Abstract
Final maturation of eukaryotic ribosomes occurs in the cytoplasm and requires the sequential removal of associated assembly factors and processing of the immature 20S pre‐RNA. Using cryo‐electron microscopy (cryo‐EM), we have determined the structure of a yeast cytoplasmic pre‐40S particle in complex with Enp1, Ltv1, Rio2, Tsr1, and Pno1 assembly factors poised to initiate final maturation. The structure reveals that the pre‐rRNA adopts a highly distorted conformation of its 3′ major and 3′ minor domains stabilized by the binding of the assembly factors. This observation is consistent with a mechanism that involves concerted release of the assembly factors orchestrated by the folding of the rRNA in the head of the pre‐40S subunit during the final stages of maturation. Our results provide a structural framework for the coordination of the final maturation events that drive a pre‐40S particle toward the mature form capable of engaging in translation.
Keywords: cryo‐EM, pre‐40S ribosome, ribosome, ribosome assembly, ribosome biogenesis
Subject Categories: Protein Biosynthesis & Quality Control, RNA Biology, Structural Biology
Introduction
The ribosome performs the essential task of decoding genetic information into proteins. While structures of the mature ribosomal subunits in different functional states provide detailed insights into the mechanisms of translation, our knowledge regarding ribosome assembly and quality control at atomic resolution is only emerging.
In eukaryotes, ribosome assembly is a complex process that stretches across the nucleolus, nucleoplasm and cytoplasm and requires the concerted actions of > 200 assembly factors and several energy‐consuming enzymes (Woolford & Baserga, 2013; Konikkat & Woolford, 2017; Kressler et al, 2017; Pena et al, 2017). The process is initiated in the nucleolus with the RNA Pol‐I‐driven synthesis of the 35S pre‐rRNA. The emerging pre‐rRNA undergoes co‐transcriptional folding and modification and recruits ribosomal proteins (r‐proteins) and assembly factors specific for the small ribosomal subunit to form the earliest ribosome precursor, the 90S (Dragon et al, 2002; Kornprobst et al, 2016; Barandun et al, 2017; Cheng et al, 2017). Cleavage within the pre‐rRNA releases the earliest small ribosomal subunit 40S precursor (pre‐40S). Pre‐60S large ribosomal subunits and pre‐40S particles mature independently by transiently interacting with assembly factors (AFs) and are separately transported by export receptors through the nuclear pore complexes into the cytoplasm. Cytoplasmic maturation events are intimately intertwined with quality control and proofreading of functional centers, which ensures that only correctly assembled ribosomal subunits are committed to the translating pool (Lo et al, 2010; Bussiere et al, 2012; Karbstein, 2013; Greber, 2016; Greber et al, 2016; Ma et al, 2017; Malyutin et al, 2017).
Exported pre‐40S ribosomal particles undergo a series of cytoplasmic maturation steps before engaging in translation, including the sequential release of assembly factors and export receptors, as well as the assembly and stable incorporation of remaining ribosomal proteins (Karbstein, 2013). During the final steps of cytoplasmic maturation of the pre‐40S, the 3′ end of the 20S rRNA is cleaved off by the nuclease Nob1 at cleavage site “D” to generate translation‐competent 40S subunits (Fatica et al, 2004; Lamanna & Karbstein, 2009; Lebaron et al, 2012). Prior to cleavage of the 20S rRNA, the pre‐40S contains several AFs including Ltv1 and Enp1 bound to the pre‐40S head and Tsr1, Rio2, Dim1, Pno1, and Nob1 in the vicinity of the pre‐40S mRNA binding channel (Strunk et al, 2011, 2012), which are exported from the nucleus in complex with the pre‐40S and released in the cytoplasm (Schafer et al, 2003).
The release of Ltv1 and Enp1 from the pre‐40S is expected to induce a major structural rearrangement during final cytoplasmic maturation that leads to the formation of a mature beak, a universal structural landmark on the head of the 40S subunit critical during translation (Mitterer et al, 2016). Beak formation also involves stable incorporation of the r‐protein uS3 into the 40S and is initiated by the kinase Hrr25 in the cytoplasm (Schafer et al, 2006), which phosphorylates Ltv1 and Enp1 leading to their release from the pre‐40S beak. However, the molecular basis underlying stable uS3 incorporation remains unclear, due to the lack of high‐resolution structural information on pre‐40S particles.
Dim1, Tsr1, and the atypical kinase/ATPase Rio2 bind to the subunit interface of the pre‐40S, where they prevent the access of translation initiation factors and the 60S subunits to the immature 40S subunit (Strunk et al, 2011). Tsr1 is a GTPase‐like protein composed of four domains, where the first three domains form a fold similar to translational GTPases such as EF‐Tu and eIF5B, but lack the residues to bind and hydrolyze GTP (McCaughan et al, 2016). Release of Rio2 is triggered by its own ATPase activity, which has been suggested to induce a conformational change in the 40S subunit (Ferreira‐Cerca et al, 2012).
Endonucleolytic cleavage of immature 20S rRNA into mature 18S rRNA by Nob1 is an essential cytoplasmic maturation event that renders the pre‐40S subunit translation‐competent (Fatica et al, 2003; Pertschy et al, 2009). The regulation of the rRNA cleavage involves the KH domain‐containing protein Pno1 (Partner of Nob1), which co‐localizes with Nob1 to the pre‐40S platform. The encounter between a mature 60S subunit with a pre‐40S particle is aided by the cytoplasmic GTPase Fun12 (eIF5B) and marks the point at which Pno1 is displaced and/or released and Nob1‐mediated 20S pre‐rRNA cleavage is triggered (Turowski et al, 2014). However, the molecular details of how Pno1 regulates 20S pre‐rRNA cleavage remains to be clarified.
Recent cryo‐EM structures of the 90S are beginning to shed light on nucleolar events of the 40S assembly (Kornprobst et al, 2016; Barandun et al, 2017; Chaker‐Margot et al, 2017; Cheng et al, 2017; Sun et al, 2017). In contrast, high‐resolution structural information of cytoplasmic pre‐40S particles has remained inaccessible in part due to their structural flexibility and heterogeneity (Strunk et al, 2011; Johnson et al, 2017). We used a catalytically inactive mutant of Nob1 in budding yeast to trap and purify a cytoplasmic pre‐40S particle poised for final cytoplasmic maturation. This approach permitted the structural determination of a cytoplasmic pre‐40S particle at a resolution of 3.4 Å by cryo‐EM. The structure provides critical insights into mechanisms that drive 40S beak formation and final pre‐rRNA folding and processing and provides a mechanistic framework for the series of events that take place during pre‐40S maturation.
Results
Isolation of cytoplasmic pre‐40S poised for 20S pre‐rRNA cleavage
In order to trap a pre‐40S particle poised to undergo the final 20S pre‐rRNA cleavage step, we exploited the catalytically inactive Nob1‐D15N allele. The D15N mutation is located in the conserved PIN domain (PilT N terminus) of Nob1 and exhibits a dominant‐negative phenotype (Fatica et al, 2004; Granneman et al, 2010), indicating that the nuclease activity is not required to bind pre‐40S ribosomes.
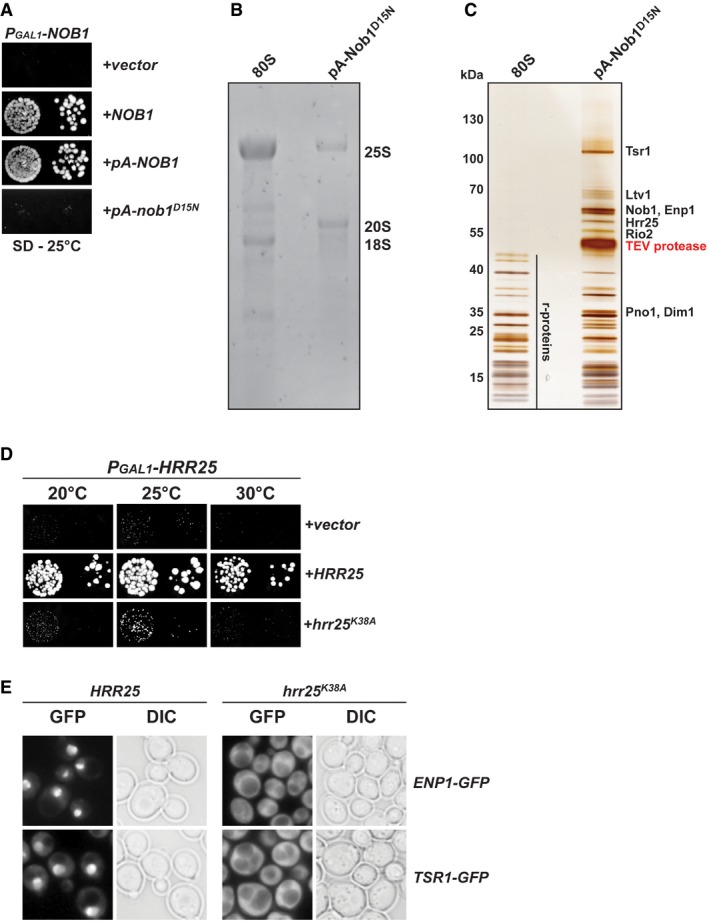
We generated a conditional Nob1 mutant strain in which the endogenous NOB1 gene was placed under the control of the GAL1 promoter (PGAL1‐NOB1; Appendix Table S1). On repressive glucose media, the PGAL1‐NOB1 strain was severely impaired in growth compared to PGAL1‐NOB1 complemented with plasmid‐encoded Nob1 (Fig 1A). Next, we introduced Protein A‐tagged N‐terminal fusions of WT Nob1 (pA‐Nob1) and the catalytically inactive Nob1‐D15N into the PGAL1‐NOB1 strain and analyzed growth of the resultant strains on glucose media (Fig 1A and Appendix Table S2). Consistent with previous studies (Granneman et al, 2010), the yeast cells expressing the pA‐Nob1 fusion protein grew at WT rates, indicating that an N‐terminal tag does not interfere with Nob1 function (Fig 1A). In contrast, the strain expressing the catalytically inactive pA‐Nob1‐D15N fusion showed a severe growth defect.
