Niewidok et al. analyze the distribution and dynamics of RNA-binding proteins (RBPs) with single-molecule resolution in living neuronal cells, providing direct support for liquid droplet behavior of stress granules in living cells and revealing transient binding of RBPs in nanocores.
Abstract
Stress granules (SGs) are cytosolic, nonmembranous RNA–protein complexes. In vitro experiments suggested that they are formed by liquid–liquid phase separation; however, their properties in mammalian cells remain unclear. We analyzed the distribution and dynamics of two paradigmatic RNA-binding proteins (RBPs), Ras GTPase-activating protein SH3-domain–binding protein (G3BP1) and insulin-like growth factor II mRNA-binding protein 1 (IMP1), with single-molecule resolution in living neuronal cells. Both RBPs exhibited different exchange kinetics between SGs. Within SGs, single-molecule localization microscopy revealed distributed hotspots of immobilized G3BP1 and IMP1 that reflect the presence of relatively immobile nanometer-sized nanocores. We demonstrate alternating binding in nanocores and anomalous diffusion in the liquid phase with similar characteristics for both RBPs. Reduction of low-complexity regions in G3BP1 resulted in less detectable mobile molecules in the liquid phase without change in binding in nanocores. The data provide direct support for liquid droplet behavior of SGs in living cells and reveal transient binding of RBPs in nanocores. Our study uncovers a surprising disconnect between SG partitioning and internal diffusion and interactions of RBPs.
Introduction
Stress granules (SGs) are cytosolic, nonmembranous RNA–protein (RNP) complexes that form in the cytosol during various stress responses. They contain mRNAs stalled in translation initiation and are thought to be involved in regulation and mediating mRNA stability as a response to environmental challenges (Anderson and Kedersha, 2008; Buchan and Parker, 2009; Protter and Parker, 2016; Rabouille and Alberti, 2017). Although physiological SG formation appears to be an adaptive and survival-promoting mechanism, inappropriate formation or chronic persistence of SGs has been implicated in aging and pathological processes during several neurodegenerative diseases such as amyotrophic lateral sclerosis (ALS), Parkinson’s disease, and Alzheimer’s disease (Wolozin, 2012; Ramaswami et al., 2013; Buchan, 2014; Alberti and Hyman, 2016).
Cytosolic RNP complexes such as SGs may form in the cell by a process called liquid–liquid phase separation (LLPS), which increases the local concentration of RNA and protein, thereby generating a concentrated subcellular microcompartment that favors downstream interactions (Brangwynne et al., 2009; Li et al., 2012; Alberti, 2017; Banani et al., 2017). Experiments in yeast indicated that SGs behave like unstructured, solid storage depots for RNA and proteins. In contrast, mammalian SGs were more liquid-like, as would be expected from structures derived by LLPS (Kroschwald et al., 2015). However, mammalian SGs appeared to be less uniform than expected. In particular, substructures, which have been referred to as “cores” with higher concentrations of proteins and mRNAs, surrounded by a less concentrated “shell” have been described (Souquere et al., 2009; Jain et al., 2016; Wheeler et al., 2016). It should be noted, though, that the presence of a more stable core was mainly concluded from biochemical fractionation experiments, and the heterogeneous nature of SGs was inferred from microscopic images of fixed material. Thus, it is unclear whether authentic SGs are heterogeneous in living cells, how persistent their potential core structures are, and how the material state of more core-like and more shell-like regions differs.
A particular feature of SGs that distinguishes them from membrane-surrounded microcompartments is that they are dynamic structures, where at least some components frequently shuttle in and out. This would also be consistent with a role of SGs as dynamic sorting stations for mRNAs and associated proteins and not merely sites of stalled mRNA translation. In fact, FRAP experiments revealed that many SG components exchange rapidly with half-lives of less than 30s, whereas other components were more immobile (Buchan and Parker, 2009). However, it is unclear whether the difference is related to the distribution of the respective components within SGs. For example, an obvious possibility would be that components in a potential shell can exchange rapidly, whereas SG components in the core are less dynamic (Jain et al., 2016). To test this hypothesis, it would be required to correlate the dynamicity of individual SG components (e.g., their shuttling frequency between granules) with their behavior and distribution within SGs.
The RNA-binding protein (RBP) Ras GTPase-activating protein SH3-domain–binding protein 1 (G3BP1) is considered to be a constitutive SG protein and has been implicated in the regulation of SG assembly and function (Kedersha et al., 2016). G3BP1 contains multiple protein-protein interaction domains and a single RNA-binding region, and it appears to dynamically shuttle in and out of granules (Moschner et al., 2014). G3BP1 is a target of transactive response DNA-binding protein 43 (TDP-43), the major pathological protein in sporadic ALS (Aulas et al., 2012, 2015), which may link it to neurodegenerative processes. Also insulin-like growth factor II mRNA-binding protein 1 (IMP1) is an RBP that can be present in SGs, but differs from G3BP1 by containing multiple RNA-binding domains and only a single protein-protein interaction module (Fig. 1 A). IMP1 is known to regulate RNA processing at several levels and is also involved in neuronal mRNA transport (Degrauwe et al., 2016). We have previously shown that exogenous expression of G3BP1 and IMP1 induces SG formation in neuronally differentiated cells and affects mRNA translation (Moschner et al., 2014). However, IMP1 appeared to be more stationary than G3BP1, which may suggest different localization in SGs; e.g., in potential core versus shell structures.
Figure 1.
G3BP1 and IMP1 exhibit different dynamics of protein exchange between SGs. (A) Protein interaction (gray) and RNA-binding domains (black) of human G3BP1 and IMP1 according to SMART analysis for identification of signaling domains (Schultz et al., 1998). RRM, RNA recognition motif; KH, K homology domain. Right: Western analysis of cellular lysates after transfection with PAGFP-tagged G3BP1 and IMP1 (arrowhead). Lysates were analyzed using size-based capillary electrophoresis, and electropherograms are represented as pseudoblots as described in Materials and methods. Molecular mass standards are indicated. The respective endogenous proteins are indicated by an arrow. Please note that analysis by size-based capillary electrophoresis can yield protein mobilities that differ from separation by standard SDS-PAGE. (B) Colocalization of exogenously expressed mCherry-G3BP1 with the SG marker TIA-1. Bar, 10 µm. (C) Colocalization and dynamic exchange of fluorescence-tagged G3BP1 and IMP1 in SGs of living PC12 cells. Granules were labeled with mCherry-IMP1 and PAGFP-G3BP1 was activated in one granule (dashed square). The outline of the cell and the nucleus are indicated in the red fluorescent micrographs. Fluorescence distribution was followed over time. After some seconds, photoactivated PAGFP-G3BP1 appears in SGs outside of the activated region indicating dynamic exchange of G3BP1 between SGs. Bar, 10 µm. (D) Bar plot showing the size distribution of SGs as determined from the area of mCherry-IMP1 positive granules. The box represents 50% of the population, whiskers range from 5% to 95% and crosses correspond to the minimal and maximal values (n = 34). (E) Schematic representation showing the FDAP approach to determine dynamics and binding of PAGFP-tagged G3BP1 and IMP1 in granules. Photoactivation and fluorescence recording was performed in a 3 × 5 µm region containing cytosol and granules between nucleus and plasma membrane (red box). The decay curves were fitted with model FDAP functions. (F) FDAP curves for PAGFP-IMP1, PAGFP-G3BP1, and 3×PAGFP (mean ± SEM, n = 13 [PAGFP-IMP1], n = 17 [PAGFP-G3BP1], n = 20 [3×PAGFP]) showing the different dynamics of IMP1 and G3BP1. (G) Residence time of PAGFP-tagged IMP1 and G3BP1 in granules as determined by the model FDAP function (mean ± SEM, n = 19 [PAGFP-IMP1], n = 20 [PAGFP-G3BP1], n = 20 [3×PAGFP] from two [3×PAGFP], four [PAGFP-G3BP1], and five [PAGFP-IMP1] independent experiments). Comparison between the constructs involved one-way ANOVA followed by post-hoc Tukey's test. **, P < 0.01; ***, P < 0.001 (compared with control [3×PAGFP]); +, P < 0.05. For all experiments, stress had been induced by a 20-min treatment with 0.5 mM sodium arsenite before imaging.
To follow the distribution and dynamics of G3BP1 and IMP1 in stressed neuronal cells, we performed fluorescence decay after photoactivation (FDAP) measurements and single-molecule localization microscopy. These complementary approaches allow for comparing the dynamics of protein exchange between SGs and the cytoplasm and the local dynamics of protein diffusion and interaction within SGs.
Results
G3BP1 and IMP1 exhibit different dynamics of protein exchange between SGs
To analyze the behavior of individual SG components, we chose the two RBPs, G3BP1 and IMP1, as representative proteins of RNP granules. Both proteins are known to be present in SGs and both contain protein- and RNA-binding domains that mediate hetero- and homotypic interactions between proteins and RNAs (Fig. 1 A, left). We have shown previously that G3BP1 and IMP1 colocalize in the same granules of neuronal cells after induction of stress (Moschner et al., 2014).
