The facilitated aggregation of misfolded proteins is a proteostasis strategy important for cell function and viability, but the molecular mechanisms are poorly understood. Grousl et al. reveal how the intrinsically disordered domains of the small heat shock protein Hsp42 promote and control the aggregation of misfolded proteins during stress conditions in yeast.
Abstract
Chaperones with aggregase activity promote and organize the aggregation of misfolded proteins and their deposition at specific intracellular sites. This activity represents a novel cytoprotective strategy of protein quality control systems; however, little is known about its mechanism. In yeast, the small heat shock protein Hsp42 orchestrates the stress-induced sequestration of misfolded proteins into cytosolic aggregates (CytoQ). In this study, we show that Hsp42 harbors a prion-like domain (PrLD) and a canonical intrinsically disordered domain (IDD) that act coordinately to promote and control protein aggregation. Hsp42 PrLD is essential for CytoQ formation and is bifunctional, mediating self-association as well as binding to misfolded proteins. Hsp42 IDD confines chaperone and aggregase activity and affects CytoQ numbers and stability in vivo. Hsp42 PrLD and IDD are both crucial for cellular fitness during heat stress, demonstrating the need for sequestering misfolded proteins in a regulated manner.
Introduction
Cells are continuously endangered by fluctuations in their environment and need to adapt to these extra- and intracellular changes. External stress conditions like heat shock cause accumulation of misfolded proteins that endanger cellular homeostasis. To counteract the impact of stress conditions, cells harbor protein quality control systems that either refold or degrade misfolded proteins as central activities. The regulated, factor-driven aggregation of misfolded proteins has been recently identified as an additional proteostasis strategy (Tyedmers et al., 2010; Chen et al., 2011; Sontag et al., 2017). The molecular mechanism of facilitated protein aggregation, however, remains poorly understood. In Saccharomyces cerevisiae, cytosolic (CytoQ or Q-bodies) and nuclear protein aggregates (intranuclear quality control compartments; INQs) form during proteotoxic stress under control of the compartment-specific aggregases Hsp42 and Btn2, respectively (Specht et al., 2011; Malinovska et al., 2012; Miller et al., 2015). The role of Hsp42 and Btn2 aggregases in promoting protein aggregation is not restricted to protein misfolding conditions. Btn2 promotes formation of noncanonical DNA stress foci during DNA replication stress (Tkach et al., 2012; Gallina et al., 2015; Miller et al., 2015). Hsp42 is crucial for the formation of several different protein aggregates in aging yeast cells (Saarikangas and Barral, 2015) including stationary-phase granules in quiescent yeast (Liu et al., 2012) and granules composed of inactive proteasome subunits (Peters et al., 2015; Marshall et al., 2016).
Hsp42 belongs to the chaperone family of small heat shock proteins (Hsps; sHsps), which are defined by the conserved α-crystallin domain (ACD; Haslbeck et al., 2004a). sHsps act as a first line of defense during unfolding stress and associate first with a broad range of misfolding proteins (Haslbeck and Vierling, 2015; Treweek et al., 2015). They sequester these proteins in near-native conformations and facilitate their subsequent refolding by ATP-dependent Hsp70-Hsp100 disaggregases (Mogk and Bukau, 2017).
sHsps gain functional diversity by N- and C-terminal extensions (NTEs and CTEs, respectively) of variable length and sequence flanking the ACD. Flexible NTEs provide the majority of substrate interaction sites, but exposed sites in ACDs and CTEs also contact substrates (Haslbeck et al., 2004b; Basha et al., 2006; Jaya et al., 2009; Fu et al., 2013; Ungelenk et al., 2016). Hsp42 harbors an unusual long NTE that is essential for inducing the aggregation of misfolded protein species, leading to CytoQ formation (Specht et al., 2011; Marshall et al., 2016). The molecular basis of this activity, however, remains unexplored. Intriguingly, Hsp42 was among the 200 yeast proteins suggested to harbor a putative prion-like domain (PrLD) as part of its NTE (Alberti et al., 2009). Furthermore, the Hsp42 NTE includes a second intrinsically disordered subdomain (IDD).
Proteins harboring IDDs promote or modulate the formation of membraneless compartments of various nature, including processing bodies or stress granules, among others (Gilks et al., 2004; Protter and Parker, 2016). These compartments form by phase separation processes and can play a role in cellular adaptations to stress conditions (Rabouille and Alberti, 2017; Riback et al., 2017). IDDs do not adopt stable secondary and tertiary structures and are dynamic and heterogeneous in conformation. The ability to self-interact and to undergo multiple weak interactions with other components represents the molecular basis of their scaffolding function (March et al., 2016). IDDs typically include regions that exhibit low hydrophobicity, high mean charge, and often low sequence complexity (Malinovska et al., 2013; van der Lee et al., 2014). For example, the IDD of Ddx4, harboring multiple F/RG repeats, is crucial for formation of nuage granules (Nott et al., 2015). A subtype of intrinsically disordered proteins harbors PrLDs, identified by an amino acid composition similar to those causing yeast prion formation (Alberti et al., 2009; March et al., 2016). PrLDs are typically rich in uncharged amino acids (Q, N, Y, S, and G) and devoid of charges. They are frequently present in proteins harboring RNA-binding domains, where they play crucial roles in RNA granule assembly (Gilks et al., 2004; Decker et al., 2007; Reijns et al., 2008; Cherkasov et al., 2015; Molliex et al., 2015; Jain et al., 2016).
In this study, we dissect the role of each type of disordered domain of yeast Hsp42 and elucidate the mechanism by which Hsp42 promotes the formation of cytosolic aggregates of misfolded proteins. We determined the activities of Hsp42 deletion variants lacking either PrLD or IDD in vivo and in vitro. We show the PrLD is essential for CytoQ formation, whereas the IDD controls the chaperone activity of Hsp42 and modulates the numbers and stability of CytoQ aggregates in vivo. The combined activities of both Hsp42 domains are required for fitness of yeast cells during heat stress, underlining the need for tightly regulated sequestration of misfolded proteins.
Results
Protein translation is crucial for the formation of large Hsp42-dependent CytoQ deposits
We aimed to assess the molecular features of Hsp42-mediated sequestration of misfolded proteins. We initially set out to define the substrate species that interact with Hsp42 for CytoQ formation under thermal stress. These species might comprise preexisting proteins that unfold during stress or require storage in a native-like state. Alternatively, they may comprise newly synthesized polypeptides including nascent chains, which misfold before reaching the native state.
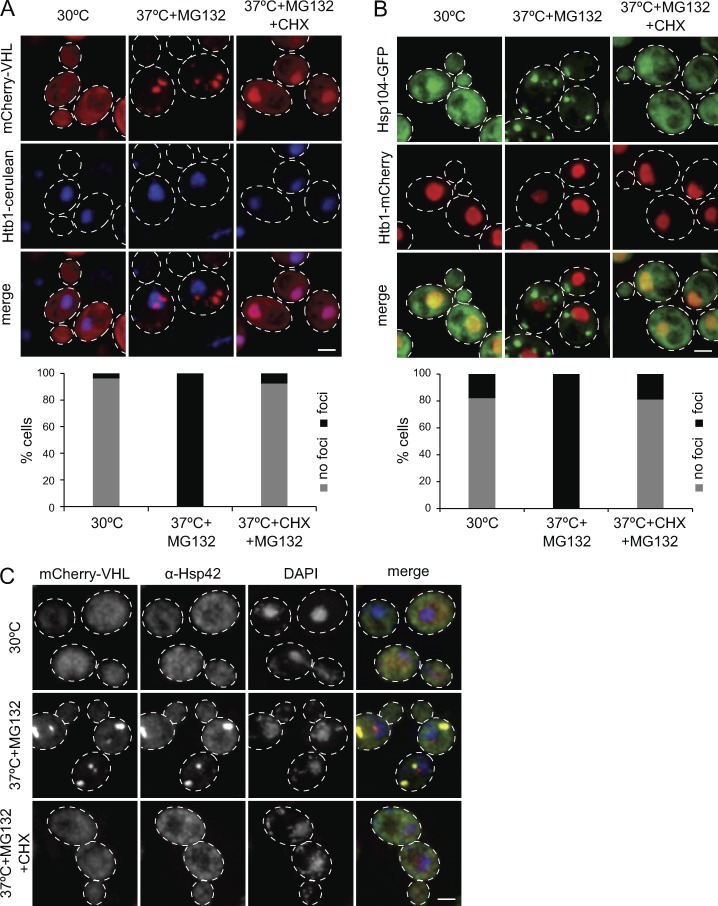
To analyze whether newly synthesized proteins constitute a source of Hsp42 substrates sequestered into microscopically visible foci during thermal stress, we stopped protein synthesis by cycloheximide (CHX) addition before heat shock and furthermore inhibited proteasomal activity by MG132 treatment. As established previously (Specht et al., 2011; Miller et al., 2015), proteasome inhibition increases the stability of forming foci, probably by adding a burden to the proteostasis network including disaggregating Hsp70-Hsp100 chaperones. This experimental design makes the analysis of generated protein aggregates experimentally more accessible without overruling the dependence of protein aggregation on Hsp42. The formation of Hsp42-dependent CytoQ deposits was monitored in three ways. In the first way, we used mCherry-VHL expressed from a galactose-driven promoter as misfolded reporter. In the second way, we used Hsp104-GFP, which labels endogenous protein aggregates induced by proteotoxic stress, including CytoQ foci, by binding to aggregate surfaces (Specht et al., 2011). In the third way, we monitored Hsp42 localization via immunofluorescence as Hsp42 represents an integral part of CytoQs. Translational arrest (+CHX) largely abrogated mCherry-VHL, Hsp104-GFP, and Hsp42 foci formation upon proteotoxic stress (Fig. 1, A–C). mCherry-VHL accumulated in the nucleus of yeast cells upon CHX addition (Fig. 1 A). Nuclear accumulation of mCherry-VHL was also observed in heat-stressed hsp42Δ btn2Δ cells, which lack cellular aggregases and are deficient in facilitated protein aggregation (Miller et al., 2015). This can be explained by shuttling of soluble misfolded proteins between cytosol and the nucleus, with nuclear import or retention being preferred (Park et al., 2013; Miller et al., 2015). This suggests that newly synthesized proteins strongly affect the formation of large CytoQ inclusions traceable by fluorescence microscopy. We noted that preexisting mCherry-VHL, appearing as diffuse fluorescence before heat treatment, also became part of foci. This indicates that the deposits can include both newly synthesized proteins and preexisting misfolded proteins (Fig. 1 A). To test whether sequestration of preexisting mCherry-VHL requires ongoing protein synthesis in general or specifically of the reporter, we first stopped reporter expression and synthesis by shifting yeast cells from galactose to glucose before applying a stress condition (37°C + MG132) in the absence and presence of CHX (Fig. S1 A). Sequestration of the preexisting mCherry-VHL pool was still observed and relied on ongoing protein synthesis as it was abrogated by CHX addition. Collectively, this suggests that newly synthesized and nascent polypeptides and/or the active translation machinery as a whole are particularly vulnerable to proteotoxic stress and might act as a seed, promoting aggregation of preexisting misfolded proteins. Identical findings were observed when only applying a heat shock (38°C) without MG132 addition, demonstrating that translation is crucial for Hsp42-dependent sequestration of misfolded proteins into large CytoQ inclusions irrespective of proteasome inhibition (Fig. S1, B and C).
Figure 1.
The formation of large protein aggregates depends on translation. (A–C) Localization patterns of mCherry-VHL (A), Hsp104-GFP (B), and Hsp42 (C) in S. cerevisiae cells and their superposition with the nuclear marker Htb1-cerulean (A and B) or DNA stained by DAPI (C) upon control conditions (30°C), and proteotoxic stress in the absence or presence of CHX (37°C for 30 min + MG132 ± CHX) are shown. Maximal projections of widefield z stack images with corresponding quantifications (n > 50/sample) are presented. Bars, 2 µm.
The absence of CytoQ foci formation upon translational arrest does not exclude the formation of smaller aggregate species that are nontraceable by microscopy. Indeed, CHX addition was shown previously to only reduce but not to prevent protein aggregation in yeast cells heat shocked at 46°C, resulting in the formation of smaller aggregates that can be pelleted by centrifugation (Wallace et al., 2015). We therefore monitored the distribution of mCherry-VHL and Hsp42 in the soluble fraction and two insoluble fractions, reporting on large (P20 fraction) or smaller (P100 fraction) aggregates in stressed yeast cells (with or without CHX; Fig. S1 D). We found ∼50% of mCherry-VHL and Hsp42 in the P20 fraction after stress application, reflecting the formation of large aggregates and consistent with formation of microscopically detectable foci. When stress was applied in presence of CHX, the mCherry-VHL and Hsp42 profiles looked similar to nonstressed control cells, and only a slightly increased P100 fraction was observed for Hsp42 (Fig. S1 D). We therefore did not exclude the formation of some small-sized aggregates, but we concluded that active protein translation is required for the formation of large-sized protein aggregates.
We considered the possibility that CHX addition also prevents upregulation of Hsps including Hsp42 and therefore might affect CytoQ formation indirectly. Levels of Hsp42 are, however, already high before heat treatment (20,000 molecules/cell) and increase only twofold after stress (Miller et al., 2015; Mackenzie et al., 2016). We therefore consider it unlikely that inhibition of protein synthesis affects protein aggregation by limiting stress-induced Hsp42 up-regulation.
