The yeast cell wall is digested to allow cell fusion during sexual reproduction. How cells coordinate this process with cell–cell contact to prevent lysis is unclear. Merlini et al. show that the Ras GAP protein Gap1, which is recruited to sites of Ras-GTP, restricts Ras activity and protects cells from lysis due to premature fusion attempts.
Abstract
In the fission yeast Schizosaccharomyces pombe, pheromone signaling engages a signaling pathway composed of a G protein–coupled receptor, Ras, and a mitogen-activated protein kinase (MAPK) cascade that triggers sexual differentiation and gamete fusion. Cell–cell fusion requires local cell wall digestion, which relies on an initially dynamic actin fusion focus that becomes stabilized upon local enrichment of the signaling cascade on the structure. We constructed a live-reporter of active Ras1 (Ras1–guanosine triphosphate [GTP]) that shows Ras activity at polarity sites peaking on the fusion structure before fusion. Remarkably, constitutive Ras1 activation promoted fusion focus stabilization and fusion attempts irrespective of cell pairing, leading to cell lysis. Ras1 activity was restricted by the guanosine triphosphatase–activating protein Gap1, which was itself recruited to sites of Ras1-GTP and was essential to block untimely fusion attempts. We propose that negative feedback control of Ras activity restrains the MAPK signal and couples fusion with cell–cell engagement.
Introduction
Inhibitory mechanisms commonly regulate signaling pathways. Negative feedbacks drive the oscillation of the cell cycle, circadian clocks, or synthetic networks (Elowitz and Leibler, 2000; Novák and Tyson, 2008; Ferrell, 2013) and serve to produce spatial patterns during multicellular development (Ribes and Briscoe, 2009) and in single cells (Wu and Lew, 2013). In cell polarization, negative controls counteract positive feedbacks that promote the formation of single polarity sites, thus providing dynamic adaptation (Ozbudak et al., 2005; Das et al., 2012), limiting the size of the growth zone (Hwang et al., 2008), promoting robustness against variation in polarity factor concentration (Howell et al., 2012), or promoting a morphogenetic transition (Okada et al., 2013). Here, we describe a negative control that coordinates the achievement of a stable cell polarization state with that of the partner cell for fusion.
We are interested in understanding the cellular events driving cell–cell fusion, a process that strongly relies on positive feedbacks. We recently described the course of events leading two haploid fission yeast (Schizosaccharomyces pombe) cells of opposite mating type to form a diploid zygote. Early during the mating process, the haploid partners exhibit a polarity patch of active Cdc42 GTPase that dynamically forms and disassembles, exploring multiple sites at the cell cortex (Bendezú and Martin, 2013). This dynamic patch is a site of both own-pheromone secretion and sensing of the opposite-type pheromone (Merlini et al., 2016). Because patch lifetime is prolonged upon higher pheromone perception, adjacent patches in cells of opposite mating type positively feedback to stabilize each other. This leads to cell pairing and growth of the two cells toward their partner.
This positive feedback is maintained and amplified during the fusion process, which requires a dedicated actin-based aster—the fusion focus—nucleated by the formin Fus1 (Petersen et al., 1995; Dudin et al., 2015). The fusion focus underlies the concentration of pheromone secretion and perception machineries at a focal point. Reciprocally, local activation of the downstream MAPK cascade spatially constrains the focus, leading to its immobilization at facing locations in the two partner cells, now committed to fusion (Dudin et al., 2016). Focus immobilization drives fusion because it allows the type V myosin motor Myo52 to deliver cell wall digestive enzymes at a precise location to locally pierce through the cell wall for plasma membrane merging (Dudin et al., 2015, 2017). Fusion commitment, and thus cell wall digestion, do not directly require cell–cell contact. Indeed, forcing the positive feedback to occur in single cells, by engineering autocrine cells that respond to self-produced pheromones, leads to fusion attempts without a partner. This causes cell lysis because the locally digested cell wall no longer resists the strong internal turgor pressure (Dudin et al., 2016). Yet lysis is extremely rare during WT cell fusion, suggesting the existence of mechanisms to couple fusion commitment with the formation of cell pairs.
A Ras-MAPK signaling pathway, functionally homologous to the mammalian Ras–Raf–MEK–ERK mitogenic pathway (Hughes et al., 1993), is intimately involved in controlling fission yeast mating. The single S. pombe Ras protein, Ras1, whose activated form directly binds the MAP3K Byr2 (Masuda et al., 1995), is an essential activator of the MAPK cascade that transduces the pheromone signal (Fukui et al., 1986; Wang et al., 1991b). Two guanine nucleotide exchange factors (GEFs) promote Ras1 activation: a constitutively expressed GEF Efc25, which activates Ras1 for cell polarization during mitotic growth (Papadaki et al., 2002), and a pheromone-induced GEF Ste6, required for Ras1 activation during mating (Hughes et al., 1994). Dependence of Ste6 transcription on the MAPK signal creates an additional positive feedback (Hughes et al., 1994; Mata and Bähler, 2006). A single GTPase-activating protein (GAP), Gap1, is predicted to promote GTP hydrolysis and return Ras1 to its inactive state (Imai et al., 1991; Wang et al., 1991a; Weston et al., 2013). Interestingly, hyperactivation of Ras1, like its deletion, causes sterility, but with a distinct phenotype (Fukui et al., 1986; Nadin-Davis et al., 1986): although ras1Δ cells do not engage in mating, GTP-locked alleles of Ras1 or deletion of gap1 provokes cell death during mating, which was proposed to result from unsustainable cell elongation from multiple sites (Weston et al., 2013). We made the alternative hypothesis that this phenotype is caused by premature fusion attempts. Here, we show that the Ras GAP Gap1 is recruited to sites of Ras1 activity to restrict Ras1 activation to sites of pheromone signaling, drive dynamic polarization, and prevent fusion commitment during early mating stages to couple it with cell–cell pairing.
Results
Constitutive Ras activation promotes untimely fusion attempts
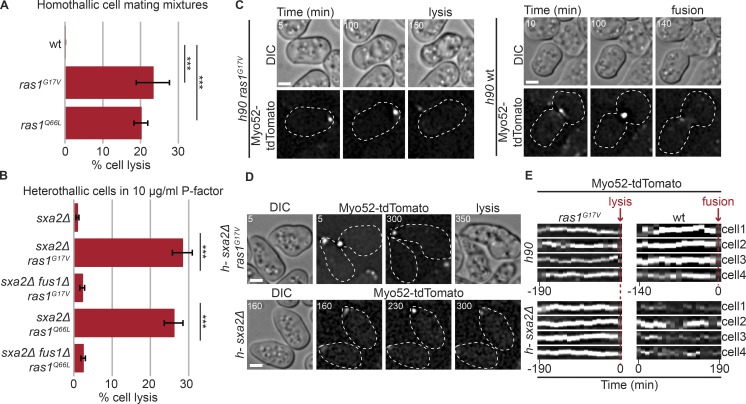
As previously shown, cells carrying a GTP-locked Ras1 allele (ras1G17V or ras1Q66L) at the native locus extended elongated single or multiple mating projections at apparent aberrant locations and often lysed (Fig. 1 A and Video 1; Nadin-Davis et al., 1986; Weston et al., 2013). Similarly, single mating type h-ras1G17V or ras1Q66L cells exposed to synthetic P-factor readily extended mating projections and lysed, whereas WT cells did not lyse, as shown previously (Fig. 1 B and Video 2; note these cells also lack the P-factor protease Sxa2 to prevent P-factor degradation; Weston et al., 2013; Dudin et al., 2016). Importantly, cell lysis was suppressed by fus1 deletion, suggesting lysis may arise from an untimely fusion attempt (Fig. 1 B).
Figure 1.
Constitutive Ras activation promotes untimely fusion attempts. (A) Percentage of cell lysis of homothallic (h90) WT and indicated ras mutants after 14 h in MSL-N (n > 500 for three independent experiments); ***, 5.85 × 10−6 ≤ P ≤ 1.1× 10−5. (B) Percentage of cell lysis of h-sxa2Δ, h-sxa2Δ ras1G17V, and h-sxa2Δ ras1Q66L cells, with or without fus1 deletion, 14 h after 10 µg/ml synthetic P-factor addition (n > 500 for three independent experiments); ***, 4.58 × 10−6 ≤ P ≤ 1.43 × 10−5. (C) Differential interference contrast (DIC) and Myo52-tdTomato time-lapse images of h90 ras1G17V and WT cells during mating. Myo52 focus persists until cell lysis in the unpaired ras1G17V cell, but only occurs in cell pairs during fusion in WT. Cell lysis (ras1G17V) and fusion (WT) are indicated. (D) DIC and Myo52-tdTomato time-lapse images of h-sxa2Δ ras1G17V and h-sxa2Δ cells treated with 10 µg/ml P-factor. Note persistent Myo52 focus and cell lysis in ras1G17V cells and unstable Myo52 signal in WT cells. (E) Kymographs of four cell tips showing a stable Myo52 focus in h90 ras1G17V mating cells and h-sxa2Δ ras1G17V cells exposed to 10 µg/ml P-factor. The kymographs are aligned to lysis time. ras1+ cells form a focus late in the fusion process (in h90 cells, kymographs aligned to fusion time) or only transiently (in h-sxa2Δ exposed to P-factor; no kymographs alignment). Bars, 2 µm. Error bars, SD. Time in minutes from the start of imaging.
Consistent with this hypothesis, cells with constitutive Ras1 activation displayed a strong, focal signal of Myo52-tdTomato, reminiscent of the fusion focus of WT cell pairs (Dudin et al., 2015). This signal formed and remained stable over long time periods in unpaired cells before cell lysis (Fig. 1, C and E; and Fig. S1 A). In contrast, WT cells formed a fusion focus only after pairing (Fig. 1, C and E). Similarly, in heterothallic sxa2Δ cells exposed to synthetic pheromone, a stable Myo52 focus was formed upon constitutive Ras1 activation, whereas the Myo52 signal was broad and only transiently focalized in ras1+ cells (Fig. 1, D and E; and Video 2). Over 97% of lysing cells showed a focalized Myo52 signal (118 of 121 ras1G17V and 84 of 86 ras1Q66L cells). These observations suggest Ras1 activation promotes fusion focus stabilization. Note that constitutive Ras1 activation did not lead to fusion attempts during mitotic growth, consistent with pheromone signaling being required for Fus1 expression (Petersen et al., 1995).
RasAct: A probe for in situ labeling of Ras-GTP
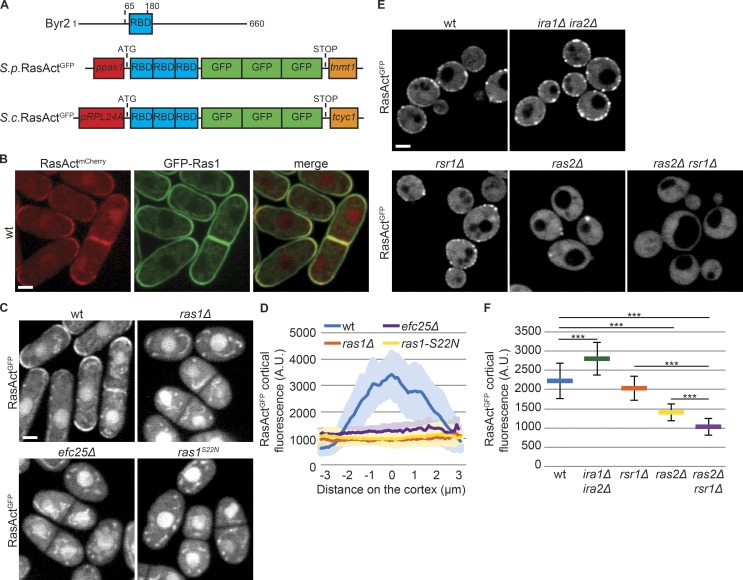
To define the cellular location of Ras activity, we developed a fluorescent probe detecting Ras1-GTP. The structure of the Byr2 Ras-GTP binding domain (RBD) has been solved (Gronwald et al., 2001). We cloned three tandem repeats of the Byr2 RBD followed by three GFPs (or mCherry) and constitutively expressed this probe, called RasActGFP (or RasActmCherry; Fig. 2 A). During mitotic growth, RasAct localized to cell poles and septa, a localization abolished in cells carrying a deletion of ras1 or the GEF efc25, or a GDP-locked ras1S22N allele (Fig. 2, B–D). RasAct also accumulated in the nucleus, but this localization was not affected by Ras1 activity state, suggesting this is a spurious localization (Fig. 2 C). In contrast to pole-restricted RasAct, GFP-Ras1 decorated the entire cell cortex independently of its activation and was only weakly enriched at cell poles in WT cells (Fig. 2 B and Fig. S1 B). We conclude that cortical RasAct localization reports on sites of Ras1-GTP, which represents only a fraction of total Ras1.
