Mitochondrial stress induces PARL-mediated cleavage and cytosolic release of the mitochondrial phosphatase Pgam5. In the cytosol, Pgam5 interacts with the Wnt pathway component axin and dephosphorylates axin-bound β-catenin, thereby cell-intrinsically activating Wnt/β-catenin signaling to induce mitochondrial biogenesis.
Abstract
Mitochondrial abundance is dynamically regulated and was previously shown to be increased by Wnt/β-catenin signaling. Pgam5 is a mitochondrial phosphatase which is cleaved by the rhomboid protease presenilin-associated rhomboid-like protein (PARL) and released from membranes after mitochondrial stress. In this study, we show that Pgam5 interacts with the Wnt pathway component axin in the cytosol, blocks axin-mediated β-catenin degradation, and increases β-catenin levels and β-catenin–dependent transcription. Pgam5 stabilized β-catenin by inducing its dephosphorylation in an axin-dependent manner. Mitochondrial stress triggered by carbonyl cyanide m-chlorophenyl hydrazone (CCCP) treatment led to cytosolic release of endogenous Pgam5 and subsequent dephosphorylation of β-catenin, which was strongly diminished in Pgam5 and PARL knockout cells. Similarly, hypoxic stress generated cytosolic Pgam5 and led to stabilization of β-catenin, which was abolished by Pgam5 knockout. Cells stably expressing cytosolic Pgam5 exhibit elevated β-catenin levels and increased mitochondrial numbers. Our study reveals a novel mechanism by which damaged mitochondria might induce replenishment of the mitochondrial pool by cell-intrinsic activation of Wnt signaling via the Pgam5–β-catenin axis.
Introduction
The Wnt/β-catenin pathway is an evolutionary conserved signaling pathway involved in the regulation of fundamental processes such as patterning of body axis during development or maintenance of stem cells (Clevers and Nusse, 2012). Inappropriate activation of the Wnt pathway can cause various cancers, best characterized in colorectal cancer. In the absence of Wnt ligands, β-catenin is phosphorylated by a destruction complex consisting of the scaffold proteins axin and conductin (axin2), the tumor suppressor adenomatous polyposis coli, and the kinases casein kinase 1α (CK1α) and glycogen synthase kinase 3 (GSK3; van Kappel and Maurice, 2017). Phosphorylated β-catenin is recognized by the β-transducin repeat–containing protein E3 ubiquitin ligase, ubiquitinated, and proteasomally degraded (Aberle et al., 1997). Binding of Wnt ligands to receptor pairs of frizzled and low-density lipoprotein receptor–related protein 5 or 6 inhibits the destruction complex, resulting in β-catenin stabilization (MacDonald and He, 2012). Stabilized β-catenin interacts with T cell factor/lymphoid enhancer–binding factor transcription factors in the nucleus to induce transcription of its target genes (Behrens et al., 1996; Molenaar et al., 1996).
Pgam5 belongs to the phosphoglycerate mutase family. In contrast with other family members, Pgam5 functions as an atypical serine/threonine protein phosphatase instead of a phosphoglycerate mutase (Takeda et al., 2009). The N-terminal 35 amino acids including a transmembrane α-helix target Pgam5 to mitochondria (Lo and Hannink, 2008). However, the submitochondrial localization of Pgam5 remains controversial. Pgam5 has been reported to localize to the outer mitochondrial membrane (Lo and Hannink, 2008; Wang et al., 2012; Wu et al., 2014; Panda et al., 2016), the inner mitochondrial membrane (Sekine et al., 2012), or both (Chen et al., 2014). Of note, several described Pgam5 functions require its interaction with cytosolic or mitochondrial outer membrane proteins (Lo and Hannink, 2008; Wang et al., 2012; Chen et al., 2014; Wu et al., 2014; Kang et al., 2015; Panda et al., 2016). Upon loss of the mitochondrial membrane potential, Pgam5 is cleaved by the intramembrane-cleaving protease presenilin-associated rhomboid-like protein (PARL), leading to the release of the larger C-terminal part including the phosphatase domain from mitochondrial membranes (Sekine et al., 2012). Several mitochondrial stressors such as the chemical inhibitor of oxidative phosphorylation carbonyl cyanide m-chlorophenyl hydrazone (CCCP) can cause loss of the mitochondrial membrane potential, thereby inducing Pgam5 cleavage (Sekine et al., 2012; Wai et al., 2016). Pgam5 is involved in regulating cell death pathways such as apoptosis and necroptosis as well as mitochondrial turnover by inducing mitophagy after mitochondrial damage (Wang et al., 2012; Chen et al., 2014; Wu et al., 2014; He et al., 2017). It was recently shown that mitochondrial uncleaved Pgam5 can act as a negative regulator of Wnt/β-catenin signaling and that it dephosphorylates disheveled (Dvl), a positive regulator of Wnt signaling (Rauschenberger et al., 2017).
In this study, we characterize cytosolic Pgam5 as novel activator of Wnt/β-catenin signaling in contrast to its suppressive role in the pathway when localized to mitochondria, thereby establishing a dual role for Pgam5 in regulating Wnt/β-catenin signaling. We show that cleaved Pgam5 interacts with axin, the central scaffold protein in the destruction complex, in the cytosol. Binding of Pgam5 to axin results in dephosphorylation and therefore stabilization of β-catenin, and finally in the activation of β-catenin–dependent transcription. In addition, cytosolic Pgam5 increases the number of mitochondria, most likely by activating Wnt/β-catenin signaling. Thus, we identify Pgam5, which is released from dysfunctional mitochondria upon the loss of mitochondrial membrane potential and activates biogenesis of new functional mitochondria, as part of a feedback loop regulating mitochondrial homeostasis.
Results
The phosphatase Pgam5 interacts with the β-catenin destruction complex component axin
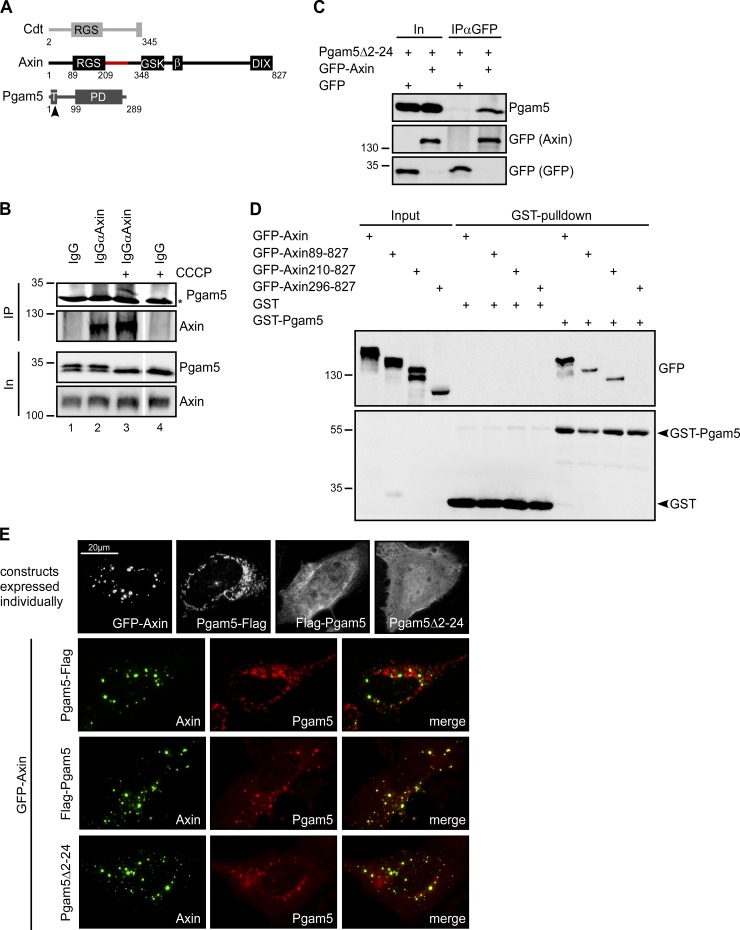
Using proteomic analysis, we found Pgam5 to coprecipitate with an N-terminal fragment of the axin family member axin2/conductin. This fragment encompassing the first 345 aa is depicted in Fig. 1 A. To confirm the interaction of Pgam5 with axin proteins, immunoprecipitation (IP) experiments were performed. Endogenous complexes of Pgam5 with axin did not coimmunoprecipitate efficiently using antiaxin antibodies (Fig. 1 B, lanes 1 and 2). We reasoned that Pgam5 bound to mitochondria might be poorly accessible to cytosolic axin under endogenous conditions. Therefore, we treated cells with the chemical uncoupler CCCP before IP, which induces cleavage of Pgam5 at the transmembrane region between amino acids 24 and 25, releasing the major part of the protein from the mitochondrial membrane (Fig. 1 A; Sekine et al., 2012). In Western blots, cleaved Pgam5 is represented by the lower of two Pgam5 bands (Fig. 1 B, In). Importantly, endogenous Pgam5 coprecipitated with endogenous axin after CCCP-induced cleavage (Fig. 1 B, lanes 3 and 4), indicating that axin interacts with Pgam5 after its release into the cytosol. Pgam5Δ2–24, which lacks the transmembrane region and localizes constitutively in the cytosol (Figs. 1 E and S1), also coprecipitated with axin (Fig. 1 C). To map the Pgam5 binding site in axin, pulldown experiments were performed with recombinant GST-tagged Pgam5 as bait. Full-length axin and N-terminally truncated axin constructs starting from amino acid 89 or 210 were pulled down with GST-Pgam5, whereas a construct starting from amino acid 296 was not (Fig. 1 D). Therefore, amino acids 210–295 of axin are critical for binding to Pgam5 (Fig. 1 A).
