Abstract
Integrin-beta 1 (ITGB1) is aberrantly overexpressed or downregulated in solid cancers; however, its prognostic value remains controversial. Therefore, we conducted a meta-analysis to explore whether ITGB1 expression is correlated with overall survival (OS) and the clinicopathological characteristics of patients with solid cancers. We systematically searched the PubMed, Embase, and Web of Science databases for eligible studies published up to June 1, 2017. In total, 22 studies involving 3,666 patients were included. A sensitivity analysis was performed to assess the validity and reliability of the pooled OS. Among the 22 studies, 7 focused on lung cancer, 3 focused on colorectal cancer, 6 focused on breast cancer, 3 involved melanoma, and 3 involved pancreatic cancer. The pooled results showed that high ITGB1 expression was significantly associated with worse OS in lung cancer (pooled hazard ratio [HR]=1.78, 95% CI: 1.19–2.65, p<0.05) and breast cancer (pooled HR=1.88, 95% CI: 1.46–2.42, p<0.01). In addition, a significant association was observed between high ITGB1 expression and disease-free survival in breast cancer (pooled HR=1.63, 95% CI: 1.17–2.25, p<0.001) and pancreatic cancer (pooled HR=2.49, 95% CI: 1.35–4.61, p<0.001). However, high ITGB1 expression was not related to OS in colorectal cancer, pancreatic cancer, or melanoma. The pooled HRs used to evaluate the prognostic value of increased ITGB1 expression in lung cancer, breast cancer, and pancreatic cancer were not significantly altered, which indicates that the pooled results were robust. The results of this study indicate that the prognostic value of decreased ITGB1 expression varies among solid cancers.
Keywords: ITGB1, solid cancer, prognosis, meta-analysis
Introduction
Cancer is a major cause of death worldwide; in 2012, 14.1 million new cancer cases and 8.2 million cancer-related deaths were reported across the world.1 Currently, several therapeutic strategies, including radical operation, chemotherapy, and radiotherapy, are available for primary solid tumors and metastatic cancers. Nevertheless, the therapeutic response differs significantly in patients. Therefore, it is imperative to develop effective and clinically applicable biomarkers to accurately evaluate the therapeutic effect and prognosis of patients with cancer.
Integrins are a family of transmembrane receptors that generally consist of non-covalently linked alpha and beta subunits. Integrins have a variety of functions in cell adhesion and contact, and anchorage-dependent cell survival, and regulate various cellular processes including tissue healing, hemostasis, immune response, cell differentiation, division, growth, recognition, and migration.2–4 Recent evidence suggests that integrins are involved in many processes associated with tumor cell adhesion to the extracellular matrix, including migration, invasion, and metastasis.5–7 Moreover, previous studies have shown that integrins play an important role in the regulation of tumor angiogenesis.8,9 Additional investigations indicate that integrins can interact with tyrosine kinase receptors, such as epidermal growth factor receptor and vascular epidermal growth factor receptor, to promote cancer cell proliferation, survival, and differentiation.10
Integrin-beta 1 (ITGB1), also known as CD29, is a member of the integrin family and is composed of alpha and beta transmembrane subunits that form at least 24 distinct heterodimeric receptors.7,11 The role of ITGB1 in malignant phenotypes of cancers has gained a lot of attention.12 Previous studies have suggested that ITGB1 is the predominantly expressed integrin in normal and tumor cells and controls various developmental processes including angiogenesis, tumor progression, and metastasis.13–17 Other studies have also shown that, in many cancer types, ITGB1 may induce resistance to radiotherapies, chemotherapies, and targeted therapies.18–22 On the basis of these findings, ITGB1 has been studied extensively in terms of the biology of solid tumors, and its aberrant expression at the protein or RNA level has been shown to correlate with poor prognosis in several types of cancer, including lung cancer,23–26 gastric cancer,27 breast cancer,28–30 prostate cancer,31 pancreatic carcinoma,32,33 and colorectal cancer,34,35 making it a potential target for antitumor therapies. Nevertheless, the prognostic significance of ITGB1 expression in patients with cancer remains inconsistent. For instance, some studies have reported that decreased ITGB1 protein expression is associated with more aggressive breast cancer types,30,36 but the conclusion was different in other studies.28,37 Some studies could not verify a significant correlation between ITGB1 protein expression and survival of patients with breast carcinoma.29,38
Thus far, no meta-analyses of studies focusing on the investigation of the prognostic and clinicopathological significance of ITGB1 in patients with solid tumors have been performed. Furthermore, conclusions made from most studies published so far are limited by small sample sizes. Therefore, we conducted a meta-analysis to estimate the prognostic and clinicopathological value of ITGB1 in patients with solid cancers.
Materials and methods
Literature search strategy and study selection
We searched the PubMed, Embase, and Web of Science databases to identify all relevant studies that assessed the association between ITGB1 expression and survival outcome in patients with solid cancers published up to June 1, 2017. The search terms included the following terms: (“ITGB1” or “Integrin β1”), (“cancer” or “tumor” or “malignancy” or “carcinoma”), and (“prognos*”). The publication language was limited to English.
Studies were included in the analysis if they met the following inclusion criteria: 1) the studies investigated the association between ITGB1 and overall survival (OS) or disease-free survival (DFS) among patients with primary solid cancers or metastasis; 2) relevant clinicopathological characteristics were presented; 3) tumor tissues from patients with solid cancers were used for the determination of ITGB1 expression; 4) patients were grouped into high and low expression groups according to the ITGB1 protein or RNA level; and 5) sufficient information and data were available to calculate hazard ratios (HRs) with 95% CIs. The exclusion criteria were as follows: 1) studies that were published as reviews, abstracts, case reports, letters, or comments, as well as duplicate studies; 2) studies in which human cell lines or animals were used; and 3) studies that failed to provide the HRs with 95% CIs or Kaplan–Meier survival curves used to calculate the OS and DFS.
Data extraction and quality assessment
All the candidate publications were reviewed, and the data were extracted by 2 independent investigators. The third investigator was responsible for reconciling disagreements when the results were controversial. The following information was extracted: cancer type, first author’s name, publication year, region, number of patients, patients’ ages, test method, rate of high ITGB1 expression, clinical and pathological features, follow-up duration, OS, and DFS. If the results of both the univariate and multivariate analyses were provided in the studies, only the latter were extracted owing to their higher accuracy, since multivariate analyses account for confounding factors. Moreover, the selection of participants, comparability, and ascertainment of outcomes were assessed. The study quality was assessed using the standard Newcastle–Ottawa Scale, which ranges from 0 (minimum) to 9 (maximum). If the final score of a study was higher, it indicated that the study’s methodological quality was better. A study with a score of 6 or higher was defined as “high-quality.”
Statistical analysis
The meta-analysis was performed using Stata SE12.0 (Stata Corp., College Station, TX, USA). HRs and 95% CIs were used to assess the prognostic value of ITGB1, and odds ratios (ORs) with 95% CIs were used to evaluate the association between ITGB1 expression and clinicopathological features of solid cancers. A sensitivity analysis was performed to assess the validity and reliability of the pooled OS in patients with lung cancer, breast cancer, and pancreatic cancer. Chi-square-based Q-tests and I2 statistics were applied to evaluate study heterogeneity, with I2>50% and p<0.05 indicating statistical heterogeneity. If no large statistical heterogeneity was detected, a fixed-effects model was used to assess the pooled HRs; otherwise, a random-effects model was used. Egger’s test and Begg’s test should have been applied to investigate publication bias, but the tests were not conducted owing to the small number of included studies used to assess the specific outcomes of interest.