Figure 2.
A probe for the visualization of Ras-GTP. (A) Schematic representation of MAP2K Byr2 (top) and probes to visualize Ras-GTP (RasActGFP) in S. pombe (S.p.; middle) and S. cerevisiae (S.c.; bottom). Promoters and terminators used for gene expression are indicated. (B and C) Localization of RasActmCherry and GFP-Ras1 (B), and RasActGFP (C), during vegetative growth of S. pombe cells. Note that the nuclear and dotted (mitochondrial) localization of RasActGFP are nonspecific, as they do not depend on ras1. (D) Cortical tip profiles of RasActGFP fluorescence in strains as in C; n = 25. Thick line, mean; shaded area, SD. (E) Localization of RasActGFP during vegetative growth of S. c. cells. (F) Mean total RasActGFP cortical fluorescence in strains as in E; n = 25; ***, 3.9 × 10−13 ≤ P ≤ 3.7 × 10−5. Error bars, SD. Bars, 2 µm.
We tested whether RasAct is able to detect Ras-GTP in other organisms. RasActGFP was constitutively expressed in Saccharomyces cerevisiae, integrated as single genomic copy (Fig. 2 A). In these cells, RasAct localized in the cytoplasm and decorated regions of the cell cortex (Fig. 2 E). S. cerevisiae encodes two Ras isoforms, Ras1 and Ras2 (Kataoka et al., 1984), implicated in glucose sensing (Conrad et al., 2014), negatively regulated by two GAPs, Ira1 and Ira2 (Tanaka et al., 1990), and localized to the plasma membrane (Manandhar et al., 2010). A third Ras-like protein, Rsr1, is involved in cell polarization (Bi and Park, 2012). Interestingly, deletion of ira1 and ira2 increased RasAct cortical levels (Fig. 2, E and F), suggesting RasAct reports on Ras-GTP levels. In cells lacking Ras2, RasAct was poorly recruited to the cell cortex (Fig. 2, E and F). The residual cortical localization was dependent on Rsr1, as RasAct did not label the cell cortex in double ras2Δ rsr1Δ mutants. We conclude that RasAct reports on both Rsr1-GTP and Ras2-GTP (Fig. 2, E and F). These results establish RasAct as a tool to detect local levels of active Ras in both fission and budding yeast cells.
Ras activity at the fusion site peaks before cell fusion
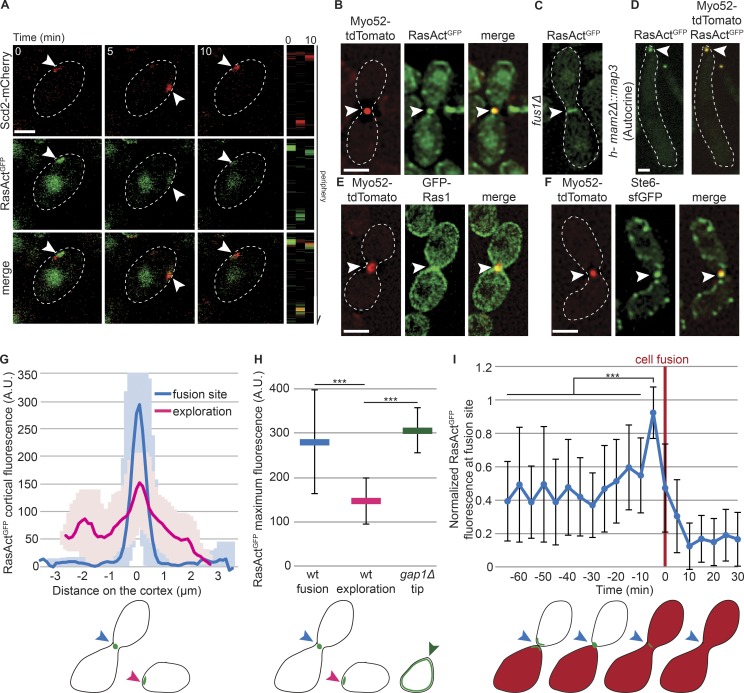
We used RasAct to probe where Ras1 is active in mating fission yeast cells. During dynamic polarization, RasAct localized to dynamic sites at the cortex, which overlapped with sites of Cdc42 activity, labeled by the scaffold protein Scd2 (Fig. 3 A and Fig. S2, A and B). GFP-Ras1 weakly accumulates at these sites, but is also present broadly at the cell cortex (Merlini et al., 2016). Time-lapse imaging showed simultaneous accumulation and loss of RasActGFP and Scd2-mCherry signals at dynamic cortical sites, in agreement with Ras1 acting as activator of Cdc42 signaling (Chang et al., 1994; Merlini et al., 2016). Thus, Ras1-GTP, like Cdc42-GTP, exhibits oscillatory polarization dynamics during early mating.
Figure 3.
Ras1 is active at polarity sites and the fusion focus. (A) h90 WT strains showing colocalization of Scd2-mCherry (red) and RasActGFP (green) during early mating. Right: Kymographs of the cell periphery. Arrowheads highlight dynamic zones of colocalization. (B) Colocalization of RasActGFP (green) and Myo52-tdTomato (red) at the fusion focus in h90 WT cells. (C) RasActGFP in fus1Δ cell pair. (D) Colocalization of RasActGFP (green) and Myo52-tdTomato (red) at the fusion focus in mam2Δ::map3 autocrine M-cells. (E) Broader localization of GFP-Ras1 (green) than Myo52-tdTomato (red) at the fusion site in h90 WT strains. (F) Colocalization of Ste6-sfGFP (green) and Myo52-tdTomato (red) at the fusion focus in h90 WT cells. (G) Cortical profiles of RasActGFP fluorescence in h90 WT strains during early (exploration; pink) or late (fusion site; blue) mating; n = 20. Thick line, mean; shaded area, SD. (H) Maximal RasActGFP cortical fluorescence in h90 WT cells during early (exploration; pink) or late (fusion; blue) mating and at the mating projections of h90 gap1Δ cells (green). Mean values of the five brightest pixels are shown; n = 20; ***, 9.7 × 10−34 ≤ P ≤ 2.8 × 10−13. (I) Normalized value of RasActGFP cortical fluorescence over time at the fusion site in h90 WT cells. Fluorescence profiles were aligned to fusion time (t = 0) and normalized to maximal value; n = 22. ***, 8.7 × 10−12 ≤ P ≤ 1.1 × 10−5. The schemes indicate where fluorescence was quantified. Error bars, SD. Bars, 2 µm.
During fusion, RasAct strongly accumulated at the fusion focus, labeled by Myo52, and its restriction to a focal site depended on Fus1 (Fig. 3, B and C; and Fig. S2 C). RasAct was also concentrated at the fusion focus in autocrine M-cells attempting fusion in the absence of a partner (Fig. 3 D). In contrast, GFP-Ras1 was enriched at the mating projection over a broader zone (Fig. 3 E), indicating that Ras1-GTP is restricted to the fusion site with Ras1-GDP also present in surrounding regions. During mating, Ras1 activation is thought to occur downstream of pheromone receptor signaling, which takes place locally first at dynamic polarization sites (Merlini et al., 2016) and then at the fusion focus (Dudin et al., 2016). Unfortunately, tagging of the pheromone-induced Ras GEF Ste6 with GFP or superfolder GFP (sfGFP) at the N or C terminus produced a protein with reduced function, as these cells exhibited only 25% mating efficiency (compared with 75% for WT cells in our assay conditions), and we were unable to detect it during dynamic polarization. However, partly functional Ste6-sfGFP colocalized with the fusion focus (Fig. 3 F), consistent with local activation of Ras1. Efc25-GFP was undetectable during mating. We conclude that Ras1 is locally activated first at dynamic polarization sites and then at the fusion site.
The spatial distribution of cortical RasAct was distinct at exploratory zones, where it formed low-intensity, broad peaks, and the fusion focus, where it formed higher-intensity, sharper ones (Fig. 3, G and H). Time-lapse data alignment to the fusion time, assessed by transfer of a cytosolic marker from one cell to its partner, further revealed that RasAct levels were highest at the fusion focus immediately before the fusion event (Fig. 3 I). Thus, local Ras1-GTP concentration peaks at the fusion site just before fusion.
Gap1 GTPase activating protein restricts Ras1 activity
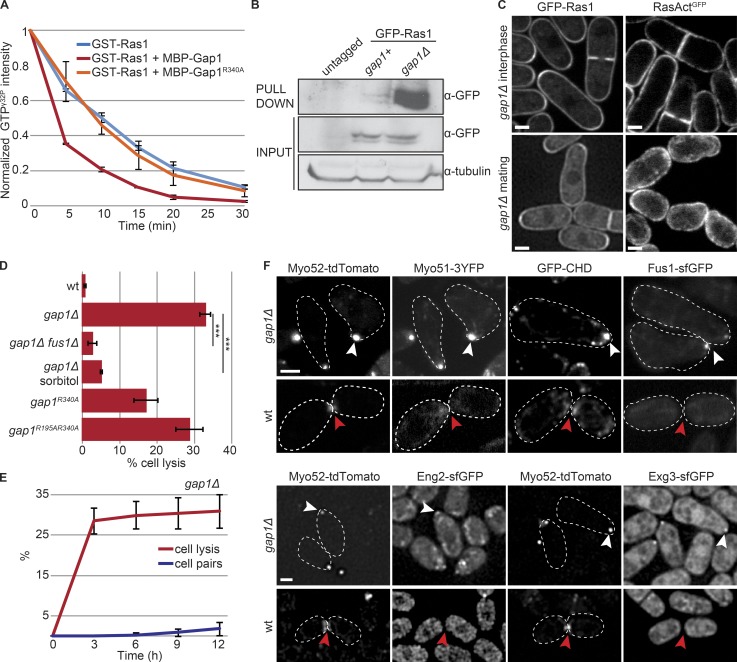
Because Ras1 activation peaks before fusion and constitutive Ras1 activation promotes untimely fusion attempts, we asked how Ras activity is controlled to induce fusion. Ras1-GTP hydrolysis is likely catalyzed by the GAP Gap1 (Imai et al., 1991; Wang et al., 1991a; Weston et al., 2013). Indeed, MBP-Gap1 accelerated GTP hydrolysis on Ras1 in vitro, whereas MBP-Gap1R340A carrying a point mutation in the GAP domain predicted to affect GTP hydrolysis (Sermon et al., 1998) had no effect (Fig. 4 A). Recombinant GSTRBD (a derivative of RasAct, containing just one copy of RBD fused to GST; Kae et al., 2004) pulled down excess Ras1-GTP from gap1Δ extracts, as compared with WT, indicating that Gap1 promotes Ras1-GTP hydrolysis also in vivo (Fig. 4 B). In cells lacking Gap1, RasAct, like GFP-Ras1, decorated the entire cortex of vegetative growing cells and was absent from the nucleus (Fig. 4 C). Thus, a large fraction of Ras1 is active and recruits RasAct to the plasma membrane. During mating, RasAct did not form discrete zones, but was distributed homogeneously at the cortex in unpolarized cells. In cells extending a growth projection, RasAct was broadly distributed around the projection tip and largely excluded from the back of the cell. This localization mimics that of GFP-Ras1, again suggesting that most Ras1 molecules are active in this mutant (Fig. 4 C). Fluorescence intensity measurements further showed that RasAct levels at gap1Δ projection tips were significantly higher than those observed during early mating in WT cells and similar to those at the fusion focus of WT cells during fusion (Fig. 3 H). Thus, Gap1 is a Ras1 GAP and restricts Ras1 activity.
Figure 4.
Gap1 is a GTPase Activating Protein for Ras1. (A) In vitro GAP assays: MBP-Gap1, but not catalytically inactive MBP-Gap1R340A, increases the rate of GST-Ras1-GTPγ32P hydrolysis; n = 3. (B) GSTRBD pulldown of extracts from h-cells of indicated genotypes. (C) GFP-Ras1 (left) and RasActGFP (right) in gap1Δ strains during vegetative growth (top) and mating (bottom). Arrowheads indicate shmoo tips. (D) Percentage of cell lysis of h90 WT and gap1 mutant (gap1R340A, gap1R195A R340A, and gap1Δ) cells treated with 1.2 M sorbitol or lacking fus1 after 14 h in MSL-N (n > 500 for three independent experiments); ***, 9.34 × 10−7 ≤ P ≤ 6.54 × 10−6. (E) Time course of cell lysis and cell pair formation in h90 gap1Δ cells placed in MSL-N (n > 200 for three independent experiments). (F) Type V myosins (Myo52-tdTomato and Myo51-3YFP), actin (GFP-CHD), formin Fus1-sfGFP, and cell wall glucanases (Eng2-sfGFP and Exg3-sfGFP) are focalized in unpaired gap1Δ cells (white arrowheads) but either not detectable or not focalized (red arrowheads) in WT cells not yet engaged in fusion (early paired cells shown). Bars, 2 µm. Plots show means and SDs.