Figure 1. Purification of trapped pre‐40S ribosomes and Hrr25 kinase activity.
-
AN‐terminally tagged Protein A (pA)‐Nob1 is functional, but requires endonuclease activity for viability. The PGAL1‐NOB1 strain was transformed with indicated plasmids and spotted in 10‐fold dilutions on selective and repressive glucose‐containing plates and grown at 25°C for 4 days.
-
BRNA composition of 80S ribosomes and purified Protein A (pA)‐tagged Nob1‐D15N particles. RNA was separated by agarose gel electrophoresis and stained with GelRed.
-
CProtein composition of 80S ribosomes and purified Protein A (pA)‐tagged Nob1‐D15N particles. Proteins were separated by SDS–PAGE and visualized by silver staining. Indicated protein bands were identified by mass spectrometry.
-
D, EThe kinase activity of Hrr25 is essential for cell viability and is required for cytoplasmic release and recycling of Enp1 and Tsr1. (D) The PGAL1‐HRR25 strain was transformed with indicated plasmids and spotted in 10‐fold dilutions on selective and repressive glucose‐containing plates and grown at indicated temperatures for 3–7 days. (E) Yeast strains expressing endogenous Enp1‐GFP or Tsr1‐GFP were transformed with plasmids carrying either a galactose‐inducible wild‐type HRR25 gene or a dominant‐negative hrr25‐K38A kinase dead mutant gene. Strains were then grown on galactose‐containing medium at 25°C, and GFP constructs were visualized by fluorescence microscopy.
We affinity‐purified the pA‐Nob1‐D15N containing ribosomal subunits from yeast cells grown in glucose‐containing medium to exclusively express the mutant protein. As expected, the affinity‐purified particles contained 20S pre‐rRNA in contrast to the mature 18S rRNA present in 80S ribosomes (Fig 1B). In addition, we found that the isolated pre‐40S particle also contained sub‐stoichiometric levels of mature 25S rRNA, indicating the presence of cytoplasmic 80S‐like complexes that contain mature 60S subunits (Fig 1B). Mass spectrometric analysis of the purified sample revealed that the eight pre‐40S assembly factors Hrr25, Ltv1, Enp1, Rio2, Tsr1, Dim1, Pno1, and Nob1 co‐purified to nearly stoichiometric levels (Fig 1C).
These AFs are released in a sequential manner once Ltv1 is removed (Strunk et al, 2012). In human cells, CK1δ/ε, the homologs of Hrr25, were reported to be the factors responsible for starting the cascade of AFs release (Fassio et al, 2010; Zemp et al, 2014; Ghalei et al, 2015). To investigate an analogous function of Hrr25 in yeast (Schafer et al, 2006), we generated conditional Hrr25 strains in which the endogenous HRR25 gene or previously characterized dominant‐negative mutants of Hrr25 lacking the kinase activity (Hoekstra et al, 1991; hrr25‐K38A) are placed under control of a GAL1 promoter (Fig 1D and Appendix Tables S1 and S2). Plasmids containing the galactose‐inducible Hrr25 variants were transformed in yeast strains expressing endogenous GFP‐tagged constructs of Enp1 or Tsr1 (Fig 1E and Appendix Tables S1 and S2). At steady‐state levels, wild‐type Enp1‐GFP and Tsr1‐GFP localized to the nucleus. In contrast, upon overexpressing the catalytically inactive hrr25‐K38A mutant in galactose‐containing medium, both Enp1‐GFP and Tsr1‐GFP failed to release and recycle back to the nucleus and were therefore mislocalized to the cytoplasm. This suggests that yeast Hrr25 also acts upstream of Enp1 and Tsr1 releases, as observed previously for human proteins (Zemp et al, 2014), and is likely involved in starting the cascade of AF release (Schafer et al, 2006).
Cytoplasmic pre‐40S structure determination
The affinity‐purified pre‐40S particles were analyzed by single‐particle cryo‐EM. The micrographs showed a mixture of particles that resemble 40S ribosomal subunits and 80S ribosomes in size and shape. Single‐particle images were selected from the micrographs and split into subsets showing 40S‐like and 80S‐like particles by 2D classification. The subset of 80S‐like particles was used to refine a structure at a resolution of 3.1 Å (Table EV1 and Fig EV1). The 3D reconstruction revealed a mature 80S (Appendix Fig S1A and B; Ben‐Shem et al, 2011) with additional density visible at the 3′ end of the 18S rRNA at lower local resolution (Appendix Fig S1A and B), which probably represents partial density for the uncleaved internal transcribed spacer 1 (ITS1) protruding flexibly from the 40S subunit (Appendix Fig S1B). However, no density for any assembly factor could be observed.
Figure EV1. Classification and refinement scheme for the pre‐40S complex, the 80S‐like particle, and the Dim1‐containing complex.
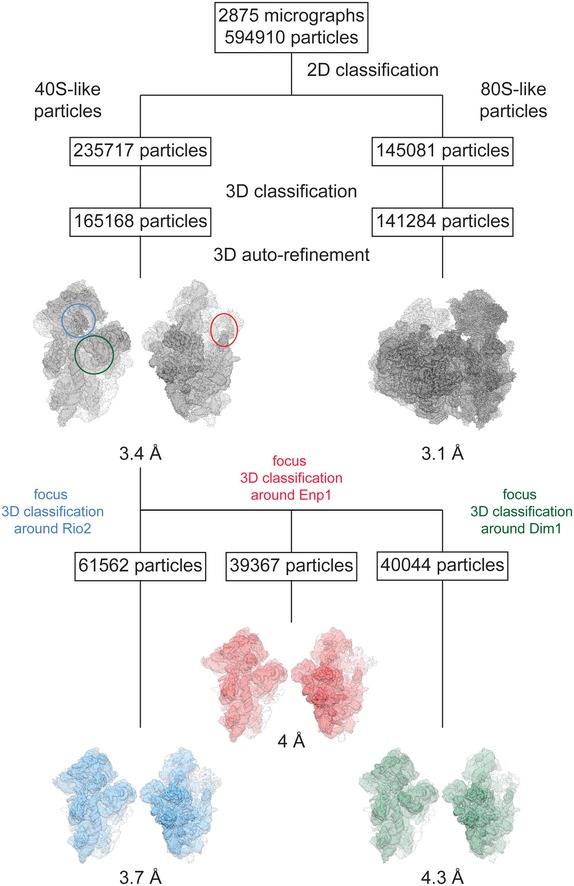
2D classification from 594,910 particles that had been picked from 2,875 micrographs revealed two particle subpopulations of “40S‐like” and “80S‐like” particles, which where separately processed and subjected to 3D classification and auto‐refinement yielding a 3D reconstruction of the pre‐40S complex at an overall resolution of 3.4 Å and a 3D reconstruction of the 80S‐like particle at an overall resolution of 3.1 Å. Subsequent rounds of focused 3D classification from this final set of pre‐40S particles focusing on either Rio2 (blue circle), Enp1 (red circle), or Dim1 (green circle) yielded subpopulations of the pre‐40S complex at overall lower resolution but with higher local resolution in the area of focus, further facilitating the model building and docking in these areas.
The 3D reconstruction from the subset of 40S‐like particles was refined reaching an overall resolution of 3.4 Å (Table EV1, Figs EV1 and EV2A). The density showed an immature 40S ribosomal subunit similar to the previously described cytoplasmic pre‐40S ribosome (Fig 2A; Strunk et al, 2011; Johnson et al, 2017).
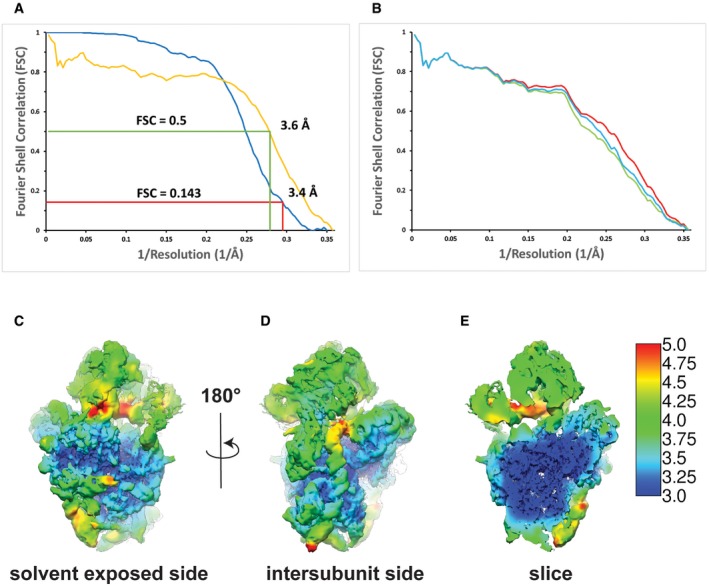
Figure EV2. Local resolution heat map of the high‐resolution pre‐40S complex and comparison of the Fourier shell correlation curves calculated for the experimental 3D‐EM reconstruction and the refined model of the pre‐40S complex.
-
AFourier shell correlation (FSC) curves for the high‐resolution EM map (blue) and the one calculated from the model versus the map (yellow). At the FSC = 0.143 criterion, the overall resolution of the high‐resolution EM map was estimated to be 3.4 Å, which coincides well with the 3.6 Å resolution obtained for the model versus map FSC at a value of 0.5.
-
BFSC curves calculated between the model refined in the density of the halfset1 of the data and the map calculated with the full dataset (red), the halfset1 map (green, half used for refinement), or the halfset2 map (blue, half not used for refinement). The agreement between the three FSCs curves indicates the absence of overfitting (Fernandez et al, 2014; Greber et al, 2014).
-
C, DLocal resolution heat map of two different orientations of the reconstruction of the full pre‐40S complex. The local resolution varies between 3 Å for the core of the ribosomal subunit and 4–5 Å for the peripheral parts of the rRNA expansion segments as well as the head of the pre‐40S and the assembly factors. Local resolution was calculated using the “relion_postprocess” from Relion 2.1 (Kimanius et al, 2016).
-
ECut‐through of the view in (D).