Results
Study selection and study characteristics
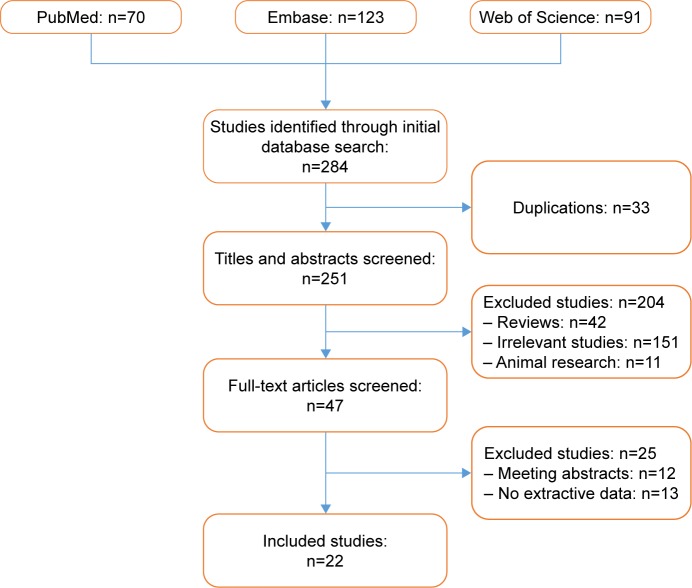
A total of 284 articles were primarily identified, 70 of which were from PubMed, 123 from Embase, and 91 from Web of Science. After duplicate publications were removed and the remaining abstracts and full texts meticulously reviewed, 22 publications were finally determined to be eligible for the present pooled analysis;24–26,28,29,33–35,39–52 the inclusion of the publications in the analysis was based on the selection criteria mentioned above. The detailed selection process is shown in Figure 1.
Figure 1.
Flow diagram of study selection.
The basic characteristics of the included studies are summarized in Table 1. In all, 22 studies involving 3,666 patients were included in the current meta-analysis, the sample size of which ranged from 30 to 959. All the included studies were published in English. The recruitment period of patients ranged from 1974 to 2014. Among the 22 studies, 7 focused on lung cancer,24–26,39–42 3 focused on colorectal cancer,34,35,43 6 focused on breast cancer,28,29,44–47 3 involved melanoma,48–50 and 3 involved pancreatic cancer.33,51,52 Moreover, 7 studies were performed in China and 3 studies were performed in the USA. The majority of the studies used immunohistochemistry to detect ITGB1 protein levels, while 3 studies used real-time quantitative reverse transcription polymerase chain reaction (Table 1). The study quality scores ranged from 5 to 7, which indicated that the quality of the included studies was moderate to high (Table 2).
Table 1.
Baseline characteristics of studies included in the meta-analysis
| Cancer type | First author (year) | Region | Time of recruitment | No of patients | Age (years), median (range) | Test method | Rate of high ITGB1 expression (%) | Survival outcomes (HR, 95% CI), H/L | Follow-up (months), median | NOS score |
|---|---|---|---|---|---|---|---|---|---|---|
| Small cell lung cancer | Chang et al25 (2012) | Korea | 2000–2008 | 112 | 66 (42–88) | IHC | 57.1 | OS: 0.98 (0.66–1.45) | 61 | 7 |
| Non-small cell lung cancer | Dingemans et al26 (2010) | the Netherlands | 1995–1999 | 68 | 69.1 (44.5–90.2) | Real-time qPCR | NR | OS: 2.11 (1.21–3.69); DFS: 1.53 (0.87–2.70) |
93 | 6 |
| Small cell lung cancer | Lawson et al39 (2010) | UK | 1998–2005 | 151 | 67.34 (9.93) | IHC | 70.2 | OS: 1.67 (1.09–2.56) | 72 | 7 |
| Non-small cell lung | Liang et al40 | China | NR | 134 | NR | IHC | NR | OS: 1.56 (0.96–2.53) | NR | 5 |
| cancer | (2017) | |||||||||
| Non-small cell lung cancer | Okamura et al41 (2007) | Japan | 1997–2000 | 118 | 65 (41–79) | IHC | 66.9 | OS: 1.41 (0.80–2.49) | 71.0 | 7 |
| Lung adenocarcinoma | Zhang et al42 (2013) | China | 2004–2006 | 108 | NR | IHC | 32.4 | RFS: 6.592 (4.106–10.584); OS: 6.183 (3.851–9.929) |
60 | 7 |
| Non-small cell lung cancer | Zheng et al24 (2016) | China | NR | 959 | 67 (38–90) | TCGA RNASeq database | NR | OS: 1.31 (1.10–1.55) | NR | 5 |
| Colorectal cancer | Langan et al43 (2012) | USA | NA | 30 | 56 (35–76) | IHC | NR | OS: 0.28 (0.06–1.27) | 113 | 6 |
| Colorectal cancer | Liu et al35 (2015) | China | 2001–2013 | 582 | NR | IHC | 52.6 | OS: 1.537 (1.147–2.059) | 100 | 6 |
| Colorectal liver metastases | Vassos et al34 (2014) | Germany | 1995–2010 | 81 | NR | IHC | 45.7 | OS: 1.23 (0.44–3.43); DFS: 0.95 (0.44–2.01) |
55 | 6 |
| HER-2-positive metastatic breast cancer | Lesniak et al44 (2009) | Canada | 1998–2002 | 78 | NR | IHC | 29.5 | OS: 2.82 (1.52–4.24) | 28 | 6 |
| Breast cancer | McSherry et al45 (2009) | Ireland | 1988–1992 | 272 | NR | IHC | NR | OS: 1.16 (0.62–2.19) | NR | 6 |
| Breast cancer | Petricevic et al29 (2012) | Croatia | 2002–2003 | 133 | NR | IHC | 36.4 | NR | 60 | 7 |
| Breast cancer | dos Santos et al46 (2012) | Brazil | 1994–2010 | 225 | 55 | IHC | 32.9 | DSS: 2.51 (1.24–3.7); DFS: 1.38 (0.86–2.23); MFS: 1.94 (1.07–3.52) |
NR | 7 |
| Breast cancer | Yao et al28 (2007) | USA | 1974–1999 | 149 | NR | IHC | 10.1 | OS: 1.73 (1.22–2.44); DFS: 1.88 (1.20–2.95) |
60 | 7 |
| Breast cancer | Yin et al47 (2016) | China | NR | 67 | 51 | IHC | 43.3 | OS: 2.772 (1.011–7.600) | NR | 6 |
| Melanoma | Hieken et al48 (1999) | USA | 1976–1995 | 111 | 53 | IHC | 32.4 | OS: 3.10 (0.9–10.73) | 105 | 7 |
| Melanoma | Nikkola et al49 (2004) | Finland | 1989–1995 | 55 | 47 | IHC | 10.7 | DFS: 3.36 (2.03–5.54) | NR | 6 |
| Melanoma | Vihinen et al50 (2000) | Finland | NR | 38 | NR | IHC | 28.9 | OS: 0.43 (0.12–1.59); DFS: 0.21 (0.07–0.60) |
NR | 5 |
| Pancreatic cancer | Sawai et al51 (2006) | Japan | NR | 78 | 65.1 | IHC | NR | Training set OS: 0.909 (0.569–1.410); validation set OS: 0.782 (0.505–1.355) | 17.6 | 7 |
| Pancreatic ductal adenocarcinoma | Yang et al52 (2016) | China | 2012–2014 | 63 | NR | IHC | 54.0 | OS: 2.375 (1.186–5.194); RFS: 2.488 (1.197–5.170) |
NR | 6 |
| Pancreatic cancer | Zhou et al33 (2013) | China | 2008–2012 | 54 | 56 | RT-PCR | 50 | OS: 3.2 (1.2–6.7); DFS: 2.5 (0.7–6.8) |
NR | 6 |
Note: H/L is high ITGB1 expression vs low ITGB1 expression.