Multiple lines of evidence showed that cells lacking Gap1 or its GAP activity engaged in untimely fusion attempts. We have recently described that gap1Δ cells display diminished dynamic Cdc42 polarization and stabilize a site of growth at reduced pheromone concentrations, leading to misoriented growth projections (Merlini et al., 2016), thus reducing pairing efficiency (Fig. 4 E). Remarkably, homothallic cells with a gap1 deletion or point mutation abolishing the GAP activity (gap1R340A, gap1R195A R340A) often lysed (Weston et al., 2013; Fig. 4 D). Cell lysis was slightly less frequent in cells carrying the mutation gap1R340A, predicted to block GTP hydrolysis, than gap1R195A R340A, predicted to also block Ras1-GTP binding (Scheffzek et al., 1997; Sermon et al., 1998), suggesting that both Ras1-GTP binding and hydrolysis contribute to Gap1 function in vivo; notably, 34.6 ± 4.1% of gap1R340A cells were able to mate. Lysis of gap1Δ cells was an early event, with almost maximal levels reached 3 h after mating induction (Fig. 4 E). Cells that did not lyse grew further, with a few forming more than one cell projection (Fig. S3 A and Video 3). In almost all cases, these growth projections formed consecutively (Fig. S3 B and Video 3). Importantly, osmostabilization by addition of 1.2 M sorbitol prevented lysis (Fig. 4 D and Fig. S3 C), suggesting lysis was caused by cell wall weakening. Furthermore, fus1 deletion suppressed lysis, though it did not prevent misoriented growth projections (Fig. 4 D and Fig. S3 C). This suggests that lysis is not a consequence of excessive growth, but represents a precocious fusion attempt.
In WT cells, the actin fusion focus forms only after cells are paired, with single cells or even cells that have just paired not yet exhibiting a tight concentration of type V myosins (Fig. 4 F; Dudin et al., 2015). Strikingly, the projection tip of unpaired gap1Δ cells displayed a focal structure labeled by markers of the fusion focus: the formin Fus1, GFP-CHD–labeled F-actin, type V myosins Myo52 and Myo51, and the glucanases Eng2 and Exg3 for cell wall digestion (Fig. 4 F and Videos 3, 4, and 5). In addition, heterothallic gap1Δ cells exposed to synthetic P- or M-factor (and lacking the respective protease) also readily assembled a stable fusion focus and lysed (Fig. S3, D–G). Again, lysis was prevented by fus1 deletion (Fig. S3, E and G). Finally, gap1 deletion increased the percentage of lysing autocrine M-cells, in which pheromone signaling focalization occurs through a cell-autonomous positive feedback (Dudin et al., 2016; Fig. S3 H). gap1Δ cells that did not lyse eventually defocused their polarity patch. We conclude that, by promoting Ras1-GTP hydrolysis, Gap1 protects cells against untimely fusion attempts.
Ras1 activity promotes MAPK focalization
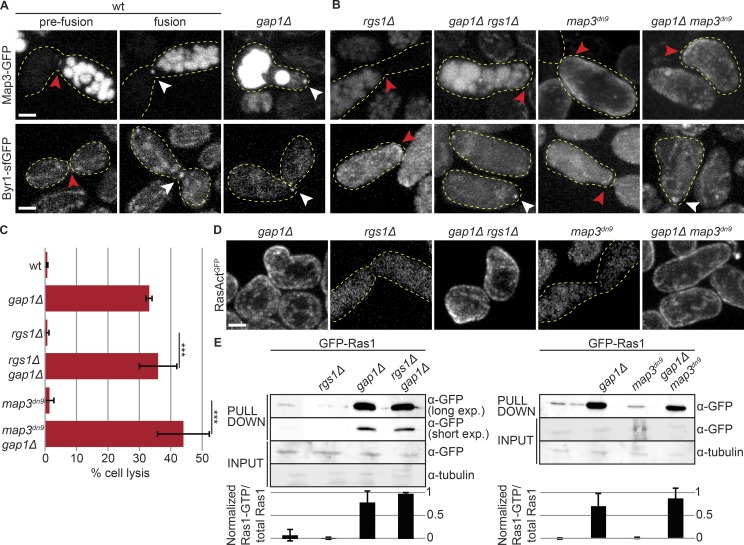
Focalization of pheromone signaling—including pheromone receptors, coupled Gα, and MAPK cascade—at the fusion focus promotes cell fusion by stabilizing the focus (Dudin et al., 2016). Because gap1Δ cells engage in precocious fusion events, we tested the spatial organization of these signaling molecules. Similar to other components of the fusion focus, the MAP2K Byr1 and the M-factor pheromone receptor Map3 prematurely localized to the fusion focus in unpaired gap1Δ cells (Fig. 5 A). Thus, constitutive Ras1 activation engages the positive feedback leading to focalization of the pheromone signaling cascade and stabilization of the fusion focus.
Figure 5.
Ras activity promotes MAPK focalization. (A and B) Pheromone receptor Map3-GFP (top) and MAP2K Byr1-sfGFP (bottom) in WT cell pairs before or during the fusion process (left), and at the projection tip of mutant strains (right). White arrowheads highlight focalization and red arrowheads highlight zones of broad or undetectable localization. Deletion of gap1 leads to Byr1 MAP2K focalization whether Map3 is focalized or not. (C) Percentage of cell lysis of h90 WT, rgs1Δ, and map3dn9 mutants, with or without gap1 deletion after 14 h in MSL-N (n > 500 for three independent experiments). ***, P ≤ 2.6 × 10−5. (D) RasActGFP during mating in mutant strains. Note undetectable local Ras activation when the pheromone receptor is unfocalized and quasiuniform cortical activation upon gap1 deletion. (E) GSTRBD pulldown of protein extracts from h90 cells with indicated genotypes shifted for 4 h to mating conditions (top). Mean intensity from three independent experiments is shown (bottom). Long (20 min) and short (5 min) exposure times of pulldown samples are shown on left. *, band caused by overflow in an empty lane. Error bars, SD. Bars, 2 µm.
To understand the relationship between Ras1 activation and engagement of the positive feedback, we used two mutants that impair receptor focalization, map3dn9 (a C-terminal truncation blocking receptor endocytosis; Hirota et al., 2001) and rgs1Δ (a deletion of the receptor-associated Gα GAP; Watson et al., 1999; Pereira and Jones, 2001), which fail to focalize Map3 receptor and Byr1 MAP2K and are fusion defective (Fig. 5 B; Dudin et al., 2016). In these mutants, RasAct did not reveal local Ras activity at the cell–cell contact site (Fig. 5 D). A small proportion of map3dn9 cells (13 of 102 cells) localized RasAct over a broad zone at the cell–cell contact. However, pulldown assays with GSTRBD detected Ras1-GTP at levels similar to those in WT in these mutants (Fig. 5 E), suggesting Ras1-GTP is present but distributed over a large area and thus not detected by microscopy. These results are consistent with the view that the narrow distribution of Ras1-GTP follows from the concentration of active pheromone signaling at the fusion focus.
Interestingly, deletion of gap1 in map3dn9 and rgs1Δ cells led to formation of mating projections at aberrant sites and lysis-provoking precocious fusion attempts, like gap1Δ single mutants (Fig. 5, C and D). Thus, constitutive Ras1 activation on the entire cell cortex can bypass the normal focalization of the pheromone receptor. Indeed, in these double mutants, the pheromone receptor was still not enriched at the fusion focus, but the downstream MAP2K cascade component Byr1 was (Fig. 5 B). These data suggest that Ras1-GTP can promote MAPK localization to the actin fusion structure independently of the localization of upstream signaling components and does not need to be restricted to the fusion focus to fulfill this function. These data are also consistent with the notion that MAPK focalization is the critical signal for fusion focus stabilization (Dudin et al., 2016).
Altogether, the aforementioned results indicate that elevated Ras1 activity levels at the mating projection promote the local accumulation of the MAPK cascade at the fusion focus for fusion commitment and that Gap1 restricts Ras1-GTP levels until cells are ready for fusion.
Gap1 locally restricts Ras1 activity
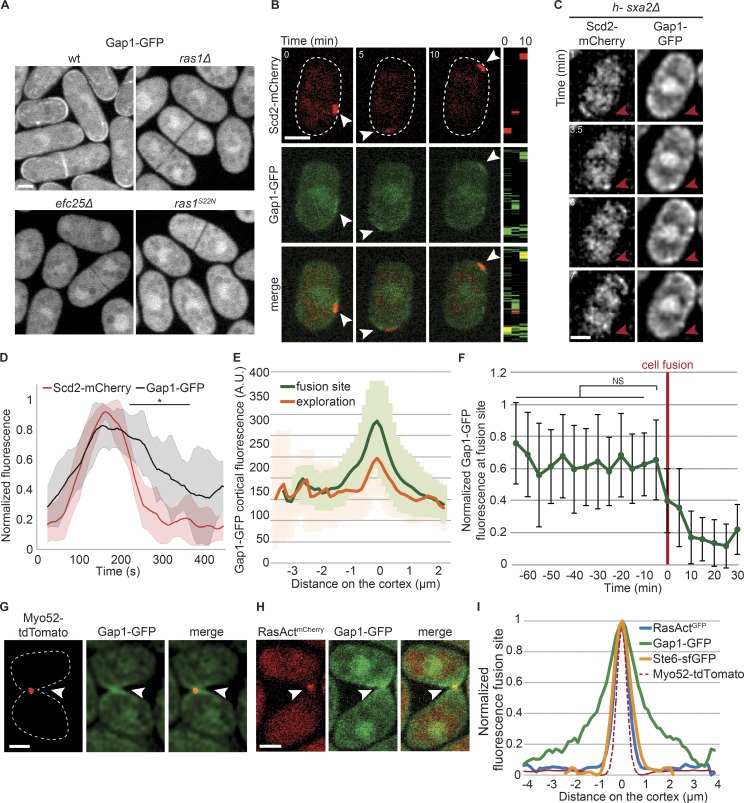
To understand how Gap1 spatially controls Ras1 activity, we examined Gap1 localization. Previous data, using overexpressed constructs, showed that Gap1 is recruited to the cell cortex by Ras1-GTP after pheromone stimulation (Weston et al., 2013). Gap1-GFP tagged as sole copy at the native locus associated with cell poles and septa in vegetative cells, and was also present in the cytosol. Gap1-GFP localization was abolished in ras1Δ, efc25Δ, or ras1S22N cells, similar to RasActGFP, in agreement with its recruitment to the cell cortex by Ras1-GTP (Fig. 6 A). During early mating, Gap1-GFP localized to sites of Cdc42 and Ras1 activation, labeled with Scd2-mCherry (112 of 139 cells with Scd2-mCherry signal; Fig. 6 B and Fig. S4 A). Short-interval time-lapse imaging revealed that Gap1-GFP was recruited to Scd2 sites and remained associated beyond their disassembly (Fig. 6, C and D; and Fig. S4, B–E). Gap1-GFP also localized to the fusion site, with a wide distribution similar to earlier time points (Fig. 6 E). In contrast to RasAct, Gap1 levels did not peak before fusion and its distribution at the fusion site was broader than that of RasAct, Ste6, or Myo52 (Fig. 6, F–I). These observations suggest Gap1 may be recruited by Ras1-GTP, but remain associated with the cell cortex beyond GTP hydrolysis.
Figure 6.
Gap1 is recruited to sites of Ras1 activity. (A) Gap1-GFP during vegetative growth in WT and mutant strains. (B) Colocalization of Scd2-mCherry (red) and Gap1-GFP (green) during early mating in h90 WT cells. Right: Kymographs of the cell periphery. Arrowheads highlight dynamic zones of colocalization. (C) Colocalization of Scd2-mCherry and Gap1-GFP in a dynamic polarity patch of h-sxa2Δ cells treated with 0.01 µg/ml P-factor. Red arrowhead highlights one Scd2-mCherry patch that appears and disappears in the course of the time lapse. For more examples, see Fig. S4. (D) Cortical profiles of Scd2-mCherry and Gap1-GFP normalized fluorescence intensity at the exploratory patch, showing that Gap1-GFP signal persists after Scd2-mCherry signal has disappeared. n = 10 patches from 10 different cells. Thick line, mean; shaded area, SD. Two-samplet test was calculated for each time point and resulted in *, P ≤ 1.3 × 10−2 between 224 and 365 s. (E) Cortical profiles of Gap1-GFP fluorescence in h90 WT strains during early (exploration) or late (fusion site) mating; n = 20. Thick line, mean; shaded area, SD. (F) Normalized value of Gap1-GFP cortical fluorescence over time at the fusion site in h90 WT cells. Fluorescence profiles were aligned to the fusion time (t = 0) and normalized to maximal value; n = 22. Error bars, SD. (G and H) Myo52-tdTomato (red) and Gap1-GFP (green; G), and RasActmCherry (red) and Gap1-GFP (green; H) in h90 WT strains during fusion. Gap1-GFP signal is broader than Myo52-tdTomato or RasActmCherry (arrowheads). (I) Cortical profiles of Gap1-GFP, RasActGFP, and Ste6-sfGFP at the fusion site in h90 WT strains as in G and Fig. 3 (B and F). The profiles were aligned to the Myo52-tdTomato signal coimaged in the same cells (dashed line); n = 20. Bars, 2 µm.
