Abstract
Objective
Vascular extracellular matrix (ECM) stiffening is a risk factor for aortic and coronary artery disease. How matrix stiffening regulates the transcriptome profile of human aortic (Ao) and coronary (Co) vascular smooth muscle cells (VSMCs) is not well understood. Furthermore, the role of long non-coding RNAs (lncRNAs) in the cellular response to stiffening has never been explored. This study characterizes the stiffness-sensitive transcriptome of human Ao and Co VSMCs and identify potentially key lncRNA regulators of stiffness-dependent VSMC functions.
Approach and Results
Ao and Co VSMCs were cultured on hydrogel substrates mimicking physiologic and pathologic ECM stiffness. Total RNA-seq was performed to compare the stiffness-sensitive transcriptome profiles of Ao and Co VSMCs. We identified 3098 genes (2842 protein coding and 157 lncRNA) that were stiffness-sensitive in both Ao and Co VSMCs (FDR<1%). Hierarchical clustering revealed that Ao and Co VSMCs grouped by stiffness rather than cell-origin. Conservation analyses also revealed that stiffness-sensitive genes were more conserved than stiffness-insensitive genes. These VSMC stiffness-sensitive genes were less tissue-type specific and expressed in more tissues than stiffness-insensitive genes. Using unbiased systems analyses, we identified MALAT1 as a stiffness-sensitive lncRNA that regulates stiffness-dependent VSMC proliferation and migration in vitro and in vivo.
Conclusions
This study provides the first transcriptomic landscape of human Ao and Co VSMCs in response to ECM stiffness and identifies novel stiffness-sensitive human lncRNAs. Our data suggest that the stiffness-sensitive transcriptome is evolutionarily important to VSMCs function, and that stiffness-sensitive lncRNAs can act as regulators of stiffness-dependent phenotypes.
Keywords: Extracellular matrix stiffness, Vascular smooth muscle cells, Long non-coding RNAs, Transcriptome
Subject Codes: Smooth Muscle Proliferation and Differentiation, Vascular Biology, Gene Expression and Regulation, Vascular Disease, Coronary Artery Disease
INTRODUCTION
Vascular stiffening is associated with aging, hypertension, diabetes, and several genetic disorders and is recognized as a risk factor for cardiovascular disease1–4. Several studies demonstrate that vascular smooth muscle cells (VSMCs) respond to extracellular matrix (ECM) stiffness by proliferating, migrating, and further remodeling the vascular ECM, thus contributing to atherosclerosis and hypertension in animal models5–7. However, the detailed transcriptomic response of VSMCs to experimental variation in ECM stiffness has never been investigated.
Furthermore, VSMCs are diverse and heterogeneous across the vasculature resulting from distinct embryologic origins and are exposed to diverse microenvironmental, mechanical, and biochemical cues. Although some studies have shown that vascular cells from different developmental origins have different transcriptome programs and can respond differently to mechanical and biochemical factors8–17, other studies suggest vascular cells from differing origins are stable and maintain their properties when transplanted to different microenvironments12,16,18,19. In particular, the VSMCs of the proximal aorta (Ao) and coronary (Co) arteries arise separately from the neural crest and proepicardium, respectively12,20 and thus may respond differently to pathologic ECM stiffening, contributing to differences in aortic and coronary artery disease. To date, most studies have predominantly utilized non-coronary and non-human models to elucidate VSMC responses to ECM stiffness.5,7,21–27.
Long noncoding RNAs (lncRNAs) contribute to transcriptome complexity and may account for cell-type and functional diversity. Several lncRNAs have been implicated in biological, developmental, and pathological processes and act through mechanisms such as chromatin modifications, cis regulation of target genes, and post-transcriptional regulation of mRNA processing28–30. Several lncRNAs such as SENCR, lincRNA-p21, MYOSLID, and SMILR have been linked to VSMC functions31–35. While stiffness-regulated transcription factors and microRNAs have been identified as coordinators of stiffness-mediated gene expression36–38, the role of lncRNAs in pathologic stiffening is unknown. Understanding the stiffness-sensitive non-coding transcriptome may shed light into the gene regulatory networks that control stiffness-mediated functions and underlie different disease phenotypes along the vascular bed.
Here, we present a comprehensive analysis of the stiffness-sensitive transcriptome in human Ao and Co VSMCs. We used engineered hydrogel substrates tuned to physiologic and pathologic arterial stiffness and profile donor-matched Ao and Co VSMCs. Our data suggest that the stiffness-sensitive transcriptome is evolutionarily important to VSMCs function and that stiffness-sensitive lncRNAs can act as regulators of stiffness-dependent phenotypes.
MATERIALS AND METHODS
Materials and Methods are available in the online-only Data Supplement
RESULTS
The Stiffness Transcriptome Landscape of Co and Ao SMCs in Culture
We implemented deep RNA-sequencing to comprehensively define gene expression in donor-matched aortic and coronary smooth muscle cells in response to physiologically soft and pathologically stiff hydrogel matrices (Fig 1A). Using GENCODE annotations, we identified a total of 17,716 genes expressed in either Ao or Co VSMCs and in either soft or stiff conditions (Fig 1B); 93.2% were expressed in both cell lineages (DESeq2 mean normalized counts > 8). Of all expressed genes, 13,574 were protein-coding genes, and 2,379 were long non-coding RNAs. Further inspection revealed protein-coding genes are more commonly expressed in both lineages of VSMCs than lncRNAs (95.7% v 82.9%). These proportions are consistent with expression data obtained from whole human coronary and aortic tissues in the GTEx database (Fig IA). The uniquely expressed lncRNAs (17.1%) and protein coding genes (4.3%) may represent contributions from the unique developmental origins of Ao and Co VSMCs.
Figure 1. The stiffness transcriptome landscape of coronary and aortic VSMCs.
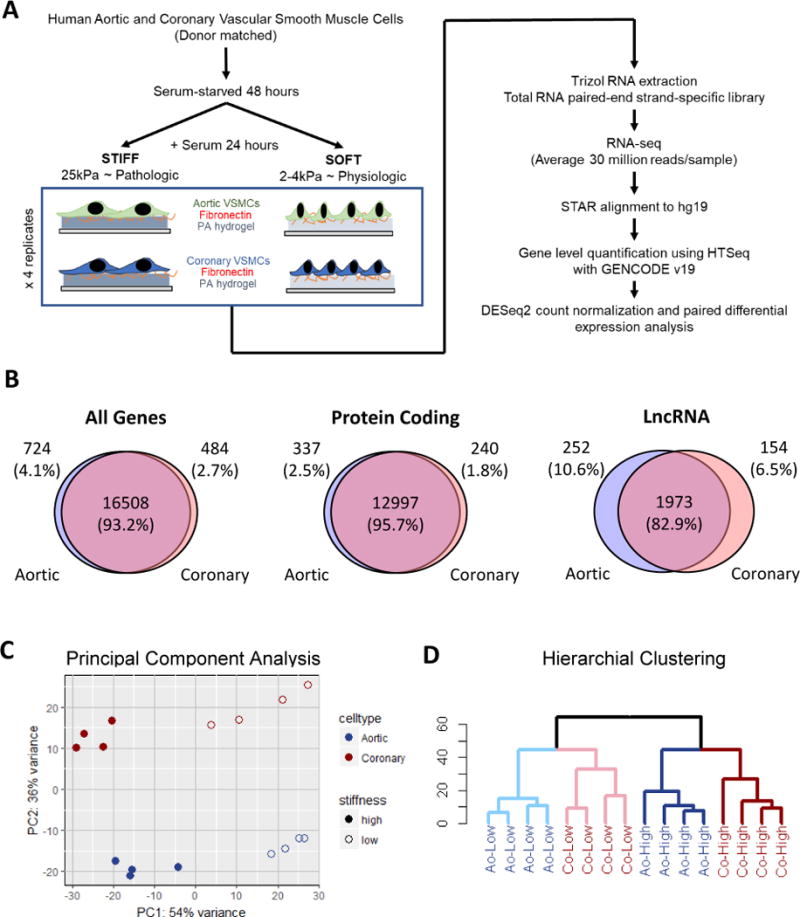
(A) Experimental and computational flow chart for interrogating the stiffness-mediated transcriptome. (B) Venn diagrams depicting the overlap of expressed genes (defined as normalized counts > 8) in Ao VSMCs and Co VSMCs by type. (C) Principal component analysis plot of the first two principal components using the top 500 variant genes (D) Hierarchical clustering of all Ao and Co VSMC samples using the top 500 variant genes
Principal component analysis of the top 500 most variant genes revealed that matrix stiffness is a major contributor to expression differences and accounts for approximately 54% of variation between transcriptomes. VSMC origin is also a major contributor accounting for about 36% of the expression variance (Fig 1C). Consistent with this analysis, unsupervised hierarchical clustering with the 500 most variant genes revealed that Ao and Co VSMCs cultured on the same matrix stiffness clustered together (Fig 1D). This clustering is similar when clustering based only on protein coding genes or even lncRNAs despite their reduced overlap in expression between Ao and Co VSMCs (Fig IB).