Hsp42 NTE harbors two distinct IDDs that differently control CytoQ formation and stability
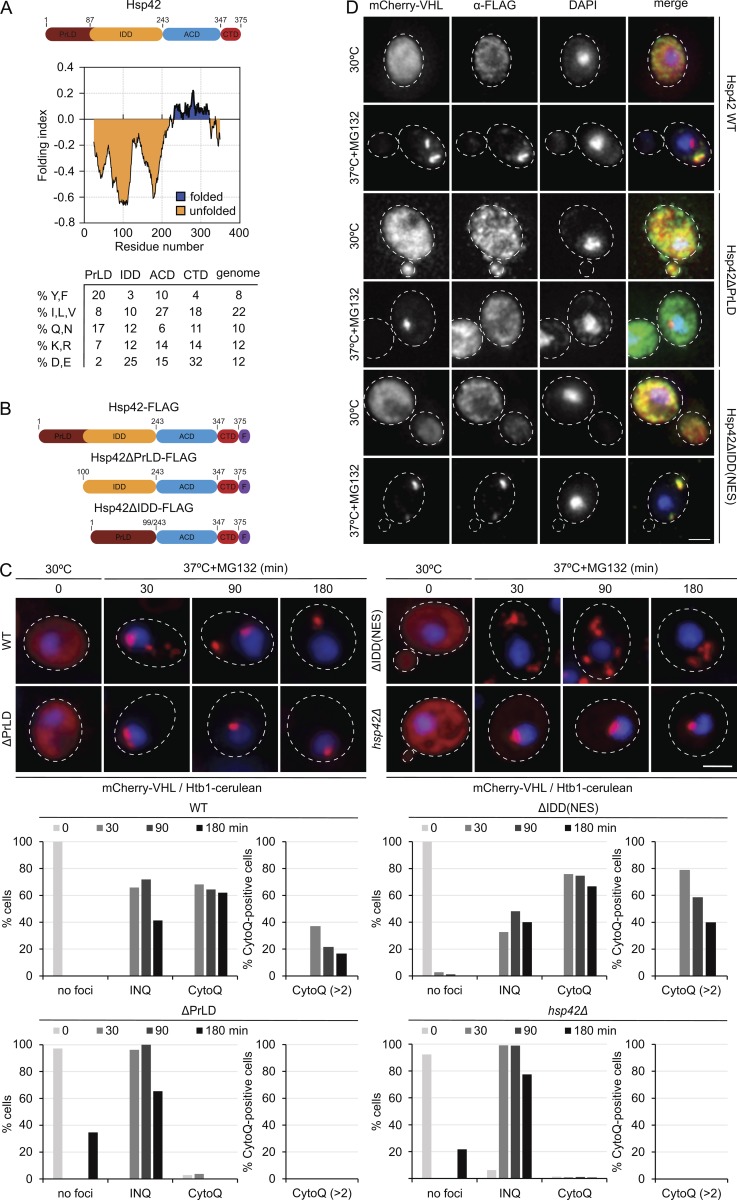
Hsp42 consists of the conserved α-crystallin scaffold domain, which is essential for sHsp activity (Haslbeck and Vierling, 2015) and flanked by NTEs and CTEs. Since the Hsp42 NTE but not CTE is essential for CytoQ formation (Specht et al., 2011), we focused our analysis on this domain, which is also unique among sHsps because of its extraordinary length. Based on sequence features, the NTE can be further subdivided into two subdomains, each predicted to be intrinsically disordered yet representing a different prototype of disordered protein segments (Figs. 2 A and S2 A; Prilusky et al., 2005). Subdomain 1 (M1-Y86) forms a putative PrLD particularly enriched in tyrosine residues (17.4% of total) and partially in glutamines and asparagines (17%; Alberti et al., 2009). Subdomain 2 (D87-F242) is strongly enriched for acidic (24.7%) and disenriched for large hydrophobic and aromatic residues (13.6% total), representing classical sequence features of IDDs (Figs. 2 A and S2 A). The roles of the disordered Hsp42 subdomains in misfolded protein sequestration are unknown. Hsp42 thereby constitutes an ideal system to investigate the impact of the different classes of disordered domains, PrLDs and IDDs, on the facilitated aggregation of misfolded proteins.
Figure 2.
The Hsp42 PrLD is essential for CytoQ formation. (A) Hsp42 domain organization. Hsp42 consists of an NTE composed of two IDDs (PrLD and IDD) followed by the ACD and a CTE. Prediction of intrinsically unfolded protein segments according to Foldindex (Prilusky et al., 2005) and the relative abundance of specific amino acids within each Hsp42 domain are shown. The average amino acid abundance of the yeast proteome is included as reference. (B) Schematic diagrams of Hsp42 WT and deletion constructs. F, FLAG tag. (C) mCherry-VHL localization patterns in S. cerevisiae hsp42Δ cells expressing Hsp42 WT or the indicated Hsp42 deletion mutants and their superposition with the nuclear marker Htb1-cerulean upon control conditions (30°C) and proteotoxic stress (37°C + MG132) are shown. The number of cells showing CytoQ (mCherry-VHL foci distant from the Htb1-cerulean signal) or INQ (mCherry-VHL foci adjacent or overlapping with the Htb1-cerulean signal) inclusions was quantified (n > 50/sample). (D) Immunofluorescence images of Hsp42 WT and deletion mutants and their superposition with mCherry-VHL and DAPI upon control conditions (30°C) and proteotoxic stress (37°C for 30 min + MG132) are shown. Maximal projections of widefield z stack images are presented. Bars, 2 µm.
To analyze the roles of the NTE subdomains for Hsp42 function, we generated Hsp42 deletion variants lacking either PrLD or IDD subdomains (Fig. 2 B). The constructs additionally harbored a C-terminal FLAG-tag, allowing us to compare Hsp42 levels by anti-FLAG Western blotting. Hsp42-FLAG was fully functional in CytoQ formation (Fig. 2 C; Specht et al., 2011). An Hsp42Δ1–86 variant only accumulated to very low levels, suggesting instability of the deletion construct (unpublished data). We therefore expressed Hsp42Δ1–99 (referred to as Hsp42ΔPrLD), additionally excluding an adjacent highly acidic peptide stretch. Hsp42ΔPrLD accumulated to Hsp42 WT levels, indicating that the deletion mutant is not sensitive toward proteolysis (Fig. S2 B). As a corresponding deletion variant, we expressed Hsp42Δ100–242 (referred to as Hsp42ΔIDD) lacking almost the entire IDD. When monitoring the localization of the Hsp42 variants in yeast cells at 30°C, we observed that Hsp42ΔIDD accumulated in the nucleus (Fig. S2 C). This contrasted with Hsp42 WT, which was largely excluded from the nucleus (Fig. 2 D; Miller et al., 2015), suggesting that the IDD is responsible for nuclear exclusion, yet without harboring an obvious nuclear export signal (NES). To ensure a proper cytosolic localization, we therefore C-terminally fused a NES to Hsp42ΔIDD, creating Hsp42ΔIDD-NES, which resided in the cytosol (Fig. 2 D) and accumulated to Hsp42 WT levels (Fig. S2 B).
We tested for the ability of the different Hsp42 constructs to restore CytoQ formation in hsp42Δ cells by using misfolded mCherry-VHL as fluorescent reporter (Fig. 2 C; Specht et al., 2011). Upon proteotoxic stress (heat shock to 37°C plus MG132 addition), mCherry-VHL forms cytosolic (CytoQ) and nuclear (INQ) foci in cells expressing Hsp42 WT, but only a single INQ focus that is adjacent to stained chromatin (Htb1-cerulean) in hsp42Δ cells (Specht et al., 2011; Miller et al., 2015). Accordingly, Hsp42 colocalized with mCherry-VHL foci except for one located next to the DNA (DAPI) signal, which corresponded with nuclear INQ (Fig. 2 D; Specht et al., 2011). CytoQ numbers reduce upon prolonged stress application, giving rise to the formation of only one or two CytoQ deposits in most Hsp42 WT cells (Fig. 2 C; Kaganovich et al., 2008; Specht et al., 2011).
Hsp42ΔPrLD expression did not restore CytoQ formation, and mCherry-VHL exclusively localized to INQ (Fig. 2 C). Nuclear localization of mCherry-VHL foci was confirmed by Nsp1 immunofluorescence, staining the nuclear envelope (Fig. S2 D). Hsp42ΔPrLD did not change cellular localization upon stress application but remained diffusely distributed, consistent with its inability to trigger cytosolic mCherry-VHL aggregation (Fig. 2 D). We conclude the PrLD of Hsp42 is therefore essential for aggregase function.
The second deletion construct Hsp42ΔIDD-NES restored formation of cytosolic mCherry-VHL aggregates in hsp42Δ cells. However, we noticed a more than twofold increased number of smaller mCherry-VHL foci (more than two CytoQ/cell) at all time points after stress application when compared with Hsp42 WT (Fig. 2 C). A large fraction (40%) of Hsp42ΔIDD-NES–expressing cells still exhibited multiple CytoQs even after prolonged stress application (180 min) as compared with 18% of Hsp42 WT cells (Fig. 2 C). Hsp42ΔIDD-NES, like Hsp42 WT, was an integral part of the aggregates as revealed by colocalization with mCherry-VHL foci (Fig. 2 D). We observed very similar mCherry-VHL foci formation in the presence of Hsp42ΔIDD (Fig. S2 E), suggesting that the construct shuttles between cytosolic and nuclear compartments and becomes trapped in the cytosol upon inducing CytoQ formation. This finding excludes that NES fusion in Hsp42ΔIDD-NES has an impact on aggregase activity and confirms the former observations. The IDD is therefore not required for CytoQ formation; however, it modulates CytoQ numbers per cell.
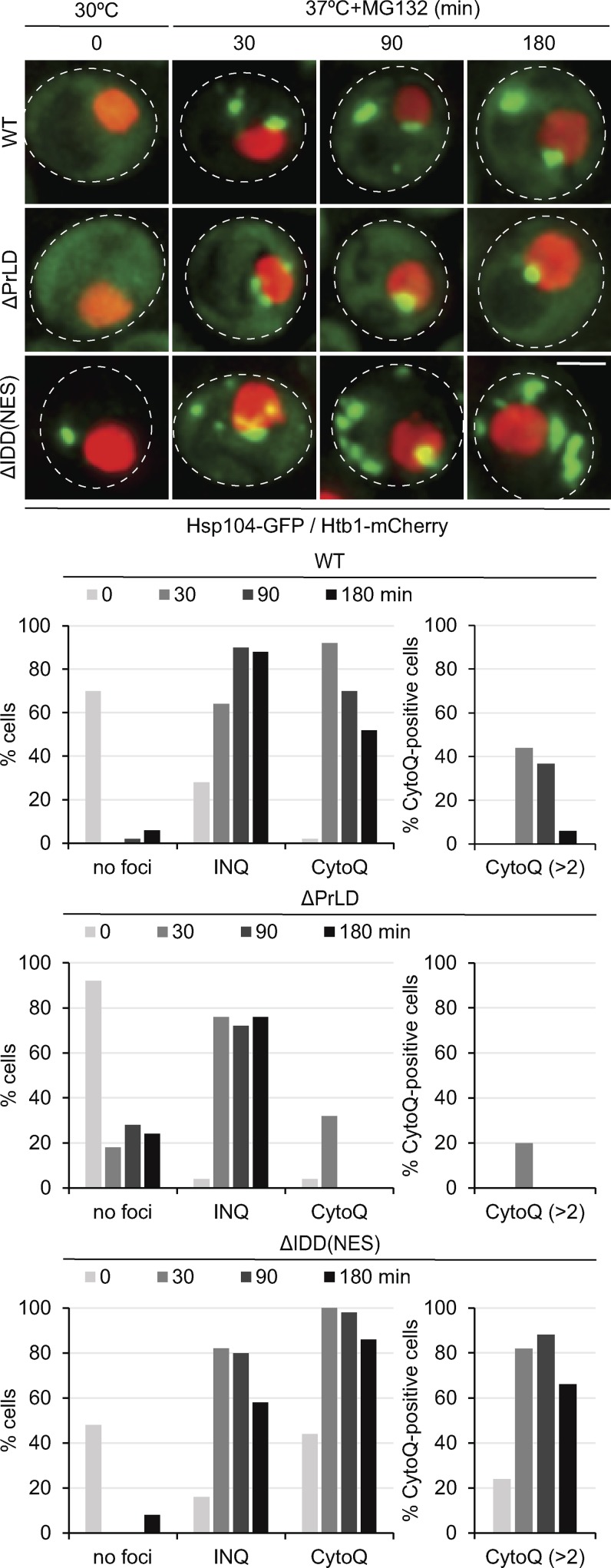
To generalize the determined defects of Hsp42ΔPrLD and Hsp42ΔIDD-NES in CytoQ formation, we monitored endogenous aggregates in respective yeast cells expressing fluorescent Hsp104-GFP disaggregase as a reporter. Yeast cells expressing Hsp42 WT frequently harbored multiple Hsp104-GFP foci during the acute stress period (30 min) but typically only one or two enlarged Hsp104-GFP foci upon prolonged (180 min) stress application (Fig. 3). Hsp42ΔPrLD-expressing cells showed only nuclear Hsp104-GFP foci (INQs next to or overlapping with Htb1-mCherry staining chromatin) and were deficient in CytoQ formation (Fig. 3). Increased numbers of cytosolic Hsp104-GFP foci were observed throughout the stress period in the presence of Hsp42ΔIDD-NES. In particular, the majority (63%) of Hsp42ΔIDD-NES–expressing cells exhibited more than two CytoQ deposits, whereas the corresponding fraction was negligible (4%) in Hsp42 WT cells after prolonged stress application (Fig. 3). We also noticed cytosolic Hsp104-GFP foci in nonstressed cells expressing Hsp42ΔIDD-NES, suggesting an increased propensity of the Hsp42 deletion variant to trigger protein aggregation even under physiological conditions. Collectively, these findings demonstrate that the defects of Hsp42 mutants observed with the mCherry-VHL reporter also hold true and are even more pronounced for CytoQs formed by endogenous yeast proteins. We infer that the disordered subdomains of Hsp42 have different functions in CytoQ formation. Although PrLD was essential for CytoQ formation, IDD impacted CytoQ numbers.
