Abstract
The Mis18 complex is a critical player in determining when and where centromeres are built. A new study identifies Polo-like kinase (Plk1) as a positive regulator required for the localization of Mis18 to centromeres. This is a critical step that is essential for proper centromere function and maintaining the integrity of the genome.
In order to accurately transmit genetic information to daughter cells during mitotic division, vertebrate cells must maintain a single centromere on each chromosome. The centromere is the chromatin site on which the kinetochore will assemble during mitosis and will attach the chromosome to the mitotic spindle. The key determinant of centromere position in most eukaryotes is the presence of the centromere-specific histone H3 variant CENP-A. The site of centromere formation and CENP-A deposition is determined epigenetically in higher eukaryotes, depending on the proteins present at the centromere but not the underlying DNA sequence [1–3]. A new study by McKinley et al. [4] published recently in Cell provides important insight into how centromere assembly is controlled by demonstrating that the mitotic kinase Plk1 is a positive regulator of new CENP-A deposition.
Centromeric CENP-A nucleosomes are highly stable and are quantitatively retained during replication of centromeric DNA in S-phase [5,6]. The redistribution of CENP-A nucleosomes between the two new DNA strands is necessary to maintain the epigenetic mark of the centromere and leads to the dilution of CENP-A nucleosomes. Therefore, new CENP-A nucleosomes must be assembled during each cell cycle to maintain CENP-A at centromeres, and to ensure that the epigenetic mark is not lost over multiple generations. Canonical histone H3.1 nucleosomes contained within general chromatin are restored to their full complement on each daughter strand during DNA replication [3]. However, CENP-A nucleosomes are not deposited concurrently with DNA replication. Instead, new CENP-A nucleosomes are deposited in early G1 in vertebrate cells [5].
Centromere assembly is thought to be controlled by a process of licensing that restricts the assembly of new CENP-A to the site of the existing centromere. This process relies on temporal control of new CENP-A deposition restricting it to G1. The unique timing of CENP-A deposition suggests a novel temporal control mechanism that is linked to the progression of cells through mitosis. Two kinases appear to provide positive and negative regulation of new CENP-A deposition to ensure it occurs exclusively in G1 phase. The new findings by McKinley et al. [4] reveal that the mitotic Polo-like kinase 1 (Plk1) is required for new CENP-A deposition in early G1. This complements earlier studies where CDK1 activity was shown to negatively regulate CENP-A deposition [7]. CDK1 activity prevents deposition from occurring prior to completion of mitosis, after which time Plk1 takes over to activate new CENP-A deposition.
Two of the factors known to be required for orchestrating CENP-A deposition in human cells are the Mis18 complex and the CENP-A-specific chaperone, Holliday junction recognition protein (HJURP) [8–11]. The Mis18 complex is composed of Mis18α, Mis18β, and M18BP1 (also known as Mis18BP1 or hsKNL2) in human cells, and is recruited to centromeres beginning in late telophase and persists through early G1 [8]. Mis18 localizes to centromeres just prior to the pre-nucleosomal HJURP/CENP-A/H4 complex and is absolutely required for HJURP to reach centromeres [12,13].
Work by the Cheeseman and Jansen labs together demonstrated that a key event in controlling the timing of CENP-A deposition is the regulation of Mis18 complex localization by phosphorylation. Silva et al. demonstrated that phosphorylation of M18BP1 by CDK1 and CDK2 negatively regulates M18BP1’s ability to localize to centromeres. Inhibiting CDK activity caused premature Mis18 complex loading onto centromeres in G2 and resulted in early CENP-A deposition [7]. The negative regulation of Mis18 complex localization by CDK phosphorylation agrees with the observed Mis18 complex localization at centromeres only after anaphase onset when CDKs are rapidly degraded and no longer present to phosphorylate Mis18.
The new work from McKinley et al. [4] reveals a complementary regulatory arm in the Mis18 complex localization pathway. Affinity purification of the Mis18 complex reveals a new kinase interacting with the CENP-A deposition machinery. Polo-like kinase 1 Plk1 purified with the Mis18 complex from cells synchronized in the early G1 CENP-A deposition timeframe. Although Plk1 is frequently considered to be a mitotic kinase, McKinley et al. found that Plk1 localized to centromeres and was active during early G1 phase. Its localization was dependent on Mis18. Strikingly, inhibiting Plk1 abolished CENP-A deposition during early G1, verifying its role in the centromere maintenance pathway.
