Despite their shared ability to regulate microtubule polymerization dynamics, kinesin-4 motors display dramatically different motility properties ranging from fast processive motility to no movement. Yue et al. demonstrate that for KIF7 and KIF27, altered chemomechanical coupling results in immotile behavior and slow processive movement, respectively.
Abstract
Kinesin-4 motors play important roles in cell division, microtubule organization, and signaling. Understanding how motors perform their functions requires an understanding of their mechanochemical and motility properties. We demonstrate that KIF27 can influence microtubule dynamics, suggesting a conserved function in microtubule organization across the kinesin-4 family. However, kinesin-4 motors display dramatically different motility characteristics: KIF4 and KIF21 motors are fast and processive, KIF7 and its Drosophila melanogaster homologue Costal2 (Cos2) are immotile, and KIF27 is slow and processive. Neither KIF7 nor KIF27 can cooperate for fast processive transport when working in teams. The mechanistic basis of immotile KIF7 behavior arises from an inability to release adenosine diphosphate in response to microtubule binding, whereas slow processive KIF27 behavior arises from a slow adenosine triphosphatase rate and a high affinity for both adenosine triphosphate and microtubules. We suggest that evolutionarily selected sequence differences enable immotile KIF7 and Cos2 motors to function not as transporters but as microtubule-based tethers of signaling complexes.
Introduction
Microtubule-based motors of the kinesin superfamily play essential roles in cell division, cell motility, intracellular trafficking, control of microtubule dynamics, and ciliary function (Hirokawa et al., 2009; Verhey and Hammond, 2009). Kinesins are defined by the presence of a kinesin motor domain and use divergent sequences outside of this domain for each motor’s unique regulatory and functional outputs. However, recent work demonstrated that sequence differences within the core motor domain are also critical for the specific functions of each kinesin motor. Indeed, substituting a kinesin’s motor domain with one from a different family cannot replicate the functional output of that kinesin (Kim et al., 2014; Ravindran et al., 2017).
Understanding how patterns of residue conservation and divergence within protein families relate to the evolution of distinct functional properties is of wide biological significance. For actin-based motors of the myosin superfamily (Richards and Cavalier-Smith, 2005), the core myosin motor domain was thought to endow myosins with equivalent chemomechanical properties to that of the “conventional” myosin-2, with family-specific tail domains giving rise to specific cellular functions. Yet recent work has demonstrated that sequence divergence within the myosin motor domain itself dictates alternative functions as force sensors, tethers, and F-actin organizers (Woolner and Bement, 2009).
The kinesin-4 family of motors provides a unique opportunity to understand how sequence divergence of a core domain leads to different functional outputs. The best-known family members, mammalian KIF4 and Xenopus laevis Xklp1, undergo typical kinesin-type processive motility and suppress microtubule dynamics upon reaching the plus end (Bringmann et al., 2004; Bieling et al., 2010; Subramanian et al., 2013). The mammalian KIF21A and KIF21B kinesins also undergo plus end–directed motion and suppress microtubule dynamics (Huang and Banker, 2012; van der Vaart et al., 2013; Cheng et al., 2014; Bianchi et al., 2016; Ghiretti et al., 2016; Muhia et al., 2016; van Riel et al., 2017). In contrast, the Arabidopsis thaliana motor FRA1 undergoes processive motility and mediates trafficking of cell wall material along cortical microtubules but does not alter plus end dynamics or microtubule organization (Zhu and Dixit, 2011; Kong et al., 2015; Zhu et al., 2015; Ganguly et al., 2017), whereas the mammalian motor KIF7 displays no microtubule-based motility but is able to suppress microtubule dynamics (He et al., 2014). The Drosophila melanogaster motor Costal2 (Cos2) has been suggested to be a homologue of KIF7, and the mammalian motor KIF27 has been suggested to be a paralog, but their motility properties and effects on microtubule dynamics have not been determined.
In this study, we systematically analyzed the motility properties of members of the kinesin-4 family using in vitro and cellular assays. We find that MmKIF27 is a slow processive motor, whereas MmKIF7 and DmCos2 show no motility at the single molecule level. Furthermore, we show that KIF27 and KIF7 are not able to cooperate for enhanced motility in a multimotor context. Unlike "conventional" kinesin motors, which alter their microtubule binding in response to their nucleotide state, KIF27 and KIF7 show tight microtubule binding regardless of nucleotide condition. The mechanistic basis of KIF27’s slow processive motility is found in its slow ATPase rate and its high affinity for both ATP and microtubules. The immotile behavior of KIF7 is explained by its defective chemomechanical coupling: KIF7 is unable to release ADP in response to microtubule binding. We suggest that differences in intrinsic catalytic domain behavior (motility, kinetics, and chemomechanical coupling) are derived from variances in catalytic domain sequence and allow kinesin-4 motors to carry out distinct cellular functions as transporters or tethers.
Results
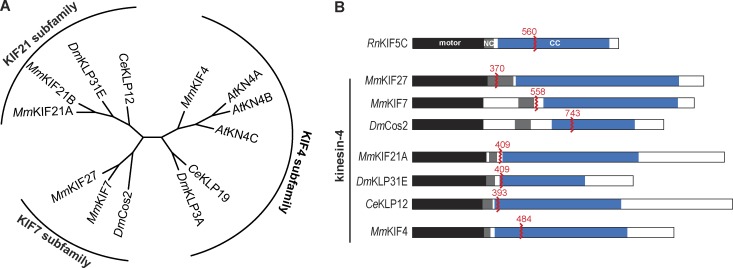
The kinesin-4 family can be divided into three subfamilies in mammals
We first set out to define the phylogenetic relationship between members of the kinesin-4 family. We focused our analysis on kinesin-4 motors in commonly studied genetic organisms (fly and worm) and/or for which motility data exist (Figs. 1 A and S1). This analysis suggests that the kinesin-4 family can be divided into three subfamilies, referred to in this study by the best-known mammalian member of each subfamily. The KIF7 subfamily contains two paralogous motors from the mammalian genome (MmKIF7 and MmKIF27) and one motor in flies (DmCos2). The KIF21 subfamily contains two paralogous motors from the mouse genome (MmKIF21A and MmKIF21B) and one member each from the fly (DmKLP31E) and worm (CeKLP12) genomes. The KIF4 subfamily contains one motor each from mammalian (MmKIF4), fly (DmKLP3A), and worm (CeKLP19) genomes and three motors in Arabidopsis (AtKN4A, also known as FRA1, AtKN4B, and AtKN4C). Although CeKLP19 and DmKLP3A are separated from the rest of the KIF4 subfamily in our unrooted tree, we place these motors in this subfamily based on the following points. Our analysis used kinesin sequences trimmed to the core motor domain, similar to the analysis of Wickstead et al. (2010), whereas a previous phylogenetic analysis that included sequences of each motor’s neck region placed CeKLP19 as most similar to KIF4 and DmKLP3A (Powers et al., 2004). In addition, a recent study used a maximum likelihood coestimating algorithm that performs multiple sequence alignment (MSA) and phylogeny calculations in tandem and placed both CeKLP19 and DmKLP3A in the KIF4 subfamily (Richard et al., 2016).
Figure 1.
Members of the kinesin-4 family. (A) Phylogenetic tree of selected members of the kinesin-4 family. (B) Domain organization of the kinesin-4 motors analyzed in this study. Black, motor domain; blue, coiled-coil (CC); gray, neck coil (NC). The red zigzags indicate the position of truncation, and the red numbers indicate the last aa in the truncated protein. The motility of the kinesin-1 motor RnKIF5C was used as control in this study. Rn, Rattus norvegicus; Mm, Mus musculus; Dm, Drosophila melanogaster; Ce, Caenorhabditis elegans; At, Arabidopsis thaliana.
The KIF27 motor domain can regulate microtubule growth
To investigate whether KIF27 is a kinesin-4 motor capable of influencing microtubule dynamics, we purified a truncated Flag-tagged version of KIF27 and compared its activity to that of its closest relative, KIF7 (Fig. 1 B). For KIF27, we generated a truncated version containing the minimum neck coil segment but found that this construct (aa 1–370; Fig. 1 B) was not a stable dimer. We thus appended the leucine zipper (LZ) of GCN4, similar to a previous strategy with kinesin-3 family motors (Tomishige et al., 2002; Huckaba et al., 2011; Soppina et al., 2014), resulting in the construct KIF27(1–370)-LZ-Flag. For KIF7, we used a truncated version consisting of aa 1–558 (Fig. 1 B) based on previous work (He et al., 2014).
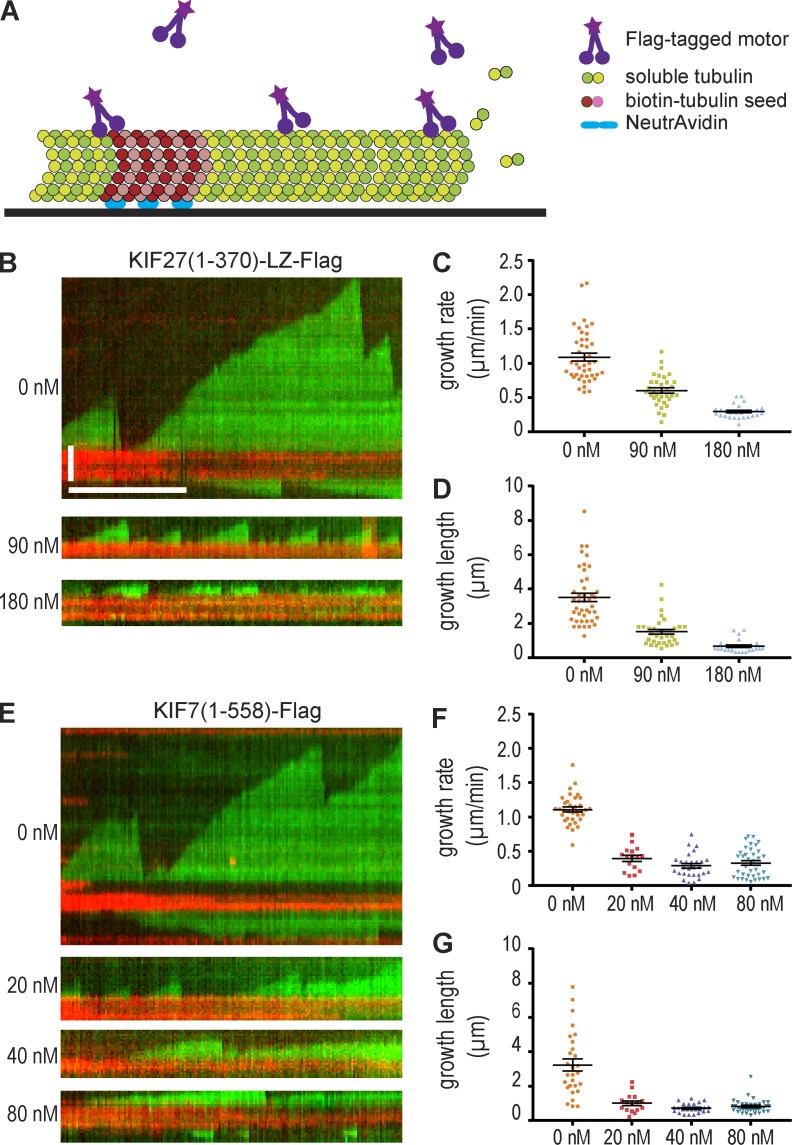
We measured the growth rates of dynamic microtubules in an in vitro assay (Fig. 2 A). In the absence of motor, the mean microtubule growth rate was 1.11 ± 0.04 µm/min. Increasing amounts of KIF7 resulted in inhibition of microtubule growth, with mean rates of 0.40 ± 0.04, 0.29 ± 0.03, and 0.33 ± 0.04 µm/min at KIF7(1–558)-Flag concentrations of 20, 40, and 80 nM, respectively (Fig. 2, E and F), similar to a previous study (He et al., 2014). Increasing concentrations of KIF7(1–558)-Flag also resulted in a decrease in the mean length of microtubule growth before catastrophe, from 3.24 ± 0.35 µm in the absence of motors to 1.01 ± 0.13, 0.72 ± 0.06, and 0.82 ± 0.07 µm at concentrations of 20, 40, and 80 nM (Fig. 2 G). In a similar manner, increasing amounts of KIF27(1–370)-LZ-Flag resulted in a decrease in the microtubule growth rate, with mean rates of 0.60 ± 0.04 µm/min at 90 nM and 0.29 ± 0.02 µm/min at 180 nM KIF27(1–370)-LZ-Flag (Fig. 2, B and C) and a decrease in the mean length of microtubule growth to 1.52 ± 0.14 and 0.67 ± 0.07 µm at concentrations of 90 and 180 nM KIF27(1–370)-LZ-Flag, respectively (Fig. 2 D). Both motors caused a decrease in the catastrophe frequency (Fig. S2 B), but this could largely be explained by a concomitant decrease in the frequency of microtubule growth events (Fig. S2 A). The primary effect of KIF7 and KIF27 motors thus appears to be, like other members of the kinesin-4 family, a suppression of microtubule growth rates as measured in in vitro assays.
Figure 2.
