We investigated the genetic diversity and origins of a long-term cultivar. Dyer’s woad has been used as a dye source for at least eight centuries in Eurasia. It was introduced to eastern USA in the 1600s, and is now considered invasive in the western USA. Our analysis of plants from the USA and Eurasia suggests that there are two distinct invasions in western USA that most likely originate from Switzerland, Ukraine and Germany. This information assists in finding effective biological control agents, and continued combination of ecological and molecular data helps bring us closer to sustainable management of plant invasions.
Keywords: AFLP, cultivation, dyer’s woad, genetic diversity, invasion, Isatis tinctoria, origins, population structure, weed
Abstract
Population structure and genetic diversity of invasions are the result of evolutionary processes such as natural selection, drift and founding events. Some invasions are also molded by specific human activities such as selection for cultivars and intentional introduction of desired phenotypes, which can lead to low genetic diversity in the resulting invasion. We investigated the population structure, diversity and origins of a species with both accidental and intentional introduction histories, as well as long-term selection as a cultivar. Dyer’s woad (Isatis tinctoria; Brassicaceae) has been used as a dye source for at least eight centuries in Eurasia, was introduced to eastern USA in the 1600s, and is now considered invasive in the western USA. Our analyses of amplified fragment length polymorphisms (AFLPs) from 645 plants from the USA and Eurasia did not find significantly lower gene diversity (Hj) in the invaded compared to the native range. This suggests that even though the species was under cultivation for many centuries, human selection of plants may not have had a strong influence on diversity in the invasion. We did find significantly lower genetic differentiation (Fst) in the invasive range but our results still suggested that there are two distinct invasions in the western USA. Our data suggest that these invasions most likely originated from Switzerland, Ukraine and Germany, which correlates with initial biological control agent survey findings. Genetic information on population structure, diversity and origins assists in efforts to control invasive species, and continued combination of ecological and molecular analyses will help bring us closer to sustainable management of plant invasions.
Introduction
Some invasive plant species have a history of cultivation before becoming invasive (e.g. Pinus spp.; Richardson 1998, Rubus fruticosus; Clark et al. 2013). Selection for desired plant phenotypes by humans over centuries can reduce genetic diversity in cultivars compared to overall diversity in wild accessions of a species (Clegg et al. 1984; Spooner et al. 2005; Miller and Schaal 2006; Dlugosch and Parker 2008; Lam et al. 2010). Purposeful movement of a limited number of cultivars to a new continent can also act to reduce genetic diversity through founding effects (Loveless and Hamrick 1984). Unintentional movement by seed contamination can likewise result in low diversity through founding effects, though multiple introductions or higher propagule pressure can mitigate loss of genetic diversity (Bossdorf et al. 2005; Genton et al. 2005). When all of these processes (cultivation, importation of cultivars to a new continent, and movement via seed contamination) occur over a centuries-long time frame, we expect that the resulting invasive populations could be exceedingly low in genetic diversity.
The genetic diversity of an invasion can give us insight into both the history and the trajectory of the invasion process. The success of an invasion is driven by the ability to tolerate and adapt to a range of environmental pressures through both plasticity and genetic diversity (Sultan 1995; Lee 2002; Ward et al. 2008). Higher levels of genetic diversity may enhance invasiveness (but see Roman and Darling 2007) and also be an indication that an invasion might be more difficult to manage due to genetically based variation in responses to tools such as herbicides (Maxwell et al. 1990; Jasieniuk et al. 1996) and highly host-specific biological control agents (Gaskin et al. 2011). Low genetic diversity might lead to failure of a population to adapt to the invasive range unless high levels of phenotypic plasticity are present.
In this study, we considered the genetic diversity and population structure of a plant species with a long history of cultivation, deliberate and accidental importation, and recent invasion; Isatis tinctoria (dyer’s woad). The current major invasions in the USA are in northern California/southern Oregon, and near the Rocky Mountains in Idaho, Utah, Montana and Wyoming (PLANTS USDA NRCS 2018). Isatis tinctoria began its interaction with humans as a source of pigment. Sporadic early use and movement in Eurasia during the Neolithic period was followed by wide use since the 1200s in Europe, especially in the dyer’s woad growing regions of England, Germany, Italy and France (Hurry 1930; Zech-Matterne and Leconte 2010). After centuries of use in Europe, dyer’s woad was purposely introduced to eastern North America in the early 1600s as a dye crop (Varga and Evans 1978), until its use was succeeded in the 1700s by a more effective pigment from true indigo, Indigofera tinctoria (Fabaceae). Despite its early introduction into the eastern USA, it is not widely naturalized there (Mack 2003; USDA NRCS 2018). There are multiple hypotheses of how dyer’s woad migrated to the western USA in the late 1800s or early 1900s; as an accidental contaminant of alfalfa seed from eastern USA (Callihan et al. 1984) or Ireland (McConnell et al. 1999) or as a contaminant of packing material (Baun 2009), dye source or ornamental from eastern USA (Kedzie-Webb et al. 1996), or perhaps a combination of these options. In the western USA, the species invades crop and rangeland, and can be locally dense and spread into sagebrush communities (Holmgren et al. 2005). Dyer’s woad is currently listed as a noxious weed in 11 western US states (USDA NRCS 2018).
Studies on the genetic diversity of dyer’s woad have been performed for the western US (Simpson 2013) and for Eurasian populations (Gilbert et al. 2002; Spataro et al. 2007), but there have been no comparisons between the US invasion and native Eurasian populations. Though we expect the invasion to contain much less genetic diversity than the native range, and for the multiple invasions to be genetically similar if they have a common origin, this remains unknown without molecular analysis. In addition, this research can provide data useful to the control of the invasion, including knowledge about genetic diversity and structuring and the Eurasian origins of the invasion, which could be used to match invasive genotypes for host-specificity testing in classical biological control. Here we used amplified fragment length polymorphisms (AFLPs) in an attempt to answer the following questions: (i) Is there a significant loss of genetic diversity overall and within regions when comparing the invasion to the native range? (ii) What are the population structures of the native and invaded ranges? (iii) What is the most probable origin of the US invasion?