Global Characteristics of Stiffness-Sensitive lncRNAs and Protein-Coding Genes
We conducted pair-wise differential expression analysis and found that a total of 5818 genes are matrix stiffness-sensitive in either Ao or Co VSMCs (FDR<1%) (Fig 2A, 2B). From here on, we will define a stiffness-sensitive (SS) gene as the following: genes identified that are unique to Ao or Co VSMCs will be referred to as UNIQUE-SS genes; genes identified in found in both Ao and Co VSMCs will be referred to as COMMON-SS genes. Genes in the union of UNIQUE-SS and COMMON-SS will be referred to as ALL-SS genes.
Figure 2. Stiffness-sensitive lncRNAs and protein coding genes.
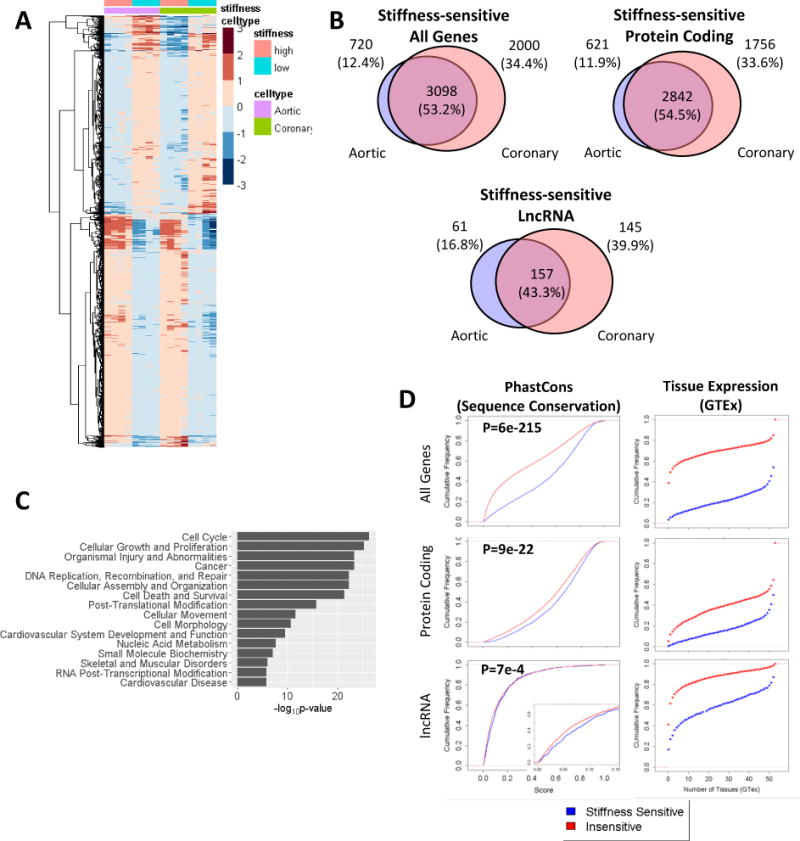
(A) Heatmap of stiffness-sensitive genes (FDR < 0.01) in both Co VSMC and Ao VSMCs. Heatmap colors correspond to log2 (normalized counts). (B) Venn diagrams depicting the overlap in stiffness-sensitive genes in Ao and Co VSMCs. (C) Top pathways enriched in shared stiffness-sensitive genes in Ao and Co VSMCs. (D) Cumulative distributions of conservation (phastCons) scores and tissue expression (GTEx) of stiffness-sensitive v. stiffness-insensitive genes. In-set figure of lncRNA distribution shows zoomed-in distribution at conservation scores 0 to 0.15. A right-shifted cumulative distribution demonstrates higher conservation or expression in more tissues. P-values determined using a Kolmogorov–Smirnov test.
Of the genes identified as stiffness-sensitive, 3098 (53.2%) are COMMON-SS, with 2842 being protein coding genes and 157 being lncRNAs. Interestingly, SS lncRNAs are more likely to be identified as UNIQUE-SS than SS protein coding genes. That is, the percentage of UNIQUE-SS in ALL-SS of lncRNAs (57%, 206/363) is greater than the percentage of UNIQUE-SS protein coding genes (45.5%, 2377/5219). We also found that ALL-SS genes were more likely to be expressed in both VSMC lineages than stiffness-insensitive genes. We observed that 93% of ALL-SS lncRNAs and 88% of UNIQUE-SS lncRNAs were expressed in both VSMC lineages compared to only 83% of stiffness-insensitive lncRNAs (Fig IIA, IIB). Similarly, 98% of ALL-SS protein-coding genes and 97% of UNIQUE protein-coding genes were expressed in both VSMC lineages compared to 96% of stiffness-insensitive protein-coding genes (Fig IIA, IIB). These observations suggest that the expression of SS genes is not cell-type specific and that the SS transcriptome may be generally conserved.
Indeed, we find that SS genes were significantly more conserved at sequence level using phastCons scores (Fig 2D). SS protein coding genes and lncRNAs both exhibited increased conservation over stiffness-insensitive genes. Furthermore, using data from the GTEx project, we find that SS genes are expressed (cutoff of RPKM>0.7) in more tissue types than stiffness-insensitive genes (Fig 2D). The SS transcriptome appears to be a conserved cellular response, which suggests that SS lncRNAs, by way of their increased conservation across species and tissue-types, may control stiffness-dependent functions.
Coordinated Expression of Stiffness-Sensitive lncRNA-Protein Coding Pairs
LncRNAs and their neighboring genes are often correlated in their gene-level expression, and several studies have demonstrated that lncRNAs may regulate the expression of neighboring coding genes through various mechanisms29,39,40. We asked whether the VSMCs transcriptome-wide stiffness response may result in part by lncRNA regulation of nearby genes. Thus, we conducted transcriptome-wide correlation analyses between all lncRNA-gene pairs within the same chromosome. We found that the frequency of statistically significantly correlated lncRNA-gene pairs increased based on the proximity of the lncRNA-gene pair (Fig V). These significant correlations were also predominantly positive. When comparing stiffness-sensitive to insensitive lncRNAs, stiffness-sensitive lncRNAs were more likely correlated to nearby genes than stiffness-insensitive genes (Fig VI).
