Abstract
The ubiquitin (Ub) system, which is centered on the conjugation and deconjugation of Ub and Ub-like (Ubls) proteins to proteins or lipids by a system of ligases and peptidases, respectively, regulates practically all aspects of eukaryotic biology. Ub/Ubls contain the beta-grasp fold (beta-GF) that is also found in Ub-related domains of several other proteins of the Ub-system and numerous proteins with biochemically distinct roles unrelated to the conventional Ub-system. Taken together, domains displaying the beta-grasp include versions performing catalytic roles, scaffolding of iron-sulfur clusters, binding of RNA and other biomolecules such as co-factors and sulfur transfer in biosynthesis of diverse metabolites. The beta-GF appears to have undergone an early radiation spawning at least seven clades prior to the divergence of extant organisms from their last universal common ancestor. The beta-GF appears to have first emerged in the context of translation-related RNA-interactions and subsequently exploded to occupy various functional niches. Most structural and biochemical diversification of the beta-GF occurred in prokaryotes, with the Ubl clade showing a dramatic expansion in the eukaryotic phase of its evolution. Hence, at least 70 distinct Ubl families are observed in eukaryotes, of which nearly 20 families were probably present in the last eukaryotic common ancestor. Diversification of Ubl families early in eukaryotic evolution played a major role in emergence of characteristic eukaryotic cellular sub-structures and systems pertaining to nucleo-cytoplasmic compartmentalization and dynamics, vesicular trafficking, autophagy and different protein degradation systems. Recent comparative genomics studies indicate that precursors of the eukaryotic Ub-system emerged first in prokaryotes. The simplest of these combine an Ubl and an E1-like enzyme involved in metabolic pathways related metallopterin, thiamine, cysteine, siderophore and perhaps modified base biosynthesis. Sampylation in archaea and Urmylation in eukaryotes appear to represent direct recruitment of such systems as simple protein-tagging apparatuses. However, other prokaryotic systems incorporated further components that more or less mirror the eukaryotic condition in possessing an E2, a RING-type E3 or both of these components. Additionally, prokaryotes have evolved conjugation systems that are independent of Ub ligases, such as the Pup system.
Keywords: Ubiquitin, Prokaryotic Ubiquitin Conjugation, Non-Ribosomal Peptide Ligases, Sumo, Rna Modification, Beta-Grasp Fold, Review
2. INTRODUCTION
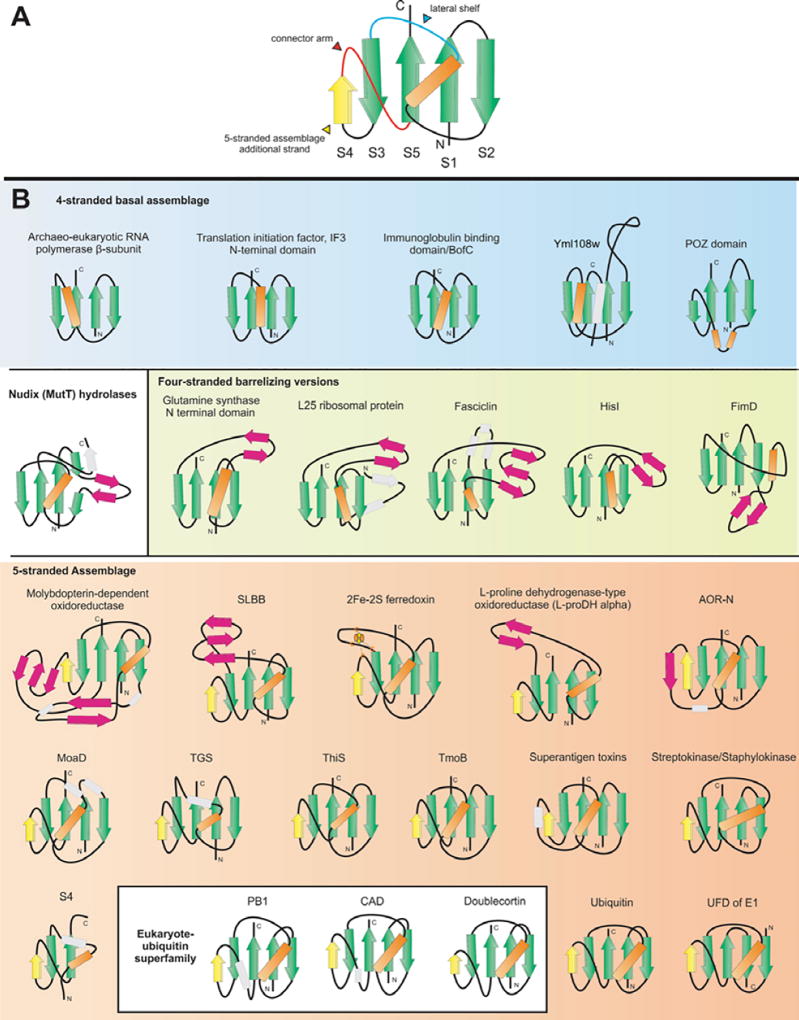
The discovery of covalent modification of eukaryotic proteins by the conjugation of ubiquitin to the epsilon-amino groups of target lysines has spawned some of the most exciting directions of research in current molecular biology (1–3). Ubiquitin (Ub) itself is a small polypeptide of 76 residues, and its crystal structure revealed a distinctive fold dominated by a beta-sheet with 5 anti-parallel beta-strands and a single helical segment (4, 5) (Figure 1A). Pioneering investigations of Kraulis, Overington and Murzin showed that this fold was not unique to Ub, but was also present in several other proteins with biologically distinct functions. These included the staphylococcal enterotoxin B, the streptococcal immunoglobulin (Ig)-binding protein G and 2Fe-2S ferredoxins (6–8). The common fold shared by these proteins was termed the beta-grasp, because the beta-sheet appears to grasp the helical segment in this domain (7). These early studies provided the first indications that, despite its small size, the beta-grasp fold (beta-GF) might serve as a multi-functional scaffold in diverse biological contexts.
Figure 1.
Topology diagrams of selected beta-GF members. A generalized representation of the beta-GF depicting the conserved secondary structure elements, and key structural features found in certain lineages of the fold, is shown in (A). Shown in (B) are idealized versions of specific lineages, the names of which are given above the diagrams. Strands are illustrated as arrows with the arrowhead pointing to the C-terminal end and helices as rectangles. Strands belonging to the 4-stranded beta-GF core are colored green, the additional strand found in the 5-stranded assemblage is colored yellow, strands forming a conserved insert within the beta-GF scaffold are colored magenta, and other strands specific to a certain lineage are colored grey and outlined with a broken line. The absolutely conserved core helix is colored orange and other helices specific to a certain lineage colored grey and outlined with a broken line. Topologies are grouped and labeled in a manner consistent with the structural classes described in the text, with members of the eukaryotic UB-like superfamily nested within other members of the 5-stranded assemblage. The 2Fe-2S cluster of the ferredoxins is shown as four small ovals bound to cysteine residues represented by the letter "C".
The centrality of Ub conjugation in eukaryotic molecular biology has led to numerous investigations on Ub and Ub-related domains (9, 10). These studies have resulted in a large body of data on the properties of the Ub-like versions of the beta-GF. One key finding has been that several other Ub-like proteins (Ubl), such as Urm1 (11), Apg12 (12), Nedd8 (13), and SUMO (14, 15) are also covalently linked to target polypeptides, just as Ub itself (16). In contrast, some Ub-related domains, like the Ubx domain or Ubl domains of IkB kinases, play adaptor roles in Ub-signaling (17–20). These studies also showed that eukaryotes possess a distinctive enzymatic apparatus for Ub-modification, comprised of a cascade of three enzymes: E1, E2 and E3. These enzymes successively activated Ub/Ubl for transfer using the free energy derived from ATP hydrolysis, relayed it via thiocarboxylate linkages involving the C-terminal residue of Ub/Ubls, and finally transferred it to the epsilon NH2 group on lysines, the amino terminal NH2 groups or on rare occasions cysteines on target polypeptides (1, 10, 21–23) (24). Eukaryotes were also shown to contain an elaborate apparatus centered primarily on thiol peptidases of the papain-like fold or JAB-superfamily metallopeptidases for removal of covalently linked Ub/Ubls and proteasomal degradation of Ub-modified proteins (25–29).
Concomitantly, structural studies also uncovered several new versions of the beta-GF in a variety of domains, greatly widening its horizon of biological functions. Examples of such beta-GF domains are: 1) the TGS domain, an RNA-binding domain found in aminoacyl tRNA synthetases and other translation regulators (PDB: 1QF6 (30, 31)). 2) The doublecortin (DCX) (PDB: 1MJD (32)), RA (PDB: 1C1Y (33)), PB1 (PDB: 1IPG (34)), and FERM N-terminal domains (PDB: 1EF1 (35)), which function as adaptors in animal signaling proteins and apoptosis regulators by mediating protein-protein interactions. 3) The soluble ligand-binding beta-GF (SLBB) domain involved in binding vitamin B12 and other solutes in animals and bacteria (PDB: 2BBC, 2FUG (36–38)). 4) Various toxins related to the staphylococcal enterotoxin B including superantigens involved in the toxic shock syndrome (PDB: 1ESF (39)). 5) Functionally obscure subunits of various enzymatic complexes, like TmoB of the aromatic monooxygenase oxygenase complex (PDB: 1T0S (40)) and RnfH of the Rnf dehydrogenases (41). 6) Conserved domains, perhaps involved in RNA binding, in the archaeo-eukaryotic RNA polymerase RPB2 subunit (42) and bacterial translation initiation factor IF3 (PDB: 1TIF (43–45)). 7) Staphylokinases and streptokinases which are fibrinolytic enzymes of low GC Gram-positive bacteria (PDB: 2SAK (46)). 8) MutT/nudix enzymes- a group of phosphohydrolases acting on diverse substrates (47). These observations suggested that the beta-GF is indeed a widely utilized structural scaffold, with an underappreciated versatility and an evolutionary history rich in adaptive radiations.
One notable evolutionary question in this regard is the origin of eukaryotic Ub and its relationships to other domains with the beta-GF. The first major advances in this direction came with the identification of the sulfur transfer proteins, ThiS and MoaD, respectively involved in thiamine and Molybdenum cofactor (MoCo) biosynthesis, which contained beta-GFs closely related to Ub (48) (49). Furthermore, it was demonstrated that their C-terminal residues formed thiocarboxylates, just like Ub, and this was catalyzed by enzymes (ThiF and MoeB) very similar to the E1 enzymes involved in Ub-conjugation (48–52). In a similar vein, the Urm1 protein, a close eukaryotic relative of the ThiS and MoaD proteins, has also recently been demonstrated to function as a sulfur carrier through thiocarboxylate formation catalyzed by the Uba4 E1-like homolog in the context of tRNA thiolation (53–56), a remarkable analogous functional role to ThiS and MoaD that was predicted in our earlier work (57).
A growing pool of evidence indicates that Ubls related to ThiS/MoaD/Urm1 not only function as sulfur carriers but are also, in apparent response to extracellular environmental cues, conjugated to target proteins similar to classical eukaryotic Ub systems. The Urm1 enzyme itself undergoes covalent attachment to target proteins in response to oxidative stress (11, 58, 59), (60). Additionally, widespread covalent conjugation of the ThiS/MoaD/Urm1-like archaeal SAMP1 and SAMP2 proteins to target proteins has recently been experimentally shown, and unlike the Urm1 protein, SAMP2 proteins appear capable of forming covalent bonds with itself to form “poly-samp” chains analogous to polyubiquitin chains (61). The available evidence indicates that these ligation reactions are solely dependent on the E1 cognate and proceed in the absence of E2 or E3 enzymes. The exact details of these E2/E3-independent ligation reactions remain poorly understood. The combination of these observations has led some to view these proteins, particularly the Urm1 protein, as potential links bridging the gap between Ubl sulfur carrier and protein modifier functions (62, 63).
In addition to these apparent E2 and E3-independent Ubl conjugation pathways, our observations showed that proteins with Ubl beta-GF domains and conjugating enzymes related to E1, E2 and deubiquitinating peptidases of the JAB domain superfamily were found in tightly-linked functional associations in diverse prokaryotic genomes. While some of these systems are likely to be involved in sulfur transfer reactions in metabolite biosynthesis, akin to ThiS, MoaD, and Urm1, others might potentially function as bona fide conjugation systems that transfer beta-GF proteins to target polypeptides (41). Finally, two very recent studies have uncovered associations between these same components with the RING-like E3 domains in both bacteria and archaea (64, 65), with the remarkable implication that the entire classical eukaryotic Ub modification system was present in prokaryotes. Hence, the eukaryotic Ub-conjugation and JAB-dependent deconjugation system might have been inherited as a single operonic unit from ancient prokaryotic precursors in the earliest phase of eukaryotic evolution.
An interesting variation in covalent attachment of a protein modifier was reported in actinobacteria, wherein the Pup protein, which is structurally unrelated to the beta-GF, is covalently ligated to target proteins. Several remarkable parallels between “pupylation” and classical, proteosomal-directing ubiquitination have been observed. Chief among these are 1) protein targets of pupylation are targeted for degradation via the action of bacterial cognates of the eukaryotic proteasomes, 2) Pup proteins are ligated to exposed lysine residues on target proteins via the action of a ligase enzyme and 3) ligation occurs at the extreme C-terminus of the Pup protein, which like ubiquitin contains a conserved diglycine motif providing flexibility necessary for the ligation reaction (66–71). However, despite these superficial similarities, the protein components of the pupylation apparatus are completely distinct from their ubiquitin counterparts (72). Despite the convergent emergence of the digylcine C-terminal residues and poorly-researched suggestions to the alternative (73), Pup does not contain a beta-GF, consisting instead of an N-terminal bi-helical unit followed by an extended C-terminal tail region housing the aforementioned diglycine motif (72, 74). Similarly, the Pup ligase (PafA) and its homologous deamidase (Dop) (72) are structurally unrelated to the Ubl E1-like ligase domain, belonging instead to the glutamine synthetase (GS)/NH2-COOH ligase fold (72). Thus, pupylation is a remarkable convergent emergence of a protein-tagging system utilized for the targeting of proteins for degradation. Subsequent studies have shown that the Pup system is widely distributed in bacteria and in some bacteria (e.g. deltaproteobacteria and planctomycetes) it might function independently of proteasomes, as a membrane-protein modifying system (72). At least one study has suggested that additional, convergently-emerged protein tagging may be present in prokaryotes (75).
The origin of Ub/Ubls and their associated biochemical networks is best understood through the study of the adaptive radiations of the beta-GF at large. Previous research provided the first comprehensive assessment of these radiations through, among other objectives, a careful comparative analysis of the structural and topological variations in the fold, determination of lineage-specific sequence-structure correlates for the varying functional adaptations of the fold, and identification of the temporal phases of adaptation leading to the construction of the first comprehensive evolutionary history incorporating the numerous distinct monophyletic families of the fold (76). This review revisits several of these structural and evolutionary themes, with an emphasis on understanding the functional shift which accompanied the emergence of the classical Ubl proteins which came to occupy a central role in a distinctive post-translational modification system that plays vital roles in several quintessentially eukaryotic systems. In the process, we will refine the previously presented evolutionary history of the beta-GF through incorporation of several novel findings relating to Ubl modification systems that have emerged since the initial characterization of the fold (76).
3. CORE CONSERVED TOPOLOGY, STRUCTURAL VARIATION, AND DERIVATIVES OF THE beta-GF
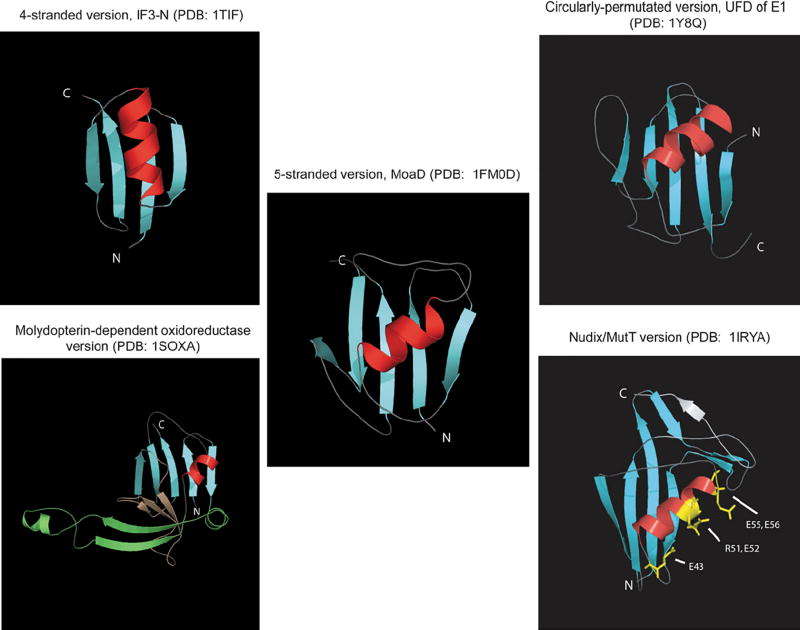
A comparison of the available beta-GF structures revealed a common core of 4 strands forming an anti-parallel sheet and a single helical region (see Table 1, Figure 1A). The characteristic topological feature is that the first and last strands are adjacent and parallel to each other, and the remaining two strands of the conserved core are anti-parallel and flank the former two strands on either side. The first and last strands are invariably located in the center of the sheet with a cross-over occurring via the single helical element. This helical region is packed against one face of the sheet, typically leaving the other face exposed. The chief interacting positions between sheet and the helical segment and the pattern of key stabilizing hydrophobic interactions are conserved throughout the fold, supporting its monophyletic origin. The beta-GF domains found in IF3 and the second largest subunit (beta-subunit orthologs) of the archaeo-eukaryotic RNA polymerase more or less correspond to this conserved core (Figure 1B). Several beta-GF domains display simple structural elaborations of this basic 4-stranded core, which can be observed in Figure 1A and are discussed below. All other versions of the beta-GF are characterized by major modifications to the 4-stranded core in the form of distinct inserts that add new secondary structure elements. The first of these insertions consists of one or more strands between the helical segment and strand 3. The conserved inserted strand seen in all domains with this version forms a hairpin with the connector segment between the helical segment and strand 3 which also assumes an extended conformation. This hairpin, together with any additional strands in the insert results in these versions of the fold assuming barrel-like structures with differing degrees of openness (Figure 1, Table 1). The most common structural elaboration in the beta-GF is typified by the presence of an additional strand that packs against the conserved third strand at the margin of the core beta-sheet. The acquisition of this additional strand has resulted in the emergence of a connector arm that joins it to the terminal conserved strand of the core sheet (Figure 1, Table 1). All ubiquitin-like beta-GF domains, including sulfur carrier proteins like MoaD and ThiS, contain this 5-stranded version of the fold. The connector arm is variable in structure and length and assumes a wide range of conformations ranging from coils to structured elements in different versions of the fold (Figure 1B, Table 1). A derivative of this Ub-like 5-stranded version is found as a C-terminal domain (UFD) in most eukaryotic E1 Ub-conjugating enzymes and their closest prokaryotic relatives (77, 78)—here a circular permutation appears to have displaced the N-terminus to the C-terminus. Given that the N- and C-terminal strands of the beta-GF are adjacent to each other, the C-terminal strand in the permuted version occupies the same position as the N-terminal strand of the classical versions, but is oriented in the opposite direction. Additional structural variations of the 5-stranded versions are depicted in Figure 1 and described below in some detail.