In order to understand why Plk1 is required for new CENP-A deposition, McKinley et al. investigated if Plk1 could directly phosphorylate the Mis18 complex. They found that Plk1 could bind and phosphorylate all three Mis18 complex components in vitro. However, mutating the Plk1 sites specifically on M18BP1 caused a severe defect in new CENP-A loading. M18BP1 with mutations in these sites failed to localize to centromeres and, complementary to this, Plk1 inhibition caused reduced Mis18α localization at centromeres. This result is consistent with the previous observation that centromere localization of the Mis18 complex members is interdependent, such that if one member is suppressed, the remaining complex members are lost from the centromere [8]. If phosphorylation of M18BP1 by Plk1 is strictly required for centromere localization of the Mis18 complex, this implies that rescuing M18BP1 localization to centromeres should also rescue the loss of CENP-A deposition seen following Plk1 inhibition. McKinley et al. elegantly show that this is the case by rescuing the non-phosphorylatable M18BP1 centromere localization by fusing it to the constitutive centromere protein CENP-C. Sure enough, artificially localizing the non-phosphorylatable M18BP1 to centromeres could rescue CENP-A deposition.
The positive regulation of Mis18 complex localization by Plk1 works alongside the negative regulation by CDKs previously documented by Silva et al. to ensure precise temporal regulation of CENP-A deposition. McKinley et al. additionally find a role for CDKs in regulating complex formation of Mis18. By mutating the CDK sites on M18BP1 and artificially targeting it to centromeres using their CENP-C fusion, they were able to induce early Mis18α recruitment during mitosis. They additionally show the early CENP-A loading following CDK inhibition originally demonstrated by Silva et al. also required Plk1, strongly suggesting that both kinases are required for CENP-A deposition.
These exciting new findings by McKinley et al. enhance our understanding of how centromere licensing is initiated. The concerted action of Plk1- and CDK-dependent phosphorylation of the Mis18 complex restricts CENP-A deposition to early G1, providing the temporal control for new centromere assembly (Figure 1). The Mis18 complex integrates temporal and spatial control of new CENP-A deposition by specifically recognizing the constitutive centromere protein CENP-C to determine the site of new CENP-A deposition [13,14]. Many new questions arise from the work by McKinley et al., including how Plk1 phosphorylation facilitates or stabilizes Mis18 complex centromere localization and how CDKs regulate Mis18 complex assembly at the centromere. While control of Mis18 is an important branch of the pathway, additional levels of control may also be at work, possibly through direct regulation of HJURP localization and activity. Other licensing processes in the cell, such as origin firing in DNA replication, are restricted to a single event in each cell cycle. Whether individual sites of new CENP-A assembly are also restricted to a single round of CENP-A deposition per cell cycle in not known. McKinley et al.’s findings of Plk1 as a novel positive regulator of Mis18 complex localization and CENP-A deposition sheds new light on the initiation of centromere licensing that ensures faithful segregation of the genome.
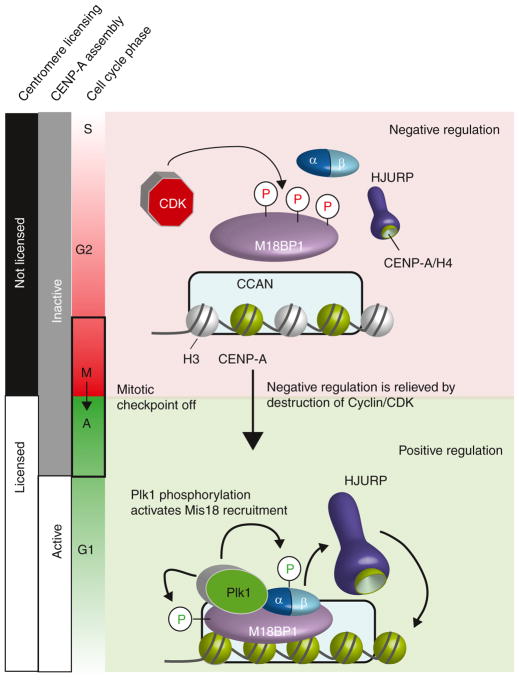
Figure 1.
Centromeric licensing is negatively and positively regulated through the phosphorylation of M18BP1.
