Abstract
Death receptor 6 (DR6) is an orphan member of the TNF receptor superfamily and controls cell death and differentiation in a cell‐autonomous manner in different cell types. Here, we report an additional non‐cell‐autonomous function for DR6 in the peripheral nervous system (PNS). DR6‐knockout (DR6 KO) mice showed precocious myelination in the PNS. Using an in vitro myelination assay, we demonstrate that neuronal DR6 acts in trans on Schwann cells (SCs) and reduces SC proliferation and myelination independently of its cytoplasmic death domain. Mechanistically, DR6 was found to be cleaved in neurons by “a disintegrin and metalloprotease 10” (ADAM10), releasing the soluble DR6 ectodomain (sDR6). Notably, in the in vitro myelination assay, sDR6 was sufficient to rescue the DR6 KO phenotype. Thus, in addition to the cell‐autonomous receptor function of full‐length DR6, the proteolytically released sDR6 can unexpectedly also act as a paracrine signaling factor in the PNS in a non‐cell‐autonomous manner during SC proliferation and myelination. This new mode of DR6 signaling will be relevant in future attempts to target DR6 in disease settings.
Keywords: ADAM10, DR6, myelin, Schwann cell, shedding
Subject Categories: Neuroscience
Introduction
Communication between cells requires cell surface membrane proteins. Their levels can be controlled by proteases, which cleave off the membrane proteins’ ectodomain in a process referred to as shedding (Lichtenthaler et al, 2011). This process shapes the cell surface, thereby terminating the function of different surface membrane proteins, such as receptors and cell adhesion proteins, and/or releasing important signaling molecules, for example, cytokines and growth factors. The contributing proteases are diverse, but are typically also membrane proteins. They include members of the “a disintegrin and metalloprotease” (ADAM) family, but also the beta‐site APP‐cleaving enzymes (BACE), the signal peptide peptidase‐like 3 (SPPL3) protease, rhomboids, and others (Reiss & Saftig, 2009; Vassar et al, 2014; Kuhn et al, 2015; Lastun et al, 2016). Among the ADAMs, particularly ADAM10 and ADAM17 contribute to membrane protein shedding. ADAM17 is predominantly involved in growth factor shedding and signaling as well as in inflammatory reactions, for example, through release of tumor necrosis factor‐α (TNF‐α). ADAM10 is a close homolog of ADAM17 and has fundamental functions during embryonic development, for instance, through Notch signaling, but also during postnatal development and adulthood. For example, in the nervous system ADAM10‐mediated membrane protein shedding is required for contact‐mediated axon repulsion, neurogenesis, and synaptic functions, but also contributes to human disorders, including Alzheimer's disease (AD; Kuhn et al, 2010; Saftig & Lichtenthaler, 2015), where it is able to prevent the generation of the neurotoxic amyloid β peptide. Different ADAM10 substrates have been identified in primary neurons using proteomics (Kuhn et al, 2016), but how ADAM10 cleavage alters their function remains unknown for most of them. This is largely due to the embryonic lethality of ADAM10‐deficient mice. Additionally, it has been little explored whether the ADAM10‐mediated cleavage products act as new signaling molecules or are merely further degraded.
Among the substrates of ADAM10 and related proteases are members of the TNF receptor superfamily (TNFRSF), which have major functions in the nervous and immune systems (Aggarwal, 2003). One family member, where proteolysis and its functional consequence have been little studied (Tam et al, 2004; DeRosa et al, 2008), is death receptor 6 (DR6, TNFRSF21). In the central nervous system (CNS) and the peripheral nervous system (PNS), DR6 is expressed in neurons. DR6 is also expressed in oligodendrocytes, which are the myelinating cells in the CNS. In neurons and oligodendrocytes, full‐length DR6 acts as a receptor that activates cell death pathways, resulting in developmental axon pruning, Wallerian degeneration, and reduced oligodendrocyte‐mediated myelination in the CNS (Nikolaev et al, 2009; Mi et al, 2011; Marik et al, 2013; Gamage et al, 2017). Consequently, DR6 has been linked to Alzheimer's disease and is considered as a drug target for multiple sclerosis (Nikolaev et al, 2009; Mi et al, 2011; Kallop et al, 2014). Additionally, DR6 controls proliferation and differentiation of immune cells (Zhao et al, 2001; Schmidt et al, 2003; DeRosa et al, 2008), and was recently shown to contribute to tumor cell‐induced endothelial cell necroptosis and autoimmunity in lupus‐prone mice (Strilic et al, 2016; Fujikura et al, 2017). In these processes, DR6 acts cell‐autonomously as a membrane‐bound receptor, similar to the central nervous system.
Whether and how proteolysis affects DR6 function and whether the cleaved soluble DR6 (sDR6) has a signaling function have not yet been investigated. While for most TNFRSF members the protease‐mediated shedding is a mechanism to terminate their receptor function (e.g. Peschon et al, 1998; McIlwain et al, 2012; Laurent et al, 2015), we now report that DR6 is a striking exception. We show that DR6 is cleaved by the metalloprotease ADAM10 and that the released sDR6 ectodomain is biologically active in the PNS, where it acts as a novel paracrine signaling factor in a non‐cell‐autonomous manner during Schwann cell (SC) proliferation and myelination.