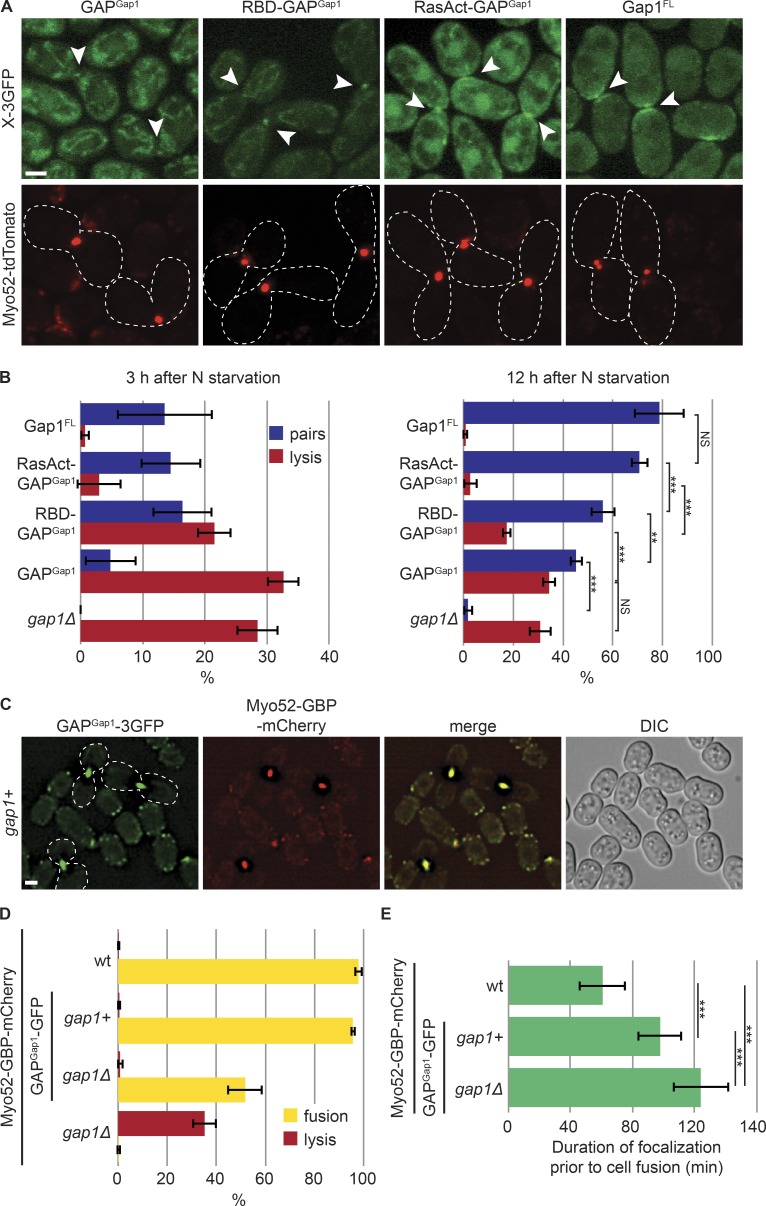
The recruitment of Gap1 to Ras1-GTP may simply reflect interaction of the GAP domain with Ras1-GTP, as previously suggested (Weston et al., 2013) and consistent with the need for a WT GAP domain (Fig. S5 D). However, the sole GAP domain (GAPGap1) only weakly localized to the Myo52 focus in both gap1Δ and gap1+ cells (Fig. 7 A). GAPGap1 expressed under control of a constitutive promoter in gap1Δ cells partly rescued cell pairing: ∼45% of cells formed pairs and eventually fused 12 h after nitrogen starvation (Fig. 7 B and Fig. S5 A). Thus, GAPGap1 retained some activity, and its weak localization likely results from the lack of N- and C-terminal localization determinants rather than a misfolding problem. Notably, GAPGap1 did not rescue the lysis phenotype of gap1Δ cells (Fig. 7 B). In contrast, full-length Gap1 (Gap1FL) expressed under the same conditions and at similar levels (Fig. S5, A and B) localized broadly at the fusion site and fully rescued the lysis and pairing defects of cells lacking gap1 (Fig. 7, A and B). Thus, the sole Gap1 GAP domain, although retaining some function, is not sufficient to prevent premature fusion attempts.
Figure 7.
Local negative regulation of Ras1 couples cell fusion with cell contact. (A) Localization of Gap1 GAP domain on its own (GAPGap1), fused to a single or triple RBD (RBD-GAPGap1, RasAct-GAPGap1), and full-length Gap1 fused to 3GFP expressed in gap1Δ cells. These constructs are detailed in Fig. S5 A. Myo52-tdTomato (red) foci indicate cell pairs in fusion (arrowheads). (B) Percentage of cell lysis and cell pair formation of h90 gap1Δ cells expressing or not the fusion constructs shown in A 3 h (left) and 12 h (right) after nitrogen (N) starvation (see also Fig. 4 E; n > 300 for three independent experiments). (C) GAPGap1-3GFP strains expressing Myo52-GBP-mCherry 8 h after nitrogen starvation. GAPGap1-3GFP is recruited to the fusion focus via interaction with the GBP moiety. (D) Percentage of cell lysis and fusion efficiency of WT, gap1Δ, GAPGap1-3GFP, and gap1Δ GAPGap1-3GFP strains expressing Myo52-GBP-mCherry 12 h after nitrogen starvation. Note that GAPGap1-3GFP recruitment to the fusion focus suppresses gap1Δ cell lysis. (E) Duration of Myo52-GBP-mCherry focalization in strains of indicated genotypes; n = 25. ***, 1.2 × 10−16 ≤ P ≤ 7.2 × 10−7. Error bars, SD. Bars, 2 µm.
Remarkably, linking GAPGap1 to one or three copies of RBD (RBD-GAPGap1 and RasAct-GAPGap1) to enhance GAPGap1 recruitment to active Ras1 suppressed the mating defects of gap1Δ cells (Fig. 7 B). These constructs were expressed as above, at similar levels as GAPGap1 (Fig. S5, A and B). This suppression relied on the GAP activity of the fusion constructs (Fig. S5, C and D). Suppression was partial for RBD-GAPGap1, but complete for RasAct-GAPGap1, which was noticeably recruited to the fusion site (Fig. 7, A and B), suggesting that the strength of the GAP recruitment is important to tune the appropriate cellular response.
Finally, we recruited GAPGap1-GFP to the fusion focus labeled with Myo52-GBP-mCherry by using the low nanomolar affinity between GFP and GBP (GFP binding protein; Rothbauer et al., 2008). Here, all detectable GAPGap1 localized to sites of polarity (Fig. 7 C). This locally recruited GAP efficiently suppressed the lysis of gap1Δ cells, though it did not fully restore fusion ability (Fig. 7 D), as these cells showed an extended fusion reaction, as measured by the duration of the Myo52-labeled fusion focus (Fig. 7 E). Recruitment of GAPGap1 to the fusion focus in otherwise gap1+ cells also significantly extended fusion duration (Fig. 7 E), though it did not have a significant effect on global fusion efficiency (Fig. 7 D). Together, these results indicate that local Gap1 enrichment is necessary to efficiently restrict Ras1 activity. As Gap1 is recruited by active Ras1, these observations are consistent with Gap1 recruitment forming a negative feedback that promotes cell pairing and coordinates the process of fusion with cell–cell contact.
Discussion
The Ras-MAPK cascade is widely used to underlie cell decisions between proliferation and differentiation or between life and death (Rauch et al., 2016). Here, we show that negative regulation of Ras1 activity forms an inherent part of the pheromone sensing system in yeast, mediating both the pairing of sexual partners and their fusion. We propose that the coupling of this inhibition with previously established positive feedback forms an integrated pheromone sensing system that coordinates cell polarization and fusion decisions.
The positive feedback that underlies the fission yeast sexual reproduction cycle is well established. At the transcriptional level, pheromone signaling promotes its own expression, such that two cells of opposite mating types stimulate each other (Merlini et al., 2013). At the cortex, pheromone signaling is coupled to cell polarization, with pheromone release occurring at polarity sites and pheromone sensing promoting polarity site stabilization (Merlini et al., 2016). This forms a positive feedback through which an initially unstable Cdc42 patch (Bendezú and Martin, 2013) becomes stabilized for cell pairing. Similar positive feedbacks likely occur, for instance, in budding yeast, where localization of pheromone receptors and transporters is polarized at the projection tip (Kuchler et al., 1993; Ayscough and Drubin, 1998) and pheromone receptor activation promotes cell polarization in the direction of the pheromone gradient (Merlini et al., 2013). Positive feedback between signaling and polarization is further enhanced during cell fusion, when the actin cytoskeleton forms a concentrated focus, immobilized in response to local pheromone–MAPK signaling (Dudin et al., 2015, 2016). Because the fusion focus is a site of cell wall digestive enzyme release (Dudin et al., 2015), its immobilization leads to local cell wall digestion, a process essential for fusion with a partner cell, but lethal if premature.
We discuss below how our data support a model in which the positive feedback is counteracted by negative regulation of Ras1, thus allowing pheromone gradient sensing to consecutively drive dynamic polarization for cell pairing and signal stabilization for fusion.
Gap1 restrains the mating process
Consistent with previous genetic analysis (Imai et al., 1991), our data show that Gap1 functions as a GAP for Ras1 both in vitro and in vivo. Indeed, its deletion leads to excessive Ras1-GTP levels and phenotypes indistinguishable from those of GTP-locked Ras1 alleles. Thus, Gap1 promotes Ras1 activity cycling, which is important to coordinate the progression of mating.
Gap1-dependent inhibition of Ras1 is critical throughout the mating process. First, gap1Δ cells exhibit higher transcriptional output upon pheromone stimulation (Weston et al., 2013). During the cell pairing process, Gap1 is necessary to destabilize polarity zones, where both Ras1-GTP and Gap1 are recruited (Merlini et al., 2016). During this stage, the formation of zones of Cdc42 activity likely relies on both spontaneous symmetry-breaking mechanisms, well described in other cells and conditions (Slaughter et al., 2009; Johnson et al., 2011; Martin, 2015), and coupling to activated pheromone receptors. Ras1 activation at sites of receptor engagement may couple local pheromone sensing with the cell polarization machinery by activating the Cdc42 GEF Scd1 (Chang et al., 1994). This spatial coupling requires Ras1 activation to be transient: in the absence of Ras1 inhibition, Ras1 activity spreads over the entire cortex such that pheromone gradient information cannot spatially bias the symmetry-breaking mechanisms that now dominate Cdc42 activation, and these cells form projections at inappropriate location (this work; Weston et al., 2013; Merlini et al., 2016). Such a mechanism is similar to that envisaged for bud site selection in budding yeast, where the cycling of the Ras-like protein Rsr1 between GTP and GDP forms couples Cdc42 to spatial landmarks (Bi and Park, 2012).
Critically, Gap1 protects against cell lysis. Several observations support the notion that lysis in gap1Δ cells results from a premature fusion attempt rather than “unsustainable elongation from multiple tips” as previously proposed (Weston et al., 2013). First, lysis occurs early after pheromone exposure, usually in cells with a single growth projection, and growth of further projections occurs later. Second, even when extending multiple projections, gap1Δ cells grow from only one cell pole at any given time (Video 3 and Fig. S3 B). Third, the lysis but not the excessive growth is suppressed by deleting fus1 and results from premature assembly of the fusion focus. This phenotype resembles that of autocrine cells (Dudin et al., 2016), suggesting that gap1Δ cells display unrestrained positive feedback. As autocrine gap1Δ cells lyse more frequently than autocrine gap1+ cells, Gap1 likely tempers the positive feedback even in these cells. We note that not all autocrine gap1Δ cells succumb to lysis, suggesting the existence of other safeguards. We conclude that Gap1 acts to dampen pheromone-dependent responses.
