Isolated gonadotropin-releasing hormone (GNRH) deficiency (IGD) is a rare highly heterogeneous disorder characterized by abnormal pubertal development and/or infertility and refers to a wide spectrum of different reproductive phenotypes, comprising GnRH deficiency with anosmia (Kallmann Syndrome, KS) and normosmic idiopathic hypogonadotrpic hypogonadism (nIHH) (2, 3), as well as a variety of non-reproductive phenotypes. This phenotypic heterogeneity is accompanied by a considerable genetic heterogeneity that has permitted >35 genes implicated in the pathogenesis of this disorder, with the genetic basis of IGD being uncovered in about 45% of patients. Several studies have estimated the prevalence of IGD genes in specific ethnic populations (1, 4, 5, 6, 7, 8) and geographically delimitated populations are enriched for specific genes that arise in the backbone of shared inherited haplotypes (4, 9, 10, 11, 12, 13, 30). With this letter, we would like to share our experience in harnessing the power of a large Greek population of IGD subjects, derived from different referral sources around Greece, to investigate the genetic basis of IGD and uncover the prevalence of gene variations in 14 major genetic players of IGD.
In this study, we analyzed 81 IGD patients of Greek origin. Detailed medical history from the proband and the extended family was obtained, including ascertainment of age at puberty, fertility and a variety of other non-reproductive features. Consent was obtained from each patient or subject after full explanation of the purpose and nature of all procedures used. The investigation was approved by the local ethical committee, functioning according to the 3rd edition of the Guidelines on the Practice of Ethical Committees in Medical Research issued by the Royal College of Physicians of London.
Sanger Sequencing for 14 well-validated genes (14) including ANOS1, previously known as KAL1, (OMIM 308700), GNRH1 (OMIM 152760), GNRHR (OMIM 138850), KISS1R (OMIM 604161), KISS1 (OMIM 603286), CHD7 (OMIM 608892), NSMF (OMIM 608137), FGF8 (OMIM 600483), FGFR1 (OMIM 136350), PROK2 (OMIM 607002), PROKR2 (OMIM 607212), HS6ST1 (OMIM 1 604846), TAC3 (OMIM 162330) and TACR3 (OMIM 162332) was performed. The PCR primers and amplification conditions for each gene have been previously published (15, 16). A rare sequence variant (RSV) was defined as (a) a variant affecting splice junctions within 10 bp of coding sequence or a protein-altering/protein-truncating non-synonymous variant and (b) present in <1% minor allele frequency (MAF) in 1000 Genome Project (1000G) (http://www.1000genomes.org/home) (17), Exome Variant Server, NHLBI GO Exome Sequencing Project (ESP), Seattle, WA (URL: http://evs.gs.washington.edu), (February 2016) including the European American (EA) sub-population, and the non-Finish European (NFE) population of the Exome Aggregation Consortium (ExAC), Cambridge, MA (URL: http://exac.broadinstitute.org) (February 2016). All coding DNA and protein changes were based on the Genome Reference Consortium, GRCh37/ hg19, and nomenclature was reported according to most updated HGSV guidelines by den Dunnen et al. (33).
Apart from the analysis of coding point sequence variation, we investigated the presence of intergenic ANOS1 deletions and/or duplication in a subset of Greek patients who met the following criteria: (i) KS phenotype, (ii) negative for ANOS1 point variation, (iii) expressing a non-reproductive phenotype consistent with ANOS1 genetic changes, such as synkinesia or URA (18). Gene dosage analysis was performed using the SALSA MLPA kit P132 Kallmann-1 (MRC Holland) designed to detect exonic deletions/duplications across the entire coding region of ANOS1 gene.
Phenotypic results of our analysis included:
Male-to-female ratio: The cohort consisted of 53 male and 28 female probands. A total of 54 probands (67%) were classified as nIHH, whereas 27 probands (33%) were diagnosed with KS. KS was strikingly predominant in men with a 3:1 male-to-female ratio (21 males and 6 females) compared to a modest 3:2 male-to-female ratio in nIHH (32 males and 22 females).