Methods
In order to obtain sufficient material for comparison across much of the range of I. tinctoria, we collected leaf material from 645 wild plants from 88 locations [seeSupporting Information—Table S1]. Two hundred and forty-five plants were from the USA and 400 were from Eurasia. This also included one location with two plants from Morocco, Africa. We designated five regions that represent: (i, ii) the two major invaded regions in western USA; (iii) the eastern US region where plants were first introduced but are not invasive; and the regions of potential origin of the species which are (iv) western Europe and (v) eastern Europe/western Asia (see Figs 1 and 2). From these regions we collected 65, 128, 33, 131 and 252 plants, respectively (Table 1). In Region 3 (eastern USA), we searched the nine locations with reputable herbarium or online location information but only found the species in four locations, and in two of those locations there were fewer than 10 plants present; thus we feel that our sampling numbers, while being low, do represent the genetic diversity that exists in that region. We were unable to include plants from the UK due to our inability to find them at ~20 historical collection sites in the UK in 2016. Plants were haphazardly collected at all 88 locations, which is an average of 7.3 plants per location, and we define a location or population as a group of plants within 10 km of each other. Young leaves of plants were stored in silica gel at ambient temperature. To broaden our possible origins in case multiple morphologically similar species are in the USA, our Eurasian collection included two species morphologically similar to I. tinctoria: 12 plants morphologically identified as I. littoralis, endemic to Ukraine, and 5 plants identified as I. glauca from Turkey, endemic to Turkey, Lebanon and Syria (Marhold 2011).
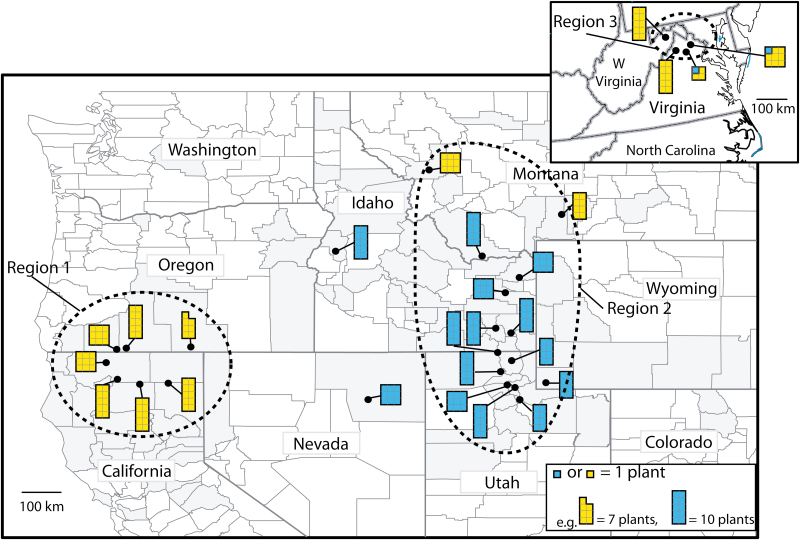
Figure 1.
Collection locations of Isatis tinctoria in the western USA. Shaded counties in the western USA have noted presence of this species at some time. The inset shows collection locations from the eastern USA. Black dots show collection location (exact coordinates are in Supporting Information—Table S1) and yellow or blue shapes indicate majority genetic cluster assignments of plant as well as number of plants collected for a location. Regions 1, 2 and 3 approximate the major invaded areas and are shown by dashed lines.
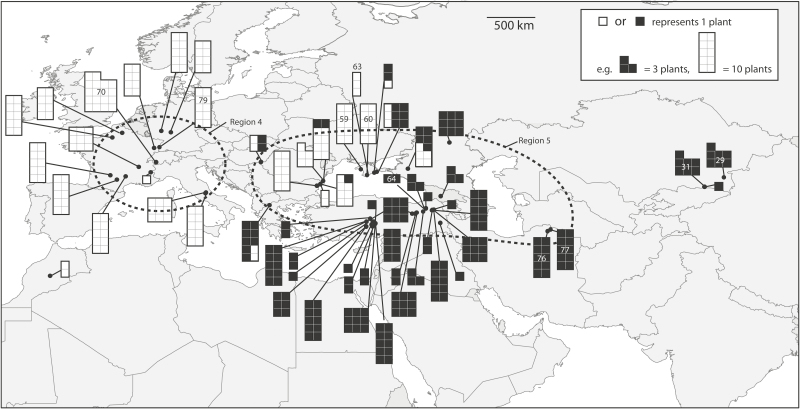
Figure 2.
Collection locations of Isatis tinctoria in Eurasia. Black dots show collection locations (exact coordinates are in Supporting Information—Table S1) and white or black shapes indicate majority genetic cluster assignments of plant as well as number of plants collected for a population. Regions 4 and 5 approximate western Europe vs. eastern Europe/western Asia locations and are shown by dashed lines. The five locations that are genetically most similar and most dissimilar to the US invasion are indicated by location numbers in black or white text, respectively.
Table 1.