KIF27 and KIF7 can inhibit microtubule growth in vitro. (A) Schematic of the microtubule dynamics assay. Microtubule seeds generated from biotin-tubulin and HiLyte647-tubulin and polymerized in GMPCPP were attached to the slide, and then microtubule polymerization was performed in the presence of 10.7 µM tubulin and 1 mM GTP. (B and E) Representative kymographs of microtubules polymerizing in the absence (0 nM) or presence of indicated concentrations of KIF27 (B) or KIF7 (E). HiLyte647 microtubule seeds are in red. X-rhodamine dynamic microtubules are in green. Time is on the x axis (bar, 5 min), and distance is on the y axis (bar, 2 µm). (C and F) Plots of microtubule growth rate measured in the absence or presence of the indicated concentrations of KIF27 (C) or KIF7 (F). (D and G) Plots of microtubule growth length before catastrophe measured in the absence or presence of indicated concentrations of KIF27 (D) or KIF7 (G). Scatterplots display means ± SEM (n = 20–43 for each condition) from two or three independent experiments.
Comparative analysis of kinesin-4 motility at the single molecule level
Some members of the kinesin-4 family have been shown to undergo processive motility along the microtubule surface and may therefore participate in cargo trafficking in cells. To examine the motility properties of dimeric motors across the kinesin-4 family, we generated constitutively active motors by truncating their sequences after the neck coil or within the subsequent coiled-coil segment (Fig. 1 B). Like MmKIF27, the minimal predicted neck coil sequence of CeKLP12 was not sufficient for stable dimer formation and we thus appended the LZ of GCN4 to the C terminus of the truncated motor. All motors were tagged with the fluorescent protein mNeonGreen (mNG). The well-studied kinesin-1 motor KIF5C(1–560) was examined as control (Fig. 1 B and Table 1).
Table 1. Summary of motility behavior and kinetics of kinesin-4 motors.
| Family | Motor | Single molecule assay (velocity) | Multimotor assay | kcat | K0.5,MT | Km,ATP | |
|---|---|---|---|---|---|---|---|
| Microtubule gliding assay (velocity) | Cellular transport ability | ||||||
| nm/s | nm/s | s−1site−1 | µM | µM | |||
| Kinesin-1 | KIF5Ca | 664 ± 10 | 728.3 ± 6.4 | Fast transportb | 20 ± 2c | 0.3 ± 0.1c | 23 ± 9c |
| Kinesin-4 | |||||||
| KIF7 | MmKIF27 | 13.51 ± 0.17 | 10.8 ± 0.4 | Slow transport | 2.5 | 0.04 | 0.6 |
| MmKIF7 | 0 | 0 | No transport | 0.02 | NA | NA | |
| DmCos2 | 0 | ND | ND | ND | ND | ND | |
| KIF21 | MmKIF21A | 391.3 ± 4.7 | 224.2 ± 4.3 | Fast transport | ND | ND | ND |
| DmKLP31E | 375.3 ± 5.2 | ND | ND | ND | ND | ND | |
| CeKLP12 | 623 ± 13 | ND | ND | ND | ND | ND | |
| KIF4 | MmKIF4 | 908 ± 15 | 836 ± 13 | Fast transport | ND | ND | ND |
NA, not applicable (could not be determined due to low kcat); ND, not determined.
For kinesin-1, the data in the table state motility behavior of RnKIF5C from this study.
Kinetics of HsKIF5C from Cochran (2015).
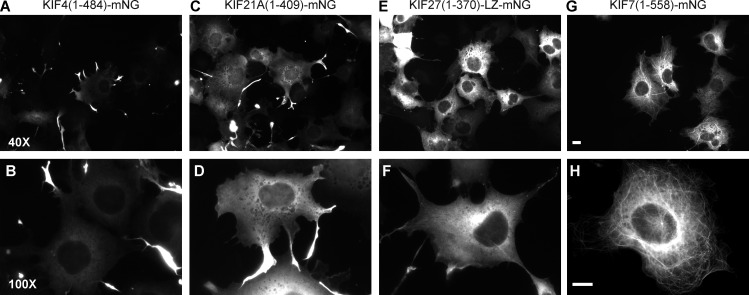
When expressed in COS-7 cells, KIF4(1–484)-mNG accumulated at the periphery of the cells, indicative of directed motion to the plus ends of the microtubules (Fig. 3, A and B). KIF21A(1–409)-mNG motors also accumulated at the periphery, although a large fraction showed a diffuse localization within the cytosol (Fig. 3, C and D). However, KIF27(1–370)-LZ-mNG displayed a largely diffuse localization (Fig. 3, E and F), suggesting that this motor may undergo infrequent and/or minimally processive motility events, and KIF7(1–558)-mNG displayed a strongly microtubule-bound state (Fig. 3, G and H) similar to “rigor” versions of other kinesin motors (Nakata and Hirokawa, 1995; Sturgill et al., 2016).
Figure 3.
Localization of truncated kinesin-4 motors expressed in COS-7 cells. (A–H) Representative images showing the localization of MmKIF4(1–484) (A and B), MmKIF21A(1–409) (C and D), MmKIF27(1–370)-LZ (E and F), and MmKIF7(1–558) (G and H) tagged at their C termini with mNG at low (top) and high (bottom) magnification. Bars, 10 µm.
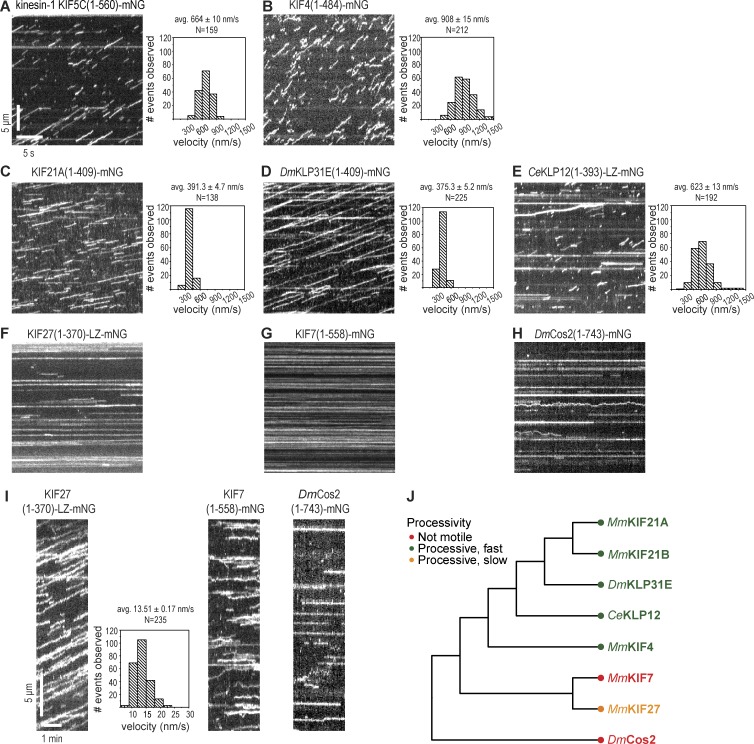
The motility properties of the truncated kinesin-4 motors along microtubules were determined in single molecule motility assays (Fig. 4). Under standard imaging conditions (2 mM ATP, room temperature, P12 buffer, and 10 Hz imaging; Norris et al., 2015; Yildiz and Vale, 2015), KIF4(1–484)-mNG showed kinesin-type motility with a mean velocity of 908 ± 15 nm/s (Fig. 4 B and Table 1). Given previous studies demonstrating processive motility for the Xenopus Xklp1 (Bringmann et al., 2004; Bieling et al., 2010) and Arabidopsis AtKN4A (Zhu and Dixit, 2011) homologues, we conclude that members of the KIF4 subfamily are processive kinesin motors (Fig. 4 J). For the KIF21 subfamily, we measured the motility properties of mammalian KIF21A and the fly (DmKLP31E) and worm (CeKLP12) homologues. All three motors showed characteristic kinesin-like motility behavior with mean velocities of 391.3 ± 4.7 nm/s for KIF21A(1–409)-mNG (Fig. 4 C and Table 1), 375.3 ± 5.2 nm/s for DmKLP31E(1–409)-mNG (Fig. 4 D and Table 1), and 623 ± 13 nm/s for CeKLP12(1–393)-LZ-mNG (Fig. 4 E and Table 1). We thus conclude that members of the KIF21 subfamily are also processive kinesin motors (Fig. 4 J).
Figure 4.
Single molecule motility properties of kinesin-4 motors. (A–H) Representative kymographs showing single molecule motility of truncated versions of the kinesin-4 motors MmKIF4 (B), MmKIF21A (C), DmKLP31E (D), CeKLP12 (E), MmKIF27 (F), MmKIF7 (G), and DmCos2 (H) as compared with the kinesin-1 motor KIF5C (A). All motors were tagged with mNG at their C termini, and videos were acquired at a fast imaging rate (10 frames per second). Time is on the x axis (bar, 5 s), and distance is on the y axis (bar, 5 µm). Histograms of the velocities for each population of motors are plotted to the right of the kymographs. (I) Representative kymographs of single molecule motility of truncated versions of the kinesin-4 motors MmKIF27, MmKIF7, and DmCos2 imaged at a slow acquisition rate (1 frame/3 s). Time is on the x axis (bar, 1 min), and distance is on the y axis (bar, 5 µm). A histogram of the velocities for the population of KIF27 motors is shown to the right of the KIF27 kymograph. Motility data are described as means ± SEM. N, number of motility events analyzed across three independent experiments. (J) Motility of kinesin-4 motors based on the single molecule imaging results depicted on the phylogenetic tree.
Unlike the characteristic kinesin-like motility observed for members of the KIF4 and KIF21 subfamilies, very different motility behavior was observed for members of the KIF7 subfamily. Under standard imaging conditions, all three motors bound to microtubules and appeared largely immotile (Fig. 4, F–H), although some diffusive movement was observed for DmCos2(1–743)-mNG (Fig. 4 H). The inability of KIF7 to undergo processive stepping along the microtubule lattice is consistent with a previous study (He et al., 2014). The inability of Cos2 to demonstrate processive motility is likely caused by sequence changes in key residues of the Cos2 motor domain (Fig. S1 A) as noted previously (Sisson et al., 1997; Matthies et al., 2001).
We considered the possibility that the standard imaging parameters of these single molecule motility assays were not optimal for observing the behavior of KIF7 subfamily motors. When we changed imaging conditions to acquire one frame every 3 s, both KIF7(1–558)-mNG and Cos2(1–743)-mNG continued to show only static binding to or diffuse movement along the microtubule surface (Fig. 4 I). Strikingly, KIF27(1–370)-LZ-mNG exhibited very slow motility, with a mean velocity of 13.51 ± 0.17 nm/s (Fig. 4 I and Table 1) under these imaging conditions. These results indicate that members of the KIF7 subfamily show dramatically different motility properties than the rest of the family (Fig. 4 J). Whereas members of the KIF4 and KIF21 subfamilies across species show processive motility characteristic of dimeric kinesin motors, Cos2 and KIF7 members of the KIF7 subfamily are immotile, and KIF27 displays very slow motility. Thus, evolutionary changes to the kinesin motor domain gave rise to dramatically different motility properties within the kinesin-4 family.
The motility of KIF7 subfamily motors is not enhanced by working in teams
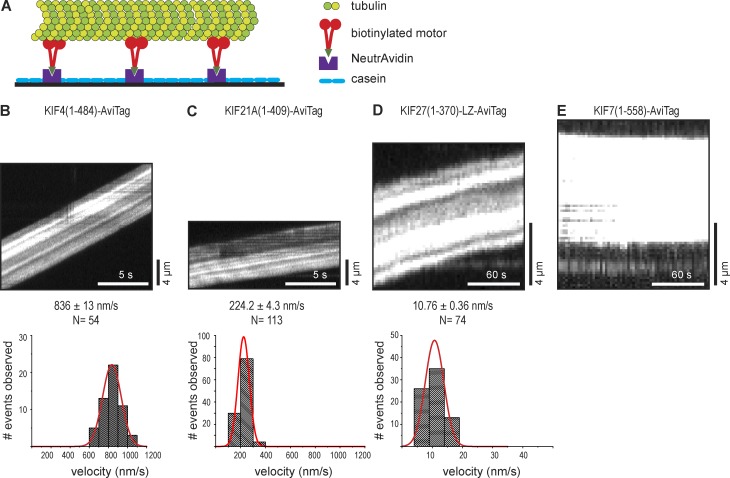
We sought to test whether immotile (KIF7 and Cos2) or slow processive (KIF27) kinesin-4 motors could work together to drive processive motility in a multimotor regime, as has been suggested for nonprocessive kinesin-14 motors (Case et al., 1997). We used two assays to test this possibility. First, we performed microtubule gliding assays with kinesin-4 motors. Truncated versions of the kinesin-4 members KIF4, KIF21A, KIF27, and KIF7 were tagged at their C termini with an AviTag sequence (Tirat et al., 2006) and biotinylated by coexpression with the bacterial biotin ligase BirA (Fig. S3; Cull and Schatz, 2000). Biotinylated motors were immobilized on NeutrAvidin-coated coverslips, and then taxol-stabilized microtubules were added to the flow chamber (Fig. 5 A). Both KIF4(1–484)-AviTag (836 ± 13 nm/s) and KIF21A(1–409)-AviTag (224.2 ± 4.3 nm/s) motors were able to drive microtubule gliding at speeds consistent with their single motor properties (Fig. 5, B and C; and Table 1). For KIF27(1–370)-LZ-AviTag motors, very slow microtubule gliding was observed (10.76 ± 0.36 nm/s; Fig. 5 D and Table 1), whereas for KIF7(1–558)-AviTag motors, no microtubule gliding activity was observed (Fig. 5 E and Table 1). These results in multimotor assays are consistent with the single-molecule motility properties of KIF27 and KIF7 (Table 1), suggesting that nonmotile and slowly processive KIF7 subfamily motors do not cooperate to increase their motility properties when working in teams.