To analyze the distribution and dynamics of G3BP1 and IMP1 in living cells, we prepared photoactivatable GFP (PAGFP)–tagged fusion proteins. The constructs were present as single polypeptides in transfected PC12 cells, indicating their integrity (Fig. 1 A, right). Compared with endogenous G3BP1 and IMP1, we observed a similar expression or a moderate overexpression of the exogenously expressed fusion proteins. To confirm that exogenously expressed fluorescence-tagged G3BP1 localizes to authentic SGs in neuronally differentiated PC12 cells we induced stress with sodium arsenite and performed immunocytochemical stainings with the SG marker TIA-1 (Kedersha et al., 2005). Indeed, all cytosolic granular structures containing mCherry-G3BP1 were also positive for TIA-1, indicating that the cytoplasmic puncta are equivalent to stress-induced granules (Fig. 1 B). To determine the dynamics of their segregation into neuronal SGs, we coexpressed PAGFP-tagged G3BP1 and mCherry-tagged IMP1 in the cells. After sodium arsenite treatment, PAGFP-G3BP1 was photoactivated in a single SG by a flash with a 407-nm laser. IMP1 and G3BP1 colocalized in the photoactivated granule, and fluorescent G3BP1 gradually appeared in IMP1-positive granules outside of the activation region after some seconds indicating dynamic exchange of G3BP1 between SGs (Fig. 1 C). Granules were heterogeneous in size with a mean area of ∼7 µm2, extending from ∼1 to 16 µm2 (Fig. 1 D), which is consistent with previous observations that SG shape and size varies significantly in cells (Buchan and Parker, 2009). Arsenite-induced SGs remained largely stationary during the observation period, probably because of anchorage to the cellular microtubule network (Nadezhdina et al., 2010; Moschner et al., 2014).
To quantify the dynamics of G3BP1 and IMP1 partitioning into neuronal SGs, we performed FDAP assays with PAGFP-tagged G3BP1 and IMP1, as well as 3×PAGFP as a cytosolic control protein of similar size. PAGFP-tagged proteins were coexpressed with mCherry-IMP1 to visualize the granules. After induction of SG formation with sodium arsenite, a small region of the cytosol containing SGs was photoactivated by a laser flash. The dissipation of the fluorescent proteins from the activated region was followed over time as schematically shown in Fig. 1 E. 3×PAGFP showed a rapid decay after photoactivation, indicating high mobility (Fig. 1 F). In contrast, PAGFP-IMP1 exhibited a very slow decay indicating strong and long-lasting interaction in SGs. PAGFP-G3BP1 showed an intermediate decay, suggesting dynamic shuttling in and out of granules. Decay curves were fitted with model FDAP functions for one (I1(t)) or two populations (I2(t); see Materials and methods). The FDAP curves for 3×PAGFP and IMP1 were fitted best by Eq. 1 (I1(t)). The decay curve of G3BP1 was better described by the model for two populations (I2(t)), indicating the presence of both a free and a granule-associated population with dynamic exchange. The decay time of the slow component (τslow) reflects the residence time of G3BP1 in the granules, and τfast reflects the mobility of G3BP1 in the cytosol.
The fit with the model FDAP functions allowed estimation of the residence time of the RBPs in the granules, which was significantly lower for G3BP1 than for IMP1 (Fig. 1 G). Assuming a first-order process, a t1/2 of ∼20 s could be estimated for G3BP1 (t1/2 = τslow × ln2), which is comparable with previous data from FRAP experiments, where half-lives of less than 30s have been reported for dynamically exchanging SG components (Buchan and Parker, 2009). τfast for G3BP1 in the presence of arsenite was the same as its decay time in the absence of arsenite (5.3 ± 1.2 s and 5.3 ± 0.6 s [mean ± SEM, n = 15 and n = 12, respectively]) and similar to τ of 3×PAGFP at control conditions (4.3 ± 0.5 s [mean ± SEM, n = 23]), indicating that G3BP1 was present in a freely diffusing form outside of granules. The data indicate that G3BP1 and IMP1 exhibit very different exchange kinetics between SGs as observed by FDAP; although G3BP1 exchanges rapidly, IMP1 is much more stationary and exchanges only slowly.
G3BP1 contains one RNA-binding domain and four protein interaction domains (low-complexity [LC] regions), whereas IMP1 contains six RNA-binding domains and only a single LC region; LC regions are defined as amino acid sequences with low information content and are thought to exhibit nonspecific weak interactions between multiple partner proteins (Coletta et al., 2010). To determine the influence of protein interaction and RNA-binding domains on the exchange kinetics between granules, we prepared deletion constructs, which contained only two protein interaction and one RNA-binding domains (G3BP1C) and four RNA-binding and one protein interaction domain (IMP1C; Fig. 2). We also prepared a construct containing two LC regions in the absence of an RNA-binding domain (G3BP1N, aa 1–154) but observed that expression of G3BP1N inhibited arsenite-induced SG formation, so that its interaction with granules could not be analyzed. G3BP1C retained the dynamic SG partitioning similar to the full-length protein (Fig. 2 A). The exchange dynamics appeared to be slightly higher than for the full-length protein, but the difference in τ did not reach significance. IMP1C, which contained two RNA-binding domains less than the full-length protein, exhibited slow exchange very similarly to IMP1 (Fig. 2 B). Thus, the data suggest that two LC regions with a RNA-binding domain regarding G3BP1 are sufficient for dynamic exchange between SGs. With respect to IMP1, the four K homology domains, responsible for RNA interactions (Siomi et al., 1994), and the LC region appear to be sufficient to mediate strong interaction with SGs similarly to the full-length protein.
Figure 2.
Effect of G3BP1 and IMP1 deletions on protein exchange between SGs. (A and B) FDAP curves for PAGFP-G3BP1C (A) and PAGFP-IMP1C (B; mean ± SEM, n = 22 and n = 13, respectively). Schematic representations of the deletion constructs are shown on top. Protein interaction (gray) and RNA-binding domains (black) are indicated. Residence time of PAGFP-tagged G3BP1C (A) and PAGFP-IMP1C (B) in granules as determined by the model FDAP function (mean ± SEM, n = 22 from four and n = 13 from six independent experiments, respectively) are shown on the right. Values for full-length PAGFP-G3BP1 and PAGFP-IMP1 from Fig. 1 are indicated for comparison by dotted lines (green and red, respectively). The deletion constructs were not showing statistically significant differences to their respective full-length counterparts, but were statistically significantly different from the control construct (3×PAGFP vs. PAGFP-G3BP1C, P < 0.05; 3×PAGFP vs. PAGFP-IMP1C, P < 0.001). Statistics involved one-way ANOVA followed by post-hoc Tukey's test. Bars, 20 µm. For all experiments, stress had been induced by a 20-min treatment with 0.5 mM sodium arsenite before imaging.
Single-molecule localization microscopy indicates the presence of distributed hotspots of G3BP1 and IMP1 within SGs
To determine the distribution of G3BP1 and IMP1 within SGs of living cells, we performed tracking and localization microscopy (TALM; Appelhans et al., 2012). TALM is based on posttranslational labeling of a small subpopulation of molecules with photostable fluorophores, which allows for localizing individual proteins over relatively long observation times. To this end, we cloned HaloTag and SNAP-tag fusion constructs of G3BP1 and IMP1 for substoichiometric covalent labeling (TMR or JF549 for HaloTag and SiR for SNAP-tag) and expressed them in PC12 cells. Cells were first neuronally differentiated with NGF, SG formation was then induced with sodium arsenite, and labeling was finally performed as described in Materials and methods. To excite fluorophores within SGs with minimal background, we used total internal reflection fluorescence (TIRF) microscopy in the highly inclined and laminated optical sheet mode (Tokunaga et al., 2008). Snapshots done before image acquisition confirmed the presence of SGs and showed that G3BP1 and IMP1 colocalized in the same granules independent of the combination of the label (Fig. 3 A, left and middle; and Fig. S1 B, left). To analyze the nanostructure of the SGs, which is rendered by the diffusing molecules, images were taken at 100 frames per second (fps), and superresolution images were achieved from localized molecules with an estimated precision of 20–30 nm (see Materials and methods). To minimize a potential bias caused by granule movement during imaging, the SiR and TMR signals were collected quasisimultaneously by switching between the corresponding channels every 100 ms. The images indicated the presence of hotspots of G3BP1 and IMP1 within the granules (Fig. 3 A, right; and Fig. S1, A and B, right). We would like to emphasize that “hotspots” within TALM images generated from such time-lapse experiments cannot be interpreted as regions where many molecules are localized but rather as regions where an individual molecule has been localized for extended time periods due to loss of mobility (You et al., 2014).
Figure 3.
G3BP1 and IMP1 are enriched in distributed nanocores within SGs. (A) Single-molecule imaging of SiR-labeled SNAP-G3BP1 and TMR-labeled HaloTag-IMP1. Snapshots before image acquisition confirmed the presence of SGs and showed that G3BP1 and IMP1 colocalized in the same granules (left). The outline of the cell and the nucleus are indicated. Localizations of single molecules in SGs of the indicated region (yellow box in the snapshot images) are shown (right). Bars: (left, middle) 10 µm; (right) 1 µm. (B) Spatial clustering of single-molecule localizations of an area within a single SG after combined expression of G3BP1 and IMP1. Raw localizations (left), the respective cluster patterns (middle), and cluster pattern overlap (right) are shown. (C) Quantitation of the fraction of the granular area, which is occupied by clusters, and weighted overlap for different combinations of expressed RBPs. The value for “n” represents the number of individual granules analyzed. Granules were obtained from 5–19 cells from three to eight independent experiments. For all experiments, stress had been induced by a 20-min treatment with 0.5 mM sodium arsenite before imaging.
To determine the distribution of the hotspots and to test whether they reflect common binding sites where different RBPs preferentially interact, we analyzed spatial clustering of the single-molecule localizations using the density-based spatial clustering of applications with noise (DBSCAN) algorithm as described in Materials and methods. The analysis revealed that the hotspots occupied a significant proportion of the granules (Figs. 3 B and Fig. S1 C) and covered up to ∼25% of the area of the SGs (Fig. 3 C). The composite of hotspots within the individual granules revealed only a moderate colocalization of G3BP1 and IMP1 (Fig. 3 B), which is probably because of the low number of sampled molecules. The weighted overlap was calculated as
where is the real overlap area and STMR and SSiR are the areas occupied by the TMR and SiR hotspots, respectively. The weighting was required to exclude the dependency of on the number of localizations for each of the dyes. The weighted overlap was between 0.12 and 0.25 for all RNP combinations including the homotypic ones (Halo-G3BP1 on a SNAP-G3BP1 background and Halo-IMP1 on a SNAP-IMP1 background; Fig. 3 C).
