF1–TRAM interaction was not altered by tyrosine phosphorylation of SLAMF1.
Regulation of the LPS-induced IFNβ response seems to differ between humans and mice. Human macrophages respond to LPS with at least 10-fold higher IFNβ mRNA expression compared with mouse BMDMs and thioglycolate-elicited peritoneal macrophages (Schroder et al., 2012). Thus, we wanted to check whether SLAMF1 and TRAM interaction was conserved across species, and we tested murine SLAMF1 and TRAM proteins for interaction. Indeed, mouse TRAMEGFP did not coprecipitate with mouse SLAMF1Flag (Fig. 6 G). Interestingly, three amino acids in human SLAMF1ct upstream of potential TRAM-binding site TNSI (321–324; UniProtKB, Q13291) are different from mouse SLAMF1ct, containing PNPT (329–332; UniProtKB, Q9QUM4; Fig. S6 A). Substitution of the TNSI sequence with PNPT in human SLAMF1 abrogated its interaction with TRAMYFP (Fig. 6 H). Thus, these amino acids are crucial for interaction. However, the sequence in human TRAM involved in the interaction with human SLAMF1 is not fully conserved in murine TRAM (Fig. S6 B), which also could explain why murine TRAM and SLAMF1 do not interact.
SLAMF1 has been shown to regulate E. coli phagosome maturation in mouse BMDMs, but it did not modify the response to ultrapure LPS (Berger et al., 2010). However, regulation of mRNA expression or secretion of type I IFNs by SLAMF1 have not been previously addressed in BMDMs. We stimulated C57BL/6 Slamf1−/− and control BMDMs with 100 ng/ml of ultrapure LPS or E. coli particles and tested for Ifnβ and Tnf mRNA expression and cytokine secretion (Fig. S6, C–E). Indeed, Slamf1−/− BMDMs showed comparable Ifnβ and Tnf mRNA levels to control BMDMs (Fig. S5 C), and the amounts of IFNβ and TNF secreted from Slamf1−/− BMDMs stimulated with LPS or E. coli were not significantly altered (Fig. S6, D and E). Thus, mouse SLAMF1 does not interact with TRAM protein. Therefore, mouse SLAMF1 did not affect TRAM-TRIF–mediated IFNβ secretion in murine macrophages.
Rab11 interacts with SLAMF1 via class I FIPs
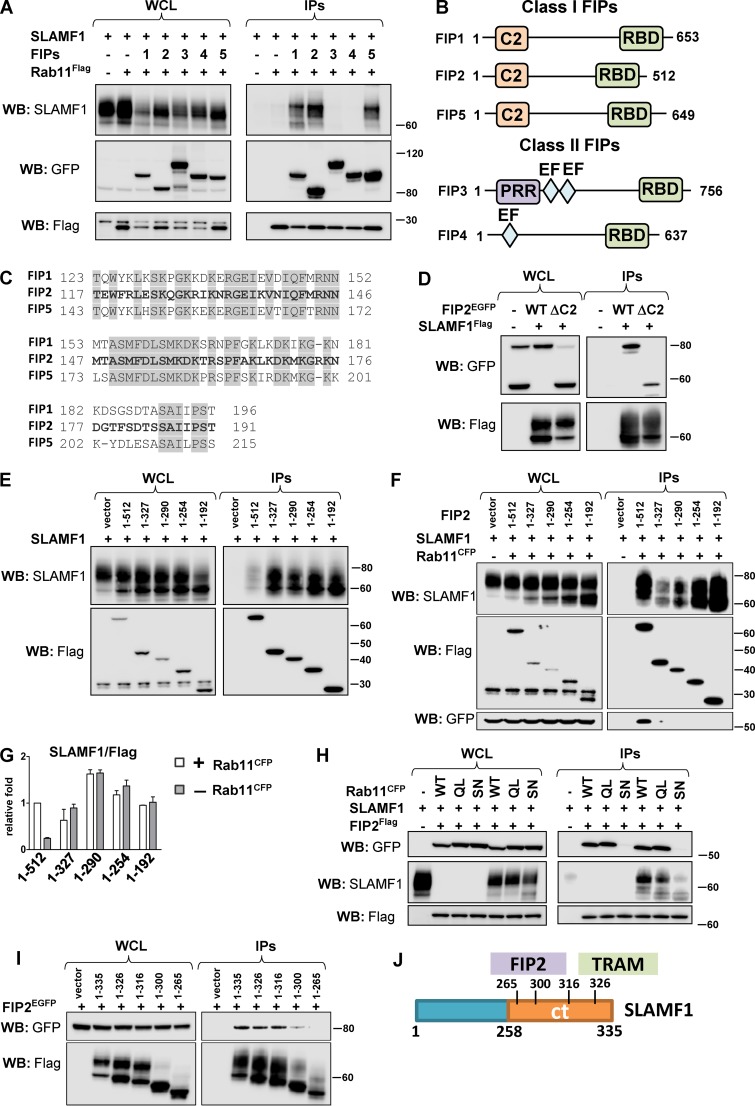
As SLAMF1 recruitment to E. coli phagosomes was found to be Rab11 dependent (Fig. 5 C), we investigated whether SLAMF1 could form a complex with Rab11 via effector proteins such as Rab11 FIPs (Horgan and McCaffrey, 2009). Individual FIPs (FIP1–5) were coexpressed together with SLAMF1 and Rab11aFlag proteins in HEK cells followed by co-IP with Rab11aFlag (Fig. 7 A). All three members of class I FIPs were found to form a complex with SLAMF1 and Rab11a (Fig. 7 A). All class I FIPs are characterized by a phospholipid-binding C2 domain (Fig. 7 B), which is located between aa 1–129 in FIP2 (Lindsay and McCaffrey, 2004). We found that the ΔC2 mutant of FIP2 (lacking aa 1–128) could still bind SLAMF1 (Fig. 7 D). Protein sequence alignment between the class I FIPs showed a highly conserved domain between aa 117–191 in FIP2 with undefined function (Fig. 7, B and C). To figure out whether this domain in FIP2 could be responsible for interaction with SLAMF1, several Flag-tagged FIP2 deletion mutants were tested with or without Rab11CFP overexpression. The 1–192-aa deletion mutant was the minimal deletion mutant found to interact with SLAMF1 (Fig. 7, D–F). Both this mutant and the ΔC2 mutant contains a common 62-aa motif that could be important for interaction with SLAMF1 (Fig. 7, D–F).
Figure 7.
SLAMF1 interacts with all class I Rab11 FIPs. (A) Anti-Flag IPs for Rab11aFlag with EGFP-tagged Rab11FIPs (1–5) and SLAMF1. (B) Schematic figure for class I and class II Rab11 FIPs domain structure. C2, phospholipid-binding C2 domain; EF, EF-hand domain; PRR, proline-rich region; RBD, Rab11 binding domain. (C) Homologous protein sequence in class I FIPs, which follow the C2 domain. Identical amino acids in all three class I FIPs are highlighted. (D) Coprecipitation of SLAMF1Flag with FIP2EGFP WT or FIP2 deletion mutant lacking the C2 domain (ΔC2). (E and F) Coprecipitation of untagged SLAMF1 with FIP2Flag (1–512 aa) and Flag-tagged FIP2 deletion mutants in anti-Flag IPs in the absence (E) or presence (F) of overexpressed Rab11CFP. (G) Quantification of coprecipitations in E and F between SLAMF1 and FIP2Flag variants correlated with the amount of Flag-tagged protein on the blot and Flag-tagged protein sizes. Error bars represent means ± SD for three independent experiments. (H) Coprecipitation of FIP2Flag with SLAMF1 and Rab11a WT, Rab11a Q70L mutant (QL), or Rab11a S25N mutant (SN). (I) Coprecipitation of SLAMF1Flag deletion mutants with FIP2EGFP. Molecular weight is given in kilodaltons. WB, Western blot; WCL, whole-cell lysate. (J) Scheme for FIP2- and TRAM-interacting domains in SLAMF1ct. The results are representative of at least three independent experiments.
The tested deletion mutants more efficiently precipitated SLAMF1 than the full-length FIP2 (Fig. 7 E), but coexpression of Rab11 with full-length FIP2 and SLAMF1 markedly increased its binding to SLAMF1 (Fig. 7, F and G). All FIP2 deletion mutants in IPs lacked a C-terminal Rab11 binding domain (Fig. 7, B, E, and F) and showed better coprecipitation with SLAMF1 without Rab11 overexpression, which suggested that Rab11 has a critical role in controlling FIP2–SLAMF1 interactions.
Next, we examined whether FIPs could coprecipitate SLAMF1 efficiently only in the presence of GTP-bound active Rab11. Rab11 GTPase functions as a molecular switch, being active in the GTP-bound state and inactive in the GDP-bound state. It has been shown that FIPs only interact with activated Rab11 (Junutula et al., 2004; Gidon et al., 2012). FIP2Flag was precipitated from cells, which coexpressed SLAMF1 with either Rab11a WT, GTP-bound Rab11Q70L, or GDP-bound Rab11a S25N (Fig. 7 G). FIP2Flag coprecipitated with SLAMF1 only in the presence of Rab11 WT and Rab11Q70L but not Rab11 serine mutant (Fig. 7 G). We also found that the interaction domain for FIP2 in SLAMF1ct was distinctly different from the TRAM interaction domain (Figs. 6 F and 7, I and J). Our results suggest that TLR4-induced activation of Rab11 is a signal for the recruitment of SLAMF1 and TRAM via the FIPs to E. coli phagosomes and that class I FIPs may link the SLAMF1–TRAM complex to Rab11.