Results
DR6 is a novel substrate for ADAM10 and γ‐secretase
Using mass spectrometry, we recently found that full‐length DR6 levels showed a trend to an increase in ADAM10‐deficient murine neurons (Kuhn et al, 2016), suggesting that DR6 may be a novel ADAM10 substrate. To test for evidence of proteolytic DR6 cleavage in vivo, we looked at the peripheral nervous system (PNS) of embryonic stage 13.5 (E13.5) wild‐type (WT) or DR6‐knockout (DR6 KO) mice (Zhao et al, 2001). Freshly prepared dorsal root ganglia (DRGs) were processed into a membrane fraction, where full‐length DR6 (DR6 FL) was detected at 98 kDa, and into a soluble fraction, where the soluble DR6 ectodomain (sDR6) was detected at around 64 kDa. Both bands were absent in DR6 KO samples (Fig 1A), demonstrating the specificity of the signal. Together, these findings reveal that DR6 undergoes shedding in vivo. To determine from which cell type in the PNS DR6 is shed, DR6 expression was analyzed in different cell types. Although in the CNS, DR6 is expressed in both neurons and oligodendrocytes (Mi et al, 2011), we surprisingly did not detect DR6 expression in SCs, at neither the protein (Fig 1B) nor RNA level (Fig 1C). However, DR6 expression was clearly detected in WT, but not in DR6 KO neurons (Fig 1B). To identify which neuronal protease may mediate DR6 shedding, neuronal cultures were used. The broad‐spectrum metalloprotease inhibitor TAPI‐I blocked 70% of sDR6 release and mildly enriched full‐length DR6 levels (Fig 1D), revealing that one or several metalloproteases mediate most of the DR6 cleavage. While membrane‐type 1 matrix metalloprotease (MT1‐MMP) was suggested as a DR6 protease in tumor cells (Tam et al, 2004; DeRosa et al, 2008), lentiviral knockdown of MT1‐MMP did not alter DR6 shedding in neurons (Fig EV1). In contrast, in ADAM10‐deficient neurons DR6 shedding was reduced by 50% (Fig 1E). To exclude any indirect effect on DR6 shedding upon ADAM10 deficiency, an in vitro cleavage assay was used where recombinant ADAM10 was incubated with full‐length DR6 and produced the same 64‐kDa sDR6 ectodomain as seen in vivo (Fig 1F). We conclude that ADAM10 is a major DR6 protease, directly cleaves DR6, and is responsible for ~50% of DR6 cleavage. The partial DR6 cleavage by ADAM10 behaves similar to other ADAM10 substrates, such as APP, which are also partly cleaved by ADAM10 and partly by other proteases, including the β‐secretase BACE1 (Hu et al, 2016; Kuhn et al, 2016).
Figure 1. DR6 ectodomain shedding.
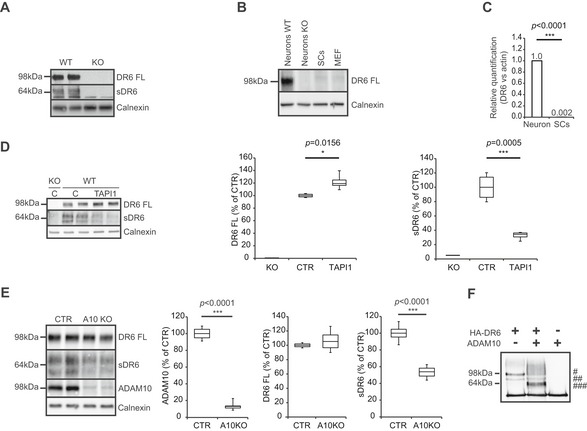
-
ADR6 shedding in vivo. DR6 full‐length (FL) and its cleavage product soluble DR6 (sDR6) were detected in WT but not in DR6 KO PNS tissue samples.
-
BWestern blot analysis of DR6 FL in WT primary neurons and SCs showing immunoreactivity only in neuronal cells. Mouse embryonic fibroblasts (MEFs) were also tested to exclude that DR6 detection resulted from a minor fibroblast contamination in DRG cultures. DR6 KO neuronal lysate was added as negative control.
-
CQuantitative real‐time PCR for murine DR6 RNA normalized to reference gene (actin) confirms absence of DR6 expression in SCs (n = 3).
-
D, EDR6 FL and sDR6 levels in primary neurons treated with broad‐spectrum metalloprotease inhibitor TAPI‐I (D, n = 4) or DMSO as a control (C, CTR) and upon ADAM10 knockout (E, n = 8) showing that ADAM10 is responsible for at least 50% of sDR6 release.
-
FIncubation of recombinant ADAM10 with recombinant DR6 FL produces a cleavage product (64 kDa) corresponding to sDR6. As additional control, ADAM10 alone was added to the assay. #: mature fully glycosylated DR6 FL; ##: immature DR6 FL; ###: sDR6.
Figure EV1. Analysis of sDR6 level in WT primary neurons upon MT1‐MMP knockdown.

Specific short hairpin RNAs targeting MT1‐MMP (sh1 and sh2) were expressed in primary neurons using a lentiviral system. A scrambled shRNA sequence was used as negative control. Levels of full‐length DR6 (DR6 FL) and its released ectodomain (sDR6) were analyzed in cell lysates and culture supernatant, respectively (n = 6 for each group), showing that upon MT1‐MMP knockdown, full‐length protein and its processing were not affected. Western blot quantifications are represented using box plots showing the first and the third quartile together with the median. The whiskers show the maximum and the minimum data point. Statistical test: unpaired Student's t‐test with two‐tailed distribution. Source data are available online for this figure.
After initial ectodomain shedding, the remaining C‐terminal fragment (CTF) of several type I transmembrane proteins can be further cleaved by the protease complex γ‐secretase within the transmembrane domain (TM) in a process referred to as regulated intramembrane proteolysis (RIP; Lichtenthaler et al, 2011; Fig EV2A). As a result, a short N‐terminal peptide—comprising the short remaining ectodomain and half of the transmembrane domain—is typically secreted from cells, whereas the intracellular domain (ICD) is released into the cytosol, where it is mostly rapidly degraded, unless proteasomal activity is blocked. γ‐Secretase cleavage can be monitored by the detection of the ICD. Inhibition of γ‐secretase blocks ICD generation and leads to increased CTF levels (for a schematic, see Fig EV2A). To test whether the type I transmembrane protein DR6 is processed by γ‐secretase, we transiently overexpressed DR6 in HEK293E cells. To detect CTF and ICD of DR6, a Flag tag was fused to the C‐terminus of DR6. Treatment of the cells with the proteasome inhibitor MG132 enabled detection of the DR6 ICD (##; Fig EV2B). Conversely, the γ‐secretase inhibitor DAPT—alone or together with MG132—blocked ICD formation and increased DR6 CTF levels (#; Fig EV2B), demonstrating that DR6 is a new substrate not only for ADAM10, but also for γ‐secretase.
Figure EV2. γ‐Secretase cleavage of DR6 CTF .