To rule out that the observed hotspots are a staining artifact and do not reflect the structure of the SGs, which is rendered by the trajectories of diffusing SG molecules, we analyzed the behavior of mEGFP-HaloTag as a negative control. As a cytosolic protein that lacks protein- and RNA-binding domains, mEGFP-HaloTag would not be expected to interact with SGs and produce trajectories. Indeed, we observed that mEGFP-HaloTag was present in the cells with no obvious localization to SGs (Fig. S2 A). Furthermore, we failed to observe trajectories within SGs, confirming that our TALM approach can be used to determine the nanostructure of granules in living cells (Fig. S2 B).
Collectively, the single-molecule localization results suggest the presence of hotspots of immobilized G3BP1 and IMP1 within SGs. The hotspots may reflect the presence of “nanocores,” where G3BP1 or IMP1 preferentially interact with other SG components. Because of the low density of labeling required for single-molecule localization, the comparably long dwell times, and the relatively fast photobleaching, we can only probe a limited number of such nanocores within each granule. We therefore assume that the number and density of nanocores are higher than suggested by the TALM images, which also explains the low overlap observed in dual color experiments.
Nanocores are relatively immobile within SGs and contain multiple binding sites
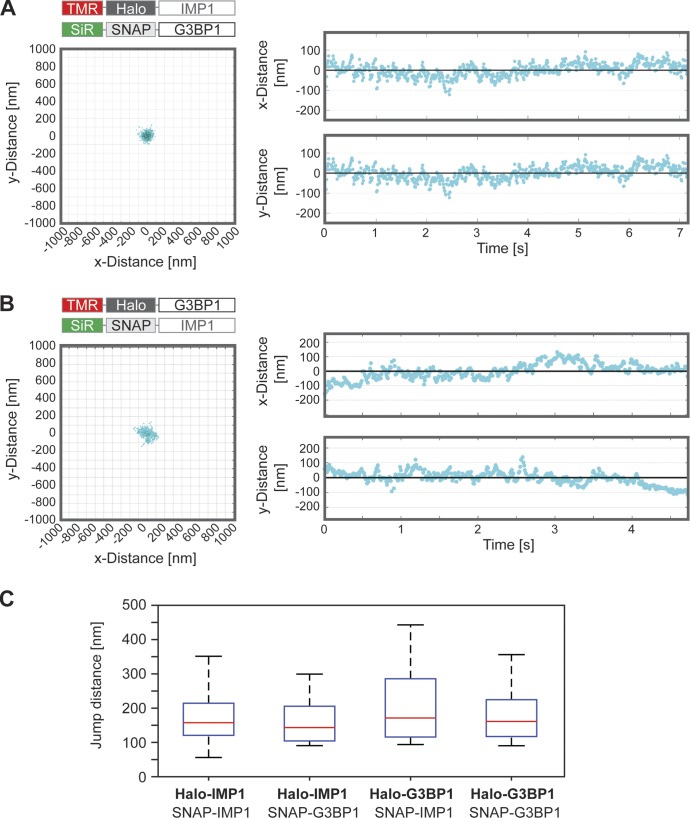
To investigate the spatial restriction of nanocores within SGs and whether they contain multiple binding sites, we applied a version of the DBSCAN algorithm adapted to group localizations that exhibit high spatiotemporal correlation as it is expected for transient binding to quasistationary multiple binding sites. The DBSCAN principle is applied within a constraint time window around each localization (see Materials and methods for further details). To assign such grouped localization sequences into an optimal number of binding sites, we made use of the step transition and state identification (STaSI) algorithm (Shuang et al., 2014). The kymographic representation of the movement of G3BP1 and IMP1 revealed that some single molecules were trapped within a single nanocore over seconds and were remarkably immobile in x and y direction (Fig. 4, A and B). In some cases a drift of the mean axis could be observed over time (see for example Fig. 4 B), which may indicate a movement of the respective nanocore within the SG. Some of the molecules exhibited a rapid transition from one position to another, probably representing jumps from one binding site to another (Fig. S3 and Video 1). We selected bona fide transition events and determined the jump distance distributions for each protein by calculating the pairwise distances between binding sites (Fig. 4 C). Jump distances were similar for G3BP1 and IMP1 in all combinations and indicated that nanocores are composed of multiple binding sites. The results also suggest that a single nanocore has a size of up to ∼150–200 nm.
Figure 4.
Nanocores are relatively immobile within SGs and contain multiple binding sites. (A and B) Top view (left) and kymographic representation (right) of a typical single binding event of Halo-IMP1 on SNAP-G3BP1 background (A) and Halo-G3BP1 on SNAP-IMP1 background (B) demonstrating spatial restriction of nanocores within SGs. (C) Bar plots showing the jump distance distributions of G3BP1 and IMP1 in different combinations indicating the presence of multiple binding sites within a nanocore. 14–27 transition events for each protein per condition were analyzed and the jump distance distributions determined by calculating the pairwise distances of all linked cluster centers. The box represents 50% of the population, whiskers range from 5% to 95%, and the horizontal line shows the median value.
Collectively, our analysis indicates that nanocores consist of multiple binding sites, which trap G3BP1 and IMP1 within a SG. The data also suggest kinetic trapping of RBPs by subsequent rebinding events of the same molecule to different binding sites within the same nanocore.
Single-molecule tracking analyses reveal alternating binding and diffusion of G3BP1 and IMP1 within SGs
To temporally and spatially resolve the local mobility of individual RBP molecules within SGs, we recorded images in the TMR channel at 100 fps and selected sequences with suitable labeling density for localizing and tracking of individual molecules (>2,000 frames per SG). Individual granules were selected based on a snapshot in the SiR channel (Fig. 5 A, left). G3BP1 could be localized within SGs, and single-molecule tracking with high fidelity was possible for extended time periods (Fig. 5 A, right; and Videos 2 and 3). Some trajectories extended for considerable distance, whereas others remained locally very restricted. Occasionally, we observed that individual molecules showed alternating phases of fast movement (Fig. 5 B, 10–60 ms) and immobile time periods (Fig. 5 B, 60–280 ms) suggesting alternating binding and diffusion within SGs (see also Video 4). To rule out that potential blinking of the fluorophore is confusing the trajectories, we performed similar experiments with fixed samples. Under these conditions, we did not observe a mobile fraction, confirming that the tracking algorithm correctly recorded the movement of individual molecules (Fig. S2, C and D).
Figure 5.
G3BP1 and IMP1 exhibit a biphasic partition in a bound and mobile fraction within SGs. (A) Example of a representative granule in a double-transfected cell. The granule was identified based on SiR staining (left) and localizations and trajectories based on TMR staining of HaloTag-G3BP1 (middle and right) are shown. The border of the SG is indicated by a dashed line. Bars, 1 µm. (B) Trajectory of a single G3BP1 molecule (as indicated by an arrowhead at time point 0) showing alternating phases of fast movement and immobile time periods. The trajectory is indicated in green. Bar, 250 nm. (C) High-mobility trajectories (mobile fraction, left) and low mobility trajectories (bound fraction, right) of HaloTag-G3BP1 within the granule shown in A. Bar, 1 µm. (D) Probability density of single-molecule diffusion coefficients from trajectories of a single granule expressing HaloTag-G3BP1 showing a biphasic partition in a bound (red) and mobile fraction (green). (E) Quantification of bound fractions of the respective Halo-tagged constructs expressed in different combinations. Calculations were performed on the total trajectories of 18–28 SGs per experimental condition as indicated by “n.” SGs were obtained from 12–15 cells from 3–10 independent experiments. For all experiments, stress had been induced by a 20-min treatment with 0.5 mM sodium arsenite before imaging.
To dissect in more detail, the local mobility of G3BP1 inside SGs, we determined diffusion coefficients from individual trajectories. The distribution of the diffusion coefficients revealed highly heterogeneous diffusion properties: next to a substantial fraction of essentially immobile molecules (log D < −2.5) a mobile fraction covering a relatively broad spectrum of diffusion coefficients was observed (Fig. 5, C and D). We interpret this biphasic mobility as RBPs alternating between a bound state (immobile) in nanocores and a diffusing state (mobile) within the liquid phase of SGs. Indeed, single-molecule trajectories of the bound fraction (Fig. 5 C, right) correlated with the hotspots observed before with negligible displacements (see Fig. 5 A, middle; and Video 3), supporting that the nanocores represent transient binding sites for the respective RBP. Analysis of the mobility of IMP1 inside granules indicated a very similar biphasic distribution in a bound and mobile fraction (Fig. S4 A).
For quantitative comparison between G3BP1 and IMP1, we determined the fraction of bound and mobile molecules by a two-component Gaussian distribution. In both cases, a similar fraction (∼60–70%) was present in the bound state (Fig. 5 E), which is consistent with our previous results showing that G3BP1 and IMP1 hotspots occupied a comparable proportion of the granules (compare Fig. 3 C).
To determine how long the proteins, on average, stay in the bound state, we first excluded mobile trajectories by setting a threshold in the respective diffusion coefficient histogram at the position where the mobile and bound fractions intersect. Then, lifetime was estimated from trajectory duration histograms for every combination of coexpressed proteins (Fig. 6 A). The histograms were fitted by single-exponential decays to extract the respective mean lifetime. For all combinations, lifetime histograms were very similar, and mean lifetimes between ∼180 and 330 ms were observed for both RBPs. Although we cannot rule out that the actual numbers are affected by photobleaching, the single molecule experiments collectively suggest very similar binding of IMP1 and G3BP1 in nanocores.