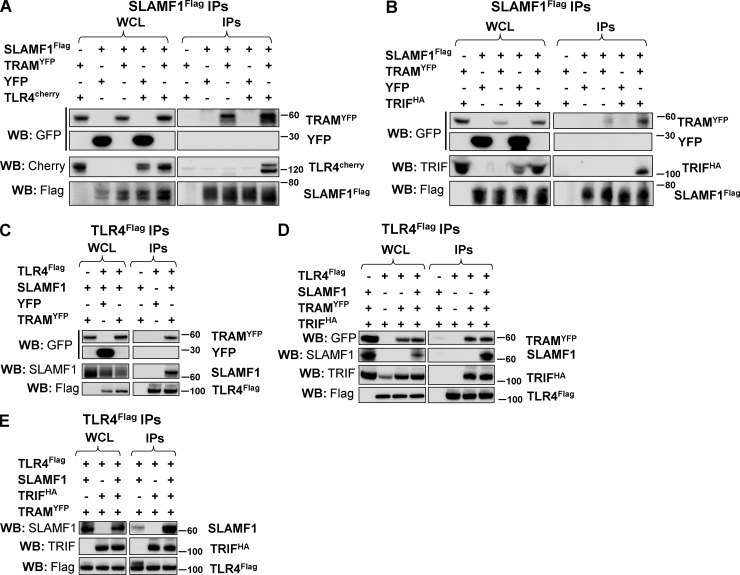
SLAMF1 is recruited to the TLR4–TRAM–TRIF complex
We defined the SLAMF1-interacting site in TRAM as the N-terminal part of TRAM TIR domain (68–95 aa; Fig. 6 C). This raised an important question of whether SLAMF1 could regulate the subsequent formation of TLR4–TRAM–TRIF complex needed for LPS-mediated signaling. To address this question, HEK cells were cotransfected by SLAMF1Flag, TRAMYFP, and TLR4Cherry or TRIFHA, and their interactions were monitored by co-IP with SLAMF1Flag. Both TLR4Cherry and TRIFHA coprecipitated with SLAMF1Flag in the presence of TRAMYFP (Fig. 8, A and B). In addition, SLAMF1 did not coprecipitate with TLR4Flag in the absence of TRAMYFP (Fig. 8 C). Overexpression of SLAMF1 did not alter the ability of TLR4 to attract TRIF via TRAM despite that SLAMF1 also coprecipitated with the complex in the presence of TRAM and TRIF (Fig. 8 D). Furthermore, TRIF overexpression strongly enhanced SLAMF1 coprecipitation with TLR4Flag (Fig. 8 E). In summary, SLAMF1 binding to TRAM seems to be unique and outside of the TIR-TIR dimerization domain as it does not interfere with TRAM–TLR4 interaction and subsequent TRIF recruitment.
Figure 8.
TRAM acts as a bridge between the SLAMF1 and TLR4 signaling complex. (A and B) Coprecipitations of SLAMF1Flag with TLR4Cherry (A) or TRIFHA (B) with or without TRAMYFP overexpression. (C) Coprecipitation of TLR4Flag with SLAMF1 with or without TRAMYFP overexpression. (D) TLR4Flag interaction with TRAMYFP and TRIFHA with or without SLAMF1 coexpression. (E) Coprecipitation of SLAMF1 with or without TRIFHA in the presence of TRAMYFP by TLR4Flag. Indicated constructs were transfected to HEK293T cells. pDuo-CD14/MD-2 vector was cotransfected to all wells (A and C–E). Anti-Flag agarose was used for IPs. At least three independent experiments were performed. Molecular weight is given in kilodaltons. WB, Western blot; WCL, whole-cell lysate.
TRAM and SLAMF1 positively regulate bacterial killing by macrophages
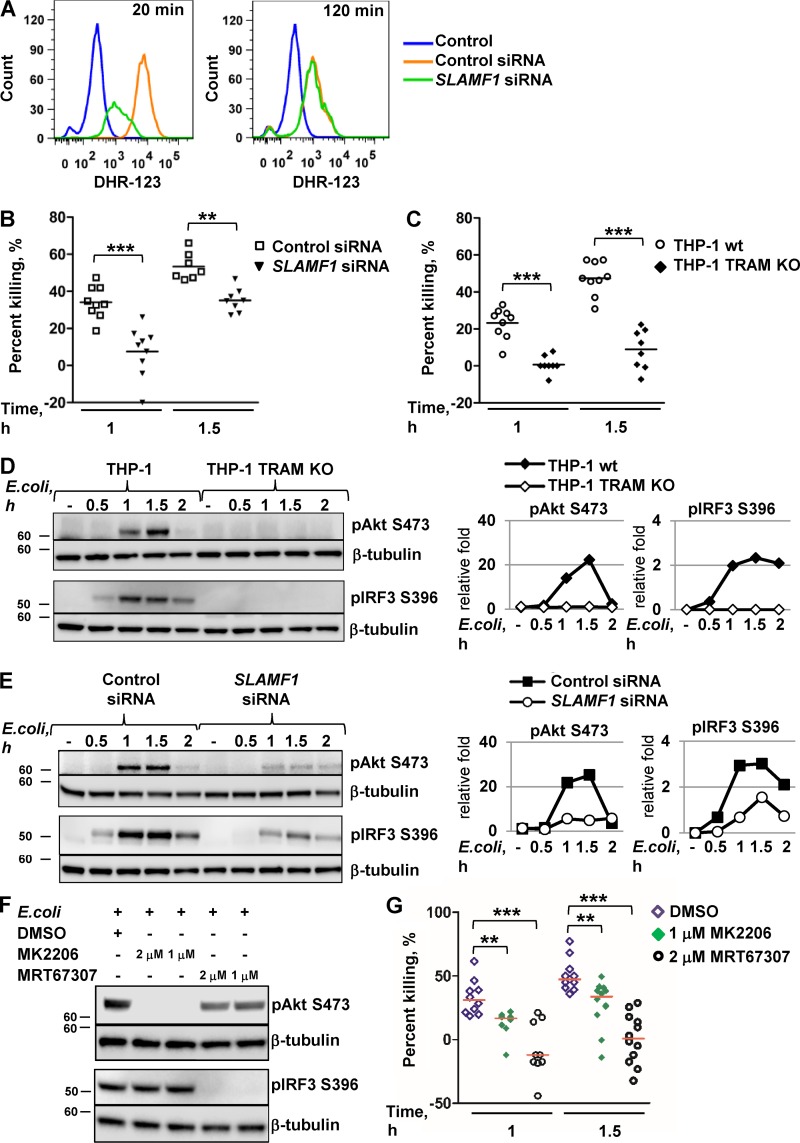
SLAMF1 controls killing of Gram-negative bacteria by mouse BMDMs through generation of ROS (Berger et al., 2010). We tested E. coli–mediated ROS generation by SLAMF1-silenced human macrophages and found that SLAMF1 also acts as a positive regulator of ROS generation in human cells (Fig. 9 A).
Figure 9.
TRAM and SLAMF1 are essential for the killing of E. coli by human macrophages. (A) Flow cytometry analysis of dihydrorhodamine 123 (DHR-123) fluorescence to access ROS activation in control siRNA or SLAMF1 siRNA human macrophages upon stimulation by E. coli red pHrodo particles. One of three experiments shown. (B and C) Bacterial killing assays by SLAMF1-silenced and control THP-1 cells (B) as well as TRAM KO and control THP-1 cells (C) infected with a DH5α strain at MOI 40. (D and E) Western blot analysis of pAkt (S473) and pIRF3 (S396) levels induced by E. coli particles in THP-1 WT and TRAM KO cells (D) as well as SLAMF1-silenced or control oligonucleotide–treated cells (E). Graphs (right) on Western blotting show quantification of protein levels relative to β-tubulin obtained with Odyssey software. (F) Western blot showing phospho-(S396) IRF3 and phospho-(S473) Akt levels in lysates of THP-1 cells coincubated with E. coli particles for 1 h in the presence or absence of TBK1-IKKε inhibitor (MRT67307), pan-Akt allosteric inhibitor (MK2206), or DMSO. Molecular weight is given in kilodaltons. (G) Bacterial killing assays by THP-1 cells with DMSO (<0.01%), 1 μM Akt inhibitor MK2206, or 2 μM TBK1-IKKε inhibitor MRT67307 upon infection by DH5α at MOI 40. Percent killing was calculated as 100 − (number of CFUs at time X/number of CFUs at time 0) × 100 for average values of technical replicates, and each dot on the graphs in B, C, and G represents a biological replicate from three independent experiments. Median values are shown by lines. Statistical significance was calculated by a Mann-Whitney nonparametric test. **, P < 0.01; ***, P < 0.001.
Slamf1−/− BMDMs demonstrated reduced bacterial killing at 6 h of E. coli infection (Berger et al., 2010). To test the effect of SLAMF1 on bacterial killing by human cells, SLAMF1-silenced THP-1 or TRAM knockout (KO) cells with respective control cells were incubated with live DH5α E. coli. Bacterial killing was strongly decreased already at early time points (1 h and 1.5 h) in SLAMF1-silenced cells (Fig. 9 B) and was almost completely abolished in TRAM KO cells (Fig. 9 C). Negative values of the percentage of killing in TRAM KO cells pointed to intracellular bacterial replication in these cells (Fig. 9 C).
TRIF-dependent signaling activates TBK1-IKKε kinases that regulate the integrity of pathogen-containing vacuoles and restrict bacterial proliferation in the cytosol (Radtke et al., 2007; Thurston et al., 2016). TRAM is a crucial adapter for TRIF recruitment to activated TLR4, leading to the activation of TBK1 and IKKε (Oshiumi et al., 2003; Yamamoto et al., 2003; Fitzgerald et al., 2004). Upon TLR4 ligation, TBK1 and IKKε phosphorylate Akt kinase (S473), resulting in Akt activation (Krawczyk et al., 2010; Everts et al., 2014). In turn, the Akt-mTORC1 signaling axis can drive phagocytosis, phagosome maturation, and ROS production, which are essential for bacterial killing (Kelly and O’Neill, 2015). As expected, TRAM KO cells had no detectable IRF3 phosphorylation in response to E. coli particles (Fig. 9 D). In control cells, E. coli–mediated Akt S473 phosphorylation underwent the similar kinetics as IRF3 phosphorylation and was completely abolished in TRAM KO cells (Fig. 9 D) and strongly decreased in SLAMF1-depleted cells (Fig. 9 E). The TBK1-IKKε inhibitor MRT67307 decreased TLR4-mediated Akt phosphorylation in THP-1 cells, whereas the Akt inhibitor MK2206 completely abrogated Akt phosphorylation (Fig. 9 F). Both compounds inhibited bacterial killing in THP-1 cells, especially at the earliest time point (Fig. 9 G). Moreover, coincubation with MRT67307 resulted in the increase of intracellular bacterial number as could be seen by negative values in the percentage of bacterial killing (Fig. 9 G). Hence, TBK1-IKKε activity and subsequent E. coli–mediated Akt phosphorylation directly correlated with the ability of cells to kill bacteria and restrict intracellular replication.