Table 1.
Secondary structure features of major β-GF structural categories.
| Higher-order Classification | Representative lineages | Secondary Structural Features Common to the β-GF Fold1 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| S1 | L1 | S2 | L2 | H | L3/LS | S3 | L4 | S4 | L5/CA | S5 | tail | notes | ||
| Basal 4-stranded versions of the β-GF | IF3-N | S1 | -- | S2 | -- | H | -- | S3 | -- | O | O | S5 | -- | |
| Archaeo-eukaryotic RNA poly. β-subunit | S1 | -- | S2 | -- | H | -- | S3 | -- | O | O | S5 | -- | ||
| Yml108w | S1 | cc | S2 | -- | H | -- | S3 | -- | O | O | S5 | h | ||
| BofC | S1 | -- | S2 | -- | H | -- | S3 | -- | O | O | S5 | -- | ||
| Immunoglobulin-binding | S1 | -- | S2 | -- | H | -- | S3 | -- | O | O | S5 | -- | ||
| POZ | S1 | -- | S2 | -- | H | h | S3 | -- | O | O | S5 | -- | ||
|
| ||||||||||||||
| Nudix superfamily | Nudix (MutT) | S1 | -- | S(ee)2 | -- | H | * | S3 | -- | O | O | S5 | e | |
|
| ||||||||||||||
| Fasciclin-like assemblage | L25 | S1 | -- | S2 | -- | H | ee* | S3 | -- | O | O | S5 | -- | 3 |
| glutamine synthetase N-terminal | S1 | -- | S2 | -- | H | eee* | S3 | -- | O | O | S5 | -- | 3 | |
| fasciclin | S1 | hhh | S2 | -- | H | ee* | S3 | -- | O | O | S5 | -- | 3 | |
| phosphoribosyl AMP cyclohydrolase (HisI) | S1 | -- | S2 | -- | H | ee* | S3 | -- | O | O | S5 | -- | 3,4 | |
|
| ||||||||||||||
| 5-stranded assemblage: classical 5-stranded clade | MoaD | S1 | H | S2 | -- | H | h** | S3 | -- | S4 | * | S5 | -- | |
| ThiS | S1 | -- | S2 | -- | H | * | S3 | -- | S4 | * | S5 | -- | ||
| TmoB | S1 | -- | S2 | -- | H | * | S3 | -- | S4 | * | S5 | -- | ||
| PHH-γ subunit | S1 | -- | S2 | e | H | h* | S3 | -- | S4 | e* | S5 | -- | ||
| Superantigen | S1 | -- | S2 | -- | H | * | S3 | -- | S4 | h* | S5 | -- | ||
| Strepto/Staphylokinase | S1 | -- | S2 | -- | H | * | S3 | -- | S4 | * | S5 | -- | ||
| YukD | S1 | -- | S2 | -- | H | * | S3 | -- | S4 | * | S5 | -- | ||
| TGS | S1 | -- | S2 | -- | H | h* | S3 | -- | S4 | * | S5 | -- | ||
| Aldehyde OR2 N-terminal domain | S1 | -- | S2 | -- | H | * | S3 | -- | S4 | eh* | S5 | -- | ||
|
| ||||||||||||||
| 5-stranded assemblage: Selected UB-like clade members | classic UB-like | S1 | -- | S2 | -- | H | * | S3 | -- | S4 | * | S5 | -- | |
| PB1 | S1 | -- | S2 | -- | H | * | S3 | -- | S4 | h* | S5 | -- | ||
| CAD/Doublecortin (DCX) | S1 | -- | S2 | -- | H | * | S3 | -- | S4 | [h]* | S5 | -- | 6 | |
| RA | S1 | -- | S2 | -- | H | * | S3 | -- | S4 | h* | S5 | -- | ||
| Elongin | S1 | -- | S2 | -- | H | * | S3 | -- | S4 | * | S5 | -- | ||
| UBX | S1 | -- | S2 | -- | H | * | S3 | -- | S4 | * | S5 | -- | ||
| E1/UFD | O | -- | S2 | -- | H | * | S3 | -- | S4 | * | S5 | S6 | 7 | |
|
| ||||||||||||||
| 5-stranded assemblage: soluble ligand binding or metal ion chelating clade | molydopterin-dependent oxidoreductase | S1 | -- | S2 | hehee | H | * | S3 | -- | S4 | eee* | S5 | -- | |
| SLBB: Nqo1-type | S1 | -- | S2 | -- | H | * | S3 | -- | S4 | hh* | S5 | -- | 5 | |
| SLBB: transcobalamin-type | S1 | -- | S2 | -- | H | eee* | S3 | -- | S4 | * | S5 | -- | ||
| 2Fe-2S ferredoxin | S1 | -- | S2 | -- | H | cc* | S3 | -- | S4 | * | S5 | -- | ||
| L-proline DH-like OR2 N-terminal domain | S1 | -- | S2 | -- | H | ee* | S3 | -- | S4 | * | S5 | -- | ||
|
| ||||||||||||||
| Miscellaneous | WWE | S1 | -- | S2 | -- | H | e* | S3 | -- | O | O | S5 | e | 8 |
| FimD N-terminal | S1 | -- | S2 | ee | H | * | S3 | -- | O | O | S5 | -- | ||
| S4 | O | O | O | O | H | h* | S3 | -- | S4 | * | S5 | -- | ||
S: Strand, L: Loop, H: Helix, LS: Lateral Shelf, CA: Connector Arm, O: absence of given feature, --: presence of a loop feature, *: presence of LS or CA, h: insert in helical conformation, e: insert in extended conformation (strand-like), cc: long coil insert.
OR: oxidoreductase.
Versions form barrel through insertion of strands at the lateral shelf.
Barrel is less pronounced in this version, strands are inserted more upstream relative to the other 3 versions.
Two small helices are present in ascending arm.
Single helix found at ascending arm in several members.
Circular permutation results in new connections between strands; the S1 strand is found at C-terminus (See Figures 1, 2).
Additional strand at tail inserted between S1 and S5; lateral shelf forms strand that also stacks with central sheet.
4. NATURAL CLASSIFICATION OF beta-GF DOMAINS
In order to address the prime evolutionary questions about the beta-GF and the emergence of the Ubls, a natural classification was constructed using structural characters and sequence features, most closely approximating the higher-order evolutionary relationships of the members of this fold (57). The small size of the majority of the versions of this domain often precludes sufficient resolution of relationships using conventional phylogenetic tree methods, sometimes even within superfamilies that display significant sequence similarity. This difficulty is further compounded by the extreme sequence divergence even between versions having highly similar tertiary structures (e.g. ubiquitin and ThiS). Hence, the reconstruction of the evolutionary history is underpinned to a great extent on structure similarity-based clustering, shared derived structural characters, and phyletic patterns of sequence superfamilies (see Ref. (76) for details). New evidence introduced by experimental and computational studies has been incorporated into this reconstruction, resulting in further refinement of a few of the higher-order relationships. The resulting classification offers the most reasonable resolution of the higher-order relationships to date, while on occasions still producing relatively flat hierarchies for lower-level clusters where existing methods cannot offer reliable resolution of relationships. A summary of this classification is presented in Table 1 and Figures 2 and 3. Monophyletic assemblages of diverse beta-GF domains are briefly discussed below to provide a broader context for the emergence of the Ubl domains before moving to a detailed description of the structural and sequences affinities of the prokaryotic and eukaryotic Ubls and their closely-allied domains.
Figure 2.
Cartoon depictions of distinct beta-GF domains. The core conserved strands and helices are colored blue and red respectively. Also shown are the critical residues in MutT responsible for catalytic activity.
Figure 3.
Reconstructed evolutionary history of the beta-grasp fold. Individual lineages, listed to the right of the figure, are grouped according to classification given in the text. The inferred evolutionary depth of the lineages is traced by solid horizontal lines across the relative temporal epochs representing major evolutionary transitional periods shown as vertical lines. Horizontal lines are colored according to their observed phyletic distributions; the key for this coloring scheme is given at the bottom of the figure. Dashed lines indicate an uncertainty in terms of the origins of a lineage, while grey ellipses group lineages of relatively restricted phyletic distribution with more broadly distributed ones, indicating that the former likely underwent rapid divergence from the latter. The major structural and functional transitions of the fold are marked by green ellipses along with a brief description. Colored, labeled squares immediately to the left of the lineage names represent broad functional categories: E, enzymatic activity; LMB, ligand or metal-binding; CO, conjugated versions; AD, mediator of protein-protein interactions; RNA, RNA metabolism-related.
4.1. Basal and other 4-stranded versions of the beta-GF
Analysis of the structural diversity of the fold suggests that the 4-stranded version is the simplest form from which all other versions could have been derived through accretion of inserts and additional secondary structure elements. Two structurally close superfamilies of the 4-stranded beta-GF domain, namely the IF3-N and the archaeo-eukaryotic RNA polymerase domain, are respectively universally conserved in the bacterial and archaeal-eukaryotic branches of life. This, taken together with their shared general functional connection to RNA metabolism, suggests that they arose from a similarly structured precursor that can be traced back to the last universal common ancestor (LUCA). This structurally simple representative of the beta-GF is likely to represent one of the most basal lineages of the fold. The remaining sequence clusters with structurally comparable, simple 4-stranded beta-GF domains show extremely limited phyletic patterns (Table 1, 2), suggesting a probable recent derivation from the more ancient versions. The eukaryote-specific POZ domain might represent another derivative of a more widely-distributed 4-stranded version, which has accreted an additional C-terminal helical bundle to form a distinctive globular structure (Figures. 1, 2 and Table 1). All remaining versions of the beta-GF fold appear to form a monophyletic clade unified by the presence of an ancestral “lateral shelf” or “flange” that forms an extended connector between the helical segment and the remaining portion of the sheet after the topological cross-over (Figure 3 and Table 1). Of these versions, the Nudix superfamily appears to be one of the early branches given that its beta-sheet retains the ancestral 4-stranded core. All members of this superfamily share an insert or “outflow” in the middle of strand 2 which forms a distinctive shelf for accommodating substrates for NDP-X binding and hydrolase activity of the domain (79, 80). Another monophyletic class of beta-GF domains features a structurally distinct insert in the lateral shelf forming a barrel-like configuration (Figure 1B) (76). This subgroup is termed the fasciclin-like assemblage which unifies the fasciclin domain (PDB: 1O70 (81)), the ribosomal protein L25 (PDB: 1B75 (82)), the FimD superfamily, and the phosphoribosyl AMP cyclohydrolase (HisI) (PDB: 1ZPS (83)) with the glutamine synthetase N-terminal domain. The WWE domain, which appears to have acquired an additional strand after the terminal strand inserted in the middle of the core sheet, is also a likely member of this assemblage (Figure 3). Of the sequence superfamilies in this assemblage, the glutamine synthetase N-terminal domain is traceable to LUCA. Hence, the fasciclin-like version of the beta-GF domain might have diverged from other major lineages of the fold prior to the LUCA.
Table 2.
Natural classification of the β-GF.
| I. Basal 4-stranded versions of the β-GF |
|
|
| A. Archaeo-eukaryotic RNA polymerase β-subunit domain superfamily (all eukaryotes, all archaea) function unknown |
| PDB: 1I6HB (residues ~573–631) |
| B. IF3-N terminal domain superfamily (all bacteria, all eukaryotes except Giardia) function unknown |
| PDB: 1TIF (residues ~9–61) |
| C. POZ superfamily (all eukaryotes) mediates dimerization and transcriptional repression and interacts with histone deacetylase co-repressor complexes |
| PDB: 1HV2, several others |
| D. BofC/IGB lineage |
| Bypass of forespore (Bof)C family (Bacillus, Geobacillus) secreted protein important in spore formation pathway |
| PDB: 2BW2 |
| Immunoglobulin-binding (Ig-binding) family (firmicutes) cell surface virulence protein |
| PDB: 1HEZ, several others |
| E. Other lineages |
| Yml108w family (budding yeast) function unknown |
| PDB: 1N6Z |
|
|
| II. Nudix (MutT) Superfamily |
|
|
| (all eukaryotes, crenarchaea, euryarchaea, all bacteria, dsDNA viruses) nucleotide-derivative phosphohydrolase, contains insert in middle of second strand |
| PDB: 1RYAA, several others |
|
|
| III. Fasciclin-like assemblage |
|
|
| A. Glutamine synthetase N-terminal domain (GS-N) superfamily (all eukaryotes except Giardia, crenarchaea, euryarchaea, all bacteria, mimivirus) two-stranded insert contributes residue to enzyme active site, forms one wall in active site |
| PDB: 2BVCA (residues ~17–100), several others |
| B. Phosphoribosyl AMP cyclohydrolase (HisI) superfamily (fungi, plants, crenarchaea, euryarchaea, all bacteria) Two enzyme active sites formed by an obligate HisI dimer |
| PDB: 1ZPS |
| C. Fasciclin I superfamily (apicomplexa, crown group, euryarchaea, actinobacteria, bacteroidetes, chlorobi, chlamydiae, chloroflexi, cyanobacteria, deinococci, acidobacteria, planctomycetes, α/β/δ/γ proteobacteria) binds sugar moieties |
| PDB: 1NYO, 1O70 |
| D. Ribosomal protein L25 superfamily (apicomplexa, plants, slime molds, Anopheles, all bacteria) binds 5S rRNA |
| PDB: 1DFU, several others |
| E. FimD N-terminal domain (FimD-N) superfamily (α/β/δ/γ proteobacteria, deinococci) interacts with FimC protein |
| PDB: 1ZE3, 1ZDV, 1ZDX |
| F. WWE superfamily (all eukaryotes except Giardia) |
| PDB: 2A90A |
|
|
| IV. 5-stranded assemblage: Classical 5-stranded clade |
|
|
| A. TGS superfamily (all eukaryotes, all archaea, all bacteria) |
| PDB: 1NYR (residues 1–62), several others |
| B. ThiS superfamily (crenarchaea, euryarchaea, all bacteria, algae, Thalassiosira, Emiliania, Phaeodactylum, Odontella). SAMP2 is conjugated |
| PDB: 1F0Z, many others |
| C. MoaD superfamily (crenarchaea, euryarchaea, all bacteria, crown group) SAMP1 is conjugated |
| PDB: 1FM0, many others |
| D. Urm-1 superfamily (all eukaryotes) conjugated version implicated in tRNA thiouridine synthesis, conjugated during oxidative stress response |
| PDB: 1XO3, 1WGK, 2AX5 |
| E. Mut7-C family (Bacteria) Often Fused to N-terminal PIN nuclease domain and C-terminal Zn-ribbon |
| F. Ribosomal protein S4 superfamily (all eukaryotes, all archaea, all bacteria) |
| PDB: 2CQJA |
| G. Aldehyde oxidoreductase N-terminal (AOR-N) domain superfamily (crenarchaea, euryarchaea, actinobacteria, chloroflexi, deinococci, firmicutes, α/β/δ/ε/γ proteobacteria) tandem repeats in same protein form dimer that recognizes metallopterins |
| PDB: 1AOR (residues 1–210) |
| H. Prokaryotic UB-like superfamily (See text for details) |
| YukD family (actinobacteria, firmicutes) |
| PDB: 2BPS |
| I. UB-like superfamily (see section VI. below) |
| J. SPK/SupAnt lineage |
| Strepto/staphylococcus kinase (SPK) family (Streptococcus, Staphylococcus, caudovirales) interacts with host plasmin protein, promoting virulence |
| PDB: 2SAK, several others |
| Superantigen (SupAnt) family (Staphylococcus, Streptococcus) interacts with T-cell receptor β-chains |
| PDB: 1TY0 (residues 104–211), several others |
| K. Other lineages of note |
| TmoB family (actinobacteria, α/β/γ proteobacteria) |
| PDB: 1T0Q, 1T0R, 1T0S |
| PHH-γ family (actinobacteria, β/γ proteobacteria) |
| PDB: 2inp-E |
| RnfH family (α/β/γ proteobacteria) |
| FliD-FlgL/K family (α/β/δ/ε proteobacteria, clostridia, planctomycetes, Thermotoga, spirochetes, acidobacteria, Mu-like phages) |
|
|
| V. 5-stranded assemblage: Soluble ligand binding or metal chelating clade |
|
|
| A. 2Fe-2S ferredoxin superfamily |
| 2Fe-2S family (all eukaryotes, all archaea, all bacteria) small insert with conserved cysteines chelates Fe ions |
| PDB: 1NEK (residues 1–106), several others |
| L-proline dehydrogenase-type oxidoreductase (L-proDH alpha) family (euryarchaea, Thermofilum, actinobacteria, firmicutes, Psychroflexus, Herpetosiphon, α/β/δ/γ proteobacteria) lack conserved cysteine residues |
| PDB: 1Y56A (residues ~10–94) |
| B. Molybdopterin-dependent oxidoreductase (SOX) superfamily (Toxoplasma, crown group, crenarchaea, euryarchaea, actinobacteria, Aquifex, bacteroidetes/chlorobi, chloroflexi, cyanobacteria, deinoccoci, acidobacteria, firmicutes, planctomycetes, α/β/δ/ε proteobacteria) two inserts in core β-sheet help facilitate enzymatic activity |
| PDB: 1SOX (residues ~150–310) |
| C. Soluble-ligand binding β-grasp (SLBB) superfamily |
| Nqo1 family (kinetoplastids, ciliates, crown group, euryarchaea, all bacteria) |
| PDB: 2FUGS (residues ~246–334) |
| Transcobalamin family (hexapoda, vertebrates, euryarchaea, firmicutes, planctomycetes) |
| PDB: 2BBC (residues ~330–415) |
|
|
| VI. UB-like superfamily members |
|
|
| A. Classic UB-like clade |
| NIP45/Reni family (crown group) |
| SUMO/SMT3 family (all eukaryotes) conjugated versions tag proteins for localization/regulation |
| PDB: 1TGZ, several others |
| ZFAND1-C family (Naegleria, apicomplexa, stramenopiles, crown group) fused to N-terminal AN1 treble-clef domains |
| UFD of E1 C-terminal UB-like domain family (all eukaryotes) C-terminal adaptor domain of E1-like enzymes that bind E2-like enzymes |
| PDB: 1Y8X |
| UBX family (all eukaryotes except Giardia) adaptor version receptor for protein processing and degradation via the ERAD system |
| PDB: 1H8C, several others |
| Rad23N family (all eukaryotes except Giardia) involved in protein recruitment to the proteosome |
| PDB : 1UEL, 1P1A |
| Sin3a/SAP18-Ddi1 family |
| Sin3a/SAP18 subfamily (Trichomonas, apicomplexa, ciliates, crown group) |
| PDB: 2HDE |
| DNA damage inducible 1 (Ddi1) subfamily (plants, fungi, animals) adaptor version regulating Ho interaction with proteosome |
| PDB: 1V5O |
| Apg8-Apg12-APG5 family |
| Apg8 subfamily (Trichomonas, Naegleria, kinetoplastids, apicomplexa, ciliates, Phytophthora, crown group, pestiviruses, Marseillevirus) target lipids to autophagy pathway |
| Apg12 subfamily (Trichomonas, Naegleria, ciliates, crown group) targets proteins to autophagy pathway |
| PDB: 1WZ3 |
| APG5 subfamily (Trichomonas kinetoplastids, apicomplexa, ciliates, entamoebidae, crown group) |
| PDB: 2DYM |
| Bmi1/Psc-Wdr48C family |