Figure 6.
G3BP1 and IMP1 have a short lifetime in the bound fraction and display anomalous subdiffusion in the mobile fraction. (A) Lifetime determination of different combinations of HaloTag- and SNAP-tagged G3BP1 and IMP1 in the bound fraction of SGs. Trajectories of the respective Halo-tagged constructs (shown in bold) are evaluated. (B) Example of the trajectories of the mobile fraction of HaloTag-G3BP1 within a single granule (left) and plot of the MSD against the elapsed time showing anomalous diffusion. Mean diffusion constants (Γ) and α values of the respective HaloTag construct in the mobile fraction as determined for the different combinations. Bar, 500 nm. Calculations were performed on the mobile trajectories of 8–11 SGs per experimental condition as indicated by n. SGs were obtained from 8–11 cells from two to eight independent experiments. For all experiments, stress had been induced by a 20-min treatment with 0.5 mM sodium arsenite before imaging.
The surprising disconnect between SG partitioning and internal diffusion and interactions of RBPs may be because of the different labels that have been used for the FDAP (PAGFP-tag) and single-molecule experiments (HaloTag). To exclude this possibility, we used HaloTag constructs also for the FDAP experiments by labeling them with the photoactivatable dye PA-JF549. We observed that the residence time in granules closely matched for PAGFP- and HaloTag-labeled constructs and that comparison between HaloTag-IMP1 and -G3BP1 revealed higher residence time for HaloTag-IMP1 (*, P < 0.05), similarly to the respective results for the PAGFP-tagged constructs (Fig. S4 B), indicating that the nature of the tag does not affect SG partitioning to a major extent.
G3BP1 and IMP1 display anomalous diffusion in the liquid phase of SGs
Determination of the diffusion behavior of G3BP1 and IMP1 within SGs can provide information about the biophysical properties of the liquid phase of SGs in living cells. To analyze diffusion of the two proteins, we excluded bound trajectories by setting a threshold in the respective diffusion coefficient histogram as described before (Fig. 5 D). For the remaining trajectories of mobile molecules, we determined the mean squared displacement (MSD) with time. We observed that G3BP1 displayed anomalous (non-Fickian) diffusive behavior in a representative SG (i.e., diffusion with a nonlinear relationship between the MSD of molecules and time; Fig. 6 B, top). In a cellular context, anomalous diffusion (characterized by α < 1) occurs as a result of macromolecular crowding, which contributes to the high viscosity of cellular compartments (Ellis, 2001). We observed α values between 0.5 and 0.7 for all combinations of G3BP1 and IMP1 (Fig. 6 B, bottom) indicative of intense molecular crowding in the liquid phase of SGs. The anomalous diffusion constants (Γ) of G3BP1 and IMP1 were very similar with values between 0.038 and 0.068 µm2/sα.
Thus, the results indicate that both G3BP1 and IMP1 undergo anomalous diffusion in the liquid phase in SGs, which provides direct support for liquid droplet behavior of highly viscous neuronal SGs in living cells.
Reduction in the number of LC regions of G3BP1 results in less detectable mobile molecules within the liquid phase without changing binding properties in nanocores
G3BP1 contains four LC regions, which are thought to exhibit nonspecific weak interactions between multiple partner proteins (Coletta et al., 2010). To determine the influence of the number of LC regions on protein behavior in SGs, we performed TALM with the carboxy-terminal fragment of G3BP1 (G3BP1C), which contained only two LC regions in addition to the RNA-binding domain. G3BP1C exhibited high exchange kinetics between SGs, similarly to full-length G3BP1, as determined by the FDAP experiments (Fig. 2 A). However, within SGs, we observed that most of the trajectories remained locally very restricted and that the distribution of the trajectories resembled the bound fraction of the full-length protein (Fig. 7 A; compare with Fig. 5 C, right). This feature was also reflected in the distribution of the diffusion coefficients, which revealed the presence of a substantial fraction of essentially immobile molecules (log D < −2.5), whereas much less trajectories with higher diffusion coefficients could be detected (Fig. 7 B). Thus, the distribution of diffusion coefficients was different from full-length G3BP1, where we observed a biphasic mobility by a two-component Gaussian distribution. The mean lifetime of binding events as determined from the trajectory length histograms of the bound fraction was similar to full-length G3BP1 (Fig. 7 C), indicating that the binding properties to nanocores were not changed by the loss of two LC regions. The data suggests that the presence of multiple interaction domains facilitates the biphasic alternation between a bound state (immobile) in nanocores and a diffusing state (mobile) within the liquid phase of SGs and that multiple interaction domains are in particular important for the diffusion in the liquid droplet.
Figure 7.
Reduction in the number of LC regions of G3BP1 results in less detectable mobile molecules within the liquid phase without changing binding properties to nanocores. (A) Example of a representative granule after expression of a deletion fragment of G3BP1 (G3BP1C) lacking two LC domains fused to HaloTag on a SNAP-IMP1 background. The granule was identified based on the SiR staining (left) and localizations and trajectories based on TMR staining of HaloTag-G3BP1C (right) are shown. The border of the SG is indicated by a dashed line. Note that most of the trajectories remained locally very restricted. Bar, 1 µm. (B) Probability density of single-molecule diffusion coefficients from trajectories of a granule expressing HaloTag-G3BP1C showing the presence of a substantial fraction of essentially immobile molecules. (C) Lifetime determination of HaloTag-G3BP1C on SNAP-IMP1 background in the bound fraction of SGs. Quantification of granules from six cells from two independent experiments was performed. For all experiments, stress had been induced by a 20-min treatment with 0.5 mM sodium arsenite before imaging.
Discussion
Stress induces aggregation of proteins and RNAs in many cell types. However, the nature of the aggregates and the dynamics of their components are not entirely clear. Experiments in yeast showed that protein aggregation after heat exposure is a reversible cellular process, indicating that aggregates may not be toxic end products but represent a new layer of cellular organization (Wallace et al., 2015). However, because many of the interpretations were based on in vitro experiments or studies with microorganisms, where SGs appear to be more rigid (Jain et al., 2016), we aimed at setting up an experimental system to analyze individual SG components in living neuron-like cells.
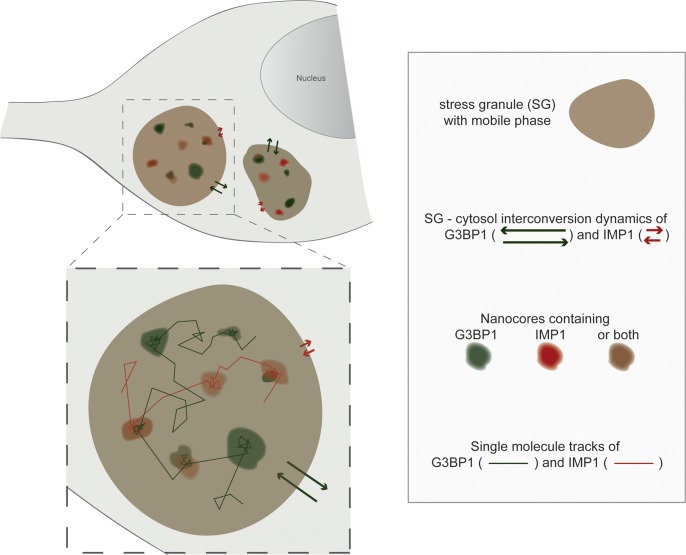
Here, we report about an approach to analyze the distribution and dynamics of two paradigmatic SG proteins, G3BP1 and IMP1, at single-molecule resolution in neuronally differentiated PC12 cells. Our major findings are as follows (see Fig. 8 for a schematic representation): (1) The two RBPs, G3BP1 and IMP1, exhibit markedly different dynamics of protein exchange between SGs (red and green arrows). (2) Within SGs, single-molecule localization microscopy revealed the presence of distributed hotspots of immobilized G3BP1 and IMP1, which reflect the presence of nanocores, where G3BP1 or IMP1 preferentially interact with other SG components. (3) Nanocores are relatively immobile within SGs and contain multiple binding sites. (4) G3BP1 and IMP1 exhibit alternating binding between a bound, nanocore-associated phase and a mobile, liquid phase with very similar characteristics (red and green single-molecule tracks). (5) Both proteins display anomalous diffusion in the mobile phase indicative of a liquid droplet environment with high concentration of molecules. (6) The number of LC regions in G3BP1 appears to be important for the anomalous diffusion in the liquid phase of SGs. Notably, the results show a highly dynamic binding and diffusion of RBPs in mammalian SGs and a remarkable disconnect between SG partitioning and internal interactions.
Figure 8.
Schematic representation visualizing the major findings of the study. The study reveals the presence of distributed nanocores within the mobile, liquid droplet-like phase of SGs. The two RBPs, G3BP1 (green arrows) and IMP1 (red arrows), exhibit markedly different SG–cytosol interconversion dynamics. Within granules, both RBPs exhibit alternating binding in the nanocores and liquid droplet-like diffusion in the mobile phase.