Diversity and population structure of Isatis tinctoria in the invasive and native ranges. Dissimilar letters following values within a row indicate statistically significant differences (P < 0.05). PLP = proportion of loci that are polymorphic; Hj = gene diversity; Fst = genetic differentiation.
| USA | Eurasia | ||||||
|---|---|---|---|---|---|---|---|
| Region 1 | Region 2 | Region 3 | Region 4 | Region 5 | |||
| Number of plants | 245 | 65 | 128 | 33 | 400 | 131 | 252 |
| Number of locations | 27 | 7 | 14 | 4 | 61 | 15 | 42 |
| PLP | 38.2 | 34.4 | 35.1 | 42.7 | 50.8 | ||
| PLP | 37 | 49.6 | |||||
| H j | 0.136a | 0.166a | |||||
| H j | 0.139a | 0.126b | 0.124b | 0.143a | 0.168a | ||
| F st | 0.098a | 0.119b | |||||
Genomic DNA was extracted from ~20 mg of silica-dried leaf material using a modified CTAB method (Hillis et al. 1996). The AFLP method followed Vos et al. (1995) with modifications as in Gaskin and Kazmer (2009). All 15 selective primer combinations of MseI + CAA, CAC, CAT, CTA, or CTA and EcoRI + AAG, ACC, or ACT were pre-screened for PCR product quality and number of variable loci using eight samples, and the two most polymorphic primer pairs were chosen (MseI + CTC/EcoRI + AAG and MseI + CAT/EcoRI + ACT). Reliable AFLP bands (loci) were determined as in Ley and Hardy (2013). Briefly, we generated all AFLP data on an Applied Biosystems (ABI, Foster City, CA, USA) 3130 Genetic Analyzer, including 155 repeats (24 % of all 645 samples), and omitted any plants that did not produce a typical electropherogram pattern (i.e. noise > 20 relative fluorescence units (rfu) or failure to produce sufficient number of peaks). All repeats and their matching original.fas files were placed in ABI PeakScanner Software 2 and analysed at a minimum peak height of 20 rfu. The resultant.csv file from the PeakScanner sizing table was prepared with a header for tinyFLP v1.22 (Arthofer 2010) automatic peak scoring and selection using settings of: minimum peak height = 20, maximum peak width = 2, minimum size = 50, maximum size = 500, size tolerance range = 0.5, minimum peak–peak distance = 1, peak height difference = 0, minimum frequency = 0.3, maximum frequency 99.7. The tinyFLP output file was prepared for SPAGeDi v1.4 (Hardy and Vekemans 2002) to test for reproducibility of peaks, using broad sense heritability (H2) and its significance, calculated as Fst of Weir and Cockerham (1984). Peaks (loci) with an H2 value of >0 and P < 0.05 were considered heritable for this study (significant H2 was always >0.26). Final allele calls for heritable loci were made with GeneMapper (ABI) at >50 rfu; bin width of 1 bp.
To compare genetic diversity between continents and between Regions 1 through 5, we calculated proportion of loci that are polymorphic at the 5 % level (PLP) and Nei’s (1987) gene diversity (Hj; equivalent to expected heterozygosity; He) in AFLP-SURV 1.0 (Vekemans 2002). Significance differences for Hj were calculated using 95 % confidence intervals derived from standard errors.
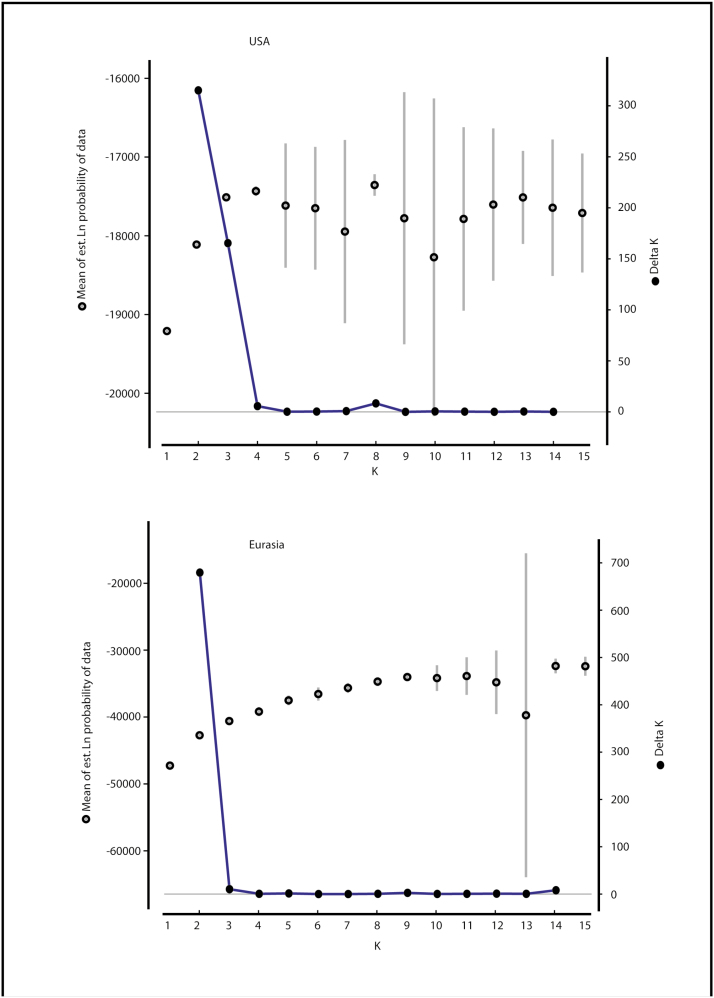
To analyse population structure, we investigated genetic differentiation (Fst) and clustering and assignment tests. Fst was compared for the USA and Eurasia in AFLP-SURV, with 95 % confidence intervals around Fst values generated from 1000 permutations. Clustering and assignment tests were performed using the software STRUCTURE v 2.3.3 (Pritchard et al. 2000; Falush et al. 2003; Falush et al. 2007). To determine the number of genetic clusters (K) represented in the genotypes, AFLP results were diploidized (Falush et al. 2007), no population or geographic location information was included, admixture was assumed as possible, allele frequencies were considered to be independent, and a 50000 run burn-in (α stabilized at ~1000 runs) and 100000 run length were used. We tested for number of genetic clusters (K = 1 to 15) with 20 repetitions for each value of K. Selection of K from this output data was done with the criterion ΔK suggested by Evanno et al. (2005), and results were visualized in the software STRUCTURE HARVESTER webv0.6.92 (Earl 2012). Because we were not convinced that allele frequencies should be considered independent, we also ran the final STRUCTURE analysis with the allele frequencies correlated option.