| Bmi1/Psc subfamily (apicomplexa, ciliates, plants, animals) |
| Wdr48 C-terminal UB-domain subfamily (crown group) |
| BIPOSTO/ARF-PB1 family |
| BIPOSTO/ARF subfamily (plants) plant transcription factor |
| PB1 subfamily (all eukaryotes except Giardia and Trichomonas) adaptor that regulates localization |
| PDB: 2BKF, several others |
| MUBs family (plants, fungi, animals) versions anchored to plasma membrane via prenylation |
| PDB: 1WHG, 1SEH9 |
| Nedd8 family (crown group) |
| PDB: 1NDD, others |
| BAG N-terminal domain family (plants, animals) adaptor version mediating proteosome and Hsc70/Hsp70 chaperone system interaction |
| PDB: 1WXV |
| ANKRD40 C-terminal domain family (animals,plants, slime molds) adaptor version fused to N-terminal ankyrin repeats or MJ1566-like domains |
| CP2 C-terminal domain family (fungi, animals) adaptor version fused to N-terminal P53-like DNA binding domains of the cytochrome F fold |
| Splicing factor 3a (Sf3a)/prp21 family (plants, animals) |
| PDB: 1WE7, 1ZKH |
| UBP11/Usp40N-GGNB1 family |
| Usp40 N-terminal UB domain subfamily (ciliates, slime molds, animals) |
| UBP11-GGNB1 UB domain subfamily (plants, slime molds, vertebrates) |
| Hepatocyte odd protein shuttling (HOPSP) family (animals) |
| PDB: 1WIA |
| Parkin family (entamoebidae, animals) adaptor that binds Rpn10 subunit of 26S proteosomal subunit |
| PDB: 1IYF, 1MG8 |
| S30 N-terminal fusion ribosomal protein (S30-N) family (animals) adaptor that associates with Bcl-G and histone 2A |
| Midnolin family (animals) regulates genes related to neurogenesis in the nucleolus |
| Bone marrow stromal cell-derived (BMSC) UB family (animals) may regulate BMSC function in cell differentiation |
| PDB: 1X1M |
| Dendritic cell-derived C-terminal (DC-C) UB domain family (animals) implicated in cell differentiation and apoptosis |
| TRS4 N-terminal domain UB family (animals) |
| PDB: 2DAF |
| IkappaB kinase beta (IKK) UB-like domain family (animals) domain required for kinase activity |
| GDX N-terminal domain family (fungi, animals) |
| Homocysteine-inducible, endoplasmic reticulum stress-inducible protein (Herp-1) UB domain family (animals) |
| AN1 UB-like domain family (animals) |
| Interferon-inducible protein (ISG-15) UB domain family (vertebrates) conjugated version that tags proteins as part of antiviral response pathway |
| PDB: 1Z2M |
| 2'–5' oligoadenylate synthetase-like protein C-terminal (Oasl2-C) UB domain family (vertebrates) interacts with MBD1 transcriptional repressor |
| PDB: 1WH3 |
| Classic UB family (all eukaryotes, Candidatus 'Caldiarchaeum subterraneum’ and certain viruses) conjugated versions modulating protein stability and interactions |
| PDB: 1XD3, many others |
| Sacsin UB-domain family (vertebrates) |
| Rb1cc1 family (animals) |
| FAT10/Diubiquitin family (slime molds, vertebrates) conjugated version that tags proteins for proteosomal degradation |
| Np95-like ring finger protein N-terminal (NIRF-N) UB domain family (vertebrates) |
| PDB: 1WY8, 2FAZ |
| HOIL-1 UB-like domain family (animals) adaptor protein that regulates degradation of suppressor of cytokine signaling (SOCS) proteins |
| Transcription elongation factor B (Elongin B) family (animals) positive regulator of RNA pol II elongation factor A, possible tumor and cytokine signaling complex suppressor, and hypoxia-inducible gene regulator |
| PDB: 1VCB, several others |
| Nedd8 ultimate buster-1 (NUB1L) N-terminal UB domain family (animals) adaptor version linking FAT10 with the 26S proteosome |
| PDB: 1WJU |
| Ubiquilin N-terminal UB domain family (animals) adaptor version that interacts with PDI and mediates delivery of tagged proteins to the proteosome |
| USP48/USP26 C terminal UB domain family (plants, animals) |
| U11/U12 snRNP 25K family (plants, animals) |
| PDB: 1V2Y |
| Glycoprotein, synaptic 2 N-terminal (Gpsn2-N) domain family (apicomplexa, plants, slime molds, animals) |
| Ubl5-HubA family |
| Ubl5 subfamily (Trichomonas, apicomplexa, crown group) possibly a conjugating version, although target unknown |
| HubA subfamily (Tetrahymena, fungi) appears to modify Snu66 in pre-mRNA splicing and localization, conjugates with cell polarity factors Sph1 and Hbt1 |
| TUG-UBL1 N-terminal UB domain family (Naegleria, slime molds, fungi, animals) |
| PDB: 2AL3 |
| CLU1/eIF-3 family (Naegleria, ciliates, crown group) |
| VCPIP1- HIP7PN family |
| Valosin-containing protein (p97)/p47 complex-interacting protein p135 (VCPIP1) UB domain subfamily (vertebrates) |
| HIV-induced protein-7-like protease N-terminal (HIP7P-N) UB-like domain subfamily (all eukaryotes except Giardia) |
| UBP7/UBP14N-Ublcp1-Atg35690N family |
| UBP7/UBP14 N-terminal UB domain subfamily (all eukaryotes except Giardia) likely adaptor protein binding to p53 |
| PDB: 1V86, 1WGG |
| Ublcp1 UB domain subfamily (crown group) |
| Atg35690 N-terminal UB domain subfamily (fungi, plants) |
| Bat3/DsK family (all eukaryotes) adaptor version that binds with Hsp70-like Stch |
| PDB: 1WX9, 2BWF, 2BWE |
| AT23465p C-terminal domain family (Trichomonas, Giardia, kinetoplastids, ciliates, animals) fused to cytochrome b5-like heme/steroid binding domain |
| TbcB-TbcE family |
| Tubulin binding cofactor B (TbcB) UB-like domain subfamily (all eukaryotes except Trichomonas) |
| PDB: 1T0Y, 1V6E |
| Tubulin binding cofactor E (TbcE) UB-like domain subfamily (kinetoplastids, ciliates, plants, animals) |
| PDB: 1WJN |
| B. RA/FERM/PI3KN/DWNN clade |
| Ras-associating (RA) family (slime molds, fungi, animals) |
| PDB: 1RAX, several others |
| FERM UB-like domain family(ciliates, plants, slime molds, entamoebidae, animals) |
| PDB: 1EF1 (residues 4–87), several others |
| Phosphoinositide 3-kinase N-terminal (PI3K-N) domain family (all eukaryotes) |
| PDB: 1E8Y (residues ~217–310), several others |
| DWNN (RBBP6 N-terminal domain) family (all eukaryotes except Giardia and kinetoplastids) |
| PDB: 27CH |
| C. CAD/DCX clade |
| CAD domain family (animals) adaptor version that inhibits DNAse activity |
| PDB: 1IBX, several others |
| Double cortin (DCX) domain family (all eukaryotes except Giardia) occur in tandem, function as microtubule-binding domains |
| D. Additional 5-stranded classical UB-like lineages |
| BM-002/Ufm1 family (all eukaryotes except Trichomonas) conjugated version |
| PDB: 1J0G |
| NPL4 N-terminal (NPL4-N) UB-like domain family (all eukaryotes) |
| PDB: 1WF9 |
4.2. The 5-stranded assemblage
The 5-stranded assemblage is unified by the addition of the fifth strand to the core sheet and the consequent emergence of the “connector arm” linking the additional strand to the terminal strand (Figure 1A). The strong conservation of this unique structural feature, in conjunction with the exclusive grouping of these versions in structure similarity-based clustering, suggests that they form a monophyletic assemblage. This clade is also supported by the presence of a highly conserved alcoholic residue (S or T) at the transition between the N-terminal hairpin and the helical segment of the fold (41). The Ub-like beta-GF domains are derived from the ThiS and MoaD-like versions and comprise the most diverse clade within the classical 5-stranded clade. This version of the fold is most prevalent, both in terms of number of distinct clades contained within it and universal representation found across all life forms. At least 4 monophyletic lineages of this assembly, namely the TGS domain, the ThiS and MoaD proteins, and the 2Fe-2S ferredoxins can be traced to the LUCA. Beyond these, there are several lineages that are conserved in a single superkingdom or distributed more sporadically within a superkingdom. On the whole, two major clades can be recognized within the 5-stranded assemblage. The first of these, termed the classical 5-stranded clade, unites the three ancient lineages TGS, ThiS, and MoaD and several other closely-related versions, notably including the diverse prokaryotic and eukaryotic Ubl domains and the prokaryotic YukD domains.
The second major clade of the 5-stranded assemblage unifies a group of beta-GF domains by the presence of a set of inserts poorly conserved in sequence but similar in terms of their position in the structure. These domains are associated with binding soluble ligands or chelating metal ions and comprise the soluble ligand or metal-binding clade. The main sequence superfamilies in this clade are the 2Fe-2S ferredoxins/L-proline dehydrogenase-type oxidoreductase domain (PDB: 1Y56 (84)), the SLBB domains (36), and the molybdopterin-dependent oxidoreductase domains. A single version of this clade, a representative of the 2Fe-2S ferredoxins, was likely present in the LUCA and all other versions were later derived from this version. Another distinctive superfamily of the 5-stranded assemblage, the N-terminal module of the aldehyde oxidoreductase (AOR-N) (PDB: 1AOR (85)) consists of two tandem, distantly related copies of the beta-GF, which are unified by the modified structure of their connector arm, ligand-binding and dimerization pattern and do not show strong affinities to other members of the 5-stranded assemblage. A final superfamily is the universally distributed S4-RNA binding domain. The S4 domain appears to be a degenerate variant of the 5-stranded TGS-like beta-GF domain, which has emerged through partial loss of the N-terminal part of the domain including the first two strands prior to the LUCA (76).
Bacterial representatives of the classical 5-stranded assemblage. As the structure of these bacterial members tend to be very similar relative to other beta-GF assemblages (Figure 1B), distinct monophyletic clades of domains are typically determined through comparison of subtle variations in sequence composition. The following lineages have been clearly distinguished to date: the ThiS, MoaD, and Urm1 sulfur-carrier families, the YukD family, the fibrinolytic adapters of several Gram-positive bacteria (e.g. streptokinase), the superantigen/toxin domains, the RnfH family, the aromatic compound monooxygenase TmoB subunit family (40), the RNA-binding TGS family (86), the Mut7-C fused family (41), TAPI phage-tail assembly component Ubls, and the sporadically distributed but related assemblage of various prokaryotic Ubl families, many of which are predicted to act as modifiers (41). In addition, the solved structure of the phenol hydroxylase (PHH) gamma-subunit recently revealed itself as a member of this assemblage, albeit with a substantially divergent sequence precluding its earlier detection (87). Thus, it now appears that on two independent occasions members of the beta-GF were recruited to bacterial multicomponent monooxygenase (BMM) complexes (87),(88). While the high sequence divergence coupled with close structural similarity makes it difficult to ascertain relationships between some of these families, there is a degree of clarity concerning the general picture of their evolution. The superantigen/toxin and streptokinase families form a unified clade, as do the Urm1, ThiS, and MoaD families from which the Mut7-C and the phage tail TAP1 families were likely later derivations. The RnfH family is also likely derived from a TGS precursor at some point early in the evolution of the proteobacteria, within which it was widely adopted. Finally, combining sequence affinities with functional connections suggest that the various prokaryotic Ubl families predicted to act in modification pathways, the YukD family, and the eukaryotic set of Ubl domains form an additional higher-order clade to the exclusion of other lineages in the 5-stranded assemblage.
ThiS/MoaD/Urm-1 clade. This clade is centered on the sulfur-carrying beta-GF members, which play integral roles in cofactor biosynthesis pathways. Despite the generally poor annotation in public databases (which often confuse membership, particularly between the ThiS and MoaD clade), they can be readily delineated into the three primary clades based on structural and sequence similarities. The ThiS clade is the structurally simplest 5-stranded domain; the lack of structural elaboration tends to yield a shorter domain in absolute amino acid length relative to the MoaD and Urm1 clades. The ThiS domain is widespread across bacteria and while the archaeal ThiS members are relatively few in number, they form strong sub-groupings indicative of ancestral representation and not horizontal transfer from bacterial. Interestingly, the stramenopile eukaryotic lineage also appears to have acquired a copy of the ThiS gene via a horizontal gene transfer (HGT) event. In contrast to the ThiS clade, both the Urm1 the MoaD clades are characterized by smaller extended regions often taking the form of helix-like inserts in the lateral shelf and between the first two core beta-strands. The MoaD clade is widely distributed across all three superkingdoms of Life, while the Urm1 is found across all eukaryotic lineages including the basal lineages suggesting it was likely present in the First Eukaryotic Common Ancestor (FECA). It was most likely derived from a MoaD-like precursor in FECA. Unlike the Urm1 and ThiS clades, which are often observed in a single copy per genome, the MoaD clade can contain several representatives per genome. Discerning the finer relationships within the MoaD clade is a difficult task given the high level of transfer and lineage-specific diversification that appears to have occurred within the family and across prokaryotic superkingdoms (unpublished observations), yielding many subfamilies with varying degrees of affinity. Consistent with this observation, while the MoaD clade is widely assumed to primarily play a well-characterized role in the biosynthesis of Molybdenum/Tungsten (Mo/W) cofactors, in reality this clade appears to have been adapted to a wide range of functional niches in prokaryotes including, but likely not limited to, proteasomal-mediated destruction of proteins through covalent conjugation of the SAMP1 proteins to target substrates (61), assembly of tungsten cofactors for reductase reactions, assembly of siderophore-like compounds (89), and cysteine synthesis (90). Several functionally-specialized domains belonging to or closely related to the ThiS/MoaD/Urm-1 clade are described in additional detail below.
SAMP1 and SAMP2 domains. Both SAMP proteins were initially identified as modification proteins conjugated to targets in response to nitrogen depletion in the archaeon Haloferrax (61). The SAMP2 protein is a representative of one of the small, ancestral archaeal ThiS sub-clades. Despite its divergence, its minimal structure and sequence affinities clearly establish it as a member of the ThiS clade. The SAMP2 proteins are restricted to euryarchaeota, with substantial representation in the haloarchaea as well as some methanoarchaeal representation. In addition to being conjugated to target proteins, SAMP2 is also involved in tRNA thiolation, a function likely independent of its penchant for conjugation (91). SAMP1 domains, on the other hand, are clearly members of the MoaD clade; however, the phylogenetic relationships of the MoaD family are complicated by the repeated occurrence of gene duplications and HGT events yielding a generally poor picture of the incursion of the conjugating functional role of SAMP1 into the MoaD clade at large. Indeed, a recent study has demonstrated that the SAMP1 protein is indispensible for MoCo/WCo biosynthesis in Haloferrax (91), suggesting the primary role for SAMP1 is akin to the classical MoaDs. Unlike SAMP2, which has been demonstrated to form conjugates with substrate proteins (and itself) across a spectrum of perturbed conditions (61) (91), observation of SAMP1 conjugation is largely restricted to substantial nitrogen depletion (61). Thus, while SAMP2-ylation appears to be a genuine adaptation restricted to a small group of archaea, the extent to which SAMP1 is used as a modifier is still a question of great interest. Further analysis of the MoaD clade is required to precisely define the conditional and functional extent of SAMP1-ylation activity amongst MoaD proteins.
Aldehyde ferredoxin oxidoreductase (AOR)-associating domains. A distinct MoaD sub-clade found strictly adjacent to genes encoding an aldehyde ferredoxin oxidoreductase (AOR) was previously characterized (41) and is present in a sporadic group of phylogenetically distant archaea and bacteria, suggesting that they might constitute a mobile gene cluster. Analogous to SAMP1-like MoaD domains, the affinities for AOR-associating MoaDs are difficult to distinguish from other members of the clade, and require further detailed investigation to define its precise functional role. Gene neighborhoods for these domains often include MoeB and occasionally other cofactor biosynthesis genes such as MoaA and MoaE, and a pyridine disulfide oxidoreductase in close vicinity to MoaD-like and the AOR genes. In some organisms this gene cluster is distinct from the MoCo biosynthesis operon found elsewhere in the genome of the same organism. Experimentally characterized versions of these AORs have been shown to utilize a tungsten-containing variant of the cofactor (92). Taken together, these observations suggest that these AOR linked MoaD-like genes might specifically participate in the synthesis of the pterin moiety for WCo generation for the AORs, another probable, to this point overlooked, functional offshoot of the MoaD-like Ubl clade.