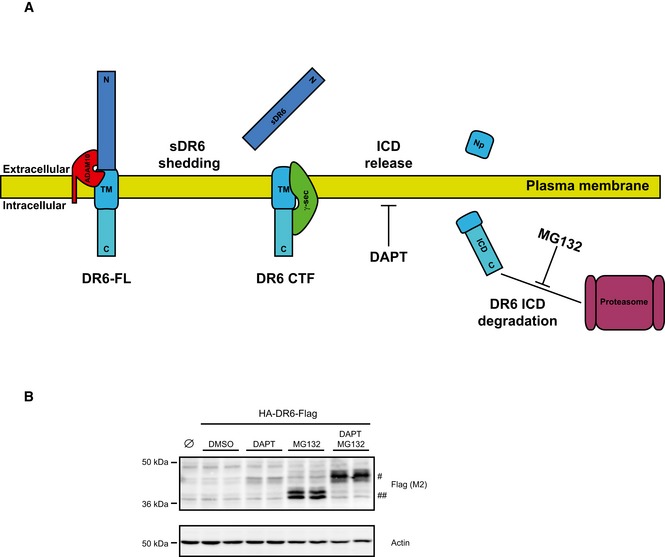
- Regulated intramembrane proteolysis (RIP) scheme. Type I transmembrane proteins such as DR6 can be cleaved by a protease (e.g., ADAM10), thus releasing their soluble ectodomain (sDR6) into the extracellular environment. The resulting C‐terminal fragment (CTF) can be further cleaved by the protease γ‐secretase within the transmembrane domain (TM). As a result, a short N‐terminal peptide (Np) is typically secreted from cells, while the intracellular domain (ICD) is released into the cytosol. The latter is mostly rapidly degraded, unless proteasomal activity is blocked through specific inhibition with MG132. Typically, the γ‐secretase inhibitor DAPT is used to induce accumulation of the CTFs and to validate candidates as γ‐secretase substrates.
- HEK293E cells overexpressing hDR6 with a Flag tag at the C‐terminus were treated for 24 h with either the γ‐secretase inhibitor DAPT (1 μM) or the proteasome inhibitor MG132 (1 μM). Cells transfected with an empty vector were used as control (Ø). Western blot analysis of cell lysates showed that DAPT treatment, alone or together with MG132, blocked ICD formation and a 45‐kDa fragment (#) corresponding to DR6 CTF was accumulating, while proteasome blockade induces accumulation of a smaller fragment (##) corresponding to DR6 ICD. Together, our data demonstrate that DR6, after initial ectodomain shedding, is further processed by the γ‐secretase complex.
Source data are available online for this figure.
DR6 controls PNS myelination
Proteolysis of membrane proteins may serve different functions. It may inactivate a cell surface receptor, as it has been shown for several TNFRSF members, but may also release a biologically active protein ectodomain, for example, of neuregulin‐1 (Luo et al, 2011, 2014; Fleck et al, 2013). To determine whether sDR6 may have a function in the PNS, we first looked whether DR6 is involved in peripheral myelination. DRG cultures were prepared from WT or DR6 KO littermate mice carrying a SC‐specific reporter gene (plp‐GFP; Zhao et al, 2001; Mallon et al, 2002). Myelination was induced with ascorbic acid. While the gross morphology and amount of axons appeared independent of the genotype (based on β3‐tubulin staining), the number of myelin segments (stained with an antibody against myelin basic protein [MBP]) was two times higher in DR6 KO compared to WT littermates (Fig 2A). The myelin sheath length of each segment was not altered. To test whether the increased number of myelin segments in DR6 KO DRG cultures was due to increased SC proliferation, a time‐course BrdU proliferation assay was used. In fact, the ratio of proliferating SCs (double‐positive for GFP and BrdU) relative to the total number of SCs in DR6 KO was higher during the first week in vitro compared to controls, with the increases ranging from 1.4‐fold to 2.5‐fold (Fig 2B). As expected, the number of immature, proliferating SCs decreased in a time‐dependent manner for both WT and KO cultures, consistent with increased maturation and the onset of myelination (Figs 2B and EV3).
Figure 2. DR6 negatively regulates Schwann cell number and myelination in the PNS in vitro .

- Increased myelination in DR6 KO DRG cultures. DR6 KO cultures showed an increased number of myelin segments (MBP‐positive segments (seg.), green) per DRG. In contrast, the length of the myelin segment (MBP segment length) was unchanged between the two genotypes. Data are normalized to the mean of WT control. Neuronal marker β3‐tubulin is shown in red (WT, n = 5; KO, n = 4; scale bar = 500 μm).
- DR6 negatively regulates SC proliferation in vitro. SC proliferation was assessed by determining the number of cells double‐positive for plp‐GFP (green) and BrdU (magenta) divided by the number of total plp‐GFP‐positive cells. Quantified numbers and ratios of the representative pictures are shown at the upper left corner. Neuronal marker β3‐tubulin is shown in red (WT, n = 5; KO, n = 4; scale bar = 100 μm).
Figure EV3. Time course of SC proliferation in DR6 KO cultures.

BrdU proliferation assay of SCs in WT and DR6 KO DRGs along the myelination period (t = days of ascorbic acid (AA) treatment to induce myelination). During the first week of AA treatment, quantification of plp‐GFP (green)‐ and BrdU (magenta)‐double‐positive cells shows a 1.4‐ to 2.5‐fold increase in proliferating SC in KO cultures in comparison with WT controls. Neurons are labeled with β3‐tubulin (WT, n = 5; KO, n = 4; scale bar = 100 μm, quantification shown in Fig 2B; t = 0, the same as in Fig 2B).