Figure 5.
Multimotor properties of kinesin-4 motors in microtubule gliding assays. (A) Schematic of microtubule gliding assay. Truncated dimeric kinesin-4 motors were tagged at their C termini with the AviTag and biotinylated by coexpression with the biotin ligase BirA. The biotinylated motors were bound to the surface of a NeutrAvidin-coated flow cell, and fluorescent microtubules were added. (B–E) Representative kymographs show microtubule gliding driven by the indicated motors (top). Time is on the x axis, and distance is on the y axis. The velocity of each microtubule gliding event was determined via kymograph analysis and plotted as a histogram for each population (bottom). The curves were fit with a normal distribution. Gliding speeds are described as means ± SEM. N, number of motility events analyzed across three independent experiments.
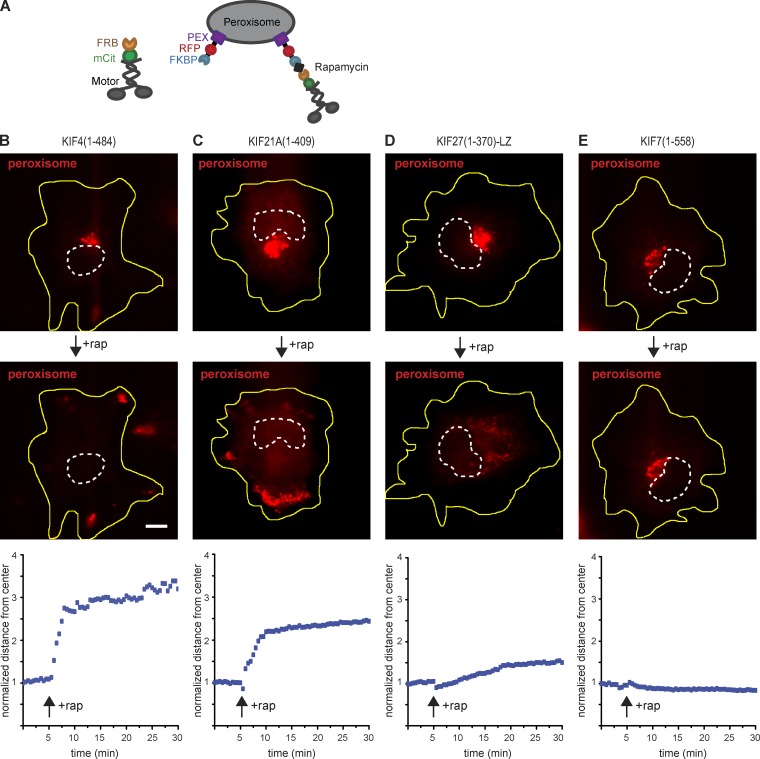
Second, we used a peroxisome dispersion assay to investigate whether KIF7 subfamily motors can act as processive cargo transporters in a cellular multimotor regime. The kinesin-4 family motors were tagged with mCitrine (mCit) and an FRB domain, whereas the peroxisomes were labeled by expressing PEX3-mRFP-FKBP. Addition of rapamycin results in the recruitment of kinesin motors to the peroxisome surface, and recruitment of active motors drives the dispersion of peroxisomes to the cell periphery (Fig. 6 A; Kapitein et al., 2010; Engelke et al., 2016). COS-7 cells coexpressing kinesin-4 motor-mCit-FRB and PEX3-mRFP-FKBP fusion constructs were observed by live-cell imaging. Before rapamycin addition, most of the peroxisomes exhibited a perinuclear distribution (Fig. 6, B–E, top; and Fig. S4). After rapamycin addition, the peroxisomes in cells expressing KIF4(1–484)-mCit-FRB or KIF21A(1–409)-mCit-FRB motors displayed rapid and robust accumulation at the cell periphery (Fig. 6, B and C; and Fig. S4). In contrast, peroxisomes in cells expressing KIF27(1–370)-LZ-mCit-FRB were moved only a short distance in the 25-min time period (Figs. 6 D and S4), whereas peroxisomes in cells expressing KIF7(1–558)-mCit-FRB remained in a perinuclear cluster (Figs. 6 E and S4). Together with the in vitro microtubule gliding results (Fig. 5), these findings indicate that KIF27 is capable of very slow motility both as single motors and in teams, whereas KIF7 lacks detectable motility in both single- and multimotor assays (Table 1).
Figure 6.
Multimotor properties of kinesin-4 motors in a cellular transport assay. (A) Schematic of the inducible peroxisome redistribution assay. Truncated dimeric kinesin motors tagged with the fluorescent protein mCit and FRB were coexpressed with the peroxisome-targeted PEX-mRFP-FKBP fusion protein in COS-7 cells. Addition of rapamycin causes heterodimerization of FRB and FKBP and thereby recruits kinesin motors to the peroxisome surface. Recruitment of active motors causes redistribution of peroxisomes to the cell periphery. (B–E) Representative images of peroxisome distribution in COS-7 cells expressing the indicated kinesin motors before (0 min; top) and after (25 min; middle) addition of rapamycin (rap). Yellow lines indicate the periphery of each cell; white dotted lines indicate the nucleus. Bar, 10 µm. Representative graphs (bottom) show the peroxisome distribution over time upon recruitment of the indicated kinesin-4 motors in a representative cell. Each blue dotted line represents the peroxisome distribution at that time point. The value was normalized by the peroxisome distance from center in the first frame. The black arrow indicates the time point of rapamycin addition.
KIF7 subfamily members KIF27 and KIF7 are defective in their chemomechanical coupling
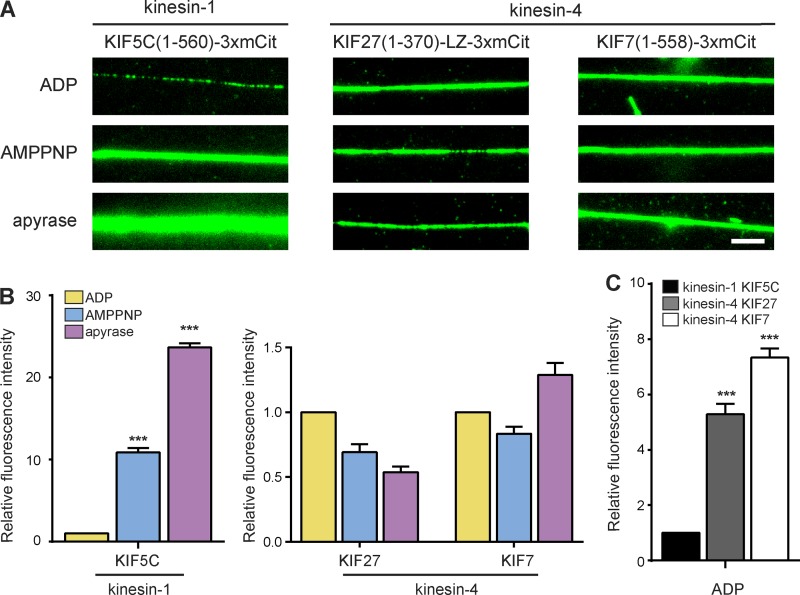
To determine the mechanisms that underlie the nonmotile association of KIF7 with microtubules and the very slow motility of KIF27, we examined the chemomechanical coupling of these motors. For processive motors like kinesin-1, chemomechanical coupling enables the motor to alter its affinity for the microtubule in response to its nucleotide state (Crevel et al., 1996). To assess the ability of kinesin-4 motors to respond to nucleotide conditions with a change in microtubule affinity, we used a fluorescence-based microtubule binding assay. The control kinesin-1 motor KIF5C(1–560)-mNG showed weak microtubule binding in the presence of ADP and strong microtubule binding in the presence of either AMPPNP (a nonhydrolyzable ATP analogue) or apyrase (which generates a no-nucleotide condition; Fig. 7 A, left). Specifically, AMPPNP and apyrase resulted in 11-fold and 23-fold increases in KIF5C(1–560)’s interaction with microtubules as compared with motors in the ADP state, respectively (Fig. 7 B), consistent with previous data (Crevel et al., 1996; Case et al., 1997; Pierce et al., 1999). Likewise, the processive kinesin-4 motors KIF4 and KIF21A displayed similar changes in their microtubule association in response to their nucleotide state. For KIF4(1–484), microtubule association was increased fourfold in the presence of AMPPNP and 49-fold in the presence of apyrase when compared with the ADP state (Fig. S5, A and B), and for KIF21A(1–409), microtubule association was increased fivefold in the presence of AMPPNP and 78-fold in the presence of apyrase when compared with the ADP state (Fig. S5, A and B).
Figure 7.
Defective chemomechanical coupling of the kinesin-4 motors KIF27 and KIF7. (A–C) Truncated dimeric motors versions of the kinesin-1 KIF5C(1–560) or the kinesin-4s KIF27(1–370)-LZ or KIF7(1–558) were tagged at their C termini with 3×mCit. Cell lysates expressing equivalent amounts of motors were added to flow cells containing taxol-stabilized microtubules in the presence of the indicated nucleotides. (A) Representative images of motor (green) binding to microtubules in the presence of ADP, AMPPNP, or apyrase. Bar, 5 µm. (B) The fluorescence intensity of each motor along microtubules was quantified for each nucleotide condition. The mean fluorescence intensity of each motor in AMPPNP and apyrase was normalized to the mean fluorescence intensity in the ADP state. ***, P < 0.001 as compared with the ADP state (two-tailed t test). (C) The fluorescence intensity of each motor in the presence of ADP was compared with that of the kinesin-1 KIF5C. ***, P < 0.001 as compared with KIF5C (two-tailed t test). Data indicate means ± SEM of more than five microtubules from one representative experiment.
In contrast, we observed that the slow processive kinesin-4 motor KIF27 and the immotile kinesin-4 motor KIF7 uniformly decorated microtubules regardless of nucleotide condition (Figs. 7 A and S5 C). For KIF27(1–370)-LZ, addition of AMPPNP and apyrase resulted in levels of microtubule association (0.69-fold and 0.54-fold), respectively, comparable with the ADP state (Fig. 7, A and B; and Fig. S5 D). For KIF7(1–558), addition of AMPPNP and apyrase resulted in levels of microtubule association (0.83-fold and 1.29-fold), respectively, comparable with the ADP state (Fig. 7, A and B; and Fig. S5 D). When compared with kinesin-1, both KIF27 and KIF7 showed a higher microtubule affinity in the ADP state; the microtubule association of KIF27 was fivefold higher than kinesin-1, and that of KIF7 was sevenfold higher (Fig. 7 C). Collectively, these results indicate the KIF7 subfamily members are defective in their chemomechanical coupling. Their inability to respond to the nucleotide state with a change in microtubule affinity is likely the underlying explanation for their defective motility.
Characterization of KIF7 and KIF27 kinetic parameters
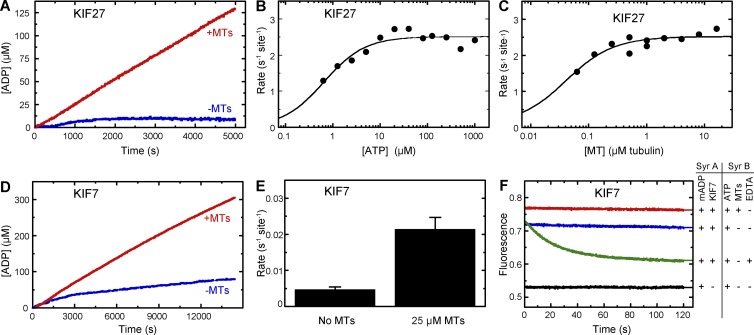
To further explore whether defective chemomechanical coupling of KIF7 and KIF27 can explain the observed motility defects, we performed a series of kinetic assays. We first measured each motor’s ATPase activity in the absence and presence of microtubules. For processive motors like kinesin-1, microtubule binding causes a >1,000-fold increase in ATPase activity over the basal level (Ma and Taylor, 1995). For the slow processive KIF27 motor, we found that the addition of microtubules resulted in a 25–50-fold increase in ATPase activity (Fig. 8 A), which was comparable to other slow processive kinesins such as human kinesin-5 (Cochran, 2015). Under steady-state kinetics conditions, KIF27 displayed a kcat = 2.5 s−1site−1 with a high affinity for both ATP (Km,ATP = 0.6 µM; Fig. 8 B and Table 1) and microtubules (K0.5,MT = 0.04 µM; Fig. 8 C and Table 1). These constants represent the tightest ATP affinity for a dimeric kinesin motor measured to date (Klumpp et al., 2004; Auerbach and Johnson, 2005; Krzysiak and Gilbert, 2006; Pan et al., 2006; Albracht et al., 2014). These results were surprising as processive kinesins typically have a reciprocal relationship between ATP binding and microtubule binding such that tight ATP binding correlates with weak microtubule binding and vice versa (Albracht et al., 2014).
Figure 8.