Figure 3.
Distinct functions of Hsp42 PrLD and IDD in CytoQ formation. Localization patterns of Hsp104-GFP and their superposition with the nuclear marker Htb1-mCherry in S. cerevisiae cells expressing Hsp42 WT or deletion mutants upon control conditions (30°C) and proteotoxic stress (37°C + MG132). The number of cells showing CytoQ (mCherry-VHL foci distant from the Htb1-cerulean signal) or INQ (mCherry-VHL foci adjacent or overlapping with the Htb1-cerulean signal) inclusions was quantified at the indicated time points (n > 50 cells). Maximal projections of widefield z stack images are presented. Bar, 2 µm.
We also tested whether the Hsp42 IDD affects the stability of the formed CytoQs during a recovery phase. We therefore determined the fraction of cells still harboring mCherry-VHL foci during a recovery period at 30°C after MG132 washout (Fig. S2 F). mCherry-VHL foci were removed in the majority (71%) of Hsp42 WT–expressing cells 120 min after stress treatment. In contrast, 81% of Hsp42ΔIDD-expressing cells still retained mCherry-VHL foci, indicating that the Hsp42 IDD also affects aggregate stabilities (Fig. S2 F).
The Hsp42 PrLD constitutes the major substrate-binding site
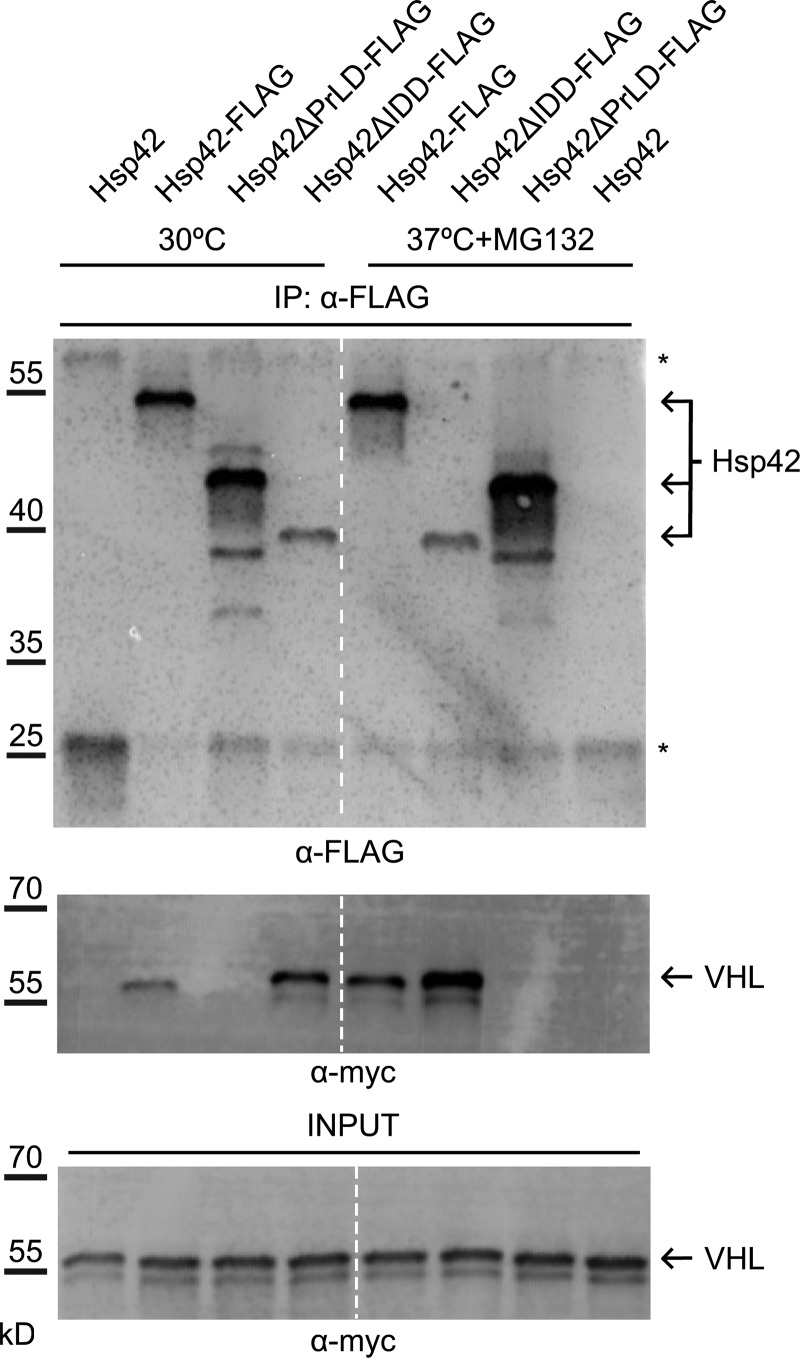
To test whether the determined defects of Hsp42 variants on CytoQ formation correlate with altered substrate-binding capacities, we performed pulldown experiments using yeast cells expressing FLAG-tagged Hsp42 (WT and mutants) and mCherry-Myc-VHL harboring an additional Myc tag. Hsp42 was isolated before and after stress treatment, and the amount of coprecipitated mCherry-Myc-VHL was determined. The amounts of precipitated Hsp42 WT–FLAG and Hsp42ΔPrLD-FLAG were comparable, whereas less Hsp42ΔIDD-FLAG was isolated (Fig. 4). mCherry-Myc-VHL was in complex with Hsp42-FLAG already before heat shock as VHL is constantly misfolded upon expression in yeast cells because of the absence of stabilizing partner proteins. The amount of Hsp42-bound mCherry-Myc-VHL increased after stress (Fig. 4). In contrast, no mCherry-Myc-VHL could be isolated in complex with Hsp42ΔPrLD-FLAG, indicating that PrLD deletion abrogates substrate interaction and explaining the deficiency of Hsp42ΔPrLD in sequestering mCherry-VHL at CytoQs (Fig. 4). An increased amount of substrate was found under all conditions associated with Hsp42ΔIDD-FLAG as compared with Hsp42-FLAG; despite this, the variant was precipitated to a lower extent (Fig. 4). Hsp42 lacking the IDD thus exhibits strongly increased substrate-binding capacity.
Figure 4.
Hsp42 PrLD is essential for mCherry–VHL interaction. Coimmunoprecipitation of Hsp42 WT and deletion mutants (all FLAG-tagged) and mCherry-myc-VHL. Cells were grown at control conditions (30°C) or stressed (37°C for 45 min + MG132) before extract preparation. Hsp42 proteins were precipitated using anti-FLAG agarose, and the amount of bound Hsp42 and mCherry-myc-VHL was quantified by immunoblot analysis using anti-FLAG and anti-myc antibodies. Fractions were analyzed for the content of FLAG-tagged proteins and mCherry–VHL interaction (anti-myc). The sizes of Hsp42 proteins are indicated (arrows). Asterisks indicate coeluting mouse IgG heavy and light chains. VHL input controls are provided. The dashed line indicates intervening lanes have been spliced out. IP, immunoprecipitation.
To provide further evidence for a central role of the PrLD in substrate interaction, we purified the Hsp42 deletion variants and characterized key properties including chaperone activity in vitro. We first analyzed for oligomer formation and determined the masses of Hsp42, Hsp42ΔPrLD, and Hsp42ΔIDD complexes by static light scattering coupled with size exclusion chromatography. The molecular weight of Hsp42 WT oligomers varied from 325–575 kD, corresponding with 8–14 subunits and with decamers representing the most populated state (Fig. S3 A). This size was similar to a previously reported one (12–16 subunits) that estimated Hsp42 particle size based on gel filtration (Haslbeck et al., 2004a). Hsp42ΔPrLD formed smaller oligomers (tetramer as most populated state), whereas Hsp42ΔIDD oligomers were larger (36-mer as most populated state; Fig. S3 A). Thus, both disordered subdomains of the Hsp42 NTE affect oligomer formation, with PrLD and IDD deletions having opposite consequences on oligomer size.
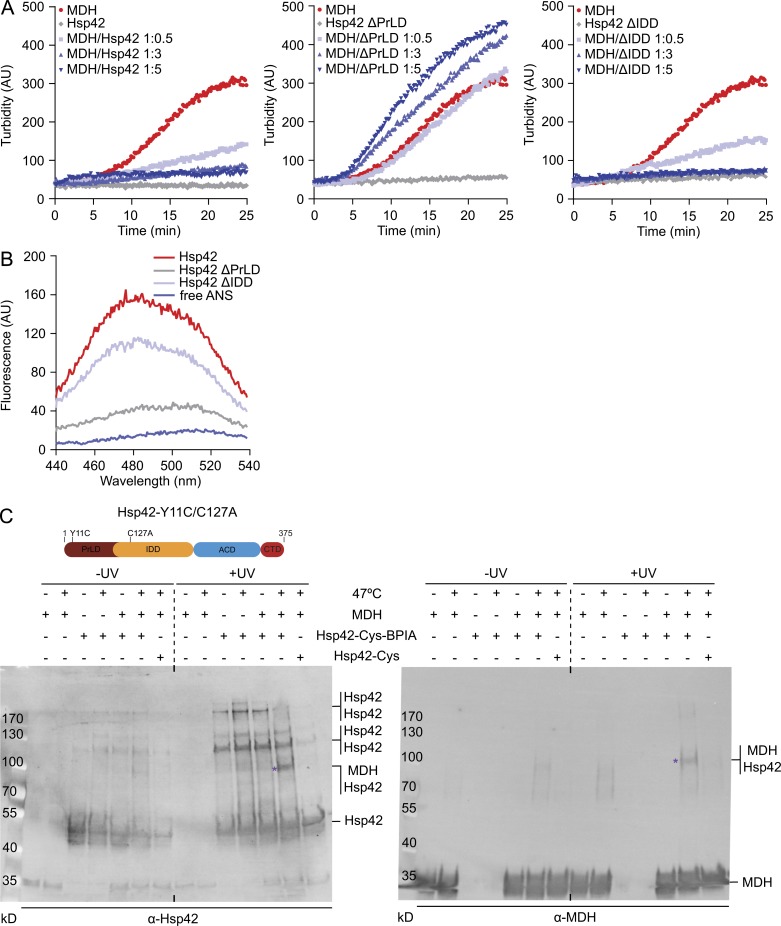
The chaperone activities of Hsp42 deletion constructs were first analyzed by monitoring aggregation of thermolabile malate dehydrogenase (MDH) in the absence and presence of Hsp42 (WT and mutants) at 47°C. Hsp42 WT associates with aggregating MDH and entirely suppresses the formation of turbid MDH aggregates if present in threefold molar excess (protomers; Fig. 5 A). In contrast, excess of Hsp42ΔPrLD did not prevent MDH aggregation as indicated by an almost unaltered MDH turbidity increase even when present in fivefold excess (Fig. 5 A). Hsp42ΔIDD exhibited a chaperone activity that appeared comparable to Hsp42 WT (Fig. 5 A). The association of sHsps with substrates facilitates the refolding of bound substrates by disaggregating Hsp70-Hsp100 (S. cerevisiae Ssa1/Ydj1/Sse1/Hsp104) chaperones during recovery phases (Mogk et al., 2003; Haslbeck et al., 2005). We therefore monitored MDH reactivation after 47°C denaturation in the absence or presence of Hsp42 WT and mutants. Hsp42 WT and Hsp42ΔIDD facilitated MDH reactivation by Hsp70-Hsp100 chaperones, whereas MDH refolding kinetics remained unchanged in the presence of Hsp42ΔPrLD as compared with aggregated MDH (Fig. S3 B). Collectively, these findings demonstrate a crucial role of the Hsp42 PrLD for chaperone activity.
Figure 5.
PrLD is the major Hsp42 substrate interaction site. (A) MDH was denatured for 30 min at 47°C in the absence or presence of Hsp42 WT and deletion mutants at indicated ratios. All control Hsp42 proteins were heated alone. The formation of MDH aggregates was followed by turbidity measurements. (B) Overall surface hydrophobicity of Hsp42 WT and deletion mutants probed by ANS fluorescence. Fluorescence of free ANS is provided as reference. (C) Cross-linking of BPIA-labeled Hsp42-Y11C/C127A (Hsp42-Cys) to unfolded MDH. MDH and Hsp42 alone or MDH/Hsp42-Cys (unlabeled and BPIA-labeled) mixtures (1:5 ratio) were incubated at 30°C or 47°C. Samples were afterward exposed to UV (+/− UV), and cross-link products were analyzed by immunoblot analysis using MDH and Hsp42-specific antibodies. Asterisks indicate Hsp42-MDH cross-link products. A schematic diagram of Hsp42-Y11C/C127A is shown at top.