To corroborate our in vitro findings from the above also in vivo, we analyzed whether an increased number of myelinated segments and SCs is also detected in the PNS of DR6 KO mice. First, we analyzed the number of myelinated fibers using toluidine blue staining in sciatic nerve sections at three different postnatal stages, that is, postnatal day 1 (P1, neonatal), P7 (young), and P21 (adolescent; Fig 3A). At P1, the number of myelinated fibers per area was increased around twofold in the DR6 KO nerve, which is in agreement with the DRG experiments. At P7 and P21, the number of myelinated fibers per area was increased compared to P1, but was no longer different between WT and DR6 KO, indicating that DR6 deficiency induces a precocious myelination in early postnatal development. Importantly, using electron microscopy in sciatic nerve sections at P7 (Fig 3B), the overall axon diameter and averaged g‐ratios were not significantly altered at P7 between WT and DR6 KO sciatic nerves (Fig 3C), demonstrating that DR6 deficiency does not induce hypermyelination. The mild, but not significant increase of the averaged g‐ratio (0.718 in WT versus 0.734 in DR6 KO) was particularly seen for axons with large diameters (> 3 μm) but not for axons with smaller diameters (Fig 3D). Additionally, there was a mild increase in the percentage of axons with larger diameters (> 3 μm) among the myelinated axons in DR6 KO as compared to WT (Fig 3E). It is possible that the axon diameters increase even further in adulthood as recently described (Gamage et al, 2017).
Figure 3. DR6 deficiency leads to precocious myelination in the PNS in vivo .

-
ASemithin sections of P1, P7, and P21 WT and DR6 KO sciatic nerves (scale bar = 10 μm). The number of myelinated fibers per area significantly increased in P1 KO mice (n = 8) as compared to littermate WT mice (n = 4). Examples of myelinated fibers in P1 mice are indicated by magenta arrowheads. Differences in the number of myelinated fibers were no longer seen in later developmental stages at P7 (WT, n = 8; KO, n = 6) or at P21 (WT, n = 3; KO, n = 4). Data were normalized to the mean of WT at P1.
-
B, CElectron micrographs of P7 WT and DR6 KO sciatic nerves (scale bar = 3 μm). The averaged axon diameter and g‐ratio were not altered in P7 DR6 KO mice (n = 7) as compared to littermate WT mice (n = 8). Data were obtained from more than 700 myelinated axons per genotype.
-
Dg‐ratio (y‐axis) was plotted against axon diameter (x‐axis). Lines represent the linear regression of obtained data points and show a significant difference in slopes between WT and DR6 KO (P = 0.0378). At P7, DR6 KO mice have increased g‐ratios for axons with larger diameters.
-
EThe distribution of myelinated fibers in P7 WT and DR6 KO mice. KO mice showed an increased number of myelinated axons with larger diameters (> 3 μm) as compared to WT mice.
Next, we sought to determine alterations in the number of SCs and their morphology during the early myelination phase. To this end, we took advantage of the neuromuscular junction (NMJ), which is a large PNS synapse with non‐myelinating, terminal SCs on the NMJ and myelinating SCs along the axons (axonal SCs) and a well‐established model to detect differences in the onset of myelination and glial coverage (Brill et al, 2011). Using plp‐GFP mice, where all SCs are labeled, we quantified the number of terminal SCs located on the NMJ, the length of axonal SCs—which is a measure of the internodal length—and the onset of myelination on terminal motor axon branches in DR6 KO versus WT littermate controls. Because myelination of the terminal branches at the NMJ starts in the second postnatal week, we omitted the neonatal time point and chose P9/P11 as the time point of the young age. P21 was used as the adolescent time point, which is identical to the toluidine blue staining in the sciatic nerve. At this time point, myelination of the terminal branches at the NMJ is completed. We found that in DR6 KO mice at the young time point P11, the number of terminal SCs was increased by about 20% per NMJ (the latter were not changed in size; Fig 4A and B). At the same time, the length of axonal SCs on terminal branches was decreased by about 25% in DR6 KO mice (Fig 4C), presumably as a consequence of higher SC proliferation on the limiting axonal space available for a larger SC population. Notably, this difference was no longer seen at the adolescent time point (P21; Fig 4C). Moreover, immunostainings for contactin‐associated protein 1 (Caspr1), which labels differentiated paranodal regions around the nodes of Ranvier, revealed that significantly more terminal axon branches showed Caspr1‐positive paranodes at the young time point (P9) in DR6 KO mice (Fig 4D and E), suggesting premature node formation. Yet, at the later adolescent stage (P21), all SCs on terminal axon branches showed Caspr1‐positive paranodes in both WT and DR6 KO mice (Fig 4F). Taken together, these findings indicate that DR6 negatively regulates SC proliferation at early developmental stages in the PNS.
Figure 4. DR6 negatively regulates Schwann cell number and myelination in the PNS in vivo .
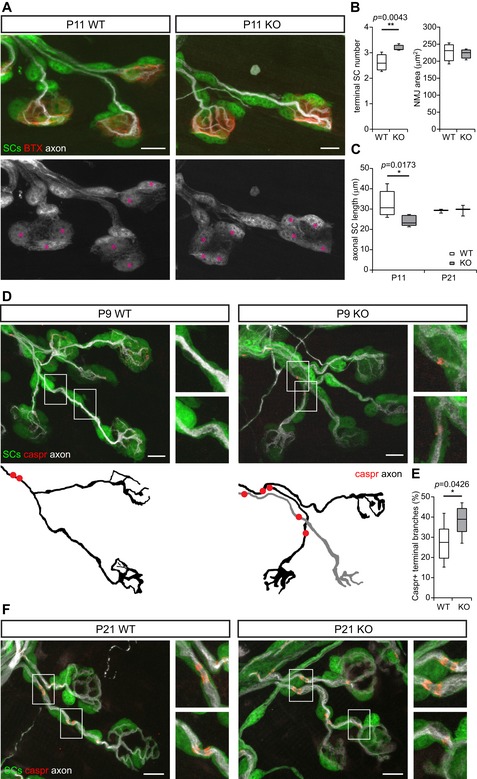
-
ADR6 negatively regulates SC proliferation in vivo. P11 NMJ in a plp‐GFP (SCs, green in top panels, gray below) × DR6 KO or WT triangularis sterni muscle with axons labeled with β3‐tubulin (white) and postsynaptic membrane labeled with α‐bungarotoxin (red). Terminal SCs are marked with magenta asterisks. Scale bars = 10 μm
-
BTerminal SC numbers (left panel) were increased in P11 DR6 KO mice (n = 5) as compared to littermate WT controls (n = 6), whereas the sizes of NMJ (right panel) were not altered. More than 30 NMJs were analyzed per animal.