Activation of PI3K and subsequent Akt phosphorylation downstream of TLR2 and TLR4 has been extensively explored in many model systems (Laird et al., 2009; Troutman et al., 2012). It was previously reported that TLR2- and TLR4-mediated Akt S473 phosphorylation is MyD88 dependent in murine model systems (Laird et al., 2009). Our data on MyD88 silencing in primary human macrophages showed that E. coli–induced Akt S473 phosphorylation was not dependent on MyD88 but was dependent on TRAM (Fig. S7 A). In human macrophages, the kinetics of Akt phosphorylation induced by E. coli particles were much faster and robust than those induced by the TLR2 ligand FSL-1 or the TLR4 ligand LPS (Fig. S7 B). TLR2 and TLR4 ligands were not inducing pAkt much over the background level in THP-1 cells, which were used for bacterial killing assays, and pAkt levels were only modestly affected by SLAMF1 or TRAM silencing (Fig. S7 C). In contrast, THP-1 cells stimulated with E. coli particles showed a 15–20-fold increase in pAkt that were almost lost in cells depleted for TRAM or SLAMF1 (Fig. 9, D and E). Thus, TRAM and SLAMF1 are involved in regulation of E. coli–mediated but not pure TLR ligand–mediated Akt phosphorylation in THP-1 cells.
Discussion
Despite that SLAMF1 has been reported to control inflammatory responses and defense against Gram-negative bacteria in mice, the underlying mechanisms are elusive (Theil et al., 2005; van Driel et al., 2012, 2016). Moreover, little has been shown about the role of SLAMF1 in modulating the inflammatory response against Gram-negative bacteria in human macrophages. In this study, we show for the first time that human SLAMF1 regulates TLR4-mediated TRAM-TRIF–dependent signaling by the unique interaction with the signaling adapter TRAM.
Mouse BMDMs express high levels of SLAMF1 on the plasma membrane, whereas resting human monocytes and human monocyte–derived macrophages have been considered to be SLAMF1 negative (Farina et al., 2004; Romero et al., 2004). In contrast, we found that human monocytes and macrophages largely expressed SLAMF1 in the intracellular Rab11+ ERC compartment; however, they had weak or no expression on the cell surface. After stimulation by E. coli, SLAMF1 relocalized from ERCs to E. coli– or LPS-containing phagosomes that resembled the previously reported Rab11a-dependent transport of TLR4 and TRAM from ERCs to E. coli phagosomes (Husebye et al., 2010). Endogenous SLAMF1 was already bound to TRAM before stimulation, and upon E. coli phagocytosis, both proteins were recruited to phagosomes by Rab11 GTPases, with class I Rab11 FIPs as effector molecules. It is known that Rab11 functions as a molecular switch, which cycles between two conformational states: a GTP-bound “active” form and a GDP-bound “inactive” form (Guichard et al., 2014). Surface TLR4 interaction with LPS on the E. coli outer membrane induces fast intracellular complex formation, resulting in multiple posttranslational modifications of signaling molecules (Mogensen, 2009). We hypothesize that TLR4 signaling results in a shift from a Rab11 GDP-bound to a Rab11 GTP-bound active state. Moreover, previous research has demonstrated that FIPs prefer binding to GTP-bound Rab11 (Junutula et al., 2004). Thus, after the Rab11 GDP/GTP ratio shifts to the GTP-bound state, FIPs would connect cargo to Rab11 vesicles, which would enhance delivery of SLAMF1 and TRAM via class I FIPs from ERCs to E. coli phagosomes.
Mouse SLAMF1 was shown to be a bacterial sensor by itself, recognizing porins in the outer bacterial membrane (Berger et al., 2010). The regulatory role of mouse SLAMF1 upon TLR4 ligation by LPS is directly dependent on the porins present in crude LPS preparations or porins in the bacterial outer membrane (Berger et al., 2010). Unlike its mouse orthologue, human SLAMF1 did not require interaction with bacterial porins to elicit its effects on TLR4-mediated IFNβ production as similar data were obtained with both E. coli bioparticles and ultrapure LPS.
The delivery of TRAM to endosomes and phagosomes is crucial for the activation of the IRF3 signaling pathway and IFNβ induction (Kagan et al., 2008; Husebye et al., 2010). We found that SLAMF1 silencing caused a significant decrease in TRAM accumulation around E. coli phagosomes. Moreover, endogenous TRAM coimmunoprecipitated with SLAMF1. These data demonstrate that SLAMF1 is a critical regulator of TRAM recruitment to the phagosomes. We were able to map the domain in SLAMF1 involved in the interaction with TRAM to 15 C-terminal amino acids as both deletion of five C-terminal amino acids and substitution of amino acids at positions 321–324 abrogated interaction of SLAMF1 protein with TRAM. The interaction domain in TRAM was located outside the BB loop as TIR-TIR dimerization is not affected, and it was mapped to the N-terminal part of TRAM TIR domain between aa 68–95. We found that mouse SLAMF1ct contains different amino acid sequences at positions corresponding with human 321–324 residues. This resulted in the absence of interaction between mouse SLAMF1 and TRAM, and consequently, LPS- or E. coli–induced IFNβ expression was not altered in Slamf1−/− BMDMs when compared with WT cells.
The failure to activate TBK1-IKKε kinase observed upon SLAMF1 silencing may affect the antibacterial functions of TBK1-IKKε (Radtke et al., 2007; Thurston et al., 2016). The Akt kinase, which is activated by TBK1-IKKε upon TLR4 ligation, is directly involved in the TLR4-mediated switch to glycolysis by phosphorylating crucial downstream target proteins (Krawczyk et al., 2010; Kelly and O’Neill, 2015). Moreover, Akt is involved in the activation of NADPH oxidase by the phosphorylating p47 subunit (Chen et al., 2003; Hoyal et al., 2003), which may result in ROS generation needed for bacterial killing (West et al., 2011). We demonstrate that SLAMF1 and TRAM were required for E. coli–mediated Akt phosphorylation via TBK1-IKKε as well as for the efficient bacterial killing. It is known that TLR2 and TLR4 ligands activate Akt S473 in a MyD88-dependent manner (Laird et al., 2009; Troutman et al., 2012). It should be noted that in contrast with these studies, we have used E. coli particles and found that Akt S473 phosphorylation was TRAM and SLAMF1 dependent. Akt phosphorylation induced by pure TLR2 and TLR4 ligands could be more dependent on MyD88, but it was not dependent on SLAMF1 or TRAM in THP-1 macrophages.
There is accumulating evidence on divergent regulation of TLR4 signaling and gene expression in different species (Schroder et al., 2012; Vaure and Liu, 2014). It is known that humans and old-world monkey species are highly sensitive to LPS with physiological changes induced by a dose at nanograms per kilogram, whereas rodents are highly insensitive to LPS with physiological changes only induced by a dose at milligrams per kilogram (Vaure and Liu, 2014). Many therapeutic agents that reduce inflammation and mortality in mouse septic shock models show no clinical benefit for humans (Poli-de-Figueiredo et al., 2008). Human monocyte-derived macrophages express much higher levels of IFNβ mRNA in response to LPS than mouse BMDM or thioglycolate-elicited peritoneal macrophages (Schroder et al., 2012). Our findings support higher LPS-induced secretion of IFNβ by human macrophages compared with BMDMs. Thus, during evolution, human macrophages must have acquired mechanisms to enhance TLR4-mediated IFNβ production in response to LPS or, vice versa, mouse cells developed less sensitive response to bacterial LPS. We suggest that SLAMF1-regulated transport of TRAM to the TLR4 signaling complex on bacterial phagosomes could be one of the features specific for human cells, which amplifies the IFNβ secretion. Thus, human SLAMF1 could potentially be targeted to regulate TLR4-mediated cytokine production in inflammatory conditions.
Materials and methods
Primary cells and cell lines
Use of human monocytes from blood donors was approved by the Regional Committees from Medical and Health Research Ethics at the Norwegian University of Science and Technology. Human monocytes were isolated from buffycoat by adherence as previously described (Husebye et al., 2010). In brief, freshly prepared buffycoat (St. Olavs Hospital) was diluted by 100 ml of PBS and applied on top of Lymphoprep (Axis-Shield) according to the manufacturer’s instructions. PBMCs were collected and washed by HBSS (Sigma-Aldrich) four times with low-speed centrifugation (150–200 g). Cells were counted using Z2 Coulter particle count and size analyzer (Beckman Coulter) on program B, resuspended in RPMI 1640 (Sigma-Aldrich) supplemented with 5% of pooled human serum at a concentration of 8 × 106 per ml, and seeded to six-well (1 ml per well) or 24-well (0.5 ml per well) cell culture dishes. After a 45-min incubation allowing surface adherence of monocytes, the dishes were washed three times by HBSS to remove nonadherent cells. Monocytes were maintained in RPMI 1640 supplemented with 10% pooled human serum (St. Olavs Hospital) and used within 24 h after isolation. Monocyte-derived macrophages were obtained by differentiating cells for 8–10 d in RPMI 1640 with 10% human serum and 25 ng/ml rhM-CSF (216-MC-025; R&D Systems). THP-1 cells (ATCC) were cultured in RMPI 1640 supplemented by 10% heat-inactivated FCS, 100 nM penicillin/streptomycin (Thermo Fisher Scientific), and 5 μM β-mercaptoethanol (Sigma-Aldrich). THP-1 cells were differentiated with 50 ng/ml of PMA (Sigma-Aldrich) for 72 h, followed by 48 h in medium without PMA. HEK293T cells (ATCC) were cultured in DMEM with 10% FCS. For making the TRAM KO THP-1 cell line, LentiCRISPRv2 plasmid (a gift from F. Zhang; 52961; Addgene; Sanjana et al., 2014) was ligated with 5′-CACCGATGACTTTGGTATCAAACC-3′ and 5′-AAACGGTTTGATACCAAAGTCATC-3′ for TRAM. Packaging plasmids pMD2.G and psPAX2 were used for producing lentivirus (provided by D. Trono, École Polytechnique Fédérale de Lausanne, Lausanne, Switzerland; 12260 and 12259; Addgene). HEK293T cells were cotransfected with the packaging and lentiCRISPRv2 plasmids and then washed after 16 h. The lentivirus-containing supernatants were collected after 48 h and used for transduction of THP-1 cells along with 8 µg/ml protamine sulphate. Transduced THP-1 cells were then selected with puromycin (1 μg/ml) for 1 mo and tested for TRAM protein expression by Western blotting. All cell lines were regularly checked for mycoplasma contamination.