Chaperones typically use hydrophobic surfaces or segments to interact with misfolded proteins (Balchin et al., 2016). We analyzed for the exposure of hydrophobic patches serving as potential substrate-binding sites in Hsp42 WT and mutants by incubating the proteins with the fluorescent reporter 1-anilino-8-napthalene-sulfonate (ANS; Fig. 5 B). ANS fluorescence strongly increases upon binding to hydrophobic surfaces and also shows a blue shift of fluorescence emission maxima (Gasymov and Glasgow, 2007). ANS exhibited blue-shifted high fluorescence intensity when incubated with Hsp42 WT. A similar increase in ANS fluorescence as well as a blue shift was noticed upon incubation with Hsp42ΔIDD (Fig. 5 B). Notably, Hsp42ΔIDD oligomers (36-mer) have a lower surface/volume ratio as compared with Hsp42 WT (10-mer), assuming dense packing of subunits. Therefore, despite reduced surface accessibility, Hsp42ΔIDD bound a similar amount of ANS, suggesting that Hsp42ΔIDD exposes more hydrophobic binding sites. In contrast, ANS fluorescence remained low in the presence of Hsp42ΔPrLD despite an increased surface exposure of the tetrameric mutant as compared with Hsp42 WT. This shows a strong reduction in accessible hydrophobic surfaces for Hsp42ΔPrLD, supporting a major role of PrLD in substrate interaction.
To provide direct evidence for interactions between the Hsp42 PrLD and substrates, we used a cross-linking approach using UV-activable benzophenone-4-iodoacetamide (BPIA), which can be site-specifically introduced at cysteine residues (Seyffer et al., 2012). BPIA has a linker length of 10 Å, and cross-link products therefore report on intimate contacts between interacting proteins. We surmised that the highly abundant tyrosine residues of the PrLD are key players in substrate binding. Indeed, replacing all tyrosines of the PrLD with serine residues (Hsp42-Y/S) abrogated CytoQ formation in vivo (Fig. S3, C and D). Accordingly, staining of Hsp42-Y/S stayed diffuse after stress application and did not colocalize with the remaining nuclear mCherry-VHL foci (Fig. S3 E). We therefore generated Hsp42-Y11C/C127A, allowing for specific labeling of the PrLD at position 11 by additionally mutating the single endogenous Cys127 (Fig. 5 C). Hsp42-Y11C/C127A was fully active in preventing MDH aggregation ensuring functionality (Fig. S3 F). Cross-linking of BPIA-labeled Hsp42-Y11C/C127A upon UV exposure in the absence of substrate led to formation of dimeric and trimeric cross-link products (Fig. 5 C), indicating intersubunit contacts of the PrLD. Incubation with MDH at 30°C did not alter this cross-link pattern and did not yield Hsp42-MDH cross-links. However, triggering MDH unfolding at 47°C led to the appearance of a specific Hsp42-Y11C/C127-MDH cross-link product, which was confirmed by Western blot analysis using MDH-specific antibodies (Fig. 5 C). This demonstrates that Tyr11 of the PrLD is at a distance of ≤10 Å from the substrate bound in Hsp42–MDH complexes formed upon MDH unfolding. We conclude that the PrLD constitutes the major substrate-binding site of Hsp42 and is therefore essential for CytoQ formation.
Hsp42 lacking IDD exhibits superior chaperone activity
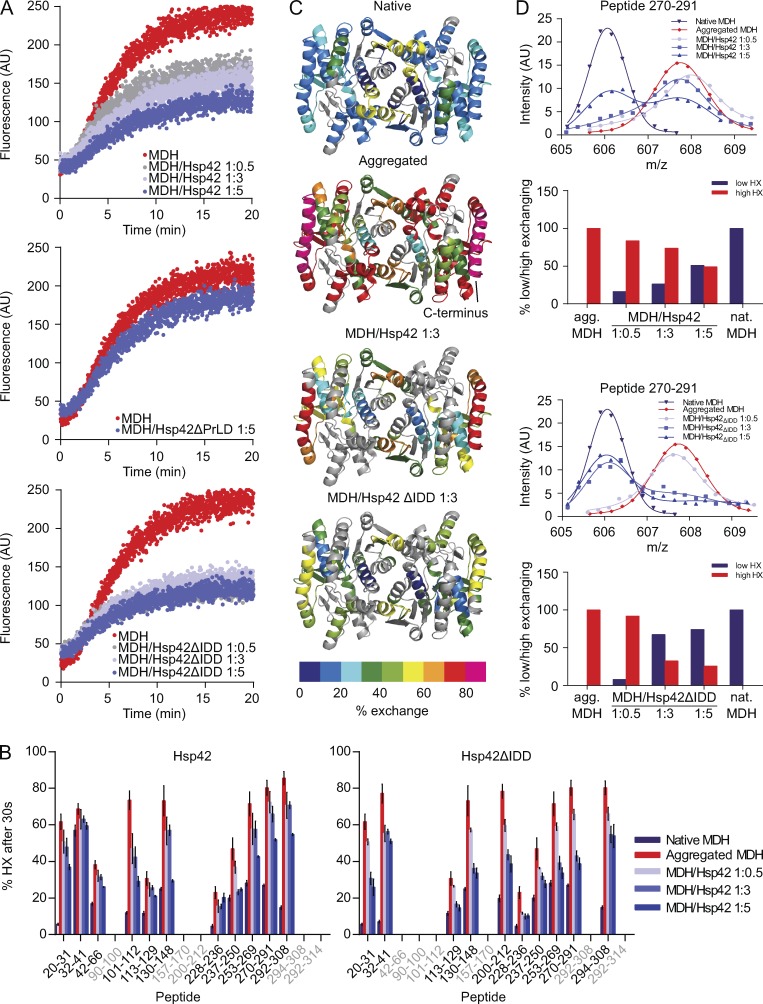
Hsp42ΔIDD exhibits increased substrate binding capacity in vivo, yet its chaperone activity in vitro seems comparable to Hsp42 WT when monitoring protein aggregation by turbidity measurements (Fig. 5 A). We therefore used a more sensitive chaperone assay in which we monitored MDH aggregation by Förster resonance energy transfer (FRET) using a mixture of MDH-YFP and MDH labeled with 7-diethylcoumarin-3-carboxylic acid as FRET pair (Ungelenk et al., 2016). FRET was observed at 47°C but not at 30°C, demonstrating that FRET reports on coaggregation of MDH donor and acceptor molecules (Ungelenk et al., 2016). An increase in FRET signal was almost immediately observed upon incubation at 47°C (Fig. 6 A), whereas turbid MDH aggregates became only detectable after a lag phase of 5 min (Fig. 5 A), demonstrating higher sensitivity of the FRET assay. Hsp42ΔPrLD presence did hardly affect FRET kinetics and efficiencies between aggregating MDH species in contrast with Hsp42 WT (Fig. 6 A). In this study, FRET efficiencies were continuously declining with increasing Hsp42 concentrations. Efficient FRET suppression was already detected in presence of substoichiometric ratios of Hsp42ΔIDD (Fig. 6 A). This suggests that Hsp42ΔIDD has an increased chaperone activity causing changes in the organization of Hsp42–MDH complexes, which are only reached in the presence of higher Hsp42 WT levels. We tested whether such higher Hsp42ΔIDD chaperone activity also changes the sizes of sHsp–MDH complexes by electron microscopy. Indeed, Hsp42ΔIDD–MDH complexes appeared smaller as compared with Hsp42–MDH complexes (Fig. S4 A). Collectively, these findings suggest that the IDD negatively regulates Hsp42 activity and that its deletion increases substrate-binding capacities.
Figure 6.
Hsp42ΔIDD exhibits superior chaperone activity. (A) Coaggregation of MDH-YFP (FRET donor) and MDH labeled with 7-diethylcoumarin-3-carboxylic acid (FRET acceptor) was monitored by FRET (increased acceptor fluorescence) at 47°C in the absence or presence of Hsp42 WT and deletion mutants. (B) Relative proton/deuteron exchange in native or aggregated MDH and MDH coaggregated with Hsp42 WT and Hsp42ΔIDD (at indicated MDH/sHsp ratios) after 30 s incubation in D2O. Aggregated MDH and MDH–sHsp complexes were formed by incubation for 30 min at 47°C. Error bars denote SD for each point based on three repetitions. All data were corrected for deuteron losses caused by back exchange using a 100% deuterated control. Grey regions could not be detected. (C) HX heat maps of native, aggregated, and Hsp42-complexed states (threefold excess of Hsp42) of the MDH dimer structure (PDB ID: 1MLD) are shown. Peptic peptides are colored according to their exchange behavior (% exchange). Grey regions could not be detected. (D) Bimodal distribution of isotope peaks of indicated MDH peptides derived from MDH–Hsp42 (WT or ΔIDD) complexes. Top: intensity versus m/z diagrams for the indicated peptic MDH fragment after 30 s HX at 30°C. Bottom: fractions of native-like (low HX) and aggregate-like (high HX) populations.
To compare the chaperone activities of Hsp42 WT and Hsp42ΔIDD at higher resolution, we used time-resolved amide hydrogen exchange (HX) combined with mass spectrometry. This approach allowed us to determine the conformational states of MDH when complexed with Hsp42. HX determines the solvent accessibility of backbone amide hydrogen atoms as a measure of structural flexibility and the conformational state of MDH. Amide hydrogens are protected from HX if engaged in (a) hydrogen bonds as a result of secondary and tertiary structure formation or (b) protein–protein interactions. We recently demonstrated that sHsps including Hsp42 stabilize MDH in a native-like conformation if present in excess over substrate (Ungelenk et al., 2016). In this study, we first determined the averaged HX profile of MDH denatured in the presence of varying Hsp42ΔIDD and Hsp42 WT concentrations and compared those with aggregated and native MDH (Fig. 6, B and C; and Fig. S4 B). MDH complex formation with Hsp42ΔIDD caused more pronounced HX protection when compared with Hsp42 WT (Fig. 6, B and C). This difference in protective capacity can be well seen for the C-terminal α helix of MDH (F287-M313), which showed greatest HX in the aggregated state and was shown to serve as major sHsp interaction site upon initial unfolding (Fig. 6 C; Ungelenk et al., 2016). The higher activity of Hsp42ΔIDD became also apparent when analyzing HX data for bimodal peak distributions of MDH peptides, which report on the degree of structural heterogeneity of MDH when present in complexes with sHsps (Figs. 6 D and S4 C). We recently showed that MDH peptides when part of an sHsp–MDH complex exist in two structural states that indicate either aggregated (high HX) or native-like (low HX) MDH conformations. Increasing the ratio of sHsps versus MDH shifted the ensemble of MDH peptide conformations toward native-like states, indicating that sHsp binding stabilizes MDH in near-native conformation and protects the bound substrate from further unfolding (Ungelenk et al., 2016). We almost exclusively observed native-like states for all analyzed MDH peptides in the presence of a threefold Hsp42ΔIDD excess (Figs. 6 D and S4 C). For comparison, a fivefold excess of Hsp42 WT was necessary to obtain a similar trend, yet a substantial fraction of some MDH peptides (113–129, 270–291, and 292–308) still exhibited aggregate-like HX under these conditions (Figs. 6 D and S4 C). We infer that our highly sensitive assays monitoring MDH aggregation and its conformational state upon association with Hsp42 variants demonstrate an increased chaperone activity of Hsp42ΔIDD, qualifying the IDD as regulatory domain.
To account for the superior chaperone activity of Hsp42ΔIDD, we considered alterations in the dynamics of Hsp42 WT and Hsp42ΔIDD subunits within the sHsp oligomers because high exchange rates correlate with high chaperone activity (Haslbeck and Vierling, 2015; Rajagopal et al., 2015). We tested for subunit dynamics by FRET using 7-diethylaminocoumarin-3-carboxylic acid and 6-(N-(7-nitrobenz-2-oxa-1,3-diazol-4-yl)amino)hexanoate (NBD-X)–labeled Hsp42 (WT or ΔIDD) as the FRET donor and acceptor (Fig. S5 A). Regain of donor fluorescence upon addition of an excess of unlabeled Hsp42 (WT or ΔIDD) enabled determination of exchange rates, which were similar for Hsp42 WT and Hsp42ΔIDD (Fig. S5 A). Differences in oligomer dynamics are therefore not causative for increased Hsp42ΔIDD activity. We therefore speculate that the increase in Hsp42ΔIDD oligomer size is linked to its higher activity, eventually reflecting altered subunit organization facilitating substrate interaction as suggested for heat-induced larger oligomers of pea Hsp18.1 (Stengel et al., 2010).