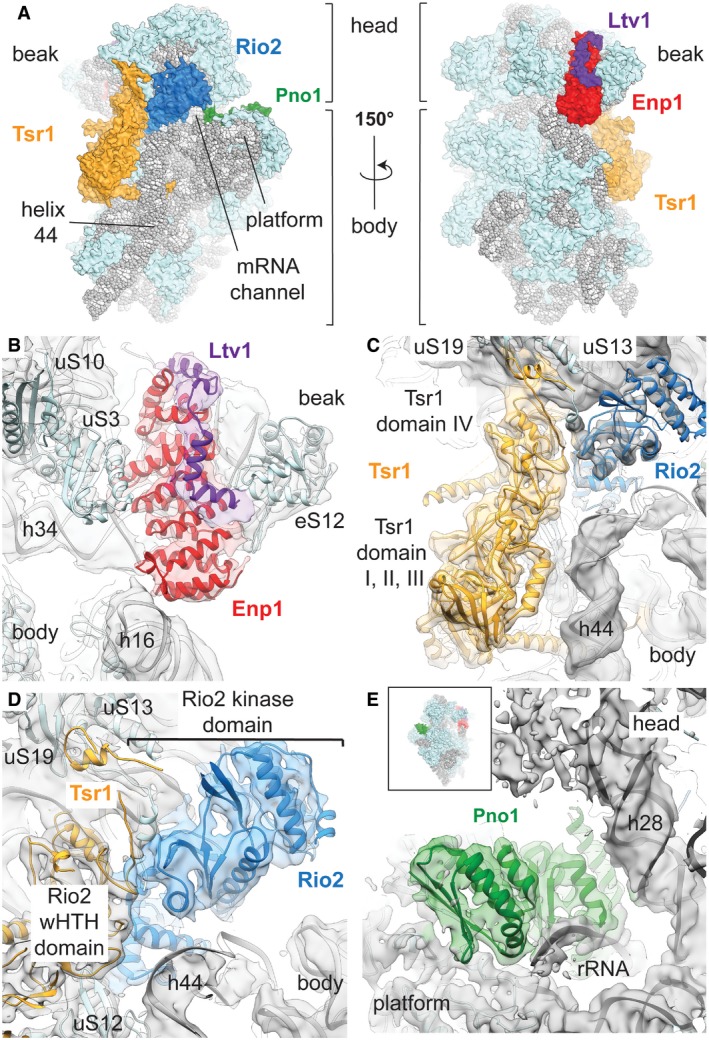
Figure 2. Structure of the cytoplasmic pre‐40S ribosome.
-
AOverview of the front and back of the structure of the pre‐40S. The 20S rRNA is depicted in gray, the r‐proteins in light blue, Tsr1 in yellow, Rio2 in blue, Pno1 in green, Enp1 in red, and Ltv1 in purple.
-
B–EOverview of the atomic models of the AFs in their corresponding densities. Densities were colored according to the models in a radius of 4 Å around the model atoms. For each panel, the shown densities correspond to the locally classified 3D reconstruction used for model building. (B) The density of Ltv1 is shown at a different contour level (1.5σ for Ltv1 and 2.5σ for ribosome and Enp1) to highlight the visible secondary structure elements of both Enp1 and Ltv1. (C) Tsr1 and Rio2 bind to the subunit interface and close to the decoding center (indicated with an asterisk at the top of helix 44). Tsr1 domain IV binds to the pre‐40S head, while domains I–III are in contact with the body. (D) Rio2 is also in contact with ribosomal proteins uS13 and uS19. (E) Pno1 binds to the “neck” helix 28 and in vicinity of the 3′ end of the rRNA to the platform of the pre‐40S particle
The local resolution of this 3.4 Å 3D reconstruction ranges from 2.8 Å to 4.9 Å for the ribosomal proteins and the rRNA (Fig EV2C–E and Appendix Table S3), allowing us to build and refine a model of the pre‐40S subunit based on the 40S subunit from the 80S structure of the yeast ribosome (Ben‐Shem et al, 2011) from which proteins eS10, eS26, eS31, and STM1 that are not present in our structure were removed. The reconstruction clearly revealed additional features that were assigned to the AFs by fitting previously solved crystal structures of individual yeast factors and their homologs (Appendix Table S4). The distribution of the maturation factors on the surface of the pre‐40S subunit forms three groups, in agreement with previously published low‐resolution reconstructions of cytoplasmic pre‐40S ribosomes (Strunk et al, 2011; Johnson et al, 2017). The Enp1 and Ltv1 factors are positioned between the head and the body of the subunit close to the mRNA entry channel on the solvent‐exposed side (Fig 2A and B). Tsr1 and Rio2 are localized on the side of the 40S subunit that interacts with the 60S subunit (Fig 2A), with Tsr1 occupying the position where eIF5B binds during translation initiation as previously reported (Fig 2C; Strunk et al, 2011; McCaughan et al, 2016; Johnson et al, 2017), while Rio2 is positioned in the middle of the mRNA channel bridging the head and the body of the 40S subunit (Fig 2D). Pno1 binds on the soluble side of the subunit behind the platform where the 3′ end of the 18S rRNA is located in the mature 40S subunit (Fig 2E). The average local resolution of the densities corresponding to Tsr1 (3.7 Å), Pno1 (3.6 Å), and Rio2 (4.0 Å) allowed us to rebuild their structures in complex with the ribosome, whereas the density corresponding to Enp1 and Ltv1 was resolved at lower local resolution (4.2 Å) and was interpreted by rigid‐body fitting the crystal structure of the partial Enp1‐Ltv1 complex from yeast (PDB: 5WWO; Sun et al, 2017; Figs 2B–E and EV2C–E and Appendix Tables S3 and S4).
The Dim1 rRNA dimethylase reported to be part of the pre‐40S could only be identified in a small fraction of the final set of 40S‐like particles after additional local classification (Table EV1, Figs EV1 and EV3A and B, and Appendix Table S4). The reconstruction of this subset of particles reached an overall resolution of 4.3 Å and showed a pre‐40S subunits containing Dim1 and helix 45 in its mature position (Fig EV3B), opposed to the displaced conformation described previously (Johnson et al, 2017). In our conformation, Dim1 (average local resolution of 6.1 Å) is at a position too distant from its target helix 45 for methylation (Boehringer et al, 2012; Johnson et al, 2017; Fig EV3A and B). This suggests that Dim1 might have already methylated helix 45 in this particle, as shown by previous primer extension analysis on the Nob1‐D15N particles (Lebaron et al, 2012), and remains only weakly attached to the rRNA.
Figure EV3. 3D reconstruction of the Dim1‐containing subclass of cytoplasmic pre‐40S particles.
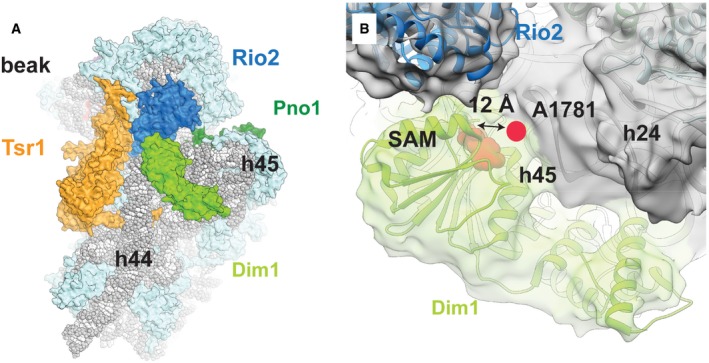
- Overview over the Dim1‐containing complex (r‐proteins in light blue, rRNA in gray) from the intersubunit face. The rRNA methylase Dim1 (bright green) binds to the complex between the platform and the top of the displaced helix 44 in close vicinity of Rio2 (blue), Tsr1 (orange), and the decoding center.
- Dim1 (bright green) binds not only to helix 44, but also helices 45 and 24. Rigid‐body fitting of the available crystal structure of human Dim1 in complex with S‐adenosyl methionine (SAM) (pink) (PDB: 1ZQ9) into the cryo‐EM density shows that the previously described active site of Dim1 would be located too distant from its methylation target, A1781 (pink circle), to allow the methylation reaction to take place. Additionally, the unmethylated helix 45 has been reported to be rotated upward to allow access for Dim1 (Johnson et al, 2017). Thus, Dim1 has likely already methylated A1781 and A1782, as shown previously by primer extension analysis of the Nob1‐D15N particle (Lebaron et al, 2012).
Configuration of Enp1, Ltv1, and uS3 on the pre‐40S head
On the solvent‐exposed side of the pre‐40S ribosome, densities corresponding to Enp1 and Ltv1 connect the beak of the highly rotated pre‐40S head (Fig 3A and B) with rRNA helix 16 on the pre‐40S body (Fig 2A and B). The superhelical fold of Enp1 fits well into the density, and an additional L‐shaped density on top of Enp1 could be attributed to residues 362–405 of Ltv1 in agreement with the high‐resolution Enp1‐Ltv1 crystal structure (Fig 2B; Sun et al, 2017). The large positively charged surface of Enp1 contacts the three‐way junction of rRNA helices 32, 33, and 34 in the beak (Appendix Fig S2A and B), where eS10 would bind in the mature 40S (Ben‐Shem et al, 2011), and is likely stabilizing these helices in an immature conformation (Fig 3B). The three helices are highly distorted compared to the mature 40S, changing the overall shape of the beak.
Figure 3. Enp1 and Ltv1 interact with the distorted rRNA in the head of the pre‐40S.
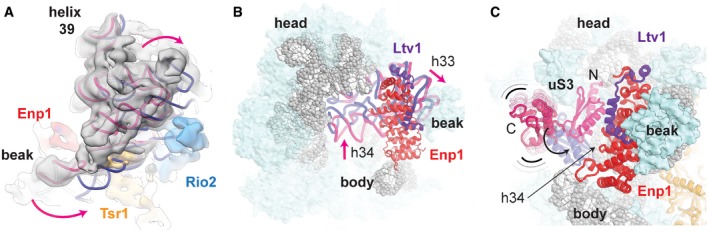
- Top view of the pre‐40S head rRNA (pink) in the cryo‐EM density (gray), low‐pass‐filtered to 6 Å for clarity. Compared to the mature head position (rRNA in dark blue, aligned to the body rRNA, PDB 4V88; Ben‐Shem et al, 2011), the pre‐40S head is highly rotated toward the solvent‐exposed side.