To further identify origins we calculated smallest pairwise Nei’s (1972) genetic distances (D) between locations in the USA and Eurasia using AFLP-SURV. Locations must have at least two plants in order to calculate D, so some locations were omitted from this analysis (seeSupporting Information—Table S1 for number of samples per location). To visually estimate origins of the US invasion, we used D values in T-REX online (Boc et al. 2012) to create a neighbor-joining tree. Bootstrap values for tree branches were created using 1000 repetitions of D in AFLP-SURV and running those with the Consense function in Phylip (Felsenstein 1989). Though our location population sizes were low in some cases, we included this analysis because: (i) the comparison of population genetic distances does not in itself create type 1 errors (false positive); and (ii) though type 2 errors (false negative) are possible, emerging patterns of origins using a high number of locations is useful information for control efforts, even if more precise information could be drawn from further sampling of more individuals or locations. To infer if the western US plants were more likely to have come from eastern USA or directly from Eurasia, we performed a Student’s t-test on the average Nei’s genetic distance (D) between locations comparing western USA (23 populations) and eastern USA (four locations; 92 pairwise comparisons) or western USA and Eurasia (four locations that were the closest genetic matches to western USA; 92 pairwise comparisons).
Results
Screening of repeatability of loci described above provided 262 significantly heritable, variable loci for MseI + CTC/EcoRI + AAG and MseI + CAT/EcoRI + ACT combined (127 and 135 loci, respectively). AFLP data are presented in Supporting Information—Table S1.
Genetic diversity and population structure
The proportion of loci that were polymorphic (PLP) was lower for the USA than for Eurasia, at both the continent and region levels (Table 1). Nei’s (1987) gene diversity (Hj) was not significantly different in the USA compared to Eurasia. Hj was significantly higher in Regions 1, 4 and 5 than in 2 and 3 (Table 1 and see Figs 1 and 2 for region designations).
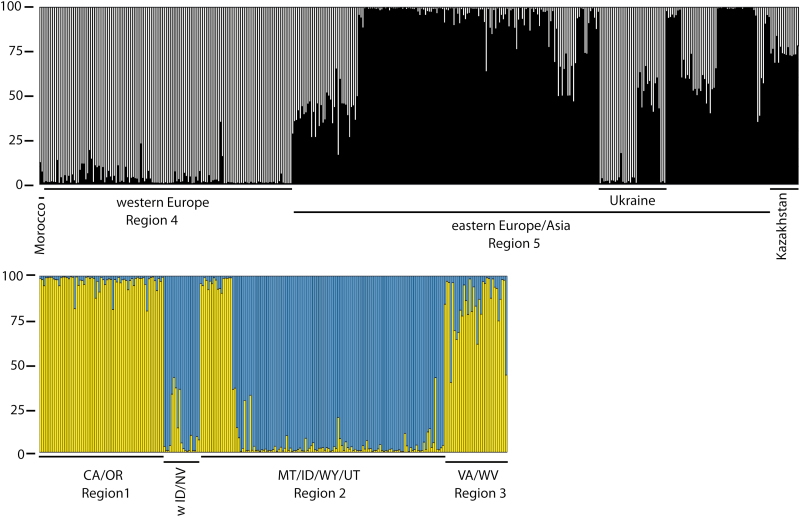
Genetic differentiation (Fst) was significantly lower in the USA compared to Eurasia. Clustering analysis within Eurasia indicated two genetic clusters (ΔK = 2, coloured black or white in Figs 2 and 3). When we analysed USA and Eurasia together under K = 2, all 245 US plants assigned to the white cluster (Fig. 2). When we analysed the US plants alone, these were further sub-clustered into two groups (Fig. 3) indicated by two colours, yellow and blue, on Fig. 1. According to STRUCTURE analysis, there was geographic structuring in western USA, with populations in each region belonging to the same cluster (blue or yellow), except for two populations in Montana (Populations 1 and 2, Fig. 1). Assignment of plants to each cluster in the USA and Eurasia was generally strong, with 93 % and 80 % of plants assigning at >90 % to a cluster, respectively [seeSupporting Information—Table S2]. When we assumed allele frequencies to be correlated in the STRUCTURE analysis, all plants assigned to the same cluster as when we assumed allele frequencies to be independent.
Figure 3.
The bar charts show assignment levels of Isatis tinctoria plants to the genetic clusters determined by STRUCTURE analysis. Bar height within one column (there are 645 columns, one for each plant) can vary from 0 to 100 % assignment value. Mixed colours within a column (plant) indicate assignment to multiple genetic clusters. CA = California; ID = Idaho; MT = Montana; OR = Oregon; NV = Nevada; UT = Utah; WY = Wyoming.
Origins
STRUCTURE analysis assigned all US plants to the cluster of populations that included western Europe/Morocco/Ukraine [seeSupporting Information—Table S2]. In the neighbor-joining tree [seeSupporting Information—Fig. S1], the US locations were nested within branches that included populations from Morocco, France, Germany, Switzerland and Ukraine (Crimean Peninsula). Average Nei’s genetic distance (D) between Eurasian locations and US locations ranged from 0.021 to 0.157 and the lowest five average genetic distances to US locations (D = 0.021–0.031) were Eurasian locations 70, 59, 60, 79, 63 (Switzerland, Ukraine, Ukraine, Germany and Ukraine, respectively; marked on Fig. 2). The same five Eurasian locations were also genetically most similar to the invasion when comparing with US Regions 1, 2 or 3 separately. The Eurasian locations least genetically similar to US locations on average (D = 0.133–0.157) were 29, 31, 77, 64, 76 (Kazakhstan, Kazakhstan, Iran, Turkey, Iran, respectively; marked on Fig. 2). The locations identified as I. littoralis were below the average pairwise genetic distance between all populations (D = 0.065 and 0.053). The locations identified as I. glauca were among the most genetically distinct (D = 0.096–0.125). Plants from Eurasia morphologically identified as I. littoralis or I. glauca had average genetic distances to US locations of D = 0.058–0.138.