The top 20 correlated stiffness-sensitive lncRNA-protein coding gene pairs are presented in Table 1. The full list is presented in Supplemental Table III. Notably, we find that the lncRNA, RP5-973M2.2 (PACER) is highly associated with the gene expression of PTGS2. PACER has been shown to positively regulate PTGS2 expression in several cell types41,42, and we and others have shown that PTGS2 is important in stiffness-sensing and contributes to vascular stiffening5,43. Furthermore, several lncRNAs are significantly correlated to matrix and cytoskeletal proteins that may potentially be involved in VSMC stiffness-sensing. EDIL3 (Del-1) is an extracellular matrix protein that regulates VSMC adhesion, migration, and proliferation44. Two nuclear membrane associated proteins are also highly correlated to stiffness-sensitive lncRNAs. SYNE3 (aka Nesprin-3), a component of the LINC (linker of the nucleoskeleton and cytoskeleton) complex which transmits mechanical stress from cytoskeleton to the nucleus, may be important in the stiffness-mediated transcriptional response45–48. TMPO (a.k.a lamin-associated polypeptide 2-alpha, LAP2α) is a LEM-domain containing nuclear membrane protein which binds Lamin A, much like Emerin, a critical regulator of nuclear mechanotransduction49–51.
Table 1. Top correlated stiffness-sensitive IncRNA-protein coding gene pairs.
Table 1 depicts the top significantly correlated stiffness-sensitive lncRNAs-stiffness-sensitive protein coding gene pairs. Protein coding genes and lncRNA genes are listed along with their log2 fold change (log2FC > 0 represents decreased expression by stiffness). The distance between gene pairs, Spearman’s correlation coefficient, and p-value are listed.
| LncRNA | Log2 FC | Distance (kB) | PCG | Log2 FC | Spearman R | Correlation p-value |
|---|---|---|---|---|---|---|
| LINC00341 | 1.41 | −65.7 | SYNE3 | 1.00 | 0.98 | 4.00E-11 |
| RP11-54O7.1 | 1.05 | −13.4 | SAMD11 | 1.06 | 0.95 | 1.20E-08 |
| AC002116.7 | −1.66 | −40.4 | WDR62 | −.17 | 0.95 | 2.70E-08 |
| RP11-800A3.4 | 2.85 | 20.7 | P2RY2 | 2.26 | 0.94 | 3.90E-08 |
| TMPO-AS1 | −1.97 | 0.9 | TMPO | −.72 | 0.94 | 3.90E-08 |
| RP11-401P9.4 | 2.32 | 97.5 | NKD1 | 2.06 | 0.94 | 7.80E-08 |
| RP11-386G11.10 | −1.97 | −61 | TUBA1C | −.21 | 0.94 | 1.10E-07 |
| CTD-2269F5.1 | 1.17 | −0.4 | EDIL3 | 1.64 | 0.93 | 1.90E-07 |
| CTB-92J24.2 | −1.61 | 12.9 | ZNF726 | −.73 | 0.92 | 4.30E-07 |
| CTD-2298J14.2 | 1.37 | −2.7 | LRFN5 | 1.98 | 0.92 | 4.30E-07 |
| JHDM1D-AS1 | 1.66 | 0.2 | JHDM1D | 1.62 | 0.92 | 4.30E-07 |
| RP11-303E16.2 | −1.5 | 24.3 | CENPN | −.56 | 0.91 | 8.80E-07 |
| RP11-110I1.12 | −1.03 | −97.5 | H2AFX | −.11 | 0.90 | 1.70E-06 |
| RP5-973M2.2 | 1.61 | 0.2 | PTGS2 | 2.23 | 0.90 | 1.70E-06 |
| RP11-386G11.10 | −1.97 | −3.6 | TUBA1B | −.01 | 0.90 | 2.10E-06 |
| RP3-462E2.3 | 1.78 | 45.9 | ALDH2 | 1.08 | 0.90 | 2.50E-06 |
| AC009133.12 | 1.18 | 30.5 | KIF22 | −.94 | −.89 | 3.60E-06 |
| CASC15 | 1.63 | 71 | SOX4 | 1.74 | 0.89 | 5.10E-06 |
| RP1-151F17.2 | 1.4 | 2.9 | ATXN1 | 1.08 | 0.89 | 5.10E-06 |
Identification of lncRNA Regulators of Stiffness-dependent Cell Responses
Most lncRNAs have no known function, and none have been implicated in the response to ECM stiffening. To identify lncRNAs that may play a role in stiffness-mediated cellular responses, we performed unbiased enrichment analyses (Fig 2C) and identified several highly enriched cellular processes in both Ao and Co VSMCs: cell cycle, growth, death, motility, morphology, and RNA post-transcriptional modifications (splicing). Among these categories, MALAT1 was the most overrepresented lncRNA annotated to these functions (Supplemental Table I and Supplemental Table II). MALAT1, a highly conserved lncRNA, has been primarily implicated in cancer biology, and, more recently, in endothelial cell function. Whether MALAT1 plays a role in VSMCs or in stiffness-dependent functions is unknown.
RT-PCR validated the RNA-seq finding that matrix stiffening reduces MALAT1 expression in both Ao and Co VSMCs (Fig 3A, IIIA). We then focused on using primary and hTERT-immortalized Co VSMCs. Knockdown of MALAT1 with two different LNA-GapmeRs resulted in significant reduction in MALAT1 expression (Fig 3B, IIIB). Upon MALAT1 knockdown, Co VSMCs lost their elongated cell morphology (Fig 3C, IIIC). MALAT1 knockdown also resulted in reduction in stiffness-dependent S-phase entry in Co VSMCs measured by EdU incorporation (Fig 3D, IIID, IVA). Reduced MALAT1 also decreased stiffness-induced Co VSMCs migration speed, total distance, and displacement (Fig 3E, IIIE), and slowed collective migration on a wound closure assay on stiff tissue culture plastic (Fig IVB). Taken together, these data show that MALAT1 is a positive regulator of VSMC proliferation and migration in response to ECM stiffness. ECM stiffness negatively regulates MALAT1, and this may represent a homeostatic negative feedback mechanism in which the reduction in MALAT1 limits stiffness-induced proliferation and migration.
Figure 3. MALAT1, a lncRNA regulator of stiffness-dependent cell responses.
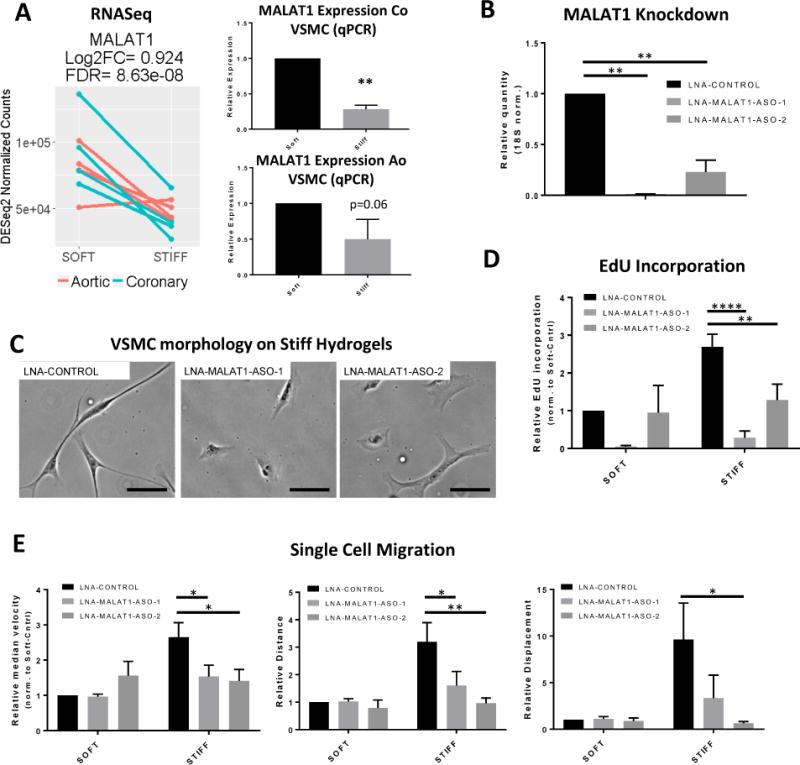
(A) Left, MALAT1 RNA-seq expression from donor-matched Ao and Co VSMCs. Right, MALAT1 expression validation by quantitative-PCR in primary Co and Ao VSMCs (n = 5 Co VSMCs, n =4 Ao VSMCs, **, p < 0.005). (B) MALAT1 knockdown using LNA-GapmeRs in primary Co VSMCs. (C) Phase image of primary Co VSMCs cultured on tissue culture plastic for 48 hrs in SmGm2 after MALAT1 knockdown. Scale bar = 100um. (D) Primary Co VSMCs cultured on soft and stiff hydrogels for 48 hours with serum containing EdU. Values are normalized to non-targeting control LNA-GapmeR on soft hydrogels (n = 4, ** p<0.005; ****p<0.0001). (E) Normalized median velocity, total distance (path-dependent) migrated, and total displacement (path-independent) migrated in primary Co VSMCs over 5 hours. Values are normalized to velocity and distance in non-targeting control LNA-GapmeR on soft hydrogels. (n = 4, * p<0.05, **p<0.005, Two-way ANOVA; Holm-Sidak’s multiple comparisons test).