- Overview of the head rRNA with the r‐proteins shown as light‐blue surfaces, Enp1 and Ltv1 as cartoon, and rRNA as gray spheres and cartoon. uS10, uS3, and uS14 were removed for clarity. The rRNA corresponding to helix 32, 33, and 34 is shown as cartoon to highlight the change in conformation between our model (pink) and the mature 40S rRNA (dark blue, aligned to helix 42 and 43 of our model PDB 4V88; Ben‐Shem et al, 2011).
- Position of the flexibly bound uS3 C‐terminal domain in our structure (pink) and the stably incorporated uS3 N‐terminal domain in the context of the mature 40S subunit (dark blue, PDB 4V88; Ben‐Shem et al, 2011). Within the pre‐40S particle, the uS3 C‐terminal domain cannot adopt its mature position, as the displaced helix 34 is in its way.
Enp1 contacts the ribosomal protein uS3 N‐terminal domain, which occupies its mature position with respect to helix 41 and uS10 (Fig 3C). On the other hand, the uS3 C‐terminal domain does not localize to its mature position, which is occupied by the displaced h34. It is only visible at low contour level as a density disconnected from the core of the pre‐40S, and therefore, this domain is likely to be integrated once h34 withdraws to its final conformation (Fig 3C). This provides a molecular explanation for the previously observed lower affinity of uS3 for pre‐40S particles (Schafer et al, 2006). Moreover, this finding implies that the Hrr25‐dependent stable incorporation of uS3 into the pre‐40S is linked to a large‐scale conformational rearrangement of rRNA in the beak upon departure of Enp1.
Tsr1 and Rio2 contact multiple functionally important sites on the 40S
The crystal structure of Tsr1 (PDB 5IW7; McCaughan et al, 2016) could be fitted and refined into an extra density located between the head and the body of the cytoplasmic pre‐40S, with only minimal adjustments of domain IV (Fig 2C). Tsr1 binds close to a displaced helix 44 (Fig 4A) on top of uS12 (Fig 4B), a highly conserved ribosomal protein linked to translational accuracy (Alksne et al, 1993; Triman, 2007). At the ribosomal P‐site close to domain IV of Tsr1, we observe density for Rio2, which could be interpreted by separately fitting homologous crystal structures of the N‐ and C‐terminal halves of Rio2 from A. fulgidus (LaRonde‐LeBlanc et al, 2005; Fig 2D). Interestingly, the eukaryote‐specific C‐terminal extension segment that contains the autoinhibitory helix (Ferreira‐Cerca et al, 2012) is disordered in our structure, as expected for Rio2 in an active conformation. Together, Tsr1 and Rio2 are blocking the A‐ and P‐site on the 40S subunit where tRNAs and initiation factors bind.
Figure 4. Tsr1 and Rio2 interact with functionally important regions of the 40S subunit.
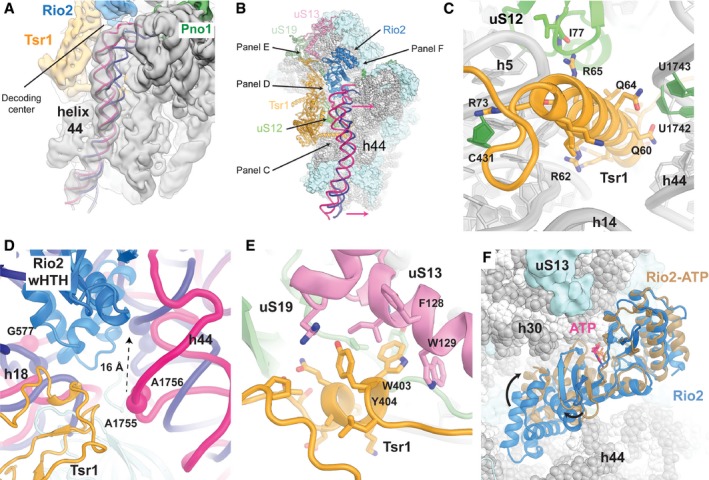
- Close‐up view of the pre‐40S rRNA helix 44 (pink) in the cryo‐EM map (gray), which was low‐pass‐filtered to 6 Å for clarity, superposed with the mature helix 44 (dark blue, PDB 4V88; Ben‐Shem et al, 2011). The immature helix 44 is highly distorted, mainly from the interaction with Tsr1 (panel C and Fig EV4A) up to the decoding center (panel D and Fig EV4B).
- Overview of the pre‐40S ribosome. The rRNA is represented as spheres (gray) and the r‐proteins as surfaces (light blue). Rio2 (blue), Tsr1 (orange), uS12 (green), uS13 (pink), and uS19 (light green) are represented as cartoon. To help with orientation arrows indicate the viewing perspectives used to generate panels (C–F). The pre‐40S helix 44 (cartoon in pink) is highly distorted compared to the mature one (dark blue, PDB 4V88; Ben‐Shem et al, 2011).
- The conserved N‐terminal α‐helix of Tsr1 (residues 51–76; Fig EV4A and Appendix Fig S3A) is wedged between the body of the pre‐40S and helix 44 causing its displacement (panels A and B).
- The highly conserved stretch of Tsr1 residues 391–406 (Fig EV4C and Appendix Fig S3B) interacts with uS13 and uS19. Tsr1 W403 is inserted into a hydrophobic pocket formed by uS13 F128, W129, and Tsr1 Y404.
- Comparison of the yeast pre‐40S Rio2 conformation (blue) and the crystal structure of ATP‐bound Rio2 from A. fulgidus (brown, PDB 1ZAO; LaRonde‐LeBlanc et al, 2005). The structures are superimposed relative to their kinase domains. The ATP visualized in the crystal structure is shown in pink to highlight the binding pocket. The arrows represent the movements of the Rio2 domains needed to reach the close conformation of the crystal structure, which is able to cleave ATP.
Remarkably, the Tsr1 density forms a helical structure that inserts between the body of the small subunit and helix 44 (Figs 4C and EV4A). This α‐helix corresponds to residues 51–75 of Tsr1 that were not resolved in the previously reported Tsr1 crystal structure (McCaughan et al, 2016). The conserved positively charged amino acids (Appendix Fig S3A) of this Tsr1 α‐helix directly interact with the minor groove of rRNA helix 44, as well as with rRNA helices 5, 11, and 24, and protein uS12 (Figs 4C and EV4A).
Figure EV4. Details of the functionally important assembly factors’ interactions in the cryo‐EM map of the cytoplasmic pre‐40S.
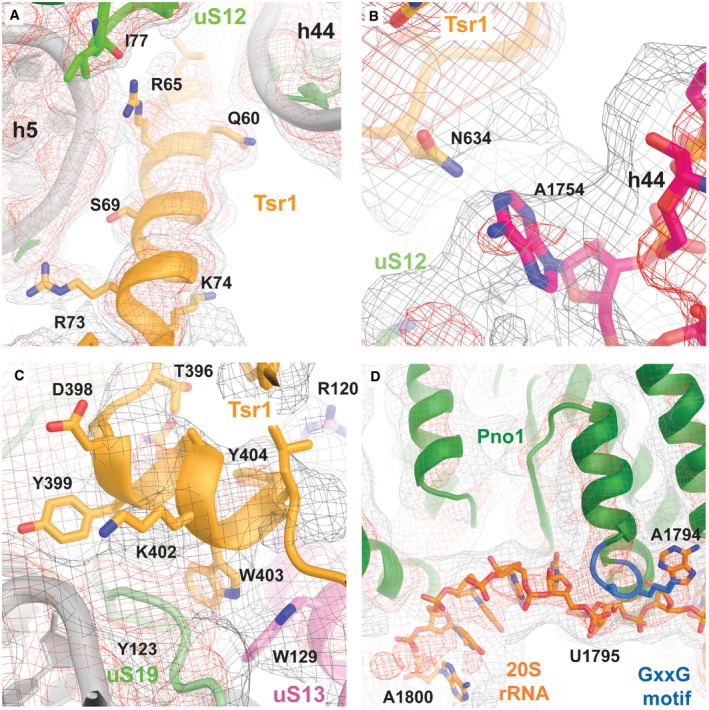
- N‐terminal α‐helix of Tsr1 (orange) interacting with helix 44.
- Visualization of the flipped out position of the decoding center base A1754 at the top of helix 44 (pink).
- Interactions of the conserved loop of Tsr1 with uS19 (green) and uS13 (pink).
- Interaction of the Pno1 (green) C‐terminal KH domain (containing a GxxG RNA binding motif shown in blue) with the 3′ terminal bases of the rRNA (orange).
In our cytoplasmic pre‐40S reconstruction, rRNA helix 44 is considerably displaced compared to its position in the mature 40S (Fig 4A and B) and its conformation would be incompatible with the joining of the 60S subunit. The conformational rearrangements also propagate to the top of helix 44, which forms the ribosomal A‐site, and result in a displacement of the decoding center bases 1755 and 1756 by approximately 16 Å (Figs 4A, B and D, and EV4B). This displacement indicates a possible role for Tsr1 in preventing the premature formation of the decoding center. Additionally, the shift of helix 44 at the decoding center allows the insertion of Rio2 (Fig 4D).
Furthermore, we could assign extra densities to residues 380–409 of Tsr1 belonging to a long linker between Tsr1 domain II and domain III, which had remained unresolved in the Tsr1 crystal structure due to conformational flexibility. The linker first contacts Rio2 and the C‐terminus of uS19 and extends along Tsr1 domain IV in the direction of the pre‐40S head. The last visible residues of the linker form specific contacts with the displaced rRNA helix 31 as well as with proteins uS13 and uS19 (Figs 4E and EV4C). These residues, 392–408, are highly conserved in eukaryotes (Appendix Fig S3B), pointing toward a key role of this interaction with these two universally conserved ribosomal proteins, whose prokaryotic homologs are important for intersubunit bridging (Jenner et al, 2010; Bowen et al, 2015) as well as for tRNA binding (Lomakin & Steitz, 2013; Graifer & Karpova, 2015).