The kinetic properties of KIF27 and KIF7 are consistent with their motility defects. (A) ATPase activity of purified KIF27(1–370)-LZ-Flag was measured in the absence and presence of microtubules (MTs) using the NADH-coupled assay. Final conditions: 0.01 µM KIF27, 0 or 0.5 µM microtubules, 20 µM taxol, and 1 mM ATP. (B) ATP-dependent ATPase activity of KIF27 in the presence of excess microtubules. Final conditions: 0.01 µM KIF27, 2 µM microtubules, 20 µM taxol, and 0.5–1000 µM ATP. Fit to the Michaelis-Menten equation yields: kcat = 2.5 ± 0.1 s−1site−1 and Km,ATP = 0.6 ± 0.1 µM. (C) Microtubule-dependent ATPase activity in the presence of excess ATP. Final conditions: 0.01 µM KIF27, 0–16 µM microtubules, 20 µM taxol, and 100 µM ATP. Fit to the quadratic equation yields: kcat = 2.4 ± 0.1 s−1site−1, K0.5,MT = 0.04 ± 0.01 µM, and kbasal = 0.08 ± 0.05 s−1. (D) ATPase activity of purified KIF7(1–558)-Flag was measured in the absence and presence of microtubules using the NADH-coupled assay. Final conditions: 1 µM KIF7, 0 or 25 µM microtubules, 40 µM taxol, and 1,000 µM ATP. (E) ATPase rates from D indicating means ± SEM from three independent experiments. (F) Mant-ADP release kinetics for KIF7 in the absence and presence of microtubules. Final conditions: 0.25 µM mant-ADP, 0.5 µM KIF7, 2 µM microtubules, 10 µM taxol, 500 µM ATP, and 5 mM EDTA.
For immotile KIF7, very little microtubule-dependent stimulation of ATPase activity was observed. Even at a high microtubule concentration (25 µM) rate, the ATPase rate (kcat = 0.02 s−1site−1; Fig. 8, D and E; Table 1) did not exceed the basal activity of most other kinesins studied to date (Cochran, 2015) and reflected only a fivefold enhancement of KIF7’s basal ATPase rate (Fig. 8, D and E). The low levels of basal and microtubule-stimulated ATPase activity for KIF7 could be caused by an inability of the motor to release ADP in response to microtubule binding or an inability to hydrolyze ATP. To distinguish these possibilities, we measured the microtubule-stimulated ADP release for KIF7 using a mant-ADP release assay. When bound in the nucleotide pocket of a kinesin motor, mant-ADP displays a higher fluorescence than when free in solution (Cochran et al., 2013), and the microtubule-stimulated mant-ADP release rate can thus be measured as a decrease in fluorescence (Nitta et al., 2008; Cochran et al., 2013; Chen et al., 2017). We found that KIF7 was capable of binding to mant-ADP as the initial fluorescence intensity was higher in the presence of KIF7 (Fig. 8 F, blue trace) than in the absence of KIF7 (Fig. 8 F, black trace). However, KIF7 was incapable of microtubule-stimulated ADP release as the addition of microtubules did not result in a decrease in mant-ADP fluorescence intensity over time (Fig. 8 F, red trace). We verified that mant-ADP could be released from the KIF7 motor domain in a control reaction (Fig. 8 F, green trace) where EDTA chelates the free Mg2+ from solution (Nitta et al., 2008). These results indicate that the underlying mechanism for the immotile behavior of the KIF7 motor is an inability to stimulate ADP release from the nucleotide pocket in response to microtubule binding.
Discussion
Sequence divergence within the kinesin superfamily resulted in motors with distinct mechanochemical properties matched to their functions during intracellular trafficking, microtubule organization, and cell division. In this study, we used single-molecule, cellular, and kinetic assays to characterize motility properties of motors across the kinesin-4 family to elucidate a molecular understanding of their cellular roles. We show that although all kinesin-4 motors examined to date are capable of influencing microtubule dynamics, these motors showed large variations in their ability to undergo processive motility. We describe the kinetic basis for the immotile behavior of mammalian KIF7 and suggest that this motor and its Drosophila homologue Cos2 function not as transporters but as tethers for signaling complexes in the Hedgehog pathway. These findings support the idea that sequence divergence has led to a diversity of mechanochemical and motility properties within the kinesin superfamily.
Kinesin-4 motors share the ability to influence microtubule dynamics but differ dramatically in their motility properties
Several members of the kinesin-4 family have been shown to regulate microtubule dynamics in in vitro assays, including the KIF4 subfamily members human KIF4, Xenopus Xklp1, and Physcomitrella patens Kin4-Ic (Bieling et al., 2010; Hu et al., 2011; Subramanian et al., 2013; de Keijzer et al., 2017), the KIF21 subfamily members KIF21A and KIF21B (van der Vaart et al., 2013; Ghiretti et al., 2016; Muhia et al., 2016; van Riel et al., 2017), and the KIF7 subfamily member KIF7 (He et al., 2014). We show that the kinesin-4 family member KIF27 reduces microtubule growth rates and the length of microtubule growth before catastrophe. It thus appears that the ability to regulate microtubule dynamics is shared across the kinesin-4 family.
In contrast, members of the kinesin-4 family display dramatically different motility properties. We found that individual dimeric KIF4 motors are processive motors, similar to previous work on members of this subfamily (Bieling et al., 2010; Zhu and Dixit, 2011; Subramanian et al., 2013) and consistent with their proposed roles in intracellular transport (Peretti et al., 2000; Zhu and Jiang, 2005; Martinez et al., 2008; Bisbal et al., 2009; Kong et al., 2015; Zhu et al., 2015). We also found that KIF21 subfamily motors across species (MmKIF21A, DmKLP31E, and CeKLP12) are processive motors and that this is likely a critical feature for their participation in intracellular transport events (Lee et al., 2012; Labonté et al., 2014; Ghiretti et al., 2016; Muhia et al., 2016). In contrast, we found very different motility behaviors for members of the KIF7 subfamily: DmCos2 is immotile and shows both static and diffusive interactions with the microtubule, MmKIF7 is immotile, and MmKIF27 is a slow processive motor. This variation highlights the diversity of ways that the core kinesin motor domain can be adapted for different functional outputs even within a kinesin family.
The immotile (DmCos2 and MmKIF7) or slowly motile (MmKIF27) behavior in single molecule assays does not a priori preclude the ability of these motors to drive intracellular trafficking events. One possibility is that KIF7 subfamily motors drive processive motion when working in teams. This has previously been demonstrated for nonprocessive kinesin-14 family motors, which undergo only short runs or diffusive movement as individual dimeric motors but drive processive motion when working in teams (Case et al., 1997; Furuta et al., 2013; Jonsson et al., 2015; Walter et al., 2015). However, we find that KIF7 and KIF27 are unable to drive processive motion when working as a team in a classic multimotor microtubule gliding assay or when transporting an exogenous cargo in cells (Table 1).
Our data provide strong support for the finding that a motor’s ability to influence polymerization dynamics at microtubule ends is not linked to its ability to undergo processive motion to the end of the microtubule. This was first noted for the kinesin-13 motor mitotic centromere-associated kinesin (MCAK), which diffuses along the microtubule lattice to reach both ends of a microtubule, where it promotes depolymerization (Hunter et al., 2003; Helenius et al., 2006). The ability to influence microtubule dynamics also does not require ATP hydrolysis, at least for the kinesin-8 motor Kip3p (Arellano-Santoyo et al., 2017) and the mammalian kinesin-4 motor KIF7 (this study). Rather, the ability to influence microtubule polymerization appears to be related to a motor’s ability to stabilize the curvature of tubulin subunits at the plus end of the microtubule. For the kinesin-8 motors Kip3 and KIF19A and the kinesin-13 motor MCAK, binding to curved tubulin at the microtubule end promotes microtubule depolymerization (Asenjo et al., 2013; Wang et al., 2016, 2017; Arellano-Santoyo et al., 2017).
The underlying mechanisms of impaired motility
DmCos2 motors bind microtubules in a stationary manner with a long dwell time regardless of nucleotide condition. The defect in chemomechanical coupling may arise from deletions of key surface loops of the motor domain, e.g., loops 8, 9, and 11 and/or mutation of the invariant kinesin switch-1 motif (SSRSH) to SPLAH (Fig. S1 A). The immotile behavior of Cos2 is consistent with previous work demonstrating an ATP-independent interaction of Cos2 with microtubules (Sisson et al., 1997) but is in contrast with a recent study suggesting that Cos2 functions as a transport motor (Farzan et al., 2008). Although GFP-Cos2 localized to mobile punctate structures in S2 cells in the latter study, the motility of these structures was not consistent with kinesin-driven movement as it was spatially limited, not directional, and similar for full-length Cos2 and a construct lacking the motor domain (Farzan et al., 2008). We thus believe that the immotile behavior in single-molecule assays and the mutation of key motility residues preclude this motor’s participation in intracellular transport events.
MmKIF7 motors also bind microtubules in a stationary manner with a long dwell time regardless of nucleotide condition. An analysis of the kinetics revealed that KIF7 is defective in the first step in the kinesin chemomechanical cycle as it fails to release ADP from the nucleotide pocket in response to microtubule binding. Although the kinesin-13 motor MCAK (KIF2C) shows little ADP release in response to binding of the microtubule lattice, ADP release is stimulated >20-fold by the ends of the microtubule (Hunter et al., 2003; Friel and Howard, 2011; Patel et al., 2016). KIF7 is thus to our knowledge the only motor that fails to release nucleotides upon microtubule interaction. The mechanism by which KIF7 is unable to couple microtubule binding to changes in nucleotide state is not apparent at this time. MmKIF7 contains the necessary residues for ATP and microtubule binding (Fig. S1), and a crystal structure of the KIF7 motor domain is superimposable on that of kinesin-1 (Klejnot and Kozielski, 2012). In addition, human KIF7 shares an overall sequence identity of 44% with KIF27 and a higher sequence identity of 61% within the core motor domain (Klejnot and Kozielski, 2012). Thus, a molecular explanation for the defective chemomechanical coupling and immotile behavior of KIF7 requires further exploration.
Although MmKIF27 appeared at first glance to also be an immotile kinesin, imaging at a slower acquisition rate demonstrated that KIF27 is a slow motor that moves directionally with a long dwell time. Analysis of the kinetic cycle of KIF27 revealed a low kcat of 2.5 s−1 in a steady-state ATPase assay (Table 1). The correlation between a low ATPase rate and slow processive motility measured in single molecule and/or microtubule gliding assays has been noted for members of the kinesin-5 family (100 nm/s and kcat = 0.48 s−1; Krzysiak and Gilbert, 2006; Valentine and Gilbert, 2007), kinesin-7 family (11 nm/s and kcat = 0.9 s−1; Sardar et al., 2010; Sardar and Gilbert, 2012), and kinesin-14 family (97 nm/s and kcat = 1.6 s−1; Furuta and Toyoshima, 2008). Surprisingly, however, KIF27 showed very high affinity for both microtubules (K0.5,MT = 0.04 µM; Table 1) and ATP (Km,ATP = 0.6 µM; Table 1). Most kinesin motors have a reciprocal relationship between microtubule and ATP binding, including the slow processive kinesin-5 (Eg5; K0.5,MT = 1.8 µM and Km,ATP = 7.9 µM; Krzysiak and Gilbert, 2006; Valentine and Gilbert, 2007) and kinesin-7 (CENP-E; K0.5,MT = 1.5 µM and Km,ATP = 18.3 µM; Sardar et al., 2010; Sardar and Gilbert, 2012; Gudimchuk et al., 2013) motors. This is the highest ATP binding affinity for a dimeric kinesin motor to date and suggests that KIF27’s motility is tuned differently from the other slow processive motors.
KIF7 and KIF27 motility properties are tuned for their cellular functions
Collectively, these results provide key insights into the basic motility properties of the kinesin-4 motors DmCos2, MmKIF7, and MmKIF27. We propose that sequence changes were evolutionarily selected to provide these motors with distinct properties for their cellular functions. All three motors have been implicated in the proper formation of cilia and/or in the proper function of signaling pathways that use the cilium for compartmentalization. KIF27 localizes to the base of motile cilia in mouse tracheal epithelial cells and interacts with Fused for assembly of the central pair of microtubules in 9+2 motile cilia (Wilson et al., 2009; Vogel et al., 2011; Nozawa et al., 2013). Whether KIF27’s slow motility and/or ability to regulate microtubule dynamics are critical for proper formation and/or function of motile cilia requires further analysis.
Although KIF27 and KIF7 are orthologues of Cos2, KIF7 is thought to be the mammalian homologue as Cos2 can functionally replace KIF7 in the zebrafish embryo (Maurya et al., 2013) and KIF7 can replace some aspects of Cos2 function in flies (Marks and Kalderon, 2011). Both Cos2 and KIF7 have been shown to complex with transcription factors of the Hedgehog pathway (Cubitus interruptus [Ci] and Gli proteins, respectively) and regulate their proteolytic processing to a repressor form. Pathway activation by Hedgehog ligand results in the release of full-length Ci/Gli and transcriptional activation (Aikin et al., 2008; Ingham et al., 2011; Lee et al., 2016). Cos2 and KIF7 can also localize to cilia and influence the ciliary localization of transmembrane signaling proteins (Maurya et al., 2013; He et al., 2014; Kuzhandaivel et al., 2014; Sanchez et al., 2016). Given our results, it seems likely Cos2 and KIF7 use a static interaction with the microtubule lattice to regulate the proteolytic processing of Ci/Gli and the localization of receptors and other pathway components to the cilium. In contrast, KIF7’s ability to influence microtubule dynamics is likely required for its proposed role in regulating cilium tip structure (He et al., 2014). We thus propose that Cos2 and KIF7 function not as conventional transport kinesin motors in cells but rather as microtubule-based tethers. In this regard, it is interesting to note a parallel situation in the myosin superfamily where myosin-18A has very low enzymatic activity and binds actin filaments regardless of nucleotide conditions (Guzik-Lendrum et al., 2013). It thus appears that during the evolutionary selection of both the kinesin and myosin superfamilies, motor sequences with uncoupled filament and nucleotide binding were selected to provide a mechanism for tethering components to the underlying cytoskeleton.