-
CAxonal SC length was decreased in NMJ of P11 DR6 KO mice (n = 5) as compared to littermate WT controls (n = 6). This difference was no longer significant at P21 (n = 3 for both genotypes).
-
D, EP9 terminal branches in WT or DR6 KO triangularis sterni with paranodes stained for Caspr1 (red) and axons with β3‐tubulin (white). SCs are labeled with plp‐GFP (green). The higher magnification of the regions in white boxes and the outlines of axonal branches are shown on the right and below (D). Quantification of Caspr1‐positive terminal branches showed an increased number in P9 KO samples (E), suggesting an advanced myelination in these mice (WT, n = 6; KO, n = 8; more than 24 NMJs were analyzed per animal; scale bars = 10 μm).
-
FP21 terminal branches in WT or DR6 KO triangularis sterni with paranodes stained for Caspr1 (red) and axons with β3‐tubulin (white). SCs are labeled with plp‐GFP (green). Note that at P21, all branches were positive for Caspr1 (scale bars = 10 μm).
DR6 acts in trans on SCs
That we did not detect DR6 expression in SCs but in neurons (Fig 1B and C) suggests that the neuronally expressed DR6 acts in trans on SCs regulating their proliferation and myelination. This is in clear contrast to the CNS, where DR6 acts as a receptor in a cell‐autonomous fashion in both neurons and oligodendrocytes (Nikolaev et al, 2009; Mi et al, 2011). To confirm our hypothesis that neuronal DR6 can act in trans on SCs, full‐length DR6 was lentivirally transduced into neurons of DR6 KO DRG cultures (Figs 5A and EV4) driven by the neuron‐specific synapsin promoter. This approach reduced the increased number of myelinated segments in DR6 KO DRGs (Fig 5A), demonstrating that neuronally expressed DR6 is sufficient to rescue the KO phenotype. Strikingly, neuronal expression of a DR6 mutant, which lacks the cytoplasmic death domain (DR6 ΔC) required for the previously described cell‐autonomous receptor function of DR6, also sufficed to rescue the phenotype of DR6 KO DRGs (Fig 5B). This result indicates that the ectodomain of DR6 is the main functional element to regulate SC proliferation in the PNS.
Figure 5. DR6 acts in trans on SCs.

- Neuronal DR6 expression negatively regulates myelination in DR6 KO DRGs. DR6 KO cultures transduced with a lentiviral vector expressing full‐length DR6 (DR6, n = 5) tagged with DsRed under a neuronal promoter (synapsin) showed a significantly reduced number of myelinated segments (seg.). A vector expressing DsRed only was used as control (CTR, n = 5).
- Neuronal expression of mutant DR6 lacking the cytoplasmic domain (DR6 ΔC, n = 15) still reduces the number of myelinated segments (MBP in magenta) in DR6 KO DRGs similar to the effect of full‐length DR6 (DR6 FL, n = 11). A vector expressing DsRed only was used as control (CTR, n = 15). Scale bars = 200 μm.
Figure EV4. Neuronally expressed DR6 rescues myelin phenotype.

- DRG cultures transduced with lentiviral vector encoding for DsRed (red) under a neuronal‐specific promoter (synapsin), showing that the construct is expressed in neurons (labeled with β3‐tubulin, upper panels, green) but not in SCs (expressing plp‐GFP, lower panels, green). Scale bar = 10 μm.
- DRG cultures without viruses (upper panels) or transduced with lentiviral vectors encoding DR6 ΔC‐DsRed (middle panels) and DR6 FL‐DsRed (bottom panels) under the synapsin promoter. Note that both DR6 ΔC‐DsRed and DR6 FL‐DsRed (red) are expressed in axons (labeled with β3‐tubulin, green). Scale bar = 10 μm.
- Western blot analysis of lentivirally transduced DRG cultures from WT and DR6 KO mice. DR6 ΔC‐DsRed (lysates), DR6 FL‐DsRed (lysates), and sDR6 (media) were detectable in both genotypes with the 6B6 antibody. Additionally, WT samples showed bands corresponding to endogenous DR6 FL and its shed ectodomain. DsRed construct alone was used as control.
Source data are available online for this figure.
sDR6 is a novel paracrine molecule regulating SC proliferation
The findings that (i) DR6 acts in trans on SCs and (ii) DR6 is converted to sDR6 suggest that the soluble DR6 ectodomain may act as a novel paracrine molecule and is sufficient to rescue the increased number of myelinated segments seen in the DR6 KO DRGs in vitro. To test this possibility, we incubated DRG cultures with murine recombinant sDR6. Indeed, sDR6 reduced the number of myelinated segments in DR6 KO DRGs to WT levels (Fig EV5), consistent with a reduced number of myelinating SCs upon sDR6 treatment. Interestingly, sDR6 also further reduced the number of myelinated segments in the WT DRGs, demonstrating that the endogenously produced sDR6 levels control, but do not fully block the level of myelination. To further verify that sDR6 is sufficient to rescue the increased number of myelinated segments in the DR6 KO condition, DR6 KO DRGs were incubated with conditioned medium from WT (containing endogenous sDR6; see Fig EV4C) or DR6 KO DRGs (Fig 6A). Importantly, only the WT conditioned medium restored the WT phenotype in the DR6 KO DRGs by reducing the number of myelinated segments. This function of sDR6 was also confirmed by analyzing the number of proliferating SCs in the BrdU assay (Fig 6B), where again the WT, but not the DR6 KO, conditioned medium restored the number of proliferating SCs. Taken together, these results demonstrate that sDR6 is sufficient for the function of DR6 as a negative regulator of SC proliferation and myelination.
Figure EV5. Rescue of knockout phenotype by sDR6.

In order to test whether sDR6 is able to rescue the myelination phenotype, WT and DR6 KO cultures were incubated with complete medium supplemented with 1 μg/ml murine recombinant sDR6. Quantification of myelin segments (seg., MBP‐positive, green) showed that the treatment with sDR6 was sufficient to downregulate myelination. Neurons were labeled with β3‐tubulin (red). Myelin segment quantification is represented using box plots showing the first and the third quartile together with the median. The whiskers show the maximum and the minimum data point. n = 6 for each group; scale bar = 500 μm.