Detailed phenotypic characteristics: Patients with or without anosmia displayed a wide spectrum of reproductive phenotypic features, including cryptorchidism and microphallus, with no statistically significant difference between KS and nIHH (36% in KS vs 35.7% in nIHH with a P value of 1 for cryptorchidism and 31.5% in KS vs 21.4% in nIHH with a P value of 0.6982 for microphallus). Of the assessed non-reproductive phenotypes, Unilateral Renal Agenesis (URA) and Synkinisia (S) were statistically enriched in KS (25% in KS vs 3.4% in IHH, P: 0.0377, for URA and 25% in KS vs 0% in nIHH, P: 0.0059, for synkinisia) highlighting the need for closer evaluation of patients with both KS and nIHH for non-reproductive features. Additionally, a familial case with a most interesting segregation of psychiatric disorders pointed toward further psychiatric evaluation in IGD cases.
Patterns of inheritance: A vast majority of the cases assembled were found to be sporadic (63/81, 78%) with the rest being familial, displaying different patterns of inheritance including X-linked (2/81, 2.4%), autosomal recessive (N 5/81, 6%) and autosomal dominant (N: 11/81, 13.2%) mode of inheritance.
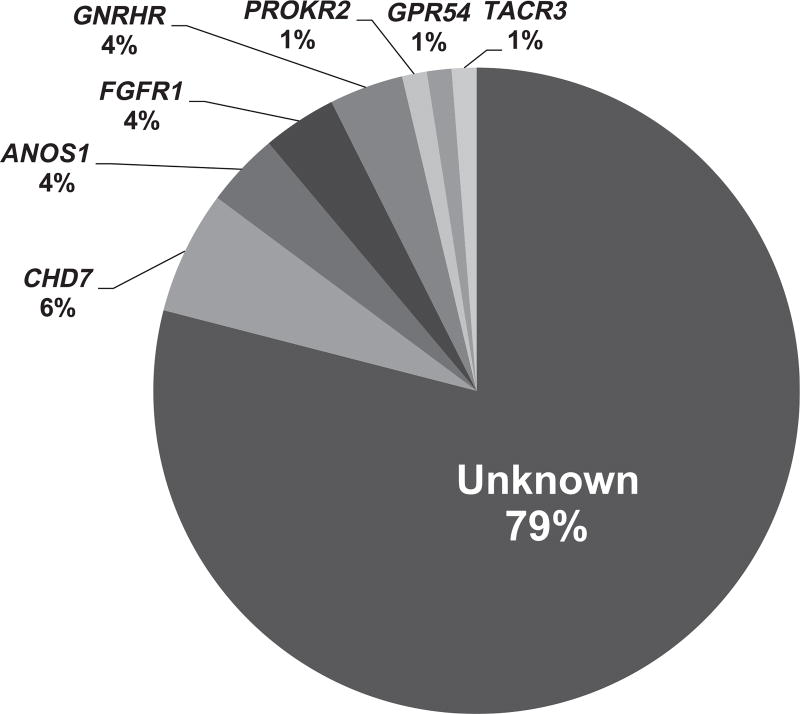
Genetic screening for a large number of IGD genes identified genetic changes in 21% of the Greek population (as illustrated in Fig. 1). A total of 15 genetic alterations were harbored by 17 IGD probands in the following genes: FGFR1, GNRHR, ANOS1, KISS1R, TACR3, CHD7, PROKR2 including:
Variants in autosomal dominant genes: From the detected variation in CHD7, the 2 missense RSVs c.7579A > C p.(Met2527Leu) (MAF of 0.1%/0.5%/0.31% in 1000G, ESP-EA and ExAC-NFE, found in 1 nIHH proband), c.1018A > G p.(Met340Val) (MAF 0.3%/0.74%/0.63% in the aforementioned databases, found in 2 probands with nIHH and KS) as well as 1 splice variant c.5051-4C > T (MAF: 0%, found in 1 KS proband) were previously found to be benign. One frame-shift deletion in the same gene, CHD7, c.8962_8963insG p.(Asp2988Glyfs*2) (MAF: 0%, found in 1 nIHH proband) has been predicted to be deleterious (19). Additionally, heterozygous RSVs in the gene of FGFR1 c.760C > T p.(Arg254Trp) (MAF: 0%, carried by 1 nIHH proband), c.1408C > T p.(Arg470Cys) (found only in ExAC-NFE with a MAF 0.0015% in 1 nIHH proband with cleft lip and palate) and c.710G > A p.(Gly237Asp) (MAF 0%, harbored by 1 KS proband) were previously tested and proven to be disease-causative (20, 21, 22).