Materials and methods
Plasmids
The constitutively active kinesin-1 constructs rat KIF5C(1–560) fused to mNG or 3×mCit have been described previously (Cai et al., 2009; Norris et al., 2015). Full-length MmKIF4 expression plasmid (Morris et al., 2014) was a gift from G. Gundersen (Columbia University, New York, NY). Full-length KIF21A expression plasmid (Huang and Banker, 2012) was a gift from G. Banker (Oregon Health Sciences, Portland, OR). Full-length DmCos2 was purchased from the Drosophila Genome Resource Center. For truncated MmKIF4(aa 1–484), MmKIF21A(aa 1–409), MmKIF27(aa 1–370), MmKIF7(aa 1–558), DmKLP31E(aa 1–409), and CeKLP12(aa 1–393), the coiled-coil regions were identified by prediction software (MARCOIL), and sequences encoding dimeric truncated motors were amplified by PCR or synthesized by GeneArt (Invitrogen) and subcloned into mNG-N1 or 3×mCit-N1 vectors (based on Takara Bio Inc.’s EYFP-N1 vectors). The dimeric state of each 3×mCit-tagged motor was tested by photobleaching analysis (Hammond et al., 2009, 2010; Soppina et al., 2014). Although MmKIF4(aa 1–484), MmKIF21A(aa 1–409), MmKIF7(aa 1–558), and DmKLP31E(aa 1–409) displayed characteristics of a stable dimer state, the motors MmKIF27(aa 1–370) and CeKLP12(aa 1–393) were largely monomeric. To ensure analysis of dimeric motors, the LZ sequence of GCN4 (the aa sequence KQLEDKVEELASKNYHLENEVARLKKLV) was appended to the C termini of MmKIF27(aa 1–370) and CeKLP12(aa 1–393) as described previously (Tomishige et al., 2002; Huckaba et al., 2011; Soppina et al., 2014).
For expression in S2 cells, pMT-DmKLP31E(1–409)-mNG and pMT-CeKLP12(1–393)-mNG were generated by using appropriate restriction digestion to insert motor sequences into pMT-mNG vector. For protein expression and purification from insect cells, truncated KIF27 and KIF7 motors fused with Flag tag were generated by restriction digestion and subcloning into the pFastBac1 vector. For microtubule gliding assays, KIF5C(1–560)-AviTag was generated by synthesizing AviTag to replace mNG tag of KIF5C(1–560)-mNG. The other motors were subcloned to replace KIF5C using appropriate restriction digestion or PCR amplification. HA-BirA and the peroxisome targeting construct PEX3-mRFP-2×FKBP (Kapitein et al., 2010), denoted in this study PEX3-mRFP-FKBP for simplicity, were gifts from C. Hoogenraad (Utrecht University, Utrecht, Netherlands). KIF5C (1–560)-mCit-FRB has been described previously (Engelke et al., 2016). All plasmids were verified by DNA sequencing.
Phylogenetic tree
Sequences of select members of the kinesin-4 family were collected from the UniProtKB database (Fig. S1 B) with a total of 14 sequences. Mouse sequences over human homologues were selected where applicable. The sequences were trimmed to the minimal motor domain using Pfam seed alignment (Kull et al., 1996; Finn et al., 2014) with the protein family ID PF00225. The sequence of the prototypical human kinesin-1 motor KIF5B was added to permit rooted phylogeny analysis. An MSA was performed with Bio3D (2.3; Skjærven et al., 2014) using default MUSCLE parameters for proteins (Edgar, 2004). Both rooted and unrooted phylogenetic trees were calculated with this MSA using BioNJ (Gascuel, 1997) and Kimura protein distance as the distance metric (Kimura, 1983). Bootstrapping was performed using Seaview (4.4.2; Gouy et al., 2010) with 1,000 iterations using a single central processing unit. Phylogenies were annotated and plotted with the R packages ggtree (Yu et al., 2017) and APE (Paradis et al., 2004). The alignment annotation of kinesin-4 family motor domain was generated by STRAP (Gille and Frömmel, 2001).
Cell culture, transfection, and lysis
Drosophila S2-DRGC cells obtained from the Drosophila Genome Resource Center (RRID: CVCL_Z232) were cultured at 25°C in Schneider’s Drosophila medium (Gibco) supplemented with 10% (vol/vol) FBS (HyClone). Plasmids for expression of truncated DmKLP31E, DmCos2, or CeKLP12 motors in the pMT vector were transfected into S2 cells using Cellfectin II (Invitrogen). Protein expression was induced by adding 1 mM CuSO4 to the medium 4–5 h after transfection. The cells were harvested after 48 h.
COS-7 (monkey kidney fibroblast) cells obtained from ATCC (RRID: CVCL_0224) were cultured in DMEM (Gibco) with 10% (vol/vol) Fetal Clone III (HyClone) and GlutaMAX (Gibco) at 37°C with 5% CO2. For single molecule assays, plasmids for expression of truncated MmKIF4, MmKIF21A, MmKIF27, MmKIF7, or RnKIF5C motors tagged with mNG or 3×mCit were transfected into cells using Trans-IT LT1 (Mirus). The cells were harvested after ∼16 h. For microtubule gliding assays, plasmids for expression of the indicated motors fused with AviTag were cotransfected with a plasmid for expression of HA-BirA at a ratio of 1:1 into using Trans-IT LT1 (Mirus). The cells were harvested or fixed after ∼16 h. For the inducible peroxisome dispersion assay, plasmids for expression of the indicated motors tagged with mCit and FRB were cotransfected with a plasmid for expression of PEX3-mRFP-FKBP at a ratio of 15:1 using Lipofectamine 2000 (Thermo Fisher Scientific), and the cells were imaged after ∼16 h. All transfections were performed according to the manufacturer’s instructions.
To prepare cell lysates, COS-7 cells 16 h after transfection or S2 cells 48 h after transfection were harvested and centrifuged at low speed at 4°C. The cell pellet was washed with PBS and resuspended in ice-cold lysis buffer (25 mM Hepes/KOH, pH 7.4, 115 mM potassium acetate, 5 mM sodium acetate, 5 mM MgCl2, 0.5 mM EGTA, and 1% [vol/vol] Triton X-100) freshly supplemented with 1 mM ATP, 1 mM PMSF, and protease inhibitor cocktail (P8340; Sigma-Aldrich). 1 mM DTT and 10% glycerol were added for S2 cell lysates. After clarifying the lysate by centrifugation for 10 min at 20,000 g at 4°C, aliquots were snap frozen in liquid nitrogen and stored at −80°C until further use.
Protein expression and purification
Sf9 cells obtained from Thermo Fisher Scientific were cultured in suspension with serum-free sf900 II SFM medium (Thermo Fisher Scientific) supplemented with antibiotic antimycotic (Gibco) in flasks at 28°C in a non-CO2 nonhumidified incubator with an orbital shaker platform set at 130 rpm. The cells were infected with baculovirus generated according to the Bac-to-Bac system (Invitrogen). In brief, plasmids encoding truncated Flag-tagged motors in the pFastBac1 vector were transformed into DH10Bac Escherichia coli to generate recombinant bacmids. Bacmid DNA was isolated with the HiPure Plasmid DNA miniprep kit (Invitrogen) and confirmed by PCR analysis. Recombinant bacmid DNA was transfected into Sf9 cells using Cellfectin II (Invitrogen) to produce the P1 recombinant baculovirus. 4 or 5 d after transfection, the supernatant containing P1 baculovirus was collected by centrifugation at 3,000 rpm for 3 min at 4°C. The baculovirus was amplified by successive infection of Sf9 cells to generate P2 and P3 baculoviruses. Baculovirus-containing supernatants were stored at 4°C in the dark.
For protein purification, Sf9 cells were infected with 10% P3 baculovirus (vol/vol). 1–2 d after infection, the cells were harvested by centrifugation for 3 min at 3,000 rpm at 4°C. The pellet was washed once with PBS and resuspended in ice-cold lysis buffer (200 mM NaCl, 4 mM MgCl2, 0.5 mM EDTA, 1 mM EGTA, 0.5% igepal, 7% sucrose, and 20 mM imidazole-HCl, pH 7.5) supplemented with 2 mM ATP, 1 mM PMSF, 5 mM DTT, and protease inhibitor cocktail. After 30 min incubation on ice, the lysates were clarified by ultracentrifugation for 30 min at 60,000 rpm in TLA-100.3 rotor (Beckman Coulter), and the supernatants were incubated with anti-FLAG M2 agarose (Sigma-Aldrich) for 1.5 h at 4°C with rotation. Bound proteins were washed with wash buffer (150 mM KCl, 25 mM imidazole-HCl, pH 7.5, 5 mM MgCl2, 1 mM EDTA, and 1 mM EGTA) supplemented with 1 mM PMSF, 3 mM DTT, 3 mM ATP, and protease inhibitor cocktail and wash buffer supplemented with 1 mM PMSF, 3 mM DTT, and protease inhibitor cocktail, subsequently. Bound proteins were eluted by incubation with elution buffer (25 mM KCl, 25 mM imidazole-HCl, pH 7.5, 5 mM EGTA, 2 mM MgCl2, 2 mM DTT, and 0.1 mM ATP) supplemented with 0.15 mg/ml 3×FLAG peptide (Sigma-Aldrich) overnight at 4°C. Supernatants were collected by centrifugation, and aliquots were snap frozen in liquid nitrogen and stored in −80°C until further use.
Imaging of live or fixed cells
Cells were rinsed with PBS and fixed in 3.7% (vol/vol) paraformaldehyde (Thermo Fisher Scientific) in PBS for 10 min at room temperature. Cells were imaged after mounting in Prolong Gold (Invitrogen) or were further processed for immunostaining. Fixed cells were permeabilized in 0.2% Triton X-100 in PBS for 5 min and blocked with 0.2% fish skin gelatin in PBS for 5 min. Primary (rabbit anti-HA; 1:500; Sigma-Aldrich) and secondary (594 nm anti–rabbit or 488 nm streptavidin; 1:500; Jackson ImmunoResearch Laboratories, Inc.) antibodies were applied in blocking buffer for 1 h at room temperature in the dark. The coverslips were mounted using Prolong Gold (Invitrogen). Images were acquired on an inverted epifluorescence microscope (TE2000E; Nikon) with a 40× 0.75 NA objective or a 100× 1.40 NA objective and a CoolSnap HQ camera (Photometrics). The fluorescence images were analyzed using ImageJ (National Institutes of Health).
For the inducible peroxisome dispersion assays, COS-7 cells were seeded onto glass-bottomed dishes (MatTek Corporation) and cotransfected with the indicated motor-mCit-FRB and PEX3-mRFP-FKBP plasmids. After ∼16 h, the cells were washed once and then incubated in Leibovitz’s l-15 medium (Gibco) and imaged at 37°C in a temperature-controlled and humidified live-imaging chamber (Tokai Hit). Live-cell imaging was performed on an inverted total internal reflection fluorescence (TIRF) microscope Ti-E/B (Nikon) equipped with the perfect focus system, a 100× 1.49 NA oil immersion TIRF objective (Nikon), three 20-mW diode lasers (488 nm, 561 nm, and 640 nm), and an electron-multiplying charge-coupled device detector (iXon X3DU897; Andor). The angle of illumination was adjusted for maximum penetration of the evanescent field into the cell. Image acquisition was controlled with Elements software (Nikon). Images were acquired in both 488- and 561-nm channels every 30 s for the first 5 min. Then, rapamycin (EMD Millipore) was added to a final concentration of 44 nM, and the cells were imaged every 30 s for the next 25 min. Images were analyzed with a custom MATLAB (MathWorks) script described by Engelke et al. (2016). In brief, first peroxisome objects were detected in each image in the 561-nm channel by a local adaptive thresholding algorithm (Guanglei Xiong; Tsinghua University; http://www.mathworks.com/matlabcentral/fileexchange/8647-local-adaptive-thresholding). Subsequently, the mean distance of all peroxisome object pixels from a manually determined cell center was reported for each frame to monitor peroxisome movement over time. Graphs were generated by Origin (OriginLab).
In vitro fluorescence microscopy assays
All in vitro imaging assays were performed on an inverted Ti-E/B TIRF microscope equipped with a perfect focus system, a 100× 1.49 NA oil immersion TIRF objective, three 20-mW diode lasers (488 nm, 561 nm, and 640 nm) and an electron-multiplying charge-coupled device detector. Image acquisition was controlled with Elements software.