In determining if the western US plants were more likely to have come from eastern USA or directly from Eurasia, the average genetic distances between western USA and the four genetically closest Eurasian locations (D = 0.027) were significantly closer than those of western USA and eastern USA (D = 0.0315) at the 95 % confidence level, suggesting that the western US invasion more likely came directly from Eurasia than eastern USA, though the paucity of populations we could find in eastern USA lowered our ability to support this claim.
Discussion
Population structure and genetic diversity of plants are affected by many factors, including mode of reproduction and dispersal. Dyer’s woad is a tetraploid biennial, typically outcrossing herb (2n = 4x = 28) (Al-Shehbaz et al. 2006). Selfing is possible, but leads to fewer siliques with lower weight and seed germinability (Spataro and Negri 2008a). Seeds are not adapted to wind dispersal and usually fall to the ground within a few meters of the plant (Evans 1991). Longer distance dispersal relies on movement by wildlife and contaminated crop products (Farah 1987). Invasions can also be found along roadsides and railroads, suggesting transportation vectors (Evans 1991).
Genetic diversity
Introduced populations typically show losses in genetic diversity (Allendorf and Lundquist 2003; Dlugosch and Parker 2008), but we did not see statistically significant losses in the USA compared to Eurasia (Table 1). Proportion of loci that are polymorphic was lower in the USA, but may have been influenced by our unequal sample sizes at the continent and region levels. Hj was lower for the USA but not significantly. Region 1 was statistically similar to Regions 4 and 5 of Eurasia for Hj, even though sampling size was smaller in Region 1, further suggesting a comparatively high level of genetic diversity in Region 1. This emerging pattern of retention of significant amounts of genetic diversity in the USA could be the result of mixing of genotypes from different population sources as they were moved from Europe to eastern USA and/or to western USA and the facultative outcrossing of this species. The high diversity within the USA also suggests that before introduction to the USA there was not strong selection for a single or a few cultivars that provided the best dye product, as historically recorded (Hurry 1930), and that maybe most individuals of the species could produce useful dye, though research by Spataro and Negri (2008b) suggests that in Europe there are now preferred lineages or varieties used for artisanal dyeing.
Population structure
Genetic differentiation (Fst) was significantly lower in the USA than in Eurasia (Table 1). This may be due to the much longer history of I. tinctoria in Europe and Asia, giving more opportunities for natural selection, mutation and drift to create population structure and offset any losses in fixation due to migration and gene flow.
Our western US invasion had two genetic clusters (blue and yellow, Fig. 1) which, except for two yellow populations in Montana, segregated roughly as California/Oregon vs. Rocky Mountains. Simpson (2013) found three genetic clusters in the western USA, and populations along the Wyoming/Idaho border formed their own cluster, separate from nearby Utah and all other western US populations. Our analysis of ΔK values (Fig. 4) suggested two clusters, but when we forced the clustering program to find three clusters as Simpson (2013) did, the clusters still segregated California/Oregon from Idaho/Wyoming/Utah/Nevada, with a third cluster containing eastern USA and some Montana populations [seeSupporting Information—Fig. S2], which is unlike the Simpson (2013) result. Also unlike our study, Simpson (2013) found less geographic clustering, with two of three clusters represented in the far west (California) and all three clusters in the Rocky Mountains. The disparity between the two studies may be due to collection from different locations in a region, or quality of DNA extracted for AFLP data, which was claimed to be poor in some populations in Simpson’s study (2013). Our study and that of Simpson (2013) included 21 compared to 15 locations and 193 compared to 268 samples, respectively, for the western USA. Our evidence of geographic structure suggested a lack of gene flow between California/Oregon and the blue coloured locations in the Rocky Mountain region, and for management, we suggest including representative plants from both yellow and blue clusters (Fig. 1) in biological control agent efficacy studies. This genetic differentiation between California/Oregon and the Rocky Mountain region might also explain why the dyer’s woad strain of the North American rust Puccinia thlaspeos infects dyer’s woad in Idaho/Utah, but fails to establish when applied to plants in California/Oregon (M. Schwarzländer, pers. comm.).
Figure 4.
Support for defining the number of genetic clusters of US and Eurasian Isatis tinctoria. Mean of estimated Ln probability of the data ± SD for each K (number of clusters) and ΔK values over 15 runs as a function of K (Evanno et al. 2005).
Origins
The claimed native range of I. tinctoria was historically unclear and debatable. Origin hypotheses range from south-eastern Asia (Sales et al. 2006), Central Asia (Spataro et al. 2007), the Caucasus and Middle East to eastern Siberia (Conert et al. 1986) to south-west Asia and south-eastern Europe (Ball and Akeroyd 2010). The Euro+Med PlantBase (Marhold 2011) listed native/introduced status for Europe, and cited references in Marhold (2011) often disagreed about native status in certain western European countries (e.g. Germany, Switzerland, Spain) but agreed on native status in other countries (e.g. UK, France, Italy). Germany, France and the UK were once big growing centres for I. tinctoria (Zech-Matterne and Leconte 2010), and in some regions naturalized populations persist in good numbers (e.g. eastern France) while other areas seem to have lost most populations (e.g. UK; J. F. Gaskin, H. Hinz, pers. comm.). Our data show that except for Ukraine, Asian populations were genetically distinct in clustering analysis from western European populations, supporting native status of this species in western Europe, or at least very old establishment and selection of dyer’s woad in Europe. However, it is also possible that western European populations are derived from Asia and our limited Asian sampling did not sample the populations that would demonstrate that connection.