Abbreviations: ITGB1, integrin-beta 1; NOS, Newcastle–Ottawa Scale; HER, human epidermal growth factor receptor; IHC, immunohistochemistry; qPCR, quantitative polymerase chain reaction; TCGA, The Cancer Genome Atlas; RT-PCR, reverse transcription polymerase chain reaction; OS, overall surviva ; DFS, disease-free survival; RFS, recurrence-free survival; DSS, disease-specific survival; MFS, metastatic-free survival; NR, not reported; NA, not applicable; HR, hazard ratio.
Table 2.
Methodological quality assessment (risk of bias) of included studies by the Newcastle–Ottawa Scale
| Study | Selection
|
Comparability | Outcome
|
Total | |||||
|---|---|---|---|---|---|---|---|---|---|
| Exposed cohort | Non-exposed cohort | Ascertainment of exposure | Outcome of interest | Assessment of outcome | Length of follow-up | Adequacy of follow-up | |||
| Chang et al25 (2012) | ★ | ★ | ★ | ★ | ★☆ | ★ | ★ | ☆ | 7 |
| Dingemans et al26 (2010) | ★ | ★ | ★ | ☆ | ★☆ | ☆ | ★ | ★ | 6 |
| Lawson et al39 (2010) | ★ | ★ | ★ | ★ | ★☆ | ☆ | ★ | ★ | 7 |
| Liang et al40 (2017) | ☆ | ★ | ★ | ☆ | ★☆ | ☆ | ★ | ★ | 5 |
| Okamura et al41 (2007) | ★ | ★ | ★ | ★ | ★☆ | ☆ | ★ | ★ | 7 |
| Zhang et al42 (2013) | ☆ | ★ | ★ | ★ | ★☆ | ★ | ★ | ★ | 7 |
| Zheng et al24 (2016) | ☆ | ★ | ★ | ☆ | ★☆ | ☆ | ★ | ★ | 5 |
| Langan et al43 (2012) | ★ | ★ | ★ | ☆ | ★☆ | ☆ | ★ | ★ | 6 |
| Liu et al35 (2015) | ☆ | ★ | ★ | ★ | ★☆ | ☆ | ★ | ★ | 6 |
| Vassos et al34 (2014) | ☆ | ★ | ★ | ★ | ★☆ | ☆ | ★ | ★ | 6 |
| Lesniak et al44 (2009) | ★ | ★ | ★ | ★ | ★☆ | ☆ | ☆ | ★ | 6 |
| McSherry et al45 (2009) | ★ | ★ | ★ | ★ | ★☆ | ★ | ☆ | ☆ | 6 |
| Petricevic et al29 (2012) | ★ | ★ | ☆ | ★ | ★☆ | ★ | ★ | ★ | 7 |
| dos Santos et al46 (2012) | ★ | ★ | ★ | ★ | ★☆ | ★ | ★ | ☆ | 7 |
| Yao et al28 (2007) | ★ | ★ | ☆ | ★ | ★☆ | ★ | ★ | ★ | 7 |
| Yin et al47 (2016) | ☆ | ★ | ★ | ★ | ★☆ | ★ | ★ | ☆ | 6 |
| Hieken et al48 (1999) | ★ | ★ | ☆ | ★ | ★☆ | ★ | ★ | ★ | 7 |
| Nikkola et al49 (2004) | ☆ | ★ | ★ | ★ | ★☆ | ★ | ☆ | ★ | 6 |
| Vihinen et al50 (2000) | ☆ | ☆ | ★ | ★ | ★☆ | ★ | ☆ | ★ | 5 |
| Sawai et al51 (2006) | ★ | ★ | ★ | ★ | ★☆ | ★ | ☆ | ★ | 7 |
| Yang et al52 (2016) | ☆ | ★ | ★ | ★ | ★☆ | ★ | ★ | ☆ | 6 |
| Zhou et al33 (2013) | ☆ | ★ | ★ | ★ | ★☆ | ★ | ★ | ☆ | 6 |
Notes: For each domain, either a “star” or “white star” is assigned, with a “star” indicating that the study design element was considered adequate and less likely to introduce bias. A maximum of 2 stars could be given for Comparability. A study could receive a maximum of 10 stars.
High ITGB1 expression and OS in solid cancers
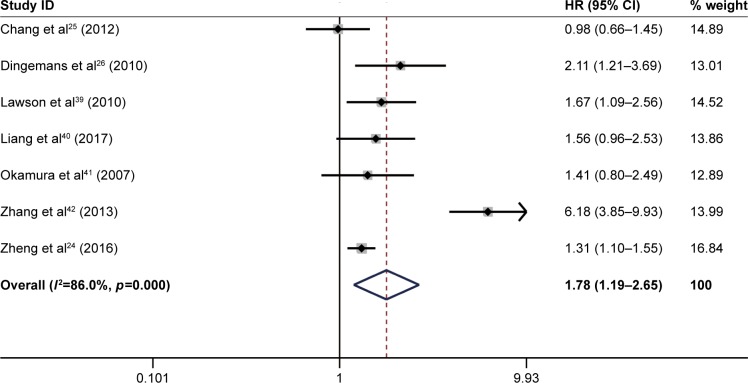
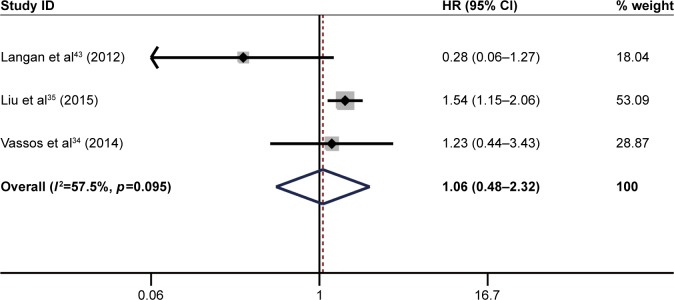
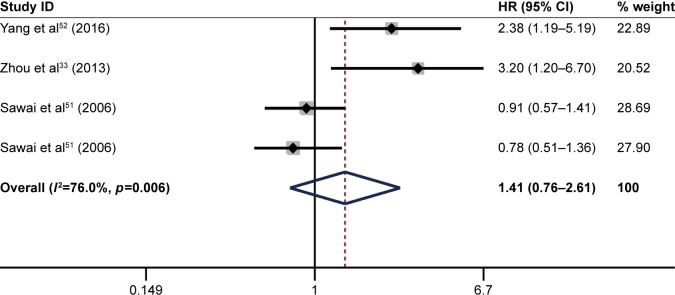
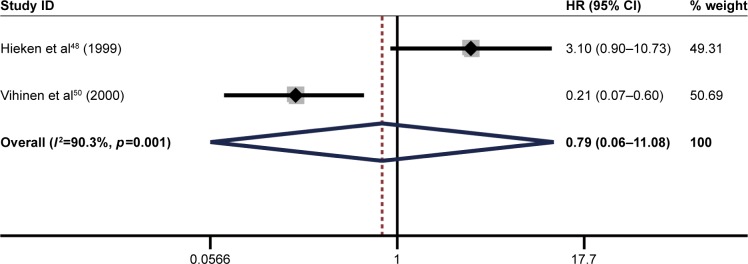
The pooled result revealed that high ITGB1 expression was significantly associated with worse OS in patients with lung cancer (pooled HR=1.78, 95% CI: 1.19–2.65, p<0.05) (Figure 2) and breast cancer (pooled HR=1.88, 95% CI: 1.46–2.42, p<0.01) (Figure 3). In addition, no significant association was found between high ITGB1 expression and OS in colorectal cancer (pooled HR=1.06, 95% CI: 0.48–2.32, p<0.001) (Figure 4), pancreatic cancer (pooled HR=1.41, 95% CI: 0.76–2.61, p<0.0001) (Figure 5), or melanoma (pooled HR=0.79, 95% CI: 0.06–11.08, p=0.01) (Figure 6).