MALAT1 is a key lncRNA regulator of the in vivo vascular injury response
We next asked whether MALAT1 plays a role during in vivo pathologic vascular stiffening by utilizing the mouse femoral artery wire injury model. In this model, wire-mediated endothelial injury leads to vascular matrix stiffening, neointima formation, and luminal stenosis largely driven by VSMC proliferation and migration27,52. Uninjured arteries in the Malat1-knockout mice were largely similar to wild-type uninjured arteries – no appreciable neointima and comparable medial areas (Fig 4A, B). This is consistent with previous reports that Malat1-knockout mice develop normally53–55. However, after vascular injury, Malat1-knockout mice had significantly reduced neointima formation, neointima/media ratios, and percent luminal stenosis compared to wild-type mice (Fig 4). These results demonstrate that MALAT1 is an important regulator of the vascular injury response, and may prevent in vivo VSMC proliferation and migration during vascular stiffening.
Figure 4. MALAT1 regulates neointima formation during vascular injury.
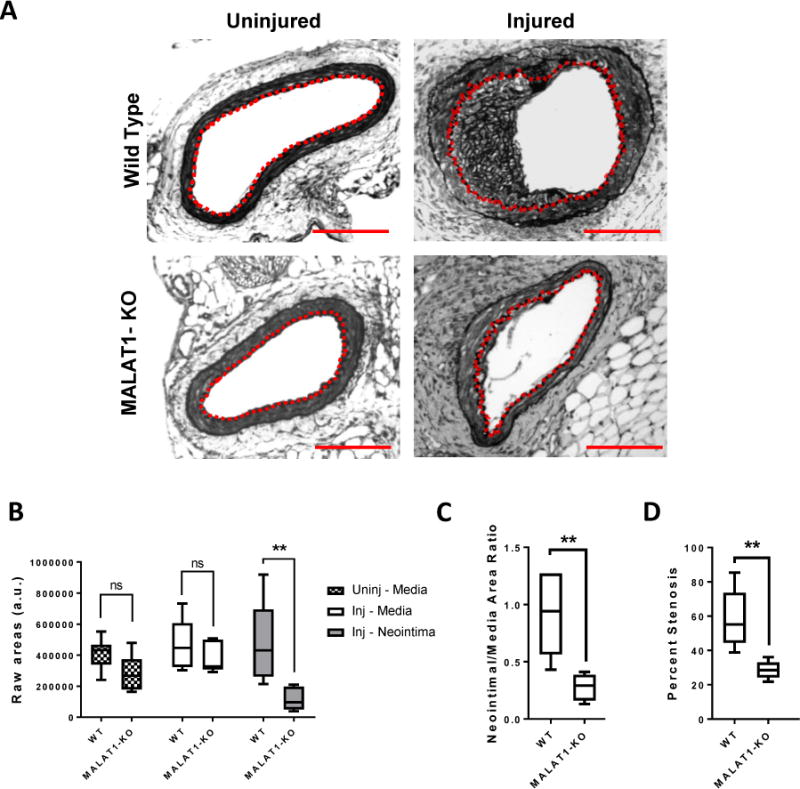
(A) Cross-sections of injured and uninjured (contralateral) femoral arteries of wild-type (WT) and MALAT1-knockout (KO) adult mice were stained for elastin to visualize the internal and external elastic lamina. The dashed red line delineates the internal elastic lamina, which circumscribes the neointima. Scale bar = 100μm. (B) Areas of the media and neointima from injured femoral arteries (inj – media, inj – neointima) and uninjured femoral arteries (uninj – media) of WT and KO mice. (C) Neointimal-Media area ratios of injured WT and KO femoral arteries. (D) Percent stenosis was calculated by dividing the neointimal area by the area circumscribed by the internal elastic lamina × 100%. (n = 7 WT, n = 5 MALAT1-KO, ** p<0.005, ns = not significant, Mann-Whitney Test).
DISCUSSION
Although vascular stiffness has been identified as an important risk factor for cardiovascular disease, our understanding of how vascular stiffness alters vascular cellular behavior is incomplete. This study provides the first comprehensive analysis comparing the stiffness-sensitive transcriptome in human aortic and coronary vascular smooth muscle cells with a focus on lncRNAs. We found that Ao and Co VSMC transcriptome profiles are significantly dictated by matrix stiffness, and that stiffness-sensitive genes (whether protein coding or lncRNA) are more commonly co-expressed in Ao and Co VSMCs compared to stiffness-insensitive genes. We also found that VSMC stiffness-sensitive genes are overall more species conserved and expressed in more tissue types than stiffness-insensitive genes. Furthermore, we observed that stiffness-sensitive lncRNAs may be more cell lineage specific (between Ao and Co VSMCs) than stiffness-sensitive protein coding genes. Using unbiased bioinformatic approaches, we predicted and confirmed that MALAT1 is a stiffness-sensitive lncRNA that regulates stiffness-mediated VSMC proliferation and migration. These insights expand our knowledge of the basic cellular responses to pathologic stiffening and human vascular disease mechanisms.
We find that ECM stiffness significantly dictates the transcriptomic identity of Ao and Co VSMCs. Our clustering analysis suggests that ECM stiffness may override the embryologic-lineage contributions to the transcriptome; the transcriptome of Co VSMCs/stiff matrix is more similar to Ao VSMCs/stiff matrix than Co VSMCs/soft matrix. Prior studies have suggested that VSMC from different embryologic origins are rather phenotypically stable even when transplanted into disparate vascular microenvironments16–18; however, these transplantation studies are uncontrolled for any particular microenvironmental parameter. By using donor-matched Ao and Co VSMCs, any differences we observe in the transcriptome of Ao and Co VSMCs arise from contributions from embryologic lineage programs or the effect of ECM stiffness.
Several studies support this notion that ECM stiffness may significantly dictate cell transcriptomic identity. ECM stiffness and other mechanical parameters are critical in tissue and organ development56. Early studies demonstrated that ECM stiffness can dictate stem cell fate and that different stiffness can promote different lineages57–60. ECM stiffness has also been shown to alter nuclear mechanics, chromatin organization and the activity of a variety of ubiquitous transcription factors in multiple cell types, leading to global transcription changes61–65.
