Liu and Tjian review how superresolution and live-cell imaging are providing new insights into transcription factor dynamics and genome organization.
Abstract
The assembly of sequence-specific enhancer-binding transcription factors (TFs) at cis-regulatory elements in the genome has long been regarded as the fundamental mechanism driving cell type–specific gene expression. However, despite extensive biochemical, genetic, and genomic studies in the past three decades, our understanding of molecular mechanisms underlying enhancer-mediated gene regulation remains incomplete. Recent advances in imaging technologies now enable direct visualization of TF-driven regulatory events and transcriptional activities at the single-cell, single-molecule level. The ability to observe the remarkably dynamic behavior of individual TFs in live cells at high spatiotemporal resolution has begun to provide novel mechanistic insights and promises new advances in deciphering causal–functional relationships of TF targeting, genome organization, and gene activation. In this review, we review current transcription imaging techniques and summarize converging results from various lines of research that may instigate a revision of models to describe key features of eukaryotic gene regulation.
Introduction
Early research established that transcription in prokaryotes is primarily controlled by proximal promoter-bound factors (e.g., repressors and σ factors) that often directly interact with RNA polymerase (Ptashne and Gann, 1997). However, as the complexity of organism increases, a key feature of higher eukaryotes (metazoans) emerged as a dramatic expansion of genome size and especially the intergenic regions that contain widely dispersed cis-regulatory elements such as enhancers and insulators (Levine et al., 2014). Specifically, enhancers communicate with promoters to regulate gene activities, whereas insulators block the communication between enhancers and promoters (Heintzman and Ren, 2009). In the past decades, functional components of the gene-regulatory machinery have been extensively characterized by classical in vitro biochemistry and in vivo genetic research (Levine et al., 2014). Moreover, the recent rapid development of next-generation sequencing–based genomewide high-throughput assays (e.g., chromatin immunoprecipitation sequencing, assay for transposase-accessible chromatin sequencing, and Hi-C) have provided additional insights into transcription factor (TF)-binding patterns as well as chromatin architecture and genome organization at the cell population level with rich sequence information (Barski et al., 2007; Lieberman-Aiden et al., 2009; Consortium and ENCODE Project Consortium, 2012; Buenrostro et al., 2013). However, these end point assays are not able to reveal 3D molecular structures or intrinsic dynamics associated with gene-regulatory activities within individual living cells. For example, how quickly and by what means does a TF find its target site inside the nucleus of a live cell? How do multiple TFs dynamically assemble at enhancer DNA? How are cis-regulatory elements organized inside the 3D space of the nucleus? Most importantly, what are the fundamental structural basis and dynamics underlying enhancer–promoter communication? Recent advances in fluorescence labeling techniques in combination with superresolution microscopy modalities have overcome several technical barriers to visualizing and tracking the dynamic movement of individual TFs in single living cells (Mazza et al., 2012; Gebhardt et al., 2013; Chen et al., 2014b), even within developing embryos (Mir et al., 2017). These studies reveal hitherto unknown intranuclear structures and TF kinetic features in live cells with unprecedented spatiotemporal resolution, providing unique opportunities to dissect transcription dynamics and genome organization. In this review, we first outline emerging transcription imaging methods and then discuss recent studies that converge on a gene regulation model that integrates TF dynamics, genome organization, and gene activity in live cells.
Imaging and labeling strategies to probe TF dynamics
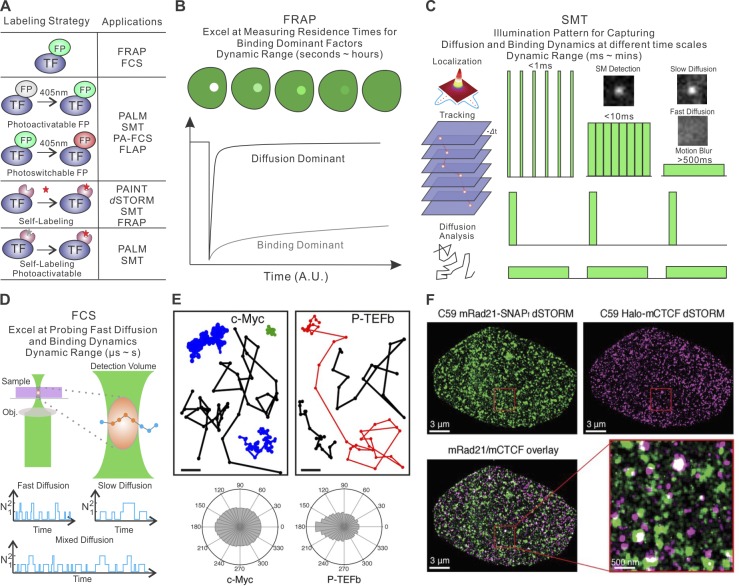
The successful development of superresolution TF imaging strategies became possible largely as a consequence of dual advances in new biomolecular labeling techniques and advanced imaging modalities (Fig. 1 A). Early pioneering studies (McNally et al., 2000; Stenoien et al., 2001) to measure TF diffusion and binding dynamics relied on fluorescent proteins (FPs; Tsien, 1998) and the use of FRAP assays (Fig. 1 B; Axelrod et al., 1976). In a typical FRAP experiment, a focused laser beam rapidly bleaches FPs within a selected region in the nucleus. The subsequent rate of fluorescence recovery in the bleached region is dependent on the diffusive and binding kinetics (Kon and Koff) of labeled molecules outside the target area as well as the dissociation rate (Koff) and diffusion kinetics of the bleached molecules within the target area. FRAP has proven to be an effective tool for studying TFs with relatively long-lived binding residence times (several seconds to hours), and as such, it provides valuable information about the behavior of some TFs in live cells. For example, it was found that core histone subunits are generally very stable with little exchange occurring even after ∼1–2 h. Somewhat surprisingly, a small fraction of H2B molecules displayed a considerably more dynamic behavior and exchanged within minutes. These results suggest that the core of the nucleosome is very stable, whereas H2B on the surface of active nucleosomes may exchange more frequently (Kimura and Cook, 2001). Strikingly, heterochromatin protein 1 (HP1) displayed remarkably transient residence times (∼10–20 s) in live cells compared with core histones (approximately hours; Cheutin et al., 2003), suggesting a rather dynamic maintenance of heterochromatin in live cells. In addition, FRAP assays targeting artificially amplified gene arrays suggested that nuclear receptor TFs (e.g., estrogen receptors and glucocorticoid receptors [GRs]) have short residence times (approximately seconds) even at cognate target sites (McNally et al., 1999, 2000; Voss et al., 2011), suggesting rapid dynamic exchange of TFs at enhancer sites in live cells in the time scales of several seconds. Until now, FRAP assays offered the best available method to estimate residence times for proteins that predominantly operated in a binding-dominant mode. However, FRAP measurements rely on averaging over a large, often heterogeneous population of molecules, and the resulting data analysis is a priori highly model dependent (Mueller et al., 2010; Mazza et al., 2012). As a result, FRAP has limited power to resolve complex fast-diffusion dynamics or to measure residence times for minor subpopulations of bound molecules.
Figure 1.
Methods for imaging TF dynamics and subnuclear structures in single cells. (A) Current TF labeling methods (left) and their suitable imaging applications (right). Specifically, three common strategies are schematized: (1) labeling with regular FPs, (2) labeling with photoactivatable/photoswitchable proteins, or (3) self-labeling tags such as HaloTag, SNAPTag, and TMPTag. Self-labeling tags can be conjugated with organic dyes for subsequent imaging applications. FLAP, fluorescence localization after photobleaching (Dunn et al., 2002); PAINT, points accumulation for imaging in nanoscale topography (Sharonov and Hochstrasser, 2006). (B) FRAP technique. A high-intensity–focused laser illumination pulse generates a bleached spot in the cell. The fluorescence recovery curve is the result of dissociation of bleached fluorescent molecules and diffusion-in of unbleached molecules. (C) SMT technique. Left: Sparsely labeled TF molecules appear to be diffraction-limited spots on the camera chip. The position of the TF molecule is determined by localization of the centroid of the spot by Gaussian fitting. The positions of one molecule across multiple frames are linked to form single-molecule trajectories. TF diffusion and binding dynamics can be extracted from single-molecule trajectories. Right: Distinct illumination patterns can be deployed to examine TF diffusion and binding dynamics at different time scales by limiting or using motion blur effect. Specifically, intensive stroboscopic illumination is an efficient way to reduce motion blurring for capturing fast diffusion events, whereas long acquisition times and low laser illumination are used for selectively imaging stable binding events. Long lapse times can also be introduced to reduce photobleaching and to specifically study long-lived binding events up to several minutes. (D) FCS estimates the diffusion dynamics of molecules by recording photon bursting of molecules diffusing through a diffraction-limited focal spot. Fast diffusion gives rise to bursts with shorter temporal widths and vice versa. (E) SMT analysis reveals distinct diffusion dynamics of c-Myc and positive transcription elongation factor (P-TEFb) in live cells. Specifically, c-Myc diffusion is less constrained in space than that of P-TEFb as highlighted by representative single-molecules trajectories (top) and the distribution histograms of the angle formed between the vectors of two consecutive translocation steps (bottom). The data suggest that local protein–protein and protein–DNA interactions alter the diffusion kinetics of P-TEFb molecules. This panel is adapted from Izeddin et al. (2014) with permission. (F) Two-color PALM experiments reveal distinct localization of CTCF (magenta) and the cohesin subunit Rad21 (green) in an interphase cell. Interestingly, CTCF and Rad21 bind to chromatin with different residence times, suggesting their distinct role in establishing chromatin loops in the nucleus. This panel is adapted from Hansen et al. (2017a) with permission. dSTORM, direct STORM.