Based on results from FDAP assays, we report that G3BP1 exhibits dynamic shuttling between SGs, whereas IMP1 exchanges only slowly. G3BP1 and IMP1 have a similar molecular mass, and both contain protein- and RNA-binding domains that mediate hetero- and homotypic interactions between proteins and RNAs, thereby regulating the fate of specific mRNAs. For example, both proteins have previously been shown to associate with the neuronal tau mRNA and regulate its expression (Atlas et al., 2004; Moschner et al., 2014). However, although IMP1 contains a total of six RNA-binding domains (two RNA recognition motifs and four K homology domains; see Fig. 1 A), G3BP1 harbors only a single RNA recognition motif. On the other hand, G3BP1 contains four LC regions, whereas IMP1 has only one. LC regions are defined as amino acid sequences with low information content and are thought to exhibit nonspecific weak interactions between multiple partner proteins (Coletta et al., 2010). We observed that a G3BP1 deletion construct with only two LC regions in addition to the RNA-binding domain (G3BP1C) still exhibited dynamic exchange between granules similar to the full-length protein, whereas we have previously observed that such a construct lost its ability to nucleate granules in the absence of stress (Moschner et al., 2014). This suggests that more than two LC domains are required to mediate the condensation of SGs, whereas the structural requirements for interaction with existing SGs are less strict. In TALM experiments, we observed that most of the trajectories of G3BP1C remained locally very restricted. It appears that, in particular, the anomalous diffusion in the liquid phase of SGs is supported by the presence of multiple protein- and RNA-binding domains that mediate hetero- and homotypic interactions. This is also consistent with the observation that a control protein (mEGFP-HaloTag), which lacks such interaction domains, is not detectable in the liquid phase despite the fact that it is not excluded from SGs. Thus, the data suggest that the anomalous diffusion of RBPs in the liquid phase is a feature that depends on multiple interaction domains and is not just a consequence of the RNA- and protein-enriched environment in this compartment.
Previously, substructures within SGs, which have been referred to as cores with higher concentrations of proteins and mRNAs, surrounded by a less concentrated shell have been described, suggesting inhomogeneous distribution of individual SG components (Souquere et al., 2009; Jain et al., 2016). In particular, G3BP1-containing SG substructures have been visualized by superresolution microscopy (Wheeler et al., 2016). However, the cells were fixed before imaging, and the protein dynamics could not be assessed. Our approach has allowed us to follow the movement of G3BP1 and IMP1 in living cells at single molecule resolution. Our data indicate the presence of distributed hotspots, where individual molecules have been immobilized for extended time periods and which reflect the presence of nanocores. The nanocores are relatively immobile within SGs, contain multiple binding sites and have, at least in our experimental setup, a size of up to ∼150–200 nm. Previously, it has been suggested that SG components in a potential shell may exchange rapidly, whereas SG components in the core are less dynamic (Jain et al., 2016). At least with respect to acute arsenite-induced stress, our data do not support such a hypothesis. Instead, our results argue for the presence of transient interactions of RBPs in distributed nanocores in the absence of a stable core structure. Such an organization may facilitate the function of SGs as dynamic sorting stations at conditions of acute stress.
Within SGs, G3BP1 and IMP1 exhibit a very similar biphasic partition into a bound and mobile phase (∼60–70% bound with a fast transition between these phases [half-lives of the bound fraction of ∼100–200 ms]). The positions where G3BP1 and IMP1 are in a bound state correlate well with the positions of the hotspots indicating that nanocores represent positions, where the RBPs transiently interact with other SG components. Interestingly, our studies uncover a surprising disconnect between SG partitioning, and internal diffusion and interactions of G3BP1 and IMP1, which indicates that the distribution behavior of G3BP1 and IMP1 within SGs is not related to their interconversion dynamics between granules. In fact, the lifetime of G3BP1 in the bound state is much shorter than the residence time of G3BP1 in a granule (∼20 s) indicating that the partition within SGs is not the major factor in determining the different granule–cytosol interconversion dynamics of G3BP1 and IMP1. It should, however, be noted that our data provided evidence that nanocores consist of multiple binding sites, which may suggest kinetic trapping of G3BP1 and IMP1 by subsequent rebinding events of the same molecule to different binding sites in the nanocore. By taking such a feature into account and applying the DBSCAN algorithm adapted to group localizations also for lifetime determination, we obtained values between 1.0 and 1.6 s for all combinations (Fig. S5 D). We still did not observe a difference between G3BP1 and IMP1, supporting that the residence time in SGs is not temporally correlated with the binding to nanocores.
Our data provide direct support for the LLPS model of neuronal SG formation, where macromolecules are kept together solely by weak intermolecular interactions, which distinguishes them from rigid oligomeric structures. As a consequence, molecules can enter SGs, get concentrated, diffuse within, and exit easily. Notably, despite their different granule–cytosol interconversion dynamics and the different number of protein- and RNA-binding domains, G3BP1 and IMP1 exhibit a very similar anomalous diffusion in the mobile phase of SGs with an anomality factor α of 0.5–0.7. Because the anomality factor decreases with increasing molecular crowding, it has been suggested that the anomality of the diffusion can be used as a quantifiable measure for the crowdedness of a medium (Weiss et al., 2004). Thus, the data are consistent with the view that RNP granules have a much higher protein concentration than the surrounding cytoplasm (Handwerger et al., 2005; Souquere et al., 2009), with estimated protein concentrations of ∼300 mM (Lin et al., 2015). The diffusion constant Γ represents a measure for the mobility of the protein inside SGs, which is modulated by the extent and strength of the homotypic and heterotypic interactions between the SG proteins and RNA, which may also tune the liquid droplet viscoelasticity.
The material state of SGs in neuronal cells could be relevant for neurodegenerative diseases such as ALS, Huntington’s disease, and some spontaneous cases of Alzheimer’s disease, where pathological and stable RNP inclusions form. It is conceivable that “healthy” RNP granules (with weaker interactions and more liquid droplet-like features) switch to a disease-causing insoluble state (with stronger interactions and a more solid state) during disease development that may be accelerated by disease-associated mutations or the formation of misfolded proteins (Patel et al., 2015; Mateju et al., 2017). In fact, macromolecular crowding favors the association of macromolecules, which may result in pathological aggregation of SG components. Our live-cell approach opens possibilities to further investigate central questions concerning the composition and subgranular organization of SGs. In particular, it may permit to analyze the potential switch in the material state of SGs to a potential disease-causing state and to identify conditions and factors that would be involved in such a switch.
Materials and methods
Constructs and materials
Eukaryotic expression plasmids for human G3BP1 and human IMP1 with amino-terminally fused HaloTag and SNAP-tag were constructed from pRc/cytomegalovirus (CMV)–based expression vectors coding for PAGFP- and mCherry-tagged G3BP1 and IMP1 (Moschner et al., 2014). The coding sequence for human G3BP1 was PCR amplified from pRc/CMV PAGFP-hG3BP1 and modified to include a SbfI and a NotI restriction site using the primers Sbf1_G3BP1_FW and G3BP1_NotI_BW (5′-GGCCCCTGCAGGGATGGTGATGGAGAAG-3′/5′-GTTATCTAGATGCGGCCGCTCACTGCCGTG-3′). The PCR product as well as the vector plasmid pSems HaloTag-hTau441wt were digested using SbfI and NotI endonucleases, followed by ligation. Cloning of SNAP-tag-hG3BP1 was performed essentially the same using pSems fSNAP-tag-Rab5a as vector. The coding sequence for IMP1 was PCR amplified from pRc/CMV PAGFP-IMP1 and modified to include a XhoI and a NotI restriction site using the primers XhoI_IMP1_FW and IMP1_NotI_BW (5′-CGCGGGCTCGAGATATCCATGAACAAG-3′/5′-CCGGCGGCCGCTCACTTCCTCCG-3′). The PCR product and the vector plasmid pSems haloTag-hTau441wt were digested using XhoI and NotI endonucleases, followed by ligation. To clone pSems SNAP-tag-IMP1, the vector pSems fSNAP-tag-Rab5a and the plasmid pSems HaloTag-IMP1 were linearized using XhoI and NotI endonucleases, followed by ligation. The pCMV-3×PAGFP plasmid (Weissmann et al., 2009) and the construct coding for mEGFP-HaloTag (Wedeking et al., 2015) were described previously. The deletion constructs G3BP1N, G3BP1C and IMP1C were prepared as described previously (Moschner et al., 2014). Sequences that were introduced by PCR were verified by DNA sequencing (Seqlab-Microsynth). Chemicals were obtained from Sigma-Aldrich, and cell culture media, supplements, culture flasks, plates, and dishes were obtained from Sigma-Aldrich and ThermoFisher Scientific, unless stated otherwise. TMR-HTL was obtained from Promega, JF549-HTL and PA-JF549-HTL from Janelia Research Campus (Ashburn, VA), and SiR (SNAP-Cell 647-SiR) from New England Biolabs. The following antibodies were used: anti–TIA-1 (G-3; mouse monoclonal; Santa Cruz Biotechnology), anti-G3BP1 (NBP2; rabbit polyclonal; Novus Biologicals), anti-IMP1 (D-9; mouse monoclonal; Santa Cruz Biotechnology). As secondary antibodies, anti–mouse Alexa488 (Jackson ImmunoResearch Laboratories) or anti–rabbit and anti–mouse detection modules for Wes (ProteinSimple Wes; ProteinSimple) were used.
Cell culture and transfections
PC12 cells were cultured in 15% serum/DMEM as described previously (Fath et al., 2002). For induction of neuronal differentiation, the medium was switched to 1% serum/DMEM with 100 ng/ml 7 S mouse NGF (Alomone Laboratories) for 4 d. Transfections of PC12 cells were performed with Lipofectamine 2000 (Invitrogen) as described previously (Fath et al., 2002). For FDAP analysis, cells were plated on 35-mm polylysine- and collagen-coated glass-bottom culture dishes (MatTek). For TIRF imaging, cells were plated on glass coverslips (24 mm, No. 1; VWR) that were coated with poly-l-lysine and collagen or poly-l-lysine–graft (polyethylene glycol)–copolymer functionalized with RGD as described previously (Wedeking et al., 2015). Before imaging, the medium was exchanged against DMEM without phenol red. For the induction of stress, 0.5 mM sodium arsenite was added to the medium. After 20 min, the medium was replaced with fresh DMEM without phenol red. Labeling of cells for imaging was performed by incubation with serum-DMEM containing 0.5–5 nM TMR-HTL, 0.25 nM JF549-HTL, and 25 nM SiR for 20 min at 37°C. For photoactivation experiments with HaloTag constructs, PA-JF549-HTL was used at 100 nM. Subsequently, cells were washed 3 × 5 min with serum-DMEM without phenol red and transferred to the microscope.