Microtubule dynamics assay
A flow cell (∼10 µl volume) was assembled by attaching a clean #1.5 coverslip (Thermo Fisher Scientific) to a glass slide (Thermo Fisher Scientific) with two stripes of double-sided tape. Microtubules were assembled as previously described (Norris et al., 2015). Microtubule seeds containing 10% HiLyte647- and 10% biotin-labeled tubulin (Cytoskeleton Inc.) were generated by polymerization in the presence of the nonhydrolyzable GTP analogue GMPCPP (Jena Bioscience) and then immobilized on coverslips incubated sequentially with the following solutions: (A) 1 mg/ml BSA-biotin (A8549; Sigma-Aldrich), (B) blocking buffer (1 mg/ml BSA in BRB80 [80 mM Pipes/KOH, pH 6.8, 1 mM MgCl2, and 1 mM EGTA]), (C) 0.5 mg/ml NeutrAvidin (31000; Thermo Fisher), (D) blocking buffer, (E) short GMPCPP-stabilized microtubule seeds, and (F) blocking buffer. Microtubule growth was then initiated by flowing in 10.7 µM tubulin containing 7% X-rhodamine–labeled tubulin (Cytoskeleton Inc.) together with motor proteins at the indicated concentrations in the reaction buffer (1 mM GTP, 2.5 mM ATP, 0.1 mg/ml BSA, 1 mg/ml casein, 1 mM MgCl2, and 0.1% methylcellulose [Sigma-Aldrich] and oxygen scavenging [1 mM DTT, 10 mM glucose, 0.1 mg/ml glucose oxidase, and 0.08 mg/ml catalase] in BRB80). The flow cells were sealed with molten paraffin wax and imaged by TIRF microscopy. Time-lapse images were acquired in both 561-nm and 640-nm channels at a rate of every 5 s for 15 min. The temperature was set at 37°C in a temperature-controlled chamber (Tokai Hit). To determine of the growth rate and growth length of microtubule plus ends, maximum-intensity projections were generated, and kymographs (width = 3 pixels) were generated using ImageJ and displayed with time on the x axis and distance on the y axis. The growth length was defined as the vertical component of the kymograph between adjacent catastrophe events. The growth rate was defined as the growth length divided by the time (horizontal component) between adjacent catastrophe events.
Single molecule motility assays
A flow cell was prepared, and microtubules were assembled as described in the previous section. Polymerized microtubules were diluted in P12 buffer (12 mM Pipes/KOH, pH 6.8, 2 mM MgCl2, and 1 mM EGTA) containing 10 µM taxol and then were infused into flow cells and incubated for 5 min at room temperature for nonspecific adsorption to the coverslips. Sequentially, the flow cells were incubated with (A) blocking buffer (15 mg/ml BSA in P12 with 10 µM taxol) and then (B) kinesin motors in the motility mixture (0.5–1 µl cell lysate, 2 mM ATP, 6 mg/ml BSA, 0.5 mg/ml casein, 10 µM taxol, 1 mM MgCl2, and oxygen scavenging in P12). The flow cells were sealed with molten paraffin wax and imaged by TIRF microscopy. The fast imaging rate was set for continuous acquisition of one frame every 0.1 s for 30 s. The slow imaging rate was one frame every 3 s for 3 min. Maximum-intensity projections were generated, and kymographs were produced by drawing along these tracks (width = 5 pixels) using Elements software. The velocity was defined as the distance on the y axis of the kymograph divided by the time on the x axis of the kymograph.
Fluorescence-based microtubule affinity
The amount of motors across COS-7 lysates was determined by Western Blot using an anti-GFP antibody (ProteinTech). Digital images of the blots were analyzed by ImageJ. Equal amounts of motors in the final motility mixture (6 mg/ml BSA, 0.5 mg/ml casein, 10 µM taxol, 1 mM MgCl2, and oxygen scavenging in P12) supplemented with either 5 U/ml apyrase or 2 mM AMPPNP or 2 U/ml hexokinase with 2 mM ADP were added to flow cells containing polymerized microtubules. The coverslips were sealed and imaged at room temperature by TIRF microscopy. All the images were acquired and analyzed using the same conditions. For each motor in each nucleotide condition, the fluorescence intensities along the microtubules were measured using ImageJ, and the fluorescence intensity of an adjacent region was subtracted as background. The experiment was repeated two times for each motor with similar results. The data from two experiments are shown separately in Figs. 7 and S5 because the different protein expression levels between the two independent experiments made it difficult to pool all the data.
Microtubule gliding assays
A flow cell was prepared, and microtubules were assembled as described in the Microtubule dynamics assay section. Biotinylated motors were generated by coexpression of motors tagged with the 15-aa AviTag and the bacterial biotin ligase BirA fused with HA tag (HA-BirA) in COS-7 cells. Biotinylated motors were attached to the coverslip surface by sequential incubation of flow cells with (A) 1 mg/ml BSA-biotin, (B) blocking buffer (0.5 mg/ml casein and 10 µM taxol in BRB80), (C) 0.5 mg/ml NeutrAvidin, (D) blocking buffer, and (E) cell lysates with 2 mM ATP and 0.5 mg/ml casein in BRB80 and blocking buffer. Taxol-stabilized HiLyte 647–labeled microtubules in motility mixture (2 mM ATP, 10 µM taxol, 2 mM MgCl2, and oxygen scavenging in BRB80) were then added, and the flow cells were sealed with molten paraffin wax and imaged by TIRF microscopy. For KIF27 and KIF7, images were acquired at one frame every 3 s for 3 min. For KIF4 and KIF21A, images were acquired continuously at 50 ms per frame for 15 s. Maximum-intensity projections were generated, and the kymographs were produced by drawing along these tracks (width = 3 pixels) using ImageJ. Stalled events were ignored. Velocity was defined as the distance on the y axis of the kymograph divided by the time on the x axis of the kymograph.
Statistical tests
Statistical analysis and plots were generated by Origin or Prism (GraphPad Software). The error bars indicate SEM.
Kinetic assays
Kinetic experiments were conducted at 298 K in ATPase buffer (10 mM Hepes, pH 7.2, using sodium hydroxide, 2 mM magnesium chloride, 10 mM potassium chloride, and 5% [vol/vol] glycerol) unless otherwise noted.
Steady-state ATPase measurements
The basal and microtubule-stimulated ATPase activities of KIF27 and KIF7 were measured by the NADH-coupled assay (Hass et al., 1961; Imamura et al., 1966; Trentham et al., 1972; Furch et al., 1998; De La Cruz et al., 2000). In brief, KIF only or microtubule • KIF was diluted to 2× final concentration in ATPase buffer. ATP was also diluted to 2× final concentration in ATPase buffer plus 2× NADH cocktail (final concentrations: 1 mM phosphoenolpyruvate, 0.8 mM NADH, 10 U/ml rabbit pyruvate kinase [Roche], and 10 U/ml lactic dehydrogenase [Sigma-Aldrich]). To initiate the reaction, equal volumes (80 µl) of KIF only or microtubule • KIF complex as well as ATP and NADH cocktail were thoroughly mixed by pipetting, and 150 µl of each reaction was transferred to a 96-well microplate for absorbance reading at 340 nm using a microplate spectrophotometer (BioTek). A standard curve from 0–800 µM NADH was used to convert A340 to ADP product concentration.
Two control reactions were performed: control 1, no kinesin with all the components of the assay at high microtubule concentration (10 µM) and high ATP concentration (1 mM); and control 2, no kinesin and no ATP with all the components of the assay. Both control reactions gave similar results, and the mean observed velocity was subtracted from the velocities of the KIF27 reactions in Fig. 8 (B and C).
For ATP-dependent experiments, the initial ATPase velocity was plotted against the ATP concentration, and the data were fit to the Michaelis-Menten equation:
where [E0] is the total concentration of KIF, kcat is the maximum rate of ATP turnover per KIF active site at infinite [ATP], and Km,ATP is the Michaelis constant, which is defined as the [ATP] that yields 1/2 [E0]kcat. For microtubule-dependent experiments, the initial ATPase velocity was plotted against the microtubule (MT) concentration, and the data were fit to the quadratic equation:
where [E0] is the total KIF concentration, kcat is the maximum rate of ATP turnover per KIF active site at infinite [MT], K0.5,MT is the [MT] that yields 1/2 [E0]kcat, and kbasal is the experimentally determined basal rate of ATP turnover at zero [MT]. The basal rate was determined using NADH-coupled and malachite green assays; both assays gave similar rates, and this data point (i.e., zero [MT]) was included with the fit as the y intercept.
Presteady-state kinetic experiments
Stopped-flow measurements were performed at 298 K using a SF-300X stopped-flow apparatus (KinTek Corp.) equipped with a Xenon arc lamp (Hamamatsu Photonics). Kinetics of the interaction of mant-ADP with KIF7 was measured by equilibrating the KIF7 mant-ADP (0.5 µM:0.25 µM) complex followed by rapid mixing with 500 µM ATP to chase the mant-ADP from the active site and with or without 2 µM microtubules or with or without 5 mM EDTA as described previously (Woodward et al., 1991). Mant fluorescence was monitored over time, λex,max equaled 356 nm, and λem,max equaled 448 nm (400 nm long-pass filter). Each averaged transient was fit to a single exponential equation.
Online supplemental material
Fig. S1 shows sequence alignment and rooted phylogenetic tree of the motor domains of kinesin-4 family motors. Fig. S2 shows plots of the effects of KIF27 and KIF7 on the frequencies of microtubule growth and catastrophe in the in vitro microtubule dynamics assay. Fig. S3 shows biotinylation of AviTag motors in COS-7 cells. Fig. S4 shows the localization of kinesin-4 motors and peroxisomes before and after addition of rapamycin in COS-7 cells. Fig. S5 shows fluorescence-based microtubule affinity across kinesin-4 motors in different nucleotide states.
Supplementary Material
Acknowledgments
We thank members of the Cochran, Grant, and Verhey laboratories for helpful discussions. We thank the Drosophila Genomics Resource Center, supported by National Institutes of Health grant 2P40OD010949, for plasmids and cells. We thank Gary Banker, Greg Gundersen, and Casper Hoogenraad for plasmids.
This work was funded by grants from the National Institutes of Health to K.J. Verhey and B.J. Grant (R01GM070862) and from the National Science Foundation to J.C. Cochran (MCB 1614514).
The authors declare no competing financial interests.
Author contributions Y. Yue, B.J. Grant, J.C. Cochran, and K.J. Verhey designed the research. Y. Yue, T.L. Blasius, S. Zhang, S. Jariwala, and B. Walker performed the research. Y. Yue, S. Jariwala, B.J. Grant, and J.C. Cochran analyzed data. Y. Yue and K.J. Verhey wrote the paper with input from all authors.