Local high MAPK signaling at the fusion site
Lysis of gap1Δ cells occurs upon premature stabilization of the fusion focus that leads to cell wall digestion. We previously showed that forced recruitment of the MAP2K Byr1 to the fusion focus stabilizes the structure and induces fusion attempts (Dudin et al., 2016). Our data now show that constitutive Ras1 activation promotes Byr1 localization to the fusion focus even in the absence of pheromone receptor localization. Although this is consistent with Ras1-GTP recruiting the MAP3K Byr2 (Bauman et al., 1998), additional inputs must restrict the localization of Byr1 on the focus because spatial MAPK restriction happens even when both pheromone receptor and Ras1 are active on a broad cortical zone. The observations that scd1 deletion suppresses gap1Δ cell lysis (Weston et al., 2013) and that Byr1 focal localization requires Fus1 (Dudin et al., 2016) suggest a possible role for Cdc42 and the actin fusion focus.
Recent work indicated that elevated MAPK signaling also promotes cell fusion in S. cerevisiae (Conlon et al., 2016). This is noteworthy because the molecular wiring of the pheromone–MAPK pathways differs significantly in the two yeasts, with the S. cerevisiae pathway signaling independently of Ras and using a MAPK scaffold, Ste5, absent in S. pombe. In S. cerevisiae, the MAPK cascade accumulates at the shmoo tip through mechanisms that involve direct transport of Ste5 scaffold along formin-nucleated actin cables (Qi and Elion, 2005), enzyme-substrate interactions (Maeder et al., 2007), or binding to the polarizome component Spa2 (Sheu et al., 1998). Despite the distinct molecular wiring, the concentration of the MAPK cascade at the fusion site may be a common switch decision to induce cell fusion.
Gap1 may form a negative feedback on Ras1 activity
Two pieces of data suggest that Gap1 could form a negative feedback. First, Ras1-GTP recruits its own inhibitor. Though this localization is expected from the known interaction of GAP domains with the GTP form of their cognate GTPase, additional determinants may contribute to Gap1 localization: the prolonged localization of Gap1 beyond polarity patch disassembly and its broader distribution around fusion foci, distinct from RasAct, suggest that Gap1 localization is more complex. Upon Ras1-GTP hydrolysis, Gap1 may be handed over to other binding partners at the membrane, or simply remain bound to Ras1-GDP. Second, synthetic recruitment of the GAP domain to Ras1-GTP, but not GAP alone, rescues the gap1Δ phenotypes, suggesting that local enrichment of the negative regulator contributes to its efficiency. These data are consistent with the idea that Ras1 activity nonlinearly recruits its own inhibitor, thus promoting its own inhibition to favor dynamic early polarization, cell pairing, and coordination of fusion with cell–cell contact.
Nonlinearity is consistent with several negative feedbacks proposed and/or demonstrated to promote spatiotemporal oscillations in other polarized systems, across eukaryotes. These include oscillations in Cdc42 GTPase activity during yeast polarization (Wu and Lew, 2013; Martin, 2015), growth-mediated recruitment of a Rho GAP in pollen tubes to promote oscillatory growth dynamics (Hwang et al., 2005), or negative feedbacks that keep migratory cells such as Dictyostelium in an excitable state (Devreotes and Horwitz, 2015). The parallel with Dictyostelium migration is particularly interesting because activation of one of the nine Ras homologues, RasG, is one of the earliest polarized responses to the cAMP chemoattractant (Sasaki et al., 2004), and deletion of its GAP DdNF1 leads to near homogeneous RasG activation at the cell cortex and loss of directional sensing (Zhang et al., 2008), similar to the case of gap1Δ in S. pombe. Its mammalian homologue, the neurofibromin NF1, has also been proposed to function in Ras feedback inhibition (Hennig et al., 2016). Thus, by promoting Ras activity cycling, inhibition of Ras activity contributes to spatial patterning across eukaryotic species.
Ordering mating progression
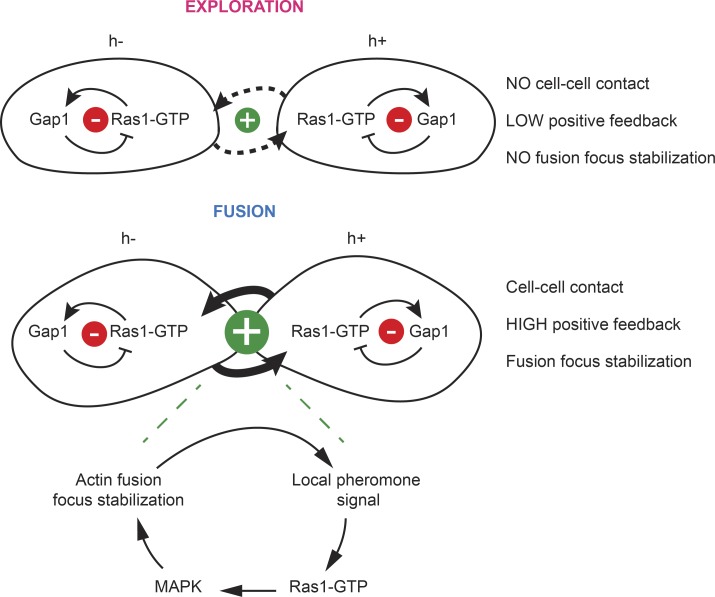
As discussed in the previous paragraphs, inhibition of Ras1 activity is critical to both promote dynamic polarization for cell pairing and prevent premature fusion events. This raises the important question of what provides order to the mating process. One possibility would be that Gap1 activity is down-regulated for fusion, though our data do not provide support for this hypothesis. Instead, we propose that the physical distance between two partner cells inherently regulates the strength of the pheromone-dependent positive feedback signal and thus orders morphological responses (Fig. 8). In support for this idea, pheromones are released locally at sites of polarity (Merlini et al., 2016), and signal perception promotes local polarization, which enhances local secretion. Thus, as the proximity between cells increases, the site of pheromone release narrows, with the consequence that the profile of the perceived pheromone gradient becomes progressively sharper and its amplitude higher. As the polarity site narrows to a tight focus, the local concentration of secreted cell wall hydrolases dominates that of cell wall synthases, leading to local cell wall digestion (Dudin et al., 2015). By constitutively inhibiting Ras1, Gap1 destabilizes the polarity site, thus extending the range of pheromone concentrations supporting dynamic polarization, while still allowing high pheromone concentrations to overcome its effect and trigger cell–cell fusion (Fig. 8). The observation that Gap1 is distributed over broad cortical regions at both exploratory and fusion sites also support this hypothesis, with RasAct zones getting narrower when local pheromone concentration rises. Thus one way to think about the Gap1-dependent inhibition of Ras1 is setting a cell-to-cell distance threshold to ensure that fusion focus stabilization happens only once the local pheromone signal is high enough, and thus couple fusion with cell–cell contact.
Figure 8.
Model for the role of Ras1 activity inhibition for mating progression. During the exploratory phase (top), when cells are not yet in contact, Gap1 recruitment to Ras1-GTP sites promotes the disassembly of the polarity patch. As cell proximity increases, the positive entrainment between partner cells increases. This consists in local pheromone release from one cell leading to local signal activation in its partner, and results in local Ras activity increase, which overcomes the negative regulation by Gap1, and promotes MAPK recruitment to the fusion focus, leading to its stabilization. When Ras1 is constitutively activated, the fusion focus is stabilized early, before cells are in contact, thus resulting in cell lysis.
Materials and methods
Strains, media, and growth conditions
Strains used in this study are listed in Table S1. S. cerevisiae strains were received from B. Hegemann (ETH, Zurich, Switzerland), S. Pelet (University of Lausanne, Lausanne, Switzerland), and S. Piatti (Cell Biology Research Institute of Montpellier–Centre National de la Recherche Scientifique, Montpellier, France). Standard genetic manipulation methods for S. pombe and S. cerevisiae transformation and tetrad dissection were used. For microscopy of fission yeast cells during exponential growth, cells were grown in Edinburgh minimal medium supplemented with amino acids as required. For biochemistry experiments, cells were grown in rich yeast extract (YE) medium or minimal sporulation medium with nitrogen (MSL+N). For assessing cells during the mating process, liquid or agar minimal sporulation medium without nitrogen (MSL-N) was used (Egel et al., 1994; Vjestica et al., 2016). All live-cell imaging during the sexual lifecycle was performed on MSL-N agarose pads (Vjestica et al., 2016). Mating assays were performed as in Vjestica et al. (2016). For microscopy of budding yeast cells, cells were grown in synthetic defined medium supplemented with amino acids as required.
Gene tagging was performed at endogenous genomic locus at the 3′ end, yielding C-terminally tagged proteins, as described previously (Bähler et al., 1998). Tagging with sfGFP was performed as in Dudin et al. (2015). Tagging with GBP-mCherry was performed as in Dudin et al. (2016). N-terminal tagging of Ras1 with GFP was performed as in Merlini et al. (2016). Gene deletion was performed as described (Bähler et al., 1998). Gene tagging and deletion were confirmed by diagnostic PCR for both sides of the gene.
Construction of fission yeast strains expressing RasActGFP (3x-Byr2RBD-3xGFP) was done by integration of RasActGFP under pak1 promoter at the leu1+ locus. A fragment encoding the RBD domain of Byr2 (Fig. 2 A) was cloned in three tandem repeats into pAV49 (pJK210; a gift from A. Vjestica, University of Lausanne, Lausanne, Switzerland). First, Byr2-RBD was amplified from genomic DNA with primers osm2930 (5′-TCCCCCCGGGCGAGAGTTTCCACGTCCATG-3′) and osm2932 (5′-CGGGATCCAGGAGAAAGGGAGGACTGTG-3′), digested with XmaI and BamHI, and ligated to similarly treated pAV49 to generate plasmid pJK210-1x-byr2-RBD. Second, Byr2-RBD was amplified from genomic DNA with primers osm2933 (5′-CGGGATCCCGAGAGTTTCCACGTCCATG-3′) and osm2934 (5′-GCTCTAGAAGGAGAAAGGGAGGACTGTG-3′), digested with BamHI and XbaI, and ligated to similarly treated pJK210-1x-byr2-RBD to generate plasmid pJK210-2x-byr2-RBD. Third, Byr2-RBD was amplified from genomic DNA with primers osm2935 (5′-GCTCTAGACGAGAGTTTCCACGTCCATG-3′) and osm3082 (5′-TCCCCGCGGTTAATTAAAGGAGAAAGGGAGGACTGTG-3′), digested with XbaI and SacII, and ligated to similarly treated pJK210-2x-byr2-RBD to generate plasmid pJK210-3x-byr2-RBD (pSM1627). The 3x-byr2-RBD fragment was then excised from plasmid pSM1627 through digestion with XmaI and PacI and ligated into similarly treated pAV55 (a pJK148-based vector containing the pak1 promoter; a gift from A. Vjestica) to generate plasmid pINT-Ppak1-3x-byr2-RBD-3xGFP-kanMX-leu1+ (pSM1628). Finally, pSM1628 digested with AfeI was stably integrated as a single copy at the leu1+ locus in the yeast genome. In primer sequences, restriction sites are underlined.
Construction of fission yeast strains expressing RasActmCherry (3x-Byr2RBD-3xmCherry) was done by integration of RasActmCherry under pak1 promoter at the leu1+ locus. The 3xmCherry fragment was excided from pSM2060 (pFa6-3xmCherry-natMX) by enzymatic digestion with PacI and AscI and ligated to similarly treated pSM1628 to generate plasmid pSM2094 (pINT-Ppak1-3x-byr2-RBD-3xmCherry-kanMX-leu1+).
Construction of budding yeast strains expressing RasActGFP was done by integration of RasActGFP under pRPL24A promoter at the ura3 locus. First, RasActGFP was amplified from pSM1628 with primers osm3900 (5′-GGACTAGTATGGGAGGTCCCGGGCGA-3′) and osm3901 (5′-ACGCGTCGACGGCGCGCCGATATTAAAG-3′), digested with SpeI and SalI and ligated to similarly treated pSM1890 (pRS413; a gift from S. Piatti) to generate plasmid pSM1898 (pRS413-Pgpd-RasActGFP-Tcyc1). Second, the RasActGFP-Tcyc1 fragment was excised from plasmid pSM1898 through digestion with SpeI and KpnI and ligated into similarly treated pSM1974 (pDA133-pRPL24A; a gift from S. Pelet) to generate plasmid pSM1977 (pRPL24A-RasActGFP-Tcyc1). Finally, pSM1977 digested with BstBI was stably integrated as a single copy at the ura3 locus in the yeast genome. In primer sequences, restriction sites are underlined.