Reagents and cell stimulation
pHrodo red E. coli and S. aureus as well as AF488-conjugated E. coli bioparticles were purchased from Thermo Fisher Scientific. Ultrapure 0111:B4, K12 LPS from E. coli, poly I:C, imidazoquinoline compound R848 (Resiquimod), thiazoloquinoline compound CL075, and synthetic diacylated lipoproteins FSL-1 (Pam2CGDPKHPKSF) and Pam3CSK4 (P3C) were from InvivoGen. Ultrapure K12 LPS or 0111:B4 LPS (InvivoGen) were used at concentrations of 100 ng/ml. E. coli bioparticles were reconstituted in 2 ml PBS, and 50 µl/well (1.5 × 107 particles) in 1 ml of media was used for cells in six-well plates (Nunc) or 35-mm glass-bottomed tissue cell dishes (MatTek Corporation), and 15 µl/well (0.45 × 107 particles) in 0.5 ml of media were used for 24-well plates (Nunc). The pan-Akt inhibitor MK2206 (1032350-13-2; Axon Medchem) and the TBK1-IKKε inhibitor MRT67307 (from P. Cohen, University of Dundee, Dundee, Scotland, UK; Clark et al., 2011) were diluted in DMSO at a concentration of 20 mM and stored at −80°C, and working solutions were prepared in cell culture media immediately before use.
Antibodies
The following primary antibodies were used: rabbit anti–TICAM-2/TRAM (GTX112785) from Genetex; rabbit mAb anti–human SLAMF1/SLAMF1 (10837-R008-50) from Sino Biological Inc.; mouse anti-GAPDH (ab9484) and rabbit anti–phospho-IRF3 Ser386 (ab76493) from Abcam; rabbit anti–phospho-Akt Ser473 (D9E; 4060), phospho-IRF3 Ser396 (4D4G; 4947), IkB-α (44D4; 4812), phospho–IkB-α (14D4; 2859), p38MAPK (9212), phospho-p38MAPK (Thr180/Tyr182; D3F9; 4511), TBK1/NAK (D1B4; 3504), phospho-TBK1/NAK (Ser172; D52C2; 5483), phospho-TAK1 (T184/187; 90C7; 4508), TAK1 5206, phospho–stress-activated protein kinase (SAPK)/JNK (Thr183/Tyr185; 81E11; 4668), anti-DYKDDDDK tag (D6W5B)/Flag tag (14793), anti-MyD88 (D80F5; 4283), and phospho-STAT1 (Tyr701; D4A7; 7649) from Cell Signaling Technology; rabbit anti–total IRF3 (FL-425; sc-9082) and proliferating cell nuclear antigen (PCNA; FL-261; sc-7907) were from Santa Cruz Biotechnology, Inc.; Living Colors rabbit anti–full-length GFP polyclonal antibodies (632592) from Takara Bio Inc.; 4G10 platinum antiphosphotyrosine antibody biotin conjugated (16-452) from EMD Millipore; and mouse anti-GST antibodies (SAB4200237) and monoclonal mouse ANTI-FLAG M2 antibodies (F1804-200UG) from Sigma-Aldrich. Secondary antibodies (HRP linked) for Western blotting were swine anti–rabbit (P039901-2) and goat anti–mouse (P044701-2) from Dako/Agilent Technologies. The following antibodies were used for staining and/or IPs: rabbit anti-LAMP1 (ab24170) and GM130 antibody (EP892Y) cis-Golgi marker (ab52649) from Abcam; rabbit anti-EEA1 (H-300; sc-33585), TICAM2/TRAM (H-85), TLR4 (H-80; sc-10741), normal rabbit IgG (sc-2027), and normal mouse IgG (sc-2025) were from Santa Cruz Biotechnology, Inc.; rabbit anti-Rab11, low endotoxin, azide-free (LEAF)-purified mouse IgG1 isotype control (MOPC-21; 400124) and LEAF-purified mouse anti-CD150 (SLAMF1) A12 (7D4; 306310) were from BioLegend; and anti-SLAMF1 IgG1 (IPO-3) was provided by S.P. Sidorenko (Natural Academy of Sciences, Kiev, Ukraine; Sidorenko and Clark, 1993). Secondary antibodies for confocal microscopy were goat anti–mouse IgG (H+L) Alexa Fluor 405 conjugate (A-31553), Alexa Fluor 488 conjugate (A-11001), Alexa Fluor 647 conjugate (A-21235), goat anti–rabbit IgG (H+L) Alexa Fluor 405 conjugate (A-31556), Alexa Fluor 488 conjugate (A-11008), and DNA stain Hoechst 33342 (62249) from Thermo Fisher Scientific.
Imaging and image analysis
Confocal images were captured using either an LSM510 META (ZEISS) equipped with a Plan Apochromat 63× 1.4 NA oil immersion objective (images presented in Fig. 1 A and used for 3D modeling in Fig. 1 B) or a TCS SP8 (Leica Microsystems) equipped with a high-contrast Plan Apochromat 63× 1.40 NA oil CS2 objective. Fluorescence was captured by standard photomultiplier tube detectors (LSM 510 META; ZEISS), stimulated emission depletion hybrid detector, or photomultiplier tube detectors (TCS SP8). Acquisition software for the LSM 510 META was Zen microscope software (2012; ZEISS) and for the TCS SP8 was LAS AF software (4.0.0.11706; Leica Microsystems). Before imaging, cells were fixed with 2% paraformaldehyde in PBS on ice, and then immunostaining was performed as described previously (Husebye et al., 2010). In brief, upon fixation, the cells were permeabilized with PEM buffer (80 mM K-Pipes, pH 6.8, 5 mM EGTA, 1 mM MgCl2, and 0.05% saponin) for 15 min on ice, quenched of free aldehyde groups in 50 mM NH4Cl with 0.05% saponin for 5 min, and blocked in PBS with 20% human serum and 0.05% saponin. The cells were incubated with primary antibody in PBS with 2% human serum and 0.05% saponin overnight at 4°C or for 2 h at RT. Alexa Fluor–labeled secondary antibodies (Invitrogen/Thermo Fisher Scientific) were incubated for 15 min at RT after three washes in PBS with 0.05% saponin. If double staining was made, cells were sequentially stained by primary antibodies, specific secondary Alexa Fluor–conjugated antibodies, second primary antibodies, and then specific secondary Alexa Fluor–conjugated antibodies. Images of stained cells, washed in PBS with 0.05% saponin and left in PBS, were captured at RT. 3D data were captured with identical settings, which were also adjusted to avoid saturation of voxel (3D pixels) intensities. For colocalization analysis, the Coloc 2 plugin with thresholds in Fiji (ImageJ; National Institutes of Health) application was applied (Schindelin et al., 2012). The pHrodo fluorescence was used to spot or surface render the volume of individual phagosomes when E. coli pHrodo, S. aureus pHrodo red, or AF488-conjugated E. coli particles were used. A binary mask was created around bacterial particles (Process/Make Binary function) and used to define the regions for quantification of MIs for TRAM and SLAMF1 voxels in original images and to quantify E. coli pHrodo particles MI when redirected to the original image. E. coli pHrodo MI was evaluated to quantify acidification of E. coli–containing phagosomes in cells treated by control or SLAMF1 siRNA. For analysis of the sum of voxel intensities of SLAMF1 inside Golgi rings, GM130 staining was used to define the region to evaluate SLAMF1 intensities for individual 3D Golgi ring structure. Using ImageJ/Fiji software, 3D Golgi ring structures were selected as a region of interest (ROI) and used as a mask to obtain a numerical value of the relative amount of SLAMF1 as a sum of voxel intensities for SLAMF1 staining in Golgi ring ROIs from original image. ImarisXT software (Bitplane) was used to surface render the imaged GM130-positive structures, giving one surface for each. The values for voxel intensities did not follow a Gaussian distribution, and therefore we used median as a measure of average intensities and the nonparametric Mann-Whitney test to evaluate statistical significance in Prism (5.03; GraphPad Software).
siRNA treatment
Oligonucleotides used for silencing were AllStars negative control siRNA (SI03650318), FlexiTube siRNA Hs_SLAMF1_2 (SI00047250), Hs_MYD88_2 (SI00038297), Hs_TICAM2_2 (SI00130893), Hs_RAB11A_5 (SI00301553), and Hs_RAB11B_6 (SI02662695; QIAGEN). On day 7, cells were transfected by silencing oligonucleotides (20 nM final concentration) using Lipofectamine 3000 (L3000008) from Invitrogen/Thermo Fisher Scientific as suggested by the manufacturer. Cells were stimulated by LPS or E. coli particles 96 h after transfection. For THP-1 cells, cells were seeded in six-well plates (Nunc) 0.4 × 106 per well in antibiotic-free media supplemented by 40 ng/ml of PMA. Transfection of siRNA was performed for 24 h, media was changed to PMA-free for 72 h, and cells were kept for another 48 h before stimulation by LPS or E. coli particles.
Quantitative PCR (qPCR)
Total RNA was isolated form the cells using Qiazol reagent (79306; QIAGEN), and chloroform extraction followed by purification was performed on RNeasy Mini columns with DNase digestion step (QIAGEN). cDNA was prepared with the Maxima first strand cDNA synthesis kit for RT-qPCR (Thermo Fisher Scientific) according to the manufacturer’s protocol. qPCR was performed using the PerfeCTa qPCR FastMix (Quanta Biosciences) in replicates and was cycled in a StepOnePlus real-time PCR cycler. The following TaqMan gene expression assays (Applied Biosystems) were used: IFNβ (Hs01077958_s1), TNF (Hs00174128_m1), SLAMF1 (Hs00900288_m1), TBP (Hs00427620_m1), CXCL10 (Hs01124251_g1), Rab11a (Hs00366449_g1), and Rab11b (Hs00188448_m1) for human cells; and Ifnβ (Mm00439552_s1), Tnf (Mm00443258_m1), and Tbp (Mm01277042_m1) for mouse cells. No–reverse transcription controls were negative. The level of TBP mRNA was used for normalization, and results are presented as relative expression compared with the control untreated sample. Relative expression was calculated using the Pfaffl’s mathematical model (Pfaffl, 2001). Results are presented as means and SD of expression fold change for biological replicates relative to nonstimulated cells. Statistical significance was evaluated with Prism software. Data distribution was assumed to be normal, but this was not formally tested. The difference between the two groups was determined by the two-tailed t test.