Photoactivation and live cell imaging
Live cell imaging for FDAP experiments was performed on a laser scanning microscope (Eclipse TE2000-U inverted; Nikon) equipped with argon (488 nm), helium/neon (543 nm), and violet diode (407 nm) lasers. The microscope was enclosed in an incubation chamber maintained at 37°C and 5% CO2 (Solent Scientific). A 60× magnification objective with NA 1.40 (oil, Plan Apo VC; Nikon) was used. Photoactivation was performed with the violet diode in a region between the nucleus and the cell membrane with a size of 3 × 5 µm. Automated image acquisition after photoactivation was essentially performed as described previously (Weissmann et al., 2009). Frames were obtained at a frequency of 1 frame per second, and 112 frames were collected per experiment. Standard series were collected at a resolution of 256 × 256 pixels.
FDAP data analysis and fitting
For determination of fluorescence decay, individual image frames from FDAP experiments were extracted from raw images using Fiji software (Schindelin et al., 2012). From all frames, the zero-level (“preactivation intensity”, t = −1 s) was subtracted to exclude the preactivation fluorescence and normalized to 1 by the maximal value at t = 0 s. Data fitting was performed using the Levenberg–Marquardt algorithm implemented in the Origin Pro 8 software package. To test for different fluorescence populations in the FDAP curves, two different model FDAP functions were used:
| (1) |
and
| (2) |
In Eq. 2, Aslow and Afast denote the relative fractions ranging from 0 to 1 of a slow and fast fraction, τslow and τfast represent the corresponding characteristic decay times of those fractions, and F∞ and F0 are auxiliary offset parameters. Eq. 1 is a simplification of Eq. 2, wherein the fraction of one of the two populations is set to 0. We underline that both models do not include information on either the geometry of the activation region or the molecular kinetics underlying the decay. Because there was no a priori knowledge whether a FDAP curve reflects the dynamic of one or two populations, both models were applied to fit every curve. The resulting fit parameters were averaged to produce a mean and SEM for each construct. By using a χ2 statistical test to compare the results of fitting, it became clear that the FDAP curves for 3×PAGFP and PAGFP-IMP1 were better described by Eq. 1 and the curves for PAGFP-G3BP1 by Eq. 2.
Single-molecule microscopy, localization, and tracking
For recording single molecules, TIRF microscopy was performed using an IX81 microscope (Olympus) with a four-line motorized TIR condenser (cellTIRF) and 405-nm (200 mW), 488-nm (200 mW), 561-nm (200 mW), and 642-nm (140 mW) lasers (Olympus). High-speed single-molecule tracking was performed using a digital scientific complementary metal–oxide–semiconductor camera (sCMOS, ORCA-Flash4.0 V2 C11440-22CU; Hamamatsu). A 150× magnification objective with NA 1.45 (oil, UAPON 150×/1.45; Olympus) was used for TIR illumination in the highly inclined and laminated optical sheet mode. The emitted fluorescence from the sample was filtered using a quad-band bandpass filter (FF01 446/523/600/677; Semrock) and a secondary single bandpass filter: BrightLine HC 525/50 (Semrock) for meGFP, BrightLine HC 600/37 (Semrock) for TMR/JF549, and BrightLine HC 697/58 (Semrock) for SiR. The microscope was enclosed in an incubation chamber maintained at 37°C and 10% CO2 (cellVivo; Olympus). Time series were recorded using Olympus CellSens 1.14 software. Cell vitality was confirmed by a bright field snapshot and double transfection verified with a snapshot in the SiR channel. Time series were performed in the TMR/JF549 channel with an exposure time of 10 ms and 2,000–8,000 frames were recorded per cell. Image stacks were imported using FIJI software (Schindelin et al., 2012) and regions of interest containing RNP granules were cropped and saved as separate TIFF stacks that were further analyzed (∼2,000–6,000 frames, where individual molecules could be localized). Localization of single molecules and single-molecule tracking as well as further data processing were performed in MATLAB (MathWorks) using well-established localization and tracking algorithms as previously described (Jaqaman et al., 2008; Sergé et al., 2008) implemented in a custom-written graphical user interface, which we call SLIMfast (software for localization-based imaging in MATLAB). The estimated localization precision achieved 20–30 nm in our experiments. Trajectory linking proceeded in parallel with the localization step and used a local criterion (a search radius determined by a hypothetical diffusion process) to build trajectories in each time step. Past statistics (instantaneous diffusion coefficient, mean intensity of spots, and blinking statistics) were taken into account when assigning new localized spots to trajectories to resolve local ambiguities such as crossing trajectories or dye blinking.
Lifetime determination and diffusion in granules
The trajectory set produced by SLIMfast contained, on the one hand, trajectories that existed for hundreds of frames and explored compact areas within granules. On the other hand, there were trajectories being rather short-lived but exploring wider areas. To separate those fractions from one another and estimate their relative abundances, the MSD for each trajectory longer than 10 frames was calculated and instantaneous diffusion coefficients were obtained by linearly fitting those MSDs between time lag 2 and 10. The distribution of diffusion constants was analyzed using a two-component Gaussian function to estimate the different populations. The Gaussians for the two subpopulations cross at a point (Dthreshold) that we defined as a border between those subpopulations. For trajectories that satisfied the inequality D < Dthreshold (defined as bound fraction), lifetime histograms were plotted and the mean lifetimes (τlife) were determined by fitting single exponential functions to them. The resulting lifetimes were corrected by subtracting an estimated bleaching rate γ (0.040 ± 0.002 s−1) from the corresponding dissociation rate 1/τlife. For trajectories with instantaneous diffusion coefficients that satisfied the inverse inequality D > Dthreshold (defined as mobile fraction), MSD plots for time lags 2 to 20–40 were constructed. The MSDs were fitted by two models: simple and anomalous diffusion. In most cases, the MSD were better described by anomalous diffusion, which allowed us to determine the anomaly exponent α and diffusion constant Γ (µm2/tα) from the fits.
Cluster analysis
Clustering of localized molecule positions was performed using the DBSCAN algorithm (Ester et al., 1996). This algorithm, given a set of points in 2D space, splits closely located points into groups (clusters) and rejects all unclustered points in low-density regions as “noise.” DBSCAN uses two parameters to define the critical density of points in a certain region to belong to a cluster, ε and m. A chosen point is a core sample when there exists at least m points within a circle of radius ε around a randomly chosen point in the set. The core (a set of closely packed core samples) is then recursively expanded by ensuring the validity of the aforesaid criterion for all neighboring points unless the local density of points drops significantly. Every detected cluster is surrounded by noncore samples that do not yet belong to noise but contain at least one core sample within their ε-neighborhood. To prepare single-molecule data for the cluster analysis, we performed the localization of TMR and SiR stacks using SLIMfast without any subsequent tracking (see Single-molecule microscopy, localization, and tracking). The coordinates of the localized positions of every granule were saved in ASCII format and were further analyzed by self-written Python scripts (Oliphant, 2007) implementing the scikit-learn library for machine learning (Pedregosa et al., 2011). Before working with experimental data, the cluster detection procedure was tested on simulated data to find optimal DBSCAN parameters (Fig. S5 A), which depend on factors such as the mean distance between points, the mean distance between clusters and the amount of noise in low-density regions. A general idea about the spatial scales present in a set of points can be given by the radial distribution function (RDF) which describes how the density of points changes as a function of distance from an arbitrary reference particle. As the test set is rather inhomogeneous, the RDF is expected to have a local maximum at low values of ε, which corresponds to the mean half-radius of high-density regions (future clusters). It is clear that the value of ε during the DBSCAN cluster detection should lie below this critical value ε*. In the opposite case, DBSCAN will ignore fine intracluster details and underestimate the number of detected clusters. Integrating the RDF over ε and multiplying it by the total number of points in the set yields the mean number of points within a circle of radius ε, <N>(ε). This information is required to estimate the optimal m parameter for the DBSCAN cluster detection. Choosing m close to <N>(ε) ensures that the density of points for the chosen optimal value of ε is directly related to the composition of the given set of points. Given an optimal area in the (ε, m)-space for the DBSCAN cluster detection (0 < ε < ε* and m = <N>(ε)), one finally has to choose an appropriate pair of the parameters to perform the final clustering with. For every pair (εi, <N>(εi)), the number of clusters Nclusters detected by DBSCAN was calculated (Fig. S5 A). Nclusters decayed with the increase of ε, but not monotonously. There was, as expected, a sharp drop in Nclusters when ε crossed the critical value ε*. This drop corresponds to a sort of “phase transition” as the fine intracluster details are not properly taken into account anymore. The sharp drop observed at small values of ε reflected an overestimation of the number of clusters because of the too high density required to classify points of clusters, which resulted in splitting of the actual clusters into smaller subclusters. The sought optimal parameter pair was therefore located in the plateau between the two drops of Nclusters. Cluster detection with these parameters yield the best result for the given set of points (Fig. S5 B). To calculate the overlap area between two cluster sets (e.g., 1 and 2), we performed the binarization of the 2D space (Fig. S5 C). For this to be done, a 2D grid with a certain cell size (d) was created and put on the 2D space containing the cluster sets. The grid corresponded to a matrix whose elements were assigned 1 depending on whether the respective cell in the 2D grid contained core elements of both cluster sets. In all other cases, the elements of the matrix were assigned 0. The overlap area was then calculated as the number of all nonzero elements of the grid matrix n times the area of a single binarization cell d2. To eliminate the dependence of the overlap area on the cell size and account for different numbers of points in the cluster sets, a unitless measure s was introduced:
where S1 and S2 are the areas taken by the cluster set 1 and 2, respectively. To validate the values of the weighted overlap, we performed two types of calculations. First, using the datasets for TMR-G3PB1 and SiR-G3BP1 (Fig. 3 C, first line), we selected signals that did not belong to any cluster detected with DBSCAN. Those signals were hence interpreted as noise. Calculating the weighted overlap for these noise contributions from the TMR and SiR channels yielded a value of 0.020 ± 0.001 (mean ± SEM, n = 8). Hence, noise detected in the different channels was spatially uncorrelated. Second, we calculated the weighted overlap between images, where the signals were generated numerically using a 2D uniform distribution and the total number of signal in either channel was comparable to that observed experimentally. This yielded 0.001 ± 0.000 (mean ± SEM, n = 8). This suggested that the measured overlaps in Fig. 3 C were significantly different from noise.