PrLD and IDD differently affect Hsp42 aggregase function in vitro
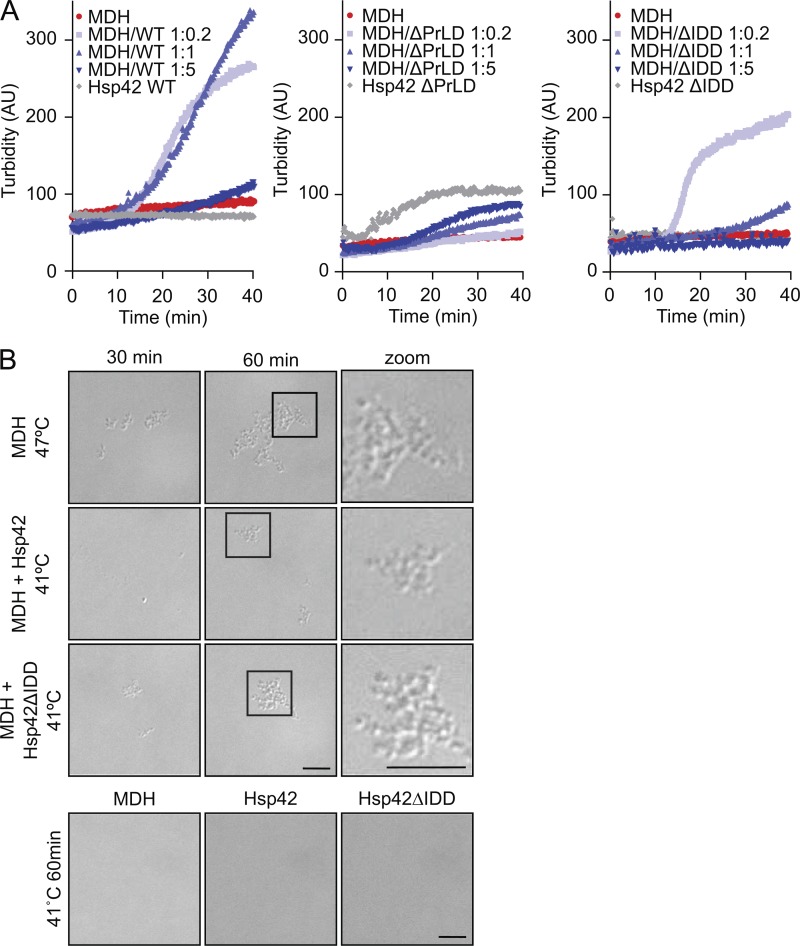
We next tested how the altered chaperone activities of Hsp42 deletion variants affect aggregase function in a reconstituted system. We recently established an aggregase assay in vitro by exposing MDH to mild denaturing conditions (41°C), which is well below its melting temperature (50.9°C; Ungelenk et al., 2016). This treatment caused slow MDH unfolding but did not result in the formation of large, turbid MDH aggregates. Presence of substoichiometric and stoichiometric Hsp42 levels triggered the formation of turbid MDH aggregates, whereas this effect is reverted in the presence of Hsp42 excess (Fig. 7 A; Ungelenk et al., 2016). The ratio of Hsp42 and MDH present in the sHsp–substrate assembly is therefore an important feature controlling complex size. Increased incorporation of Hsp42 will cause formation of smaller, nonturbid substrate complexes.
Figure 7.
Aggregase function of Hsp42 deletion mutants. (A) MDH was denatured for 60 min at 41°C in the absence or presence of Hsp42 WT and deletion mutants at the indicated ratios. As a control, Hsp42 proteins were heated alone. The formation of MDH aggregates was followed by turbidity measurements. (B) Complexes of MDH with Hsp42 WT or Hsp42ΔIDD formed at 41°C at equimolar ratios were analyzed by light microscopy. MDH aggregated at 47°C served as reference. As control, MDH and Hsp42 proteins were heated alone. Widefield images are presented. Bars, 5 µm.
Hsp42ΔPrLD hardly affected MDH aggregation (Fig. 7 A), in agreement with its deficiency in supporting CytoQ formation (Figs. 2 C and 3) and interacting with MDH in chaperone assays (Figs. 5 and 6). Hsp42ΔIDD exhibited aggregase function that was highly concentration dependent (Fig. 7 A). When Hsp42ΔIDD was present at substoichiometric ratios (MDH/Hsp42ΔIDD–MDH of 1:0.2), it exhibited aggregase activity, and the time course of Hsp42ΔIDD–MDH complex turbidity was slightly steeper as compared with Hsp42–MDH complexes, suggesting a faster aggregation process. Stoichiometric levels of Hsp42ΔIDD already reverted the aggregase effect in contrast with Hsp42 WT, and Hsp42ΔIDD excess did not lead to formation of large turbid assemblies any longer (Fig. 7 A). These findings can be explained by a higher substrate-binding capacity of Hsp42ΔIDD, thereby changing the ratio of substrate and sHsps present in the formed complexes.
In an independent approach, we monitored Hsp42-facilitated MDH aggregation by light microscopy as pure qualitative readout (Fig. 7 B). As a reference, we used MDH aggregates formed at 47°C, which appeared as particles of 500-nm diameter. Many of these particles were found sticking together, forming larger clumps. Incubation of MDH at 41°C did not result in the formation of detectable particles, consistent with the absence of MDH aggregation in turbidity measurements (Fig. 7 B). In contrast, incubation of MDH with stoichiometric amounts of Hsp42ΔIDD or Hsp42 WT triggered formation of detectable particles at 41°C, which appeared similar in size and shape as compared with MDH aggregates formed at 47°C (Fig. 7 B). Particles were not observed when Hsp42 WT or Hsp42ΔIDD were incubated without MDH, documenting aggregase function. In summary, Hsp42ΔPrLD and Hsp42ΔIDD activities determined in vivo concur well with their activities in aggregase assays in vitro.
Hsp42 PrLD and IDD provide fitness to yeast cells during repetitive stress cycles
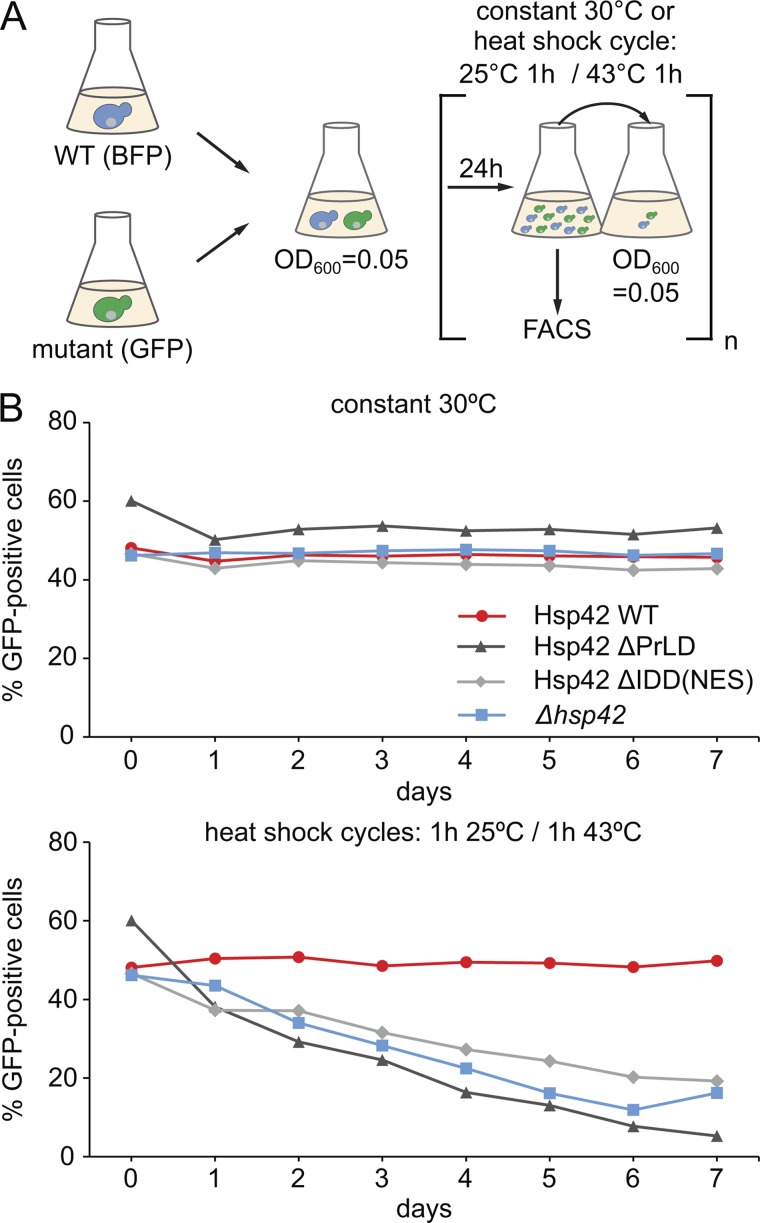
We finally tested whether the distinct consequences of Hsp42 subdomain deletions on chaperone and aggregase activities (Hsp42ΔPrLD, loss of activity; Hsp42ΔIDD, superior activity) impacts the fitness of yeast cells during heat stress conditions. We recently unraveled by growth competition assays involving coincubation of S. cerevisiae WT and hsp42Δ cells that Hsp42 aggregase function is critical for cellular fitness upon repetitive heat stress (Ungelenk et al., 2016). When subjected to heat shock cycles (25–43°C), hsp42Δ cells were continuously outcompeted by WT cells, whereas no competition was observed when cells were constantly incubated at 30°C. We compared the competitive capacity of yeast cells expressing either Hsp42ΔPrLD or Hsp42ΔIDD-NES at 30°C and during heat shock cycles (Fig. 8). Expression of BFP and GFP in WT and hsp42 mutant cells allowed for precise calculation of the respective cell population by FACS analysis after each day (Fig. 8 A). We found that Hsp42ΔPrLD–expressing cells, which are deficient in CytoQ formation, were outcompeted by yeast WT cells under stress conditions, confirming cytoprotective functions of Hsp42-dependent CytoQ formation (Fig. 8 B). Notably, Hsp42ΔIDD-NES–expressing cells were displaced by WT cells from the total cell population with similar kinetics (Fig. 8 B). Almost identical results were obtained for Hsp42ΔIDD–expressing cells excluding a detrimental effect of the fused NES (Fig. S5 B). These findings indicate that Hsp42 PrLD–mediated CytoQ formation is not sufficient for cellular protection but that Hsp42 IDD–mediated control of CytoQ formation and reversibility is additionally required. The regulatory role of Hsp42 IDD is therefore crucial for fitness of yeast cells during stress conditions.
Figure 8.
PrLD and IDD are crucial for cytoprotective Hsp42 activity during heat stress. (A) Schematic diagram of the growth competition assay. S. cerevisiae WT cells expressing BFP and indicated hsp42 mutant cells expressing GFP were mixed 1:1. Mixtures were either grown at 30°C or were subjected to heat shock cycles, switching repetitively between 25°C and 43°C. Each day, the proportion of WT and mutant cells was determined by FACS, and cell mixtures were diluted to OD600 0.05. (B) Fractions of Hsp42 WT and Hsp42 deletion mutants or hsp42Δ deletion strain in the mixed cultures upon constant growth at 30°C or repetitive heat stress are plotted. The representative result of one out of three biological replicates is shown.
Discussion
Proteins harboring intrinsically disordered domains play important roles in the formation of membrane-free RNA sequestering compartments because of their ability to undergo homo- and heterotypic multivalent interactions. In this study, we have analyzed the mechanism of organized, factor-dependent sequestration of misfolded proteins. Misfolded protein sequestration into cytosolic aggregates (CytoQs) during physiological heat stress requires the chaperone Hsp42 (Specht et al., 2011; Escusa-Toret et al., 2013; Song et al., 2014; Saarikangas and Barral, 2015). The long NTE of Hsp42 is essential for CytoQ formation (Specht et al., 2011; Marshall et al., 2016). It harbors two prototypes of IDDs, a PrLD and a highly charged segment with low hydrophobicity. This enabled us to dissect the role of each type of disordered domain in the organized aggregation of misfolded proteins.
We identified the Hsp42 PrLD as key factor in CytoQ formation, expanding the established role of PrLDs in the formation of RNA granules to organized protein aggregates. We show that ongoing translation is required for sequestration of misfolded proteins at large deposition sites upon physiological heat stress (e.g., 38°C), suggesting that newly synthesized proteins are particularly vulnerable toward stress application and represent a seed required for formation of large aggregates. These microscopic findings confirm and extend previous findings using more severe heat shock (42°C) to induce the formation of large protein aggregates in yeast cells (Zhou et al., 2014).
Our findings are partially different from a previous study, which showed that smaller protein aggregates, which cannot be detected by fluorescence microscopy, still form in heat-stressed yeast cells (Wallace et al., 2015). We would like to point out that stress conditions applied here (37°C + MG132) were less severe as compared with Wallace et al. (2015; 42–46°C), potentially explaining different outcomes. These more severe heat shock conditions also cause Hsp42-independent protein aggregation (Specht et al., 2011). We do not, however, exclude the possibility that small-sized aggregates, which are not traceable by the methods applied in this study, form upon translation stop.
RNA granule assembly factors harbor PrLDs that are fused to RNA binding domains. This separates the two obligatory activities for RNA granule formation, the abilities for self-assembly and substrate binding, into distinct domains. In contrast, the PrLD of Hsp42 is bifunctional as it mediates homo- and heterotypic multivalent interactions. The PrLD directly binds substrates but also interacts with other Hsp42 subunits. This is supported by demonstrating the direct contacts of the Hsp42 PrLD to substrate and other Hsp42 subunits by cross-linking (Fig. 5 C) as well as reduced oligomeric sizes of Hsp42ΔPrLD (Fig. S3 A). We suggest this dual activity of the Hsp42 PrLD constitutes the central mechanistic element for CytoQ formation. Accordingly, removal of the PrLD abrogates substrate binding and leads to deficiencies in all chaperone assays and CytoQ formation in yeast cells. Attempts to purify and further characterize the isolated Hsp42 PrLD failed because of its high aggregation propensity (not depicted), a feature shared by many PrLDs from other proteins (Malinovska et al., 2013).