CDK phosphorylation during G2 and mitosis, prior to the metaphase-to-anaphase transition, limits the ability of M18BP1 to interact with Mis18α/β and to be recruited to existing centromeres through its interactions with the CCAN (constitutive centromere associated network). Satisfaction of the mitotic checkpoint, leading to CDK degradation, relieves negative inhibition of centromere licensing, placing it under the control of positive regulation by Plk1 phosphorylation. Plk1 is recruited to centromeres through an interaction with the Mis18 complex. Plk1 phosphorylation of Mis18BP1 positively regulates M18BP1’s localization at centromeres. HJURP recruitment and new CENP-A assembly occurs in early G1 following the licensing of existing centromeres by Mis18 complex recruitment.
References
- 1.Stellfox ME, Bailey AO, Foltz DR. Putting CENP-A in its place. Cell Mol Life Sci. 2012;70:387–406. doi: 10.1007/s00018-012-1048-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Probst AV, Almouzni G. Pericentric heterochromatin: dynamic organization during early development in mammals. Differentiation. 2008;76:15–23. doi: 10.1111/j.1432-0436.2007.00220.x. [DOI] [PubMed] [Google Scholar]
- 3.Probst AV, Dunleavy E, Almouzni G. Epigenetic inheritance during the cell cycle. Nat Rev Mol Cell Biol. 2009;10:192–206. doi: 10.1038/nrm2640. [DOI] [PubMed] [Google Scholar]
- 4.McKinley KL, Cheeseman IM. Polo-like kinase 1 licences CENP-A deposition at centromeres. Cell. 2014;158:397–411. doi: 10.1016/j.cell.2014.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jansen LE, Black BE, Foltz DR, Cleveland DW. Propagation of centromeric chromatin requires exit from mitosis. J Cell Biol. 2007;176:795–805. doi: 10.1083/jcb.200701066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bodor DL, Valente LP, Mata JF, Black BE, Jansen LE. Assembly in G1 phase and long-term stability are unique intrinsic features of CENP-A nucleosomes. Mol Biol Cell. 2013;24:923–932. doi: 10.1091/mbc.E13-01-0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Silva MC, Bodor DL, Stellfox ME, Martins NM, Hochegger H, Foltz DR, Jansen LE. Cdk activity couples epigenetic centromere inheritance to cell cycle progression. Dev Cell. 2012;22:52–63. doi: 10.1016/j.devcel.2011.10.014. [DOI] [PubMed] [Google Scholar]
- 8.Fujita Y, Hayashi T, Kiyomitsu T, Toyoda Y, Kokubu A, Obuse C, Yanagida M. Priming of centromere for CENP-A recruitment by human hMis18alpha, hMis18beta, and M18BP1. Dev Cell. 2007;12:17–30. doi: 10.1016/j.devcel.2006.11.002. [DOI] [PubMed] [Google Scholar]
- 9.Foltz DR, Jansen LE, Bailey AO, Yates JR, 3rd, Bassett EA, Wood S, Black BE, Cleveland DW. Centromere-specific assembly of CENP-a nucleosomes is mediated by HJURP. Cell. 2009;137:472–484. doi: 10.1016/j.cell.2009.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dunleavy EM, Roche D, Tagami H, Lacoste N, Ray-Gallet D, Nakamura Y, Daigo Y, Nakatani Y, Almouzni-Pettinotti G. HJURP is a cell-cycledependent maintenance and deposition factor of CENP-A at centromeres. Cell. 2009;137:485–497. doi: 10.1016/j.cell.2009.02.040. [DOI] [PubMed] [Google Scholar]
- 11.Maddox PS, Hyndman F, Monen J, Oegema K, Desai A. Functional genomics identifies a Myb domain-containing protein family required for assembly of CENP-A chromatin. J Cell Biol. 2007;176:757–763. doi: 10.1083/jcb.200701065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barnhart MC, Kuich PH, Stellfox ME, Ward JA, Bassett EA, Black BE, Foltz DR. HJURP is a CENP-A chromatin assembly factor sufficient to form a functional de novo kinetochore. J Cell Biol. 2011;194:229–243. doi: 10.1083/jcb.201012017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Moree B, Meyer CB, Fuller CJ, Straight AF. CENP-C recruits M18BP1 to centromeres to promote CENP-A chromatin assembly. J Cell Biol. 2011;194:855–871. doi: 10.1083/jcb.201106079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dambacher S, Deng W, Hahn M, Sadic D, Frohlich J, Nuber A, Hoischen C, Diekmann S, Leonhardt H, Schotta G. CENP-C facilitates the recruitment of M18BP1 to centromeric chromatin. Nucleus. 2012;3:101–110. doi: 10.4161/nucl.18955. [DOI] [PMC free article] [PubMed] [Google Scholar]