Phage tail Ubls. Genomes of lambdoid and T1-like phages contain related tail assembly gene complexes (93). In a large number of phages this complex encodes a protein TAPI that contains a beta-GF. Past analyses have indicated that this domain is most closely related to the ThiS/MoaD/Urm1 Ubl clade (41). Accordingly, the TAP1 Ubl strongly conserves C-terminal small amino acid residues characteristic of the ThiS/MoaD/Urm1 domains. An unusual lineage-specific character is the predicted presence of an extended lateral shelf region that is unique within other domains within the 5-stranded assemblage (41). Analysis of TAPI gene neighborhoods revealed that it is most often flanked by the genes encoding the TAPK protein, with JAB and NlpC/P60 peptidase domains, and the TAPJ protein, which is required for host specificity (41). The JAB domains found in these gene associations are also a part of the monophyletic clade, including those from the above-described class of gene neighborhoods. Variants of this organization lacking either of the two flanking genes are seen in a few phages/prophages, and in a small group of phages TAPI is flanked by a version of TAPK containing only an NlpC/P60 peptidase domain. The association of these JAB peptidases with an Ubl domain with a C-terminal glycine in the phage tail assembly operons strongly implies that the two domains form a functional unit. It is quite probable that the phage TAPI is processed by the peptidase domains of TAPK, with the JAB probably releasing the Ubl domain by cleaving at the point of the C-terminal-most glycine of the Ub-domain. Though there no evidence for this Ubl being incorporated into the mature phage tail, it is possible that it plays a role in assembly of the tail.
Mut7C-fused clade. A small yet phyletically diverse superfamily of classical bacterial Ubls is fused to the Mut7-C RNAse domain, a member of the PilT N-terminal (PIN) RNAse fold. This family is very similar in phyletic distribution to the predicted prokaryotic Ubl modifier domains, suggesting a similar mode of emergence and subsequent dispersion (see below). The specific enrichment observed for this family in beta-proteobacteria suggests it may have initially emerged in this lineage before distribution via HGT. This family appears to show closest affinity to the ThiS/MoaD/Urm1 clade and the TGS domains to the exclusion of others in the 5-stranded assemblage.
YukD clade. The YukD clade was initially identified in a bacteriophage receptor operon in Bacillus and homologs were subsequently observed in several low GC content Gram-positive bacteria (94),(95). Further analysis identified additional homologs in several actinobacteria and distant YukD homologs in bacterial lineages including planctomycetes and chloroflexi (65). In the Low GC gram positive bacteria, many of these domains appear as stand-alone versions (95). In the actinobacteria, the YukD-like Ub domain is fused to an integral membrane domain with 12 transmembrane helices. In both groups, the YukD protein is found in the neighborhood of the ESAT-6 export system, which at its core consists of a a-helical polypeptide, the virulence protein ESAT-6, and an FtsK-like ATPase that pumps these polypeptides outside the cell (96),(97),(98). Additionally, the actinobacterial operons contain a subtilisin-like protease (mycosin), members of the a-helical PE family and the membrane-associated PPE family of proteins. The operons of the low-GC Gram-positive bacteria, in contrast, encode a membrane-associated enzyme with a domain related to the protein serine/threonine kinase domain (YukC/EssB/Ukp) and a membrane protein prototyped by the Bacillus YueB protein. Given that such kinase domains have been shown to function as peptide ligases in several non-ribosomal peptide biosynthesis systems (e.g. pyoverdin and vibrioferrin synthetases) (75), it is conceivable that the YukC/EssB proteins could act as peptide ligases that help conjugate the YukD in these bacteria to specific targets such as the large membrane protein encoded by the same operon. As an added wrinkle, recently some YukD members have been found in association with components of prokaryotic Ubl conjugation systems (65). The YukD domains thus appear to have striking parallels with the eukaryotic Ubl domains, with some representatives involved in structural roles likely mediating protein-protein contacts and some representatives conjugated to target proteins. This points to the kinship of the YukD domains with the eukaryotic Ubl domains and their prokaryotic predecessors and could even suggest that the classical conjugated eukaryotic Ub descended from a YukD-like lineage. It is worth noting that despite the lack of experimental evidence, some of the standalone YukD versions could also be attached to target proteins given the conservation of small, C-terminal residues. Perhaps, as is in the case of the conjugated SAMPs and Urm1s, conjugation occurs in a condition-specific manner that has yet to be determined.
Prokaryotic Ubl modifier domains. Four distinct families of Ubls are found in conserved gene neighborhood associations with JAB domain peptidases and E1 and E2 Ub-ligases. All of these families are sporadically distributed across multiple bacterial lineages, and to this point have not been observed in archaea. These observed phyletic distributions are suggestive of a dispersion across diverse lineages through HGT. Three of these families are found fused to each other in the same polypeptides, forming bacterial genes resembling eukaryotic polyubiquitin-like genes (41). Interestingly, the order and frequency of the three families within a gene can vary, further suggesting the individual families show high evolutionary mobility. With the exception of the first of these three families, the small, conserved C-terminal residues typical of conjugating Ubls are absent, leaving open the question of how the individual components of these prokaryotic Ubl systems are interacting. Alignments previously constructed for the final family, a stand-alone domain, reveal conserved C-terminal small residues as well as a possibly abbreviated connector-arm region (41).
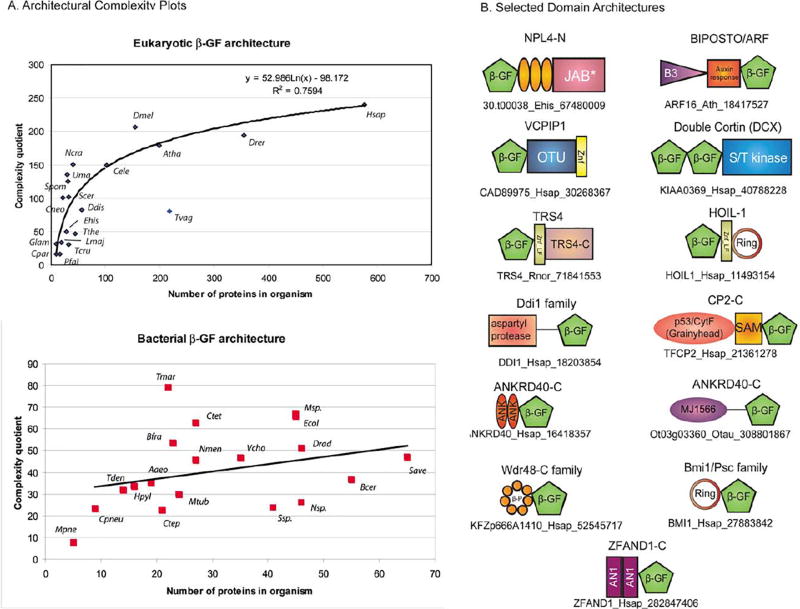
Eukaryotic representatives of the Ubl clade of beta-GF domains. In eukaryotes, this clade has undergone an explosive diversification with at least 19–20 distinct families which can be traced back to the last eukaryotic common ancestor (LECA). These families include six conjugated versions (ubiquitin, Urm1, Apg8/Aut7, Apg12, Ufm1 and SUMO/SMT3) (99, 100) and several versions that are known or predicted to function as adapters in multi-domain proteins, like the tubulin cofactor B (TBCB) (101,77,78) and phosphatidyl-inositol 3 kinase (PI3K) (102). Overall, in the course of eukaryotic evolution, at least 70 distinct sequence families appear to have emerged within this clade with some restricted to particular eukaryotic kingdoms like animals or plants. Some of these include poorly characterized families such as NPL4p, the Ubl domains of the BMI1/Posterior Sex Combs family of chromatin associated E3 ligases, a family with the Ubl domain fused to a cytochrome b5 domain, and the auxin response factor (BIPOSTO) in plants (Figure 5, Table 2). On the whole, comparisons of sequence conservation profiles showed that beta-GF domains related to the classical ubiquitin domain form a large monophyletic assemblage within the clade, including several distinct families such as Nedd8, SUMO, ubiquitin, NPL4, BAG, the Ubx domain, the tubulin cofactors or chaperones (TBCB and TBCE), Bat3/Dsk and Apg12/Gate16 (Figure 3, Table 2). The circularly permuted C-terminal UFD of eukaryote-type E1s also appears to have been derived from this lineage, though the recent evidence from the Caldiarchaeum Ub system seen in archaea suggests that this event occurred prior to the origin of the eukaryotes (64). Sequence comparisons also showed that the RA, FERM N-terminal module, and PI3K adapter domain families form another distinct higher-order monophyletic lineage within the eukaryotic radiation. The remaining lineages typified by ECR1/UBA1 and BM-002, while structurally close to the rest, form distinct sequence families that could not be placed into the any of the above larger assemblages of families (Table 2).
Figure 5.
A) Architectural complexity plot of beta-grasp domains found in eukaryotes and prokaryotes. The complexity quotient (CQ) for a given species (y-axis) is plotted against the total number of beta-grasp domain containing proteins in the same species. Species abbreviations are given next to plot points. B) Domain architectures of selected beta-grasp domains. Proteins are denoted by their gene names, species abbreviations and genbank index numbers. Proteins are not drawn to scale. The conserved cysteine clusters observed in the NPL4-N family are shown as orange ellipses. Expansion of domain abbreviations: B3, DNA-binding domain; Auxin response, auxin-responsive transcription factor domain, also called Aux-RF; OTU, OTU-like family of cysteine proteases; Znf, zinc-finger; Znf_LF, little finger family of zinc finger domains; beta-P, beta-propeller domain; X, previously uncharacterized BofC C-terminal domain. Species abbreviations are as follows: Aaeo, Aquifex aeolicus; Ath, Arabidopsis thaliana; Bfra, Bacteroides fragilis; Cele, Caenorhabditis elegans; Cneo, Cryptococcus neoformans; Cpneu, Chlamydophila pneumoniae.; Cpar, Cryptosporidium parva; Ctet, Clostridium tetani; Ctep, Chlorobium tepidum; Ddis, Dictyostelium discoideum; Dmel, Drosophila melonogaster; Drad, Deinococcus radiodurans; Drer, Danio rerio; Ecol, Escherichia cioli; Ehis, Entamoeba histolytica; Glam, Giardia lamblia; Hpyl, Helicobacter pylori; Hsap, Homo sapiens; Lmaj, Leishmania major; Mpne, Mycoplasma pneumoniae; Msp., Mesorhizobium sp.; Mtub, Mycobacterium tuberculosis; Ncra, Neurospora crassa; Nmen, Neisseria meningitides; Nsp., Nostoc sp.; Otau, Ostreococcus tauri; Pfal, Plasmodium falciparum; Rnor, Rattus norvegicus; Save, Streptomyces avermitilis; Scer, Saccharomyces cerevisiae; Spom, Schizosaccharomyces pombe; Ssp., Synecococcus sp.; Tcru, Trypanosoma cruzi; Tden, Treponema denticola; Tmar, Thermotoga maritima; Tthe, Tetrahymena thermophila; Tvag, Trichomonas vaginalis; Uma, Ustilago maydis; Vcho, Vibrio cholerae.
5. THE RELATIVE TIMELINE OF MAJOR ADAPTIVE RADIATIONS AND FUNCTIONAL TRANSITIONS OF THE beta-GF DOMAINS
5.1. The pre-LUCA phase and inference of the ancestral function of the beta-GF
The inference of at least 7 beta-GF or beta-GF-derived lineages (the S4 domain) in the LUCA suggests that there was a major diversification of the fold even before the LUCA (Figure 3). In structural terms, the inferred representatives in the LUCA span all major variants of the fold, from the simplest 4-stranded versions to the barrel-like forms (GS-N domain) to simple and elaborated versions the 5-stranded form. This suggests that the major structural variations were already in place as a result of the early diversification events of the pre-LUCA phase. In functional terms, versions close to the primitive state of both the 4- and 5-stranded forms, the RNA polymerase/IF3-N domain and the TGS domain, respectively, as well as the possible beta-GF derivative, the S4 domain, have functions related to RNA metabolism or RNA-binding (31, 45, 103). Even members of the Nudix clade are known to interact with nucleic acids or chemically-related molecules such as nucleoside diphosphate derivatives (79). In the eukaryotic lineage the Urm1 has been demonstrated to play a role in tRNA modification, whereas archaeal members of the ThiS/MoaD/Urm1 clade are predicted to be involved in tRNA-linked amino acid biosynthesis (57). RNA metabolism-associated functions are also sporadically observed in later-derived lineages such as the L25 ribosomal proteins in the fasciclin-like assemblage, the family of prokaryotic Ub-related domains fused to the Mut-7C-like RNAses (41), and several eukaryotic Ubl domains like those found in eIF3 p135/Clu-1 (supplementary material), RBBP6 (DWNN domain) (104), and Prp21/Splicing factor 3 (105). Given that the at least 4 of the seven main lineages traceable to the LUCA, including some of the inferred basal lineages, have a RNA/ribonucleoprotein associated role, it appears likely that the ancestral version of the beta-GF was probably involved in RNA-binding. The distribution of RNA-related roles (Figure 3, 4) implies that this function seems to have been retained or re-acquired in some sense in several later derived versions of the fold.
Figure 4.
Reconstructed evolutionary history of the eukaryotic ubiquitin superfamily. In contrast to Figure 3, major evolutionary transitions are now shown as horizontal lines and the maximum depth to which these individual lineages can be traced is shown with solid vertical lines. The rest of the details that includes functional categories are as in Figure 3.
A corollary to the inference of the ancestral function of the fold is that there were major functional innovations even in the pre-LUCA period. These are most prominently seen in the 5-stranded assemblage, and appear to be associated with the emergence of distinctive roles in sulfur delivery and scaffolding of Fe-S clusters. Previous observations have shown biochemical links between the formation of metal-sulfur clusters and sulfur transfer, including pathways in which ThiS and MoaD-like proteins participate (106). This observation raises the intriguing possibility that the earliest functional shift involved recruitment of a 5-stranded beta-GF domain for a shared general role in both sulfur transfer and generation of Fe-S clusters. It is quite possible that the subsequent specialization of such a generic precursor spawned the MoaD/ThiS/Urm1 precursor related to sulfur transfer on one hand and the 2Fe-2S ferredoxins on the other. The former function is consistent with the inferred presence of an E1-like enzyme similar to MoeB in the LUCA (57), which adenylates the Ubl protein prior to sulfotransfer. Further, the presence of molybdopterin, thiamine and different thiouridines in tRNA across the three superkingdoms of life suggests that sulfur transfer for at least a subset of these metabolites was already being catalyzed by an E1-like enzyme/Ubl-dependent system in the LECA. The rise of the 2Fe-2S ferredoxins probably coincided with the emergence of the precursors of the electron transfer chains of respiratory metabolism.
5.2. The post-LUCA phase: the prokaryotic superkingdoms
The emergence of the two prokaryotic superkingdoms, the archaea and bacteria, was marked by numerous superkingdom-specific innovations. Several of these innovations appear to have happened early in the history of the bacteria followed by multiple lateral transfers to the archaea. Likewise, innovations occurring in bacteria were also transferred to eukaryotes both during the primary endosymbiotic event and sporadically through later transfers. Members performing some form of most of the biochemical functions observed in extant representatives of the fold emerged in course of the post-LUCA diversification in bacteria. In certain cases there were no major shifts in basic biochemical activity but only an expansion of the range of specific biological contexts in which these activities were deployed. These included new RNA-binding/ribonucleoprotein-related functions emerging within diverse branches of the clade or adaptation of ThiS/MoaD-type proteins in sulfur transfer systems related to synthesis of lineage-specific metabolites (107). The principal, early functional innovations in the prokaryotic radiations were the independent acquisition of multiple small molecule/solute-binding capabilities across distant members of the fold and the emergence of catalytic versions, which might have in turn emerged from ligand-binding precursors (Figure 3). This phase also saw the recruitment of several forms of the beta-GF domain for mediating specific protein-protein interactions in the assembly or stabilization of multi-protein complexes, as evidenced by incorporation into flagella/pili structures (108–110), plasmin-interacting strepto/staphlyokinases, vertebrate T-cell interacting superantigens (111), and immunoglobulin-binding domains (112). The classical 5-stranded clade in particular appears to have given rise to several lineages that seem to function as protein interaction adapters, assembly or stability factors in very different biochemical contexts. For example, the TmoB and PHH gamma-subunit families might function in stabilizing the proteobacterial aromatic monooxygenase and the phenol hydroxylase complexes, respectively (87) (40), different members of the RnfH family might play roles in protein stability or assembly of the Rnf oxidoreductase complex, and some YukD members in the assembly of the ESAT-type export systems of Gram-positive bacteria (41).
5.3. The post-LUCA phase: covalently-attached protein modifiers emerge from sulfur carriers
The emergence of an E1-mediated covalent attachment of beta-GF domains to target proteins in the sulfur-carrying MoaD and ThiS-like clades, as evidenced by the SAMP1 and SAMP2 proteins appears to be a major functional shift in the biochemical activity that occurred post-LUCA. In functional terms, this shift to a protein modifier role represents a thematic collusion of the sulfur-transfer aspect with the protein interaction function, which were simultaneously expanding in members of the fold. Sampylation itself is currently thought to be restricted to a subset of archaea, however, detailed experimental studies probing the boundaries (and specifics) of this function have yet to be performed. This attachment mechanism appears to have either persisted in the eukaryotic Urm1 lineage or re-emerged independently; better characterization of the complete scope of Sampylation-like modifications in prokaryotes will assist in determining which scenario is more likely. An additional possible emergence of a protein modifier functional role is observed in the potential conjugation systems covalently linking ubiquitin-like beta-GF domains to other proteins through the E1, E2, and E3 ligase enzymes (predecessors of the eukaryotic conjugation systems). Better characterization of the E2 and E3-independent ligation mechanism will provide a stronger understanding of how this relates to the E2- and E3-dependent mechanism.
At least two systems found across a broad range of sporadically-distributed bacteria link the E1 and E2 ligases (along with the JAB domain, the primary proteasomal isopeptidase) to an Ubl. Several additional systems contain different combination of linkages between E1, E2, and JAB domains in the absence of Ubls (41). These systems may have emerged first through association of the JAB domain with the E1-Ubl interaction, which has two related, experimentally characterized functions in prokaryotes: the pre-processing of Ubl-containing peptides to expose small residues at the C-terminus (89) and ubiquitanase-like removal of Ubls from amino acid metabolism intermediates (90). Finally, a limited number of systems appear to have acquired the E3 RING-like ligase domain. Despite the limited number, systems containing the complete Ubl ligase complement are currently known from both bacteria and archaea, specifically in the archaeon Candidatus 'Caldiarchaeum subterraneum’ (64), the planctomycete Pirellula staleyi, the acidobacterium Acidobacteria sp. MP5ACTX8, the actinobacterium Franki alni, and the planctomycete Isosphaera pallida (65).