A more recently developed and complementary imaging technology for dissecting TF dynamics is direct observation of individual molecules in motion within live cells (Fig. 1 C). Although single-particle tracking is an ancient technique that was first applied to study random movements (Brownian motion) of tiny objects (e.g., pollen) under the microscope as early as 1820s (Brown, 1828), single-molecule tracking (SMT) in live cells was not possible until the realization of fluorescence microscopy (Fluoreszenzmikroskop., 1911) and subsequent development of suitable protein-labeling techniques. For example, SMT was first applied to study the dynamics of membrane proteins by antibody labeling more than two decades ago (Ghosh and Webb, 1994). However, because of the high packing density of intracellular, and particularly intranuclear, proteins and the lack of tools for sparse labeling, reliable intracellular single-molecule imaging even in fixed cells was not possible until the discovery of photoactivatable or photoswitchable FPs (or dyes; Patterson and Lippincott-Schwartz, 2002). These are proteins whose fluorescence upon excitation at certain wavelengths (usually 405 nm) can be either switched on or off or modified to a different emission spectrum (Lukyanov et al., 2005). These advanced labeling strategies, when combined with the subsequent development of photoactivated localization microscopy (PALM; Betzig et al., 2006) and stochastic optical reconstruction microscopy (STORM; Rust et al., 2006), ushered in a new era of superresolution imaging. With photoactivation, sparse labeling can be achieved by tunable stochastic activation of fluorophores, separating the appearance of individual molecules temporally. However, FPs are generally not very photostable and thus cannot support long-term single-molecule observation (Manley et al., 2008; Xia et al., 2013). Recent development of self-labeling tags (e.g., HaloTag and SNAPTag) and a new suite of photostable cell-permeable organic dyes (e.g., JF549 and JF646; Grimm et al., 2015) further expanded the spatiotemporal length scales of single-molecule measurements in live cells, enabling high signal-to-noise single-molecule detection in individual living cells. The 2D and 3D imaging modalities and requirements for a specific SMT application have been extensively discussed previously; for detailed reviews, we refer the readers to Liu et al. (2015), Manzo and Garcia-Parajo (2015), Presman et al. (2017), and Shen et al. (2017). The next major challenge of labeling was to devise means to sparsely label biomolecules in live cells for reliable single-molecule imaging and SMT under physiologically relevant conditions. Encouragingly, the rapid development of genome-editing techniques (Doudna and Charpentier, 2014) enabled functional labeling and imaging of endogenous proteins as reported in several recent studies (Teves et al., 2016; Hansen et al., 2017a).
Because of its ability to monitor the dynamics of individual biomolecules, SMT has the resolving power to monitor complex TF diffusion and binding kinetics (Fig. 1 E; Mazza et al., 2012; Gebhardt et al., 2013; Chen et al., 2014b; Izeddin et al., 2014; Ball et al., 2016; Paakinaho et al., 2017), investigate subpopulation-associated structures (Fig. 1 F; Liu et al., 2014; Mir et al., 2017; Wollman et al., 2017), and determine the order of multistep molecular binding events in live cells (Chen et al., 2014b; Xie et al., 2017). Moreover, robust computational pipelines have been devised to achieve accurate data analysis (Jaqaman et al., 2008; Sergé et al., 2008; Hansen et al., 2017b).
An important technical aspect to consider for SMT experiments in live cells is the effect of motion blurring. In fixed samples, molecules are by definition stationary. Thus, image acquisition times do not significantly influence single-molecule detection. However, in live cells, rapid molecular diffusion introduces motion blur during image acquisition, limiting localization precision. Various techniques have been deployed to counteract or, in some cases, take advantage of the motion blur effect (Fig. 1 C). At one end of the spectrum, high-intensity stroboscopic illumination pulses (<1 ms) have been used to minimize motion blur and capture rapid binding and unbinding events in the range of milliseconds (Elf et al., 2007). At the other extreme, low laser power and long acquisition times can be deployed to image relatively stable TF–chromatin binding events by blending blurred images of fast-diffusing unbound molecules into the background while also preserving in clear relief stable binding events as bright diffraction-limited spots for measuring binding events that last for seconds (Chen et al., 2014b; Swinstead et al., 2016; Hansen et al., 2017a; Xie et al., 2017). In addition, long dark periods between acquisitions can be introduced to reduce photobleaching, thereby increasing the dynamic range of single-molecule detection up to ∼10–20 min (Gebhardt et al., 2013; Normanno et al., 2015; Paakinaho et al., 2017; Liu et al., 2018). In essence, because the number of photons (photon budget) that can be emitted from any FP or dye molecule is finite, distinct illumination patterns may be used to capture desired TF dynamics occurring at widely different time scales. Because of this reason, SMT-based residence time measurements are highly sensitive to changes in imaging setups (e.g., the laser power, the objective NA, the camera sensitivity, the photostability of the label, and the imaging acquisition strategy). For example, it was reported that dimeric GR has a residence time of ∼1.45 s by using a EOS2 fusion protein (Gebhardt et al., 2013), whereas the GR-stable binding residence time measured by using the more photostable HaloTag is several-fold longer (∼7.4 s; Swinstead et al., 2016). Similarly, CTCF residence times estimated by different SMT imaging strategies showed drastic differences (∼1–2 min [Hansen et al., 2017a] versus >15 min [Agarwal et al., 2017]). These results highlight that specific imaging regimes are only optimized to detect the TF dwell times in particular temporal domains. Therefore, the best practice to avoid bias in data interpretation is to derive conclusions based on relative changes of residence times followed by well-controlled perturbations.
Another useful technique to probe fast TF diffusion dynamics is fluorescence correlation spectroscopy (FCS; Magde et al., 1972; Chen et al., 2008). Specifically, in FCS, a high-sensitivity point detector is used to record motion-induced fluorescence fluctuations at the focal spot (Fig. 1 D). Because molecules with different diffusion kinetics give rise to distinct photon burst patterns, the molecular diffusion characteristics can be estimated from subsequent autocorrelation function calculations. This technique can be quite useful for resolving fast diffusion dynamics. One limitation, however, is that the concentration of molecules in cells must be low enough (<10 nM) to generate robust fluctuations for detection. Combining FCS with photoactivatable GFPs (PA-FCS) can overcome this problem and allow FCS measurement in densely labeled samples (White et al., 2016). Another limitation of FCS is that immobile fluorophores in the focus region do not generate fluorescence fluctuations and can be bleached quickly (Stasevich et al., 2010). Thus, this technique is not suitable for studying stable TF binding events. In addition, FCS suffers from the same model-fitting problem as FRAP: mechanistically distinct models can often fit the same FCS curve equally well (Mazza et al., 2012).
TF dynamics
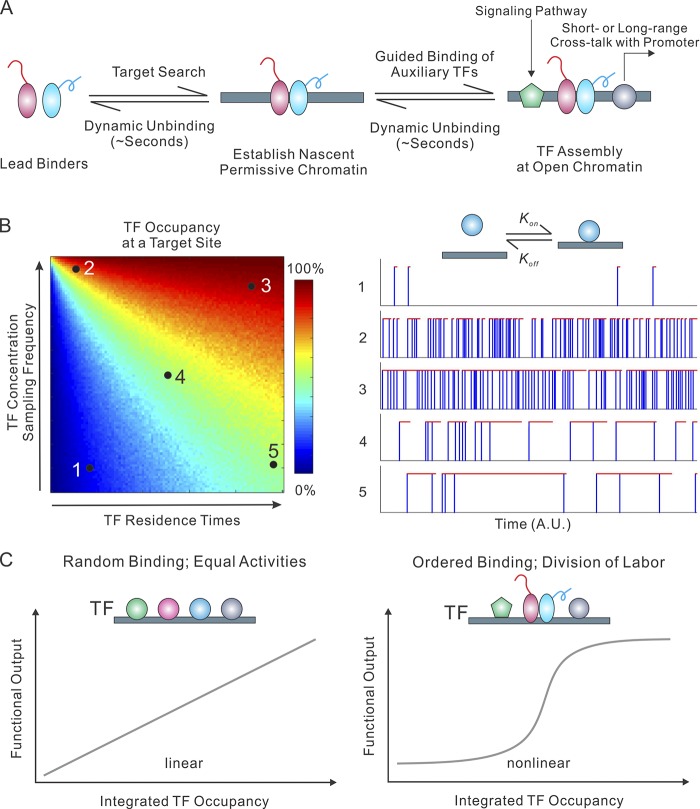
The assembly of multiple TFs at distal enhancers is a signature feature of gene regulation in metazoans. However, one longstanding unresolved question in the field (Halford, 2009; Mirny et al., 2009) is how a TF molecule efficiently navigates through the complex and crowded nuclear environment, searching through several billion base pairs of DNA in the form of chromatin before locating a cognate target site. Early FRAP studies found that site-specific TFs show fast recovery after photobleaching, suggesting that a large fraction of TF molecules are in fast diffusion states (McNally et al., 2000; Stenoien et al., 2001; Sprague et al., 2004). The recent development of higher quantum yield and more stable self-labeling dyes enabled long-term tracking of TFs at the single molecule level. A study focused on imaging Sox2 and Oct4 dynamics in live embryonic stem (ES) cells observed that Sox2 and Oct4 molecules use a 3D diffusion–dominant trial-and-error target search mechanism (Chen et al., 2014b). Specifically, Sox2 molecules spend most of the time (∼97%) in stochastic diffusion and collision with nonspecific DNA before engaging a cognate-binding site. It is estimated that only a small fraction of Sox2 molecules (∼3%) in ES cells are bound to specific recognition sites in the equilibrium state. Consistent with previous studies on nuclear receptors (McNally et al., 1999, 2000; Voss et al., 2011; Paakinaho et al., 2017), Sox2 also displayed short mean residence times on the scale of ∼12 s at specific target sites and even more fleeting dwell times (0.7 s) at nonspecific sites. Interestingly, recent SMT studies also show that in live cells, Sox2 interacts with the mitotic chromosomes for bookmarking (Deluz et al., 2016; Teves et al., 2016). Depletion of Sox2 at the M–G1 transition abolishes the ability of ES cells to differentiate into neuroectodermal lineages but does not interfere with reprogramming toward induced pluripotent stem cells, suggesting that dynamic interaction of Sox2 with mitotic chromosome is critical for poised activation of neural gene expression programs (Deluz et al., 2016). Surprisingly, one recent study reveals that the FOXA1 pioneer TF fails to generate significant DNA footprints at chromatin-binding sites and likely only has a short specific binding residence time in the range of seconds (Swinstead et al., 2016). These results highlight a highly dynamic turnover of TFs at enhancer sites in live cells. Simulation experiments suggest that one important feature of rapid and dynamic TF binding and unbinding is that TF occupancy at the target site is highly sensitive to TF concentrations in the nucleus (Fig. 2 B; Chen et al., 2014b). Specifically, to compensate for short dwell times, high TF concentrations, at least locally, are required to increase TF sampling frequency at the enhancer to maintain functionally relevant TF temporal occupancy. However, if a TF binds DNA with very long residence times, its occupancy at a target site remains high regardless of its concentration in the cell and thus, the system becomes much less tunable (Fig. 2 B). It seems plausible that during evolution, there has been a tradeoff between TF DNA binding affinity and a more tunable on/off system. Rapid TF unbinding is likely desirable at the enhancers of those genes that need precise spatiotemporal expression patterns. Indeed, one study found that even a slight enhancement of Hox TF DNA binding affinity disrupts normal Drosophila melanogaster developmental programs (Crocker et al., 2015).