Figure 1.
Axin interacts with Pgam5. (A) Schematic presentation of conductin (Cdt) amino acids 2–345 used as a bait to identify new interaction partners by proteomics, axin, and Pgam5. β, β-catenin binding site; DIX, disheveled and axin (DIX) domain; GSK, GSK3 binding site; PD, PGAM domain; RGS, regulator of G protein signaling domain; T, transmembrane region. The arrowhead indicates the PARL cleavage position between amino acids 24 and 25. Red highlighting in axin marks the mapped Pgam5 binding site. The white line indicates that intervening lanes have been spliced out. (B) Western blotting for Pgam5 and axin after IP with IgGαAxin or control IgG from lysates of HEK293T cells treated with 25 µM CCCP for 2 h or left untreated. Input (In) shows similar levels of axin and Pgam5 as well as CCCP-induced Pgam5 cleavage indicated by the loss of the top Pgam5 band. The asterisk indicates an unspecific band. (C) Western blotting for Pgam5 and GFP in lysates of HEK293T cells transfected with Pgam5Δ2–24 together with GFP or GFP-axin (In) and after IP with a GFP antibody (IPαGFP) from these lysates. (D) Western blotting for GFP and GST in lysates of HEK293T cells transfected with indicated axin constructs (input) and after GST pulldown from these lysates using GST (control) or GST-Pgam5. Molecular masses are given in kilodaltons. (E) GFP fluorescence (axin) and immunofluorescence staining of Pgam5 in U2OS cells expressing GFP-axin, Pgam5-Flag, Flag-Pgam5, and Pgam5Δ2–24 either individually or in combinations as indicated on the left.
We next determined the intracellular localization of axin and Pgam5 when expressed alone and in combination. Transiently expressed Pgam5 (Pgam5; Pgam5-Flag) localized at mitochondria similar to endogenous Pgam5 (Fig. S1, top three rows). When the N-terminal mitochondrial localization sequence was deleted (Pgam5Δ2–24) or fused to an N-terminal tag (Flag-Pgam5 or Myc-Pgam5), Pgam5 localized in the cytosol and occasionally in the nucleus (Fig. S1, bottom three rows). Axin is typically located in round puncta representing β-catenin destruction complexes within the cytosol (Fig. 1 E; Fagotto et al., 1999; Fiedler et al., 2011). When coexpressed, mitochondria-associated Pgam5-Flag and axin did not colocalize. In contrast, cytosolic Pgam5 (Flag-Pgam5; Pgam5Δ2–24) almost completely colocalized with axin at puncta (Fig. 1 E).
Pgam5 is released from mitochondria into the cytosol upon mitochondrial stress
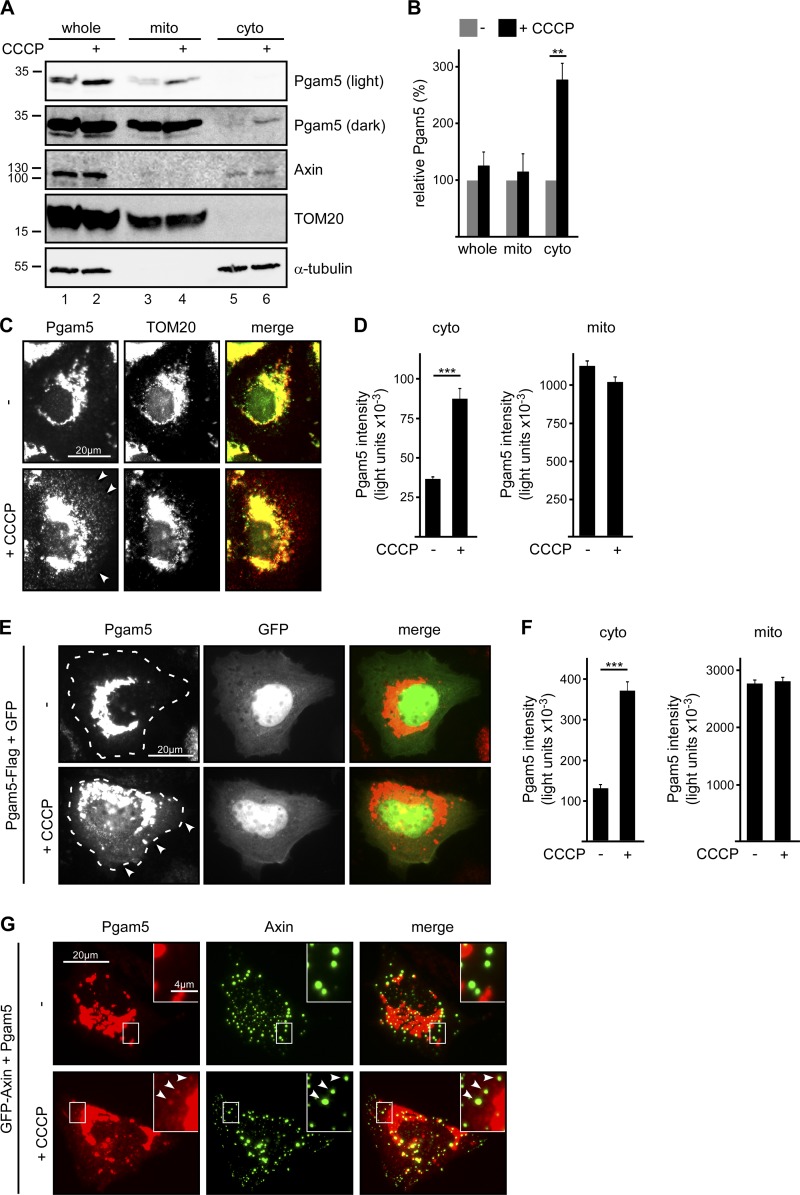
Because our data show interaction of cytosolic Pgam5 with axin, we hypothesized that mitochondrial Pgam5 might get released into the cytosol upon certain conditions. To test whether mitochondrial stress, which induces Pgam5 cleavage, also induces cytosolic release of Pgam5, cells were treated with the mitochondrial stressor CCCP. Biochemically, subcellular fractionation showed that a small but considerable fraction of cleaved Pgam5 is released into the cytosol upon CCCP treatment, increasing the amounts of cytosolic Pgam5 by about threefold (Fig. 2 A, lanes 5 and 6, and Fig. 2 B). The presence of Pgam5 in the cytosol of CCCP-treated cells was also shown by immunofluorescence staining of endogenous and transiently expressed Pgam5 (Fig. 2, C–G): A distinct fraction of endogenous Pgam5 was observed outside mitochondria in CCCP-treated cells as revealed by Pgam5 staining beyond staining of the mitochondrial-restricted translocase of outer membrane 20 (TOM20; Fig. 2, C and D). Similarly, Pgam5-Flag, which was largely restricted to mitochondrial structures in control cells, acquired an additional diffuse cytosolic localization upon CCCP treatment (Fig. 2, E and F). Moreover, untagged Pgam5 interacted with coexpressed GFP-axin in the cytosol of CCCP-treated cells as revealed by colocalization in axin puncta (Fig. 2 G).
Figure 2.
Pgam5 is released into the cytosol upon CCCP treatment. (A) Western blotting for endogenous Pgam5 and axin in whole-cell extracts (whole), mitochondrial fractions (mito), and cytosolic fractions (cyto) of U2OS cells that were treated with 100 µM CCCP for 2 h or left untreated. The mitochondrial protein TOM20 and the cytosolic protein α-tubulin served as loading controls and show fraction purity. The Pgam5 blot is shown in two versions of different brightness (light and dark). Molecular masses are given in kilodaltons. (B) Pgam5 band intensities were determined by 2D densitometry of four independent experiments as in A and normalized to TOM20 (whole and mitochondrial fractions) or α-tubulin (cytosolic fractions). Normalized Pgam5 intensities of extracts/fractions from untreated cells were set to 100%. n = 4. (C) Immunofluorescence staining of endogenous Pgam5 (red) and TOM20 (green) in U2OS cells that were treated with 100 µM CCCP for 4 h or left untreated. Arrowheads point to cytosolic Pgam5. (D) Quantification of Pgam5 intensity from four measuring sites per cell, two of which were randomly positioned within the cytosol and two within mitochondria of individual U2OS cells. n = 120 cells. (E) Immunofluorescence staining of Pgam5 (red) in U2OS cells transfected with Pgam5-Flag and GFP (green) that were treated with 100 µM CCCP for 4 h or left untreated. Dotted lines indicate cell borders according to GFP distribution. Arrowheads point to cytosolic Pgam5. (F) Quantification of cytosolic and mitochondrial Pgam5 of experiments described in E as described in D. Results are means ± SEM of four independent experiments. **, P < 0.01; ***, P < 0.001 (Student’s t test). (G) Immunofluorescence staining of Pgam5 (red) in U2OS cells transfected with untagged Pgam5 together with GFP-axin (green) that were treated with 100 µM CCCP for 4 h or left untreated. Insets are magnified in the top right corner. Arrowheads point to Pgam5 colocalizing with axin puncta.