Analysis of quasistationary multiple binding sites
Nanocore binding sites within the SGs are expected to be quasistationary within the observed time period. Therefore binding events to such sites correspond to transient immobilization of G3BP1 and IMP1 and were detected using the DBSCAN principle as described above but adapted to work only within a constraint time window around each localization. Before localization, raw image stacks were preprocessed using a highly localized 3 × 3 × 3 median filter (width [pixels] × height [pixels] × depth [frames]) to specifically increase the signal to noise ratio of the immobilized particles. We then scanned around each localization within a radius ε of 225 nm (five times the apparent localization precision as determined from manually preselected binding events) tolerating a 50% probability to not observe the respective molecule because of blinking. We started with a time window of 30s (3,000 frames) to pick up prolonged binding events and sequentially reduced the time window to 1s (100 frames) to be able to detect more transient events but still discriminate mobile particles. Precise detection of the starting point and endpoint of the immobilization events was realized by initially clustering the localizations in a pure forward- or backward-looking sweep (for a point localized at time t0 only those points localized at t > t0 are considered in the forward sweep and vice versa in the backward sweep) and subsequent fusion of corresponding cluster via maximal overlap. Spurious start and end points caused by noise peaks were removed by enforcing at least three subsequent localizations to be present at these points. If the endpoint of one cluster coincided with the starting point of a second cluster within a radius of 500 nm and 1 s, then these clusters were subsequently merged into one. To determine the number of binding sites present, we extracted for each cluster the x-t- and y-t-kymographs and applied the STaSI algorithm to each coordinate (Shuang et al., 2014). This algorithm decomposed the observed kymographs into an optimal number of piece-wise constant periods. The averages of these piece-wise constant periods in both dimensions then represent the position of the respective binding sites and their change points the sequential hopping of the RBPs from one binding site to the next. Because the binding sites showed some form of additional fluctuation, we set the minimum accepted jump distance to 90 nm (two times the apparent localization precision). To approximate the size of the nanocore, we calculated the pairwise distances between binding sites within one nanocore among selected samples.
Other methods
PC12 cells transfected with PAGFP-G3BP1 or PAGFP-IMP1 constructs were lysed with RIPA buffer containing protease inhibitors as described previously (Moschner et al., 2014). Lysates were analyzed in the Simple Western size-based capillary electrophoresis system (ProteinSimple Wes) following the manufacturer’s instructions. Electropherograms were represented as pseudo-blots, generated using the inbuilt Compass software (ProteinSimple).
Statistical analysis
Statistical analysis among experimental groups was performed using Student’s t test. One-way ANOVA followed by post-hoc Tukey’s test for multiple comparisons was used for FDAP analysis (*, P < 0.05; **, P < 0.01; ***, P < 0.001).
Online supplemental material
Fig. S1 shows single-molecule imaging of G3BP1 and IMP1 in different combinations and examples of spatial clustering of single molecule localizations. Fig. S2 shows single-molecule imaging of mEGFP-HaloTag in living cells and single molecule imaging of fixed samples after expression of SiR-labeled SNAP-IMP1 and TMR-labeled HaloTag-G3BP1. Fig. S3 shows RNP-binding dynamics on a single-nanocore level. Fig. S4 shows localizations, trajectories, and biphasic partitioning of IMP1 within a SG and comparison of residence times for PAGFP-G3BP1 and PAGFP-IMP1 with the respective HaloTag constructs. Fig. S5 shows determination of optimal DBSCAN parameters for cluster detection using simulated data and lifetime determination of different combinations of HaloTag- and SNAP-tagged G3BP1 and IMP1 using the DBSCAN algorithm adapted to group localizations. Video 1 shows a visualization of DBSCAN algorithm adapted to group localizations that exhibit high spatiotemporal correlation. Video 2 shows single-molecule tracking of G3BP1 molecules in a SG. Video 3 shows TALM visualizing immobile hot spots of G3BP1 binding events. Video 4 shows single-molecule track of a G3BP1 molecule in a SG. Source code 1 shows custom-made Python scripts for detection of optimal DBSCAN parameters and analysis of G3BP1 and IMP1 clusters and colocalization. Source code 2 (available at https://github.com/CPaoloR/smDBSCAN/releases/tag/1.0-JCB) shows MATLAB scripts for image smoothing, molecule localization, cluster analysis, kymograph extraction, and subsequent STaSI analysis for scrutinizing quasi-stationary multiple binding sites in SGs.
Supplementary Material
Acknowledgments
We thank Luke Lavis (Janelia Research Campus, Ashburn, VA) for providing Janelia Fluor dyes, Mohammad Ashikur Rahman and Michael Peters for help with imaging, and Lidia Bakota for critical reading, helpful suggestions, and preparation of the schematic model.
This work was supported by Deutsche Forschungsgemeinschaft (grant SFB 944, project P1, to R. Brandt and J. Piehler).
The authors declare no competing financial interests.
Author contributions: Conceptualization, J. Piehler and R. Brandt; Methodology, B. Niewidok, M. Igaev, C.P. Richter, R. Kurre, J. Piehler, and R. Brandt; Investigation, B. Niewidok, M. Igaev, A. Pereira da Graca, A. Strassner, C.P. Richter, and C. Lenzen; Resources, J. Piehler and R. Brandt; Writing – original draft, B. Niewidok, M. Igaev, C.P. Richter, R. Kurre, J. Piehler, and R. Brandt; Writing – review and editing, J. Piehler and R. Brandt; Funding Acquisition, R. Brandt; Supervision, R. Brandt.
References
- Alberti S. 2017. The wisdom of crowds: regulating cell function through condensed states of living matter. J. Cell Sci. 130:2789–2796. 10.1242/jcs.200295 [DOI] [PubMed] [Google Scholar]
- Alberti S., and Hyman A.A.. 2016. Are aberrant phase transitions a driver of cellular aging? BioEssays. 38:959–968. 10.1002/bies.201600042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson P., and Kedersha N.. 2008. Stress granules: the Tao of RNA triage. Trends Biochem. Sci. 33:141–150. 10.1016/j.tibs.2007.12.003 [DOI] [PubMed] [Google Scholar]
- Appelhans T., Richter C.P., Wilkens V., Hess S.T., Piehler J., and Busch K.B.. 2012. Nanoscale organization of mitochondrial microcompartments revealed by combining tracking and localization microscopy. Nano Lett. 12:610–616. 10.1021/nl203343a [DOI] [PubMed] [Google Scholar]
- Atlas R., Behar L., Elliott E., and Ginzburg I.. 2004. The insulin-like growth factor mRNA binding-protein IMP-1 and the Ras-regulatory protein G3BP associate with tau mRNA and HuD protein in differentiated P19 neuronal cells. J. Neurochem. 89:613–626. 10.1111/j.1471-4159.2004.02371.x [DOI] [PubMed] [Google Scholar]
- Aulas A., Stabile S., and Vande Velde C.. 2012. Endogenous TDP-43, but not FUS, contributes to stress granule assembly via G3BP. Mol. Neurodegener. 7:54 10.1186/1750-1326-7-54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aulas A., Caron G., Gkogkas C.G., Mohamed N.V., Destroismaisons L., Sonenberg N., Leclerc N., Parker J.A., and Vande Velde C.. 2015. G3BP1 promotes stress-induced RNA granule interactions to preserve polyadenylated mRNA. J. Cell Biol. 209:73–84. 10.1083/jcb.201408092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banani S.F., Lee H.O., Hyman A.A., and Rosen M.K.. 2017. Biomolecular condensates: organizers of cellular biochemistry. Nat. Rev. Mol. Cell Biol. 18:285–298. 10.1038/nrm.2017.7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brangwynne C.P., Eckmann C.R., Courson D.S., Rybarska A., Hoege C., Gharakhani J., Jülicher F., and Hyman A.A.. 2009. Germline P granules are liquid droplets that localize by controlled dissolution/condensation. Science. 324:1729–1732. 10.1126/science.1172046 [DOI] [PubMed] [Google Scholar]
- Buchan J.R. 2014. mRNP granules. Assembly, function, and connections with disease. RNA Biol. 11:1019–1030. 10.4161/15476286.2014.972208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchan J.R., and Parker R.. 2009. Eukaryotic stress granules: the ins and outs of translation. Mol. Cell. 36:932–941. 10.1016/j.molcel.2009.11.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coletta A., Pinney J.W., Solís D.Y., Marsh J., Pettifer S.R., and Attwood T.K.. 2010. Low-complexity regions within protein sequences have position-dependent roles. BMC Syst. Biol. 4:43 10.1186/1752-0509-4-43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degrauwe N., Suvà M.L., Janiszewska M., Riggi N., and Stamenkovic I.. 2016. IMPs: an RNA-binding protein family that provides a link between stem cell maintenance in normal development and cancer. Genes Dev. 30:2459–2474. 10.1101/gad.287540.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis R.J. 2001. Macromolecular crowding: obvious but underappreciated. Trends Biochem. Sci. 26:597–604. 10.1016/S0968-0004(01)01938-7 [DOI] [PubMed] [Google Scholar]