Variants in autosomal recessive genes: Heterozygous and compound heterozygous RSVs in GNRHR c.436C > T p.(Pro146Ser) (MAF 0% in 1000G, 0.069% and 0.11% in ESP-EA and ExAC-NFE respectively, found in 1 nIHH proband) c.317A > G p.(Gln106Arg) (MAF 0.2%/0.3488%/0.3% in the aforementioned databases) and c.785G > A p.(Arg262Gln) (0.1%/0.22%/0.2% in the same databases) were detected in both sporadic and familial nIHH cases. The p.(Gln106Arg) and p.(Arg262Gln) were previously reported in nIHH individuals and were functionally validated in vitro (23, 24, 25, 26). Segregation of the variants in one familial case revealed recessive compound heterozygous mode of inheritance with the proband and her sister both diagnosed with partial nIHH, whereas the family history revealed the presence of 2 paternal aunts with delayed puberty and/or infertility. Additionally, a heterozygous variation in a traditional autosomal gene of PROKR2 was identified. The rare RSV c.1069C > T p.(Arg357Trp) was only found in ExAC-NFE with a MAF of 0.089% and was carried by a KS patient. However, this substitution of the Arginine to Tryptophan in the position 357 has been previously functionally validated as benign (27).
Intergenic deletions in ANOS1: 2 cases with ANOS1 deletions were identified, including one most interesting familial case. In particular, an ANOS1 hemizygous deletion was detected in 2 brothers diagnosed with KS, URA, synkinisia and icthyosis. Importantly, the reproductively unaffected mother, who presented as the obligate carrier of the Xp22 deletion, displayed URA. The role of ANOS1 and its encoded protein, anosmin 1, in the renal mesenchyme is known (28). The presence of the isolated URA in the female obligate carrier of the ANOS1 deletion can be explained by either (i) the presence of a separate cause of the URA that is transmitted from the mother to both affected sons or (ii) a variable pattern of X-inactivation of ANOS1 in different tissues. Further studies are required to identify the role and differential expression of ANOS1 in different tissues.
Novel variants: We were able to detect novel heterozygous variants in KISS1R, c.458C > T p.(Arg153Cys) in 1 KS proband, TACR3, c.1305T > A p.(Ser435Arg) in 1 nIHH proband and ANOS1 c.1532C > A p.(Ser511Tyr) harbored by 1 KS proband. The novel variants were not found in the normative databases apart from the TACR3 RSV that has a MAF of 0.0029% in ExAC-NFE dataset. Further validation is required to assess the role of the functionality of the discovered RSVs. Thus, even though the phenotype of the majority of probands carrying a RSV can be explained by genetic screening, further validation of the detected genetic alterations, as well as deep sequencing for detection of additional genes or modifiers that could synergistically act with the known IGD genes for the expression of the phenotype, will be crucial for some further understanding of the GnRH biology and the pathophysiology of the disease.
Figure 1.
A putative genetic change was discovered in 21% of the cohort comprising RSVs in the genes CHD7, ANOS1, FGFR1, PROKR2, GPR54, TACR3 and 2 intergenic deletions in the gene of ANOS1.
In this study, we explored the genotypic and phenotypic features of the largest cohort of IGD patients of Greek origin, which has so far been reported. Our patient cohort was represented by a larger number of nIHH patients compared to patients with KS, suggestive of a phenotypic enrichment of the normosmic feature, even though evaluation of the olfactory function of a subset of nIHH patients was based only on data given by our referring collaborators. Previous reports of ethnically mixed IGD patients have shown only a small excess of patients with nIHH phenotypes (29). However, studies in ethnically homogeneous populations postulated supremacy of one of the 2 sub-phenotypes (4, 30). Thus, the higher percentage of the nIHH phenotype in our population, may reflect a common genetic background among those nIHH patients, which is yet undiscovered.
Similar to previous studies in other ethnic groups, we report a higher male-to-female ratio in the total number of IGD patients with an enrichment for higher male-to-female ratio in KS form of IGD compared to nIHH patients. The precise reason for the male predominance in KS has hitherto been considered to be secondary to the X-linked form of KS. However, the presence of ANOS1 variations was only seen in a minority of KS patients in this study, and this observation suggests either a potential sex-dependent penetrance of the phenotype or a non-genetic cause or a possible bias of ascertainment. A more precise estimation of the KS-to-nIHH ratio in males could also be addressed by studying males registering in the Greek military service.