The recent characterization of the additional bacterial systems with RING-like E3 domains has considerable significance in elucidating the possible events leading to the inheritance of the eukaryotic Ub system (65). These systems, while more distantly related to their eukaryotic counterparts than the recently discovered Caldiarchaeum system (64) (but notably closer in their respective affinities than the systems lacking RING-like E3 domains), suggests that the Caldiarchaeum system is merely one of a larger range of such systems that are present in prokaryotes. Careful analysis of the complete complement of prokaryotic systems described above reveals a sequence and organizational diversity that is much higher than that seen in their eukaryotic cognates. This strongly suggests that systems resembling eukaryotic Ub-conjugation systems to different degrees were put together in prokaryotes during the diversification of various systems containing Ubls, E1s, E2s, RINGs and JAB peptidases. For example, the cysteine, molybdopterin, thiamin and siderophore biosynthesis systems merely contain Ubls, E1 and JAB peptidases in adenylation and sulfur transfer reactions (41). The more complex systems including an E2 component are likely to serve as regular Ub/Ubl-conjugation-like systems. Finally, there are those with RINGs that are likely to be close to the eukaryotic systems in every sense (64). Together, this lends strong support for a primarily prokaryotic origin for the complete Ub-system in the form of an operonic assembly linking all the key components that was acquired by the eukaryotic progenitor. Such operons are present across phylogenetically distant prokaryotes, and often missing in close relatives of the forms that display such systems. Hence, these prokaryotic Ub/Ubl-related systems are apparently highly mobile and widely disseminated through lateral transfer, analogous to the restriction-modification and secondary metabolite biosynthesis gene clusters (41),(75),(113). Therefore, we cannot yet be certain if the eukaryotic Ub-system emerged from a Caldiarchaeum-like system in the archaeal symbiont during eukaryogenesis. Indeed, such systems might be present in as yet unsampled bacteria suggesting that it is not unlikely that eukaryotes acquired such a system from the primary bacterial symbiont or even via an independent lateral transfer of the operon from yet another prokaryote.
5.4. Emergence of other possible links between protein stability and Ubl domains in prokaryotes
Two additional sets of prokaryotic Ubl domains belonging to the RnfH and ThiS/MoaD-like clades contain linkages to systems involved in protein stability. The RnfH protein is highly conserved across the beta/gamma proteobacteria and is found in two conserved gene neighborhood themes. The first conserved gene neighborhood containing an RnfH gene is found sporadically in a few proteobacteria, where it is linked to a group of Rnf genes whose products form a membrane associated complex involved in transporting electrons for various reductive reactions such as nitrogen fixation (114). In this system, it appears likely that the RnfH domain is acting as a subunit required for structural organization or assembly of the catalytic complex. However, in the second strongly conserved gene neighborhood theme, the RnfH domain associates with genes for a START domain protein (115), the tmRNA-binding protein SmpB and a small poorly understood membrane protein SmpA. Within this conserved neighborhood, the genes for the SmpB, the START domain protein and RnfH appear to share a common transcriptional regulatory region with the former gene being transcribed in the opposite direction to the latter two. This neighborhood is of particular interest given that the SmpB-tmRNA complex is used in bacteria to tag proteins from mRNAs lacking stop codons with small peptide. This tag targets proteins for degradation analogous to the eukaryotic Ub-system (116). This suggests a tantalizing functional link between these RnfH Ubls and the tmRNA-based regulation of protein stability in certain organisms, which might also additionally involve recognition of ligands by the START domain protein in this system. The second case with a linkage to protein stability features an Ubl domain of the ThiS/MoaD clade, which is encoded in a conserved operon that also displays genes for a JAB domain protein and ClpS (41). The ClpS domain recognizes the N-terminal domain of proteins targeted for destruction and links them to the protein-degrading ClpAP machine in bacteria and the RING finger E3 ligase of the eukaryotic N-recognins (117),(118). In light of these observations, it remains to be seen if this system might be involved in modification of proteins by an Ubl modification prior to recruitment by ClpS for degradation.
5.5. The eukaryotic phase of beta-GF evolution: expansion of the ubiquitin-like domains
Genomic and cell biological evidence suggests that the eukaryotes emerged as a result of a basic endosymbiotic event between a proteobacterium and an archaeon (perhaps related to the thaumarchaeal lineage) (119–121). Consequently, eukaryotes inherited several versions of the beta-GF domain found in both their archaeal and bacterial (mitochondrial) precursors (see Figure 2) (76). Eukaryotes showed an explosive development of the ubiquitin-like lineage resulting in forms that occupied biological functional niches across the entire cell, after inheritance of the core Ub system from one of their prokaryotic progenitors (see above). Most of these functions depend on the ancient property of the classical ubiquitin-like 5-stranded version to mediate protein-protein interactions, particularly in relation to the assembly or stabilization of complexes. These functions were performed either via conjugation of Ub/Ubls to target proteins and phosphatidylethanolamine, or as domains within multi-domain proteins. The biochemical diversification of the Ubl clade to perform multiple biological roles appears to have been notable even in the LECA (Figure 4). These adaptations include: 1) conjugation to proteins destined for degradation (classical Ub). 2) Tagging of proteins for altering interactions and localization (e.g. SUMO/SMT3) (14, 15) 3) conjugation to both a protein target (Apg5p) and the amino group of the lipid phosphatidylethanolamine (Agp8p/Aut7p) in regulation of the distinctly eukaryotic process of autophagy. 4) Possible recognition of proteins with conjugated Ub moieties (e.g. NPL4) (122). 5) Assembly of tubulin polymers (TBCB) (101) and microtubule-binding (DCX domains (32)). 6) Protein-protein interactions in Ub-modification (e.g. Ubl domains in Ub-deconjugating enzymes like Ubp7/Ubp14 and the Bmi1/Posterior Sex Combs-like E3s) and other signaling pathways (e.g. PI3K N-terminal domain) (102). The ancestral member of the eukaryotic Ubl clade is likely to have been a conjugated version because: 1) conjugated forms are seen across the entire diversity of the eukaryotic Ubl clade, which includes at least 5 versions traceable to the LECA and 2) they preserve the basic thiocarboxylate-forming chemistry seen in their even more ancient precursors like ThiS or MoaD. Given the inferred presence of multiple non-conjugated forms in the LECA, multiple early functional shifts resulting in non-conjugated forms appear to have occurred prior to the divergence of extant eukaryotes from the LECA, but after the emergence of the first eukaryotic cell. Of these, the UFD domain of E1s appears to have emerged in prokaryotes themselves from a conjugated Ub-like precursor and was recruited to a role in mediating E1 contacts during Ubl transfer, foreshadowing the recruitment of many Ubl families to protein-protein interaction roles later in eukaryotic evolution.
The diversification of the conjugated members of the eukaryotic Ubl radiation might have played a role in the emergence of distinct sub-cellular compartments in eukaryotes. While Ub and SUMO are linked to both cytoplasmic and nuclear proteins the available data points to a strong signal for the preferential nuclear enrichment of SUMO targets compared to the cytoplasmic enrichment of Ub targets, especially in the context of vesicular, vacuolar and ER complexes (24). Even the SUMO E3s show predominantly nuclear localization and nuclear interaction partners. This suggests that divergence of Ub and SUMO was probably correlated and coeval with the emergence of the nucleus as a separate compartment from the cytoplasmic ER network, with SUMO acquiring a dominant nuclear role and Ub a dominant cytoplasmic role. Sumoylation has been shown to exhibit a preference for lysine occurring in the signature sequence hKx[ED] (where h is a hydrophobic residue and×any residue)(123). Analysis of the extensive yeast dataset identifying individual modified lysines on Ub targets (124) revealed a preference for a motif of the form [ED]Kx4 [ED] spanning the modified lysine, and a mild general enrichment for acidic residues for around five positions on either side of the modified K (24). This suggests that in addition to divergence of the SUMO and Ub modifiers themselves, even their target site preferences differentiated to a certain extent. Consistent with this, the E1, E2 and E3 enzymes for Ub and SUMO appear to have diverged considerably in the interval between the FECA ancestor and the LECA, with distinct SUMO- and Ub-specific E3s by the time of the LECA. Further, specific nucleolar enrichment and function suggest that the divergence of SUMO might be related to the emergence of this key subcompartment within the nucleus (24).
Likewise, the emergence of the eukaryotic Ubx family of Ubl domains might have played an important role in the emergence of the eukaryote-specific endoplasmic reticulum (ER) associated degradation system (ERAD), which is involved in degradation or processing of proteins associated with the ER system (24). In this system, the Ubx domains function as receptors for recognition of the target proteins. This system also includes the Cdc48 ATPase of the AAA+ superclass which is of archaeal origin, and the membrane-embedded rhomboid-like peptidases (Der1 and Dfm1) of possibly bacterial origin. Thus, the ERAD system appears to have been pieced together in eukaryotes from systems drawn from both the archaeal and bacterial progenitors, as well as the Ubx domains innovated in the eukaryotes. Network analysis revealed that the ERAD system protein also includes an uncharacterized protein, ZFAND1/Ynl155w that contains an amino-terminal An1-finger combined with a distinct carboxy-terminal Ubl domain (ZFAND1-C family). The phyletic distribution of this domain (Table 2) suggests that it emerged relatively early in eukaryotic evolution, prior to the divergence of the heteroloboseans. This suggests that multiple distinct families of Ubl domains were recruited to mediate potential interactions with target proteins in course of evolution of the ERAD system. Finally, Ub conjugation plays a central role in processes such as vesicular trafficking, lysosomal targeting of proteins and cell-cycle progression, which are defining features of the eukaryotic step (e.g. see Ref (125). In each case tagging of proteins with Ub is necessary for the further processing of proteins through each of these systems, and might involve other proteins with Ubl domains. This suggests that the emergence of these key eukaryotic features was dependent on the Ub-system being in place.
Subsequently in eukaryotic evolution, there appear to have been several innovations of non-conjugated versions. Many of these continued to function in contexts related to Ub signaling, presumably by recognizing conjugated Ub moieties and target proteins (Figure 4, Table 2). However, some seem to have acquired apparently unrelated functions depending on the more general protein-protein interaction capabilities of the domain; for example, the RA domain in RAS signaling (33) and the CAD domain in apoptotic signaling (126–129). In temporal terms, a major pre-LECA expansion resulted in at least 19–20 distinct families in the ancestor of extant eukaryotes, followed by new families, like the PB1 and ZFAND1-C domains, appearing a little later in eukaryotic evolution. A notable phase of new innovation through sequence diversification resulted in several new families (e.g. Nedd8) prior to the radiation of the eukaryotic crown clade comprised of plants, slime molds, fungi, and animals. Interestingly, in the animal lineage alone, there appears to have been another massive round of diversification resulting in more than 10 distinct sequence families. The plants show a lineage expansion of a group of Ubl domains in the BIPOSTO/ARF transcriptional regulators (Table 2) which emerged from the more ancient PB1 family. Thus, in general, there appears to be a correlation between the emergence of new Ubl families and that of multi-domain proteins in the signaling systems of crown group eukaryotes, especially animals (130). Parallel to this expansion of Ubl domains in eukaryotes, there was also an expansion of other components of the Ub-conjugation system such as E1, E2, and E3 enzymes, F-box and UBA domains, and deubiquitinating peptidases (21, 25, 28).
Finally, on a few occasions eukaryotic Ubls appear to have been acquired by certain bacterial lineages. The best examples of these are seen in the plant pathogen Acidovorax citrulli (Aave_4710; gi: 120591805) and the vertebrate commensal Bacteroides fragilis (BF3883; gi: 60683320). The Acidovorax Ubl has a predicted signal peptide and is likely to be secreted into the host. While the Bacteroides lacks a secretory signal, it could be potential delivered to the host via other secretory mechanisms. Both these Ubls are closely related to Ub itself and are likely to have been derived from it; however, they lack the C-terminal glycines typical of Ub. Hence, they might interact with the host Ub-systems proteins to interfere with the transfer or the removal of endogenous Ub adducts, and thereby regulate host behavior. In a similar vein, certain RNA viruses of the pestivirus family have acquired domains related to Ub, NEDD8 and Apg8. These appear to be independent acquisitions from the host with different strains of viruses having acquired different Ubls. However, the potential role of all these Ubl inserts in interacting with the host Ub-system by possible targeting host proteins is supported by their requirement for enhanced pathogenicity of the bovine viral diarrhea virus (131, 132). Apg8 has also been acquired by a larger nucleocytoplasmic DNA virus, the Marseillevirus (gi: 284504416; misannotated as ubiquitin-like protein) and might represent a strategy used more widely across different viral groups.
6. EVOLUTIONARY TRENDS IN THE DOMAIN ARCHITECTURES OF beta-GF DOMAINS
6. 1. General architectural themes in the beta-GF
Previous studies on domains occurring in diverse architectural contexts in multi-domain proteins have hinted at a strong relationship between domain architectures and functional constraints (133). A systematic analysis of the domain architectures of the beta-GF domains and their conservation across evolution has assisted in the identification of these constraints. Both the sulfur-carrier and attachment to other proteins as a modifier functionalities require the free carboxy-terminus of the standalone beta-GF domain. As a result, the standalone copies of the 5-stranded Ub-like version have been preserved across all three superkingdoms since the LUCA. But an alternative strategy to this, observed primarily in eukaryotes, is the generation of free C-termini through post-translational proteolytic cleavage as seen in the polyubiquitins, APG8p (Aut7p), and even prokaryotic sulfur-carrying Ubls like those involved in siderophore biosynthesis (89). This raises that possibility that there might be other as yet undiscovered versions which are released for conjugation by proteolytic processing, as has been previously proposed for the DWNN domain (104). In this context, it still remains to be seen if the Ubl domain in the eukaryotic DDI1p-like proteins (41), which is connected via a glycine-rich linker to the rest of the protein (Figure 5) might be processed by the C-terminal aspartyl peptidase domain to release a free Ubl polypeptide.
In contrast, versions involved in protein and nucleic acid interactions are under no major constraints to remain as standalone forms of the domain. Hence, numerous instances of beta-GF domains involved in this function occur in multi-domain architectures (Figure 5). In most cases, the multi-domain architectures of RNA metabolism-related proteins are well-conserved across entire superkingdoms or even the three superkingdoms of Life because of the universality of these functions in their respective phyletic ranges. Multi-domain architectures associated with signaling or small-molecule interactions are often more restricted in their phyletic range and show lineage-specific diversity (130, 134).
The complexity quotient (CQ) (20), which measures the complexity of domain architectures for a given domain can be used to objectively assess the trends in domain architectural complexity of proteins (76) (Figure 5). This was done for 19 completely sequenced species of prokaryotes and 19 eukaryotic. In the case of prokaryotes, the plot reveals a more or less flat line, with an approximately constant domain architectural complexity across all species, irrespective of the number of beta-GF proteins they possessed (Figure 5). The plot only showed a few anomalous points: there was a greater than expected paucity of beta-GF proteins in the highly reduced genome of Mycoplasma and an inexplicably high architectural complexity in Thermotoga maritima. Thus, barring very few exceptions, the main tendency in prokaryotes is a wide variability in the number of proteins with beta-GF domains rather than any concerted increase in architectural complexity.
Eukaryotes not only have greater numbers of beta-GF domain proteins, but also appear to display greater diversity of domain architectures relative to the prokaryotes. The complexity of the beta-GF proteins as well as their numbers appear to increase throughout eukaryotic evolution with the highest figures observed in multicellular organisms of the eukaryotic crown group. However, the increase in architectural complexity is not linear across eukaryotes, with a tendency to plateau in animals. The only exception to the strong trend is Trichomonas vaginalis, a basal eukaryote, which appears to have undergone a massive, relatively recent proliferation across most protein families (135). As a result it possesses an unexpectedly large number of beta-GF proteins, but low architectural complexity comparable to other basal eukaryotes with similar numbers of beta-GF-containing proteins (Figure 5). In terms of actual architectures, the multicellular eukaryotes show numerous lineage-specific multi-domain proteins with different beta-GF domains, which are often involved in specific signaling pathways that correspond to unique aspects of the biology of these organisms. Typically, many of the eukaryotic multi-domain architectures, both ancient and lineage-specific, tend to combine the Ubl domains with other signaling domains, typically those involved in Ub-signaling. These combinations include those with deubiquitinating peptidases (e.g. of the OTU family), E3 ligases usually of the RING superfamily (e.g. HOIL1/RBCK1; Figure 5), and other Ub-binding domains like Uba, or other kinds of signaling domains like kinases as seen in the IKKs and Doublecortin. Another feature seen in eukaryotic architectures is the architectural variability through domain loss or accretion that is seen even in highly conserved orthologous proteins. One example is the Npl4p family (136) of Ubls which is conserved throughout eukaryotes and might play a role as a novel E3 in degradation of proteins in the endoplasmic reticulum.
6.2. Structural correlates for functional diversity in the beta-GF
The availability of multiple crystal and NMR structures has allowed exploration of the relationship between functional diversification of the beta-GF and its structural elaborations of the fold has been investigated in depth (see Figure 6 and (76) for methods). We briefly summarize below some of the highlights of these findings, including the structural correlates influencing the diversification of the eukaryotic Ubls.
Figure 6.
Relative location of beta-grasp interacting partners. The strands and core helix of an idealized beta-GF domain have been sectored into interaction zones, and the names of representatives of the beta-GF that interact using each of these zones are listed. The top view depicts the exposed face while the bottom view depicts the obscured face.
The apparent rarity of the simple 4-stranded versions suggests that there appears to be a tendency to elaborate the core sheet to provide an increased interface for interactions. On the whole, the exposed face (i.e. the face of the domain without the conserved alpha-helix) mediates more interactions across the beta-GF fold compared to the obscured face (i.e. the face “obscured” by the packing helical segment, lateral shelf, and connector arm). An example of soluble ligand binding is observed in the SLBB/ferredoxin/molbdopterin-dependent clade, wherein the unifying inserts of the clade typically occur in the region prior to strand 3 and in the region associated with the connector arm or the additional strand of the 5-stranded core. However, there is considerable diversity in the means by which these inserts mediate specific interactions, both between and within different families of this clade. The 2Fe-2S ferredoxins contain a characteristic set of four cysteines, three of which come from the pre-strand 3 insert and one from the connector arm-associated insert which help in coordination of the 2Fe-2S cluster (137). The subsequent diversification of this clade appears to have involved extensive adaptation of the binding site that originally contained the 2Fe-2S cluster for accommodating a diverse set of new ligands, including transcobalamin and related B12-binding proteins via a conserved aromatic residue in the pre-strand 3 insert of the SLBB domains (36), and a molybdopterin ligand via a conserved cysteine in a distinct pre-strand-3 barrel-like insert (Figure 6). Additionally, the exposed face in most of these cases remains available for interaction with other domains or polypeptides to recruit the beta-GF domain to larger complexes. This has been extensively demonstrated in the case of the 2Fe-2S ferredoxins (138, 139).