References
- Aikin R.A., Ayers K.L., and Thérond P.P.. 2008. The role of kinases in the Hedgehog signalling pathway. EMBO Rep. 9:330–336. 10.1038/embor.2008.38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albracht C.D., Rank K.C., Obrzut S., Rayment I., and Gilbert S.P.. 2014. Kinesin-2 KIF3AB exhibits novel ATPase characteristics. J. Biol. Chem. 289:27836–27848. 10.1074/jbc.M114.583914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arellano-Santoyo H., Geyer E.A., Stokasimov E., Chen G.-Y., Su X., Hancock W., Rice L.M., and Pellman D.. 2017. A Tubulin Binding Switch Underlies Kip3/Kinesin-8 Depolymerase Activity. Dev. Cell. 42:37–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asenjo A.B., Chatterjee C., Tan D., DePaoli V., Rice W.J., Diaz-Avalos R., Silvestry M., and Sosa H.. 2013. Structural model for tubulin recognition and deformation by kinesin-13 microtubule depolymerases. Cell Reports. 3:759–768. 10.1016/j.celrep.2013.01.030 [DOI] [PubMed] [Google Scholar]
- Auerbach S.D., and Johnson K.A.. 2005. Alternating site ATPase pathway of rat conventional kinesin. J. Biol. Chem. 280:37048–37060. 10.1074/jbc.M502984200 [DOI] [PubMed] [Google Scholar]
- Bianchi S., van Riel W.E., Kraatz S.H., Olieric N., Frey D., Katrukha E.A., Jaussi R., Missimer J., Grigoriev I., Olieric V., et al. 2016. Structural basis for misregulation of kinesin KIF21A autoinhibition by CFEOM1 disease mutations. Sci. Rep. 6:30668 10.1038/srep30668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieling P., Telley I.A., and Surrey T.. 2010. A minimal midzone protein module controls formation and length of antiparallel microtubule overlaps. Cell. 142:420–432. 10.1016/j.cell.2010.06.033 [DOI] [PubMed] [Google Scholar]
- Bisbal M., Wojnacki J., Peretti D., Ropolo A., Sesma J., Jausoro I., and Cáceres A.. 2009. KIF4 mediates anterograde translocation and positioning of ribosomal constituents to axons. J. Biol. Chem. 284:9489–9497. 10.1074/jbc.M808586200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bringmann H., Skiniotis G., Spilker A., Kandels-Lewis S., Vernos I., and Surrey T.. 2004. A kinesin-like motor inhibits microtubule dynamic instability. Science. 303:1519–1522. 10.1126/science.1094838 [DOI] [PubMed] [Google Scholar]
- Cai D., McEwen D.P., Martens J.R., Meyhofer E., and Verhey K.J.. 2009. Single molecule imaging reveals differences in microtubule track selection between Kinesin motors. PLoS Biol. 7:e1000216 10.1371/journal.pbio.1000216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Case R.B., Pierce D.W., Hom-Booher N., Hart C.L., and Vale R.D.. 1997. The directional preference of kinesin motors is specified by an element outside of the motor catalytic domain. Cell. 90:959–966. 10.1016/S0092-8674(00)80360-8 [DOI] [PubMed] [Google Scholar]
- Chen G.-Y., Kang Y.J., Gayek A.S., Youyen W., Tüzel E., Ohi R., and Hancock W.O.. 2017. Eg5 inhibitors have contrasting effects on microtubule stability and metaphase spindle integrity. ACS Chem. Biol. 12:1038–1046. 10.1021/acschembio.6b01040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng L., Desai J., Miranda C.J., Duncan J.S., Qiu W., Nugent A.A., Kolpak A.L., Wu C.C., Drokhlyansky E., Delisle M.M., et al. 2014. Human CFEOM1 mutations attenuate KIF21A autoinhibition and cause oculomotor axon stalling. Neuron. 82:334–349. 10.1016/j.neuron.2014.02.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cochran J.C. 2015. Kinesin Motor Enzymology: Chemistry, Structure, and Physics of Nanoscale Molecular Machines. Biophys. Rev. 7:269–299. 10.1007/s12551-014-0150-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cochran J.C., Thompson M.E., and Kull F.J.. 2013. Metal switch-controlled myosin II from Dictyostelium discoideum supports closure of nucleotide pocket during ATP binding coupled to detachment from actin filaments. J. Biol. Chem. 288:28312–28323. 10.1074/jbc.M113.466045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crevel I.M., Lockhart A., and Cross R.A.. 1996. Weak and strong states of kinesin and ncd. J. Mol. Biol. 257:66–76. 10.1006/jmbi.1996.0147 [DOI] [PubMed] [Google Scholar]
- Cull M.G., and Schatz P.J.. 2000. Biotinylation of proteins in vivo and in vitro using small peptide tags. Methods Enzymol. 326:430–440. 10.1016/S0076-6879(00)26068-0 [DOI] [PubMed] [Google Scholar]
- de Keijzer J., Kieft H., Ketelaar T., Goshima G., and Janson M.E.. 2017. Shortening of microtubule overlap regions defines membrane delivery sites during plant cytokinesis. Curr. Biol. 27:514–520. 10.1016/j.cub.2016.12.043 [DOI] [PubMed] [Google Scholar]
- De La Cruz E.M., Sweeney H.L., and Ostap E.M.. 2000. ADP inhibition of myosin V ATPase activity. Biophys. J. 79:1524–1529. 10.1016/S0006-3495(00)76403-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R.C. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32:1792–1797. 10.1093/nar/gkh340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelke M.F., Winding M., Yue Y., Shastry S., Teloni F., Reddy S., Blasius T.L., Soppina P., Hancock W.O., Gelfand V.I., and Verhey K.J.. 2016. Engineered kinesin motor proteins amenable to small-molecule inhibition. Nat. Commun. 7:11159 10.1038/ncomms11159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farzan S.F., Ascano M. Jr., Ogden S.K., Sanial M., Brigui A., Plessis A., and Robbins D.J.. 2008. Costal2 functions as a kinesin-like protein in the hedgehog signal transduction pathway. Curr. Biol. 18:1215–1220. 10.1016/j.cub.2008.07.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn R.D., Bateman A., Clements J., Coggill P., Eberhardt R.Y., Eddy S.R., Heger A., Hetherington K., Holm L., Mistry J., et al. 2014. Pfam: the protein families database. Nucleic Acids Res. 42(D1):D222–D230. 10.1093/nar/gkt1223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friel C.T., and Howard J.. 2011. The kinesin-13 MCAK has an unconventional ATPase cycle adapted for microtubule depolymerization. EMBO J. 30:3928–3939. 10.1038/emboj.2011.290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furch M., Geeves M.A., and Manstein D.J.. 1998. Modulation of actin affinity and actomyosin adenosine triphosphatase by charge changes in the myosin motor domain. Biochemistry. 37:6317–6326. 10.1021/bi972851y [DOI] [PubMed] [Google Scholar]
- Furuta K., and Toyoshima Y.Y.. 2008. Minus-end-directed motor Ncd exhibits processive movement that is enhanced by microtubule bundling in vitro. Curr. Biol. 18:152–157. 10.1016/j.cub.2007.12.056 [DOI] [PubMed] [Google Scholar]
- Furuta K., Furuta A., Toyoshima Y.Y., Amino M., Oiwa K., and Kojima H.. 2013. Measuring collective transport by defined numbers of processive and nonprocessive kinesin motors. Proc. Natl. Acad. Sci. USA. 110:501–506. 10.1073/pnas.1201390110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganguly A., DeMott L., and Dixit R.. 2017. The Arabidopsis kinesin-4, FRA1, requires a high level of processive motility to function correctly. J. Cell Sci. 130:1232–1238. 10.1242/jcs.196857 [DOI] [PubMed] [Google Scholar]
- Gascuel O. 1997. BIONJ: an improved version of the NJ algorithm based on a simple model of sequence data. Mol. Biol. Evol. 14:685–695. 10.1093/oxfordjournals.molbev.a025808 [DOI] [PubMed] [Google Scholar]
- Ghiretti A.E., Thies E., Tokito M.K., Lin T., Ostap E.M., Kneussel M., and Holzbaur E.L.F.. 2016. Activity-Dependent Regulation of Distinct Transport and Cytoskeletal Remodeling Functions of the Dendritic Kinesin KIF21B. Neuron. 92:857–872. 10.1016/j.neuron.2016.10.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gille C., and Frömmel C.. 2001. STRAP: editor for STRuctural Alignments of Proteins. Bioinformatics. 17:377–378. 10.1093/bioinformatics/17.4.377 [DOI] [PubMed] [Google Scholar]
- Gouy M., Guindon S., and Gascuel O.. 2010. SeaView version 4: A multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol. Biol. Evol. 27:221–224. 10.1093/molbev/msp259 [DOI] [PubMed] [Google Scholar]
- Gudimchuk N., Vitre B., Kim Y., Kiyatkin A., Cleveland D.W., Ataullakhanov F.I., and Grishchuk E.L.. 2013. Kinetochore kinesin CENP-E is a processive bi-directional tracker of dynamic microtubule tips. Nat. Cell Biol. 15:1079–1088. 10.1038/ncb2831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzik-Lendrum S., Heissler S.M., Billington N., Takagi Y., Yang Y., Knight P.J., Homsher E., and Sellers J.R.. 2013. Mammalian myosin-18A, a highly divergent myosin. J. Biol. Chem. 288:9532–9548. 10.1074/jbc.M112.441238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond J.W., Cai D., Blasius T.L., Li Z., Jiang Y., Jih G.T., Meyhofer E., and Verhey K.J.. 2009. Mammalian Kinesin-3 motors are dimeric in vivo and move by processive motility upon release of autoinhibition. PLoS Biol. 7:e1000072 10.1371/journal.pbio.1000072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond J.W., Blasius T.L., Soppina V., Cai D., and Verhey K.J.. 2010. Autoinhibition of the kinesin-2 motor KIF17 via dual intramolecular mechanisms. J. Cell Biol. 189:1013–1025. 10.1083/jcb.201001057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hass L.F., Boyer P.D., and Reynard A.M.. 1961. Studies on possible phosphoryl enzyme formation in catalysis by hexokinase, pyruvate kinase, and glucose 6-phosphatase. J. Biol. Chem. 236:2284–2291. [PubMed] [Google Scholar]
- He M., Subramanian R., Bangs F., Omelchenko T., Liem K.F. Jr., Kapoor T.M., and Anderson K.V.. 2014. The kinesin-4 protein Kif7 regulates mammalian Hedgehog signalling by organizing the cilium tip compartment. Nat. Cell Biol. 16:663–672. 10.1038/ncb2988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helenius J., Brouhard G., Kalaidzidis Y., Diez S., and Howard J.. 2006. The depolymerizing kinesin MCAK uses lattice diffusion to rapidly target microtubule ends. Nature. 441:115–119. 10.1038/nature04736 [DOI] [PubMed] [Google Scholar]
- Hirokawa N., Noda Y., Tanaka Y., and Niwa S.. 2009. Kinesin superfamily motor proteins and intracellular transport. Nat. Rev. Mol. Cell Biol. 10:682–696. 10.1038/nrm2774 [DOI] [PubMed] [Google Scholar]
- Hu C.-K., Coughlin M., Field C.M., and Mitchison T.J.. 2011. KIF4 regulates midzone length during cytokinesis. Curr. Biol. 21:815–824. 10.1016/j.cub.2011.04.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C.F., and Banker G.. 2012. The translocation selectivity of the kinesins that mediate neuronal organelle transport. Traffic. 13:549–564. 10.1111/j.1600-0854.2011.01325.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huckaba T.M., Gennerich A., Wilhelm J.E., Chishti A.H., and Vale R.D.. 2011. Kinesin-73 is a processive motor that localizes to Rab5-containing organelles. J. Biol. Chem. 286:7457–7467. 10.1074/jbc.M110.167023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter A.W., Caplow M., Coy D.L., Hancock W.O., Diez S., Wordeman L., and Howard J.. 2003. The kinesin-related protein MCAK is a microtubule depolymerase that forms an ATP-hydrolyzing complex at microtubule ends. Mol. Cell. 11:445–457. 10.1016/S1097-2765(03)00049-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imamura K., Tada M., and Tonomura Y.. 1966. The pre-steady state of the myosin--adenosine triphosphate system. IV. Liberation of ADP from the myosin--ATP system and effects of modifiers on the phosphorylation of myosin. J. Biochem. 59:280–289. [PubMed] [Google Scholar]
- Ingham P.W., Nakano Y., and Seger C.. 2011. Mechanisms and functions of Hedgehog signalling across the metazoa. Nat. Rev. Genet. 12:393–406. 10.1038/nrg2984 [DOI] [PubMed] [Google Scholar]
- Jonsson E., Yamada M., Vale R.D., and Goshima G.. 2015. Clustering of a kinesin-14 motor enables processive retrograde microtubule-based transport in plants. Nat. Plants. 1:15087 10.1038/nplants.2015.87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapitein L.C., Schlager M.A., van der Zwan W.A., Wulf P.S., Keijzer N., and Hoogenraad C.C.. 2010. Probing intracellular motor protein activity using an inducible cargo trafficking assay. Biophys. J. 99:2143–2152. 10.1016/j.bpj.2010.07.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H., Fonseca C., and Stumpff J.. 2014. A unique kinesin-8 surface loop provides specificity for chromosome alignment. Mol. Biol. Cell. 25:3319–3329. 10.1091/mbc.E14-06-1132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M. 1983. The neutral theory of molecular evolution. Cambridge University Press, Cambridge, England, UK. 367 pp. 10.1017/CBO9780511623486 [DOI] [Google Scholar]
- Klejnot M., and Kozielski F.. 2012. Structural insights into human Kif7, a kinesin involved in Hedgehog signalling. Acta Crystallogr. D Biol. Crystallogr. 68:154–159. 10.1107/S0907444911053042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klumpp L.M., Hoenger A., and Gilbert S.P.. 2004. Kinesin’s second step. Proc. Natl. Acad. Sci. USA. 101:3444–3449. 10.1073/pnas.0307691101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong Z., Ioki M., Braybrook S., Li S., Ye Z.-H., Julie Lee Y.R., Hotta T., Chang A., Tian J., Wang G., and Liu B.. 2015. Kinesin-4 functions in vesicular transport on cortical microtubules and regulates cell wall mechanics during cell elongation in plants. Mol. Plant. 8:1011–1023. 10.1016/j.molp.2015.01.004 [DOI] [PubMed] [Google Scholar]