Cloning, expression vectors, and DNA transfection
Phusion high-fidelity DNA polymerase and respective Fast Digest enzymes (Thermo Fisher Scientific) were used for cDNA recloning. Plasmids we purified by the Endofree plasmid maxi kit (QIAGEN). Sequencing of plasmids was done at the Eurofins genomics facility. Primers for cloning are listed below. SLAMF1 coding sequence was subcloned from retroviral vector (from A. Taranin, Siberian Branch of the Russian Academy of Sciences, Novosibirsk, Russia) to pcDNA3.1 (Invitrogen), C-terminal DYKDDDDK (Flag tag) vector (Takara Bio Inc.), deletion mutants of SLAMF1 made in pcDNA3.1 vector, or C-terminal DYKDDDDK vector. Human TRIFHA and TRAMYFP from K. Fitzgerald (University of Massachusetts Medical School, Worcester, MA) were used for transfections or as templates for subcloning and making TRAM deletion mutants. Rab11aFlag coding construct was described previously (Klein et al., 2015); Rab11FIP1, Rab11FIP2 ΔC2, Rab11FIP2, Rab11FIP3, Rab11FIP4, and Rab11FIP5 in pEGFPC1 vector were from M. McCaffrey (University College Cork, Cork, Ireland). Rab11aQ70L and Rab11aS25N were PCR amplified from pEGFP-Rab11Q70L and pEGFP-Rab11aS25N (Husebye et al., 2010), respectively. The amplified fragments were inserted into SalI- and BamHI-restricted pECFP-C1 vector. TLR4 was subcloned from a TLR4Cherry construct (Husebye et al., 2010) to a C-terminal DYKDDDDK vector. Mouse SLAMF1 was subcloned to a C-terminal DYKDDDDK vector, and mouse TRAM was subcloned from a GeneScript ORF clone (OMu22478D) to pEGFP-N1 vector (Takara Bio Inc.). pDUO-hMD-2/CD14 (Invivogen) was coexpressed with TLR4 to ensure TLR4 dimer formation. HEK 293T cells in six-well plates were transfected by 0.2–0.4 µg of vector/well using Genejuice transfection reagent (EMD Millipore). Lysates were prepared 48 h after transfection. Primers used for cloning are listed in Table 1.
Table 1. Primers used for cloning of full-size or deletion mutants of SLAMF1, TRAM, TLR4, and Rab11 FIP2.
| Construct | For/Rev | Primer sequence (5′–3′) | Restriction enzyme | PCR product mapped on nucleotide sequence |
|---|---|---|---|---|
| SLAMF1 ( NM_003037.3 ) | ||||
| pcDNA3.1 and pLVX-EF1α-IRES-ZsGreen1 | For | TTCGAATTCTGATGGGATCCCAAGGGGCTCC | EcoRI | 367–1,374 |
| Rev | GCAGCGGCCGCTCAGCTCTCTGGAAGTGTCA | NotI | ||
| C-terminal DYKDDDDK | For | TTAGAATTCATGGATCCCAAGGGGCTCC | EcoRI | 367–1,371 |
| Rev | GGTACCTCGAGAGCTCTCTGGAAGTGTCACAC | XhoI | ||
| pGEX-2TK | For | TATGGATCCCAGTTGAGAAGAAGAGGTAAAACG | BamHI | 1,141–1,374 |
| Rev | ATAGAATTCTCAGCTCTCTGGAAGTGTCAC | EcoRI | ||
| SLAMF1 ( NM_003037.3 ) deletion mutants to C-terminal DYKDDDDK | ||||
| All deletion mutants | For | TTAGAATTCATGGATCCCAAGGGGCTCC | EcoRI | N/a |
| 1–265 aa | Rev | GGTACCTCGAGATTTACCTCTTCTTCTCAACTGTAG | XhoI | 367–1,161 |
| 1–326 aa | Rev | GTACCTCGAGAGACTGTGATGGAATTTGTTTCCTG | XhoI | 367–1,344 |
| 1–330 aa | Rev | GTACCTCGAGACACACTAGCATAGACTGTGATG | XhoI | 367–1,356 |
| SLAMF1 ( NM_003037.3 ) deletion mutant to pcDNA3.1 | ||||
| 1–265 aa | For | TTCGAATTCTGATGGGATCCCAAGGGGCTCC | EcoRI | 367–1,161 |
| Rev | TTAAGCGGCCGCTCATTTACCTCTTCTTCTCAACTG | NotI | ||
| SLAMF1 ( NM_003037.3 ) mutants with amino acid substitutions to C-terminal DYKDDDDK | ||||
| SLAMF1 Y281F | For | TTAGAATTCATGGATCCCAAGGGGCTCC | EcoRI | 367–1,225 |
| Rev | GTTTCTGGACTTGGGCAAAGATCGTAAGGC | |||
| For | GCCTTACGATCTTTGCCCAAGTCCAGAAAC | 1,196–1,371 | ||
| Rev | GGTACCTCGAGATTTACCTCTTCTTCTCAACTGTAG | XhoI | ||
| SLAMF1 Y327F | For | TTAGAATTCATGGATCCCAAGGGGCTCC | EcoRI | 367–1,371 |
| Rev | GGTACCTCGAGAGCTCTCTGGAAGTGTCACACTAGCAAAGACTGTG | XhoI | ||
| SLAMF1 TNSI/PNPT | For | TTAGAATTCATGGATCCCAAGGGGCTCC | EcoRI | 367–1,341 |
| Rev | TGTGGTGGGGTTTGGTTCCTGGACAGACTCTGG | |||
| For | GAACCAAACCCCACCACAGTCTATGCTAGTGTGACACTTC | 1,324–1,371a | ||
| Revb | CTTGTCATCGTCGTCCTTG | |||
| TRAM/TICAM-2 ( NM_021649.7 ) | ||||
| C-terminal DYKDDDDK | For | CATGAATTCATGGGTATCGGGAAGTCTAAA | EcoRI | 443–1,147 |
| Rev | TTAACTCGAGCGGCAATAAATTGTCTTTGTACC | XhoI | ||
| TRAM deletion mutants to C-terminal DYKDDDDK | ||||
| 1–68 aa | For | CATGAATTCATGGGTATCGGGAAGTCTAAA | EcoRI | 443–646 |
| Rev | TTAACTCGAGCCATCTCTTCCACGCTCTGAGC | XhoI | ||
| 1–79 aa | For | CATGAATTCATGGGTATCGGGAAGTCTAAA | EcoRI | 443–679 |
| Rev | TTACCTCGAGAGAGGAACACCTCTTCTTCAGC | XhoI | ||
| 1–90 aa | For | CATGAATTCATGGGTATCGGGAAGTCTAAA | EcoRI | 443–712 |
| Rev | TTACCTCGAGATGTGTCATCTTCTGCATGCAATATC | XhoI | ||
| 1–100 aa | For | CATGAATTCATGGGTATCGGGAAGTCTAAA | EcoRI | 443–742 |
| Rev | TTACCTCGAGATAGCAGATTCTGGACTCTGAGG | XhoI | ||
| 1–120 aa | For | CATGAATTCATGGGTATCGGGAAGTCTAAA | EcoRI | 443–802 |
| Rev | TTACCTCGAGACTGTCTGCCACATGGCATCTC | XhoI | ||
| 68–235 aa | For | CATGAATTCATGTTTGAAGAAGAAGCTGAA | EcoRI | 644–1,147 |
| Rev | TTAACTCGAGCGGCAATAAATTGTCTTTGTACC | XhoI | ||
| 68–176 aa | For | CATGAATTCATGTTTGAAGAAGAAGCTGAA | EcoRI | 644–970 |
| Rev | TTAACTCGAGCCAGGGGCCGCATGGGTATAACAG | XhoI | ||
| 158–235 aa | For | CATGAATTCATGAACTCCGTTAACAGGCAGC | EcoRI | 914–1,147 |
| Rev | TTAACTCGAGCGGCAATAAATTGTCTTTGTACC | XhoI | ||
| TLR4 ( NM_138554.4 ) | ||||
| C-terminal DYKDDDDK | For | CGGTCGACCGAGATCTCATGATGTCTGCCTCGCGCCTGG | N/a | 299–2,815 |
| Rev | CTTGTAGTCGCCGGTACCGATAGATGTTGCTTCCTGCCAATTG | N/a | ||
| Mus Musculus TRAM/Ticam-2 ( NM_173394.3 ) to EGFP-N1 | ||||
| 1–232 aa | For | ACTAAGCTTATGGGTGTTGGGAAGTCTAAAC | HindIII | 477–1,175 |
| Rev | ATATGGATCCCGGGCAATGAACTGTTTCTGCGAC | BamHI | ||
| M. Musculus Slamf1 ( NM_013730.4 ) from pDisplay vector to C-terminal DYKDDDDK | ||||
| 30–343 aa | For | TTGGAATTCGGCTTGGGGATATCCACCATGG | EcoRI | 183–1,127 |
| Rev | TATACCTCGAGAGCTCTCTGGCAGTGTCACACTG | XhoI | ||
| Rab11 FIP2 ( NM_014904.2 ) and deletion mutants to N-terminal DYKDDDDK | ||||
| All constructs | For | GCCCGAATTCGGCTGTCCGAGCAAGCCCAAAAG | EcoRI | |
| 1–512 aa | Rev | ATAGCGGCCGCTCATTAACTGTTAGAGAATTTGCCAGC | NotI | 446–1,980 |
| 1–327 aa | Rev | ATAGCGGCCGCTCATTCGCTGCTTTCTTCAAATGG | NotI | 446–1,429 |
| 1–290 aa | Rev | ATAGCGGCCGCTTACACAATGCTGTCAGGTTGG | NotI | 446–1,310 |
| 1–254 aa | Rev | ATAGCGGCCGCTTATCCGAGAAGATGTGTTTGACC | NotI | 446–1,199 |
| 1–192 aa | Rev | ATAGCGGCCGCTTAGTGAGTACTTGGAATGATTGC | NotI | 446–1,016 |
| Rab11a ( NM_004663.4 ) QL and SN mutants to pECFP-C1 | ||||
| 1–216 aa | For | ATCAGTCGACATGGGCACCCGCGACGAC | SalI | 128–145 |
| Rev | TTAAGGATCCTTATATGTTCTGACAGCACTG | BamHI | 758–774 |
For, forward; QL, Rab11a Q70L mutant; Rev, reverse; SN, Rab11a S25N mutant.