- Ester M., Kriegel H.P., Sander J., and Xu X.. 1996. A Density-Based Algorithm for Discovering Clusters in Large Spatial Databases with Noise. AAAI Press, Palo Alto, CA. 226–231. [Google Scholar]
- Fath T., Eidenmüller J., and Brandt R.. 2002. Tau-mediated cytotoxicity in a pseudohyperphosphorylation model of Alzheimer’s disease. J. Neurosci. 22:9733–9741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handwerger K.E., Cordero J.A., and Gall J.G.. 2005. Cajal bodies, nucleoli, and speckles in the Xenopus oocyte nucleus have a low-density, sponge-like structure. Mol. Biol. Cell. 16:202–211. 10.1091/mbc.E04-08-0742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain S., Wheeler J.R., Walters R.W., Agrawal A., Barsic A., and Parker R.. 2016. ATPase-Modulated Stress Granules Contain a Diverse Proteome and Substructure. Cell. 164:487–498. 10.1016/j.cell.2015.12.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaqaman K., Loerke D., Mettlen M., Kuwata H., Grinstein S., Schmid S.L., and Danuser G.. 2008. Robust single-particle tracking in live-cell time-lapse sequences. Nat. Methods. 5:695–702. 10.1038/nmeth.1237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha N., Stoecklin G., Ayodele M., Yacono P., Lykke-Andersen J., Fritzler M.J., Scheuner D., Kaufman R.J., Golan D.E., and Anderson P.. 2005. Stress granules and processing bodies are dynamically linked sites of mRNP remodeling. J. Cell Biol. 169:871–884. 10.1083/jcb.200502088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha N., Panas M.D., Achorn C.A., Lyons S., Tisdale S., Hickman T., Thomas M., Lieberman J., McInerney G.M., Ivanov P., and Anderson P.. 2016. G3BP-Caprin1-USP10 complexes mediate stress granule condensation and associate with 40S subunits. J. Cell Biol. 212:845–860. 10.1083/jcb.201508028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroschwald S., Maharana S., Mateju D., Malinovska L., Nüske E., Poser I., Richter D., and Alberti S.. 2015. Promiscuous interactions and protein disaggregases determine the material state of stress-inducible RNP granules. eLife. 4:e06807 10.7554/eLife.06807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P., Banjade S., Cheng H.C., Kim S., Chen B., Guo L., Llaguno M., Hollingsworth J.V., King D.S., Banani S.F., et al. . 2012. Phase transitions in the assembly of multivalent signalling proteins. Nature. 483:336–340. 10.1038/nature10879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y., Protter D.S., Rosen M.K., and Parker R.. 2015. Formation and Maturation of Phase-Separated Liquid Droplets by RNA-Binding Proteins. Mol. Cell. 60:208–219. 10.1016/j.molcel.2015.08.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mateju D., Franzmann T.M., Patel A., Kopach A., Boczek E.E., Maharana S., Lee H.O., Carra S., Hyman A.A., and Alberti S.. 2017. An aberrant phase transition of stress granules triggered by misfolded protein and prevented by chaperone function. EMBO J. 36:1669–1687. 10.15252/embj.201695957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moschner K., Sündermann F., Meyer H., da Graca A.P., Appel N., Paululat A., Bakota L., and Brandt R.. 2014. RNA protein granules modulate tau isoform expression and induce neuronal sprouting. J. Biol. Chem. 289:16814–16825. 10.1074/jbc.M113.541425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadezhdina E.S., Lomakin A.J., Shpilman A.A., Chudinova E.M., and Ivanov P.A.. 2010. Microtubules govern stress granule mobility and dynamics. Biochim. Biophys. Acta. 1803:361–371. 10.1016/j.bbamcr.2009.12.004 [DOI] [PubMed] [Google Scholar]
- Oliphant T.E. 2007. Python for Scientific Computing. Comput. Sci. Eng. 9:10–20. 10.1109/MCSE.2007.58 [DOI] [Google Scholar]
- Patel A., Lee H.O., Jawerth L., Maharana S., Jahnel M., Hein M.Y., Stoynov S., Mahamid J., Saha S., Franzmann T.M., et al. . 2015. A Liquid-to-Solid Phase Transition of the ALS Protein FUS Accelerated by Disease Mutation. Cell. 162:1066–1077. 10.1016/j.cell.2015.07.047 [DOI] [PubMed] [Google Scholar]
- Pedregosa F., Varoquaux G., Gramfort A., Michel V., Thirion B., Grisel O., Blondel M., Prettenhofer P., Weiss R., Dubourg V., et al. . 2011. Scikit-learn: Machine Learning in Python. J. Mach. Learn. Res. 12:2825–2830. [Google Scholar]
- Protter D.S., and Parker R.. 2016. Principles and Properties of Stress Granules. Trends Cell Biol. 26:668–679. 10.1016/j.tcb.2016.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabouille C., and Alberti S.. 2017. Cell adaptation upon stress: the emerging role of membrane-less compartments. Curr. Opin. Cell Biol. 47:34–42. 10.1016/j.ceb.2017.02.006 [DOI] [PubMed] [Google Scholar]
- Ramaswami M., Taylor J.P., and Parker R.. 2013. Altered ribostasis: RNA-protein granules in degenerative disorders. Cell. 154:727–736. 10.1016/j.cell.2013.07.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., et al. . 2012. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 9:676–682. 10.1038/nmeth.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz J., Milpetz F., Bork P., and Ponting C.P.. 1998. SMART, a simple modular architecture research tool: identification of signaling domains. Proc. Natl. Acad. Sci. USA. 95:5857–5864. 10.1073/pnas.95.11.5857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sergé A., Bertaux N., Rigneault H., and Marguet D.. 2008. Dynamic multiple-target tracing to probe spatiotemporal cartography of cell membranes. Nat. Methods. 5:687–694. 10.1038/nmeth.1233 [DOI] [PubMed] [Google Scholar]
- Shuang B., Cooper D., Taylor J.N., Kisley L., Chen J., Wang W., Li C.B., Komatsuzaki T., and Landes C.F.. 2014. Fast Step Transition and State Identification (STaSI) for Discrete Single-Molecule Data Analysis. J. Phys. Chem. Lett. 5:3157–3161. 10.1021/jz501435p [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siomi H., Choi M., Siomi M.C., Nussbaum R.L., and Dreyfuss G.. 1994. Essential role for KH domains in RNA binding: impaired RNA binding by a mutation in the KH domain of FMR1 that causes fragile X syndrome. Cell. 77:33–39. 10.1016/0092-8674(94)90232-1 [DOI] [PubMed] [Google Scholar]
- Souquere S., Mollet S., Kress M., Dautry F., Pierron G., and Weil D.. 2009. Unravelling the ultrastructure of stress granules and associated P-bodies in human cells. J. Cell Sci. 122:3619–3626. 10.1242/jcs.054437 [DOI] [PubMed] [Google Scholar]
- Tokunaga M., Imamoto N., and Sakata-Sogawa K.. 2008. Highly inclined thin illumination enables clear single-molecule imaging in cells. Nat. Methods. 5:159–161. 10.1038/nmeth1171 [DOI] [PubMed] [Google Scholar]
- Wallace E.W., Kear-Scott J.L., Pilipenko E.V., Schwartz M.H., Laskowski P.R., Rojek A.E., Katanski C.D., Riback J.A., Dion M.F., Franks A.M., et al. . 2015. Reversible, Specific, Active Aggregates of Endogenous Proteins Assemble upon Heat Stress. Cell. 162:1286–1298. 10.1016/j.cell.2015.08.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wedeking T., Löchte S., Richter C.P., Bhagawati M., Piehler J., and You C.. 2015. Single Cell GFP-Trap Reveals Stoichiometry and Dynamics of Cytosolic Protein Complexes. Nano Lett. 15:3610–3615. 10.1021/acs.nanolett.5b01153 [DOI] [PubMed] [Google Scholar]
- Wedeking T., Löchte S., Birkholz O., Wallenstein A., Trahe J., Klingauf J., Piehler J., and You C.. 2015. Spatiotemporally Controlled Reorganization of Signaling Complexes in the Plasma Membrane of Living Cells. Small. 11:5912–5918. 10.1002/smll.201502132 [DOI] [PubMed] [Google Scholar]
- Weiss M., Elsner M., Kartberg F., and Nilsson T.. 2004. Anomalous subdiffusion is a measure for cytoplasmic crowding in living cells. Biophys. J. 87:3518–3524. 10.1529/biophysj.104.044263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissmann C., Reyher H.J., Gauthier A., Steinhoff H.J., Junge W., and Brandt R.. 2009. Microtubule binding and trapping at the tip of neurites regulate tau motion in living neurons. Traffic. 10:1655–1668. 10.1111/j.1600-0854.2009.00977.x [DOI] [PubMed] [Google Scholar]
- Wheeler J.R., Matheny T., Jain S., Abrisch R., and Parker R.. 2016. Distinct stages in stress granule assembly and disassembly. eLife. 5:5 10.7554/eLife.18413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolozin B. 2012. Regulated protein aggregation: stress granules and neurodegeneration. Mol. Neurodegener. 7:56 10.1186/1750-1326-7-56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- You C., Richter C.P., Löchte S., Wilmes S., and Piehler J.. 2014. Dynamic submicroscopic signaling zones revealed by pair correlation tracking and localization microscopy. Anal. Chem. 86:8593–8602. 10.1021/ac501127r [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.