Figure 2.
TF binding dynamics at enhancers. (A) TF binding kinetics at cis-regulatory elements is dynamic in live cells. Accumulating evidence suggests that a small group of site-specific TFs act as lead or pioneer binders that efficiently scan and engage with silent chromatin, establishing a permissive chromatin for the subsequent binding of auxiliary TFs, which in turn reinforces an open-chromatin state. TFs bound within the assembly mediate distinct functions (e.g., signal transduction and interplay with core promoter), suggesting a functional division of labor for TFs. (B) The TF temporal occupancy pattern at a specific target site is exquisitely regulated by TF concentration in the nucleus and TF residence times at the target site (left). For this numerical simulation, random TF residence times (R; AU) and sampling intervals (I; AU) were generated based on a log-normal distribution. We set the σ to 1/4 Log(R) or Log(I). The selection of this distribution is based on the observation that the log-scale value of Sox2 residence times on specific DNA probes roughly follows a normal distribution (Chen et al., 2014b). For each pair of mean TF residence times (R) and sampling intervals (I), 1,000 continuous binding events were simulated, and the TF occupancy (color-encoded) of the target site was calculated based on the total binding on/off durations. Representative binding traces of a TF at the target site with defined parameters (1–5) are shown on the right. Blue vertical lines represent TF sampling events. Red horizontal lines reflect TF dwelling events at the target site. (C) The relationship between TF binding dynamics and the functional output of an enhancer. If multiple TFs have an equal ability to bind to the target site and activate gene expression, the functional output of an enhancer should be linearly proportional to integrated TF occupancy at the enhancer. An ordered TF assembly and functional division-of-labor mechanism could potentially generate rapid and nonlinear functional outputs.
Another interesting question is whether multiple TFs bind to enhancers via a symmetric (random order) or asymmetric (hierarchically ordered) manner. One model is that TFs are symmetrically required for the binding and establishment of open chromatin at the target site. However, this model was quickly excluded by the finding that the FoxA1 factor binding in the genome opens up the chromatin and is required for the binding of other TFs to the target sites (Cirillo and Zaret, 1999; Cirillo et al., 2002). However, it was recently shown that the binding of FoxA1 to chromatin in many cases is dependent on steroid receptors (Swinstead et al., 2016). These results suggest that although the assembly of multiple TFs at enhancer sites is hierarchical, the order of assembly at particular enhancer sites could be highly context dependent. Supporting this model, single-molecule analysis found that in ES cells, Sox2 is kinetically favored to engage with a target site first and assists the subsequent binding of Oct4 (Chen et al., 2014b) in a hierarchically ordered fashion. Consistent with this observation, it was revealed by PA-FCS that the binding dynamics of Sox2 rather than that of Oct4 predicts the lineage fate of a cell in early embryos (White et al., 2016). Interestingly, a lead binder, Zelda, was identified in Drosophila with similar functions (Harrison et al., 2011; Mir et al., 2017). In the analysis of a more complex system, deletion of a single Oct4/Sox2 composite site at a distal Klf4 enhancer abolishes chromatin accessibility and the binding of Esrrb and Stat3 to the enhancer (Xie et al., 2017). In contrast, depletion of Esrrb and Stat3 had no significant effect on the binding of Sox2 and Oct4 to the enhancer, adding further support for an ordered assembly process at least for some enhancers in ES cells. Interestingly, Stat3 is the downstream component of the leukemia inhibitory factor signaling pathway (Dahéron et al., 2004), whereas Esrrb has been shown to interact with core promoter factors and Pol II (Percharde et al., 2012), suggesting a functional division of labor among individual TFs assembling at complex enhancers. Specifically, this model envisions that a small subset of lead binders (e.g., Sox2) are required to scan the genome, find its target site, and establish a permissive chromatin environment to assist other auxiliary TFs to bind subsequently (Fig. 2 A). The advantage for such a hierarchical and modular system is that TFs have overlapping as well as differentiating functions, which provides more flexibility and greater specificity, enabling the generation of rapid, nonlinear spatiotemporal outputs (Fig. 2 C). In contrast, in a symmetric system where partner TFs exhibit the same target search efficiency and equal ability to open up chromatin to initiate transcription, gene activity output would be a linear function of TF binding occupancy at the enhancer (Fig. 2 C).
Genome organization
Roughly 2 m (approximately six billion base pairs) of linear DNA must be packed into the nucleus of each diploid human cell (∼5–10 µm in diameter). With the same packaging density, it is possible to put a strand of DNA >6,000 times the Earth’s circumference inside a chicken egg. Effective DNA packing into the cell nucleus is partially achieved by wrapping DNA around histones into nucleosomes. Currently, we have crystal structures of the nucleosome (Luger et al., 1997). Based on in vitro structural work (Schalch et al., 2005; Woodcock and Ghosh, 2010; Song et al., 2014), it was proposed that nucleosome polymers can assemble into 30-nm fibers. Recently, a new DNA EM dye has been developed to image detailed high-resolution chromatin fiber organization in single fixed cells. These studies revealed very disordered structures, and fibers of ∼5–24 nm were found in situ (Ou et al., 2017), suggesting that the 30-nm fiber is probably not the dominant form of chromatin in the nucleus. Another finding from this study suggests that 3D subnuclear domains are assembled with distinct chromatin densities, and this feature likely determines the global accessibility and activity of DNA in the nucleus.
Currently, large subnuclear compartments such as heterochromatin, nuclear speckles, Cajal bodies, histone locus clusters, and nucleoids can be visualized in live cells with light microscopy (Mao et al., 2011). However, because of the high packing density of native chromatin fibers and a lack of noninvasive tools for specific gene locus labeling, the structural information underlying enhancer–promoter communication in the cell remains out of reach by conventional structural biology or light microscopy. The current information regarding potential structures driving enhancer–promoter communication is mainly derived from indirect methods such as chromosome conformation capture (3C)-based assays (de Wit and de Laat, 2012). Specifically, chemically cross-linked chromatin fibers are digested by restriction enzymes, and the contact frequency between chromatin fragments is inferred from proximity ligation reactions performed with mixtures of large populations of cells. In the past decade, extensive genomewide studies (4C, 5C, and Hi-C) have provided a model for chromatin folding, topological domains, and potential 3D genome organization (Dekker et al., 2013; Dekker and Mirny, 2016). The two main features extracted from these studies are: (1) as proposed originally from the earliest enhancer research (Su et al., 1991), enhancers and promoters likely communicate with each other via chromatin loops, and (2) the genome is organized into topologically associated domains (TADs) that might compartmentalize local gene activities. However, the data from imaging and genomics sometimes show significant discrepancies (Belmont, 2014; Williamson et al., 2014; Boettiger et al., 2016; Wang et al., 2016b), suggesting that 3C-based assays might have limited ability to capture the true 3D structures and physical proximity of elements in single live cells. There is also the added possibility that chemical cross-linking may introduce some unintended consequences not easily anticipated or interpreted. For example, recent SMT studies show that in live cells, Sox2 interacts with the mitotic chromosomes for bookmarking (Deluz et al., 2016; Teves et al., 2016), whereas most strikingly, chemical cross-linking leads to exclusion of Sox2 from mitotic chromosomes (Teves et al., 2016). In addition, a recent study on CTCF and cohesin with SMT reveal that chromatin loop formation might be a highly dynamic (~min) and regulated event in the cell (Hansen et al., 2017a). Results from such imaging studies using live cells suggest that the interaction frequency detected by 3C assays more likely reflect a complex convolution of physical proximity, DNA looping frequency, local chromatin environment, and cellular heterogeneity (Fudenberg and Imakaev, 2017). Consistent with this notion, single-cell Hi-C experiments reveal that stochastic clusters of contacts can occur across TAD boundaries in single cells but average into TADs in ensemble assays (Flyamer et al., 2017), suggesting that TADs might be the result of computational normalization of cross-linking events over millions of cells.