Partially on vector sequence.
On C-terminal DYKDDDDK vector.
IPs
HEK293T cells expressing Flag-tagged proteins or macrophages for endogenous IPs were lysed using 1× lysis buffer (150 mM NaCl, 50 mM Tris-HCl, pH 8.0, 1 mM EDTA, and 1% NP-40) or 1× lysis buffer with high salt (400 mM NaCl, 50 mM Tris-HCl, pH 7.5, 1% Triton X-100, and 5 mM EDTA) for antiphosphotyrosine IPs supplemented with EDTA-free Complete mini protease inhibitor cocktail tablets and PhosSTOP phosphatase inhibitor cocktail (Roche), 50 mM NaF, and 2 mM Na3VO3 (Sigma-Aldrich). IPs were carried out by rotation at 4°C for 2 h of cell lysates with either anti-Flag (M2) agarose (Sigma-Aldrich) or specific antibodies coupled to Dynabeads (M-270 Epoxy; Thermo Fisher Scientific) or phosphotyrosine-biotinylated antibodies on streptavidin beads (Invitrogen). Agarose, Sepharose, or Dynabeads were washed five times by respective lysis buffers and heated for 5 min with 1× NuPAGE LDS sample buffer (Thermo Fisher Scientific) for agarose and Sepharose beads or eluted by elution buffer (from Dynabeads co-IP kit; 14321D; Thermo Fisher Scientific) for Dynabeads before analysis by Western blotting.
Western blotting
Cell lysates other than those used as controls in IPs were prepared by simultaneous extraction of proteins and total RNA using Qiazol reagent as suggested by the manufacturer. Protein pellets were dissolved by heating protein pellets for 10 min at 95°C in buffer containing 4 M urea, 1% SDS (Sigma-Aldrich), and NuPAGE LDS sample buffer (4×; Thermo Fisher Scientific). Otherwise, lysates were made using 1× RIPA lysis buffer (150 mM NaCl, 50 mM Tris-HCl, pH 7.5, 1% Triton X-100, 5 mM EDTA, protease inhibitors, and phosphatase inhibitors). For Western blot analysis, we used precast protein gels NuPAGE, Novex, iBlot transfer stacks, and the iBlot gel transfer device (Thermo Fisher Scientific).
Lentiviral transduction
For making the TRAM KO cell line, LentiCRISPRv2 plasmid (gift from F. Zhang; 52961; Addgene; Sanjana et al., 2014) was ligated with 5′-CACCGATGACTTTGGTATCAAACC-3′ and 5′-AAACGGTTTGATACCAAAGTCATC-3′ for TRAM. The second-generation packaging plasmids pMD2.G and psPAX2 were used for producing lentivirus (provided by D. Trono; 12260 and 12259; Addgene). HEK293T cells were cotransfected with the packaging and lentiCRISPRv2 plasmids and then washed after 16 h. The lentivirus-containing supernatants were collected after 48 h and used for transduction of THP-1 WT cells along with protamine sulphate (8 µg/ml final concentration). Transduced THP-1 cells were selected with puromycin (1 μg/ml) for 1 mo and then tested for TRAM protein expression by Western blotting. Lentivirus construct of SLAMF1 was prepared by cloning full-size SLAMF1 with or without Flag tag to the bicistronic lentiviral expression vector pLVX-EF1α-IRES-ZsGreen1 (Takara Bio Inc.; primers are listed in Table 1). The construct was sequenced and cotransfected with packaging plasmids (psPAX2 and pMD2.G provided by D. Trono; 12260 and 12259; Addgene) to produce pseudoviral particles in HEK293T cells. Supernatants were collected at 48 and 72 h, combined, and concentrated using Lenti-X concentrator (631231; Takara Bio Inc.). Viral particles were titrated in HEK293T cells. Titrated virus particles, which gave 90–100% of transduction efficiencies, were subsequently used for transduction of primary human macrophages (resulting in 30–40% of ZsGreen-positive cells). Macrophages were infected on day 6 of differentiation, media was changed after 24 h, and stimulation by LPS (100 ng/ml) was performed 72 h after transduction. Cell lysates for simultaneous RNA/protein isolation were prepared using Qiazol reagent.
Flow cytometry
Untreated or LPS-stimulated monocyte-derived macrophages were detached using accutase (A6964; Sigma-Aldrich) and stained with a cocktail of antibodies against human CD14 (MΦP9) FITC conjugated from BD and SLAMF1 IgG1 (IPO-3) mAbs labeled by AF647 using the Alexa Fluor 647 protein labeling kit (A20173) for 30 min on ice. Flow cytometry was performed using LSR II (BD) with FACS Diva software (BD). Samples were analyzed with FlowJo software (7.6; TreeStar).
ELISA and multiplex cytokine assay
TNF in supernatants was detected using a human TNF-α DuoSet ELISA (DY210-05; R&D Systems), and IFNβ level was detected using VeriKine-HS human IFNβ serum ELISA kit (41415; PBL Assay Science). Supernatants were also analyzed by multiplex cytokine assay (Bio-Plex; Bio-Rad Laboratories) for IL-1β, IL-6, IL-8, and CXCL-10/IP-10. TNF in BMDM supernatants was detected using mouse TNF-α DuoSet ELISA (DY410-05; R&D Systems), and IFNβ level was assessed using a VeriKine-HS mouse IFNβ serum ELISA kit (42410-1; PBL Assay Science). Results are presented as means and SD for biological replicates for representative donors (primary human macrophages) or at least three independent experiments for model cell line THP-1. Statistical significance was evaluated in Prism software. Data distribution was assumed to be normal, but this was not formally tested. The difference between the two groups was determined by a two-tailed t test.
Blocking IFN receptors by specific antibodies
Differentiated THP-1 cells in six-well plates were incubated for 30 min at 37oC with 2.5 μg/ml anti-IFNAR chain 2 antibodies, clone MMHAR-2 isotype IgG2a (MAB1155; EMD Millipore), or control mAbs LEAF-purified mouse IgG2a κ isotype MOPC-173 (400224; BioLegend). After preincubation with mAbs, cells were stimulated with LPS (100 ng/ml) and lysed using Qiazol reagent for simultaneous extraction of proteins and total RNA.
GST pulldown assays
The GST fusion protein construct of SLAMF1ct (GST-SLAMF1ct) was prepared by cloning SLAMF1ct (corresponding with 259–335 aa of SLAMF1 protein; UniProtKB, Q13291) to pGEX-2TK vector (GE Healthcare) with the help of V. Kashuba (Karolinska Institute, Stockholm, Sweden). The sequenced plasmid was transformed into the BL21 DE3 bacterial strain (New England Biolabs, Inc.) or to the TKX1 strain (Agilent Technologies) for production of tyrosine-phosphorylated GST-SLAMF1ct-antiphosphotyrosine. Expression and purification of GST fusion proteins were performed as described previously (Shlapatska et al., 2001). For protein purification and pulldown assays, we used Glutathione high-capacity magnetic agarose beads (G0924; Sigma-Aldrich). Cell lysates of untreated and LPS-stimulated macrophages were prepared in 1× lysis buffer (0.5% NP-40, 150 mM NaCl, and 50 mM Tris-HCl, pH 8.0). Pulldowns were performed as described previously (Shlapatska et al., 2001).
Mouse BMDM differentiation and stimulation
All protocols on animal work were approved by the Norwegian National Animal Research Authorities and were carried out in accordance with Norwegian and European regulations and guidelines. BMDM cultures were generated from bone marrow aspirates extracted from the femurs of C57BL/6 8–10-wk-old male control mice or from Slamf1−/− C57BL/6 mice (Wang et al., 2004). Cells were cultured in complete RPMI 1640 medium containing 20% of L929-conditioned media produced in-house for 8–10 d in sterile Petri dishes; cells were counted and seeded to 24-well cell culture plates in RPMI 1640 with 10% FCS at concentrations of 0.3 × 106 per well in triplicate, left overnight, and treated the next day in fresh media by 100 ng/ml ultrapure LPS (Invivogen) or 50 µg/well for six-well plates or 20 µg/well for 24-well plates of E. coli particles. Cell lysates for RNA isolation were made using Qiazol reagent.
ROS activation assay
Primary human macrophages (in six-well plates) were treated by siRNA as described in the siRNA treatment section, and treated by E. coli red pHrodo bacterial particles (excitation wavelength, 561 nm) in 0.5 ml RPMI 1640 containing 10% human serum on water bath (for 20 min incubation) or in a CO2 incubator (for 120 min incubation). Freshly dissolved in washing buffer (reagent A) or dihydrorhodamine 123 (DHR-123; excitation wavelength, 488 nm; reagent E), both from the PHAGOBURST kit (Glycotope Biotechnology), was added to the wells (except for control well, to which washing buffer was added) for the last 10 min of incubation. After stimulation, cells were placed on ice, washed by cold PBS, incubated with accutase (Sigma-Aldrich) on ice for 5 min, and scraped using cell scrapers. Cells were washed by flow wash (PBS with 0.5% FCS), fixed by fixation buffer (BD), washed by PBS, and analyzed by flow cytometry using LSR II with FACS Diva software. In FlowJo software, cells were gated for pHrodo-positive cells, and DHR-123 fluorescence was presented on the graphs for this gate.