- Krzysiak T.C., and Gilbert S.P.. 2006. Dimeric Eg5 maintains processivity through alternating-site catalysis with rate-limiting ATP hydrolysis. J. Biol. Chem. 281:39444–39454. 10.1074/jbc.M608056200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kull F.J., Sablin E.P., Lau R., Fletterick R.J., and Vale R.D.. 1996. Crystal structure of the kinesin motor domain reveals a structural similarity to myosin. Nature. 380:550–555. 10.1038/380550a0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzhandaivel A., Schultz S.W., Alkhori L., and Alenius M.. 2014. Cilia-mediated hedgehog signaling in Drosophila. Cell Reports. 7:672–680. 10.1016/j.celrep.2014.03.052 [DOI] [PubMed] [Google Scholar]
- Labonté D., Thies E., and Kneussel M.. 2014. The kinesin KIF21B participates in the cell surface delivery of γ2 subunit-containing GABAA receptors. Eur. J. Cell Biol. 93:338–346. 10.1016/j.ejcb.2014.07.007 [DOI] [PubMed] [Google Scholar]
- Lee K.-H., Lee J.S., Lee D., Seog D.-H., Lytton J., Ho W.-K., and Lee S.-H.. 2012. KIF21A-mediated axonal transport and selective endocytosis underlie the polarized targeting of NCKX2. J. Neurosci. 32:4102–4117. 10.1523/JNEUROSCI.6331-11.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee R.T.H., Zhao Z., and Ingham P.W.. 2016. Hedgehog signalling. Development. 143:367–372. 10.1242/dev.120154 [DOI] [PubMed] [Google Scholar]
- Ma Y.-Z., and Taylor E.W.. 1995. Kinetic mechanism of kinesin motor domain. Biochemistry. 34:13233–13241. 10.1021/bi00040a039 [DOI] [PubMed] [Google Scholar]
- Marks S.A., and Kalderon D.. 2011. Regulation of mammalian Gli proteins by Costal 2 and PKA in Drosophila reveals Hedgehog pathway conservation. Development. 138:2533–2542. 10.1242/dev.063479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez N.W., Xue X., Berro R.G., Kreitzer G., and Resh M.D.. 2008. Kinesin KIF4 regulates intracellular trafficking and stability of the human immunodeficiency virus type 1 Gag polyprotein. J. Virol. 82:9937–9950. 10.1128/JVI.00819-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthies H.J., Baskin R.J., and Hawley R.S.. 2001. Orphan kinesin NOD lacks motile properties but does possess a microtubule-stimulated ATPase activity. Mol. Biol. Cell. 12:4000–4012. 10.1091/mbc.12.12.4000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurya A.K., Ben J., Zhao Z., Lee R.T.H., Niah W., Ng A.S.M., Iyu A., Yu W., Elworthy S., van Eeden F.J., and Ingham P.W.. 2013. Positive and negative regulation of Gli activity by Kif7 in the zebrafish embryo. PLoS Genet. 9:e1003955 10.1371/journal.pgen.1003955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris E.J., Nader G.P., Ramalingam N., Bartolini F., and Gundersen G.G.. 2014. Kif4 interacts with EB1 and stabilizes microtubules downstream of Rho-mDia in migrating fibroblasts. PLoS One. 9:e91568 10.1371/journal.pone.0091568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhia M., Thies E., Labonté D., Ghiretti A.E., Gromova K.V., Xompero F., Lappe-Siefke C., Hermans-Borgmeyer I., Kuhl D., Schweizer M., et al. 2016. The kinesin KIF21B regulates microtubule dynamics and is essential for neuronal morphology, synapse function, and learning and memory. Cell Reports. 15:968–977. 10.1016/j.celrep.2016.03.086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakata T., and Hirokawa N.. 1995. Point mutation of adenosine triphosphate-binding motif generated rigor kinesin that selectively blocks anterograde lysosome membrane transport. J. Cell Biol. 131:1039–1053. 10.1083/jcb.131.4.1039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nitta R., Okada Y., and Hirokawa N.. 2008. Structural model for strain-dependent microtubule activation of Mg-ADP release from kinesin. Nat. Struct. Mol. Biol. 15:1067–1075. 10.1038/nsmb.1487 [DOI] [PubMed] [Google Scholar]
- Norris S.R., Núñez M.F., and Verhey K.J.. 2015. Influence of fluorescent tag on the motility properties of kinesin-1 in single-molecule assays. Biophys. J. 108:1133–1143. 10.1016/j.bpj.2015.01.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nozawa Y.I., Lin C., and Chuang P.-T.. 2013. Hedgehog signaling from the primary cilium to the nucleus: an emerging picture of ciliary localization, trafficking and transduction. Curr. Opin. Genet. Dev. 23:429–437. 10.1016/j.gde.2013.04.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan X., Ou G., Civelekoglu-Scholey G., Blacque O.E., Endres N.F., Tao L., Mogilner A., Leroux M.R., Vale R.D., and Scholey J.M.. 2006. Mechanism of transport of IFT particles in C. elegans cilia by the concerted action of kinesin-II and OSM-3 motors. J. Cell Biol. 174:1035–1045. 10.1083/jcb.200606003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paradis E., Claude J., and Strimmer K.. 2004. APE: analyses of phylogenetics and evolution in R language. Bioinformatics. 20:289–290. 10.1093/bioinformatics/btg412 [DOI] [PubMed] [Google Scholar]
- Patel J.T., Belsham H.R., Rathbone A.J., Wickstead B., Gell C., and Friel C.T.. 2016. The family-specific α4-helix of the kinesin-13, MCAK, is critical to microtubule end recognition. Open Biol. 6:160223 10.1098/rsob.160223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peretti D., Peris L., Rosso S., Quiroga S., and Cáceres A.. 2000. Evidence for the involvement of KIF4 in the anterograde transport of L1-containing vesicles. J. Cell Biol. 149:141–152. 10.1083/jcb.149.1.141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierce D.W., Hom-Booher N., Otsuka A.J., and Vale R.D.. 1999. Single-molecule behavior of monomeric and heteromeric kinesins. Biochemistry. 38:5412–5421. 10.1021/bi9830009 [DOI] [PubMed] [Google Scholar]
- Powers J., Rose D.J., Saunders A., Dunkelbarger S., Strome S., and Saxton W.M.. 2004. Loss of KLP-19 polar ejection force causes misorientation and missegregation of holocentric chromosomes. J. Cell Biol. 166:991–1001. 10.1083/jcb.200403036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravindran M.S., Engelke M.F., Verhey K.J., and Tsai B.. 2017. Exploiting the kinesin-1 molecular motor to generate a virus membrane penetration site. Nat. Commun. 8:15496 10.1038/ncomms15496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richard J., Kim E.D., Nguyen H., Kim C.D., and Kim S.. 2016. Allostery Wiring Map for Kinesin Energy Transduction and Its Evolution. J. Biol. Chem. 291:20932–20945. 10.1074/jbc.M116.733675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards T.A., and Cavalier-Smith T.. 2005. Myosin domain evolution and the primary divergence of eukaryotes. Nature. 436:1113–1118. 10.1038/nature03949 [DOI] [PubMed] [Google Scholar]
- Sanchez G.M., Alkhori L., Hatano E., Schultz S.W., Kuzhandaivel A., Jafari S., Granseth B., and Alenius M.. 2016. Hedgehog signaling regulates the ciliary transport of odorant receptors in Drosophila. Cell Reports. 14:464–470. 10.1016/j.celrep.2015.12.059 [DOI] [PubMed] [Google Scholar]
- Sardar H.S., and Gilbert S.P.. 2012. Microtubule capture by mitotic kinesin centromere protein E (CENP-E). J. Biol. Chem. 287:24894–24904. 10.1074/jbc.M112.376830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sardar H.S., Luczak V.G., Lopez M.M., Lister B.C., and Gilbert S.P.. 2010. Mitotic kinesin CENP-E promotes microtubule plus-end elongation. Curr. Biol. 20:1648–1653. 10.1016/j.cub.2010.08.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sisson J.C., Ho K.S., Suyama K., and Scott M.P.. 1997. Costal2, a novel kinesin-related protein in the Hedgehog signaling pathway. Cell. 90:235–245. 10.1016/S0092-8674(00)80332-3 [DOI] [PubMed] [Google Scholar]
- Skjærven L., Yao X.-Q., Scarabelli G., and Grant B.J.. 2014. Integrating protein structural dynamics and evolutionary analysis with Bio3D. BMC Bioinformatics. 15:399 10.1186/s12859-014-0399-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soppina V., Norris S.R., Dizaji A.S., Kortus M., Veatch S., Peckham M., and Verhey K.J.. 2014. Dimerization of mammalian kinesin-3 motors results in superprocessive motion. Proc. Natl. Acad. Sci. USA. 111:5562–5567. 10.1073/pnas.1400759111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturgill E.G., Norris S.R., Guo Y., and Ohi R.. 2016. Kinesin-5 inhibitor resistance is driven by kinesin-12. J. Cell Biol. 213:213–227. 10.1083/jcb.201507036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian R., Ti S.-C., Tan L., Darst S.A., and Kapoor T.M.. 2013. Marking and measuring single microtubules by PRC1 and kinesin-4. Cell. 154:377–390. 10.1016/j.cell.2013.06.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirat A., Freuler F., Stettler T., Mayr L.M., and Leder L.. 2006. Evaluation of two novel tag-based labelling technologies for site-specific modification of proteins. Int. J. Biol. Macromol. 39:66–76. 10.1016/j.ijbiomac.2006.01.012 [DOI] [PubMed] [Google Scholar]
- Tomishige M., Klopfenstein D.R., and Vale R.D.. 2002. Conversion of Unc104/KIF1A kinesin into a processive motor after dimerization. Science. 297:2263–2267. 10.1126/science.1073386 [DOI] [PubMed] [Google Scholar]
- Trentham D.R., Bardsley R.G., Eccleston J.F., and Weeds A.G.. 1972. Elementary processes of the magnesium ion-dependent adenosine triphosphatase activity of heavy meromyosin. A transient kinetic approach to the study of kinases and adenosine triphosphatases and a colorimetric inorganic phosphate assay in situ. Biochem. J. 126:635–644. 10.1042/bj1260635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valentine M.T., and Gilbert S.P.. 2007. To step or not to step? How biochemistry and mechanics influence processivity in Kinesin and Eg5. Curr. Opin. Cell Biol. 19:75–81. 10.1016/j.ceb.2006.12.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Vaart B., van Riel W.E., Doodhi H., Kevenaar J.T., Katrukha E.A., Gumy L., Bouchet B.P., Grigoriev I., Spangler S.A., Yu K.L., et al. 2013. CFEOM1-associated kinesin KIF21A is a cortical microtubule growth inhibitor. Dev. Cell. 27:145–160. 10.1016/j.devcel.2013.09.010 [DOI] [PubMed] [Google Scholar]
- van Riel W.E., Rai A., Bianchi S., Katrukha E.A., Liu Q., Heck A.J., Hoogenraad C.C., Steinmetz M.O., Kapitein L.C., and Akhmanova A.. 2017. Kinesin-4 KIF21B is a potent microtubule pausing factor. eLife. 6:e24746 10.7554/eLife.24746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhey K.J., and Hammond J.W.. 2009. Traffic control: regulation of kinesin motors. Nat. Rev. Mol. Cell Biol. 10:765–777. 10.1038/nrm2782 [DOI] [PubMed] [Google Scholar]
- Vogel P., Read R., Hansen G., Wingert J., Dacosta C.M., Buhring L.M., and Shadoan M.. 2011. Pathology of congenital generalized lipodystrophy in Agpat2-/- mice. Vet. Pathol. 48:642–654. 10.1177/0300985810383870 [DOI] [PubMed] [Google Scholar]
- Walter W.J., Machens I., Rafieian F., and Diez S.. 2015. The non-processive rice kinesin-14 OsKCH1 transports actin filaments along microtubules with two distinct velocities. Nat. Plants. 1:15111 10.1038/nplants.2015.111 [DOI] [PubMed] [Google Scholar]
- Wang D., Nitta R., Morikawa M., Yajima H., Inoue S., Shigematsu H., Kikkawa M., and Hirokawa N.. 2016. Motility and microtubule depolymerization mechanisms of the Kinesin-8 motor, KIF19A. eLife. 5:e18101 10.7554/eLife.18101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Cantos-Fernandes S., Lv Y., Kuerban H., Ahmad S., Wang C., and Gigant B.. 2017. Insight into microtubule disassembly by kinesin-13s from the structure of Kif2C bound to tubulin. Nat. Commun. 8:70 10.1038/s41467-017-00091-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickstead B., Gull K., and Richards T.A.. 2010. Patterns of kinesin evolution reveal a complex ancestral eukaryote with a multifunctional cytoskeleton. BMC Evol. Biol. 10:110 10.1186/1471-2148-10-110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson C.W., Nguyen C.T., Chen M.-H., Yang J.-H., Gacayan R., Huang J., Chen J.-N., and Chuang P.-T.. 2009. Fused has evolved divergent roles in vertebrate Hedgehog signalling and motile ciliogenesis. Nature. 459:98–102. 10.1038/nature07883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodward S.K., Eccleston J.F., and Geeves M.A.. 1991. Kinetics of the interaction of 2‘(3’)-O-(N-methylanthraniloyl)-ATP with myosin subfragment 1 and actomyosin subfragment 1: characterization of two acto∙S1∙ADP complexes. Biochemistry. 30:422–430. 10.1021/bi00216a017 [DOI] [PubMed] [Google Scholar]
- Woolner S., and Bement W.M.. 2009. Unconventional myosins acting unconventionally. Trends Cell Biol. 19:245–252. 10.1016/j.tcb.2009.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yildiz A., and Vale R.D.. 2015. Tracking movements of the microtubule motors kinesin and dynein using total internal reflection fluorescence microscopy. Cold Spring Harb. Protoc. 10.1101/pdb.prot086355 [DOI] [PubMed] [Google Scholar]
- Yu G., Smith D.K., Zhu H., Guan Y., and Lam T.T.-Y.. 2017. ggtree: an R package for visualization and annotation of phylogenetic trees with their covariates and other associated data. Methods Ecol. Evol. 8:28–36. 10.1111/2041-210X.12628 [DOI] [Google Scholar]
- Zhu C., and Dixit R.. 2011. Single molecule analysis of the Arabidopsis FRA1 kinesin shows that it is a functional motor protein with unusually high processivity. Mol. Plant. 4:879–885. 10.1093/mp/ssr077 [DOI] [PubMed] [Google Scholar]
- Zhu C., and Jiang W.. 2005. Cell cycle-dependent translocation of PRC1 on the spindle by Kif4 is essential for midzone formation and cytokinesis. Proc. Natl. Acad. Sci. USA. 102:343–348. 10.1073/pnas.0408438102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu C., Ganguly A., Baskin T.I., McClosky D.D., Anderson C.T., Foster C., Meunier K.A., Okamoto R., Berg H., and Dixit R.. 2015. The fragile Fiber1 kinesin contributes to cortical microtubule-mediated trafficking of cell wall components. Plant Physiol. 167:780–792. 10.1104/pp.114.251462 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.