Figure 2.
Results of pooled hazard ratios of overall survival of patients with high integrin-beta 1 (ITGB1) expression level in lung cancer.
Note: Weights are from random effects analysis.
Abbreviation: HR, hazard ratio.
Figure 3.
Results of pooled hazard ratios of overall survival of patients with high integrin-beta 1 (ITGB1) expression level in breast cancer.
Abbreviation: HR, hazard ratio.
Figure 4.
Results of pooled hazard ratios of overall survival of patients with high integrin-beta 1 (ITGB1) expression level in colorectal cancer.
Note: Weights are from random effects analysis.
Abbreviation: HR, hazard ratio.
Figure 5.
Results of pooled hazard ratios of overall survival of patients with high integrin-beta 1 (ITGB1) expression level in pancreatic cancer.
Note: Weights are from random effects analysis.
Abbreviation: HR, hazard ratio.
Figure 6.
Results of pooled hazard ratios of overall survival of patients with high integrin-beta 1 (ITGB1) expression level in melanoma.
Note: Weights are from random effects analysis.
Abbreviation: HR, hazard ratio.
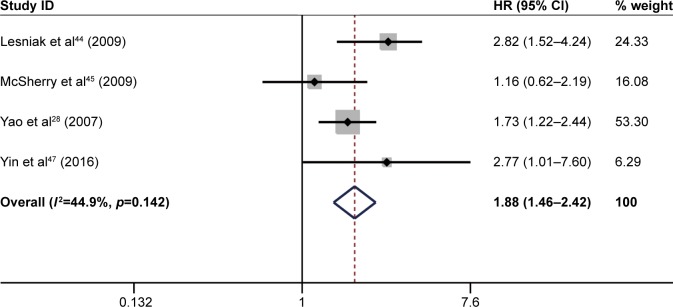
High ITGB1 expression and DFS in solid cancers
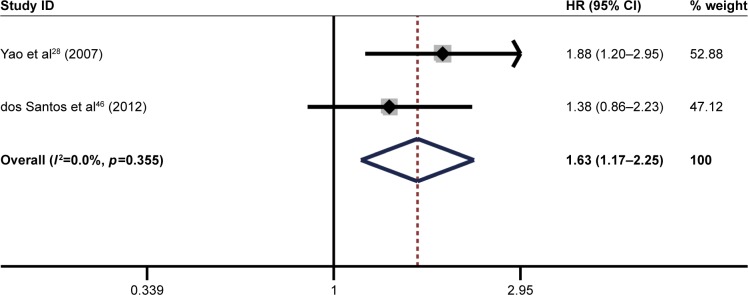
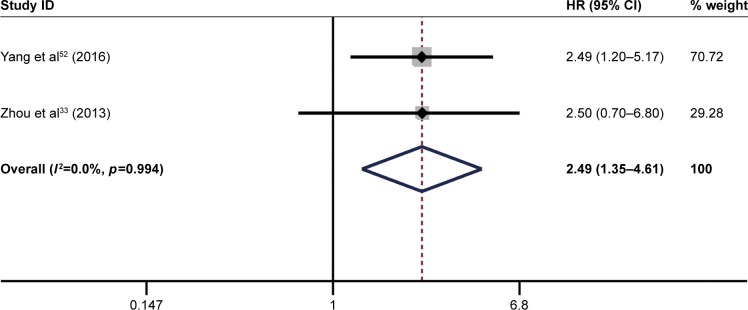
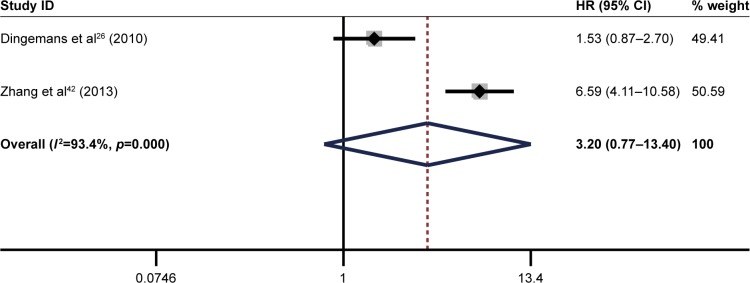
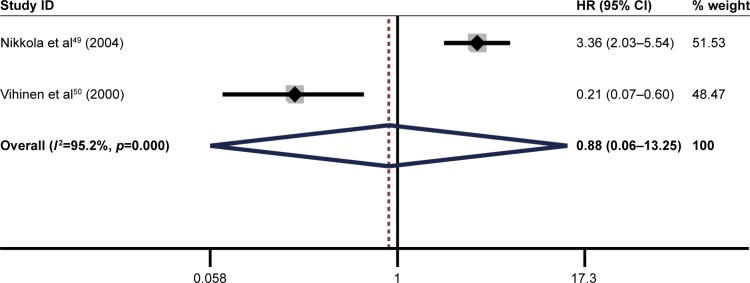
The pooled result revealed that high ITGB1 expression was significantly associated with worse DFS in patients with breast cancer (pooled HR=1.63, 95% CI: 1.17–2.25, p<0.001) (Figure 7) and pancreatic cancer (pooled HR=2.49, 95% CI: 1.35–4.61, p<0.001) (Figure 8). In addition, no significant association was found between high ITGB1 expression and DFS in lung cancer (pooled HR=3.20, 95% CI: 0.77–13.40, p<0.01) (Figure 9) or melanoma (pooled HR=0.88, 95% CI: 0.06–13.25, p<0.05) (Figure 10).
Figure 7.
Results of pooled hazard ratios of disease-free survival of patients with high integrin-beta 1 (ITGB1) expression level in breast cancer.
Abbreviation: HR, hazard ratio.
Figure 8.
Results of pooled hazard ratios of disease-free survival of patients with high integrin-beta 1 (ITGB1) expression level in lung cancer.
Note: Weights are from random effects analysis.
Abbreviation: HR, hazard ratio.
Figure 9.
Results of pooled hazard ratios of disease-free survival of patients with high integrin-beta 1 (ITGB1) expression level in pancreatic cancer.
Note: Weights are from random effects analysis.
Abbreviation: HR, hazard ratio.
Figure 10.
Results of pooled hazard ratios of disease-free survival of patients with high integrin-beta 1 (ITGB1) expression level in melanoma.
Note: Weights are from random effects analysis.
Abbreviation: HR, hazard ratio.