Cytosolic Pgam5 activates Wnt/β-catenin signaling
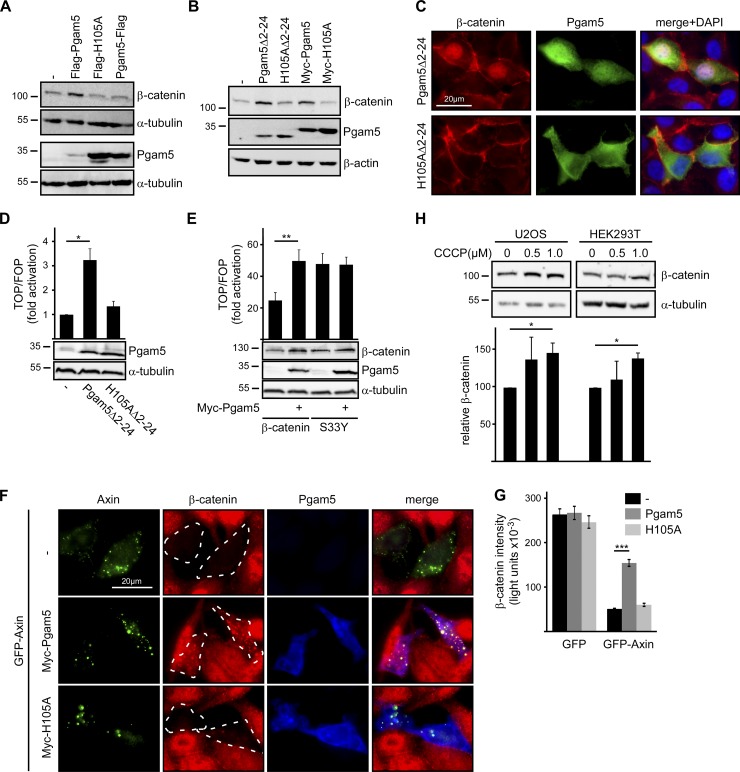
After validating the Pgam5–axin interaction, we investigated whether binding of cytosolic Pgam5 to axin alters Wnt/β-catenin pathway activity. Importantly, transient expression of cytosolic Pgam5 (Flag-Pgam5, Pgam5Δ2–24, and Myc-Pgam5) increased β-catenin levels, whereas mitochondria-associated Pgam5 (Pgam5-Flag) did not (Fig. 3, A and B). Stabilization of β-catenin by Pgam5 is dependent on its phosphatase activity as phosphatase-dead H105A mutants of Pgam5 did not increase β-catenin (Fig. 3, A and B). In addition to stabilization of β-catenin, cytosolic Pgam5 induced nuclear accumulation of β-catenin, which was observed by immunofluorescence staining in Pgam5Δ2–24-transfected cells but not in cells transfected with the phosphatase-inactive H105AΔ2–24 mutant (Fig. 3 C). Consistent with stabilization and nuclear accumulation of β-catenin, cytosolic Pgam5 (Pgam5Δ2–24) increased the transcriptional activity of the Wnt/β-catenin pathway as measured by the TCF optimal reporter (TOP)-FLASH reporter assay (Fig. 3 D). This increase again required the phosphatase activity of Pgam5. In addition, TOP-FLASH reporter activity induced by WT β-catenin expression was enhanced by coexpression of cytosolic Pgam5 (Myc-Pgam5), whereas TOP-FLASH activity induced by expression of the unphosphorylatable stable β-catenin mutant S33Y was not (Fig. 3 E). This indicates that activation of β-catenin–dependent transcription by Pgam5 depends on counteracting phosphorylation and degradation of β-catenin. Overexpression of axin in SW480 cells decreases the high β-catenin level present in this colorectal cancer cell line, which can be studied by immunofluorescence microscopy (Bernkopf et al., 2015). Degradation of β-catenin induced by axin was strongly attenuated by coexpression of cytosolic Pgam5 but not the H105A mutant (Fig. 3, F and G). Thus, Pgam5 interferes with axin-mediated degradation of β-catenin. Residual β-catenin that was not degraded in the presence of Pgam5 colocalized with axin in puncta, indicating that Pgam5 does not prevent axin–β-catenin interaction (Fig. 3 F). We next analyzed whether cytosolic release of Pgam5 by CCCP (Fig. 2) would also stabilize β-catenin levels. Indeed, treatment with low doses of CCCP for 72 h significantly increased β-catenin levels in two different cell lines (Fig. 3 H).
Figure 3.
Cytosolic Pgam5 activates Wnt/β-catenin signaling. (A) Western blotting for β-catenin in hypotonic lysates and for Pgam5 in detergent lysates of HEK293T cells transfected with plasmids indicated above the blots. α-Tubulin was used as a loading control. (B) Western blotting for β-catenin and Pgam5 in hypotonic lysates of HEK293T cells transfected with indicated plasmids. β-Actin was used as a loading control. (C) Immunofluorescence staining of endogenous β-catenin (red) and exogenous Pgam5 (green) in HEK293T cells transfected with Pgam5Δ2–24 or H105AΔ2–24. DAPI-stained nuclei are shown in blue in merge pictures. (D and E) Luciferase activity (TOP/TCF far-from-optimal reporter [FOP]) in HEK293T cells transfected with plasmids indicated below the blots. Results are means ± SEM of four independent experiments. Western blots show expression of transfected proteins. (F) Immunofluorescence staining of β-catenin (red) in SW480 cells transfected with GFP-axin (green) alone or together with either Myc-Pgam5 or Myc-H105A (blue). Dotted lines mark transfected cells. (G) Quantification of β-catenin fluorescence intensities in transfected SW480 cells from three independent experiments as described in F. Results are means ± SEM (n = 60). (H) Top: Western blotting for β-catenin and α-tubulin (loading control) in hypotonic lysates of U2OS or HEK293T cells treated with indicated CCCP concentrations for 72 h. Bottom: 2D densitometry quantification of β-catenin normalized to α-tubulin of four independent experiments. Results are means ± SEM (n = 4). *, P < 0.05; **, P < 0.01; ***, P < 0.001 (Student’s t test). Molecular masses are given in kilodaltons.
Pgam5 dephosphorylates axin-bound β-catenin
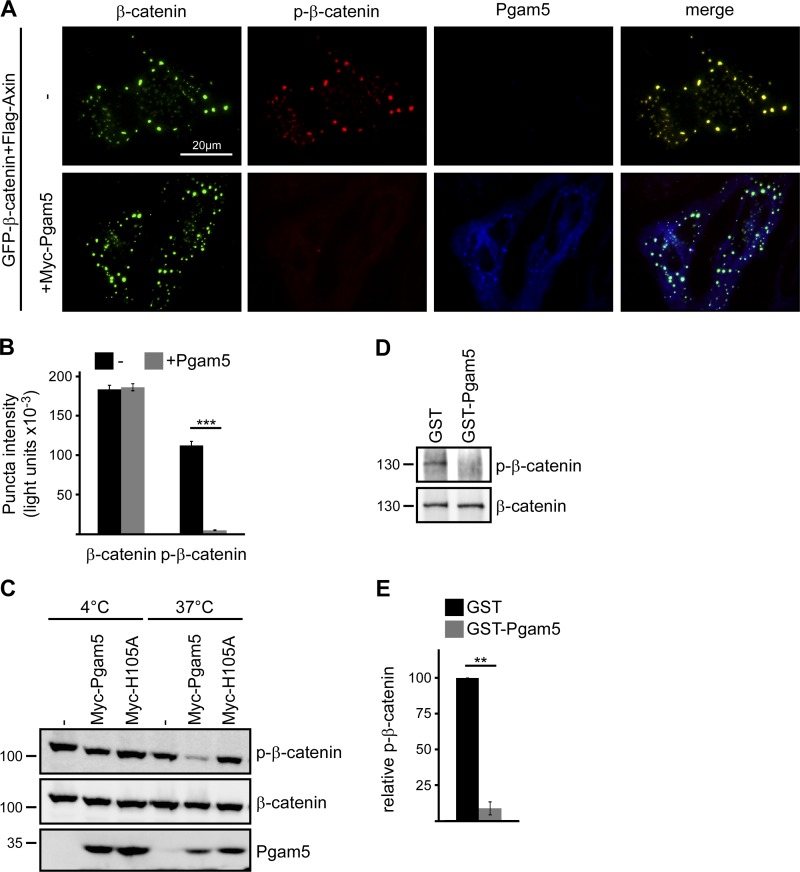
Because Pgam5 regulates β-catenin levels in a phosphatase activity–dependent manner, we investigated whether Pgam5 affects β-catenin phosphorylation. Expression of cytosolic Pgam5 significantly reduced phosphorylation of axin-bound β-catenin in axin puncta (Fig. 4, A and B). Furthermore, when lysates of U2OS cells exhibiting high phospho–β-catenin levels were mixed with lysates of Pgam5-transfected cells, phospho–β-catenin levels decreased during incubation at 37°C, again indicating Pgam5-mediated β-catenin dephosphorylation (Fig. 4 C). β-Catenin dephosphorylation depends on the phosphatase activity of Pgam5 as it was not observed with the H105A mutant (Fig. 4 C). Recombinant GST-Pgam5 also dephosphorylated immunoprecipitated β-catenin, indicating that phospho–β-catenin is a direct substrate of Pgam5 (Fig. 4, D and E). Our data suggest that Pgam5 stabilizes β-catenin by directly dephosphorylating phospho–β-catenin.
Figure 4.