An additional evolutionary trend is observed in the proliferation and widespread utilization of the 5-stranded version, which might be associated with the availability of a larger surface on the exposed face for mediating contacts. This is manifested in the diverse range of protein-protein interactions by both prokaryotic and eukaryotic members of the 5-stranded assemblage, including those with the E1, E2 and E3 enzymes or their prokaryotic counterparts. The structure of the complex of Nedd8 with its E1 and E2 enzymes (77), in conjunction with the data accumulated from several other structures and mutagenesis experiments helps in deciphering the key modes of interaction prevalent in the 5-stranded clade. Nedd8 interacts via the exposed face with the sheet of the Rossmann fold domain of the adenylating domain of the E1, as in the case of the ThiS/MoaD clade (50, 51). Similarly the exposed face is also used by the beta-GF of the C-terminal UFD of the E1 to recruit the E2. More generally, different parts of the exposed face of the sheet mediate interactions specific to particular representatives of the 5-stranded assemblage (Figure 6). In particular, zones corresponding to the C-termini of the first and last strands which lie in the center of the sheet are utilized for protein interactions by all studied members of the classical 5-stranded clade. The structures of the eukaryotic members of the classical 5-stranded clade show that many of the interaction positions on the exposed face are shared, though the actual residues at those positions might not be conserved. Hence, the interaction specificity of different members has mainly arisen via sequence diversity at spatially congruent interaction sites, as opposed to acquisition of entirely new modes of interaction. The availability of the exposed face that provides an extended surface for interaction appears to be the primary factor for the pervasive use of this fold as mediator of protein-protein interactions across biologically disparate contexts. In a few instances, the obscured face of the RA (PDB: 1LFD (140)) and elongin domains (PDB: 1VCB (141)) might mediate specific interactions suggesting that their adapter function might depend on using both faces to mediate different sets of specific interactions.
In the complex of Nedd8 with its conjugating enzymes, the Nedd8 moiety covalently linked to the cysteine in the thioester-forming a-helical domain of the E1 protein also serves to recruit its specific E2 (77). This occurs via a unique interaction involving the cleft formed between then sheet and the helix of the beta-GF, which constitutes the “open-end” of the barrel-like form of the fold in Nedd8. From the side of the E2, the interaction is mediated via the conserved C-terminal helix. The high diversity of the residues in the E2 helix as well as the cleft of the Ub/Ubls suggests that this interaction is required for the specificity of E2-Ubl association. This interaction is representative of the more generic tendency of peripheral locations on the fold to be deployed in specific interactions that might be required only for the unique function performed by a particular clade (Figure 6). In the sulfur carrier and conjugated versions, the C-terminal tail plays a specific role in interaction with the active site of enzymes performing the adenylation or thioesterification (49–51, 77). The conserved presence of two small C-terminal residues in the tail of sulfur-carrier and conjugated versions strongly suggests this is a structural pre-adaptation for the emergence of the conjugation function. The convergent presence of small residues in the C-terminal tail of the bacterial Pup modifiers, in addition to extensive experimental studies on the role of these residues, supports this proposal (142),(143),(69). The role of the exposed face in protein-protein interactions appears to be a conservative aspect of the entire 5-stranded assemblage, which has been preserved from a period predating the LUCA. The apparently complex multiple protein-protein interactions in the eukaryotic Ub-conjugation process also appear to have emerged from the repeated use of the exposed face for interaction with E1, E2 and E3 partners.
7. DISCUSSION AND GENERAL CONCLUSIONS
Reconstruction of the evolution of the beta-GF fold suggests that the major structural variants and some of the basic biochemical features and modes of interaction had emerged prior to the LUCA. The evolutionary scenario emerging from the currently available structural and genomic data suggests that the earliest reconstructed function of the beta-GF domain was in the context of ribonucleoprotein complexes, probably as an RNA-binding domain. Based on the functions of extant versions of the domain, like the TGS domain, the IF3-N domain, and early structural derivatives such as the S4 superfamily, it is quite possible that the earliest versions of the fold played a generic role in a primitive pre-LUCA translation system. Amongst the major pre-LUCA functional shifts were those relating to the biosynthesis of sulfur-containing compounds and scaffolding of Fe-S clusters. On the face, such functional shifts from earlier roles in translation-associated RNPs appear drastic and puzzling. However, it should be noted that there is a functional connection between the sulfur incorporation pathways of thiamine biosynthesis and thiouridine synthesis in RNA (106, 144). Hence, it is possible that these shifts might have occurred in the context of 5-stranded versions of the beta-GF providing scaffolds for the synthesis of thio-base containing RNAs, a function preserved or re-emergent in Urm1- and SAMP2-mediated sulfur transfer (54),(56),(53),(57) (91). The reconstruction also implies that the versions of the beta-GF associated with major metabolic functions, including respiratory metabolism, radiated from the ancestral RNA-binding versions.
The post-LUCA phases of the evolutionary history of the beta-GF fold saw two major spurts of innovation. The first, occurring primarily in the bacteria, was accompanied by an extensive exploration of the biochemical function and interaction space by different versions of the fold. Most notably, the scaffold on at least 3 independent occasions acquired very different enzymatic activities even though the beta-GF fold does not seem to have ancestrally supported catalytic activities. The eukaryotic phase did not see extensive innovation in terms of fundamentally different biochemical functions, but the diversity of protein-protein interactions within the Ubl clade of the 5-stranded assemblage was vastly expanded through extensive sequence divergence of the primary interaction surfaces. In particular the diversification of the conjugated members of the eukaryotic Ubl radiation might have had an important role in the emergence of quintessential features of the cell such as the nucleo-cytoplasmic compartmentalization. This phase was also accompanied by ongoing innovation of new multi-domain architectures associated with the eukaryotic expansions of Ubl signaling domains (Figure 5).
Of primary interest when examining the evolution of the beta-GF is the understanding of the emergence of the eukaryotic Ub/Ubl modification system from the sulfur-carrying versions. The emerging genomic evidence together with certain experimental studies indicates that entire Ubl-systems are present and function as a regulated unit in prokaryotes. These systems show remarkable diversity in terms of domain content and even architectural variation. In large part this appears to have been driven by continual horizontal transfer and recombination across prokaryotic lineages as evidenced by the sporadic phyletic distributions for these systems. Observed from a general perspective, variation in domain content indicates a “piecemeal” construction of the Ub system in prokaryotes leading to increasing complexity until systems containing a tri-ligase complement of E1-like, E2-like, and RING-like E3 domains evolved. The general steps in the assembly are as follows: 1) the first association of a sulfur carrier Ubl with an E1-like domain in the LUCA. From this beginning, Ubls of the ThiS/MoaD/Urm1 clade, in collaboration with their cognate E1-like domains, diversified to occupy functional niches related to sulfur transfer primarily in the contexts of metabolic biosynthetic processes. The E1-domain itself was further recruited for adenylating activities as a peptide-ligase in Ubl-independent systems producing peptide antibiotics (145),(146),(147). Given the sequence and operonic diversification observed in the ThiS/MoaD/Urm1 clade, there is likely to be as-yet-uncharacterized, perhaps phyletically limited, functions pertaining to sulfotransfer and protein tagging derived from the ThiS/MoaD domains. 2) At some point these Ubl-E1-containing systems became associated with JAB domains, which in some cases functioned in pre-processing of Ubls as in siderophore biosynthesis (89) and in other cases functioned in removal of Ubls from amino acid metabolic intermediates (90). In parallel, in some phage tail assembly systems the Ubl associated with a JAB domain independent of an E1-like domain. Possible adaptations of these domains related to protein stability are suggested by the conserved gene neighborhood association of an Ubl, JAB domain, and ClpS domain. 3) Several of the above systems added an association with an E2-like ligase. While the functional roles of these await detailed experimental characterization, it appears likely that at least a few are involved in covalent attachment of Ubls to target proteins. 4) In relatively rare instances, a RING-like E3 ligase was added to systems with Ubls, E1-like, E2-like, and JAB domain peptidases (64, 65), which are likely to serve very similar functional roles to their eukaryotic counterparts. Remarkably, the components of these RING-containing systems (including the Ubl, E1, E2 and JAB peptidase) display by far the strongest affinities of any of the above to the eukaryotic Ubl system components, suggesting they were the forerunners of the classical eukaryotic Ub ligation systems. The mobility and diversity of these tri-ligase systems, evident in their percolation across distant bacterial and archaeal lineages, suggests a fundamental strength of these systems is their adaptability to different functional contexts. This attribute could have favored their selection as the founder of the eukaryotic Ub-system.
In contrast, Urmylation in eukaryotes (58) and Sampylation in archaea (61) are apparently E1-only Ubl conjugation systems deployed in certain specific functional contexts. Mechanistically these could be interpreted as resembling the most rudimentary form of the Ubl-conjugation system. However, this does not necessarily mean that they should be considered evolutionary precursors of the classical Ub-system of the eukaryotes. The characterization of the pupylation system and its coupling with the proteasomal system as well as the prediction of other prokaryotic peptide ligase systems (75) (see also section on YukD above) suggests that protein ligation emerged several times in prokaryotic evolution from preexisting metabolic pathways. On more than one occasion they were combined with the proteasomal system for modulating protein stability. Thus, it is possible that Sampylation and Urmylation via E1-only conjugation systems represent separate developments that emerged parallel to the elaboration the E2 and E3 containing systems, rather than being precursors of the latter.
We hope that the summary presented here renews the interest of researchers regarding both eukaryotic and prokaryotic Ubl systems and spurs the detailed investigation of the poorly understood versions of the beta-GF fold.
Supplementary Material
Acknowledgments
Work by LMI and LA is supported by the intramural funds of the National Library of Medicine at the National Institutes of Health, USA. Supplementary material can be found at: ftp://ftp.ncbi.nih.gov/pub/aravind/UB/Ubls.html
Abbreviations
- beta-GF
beta-grasp fold
- Ub
Ubiquitin
- Ubl
Ubiquitin-like
- Ig
Immunoglobulin
- DCX
Doublecortin
- SLBB
soluble ligand binding beta-grasp fold
- MoCo
Molybdenum cofactor
- GS
glutamine synthetase
- LUCA
Last Universal Common Ancestor
- AOR
aldehyde oxidoreductase
- PHH
phenol hydroxylase
- BMM
bacterial multicomponent monooxygenase
- HGT
horizontal gene transfer
- FECA
First Eukaryotic Common Ancestor
- PIN
PilT N-terminal
- LECA
Last Eukaryotic Common Ancestor
- TBCB
tubulin cofactor B
- PI3K
phosphatidyl-inositol 3 kinase
- ER
endoplasmic reticulum
- ERAD
endoplasmic reticulum-associated degradation system
- CQ
complexity quotient
Footnotes
Statement on Deposited Manuscripts, as Required by Frontiers in Bioscience: This is an un-copyedited author manuscript that has been accepted for publication in the Frontiers in Bioscience. Cite this article as it appears in the Journal of Frontiers in Bioscience. Full citation can be found by searching the Frontiers in Bioscience (Search for articles) following publication and at PubMed (http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?CMD=search&DB=pubmed) following indexing. This article may not be duplicated or reproduced, other than for personal use or within the rule of "Fair Use of Copyrighted Materials" (section 107, Title 17, U.S. Code) without permission of the copyright holder, the Frontiers in Bioscience. From the time of acceptance following peer review, the full final copy edited article of this manuscript will be made available at https://www.bioscience.org/. The Frontiers in Bioscience disclaims any responsibility or liability for errors or omissions in this version of the un-copyedited manuscript or in any version derived from it by the National Institutes of Health or other parties.
References
- 1.Glickman MH, Ciechanover A. The ubiquitin-proteasome proteolytic pathway: destruction for the sake of construction. Physiol Rev. 2002;82(2):373–428. doi: 10.1152/physrev.00027.2001. [DOI] [PubMed] [Google Scholar]
- 2.Goldstein G, Scheid M, Hammerling U, Schlesinger DH, Niall HD, Boyse EA. Isolation of a polypeptide that has lymphocyte-differentiating properties and is probably represented universally in living cells. Proc Natl Acad Sci U S A. 1975;72(1):11–5. doi: 10.1073/pnas.72.1.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wilkinson KD. The discovery of ubiquitin-dependent proteolysis. Proc Natl Acad Sci U S A. 2005;102(43):15280–2. doi: 10.1073/pnas.0504842102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vijay-Kumar S, Bugg CE, Cook WJ. Structure of ubiquitin refined at 1.8 A resolution. J Mol Biol. 1987;194(3):531–44. doi: 10.1016/0022-2836(87)90679-6. [DOI] [PubMed] [Google Scholar]
- 5.Vijay-Kumar S, Bugg CE, Wilkinson KD, Cook WJ. Three-dimensional structure of ubiquitin at 2.8 A resolution. Proc Natl Acad Sci U S A. 1985;82(11):3582–5. doi: 10.1073/pnas.82.11.3582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Murzin AG, Brenner SE, Hubbard T, Chothia C. SCOP: a structural classification of proteins database for the investigation of sequences and structures. J Mol Biol. 1995;247(4):536–40. doi: 10.1006/jmbi.1995.0159. [DOI] [PubMed] [Google Scholar]
- 7.Overington JP. Comparison of three-dimensional structures of homologous proteins. Curr Opin. Struct. Biol. 1992;2(3):394–401. [Google Scholar]
- 8.Kraulis PJ. Similarity of protein G and ubiquitin. Science. 1991;254(5031):581–2. doi: 10.1126/science.1658931. [DOI] [PubMed] [Google Scholar]
- 9.Schwartz DC, Hochstrasser M. A superfamily of protein tags: ubiquitin, SUMO and related modifiers. Trends Biochem Sci. 2003;28(6):321–8. doi: 10.1016/S0968-0004(03)00113-0. [DOI] [PubMed] [Google Scholar]
- 10.Weissman AM. Themes and variations on ubiquitylation. Nat Rev Mol Cell Biol. 2001;2(3):169–78. doi: 10.1038/35056563. [DOI] [PubMed] [Google Scholar]
- 11.Furukawa K, Mizushima N, Noda T, Ohsumi Y. A protein conjugation system in yeast with homology to biosynthetic enzyme reaction of prokaryotes. J Biol Chem. 2000;275(11):7462–5. doi: 10.1074/jbc.275.11.7462. [DOI] [PubMed] [Google Scholar]
- 12.Mizushima N, Noda T, Yoshimori T, Tanaka Y, Ishii T, George MD, Klionsky DJ, Ohsumi M, Ohsumi Y. A protein conjugation system essential for autophagy. Nature. 1998;395(6700):395–8. doi: 10.1038/26506. [DOI] [PubMed] [Google Scholar]
- 13.Kamitani T, Kito K, Nguyen HP, Yeh ET. Characterization of NEDD8, a developmentally down-regulated ubiquitin-like protein. J Biol Chem. 1997;272(45):28557–62. doi: 10.1074/jbc.272.45.28557. [DOI] [PubMed] [Google Scholar]
- 14.Dohmen RJ. SUMO protein modification. Biochim Biophys Acta. 2004;1695(1–3):113–31. doi: 10.1016/j.bbamcr.2004.09.021. [DOI] [PubMed] [Google Scholar]
- 15.Hay RT. SUMO: a history of modification. Mol Cell. 2005;18(1):1–12. doi: 10.1016/j.molcel.2005.03.012. [DOI] [PubMed] [Google Scholar]
- 16.Hochstrasser M. Origin and function of ubiquitin-like proteins. Nature. 2009;458(7237):422–9. doi: 10.1038/nature07958. doi:nature07958 (pii) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.May MJ, Larsen SE, Shim JH, Madge LA, Ghosh S. A novel ubiquitin-like domain in IkappaB kinase beta is required for functional activity of the kinase. J Biol Chem. 2004;279(44):45528–39. doi: 10.1074/jbc.M408579200. [DOI] [PubMed] [Google Scholar]