Figure 6. Soluble DR6 ectodomain is sufficient to rescue the DR6 KO phenotype.
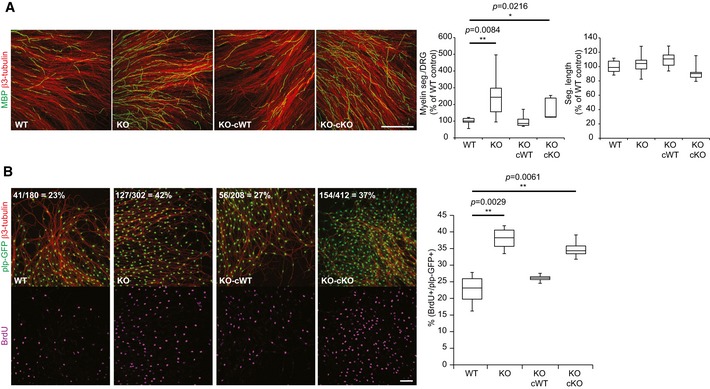
-
A, BIncubation of DR6‐deficient (KO) DRGs with WT, but not KO, conditioned medium reduced the number of myelin segments (seg.) (A; MBP in green; WT, n = 7; KO, n = 8; KO cWT, n = 9; KO cKO, n = 5) and proliferating SCs (B; plp‐GFP in green; BrdU in magenta, n = 4 for each group) compared to WT controls. β3‐Tubulin (red) was used as a neuronal marker (scale bar: 500 μm in A and 100 μm in B). Quantification results of the representative pictures are shown at the upper left corner.
Discussion
Death receptor 6 is established as a receptor controlling cell death and differentiation in a cell‐autonomous manner in different cell types including neurons, oligodendrocytes, endothelial cells, and immune cells (Zhao et al, 2001; Schmidt et al, 2003; DeRosa et al, 2008; Nikolaev et al, 2009; Mi et al, 2011; Strilic et al, 2016). Our study demonstrates for the first time an additional, but non‐cell‐autonomous function for DR6, where the soluble DR6 ectodomain (sDR6) is released from neurons and acts in trans on SCs to suppress their proliferation and thereby myelination in the PNS (Fig 7). Thus, in this setting sDR6 acts in a manner similar to cytokines (e.g., TNF), which also activate other cells in trans. Whether sDR6 acts through a receptor on SCs or by sequestering a ligand which otherwise would stimulate SC proliferation requires further studies.
Figure 7. sDR6 negatively modulates SC proliferation and myelination.

Neuronally expressed full‐length DR6 (DR6 FL) can be cleaved by the metalloprotease ADAM10, thus releasing its soluble ectodomain (sDR6) into the extracellular environment (1). sDR6 can act in trans on SCs inhibiting their proliferation in an early developmental stage and consequently negatively regulates the onset of myelination. In DR6 KO mice, a higher SC number in an early developmental stage leads to precocious myelination (2). Re‐introduction of sDR6 alone into DR6 KO cultures is sufficient to rescue the phenotype (3).
The novel signaling function is based on several experimental findings. First, DR6 works as negative regulator of PNS myelination both in vitro and in vivo. Second, we found DR6 to be expressed in neurons, but not in SCs. Third, DR6 expression selectively in neurons of DRG cultures deficient in DR6 was sufficient to restore the WT phenotype. Fourth, the same result was obtained when a deletion mutant of DR6 was expressed which lacks the cytoplasmic death domain required for the previously established receptor function. Thus, even if low amounts of DR6 were expressed in SCs, the signaling mechanism would be independent of the death domain and consequently of the receptor function, consistent with the novel, non‐cell‐autonomous signaling mechanism. Fifth, the soluble DR6 ectodomain, which we found to be physiologically released by ADAM10 and other proteases, was sufficient to rescue the increased number of myelinated segments and enhanced SC proliferation of DR6 KO DRGs. This effect was seen both with recombinant sDR6 and in a medium‐transfer experiment, where endogenous sDR6 was transferred.
Our study establishes that DR6 deficiency leads to precocious myelination in the PNS. While this phenotype is similar to the CNS, as reported previously (Mi et al, 2011), the underlying molecular mechanisms are clearly different. For the PNS, we report a non‐cell‐autonomous mechanism of DR6 signaling, whereas in the CNS, DR6 acts in cis in a cell‐autonomous manner in oligodendrocytes (Mi et al, 2011). There are additional differences between the PNS and the CNS with regard to early stages of myelination. DR6 deficiency did not alter the g‐ratio in the PNS of very young mice at P7 in our study, but lowered the g‐ratio in the CNS by 15% at P15 (Mi et al, 2011). Notably, this reduction of the g‐ratio is not anymore seen in adult DR6 KO mice both in the CNS (Mi et al, 2011) and in the PNS (Gamage et al, 2017).
The DR6 signaling mechanism being different between PNS and CNS is reminiscent of neuregulin‐1, which is required for myelination in the PNS, but not in the CNS (Brinkmann et al, 2008; Luo et al, 2011; Fleck et al, 2013). Given that there is not yet an extensive literature on DR6 function and signaling, it is possible that the non‐cell‐autonomous signaling function of DR6 will also be found in additional tissues besides the PNS.
The finding that ADAM10 is the major, but not the only DR6 protease is consistent with previous reports that myelination in vivo or in DRG cultures is not significantly altered upon inhibition or deficiency of ADAM10 (Freese et al, 2009; Jangouk et al, 2009; Luo et al, 2011). In particular, g‐ratios were unchanged in sciatic nerves of both newborn and adult mice upon ADAM10 deletion in either neurons or Schwann cells (Meyer zu Horste et al, 2015). We found that ADAM10 mediates 50% of total sDR6 release, while release of the remaining 50% must be catalyzed by other proteases upon ADAM10 deficiency. These 50% may be sufficient to allow a relatively normal PNS myelination.