Construction of strains expressing the constitutively active ras1G17V and ras1Q66L or the GDP-bound ras1S22N alleles was done by integration at the endogenous ras1 locus. First, a fragment including ras1 coding region, 5′- and 3′-extensions, was amplified from genomic DNA with primers osm2148 (5′-ACGCGTCGACCACATTTTAACGAGCTTAAGACC-3′) and osm2149 (5′-TCCCCCGGGGCTGCTAATAATTGTGTTAAATG-3′), digested with SalI and XmaI and ligated to similarly treated pSM1232 to generate plasmid pSM1316 (pSP72-ras1). Second, pSM1316 was subjected to site-directed mutagenesis with primers osm2163 (5′-GGTAGTTGTAGGAGATGTTGGTGTTGGTAAAAGTG-3′) and osm2164 (5′-CACTTTTACCAACACCAACATCTCCTACAACTACC-3′) to generate plasmid pSM1320 (pSP72-ras1G17V), with primers osm2167 (5′-GTATTGGACACGGCCGGTCTAGAGGAATATTCCGCTATG-3′) and osm2168 (5′-CATAGCGGAATATTCCTCTAGACCGGCCGTGTCCAATAC-3′) to generate plasmid pSM1322 (pSP72-ras1Q66L) or with primers osm2165 (5′-GGTGGTGTTGGTAAAAATGCTTTGACAATTCAAT-3′) and osm2166 (5′-ATTGAATTGTCAAAGCATTTTTACCAACACCACC-3′) to generate plasmid pSM1321 (pSP72-ras1S22N). Finally, pSM1320, pSM1321, or pSM1322 digested with SalI and XmaI were stably integrated as single copy at the ras1 locus in the yeast genome, through transformation of a ras1::ura4+ strain and selection on agar plates containing 5-fluoroorotic acid. In primer sequences, restriction sites are underlined, and inserted mutations are bold.
Construction of strains expressing the gap1R340A or gap1R195AR340A alleles was done by integration at the endogenous gap1 locus. First, a fragment including gap1 coding region, 5′- and 3′-extension was amplified from genomic DNA with primers osm2232 (5′-ACGCGTCGACCTTAGTATAATATCCATCCTTG-3′) and osm2233 (5′-TCCCCCGGGGACGATTAATGTATAAGAAAC-3′), digested with SalI and XmaI and ligated to similarly treated pSM1232 to generate plasmid pSM1348 (pSP72-gap1). Second, pSM1348 was subjected to site-directed mutagenesis with primers osm2567 (5′-GATTTTTCTTTCTTGCTTTCGTTAATCCAGC-3′) and osm2568 (5′-GCTGGATTAACGAAAGCAAGAAAGAAAAATC-3′) to generate plasmid pSM1513 (pSP72-gap1R340A). Third, pSM1513 was subjected to site-directed mutagenesis with primers osm2571 (5′-GTTTTGTCTCTGCTTGCGGCTAATACTCCGG-3′) and osm2572 (5′-CCGGAGTATTAGCCGCAAGCAGAGACAAAAC-3′) to generate plasmid pSM1645 (pSP72-gap1R195AR340A). Finally, pSM1513 or pSM1645 digested with SalI and XmaI was stably integrated as a single copy at the gap1 locus in the yeast genome, through transformation of a gap1::ura4+ strain and 5-fluoroorotic acid selection. In primer sequences, restriction sites are underlined, and inserted mutations are bold.
Construction of strains in Fig. 6 expressing Gap1FL, GAPGap1, or the fusion constructs RBD-GAPGap1 and RasAct-GAPGap1 was done by integration under pak1 promoter at the leu1+ locus. Gap1 gene was amplified from genomic DNA with primers osm4122 (5′-TCCCCCCGGGACTAAGCGGCACTCTGGTACCC-3′) and osm4123 (5′-CCTTAATTAACTTTCGTAAAAACAATTGTTC-3′), digested with XmaI and PacI, and ligated to similarly treated pSM1628 to generate plasmid pSM1984 (pINT-Ppak1-Gap1FL-3XGFP-kanMX-leu1+). The GAP domain of Gap1 was amplified from genomic DNA with primers osm3922 (5′-TCCCCCCGGGCTTCAGTTGTATGGAGCGTTG-3′) and osm3921 (5′-CCTTAATTAATAAATCAGGAATTGAAGAATCCCATCG-3′), digested with XmaI and PacI, and ligated to similarly treated pSM1628 to generate plasmid pSM1954 (pINT-Ppak1-GAPGap1-3XGFP-kanMX-leu1+). The GAP domain of Gap1 was amplified from genomic DNA with primers osm3922 and osm3923 (5′-TCCCCCCGGGTAAATCAGGAATTGAAGAATCCCATCG-3′) digested with XmaI and ligated to similarly treated pSM1628 to generate plasmid pSM1896 (pINT-Ppak1-RasAct-GAPGap1-kanMX-leu1+) or pSM1603 (pINT-Ppak1-1xbyr2-RBD-3xGFP-kanMX-leu1+) to generate plasmid pSM1897 (pINT-Ppak1-RBD-GAPGap1-kanMX-leu1+). Plasmids carrying mutations in the Gap domain of Gap1 were generated by site directed mutagenesis as explained in the previous paragraph. Finally, plasmids digested with AfeI were stably integrated as a single copy at the leu1+ locus in the yeast genome. In primer sequences, restriction sites are underlined.
For recombinant protein production, ras1 was amplified from a cDNA library with primers osm1941 (5′-CGGGATCCATGAGGTCTACCTACTTAAGAGAGTAC-3′) and osm1942 (5′-CCGCTCGAGCTAACATATAACACAACATTTAGTTG-3′) digested with BamHI and XhoI and ligated to similarly treated pSM394 (pGEX-4T-1) to generate plasmid pSM1282 (pGEX-4T-1-ras1). RBD fragment was amplified from genomic DNA with primers osm2933 and osm3232 (5′-TCCCCCCGGGTTAAGGAGAAAGGGAGGACTGTG-3′) digested with BamHI and XmaI and ligated to similarly treated pSM394 to generate plasmid pSM1713 (pGEX-4T-1-RBD). gap1 was amplified from a cDNA library with primers osm2152 (5′-AAGGAAAAAAGCGGCCGCCATGCTTCAGTTGTATGGAGCGTTG-3′) and osm2311 (5′-CCGCTCGAGTTACTTTCGTAAAAACAATTGTTC-3′) digested with NotI and XhoI and ligated to similarly treated pSM819 (pMAL-TEV) to generate plasmid pSM1401 (pMAL-TEV-gap1). Note that we were not able to purify the full-length Gap1 recombinant protein, and a protein lacking the first 116 aa was used for the in vitro GAP experiments. In primer sequences, restriction sites are underlined, and stop codon is bold.
Mating assays
Mating assays were performed as in Vjestica et al. (2016). In brief, precultures of cells were grown overnight in MSL+N at 25°C to reach an optical density measured at a wavelength of 600 nm (OD600) of between 0.5 and 1. Cultures were then diluted to an OD600 of 0.025 in MSL+N and grown for 16–20 h to an OD600 of between 0.5 and 1 at 30°C. Cells were washed three times with MSL-N, diluted to an OD600 of 1.5 in 1 ml MSL-N, and incubated at 30°C for 1–4 h (depending on the mating stage to be visualized). Cells were mounted onto MSL-N agarose pads (2% agarose) before imaging in overnight videos or incubated at 18°C or at 25°C overnight before imaging. Fusion efficiency, mating efficiency, and percentage of cell lysis were measured as in Dudin et al. (2015, 2016. For pheromone treatments, P-factor pheromone was purchased from Pepnome and used from a stock solution of 1 mg/ml in methanol. M-factor was synthetized and purchased from Schafer-N and used from a stock solution of 2 mg/ml in methanol. Different concentrations of pheromones were directly added to the melted MSL-N agarose before mounting cells on the pads. Cells were then imaged overnight or incubated at 25°C before imaging. Methanol was used as a control.
Microscopy and image analysis
The DeltaVision platform (Applied Precision) described previously (Bendezú and Martin, 2011) was used for time-lapse imaging overnight, quantitative analyses of mating efficiency, fusion efficiency and cell lysis, short time-lapse imaging, and quantification of cortical signal (Fig. 1; Fig. 3, G–I; Fig. 4, C–F; Fig. 6, C and E; Fig. 7, B–E; Fig. S1 A; Fig. S3; Fig. S4; and Fig. S5 C). Images in Fig. 1 and Fig. 4 C (bottom images), Fig. 4 F (bottom images), Fig. 6 C, Fig. 7 C, Fig. S1 A, and Fig. S4 B have been deconvolved. Spinning disk microscopy previously described (Bendezú and Martin, 2011; Dudin et al., 2015) was used for high-temporal resolution and z-stack maximal projection images, time-lapse imaging, and quantification of cortical signal (Fig. 2, B–F; Fig. 3, A–F; Fig. 4, C–F; Fig. 5; Fig. 6, A, B, F, and G; Fig. 7 A; Fig. S1 B; Fig. S2, A and B; Fig. S4 A; and Fig. S5 D). Spinning disk confocal projections are shown in Fig. 3, A–F; Fig. 4 F (top images); Fig. 6, B, G, and H; Fig. S2, A and B; and Fig. S4 A. Kymographs in Fig. 1, Fig. 3, Fig. 6, and Fig. S3 were constructed in ImageJ version 1.47 (National Institutes of Health) by drawing a 3-pixel-wide line at the cell tip or around the cell cortex.
Quantification of cortical fluorescence at the cell tip in Fig. 2 D and whole cortical fluorescence in Fig. 2 F were done by using the sum projection of five consecutive images. The intensity of a 3-pixel-wide segment was collected. Images were corrected for the external background. Quantification of cortical fluorescence at the shmoo or fusion tip in Fig. 3 (G and H) and Fig. 6 E was done by drawing a 3-pixel-wide line at the cell tip. The curves in Fig. 3 G and Fig. 6 E were centered on the maximum pixel values for the Myo52-tdTomato channel (not depicted). Quantifications in Fig. 3 H were done on the five brightest pixels in the selected area. Images were corrected for the external background. Quantification of RasActGFP and Gap1-GFP fluorescence over time during the fusion process in Fig. 3 I and Fig. 6 F was done by drawing a 3-pixel-wide line at the cell tip. The curves were aligned to the time of fusion visualized by observing the diffusion of a cytosolic marker (pmap3-mCherry) from one cell to the other (Vjestica et al., 2016). Images were corrected for the internal background by substracting the mean intensity of a reference cell in the same field at each time point. A Lilliefors test was used to check that data were not significantly different from normal. A t test with Bonferroni correction for comparison with each of the first 12 time points was used for statistical analysis. The levels of Gap1-GFP fluorescence were found not to change significantly before fusion, as comparison with each of the first 12 time points gave P ≥ 0.09, whereas a significance level of 0.05 would require P < 0.04 after Bonferroni correction. Quantification of Gap1-GFP, Ste6-sfGFP, and RasActGFP fluorescence at the fusion focus in Fig. 6 F was done by drawing a 3-pixel-wide line at the fusion site. The curves were centered on the maximum pixel values for the Myo52-tdTomato channel (expressed in each strain). Images were corrected for the external background. Duration of Myo52-GBP-mCherry focalization in Fig. 7 E was measured between the first formation of the Myo52 focus and its postfusion disassembly.
To analyze the lifetime of the Scd2-mCherry/Gap1-GFP patch during exploration in Fig. 6 (C and D), h-sxa2Δ cells coexpressing Gap1-GFP and Scd2-mCherry were imaged in the presence of 0.01 µg/ml P-factor (to promote exploration, but not stabilization of the patch; Bendezú and Martin, 2013) every 30 s for 10 min. To follow the entire processes of Scd2-mCherry/Gap1-GFP patch formation and disassembly, we selected cells in which the Scd2-mCherry patch formed within the first half of the video. We further selected cells in which the exploring patch did not reappear at the same location. To measure photo bleaching, we fitted an exponential decay function with decay constant rPB to the whole cell signal after subtracting the out-of-cell background. To correct for photo bleaching, the pixel intensity at every time point was corrected by multiplying by after subtracting the out-of-cell background. For some of the videos in which the slide was drifting, the translation function from StackReg plugin for ImageJ (Thévenaz et al., 1998) was used to align the slices. The GFP channel was used to manually draw the cell boundary around the cell cortex before recording the intensity of Gap1-GFP and Scd2-mCherry for all time points. We defined the width of the Scd2 patch as the region over which the Scd2-mCherry signal exceeded the mean cortical signal in neighboring regions and obtained mean intensities of Gap1-GFP and Scd2-mCherry for this region. Patch width ranged between 11–16 pixels, equivalent to 1.4–2.0 µm. We also did the same analysis using the full width at half maximum signal of the Scd2-mCherry, which led to the same conclusions. Profiles were normalized, after subtraction of the value in a neighboring zone of similar size, to the maximal value at the patch. Examples of such traces are shown in Fig. S4 (C–E). To plot the mean of these traces, we aligned the Scd2-mCherry intensity traces using the continuous alignment method of Berro and Pollard (2014) and coaligned the Gap1-GFP traces. All statistical comparisons were performed using the t test assay, with the obtained p-value reported in the figure legends, unless reported otherwise.