In further support of this idea, we found that stiffness-sensitive genes identified as stiffness-sensitive in either Ao or Co VSMCs tended to be expressed in both VSMC lineages and across more tissue types than stiffness-insensitive genes. These stiffness-sensitive genes were also more conserved across species at the sequence level. This suggests that the stiffness-sensitive gene set may represent a critical response module across many cell and tissue types that regulates cell-ubiquitous processes, as predicted by pathway enrichment analysis.
In this study, we found that stiffness-sensitive lncRNAs were significantly correlated with more of their nearby genes when compared to stiffness-insensitive lncRNAs. Suggesting that stiffness-sensitive lncRNAs may coordinate stiffness-dependent protein coding gene expression. Indeed, some of the lncRNAs in Table 1 may not be functionally linked to their correlated protein coding gene, while others may represent true cis-regulatory lncRNAs and directly control the gene expression of nearby protein coding genes.
Among the top 20 correlated lncRNAs in Table 1, only two have been previously studied: CASC15 and RP5-973M2.2 (PACER), and both have been shown to regulate coding gene expression and in particular PACER was shown to be a cis-regulator of its neighboring coding gene PTGS241,66,67.
Analysis of stiffness-enriched pathways and gene sets for stiffness-sensitive lncRNAs allowed us to identify MALAT1 as a lncRNA regulator of VSMC stiffness-dependent proliferation and migration. To our knowledge, this is the first study implicating a role for MALAT1 in VSMCs and its interplay with matrix stiffness. MALAT1 had previously been linked to tumor aggressiveness in in vitro and clinical studies68–74. However, its importance in vascular disease is relatively unknown, with only one study implicating MALAT1 in endothelial cell function75. While we found that MALAT1-dependent proliferation is similar in both VSMCs and endothelial cells, MALAT1 knockdown enhances migration in endothelial cells while reducing migration in VSMCs. Our study also identified splicing as a significant stiffness-mediated pathway for which MALAT1 has a documented role. Whether stiffness regulates RNA splicing and whether MALAT1 regulates splicing in VSMCs will require future work.
Interestingly, despite many studies linking MALAT1 to critical cell functions, MALAT1 whole-body knockout mice develop normally53–55. We hypothesize that MALAT1 is dispensable during physiologic/normal conditions, but is necessary for critical cell functions such as proliferation and migration during pathologic/abnormal stiffening conditions. In support of this, most in vitro studies have interrogated MALAT1 using only supra-physiologically stiff tissue culture plastic, while our data further demonstrates that MALAT1 is necessary for VSMC proliferation and migration in stiff context (stiff hydrogels and vascular injury), and is less important in physiologically soft context (soft hydrogels and uninjured arteries). These findings also suggest that MALAT1 may serve as a potential therapeutic target as the loss of MALAT1 prevented neointimal formation and luminal stenosis in injured arteries with limited effects on healthy/uninjured arteries.
We also find that MALAT1 expression is reduced by ECM stiffness in VSMCs and perhaps acts in a negative feedback mechanism to limit stiffness-mediated proliferation and migration. However, this effect may not necessarily be universal. RT-PCR validated the stiffness-mediated reduction in MALAT1 in the donor-matched Ao and Co VSMCs used for the RNAseq. We also validated this finding in several different donors of Ao and Co VSMCs, but not in all donors. We believe there may be a level of variability in VSMCs from specific donors that exhibit stiffness-mediated MALAT1 reduction while others do not. This potential variability may reflect differences in the risk for cardiovascular disease from vascular stiffness in various populations76,77.
In summary, pathologic matrix stiffening is a major regulator of both human Ao and Co VSMC transcriptomes and function. Our study demonstrates that lncRNAs respond to ECM stiffening and regulate VSMC cellular functions. We also identify stiffness-sensitive lncRNAs that may coordinate the expression of nearby protein coding genes involved in the response to ECM stiffening. Yet, many stiffness-sensitive lncRNAs identified in this study have no known or predicted function. Further studies to understand the complex response to ECM stiffening may help identify novel diagnostic and therapeutic targets for vascular stiffness.
Supplementary Material
HIGHLIGHTS.
Extracellular matrix stiffness is a significant determinant of human aortic and coronary vascular smooth muscle cell transcriptome identity.
Stiffness-sensitive genes are conserved across species and cell-types suggesting that the stiffness-sensitive transcriptome is an evolutionarily important response
This is the first study to characterize the long non-coding RNA response to extracellular matrix stiffening and in vascular smooth muscle cells
We identify a novel role for MALAT1 in regulating stiffness-dependent vascular smooth muscle cell proliferation and migration in vitro and in vivo.
Acknowledgments
We would like to thank the UPenn Next Gen Sequencing Core for help in conducting the RNAseq, the Biomechanics Core Facility at Institute for Translational Medicine and Therapeutics at Penn for assistance in fabricating hydrogels of tunable stiffness, and the Penn Molecular Profiling Facility. We would also like to acknowledge the following for helpful scientific discussion: Nathan D. Bade, Rachel Ballantyne, Paola Castagnino, Nicholas J. Hand, Sue I. Lee, Keeley Mui, Kristy Ou, Ziba Razinia, and Chenyi Xue. Genotype-Tissue Expression (GTEx) Project data was used for analysis in this manuscript obtained from the GTEx portal.
SOURCES OF FUNDING
This work was supported by the National Institutes of Health (HL119346 and T32HL007954-17). Center for Engineering MechanoBiology (CEMB), an NSF Science and Technology Center, under grant agreement CMMI: 15-48571.
ABBREVIATIONS
- VSMC
Vascular Smooth Muscle Cells
- ECM
Extracellular Matrix
- Ao
Aorta
- Co
Coronary
- lncRNAs
Long noncoding RNAs
- SS
Stiffness-sensitive
- GTEX
Genotype-Tissue Expression
Footnotes
DISCLOSURES
None
References
- 1.Gepner AD, Korcarz CE, Colangelo LA, Hom EK, Tattersall MC, Astor BC, Kaufman JD, Liu K, Stein JH. Longitudinal effects of a decade of aging on carotid artery stiffness: the multiethnic study of atherosclerosis. Stroke. 2014;45(1):48–53. doi: 10.1161/STROKEAHA.113.002649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.van Popele NM, Grobbee DE, Bots ML, Asmar R, Topouchian J, Reneman RS, Hoeks AP, van der Kuip DA, Hofman A, Witteman JC. Association between arterial stiffness and atherosclerosis: the Rotterdam Study. Stroke. 2001;32(2):454–460. doi: 10.1161/01.str.32.2.454. [DOI] [PubMed] [Google Scholar]