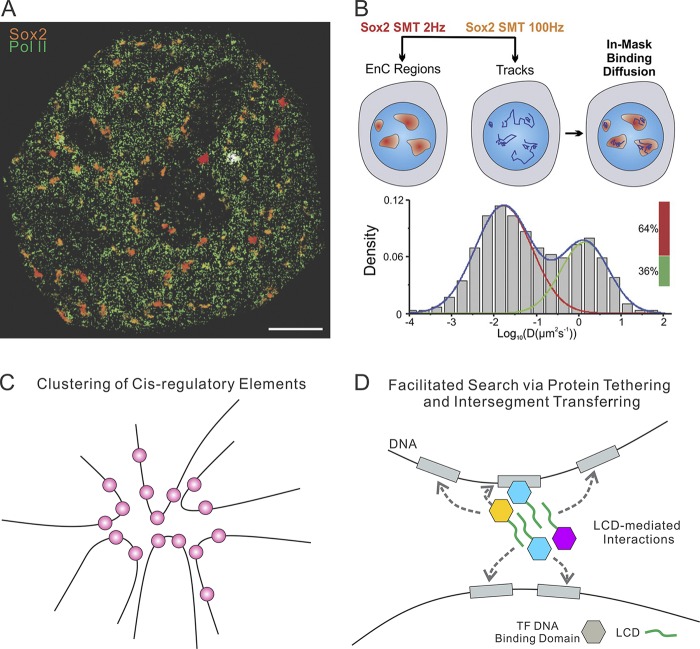
Despite some room for debate, live-cell imaging data do converge with genomic studies on the presence of topological structures in the nucleus that likely also shape local gene-regulatory activities. What is the reciprocal relationship between these topological structures and the spatial distribution of various TFs in the nucleus? By imaging long-lived Sox2 binding events in live cells with lattice light-sheet microscopy, it was possible to systematically map 3D Sox2 enhancer organization in ES cells. It was observed that Sox2 stable binding sites (most likely enhancers) form spatially restricted clusters in the nucleus of live ES cells (Liu et al., 2014). These enhancer clusters are spatially segregated from heterochromatic regions but overlap with a subset of Pol II–enriched clusters (Fig. 3 A). Furthermore, SMT experiments revealed that inside enhancer clusters, Sox2 displays significantly faster forward association rates (kon), thereby increasing local TF concentrations and allowing rapid TF rebinding to stretches of open chromatin (Fig. 3 B). Consistent with this early observation, SMT experiments confirmed that TF also forms clusters in yeast and Drosophila cells and that the TF clustering likely regulates transcriptional output from downstream genes (Mir et al., 2017; Wollman et al., 2017). In addition, detailed analysis of long-lived Sox2-binding events at the single molecule level revealed that Sox2 hops between clustered binding sites in spatially restricted subnuclear regions (Liu et al., 2018). These results support the existence of certain topological structures and TF hubs in the nucleus that can shape local TF target search dynamics and potentially fine-tune the rates of TF complex assembly at cis-regulatory elements.
Figure 3.
TF diffusion and binding dynamics. (A) Colocalization of Sox2 enhancer clusters (orange) and Pol II clusters (green) in live cells. Sox2 enhancer clusters were mapped by time-resolved, 2D single-molecule imaging/tracking. Stable binding events (>2 s) are shown. Pol II clusters are mapped by fast imaging acquisition with 100-Hz frame rates. Bar, 2 µm. This panel is adapted from Liu et al. (2014) with permission. (B) Increased Sox2 forward association with chromatin in the nucleus. Two-color single-molecule imaging to probe Sox2 binding and diffusion dynamics in enhancer clusters (EnCs). Enhancer cluster regions were first mapped by the low-excitation, long-acquisition time condition. Then, the diffusion coefficient histogram of tracks within the enhancer cluster regions was calculated and displayed in the below graph. Despite fast diffusion being the dominant fraction in the whole cell, the Sox2 enhancer clusters score elevated TF-bound fractions. This panel is adapted from Liu et al. (2014) with permission. (C) The spatial clustering of cis-regulatory elements in the nucleus of the cell was proposed by several recent imaging-based studies (Liu et al., 2014; Mir et al., 2017; Wollman et al., 2017). (D) The cis-regulatory elements clustering and LCD-mediated weak interactions between TFs would facilitate the TF target search by the mechanisms of protein tethering and intersegment transferring.
What are the forces holding together such cis-regulatory element hubs? From the earliest cloning and characterization of classical sequence-specific TFs, a puzzling feature emerged: the discovery of simple repetitive, largely unstructured amino acid motifs (i.e., glycine- and proline-rich acidic repeats) that serve as activation domains (ADs) coupled with DNA-binding domains (Courey and Tjian, 1988). More recent evidence suggests that such simple repetitive amino acid motifs, now referred to as low-complexity domains (LCDs), are found to be highly prevalent in a variety of regulatory proteins including many classical TFs and other proteins such as Fused in sarcoma (FUS), TAF15, and Ewing sarcoma protein (EWS; Kwon et al., 2013) and can participate in forming dynamic phase-separated compartments in live cells (Patel et al., 2015; Shin and Brangwynne, 2017). We speculate that the in vivo enhancer clustering or “hub formation” observed in live-cell studies opens the possibility that high local concentrations of TFs may result from the formation of LCD clustering, at least transiently. Thus, it is possible that these enhancer clusters could serve as multivalent docking sites for dynamic TF recruitment via weak protein–protein interactions potentially directed by LCD-containing proteins. In turn, it is also possible that such weak protein–protein interactions reinforce and contribute to the establishment of cis-regulatory element clustering. Indeed, LCDs within the ADs of Sp1 and Foxp2 can dynamically interact with PolyQ arrays on the surface of mutant huntingtin aggregates in live cells (Li et al., 2016). Similarly, we can imagine that such hubs of weak multivalent protein–protein interactions within enhancer clusters could be assisted by stronger sequence-specific protein–DNA transactions that together influence the local chromatin environment to regulate gene activities. Interestingly, in vitro structural analysis on reconstituted Mig1 suggests that TF clusters in the yeast nucleus are stabilized by interactions between LCDs, highlighting a crucial role of LCD in mediating the formation of TF clusters (Fig. 3 C). Consistent with the increased forward association rate for Sox2 in the enhancer cluster, it is proposed that formation of these TF clusters in yeast reduces promoter search times through intersegment transfer while stabilizing gene expression (Fig. 3 D; Wollman et al., 2017). An interesting theoretical work also suggests that in the presence of TFs, DNA could undergo phase separation (Le Treut et al., 2016). Consistent with this TF hub model, other well-described phase-separated structures known as nuclear bodies (e.g., nuclear speckles, promyelocytic leukemia protein bodies, and Cajal bodies) are thought to be capable of influencing genome organization by sequestering target genes in specialized microenvironments (Brown et al., 2008; Ching et al., 2013; Wang et al., 2016a). Modeling of cell population–based genomic data also suggests clustering and higher-order organization of cis-regulatory elements in the nucleus (Canals-Hamann et al., 2013; Hnisz et al., 2013, 2017; Dai et al., 2016; Beagrie et al., 2017). However, these studies and hypothetical models do not provide spatial aspect information of the clusters at the single-cell, single-molecule level. Despite emerging evidence for TF clustering, it is important to note that the relationship between genome organization, gene positioning, and gene activities have not been clearly established. On one hand, it was shown that gene positioning in the nucleus could be potentially critical for mediating gene activities. For example, it was shown in pro-B cells and muscle cells that activation or repression of specific genes are associated with translocation of genes into new locations (Kosak et al., 2002; Moen et al., 2004). On the other hand, genomic data suggest that regions contacting GR-regulated genes are not particularly enriched for GR targeting sites or for any functional group of genes (Hakim et al., 2011), suggesting that functional genome organization does not likely respond to the clustered binding of individual TFs. The contact regions are, however, enriched for DNaseI-hypersensitive sites, indicating that the nucleus is preorganized in a conformation favorable to rapid transcriptional reprogramming, and this organization is likely orchestrated by chromatin sites accessible to diverse regulatory factors.
Core promoters and Pol II transcription
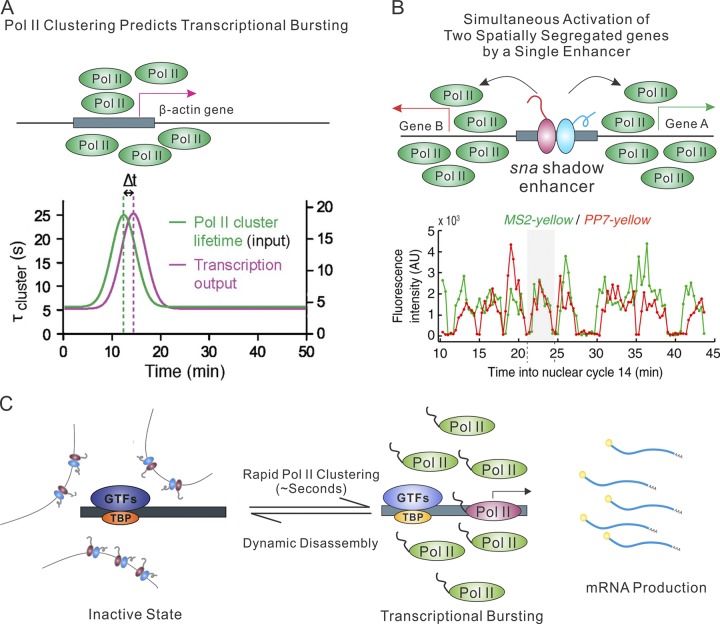
In addition to enhancer-mediated control, Pol II transcription is also extensively regulated at the core promoter by diverse sequence elements and cell type–specific core promoter factors (Goodrich and Tjian, 2010; Juven-Gershon and Kadonaga, 2010). Early fluorescence imaging and EM research led to the proposal that genes may be transcribed in “transcription factories” formed by clustered Pol II molecules in the nucleus (Cook, 1999). Later, live-cell imaging by FRAP and superresolution imaging in fixed cells showed that contrary to the “factory” hypothesis, there were no detectable stably bound and transcriptionally engaged Pol II within the clustered hot spots in the nucleus (Darzacq et al., 2007; Zhao et al., 2014). More recently, a live-cell PALM study revealed that Pol II molecules indeed form subdiffraction-size clusters (Cisse et al., 2013), but these clusters displayed lifetimes of only a few seconds incompatible with actively transcribing Pol II. These striking findings provide compelling evidence that alternative mechanisms must underlie Pol II–mediated transcription. A followup study on the β-actin locus revealed that dynamic Pol II cluster formation precedes mRNA production (Fig. 4 A; Cho et al., 2016), consistent with the notion that the observed Pol II clustering might participate in transcription initiation rather than elongation. Similarly, other research showed that low-complexity sequences within the Pol II C-terminal domain are required for phosphorylation-dependent Pol II hydrogel formation in vitro (Kwon et al., 2013). These data highlight the rapid molecular kinetics that likely drives transcription initiation. In light of observed TF enhancer clusters and Pol II hubs, one hypothesis is that multivalent weak interactions and cooperative binding within the enhancer clusters lead to increased local TF concentrations. Such TF hubs would in turn dictate local target search dynamics of key transcriptional preinitiation components including chromatin-remodeling complexes and general TFs, thereby triggering dynamic Pol II clustering and subsequent transcription initiation from nearby genes (Fig. 4 C). It is tempting to speculate that the enhancer clustering and its associated cofactors could thus form local TF hubs required for coordinated and synergistic gene regulation. Whether the enhancer clusters observed represent actively transcribed regions remains unclear, but the significant colocalization between enhancer clusters and Pol II would be consistent with such an interpretation (Liu et al., 2014).