This is the first report of the prevalence of rare genetic variants in a large number of IGD genes in this heterogeneous sample of IGD patients of Greek origin, in contrast to studies in population isolates that reflect the presence of distinct founder variations. The genetic heterogeneity which was observed could be attributed to the heterogeneity of the sample selected as patients came to the clinic from different parts of Greece. Recently, few studies in Greek population isolates, both in the Pomak villages in the North of Greece and the Mylopotamos villages in Crete, showed an enrichment of genomic loci associated with complex phenotypes (such as hematologic traits) (31), suggestive of a common genetic backbone in isolated populations of Greece. Studies in IGD patients from such populations will be crucial for establishing other novel genetic causes.
Importantly, pathogenic genetic variation with multiple inheritance patterns was observed and both single point changes as well as intergenic deletions were detected in genes affecting either the neurodevelopmental pathway of IGD, including ANOS1, FGFR1, FGF8, CHD7, PROKR2 or the neuroendocrine pathway of the disorders, including GNRHR, KISSR1 and TACR3. Additionally, the majority of the RSVs were detected in sporadic cases of IGD, and none of them was proven to be de novo; this finding is suggestive of a recessive or dominant nature of the variations with incomplete penetrance. In our study, we also had the opportunity to detect 3 novel (to our knowledge) RSVs in the genes of ANOS1, KISS1R and TACR3. All three RSVs were well conserved across species and were found in a heterozygous state, despite the recessive mode of inheritance that characterizes the genes, implying an additional genetic or non-genetic cause for the expression of the phenotype in the probands carrying the RSVs. Further in vitro analysis for testing their functional significance is required. Limitations of this study include (i) genetic screening was not performed for all known IGD genes (studies for the remaining genes are ongoing); (ii) not all detected rare variants have been functionally validated in vitro and hence some may represent neutral polymorphisms; (iii) missing phenotypic details for the subjects referred to the University of Patras.
In summary, in this study, we detected total genetic variation in ~21% of our IGD patients with a lack of enrichment for a particular genetic cause. Additionally, even though a large number of IGD genes was screened, our sample lacked the presence of oligogenicity, i.e., variations in more than 1 gene cause the expression of the phenotype, a finding present in previous studies, affecting about 2.5% of the IGD population (32). All these observations, accompanied by an association of both familial and sporadic cases with psychiatric disorders could reflect the presence of a yet unidentified genetic cause underlying the Greek cohort of IGD, ‘hidden’ variation in the regulatory regions of the known genes as well as environmental factors contributing to the expression of the phenotype.
Acknowledgments
Funding
Dr Maria I Stamou would like to thank the Alexander S Onasis Foundation for the support of her research training.
The authors would like to thank Dr William F Crowley for his contribution in the genotyping of the patient cohort in the Reproductive Endocrine Unit of Massachusetts General Hospital and his valuable and constant mentorship and Dr Ravikumar Balasubramanian from Reproductive Endocrine Unit of Massachusetts General Hospital, for his valuable advice in constructing the project and drafting the manuscript. They would also like to thank Dr K Assimakopoulos for performing the psychiatric evaluation of our IGD patients as well as the following physicians for sharing data of their IGD patients within this cohort: Dr A Dakou-Voutetaki (Athens), Dr C Kanaka (Athens), Dr A Tsatsoulis (Ioannina), Dr M Andrikoula (Ioanninna), Dr A Tsinopoulou-Galli (Thessaloniki), Dr D Panidis (Thessaloniki), Dr I Papadimas (Thessaloniki), Dr D Goulis (Thessaloniki), Dr Pantsiotou (Athens) and Dr M Anapliotou (Athens).
Footnotes
Declaration of interest
The authors declare that there is no conflict of interest that could be perceived as prejudicing the impartiality of this article.
Author contribution statement
M I S conceived the study, participated in its design and coordination, participated in the molecular genetics studies and drafted the manuscript. V P participated in the design of the study patient data, sample and consent collection. K M participated in patient data, sample and consent collection. A F participated in patient data, sample and consent collection. V A participated in patient data, sample and consent collection. J J carried out the molecular genetic studies and participated in the sequence alignment. P L participated in coordination of the project, carried out the molecular genetic studies and participated in the sequence alignment. K V participated in coordination of the project, carried out the molecular genetic studies and participated in the sequence alignment. G N conceived the study, participated in its design and coordination and helped to draft the manuscript. All authors read and approved the final manuscript.