Rio2 is bound at the decoding center of the 40S (Fig 4D). The Rio2 N‐terminal winged helix‐turn‐helix (wHTH) domain is inserted between 20S rRNA helices 1, 18, 28, 34, and 44, which form the decoding center of the mature 40S, while the bi‐lobed C‐terminal kinase domain blocks the cytoplasmic pre‐40S P‐site from tRNA binding (Fig 4B). The wHTH domain interacts with and distorts helices 18 and 44, restraining their conformation and preventing premature folding of the decoding center (Fig 4D).
Interestingly, the structure of cytoplasmic pre‐40S‐bound Rio2 shows that the two halves of the Rio2 kinase domain, which form the nucleotide binding pocket, are rotated relative to each other when compared to the crystal structures of the A. fulgidus (LaRonde‐LeBlanc & Wlodawer, 2004; LaRonde‐LeBlanc et al, 2005; Fig 4F). As a result, the residues responsible for ATP binding are separated by greater distances, which most likely render Rio2 unable to cleave ATP.
Pno1 interacts with the 3′ end of rRNA at the pre‐40S platform
Pno1 is the interaction partner of the endonuclease Nob1 that mediates the cleavage of 20S rRNA into mature 18S rRNA (Woolls et al, 2011). On the platform, close to the 40S “neck”, where eS26 is located in the mature 40S, additional density could be assigned to Pno1 (Fig 2E). The Pno1 KH‐like and KH domains were modeled using the crystal structure of the homologous P. horikoshii Pno1 protein (Jia et al, 2010; Appendix Table S4). We observed that the C‐terminal part of Pno1 would clash with the mature position of eS28 and helix 28 (Fig 5A), indicating that the tilted conformation of the cytoplasmic pre‐40S head is required for Pno1 binding. Interestingly, the KH domain is also interacting with the loop between helices 44 and 45, which is in a stretched‐out conformation due to the displaced position of helix 44 in the cytoplasmic pre‐40S.
Figure 5. Pno1 interacts with the rRNA of the platform.
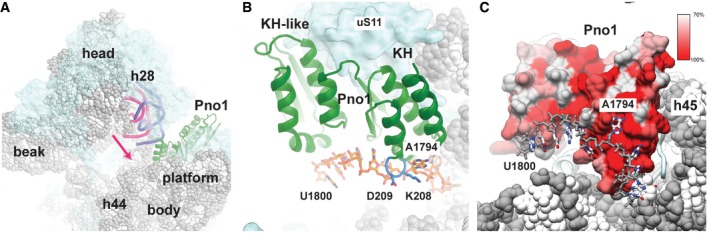
- Overview of the pre‐40S ribosome. The rRNA is represented as spheres (gray) and the r‐proteins as surfaces (light blue). In the pre‐40S particle, helix 28 (cartoon in pink) is highly distorted compared to the mature conformation (dark blue, PDB 4V88; Ben‐Shem et al, 2011) in which it would clash with Pno1 (green).
- Pno1 interacts with 20S rRNA nucleotides 1787–1800. Nucleotide A1794 is specifically recognized by the C‐terminal KH domain of Pno1, which contains a conserved GxxG RNA binding motif (blue). A1794 is flipped out, which twists the RNA such that the bases that follow bind to the surface of Pno1 (Fig EV4D).
- Surface representation of Pno1 colored by residue conservation. Highly conserved residues (red) form the surface interacting with the 20S rRNA (Appendix Fig S4).
Our density reveals that Pno1 binds the 3′ end of the rRNA at the location that would precede the final D cleavage site at nucleotide 1800 targeted by Nob1 (Figs 5B and EV4D). Particularly, nucleotides 1793–1795 are recognized in a flipped out conformation by specific interactions with the highly conserved GXXG motif of the C‐terminal KH domain of Pno1 (Appendix Fig S4), whereas the RNA up to the cleavage site is twisted such that the bases point away from the surface of the protein (Figs 5C and EV4D) and fades for the remainder of 20S pre‐rRNA sequence into featureless density.
Despite being used as the bait to isolate the pre‐40S particle, no density could be assigned to Nob1‐D15N at the D cleavage site where Nob1 was reported to be located (Strunk et al, 2011; Johnson et al, 2017) even after further focused classification (Scheres, 2016). However, it is possible that Nob1 is flexibly bound to downstream parts of the 20S rRNA (Lamanna & Karbstein, 2009; Turowski et al, 2014).
Discussion
The reconstruction of the cytoplasmic pre‐40S subunit at 3.4 Å resolution revealed how the AFs check the integrity of functionally important regions of the subunit, lock the pre‐rRNA conformation to prevent premature progression through the maturation process, and prepare the pre‐rRNA for further processing. We observe striking differences between the cytoplasmic pre‐40S state and the mature 40S ribosome exemplified by the distortion of the 3′ minor and 3′ major domains of the rRNA that is accompanied by an extreme rotation of the cytoplasmic pre‐40S head that exceeds any 40S head rotation observed during translation (Fig 3A). This stage of maturation seems to be focused on the 3′ major domain of the rRNA since it is contacted by all observed AFs except Pno1 (Fig 2A–E).
Final maturation of the pre‐40S beak
In our structure, the large positively charged surface of Enp1 directly interacts with the beak rRNA, most likely acting as a scaffold and restraining it in its observed distorted conformation (Fig 3B and Appendix Fig S2A and B). The protein kinase Hrr25 was reported to regulate the cytoplasmic maturation of the pre‐40S through the phosphorylation of Enp1 and Ltv1, leading to the release of Ltv1 (Ghalei et al, 2015), and stable incorporation of uS3 (Schafer et al, 2006). Since the displaced helix 34 prevents the formation of the mature binding pocket for the uS3 C‐terminal domain, Hrr25‐dependent incorporation of uS3 must be linked to the conformational rearrangement of helix 34 (Fig 3B and C). One possibility is that the release of Ltv1 after phosphorylation of Ltv1 and Enp1 by Hrr25 (Ghalei et al, 2015) destabilizes the binding of Enp1, which dissociates and allows the beak and uS3 to adopt their mature conformations and the head to rotate which, in turn, changes the conformation of Rio2 (Fig 6).
Figure 6. A model for cytoplasmic maturation of the pre‐40S subunit through series of interdependent conformational changes.
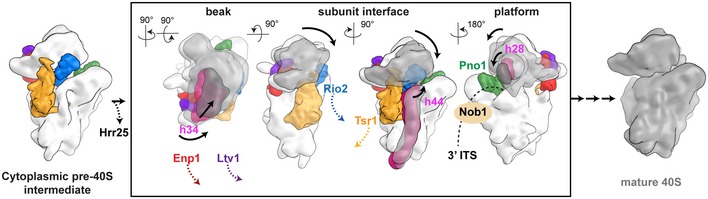
The boxed region of the scheme represents a single species and highlights the conformationally coupled changes upon the release of individual assembly factors in rRNA conformation between the pre‐40S (white, changing part in pink) and the mature 40S (gray, changed part in dark gray) without implying a sequential order. The tilt and rotation of the head as well as conformational changes in helix 34, helix 44, and helix 28 are interconnected such that conformational changes in the beak, the subunit interface, and the platform lead to the release of AFs from their respective binding sites.
Tsr1 and Rio2 probe the intersubunit interface and the decoding center
It has been suggested that the role of Tsr1 is to prevent the premature binding of 60S ribosomal subunit by sterically blocking the access of the 60S subunit (Strunk et al, 2011; McCaughan et al, 2016; Johnson et al, 2017). In the structure described here, we additionally observe that Tsr1 is responsible for stabilizing helix 44 in a shifted conformation compared to the mature 40S, which also clearly prevents the premature binding of the 60S (Figs 4A–C and EV4A).
The two domains of Rio2 are bound to the head and the body of the subunit, and therefore, their relative positioning is linked to the head rotation. In the observed conformation, Rio2 cannot be active in ATP hydrolysis since the two lobes of the kinase domain forming the nucleotide binding cleft are rotated away from each other compared to the ATP‐ or ADP‐bound structures of Rio2 homologs from A. fulgidus (Fig 4F; LaRonde‐LeBlanc & Wlodawer, 2004; LaRonde‐LeBlanc et al, 2005). Consequently, head rRNA rearrangement to its mature conformation upon dissociation of Enp1‐Ltv1 could possibly induce a closure of the Rio2 nucleotide binding cleft, thereby activating Rio2 to hydrolyze ATP that leads to autophosphorylation and to dissociation (Fig 6; LaRonde‐LeBlanc et al, 2005; Ferreira‐Cerca et al, 2012). Since both Rio2 and Tsr1 interact with helix 44 of the pre‐rRNA and they furthermore contact each other (Fig 4A, B and D), dissociation of Rio2 following autophosphorylation would lead to destabilization and dissociation of Tsr1. This allows the formation of the decoding center and accommodation of rRNA helix 44 to its mature conformation propagating conformational changes toward the platform where Pno1 interacts with the 3′ end of the rRNA (Fig 6).
The contacts of Rio2 and Tsr1 with the decoding center and important regions around it, including rRNA helices 18 and 44, and proteins uS12, uS13, and uS19 (Figs 4E and EV4C), point toward an additional function for Rio2 and Tsr1 as AFs that monitor the proper formation of this key functional region of the 40S subunit. As Rio2 and Tsr1 bind the pre‐40S in the nucleus and are needed for transport into the cytoplasm (Schafer et al, 2003), these interactions could serve as one of the “checkpoints” for nuclear export by preferentially binding only to pre‐40S particles, which contain these r‐proteins.
Repositioning of Pno1 is required for 3′ end processing
Our structure also reveals how Pno1 is bound to the 20S RNA in the region corresponding to the 3′‐end of the mature rRNA in the immediate vicinity of site D where Nob1 endonuclease binds (Fig 5B; Turowski et al, 2014). In this conformation, the close proximity of Pno1 to adjacent rRNA elements would prevent access of the Nob1 PIN domain to the D cleavage site, which explains why pre‐rRNA processing does not take place at this stage of maturation. Pno1 occupies the binding site for eS26 protein, which is not visible in our pre‐40S structure, but was readily identified by mass spectrometry. Therefore, it appears that Pno1 repositioning, or release, may permit clamping of the 3′end by eS26, a key step to trigger cleavage of the 20S pre‐rRNA by Nob1. Notably, provided that Pno1 moves from its current location, Nob1 would be able to interact with the KH‐like domain of Pno1, as observed biochemically (Woolls et al, 2011), and cleave the rRNA. In support of this notion, it was recently shown that a conformational change in Pno1 is required to allow Nob1 access for 20S pre‐rRNA cleavage (Widmann et al, 2012; Turowski et al, 2014) and that eS26 depletion impairs final cytoplasmic 20S pre‐rRNA processing (Schutz et al, 2014).