- 18.Neuber O, Jarosch E, Volkwein C, Walter J, Sommer T. Ubx2 links the Cdc48 complex to ER-associated protein degradation. Nat Cell Biol. 2005;7(10):993–8. doi: 10.1038/ncb1298. [DOI] [PubMed] [Google Scholar]
- 19.Schuberth C, Buchberger A. Membrane-bound Ubx2 recruits Cdc48 to ubiquitin ligases and their substrates to ensure efficient ER-associated protein degradation. Nat Cell Biol. 2005;7(10):999–1006. doi: 10.1038/ncb1299. [DOI] [PubMed] [Google Scholar]
- 20.Aravind L, Dixit VM, Koonin EV. Apoptotic molecular machinery: vastly increased complexity in vertebrates revealed by genome comparisons. Science. 2001;291(5507):1279–84. doi: 10.1126/science.291.5507.1279. [DOI] [PubMed] [Google Scholar]
- 21.Tanaka K, Suzuki T, Chiba T. The ligation systems for ubiquitin and ubiquitin-like proteins. Mol Cells. 1998;8(5):503–12. [PubMed] [Google Scholar]
- 22.Ardley HC, Robinson PA. E3 ubiquitin ligases. Essays Biochem. 2005;41:15–30. doi: 10.1042/EB0410015. [DOI] [PubMed] [Google Scholar]
- 23.Pickart CM. Mechanisms underlying ubiquitination. Annu Rev Biochem. 2001;70:503–33. doi: 10.1146/annurev.biochem.70.1.503. [DOI] [PubMed] [Google Scholar]
- 24.Venancio TM, Balaji S, Iyer LM, Aravind L. Reconstructing the ubiquitin network: cross-talk with other systems and identification of novel functions. Genome Biol. 2009;10(3):R33. doi: 10.1186/gb-2009-10-3-r33. doi:gb-2009-10-3-r33 (pii) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Soboleva TA, Baker RT. Deubiquitinating enzymes: their functions and substrate specificity. Curr Protein Pept Sci. 2004;5(3):191–200. doi: 10.2174/1389203043379765. [DOI] [PubMed] [Google Scholar]
- 26.Guterman A, Glickman MH. Deubiquitinating enzymes are IN/ (trinsic to proteasome function) Curr Protein Pept Sci. 2004;5(3):201–11. doi: 10.2174/1389203043379756. [DOI] [PubMed] [Google Scholar]
- 27.Iyer LM, Koonin EV, Aravind L. Novel predicted peptidases with a potential role in the ubiquitin signaling pathway. Cell Cycle. 2004;3(11):1440–50. doi: 10.4161/cc.3.11.1206. [DOI] [PubMed] [Google Scholar]
- 28.Nijman SM, Luna-Vargas MP, Velds A, Brummelkamp TR, Dirac AM, Sixma TK, Bernards R. A genomic and functional inventory of deubiquitinating enzymes. Cell. 2005;123(5):773–86. doi: 10.1016/j.cell.2005.11.007. [DOI] [PubMed] [Google Scholar]
- 29.Wing SS. Deubiquitinating enzymes--the importance of driving in reverse along the ubiquitin-proteasome pathway. Int J Biochem Cell Biol. 2003;35(5):590–605. doi: 10.1016/s1357-2725(02)00392-8. [DOI] [PubMed] [Google Scholar]
- 30.Sankaranarayanan R, Dock-Bregeon AC, Romby P, Caillet J, Springer M, Rees B, Ehresmann C, Ehresmann B, Moras D. The structure of threonyl-tRNA synthetase-tRNA (Thr) complex enlightens its repressor activity and reveals an essential zinc ion in the active site. Cell. 1999;97(3):371–81. doi: 10.1016/s0092-8674(00)80746-1. [DOI] [PubMed] [Google Scholar]
- 31.Wolf YI, Aravind L, Grishin NV, Koonin EV. Evolution of aminoacyl-tRNA synthetases--analysis of unique domain architectures and phylogenetic trees reveals a complex history of horizontal gene transfer events. Genome Res. 1999;9(8):689–710. [PubMed] [Google Scholar]
- 32.Kim MH, Cierpicki T, Derewenda U, Krowarsch D, Feng Y, Devedjiev Y, Dauter Z, Walsh CA, Otlewski J, Bushweller JH, Derewenda ZS. The DCX-domain tandems of doublecortin and doublecortin-like kinase. Nat Struct Biol. 2003;10(5):324–33. doi: 10.1038/nsb918. [DOI] [PubMed] [Google Scholar]
- 33.Nassar N, Horn G, Herrmann C, Scherer A, McCormick F, Wittinghofer A. The 2.2 A crystal structure of the Ras-binding domain of the serine/threonine kinase c-Raf1 in complex with Rap1A and a GTP analogue. Nature. 1995;375(6532):554–60. doi: 10.1038/375554a0. [DOI] [PubMed] [Google Scholar]
- 34.Ito T, Matsui Y, Ago T, Ota K, Sumimoto H. Novel modular domain PB1 recognizes PC motif to mediate functional protein-protein interactions. Embo J. 2001;20(15):3938–46. doi: 10.1093/emboj/20.15.3938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pearson MA, Reczek D, Bretscher A, Karplus PA. Structure of the ERM protein moesin reveals the FERM domain fold masked by an extended actin binding tail domain. Cell. 2000;101(3):259–70. doi: 10.1016/s0092-8674(00)80836-3. [DOI] [PubMed] [Google Scholar]
- 36.Burroughs AM, Balaji S, Iyer LM, Aravind L. A novel superfamily containing the b-grasp fold involved in binding diverse soluble ligands. Biology Direct. 2007;2:4. doi: 10.1186/1745-6150-2-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sazanov LA, Hinchliffe P. Structure of the hydrophilic domain of respiratory complex I from Thermus thermophilus. Science. 2006;311(5766):1430–6. doi: 10.1126/science.1123809. [DOI] [PubMed] [Google Scholar]
- 38.Wuerges J, Garau G, Geremia S, Fedosov SN, Petersen TE, Randaccio L. Structural basis for mammalian vitamin B12 transport by transcobalamin. Proc Natl Acad Sci U S A. 2006;103(12):4386–91. doi: 10.1073/pnas.0509099103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fraser JD, Urban RG, Strominger JL, Robinson H. Zinc regulates the function of two superantigens. Proc Natl Acad Sci U S A. 1992;89(12):5507–11. doi: 10.1073/pnas.89.12.5507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sazinsky MH, Bard J, Di Donato A, Lippard SJ. Crystal structure of the toluene/o-xylene monooxygenase hydroxylase from Pseudomonas stutzeri OX1. Insight into the substrate specificity, substrate channeling, and active site tuning of multicomponent monooxygenases. J Biol Chem. 2004;279(29):30600–10. doi: 10.1074/jbc.M400710200. [DOI] [PubMed] [Google Scholar]
- 41.Iyer LM, Burroughs AM, Aravind L. The prokaryotic antecedents of the ubiquitin-signaling system and the early evolution of ubiquitin-like beta-grasp domains. Genome Biol. 2006;7(7):R60. doi: 10.1186/gb-2006-7-7-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gnatt AL, Cramer P, Fu J, Bushnell DA, Kornberg RD. Structural basis of transcription: an RNA polymerase II elongation complex at 3.3 A resolution. Science. 2001;292(5523):1876–82. doi: 10.1126/science.1059495. [DOI] [PubMed] [Google Scholar]
- 43.Biou V, Shu F, Ramakrishnan V. X-ray crystallography shows that translational initiation factor IF3 consists of two compact alpha/beta domains linked by an alpha-helix. Embo J. 1995;14(16):4056–64. doi: 10.1002/j.1460-2075.1995.tb00077.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kycia JH, Biou V, Shu F, Gerchman SE, Graziano V, Ramakrishnan V. Prokaryotic translation initiation factor IF3 is an elongated protein consisting of two crystallizable domains. Biochemistry. 1995;34(18):6183–7. doi: 10.1021/bi00018a022. [DOI] [PubMed] [Google Scholar]
- 45.Iyer LM, Koonin EV, Aravind L. Evolutionary connection between the catalytic subunits of DNA-dependent RNA polymerases and eukaryotic RNA-dependent RNA polymerases and the origin of RNA polymerases. BMC Struct Biol. 2003;3:1. doi: 10.1186/1472-6807-3-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rabijns A, De Bondt HL, De Ranter C. Three-dimensional structure of staphylokinase, a plasminogen activator with therapeutic potential. Nat Struct Biol. 1997;4(5):357–60. doi: 10.1038/nsb0597-357. [DOI] [PubMed] [Google Scholar]
- 47.Weber DJ, Abeygunawardana C, Bessman MJ, Mildvan AS. Secondary structure of the MutT enzyme as determined by NMR. Biochemistry. 1993;32(48):13081–8. doi: 10.1021/bi00211a018. [DOI] [PubMed] [Google Scholar]
- 48.Lake MW, Wuebbens MM, Rajagopalan KV, Schindelin H. Mechanism of ubiquitin activation revealed by the structure of a bacterial MoeB-MoaD complex. Nature. 2001;414(6861):325–9. doi: 10.1038/35104586. [DOI] [PubMed] [Google Scholar]
- 49.Xi J, Ge Y, Kinsland C, McLafferty FW, Begley TP. Biosynthesis of the thiazole moiety of thiamin in Escherichia coli: identification of an acyldisulfide-linked protein--protein conjugate that is functionally analogous to the ubiquitin/E1 complex. Proc Natl Acad Sci U S A. 2001;98(15):8513–8. doi: 10.1073/pnas.141226698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Duda DM, Walden H, Sfondouris J, Schulman BA. Structural analysis of Escherichia coli ThiF. J Mol Biol. 2005;349(4):774–86. doi: 10.1016/j.jmb.2005.04.011. [DOI] [PubMed] [Google Scholar]
- 51.Lehmann C, Begley TP, Ealick SE. Structure of the Escherichia coli ThiS-ThiF complex, a key component of the sulfur transfer system in thiamin biosynthesis. Biochemistry. 2006;45(1):11–9. doi: 10.1021/bi051502y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rudolph MJ, Wuebbens MM, Rajagopalan KV, Schindelin H. Crystal structure of molybdopterin synthase and its evolutionary relationship to ubiquitin activation. Nat Struct Biol. 2001;8(1):42–6. doi: 10.1038/83034. [DOI] [PubMed] [Google Scholar]
- 53.Schlieker CD, Van der Veen AG, Damon JR, Spooner E, Ploegh HL. A functional proteomics approach links the ubiquitin-related modifier Urm1 to a tRNA modification pathway. Proc Natl Acad Sci U S A. 2008;105(47):18255–60. doi: 10.1073/pnas.0808756105. doi:0808756105 (pii) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Noma A, Sakaguchi Y, Suzuki T. Mechanistic characterization of the sulfur-relay system for eukaryotic 2-thiouridine biogenesis at tRNA wobble positions. Nucleic Acids Res. 2009;37(4):1335–52. doi: 10.1093/nar/gkn1023. doi:gkn1023 (pii) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nakai Y, Nakai M, Hayashi H. Thio-modification of yeast cytosolic tRNA requires a ubiquitin-related system that resembles bacterial sulfur transfer systems. J Biol Chem. 2008;283(41):27469–76. doi: 10.1074/jbc.M804043200. doi:M804043200 (pii) [DOI] [PubMed] [Google Scholar]
- 56.Leidel S, Pedrioli PG, Bucher T, Brost R, Costanzo M, Schmidt A, Aebersold R, Boone C, Hofmann K, Peter M. Ubiquitin-related modifier Urm1 acts as a sulphur carrier in thiolation of eukaryotic transfer RNA. Nature. 2009;458(7235):228–32. doi: 10.1038/nature07643. doi:nature07643 (pii) [DOI] [PubMed] [Google Scholar]
- 57.Burroughs AM, Iyer LM, Aravind L. Natural history of the E1-like superfamily: implication for adenylation, sulfur transfer, and ubiquitin conjugation. Proteins. 2009;75(4):895–910. doi: 10.1002/prot.22298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Van der Veen AG, Schorpp K, Schlieker C, Buti L, Damon JR, Spooner E, Ploegh HL, Jentsch S. Role of the ubiquitin-like protein Urm1 as a noncanonical lysine-directed protein modifier. Proc Natl Acad Sci U S A. 2011;108(5):1763–70. doi: 10.1073/pnas.1014402108. doi:1014402108 (pii) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Goehring AS, Rivers DM, Sprague GF., Jr Urmylation: a ubiquitin-like pathway that functions during invasive growth and budding in yeast. Mol Biol Cell. 2003;14(11):4329–41. doi: 10.1091/mbc.E03-02-0079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Goehring AS, Rivers DM, Sprague GF., Jr Attachment of the ubiquitin-related protein Urm1p to the antioxidant protein Ahp1p. Eukaryot Cell. 2003;2(5):930–6. doi: 10.1128/EC.2.5.930-936.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Humbard MA, Miranda HV, Lim JM, Krause DJ, Pritz JR, Zhou G, Chen S, Wells L, Maupin-Furlow JA. Ubiquitin-like small archaeal modifier proteins (SAMPs) in Haloferax volcanii. Nature. 2010;463(7277):54–60. doi: 10.1038/nature08659. doi:nature08659 (pii) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pedrioli PG, Leidel S, Hofmann K. Urm1 at the crossroad of modifications. 'Protein Modifications: Beyond the Usual Suspects' Review Series. EMBO Rep. 2008;9(12):1196–202. doi: 10.1038/embor.2008.209. doi:embor2008209 (pii) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Xu J, Zhang J, Wang L, Zhou J, Huang H, Wu J, Zhong Y, Shi Y. Solution structure of Urm1 and its implications for the origin of protein modifiers. Proc Natl Acad Sci U S A. 2006;103(31):11625–30. doi: 10.1073/pnas.0604876103. doi:0604876103 (pii) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nunoura T, Takaki Y, Kakuta J, Nishi S, Sugahara J, Kazama H, Chee GJ, Hattori M, Kanai A, Atomi H, Takai K, Takami H. Insights into the evolution of Archaea and eukaryotic protein modifier systems revealed by the genome of a novel archaeal group. Nucleic Acids Res. 2010 doi: 10.1093/nar/gkq1228. doi:gkq1228 (pii) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Burroughs AM, Iyer LM, Aravind L. Functional diversification of the RING finger and other binuclear treble clef domains in prokaryotes and the early evolution of the ubiquitin system. Mol Biosyst. 2011 doi: 10.1039/c1mb05061c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pearce MJ, Mintseris J, Ferreyra J, Gygi SP, Darwin KH. Ubiquitin-like protein involved in the proteasome pathway of Mycobacterium tuberculosis. Science. 2008;322(5904):1104–7. doi: 10.1126/science.1163885. doi:1163885 (pii) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sutter M, Damberger FF, Imkamp F, Allain FH, Weber-Ban E. Prokaryotic ubiquitin-like protein (Pup) is coupled to substrates via the side chain of its C-terminal glutamate. J Am Chem Soc. 2010;132(16):5610–2. doi: 10.1021/ja910546x. [DOI] [PubMed] [Google Scholar]
- 68.Imkamp F, Rosenberger T, Striebel F, Keller PM, Amstutz B, Sander P, Weber-Ban E. Deletion of dop in Mycobacterium smegmatis abolishes pupylation of protein substrates in vivo. Mol Microbiol. 2010;75(3):744–54. doi: 10.1111/j.1365-2958.2009.07013.x. doi:MMI7013 (pii) [DOI] [PubMed] [Google Scholar]
- 69.Cerda-Maira FA, Pearce MJ, Fuortes M, Bishai WR, Hubbard SR, Darwin KH. Molecular analysis of the prokaryotic ubiquitin-like protein (Pup) conjugation pathway in Mycobacterium tuberculosis. Mol Microbiol. 2010;77(5):1123–35. doi: 10.1111/j.1365-2958.2010.07276.x. doi:MMI7276 (pii) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Guth E, Thommen M, Weber-Ban E. Mycobacterial ubiquitin-like protein ligase PafA follows a two-step reaction pathway with a phosphorylated pup intermediate. J Biol Chem. 2011;286(6):4412–9. doi: 10.1074/jbc.M110.189282. doi:M110.189282 (pii) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Striebel F, Imkamp F, Sutter M, Steiner M, Mamedov A, Weber-Ban E. Bacterial ubiquitin-like modifier Pup is deamidated and conjugated to substrates by distinct but homologous enzymes. Nat Struct Mol Biol. 2009;16(6):647–51. doi: 10.1038/nsmb.1597. doi:nsmb.1597 (pii) [DOI] [PubMed] [Google Scholar]
- 72.Iyer LM, Burroughs AM, Aravind L. Unraveling the biochemistry and provenance of pupylation: a prokaryotic analog of ubiquitination. Biol Direct. 2008;3:45. doi: 10.1186/1745-6150-3-45. doi:1745-6150-3-45 (pii) 10.1186/1745-6150-3-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Liao S, Shang Q, Zhang X, Zhang J, Xu C, Tu X. Pup, a prokaryotic ubiquitin-like protein, is an intrinsically disordered protein. Biochem J. 2009;422(2):207–15. doi: 10.1042/BJ20090738. doi:BJ20090738 (pii) [DOI] [PubMed] [Google Scholar]
- 74.Chen X, Solomon WC, Kang Y, Cerda-Maira F, Darwin KH, Walters KJ. Prokaryotic ubiquitin-like protein pup is intrinsically disordered. J Mol Biol. 2009;392(1):208–17. doi: 10.1016/j.jmb.2009.07.018. doi:S0022-2836 (09)00837-7 (pii) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Iyer LM, Abhiman S, Maxwell Burroughs A, Aravind L. Amidoligases with ATP-grasp, glutamine synthetase-like and acetyltransferase-like domains: synthesis of novel metabolites and peptide modifications of proteins. Mol Biosyst. 2009;5(12):1636–60. doi: 10.1039/b917682a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Burroughs AM, Balaji S, Iyer LM, Aravind L. Small but versatile: the extraordinary functional and structural diversity of the beta-grasp fold. Biol Direct. 2007;2:18. doi: 10.1186/1745-6150-2-18. doi:1745-6150-2-18 (pii) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Huang DT, Hunt HW, Zhuang M, Ohi MD, Holton JM, Schulman BA. Basis for a ubiquitin-like protein thioester switch toggling E1-E2 affinity. Nature. 2007;445(7126):394–8. doi: 10.1038/nature05490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lois LM, Lima CD. Structures of the SUMO E1 provide mechanistic insights into SUMO activation and E2 recruitment to E1. Embo J. 2005;24(3):439–51. doi: 10.1038/sj.emboj.7600552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bessman MJ, Frick DN, O'Handley SF. The MutT proteins or "Nudix" hydrolases, a family of versatile, widely distributed, "housecleaning" enzymes. J Biol Chem. 1996;271(41):25059–62. doi: 10.1074/jbc.271.41.25059. [DOI] [PubMed] [Google Scholar]
- 80.Koonin EV. A highly conserved sequence motif defining the family of MutT-related proteins from eubacteria, eukaryotes and viruses. Nucleic Acids Res. 1993;21(20):4847. doi: 10.1093/nar/21.20.4847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Clout NJ, Tisi D, Hohenester E. Novel fold revealed by the structure of a FAS1 domain pair from the insect cell adhesion molecule fasciclin I. Structure. 2003;11(2):197–203. doi: 10.1016/s0969-2126(03)00002-9. [DOI] [PubMed] [Google Scholar]