References
- 1.Laitinen EM, Vaaralahti K, Tommiska J, Eklund E, Tervaniemi M, Valanne L, Raivio T. Incidence, phenotypic features and molecular genetics of Kallmann syndrome in Finland. Orphanet Journal of Rare Diseases. 2011;6:41. doi: 10.1186/1750-1172-6-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Balasubramanian R, Dwyer A, Seminara SB, Pitteloud N, Kaiser UB, Crowley WF., Jr Human GnRH deficiency: a unique disease model to unravel the ontogeny of GnRH neurons. Neuroendocrinology. 2010;92:81–99. doi: 10.1159/000314193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brioude F, Bouligand J, Trabado S, Francou B, Salenave S, Kamenicky P, Brailly-Tabard S, Chanson P, Guiochon-Mantel A, Young J. Non-syndromic congenital hypogonadotropic hypogonadism: clinical presentation and genotype-phenotype relationships. European Journal of Endocrinology/European Federation of Endocrine Societies. 2010;162:835–851. doi: 10.1530/EJE-10-0083. [DOI] [PubMed] [Google Scholar]
- 4.Sarfati J, Fouveaut C, Leroy C, Jeanpierre M, Hardelin JP, Dode C. Greater prevalence of PROKR2 mutations in Kallmann syndrome patients from the Maghreb than in European patients. European Journal of Endocrinology/European Federation of Endocrine Societies. 2014;169:805–809. doi: 10.1530/EJE-13-0419. [DOI] [PubMed] [Google Scholar]
- 5.Tommiska J, Kansakoski J, Christiansen P, Jorgensen N, Lawaetz JG, Juul A, Raivio T. Genetics of congenital hypogonadotropic hypogonadism in Denmark. European Journal of Medical Genetics. 2014;57:345–348. doi: 10.1016/j.ejmg.2014.04.002. [DOI] [PubMed] [Google Scholar]
- 6.Gu WJ, Zhang Q, Wang YQ, Yang GQ, Hong TP, Zhu DL, Yang JK, Ning G, Jin N, Chen K, et al. Mutation analyses in pedigrees and sporadic cases of ethnic Han Chinese Kallmann syndrome patients. Experimental Biology and Medicine. 2015;240:1480–1489. doi: 10.1177/1535370215587531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bonomi M, Libri DV, Guizzardi F, Guarducci E, Maiolo E, Pignatti E, Asci R, Persani L. New understandings of the genetic basis of isolated idiopathic central hypogonadism. Asian Journal of Andrology. 2012;14:49–56. doi: 10.1038/aja.2011.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Della Valle E, Vezzani S, Rochira V, Granata AR, Madeo B, Genovese E, Pignatti E, Marino M, Carani C, Simoni M. Prevalence of olfactory and other developmental anomalies in patients with central hypogonadotropic hypogonadism. Frontiers in Endocrinology. 2013;4:70. doi: 10.3389/fendo.2013.00070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de Roux N, Genin E, Carel JC, Matsuda F, Chaussain JL, Milgrom E. Hypogonadotropic hypogonadism due to loss of function of the KiSS1-derived peptide receptor GPR54. PNAS. 2003;100:10972–10976. doi: 10.1073/pnas.1834399100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Seminara SB, Messager S, Chatzidaki EE, Thresher RR, Acierno JS, Jr, Shagoury JK, Bo-Abbas Y, Kuohung W, Schwinof KM, Hendrick AG, et al. The GPR54 gene as a regulator of puberty. New England Journal of Medicine. 2003;349:1614–1627. doi: 10.1056/NEJMoa035322. [DOI] [PubMed] [Google Scholar]
- 11.Topaloglu AK, Reimann F, Guclu M, Yalin AS, Kotan LD, Porter KM, Serin A, Mungan NO, Cook JR, Ozbek MN, et al. TAC3 and TACR3 mutations in familial hypogonadotropic hypogonadism reveal a key role for Neurokinin B in the central control of reproduction. Nature Genetics. 2009;41:354–358. doi: 10.1038/ng.306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Topaloglu AK, Tello JA, Kotan LD, Ozbek MN, Yilmaz MB, Erdogan S, Gurbuz F, Temiz F, Millar RP, Yuksel B. Inactivating KISS1 mutation and hypogonadotropic hypogonadism. New England Journal of Medicine. 2012;366:629–635. doi: 10.1056/NEJMoa1111184. [DOI] [PubMed] [Google Scholar]