Figure 4.
Pol II clustering and enhancer function. (A) Top: Dynamic RNA Pol II clustering proceeds and predicts transcriptional bursting of β-actin gene. Bottom: Pol II cluster lifetime used as model input (green) is overlaid with a plot of the best fit for the mRNA output (magenta) from theoretical model. A time lag (delay Δt) is observed in the model. The bottom panel is adapted from Cho et al. (2016) with permission. (B) Simultaneous activation of two symmetrically localized genes by a single enhancer. Top: Single sna shadow enhancer driving the expression of the symmetric snaPr-MS2-yellow and snaPr-PP7-yellow reporters. Bottom: Transcription activities for the two reporter genes. The intensity of green (MS2-yellow) and red (PP7-yellow) false coloring is proportional to the signal strength for each transcription focus at a given time. The bottom panel is adapted from Fukaya et al. (2016) with permission. (C) Live-cell imaging data suggest that RNA Pol II functions at the core promoter by rapid assembly and disassembly. RNA Pol II clustering predicts RNA production, suggesting that the clustering is likely involved in transcription initiation processes.
This model predicts that rapid and highly dynamic weak interactions characterized by local high concentrations and physical proximity of TF and DNA elements rather than conventional stable lock-and-key protein–protein and TF–DNA contact is the driving force for enhancer–promoter communication. Results from several lines of research are consistent with this prediction. Most interestingly, when a single enhancer was used to drive the expression of two symmetric genes located upstream and downstream of the enhancer, it was observed that the two genes have synchronized transcription-bursting kinetics (Fig. 4 B; Fukaya et al., 2016). This result argues against a stable lock-and-key looping model for enhancer–promoter communication. Consistent with this finding, an early RNA FISH study showed that transcription bursting of two genes inserted into the same genomic locus is highly correlated, whereas the correlation disappears when the two genes are at separate genomic locations (Raj et al., 2006). Both examples suggest the existence of coordinated control of gene activities by local chromatin environment. Another interesting study based on CRISPR/Cas9 cis-regulatory deletion screening found that even deletions of a promoter could affect the expression of other nearby genes (Diao et al., 2016). These results strongly suggest that although enhancer elements play an important role in driving gene expression, their influence can be extensively modulated by the local chromatin environment and the level of available weak interactions in proximity.
Concluding remarks
Live-cell imaging of transcriptional regulation has the potential to greatly advance our understanding of molecular mechanisms underlying precise spatiotemporal gene expression programs. In this review, we have highlighted a few key concepts derived from recent studies linking TF dynamics, genome organization, and gene expression. Collectively, these studies reveal a highly dynamic yet regulated TF assembly process at enhancer elements. Specifically, rapid and dynamic binding and unbinding of TFs makes the enhancer system highly tunable and sensitive to TF concentration fluctuations in the cell. Hierarchically ordered TF assembly at enhancers enables rapid and flexible regulatory outputs with high specificity (Fig. 2). These live-cell studies also support the existence of topological structures in the nucleus that shape local TF target search dynamics and potentially other gene-regulatory activities. Based on these results, we propose an enhancer-mediated regulatory mechanism driven by and dependent on dynamic weak protein–protein and protein–DNA interactions (Fig. 3) that is quite distinct from prevalent “textbook” models. An important feature of this revised model is that physical proximity (i.e., local high concentration of TFs and DNA elements) rather than direct stable lock and key–type interactions between distal enhancer elements and gene-proximal promoters may be sufficient to deliver transcription activation by TFs bound to distal cis-elements communicating with core promoters. We envision that cis-regulatory element clustering with its high local TF and cofactor concentrations accompanied by altered target search features may be sufficient to serve as an alternative mechanism for achieving distal enhancer–directed transcription activation (Fig. 3 D). In the future, it will be critical to further probe mechanistic links between TF dynamics, Pol II clustering, and transcription output along with a deeper understanding of the molecular basis and functional relevance of topological structures in living cells. To address these problems, we will need to devise strategies to image functionally linked events (e.g., simultaneous measurements of TF binding, 3D single locus gene position, and active mRNA production) at single-molecule resolution and in single living cells. With continued development of noninvasive multicolor imaging modalities (Schermelleh et al., 2008; Chen et al., 2014a; Balzarotti et al., 2017), more robust live-cell single locus labeling tools (Chen et al., 2013; Ochiai et al., 2015), and large-scale FISH platforms (Beliveau et al., 2015; Shachar et al., 2015), we foresee a promising future for more accurately delineating gene-regulatory mechanisms and deciphering the dynamic behavior of key TFs at single-cell and single-molecule resolution. Indeed, we are optimistic that we can extend these types of quantitative measurements to whole organisms in vivo and perhaps even throughout the course of embryonic development with next-generation deep-tissue imaging techniques (Ji, 2017).
Acknowledgments
We thank Claudia Cattoglio, Anders Sejr Hansen, Liangqi Xie, and Mustafa Mir for proofreading the manuscript.
This work is solely funded by Howard Hughes Medical Institute.
The authors declare no competing financial interests.
References
- Agarwal H., Reisser M., Wortmann C., and Gebhardt J.C.M.. 2017. Direct Observation of Cell-Cycle-Dependent Interactions between CTCF and Chromatin. Biophys. J. 112:2051–2055. 10.1016/j.bpj.2017.04.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axelrod D., Koppel D.E., Schlessinger J., Elson E., and Webb W.W.. 1976. Mobility measurement by analysis of fluorescence photobleaching recovery kinetics. Biophys. J. 16:1055–1069. 10.1016/S0006-3495(76)85755-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ball D.A., Mehta G.D., Salomon-Kent R., Mazza D., Morisaki T., Mueller F., McNally J.G., and Karpova T.S.. 2016. Single molecule tracking of Ace1p in Saccharomyces cerevisiae defines a characteristic residence time for non-specific interactions of transcription factors with chromatin. Nucleic Acids Res. 44:e160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balzarotti F., Eilers Y., Gwosch K.C., Gynnå A.H., Westphal V., Stefani F.D., Elf J., and Hell S.W.. 2017. Nanometer resolution imaging and tracking of fluorescent molecules with minimal photon fluxes. Science. 355:606–612. 10.1126/science.aak9913 [DOI] [PubMed] [Google Scholar]
- Barski A., Cuddapah S., Cui K., Roh T.Y., Schones D.E., Wang Z., Wei G., Chepelev I., and Zhao K.. 2007. High-resolution profiling of histone methylations in the human genome. Cell. 129:823–837. 10.1016/j.cell.2007.05.009 [DOI] [PubMed] [Google Scholar]
- Beagrie R.A., Scialdone A., Schueler M., Kraemer D.C.A., Chotalia M., Xie S.Q., Barbieri M., de Santiago I., Lavitas L.M., Branco M.R., et al. 2017. Complex multi-enhancer contacts captured by genome architecture mapping. Nature. 543:519–524. 10.1038/nature21411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beliveau B.J., Boettiger A.N., Avendaño M.S., Jungmann R., McCole R.B., Joyce E.F., Kim-Kiselak C., Bantignies F., Fonseka C.Y., Erceg J., et al. 2015. Single-molecule super-resolution imaging of chromosomes and in situ haplotype visualization using Oligopaint FISH probes. Nat. Commun. 6:7147 10.1038/ncomms8147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belmont A.S. 2014. Large-scale chromatin organization: the good, the surprising, and the still perplexing. Curr. Opin. Cell Biol. 26:69–78. 10.1016/j.ceb.2013.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betzig E., Patterson G.H., Sougrat R., Lindwasser O.W., Olenych S., Bonifacino J.S., Davidson M.W., Lippincott-Schwartz J., and Hess H.F.. 2006. Imaging intracellular fluorescent proteins at nanometer resolution. Science. 313:1642–1645. 10.1126/science.1127344 [DOI] [PubMed] [Google Scholar]
- Boettiger A.N., Bintu B., Moffitt J.R., Wang S., Beliveau B.J., Fudenberg G., Imakaev M., Mirny L.A., Wu C.T., and Zhuang X.. 2016. Super-resolution imaging reveals distinct chromatin folding for different epigenetic states. Nature. 529:418–422. 10.1038/nature16496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown R. 1828. A brief account of microscopical observations made in the months of June, July and August, 1827, on the particles contained in the pollen of plants; and on the general existence of active molecules in organic and inorganic bodies. Philos. Mag. 4:161–173. 10.1080/14786442808674769 [DOI] [Google Scholar]