While we do not propose to revise the taxonomy of Isatis here, we note that our plants morphologically identified as I. littoralis fell below the average genetic distance between I. tinctoria populations, suggesting that I. littoralis is a taxon within I. tinctoria, or our morphological identification was wrong, and the individuals were actually I. tinctoria. Plants morphologically identified as I. glauca were among the most genetically distinct in our analysis supporting them as not being I. tinctoria, consistent with results from Gilbert et al. (2002). This indicates that I. littoralis be included, and I. glauca be excluded, from searches for biological control agents of I. tinctoria.
Our best estimates of Eurasian origins of the US invasion were Switzerland, Ukraine and Germany (plants were not available to address the hypothesis of a UK origin). This suggests that potential biological control candidate species from these regions found on plants genetically similar to the invasion may be better adapted to the invasive genotypes and potentially more effective control organisms compared to those from other locations (e.g. Turkey, Iran, Kazakhstan), though new host–enemy associations can also be damaging (Evans and Ellison 2004). Even though biological control agents with broad host specificity that utilize all I. tinctoria genotypes may exist, there are examples of very host-specific biological control systems (e.g. Goolsby et al. 2006), warning that agents from just one location, chosen without considering specific invasion origins, may not control all invasive genotypes, leading potentially to replacement by biological control resistant plants and lowering overall weed control efficacy, as was seen in control efforts for Chondrilla juncea in Australia (Burdon et al. 1981). Our hypothesis regarding specific origins of the western US genotypes was reinforced by the fact that all three currently studied potential biological control agents for I. tinctoria were found in western to south-western Europe (Germany and Italy) and never during surveys in eastern Europe or Asia (H. Hinz, unpubl. data). This includes the near-monophagous, root-crown mining weevil Ceutorhynchus rusticus, the near-monophagous, seed-feeding weevil C. peyerimhoffi and the mite Metaculus sp.
Our evidence that the western US invasion more likely came directly from Eurasia than the eastern USA due to average genetic similarity between locations from the two ranges still leaves other hypotheses to be tested with additional data. For example, there may be more eastern US populations to collect and increase the statistical power, though we found that many previous collection sites were now extirpated for this species. Also, we were unable to test the hypothesis of a UK origin for western US invasion due to our inability to find plants at ~20 historical collection sites in the UK.
Conclusions
We found that the dyer’s woad invasion has emerging patterns of lower, but not significantly lower, genetic diversity which was not expected from the long history of selective forces that could have had extreme effects on diversity. Unexpected retention of gene diversity in the invasion populations is likely tied to low levels of human-mediated selection for cultivars, facultative outcrossing and perhaps multiple introductions events. Our analysis demonstrated the utility of molecular data in studying invasion processes and patterns, including diversity, geographic structuring of genotypes and origins. The finding that the invasion was relatively diverse but geographically structured gave us insight into the invasion process and should facilitate management of the species. These patterns can help inform invasion control decisions at the researcher and land manager levels. Continued combination of ecological and molecular data will bring us closer to sustainable management of plant invasions.
Sources of Funding
This research was made possible in part by funding from the Bureau of Land Management (Montana, South and North Dakota) Billings Office, BLM Idaho, various Idaho and Utah counties, the Idaho State Department of Agriculture, the USDA Forest Service, Bear Lake RD&D, the Wyoming Biological Control Steering Committee and USDA-APHIS-CPHST.
Contributions by the Authors
J.F.G., H.H., M.S., R.D.G., H.S., D.L.M. and E.G. developed and planned the project and made collections. J.F.G. performed data analysis and wrote the paper with substantial input from all authors.
Conflict of Interest
None declared.
Supporting Information
The following additional information is available in the online version of this article—
Table S1. Plant collection location, region, population and amplified fragment length polymorphism (AFLP) data.
Table S2. Assignment values to K = 2 and K = 3 clusters for US and Eurasian samples.
Figure S1. Neighbor-joining tree of Isatis populations from North America and Eurasia.
Figure S2. Assignment to K = 3 clusters for USA.
Acknowledgements
Thanks to K. Mann and J. Lassey of USDA ARS, Sidney, MT, for generating AFLP data.
Literature Cited
- Allendorf FW, Lundquist LL. 2003. Introduction: population biology, evolution, and control of invasive species. Conservation Biology 17:24–30. [Google Scholar]
- Al-Shehbaz IA, Beilstein MA, Kellog EA. 2006. Systematics and phylogeny of the Brassicaceae (Cruciferae): an overview. Plant Systematics and Evolution 259:89–120. [Google Scholar]
- Arthofer W. 2010. tinyFLP and tinyCAT: software for automatic peak selection and scoring of AFLP data tables. Molecular Ecology Resources 10:385–388. [DOI] [PubMed] [Google Scholar]
- Ball PW, Akeroyd JR. 2010. Isatis L. In: Tutin TG, Burges NA, Chater AO, Edmondson JR, Heywood VH, Moore DM, Valentine DH, Walters SM, Webb DA, eds. Flora Europaea. Volume 1; Psilotaceae to Platanaceae. Cambridge, UK: Cambridge University Press, 324–325. [Google Scholar]
- Baun M. 2009. Yreka FWO: score one for the woad warriors. US Fish and Wildlife Service Journal, Region 8 https://www.fws.gov/FieldNotes/regmap.cfm?arskey=27244 (September 2017).