High ITGB1 expression and clinicopathological factors in lung cancer
Considering that the biology, pathology, clinical courses, and treatments vary enormously among different types of solid cancers, we assessed the associations between high ITGB1 expression and clinicopathological characteristics in lung cancer, breast cancer, and pancreatic cancer. However, high expression of ITGB1 was not evaluated in colorectal cancer and melanoma owing to limited data on the clinicopathological features. Six studies reported a relationship between high ITGB1 expression and clinicopathological factors in lung cancer, including 2 studies that investigated tumor differentiation, N stage, T stage, and age (Table 3). Except for T stage (I2=0, p=0.50) and age (I2=5%, p=0.305), significant heterogeneity was observed between high ITGB1 expression and tumor differentiation (I2=85%, p=0.01) and N stage (I2=74.7% p=0.047). The pooled analysis showed no significant association between high ITGB1 expression and tumor differentiation (OR=0.69, 95% CI: 0.10–4.98, p=0.712), N stage (OR=0.40, 95% CI: 0.16–1.01, p=0.053), or age (OR=0.98, 95% CI: 0.70–1.37, p=0.921) (Table 3), while high ITGB1 expression showed a strong association with worse T stage (OR=0.64, 95% CI: 0.41–0.98, p=0.041) (Table 3).
Table 3.
Meta-analysis results of the associations of integrin-beta 1 with clinicopathological parameters
| Clinicopathological parameter | No of studies | Reference no | OR (95% CI) | p-value | Heterogeneity test
|
|
|---|---|---|---|---|---|---|
| I2 (%) | p-value | |||||
| Lung cancer | ||||||
| Tumor differentiation (moderate–well vs poor) | 2 | 637 | 0.69 (0.10–4.98) | 0.712 | 85.0 | 0.010 |
| N stage (N0 vs N+) | 2 | 663 | 0.40 (0.160–1.01) | 0.053 | 74.7 | 0.047 |
| T stage (T1–2 vs T3–4) | 2 | 663 | 0.64 (0.41–0.98) | 0.041 | 0.0 | 0.5 |
| Age (<60 vs ≥60 years) | 2 | 665 | 0.98 (0.70–1.37) | 0.921 | 5.0 | 0.305 |
| Breast cancer | ||||||
| N stage (N0 vs N+) | 3 | 399 | 0.79 (0.52–1.19) | 0.262 | 2.0 | 0.360 |
| Tumor grade (I–II vs III–IV) | 3 | 424 | 1.20 (0.74–1.94) | 0.462 | 0.0 | 0.636 |
| T stage (T1 vs T2–3) | 3 | 424 | 0.78 (0.51–1.19) | 0.250 | 0.0 | 0.639 |
| Metastasis (yes vs no) | 2 | 292 | 1.99 (1.18–3.38) | 0.010 | 0.0 | 0.357 |
| ER (+ vs −) | 2 | 352 | 1.45 (0.90–2.35) | 0.126 | 14.8 | 0.279 |
| PR (+ vs −) | 2 | 352 | 1.15 (0.74–1.80) | 0.527 | 43.6 | 0.183 |
| HER (+ vs −) | 2 | 357 | 1.70 (0.98–2.93) | 0.057 | 29.9 | 0.232 |
| Pancreatic cancer | ||||||
| Age (<60 vs ≥60 years) | 2 | 117 | 1.35 (0.65–2.80) | 0.417 | 0.0 | 0.459 |
Abbreviations: ER, estrogen receptor; PR, progesterone receptor; HER, human epidermal growth factor receptor; OR, odds ratio.
High ITGB1 expression and clinicopathological factors in breast cancer
A total of 6 studies identified an association between ITGB1 expression and clinicopathological factors in breast cancer. No significant heterogeneity was identified between high ITGB1 expression and N stage (I2=2%, p=0.36), tumor grade (I2=0, p=0.636), T stage (I2=0, p=0.639), metastasis (I2=0, p=0.357), estrogen receptor (ER) (I2=14.8%, p=0.279), progesterone receptor (PR) (I2=43.6%, p=0.183), or human epidermal growth factor receptor (HER) (I2=29.9%, p=0.232); therefore, a fixed-effects model was used for the analysis (Table 3). The pooled analysis revealed no significant relationship between increased ITGB1 expression and N stage (OR=0.79, 95% CI: 0.52–1.19, p=0.262), tumor grade (OR=1.20, 95% CI: 0.74–1.94, p=0.462), T stage (OR=0.78, 95% CI: 0.51–1.19, p=0.25), ER (OR=1.45, 95% CI: 0.90–2.35, p=0.462), PR (OR=1.15, 95% CI: 0.74–1.80, p=0.527), or HER (OR=1.70, 95% CI: 0.98–2.93, p=0.057), while high ITGB1 expression demonstrated a strong association with metastasis (OR=1.99, 95% CI: 1.18–3.38, p=0.010) (Table 3).
ITGB1 expression and clinicopathological factors in pancreatic cancer
Only 2 studies reported a relationship between ITGB1 expression and age in pancreatic cancer. No significant heterogeneity was detected in the studies regarding age (I2=0, p=0.459); therefore, a fixed-effects model was applied. The pooled results showed that high ITGB1 expression was not significantly correlated with age (OR=1.35, 95% CI: 0.65–2.80, p=0.417) (Table 3).
Sensitivity analysis
Sensitivity analyses were performed to evaluate the stability of the pooled results for OS in lung cancer (Figure 11), breast cancer (Figure 12), and pancreatic cancer (Figure 13). The results of the sensitivity analyses showed that the pooled HRs for OS did not change substantially, which indicates that the conclusions from our meta-analysis were relatively reliable.
Figure 11.
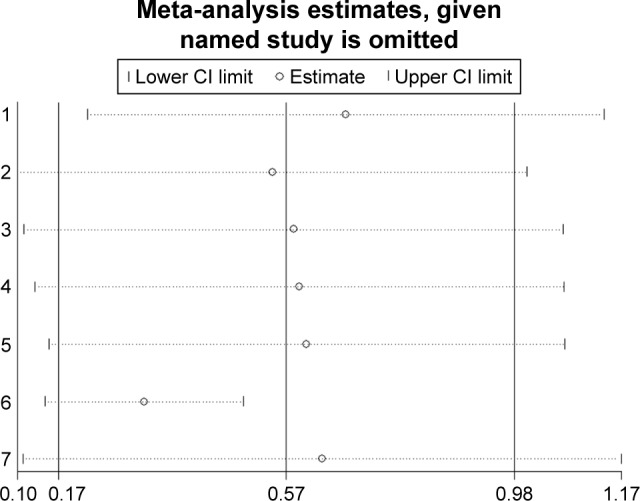
Sensitivity analysis of pooled hazard ratios of overall survival of patients with high integrin-beta 1 (ITGB1) expression level in lung cancer.
Figure 12.
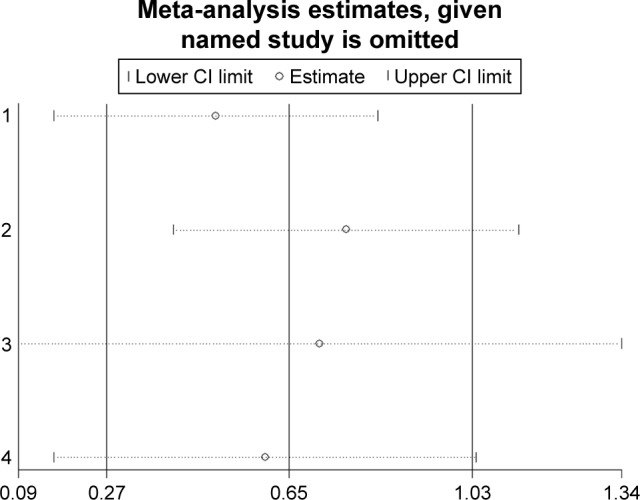
Sensitivity analysis of pooled hazard ratios of overall survival of patients with high integrin-beta 1 (ITGB1) expression level in breast cancer.