Pgam5 dephosphorylates β-catenin. (A) Immunofluorescence staining of phospho–β-catenin (p-β-catenin; red) in U2OS cells transfected with GFP–β-catenin (green) together with Flag-axin (not depicted) and with either a control plasmid (−) or Myc-Pgam5 (blue). (B) Quantification of β-catenin and phospho–β-catenin fluorescence intensities in individual puncta of transfected U2OS cells from three independent experiments as described in A. n = 100. (C) Western blotting for indicated proteins in lysates of untransfected U2OS cells mixed with lysates of U2OS cells transfected with a control plasmid (−), Myc-Pgam5, or Myc-H105A in a 1:1 ratio and either left at 4°C or incubated at 37°C for 30 min. (D) Western blotting for phospho–β-catenin and β-catenin after IP with an antibody against β-catenin and incubation with either GST or GST-Pgam5 at 37°C for 1 h. Molecular masses are given in kilodaltons. (E) 2D densitometry quantification of phospho–β-catenin band intensities normalized to β-catenin band intensities of three independent experiments as in D. n = 3. Results are means ± SEM. **, P < 0.01; ***, P < 0.001 (Student’s t test).
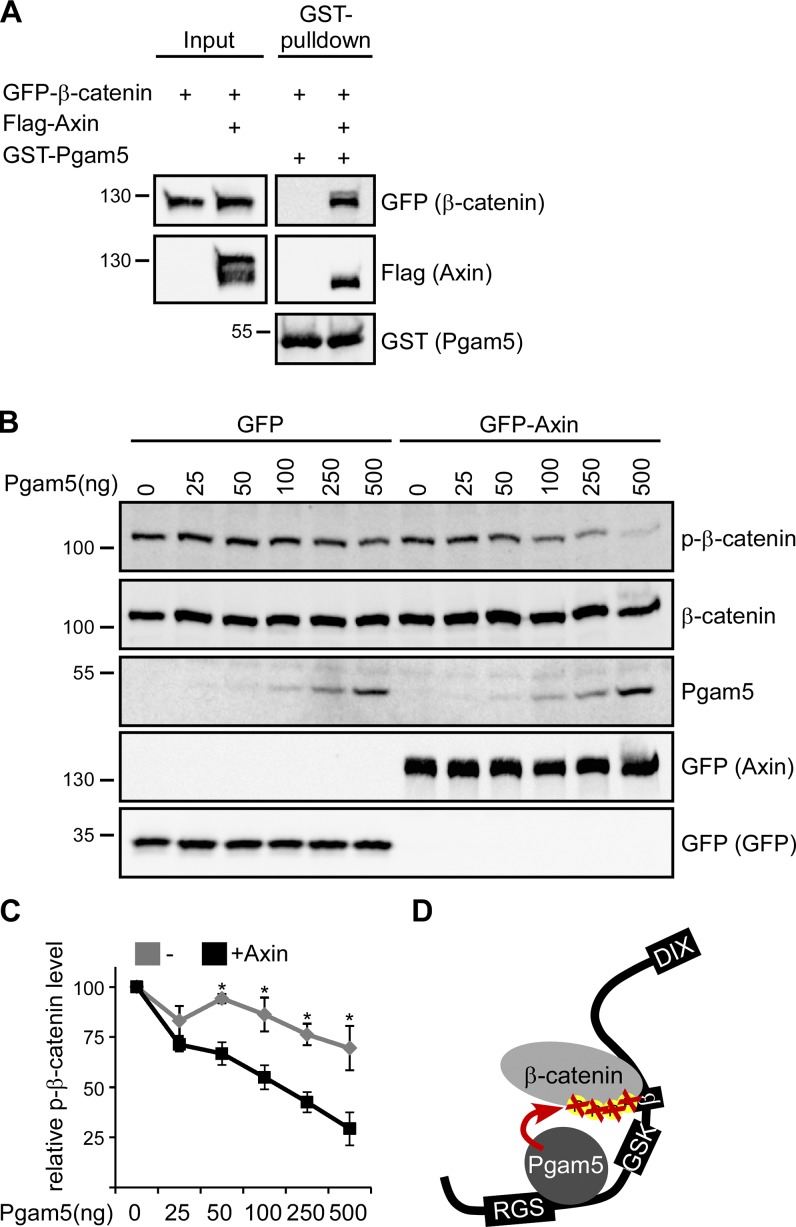
Next, we investigated whether Pgam5 binds to β-catenin. β-Catenin could not be pulled down with GST-Pgam5 from lysates of cells transiently expressing β-catenin, indicating no/weak direct interaction between Pgam5 and β-catenin. However, β-catenin was strongly pulled down with GST-Pgam5 from lysates of cells coexpressing axin (Fig. 5 A), indicating that axin connects Pgam5 with β-catenin (Fig. 5 D). Moreover, Pgam5-mediated dephosphorylation of β-catenin by recombinant GST-Pgam5 was enhanced in lysates of cells transiently expressing GFP-axin compared with lysates of GFP-expressing control cells (Fig. 5, B and C). Together, the experiments show that axin scaffolds and thereby enhances Pgam5-mediated dephosphorylation of β-catenin (Fig. 5 D).
Figure 5.
Axin scaffolds β-catenin dephosphorylation by Pgam5. (A) Western blotting for GFP, Flag, and GST in lysates of HEK293T cells transfected with indicated constructs (Input) and after GST pulldown from these lysates using GST-Pgam5. (B) Western blotting for indicated proteins in lysates of U2OS cells transfected with GFP or GFP-axin that were incubated with indicated amounts of GST-Pgam5 at 37°C for 20 min. Molecular masses are given in kilodaltons. (C) 2D densitometry quantification of phospho–β-catenin (p-β-catenin) band intensities normalized to β-catenin band intensities of three independent experiments as in B. n = 3. Results are means ± SEM. *, P < 0.05 (Student’s t test). (D) Model for axin (black)-scaffolded β-catenin dephosphorylation by Pgam5. Domains are as in Fig. 1 A. DIX, disheveled and axin domain.
Mitochondrial stress induces Pgam5-dependent dephosphorylation and stabilization of β-catenin
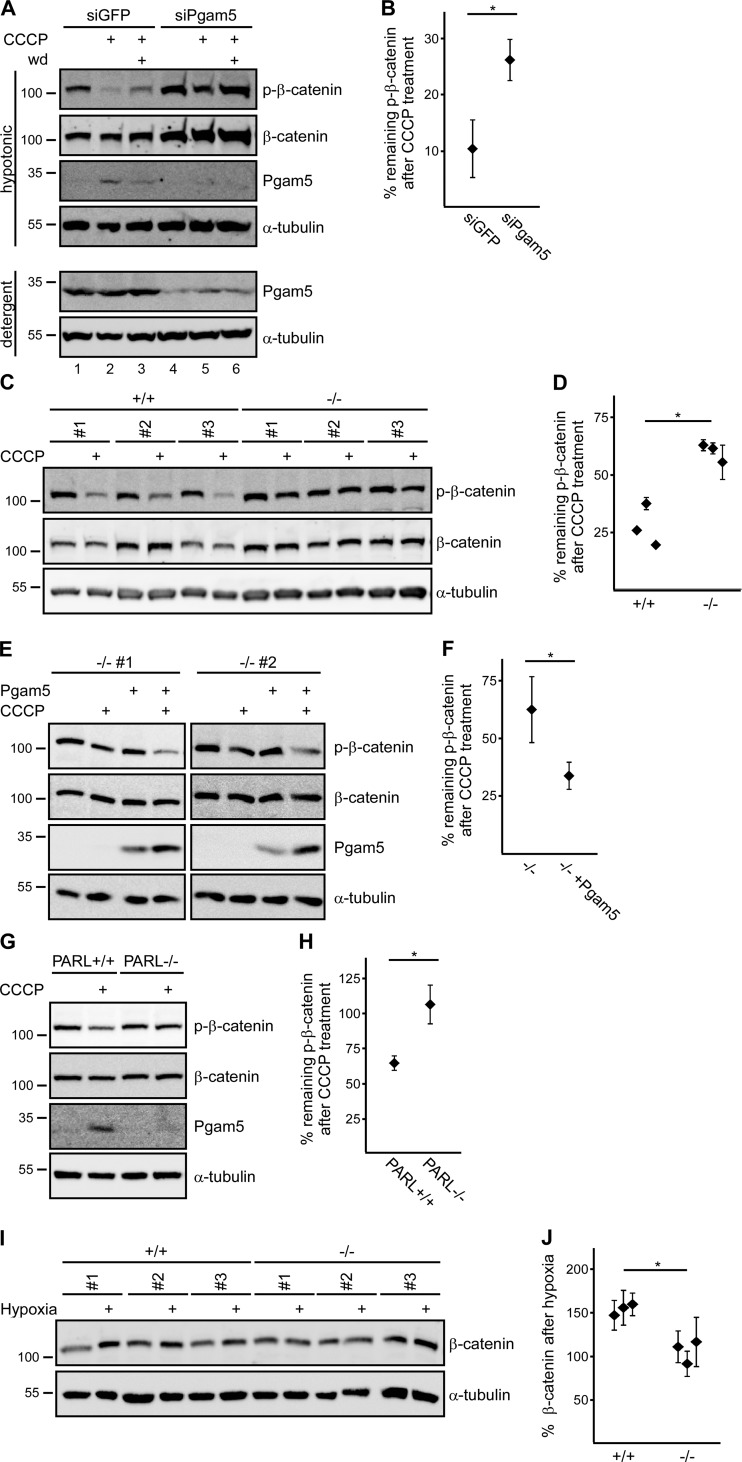
To study the impact of endogenous cytosolic Pgam5 on β-catenin, cells were treated with CCCP to induce Pgam5 cleavage and release from mitochondria followed by Western blotting for Pgam5 and phospho–β-catenin. After CCCP treatment for 2 h, cleaved Pgam5 appeared in hypotonic cell lysates, suggesting its release into the cytosolic fraction (Fig. 6 A, lane 2) in line with our subcellular fractionation results (Fig. 2 A). At the same time, phospho–β-catenin levels dropped. Amounts of released cytosolic Pgam5 were CCCP dosage–dependent and correlated with reduction of phospho–β-catenin levels (Fig. S2). After withdrawal of CCCP for another 2 h, cytosolic Pgam5 levels were reduced, whereas levels of phospho–β-catenin recovered (Fig. 6 A, lane 3). To clarify whether dephosphorylation of β-catenin upon CCCP treatment requires Pgam5, Pgam5 was knocked down by siRNA. As described previously, Pgam5 knockdown increased β-catenin levels, possibly reflecting the negative regulatory role of endogenous mitochondria-associated Pgam5 (Rauschenberger et al., 2017). Importantly, dephosphorylation of β-catenin after CCCP treatment was significantly reduced in Pgam5-knockdown cells compared with siGFP control cells (Fig. 6 A, lane 2 vs. lane 5, and Fig. 6 B). To completely eliminate Pgam5, Pgam5 knockout cells were generated using the CRISPR/Cas9 system (Fig. S3). In control clones, CCCP treatment reduced phospho–β-catenin levels on average to 27% of untreated cells, whereas phospho–β-catenin levels were only reduced to 60% in Pgam5 knockout clones (Fig. 6, C and D). These results suggest that CCCP induces β-catenin dephosphorylation at least in part via Pgam5. In line, transient reexpression of Pgam5 rescued CCCP-induced dephosphorylation of β-catenin in Pgam5−/− cells (Fig. 6, E and F). To determine whether PARL-mediated cleavage of Pgam5 is required for Pgam5-dependent β-catenin dephosphorylation, we used PARL knockout cells (Saita et al., 2017). Pgam5 present in hypotonic lysates after CCCP treatment was markedly reduced upon PARL knockout (Fig. 6 G). Importantly, CCCP-induced dephosphorylation of β-catenin was also significantly attenuated in PARL knockout cells compared with control cells, suggesting that PARL is required for Pgam5-mediated β-catenin dephosphorylation after mitochondrial stress (Fig. 6, G and H). Collectively, our data suggest that mitochondrial stressors like CCCP induce PARL-mediated cleavage of mitochondrial Pgam5, which is then released into the cytosol, where it dephosphorylates β-catenin to regulate β-catenin stability.