- Boc A, Diallo AB, Makarenkov V. 2012. T-REX: a web server for inferring, validating and visualizing phylogenetic trees and networks. Nucleic Acids Research 40:W573–W579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bossdorf O, Auge H, Lafuma L, Rogers WE, Siemann E, Prati D. 2005. Phenotypic and genetic differentiation between native and introduced plant populations. Oecologia 144:1–11. [DOI] [PubMed] [Google Scholar]
- Burdon JJ, Groves RH, Cullen JM. 1981. The impact of biological control on the distribution and abundance of Chondrilla juncea in south-eastern Australia. Journal of Applied Ecology 18:957–966. [Google Scholar]
- Callihan RH, Dewey SA, Patton JE, Thill DC. 1984. Distribution, biology and habitat of dyers woad (Isatis tinctoria L.) in Idaho. Journal of the Idaho Academy of Science 20:18–32. [Google Scholar]
- Clark LV, Evans KJ, Jasieniuk M. 2013. Origins and distribution of invasive Rubus fruticosus L. agg. (Rosaceae) clones in the Western United States. Biological Invasions 15:1331–1342. [Google Scholar]
- Clegg MT, Brown AHD, Whitfeld PR. 1984. Chloroplast DNA diversity in wild and cultivated barley: implications for genetic conservation. Genetical Research 43:339–343. [Google Scholar]
- Conert HJ, Hamann U, Schultze-Motel W, Wagenitz G. 1986. Illustrierte flora von mitteleuropa. Berlin, Hamburg: Paul Parey. [Google Scholar]
- Dlugosch KM, Parker IM. 2008. Founding events in species invasions: genetic variation, adaptive evolution, and the role of multiple introductions. Molecular Ecology 17:431–449. [DOI] [PubMed] [Google Scholar]
- Earl DA. 2012. STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conservation Genetics Resources 4:359–361. [Google Scholar]
- Evans JO. 1991. The importance, distribution, and control of dyers woad (Isatis tinctoria). In: James LF, Evans JO, Ralphs MH, Child RD, eds. Noxious range weeds. Westview special studies in agricultural science and policy. Boulder, CO: Westview Press, 387–393. [Google Scholar]
- Evans HC, Ellison CA. 2004. The new encounter concept: centres of origin, host specificity and plant pathogens. In: Cullen JM, Briese DT, Kriticos DJ, Lonsdale WM, Morin L, Scott JK, eds. XI International Symposium on Biological Control of Weeds. Canberra, Australia: CSIRO Entomology, 42–47. [Google Scholar]
- Evanno G, Regnaut S, Goudet J. 2005. Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Molecular Ecology 14:2611–2620. [DOI] [PubMed] [Google Scholar]
- Falush D, Stephens M, Pritchard JK. 2003. Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics 164:1567–1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falush D, Stephens M, Pritchard JK. 2007. Inference of population structure using multilocus genotype data: dominant markers and null alleles. Molecular Ecology Notes 7:574–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farah KO. 1987. Autecological and grazing control studies of dyers woad (Isatis tinctoria L.) on northern Utah rangelands. Dissertation, Utah State University, Logan, UT. [Google Scholar]
- Felsenstein J. 1989. PHYLIP - Phylogeny Inference Package (version 3.2). Cladistics 5:164–166. [Google Scholar]
- Gaskin JF, Bon MC, Cock MJ, Cristofaro M, De Biase A, De Clerck-Floate, Ellison CA, Hinz HL, Hufbauer RA, Julien MH, Sforza R. 2011. Applying molecular-based approaches to classical biological control of weeds. Biological Control 58:1–21. [Google Scholar]
- Gaskin JF, Kazmer DJ. 2009. Introgression between invasive saltcedars (Tamarix chinensis and T. ramosissima) in the USA. Biological Invasions 11:1121–1130. [Google Scholar]
- Genton BJ, Shykoff JA, Giraud T. 2005. High genetic diversity in French invasive populations of common ragweed, Ambrosia artemisiifolia, as a result of multiple sources of introduction. Molecular Ecology 14:4275–4285. [DOI] [PubMed] [Google Scholar]
- Gilbert K, Garton S, Karam A, Arnold M, Karp A, Edwards J, Cooke T, Barker A. 2002. A high degree of genetic diversity is revealed in Isatis spp. (dyer’s woad) by amplified fragment length polymorphism (AFLP). Theoretical and Applied Genetics 104:1150–1156. [DOI] [PubMed] [Google Scholar]
- Goolsby JA, Van Klinken RD, Palmer WA. 2006. Maximising the contribution of native‐range studies towards the identification and prioritisation of weed biocontrol agents. Austral Entomology 45:276–286. [Google Scholar]
- Hardy OJ, Vekemans X. 2002. SPAGeDi: a versatile computer program to analyse spatial genetic structure at the individual or population levels. Molecular Ecology Resources 2:618–20. [Google Scholar]
- Hillis DM, Huelsenbeck J, Mable BK, Larson A, Zimmer EA. 1996. Nucleic acids IV: sequencing and cloning. In: Hillis DM, Moritz C, Mable BK, eds. Molecular systematics, 2nd edn. Sunderland, MA: Sinauer Associates, 321–383. [Google Scholar]
- Holmgren NH, Holmgren PK, Cronquist A. 2005. Intermountain flora: vascular plants of the Intermountain West, U.S.A. Vol. 2, part B: subclass Dilleniidae. New York: The New York Botanical Garden. [Google Scholar]
- Hurry JB. 1930. The woad plant and its dye. Oxford: Oxford University Press. [Google Scholar]
- Jasieniuk M, Brûlé-Babel AL, Morrison IN. 1996. The evolution and genetics of herbicide resistance in weeds. Weed Science 44:176–193. [Google Scholar]
- Kedzie-Webb S, Sheley R, Dewey S. 1996. Dyer’s woad: a threat to rangeland in Montana. Bozeman, MT: Montana State University Cooperative Extension. Montguide MT9614. [Google Scholar]
- Lam HM, Xu X, Liu X, Chen W, Yang G, Wong FL, Li MW, He W, Qin N, Wang B, Li J, Jian M, Wang J, Shao G, Wang J, Sun SS, Zhang G. 2010. Resequencing of 31 wild and cultivated soybean genomes identifies patterns of genetic diversity and selection. Nature Genetics 42:1053–1059. [DOI] [PubMed] [Google Scholar]
- Lee CE. 2002. Evolutionary genetics of invasive species. Trends in Ecology and Evolution 17:386–391. [Google Scholar]
- Ley AC, Hardy OJ. 2013. Improving AFLP analysis of large-scale patterns of genetic variation–a case study with the Central African lianas Haumania spp. (Marantaceae) showing interspecific gene flow. Molecular Ecology 22:1984–1997. [DOI] [PubMed] [Google Scholar]
- Loveless MD, Hamrick JL. 1984. Ecological determinants of genetic structure in plant populations. Annual Review of Ecology and Systematics 15:65–95. [Google Scholar]
- Mack RN. 2003. Plant naturalizations and invasions in the eastern United States: 1634–1860. Annals of the Missouri Botanical Garden 90:77–90. [Google Scholar]
- Marhold K. 2011. Brassicaceae – In: Euro+Med Plantbase - the information resource for Euro-Mediterranean plant diversity http://ww2.bgbm.org/EuroPlusMed/ (December 2017).