Figure 13.
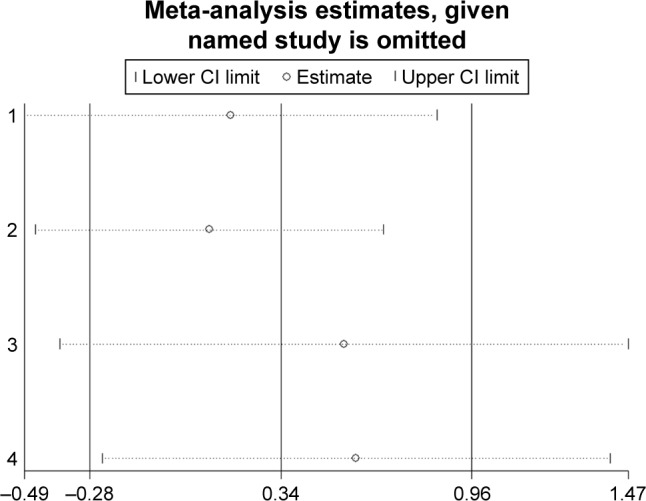
Sensitivity analysis of pooled hazard ratios of overall survival of patients with high integrin-beta 1 (ITGB1) expression level in pancreatic cancer.
Publication bias
Begg’s and Egger’s tests were used to assess publication bias in our meta-analysis. The results revealed that there was no significant bias in the pooled HRs for OS in lung cancer (Begg’s test, z=1.05, p=0.293; Egger’s test, t-bias=1.10, p=0.320). Publication bias was not observed when assessing for potential bias in the pooled HRs for the correlation between high ITGB1 expression and DFS, owing to the limited number of studies investigating this relationship.
Discussion
Studies have shown conflicting results regarding the role of ITGB1 in the carcinogenesis of solid cancers;28–30 therefore, its prognostic value in patients with solid cancers is also inconsistent and remains unknown. Although several previous studies reported that ITGB1 could induce resistance to chemotherapy and radiation in several human cancers,53,54 the impact of increased ITGB1 expression on the prognosis of patients with solid cancers has not been fully explored. Hence, we combined 22 publications involving 3,666 patients to perform the first meta-analysis that evaluated the association between ITGB1 and OS and DFS in patients with solid cancers. In addition, we explored the relationship between low ITGB1 expression and the clinicopathological features of solid cancers.
The pooled results of the meta-analysis revealed that high ITGB1 expression was significantly associated with worse OS in lung cancer and breast cancer. Moreover, the results showed that high ITGB1 expression was not related to OS in colorectal cancer, pancreatic cancer, or melanoma. In addition, the pooled result revealed that high ITGB1 expression was significantly associated with worse DFS in breast cancer and pancreatic cancer. The sensitivity analyses demonstrated that the pooled HRs used to evaluate the prognostic value of increased ITGB1 expression in lung cancer, breast cancer, and pancreatic cancer were not significantly altered, which indicates that the pooled results were robust. Considering the above findings, we believe that the prognostic value of increased ITGB1 expression varies according to cancer type in patients with solid cancer.
In addition, we investigated the relationship between ITGB1 expression and clinicopathological characteristics to further validate the pooled results of the association between OS and ITGB1 expression. Increased ITGB1 expression was significantly associated with worse T stage in lung cancer. However, the results showed no significant association between high ITGB1 expression and tumor differentiation and N stage. In addition, high ITGB1 expression strongly correlated with metastasis in breast cancer. Similarly, the results revealed no significant relationship between increased ITGB1 expression and N stage, tumor grade, T stage, ER, PR, or HER in breast cancer. This result should be interpreted with caution. On the one hand, the prognostic value and association between ITGB1 expression and the clinicopathological features of lung cancer and breast cancer may vary with the subtype of cancer. On the other hand, the included sample size was relatively small, which may have led to statistical bias. Previous studies have verified that ITGB1 overexpression promoted lymph-node metastasis in lung cancer.42,55 Bredin et al found that ITGB1 was involved in lung cancer cell migration in vitro,56 and upregulation of ITGB1 was identified as an important factor for gefitinib resistance in a lung cancer cell line.57 In addition, the overexpression of ITGB1 was found to promote the invasion and metastasis of lung cancer.25 In addition, Klahan et al reported that knockdown of ITGB1 inhibited the migration and invasion of breast cancer cells. Therefore, to date, the published literature supports the notion that the effects of ITGB1 on the biological functions of solid cancer cells vary according to the type of malignant cells, which is consistent with the results of our meta-analysis.
Our study had several significant limitations; therefore, the results should be interpreted with caution. First, only English language publications were included in this meta-analysis, which may have introduced publication bias. Second, only 3 studies on colorectal cancer, melanoma, and pancreatic cancer were included; therefore, it was difficult to accurately assess the association between ITGB1 expression and survival in these 3 cancer types. Third, although the HRs with 95% CIs in most of the included studies were calculated via a multivariate analysis, the variables added into the Cox proportional hazard models varied across studies. Fourth, all the included studies failed to carefully describe the location of the ITGB1 expression in the tumor specimens; therefore, further studies should be performed to determine whether the increased ITGB1 expression appears at cell-matrix attachment sites or in the epithelial or stromal regions. Fifth, this meta-analysis did not explore the correlation between increased ITGB1 expression and ER+/HER2+ and triple-negative breast cancers owing to the unavailability of data. Sixth, our study failed to determine whether high ITGB1 expression was associated with the type of metastasis. Last but not least, the sample size and the number of the included studies for the pooled estimate of clinicopathological parameters were rather small, which may have affected the reliability of the pooled ORs for the associations between high ITGB1 expression and clinicopathological parameters.
Conclusion
The prognostic significance of increased ITGB1 expression varies according to cancer type. Overall, high ITGB1 expression was significantly associated with a worse OS in lung cancer and breast cancer and a worse DFS in breast cancer and pancreatic cancer. However, high ITGB1 expression was not related to OS in colorectal cancer, pancreatic cancer, or melanoma. Further high-quality clinical studies that overcome the aforementioned limitations need to be performed to validate the prognostic value of increased ITGB1 expression in patients with solid cancer.