- Brown J.M., Green J., das Neves R.P., Wallace H.A.C., Smith A.J.H., Hughes J., Gray N., Taylor S., Wood W.G., Higgs D.R., et al. 2008. Association between active genes occurs at nuclear speckles and is modulated by chromatin environment. J. Cell Biol. 182:1083–1097. 10.1083/jcb.200803174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buenrostro J.D., Giresi P.G., Zaba L.C., Chang H.Y., and Greenleaf W.J.. 2013. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat. Methods. 10:1213–1218. 10.1038/nmeth.2688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canals-Hamann A.Z., das Neves R.P., Reittie J.E., Iñiguez C., Soneji S., Enver T., Buckle V.J., and Iborra F.J.. 2013. A biophysical model for transcription factories. BMC Biophys. 6:2 10.1186/2046-1682-6-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B., Gilbert L.A., Cimini B.A., Schnitzbauer J., Zhang W., Li G.W., Park J., Blackburn E.H., Weissman J.S., Qi L.S., and Huang B.. 2013. Dynamic imaging of genomic loci in living human cells by an optimized CRISPR/Cas system. Cell. 155:1479–1491. 10.1016/j.cell.2013.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B.C., Legant W.R., Wang K., Shao L., Milkie D.E., Davidson M.W., Janetopoulos C., Wu X.S., Hammer J.A. III, Liu Z., et al. 2014a Lattice light-sheet microscopy: imaging molecules to embryos at high spatiotemporal resolution. Science. 346:1257998 10.1126/science.1257998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H., Farkas E.R., and Webb W.W.. 2008. Chapter 1: In vivo applications of fluorescence correlation spectroscopy. Methods Cell Biol. 89:3–35. 10.1016/S0091-679X(08)00601-8 [DOI] [PubMed] [Google Scholar]
- Chen J., Zhang Z., Li L., Chen B.C., Revyakin A., Hajj B., Legant W., Dahan M., Lionnet T., Betzig E., et al. 2014b Single-molecule dynamics of enhanceosome assembly in embryonic stem cells. Cell. 156:1274–1285. 10.1016/j.cell.2014.01.062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheutin T., McNairn A.J., Jenuwein T., Gilbert D.M., Singh P.B., and Misteli T.. 2003. Maintenance of stable heterochromatin domains by dynamic HP1 binding. Science. 299:721–725. 10.1126/science.1078572 [DOI] [PubMed] [Google Scholar]
- Ching R.W., Ahmed K., Boutros P.C., Penn L.Z., and Bazett-Jones D.P.. 2013. Identifying gene locus associations with promyelocytic leukemia nuclear bodies using immuno-TRAP. J. Cell Biol. 201:325–335. 10.1083/jcb.201211097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho W.K., Jayanth N., English B.P., Inoue T., Andrews J.O., Conway W., Grimm J.B., Spille J.H., Lavis L.D., Lionnet T., and Cisse I.I.. 2016. RNA Polymerase II cluster dynamics predict mRNA output in living cells. eLife. 5:e13617 10.7554/eLife.13617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirillo L.A., and Zaret K.S.. 1999. An early developmental transcription factor complex that is more stable on nucleosome core particles than on free DNA. Mol. Cell. 4:961–969. 10.1016/S1097-2765(00)80225-7 [DOI] [PubMed] [Google Scholar]
- Cirillo L.A., Lin F.R., Cuesta I., Friedman D., Jarnik M., and Zaret K.S.. 2002. Opening of compacted chromatin by early developmental transcription factors HNF3 (FoxA) and GATA-4. Mol. Cell. 9:279–289. 10.1016/S1097-2765(02)00459-8 [DOI] [PubMed] [Google Scholar]
- Cisse I.I., Izeddin I., Causse S.Z., Boudarene L., Senecal A., Muresan L., Dugast-Darzacq C., Hajj B., Dahan M., and Darzacq X.. 2013. Real-time dynamics of RNA polymerase II clustering in live human cells. Science. 341:664–667. 10.1126/science.1239053 [DOI] [PubMed] [Google Scholar]
- Consortium E.P., ENCODE Project Consortium 2012. An integrated encyclopedia of DNA elements in the human genome. Nature. 489:57–74. 10.1038/nature11247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook P.R. 1999. The organization of replication and transcription. Science. 284:1790–1795. 10.1126/science.284.5421.1790 [DOI] [PubMed] [Google Scholar]
- Courey A.J., and Tjian R.. 1988. Analysis of Sp1 in vivo reveals multiple transcriptional domains, including a novel glutamine-rich activation motif. Cell. 55:887–898. 10.1016/0092-8674(88)90144-4 [DOI] [PubMed] [Google Scholar]
- Crocker J., Abe N., Rinaldi L., McGregor A.P., Frankel N., Wang S., Alsawadi A., Valenti P., Plaza S., Payre F., et al. 2015. Low affinity binding site clusters confer hox specificity and regulatory robustness. Cell. 160:191–203. 10.1016/j.cell.2014.11.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahéron L., Opitz S.L., Zaehres H., Lensch M.W., Andrews P.W., Itskovitz-Eldor J., and Daley G.Q.. 2004. LIF/STAT3 signaling fails to maintain self-renewal of human embryonic stem cells. Stem Cells. 22:770–778. 10.1634/stemcells.22-5-770 [DOI] [PubMed] [Google Scholar]
- Dai C., Li W., Tjong H., Hao S., Zhou Y., Li Q., Chen L., Zhu B., Alber F., and Jasmine Zhou X.. 2016. Mining 3D genome structure populations identifies major factors governing the stability of regulatory communities. Nat. Commun. 7:11549 10.1038/ncomms11549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darzacq X., Shav-Tal Y., de Turris V., Brody Y., Shenoy S.M., Phair R.D., and Singer R.H.. 2007. In vivo dynamics of RNA polymerase II transcription. Nat. Struct. Mol. Biol. 14:796–806. 10.1038/nsmb1280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekker J., and Mirny L.. 2016. The 3D Genome as Moderator of Chromosomal Communication. Cell. 164:1110–1121. 10.1016/j.cell.2016.02.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekker J., Marti-Renom M.A., and Mirny L.A.. 2013. Exploring the three-dimensional organization of genomes: interpreting chromatin interaction data. Nat. Rev. Genet. 14:390–403. 10.1038/nrg3454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deluz C., Friman E.T., Strebinger D., Benke A., Raccaud M., Callegari A., Leleu M., Manley S., and Suter D.M.. 2016. A role for mitotic bookmarking of SOX2 in pluripotency and differentiation. Genes Dev. 30:2538–2550. 10.1101/gad.289256.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wit E., and de Laat W.. 2012. A decade of 3C technologies: insights into nuclear organization. Genes Dev. 26:11–24. 10.1101/gad.179804.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diao Y., Li B., Meng Z., Jung I., Lee A.Y., Dixon J., Maliskova L., Guan K.L., Shen Y., and Ren B.. 2016. A new class of temporarily phenotypic enhancers identified by CRISPR/Cas9-mediated genetic screening. Genome Res. 26:397–405. 10.1101/gr.197152.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doudna J.A., and Charpentier E.. 2014. The new frontier of genome engineering with CRISPR-Cas9. Science. 346:1258096 10.1126/science.1258096 [DOI] [PubMed] [Google Scholar]
- Dunn G.A., Dobbie I.M., Monypenny J., Holt M.R., and Zicha D.. 2002. Fluorescence localization after photobleaching (FLAP): a new method for studying protein dynamics in living cells. J. Microsc. 205:109–112. 10.1046/j.0022-2720.2001.001007.x [DOI] [PubMed] [Google Scholar]
- Elf J., Li G.W., and Xie X.S.. 2007. Probing transcription factor dynamics at the single-molecule level in a living cell. Science. 316:1191–1194. 10.1126/science.1141967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flyamer I.M., Gassler J., Imakaev M., Brandão H.B., Ulianov S.V., Abdennur N., Razin S.V., Mirny L.A., and Tachibana-Konwalski K.. 2017. Single-nucleus Hi-C reveals unique chromatin reorganization at oocyte-to-zygote transition. Nature. 544:110–114. 10.1038/nature21711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fudenberg G., and Imakaev M.. 2017. FISH-ing for captured contacts: towards reconciling FISH and 3C. Nat. Methods. 14:673–678. 10.1038/nmeth.4329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukaya T., Lim B., and Levine M.. 2016. Enhancer Control of Transcriptional Bursting. Cell. 166:358–368. 10.1016/j.cell.2016.05.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebhardt J.C., Suter D.M., Roy R., Zhao Z.W., Chapman A.R., Basu S., Maniatis T., and Xie X.S.. 2013. Single-molecule imaging of transcription factor binding to DNA in live mammalian cells. Nat. Methods. 10:421–426. 10.1038/nmeth.2411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh R.N., and Webb W.W.. 1994. Automated detection and tracking of individual and clustered cell surface low density lipoprotein receptor molecules. Biophys. J. 66:1301–1318. 10.1016/S0006-3495(94)80939-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodrich J.A., and Tjian R.. 2010. Unexpected roles for core promoter recognition factors in cell-type-specific transcription and gene regulation. Nat. Rev. Genet. 11:549–558. 10.1038/nrg2847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimm J.B., English B.P., Chen J., Slaughter J.P., Zhang Z., Revyakin A., Patel R., Macklin J.J., Normanno D., Singer R.H., et al. 2015. A general method to improve fluorophores for live-cell and single-molecule microscopy. Nat. Methods. 12:244–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakim O., Sung M.H., Voss T.C., Splinter E., John S., Sabo P.J., Thurman R.E., Stamatoyannopoulos J.A., de Laat W., and Hager G.L.. 2011. Diverse gene reprogramming events occur in the same spatial clusters of distal regulatory elements. Genome Res. 21:697–706. 10.1101/gr.111153.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halford S.E. 2009. An end to 40 years of mistakes in DNA-protein association kinetics? Biochem. Soc. Trans. 37:343–348. 10.1042/BST0370343 [DOI] [PubMed] [Google Scholar]