Figure 6.
Cleaved endogenous Pgam5 dephosphorylates and stabilizes β-catenin. (A) Western blotting for indicated proteins in hypotonic or detergent lysates of U2OS cells transfected with siGFP (control) or siPgam5. Before lysis, cells were treated with 100 µM CCCP for 2 h (CCCP), incubated without CCCP for another 2 h after CCCP was washed off (withdrawal; wd), or left untreated, as indicated above the blots. (B) Phospho–β-catenin (p-β-catenin) band intensities were determined by 2D densitometry of four independent experiments as in A and normalized to β-catenin. Normalized phospho–β-catenin intensities after CCCP treatment are shown as percentages of intensities without CCCP treatment. (C) Western blotting for phospho–β-catenin, β-catenin, and α-tubulin (loading control) in lysates of the three CRISPR/Cas9 control clones and the three Pgam5 knockout clones shown in Fig. S3, which were left untreated or incubated with 100 µM CCCP for 2 h before lysis. (D) Normalized phospho–β-catenin intensities of four independent experiments as in C determined and presented as described in B. Symbols represent individual clones. (E) Western blotting for indicated proteins in hypotonic lysates of two Pgam5 knockout clones transfected with control plasmid or Pgam5, which were left untreated or incubated with 100 µM CCCP for 2 h before lysis. (F) Normalized phospho–β-catenin intensities of four independent experiments as in E with one Pgam5 knockout clone determined and presented as described in B. (G) Western blotting for indicated proteins in hypotonic lysates of HeLa WT (PARL+/+) or PARL knockout (PARL−/−) cells that were left untreated or incubated with 50 µM CCCP for 2 h before lysis. (H) Normalized phospho–β-catenin intensities of four independent experiments as in G determined and presented as described in B. (I) Western blotting for β-catenin and α-tubulin (loading control) in hypotonic lysates of the three control clones and the three Pgam5 knockout clones (Fig. S3), which were cultured under normoxic or hypoxic conditions for 24 h before lysis. Molecular masses are given in kilodaltons. (J) 2D densitometry quantification of β-catenin normalized to α-tubulin of three independent experiments as in I. Normalized β-catenin intensity after hypoxia is shown as a percentage of intensity under normoxic conditions for each clone. Symbols represent individual clones. n = 4 (B, D, F, and H) or 3 (J). Results are means ± SEM. *, P < 0.05 (Student’s t test).
Hypoxia was already described as a physiologically relevant mitochondrial stressor that can be used alternatively to CCCP to induce Pgam5-dependent processes, e.g., mitophagy of damaged mitochondria (Chen et al., 2014; Wu et al., 2014). Therefore, we wanted to analyze whether hypoxia-induced mitochondrial stress also results in Pgam5-dependent regulation of β-catenin. After culturing U2OS cells in hypoxic conditions, Pgam5 was present in hypotonic lysates (Fig. S4), again suggesting its cytosolic release. Furthermore, hypoxia increased β-catenin levels by on average 50% (Fig. 6, I and J). Importantly, this stabilization of β-catenin depended at least partially on Pgam5 because it was significantly reduced in three Pgam5 knockout clones compared with three control clones (Fig. 6, I and J). Pgam5-dependent stabilization of β-catenin after hypoxia is in line with CCCP-induced β-catenin dephosphorylation by Pgam5. These data suggest that mitochondrial stress triggers cytosolic release of Pgam5 to activate Wnt/β-catenin signaling.
Cytosolic Pgam5 increases the number of mitochondria
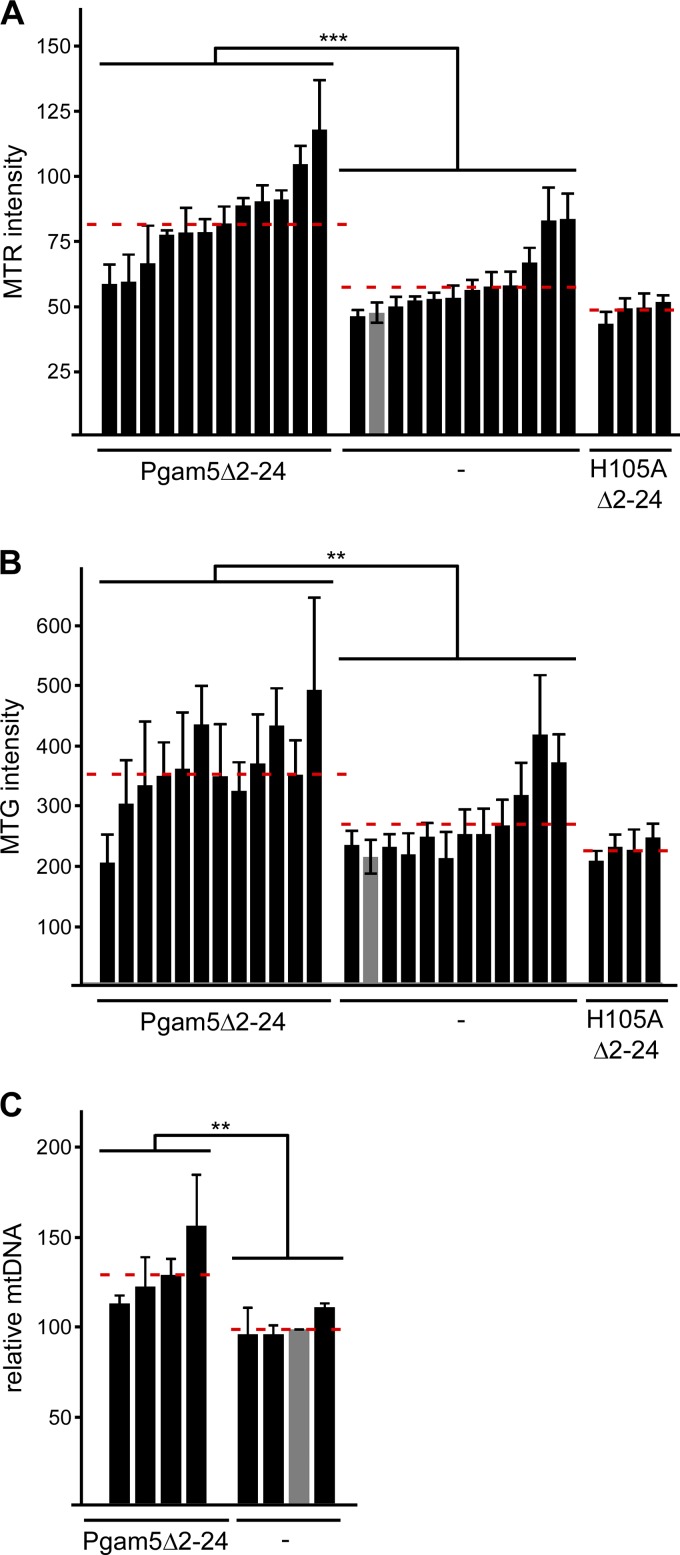
Wnt/β-catenin signaling was described to stimulate biogenesis of mitochondria e.g., in C2C12 myocytes (Yoon et al., 2010; Undi et al., 2017). As our data show that cytosolic Pgam5 activates Wnt/β-catenin signaling, we investigated whether it would also increase mitochondrial numbers. We generated C2C12 clones stably expressing cytosolic Pgam5Δ2–24, which was present in hypotonic extracts (Fig. S5 A). In line with results shown in Fig. 3, β-catenin levels were elevated in Pgam5Δ2–24 clones compared with control clones (Fig. S5 A). Mitochondria number was assessed by staining mitochondria with MitoTracker red (MTR) or MitoTracker green (MTG), which detect mitochondria with intact membrane potential only or all mitochondria, respectively. FACS-based measurement of staining intensities revealed that both MTR- and MTG-positive mitochondria were significantly increased in Pgam5Δ2–24 clones by on average 41% and 33%, respectively (Fig. 7, A and B). Furthermore, the increase of mitochondria in Pgam5Δ2–24 clones was confirmed by a significant increase of mitochondrial DNA (Fig. 7 C). Of note, stable expression of the phosphatase-inactive mutant H105AΔ2–24 did neither increase β-catenin levels (Fig. S5 B) nor MTR- or MTG-positive mitochondria (Fig. 7, A and B).