The Hsp42 PrLD was originally identified in a bioinformatic survey to identify novel yeast prion proteins (Alberti et al., 2009). The Hsp42 PrLD, however, was ranked low because of the absence of expanded Q/N stretches and the presence of structure-breaking proline residues, which interfere with highly ordered cross–β-sheet (amyloid) formation (Toombs et al., 2010). We speculate that these sequence features of Hsp42 PrLD are critical for CytoQ formation by still allowing for Hsp42 self-assembly while also promoting promiscuous interactions with a broad range of misfolded proteins. Such activity is not expected for classical prion proteins, which can form amyloids based on defined interactions between identical prion subunits (Wickner et al., 2015). Indeed, replacing the Hsp42 NTE by the prion-forming domains of the yeast prion proteins Sup35 (NM domains) and Lsm4 failed to restore CytoQ formation (Fig. S5 C). This is different from results obtained for TIA1, which is necessary for formation of arsenite-induced stress granules in human cells. In this study, replacement of the TIA-1 PrLD with the prion domain of Sup35 enabled stress granule formation (Gilks et al., 2004). We suggest that this can be explained by the separation of substrate (RNA) binding and self-assembly activities into two independent TIA-1 domains, with the latter activity being artificially executed upon fusion of a yeast prion domain. In the case of Hsp42, both activities are united in the PrLD, and the binding to misfolded proteins cannot be taken over by a fused yeast prion domain.
The Hsp42 PrLD is particularly enriched for tyrosine residues (Fig. 2 A). Tyrosines are largely spread across the PrLD sequence; however, two conserved clusters consisting of three tyrosines (Y69–Y71 and Y78–Y80) are present (Fig. S2 A). We show that PrLD tyrosine residues are essential for CytoQ formation (Fig. S3 C) and direct substrate contacts (Fig. 5 C). Tyrosines are particularly well suited for mediating multivalent interactions as they can interact with aromatic and hydrophobic but also basic residues via cation–π interactions. They might also confer binding specificity for misfolded proteins. In agreement with such function, aromatic residues are generally enriched in NTEs of sHsps (Kriehuber et al., 2010).
The second disordered domain IDD of the Hsp42 NTE is not essential for CytoQ formation; however, it influences the numbers of CytoQ foci per cell, which are strongly increased upon IDD deletion (Figs. 2 C and 3). Increased CytoQ numbers might be caused by defects in aggregate coalescence or cytosolic transport processes (Saarikangas and Barral, 2015; Hill et al., 2016), for which IDD might be required for recruitment of cellular factors involved. Our analysis, however, offers an alternative explanation, according to which the IDD directly affects CytoQ numbers by negatively modulating the Hsp42 chaperone activity. This is supported by our findings that Hsp42ΔIDD is superior in interacting with misfolded proteins in vivo (Fig. 4) and in vitro (Fig. 6). This qualifies the IDD as key regulatory domain, which is restricting substrate-binding activity of the PrLD. An increased substrate-binding capacity will change the ratio of Hsp42ΔIDD to substrate in a given complex as compared with Hsp42 WT, leading to an altered composition and organization of the Hsp42ΔIDD–substrate complexes. We suggest that the Hsp42/substrate ratio present in a complex is a key factor in controlling the aggregation process. Although low Hsp42 levels promote the formation of large Hsp42–substrate assemblies, Hsp42 excess shifts these complexes to smaller sizes. We suggest that low Hsp42 levels promote the aggregation process by increasing local concentrations of misfolded proteins at the Hsp42–substrate complex surface, thereby stimulating the nucleation of protein aggregation. In contrast, high Hsp42 levels reduce local concentrations of misfolded proteins in the respective Hsp42–substrate complexes, thereby hampering the nucleation step and reducing protein aggregation. These seemingly opposing effects are shifted to lower concentrations for Hsp42ΔIDD, consistent with its superior chaperone and aggregase activity (Figs. 6 and 7). Increased numbers of CytoQ in the presence of Hsp42ΔIDD in yeast cells implies that the Hsp42/substrate ratio in vivo is low. Deregulation of Hsp42ΔIDD activity might therefore be sufficient to explain the increase in CytoQ numbers in yeast cells. We additionally show that mCherry-VHL foci formed in Hsp42ΔIDD–expressing yeast cells are more slowly removed during a recovery phase (Fig. S2 E). We did not observe a detrimental effect of Hsp42ΔIDD on Hsp70-Hsp100–mediated refolding of complexed MDH in vitro (Fig. S3 B). This excludes an inhibitory effect of Hsp42ΔIDD on disaggregating chaperones and leaves the molecular basis of aggregate stabilization in vivo unresolved.
Importantly, we demonstrated that deregulation of Hsp42 chaperone activity by IDD deletion decreases cellular fitness during stress application (Fig. 8). Next to CytoQ formation, the numbers and stability of CytoQs, which are both altered in Hsp42ΔIDD–expressing cells, are also crucial for cellular protection. We suggest that Hsp42ΔIDD hyperactivity might cause too extensive sequestration of cellular proteins upon stress or lead to exhaustive depletion of protein quality control components that bind to CytoQs. Alternatively, increased CytoQ stability will hamper recovery of sequestered proteins and can affect growth restart during recovery periods.
Collectively, we define the crucial function of the Hsp42 PrLD and its regulation by a second disordered N-terminal subdomain (IDD) in sequestering misfolded proteins into large protein aggregates. The Hsp42 PrLD has dual activities and combines the ability for self-interaction with substrate binding, expanding the function of PrLDs in the formation of stress-induced macromolecular assemblies.
Materials and methods
Yeast strains and growth conditions
All S. cerevisiae strains used in this study are derived from BY4741 and are listed in Table S1. FLAG-tagged versions of Hsp42 WT and deletion mutants were integrated into the native genomic locus of hsp42Δ cells. Yeast cultures were cultivated in liquid Synthetic Complete media (1.7 g/liter yeast nitrogen base without ammonium sulfate with 1 g/liter monosodium glutamic acid and 2% glucose or 2% galactose and 2% raffinose supplemented with a complete or appropriate mixture of amino acids) at 30°C or at the indicated conditions. The corresponding solid media contained 2% (wt/vol) agar. To select for resistance to the antibiotic, they were added to the final concentration of 300 µg/ml (geneticin), 100 µg/ml (nourseothricin), or 300 µg/ml (hygromycin).
Proteins
If not stated otherwise, all proteins were produced in derivatives of Escherichia coli strain MC4100. Ssa1 and Sse1 were expressed with an N-terminal His6-Smt3 tag. The fusion proteins were purified by affinity chromatography using an Ni-IDA Sepharose matrix (MACHERY-NAGEL) according to the manufacturer’s protocol. Fused Smt3 was cleaved off by Ulp1 treatment. Ssa1 and Sse1 were separated from Smt3 and Ulp1 by size exclusion using a Superdex200 16/60 column (GE Healthcare). His6-tagged MDH, Sis1, and MDH-YFP were purified by affinity chromatography using an Ni-IDA Sepharose matrix according to the manufacturer’s protocol. MDH-YFP was further purified by size exclusion using a Superdex75 16/60 column (GE Healthcare). After Ni chromatography, Sis1-containing fractions were pooled and dialyzed, and then contaminants were separated on a ResourceQ column (GE Healthcare). His6-GroES and Hsp104-His6 were purified by affinity chromatography using an Ni-IDA Sepharose followed by size exclusion using a Superdex200 16/60 column. GroEL was purified by DEAE-Sepharose anion exchange chromatography followed by size exclusion using a Superdex200 16/60 column as also described previously (Weibezahn et al., 2004; Tessarz et al., 2008).
C-terminally FLAG-tagged Hsp42 WT and deletion mutants were cloned into pMal-c2E (New England Biolabs, Inc.), creating a N-terminally fused maltose-binding protein tag. The enterokinase cleavage site was changed to a PreScission cleavage site by site-directed mutagenesis, and the vector was transformed into ArcticExpress (Agilent Technologies). Cells were grown at 37°C to OD600 0.9, 0.5 mM IPTG was added, and protein was expressed at 13°C overnight. Cells were resuspended in buffer B (50 mM Tris-HCl, 200 mM NaCl, 2 mM DTT, and 10% glycerol), lysed, and centrifuged. The soluble extract was incubated with amylose resin (New England Biolabs, Inc.), and the manufacturer’s protocol was followed. Hsp42-containing fractions were pooled, and the maltose-binding protein tag was cleaved at 4°C overnight by PreScission protease (Sigma-Aldrich) followed by size exclusion using a Sephacryl S-300 HR 16/60 column (GE Healthcare) equilibrated in buffer B. Fractions containing Hsp42 were pooled and concentrated by dialysis against buffer B containing 20% (wt/vol) PEG 20,000.
His6-MDH and MDH-YFP were expressed from pDS56-MDH and pSU61 (lab collection) in E. coli XL1 blue placIq. Cells were grown at 37°C in 2× yeast extract tryptone–containing ampicillin (100 μg/ml) and spectinomycin (50 μg/ml). At OD600 0.6, cells were shifted to 20°C, and protein production was induced by adding 0.1 mM IPTG. Both proteins were purified by Ni-IDA chromatography followed by size exclusion using a Superdex75 16/60 column–equilibrated buffer A (50 mM Hepes, pH 7.6, 50 mM KCl, 5 mM MgCl2, and 2 mM DTT) supplemented with 10% (vol/vol) glycerol. MDH used for fluorescent labeling was purchased from Roche, and pyruvate kinase was obtained from Sigma-Aldrich.
Coimmunoprecipitation
Exponentially growing yeast cells were treated as indicated, harvested by centrifugation (1,300 g at 4°C for 2 min), resuspended in 1 ml TBS-I (50 mM Tris, pH 7.5, 150 mM NaCl, 1 mM DTT, and cOmplete EDTA-free protease inhibitors [Roche]), and snap-frozen in liquid nitrogen. Cells were pulverized by mixer milling (MM400; Retsch). Pulldown was subsequently performed with ANTI-FLAG M2 affinity gel (Sigma-Aldrich) according to the manufacturer’s instructions. Beads were prepared by washing two times with 1 ml TBS-I. Sedimentation of beads was always achieved by centrifugation at 5,000 g for 30 s. After centrifugation, the resin was allowed to settle for 2 min on ice before removal of supernatants. 500–600 μl of cell powder were thawed on ice and mixed with 500 μl TBS-I. These lysates were precleared by two centrifugation steps (370 g at 4°C for 5 min and 2,300 g at 4°C for 5 min). The cleared lysates were mixed with 40 μl equilibrated M2-FLAG resin and incubated for 2.5 h at RT on a turning wheel to allow binding. Beads were sedimented, and supernatants were removed and washed three times with 500 μl TBS and transferred to new reaction tubes. Bound protein was eluted by addition of 100 μl of 100-mM glycine/HCl, pH 3.5, and then was incubated for 10 min at RT. After sedimentation of the beads, supernatants were transferred into new reaction tubes and mixed with 10 μl of 0.5 M Tris, pH 7.4, and 1.5 M NaCl. Bound proteins were analyzed by SDS-PAGE and Western blotting.
Cell fractionation
Yeast cells grown at 30°C or heat shocked (37°C + MG132) in the absence or presence of CHX were harvested at 1,150 g for 5 min, washed with 1 ml of ice-cold 50 mM Tris/HCl, pH 8.5, and resuspended in 250 μl of 50-mM Tris/HCl, pH 8.5, and 500 mM NaCl supplemented with cOmplete protease inhibitor cocktail (Sigma-Aldrich). Cells were frozen in liquid nitrogen, and cell lysis was performed in 2 ml Eppendorf Safe-Lock tubes by mixer milling (MM 400; Retsch) at 30 Hz for 90 s and five cycles. The lysate was thawed and precleared at 3,000 g for 1 min at 4°C. The total cell lysate was decanted into a prechilled new reaction tube followed by separation of soluble and insoluble (P20) fractions at 16,000 g for 20 min at 4°C. The soluble fraction was removed and subjected to a second round of centrifugation at 100,000 g (P100) for 20 min at 4°C. Pellets isolated at 16,000 g (P20)/100,000 g (P100) were washed with 50 mM Tris, pH 8.5, and 150 mM NaCl supplemented with cOmplete protease inhibitor cocktail and spun down again for 20 min at the appropriate g. Pellets were resuspended in 50 mM Tris, pH 8.5, 150 mM NaCl, 8 M urea, 2% (wt/vol) SDS, and 2 mM DTT by shaking in a thermomixer (Eppendorf) at 37°C for 5 min. All fractions were mixed with 6× protein loading buffer (50 mM Tris/HCl, pH 6.8, 30% [vol/vol] glycerol, 10% [wt/vol] SDS, 5% β-mercaptoethanol, and 0.05% [wt/vol] Bromphenol blue). Distribution of mCherry-VHL and Hsp42 among soluble and insoluble fractions was monitored by SDS-PAGE and Western blotting.
Western blotting
FLAG tag (clone M2) and myc tag (clone 9E10) antibodies were obtained from Sigma-Aldrich and used at 1:1,000 dilution. Actin antibody was obtained from Merck (clone C4) and used at 1:1,650 dilution. Polyclonal antibodies against MDH, Hsp26, Hsp42, and mCherry were made by Davids Biotechnology using purified proteins and used at the following dilutions: α-MDH, 1:1,000; α-Hsp42, 1:4,000/1:10,000; and α-mCherry, 1:5,000. Antibody specificity was documented by use of purified proteins or yeast knockout cells.