- 3.Laurent S, Boutouyrie P, Asmar R, Gautier I, Laloux B, Guize L, Ducimetiere P, Benetos A. Aortic stiffness is an independent predictor of all-cause and cardiovascular mortality in hypertensive patients. Hypertension. 2001;37(5):1236–1241. doi: 10.1161/01.hyp.37.5.1236. [DOI] [PubMed] [Google Scholar]
- 4.Mitchell GF, Hwang S-J, Vasan RS, Larson MG, Pencina MJ, Hamburg NM, Vita JA, Levy D, Benjamin EJ. Arterial stiffness and cardiovascular events: the Framingham Heart Study. Circulation. 2010;121(4):505–511. doi: 10.1161/CIRCULATIONAHA.109.886655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kothapalli D, Liu S-L, Bae YH, et al. Cardiovascular protection by ApoE and ApoE-HDL linked to suppression of ECM gene expression and arterial stiffening. Cell Rep. 2012;2(5):1259–1271. doi: 10.1016/j.celrep.2012.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Weisbrod RM, Shiang T, Al Sayah L, Fry JL, Bajpai S, Reinhart-King CA, Lob HE, Santhanam L, Mitchell G, Cohen RA, Seta F. Arterial stiffening precedes systolic hypertension in diet-induced obesity. Hypertension. 2013;62(6):1105–1110. doi: 10.1161/HYPERTENSIONAHA.113.01744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu S-L, Bae YH, Yu C, Monslow J, Hawthorne EA, Castagnino P, Branchetti E, Ferrari G, Damrauer SM, Puré E, Assoian RK. Matrix metalloproteinase-12 is an essential mediator of acute and chronic arterial stiffening. Sci Rep. 2015;5:17189. doi: 10.1038/srep17189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gadson PF, Dalton ML, Patterson E, Svoboda DD, Hutchinson L, Schram D, Rosenquist TH. Differential response of mesoderm- and neural crest-derived smooth muscle to TGF-beta1: regulation of c-myb and alpha1 (I) procollagen genes. Exp Cell Res. 1997;230(2):169–180. doi: 10.1006/excr.1996.3398. [DOI] [PubMed] [Google Scholar]
- 9.Leroux-Berger M, Queguiner I, MacIel TT, Ho A, Relaix F, Kempf H. Pathologic calcification of adult vascular smooth muscle cells differs on their crest or mesodermal embryonic origin. J Bone Miner Res. 2011;26(7):1543–1553. doi: 10.1002/jbmr.382. [DOI] [PubMed] [Google Scholar]
- 10.Gittenberger-de Groot AC, DeRuiter MC, Bergwerff M, Poelmann RE. Smooth muscle cell origin and its relation to heterogeneity in development and disease. Arterioscler Thromb Vasc Biol. 1999;19(7):1589–1594. doi: 10.1161/01.atv.19.7.1589. [DOI] [PubMed] [Google Scholar]
- 11.Civelek M, Manduchi E, Riley RJ, Stoeckert CJ, Davies PF. Coronary artery endothelial transcriptome in vivo identification of endoplasmic reticulum stress and enhanced reactive oxygen species by gene connectivity network analysis. Circ Cardiovasc Genet. 2011;4(3):243–252. doi: 10.1161/CIRCGENETICS.110.958926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Majesky MW. Developmental basis of vascular smooth muscle diversity. Arterioscler Thromb Vasc Biol. 2007;27(6):1248–1258. doi: 10.1161/ATVBAHA.107.141069. [DOI] [PubMed] [Google Scholar]
- 13.Topouzis S, Majesky MW. Smooth muscle lineage diversity in the chick embryo. Two types of aortic smooth muscle cell differ in growth and receptor-mediated transcriptional responses to transforming growth factor-beta. Dev Biol. 1996;178(2):430–445. doi: 10.1006/dbio.1996.0229. [DOI] [PubMed] [Google Scholar]
- 14.Thieszen SL, Dalton M, Gadson PF, Patterson E, Rosenquist TH. Embryonic lineage of vascular smooth muscle cells determines responses to collagen matrices and integrin receptor expression. Exp Cell Res. 1996;227(1):135–145. doi: 10.1006/excr.1996.0258. [DOI] [PubMed] [Google Scholar]
- 15.Cheung C, Bernardo AS, Trotter MWB, Pedersen RA, Sinha S. Generation of human vascular smooth muscle subtypes provides insight into embryological origin-dependent disease susceptibility. Nat Biotechnol. 2012;30(2):165–173. doi: 10.1038/nbt.2107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yoshida T, Owens GK. Molecular determinants of vascular smooth muscle cell diversity. Circ Res. 2005;96(3):280–291. doi: 10.1161/01.RES.0000155951.62152.2e. [DOI] [PubMed] [Google Scholar]
- 17.Dinardo CL, Venturini G, Zhou EH, Watanabe IS, Campos LCG, Dariolli R, da Motta-Leal-Filho JM, Carvalho VM, Cardozo KHM, Krieger JE, Alencar AM, Pereira AC. Variation of mechanical properties and quantitative proteomics of VSMC along the arterial tree. Am J Physiol Heart Circ Physiol. 2014;306(4):H505–16. doi: 10.1152/ajpheart.00655.2013. [DOI] [PubMed] [Google Scholar]
- 18.Bochaton-Piallat M-L, Clowes AW, Clowes MM, Fischer JW, Redard M, Gabbiani F, Gabbiani G. Cultured Arterial Smooth Muscle Cells Maintain Distinct Phenotypes When Implanted Into Carotid Artery. Arterioscler Thromb. 2001;21(21):949–954. doi: 10.1161/01.atv.21.6.949. [DOI] [PubMed] [Google Scholar]
- 19.Haimovici H, Maier N. Fate of Aortic Homografts in Canine Atherosclerosis III. Study of Fresh Abdominal and Thoracic Aortic Implants Into Thoracic Aorta: Role of Tissue Susceptibility in Atherogenesis. Arch Surg. 1964;89:961–969. doi: 10.1001/archsurg.1964.01320060029006. [DOI] [PubMed] [Google Scholar]
- 20.Gittenberger-de Groot AC, Vrancken Peeters MP, Mentink MM, Gourdie RG, Poelmann RE. Epicardium-derived cells contribute a novel population to the myocardial wall and the atrioventricular cushions. Circ Res. 1998;82(10):1043–1052. doi: 10.1161/01.res.82.10.1043. [DOI] [PubMed] [Google Scholar]
- 21.Mui KL, Bae YH, Gao L, Liu S-L, Xu T, Radice GL, Chen CS, Assoian RK. N-Cadherin Induction by ECM Stiffness and FAK Overrides the Spreading Requirement for Proliferation of Vascular Smooth Muscle Cells. Cell Rep. 2015 Mar; doi: 10.1016/j.celrep.2015.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sazonova OVV, Lee KLL, Isenberg BCC, Rich CBB, Nugent MAA, Wong JYY. Cell-Cell Interactions Mediate the Response of Vascular Smooth Muscle Cells to Substrate Stiffness. Biophys J. 2011;101(3):622–630. doi: 10.1016/j.bpj.2011.06.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brown XQ, Bartolak-Suki E, Williams C, Walker ML, Weaver VM, Wong JY. Effect of substrate stiffness and PDGF on the behavior of vascular smooth muscle cells: Implications for atherosclerosis. J Cell Physiol. 2010;225(1):115–122. doi: 10.1002/jcp.22202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Peyton SR, Kim PD, Ghajar CM, Seliktar D, Putnam AJ. The effects of matrix stiffness and RhoA on the phenotypic plasticity of smooth muscle cells in a 3-D biosynthetic hydrogel system. Biomaterials. 2008;29(17):2597–2607. doi: 10.1016/j.biomaterials.2008.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Peyton SR, Putnam AJ. Extracellular matrix rigidity governs smooth muscle cell motility in a biphasic fashion. J Cell Physiol. 2005;204(1):198–209. doi: 10.1002/jcp.20274. [DOI] [PubMed] [Google Scholar]