Interdependent AF departure events lead to 40S maturation
The structure of the pre‐40S subunit described here reveals the molecular basis of concerted events that take place upon departure of the visualized AFs. Although it is not possible to suggest the exact temporal sequence of maturation events, we now understand the interdependence of various AFs in stabilizing a particular conformation of the pre‐rRNA (Fig 6). Based on the structure and the available biochemical and genetic evidences, the process is likely to start with the phosphorylation of Ltv1 and Enp1 by Hrr25 that destabilizes their interactions with the beak of the pre‐40S subunit (Fig 1E; Schafer et al, 2006; Zemp et al, 2014; Ghalei et al, 2015). This leads to a conformational change in the rRNA allowing accommodation of the C‐terminal domain of uS3 (Fig 3C; Schafer et al, 2006). After or during beak formation, the 40S head likely rotates to its mature position, triggering ATP hydrolysis by Rio2 and its dissociation (LaRonde‐LeBlanc & Wlodawer, 2004; LaRonde‐LeBlanc et al, 2005; Ferreira‐Cerca et al, 2012). Since both Rio2 and Tsr1 stabilize helix 44 in a distorted conformation (Fig 4A, B and D), dissociation of Rio2 will also lead to dissociation of Tsr1. Accommodation of helix 44 into its mature position will lead to conformational changes in the rRNA at the platform, allowing formation of rRNA helix 28 and rigid docking of eS26, which then repositions Pno1 and allows binding of Nob1 for pre‐rRNA cleavage. This mechanism also implies that Nob1 would only have access to Pno1 once the 40S matures to the point of being able to associate with the 60S subunit, in agreement with previous biochemical results (Fig 6; Lebaron et al, 2012; Strunk et al, 2012; Turowski et al, 2014).
In conclusion, our structure sheds new light on the cytoplasmic maturation pathway of the 40S subunit by revealing contact sites of the assembly factors Ltv1, Enp1, Rio2, Tsr1, and Pno1, and their interactions with 20S rRNA. The cytoplasmic pre‐40S exhibits a highly distorted 3′ major and 3′ minor rRNA domain conformation stabilized by the AFs, which suggests a mechanism for the concerted release of the AFs orchestrated by the folding of the rRNA in the head of the pre‐40S subunit during the final stages of maturation. These results provide the structural basis for biochemical and genetic experiments to dissect the effects of the observed interactions on the cascade of mechanistic events leading to the formation of the mature 40S subunit.
Materials and Methods
Purification of pre‐40S ribosomes
Pre‐40S ribosomes were purified from a Saccharomyces cerevisiae strain expressing the catalytically inactive Nob1‐D15N mutant fused to an N‐terminal and cleavable Protein A tag (pA‐TEV‐FLAG‐Nob1D15N). For this purpose, a NOB1 conditional mutant strain (P Gal1 ‐NOB1) was transformed with a plasmid encoding the tagged Nob1 mutant gene (pRS315‐pA‐TEV‐FLAG‐Nob1D15N). Cells were grown at 25°C in glucose‐containing synthetic medium to an optical density of 1.5 and then harvested at 4,000 × g at 4°C for 15 min. Pellets were first washed with ddH2O and then with one pellet volume of 20 mM HEPES pH 7.4 buffer supplemented with 1.2% polyvinylpyrrolidone and protease inhibitors (Roche Complete, PMSF). The final pellet was transferred to a 20‐ml syringe and pushed directly into liquid nitrogen to produce frozen yeast noodles. Cell lysis was performed by cryo‐grinding in a Planetary Ball Mill (Retsch Pulverisette 6) to generate fine yeast powder to be stored at −80°C. To purify pre‐40S ribosomes, 3–5 g of yeast powder was resuspended in 50 ml of buffer A (20 mM HEPES pH 7.4, 110 mM KOAc, 40 mM NaCl, 10 mM MgCl2, 0.5% Triton‐X 100, 0.1% Tween 20, PMSF), cleared by centrifugation at 4,000 × g for 5 min, and then filtered with a syringe first through a 2.7‐μm and then through a 1.6‐μm filter (Whatman). The cleared lysate was incubated on a nutator for 30 min at 4°C with a 100 μl slurry of magnetic Dynabeads (Epoxy M‐270, Thermo Fischer Scientific) conjugated with rabbit IgG antibodies (Sigma‐Aldrich). IgG antibodies were conjugated to Dynabeads as described (Oeffinger et al, 2007). After incubation, beads were collected with a magnet rack and washed five times in 1 ml of buffer A and then three times in 1 ml of buffer B (buffer A without Triton‐X 100 and Tween 20). Finally, beads were resuspended in 50 μl of buffer B and then incubated at 4°C overnight with 10 units of TEV protease (Sigma‐Aldrich) to elute the purified pre‐40S ribosome. Mature 80S ribosomes were purified from Saccharomyces cerevisiae as described previously (Greber et al, 2016) and using associative conditions (20 mM HEPES pH 7.4, 100 mM KCl, 20 mM MgCl2) in the final gradient.
The RNA composition of ribosomal particles was determined by gel electrophoresis (2% w/v agarose) followed by GelRed (Biotium) staining, while the protein composition was determined by SDS–PAGE (NuPAGE 4–12% v/v Bis–Tris, Thermo Fischer Scientific) followed by silver staining. For protein identification, silver‐stained bands were cut out and identified by mass spectrometry.
Yeast methods and fluorescence microscopy
The Saccharomyces cerevisiae strains used in this study are listed in Appendix Table S1. Genomic disruptions, C‐terminal tagging and promoter switches at genomic loci were performed as described previously (Janke et al, 2004).
Plasmids used in this study are listed in Appendix Table S2. Mutant constructs of NOB1, HRR25, RIO2, and TSR1 were generated using the QuikChange site‐directed mutagenesis kit (Agilent Technologies). All cloned DNA fragments and mutagenized plasmids were verified by sequencing.
To monitor subcellular localization of Enp1 and Tsr1, yeast cells expressing catalytically inactive variants of Hrr25 (Hrr25‐K38A) and GFP‐tagged Enp1 or Tsr1 constructs were grown in YPD at 25°C to log phase. Cells were visualized using a DM6000B microscope (Leica Microsystems) equipped with a HCX PL Fluotar 63×/1.25 NA oil immersion objective (Leica Microsystems). Images were acquired with a fitted digital camera (ORCA‐ER; Hamamatsu Photonics) and Openlab software (PerkinElmer).
EM sample preparation and data collection
Quantifoil R2/2 holey carbon copper grids (Quantifoil Micro Tools) were glow‐discharged for 15 s at 15 mA using an easiGlow Discharge cleaning system (PELCO) prior to coating with graphene oxide (Pantelic et al, 2010). The sample at 70 ng/μL was used for cryo‐grid preparation using a Vitrobot (FEI Company), whose chamber was set to 4°C and 100% humidity. After incubating for 90 s in the chamber with 5 μL of sample, the grids were blotted for 12–15 s and rapidly plunge‐frozen in a mixture 2:1 of propane:ethane (Carbagas) at liquid nitrogen temperature.
One grid was used for data collection in a Titan Krios cryo‐transmission electron microscope (FEI Company) operating at 300 kV and equipped with a Falcon 3EC. A dataset of 3,083 micrographs was collected using the EPU software (FEI Company) in a range of defocus between −0.6 and −2.5 μm and a magnification of 100,720× (pixel size of 1.39 Å/pixel). Each micrograph was collected in integration mode split into 38 frames using a total dose of 40 electrons/Å2.
Data processing and reconstruction
The stacks of frames were aligned to correct for beam‐induced motion and dose‐weighted using MotionCor2 (Zheng et al, 2017). The non‐dose‐weighted micrographs were used to estimate the Contrast Transfer Function (CTF) using GCTF (Zhang, 2016); 2,867 good micrographs were selected by evaluating the quality of their CTF estimation, the quality of their power spectra, and the quality of the micrographs themselves. From these, roughly 595000 particles were picked using GAutomatch (http://www.mrc-lmb.cam.ac.uk/kzhang/) using as template 2D averages of 5,000 particles picked without reference. These particles were then processed using Relion 2.1 (Kimanius et al, 2016). The particles were extracted and binned 5 times (pixel size of 6.95 Å/pixel) before being used for a reference‐free 2D classification.
After removing images representing ice crystals and carbon edges, we separately selected approximately 145,000 80S‐like particles and 236,000 40S‐like particles. The 40S‐like particles were refined using a mature 40S density as reference followed by a 3D classification without alignment of the particles. The 165,000 good particles containing Tsr1 were selected for a refinement using the full pixel size (1.39 Å/pixel). After post‐processing and sharpening, the reconstruction reached an overall resolution of 3.4 Å (Fig EV2A).
Further focused classifications were performed with different masks around densities corresponding to Enp1, Rio2, Dim1, and Pno1 generated manually using UCSF Chimera (Pettersen et al, 2004).
The 80S‐like particles that were separated after the 2D classification were refined using the full pixel size (1.39 Å/pixel). The reconstruction reached an overall resolution of 3.1 Å after post‐processing and sharpening (Fig EV1 and Appendix Fig S1A and B).