Bacterial killing assay
THP-1 cells were plated at 2 × 105 cells/well in 24-well plates and differentiated as described in the Primary cells and cell lines section for 5 d. Cells were washed and transferred to serum-free RPMI medium. Live DH5α E. coli were added at an MOI of 40. E. coli were centrifuged onto differentiated THP-1 monolayers at 2,000 rpm for 5 min at 4°C. Plates were warmed to 37°C for 15 min in a water bath. Each well was then washed 3× with ice-cold PBS and incubated with warm 10% FCS RPMI medium containing 100 μg/ml of gentamycin for 30 min at 37°C to remove extracellular bacteria. If inhibitors were used in the assay, either DMSO or inhibitors were added to the media at designated concentrations. Cells were washed again 2× with PBS. This time point (45 min after adding bacteria) was designated as time 0. To measure colony-forming units (CFUs) at the end of incubation time, triplicate wells were washed and lysed in 1 ml sterile water. Plates for time points 1 h and 1.5 h were further incubated at 37°C in a CO2 incubator in medium with 10% FCS without antibiotics and with or without kinase inhibitors. At each time point, triplicate wells were washed 3× with PBS before lysing the cells. Viable counts were determined by plating 10 μl of 10-fold dilutions, 1:100 and 1:1,000, onto Luria-Bertani agar (in triplicates to account for technical pipetting error). CFUs were counted at each time point including time 0. Percent killing was calculated as 100 − (number of CFUs at time X/number of CFUs at time 0) × 100 for average values of technical replicates. Statistical significance was calculated in Prism software for biological replicates using unpaired two-tailed tests.
Online supplemental material
Fig. S1 shows that LPS treatment induces SLAMF1 expression in human cells, resulting in its surface localization, and that the increase in SLAMF1 expression is not dependent on signaling from IFNAR. Fig. S2 shows that SLAMF1 is involved in regulation of E. coli– or LPS-mediated but not TLR3-, TLR8-, or RIG-I/MDA5-mediated IFNβ or TNF mRNA expression. Fig. S3 shows that knockdown of SLAMF1 in THP-1 cells impairs TLR4-mediated phosphorylation of TBK1, IRF3, and TAK1 in response to LPS or E. coli particles. Fig. S4 shows that SLAMF1 relocalizes from ERCs to early and late E. coli phagosomes but not to S. aureus phagosomes and that SLAMF1 is required for E. coli phagosome acidification in human cells. Fig. S5 shows that SLAMF1 interaction with TRAM is independent from SLAMF1 tyrosine phosphorylation. Fig. S6 shows that TLR4-mediated IFNβ and TNF mRNA expression and corresponding cytokine secretion are not altered in Slamf1−/− BMDMs and provides human and murine SLAMF1 and TRAM proteins sequence alignments. Fig. S7 shows that E. coli–mediated Akt phosphorylation in human macrophages is not dependent on MyD88 expression and that TLR2- and TLR4-induced phosphorylation of Akt is weak and not much dependent on SLAMF1 or TRAM expression.
Supplementary Material
Acknowledgments
We thank S. Sidorenko, V. Kashuba, and M. McCaffrey for providing reagents and V. Boyartchuk for expert advice. Confocal imaging was performed at the Cellular and Molecular Imaging Core Facility of the Norwegian University of Science and Technology.
This work was supported by the Research Council of Norway through its Centers of Excellence funding scheme grant 223255/F50 (to T. Espevik), by the Norwegian University of Science and Technology’s Onsager Fellowship (to R.K. Kandasamy), by grants from the Liaison Committee for Education, Research and Innovation in Central Norway (to T. Espevik), and by the Joint Research Committee between St. Olavs Hospital and Faculty of Medicine and Health Science of the Norwegian University of Science and Technology (to H. Husebye).
The authors declare no competing financial interests.
Author contributions: Conceptualization: M. Yurchenko, H. Husebye, and T. Espevik; Methodology: M. Yurchenko, H. Husebye, A. Skjesol, and T. Espevik; Investigation: M. Yurchenko, A. Skjesol, L. Ryan, H. Husebye, G.M. Richard, and N. Wang; Writing, original draft: M. Yurchenko; Writing, review and editing: M. Yurchenko, A. Skjesol, H. Husebye, C. Terhost, and T. Espevik; Supervision: H. Husebye and T. Espevik; Resources: R.K. Kandasamy, C. Terhorst, and N. Wang.
References
- Avota, E., Gulbins E., and Schneider-Schaulies S.. 2011. DC-SIGN mediated sphingomyelinase-activation and ceramide generation is essential for enhancement of viral uptake in dendritic cells. PLoS Pathog. 7:e1001290. 10.1371/journal.ppat.1001290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger, S.B., Romero X., Ma C., Wang G., Faubion W.A., Liao G., Compeer E., Keszei M., Rameh L., Wang N., et al. 2010. SLAM is a microbial sensor that regulates bacterial phagosome functions in macrophages. Nat. Immunol. 11:920–927. 10.1038/ni.1931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang, L., and Karin M.. 2001. Mammalian MAP kinase signalling cascades. Nature. 410:37–40. 10.1038/35065000 [DOI] [PubMed] [Google Scholar]
- Chen, Q., Powell D.W., Rane M.J., Singh S., Butt W., Klein J.B., and McLeish K.R.. 2003. Akt phosphorylates p47phox and mediates respiratory burst activity in human neutrophils. J. Immunol. 170:5302–5308. 10.4049/jimmunol.170.10.5302 [DOI] [PubMed] [Google Scholar]
- Clark, K., Peggie M., Plater L., Sorcek R.J., Young E.R., Madwed J.B., Hough J., McIver E.G., and Cohen P.. 2011. Novel cross-talk within the IKK family controls innate immunity. Biochem. J. 434:93–104. 10.1042/BJ20101701 [DOI] [PubMed] [Google Scholar]
- Cocks, B.G., Chang C.C., Carballido J.M., Yssel H., de Vries J.E., and Aversa G.. 1995. A novel receptor involved in T-cell activation. Nature. 376:260–263. 10.1038/376260a0 [DOI] [PubMed] [Google Scholar]
- Everts, B., Amiel E., Huang S.C., Smith A.M., Chang C.H., Lam W.Y., Redmann V., Freitas T.C., Blagih J., van der Windt G.J., et al. 2014. TLR-driven early glycolytic reprogramming via the kinases TBK1-IKKɛ supports the anabolic demands of dendritic cell activation. Nat. Immunol. 15:323–332. 10.1038/ni.2833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farina, C., Theil D., Semlinger B., Hohlfeld R., and Meinl E.. 2004. Distinct responses of monocytes to Toll-like receptor ligands and inflammatory cytokines. Int. Immunol. 16:799–809. 10.1093/intimm/dxh083 [DOI] [PubMed] [Google Scholar]
- Fitzgerald, K.A., McWhirter S.M., Faia K.L., Rowe D.C., Latz E., Golenbock D.T., Coyle A.J., Liao S.M., and Maniatis T.. 2003a. IKKepsilon and TBK1 are essential components of the IRF3 signaling pathway. Nat. Immunol. 4:491–496. 10.1038/ni921 [DOI] [PubMed] [Google Scholar]
- Fitzgerald, K.A., Rowe D.C., Barnes B.J., Caffrey D.R., Visintin A., Latz E., Monks B., Pitha P.M., and Golenbock D.T.. 2003b. LPS-TLR4 signaling to IRF-3/7 and NF-kappaB involves the toll adapters TRAM and TRIF. J. Exp. Med. 198:1043–1055. 10.1084/jem.20031023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzgerald, K.A., Rowe D.C., and Golenbock D.T.. 2004. Endotoxin recognition and signal transduction by the TLR4/MD2-complex. Microbes Infect. 6:1361–1367. 10.1016/j.micinf.2004.08.015 [DOI] [PubMed] [Google Scholar]
- Ford, E., and Thanos D.. 2010. The transcriptional code of human IFN-beta gene expression. Biochim. Biophys. Acta. 1799:328–336. 10.1016/j.bbagrm.2010.01.010 [DOI] [PubMed] [Google Scholar]
- Gidon, A., Bardin S., Cinquin B., Boulanger J., Waharte F., Heliot L., de la Salle H., Hanau D., Kervrann C., Goud B., and Salamero J.. 2012. A Rab11A/myosin Vb/Rab11-FIP2 complex frames two late recycling steps of langerin from the ERC to the plasma membrane. Traffic. 13:815–833. 10.1111/j.1600-0854.2012.01354.x [DOI] [PubMed] [Google Scholar]
- Guichard, A., Nizet V., and Bier E.. 2014. RAB11-mediated trafficking in host-pathogen interactions. Nat. Rev. Microbiol. 12:624–634. 10.1038/nrmicro3325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horgan, C.P., and McCaffrey M.W.. 2009. The dynamic Rab11-FIPs. Biochem. Soc. Trans. 37:1032–1036. 10.1042/BST0371032 [DOI] [PubMed] [Google Scholar]
- Hoyal, C.R., Gutierrez A., Young B.M., Catz S.D., Lin J.H., Tsichlis P.N., and Babior B.M.. 2003. Modulation of p47PHOX activity by site-specific phosphorylation: Akt-dependent activation of the NADPH oxidase. Proc. Natl. Acad. Sci. USA. 100:5130–5135. 10.1073/pnas.1031526100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Husebye, H., Halaas Ø., Stenmark H., Tunheim G., Sandanger Ø., Bogen B., Brech A., Latz E., and Espevik T.. 2006. Endocytic pathways regulate Toll-like receptor 4 signaling and link innate and adaptive immunity. EMBO J. 25:683–692. 10.1038/sj.emboj.7600991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Husebye, H., Aune M.H., Stenvik J., Samstad E., Skjeldal F., Halaas O., Nilsen N.J., Stenmark H., Latz E., Lien E., et al. 2010. The Rab11a GTPase controls Toll-like receptor 4-induced activation of interferon regulatory factor-3 on phagosomes. Immunity. 33:583–596. 10.1016/j.immuni.2010.09.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Junutula, J.R., Schonteich E., Wilson G.M., Peden A.A., Scheller R.H., and Prekeris R.. 2004. Molecular characterization of Rab11 interactions with members of the family of Rab11-interacting proteins. J. Biol. Chem. 279:33430–33437. 10.1074/jbc.M404633200 [DOI] [PubMed] [Google Scholar]