Hydrogen deuterium exchange experiments
Native His6-MDH (2 µM and 400 µl), thermally aggregated His6-MDH, or sHsp/His6–MDH complexes (both formed for 30 min at 47°C) in buffer A (50 mM Hepes, pH 7.6, 50 mM KCl, 5 mM MgCl2, and 2 mM DTT) were incubated with 50 µl MagneHis Ni particles (Promega) for 15 min at RT. His6-MDH (aggregated or sHsp-bound) was isolated by placing the reaction in a magnetic rack. The supernatant was subsequently removed, and the beads were washed once with buffer A. D2O-based buffer A was added to initiate amide proton–deuteron exchange. After 30 s, the exchange reaction was quenched by adding ice-cold low-pH quench buffer (500 mM K-phosphate buffer, pH 2.2) containing pepsin (25 µg/ml; Roche). Protein was digested from the Ni particles for 1 min on ice. Then, quenched, digested samples were injected into the HPLC setup with online peptic digest and analyzed on an electrospray ionization quadrupole time-of-flight mass spectrometer (QSTAR Pulsar; Applied Biosystems) as previously described (Rist et al., 2003). Analysis of deuteron incorporation into peptides was performed by using AnalystQS software (MDS SCIEX; Applied Biosystems). The assignment of the isotope peaks and the selection of the peptides presented were done manually. Analysis of bimodal isotope distributions was performed as described previously (Ungelenk et al., 2016).
Aggregation assays
Light scattering
For light-scattering measurements, 0.5 µM MDH in buffer A was denatured at 47°C or 41°C in the absence or presence of various Hsp42 (WT and deletion mutants) concentrations. Turbidity was measured at an excitation and emission wavelength of 550 nm (47°C) or 600 nm (41°C), respectively, using the LS50B fluorescence spectrometer (PerkinElmer).
Light microscopy
To document the morphology of MDH aggregates in vitro, 2 µM MDH and Hsp42 WT or deletion mutants were mixed in buffer A and incubated at 41°C or 47°C. 2 µl of the sample were inspected on an Olympus IX81 microscope using a Plan Apochromat 100× 1.45 NA oil objective at indicated time points, and differential interference contrast images were taken.
Disaggregation and refolding of thermally aggregated MDH
MDH (0.5 μM) was denatured for 30 min at 47°C in buffer A in the absence or presence of Hsp42 (WT or deletion mutants). Protein disaggregation and refolding was started by diluting aggregated MDH or Hsp42–MDH complexes and chaperones 1:1 (2 µM Ssa1, 1 µM Sis1, 0.1 µM Sse1, 1 µM Hsp104, 1 µM GroEL, and 1 µM GroES) in buffer A containing 0.1 mg/ml BSA and an ATP-regenerating system (3 mM phosphoenolpyruvate, 20 μg/ml pyruvate kinase, and 2 mM ATP) at 30°C. The reactivation of MDH was monitored as published previously (Mogk et al., 2003) using a Novaspec Plus spectrophotometer (GE Healthcare).
FRET during thermal aggregation of MDH
MDH (Roche) was labeled with 7-diethylcoumarin-3-carboxylic acid succinimidyl ester (Molecular Probes) according to the manufacturer’s protocol. The labeled MDH and a C-terminally YFP-tagged MDH variant (each 0.25 µM) were mixed in preheated buffer A, and the FRET signal was recorded at 527 nm in an LS50B fluorescence spectrometer at 47°C.
Subunit exchange of Hsp42
Hsp42 WT and Hsp42ΔIDD were labeled in buffer A with 7-diethylaminocoumarin-3-carboxylic-acid succinimidyl ester (SE) and succinimidyl NBD-X (Thermo Fisher Scientific) for 2 h at RT according to the manufacturer’s protocol. Unreacted dyes were separated by PD10 columns and subsequent dialysis against buffer A. Donor- and acceptor-labeled proteins (each 1 µM) were mixed and incubated at 25°C overnight. A 10-fold excess of unlabeled protein was added, and fluorescence spectra were recorded at 30°C (excitation at 420 nm) using an LS55 fluorescence spectrometer. The observed increase of donor fluorescence with time was fitted using an exponential two-phase association equation in Prism5 (GraphPad Software).
Binding of ANS
10 µM Hsp42 WT or deletion mutants were mixed in PBS buffer with 1 mM ANS and incubated for 15 min at 30°C. Fluorescence spectra were recorded (excitation at 424 nm) using an LS55 fluorescence spectrometer.
BPIA cross-linking
Hsp42-Y11C/C127A was dialyzed against buffer A lacking DTT. Next, a 10-fold excess of BPIA (100 mM stock in DMSO) was added and incubated for 2 h at RT. The reaction was quenched by addition of 2 mM β-mercaptoethanol for 15 min, and unreacted BPIA was removed by dialysis overnight. Cross-linking was performed using 2.5 µM Hsp42-Y11C/C127A and 0.5 µM MDH incubated for 30 min at 30°C or 47°C. Samples were irradiated with UV light (365 nm at 100 W) for 10 min on ice. Cross-link products were analyzed by immunoblot analysis using MDH- and Hsp42-specific antibodies.
Static light scattering
Molar masses of Hsp42 WT and deletion mutants were determined by static light scattering measurements performed at RT using a miniDawn instrument coupled with WTC-0305N5 (Wyatt Technology) size-exclusion chromatography in buffer A. 50 µM protein solutions were injected, and molar masses were determined by Astra software (Wyatt Technology).
Transmission negative-stain electron microscopy
Heat-induced aggregates of MDH alone or Hsp42-complexed state were formed as described in the Aggregation assays section. Images were recorded using an EM900 microscope (ZEISS).
Competition assays
S. cerevisiae WT cells expressing BFP and respective mutant cells expressing GFP were grown to mid-log phase. Equal amounts of both cultures were mixed and diluted to OD600 0.05. The mixed culture was split into two tubes (halves), and both were grown for 7 d with daily dilutions to OD600 0.05. One culture was constantly grown at 30°C, and the other culture was subjected to temperature cycles switching between 1 h at 25°C and 1 h at 43°C. Each day, the proportion of WT and mutant cells was measured by monitoring the fraction of GFP- and BFP-positive cells by FACS analysis (FACSCanto II; BD).
Live-cell fluorescence microscopy
To perform live-cell imaging, overnight yeast cultures were diluted into fresh medium to an OD600 of 0.1–0.15, further cultivated at 30°C to the exponential growth phase of OD600 = 0.5–0.6, and treated as indicated. An expression of mCherry-VHL was inhibited by an addition of glucose (2% final concentration) before an application of stress conditions unless stated otherwise. To inhibit global protein synthesis, CHX was added to a final concentration of 50 µg/ml. MG132 from PeptaNova (3175-v) was used at a concentration of 80 µM. Samples were taken at indicated time points, mounted on a cover glass, and coated with a slice of 1.5% (wt/vol) agarose in Synthetic Complete media. All cells were inspected immediately after the preparation on an IX81 microscope equipped with a Plan Apochromat 100× 1.45 NA oil objective and an electron-multiplying charge-coupled device Hamamatsu camera. Acquired z stack images were deconvolved using Wiener filter (eXcellence software; Olympus) and further processed with ImageJ (National Institutes of Health) software. For the purpose of CytoQ/INQ quantification, a superposition of either mCherry-VHL or Hsp104-GFP foci with the nuclear marker Htb1 was used. A focus was assigned to be INQ when overlapped or laying in the close proximity of Htb1 signal.
Immunofluorescence
For immunofluorescence imaging, cells were treated as indicated, prefixed by formaldehyde in 100 mM phosphate buffer, pH 6.5 (3.7% final concentration), and subsequently fixed by paraformaldehyde in 100 mM phosphate buffer, pH 6.5 (4% final concentration). The cell wall was digested using 500 μg/ml zymolyase T-100 in wash buffer (1.2 M sorbitol, 20 mM β-mercaptoethanol, and 100 mM phosphate buffer, pH 6.5), and spheroplasts were finally permeabilized by 1% Triton X-100 in 100 mM phosphate buffer, pH 6.5. Samples were blocked by 1% (wt/vol) BSA in 100 mM phosphate buffer, pH 6.5, and incubated with respective primary (1:500 anti-FLAG from Sigma-Aldrich; 1:1,000 anti-Nsp1 from EnCor Bio.; and 1:2,000 anti-Hsp42 from Bukau Lab) and secondary (1;1,000 Alexa Fluor 488; Invitrogen) antibodies. DNA was stained with 50 ng/ml DAPI or 20 μg/ml Hoechst 33442. Samples were inspected on an IX81 microscope equipped with a Plan Apochromat 100× 1.45 NA oil objective and an electron-multiplying charge-coupled device Hamamatsu camera. Acquired z stack images were deconvolved using Wiener filter and further processed with ImageJ software.
Online supplemental material
Fig. S1 demonstrates that protein aggregation depends on translation. Fig. S2 reports the analysis of Hsp42 deletion mutants. Fig. S3 shows that PrLD is the major Hsp42 substrate interaction site. Fig. S4 shows that Hsp42ΔIDD exhibits superior chaperone activity. Fig. S5 describes the subunit exchange kinetics of Hsp42 WT and Hsp42ΔIDD and effects on CytoQ formation and cell viability. Table S1 shows a detailed description of yeast strains used in this study.
Supplementary Material
Acknowledgments
We thank Thomas Ruppert for valuable support.
S. Ungelenk, S. Miller, and C. Ho were supported by the Universität Heidelberg Hartmut Hoffmann-Berling International Graduate School of Molecular and Cellular Biology. S. Ungelenk was additionally supported by a fellowship of the Verband der Chemischen Industrie Fonds der Chemischen Industrie, and S. Miller was supported by a fellowship of the Boehringer Ingelheim Fonds. This work was supported by the Deutsche Forschungsgemeinschaft (SFB1036 projects A8 to A. Mogk and B. Bukau, A9 to M.P. Mayer, and Z1 to T. Ruppert).
The authors declare no competing financial interests.
Author contributions: conceived and designed experiments: T. Grousl, S. Ungelenk, S. Miller, C.-T. Ho, M. Khokhrina, M.P. Mayer, B. Bukau, and A. Mogk. Performed experiments: T. Grousl, S. Ungelenk, S. Miller, C.-T. Ho, and M. Khokhrina. Analyzed the data: T. Grousl, S. Ungelenk, S. Miller, C.-T. Ho, M. Khokhrina, M.P. Mayer, B. Bukau, and A. Mogk. Wrote the manuscript: B. Bukau and A. Mogk.