- Hansen A.S., Pustova I., Cattoglio C., Tjian R., and Darzacq X.. 2017a CTCF and cohesin regulate chromatin loop stability with distinct dynamics. eLife. 6:e25776 10.7554/eLife.25776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen A.S., Woringer M., Grimm J.B., Lavis L.D., Tjian R., and Darzacq X.. 2017b Spot-On: robust model-based analysis of single-particle tracking experiments. bioRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison M.M., Li X.Y., Kaplan T., Botchan M.R., and Eisen M.B.. 2011. Zelda binding in the early Drosophila melanogaster embryo marks regions subsequently activated at the maternal-to-zygotic transition. PLoS Genet. 7:e1002266 10.1371/journal.pgen.1002266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintzman N.D., and Ren B.. 2009. Finding distal regulatory elements in the human genome. Curr. Opin. Genet. Dev. 19:541–549. 10.1016/j.gde.2009.09.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hnisz D., Abraham B.J., Lee T.I., Lau A., Saint-André V., Sigova A.A., Hoke H.A., and Young R.A.. 2013. Super-enhancers in the control of cell identity and disease. Cell. 155:934–947. 10.1016/j.cell.2013.09.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hnisz D., Shrinivas K., Young R.A., Chakraborty A.K., and Sharp P.A.. 2017. A Phase Separation Model for Transcriptional Control. Cell. 169:13–23. 10.1016/j.cell.2017.02.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izeddin I., Recamier V., Bosanac L., Cisse I.I., Boudarene L., Dugast-Darzacq C., Proux F., Benichou O., Voituriez R., Bensaude O., et al. 2014. Single-molecule tracking in live cells reveals distinct target-search strategies of transcription factors in the nucleus. eLife. 3:e02230 10.7554/eLife.02230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaqaman K., Loerke D., Mettlen M., Kuwata H., Grinstein S., Schmid S.L., and Danuser G.. 2008. Robust single-particle tracking in live-cell time-lapse sequences. Nat. Methods. 5:695–702. 10.1038/nmeth.1237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji N. 2017. Adaptive optical fluorescence microscopy. Nat. Methods. 14:374–380. 10.1038/nmeth.4218 [DOI] [PubMed] [Google Scholar]
- Juven-Gershon T., and Kadonaga J.T.. 2010. Regulation of gene expression via the core promoter and the basal transcriptional machinery. Dev. Biol. 339:225–229. 10.1016/j.ydbio.2009.08.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura H., and Cook P.R.. 2001. Kinetics of core histones in living human cells: Little exchange of H3 and H4 and some rapid exchange of H2B. J. Cell Biol. 153:1341–1353. 10.1083/jcb.153.7.1341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosak S.T., Skok J.A., Medina K.L., Riblet R., Le Beau M.M., Fisher A.G., and Singh H.. 2002. Subnuclear compartmentalization of immunoglobulin loci during lymphocyte development. Science. 296:158–162. 10.1126/science.1068768 [DOI] [PubMed] [Google Scholar]
- Kwon I., Kato M., Xiang S., Wu L., Theodoropoulos P., Mirzaei H., Han T., Xie S., Corden J.L., and McKnight S.L.. 2013. Phosphorylation-regulated binding of RNA polymerase II to fibrous polymers of low-complexity domains. Cell. 155:1049–1060. 10.1016/j.cell.2013.10.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Treut G., Képès F., and Orland H.. 2016. Phase Behavior of DNA in the Presence of DNA-Binding Proteins. Biophys. J. 110:51–62. 10.1016/j.bpj.2015.10.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine M., Cattoglio C., and Tjian R.. 2014. Looping back to leap forward: transcription enters a new era. Cell. 157:13–25. 10.1016/j.cell.2014.02.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Liu H., Dong P., Li D., Legant W.R., Grimm J.B., Lavis L.D., Betzig E., Tjian R., and Liu Z.. 2016. Real-time imaging of Huntingtin aggregates diverting target search and gene transcription. eLife. 5:e17056 10.7554/eLife.17056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieberman-Aiden E., van Berkum N.L., Williams L., Imakaev M., Ragoczy T., Telling A., Amit I., Lajoie B.R., Sabo P.J., Dorschner M.O., et al. 2009. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 326:289–293. 10.1126/science.1181369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H., Dong P., Ioannou M.S., Li L., Shea J., Pasolli A., Grimm J., Rivlin P., Lavis L.D., Koyama M., et al. 2018. Visualizing Long-Term Single-Molecule Dynamics in vivo by Stochastic Protein Labeling. Proc. Natl. Acad. Sci. USA. 115:343–348. 10.1073/pnas.1713895115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z., Legant W.R., Chen B.C., Li L., Grimm J.B., Lavis L.D., Betzig E., and Tjian R.. 2014. 3D imaging of Sox2 enhancer clusters in embryonic stem cells. eLife. 3:e04236 10.7554/eLife.04236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z., Lavis L.D., and Betzig E.. 2015. Imaging live-cell dynamics and structure at the single-molecule level. Mol. Cell. 58:644–659. 10.1016/j.molcel.2015.02.033 [DOI] [PubMed] [Google Scholar]
- Luger K., Mäder A.W., Richmond R.K., Sargent D.F., and Richmond T.J.. 1997. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 389:251–260. 10.1038/38444 [DOI] [PubMed] [Google Scholar]
- Lukyanov K.A., Chudakov D.M., Lukyanov S., and Verkhusha V.V.. 2005. Innovation: Photoactivatable fluorescent proteins. Nat. Rev. Mol. Cell Biol. 6:885–890. 10.1038/nrm1741 [DOI] [PubMed] [Google Scholar]
- Magde D., Webb W.W., and Elson E.. 1972. Thermodynamic Fluctuations in a Reacting System - Measurement by Fluorescence Correlation Spectroscopy. Phys. Rev. Lett. 29:705–708. 10.1103/PhysRevLett.29.705 [DOI] [Google Scholar]
- Manley S., Gillette J.M., Patterson G.H., Shroff H., Hess H.F., Betzig E., and Lippincott-Schwartz J.. 2008. High-density mapping of single-molecule trajectories with photoactivated localization microscopy. Nat. Methods. 5:155–157. 10.1038/nmeth.1176 [DOI] [PubMed] [Google Scholar]
- Manzo C., and Garcia-Parajo M.F.. 2015. A review of progress in single particle tracking: from methods to biophysical insights. Rep. Prog. Phys. 78:124601 10.1088/0034-4885/78/12/124601 [DOI] [PubMed] [Google Scholar]
- Mao Y.S., Zhang B., and Spector D.L.. 2011. Biogenesis and function of nuclear bodies. Trends Genet. 27:295–306. 10.1016/j.tig.2011.05.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazza D., Abernathy A., Golob N., Morisaki T., and McNally J.G.. 2012. A benchmark for chromatin binding measurements in live cells. Nucleic Acids Res. 40:e119 10.1093/nar/gks701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNally J.G., Mueller W.G., Walker D., Wolford R., and Hager G.L.. 1999. The glucocorticoid receptor undergoes rapid exchange with regulatory sites in living cells. Mol. Biol. Cell. 10:326a. [DOI] [PubMed] [Google Scholar]
- McNally J.G., Müller W.G., Walker D., Wolford R., and Hager G.L.. 2000. The glucocorticoid receptor: rapid exchange with regulatory sites in living cells. Science. 287:1262–1265. 10.1126/science.287.5456.1262 [DOI] [PubMed] [Google Scholar]
- Mir M., Reimer A., Haines J.E., Li X.Y., Stadler M., Garcia H., Eisen M.B., and Darzacq X.. 2017. Dense Bicoid hubs accentuate binding along the morphogen gradient. Genes Dev. 31:1784–1794. 10.1101/gad.305078.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirny L., Slutsky M., Wunderlich Z., Tafvizi A., Leith J., and Kosmrlj A.. 2009. How a protein searches for its site on DNA: the mechanism of facilitated diffusion. J. Phys. A Math. Theor. 42:434013. [Google Scholar]