Modeling and docking
First, atomic models of the 40S head and body obtained from the crystal structure of the mature S. cerevisiae 80S (PDB: 4V88; Ben‐Shem et al, 2011) were docked as rigid bodies into the EM density of the high‐resolution 40S after removal of eS10, eS26, eS31, and STM1, which are not visible from the pre‐40S, using UCSF Chimera (Pettersen et al, 2004). uS3 was split into its N‐ and C‐terminal domains, and each domain was docked independently. For initial interpretation of the AFs, the crystal structures of yeast Tsr1 (PDB: 5IW7; McCaughan et al, 2016) and the yeast Enp1‐Ltv1 complex (PDB: 5WWO; Sun et al, 2017) as well as homology models based on the crystal structures of A. fulgidus Rio2 (PDB: 1ZAO; LaRonde‐LeBlanc et al, 2005), human Dim1 (PDB: 1ZQ9) and Pyrococcus horikoshii Pno1 (PDB: 3AEV; Jia et al, 2010), which were generated using PHYRE2 (Kelley et al, 2015), could be unambiguously fitted into the EM densities obtained from the focused alignment runs (Figs 2B–E and EV1, and Appendix Table S4). Due to the limited resolution obtained for the densities of Dim1 and the Enp1‐Ltv1 complex, these AFs were docked as rigid bodies, whereas the remaining AFs that were resolved between 3.6 and 4.0 Å (Appendix Table S3) were manually rebuilt using COOT (Emsley et al, 2010) and O (Jones, 2004). In the r‐proteins and rRNA, structural differences between the mature 40S and the pre‐40S were observed in the vicinity of the AFs, and these areas were readjusted in areas with sufficient resolution. Major rRNA rearrangements included the beak and helix 34 in the pre‐40S head as well as helix 44 in the body. With the exception of Dim1, a final pre‐40S model was then assembled from the coordinates built into the individual EM densities obtained after focused alignment and fitted into the best resolved 3.4 Å EM density map. To regularize the geometry and optimize the fit of the final atomic model to cryo‐EM map, the structure was then refined in reciprocal space using PHENIX.REFINE (Adams et al, 2010) against structure factors back‐calculated from the high‐resolution EM density as described earlier (Greber et al, 2014; Table EV1). The refinement was stabilized in areas of weaker density by applying Ramachandran restraints and secondary structure restraints for protein α‐helices and β‐strands as well as RNA base pair restraints for the rRNA. An optimal weighting of the model geometry versus the structure factors during refinement was obtained with a wxc value of 1.2. The model was validated by comparison of the FSCs calculated for the experimental density and the model (Fig EV2A). Additionally, the pre‐40S model was refined into the density reconstructed from half of the particles (halfset1), and FSCs were calculated between this model and the density of the same half of the dataset (halfset1) and the density of the other half (halfset2). The good agreement between these two curves and the FSC calculated for the whole dataset and the model indicates that the structure is not overfitted (Fig EV2B; Fernandez et al, 2014; Greber et al, 2014).
Figure generation
Local resolutions were estimated using the “relion_postprocess” from Relion 2.1 (Kimanius et al, 2016). Figures showing cryo‐EM structures were generated using UCSF Chimera (Pettersen et al, 2004) and Pymol (The Pymol Molecular Graphics System version 1.8.4.0 Schrödinger). The electrostatic surface charge distribution was calculated using the APBS (Adaptive Poisson–Boltzmann Solver; Baker et al, 2001; Lerner & Carlson, 2006) plug‐in of Pymol. Multisequence alignments were performed using Clustal Omega (Sievers et al, 2011) and visualized with ESPript (Robert & Gouet, 2014).
Data and materials availability
The final model containing the cytoplasmic pre‐40S with Enp1, Ltv1, Tsr1, Rio2, and Pno1 is available from the Protein Data Bank (PDB) under accession number 6FAI. The cryo‐EM map of the unclassified pre‐40S is deposited in the Electron Microscopy Data Bank (EMDB) under accession number EMD‐4214, as well as the reconstructions from the focus classifications, which can be found under the accession numbers EMD‐4216 (Rio2), EMD‐4217 (Enp1), and EMD‐4218 (Dim1), respectively. The map of the empty 80S is deposited in the EMDB under accession number EMD‐4215.
Author contributions
CP and VGP conceived the project. CP designed the system to isolate the trapped pre‐40S, performed the purification and validated the preparations by negative‐stain electron microscopy. AS prepared grids for cryo‐EM, collected electron micrographs, and processed them with the aid of DB. The atomic model was interpreted by ML and MW, SG and PK‐N performed yeast in vivo experiments. All authors were involved in the interpretation of results. AS, CP, MW, DB, VGP, and NB wrote the manuscript.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting information
Appendix
Expanded View Figures PDF
Table EV1
Review Process File
Acknowledgements
We would like to thank the ETH Zürich scientific center for optical and electron microscopy (ScopeM) and Center for Microscopy and Image Analysis, University of Zurich, for access to electron microscopy equipment and are indebted to P. Tittmann for technical support. We thank all members of the Panse laboratory for enthusiastic discussions, and the Institute of Medical Microbiology and the UZH for continued support. VG Panse is supported by grants from the Swiss National Science Foundation, NCCR RNA & Disease, Novartis Foundation, Olga Mayenfisch Stiftung, and a Starting Grant Award from the European Research Council (EURIBIO260676). This work was supported by the Swiss National Science Foundation grant and via the National Centre of Excellence in RNA and Disease & project funding 138262 to N. Ban.
The EMBO Journal (2018) 37: e98499
Contributor Information
Vikram Govind Panse, Email: vpanse@imm.uzh.ch.
Nenad Ban, Email: ban@mol.biol.ethz.ch.
References
- Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse‐Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH (2010) PHENIX: a comprehensive Python‐based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr 66: 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alksne LE, Anthony RA, Liebman SW, Warner JR (1993) An accuracy center in the ribosome conserved over 2 billion years. Proc Natl Acad Sci USA 90: 9538–9541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA (2001) Electrostatics of nanosystems: application to microtubules and the ribosome. Proc Natl Acad Sci USA 98: 10037–10041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barandun J, Chaker‐Margot M, Hunziker M, Molloy KR, Chait BT, Klinge S (2017) The complete structure of the small‐subunit processome. Nat Struct Mol Biol 24: 944–953 [DOI] [PubMed] [Google Scholar]
- Ben‐Shem A, Garreau de Loubresse N, Melnikov S, Jenner L, Yusupova G, Yusupov M (2011) The structure of the eukaryotic ribosome at 3.0 A resolution. Science 334: 1524–1529 [DOI] [PubMed] [Google Scholar]
- Boehringer D, O'Farrell HC, Rife JP, Ban N (2012) Structural insights into methyltransferase KsgA function in 30S ribosomal subunit biogenesis. J Biol Chem 287: 10453–10459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowen AM, Musalgaonkar S, Moomau CA, Gulay SP, Mirvis M, Dinman JD (2015) Ribosomal protein uS19 mutants reveal its role in coordinating ribosome structure and function. Translation (Austin) 3: e1117703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bussiere C, Hashem Y, Arora S, Frank J, Johnson AW (2012) Integrity of the P‐site is probed during maturation of the 60S ribosomal subunit. J Cell Biol 197: 747–759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaker‐Margot M, Barandun J, Hunziker M, Klinge S (2017) Architecture of the yeast small subunit processome. Science 355: eaal1880 [DOI] [PubMed] [Google Scholar]
- Cheng J, Kellner N, Berninghausen O, Hurt E, Beckmann R (2017) 3.2‐A‐resolution structure of the 90S preribosome before A1 pre‐rRNA cleavage. Nat Struct Mol Biol 24: 954–964 [DOI] [PubMed] [Google Scholar]
- Dragon F, Gallagher JE, Compagnone‐Post PA, Mitchell BM, Porwancher KA, Wehner KA, Wormsley S, Settlage RE, Shabanowitz J, Osheim Y, Beyer AL, Hunt DF, Baserga SJ (2002) A large nucleolar U3 ribonucleoprotein required for 18S ribosomal RNA biogenesis. Nature 417: 967–970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, Cowtan K (2010) Features and development of Coot. Acta Crystallogr D Biol Crystallogr 66: 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fassio CA, Schofield BJ, Seiser RM, Johnson AW, Lycan DE (2010) Dominant mutations in the late 40S biogenesis factor Ltv1 affect cytoplasmic maturation of the small ribosomal subunit in Saccharomyces cerevisiae. Genetics 185: 199–209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fatica A, Oeffinger M, Dlakic M, Tollervey D (2003) Nob1p is required for cleavage of the 3′ end of 18S rRNA. Mol Cell Biol 23: 1798–1807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fatica A, Tollervey D, Dlakic M (2004) PIN domain of Nob1p is required for D‐site cleavage in 20S pre‐rRNA. RNA 10: 1698–1701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez IS, Bai XC, Murshudov G, Scheres SH, Ramakrishnan V (2014) Initiation of translation by cricket paralysis virus IRES requires its translocation in the ribosome. Cell 157: 823–831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira‐Cerca S, Sagar V, Schafer T, Diop M, Wesseling AM, Lu H, Chai E, Hurt E, LaRonde‐LeBlanc N (2012) ATPase‐dependent role of the atypical kinase Rio2 on the evolving pre‐40S ribosomal subunit. Nat Struct Mol Biol 19: 1316–1323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghalei H, Schaub FX, Doherty JR, Noguchi Y, Roush WR, Cleveland JL, Stroupe ME, Karbstein K (2015) Hrr25/CK1delta‐directed release of Ltv1 from pre‐40S ribosomes is necessary for ribosome assembly and cell growth. J Cell Biol 208: 745–759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graifer D, Karpova G (2015) Interaction of tRNA with eukaryotic ribosome. Int J Mol Sci 16: 7173–7194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granneman S, Petfalski E, Swiatkowska A, Tollervey D (2010) Cracking pre‐40S ribosomal subunit structure by systematic analyses of RNA‐protein cross‐linking. EMBO J 29: 2026–2036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greber BJ, Boehringer D, Leibundgut M, Bieri P, Leitner A, Schmitz N, Aebersold R, Ban N (2014) The complete structure of the large subunit of the mammalian mitochondrial ribosome. Nature 515: 283–286 [DOI] [PubMed] [Google Scholar]