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA: Cancer J Clin. 2015;65(2):87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 2.Hynes RO, Zhao Q. The evolution of cell adhesion. J Cell Biol. 2000;150(2):F89–F96. doi: 10.1083/jcb.150.2.f89. [DOI] [PubMed] [Google Scholar]
- 3.Chung J, Kim TH. Integrin-dependent translational control: implication in cancer progression. Microsc Res Tech. 2008;71(5):380–386. doi: 10.1002/jemt.20566. [DOI] [PubMed] [Google Scholar]
- 4.Schwartz MA, Ginsberg MH. Networks and crosstalk: integrin signalling spreads. Nat Cell Biol. 2002;4(4):E65–E68. doi: 10.1038/ncb0402-e65. [DOI] [PubMed] [Google Scholar]
- 5.Naci D, Vuori K, Aoudjit F. Alpha2beta1 integrin in cancer development and chemoresistance. Semin Cancer Biol. 2015;35:145–153. doi: 10.1016/j.semcancer.2015.08.004. [DOI] [PubMed] [Google Scholar]
- 6.Scales TM, Parsons M. Spatial and temporal regulation of integrin signalling during cell migration. Curr Opin Cell Biol. 2011;23(5):562–568. doi: 10.1016/j.ceb.2011.05.008. [DOI] [PubMed] [Google Scholar]
- 7.Hynes RO. Integrins: bidirectional, allosteric signaling machines. Cell. 2002;110(6):673–687. doi: 10.1016/s0092-8674(02)00971-6. [DOI] [PubMed] [Google Scholar]
- 8.Friedlander M, Brooks PC, Shaffer RW, Kincaid CM, Varner JA, Cheresh DA. Definition of two angiogenic pathways by distinct alpha v integrins. Science. 1995;270(5241):1500–1502. doi: 10.1126/science.270.5241.1500. [DOI] [PubMed] [Google Scholar]
- 9.Brooks PC, Clark RA, Cheresh DA. Requirement of vascular integrin alpha v beta 3 for angiogenesis. Science. 1994;264(5158):569–571. doi: 10.1126/science.7512751. [DOI] [PubMed] [Google Scholar]
- 10.Hehlgans S, Haase M, Cordes N. Signalling via integrins: implications for cell survival and anticancer strategies. Biochim Biophys Acta. 2007;1775(1):163–180. doi: 10.1016/j.bbcan.2006.09.001. [DOI] [PubMed] [Google Scholar]
- 11.Rintoul RC, Sethi T. The role of extracellular matrix in small-cell lung cancer. Lancet Oncol. 2001;2(7):437–442. doi: 10.1016/S1470-2045(00)00421-6. [DOI] [PubMed] [Google Scholar]
- 12.Kenny PA, Lee GY, Myers CA, et al. The morphologies of breast cancer cell lines in three-dimensional assays correlate with their profiles of gene expression. Mol Oncol. 2007;1(1):84–96. doi: 10.1016/j.molonc.2007.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guo G, Gong K, Wohlfeld B, Hatanpaa KJ, Zhao D, Habib AA. Ligand-independent EGFR signaling. Cancer Res. 2015;75(17):3436–3441. doi: 10.1158/0008-5472.CAN-15-0989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fujita S, Watanabe M, Kubota T, Teramoto T, Kitajima M. Alteration of expression in integrin beta 1-subunit correlates with invasion and metastasis in colorectal cancer. Cancer Lett. 1995;91(1):145–149. doi: 10.1016/0304-3835(95)03735-f. [DOI] [PubMed] [Google Scholar]
- 15.Hu C, Ni Z, Li BS, et al. hTERT promotes the invasion of gastric cancer cells by enhancing FOXO3a ubiquitination and subsequent ITGB1 upregulation. Gut. 2017;66(1):31–42. doi: 10.1136/gutjnl-2015-309322. [DOI] [PubMed] [Google Scholar]
- 16.Zhou P, Erfani S, Liu Z, et al. CD151-alpha3beta1 integrin complexes are prognostic markers of glioblastoma and cooperate with EGFR to drive tumor cell motility and invasion. Oncotarget. 2015;6(30):29675–29693. doi: 10.18632/oncotarget.4896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Williams KC, Coppolino MG. SNARE-dependent interaction of Src, EGFR and β1 integrin regulates invadopodia formation and tumor cell invasion. J Cell Sci. 2014;127(Pt 8):1712–1725. doi: 10.1242/jcs.134734. [DOI] [PubMed] [Google Scholar]
- 18.Fedorenko IV, Abel EV, Koomen JM, et al. Fibronectin induction abrogates the BRAF inhibitor response of BRAF V600E/PTEN-null melanoma cells. Oncogene. 2016;35(10):1225–1235. doi: 10.1038/onc.2015.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hirata E, Girotti MR, Viros A, et al. Intravital imaging reveals how BRAF inhibition generates drug-tolerant microenvironments with high integrin β1/FAK signaling. Cancer Cell. 2015;27(4):574–588. doi: 10.1016/j.ccell.2015.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Damiano JS. Integrins as novel drug targets for overcoming innate drug resistance. Curr Cancer Drug Targets. 2002;2(1):37–43. doi: 10.2174/1568009023334033. [DOI] [PubMed] [Google Scholar]
- 21.Cordes N, Beinke C. Fibronectin alters cell survival and intracellular signaling of confluent A549 cultures after irradiation. Cancer Biol Ther. 2004;3(1):47–53. [PubMed] [Google Scholar]
- 22.Huang C, Park CC, Hilsenbeck SG, et al. β1 integrin mediates an alternative survival pathway in breast cancer cells resistant to lapatinib. Breast Cancer Res. 2011;13(4):R84. doi: 10.1186/bcr2936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guo L, Zhang F, Cai Y, Liu T. Expression profiling of integrins in lung cancer cells. Pathol Res Pract. 2009;205(12):847–853. doi: 10.1016/j.prp.2009.07.005. [DOI] [PubMed] [Google Scholar]
- 24.Zheng W, Jiang C, Li R. Integrin and gene network analysis reveals that ITGA5 and ITGB1 are prognostic in non-small-cell lung cancer. Onco Targets Ther. 2016;9:2317–2327. doi: 10.2147/OTT.S91796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chang MH, Lee K, Lee KY, Kim YS, Kim YK, Kang JH. Prognostic role of integrin β1, E-cadherin, and rac1 expression in small cell lung cancer. APMIS. 2012;120(1):28–38. doi: 10.1111/j.1600-0463.2011.02788.x. [DOI] [PubMed] [Google Scholar]
- 26.Dingemans AM, van den Boogaart V, Vosse BA, van Suylen RJ, Griffioen AW, Thijssen VL. Integrin expression profiling identifies integrin alpha5 and beta1 as prognostic factors in early stage non-small cell lung cancer. Mol Cancer. 2010;9:152. doi: 10.1186/1476-4598-9-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhao ZS, Wang YY, Chu YQ, Ye ZY, Tao HQ. SPARC is associated with gastric cancer progression and poor survival of patients. Clin Cancer Res. 2010;16(1):260–268. doi: 10.1158/1078-0432.CCR-09-1247. [DOI] [PubMed] [Google Scholar]
- 28.Yao ES, Zhang H, Chen YY, et al. Increased beta1 integrin is associated with decreased survival in invasive breast cancer. Cancer Res. 2007;67(2):659–664. doi: 10.1158/0008-5472.CAN-06-2768. [DOI] [PubMed] [Google Scholar]
- 29.Petricevic B, Vrbanec D, Jakic-Razumovic J, et al. Expression of Toll-like receptor 4 and beta 1 integrin in breast cancer. Med Oncol. 2012;29(2):486–494. doi: 10.1007/s12032-011-9885-0. [DOI] [PubMed] [Google Scholar]
- 30.Lanzafame S, Emmanuele C, Torrisi A. Correlation of α2β1 integrin expression with histological type and hormonal receptor status in breast carcinomas. Pathol Res Pract. 1996;192(10):1031–1038. doi: 10.1016/s0344-0338(96)80045-8. [DOI] [PubMed] [Google Scholar]