- 13.Kotan LD, Hutchins BI, Ozkan Y, Demirel F, Stoner H, Cheng PJ, Esen I, Gurbuz F, Bicakci YK, Mengen E, et al. Mutations in FEZF1 cause Kallmann syndrome. American Journal of Human Genetics. 2014;95:326–331. doi: 10.1016/j.ajhg.2014.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stamou MI, Cox KH, Crowley WF., Jr Discovering genes essential to the hypothalamic regulation of human reproduction using a human disease model: adjusting to life in the ‘-omics’ era. Endocrine Reviews. 2016;2016:4–22. doi: 10.1210/er.2015-1045.2016.1.test. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 15.Shaw ND, Seminara SB, Welt CK, Au MG, Plummer L, Hughes VA, Dwyer AA, Martin KA, Quinton R, Mericq V, et al. Expanding the phenotype and genotype of female GnRH deficiency. Journal of Clinical Endocrinology and Metabolism. 96:E566–E576. doi: 10.1210/jc.2010-2292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tornberg J, Sykiotis GP, Keefe K, Plummer L, Hoang X, Hall JE, Quinton R, Seminara SB, Hughes V, Van Vliet G, et al. Heparan sulfate 6-O-sulfotransferase 1, a gene involved in extracellular sugar modifications, is mutated in patients with idiopathic hypogonadotrophic hypogonadism. PNAS. 2011;108:11524–11529. doi: 10.1073/pnas.1102284108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Abecasis GR, Auton A, Brooks LD, DePristo MA, Durbin RM, Handsaker RE, Kang HM, Marth GT, McVean GA. An integrated map of genetic variation from 1092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Costa-Barbosa FA, Balasubramanian R, Keefe KW, Shaw ND, Al-Tassan N, Plummer L, Dwyer AA, Buck CL, Choi JH, Seminara SB, et al. Prioritizing genetic testing in patients with Kallmann syndrome using clinical phenotypes. Journal of Clinical Endocrinology and Metabolism. 2013;98:E943–E953. doi: 10.1210/jc.2012-4116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bartels CF, Scacheri C, White L, Scacheri PC, Bale S. Mutations in the CHD7 gene: the experience of a commercial laboratory. Genetic Testing and Molecular Biomarkers. 2010;14:881–891. doi: 10.1089/gtmb.2010.0101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pitteloud N, Quinton R, Pearce S, Raivio T, Acierno J, Dwyer A, Plummer L, Hughes V, Seminara S, Cheng YZ, et al. Digenic mutations account for variable phenotypes in idiopathic hypogonadotropic hypogonadism. Journal of Clinical Investigation. 2007;117:457–463. doi: 10.1172/JCI29884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pitteloud N, Meysing A, Quinton R, Acierno JS, Jr, Dwyer AA, Plummer L, Fliers E, Boepple P, Hayes F, Seminara S, et al. Mutations in fibroblast growth factor receptor 1 cause Kallmann syndrome with a wide spectrum of reproductive phenotypes. Molecular and Cellular Endocrinology. 2006;254–255:60–69. doi: 10.1016/j.mce.2006.04.021. [DOI] [PubMed] [Google Scholar]
- 22.Koika V, Varnavas P, Valavani H, Sidis Y, Plummer L, Dwyer A, Quinton R, Kanaka-Gantenbein C, Pitteloud N, Sertedaki A, et al. Comparative functional analysis of two fibroblast growth factor receptor 1 (FGFR1) mutations affecting the same residue (R254W and R254Q) in isolated hypogonadotropic hypogonadism (IHH) Gene. 2013;516:146–151. doi: 10.1016/j.gene.2012.12.041. [DOI] [PubMed] [Google Scholar]
- 23.de Roux N, Young J, Misrahi M, Genet R, Chanson P, Schaison G, Milgrom E. A family with hypogonadotropic hypogonadism and mutations in the gonadotropin-releasing hormone receptor. New England Journal of Medicine. 1997;337:1597–1602. doi: 10.1056/nejm199711273372205. [DOI] [PubMed] [Google Scholar]