References
- Alberti S., Halfmann R., King O., Kapila A., and Lindquist S.. 2009. A systematic survey identifies prions and illuminates sequence features of prionogenic proteins. Cell. 137:146–158. 10.1016/j.cell.2009.02.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balchin D., Hayer-Hartl M., and Hartl F.U.. 2016. In vivo aspects of protein folding and quality control. Science. 353:aac4354 10.1126/science.aac4354 [DOI] [PubMed] [Google Scholar]
- Basha E., Friedrich K.L., and Vierling E.. 2006. The N-terminal arm of small heat shock proteins is important for both chaperone activity and substrate specificity. J. Biol. Chem. 281:39943–39952. 10.1074/jbc.M607677200 [DOI] [PubMed] [Google Scholar]
- Chen B., Retzlaff M., Roos T., and Frydman J.. 2011. Cellular strategies of protein quality control. Cold Spring Harb. Perspect. Biol. 3:a004374 10.1101/cshperspect.a004374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherkasov V., Grousl T., Theer P., Vainshtein Y., Glässer C., Mongis C., Kramer G., Stoecklin G., Knop M., Mogk A., and Bukau B.. 2015. Systemic control of protein synthesis through sequestration of translation and ribosome biogenesis factors during severe heat stress. FEBS Lett. 589:3654–3664. 10.1016/j.febslet.2015.10.010 [DOI] [PubMed] [Google Scholar]
- Decker C.J., Teixeira D., and Parker R.. 2007. Edc3p and a glutamine/asparagine-rich domain of Lsm4p function in processing body assembly in Saccharomyces cerevisiae. J. Cell Biol. 179:437–449. 10.1083/jcb.200704147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escusa-Toret S., Vonk W.I., and Frydman J.. 2013. Spatial sequestration of misfolded proteins by a dynamic chaperone pathway enhances cellular fitness during stress. Nat. Cell Biol. 15:1231–1243. 10.1038/ncb2838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu X., Shi X., Yin L., Liu J., Joo K., Lee J., and Chang Z.. 2013. Small heat shock protein IbpB acts as a robust chaperone in living cells by hierarchically activating its multi-type substrate-binding residues. J. Biol. Chem. 288:11897–11906. 10.1074/jbc.M113.450437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallina I., Colding C., Henriksen P., Beli P., Nakamura K., Offman J., Mathiasen D.P., Silva S., Hoffmann E., Groth A., et al. 2015. Cmr1/WDR76 defines a nuclear genotoxic stress body linking genome integrity and protein quality control. Nat. Commun. 6:6533 10.1038/ncomms7533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasymov O.K., and Glasgow B.J.. 2007. ANS fluorescence: potential to augment the identification of the external binding sites of proteins. Biochim. Biophys. Acta. 1774:403–411. 10.1016/j.bbapap.2007.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilks N., Kedersha N., Ayodele M., Shen L., Stoecklin G., Dember L.M., and Anderson P.. 2004. Stress granule assembly is mediated by prion-like aggregation of TIA-1. Mol. Biol. Cell. 15:5383–5398. 10.1091/mbc.E04-08-0715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haslbeck M., and Vierling E.. 2015. A first line of stress defense: small heat shock proteins and their function in protein homeostasis. J. Mol. Biol. 427:1537–1548. 10.1016/j.jmb.2015.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haslbeck M., Braun N., Stromer T., Richter B., Model N., Weinkauf S., and Buchner J.. 2004a Hsp42 is the general small heat shock protein in the cytosol of Saccharomyces cerevisiae. EMBO J. 23:638–649. 10.1038/sj.emboj.7600080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haslbeck M., Ignatiou A., Saibil H., Helmich S., Frenzl E., Stromer T., and Buchner J.. 2004b A domain in the N-terminal part of Hsp26 is essential for chaperone function and oligomerization. J. Mol. Biol. 343:445–455. 10.1016/j.jmb.2004.08.048 [DOI] [PubMed] [Google Scholar]
- Haslbeck M., Miess A., Stromer T., Walter S., and Buchner J.. 2005. Disassembling protein aggregates in the yeast cytosol. The cooperation of Hsp26 with Ssa1 and Hsp104. J. Biol. Chem. 280:23861–23868. 10.1074/jbc.M502697200 [DOI] [PubMed] [Google Scholar]
- Hill S.M., Hao X., Grönvall J., Spikings-Nordby S., Widlund P.O., Amen T., Jörhov A., Josefson R., Kaganovich D., Liu B., and Nyström T.. 2016. Asymmetric Inheritance of Aggregated Proteins and Age Reset in Yeast Are Regulated by Vac17-Dependent Vacuolar Functions. Cell Reports. 16:826–838. 10.1016/j.celrep.2016.06.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain S., Wheeler J.R., Walters R.W., Agrawal A., Barsic A., and Parker R.. 2016. ATPase-Modulated Stress Granules Contain a Diverse Proteome and Substructure. Cell. 164:487–498. 10.1016/j.cell.2015.12.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaya N., Garcia V., and Vierling E.. 2009. Substrate binding site flexibility of the small heat shock protein molecular chaperones. Proc. Natl. Acad. Sci. USA. 106:15604–15609. 10.1073/pnas.0902177106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaganovich D., Kopito R., and Frydman J.. 2008. Misfolded proteins partition between two distinct quality control compartments. Nature. 454:1088–1095. 10.1038/nature07195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriehuber T., Rattei T., Weinmaier T., Bepperling A., Haslbeck M., and Buchner J.. 2010. Independent evolution of the core domain and its flanking sequences in small heat shock proteins. FASEB J. 24:3633–3642. 10.1096/fj.10-156992 [DOI] [PubMed] [Google Scholar]
- Liu I.C., Chiu S.W., Lee H.Y., and Leu J.Y.. 2012. The histone deacetylase Hos2 forms an Hsp42-dependent cytoplasmic granule in quiescent yeast cells. Mol. Biol. Cell. 23:1231–1242. 10.1091/mbc.E11-09-0752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackenzie R.J., Lawless C., Holman S.W., Lanthaler K., Beynon R.J., Grant C.M., Hubbard S.J., and Eyers C.E.. 2016. Absolute protein quantification of the yeast chaperome under conditions of heat shock. Proteomics. 16:2128–2140. 10.1002/pmic.201500503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malinovska L., Kroschwald S., Munder M.C., Richter D., and Alberti S.. 2012. Molecular chaperones and stress-inducible protein-sorting factors coordinate the spatiotemporal distribution of protein aggregates. Mol. Biol. Cell. 23:3041–3056. 10.1091/mbc.E12-03-0194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malinovska L., Kroschwald S., and Alberti S.. 2013. Protein disorder, prion propensities, and self-organizing macromolecular collectives. Biochim. Biophys. Acta. 1834:918–931. 10.1016/j.bbapap.2013.01.003 [DOI] [PubMed] [Google Scholar]
- March Z.M., King O.D., and Shorter J.. 2016. Prion-like domains as epigenetic regulators, scaffolds for subcellular organization, and drivers of neurodegenerative disease. Brain Res. 1647:9–18. 10.1016/j.brainres.2016.02.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall R.S., McLoughlin F., and Vierstra R.D.. 2016. Autophagic Turnover of Inactive 26S Proteasomes in Yeast Is Directed by the Ubiquitin Receptor Cue5 and the Hsp42 Chaperone. Cell Reports. 16:1717–1732. 10.1016/j.celrep.2016.07.015 [DOI] [PubMed] [Google Scholar]
- Miller S.B., Ho C.T., Winkler J., Khokhrina M., Neuner A., Mohamed M.Y., Guilbride D.L., Richter K., Lisby M., Schiebel E., et al. 2015. Compartment-specific aggregases direct distinct nuclear and cytoplasmic aggregate deposition. EMBO J. 34:778–797. 10.15252/embj.201489524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mogk A., and Bukau B.. 2017. Role of sHsps in organizing cytosolic protein aggregation and disaggregation. Cell Stress Chaperones. 22:493–502. 10.1007/s12192-017-0762-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mogk A., Schlieker C., Friedrich K.L., Schönfeld H.-J., Vierling E., and Bukau B.. 2003. Refolding of substrates bound to small Hsps relies on a disaggregation reaction mediated most efficiently by ClpB/DnaK. J. Biol. Chem. 278:31033–31042. 10.1074/jbc.M303587200 [DOI] [PubMed] [Google Scholar]
- Molliex A., Temirov J., Lee J., Coughlin M., Kanagaraj A.P., Kim H.J., Mittag T., and Taylor J.P.. 2015. Phase separation by low complexity domains promotes stress granule assembly and drives pathological fibrillization. Cell. 163:123–133. 10.1016/j.cell.2015.09.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nott T.J., Petsalaki E., Farber P., Jervis D., Fussner E., Plochowietz A., Craggs T.D., Bazett-Jones D.P., Pawson T., Forman-Kay J.D., and Baldwin A.J.. 2015. Phase transition of a disordered nuage protein generates environmentally responsive membraneless organelles. Mol. Cell. 57:936–947. 10.1016/j.molcel.2015.01.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S.H., Kukushkin Y., Gupta R., Chen T., Konagai A., Hipp M.S., Hayer-Hartl M., and Hartl F.U.. 2013. PolyQ proteins interfere with nuclear degradation of cytosolic proteins by sequestering the Sis1p chaperone. Cell. 154:134–145. 10.1016/j.cell.2013.06.003 [DOI] [PubMed] [Google Scholar]
- Peters L.Z., Karmon O., David-Kadoch G., Hazan R., Yu T., Glickman M.H., and Ben-Aroya S.. 2015. The protein quality control machinery regulates its misassembled proteasome subunits. PLoS Genet. 11:e1005178 10.1371/journal.pgen.1005178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prilusky J., Felder C.E., Zeev-Ben-Mordehai T., Rydberg E.H., Man O., Beckmann J.S., Silman I., and Sussman J.L.. 2005. FoldIndex: a simple tool to predict whether a given protein sequence is intrinsically unfolded. Bioinformatics. 21:3435–3438. 10.1093/bioinformatics/bti537 [DOI] [PubMed] [Google Scholar]
- Protter D.S., and Parker R.. 2016. Principles and Properties of Stress Granules. Trends Cell Biol. 26:668–679. 10.1016/j.tcb.2016.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabouille C., and Alberti S.. 2017. Cell adaptation upon stress: the emerging role of membrane-less compartments. Curr. Opin. Cell Biol. 47:34–42. 10.1016/j.ceb.2017.02.006 [DOI] [PubMed] [Google Scholar]
- Rajagopal P., Tse E., Borst A.J., Delbecq S.P., Shi L., Southworth D.R., and Klevit R.E.. 2015. A conserved histidine modulates HSPB5 structure to trigger chaperone activity in response to stress-related acidosis. eLife. 4:e07304 10.7554/eLife.07304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reijns M.A., Alexander R.D., Spiller M.P., and Beggs J.D.. 2008. A role for Q/N-rich aggregation-prone regions in P-body localization. J. Cell Sci. 121:2463–2472. 10.1242/jcs.024976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riback J.A., Katanski C.D., Kear-Scott J.L., Pilipenko E.V., Rojek A.E., Sosnick T.R., and Drummond D.A.. 2017. Stress-Triggered Phase Separation Is an Adaptive, Evolutionarily Tuned Response. Cell. 168:1028–1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rist W., Jørgensen T.J., Roepstorff P., Bukau B., and Mayer M.P.. 2003. Mapping temperature-induced conformational changes in the Escherichia coli heat shock transcription factor sigma 32 by amide hydrogen exchange. J. Biol. Chem. 278:51415–51421. 10.1074/jbc.M307160200 [DOI] [PubMed] [Google Scholar]
- Saarikangas J., and Barral Y.. 2015. Protein aggregates are associated with replicative aging without compromising protein quality control. eLife. 4:e06197 10.7554/eLife.06197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seyffer F., Kummer E., Oguchi Y., Winkler J., Kumar M., Zahn R., Sourjik V., Bukau B., and Mogk A.. 2012. Hsp70 proteins bind Hsp100 regulatory M domains to activate AAA+ disaggregase at aggregate surfaces. Nat. Struct. Mol. Biol. 19:1347–1355. 10.1038/nsmb.2442 [DOI] [PubMed] [Google Scholar]
- Song J., Yang Q., Yang J., Larsson L., Hao X., Zhu X., Malmgren-Hill S., Cvijovic M., Fernandez-Rodriguez J., Grantham J., et al. 2014. Essential genetic interactors of SIR2 required for spatial sequestration and asymmetrical inheritance of protein aggregates. PLoS Genet. 10:e1004539 10.1371/journal.pgen.1004539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sontag E.M., Samant R.S., and Frydman J.. 2017. Mechanisms and Functions of Spatial Protein Quality Control. Annu. Rev. Biochem. 86:97–122. 10.1146/annurev-biochem-060815-014616 [DOI] [PubMed] [Google Scholar]
- Specht S., Miller S.B., Mogk A., and Bukau B.. 2011. Hsp42 is required for sequestration of protein aggregates into deposition sites in Saccharomyces cerevisiae. J. Cell Biol. 195:617–629. 10.1083/jcb.201106037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stengel F., Baldwin A.J., Painter A.J., Jaya N., Basha E., Kay L.E., Vierling E., Robinson C.V., and Benesch J.L.. 2010. Quaternary dynamics and plasticity underlie small heat shock protein chaperone function. Proc. Natl. Acad. Sci. USA. 107:2007–2012. 10.1073/pnas.0910126107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tessarz P., Mogk A., and Bukau B.. 2008. Substrate threading through the central pore of the Hsp104 chaperone as a common mechanism for protein disaggregation and prion propagation. Mol. Microbiol. 68:87–97. 10.1111/j.1365-2958.2008.06135.x [DOI] [PubMed] [Google Scholar]
- Tkach J.M., Yimit A., Lee A.Y., Riffle M., Costanzo M., Jaschob D., Hendry J.A., Ou J., Moffat J., Boone C., et al. 2012. Dissecting DNA damage response pathways by analysing protein localization and abundance changes during DNA replication stress. Nat. Cell Biol. 14:966–976. 10.1038/ncb2549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toombs J.A., McCarty B.R., and Ross E.D.. 2010. Compositional determinants of prion formation in yeast. Mol. Cell. Biol. 30:319–332. 10.1128/MCB.01140-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treweek T.M., Meehan S., Ecroyd H., and Carver J.A.. 2015. Small heat-shock proteins: important players in regulating cellular proteostasis. Cell. Mol. Life Sci. 72:429–451. 10.1007/s00018-014-1754-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyedmers J., Mogk A., and Bukau B.. 2010. Cellular strategies for controlling protein aggregation. Nat. Rev. Mol. Cell Biol. 11:777–788. 10.1038/nrm2993 [DOI] [PubMed] [Google Scholar]
- Ungelenk S., Moayed F., Ho C.T., Grousl T., Scharf A., Mashaghi A., Tans S., Mayer M.P., Mogk A., and Bukau B.. 2016. Small heat shock proteins sequester misfolding proteins in near-native conformation for cellular protection and efficient refolding. Nat. Commun. 7:13673 10.1038/ncomms13673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Lee R., Buljan M., Lang B., Weatheritt R.J., Daughdrill G.W., Dunker A.K., Fuxreiter M., Gough J., Gsponer J., Jones D.T., et al. 2014. Classification of intrinsically disordered regions and proteins. Chem. Rev. 114:6589–6631. 10.1021/cr400525m [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace E.W., Kear-Scott J.L., Pilipenko E.V., Schwartz M.H., Laskowski P.R., Rojek A.E., Katanski C.D., Riback J.A., Dion M.F., Franks A.M., et al. 2015. Reversible, Specific, Active Aggregates of Endogenous Proteins Assemble upon Heat Stress. Cell. 162:1286–1298. 10.1016/j.cell.2015.08.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weibezahn J., Tessarz P., Schlieker C., Zahn R., Maglica Z., Lee S., Zentgraf H., Weber-Ban E.U., Dougan D.A., Tsai F.T., et al. 2004. Thermotolerance requires refolding of aggregated proteins by substrate translocation through the central pore of ClpB. Cell. 119:653–665. 10.1016/j.cell.2004.11.027 [DOI] [PubMed] [Google Scholar]
- Wickner R.B., Shewmaker F.P., Bateman D.A., Edskes H.K., Gorkovskiy A., Dayani Y., and Bezsonov E.E.. 2015. Yeast prions: structure, biology, and prion-handling systems. Microbiol. Mol. Biol. Rev. 79:1–17. 10.1128/MMBR.00041-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou C., Slaughter B.D., Unruh J.R., Guo F., Yu Z., Mickey K., Narkar A., Ross R.T., McClain M., and Li R.. 2014. Organelle-based aggregation and retention of damaged proteins in asymmetrically dividing cells. Cell. 159:530–542. 10.1016/j.cell.2014.09.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.