- 31.Pontes-Júnior J, Reis ST, Bernardes FS, et al. Correlation between Beta1 integrin expression and prognosis in clinically localized prostate cancer. Int Braz J Urol. 2013;39(3):335–342. doi: 10.1590/S1677-5538.IBJU.2013.03.06. discussion 343. [DOI] [PubMed] [Google Scholar]
- 32.Böttger TC, Maschek H, Lobo M, Gottwohl RG, Brenner W, Junginger T. Prognostic value of immunohistochemical expression of beta-1 integrin in pancreatic carcinoma. Oncology. 1999;56(4):308–313. doi: 10.1159/000011984. [DOI] [PubMed] [Google Scholar]
- 33.Zhou G, Chiu D, Qin D, et al. Expression of CD44v6 and integrin-β1 for the prognosis evaluation of pancreatic cancer patients after cryosurgery. Diagn Pathol. 2013;8:146. doi: 10.1186/1746-1596-8-146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vassos N, Rau T, Merkel S, et al. Prognostic value of β1 integrin expression in colorectal liver metastases. Int J Clin Exp Pathol. 2014;7(1):288–300. [PMC free article] [PubMed] [Google Scholar]
- 35.Liu QZ, Gao XH, Chang WJ, et al. Expression of ITGB1 predicts prognosis in colorectal cancer: a large prospective study based on tissue microarray. Int J Clin Exp Pathol. 2015;8(10):12802–12810. [PMC free article] [PubMed] [Google Scholar]
- 36.Gonzalez MA, Pinder SE, Wencyk PM, et al. An immunohistochemical examination of the expression of E-cadherin, alpha- and beta/gamma-catenins, and alpha2- and beta1-integrins in invasive breast cancer. J Pathol. 1999;187(5):523–529. doi: 10.1002/(SICI)1096-9896(199904)187:5<523::AID-PATH296>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 37.Klahan S, Huang WC, Chang CM, et al. Gene expression profiling combined with functional analysis identify integrin beta1 (ITGB1) as a potential prognosis biomarker in triple negative breast cancer. Pharmacol Res. 2016;104:31–37. doi: 10.1016/j.phrs.2015.12.004. [DOI] [PubMed] [Google Scholar]
- 38.Berry MG, Gui GP, Wells CA, Carpenter R. Integrin expression and survival in human breast cancer. Eur J Surg Oncol. 2004;30(5):484–489. doi: 10.1016/j.ejso.2004.01.016. [DOI] [PubMed] [Google Scholar]
- 39.Lawson MH, Cummings NM, Rassl DM, et al. Bcl-2 and β1-integrin predict survival in a tissue microarray of small cell lung cancer. Br J Cancer. 2010;103(11):1710–1715. doi: 10.1038/sj.bjc.6605950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liang Z, Kong R, He Z, et al. High expression of miR-493-5p positively correlates with clinical prognosis of non small cell lung cancer by targeting oncogene ITGB1. Oncotarget. 2017;8(29):47389–47399. doi: 10.18632/oncotarget.17650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Okamura M, Yamaji S, Nagashima Y, et al. Prognostic value of inte-grin β1-ILK-pAkt signaling pathway in non-small cell lung cancer. Hum Pathol. 2007;38(7):1081–1091. doi: 10.1016/j.humpath.2007.01.003. [DOI] [PubMed] [Google Scholar]
- 42.Zhang PF, Zeng GQ, Yi LZ, et al. Identification of integrin β1 as a prognostic biomarker for human lung adenocarcinoma using 2D-LC-MS/MS combined with iTRAQ technology. Oncol Rep. 2013;30(1):341–349. doi: 10.3892/or.2013.2477. [DOI] [PubMed] [Google Scholar]
- 43.Langan RC, Mullinax JE, Ray S, et al. A pilot study assessing the potential role of non-CD133 colorectal cancer stem cells as biomarkers. J Cancer. 2012;3:231–240. doi: 10.7150/jca.4542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lesniak D, Xu Y, Deschenes J, et al. Beta1-integrin circumvents the antiproliferative effects of trastuzumab in human epidermal growth factor receptor-2-positive breast cancer. Cancer Res. 2009;69(22):8620–8628. doi: 10.1158/0008-5472.CAN-09-1591. [DOI] [PubMed] [Google Scholar]
- 45.McSherry EA, McGee SF, Jirstrom K, et al. JAM-A expression positively correlates with poor prognosis in breast cancer patients. Int J Cancer. 2009;125(6):1343–1351. doi: 10.1002/ijc.24498. [DOI] [PubMed] [Google Scholar]
- 46.dos Santos PB, Zanetti JS, Ribeiro-Silva A, Beltrão EI. Beta 1 integrin predicts survival in breast cancer: a clinicopathological and immunohistochemical study. Diagn Pathol. 2012;7:104. doi: 10.1186/1746-1596-7-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yin HL, Wu CC, Lin CH, et al. β1 Integrin as a prognostic and predictive marker in triple-negative breast cancer. Int J Mol Sci. 2016;17(9):1432. doi: 10.3390/ijms17091432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hieken TJ, Ronan SG, Farolan M, Shilkaitis AL, Das Gupta TK. Molecular prognostic markers in intermediate-thickness cutaneous malignant melanoma. Cancer. 1999;85(2):375–382. doi: 10.1002/(sici)1097-0142(19990115)85:2<375::aid-cncr15>3.0.co;2-1. [DOI] [PubMed] [Google Scholar]
- 49.Nikkola J, Vihinen P, Vlaykova T, Hahka-Kemppinen M, Heino J, Pyrhonen S. Integrin chains beta1 and alphav as prognostic factors in human metastatic melanoma. Melanoma Res. 2004;14(1):29–37. doi: 10.1097/00008390-200402000-00005. [DOI] [PubMed] [Google Scholar]
- 50.Vihinen P, Nikkola J, Vlaykova T, et al. Prognostic value of beta1 integrin expression in metastatic melanoma. Melanoma Res. 2000;10(3):243–251. [PubMed] [Google Scholar]
- 51.Sawai H, Okada Y, Funahashi H, et al. Integrin-linked kinase activity is associated with interleukin-1 alpha-induced progressive behavior of pancreatic cancer and poor patient survival. Oncogene. 2006;25(23):3237–3246. doi: 10.1038/sj.onc.1209356. [DOI] [PubMed] [Google Scholar]
- 52.Yang D, Shi J, Fu H, et al. Integrinβ1 modulates tumour resistance to gemcitabine and serves as an independent prognostic factor in pancreatic adenocarcinomas. Tumour Biol. 2016;37(9):12315–12327. doi: 10.1007/s13277-016-5061-7. [DOI] [PubMed] [Google Scholar]
- 53.Aoudjit F, Vuori K. Integrin signaling inhibits paclitaxel-induced apoptosis in breast cancer cells. Oncogene. 2001;20(36):4995–5004. doi: 10.1038/sj.onc.1204554. [DOI] [PubMed] [Google Scholar]
- 54.Nam JM, Chung Y, Hsu HC, Park CC. Beta1 integrin targeting to enhance radiation therapy. Int J Radiat Biol. 2009;85(11):923–928. doi: 10.3109/09553000903232876. [DOI] [PubMed] [Google Scholar]
- 55.Han JY, Kim HS, Lee SH, Park WS, Lee JY, Yoo NJ. Immunohistochemical expression of integrins and extracellular matrix proteins in non-small cell lung cancer: correlation with lymph node metastasis. Lung Cancer. 2003;41(1):65–70. doi: 10.1016/s0169-5002(03)00146-6. [DOI] [PubMed] [Google Scholar]
- 56.Bredin CG, Sundqvist KG, Hauzenberger D, Klominek J. Integrin dependent migration of lung cancer cells to extracellular matrix components. Eur Respir J. 1998;11(2):400–407. doi: 10.1183/09031936.98.11020400. [DOI] [PubMed] [Google Scholar]
- 57.Ju L, Zhou C, Li W, Yan L. Integrin beta1 over-expression associates with resistance to tyrosine kinase inhibitor gefitinib in non-small cell lung cancer. J Cell Biochem. 2010;111(6):1565–1574. doi: 10.1002/jcb.22888. [DOI] [PubMed] [Google Scholar]