In the nervous system, ADAM10‐deficient mice show several phenotypes, such as epileptic seizures, learning deficits, altered spine morphology, and defective synaptic functions (Prox et al, 2013). While the contributing substrates remain largely unknown, it is clear that ADAM10 has multiple substrates in the nervous system, as shown in a previous proteomic study using ADAM10 KO neurons (Kuhn et al, 2016). In that study, cleavage of several ADAM10 substrates, including NCAM1 and neuroligin‐1, was nearly completely abolished in the absence of ADAM10. In contrast, the soluble ectodomain of most substrates was still released to a significant extent, for example, 69% for APP (Kuhn et al, 2016), demonstrating that many membrane proteins in the nervous system undergo shedding by multiple proteases. This is in good agreement with our finding that ADAM10 mediates 50% of total DR6 cleavage, but that other proteases also contribute. Similar results for substrate cleavage by different proteases have also been reported for other proteases, including the Alzheimer's disease‐related protease BACE1 (Kuhn et al, 2012; Dislich et al, 2015), but the functional consequences of different proteolytic cleavages have in most cases not yet been explored. Importantly, substrate cleavage may occur by different proteases in a tissue‐specific manner. For example, APP and the cell adhesion proteins L1 and CHL1 are mostly cleaved by BACE1 in nervous tissue, but mostly by ADAM proteases outside of the nervous system (Naus et al, 2004; Maretzky et al, 2005; Kuhn et al, 2012). A similar tissue‐specific cleavage pattern appears to be true for DR6. We report here that in the nervous system, DR6 is a major substrate for ADAM10, but not for MT1‐MMP, whereas a previous report demonstrated DR6 shedding by MT1‐MMP in breast cancer cells, at least upon overexpression of MT1‐MMP (Tam et al, 2004; DeRosa et al, 2008).
Death receptor 6 stands out from other TNFRSF members in that it can act as a receptor or—as shown here—as a putative ligand. Another example for dual functions is APP, which can act as a putative cell adhesion protein at the NMJ or as a proteolytically released paracrine signaling molecule controlling spine density (Wang et al, 2007; Weyer et al, 2014). It is tempting to speculate that other TNFRSF members besides DR6 may have similar dual functions which, however, still need to be uncovered.
Materials and Methods
Animal procedures
Both male and female mice were used. Animals had access to water and food ad libitum in a 12/12‐h light–dark cycle in IVC System. All experimental procedures on animals were performed in accordance with the European Communities Council Directive (2010/63/EU) and in compliance with the German animal welfare law.
DR6 antibody generation
Antibodies against full‐length murine DR6 ectodomain were produced as previously published (Kuhn et al, 2016). Briefly, murine DR6 ectodomain with an N‐terminal CD5 signal peptide and a C‐terminal 2XStrepII tag was transiently expressed in HEK293T cells. Recombinant ectodomain was purified from conditioned media using Strep‐Tactin Sepharose (IBA GmbH). Purified fusion protein was injected into LOU/C rats for immunization. Hybridoma cell line supernatants were used for antibody validation and identification of positive clones in ELISA and Western blot analysis. DR6 clone 6B6 (rat IgG2b/k) was subcloned twice by limiting dilution and used in this study. While this antibody works for Western blots, it does not work in immunostainings.
DRG cultures—myelination assay
E13.5 dorsal root ganglia (DRGs) were explanted from B6‐tnfrsf21tm1Dxt (DR6 KO) × plp1‐GFP mice (expressing GFP under the plp1 promoter) and plated in IBIDI μ‐slide previously coated with collagen and poly‐d‐lysine. DR6 KO mice were kindly provided by Genentech Inc., San Francisco (Zhao et al, 2001); plp‐GFP mice were kindly provided by Wendy Macklin (Mallon et al, 2002). After explant, cells were cultured in DMEM‐high glucose plus GlutaMAX, FBS 10%, Pen/Strep 1%, and 50 ng/ml nerve growth factor (NGF). On day 3 in vitro (3 DIV), medium was changed completely to induce myelination with Neurobasal, B27 supplement 2%, 4 g/l d‐glucose, 2 mM glutamine, 50 ng/ml NGF, and 50 μg/ml ascorbic acid. On 21 DIV, cells were fixed with PFA 4%/sucrose 5%. Fixed DRG cultures were permeabilized with 100% ice‐cold methanol for 10 min. Samples were blocked with PBS/5% normal goat serum (NGS). To detect myelin, an antibody against MBP was used (#SMI‐94R; Covance), while neuronal cells were stained using an antibody against β3‐tubulin (#5568; Cell Signaling). Secondary Alexa antibodies were obtained from Invitrogen. Each well of a μ‐slide was examined by confocal microscopy on a Leica SP5 confocal microscope using a tiled mosaic acquisition protocol. Myelin was quantified by counting the number of MBP‐positive segments and measuring their length using the straight line and the cell counter plug‐ins of the ImageJ software (NIH). Quantification was performed blinded to sample genotype and based on at least four DRGs per group (n = 4) from at least two independent litters.
BrdU assay—time course
Dorsal root ganglia cultures were incubated at different time points (0, 1, 3, 7, and 14 days of myelin induction) in complete medium including 3 μg/ml BrdU reagent (Invitrogen) for 4 h. After BrdU incorporation, cells were washed and fixed with 4% PFA/5% sucrose, treated with 2 N HCl, and blocked with PBS/5% NGS. For BrdU detection, an antibody against BrdU conjugated to Alexa 647 (#B35133; Invitrogen) was used. Each well of a μ‐slide was examined by a Leica SP5 confocal microscope. To quantify proliferating SCs, the number of cells double‐positive for BrdU and plp‐GFP (proliferating SC number) was normalized to the total number of plp‐GFP‐positive cells (total SC number) using the cell counter plug‐in of the ImageJ software (NIH). Quantification was based on at least four DRGs per group from at least two independent litters.
Myelin phenotype rescue (recombinant ectodomain)
Murine recombinant full DR6 ectodomain was purified according to Kuhn et al (2016). To rescue the KO phenotype, DR6 KO and WT DRG cultures were incubated with complete medium supplemented with 1 μg/ml ectodomain during the myelination induction (from 3 DIV to 21 DIV). As negative control, the elution buffer used for the ectodomain purification was used. Myelin quantification was performed as previously described. Quantification was based on six DRGs per group from at least two independent litters.