- Maxwell BD, Roush ML, Radosevich SR. 1990. Predicting the evolution and dynamics of herbicide resistance in weed populations. Weed Technology 4:2–13. [Google Scholar]
- McConnell EG, Evans JO, Dewey SA. 1999. Dyer’s woad. In: Sheley RL, Petroff JK, eds. Biology and management of noxious rangeland weeds. Corvalis, OR: OSU Press, 231–237. [Google Scholar]
- Miller AJ, Schaal BA. 2006. Domestication and the distribution of genetic variation in wild and cultivated populations of the Mesoamerican fruit tree Spondias purpurea L. (Anacardiaceae). Molecular Ecology 15:1467–1480. [DOI] [PubMed] [Google Scholar]
- Nei M. 1987. Molecular evolutionary genetics. New York: Columbia: University Press. [Google Scholar]
- Nei M. 1972. Genetic distance between populations. The American Naturalist 106:283–292. [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P. 2000. Inference of population structure using multilocus genotype data. Genetics 155:945–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson DM. 1998. Forestry trees as invasive aliens. Conservation Biology 12:18–26. [Google Scholar]
- Roman J, Darling JA. 2007. Paradox lost: genetic diversity and the success of aquatic invasions. Trends in Ecology & Evolution 22:454–464. [DOI] [PubMed] [Google Scholar]
- Sales E, Kanhonou R, Baixauli C, Giner A, Cooke D, Gilbert K, Arrillaga I, Segura J, Ros R. 2006. Sowing date, transplanting, plant density and nitrogen fertilization affect indigo production from Isatis species in a Mediterranean region of Spain. Industrial Crops and Products 23:29–39. [Google Scholar]
- Simpson HL. 2013. Genetic diversity, local differentiation, and adaptive evolution in introduced populations of the invasive plant Isatis tinctoria L.(Brassicaceae). Doctoral dissertation, University of New Mexico, Albuquerque, NM. [Google Scholar]
- Spataro G, Negri V. 2008a. Assessment of the reproductive system of Isatis tinctoria L. Euphytica 159:229–231. [Google Scholar]
- Spataro G, Negri V. 2008b. Adaptability and variation in Isatis tinctoria L.: a new crop for Europe. Euphytica 163:89–102. [Google Scholar]
- Spataro G, Taviani P, Negri V. 2007. Genetic variation and population structure in a Eurasian collection of Isatis tinctoria L. Genetic Resources and Crop Evolution 54:573–584. [Google Scholar]
- Spooner DM, McLean K, Ramsay G, Waugh R, Bryan GJ. 2005. A single domestication for potato based on multilocus amplified fragment length polymorphism genotyping. Proceedings of the National Academy of Sciences of the United States of America 102:14694–14699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sultan SE. 1995. Phenotypic plasticity and plant adaptation. Acta Botanica Neerlandia 44:363–383. [Google Scholar]
- USDA NRCS. 2018. The PLANTS Database. Greensboro, NC: National Plant Data Team; http://plants.usda.gov (15 March 2018). [Google Scholar]
- Varga WA, Evans JO. 1978. Dyer’s woad: from cultivated to curse. Utah Science 39:87–90. [Google Scholar]
- Vekemans X. 2002. AFLP-SURV version 1.0 Belgium: Laboratoire de Génétique et Ecologie Végétale, Université Libre de Bruxelles. [Google Scholar]
- Vos P, Hogers R, Bleeker M, Reijans M, van de Lee T, Hornes M, Frijters A, Pot J, Peleman J, Kuiper M. 1995. AFLP: a new technique for DNA fingerprinting. Nucleic Acids Research 23:4407–4414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward SM, Gaskin JF, Wilson LM. 2008. Ecological genetics of plant invasion: what do we know?Invasive Plant Science and Management 1:98–109. [Google Scholar]
- Weir BS, Cockerham CC. 1984. Estimating F-statistics for the analysis of population structure. Evolution 38:1358–1370. [DOI] [PubMed] [Google Scholar]
- Zech-Matterne V, Leconte L. 2010. New archaeobotanical finds of Isatis tinctoria L. (woad) from Iron Age Gaul and a discussion of the importance of woad in ancient time. Vegetation History and Archaeobotany 19:137–142. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.