- Kagan, J.C., Su T., Horng T., Chow A., Akira S., and Medzhitov R.. 2008. TRAM couples endocytosis of Toll-like receptor 4 to the induction of interferon-beta. Nat. Immunol. 9:361–368. 10.1038/ni1569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly, B., and O’Neill L.A.. 2015. Metabolic reprogramming in macrophages and dendritic cells in innate immunity. Cell Res. 25:771–784. 10.1038/cr.2015.68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein, D.C., Skjesol A., Kers-Rebel E.D., Sherstova T., Sporsheim B., Egeberg K.W., Stokke B.T., Espevik T., and Husebye H.. 2015. CD14, TLR4 and TRAM Show Different Trafficking Dynamics During LPS Stimulation. Traffic. 16:677–690. 10.1111/tra.12274 [DOI] [PubMed] [Google Scholar]
- Krawczyk, C.M., Holowka T., Sun J., Blagih J., Amiel E., DeBerardinis R.J., Cross J.R., Jung E., Thompson C.B., Jones R.G., and Pearce E.J.. 2010. Toll-like receptor-induced changes in glycolytic metabolism regulate dendritic cell activation. Blood. 115:4742–4749. 10.1182/blood-2009-10-249540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laird, M.H., Rhee S.H., Perkins D.J., Medvedev A.E., Piao W., Fenton M.J., and Vogel S.N.. 2009. TLR4/MyD88/PI3K interactions regulate TLR4 signaling. J. Leukoc. Biol. 85:966–977. 10.1189/jlb.1208763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsay, A.J., and McCaffrey M.W.. 2004. The C2 domains of the class I Rab11 family of interacting proteins target recycling vesicles to the plasma membrane. J. Cell Sci. 117:4365–4375. 10.1242/jcs.01280 [DOI] [PubMed] [Google Scholar]
- Ma, C., Wang N., Detre C., Wang G., O’Keeffe M., and Terhorst C.. 2012. Receptor signaling lymphocyte-activation molecule family 1 (Slamf1) regulates membrane fusion and NADPH oxidase 2 (NOX2) activity by recruiting a Beclin-1/Vps34/ultraviolet radiation resistance-associated gene (UVRAG) complex. J. Biol. Chem. 287:18359–18365. 10.1074/jbc.M112.367060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makani, S.S., Jen K.Y., and Finn P.W.. 2008. New costimulatory families: signaling lymphocytic activation molecule in adaptive allergic responses. Curr. Mol. Med. 8:359–364. 10.2174/156652408785161005 [DOI] [PubMed] [Google Scholar]
- Mancuso, G., Midiri A., Biondo C., Beninati C., Zummo S., Galbo R., Tomasello F., Gambuzza M., Macrì G., Ruggeri A., et al. 2007. Type I IFN signaling is crucial for host resistance against different species of pathogenic bacteria. J. Immunol. 178:3126–3133. 10.4049/jimmunol.178.5.3126 [DOI] [PubMed] [Google Scholar]
- Mikhalap, S.V., Shlapatska L.M., Berdova A.G., Law C.L., Clark E.A., and Sidorenko S.P.. 1999. CDw150 associates with src-homology 2-containing inositol phosphatase and modulates CD95-mediated apoptosis. J. Immunol. 162:5719–5727. [PubMed] [Google Scholar]
- Mogensen, T.H. 2009. Pathogen recognition and inflammatory signaling in innate immune defenses. Clin. Microbiol. Rev. 22:240–273. 10.1128/CMR.00046-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oshiumi, H., Sasai M., Shida K., Fujita T., Matsumoto M., and Seya T.. 2003. TIR-containing adapter molecule (TICAM)-2, a bridging adapter recruiting to toll-like receptor 4 TICAM-1 that induces interferon-beta. J. Biol. Chem. 278:49751–49762. 10.1074/jbc.M305820200 [DOI] [PubMed] [Google Scholar]
- Pfaffl, M.W. 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29:e45. 10.1093/nar/29.9.e45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poli-de-Figueiredo, L.F., Garrido A.G., Nakagawa N., and Sannomiya P.. 2008. Experimental models of sepsis and their clinical relevance. Shock. 30(Suppl 1):53–59. 10.1097/SHK.0b013e318181a343 [DOI] [PubMed] [Google Scholar]
- Radtke, A.L., Delbridge L.M., Balachandran S., Barber G.N., and O’Riordan M.X.. 2007. TBK1 protects vacuolar integrity during intracellular bacterial infection. PLoS Pathog. 3:e29. 10.1371/journal.ppat.0030029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Réthi, B., Gogolák P., Szatmari I., Veres A., Erdôs E., Nagy L., Rajnavölgyi E., Terhorst C., and Lányi A.. 2006. SLAM/SLAM interactions inhibit CD40-induced production of inflammatory cytokines in monocyte-derived dendritic cells. Blood. 107:2821–2829. 10.1182/blood-2005-06-2265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romanets-Korbut, O., Najakshin A.M., Yurchenko M., Malysheva T.A., Kovalevska L., Shlapatska L.M., Zozulya Y.A., Taranin A.V., Horvat B., and Sidorenko S.P.. 2015. Expression of CD150 in tumors of the central nervous system: identification of a novel isoform. PLoS One. 10:e0118302. 10.1371/journal.pone.0118302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero, X., Benítez D., March S., Vilella R., Miralpeix M., and Engel P.. 2004. Differential expression of SAP and EAT-2-binding leukocyte cell-surface molecules CD84, CD150 (SLAM), CD229 (Ly9) and CD244 (2B4). Tissue Antigens. 64:132–144. 10.1111/j.1399-0039.2004.00247.x [DOI] [PubMed] [Google Scholar]
- Sakaguchi, S., Negishi H., Asagiri M., Nakajima C., Mizutani T., Takaoka A., Honda K., and Taniguchi T.. 2003. Essential role of IRF-3 in lipopolysaccharide-induced interferon-beta gene expression and endotoxin shock. Biochem. Biophys. Res. Commun. 306:860–866. 10.1016/S0006-291X(03)01049-0 [DOI] [PubMed] [Google Scholar]
- Sanjana, N.E., Shalem O., and Zhang F.. 2014. Improved vectors and genome-wide libraries for CRISPR screening. Nat. Methods. 11:783–784. 10.1038/nmeth.3047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin, J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., et al. 2012. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 9:676–682. 10.1038/nmeth.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroder, K., Irvine K.M., Taylor M.S., Bokil N.J., Le Cao K.A., Masterman K.A., Labzin L.I., Semple C.A., Kapetanovic R., Fairbairn L., et al. 2012. Conservation and divergence in Toll-like receptor 4-regulated gene expression in primary human versus mouse macrophages. Proc. Natl. Acad. Sci. USA. 109:E944–E953. 10.1073/pnas.1110156109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shlapatska, L.M., Mikhalap S.V., Berdova A.G., Zelensky O.M., Yun T.J., Nichols K.E., Clark E.A., and Sidorenko S.P.. 2001. CD150 association with either the SH2-containing inositol phosphatase or the SH2-containing protein tyrosine phosphatase is regulated by the adaptor protein SH2D1A. J. Immunol. 166:5480–5487. 10.4049/jimmunol.166.9.5480 [DOI] [PubMed] [Google Scholar]
- Sidorenko, S.P., and Clark E.A.. 1993. Characterization of a cell surface glycoprotein IPO-3, expressed on activated human B and T lymphocytes. J. Immunol. 151:4614–4624. [PubMed] [Google Scholar]
- Theil, D., Farina C., and Meinl E.. 2005. Differential expression of CD150 (SLAM) on monocytes and macrophages in chronic inflammatory contexts: abundant in Crohn’s disease, but not in multiple sclerosis. J. Clin. Pathol. 58:110–111. 10.1136/jcp.2004.019323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thurston, T.L., Boyle K.B., Allen M., Ravenhill B.J., Karpiyevich M., Bloor S., Kaul A., Noad J., Foeglein A., Matthews S.A., et al. 2016. Recruitment of TBK1 to cytosol-invading Salmonella induces WIPI2-dependent antibacterial autophagy. EMBO J. 35:1779–1792. 10.15252/embj.201694491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toshchakov, V., Jones B.W., Perera P.Y., Thomas K., Cody M.J., Zhang S., Williams B.R., Major J., Hamilton T.A., Fenton M.J., and Vogel S.N.. 2002. TLR4, but not TLR2, mediates IFN-beta-induced STAT1alpha/beta-dependent gene expression in macrophages. Nat. Immunol. 3:392–398. 10.1038/ni774 [DOI] [PubMed] [Google Scholar]
- Trinchieri, G. 2010. Type I interferon: friend or foe? J. Exp. Med. 207:2053–2063. 10.1084/jem.20101664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troutman, T.D., Hu W., Fulenchek S., Yamazaki T., Kurosaki T., Bazan J.F., and Pasare C.. 2012. Role for B-cell adapter for PI3K (BCAP) as a signaling adapter linking Toll-like receptors (TLRs) to serine/threonine kinases PI3K/Akt. Proc. Natl. Acad. Sci. USA. 109:273–278. 10.1073/pnas.1118579109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Driel, B., Liao G., Romero X., O’Keeffe M.S., Wang G., Faubion W.A., Berger S.B., Magelky E.M., Manocha M., Azcutia V., et al. 2012. Signaling lymphocyte activation molecule regulates development of colitis in mice. Gastroenterol. 143:1544–1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Driel, B.J., Liao G., Engel P., and Terhorst C.. 2016. Responses to Microbial Challenges by SLAMF Receptors. Front. Immunol. 7:4. 10.3389/fimmu.2016.00004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaure, C., and Liu Y.. 2014. A comparative review of toll-like receptor 4 expression and functionality in different animal species. Front. Immunol. 5:316. 10.3389/fimmu.2014.00316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, N., Satoskar A., Faubion W., Howie D., Okamoto S., Feske S., Gullo C., Clarke K., Sosa M.R., Sharpe A.H., and Terhorst C.. 2004. The cell surface receptor SLAM controls T cell and macrophage functions. J. Exp. Med. 199:1255–1264. 10.1084/jem.20031835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- West, A.P., Brodsky I.E., Rahner C., Woo D.K., Erdjument-Bromage H., Tempst P., Walsh M.C., Choi Y., Shadel G.S., and Ghosh S.. 2011. TLR signalling augments macrophage bactericidal activity through mitochondrial ROS. Nature. 472:476–480. 10.1038/nature09973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto, M., Sato S., Hemmi H., Uematsu S., Hoshino K., Kaisho T., Takeuchi O., Takeda K., and Akira S.. 2003. TRAM is specifically involved in the Toll-like receptor 4-mediated MyD88-independent signaling pathway. Nat. Immunol. 4:1144–1150. 10.1038/ni986 [DOI] [PubMed] [Google Scholar]
- Yamashiro, D.J., Tycko B., Fluss S.R., and Maxfield F.R.. 1984. Segregation of transferrin to a mildly acidic (pH 6.5) para-Golgi compartment in the recycling pathway. Cell. 37:789–800. 10.1016/0092-8674(84)90414-8 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.