- Greber BJ (2016) Mechanistic insight into eukaryotic 60S ribosomal subunit biogenesis by cryo‐electron microscopy. RNA 22: 1643–1662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greber BJ, Gerhardy S, Leitner A, Leibundgut M, Salem M, Boehringer D, Leulliot N, Aebersold R, Panse VG, Ban N (2016) Insertion of the biogenesis factor Rei1 probes the ribosomal tunnel during 60S maturation. Cell 164: 91–102 [DOI] [PubMed] [Google Scholar]
- Hoekstra MF, Liskay RM, Ou AC, DeMaggio AJ, Burbee DG, Heffron F (1991) HRR25, a putative protein kinase from budding yeast: association with repair of damaged DNA. Science 253: 1031–1034 [DOI] [PubMed] [Google Scholar]
- Janke C, Magiera MM, Rathfelder N, Taxis C, Reber S, Maekawa H, Moreno‐Borchart A, Doenges G, Schwob E, Schiebel E, Knop M (2004) A versatile toolbox for PCR‐based tagging of yeast genes: new fluorescent proteins, more markers and promoter substitution cassettes. Yeast 21: 947–962 [DOI] [PubMed] [Google Scholar]
- Jenner L, Demeshkina N, Yusupova G, Yusupov M (2010) Structural rearrangements of the ribosome at the tRNA proofreading step. Nat Struct Mol Biol 17: 1072–1078 [DOI] [PubMed] [Google Scholar]
- Jia MZ, Horita S, Nagata K, Tanokura M (2010) An archaeal Dim2‐like protein, aDim2p, forms a ternary complex with a/eIF2 alpha and the 3′ end fragment of 16S rRNA. J Mol Biol 398: 774–785 [DOI] [PubMed] [Google Scholar]
- Johnson MC, Ghalei H, Doxtader KA, Karbstein K, Stroupe ME (2017) Structural heterogeneity in Pre‐40S ribosomes. Structure 25: 329–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones TA (2004) Interactive electron‐density map interpretation: from INTER to O. Acta Crystallogr D Biol Crystallogr 60: 2115–2125 [DOI] [PubMed] [Google Scholar]
- Karbstein K (2013) Quality control mechanisms during ribosome maturation. Trends Cell Biol 23: 242–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJ (2015) The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc 10: 845–858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimanius D, Forsberg BO, Scheres SH, Lindahl E (2016) Accelerated cryo‐EM structure determination with parallelisation using GPUs in RELION‐2. Elife 5: e18722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konikkat S, Woolford JL Jr (2017) Principles of 60S ribosomal subunit assembly emerging from recent studies in yeast. Biochem J 474: 195–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornprobst M, Turk M, Kellner N, Cheng J, Flemming D, Kos‐Braun I, Kos M, Thoms M, Berninghausen O, Beckmann R, Hurt E (2016) Architecture of the 90S pre‐ribosome: a structural view on the birth of the eukaryotic ribosome. Cell 166: 380–393 [DOI] [PubMed] [Google Scholar]
- Kressler D, Hurt E, Bassler J (2017) A puzzle of life: crafting ribosomal subunits. Trends Biochem Sci 42: 640–654 [DOI] [PubMed] [Google Scholar]
- Lamanna AC, Karbstein K (2009) Nob1 binds the single‐stranded cleavage site D at the 3′‐end of 18S rRNA with its PIN domain. Proc Natl Acad Sci USA 106: 14259–14264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaRonde‐LeBlanc N, Wlodawer A (2004) Crystal structure of A. fulgidus Rio2 defines a new family of serine protein kinases. Structure 12: 1585–1594 [DOI] [PubMed] [Google Scholar]
- LaRonde‐LeBlanc N, Guszczynski T, Copeland T, Wlodawer A (2005) Autophosphorylation of Archaeoglobus fulgidus Rio2 and crystal structures of its nucleotide‐metal ion complexes. FEBS J 272: 2800–2810 [DOI] [PubMed] [Google Scholar]
- Lebaron S, Schneider C, van Nues RW, Swiatkowska A, Walsh D, Bottcher B, Granneman S, Watkins NJ, Tollervey D (2012) Proofreading of pre‐40S ribosome maturation by a translation initiation factor and 60S subunits. Nat Struct Mol Biol 19: 744–753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerner MG, Carlson HA (2006) APBS plugin for PyMOL. Ann Arbor: University of Michigan; [Google Scholar]
- Lo KY, Li Z, Bussiere C, Bresson S, Marcotte EM, Johnson AW (2010) Defining the pathway of cytoplasmic maturation of the 60S ribosomal subunit. Mol Cell 39: 196–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomakin IB, Steitz TA (2013) The initiation of mammalian protein synthesis and mRNA scanning mechanism. Nature 500: 307–311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma C, Wu S, Li N, Chen Y, Yan K, Li Z, Zheng L, Lei J, Woolford JL Jr, Gao N (2017) Structural snapshot of cytoplasmic pre‐60S ribosomal particles bound by Nmd3, Lsg1, Tif6 and Reh1. Nat Struct Mol Biol 24: 214–220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malyutin AG, Musalgaonkar S, Patchett S, Frank J, Johnson AW (2017) Nmd3 is a structural mimic of eIF5A, and activates the cpGTPase Lsg1 during 60S ribosome biogenesis. EMBO J 36: 854–868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCaughan UM, Jayachandran U, Shchepachev V, Chen ZA, Rappsilber J, Tollervey D, Cook AG (2016) Pre‐40S ribosome biogenesis factor Tsr1 is an inactive structural mimic of translational GTPases. Nat Commun 7: 11789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitterer V, Gantenbein N, Birner‐Gruenberger R, Murat G, Bergler H, Kressler D, Pertschy B (2016) Nuclear import of dimerized ribosomal protein Rps3 in complex with its chaperone Yar1. Sci Rep 6: 36714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oeffinger M, Wei KE, Rogers R, DeGrasse JA, Chait BT, Aitchison JD, Rout MP (2007) Comprehensive analysis of diverse ribonucleoprotein complexes. Nat Methods 4: 951–956 [DOI] [PubMed] [Google Scholar]
- Pantelic RS, Meyer JC, Kaiser U, Baumeister W, Plitzko JM (2010) Graphene oxide: a substrate for optimizing preparations of frozen‐hydrated samples. J Struct Biol 170: 152–156 [DOI] [PubMed] [Google Scholar]
- Pena C, Hurt E, Panse VG (2017) Eukaryotic ribosome assembly, transport and quality control. Nat Struct Mol Biol 24: 689–699 [DOI] [PubMed] [Google Scholar]
- Pertschy B, Schneider C, Gnadig M, Schafer T, Tollervey D, Hurt E (2009) RNA helicase Prp43 and its co‐factor Pfa1 promote 20 to 18 S rRNA processing catalyzed by the endonuclease Nob1. J Biol Chem 284: 35079–35091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE (2004) UCSF Chimera–a visualization system for exploratory research and analysis. J Comput Chem 25: 1605–1612 [DOI] [PubMed] [Google Scholar]
- Robert X, Gouet P (2014) Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res 42: W320–W324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schafer T, Strauss D, Petfalski E, Tollervey D, Hurt E (2003) The path from nucleolar 90S to cytoplasmic 40S pre‐ribosomes. EMBO J 22: 1370–1380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schafer T, Maco B, Petfalski E, Tollervey D, Bottcher B, Aebi U, Hurt E (2006) Hrr25‐dependent phosphorylation state regulates organization of the pre‐40S subunit. Nature 441: 651–655 [DOI] [PubMed] [Google Scholar]
- Scheres SH (2016) Processing of structurally heterogeneous cryo‐EM data in RELION. Methods Enzymol 579: 125–157 [DOI] [PubMed] [Google Scholar]
- Schutz S, Fischer U, Altvater M, Nerurkar P, Pena C, Gerber M, Chang Y, Caesar S, Schubert OT, Schlenstedt G, Panse VG (2014) A RanGTP‐independent mechanism allows ribosomal protein nuclear import for ribosome assembly. Elife 3: e03473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Soding J, Thompson JD, Higgins DG (2011) Fast, scalable generation of high‐quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol 7: 539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strunk BS, Loucks CR, Su M, Vashisth H, Cheng S, Schilling J, Brooks CL III, Karbstein K, Skiniotis G (2011) Ribosome assembly factors prevent premature translation initiation by 40S assembly intermediates. Science 333: 1449–1453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strunk BS, Novak MN, Young CL, Karbstein K (2012) A translation‐like cycle is a quality control checkpoint for maturing 40S ribosome subunits. Cell 150: 111–121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Q, Zhu X, Qi J, An W, Lan P, Tan D, Chen R, Wang B, Zheng S, Zhang C, Chen X, Zhang W, Chen J, Dong MQ, Ye K (2017) Molecular architecture of the 90S small subunit pre‐ribosome. Elife 6: e22086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Triman KL (2007) Mutational analysis of the ribosome. Adv Genet 58: 89–119 [DOI] [PubMed] [Google Scholar]
- Turowski TW, Lebaron S, Zhang E, Peil L, Dudnakova T, Petfalski E, Granneman S, Rappsilber J, Tollervey D (2014) Rio1 mediates ATP‐dependent final maturation of 40S ribosomal subunits. Nucleic Acids Res 42: 12189–12199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widmann B, Wandrey F, Badertscher L, Wyler E, Pfannstiel J, Zemp I, Kutay U (2012) The kinase activity of human Rio1 is required for final steps of cytoplasmic maturation of 40S subunits. Mol Biol Cell 23: 22–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolford JL Jr, Baserga SJ (2013) Ribosome biogenesis in the yeast Saccharomyces cerevisiae. Genetics 195: 643–681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolls HA, Lamanna AC, Karbstein K (2011) Roles of Dim2 in ribosome assembly. J Biol Chem 286: 2578–2586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zemp I, Wandrey F, Rao S, Ashiono C, Wyler E, Montellese C, Kutay U (2014) CK1delta and CK1epsilon are components of human 40S subunit precursors required for cytoplasmic 40S maturation. J Cell Sci 127: 1242–1253 [DOI] [PubMed] [Google Scholar]
- Zhang K (2016) Gctf: real‐time CTF determination and correction. J Struct Biol 193: 1–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng SQ, Palovcak E, Armache JP, Verba KA, Cheng Y, Agard DA (2017) MotionCor2: anisotropic correction of beam‐induced motion for improved cryo‐electron microscopy. Nat Methods 14: 331–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix
Expanded View Figures PDF
Table EV1
Review Process File