Figure 7.
Cytosolic Pgam5 increases number of mitochondria. (A and B) Staining intensity of MTR- (A) and MTG-stained (B) cells measured by FACS. Measurements of 12 clones stably expressing Pgam5Δ2–24, 12 control clones (−), and four clones stably expressing H105AΔ2–24 are shown as means ± SEM (n = 4). The gray bar shows parental cells. Dotted red lines indicate mean staining intensities of the 12 Pgam5Δ2–24 clones, the 12 control clones, or the four H105AΔ2–24 clones. (C) Relative mitochondrial DNA (mtDNA) normalized to nuclear DNA of four Pgam5Δ2–24-expressing C2C12 clones and four control clones (−) shown as means ± SEM (n = 3). Normalized mitochondrial DNA amount of parental cells (gray bar) was set to 100%. Dotted red lines indicate mean mitochondrial DNA amounts of the four Pgam5Δ2–24 or the four control clones. **, P < 0.01; ***, P < 0.001 (Student’s t test).
Discussion
In this study, we have identified the cytosolic form of the mitochondrial phosphatase Pgam5 as a potent novel activator of Wnt/β-catenin signaling. Cytosolic Pgam5 inhibits β-catenin degradation, leading to its stabilization, and to increased β-catenin–dependent transcription. Our data indicate that Pgam5 acts by directly dephosphorylating the N-terminal phosphoresidues that earmark β-catenin for degradation: (A) Pgam5 efficiently dephosphorylates β-catenin in vitro and in vivo, (B) stimulation of Wnt/β-catenin signaling requires the phosphatase activity of Pgam5, and (C) Pgam5 increases transcriptional activation by WT β-catenin but not β-catenin mutated at the N-terminal phosphorylation sites.
We also show that the core component of the β-catenin destruction complex, axin, promotes β-catenin dephosphorylation by Pgam5. Axin binds both β-catenin and Pgam5 via separate domains and might thereby act as a scaffold to bring these proteins into close vicinity. Obviously, this is reminiscent of the scaffolding function of axin during β-catenin phosphorylation by the β-catenin kinases CK1α and GSK3 (Ikeda et al., 1998; Liu et al., 2002). Thus, depending on its interaction with β-catenin kinases and phosphatases, axin can promote or inhibit β-catenin phosphorylation. Whereas interactions of axin with CK1α and GSK3 represent the default state in the cell, the interaction with Pgam5 becomes effective when mitochondria are stressed, leading to the cleavage and release of Pgam5. Indeed, we could show biochemically and via immunocytochemistry that CCCP treatment of cells increased cytosolic Pgam5 levels about threefold, although most Pgam5 remains in mitochondria, agreeing with previous observations (Sekine et al., 2012). As cellular axin levels are relatively low, rendering axin rate limiting for β-catenin phosphorylation (Lee et al., 2003), already low cytosolic amounts of the enzyme Pgam5 might suffice to strongly reduce β-catenin phosphorylation. In line with this, CCCP treatment induced Pgam5-dependent reduction of phospho–β-catenin levels, and hypoxia, a physiological mitochondrial stressor, induced Pgam5-dependent stabilization of β-catenin.
Our previous study characterizes mitochondrial Pgam5 as repressor of the Wnt pathway by dephosphorylating Dvl (Rauschenberger et al., 2017), whereas our data presented in this study suggest that cytosolic Pgam5 acts as an activator. This cellular localization–dependent dual role of Pgam5 in regulating Wnt/β-catenin signaling leads to the question of how mitochondrial Pgam5 specifically dephosphorylates Dvl but not β-catenin, whereas cytosolic Pgam5 is dedicated toward β-catenin dephosphorylation. The answer might arise from the lower diffusion dynamics of axin compared with Dvl (Schwarz-Romond et al., 2007). We assume that phospho–β-catenin, which is bound to axin and therefore less dynamic, is less accessible to stationary mitochondria-bound Pgam5 than dynamic Dvl and that Dvl dephosphorylation predominates. The situation is changed by mobilization of Pgam5 via PARL-mediated cleavage: mobile cytosolic Pgam5 interacts with axin and is, via this scaffold, dedicated to β-catenin dephosphorylation, which then predominates.
We can assume that activation of Wnt/β-catenin signaling by Pgam5 is independent of upstream stimulation by Wnt ligands. Instead, stimulation occurs by mitochondrial damage, leading to the activation of proteases sensitive to changes in the mitochondrial membrane potential, such as PARL, that initiate cleavage and release of Pgam5. Thus, similar to their role in cell-intrinsic triggering of apoptosis, we suggest a role for mitochondria in stimulating Wnt/β-catenin signaling in a cell-intrinsic fashion. This is of special interest because not much is known about cell-intrinsic activation of Wnt/β-catenin signaling in contrast with detailed studies on extrinsic activation via Wnt ligands. The Pgam5–β-catenin axis constitutes a novel cell-intrinsic way of activating the Wnt/β-catenin pathway, which acts independent of extrinsic Wnt ligands.
One cellular consequence of activated Wnt/β-catenin signaling is the stimulation of mitochondrial biogenesis as shown in several cell systems (Yoon et al., 2010; Undi et al., 2017). We propose that release of Pgam5 from damaged mitochondria represents a means of replenishing the mitochondria pool by activating Wnt/β-catenin signaling. In line with this, our cell clones stably expressing cytosolic Pgam5 exhibited increased mitochondrial numbers. Interestingly, Pgam5 was implicated in mitophagy to remove damaged mitochondria (Chen et al., 2014; Wu et al., 2014). It appears therefore that Pgam5 controls mitochondrial homeostasis at two levels: by depleting old mitochondria and by generating new ones.
Materials and methods
Cell culture, transfection, small chemicals, and hypoxia
HEK293T, HeLa, U2OS, SW480, and C2C12 cells were cultured in DMEM supplemented with 10% FBS and 1% penicillin-streptomycin at 37°C in a 10% CO2 atmosphere and then were subcultured according to ATCC recommendations. Plasmids were transfected with polyethylenimine (C2C12, HEK293T, and U2OS) or Lipofectamine 2000 (SW480; Invitrogen), and siRNAs were transfected with oligofectamine (Invitrogen) according to the manufacturer’s instructions. HeLa PARL knockout cells were provided by T. Langer (Saita et al., 2017). CCCP was obtained from Abcam. For hypoxic conditions, cells were cultured in an Invivo400 hypoxia workstation (Ruskin) at 0.5% O2.
Generation of stable cell lines
Pgam5−/− clones were generated using the CRISPR/Cas9 system as described elsewhere (Ran et al., 2013). In short, two top-scoring guide RNAs according to the CRISPR design tool provided by F. Zhang (Massachusetts Institute of Technology, Cambridge, MA) were cloned in the PX458 Cas9/GFP/guide RNA expression vector purchased from Addgene (48138). The guide RNAs are complementary to exon 1 of human PGAM5 and anneal shortly 3′ to the translational start site. Guide RNA sequences were (sequence 1) 5′-TACCGCCACGGCCGAGAAGA-3′ (score: 96), and (sequence 2) 5′-TGGCGGTAGGGAAGCCGCGC-3′ (score: 89). 24 h after transfection, GFP-positive U2OS cells were sorted and seeded as one cell per well in 96-well plates. Outgrowing colonies were screened by Western blotting for loss of Pgam5 expression.
To generate C2C12 myocytes stably expressing Pgam5Δ2–24 or H105AΔ2–24, C2C12 cells were transfected with a Pgam5Δ2–24 or H105AΔ2–24 expression plasmid containing a neomycin resistance gene expression cassette, replated 24 h after transfection at low density, and selected for stable integration of the Pgam5Δ2–24 or H105AΔ2–24/neomycin resistance expression plasmid in selection media containing 500 µg/ml or 800 µg/ml G418 (Sigma-Aldrich). After 20 d of selection, individual cell colonies were expanded and screened for the presence of Pgam5Δ2–24 or H105AΔ2–24 in hypotonic cell lysates by Western blotting.
Plasmids and siRNA
Expression plasmids for Pgam5-Flag, Flag-Pgam5, and Flag-Pgam5 H105A were gifts from H. Ichijo (Takeda et al., 2009), and Myc-Pgam5 and the corresponding H105A mutant were from X. Wang (Wang et al., 2012). Plasmids for expression of Flag-axin, GFP-axin, GFP-Cdt 2–345, GFP–β-catenin, and YFP–β-catenin S33Y have been described previously (Brauburger et al., 2014; Bernkopf et al., 2015; Rauschenberger et al., 2017). GFP-axin truncation constructs (89–827, 210–827, and 296–827), Pgam5, Pgam5Δ2–24, and Pgam5Δ2–24 H105A were cloned by standard molecular biology methods. SiRNA against human PGAM5 (sense: 5′-CGCCCGUGUCUCAUUGGAA-3′) was predesigned from QIAGEN.