- 26.McDaniel DP, Shaw GA, Elliott JT, Bhadriraju K, Meuse C, Chung K-H, Plant AL. The stiffness of collagen fibrils influences vascular smooth muscle cell phenotype. Biophys J. 2007;92(5):1759–1769. doi: 10.1529/biophysj.106.089003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Klein EA, Yin L, Kothapalli D, Castagnino P, Byfield FJ, Xu T, Levental I, Hawthorne E, Janmey PA, Assoian RK. Cell-cycle control by physiological matrix elasticity and in vivo tissue stiffening. Curr Biol. 2009;19(18):1511–1518. doi: 10.1016/j.cub.2009.07.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fatica A, Bozzoni I. Long non-coding RNAs: new players in cell differentiation and development. Nat Rev Genet. 2014;15(1):7–21. doi: 10.1038/nrg3606. [DOI] [PubMed] [Google Scholar]
- 29.Engreitz JM, Haines JE, Perez EM, Munson G, Chen J, Kane M, McDonel PE, Guttman M, Lander ES. Local regulation of gene expression by lncRNA promoters, transcription and splicing. Nature. 2016;539(7629):452–455. doi: 10.1038/nature20149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Iyer MK, Niknafs YS, Malik R, et al. The landscape of long noncoding RNAs in the human transcriptome. Nat Genet. 2015;47(3):199–208. doi: 10.1038/ng.3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Congrains A, Kamide K, Katsuya T, Yasuda O, Oguro R, Yamamoto K, Ohishi M, Rakugi H. CVD-associated non-coding RNA, ANRIL, modulates expression of atherogenic pathways in VSMC. Biochem Biophys Res Commun. 2012;419(4):612–616. doi: 10.1016/j.bbrc.2012.02.050. [DOI] [PubMed] [Google Scholar]
- 32.Wu G, Cai J, Han Y, et al. LincRNA-p21 regulates neointima formation, vascular smooth muscle cell proliferation, apoptosis, and atherosclerosis by enhancing p53 activity. Circulation. 2014;130(17):1452–1465. doi: 10.1161/CIRCULATIONAHA.114.011675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bell RD, Long X, Lin M, Bergmann JH, Nanda V, Cowan SL, Zhou Q, Han Y, Spector DL, Zheng D, Miano JM. Identification and Initial Functional Characterization of a Human Vascular Cell-Enriched Long Noncoding RNA. Arterioscler Thromb Vasc Biol. 2014;34(6):1249–1259. doi: 10.1161/ATVBAHA.114.303240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ballantyne MD, Pinel K, Dakin R, et al. Smooth Muscle Enriched Long Noncoding RNA (SMILR) Regulates Cell Proliferation. Circulation. 2016;133(21):2050–2065. doi: 10.1161/CIRCULATIONAHA.115.021019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhao J, Zhang W, Lin M, Wu W, Jiang P, Tou E, Xue M, Richards A, Jourd’heuil D, Asif A, Zheng D, Singer HA, Miano JM, Long X. MYOSLID Is a Novel Serum Response Factor-Dependent Long Noncoding RNA That Amplifies the Vascular Smooth Muscle Differentiation Program. Arterioscler Thromb Vasc Biol. 2016;36(10):2088–2099. doi: 10.1161/ATVBAHA.116.307879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mouw JK, Yui Y, Damiano L, Bainer RO, Lakins JN, Acerbi I, Ou G, Wijekoon AC, Levental KR, Gilbert PM, Hwang ES, Chen Y-Y, Weaver VM. Tissue mechanics modulate microRNA-dependent PTEN expression to regulate malignant progression. Nat Med. 2014;20(4):360–367. doi: 10.1038/nm.3497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dupont S, Morsut L, Aragona M, Enzo E, Giulitti S, Cordenonsi M, Zanconato F, Le Digabel J, Forcato M, Bicciato S, Elvassore N, Piccolo S. Role of YAP/TAZ in mechanotransduction. Nature. 2011;474(7350):179–183. doi: 10.1038/nature10137. [DOI] [PubMed] [Google Scholar]
- 38.Le LT-N, Cazares O, Mouw JK, Chatterjee S, Macias H, Moran A, Ramos J, Keely PJ, Weaver VM, Hinck L. Loss of miR-203 regulates cell shape and matrix adhesion through ROBO1/Rac/FAK in response to stiffness. J Cell Biol. 2016;212(6):707–719. doi: 10.1083/jcb.201507054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Touma M, Kang X, Zhao Y, Cass AA, Gao F, Biniwale R, Coppola G, Xiao X, Reemtsen B, Wang Y. Decoding the Long Noncoding RNA during Cardiac Maturation: A Roadmap for Functional Discovery. Circ Cardiovasc Genet. 2016;9(5) doi: 10.1161/CIRCGENETICS.115.001363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu SJ, Horlbeck MA, Cho SW, et al. CRISPRi-based genome-scale identification of functional long noncoding RNA loci in human cells. Science (80-) 2017;355(6320) doi: 10.1126/science.aah7111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Krawczyk M, Emerson BM. p50-associated COX-2 extragenic RNA (PACER) activates COX-2 gene expression by occluding repressive NF-κB complexes. Elife. 2014;3(3):e01776. doi: 10.7554/eLife.01776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Qian M, Yang X, Li Z, Jiang C, Song D, Yan W, Liu T, Wu Z, Kong J, Wei H, Xiao J. P50-associated COX-2 extragenic RNA (PACER) overexpression promotes proliferation and metastasis of osteosarcoma cells by activating COX-2 gene. Tumour Biol. 2016;37(3):3879–3886. doi: 10.1007/s13277-015-3838-8. [DOI] [PubMed] [Google Scholar]
- 43.Avendaño MS, Martínez-Revelles S, Aguado A, et al. Role of COX-2-derived PGE2 on vascular stiffness and function in hypertension. Br J Pharmacol. 2016;173(9):1541–1555. doi: 10.1111/bph.13457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rezaee M, Penta K, Quertermous T. Del1 mediates VSMC adhesion, migration, and proliferation through interaction with integrin alpha(v)beta(3) Am J Physiol Heart Circ Physiol. 2002;282(5):H1924–32. doi: 10.1152/ajpheart.00921.2001. [DOI] [PubMed] [Google Scholar]
- 45.Starr DA. KASH and SUN proteins. Curr Biol. 2011;21(11):R414–R415. doi: 10.1016/j.cub.2011.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lovett DB, Shekhar N, Nickerson JA, Roux KJ, Lele TP. Modulation of Nuclear Shape by Substrate Rigidity. Cell Mol Bioeng. 2013;6(2):230–238. doi: 10.1007/s12195-013-0270-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Alam SG, Zhang Q, Prasad N, Li Y, Chamala S, Kuchibhotla R, KC B, Aggarwal V, Shrestha S, Jones AL, Levy SE, Roux KJ, Nickerson JA, Lele TP. The mammalian LINC complex regulates genome transcriptional responses to substrate rigidity. Sci Rep. 2016;6(1):38063. doi: 10.1038/srep38063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stewart-Hutchinson PJ, Hale CM, Wirtz D, Hodzic D. Structural requirements for the assembly of LINC complexes and their function in cellular mechanical stiffness. Exp Cell Res. 2008;314(8):1892–1905. doi: 10.1016/j.yexcr.2008.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Guilluy C, Osborne LD, Van Landeghem L, Sharek L, Superfine R, Garcia-Mata R, Burridge K. Isolated nuclei adapt to force and reveal a mechanotransduction pathway in the nucleus. Nat Cell Biol. 2014;16(4):376–381. doi: 10.1038/ncb2927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dechat T, Korbei B, Vaughan OA, Vlcek S, Hutchison CJ, Foisner R. Lamina-associated polypeptide 2alpha binds intranuclear A-type lamins. J Cell Sci. 2000;113(Pt 19):3473–3484. doi: 10.1242/jcs.113.19.3473. [DOI] [PubMed] [Google Scholar]
- 51.Wagner N, Krohne G. LEM-Domain proteins: new insights into lamin-interacting proteins. Int Rev Cytol. 2007;261:1–46. doi: 10.1016/S0074-7696(07)61001-8. [DOI] [PubMed] [Google Scholar]