- 24.Bedecarrats GY, Linher KD, Kaiser UB. Two common naturally occurring mutations in the human gonadotropin-releasing hormone (GnRH) receptor have differential effects on gonadotropin gene expression and on GnRH-mediated signal transduction. Journal of Clinical Endocrinology and Metabolism. 2003;88:834–843. doi: 10.1210/jc.2002-020806. [DOI] [PubMed] [Google Scholar]
- 25.Caron P, Chauvin S, Christin-Maitre S, Bennet A, Lahlou N, Counis R, Bouchard P, Kottler ML. Resistance of hypogonadic patients with mutated GnRH receptor genes to pulsatile GnRH administration. Journal of Clinical Endocrinology and Metabolism. 1999;84:990–996. doi: 10.1210/jcem.84.3.5518. [DOI] [PubMed] [Google Scholar]
- 26.Gianetti E, Hall JE, Au MG, Kaiser UB, Quinton R, Stewart JA, Metzger DL, Pitteloud N, Mericq V, Merino PM, et al. When genetic load does not correlate with phenotypic spectrum: lessons from the GnRH receptor (GNRHR) Journal of Clinical Endocrinology and Metabolism. 2012;97:E1798–E1807. doi: 10.1210/jc.2012-1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cole LW, Sidis Y, Zhang C, Quinton R, Plummer L, Pignatelli D, Hughes VA, Dwyer AA, Raivio T, Hayes FJ, et al. Mutations in prokineticin 2 and prokineticin receptor 2 genes in human gonadotrophin-releasing hormone deficiency: molecular genetics and clinical spectrum. Journal of Clinical Endocrinology and Metabolism. 2008;93:3551–3559. doi: 10.1210/jc.2007-2654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hardelin JP, Julliard AK, Moniot B, Soussi-Yanicostas N, Verney C, Schwanzel-Fukuda M, Ayer-Le Lievre C, Petit C. Anosmin-1 is a regionally restricted component of basement membranes and interstitial matrices during organogenesis: implications for the developmental anomalies of X chromosome-linked Kallmann syndrome. Developmental Dynamics. 1999;215:26–44. doi: 10.1002/(SICI)1097-0177(199905)215:1<26::AID-DVDY4>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 29.Pitteloud N, Hayes FJ, Boepple PA, DeCruz S, Seminara SB, MacLaughlin DT, Crowley WF., Jr The role of prior pubertal development, biochemical markers of testicular maturation, and genetics in elucidating the phenotypic heterogeneity of idiopathic hypogonadotropic hypogonadism. Journal of Clinical Endocrinology and Metabolism. 2002;87:152–160. doi: 10.1210/jcem.87.1.8131. [DOI] [PubMed] [Google Scholar]
- 30.Gurbuz F, Kotan LD, Mengen E, Siklar Z, Berberoglu M, Dokmetas S, Kilicli MF, Guven A, Kirel B, Saka N, et al. Distribution of gene mutations associated with familial normosmic idiopathic hypogonadotropic hypogonadism. Journal of Clinical Research in Pediatric Endocrinology. 2012;4:121–126. doi: 10.4274/Jcrpe.725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Panoutsopoulou K, Hatzikotoulas K, Xifara DK, Colonna V, Farmaki AE, Ritchie GR, Southam L, Gilly A, Tachmazidou I, Fatumo S, et al. Genetic characterization of Greek population isolates reveals strong genetic drift at missense and trait-associated variants. Nature Communications. 2014;5:5345. doi: 10.1038/ncomms6345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sykiotis GP, Plummer L, Hughes VA, Au M, Durrani S, Nayak-Young S, Dwyer AA, Quinton R, Hall JE, Gusella JF, et al. Oligogenic basis of isolated gonadotropin-releasing hormone deficiency. PNAS. 2010;34:15140–15144. doi: 10.1073/pnas.1009622107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.den Dunnen JT, Dalgleish R, Maglott DR, Hart RK, Greenblatt MS, McGowan-Jordan J, Roux AF, Smith T, Antonarakis SE, Taschner PE. HGSV recommendations for the descriptions of sequence variants: 2016 update. Human Mutation. 2016;6:564–569. doi: 10.1002/humu.22981. [DOI] [PubMed] [Google Scholar]