Cell lysis, IP, and Western blotting
For Western blot analysis, cells were lysed in Triton X-100–based (150 mM NaCl, 20 mM Tris-HCl, pH 7.5, 5 mM EDTA, 1% Triton X-100, and Roche protease inhibitor cocktail) or hypotonic (20 mM Tris-HCl, pH 7.5, 1 mM EDTA, and Roche protease inhibitor cocktail) lysis buffer as indicated in figure legends. For IP, Triton X-100–containing lysates were supplemented with indicated antibodies and G/A beads (Santa Cruz Biotechnology, Inc.) and rotated at 4°C. Beads were washed with low-salt NET buffer (150 mM NaCl, 50 mM Tris-HCl, pH 8.0, 5 mM EDTA, and 1% Triton X-100), and proteins were eluted from the beads. Proteins were separated according to sizes by SDS-PAGE and blotted on a nitrocellulose membrane (VWR), which was probed with indicated antibodies. Antibodies were purchased from the following distributors: Abcam, rb α Pgam5 (ab126534); BD, m α β-catenin (610153); Cell Signaling Technology, rb α axin1 (C76H11), m α Myc (9B11), rb α phospho–β-catenin (Ser33/37); Roche, m α GFP (11814460001), Santa Cruz Biotechnology, Inc., rb α β-catenin (H-102); AbD Serotec, r α α-tubulin (MCA77G); and Sigma-Aldrich, rb α Flag (F7425), m α GST (G1160), rb and m α TOM20 (HPA011562/MABT166), and m α β-actin (A5441). Intensities of Western blot bands were quantified with AIDA 2D densitometry.
Mitochondrial fractionation
Subcellular fractionation was essentially performed as described previously (Saita et al., 2017). In short, cells were collected in homogenization buffer (220 mM mannitol, 70 mM sucrose, 20 mM Hepes/KOH, pH 7.4, 1 mM EDTA, and Roche protease inhibitor cocktail) and homogenized using an Ultra-turrax T-25 Basic (5× 45 s at setting I; IKA). The homogenate was cleared from nuclei and cell debris by centrifugation (600 g at 4°C for 5 min) before the resulting supernatant was centrifuged at 8,000 g at 4°C for 15 min to obtain the mitochondrial fraction (pellet) and a cytosolic/membrane fraction (supernatant). The cytosolic fraction was cleared from membranes by ultracentrifugation (100,000 g at 4°C for 30 min).
Mass spectrometry
GFP-Cdt 2–345 (Fig. 1 A) was immunoprecipitated from lysates of transiently transfected U2OS cells by anti-GFP antibodies (Roche) bound to G/A beads (Santa Cruz Biotechnology, Inc.). After three washes and elution from the beads, GFP-Cdt 2–345 and coprecipitated proteins were subsequently digested with trypsin and Lys-C. Samples were desalted on C-18 stage tips (Nest Group) and analyzed by nanoflow HPLC tandem mass spectrometry (2D-NanoLC; Eksigent; Orbitrap Velos mass spectrometry; Thermo Fisher Scientific; Vasilj et al., 2012). Identification of proteins was performed with Mascot (V2.2; Matrix Science), and isoform specificity of peptides was obtained from the Proteomicsdb database (Wilhelm et al., 2014).
Recombinant Pgam5 and GST pulldown
A Pgam5 deletion construct lacking amino acids 13–50, thereby removing the N-terminal transmembrane region for facilitated purification, was cloned in the pGEX4T3 expression vector (GE Healthcare). Protein expression of GST-Pgam5 was induced by IPTG in transformed Rosetta (DE3) bacteria. Bacteria were lysed, and GST-Pgam5 was precipitated on glutathione beads, washed on beads, and eluted by adding free glutathione. For GST pulldowns, lysates of cells transfected with indicated plasmids were supplemented with GST-Pgam5 and glutathione beads and then rotated at 4°C. Bound GST-Pagm5 and interacting proteins were precipitated on the beads, washed, eluted, and analyzed by Western blotting.
Immunofluorescence
Immunofluorescence staining was performed as previously described (Bernkopf et al., 2015). In short, cells were fixed, permeabilized, and stained with primary antibodies against indicated proteins (see the Western blot section) and fluorochrome-conjugated secondary antibodies (Cy2- or Cy3-conjugated antibodies; 115–225-164; 111-225-144; 115-165-146; 111-165-144; Jackson ImmunoResearch Laboratories, Inc.) or Alexa Fluor 405–conjugated antibodies (ab175660; ab175652; Abcam). When required, mitochondria were stained with MTR (Thermo Fisher Scientific). Stained cells were analyzed with an Axioplan II microscope system (ZEISS) using a Plan-NEOFLUAR 100× 1.30 NA oil objective. Images were acquired at RT with a SPOT RT Monochrome camera (DIAGNOSTIC Instruments Inc). Fluorescence intensities were quantified from images acquired at constant exposure times using MetaMorph analysis software (ZEISS; Molecular Devices).
TOP/FOP assay
To analyze changes in β-catenin–dependent transcription, cells were cotransfected with expression plasmids for proteins of interest, for β-galactosidase, and for either the TOP-FLASH (firefly luciferase under the control of a minimal promoter plus TCF/β-catenin binding sites) or FOP-FLASH (as with TOP but with mutated TCF/β-catenin binding sites) plasmid, and lysed 24 h after transfection. Luciferase activity was measured with a Centro LB 960 microplate luminometer (Berthold Technologies) as light emission upon luciferin conversion. β-Galactosidase activity was measured by cleavage of o-nitrophenyl-β-d-galactopyranosid releasing yellow o-nitrophenol. Luciferase activities from TOP and FOP lysates were normalized to corresponding β-galactosidase activities before calculating TOP/FOP ratios. TOP/FOP assays were performed in technical duplicates.
FACS measurement of MTR and MTG staining intensities
Before FACS analysis, cells were stained with 250 nM MTR or MTG (Thermo Fisher Scientific) at 37°C, trypsinized, washed with PBS, and suspended in FACS buffer (1× PBS supplemented with 2% FBS and 5 mM EDTA). Samples were analyzed at a FACSCalibur (BD): cells were gated on forward and side scatter to exclude cell debris from the analysis, and red fluorescence intensity (MTR) or green fluorescence intensity (MTG) of 10,000 individual cells was measured.
Quantitative PCR analysis of mitochondrial DNA
Total cellular DNA was isolated using the DNeasy blood and tissue kit (QIAGEN) according to the manufacturer’s guidelines. Quantitative PCR for mt-Co2 (forward, 5′-GCCGACTAAATCAAGCAACA-3′; and reverse, 5′-CAATGGGCATAAAGCTATGG-3′) and Hbb-bt (forward, 5′-GAAGCGATTCTAGGGAGCAG-3′; and reverse 5′-GGAGCAGCGATTCTGAGTAGA-3′) was performed in technical triplicates with a CFX96 real-time system (Bio-Rad Laboratories) using previously published primers (Yoon et al., 2010), and relative abundance of mitochondrial mt-Co2 was normalized to nuclear encoded Hbb-bt.
Statistical analyses
To test for statistical significance, two-sided Student’s t tests were performed for paired or nonpaired samples as appropriate. Data distribution was assumed to be normal, but this was not formally tested. In figures, statistical significance is indicated by asterisks (*, P < 0.05; **, P < 0.01; ***, P < 0.001) as also mentioned in the figure legends. Sample sizes (n) of tested datasets are stated in their respective figure legends.
Online supplemental material
Fig. S1 shows localization of endogenous Pgam5 as well as of all Pgam5 constructs used for transient expression throughout this study. Fig. S2 shows CCCP dosage-dependent release of Pgam5 and β-catenin dephosphorylation. Fig. S3 shows loss of Pgam5 protein in CRISPR/Cas9 Pgam5 knockout clones. Fig. S4 shows hypoxia-induced release of Pgam5 and increase of β-catenin levels. Fig. S5 shows the increase of β-catenin levels in C2C12 clones stably expressing Pgam5Δ2–24 and expression of Pgam5Δ2–24 and H105AΔ2–24 in C2C12 clones.
Supplementary Material
Acknowledgments
The authors are grateful to Hidenori Ichijo and Xiaodong Wang for providing Pgam5 expression plasmids as well as Thomas Langer for providing PARL knockout cells. In addition, the authors thank the core unit Cell sorting with Immunomonitoring (Friedrich-Alexander University Erlangen-Nuremberg). We thank Gabriele Daum for excellent technical assistance.
This study was funded by grants from the Friedrich-Alexander University of Erlangen-Nuremberg Interdisciplinary Center for Clinical Research to J. Behrens (D22) and D.B. Bernkopf (J58) and by a grant from the Johannes und Frieda Marohn-Stiftung to D.B. Bernkopf.
The authors declare no competing financial interests.
Author contributions: D.B. Bernkopf: conceptualization, data curation, funding acquisition, investigation, project administration, verification, visualization, and writing (draft, review, and editing). K. Jalal: investigation and methodology. M. Brückner: investigation and data curation. K.X. Knaup: methodology and resources. M. Gentzel: investigation, methodology, and resources. A. Schambony: conceptualization. J. Behrens: conceptualization, funding acquisition, project administration, supervision, verification, and writing (draft, review, and editing).