- 82.Stoldt M, Wohnert J, Gorlach M, Brown LR. The NMR structure of Escherichia coli ribosomal protein L25 shows homology to general stress proteins and glutaminyl-tRNA synthetases. Embo J. 1998;17(21):6377–84. doi: 10.1093/emboj/17.21.6377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Sivaraman J, Myers RS, Boju L, Sulea T, Cygler M, Jo Davisson V, Schrag JD. Crystal structure of Methanobacterium thermoautotrophicum phosphoribosyl-AMP cyclohydrolase HisI. Biochemistry. 2005;44(30):10071–80. doi: 10.1021/bi050472w. [DOI] [PubMed] [Google Scholar]
- 84.Tsuge H, Kawakami R, Sakuraba H, Ago H, Miyano M, Aki K, Katunuma N, Ohshima T. Crystal structure of a novel FAD-, FMN-, and ATP-containing L-proline dehydrogenase complex from Pyrococcus horikoshii. J Biol Chem. 2005;280(35):31045–9. doi: 10.1074/jbc.C500234200. [DOI] [PubMed] [Google Scholar]
- 85.Chan MK, Mukund S, Kletzin A, Adams MW, Rees DC. Structure of a hyperthermophilic tungstopterin enzyme, aldehyde ferredoxin oxidoreductase. Science. 1995;267(5203):1463–9. doi: 10.1126/science.7878465. [DOI] [PubMed] [Google Scholar]
- 86.Staker BL, Korber P, Bardwell JC, Saper MA. Structure of Hsp15 reveals a novel RNA-binding motif. Embo J. 2000;19(4):749–57. doi: 10.1093/emboj/19.4.749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Sazinsky MH, Dunten PW, McCormick MS, DiDonato A, Lippard SJ. X-ray structure of a hydroxylase-regulatory protein complex from a hydrocarbon-oxidizing multicomponent monooxygenase, Pseudomonas sp. OX1 phenol hydroxylase. Biochemistry. 2006;45(51):15392–404. doi: 10.1021/bi0618969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Merkx M, Kopp DA, Sazinsky MH, Blazyk JL, Muller J, Lippard SJ. Dioxygen Activation and Methane Hydroxylation by Soluble Methane Monooxygenase: A Tale of Two Irons and Three Proteins A list of abbreviations can be found in Section 7. Angew Chem Int Ed Engl. 2001;40(15):2782–2807. 10.1002/1521-3773 (20010803)40:15<2782::AID-ANIE2782>3.0.CO;2-P (pii) [PubMed] [Google Scholar]
- 89.Godert AM, Jin M, McLafferty FW, Begley TP. Biosynthesis of the thioquinolobactin siderophore: an interesting variation on sulfur transfer. J Bacteriol. 2007;189(7):2941–4. doi: 10.1128/JB.01200-06. doi:JB.01200-06 (pii) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Burns KE, Baumgart S, Dorrestein PC, Zhai H, McLafferty FW, Begley TP. Reconstitution of a new cysteine biosynthetic pathway in Mycobacterium tuberculosis. J Am Chem Soc. 2005;127(33):11602–3. doi: 10.1021/ja053476x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Miranda HV, Nembhard N, Su D, Hepowit N, Krause DJ, Pritz JR, Phillips C, Soll D, Maupin-Furlow JA. E1- and ubiquitin-like proteins provide a direct link between protein conjugation and sulfur transfer in archaea. Proc Natl Acad Sci U S A. 2011;108(11):4417–22. doi: 10.1073/pnas.1018151108. doi:1018151108 (pii) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Johnson JL, Rajagopalan KV, Mukund S, Adams MW. Identification of molybdopterin as the organic component of the tungsten cofactor in four enzymes from hyperthermophilic Archaea. J Biol Chem. 1993;268(7):4848–52. [PubMed] [Google Scholar]
- 93.Wietzorrek A, Schwarz H, Herrmann C, Braun V. The genome of the novel phage Rtp, with a rosette-like tail tip, is homologous to the genome of phage T1. J Bacteriol. 2006;188(4):1419–36. doi: 10.1128/JB.188.4.1419-1436.2006. doi:188/4/1419 (pii) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Sao-Jose C, Baptista C, Santos MA. Bacillus subtilis operon encoding a membrane receptor for bacteriophage SPP1. J Bacteriol. 2004;186(24):8337–46. doi: 10.1128/JB.186.24.8337-8346.2004. doi:186/24/8337 (pii) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.van den Ent F, Lowe J. Crystal structure of the ubiquitin-like protein YukD from Bacillus subtilis. FEBS Lett. 2005;579(17):3837–41. doi: 10.1016/j.febslet.2005.06.002. doi:S0014-5793 (05)00711-8 (pii) [DOI] [PubMed] [Google Scholar]
- 96.Pallen MJ. The ESAT-6/WXG100 superfamily -- and a new Gram-positive secretion system? Trends Microbiol. 2002;10(5):209–12. doi: 10.1016/s0966-842x(02)02345-4. doi:S0966842X02023454 (pii) [DOI] [PubMed] [Google Scholar]
- 97.Iyer LM, Makarova KS, Koonin EV, Aravind L. Comparative genomics of the FtsK-HerA superfamily of pumping ATPases: implications for the origins of chromosome segregation, cell division and viral capsid packaging. Nucleic Acids Res. 2004;32(17):5260–79. doi: 10.1093/nar/gkh828. doi:32/17/5260 (pii) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Brodin P, Rosenkrands I, Andersen P, Cole ST, Brosch R. ESAT-6 proteins: protective antigens and virulence factors? Trends Microbiol. 2004;12(11):500–8. doi: 10.1016/j.tim.2004.09.007. doi:S0966-842X (04)00211-2 (pii) [DOI] [PubMed] [Google Scholar]
- 99.Mahajan R, Delphin C, Guan T, Gerace L, Melchior F. A small ubiquitin-related polypeptide involved in targeting RanGAP1 to nuclear pore complex protein RanBP2. Cell. 1997;88(1):97–107. doi: 10.1016/s0092-8674(00)81862-0. [DOI] [PubMed] [Google Scholar]
- 100.Matunis MJ, Coutavas E, Blobel G. A novel ubiquitin-like modification modulates the partitioning of the Ran-GTPase-activating protein RanGAP1 between the cytosol and the nuclear pore complex. J Cell Biol. 1996;135(6 Pt 1):1457–70. doi: 10.1083/jcb.135.6.1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Grynberg M, Jaroszewski L, Godzik A. Domain analysis of the tubulin cofactor system: a model for tubulin folding and dimerization. BMC Bioinformatics. 2003;4:46. doi: 10.1186/1471-2105-4-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Walker EH, Perisic O, Ried C, Stephens L, Williams RL. Structural insights into phosphoinositide 3-kinase catalysis and signalling. Nature. 1999;402(6759):313–20. doi: 10.1038/46319. [DOI] [PubMed] [Google Scholar]
- 103.Aravind L, Koonin EV. Novel predicted RNA-binding domains associated with the translation machinery. J Mol Evol. 1999;48(3):291–302. doi: 10.1007/pl00006472. [DOI] [PubMed] [Google Scholar]
- 104.Pugh DJ, Ab E, Faro A, Lutya PT, Hoffmann E, Rees DJ. DWNN, a novel ubiquitin-like domain, implicates RBBP6 in mRNA processing and ubiquitin-like pathways. BMC Struct Biol. 2006;6:1. doi: 10.1186/1472-6807-6-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Arenas JE, Abelson JN. The Saccharomyces cerevisiae PRP21 gene product is an integral component of the prespliceosome. Proc Natl Acad Sci U S A. 1993;90(14):6771–5. doi: 10.1073/pnas.90.14.6771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Mueller EG. Trafficking in persulfides: delivering sulfur in biosynthetic pathways. Nat Chem Biol. 2006;2(4):185–94. doi: 10.1038/nchembio779. [DOI] [PubMed] [Google Scholar]
- 107.Matthijs S, Baysse C, Koedam N, Tehrani KA, Verheyden L, Budzikiewicz H, Schafer M, Hoorelbeke B, Meyer JM, De Greve H, Cornelis P. The Pseudomonas siderophore quinolobactin is synthesized from xanthurenic acid, an intermediate of the kynurenine pathway. Mol Microbiol. 2004;52(2):371–84. doi: 10.1111/j.1365-2958.2004.03999.x. [DOI] [PubMed] [Google Scholar]
- 108.Klemm P, Christiansen G. The fimD gene required for cell surface localization of Escherichia coli type 1 fimbriae. Mol Gen Genet. 1990;220(2):334–8. doi: 10.1007/BF00260505. [DOI] [PubMed] [Google Scholar]
- 109.Saulino ET, Bullitt E, Hultgren SJ. Snapshots of usher-mediated protein secretion and ordered pilus assembly. Proc Natl Acad Sci U S A. 2000;97(16):9240–5. doi: 10.1073/pnas.160070497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Saulino ET, Thanassi DG, Pinkner JS, Hultgren SJ. Ramifications of kinetic partitioning on usher-mediated pilus biogenesis. Embo J. 1998;17(8):2177–85. doi: 10.1093/emboj/17.8.2177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Papageorgiou AC, Tranter HS, Acharya KR. Crystal structure of microbial superantigen staphylococcal enterotoxin B at 1.5 A resolution: implications for superantigen recognition by MHC class II molecules and T-cell receptors. J Mol Biol. 1998;277(1):61–79. doi: 10.1006/jmbi.1997.1577. [DOI] [PubMed] [Google Scholar]
- 112.Derrick JP, Wigley DB. The third IgG-binding domain from streptococcal protein G. An analysis by X-ray crystallography of the structure alone and in a complex with Fab. J Mol Biol. 1994;243(5):906–18. doi: 10.1006/jmbi.1994.1691. [DOI] [PubMed] [Google Scholar]
- 113.Roberts RJ. Restriction endonucleases. CRC Crit Rev Biochem. 1976;4(2):123–64. doi: 10.3109/10409237609105456. [DOI] [PubMed] [Google Scholar]
- 114.Jouanneau Y, Jeong HS, Hugo N, Meyer C, Willison JC. Overexpression in Escherichia coli of the rnf genes from Rhodobacter capsulatus--characterization of two membrane-bound iron-sulfur proteins. Eur J Biochem. 1998;251(1–2):54–64. doi: 10.1046/j.1432-1327.1998.2510054.x. [DOI] [PubMed] [Google Scholar]
- 115.Iyer LM, Koonin EV, Aravind L. Adaptations of the helix-grip fold for ligand binding and catalysis in the START domain superfamily. Proteins. 2001;43(2):134–44. doi: 10.1002/1097-0134(20010501)43:2<134::aid-prot1025>3.0.co;2-i. 10.1002/1097-0134 (20010501)43-2<134::AID-PROT1025>3.0.CO;2-I (pii) [DOI] [PubMed] [Google Scholar]
- 116.Karzai AW, Roche ED, Sauer RT. The SsrA-SmpB system for protein tagging, directed degradation and ribosome rescue. Nat Struct Biol. 2000;7(6):449–55. doi: 10.1038/75843. [DOI] [PubMed] [Google Scholar]
- 117.Lupas AN, Koretke KK. Bioinformatic analysis of ClpS, a protein module involved in prokaryotic and eukaryotic protein degradation. J Struct Biol. 2003;141(1):77–83. doi: 10.1016/s1047-8477(02)00582-8. doi:S1047847702005828 (pii) [DOI] [PubMed] [Google Scholar]
- 118.Erbse A, Schmidt R, Bornemann T, Schneider-Mergener J, Mogk A, Zahn R, Dougan DA, Bukau B. ClpS is an essential component of the N-end rule pathway in Escherichia coli. Nature. 2006;439(7077):753–6. doi: 10.1038/nature04412. doi:nature04412 (pii) [DOI] [PubMed] [Google Scholar]
- 119.Cavalier-Smith T. The origin of eukaryotic and archaebacterial cells. Ann N Y Acad Sci. 1987;503:17–54. doi: 10.1111/j.1749-6632.1987.tb40596.x. [DOI] [PubMed] [Google Scholar]
- 120.Margulis L. Symbiosis in Cell Evolution. WH Freeman; New York: 1993. [Google Scholar]
- 121.Zillig W. Comparative biochemistry of Archaea and Bacteria. Curr Opin Genet Dev. 1991;1(4):544–51. doi: 10.1016/s0959-437x(05)80206-0. [DOI] [PubMed] [Google Scholar]
- 122.Bruderer RM, Brasseur C, Meyer HH. The AAA ATPase p97/VCP interacts with its alternative co-factors, Ufd1-Npl4 and p47, through a common bipartite binding mechanism. J Biol Chem. 2004;279(48):49609–16. doi: 10.1074/jbc.M408695200. [DOI] [PubMed] [Google Scholar]
- 123.Xue Y, Zhou F, Fu C, Xu Y, Yao X. SUMOsp: a web server for sumoylation site prediction. Nucleic Acids Res. 2006;34(Web Server issue):W254–7. doi: 10.1093/nar/gkl207. doi:34/suppl_2/W254 (pii) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Peng J, Schwartz D, Elias JE, Thoreen CC, Cheng D, Marsischky G, Roelofs J, Finley D, Gygi SP. A proteomics approach to understanding protein ubiquitination. Nat Biotechnol. 2003;21(8):921–6. doi: 10.1038/nbt849. [DOI] [PubMed] [Google Scholar]
- 125.Hurley JH, Lee S, Prag G. Ubiquitin-binding domains. Biochem J. 2006;399(3):361–72. doi: 10.1042/BJ20061138. doi:BJ20061138 (pii) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Enari M, Sakahira H, Yokoyama H, Okawa K, Iwamatsu A, Nagata S. A caspase-activated DNase that degrades DNA during apoptosis, and its inhibitor ICAD. Nature. 1998;391(6662):43–50. doi: 10.1038/34112. [DOI] [PubMed] [Google Scholar]
- 127.Halenbeck R, MacDonald H, Roulston A, Chen TT, Conroy L, Williams LT. CPAN, a human nuclease regulated by the caspase-sensitive inhibitor DFF45. Curr Biol. 1998;8(9):537–40. doi: 10.1016/s0960-9822(98)79298-x. [DOI] [PubMed] [Google Scholar]
- 128.Liu X, Li P, Widlak P, Zou H, Luo X, Garrard WT, Wang X. The 40-kDa subunit of DNA fragmentation factor induces DNA fragmentation and chromatin condensation during apoptosis. Proc Natl Acad Sci U S A. 1998;95(15):8461–6. doi: 10.1073/pnas.95.15.8461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Mukae N, Enari M, Sakahira H, Fukuda Y, Inazawa J, Toh H, Nagata S. Molecular cloning and characterization of human caspase-activated DNase. Proc Natl Acad Sci U S A. 1998;95(16):9123–8. doi: 10.1073/pnas.95.16.9123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Anantharaman V, Iyer LM, Aravind L. Comparative Genomics of Protists: New Insights on Evolution of Eukaryotic Signal Transduction and Gene Regulation. Annu Rev Microbiol. 2006 doi: 10.1146/annurev.micro.61.080706.093309. [DOI] [PubMed] [Google Scholar]
- 131.Tautz N, Meyers G, Thiel HJ. Processing of poly-ubiquitin in the polyprotein of an RNA virus. Virology. 1993;197(1):74–85. doi: 10.1006/viro.1993.1568. doi:S0042-6822 (83)71568-0 (pii) [DOI] [PubMed] [Google Scholar]
- 132.Baroth M, Orlich M, Thiel HJ, Becher P. Insertion of cellular NEDD8 coding sequences in a pestivirus. Virology. 2000;278(2):456–66. doi: 10.1006/viro.2000.0644. [DOI] [PubMed] [Google Scholar]
- 133.Aravind L. Guilt by association: contextual information in genome analysis. Genome Res. 2000;10(8):1074–7. doi: 10.1101/gr.10.8.1074. [DOI] [PubMed] [Google Scholar]
- 134.Anantharaman V, Koonin EV, Aravind L. Regulatory potential, phyletic distribution and evolution of ancient, intracellular small-molecule-binding domains. J Mol Biol. 2001;307(5):1271–92. doi: 10.1006/jmbi.2001.4508. [DOI] [PubMed] [Google Scholar]
- 135.Carlton JM, Hirt RP, Silva JC, Delcher AL, Schatz M, Zhao Q, Wortman JR, Bidwell SL, Alsmark UC, Besteiro S, Sicheritz-Ponten T, Noel CJ, Dacks JB, Foster PG, Simillion C, Van de Peer Y, Miranda-Saavedra D, Barton GJ, Westrop GD, Muller S, Dessi D, Fiori PL, Ren Q, Paulsen I, Zhang H, Bastida-Corcuera FD, Simoes-Barbosa A, Brown MT, Hayes RD, Mukherjee M, Okumura CY, Schneider R, Smith AJ, Vanacova S, Villalvazo M, Haas BJ, Pertea M, Feldblyum TV, Utterback TR, Shu CL, Osoegawa K, de Jong PJ, Hrdy I, Horvathova L, Zubacova Z, Dolezal P, Malik SB, Logsdon JM, Jr, Henze K, Gupta A, Wang CC, Dunne RL, Upcroft JA, Upcroft P, White O, Salzberg SL, Tang P, Chiu CH, Lee YS, Embley TM, Coombs GH, Mottram JC, Tachezy J, Fraser-Liggett CM, Johnson PJ. Draft genome sequence of the sexually transmitted pathogen Trichomonas vaginalis. Science. 2007;315(5809):207–12. doi: 10.1126/science.1132894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Bays NW, Wilhovsky SK, Goradia A, Hodgkiss-Harlow K, Hampton RY. HRD4/NPL4 is required for the proteasomal processing of ubiquitinated ER proteins. Mol Biol Cell. 2001;12(12):4114–28. doi: 10.1091/mbc.12.12.4114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Muller JJ, Muller A, Rottmann M, Bernhardt R, Heinemann U. Vertebrate-type and plant-type ferredoxins: crystal structure comparison and electron transfer pathway modelling. J Mol Biol. 1999;294(2):501–13. doi: 10.1006/jmbi.1999.3253. [DOI] [PubMed] [Google Scholar]
- 138.Truglio JJ, Theis K, Leimkuhler S, Rappa R, Rajagopalan KV, Kisker C. Crystal structures of the active and alloxanthine-inhibited forms of xanthine dehydrogenase from Rhodobacter capsulatus. Structure. 2002;10(1):115–25. doi: 10.1016/s0969-2126(01)00697-9. [DOI] [PubMed] [Google Scholar]
- 139.Yankovskaya V, Horsefield R, Tornroth S, Luna-Chavez C, Miyoshi H, Leger C, Byrne B, Cecchini G, Iwata S. Architecture of succinate dehydrogenase and reactive oxygen species generation. Science. 2003;299(5607):700–4. doi: 10.1126/science.1079605. [DOI] [PubMed] [Google Scholar]
- 140.Huang L, Hofer F, Martin GS, Kim SH. Structural basis for the interaction of Ras with RalGDS. Nat Struct Biol. 1998;5(6):422–6. doi: 10.1038/nsb0698-422. [DOI] [PubMed] [Google Scholar]
- 141.Stebbins CE, Kaelin WG, Jr, Pavletich NP. Structure of the VHL-ElonginC-ElonginB complex: implications for VHL tumor suppressor function. Science. 1999;284(5413):455–61. doi: 10.1126/science.284.5413.455. [DOI] [PubMed] [Google Scholar]
- 142.Johnson ES, Schwienhorst I, Dohmen RJ, Blobel G. The ubiquitin-like protein Smt3p is activated for conjugation to other proteins by an Aos1p/Uba2p heterodimer. EMBO J. 1997;16(18):5509–19. doi: 10.1093/emboj/16.18.5509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Wilkinson KD, Audhya TK. Stimulation of ATP-dependent proteolysis requires ubiquitin with the COOH-terminal sequence Arg-Gly-Gly. J Biol Chem. 1981;256(17):9235–41. [PubMed] [Google Scholar]
- 144.Palenchar PM, Buck CJ, Cheng H, Larson TJ, Mueller EG. Evidence that ThiI, an enzyme shared between thiamin and 4-thiouridine biosynthesis, may be a sulfurtransferase that proceeds through a persulfide intermediate. J Biol Chem. 2000;275(12):8283–6. doi: 10.1074/jbc.275.12.8283. [DOI] [PubMed] [Google Scholar]
- 145.Onaka H, Nakaho M, Hayashi K, Igarashi Y, Furumai T. Cloning and characterization of the goadsporin biosynthetic gene cluster from Streptomyces sp. TP-A0584. Microbiology. 2005;151(Pt 12):3923–33. doi: 10.1099/mic.0.28420-0. doi:151/12/3923 (pii) [DOI] [PubMed] [Google Scholar]
- 146.Gonzalez-Pastor JE, San Millan JL, Castilla MA, Moreno F. Structure and organization of plasmid genes required to produce the translation inhibitor microcin C7. J Bacteriol. 1995;177(24):7131–40. doi: 10.1128/jb.177.24.7131-7140.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Roush RF, Nolan EM, Lohr F, Walsh CT. Maturation of an Escherichia coli ribosomal peptide antibiotic by ATP-consuming N-P bond formation in microcin C7. J Am Chem Soc. 2008;130(11):3603–9. doi: 10.1021/ja7101949. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.