Biochemistry methods
Recombinant proteins were produced in BL21 cells and purified on GST-Sepharose (GE Healthcare) or amylose beads (New England Biolabs). Cells were grown overnight in 10 ml LB-Amp (lysogeny broth with 100 µg/ml ampicillin) at 37°C. The day after, 6.25 ml of the saturated culture was inoculated in 250 ml of LB-Amp, grown 3 h at 37°C, and cooled down 15 min at 4°C. Protein expression was induced by the addition of 100 µM IPTG for 5–6 h at 18°C. For purification of MBP-Gap1 and MBP-Gap1R340A, bacterial pellets were resuspended in 10 ml cold resuspension buffer (50 mM Tris-HCl, pH 8, 1 mM EDTA, 100 mM KCl, and phenylmethylsulfonyl fluoride), sonicated three times for 30 s (50% power amplitude), incubated 30 min with 1% Triton X-100 at 4°C, and centrifuged 15 min at 4°C at 10,000 g. Soluble extract was incubated with 400 µl of amylose beads for 2 h at 4°C. Finally, beads were washed three times with cold resuspension buffer and eluted in three steps in 100 µl elution buffer (50 mM Tris-HCl, pH 8, 1 mM EDTA, 100 mM KCl, and 10 mM maltose). For purification of GST-Ras1 and GST-RBD, bacterial pellets were resuspended in 5 ml cold 1× PBS containing phenylmethylsulfonyl fluoride, sonicated six times for 30 s (40% power amplitude), incubated 30 min with 1% Triton X-100 at 4°C, and centrifuged 15 min at 4°C at 10,000 g. Soluble extract was incubated with 200 µl of GST-Sepharose beads for 2 h at 4°C. Finally, beads were washed three times with 1× PBS and eluted in three steps in 100 µl elution buffer (50 mM Tris-HCl, pH 8, and 15 mM reduced glutathione).
For in vitro GAP assays (Geymonat et al., 2002), 1 mg of purified GST-Ras1 was resuspended in 70 µl of loading buffer (20 mM Tris-HCl, pH 7.5, 25 mM NaCl, 5 mM MgCl2, and 0.1 mM DTT) containing 1.5 µl γ-[32P]GTP for 10 min at 30°C. After cooling on ice, 10 µl was incubated in 50 µl reaction buffer (20 mM Tris-HCl, pH 7.5, 2 mM GTP, and 0.6 µg/µl BSA) with 18 µg of purified MBP-Gap1, MBP-Gap1R340A, or MBP. The reactions were incubated at 30°C, and every 5 min, 10 µl of reaction was diluted in 990 µl of cold washing buffer (20 mM Tris-HCl, pH 7.5, 50 mM NaCl, and 5 mM MgCl2). The samples were filtered through prewetted nitrocellulose filters (Millipore), washed three times with 4 ml of cold washing buffer, and air-dried. The amount of radioactive nucleotide bound to the protein was determined by scintillation counting.
For Ras-GTP pulldown (Soto et al., 2010), extracts from yeast cells grown in 200 ml of YE medium (vegetative growth, Fig. 4) or 200 ml MSL-N (mating conditions, Fig. 5, pregrowth in MSL+N and shift to MSL-N for 4 h) were prepared in cold Binding Buffer (25 mM Tris-HCl, pH 7.5, 1 mM DTT, 30 mM MgCl2, 40 mM NaCl, and 0.5% NP-40) containing Anti Proteolitic (Roche) and Phostop (Roche) tablets. Cell lysates were obtained at 4°C via mechanical breakeage with acid-treated glass beads (Sigma) in a BeadBeater homogenizer (10 times at 4.5 V for 30 s with 30-s break on ice every cycle) and centrifugation for 20 min at 4°C at 10,000 g. 4–6 mg of protein extract was incubated with 20 µl of GST-Sepharose beads and 25 µg of purified GST-RBD for 1 h at 4°C. Beads were washed three times with 500 µl of cold washing buffer 1 (25 mM Tris-HCl, pH 7.5, 1 mM DTT, 30 mM MgCl2, 40 mM NaCl, and 1% NP-40) and two times with 500 µl of cold washing buffer 2 (25 mM Tris-HCl, pH 7.5, 1 mM DTT, 30 mM MgCl2, and 40 mM NaCl) containing protease inhibitor cocktail. Proteins were resolved by SDS-PAGE for Western blot analysis. The mean intensity quantification from three independent experiments is shown in Fig. 5 E. Before averaging, each experiment was corrected for the GFP-Ras1 levels in the inputs for each sample and normalized to the maximum value for each experiment.
Standard protocols were used for SDS-PAGE and Western blot analysis. Antibodies used on Western blots were anti-GFP monoclonal antibody (Roche) and anti-TAT1 monoclonal antibody for α-tubulin detection.
Figures were assembled with Adobe Photoshop CS5 and Adobe Illustrator CS5. All error bars are SDs. All experiments were done a minimum of three independent times.
Online supplemental material
Fig. S1 shows constitutively active Ras1 mutant undergoing lysis and localization of GFP-Ras1 in interphase cells. Fig. S2 shows colocalization of RasActGFP and Scd2-mCherry at dynamic polarity sites. Fig. S3 shows quantification of multiple shmoos in cells lacking Gap1, suppression of gap1Δ cell lysis by fus1 deletion, and quantification of cell lysis of heterothallic sxa1Δ gap1Δ and sxa2Δ gap1Δ cells treated with synthetic pheromones. Images and quantifications in Fig. S4 show that Gap1-GFP is recruited to dynamic polarity Scd2-mCherry sites and stays associated after their disassembly. Fig. S5 shows construction of fusion strains analyzed in Fig. 7 and their expression levels, and that their activity depends on a functional GAP domain. Video 1 shows constitutively active ras mutants during mating, forming single or multiple projections. Video 2 shows single mating type (h-) constitutively active ras mutants lacking pheromone protease undergoing lysis after the assembly of stable Myo52-tdTomato foci upon synthetic pheromone treatment. Video 3 shows homothallic (h90) cells lacking Gap1 forming two consecutive mating projections. Videos 4 and 5 show homothallic (h90) cells lacking Gap1, which assemble a stable focus of Myo52-tdTomato (Video 4) or actin (GFP-CHD; Video 5) before undergoing cell lysis. Table S1 shows strains used in this study.
Supplementary Material
Acknowledgments
We thank B. Hegemann, S. Pelet, and S. Piatti for strains and plasmids, S. Mitri for help with statistics, D. Vavylonis for discussions and critical input, and S. Pelet, S. Piatti, and laboratory members for critical reading of the manuscript.
This work was supported by Swiss National Science Foundation grant 31003A_155944 and a European Research Council Consolidator Grant (CellFusion) to S.G. Martin. B. Khalili was supported by National Institutes of Health grant R01GM098430 (to D. Vavylonis).
The authors declare no competing financial interests.
Author contributions: L. Merlini and S.G. Martin designed the experimental strategy. L. Merlini, O. Dudin, L. Michon, and V. Vincenzetti performed experiments. O. Dudin performed and analyzed experiments on autocrine cells (Fig. 3 D and Fig. S3 H) and helped acquiring images for Fig. 5 B. L. Michon and V. Vincenzetti helped performing Ras-GTP pulldown in Fig. 5 E and constructing strains for Fig. S5 D. B. Khalili analyzed data in Fig. 5 D and Fig. S5 (C–E). L. Merlini performed all the other experiments. L. Merlini and S.G. Martin analyzed data and wrote the manuscript. S.G. Martin acquired funding.
References
- Ayscough K.R., and Drubin D.G.. 1998. A role for the yeast actin cytoskeleton in pheromone receptor clustering and signalling. Curr. Biol. 8:927–930. 10.1016/S0960-9822(07)00374-0 [DOI] [PubMed] [Google Scholar]
- Bähler J., Wu J.Q., Longtine M.S., Shah N.G., McKenzie A. III, Steever A.B., Wach A., Philippsen P., and Pringle J.R.. 1998. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast. 14:943–951. [DOI] [PubMed] [Google Scholar]
- Bauman P., Cheng Q.C., and Albright C.F.. 1998. The Byr2 kinase translocates to the plasma membrane in a Ras1-dependent manner. Biochem. Biophys. Res. Commun. 244:468–474. 10.1006/bbrc.1998.8292 [DOI] [PubMed] [Google Scholar]
- Bendezú F.O., and Martin S.G.. 2011. Actin cables and the exocyst form two independent morphogenesis pathways in the fission yeast. Mol. Biol. Cell. 22:44–53. 10.1091/mbc.E10-08-0720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendezú F.O., and Martin S.G.. 2013. Cdc42 explores the cell periphery for mate selection in fission yeast. Curr. Biol. 23:42–47. 10.1016/j.cub.2012.10.042 [DOI] [PubMed] [Google Scholar]
- Berro J., and Pollard T.D.. 2014. Local and global analysis of endocytic patch dynamics in fission yeast using a new “temporal superresolution” realignment method. Mol. Biol. Cell. 25:3501–3514. 10.1091/mbc.E13-01-0004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi E., and Park H.O.. 2012. Cell polarization and cytokinesis in budding yeast. Genetics. 191:347–387. 10.1534/genetics.111.132886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang E.C., Barr M., Wang Y., Jung V., Xu H.P., and Wigler M.H.. 1994. Cooperative interaction of S. pombe proteins required for mating and morphogenesis. Cell. 79:131–141. 10.1016/0092-8674(94)90406-5 [DOI] [PubMed] [Google Scholar]
- Conlon P., Gelin-Licht R., Ganesan A., Zhang J., and Levchenko A.. 2016. Single-cell dynamics and variability of MAPK activity in a yeast differentiation pathway. Proc. Natl. Acad. Sci. USA. 113:E5896–E5905. 10.1073/pnas.1610081113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad M., Schothorst J., Kankipati H.N., Van Zeebroeck G., Rubio-Texeira M., and Thevelein J.M.. 2014. Nutrient sensing and signaling in the yeast Saccharomyces cerevisiae. FEMS Microbiol. Rev. 38:254–299. 10.1111/1574-6976.12065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das M., Drake T., Wiley D.J., Buchwald P., Vavylonis D., and Verde F.. 2012. Oscillatory Dynamics of Cdc42 GTPase in the Control of Polarized Growth. Science. 337:239–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devreotes P., and Horwitz A.R.. 2015. Signaling networks that regulate cell migration. Cold Spring Harb. Perspect. Biol. 7:a005959 10.1101/cshperspect.a005959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudin O., Bendezú F.O., Groux R., Laroche T., Seitz A., and Martin S.G.. 2015. A formin-nucleated actin aster concentrates cell wall hydrolases for cell fusion in fission yeast. J. Cell Biol. 208:897–911. 10.1083/jcb.201411124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudin O., Merlini L., and Martin S.G.. 2016. Spatial focalization of pheromone/MAPK signaling triggers commitment to cell-cell fusion. Genes Dev. 30:2226–2239. 10.1101/gad.286922.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudin O., Merlini L., Bendezú F.O., Groux R., Vincenzetti V., and Martin S.G.. 2017. A systematic screen for morphological abnormalities during fission yeast sexual reproduction identifies a mechanism of actin aster formation for cell fusion. PLoS Genet. 13:e1006721 10.1371/journal.pgen.1006721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egel R., Willer M., Kjaerulff S., Davey J., and Nielsen O.. 1994. Assessment of pheromone production and response in fission yeast by a halo test of induced sporulation. Yeast. 10:1347–1354. 10.1002/yea.320101012 [DOI] [PubMed] [Google Scholar]
- Elowitz M.B., and Leibler S.. 2000. A synthetic oscillatory network of transcriptional regulators. Nature. 403:335–338. 10.1038/35002125 [DOI] [PubMed] [Google Scholar]
- Ferrell J.E., Jr 2013. Feedback loops and reciprocal regulation: recurring motifs in the systems biology of the cell cycle. Curr. Opin. Cell Biol. 25:676–686. 10.1016/j.ceb.2013.07.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukui Y., Kozasa T., Kaziro Y., Takeda T., and Yamamoto M.. 1986. Role of a ras homolog in the life cycle of Schizosaccharomyces pombe. Cell. 44:329–336. 10.1016/0092-8674(86)90767-1 [DOI] [PubMed] [Google Scholar]
- Geymonat M., Spanos A., Smith S.J., Wheatley E., Rittinger K., Johnston L.H., and Sedgwick S.G.. 2002. Control of mitotic exit in budding yeast. In vitro regulation of Tem1 GTPase by Bub2 and Bfa1. J. Biol. Chem. 277:28439–28445. 10.1074/jbc.M202540200 [DOI] [PubMed] [Google Scholar]