References
- Aberle H., Bauer A., Stappert J., Kispert A., and Kemler R.. 1997. beta-catenin is a target for the ubiquitin-proteasome pathway. EMBO J. 16:3797–3804. 10.1093/emboj/16.13.3797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behrens J., von Kries J.P., Kühl M., Bruhn L., Wedlich D., Grosschedl R., and Birchmeier W.. 1996. Functional interaction of beta-catenin with the transcription factor LEF-1. Nature. 382:638–642. 10.1038/382638a0 [DOI] [PubMed] [Google Scholar]
- Bernkopf D.B., Hadjihannas M.V., and Behrens J.. 2015. Negative-feedback regulation of the Wnt pathway by conductin/axin2 involves insensitivity to upstream signalling. J. Cell Sci. 128:33–39. 10.1242/jcs.159145 [DOI] [PubMed] [Google Scholar]
- Brauburger K., Akyildiz S., Ruppert J.G., Graeb M., Bernkopf D.B., Hadjihannas M.V., and Behrens J.. 2014. Adenomatous polyposis coli (APC) membrane recruitment 3, a member of the APC membrane recruitment family of APC-binding proteins, is a positive regulator of Wnt-β-catenin signalling. FEBS J. 281:787–801. 10.1111/febs.12624 [DOI] [PubMed] [Google Scholar]
- Chen G., Han Z., Feng D., Chen Y., Chen L., Wu H., Huang L., Zhou C., Cai X., Fu C., et al. 2014. A regulatory signaling loop comprising the PGAM5 phosphatase and CK2 controls receptor-mediated mitophagy. Mol. Cell. 54:362–377. 10.1016/j.molcel.2014.02.034 [DOI] [PubMed] [Google Scholar]
- Clevers H., and Nusse R.. 2012. Wnt/β-catenin signaling and disease. Cell. 149:1192–1205. 10.1016/j.cell.2012.05.012 [DOI] [PubMed] [Google Scholar]
- Fagotto F., Jho E., Zeng L., Kurth T., Joos T., Kaufmann C., and Costantini F.. 1999. Domains of axin involved in protein-protein interactions, Wnt pathway inhibition, and intracellular localization. J. Cell Biol. 145:741–756. 10.1083/jcb.145.4.741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiedler M., Mendoza-Topaz C., Rutherford T.J., Mieszczanek J., and Bienz M.. 2011. Dishevelled interacts with the DIX domain polymerization interface of Axin to interfere with its function in down-regulating β-catenin. Proc. Natl. Acad. Sci. USA. 108:1937–1942. 10.1073/pnas.1017063108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He G.W., Günther C., Kremer A.E., Thonn V., Amann K., Poremba C., Neurath M.F., Wirtz S., and Becker C.. 2017. PGAM5-mediated programmed necrosis of hepatocytes drives acute liver injury. Gut. 66:716–723. 10.1136/gutjnl-2015-311247 [DOI] [PubMed] [Google Scholar]
- Ikeda S., Kishida S., Yamamoto H., Murai H., Koyama S., and Kikuchi A.. 1998. Axin, a negative regulator of the Wnt signaling pathway, forms a complex with GSK-3beta and beta-catenin and promotes GSK-3beta-dependent phosphorylation of beta-catenin. EMBO J. 17:1371–1384. 10.1093/emboj/17.5.1371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang Y.J., Bang B.R., Han K.H., Hong L., Shim E.J., Ma J., Lerner R.A., and Otsuka M.. 2015. Regulation of NKT cell-mediated immune responses to tumours and liver inflammation by mitochondrial PGAM5-Drp1 signalling. Nat. Commun. 6:8371 10.1038/ncomms9371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee E., Salic A., Krüger R., Heinrich R., and Kirschner M.W.. 2003. The roles of APC and Axin derived from experimental and theoretical analysis of the Wnt pathway. PLoS Biol. 1:e10 10.1371/journal.pbio.0000010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C., Li Y., Semenov M., Han C., Baeg G.H., Tan Y., Zhang Z., Lin X., and He X.. 2002. Control of beta-catenin phosphorylation/degradation by a dual-kinase mechanism. Cell. 108:837–847. 10.1016/S0092-8674(02)00685-2 [DOI] [PubMed] [Google Scholar]
- Lo S.C., and Hannink M.. 2008. PGAM5 tethers a ternary complex containing Keap1 and Nrf2 to mitochondria. Exp. Cell Res. 314:1789–1803. 10.1016/j.yexcr.2008.02.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDonald B.T., and He X.. 2012. Frizzled and LRP5/6 receptors for Wnt/β-catenin signaling. Cold Spring Harb. Perspect. Biol. 4:a007880 10.1101/cshperspect.a007880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molenaar M., van de Wetering M., Oosterwegel M., Peterson-Maduro J., Godsave S., Korinek V., Roose J., Destrée O., and Clevers H.. 1996. XTcf-3 transcription factor mediates beta-catenin-induced axis formation in Xenopus embryos. Cell. 86:391–399. 10.1016/S0092-8674(00)80112-9 [DOI] [PubMed] [Google Scholar]
- Panda S., Srivastava S., Li Z., Vaeth M., Fuhs S.R., Hunter T., and Skolnik E.Y.. 2016. Identification of PGAM5 as a Mammalian Protein Histidine Phosphatase that Plays a Central Role to Negatively Regulate CD4(+) T Cells. Mol. Cell. 63:457–469. 10.1016/j.molcel.2016.06.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran F.A., Hsu P.D., Wright J., Agarwala V., Scott D.A., and Zhang F.. 2013. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 8:2281–2308. 10.1038/nprot.2013.143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauschenberger V., Bernkopf D.B., Krenn S., Jalal K., Heller J., Behrens J., Gentzel M., and Schambony A.. 2017. The phosphatase Pgam5 antagonizes Wnt/β-Catenin signaling in embryonic anterior-posterior axis patterning. Development. 144:2234–2247. 10.1242/dev.144477 [DOI] [PubMed] [Google Scholar]
- Saita S., Nolte H., Fiedler K.U., Kashkar H., Venne A.S., Zahedi R.P., Krüger M., and Langer T.. 2017. PARL mediates Smac proteolytic maturation in mitochondria to promote apoptosis. Nat. Cell Biol. 19:318–328. 10.1038/ncb3488 [DOI] [PubMed] [Google Scholar]
- Schwarz-Romond T., Metcalfe C., and Bienz M.. 2007. Dynamic recruitment of axin by Dishevelled protein assemblies. J. Cell Sci. 120:2402–2412. 10.1242/jcs.002956 [DOI] [PubMed] [Google Scholar]
- Sekine S., Kanamaru Y., Koike M., Nishihara A., Okada M., Kinoshita H., Kamiyama M., Maruyama J., Uchiyama Y., Ishihara N., et al. 2012. Rhomboid protease PARL mediates the mitochondrial membrane potential loss-induced cleavage of PGAM5. J. Biol. Chem. 287:34635–34645. 10.1074/jbc.M112.357509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda K., Komuro Y., Hayakawa T., Oguchi H., Ishida Y., Murakami S., Noguchi T., Kinoshita H., Sekine Y., Iemura S., et al. 2009. Mitochondrial phosphoglycerate mutase 5 uses alternate catalytic activity as a protein serine/threonine phosphatase to activate ASK1. Proc. Natl. Acad. Sci. USA. 106:12301–12305. 10.1073/pnas.0901823106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Undi R.B., Gutti U., and Gutti R.K.. 2017. LiCl regulates mitochondrial biogenesis during megakaryocyte development. J. Trace Elem. Med. Biol. 39:193–201. 10.1016/j.jtemb.2016.10.003 [DOI] [PubMed] [Google Scholar]
- van Kappel E.C., and Maurice M.M.. 2017. Molecular regulation and pharmacological targeting of the β-catenin destruction complex. Br. J. Pharmacol. 174:4575–4588. 10.1111/bph.13922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasilj A., Gentzel M., Ueberham E., Gebhardt R., and Shevchenko A.. 2012. Tissue proteomics by one-dimensional gel electrophoresis combined with label-free protein quantification. J. Proteome Res. 11:3680–3689. 10.1021/pr300147z [DOI] [PubMed] [Google Scholar]
- Wai T., Saita S., Nolte H., Müller S., König T., Richter-Dennerlein R., Sprenger H.G., Madrenas J., Mühlmeister M., Brandt U., et al. 2016. The membrane scaffold SLP2 anchors a proteolytic hub in mitochondria containing PARL and the i-AAA protease YME1L. EMBO Rep. 17:1844–1856. 10.15252/embr.201642698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Jiang H., Chen S., Du F., and Wang X.. 2012. The mitochondrial phosphatase PGAM5 functions at the convergence point of multiple necrotic death pathways. Cell. 148:228–243. 10.1016/j.cell.2011.11.030 [DOI] [PubMed] [Google Scholar]
- Wilhelm M., Schlegl J., Hahne H., Gholami A.M., Lieberenz M., Savitski M.M., Ziegler E., Butzmann L., Gessulat S., Marx H., et al. 2014. Mass-spectrometry-based draft of the human proteome. Nature. 509:582–587. 10.1038/nature13319 [DOI] [PubMed] [Google Scholar]
- Wu H., Xue D., Chen G., Han Z., Huang L., Zhu C., Wang X., Jin H., Wang J., Zhu Y., et al. 2014. The BCL2L1 and PGAM5 axis defines hypoxia-induced receptor-mediated mitophagy. Autophagy. 10:1712–1725. 10.4161/auto.29568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon J.C., Ng A., Kim B.H., Bianco A., Xavier R.J., and Elledge S.J.. 2010. Wnt signaling regulates mitochondrial physiology and insulin sensitivity. Genes Dev. 24:1507–1518. 10.1101/gad.1924910 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.