- 52.Thyberg J, Hedin U, Sjölund M, Palmberg L, Bottger BA. Regulation of differentiated properties and proliferation of arterial smooth muscle cells. Arteriosclerosis. 10(6):966–990. doi: 10.1161/01.atv.10.6.966. [DOI] [PubMed] [Google Scholar]
- 53.Zhang B, Arun G, Mao YS, Lazar Z, Hung G, Bhattacharjee G, Xiao X, Booth CJ, Wu J, Zhang C, Spector DL. The lncRNA Malat1 is dispensable for mouse development but its transcription plays a cis-regulatory role in the adult. Cell Rep. 2012;2(1):111–123. doi: 10.1016/j.celrep.2012.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nakagawa S, Ip JY, Shioi G, Tripathi V, Zong X, Hirose T, Prasanth KV. Malat1 is not an essential component of nuclear speckles in mice. RNA. 2012;18(8):1487–1499. doi: 10.1261/rna.033217.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Eißmann M, Gutschner T, Hämmerle M, Günther S, Caudron-Herger M, Groß M, Schirmacher P, Rippe K, Braun T, Zörnig M, Diederichs S. Loss of the abundant nuclear non-coding RNA MALAT1 is compatible with life and development. RNA Biol. 2012;9(8):1076–1087. doi: 10.4161/rna.21089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mammoto T, Ingber DE. Mechanical control of tissue and organ development. Development. 2010;137(9):1407–1420. doi: 10.1242/dev.024166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Engler AJ, Sen S, Sweeney HL, Discher DE. Matrix elasticity directs stem cell lineage specification. Cell. 2006;126(4):677–689. doi: 10.1016/j.cell.2006.06.044. [DOI] [PubMed] [Google Scholar]
- 58.Przybyla L, Lakins JN, Weaver VM. Tissue Mechanics Orchestrate Wnt-Dependent Human Embryonic Stem Cell Differentiation. Cell Stem Cell. 2016;19(4):462–475. doi: 10.1016/j.stem.2016.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Saha K, Keung AJ, Irwin EF, Li Y, Little L, Schaffer DV, Healy KE. Substrate modulus directs neural stem cell behavior. Biophys J. 2008;95(9):4426–4438. doi: 10.1529/biophysj.108.132217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cosgrove BD, Mui KL, Driscoll TP, Caliari SR, Mehta KD, Assoian RK, Burdick JA, Mauck RL. N-cadherin adhesive interactions modulate matrix mechanosensing and fate commitment of mesenchymal stem cells. Nat Mater. 2016;15(12):1297–1306. doi: 10.1038/nmat4725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Swift J, Ivanovska IL, Buxboim A, Harada T, Dingal PCDP, Pinter J, Pajerowski JD, Spinler KR, Shin J-WJ-W, Tewari M, Rehfeldt F, Speicher DW, Discher DE. Nuclear lamin-A scales with tissue stiffness and enhances matrix-directed differentiation. Science. 2013;341(6149):1240104. doi: 10.1126/science.1240104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Janmey PA, Wells RG, Assoian RK, McCulloch CA. From tissue mechanics to transcription factors. Differentiation. 2013;86(3):112–120. doi: 10.1016/j.diff.2013.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Discher DE, Janmey P, Wang Y-L. Tissue cells feel and respond to the stiffness of their substrate. Science. 2005;310(5751):1139–1143. doi: 10.1126/science.1116995. [DOI] [PubMed] [Google Scholar]
- 64.Dupont S. Role of YAP/TAZ in cell-matrix adhesion-mediated signalling and mechanotransduction. Exp Cell Res. 2016;343(1):42–53. doi: 10.1016/j.yexcr.2015.10.034. [DOI] [PubMed] [Google Scholar]
- 65.Le HQ, Ghatak S, Yeung C-YCC, et al. Mechanical regulation of transcription controls Polycomb-mediated gene silencing during lineage commitment. Nat Cell Biol. 2016;18(8):864–875. doi: 10.1038/ncb3387. [DOI] [PubMed] [Google Scholar]
- 66.Lessard L, Liu M, Marzese DM, et al. The CASC15 Long Intergenic Noncoding RNA Locus Is Involved in Melanoma Progression and Phenotype Switching. J Invest Dermatol. 2015;135(10):2464–2474. doi: 10.1038/jid.2015.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Russell MR, Penikis A, Oldridge DA, et al. CASC15-S Is a Tumor Suppressor lncRNA at the 6p22 Neuroblastoma Susceptibility Locus. Cancer Res. 2015;75(15):3155–3166. doi: 10.1158/0008-5472.CAN-14-3613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ji P, Diederichs S, Wang W, Böing S, Metzger R, Schneider PM, Tidow N, Brandt B, Buerger H, Bulk E, Thomas M, Berdel WE, Serve H, Müller-Tidow C. MALAT-1, a novel noncoding RNA, and thymosin beta4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene. 2003;22(39):8031–8041. doi: 10.1038/sj.onc.1206928. [DOI] [PubMed] [Google Scholar]
- 69.Arun G, Diermeier S, Akerman M, Chang KC, Wilkinson JE, Hearn S, Kim Y, MacLeod AR, Krainer AR, Norton L, Brogi E, Egeblad M, Spector DL. Differentiation of mammary tumors and reduction in metastasis upon Malat1 lncRNA loss. Genes Dev. 2016;30(1):34–51. doi: 10.1101/gad.270959.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hirata H, Hinoda Y, Shahryari V, Deng G, Nakajima K, Tabatabai ZL, Ishii N, Dahiya R. Long Noncoding RNA MALAT1 Promotes Aggressive Renal Cell Carcinoma through Ezh2 and Interacts with miR-205. Cancer Res. 2015;75(7):1322–1331. doi: 10.1158/0008-5472.CAN-14-2931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hu L, Wu Y, Tan D, Meng H, Wang K, Bai Y, Yang K. Up-regulation of long noncoding RNA MALAT1 contributes to proliferation and metastasis in esophageal squamous cell carcinoma. J Exp Clin Cancer Res. 2015;34:7. doi: 10.1186/s13046-015-0123-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yang M-H, Hu Z-Y, Xu C, Xie L-Y, Wang X-Y, Chen S-Y, Li Z-G. MALAT1 promotes colorectal cancer cell proliferation/migration/invasion via PRKA kinase anchor protein 9. Biochim Biophys Acta. 2015;1852(1):166–174. doi: 10.1016/j.bbadis.2014.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yamada K, Kano J, Tsunoda H, Yoshikawa H, Okubo C, Ishiyama T, Noguchi M. Phenotypic characterization of endometrial stromal sarcoma of the uterus. Cancer Sci. 2006;97(2):106–112. doi: 10.1111/j.1349-7006.2006.00147.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lai M, Yang Z, Zhou L, Zhu Q, Xie H, Zhang F, Wu L, Chen L, Zheng S. Long non-coding RNA MALAT-1 overexpression predicts tumor recurrence of hepatocellular carcinoma after liver transplantation. Med Oncol. 2012;29(3):1810–1816. doi: 10.1007/s12032-011-0004-z. [DOI] [PubMed] [Google Scholar]
- 75.Michalik KM, You X, Manavski Y, Doddaballapur A, Zörnig M, Braun T, John D, Ponomareva Y, Chen W, Uchida S, Boon Ra, Dimmeler S. Long noncoding RNA MALAT1 regulates endothelial cell function and vessel growth. Circ Res. 2014;114(9):1389–1397. doi: 10.1161/CIRCRESAHA.114.303265. [DOI] [PubMed] [Google Scholar]
- 76.Winston GJ, Palmas W, Lima J, Polak JF, Bertoni AG, Burke G, Eng J, Gottesman R, Shea S. Pulse Pressure and Subclinical Cardiovascular Disease in the Multi-Ethnic Study of Atherosclerosis. Am J Hypertens. 2013;26(5):636–642. doi: 10.1093/ajh/hps092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Vlachopoulos C, Aznaouridis K, Stefanadis C. Prediction of Cardiovascular Events and All-Cause Mortality With Arterial Stiffness. J Am Coll Cardiol. 2010;55(13):1318–1327. doi: 10.1016/j.jacc.2009.10.061. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