- Gronwald W., Huber F., Grünewald P., Spörner M., Wohlgemuth S., Herrmann C., and Kalbitzer H.R.. 2001. Solution structure of the Ras binding domain of the protein kinase Byr2 from Schizosaccharomyces pombe. Structure. 9:1029–1041. 10.1016/S0969-2126(01)00671-2 [DOI] [PubMed] [Google Scholar]
- Hennig A., Markwart R., Wolff K., Schubert K., Cui Y., Prior I.A., Esparza-Franco M.A., Ladds G., and Rubio I.. 2016. Feedback activation of neurofibromin terminates growth factor-induced Ras activation. Cell Commun. Signal. 14:5 10.1186/s12964-016-0128-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirota K., Tanaka K., Watanabe Y., and Yamamoto M.. 2001. Functional analysis of the C-terminal cytoplasmic region of the M-factor receptor in fission yeast. Genes Cells. 6:201–214. 10.1046/j.1365-2443.2001.00415.x [DOI] [PubMed] [Google Scholar]
- Howell A.S., Jin M., Wu C.F., Zyla T.R., Elston T.C., and Lew D.J.. 2012. Negative feedback enhances robustness in the yeast polarity establishment circuit. Cell. 149:322–333. 10.1016/j.cell.2012.03.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes D.A., Ashworth A., and Marshall C.J.. 1993. Complementation of byr1 in fission yeast by mammalian MAP kinase kinase requires coexpression of Raf kinase. Nature. 364:349–352. 10.1038/364349a0 [DOI] [PubMed] [Google Scholar]
- Hughes D.A., Yabana N., and Yamamoto M.. 1994. Transcriptional regulation of a Ras nucleotide-exchange factor gene by extracellular signals in fission yeast. J. Cell Sci. 107:3635–3642. [DOI] [PubMed] [Google Scholar]
- Hwang J.U., Gu Y., Lee Y.J., and Yang Z.. 2005. Oscillatory ROP GTPase activation leads the oscillatory polarized growth of pollen tubes. Mol. Biol. Cell. 16:5385–5399. 10.1091/mbc.E05-05-0409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang J.U., Vernoud V., Szumlanski A., Nielsen E., and Yang Z.. 2008. A tip-localized RhoGAP controls cell polarity by globally inhibiting Rho GTPase at the cell apex. Curr. Biol. 18:1907–1916. 10.1016/j.cub.2008.11.057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai Y., Miyake S., Hughes D.A., and Yamamoto M.. 1991. Identification of a GTPase-activating protein homolog in Schizosaccharomyces pombe. Mol. Cell. Biol. 11:3088–3094. 10.1128/MCB.11.6.3088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson J.M., Jin M., and Lew D.J.. 2011. Symmetry breaking and the establishment of cell polarity in budding yeast. Curr. Opin. Genet. Dev. 21:740–746. 10.1016/j.gde.2011.09.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kae H., Lim C.J., Spiegelman G.B., and Weeks G.. 2004. Chemoattractant-induced Ras activation during Dictyostelium aggregation. EMBO Rep. 5:602–606. 10.1038/sj.embor.7400151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kataoka T., Powers S., McGill C., Fasano O., Strathern J., Broach J., and Wigler M.. 1984. Genetic analysis of yeast RAS1 and RAS2 genes. Cell. 37:437–445. 10.1016/0092-8674(84)90374-X [DOI] [PubMed] [Google Scholar]
- Kuchler K., Dohlman H.G., and Thorner J.. 1993. The a-factor transporter (STE6 gene product) and cell polarity in the yeast Saccharomyces cerevisiae. J. Cell Biol. 120:1203–1215. 10.1083/jcb.120.5.1203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeder C.I., Hink M.A., Kinkhabwala A., Mayr R., Bastiaens P.I., and Knop M.. 2007. Spatial regulation of Fus3 MAP kinase activity through a reaction-diffusion mechanism in yeast pheromone signalling. Nat. Cell Biol. 9:1319–1326. 10.1038/ncb1652 [DOI] [PubMed] [Google Scholar]
- Manandhar S.P., Hildebrandt E.R., Jacobsen W.H., Santangelo G.M., and Schmidt W.K.. 2010. Chemical inhibition of CaaX protease activity disrupts yeast Ras localization. Yeast. 27:327–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin S.G. 2015. Spontaneous cell polarization: Feedback control of Cdc42 GTPase breaks cellular symmetry. BioEssays. 37:1193–1201. 10.1002/bies.201500077 [DOI] [PubMed] [Google Scholar]
- Masuda T., Kariya K., Shinkai M., Okada T., and Kataoka T.. 1995. Protein kinase Byr2 is a target of Ras1 in the fission yeast Schizosaccharomyces pombe. J. Biol. Chem. 270:1979–1982. 10.1074/jbc.270.5.1979 [DOI] [PubMed] [Google Scholar]
- Mata J., and Bähler J.. 2006. Global roles of Ste11p, cell type, and pheromone in the control of gene expression during early sexual differentiation in fission yeast. Proc. Natl. Acad. Sci. USA. 103:15517–15522. 10.1073/pnas.0603403103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merlini L., Dudin O., and Martin S.G.. 2013. Mate and fuse: how yeast cells do it. Open Biol. 3:130008 10.1098/rsob.130008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merlini L., Khalili B., Bendezú F.O., Hurwitz D., Vincenzetti V., Vavylonis D., and Martin S.G.. 2016. Local Pheromone Release from Dynamic Polarity Sites Underlies Cell-Cell Pairing during Yeast Mating. Curr. Biol. 26:1117–1125. 10.1016/j.cub.2016.02.064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadin-Davis S.A., Nasim A., and Beach D.. 1986. Involvement of ras in sexual differentiation but not in growth control in fission yeast. EMBO J. 5:2963–2971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novák B., and Tyson J.J.. 2008. Design principles of biochemical oscillators. Nat. Rev. Mol. Cell Biol. 9:981–991. 10.1038/nrm2530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada S., Leda M., Hanna J., Savage N.S., Bi E., and Goryachev A.B.. 2013. Daughter cell identity emerges from the interplay of Cdc42, septins, and exocytosis. Dev. Cell. 26:148–161. 10.1016/j.devcel.2013.06.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozbudak E.M., Becskei A., and van Oudenaarden A.. 2005. A system of counteracting feedback loops regulates Cdc42p activity during spontaneous cell polarization. Dev. Cell. 9:565–571. 10.1016/j.devcel.2005.08.014 [DOI] [PubMed] [Google Scholar]
- Papadaki P., Pizon V., Onken B., and Chang E.C.. 2002. Two ras pathways in fission yeast are differentially regulated by two ras guanine nucleotide exchange factors. Mol. Cell. Biol. 22:4598–4606. 10.1128/MCB.22.13.4598-4606.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira P.S., and Jones N.C.. 2001. The RGS domain-containing fission yeast protein, Rgs1p, regulates pheromone signalling and is required for mating. Genes Cells. 6:789–802. 10.1046/j.1365-2443.2001.00465.x [DOI] [PubMed] [Google Scholar]
- Petersen J., Weilguny D., Egel R., and Nielsen O.. 1995. Characterization of fus1 of Schizosaccharomyces pombe: a developmentally controlled function needed for conjugation. Mol. Cell. Biol. 15:3697–3707. 10.1128/MCB.15.7.3697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi M., and Elion E.A.. 2005. Formin-induced actin cables are required for polarized recruitment of the Ste5 scaffold and high level activation of MAPK Fus3. J. Cell Sci. 118:2837–2848. 10.1242/jcs.02418 [DOI] [PubMed] [Google Scholar]
- Rauch N., Rukhlenko O.S., Kolch W., and Kholodenko B.N.. 2016. MAPK kinase signalling dynamics regulate cell fate decisions and drug resistance. Curr. Opin. Struct. Biol. 41:151–158. 10.1016/j.sbi.2016.07.019 [DOI] [PubMed] [Google Scholar]
- Ribes V., and Briscoe J.. 2009. Establishing and interpreting graded Sonic Hedgehog signaling during vertebrate neural tube patterning: the role of negative feedback. Cold Spring Harb. Perspect. Biol. 1:a002014 10.1101/cshperspect.a002014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothbauer U., Zolghadr K., Muyldermans S., Schepers A., Cardoso M.C., and Leonhardt H.. 2008. A versatile nanotrap for biochemical and functional studies with fluorescent fusion proteins. Mol. Cell. Proteomics. 7:282–289. 10.1074/mcp.M700342-MCP200 [DOI] [PubMed] [Google Scholar]
- Sasaki A.T., Chun C., Takeda K., and Firtel R.A.. 2004. Localized Ras signaling at the leading edge regulates PI3K, cell polarity, and directional cell movement. J. Cell Biol. 167:505–518. 10.1083/jcb.200406177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheffzek K., Ahmadian M.R., Kabsch W., Wiesmüller L., Lautwein A., Schmitz F., and Wittinghofer A.. 1997. The Ras-RasGAP complex: structural basis for GTPase activation and its loss in oncogenic Ras mutants. Science. 277:333–338. 10.1126/science.277.5324.333 [DOI] [PubMed] [Google Scholar]
- Sermon B.A., Lowe P.N., Strom M., and Eccleston J.F.. 1998. The importance of two conserved arginine residues for catalysis by the ras GTPase-activating protein, neurofibromin. J. Biol. Chem. 273:9480–9485. 10.1074/jbc.273.16.9480 [DOI] [PubMed] [Google Scholar]
- Sheu Y.J., Santos B., Fortin N., Costigan C., and Snyder M.. 1998. Spa2p interacts with cell polarity proteins and signaling components involved in yeast cell morphogenesis. Mol. Cell. Biol. 18:4053–4069. 10.1128/MCB.18.7.4053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slaughter B.D., Smith S.E., and Li R.. 2009. Symmetry breaking in the life cycle of the budding yeast. Cold Spring Harb. Perspect. Biol. 1:a003384 10.1101/cshperspect.a003384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soto T., Villar-Tajadura M.A., Madrid M., Vicente J., Gacto M., Pérez P., and Cansado J.. 2010. Rga4 modulates the activity of the fission yeast cell integrity MAPK pathway by acting as a Rho2 GTPase-activating protein. J. Biol. Chem. 285:11516–11525. 10.1074/jbc.M109.071027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka K., Nakafuku M., Satoh T., Marshall M.S., Gibbs J.B., Matsumoto K., Kaziro Y., and Toh-e A.. 1990. S. cerevisiae genes IRA1 and IRA2 encode proteins that may be functionally equivalent to mammalian ras GTPase activating protein. Cell. 60:803–807. 10.1016/0092-8674(90)90094-U [DOI] [PubMed] [Google Scholar]
- Thévenaz P., Ruttimann U.E., and Unser M.. 1998. A pyramid approach to subpixel registration based on intensity. IEEE Trans. Image Process. 7:27–41. 10.1109/83.650848 [DOI] [PubMed] [Google Scholar]
- Vjestica A., Merlini L., Dudin O., Bendezu F.O., and Martin S.G.. 2016. Microscopy of Fission Yeast Sexual Lifecycle. J. Vis. Exp. e53801 10.3791/53801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Boguski M., Riggs M., Rodgers L., and Wigler M.. 1991a sar1, a gene from Schizosaccharomyces pombe encoding a protein that regulates ras1. Cell Regul. 2:453–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Xu H.P., Riggs M., Rodgers L., and Wigler M.. 1991b byr2, a Schizosaccharomyces pombe gene encoding a protein kinase capable of partial suppression of the ras1 mutant phenotype. Mol. Cell. Biol. 11:3554–3563. 10.1128/MCB.11.7.3554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson P., Davis K., Didmon M., Broad P., and Davey J.. 1999. An RGS protein regulates the pheromone response in the fission yeast Schizosaccharomyces pombe. Mol. Microbiol. 33:623–634. 10.1046/j.1365-2958.1999.01510.x [DOI] [PubMed] [Google Scholar]
- Weston C., Bond M., Croft W., and Ladds G.. 2013. The coordination of cell growth during fission yeast mating requires Ras1-GTP hydrolysis. PLoS One. 8:e77487 10.1371/journal.pone.0077487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C.F., and Lew D.J.. 2013. Beyond symmetry-breaking: competition and negative feedback in GTPase regulation. Trends Cell Biol. 23:476–483. 10.1016/j.tcb.2013.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S., Charest P.G., and Firtel R.A.. 2008. Spatiotemporal regulation of Ras activity provides directional sensing. Curr. Biol. 18:1587–1593. 10.1016/j.cub.2008.08.069 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.