- Moen P.T. Jr., Johnson C.V., Byron M., Shopland L.S., de la Serna I.L., Imbalzano A.N., and Lawrence J.B.. 2004. Repositioning of muscle-specific genes relative to the periphery of SC-35 domains during skeletal myogenesis. Mol. Biol. Cell. 15:197–206. 10.1091/mbc.E03-06-0388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller F., Mazza D., Stasevich T.J., and McNally J.G.. 2010. FRAP and kinetic modeling in the analysis of nuclear protein dynamics: what do we really know? Curr. Opin. Cell Biol. 22:403–411. 10.1016/j.ceb.2010.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Normanno D., Boudarène L., Dugast-Darzacq C., Chen J., Richter C., Proux F., Bénichou O., Voituriez R., Darzacq X., and Dahan M.. 2015. Probing the target search of DNA-binding proteins in mammalian cells using TetR as model searcher. Nat. Commun. 6:7357 10.1038/ncomms8357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochiai H., Sugawara T., and Yamamoto T.. 2015. Simultaneous live imaging of the transcription and nuclear position of specific genes. Nucleic Acids Res. 43:e127 10.1093/nar/gkv624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou H.D., Phan S., Deerinck T.J., Thor A., Ellisman M.H., and O’Shea C.C.. 2017. ChromEMT: Visualizing 3D chromatin structure and compaction in interphase and mitotic cells. Science. 357:eaag0025 10.1126/science.aag0025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paakinaho V., Presman D.M., Ball D.A., Johnson T.A., Schiltz R.L., Levitt P., Mazza D., Morisaki T., Karpova T.S., and Hager G.L.. 2017. Single-molecule analysis of steroid receptor and cofactor action in living cells. Nat. Commun. 8:15896 10.1038/ncomms15896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel A., Lee H.O., Jawerth L., Maharana S., Jahnel M., Hein M.Y., Stoynov S., Mahamid J., Saha S., Franzmann T.M., et al. 2015. A Liquid-to-Solid Phase Transition of the ALS Protein FUS Accelerated by Disease Mutation. Cell. 162:1066–1077. 10.1016/j.cell.2015.07.047 [DOI] [PubMed] [Google Scholar]
- Patterson G.H., and Lippincott-Schwartz J.. 2002. A photoactivatable GFP for selective photolabeling of proteins and cells. Science. 297:1873–1877. 10.1126/science.1074952 [DOI] [PubMed] [Google Scholar]
- Percharde M., Lavial F., Ng J.H., Kumar V., Tomaz R.A., Martin N., Yeo J.C., Gil J., Prabhakar S., Ng H.H., et al. 2012. Ncoa3 functions as an essential Esrrb coactivator to sustain embryonic stem cell self-renewal and reprogramming. Genes Dev. 26:2286–2298. 10.1101/gad.195545.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Presman D.M., Ball D.A., Paakinaho V., Grimm J.B., Lavis L.D., Karpova T.S., and Hager G.L.. 2017. Quantifying transcription factor binding dynamics at the single-molecule level in live cells. Methods. 123:76–88. 10.1016/j.ymeth.2017.03.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ptashne M., and Gann A.. 1997. Transcriptional activation by recruitment. Nature. 386:569–577. 10.1038/386569a0 [DOI] [PubMed] [Google Scholar]
- Raj A., Peskin C.S., Tranchina D., Vargas D.Y., and Tyagi S.. 2006. Stochastic mRNA synthesis in mammalian cells. PLoS Biol. 4:e309 10.1371/journal.pbio.0040309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rust M.J., Bates M., and Zhuang X.. 2006. Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM). Nat. Methods. 3:793–796. 10.1038/nmeth929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schalch T., Duda S., Sargent D.F., and Richmond T.J.. 2005. X-ray structure of a tetranucleosome and its implications for the chromatin fibre. Nature. 436:138–141. 10.1038/nature03686 [DOI] [PubMed] [Google Scholar]
- Schermelleh L., Carlton P.M., Haase S., Shao L., Winoto L., Kner P., Burke B., Cardoso M.C., Agard D.A., Gustafsson M.G.L., et al. 2008. Subdiffraction multicolor imaging of the nuclear periphery with 3D structured illumination microscopy. Science. 320:1332–1336. 10.1126/science.1156947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sergé A., Bertaux N., Rigneault H., and Marguet D.. 2008. Dynamic multiple-target tracing to probe spatiotemporal cartography of cell membranes. Nat. Methods. 5:687–694. 10.1038/nmeth.1233 [DOI] [PubMed] [Google Scholar]
- Shachar S., Voss T.C., Pegoraro G., Sciascia N., and Misteli T.. 2015. Identification of Gene Positioning Factors Using High-Throughput Imaging Mapping. Cell. 162:911–923. 10.1016/j.cell.2015.07.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharonov A., and Hochstrasser R.M.. 2006. Wide-field subdiffraction imaging by accumulated binding of diffusing probes. Proc. Natl. Acad. Sci. USA. 103:18911–18916. 10.1073/pnas.0609643104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen H., Tauzin L.J., Baiyasi R., Wang W., Moringo N., Shuang B., and Landes C.F.. 2017. Single Particle Tracking: From Theory to Biophysical Applications. Chem. Rev. 117:7331–7376. 10.1021/acs.chemrev.6b00815 [DOI] [PubMed] [Google Scholar]
- Shin Y., and Brangwynne C.P.. 2017. Liquid phase condensation in cell physiology and disease. Science. 357:eaaf4382 10.1126/science.aaf4382 [DOI] [PubMed] [Google Scholar]
- Song F., Chen P., Sun D., Wang M., Dong L., Liang D., Xu R.M., Zhu P., and Li G.. 2014. Cryo-EM study of the chromatin fiber reveals a double helix twisted by tetranucleosomal units. Science. 344:376–380. 10.1126/science.1251413 [DOI] [PubMed] [Google Scholar]
- Sprague B.L., Pego R.L., Stavreva D.A., and McNally J.G.. 2004. Analysis of binding reactions by fluorescence recovery after photobleaching. Biophys. J. 86:3473–3495. 10.1529/biophysj.103.026765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stasevich T.J., Mueller F., Michelman-Ribeiro A., Rosales T., Knutson J.R., and McNally J.G.. 2010. Cross-validating FRAP and FCS to quantify the impact of photobleaching on in vivo binding estimates. Biophys. J. 99:3093–3101. 10.1016/j.bpj.2010.08.059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenoien D.L., Patel K., Mancini M.G., Dutertre M., Smith C.L., O’Malley B.W., and Mancini M.A.. 2001. FRAP reveals that mobility of oestrogen receptor-alpha is ligand- and proteasome-dependent. Nat. Cell Biol. 3:15–23. [DOI] [PubMed] [Google Scholar]
- Su W., Jackson S., Tjian R., and Echols H.. 1991. DNA looping between sites for transcriptional activation: self-association of DNA-bound Sp1. Genes Dev. 5:820–826. 10.1101/gad.5.5.820 [DOI] [PubMed] [Google Scholar]
- Swinstead E.E., Miranda T.B., Paakinaho V., Baek S., Goldstein I., Hawkins M., Karpova T.S., Ball D., Mazza D., Lavis L.D., et al. 2016. Steroid Receptors Reprogram FoxA1 Occupancy through Dynamic Chromatin Transitions. Cell. 165:593–605. 10.1016/j.cell.2016.02.067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teves S.S., An L., Hansen A.S., Xie L., Darzacq X., and Tjian R.. 2016. A dynamic mode of mitotic bookmarking by transcription factors. eLife. 5:e22280 10.7554/eLife.22280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsien R.Y. 1998. The green fluorescent protein. Annu. Rev. Biochem. 67:509–544. 10.1146/annurev.biochem.67.1.509 [DOI] [PubMed] [Google Scholar]
- Voss T.C., Schiltz R.L., Sung M.H., Yen P.M., Stamatoyannopoulos J.A., Biddie S.C., Johnson T.A., Miranda T.B., John S., and Hager G.L.. 2011. Dynamic exchange at regulatory elements during chromatin remodeling underlies assisted loading mechanism. Cell. 146:544–554. 10.1016/j.cell.2011.07.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Sawyer I.A., Sung M.H., Sturgill D., Shevtsov S.P., Pegoraro G., Hakim O., Baek S., Hager G.L., and Dundr M.. 2016a Cajal bodies are linked to genome conformation. Nat. Commun. 7:10966 10.1038/ncomms10966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Su J.H., Beliveau B.J., Bintu B., Moffitt J.R., Wu C.T., and Zhuang X.. 2016b Spatial organization of chromatin domains and compartments in single chromosomes. Science. 353:598–602. 10.1126/science.aaf8084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White M.D., Angiolini J.F., Alvarez Y.D., Kaur G., Zhao Z.W., Mocskos E., Bruno L., Bissiere S., Levi V., and Plachta N.. 2016. Long-Lived Binding of Sox2 to DNA Predicts Cell Fate in the Four-Cell Mouse Embryo. Cell. 165:75–87. 10.1016/j.cell.2016.02.032 [DOI] [PubMed] [Google Scholar]
- Williamson I., Berlivet S., Eskeland R., Boyle S., Illingworth R.S., Paquette D., Dostie J., and Bickmore W.A.. 2014. Spatial genome organization: contrasting views from chromosome conformation capture and fluorescence in situ hybridization. Genes Dev. 28:2778–2791. 10.1101/gad.251694.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wollman A.J., Shashkova S., Hedlund E.G., Friemann R., Hohmann S., and Leake M.C.. 2017. Transcription factor clusters regulate genes in eukaryotic cells. eLife. 6:e27451 10.7554/eLife.27451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodcock C.L., and Ghosh R.P. (2010). Chromatin Higher-order Structure and Dynamics. Csh Perspect Biol. 2. [DOI] [PMC free article] [PubMed]
- Xia T., Li N., and Fang X.. 2013. Single-molecule fluorescence imaging in living cells. Annu. Rev. Phys. Chem. 64:459–480. 10.1146/annurev-physchem-040412-110127 [DOI] [PubMed] [Google Scholar]
- Xie L., Torigoe S.E., Xiao J., Mai D.H., Li L., Davis F.P., Dong P., Marie-Nelly H., Grimm J., Lavis L., et al. 2017. A dynamic interplay of enhancer elements regulates Klf4 expression in naïve pluripotency. Genes Dev. 31:1795–1808. 10.1101/gad.303321.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Z.W., Roy R., Gebhardt J.C., Suter D.M., Chapman A.R., and Xie X.S.. 2014. Spatial organization of RNA polymerase II inside a mammalian cell nucleus revealed by reflected light-sheet superresolution microscopy. Proc. Natl. Acad. Sci. USA. 111:681–686. 10.1073/pnas.1318496111 [DOI] [PMC free article] [PubMed] [Google Scholar]