Myelin phenotype rescue (conditioned media)
To rescue the KO phenotype, DR6 KO DRG cultures were incubated with 48‐h conditioned media from DR6 WT cultures during the myelination induction (from 3 DIV to 21 DIV). As negative control, media from DR6 KO cultures were used. Myelin quantification was performed as described above. Quantification was based on at least five DRGs per group from at least two independent litters.
For the BrdU assay, myelin induction was stopped after 3 days (t = 3 d) and cultures were analyzed in order to obtain a reasonable number of BrdU‐positive SCs. Proliferating SCs were quantified as previously described. Quantification was based on at least four DRGs per group from at least two independent litters.
Myelin phenotype rescue (DR6 overexpression in neurons)
DsRed, DR6 full‐length, or DR6 delta cytoplasmic mutant (UniProtKB: Q9EPU5 aa 1–383) C‐terminally tagged with DsRed was overexpressed in DRG neurons under the human synapsin promoter using the lentivirus system. Murine DR6 full‐length cDNA was PCR‐amplified and cloned into the BamHI site of pLV‐hSyn‐DsRed vector (Addgene #22909; Nathanson et al, 2009). cDNA of the DR6 delta cytoplasmic mutant (ΔCyto DR6) was cloned into the EcoRI/BamHI sites of pLV‐hSyn‐DsRed vector. Lentiviral particles were purified as previously described (Kuhn et al, 2010) with minor modifications. To increase the viral titer, the medium of HEK293T cells was replaced with packaging medium (DMEM + 2% FCS, 10 mM sodium butyrate) 24 h after transfection and the conditioned medium was collected 48 h after transfection. DRG explants were infected with lentivirus at 7 DIV. At 21 DIV, myelinating SCs were quantified as previously described. Quantification was based on at least five DRGs per group from at least two independent litters.
PNS DR6 shedding ex vivo analysis
E13.5 dorsal root ganglia together with peripheral nerves (n = 4) were dissected and subsequently homogenized in order to obtain soluble and membrane fractions as previously described (Kuhn et al, 2012). Briefly, the soluble fraction was generated by homogenizing fresh tissue in ice‐cold diethyl amine (DEA) buffer. After centrifugation, supernatant (soluble fraction) underwent clarifying ultracentrifugation. Pellets (membrane fraction) were further lysed in lysis buffer (150 mM NaCl, 50 mM Tris pH 7.5, 2 mM EDTA) with 1% Triton X‐100. Both fractions were analyzed on 8% acrylamide gels in non‐reducing conditions and blotted on PVDF membrane. DR6 full‐length and ectodomain were detected using an antibody against DR6 N‐terminus (clone 6B6). Anti‐rat HRP‐conjugated secondary antibodies were purchased from Santa Cruz Biotech. DR6 KO samples were used as negative controls for antibody validation.
Endogenous DR6 sheddase identification
Embryonic primary cortical neurons from WT and ADAM10 conditional KO mice (A10cKO; kindly provided by Howard Carwford; Gibb et al, 2010) were prepared as previously described (Colombo et al, 2007). Briefly, cortex samples from E16.5 embryos were collected and enzymatically dissociated with papain (Sigma‐Aldrich). Plating medium was B27/Neurobasal (Gibco) supplemented with 0.5 mM glutamine and 1% penicillin–streptomycin. Knockout of ADAM10 was induced in A10cKO neurons through infection with lentiviral vector expressing Cre recombinase as previously described (Colombo et al, 2013; n = 8 from four independent cultures). To knock down MT1‐MMP in WT primary neurons, specific short hairpin RNAs (Sh385: CCCCGGACTGAGATCAAGGCCAATGTTCAAGAGACATTGGCCTTGATCTCAGTCCTTTTT; and sh386: CCCCGGATGGACACAGAGAACTTCGTTCAAGAGACGAAGTTCTCTGTGTCCATCCTTTTT) were expressed in neuronal cells using lentiviral vectors as previously described (Kuhn et al, 2010; n = 6 from three independent cultures). An expression vector containing a scrambled shRNA sequence was used as control.
Additionally, WT neurons were treated with the broad‐spectrum metalloprotease inhibitor TAPI‐I (50 μM; Peptides International) for 24 h before harvesting (n = 4 from two independent cultures). At 8 DIV, cells were lysed in lysis buffer and the 48‐h conditioned media were collected. Samples were analyzed by Western blotting, and full‐length DR6 was detected in the lysates using the 6B6 antibody. In order to detect DR6 ectodomains released in the media, samples were enriched for glycosylated proteins using concanavalin A (Sigma‐Aldrich) and then detected by Western blotting using the 6B6 antibody. Downregulation of secretases was confirmed in lysates using specific antibodies against ADAM10 (#MAB1427; R&D) and MT1‐MMP (#EP1264Y; Abcam). Blots were developed using horseradish peroxidase‐conjugated secondary antibodies and the ECL chemiluminescence system. An antibody against calnexin (#ADI‐SPA‐860; Stressgen) was used as loading control.
Cell‐free ADAM10 cleavage assay
Human full‐length DR6 with HA tag at N‐terminus (HA‐DR6) was stably overexpressed in HEK293E cells (ATCC) and subsequently purified from cell lysates using HA‐conjugated beads (clone HA‐7; Sigma‐Aldrich). Human recombinant ADAM10 (R&D Systems) was incubated O/N with HA‐DR6 at 37°C in the assay buffer (25 mM Tris, 2 μM ZnCl2, 0.005% Brij‐35, pH 9.0). Samples were analyzed on 8% acrylamide gels and blotted on PVDF membrane. DR6 full‐length and ectodomain were detected using antibody against HA tag (clone HA‐11; Covance). ADAM10 alone was used as an additional negative control. Similar results were obtained in three independent experiments.
DR6 expression analysis
Western blot analysis was performed on cell lysates of WT primary cortical neurons and SCs using the 6B6 antibody as described above. Additionally, murine embryonic fibroblast (MEF) lysate was included to exclude that DR6 detection was due to a minor population of MEFs in DRG cultures. MEFs were isolated from WT E13.5 C57BL/6 embryos, cultured, and immortalized as described earlier (Xu, 2005). DR6 KO neuronal lysates were used as the negative control to validate the specificity of the DR6 band. Similar results were obtained in four independent experiments.
Moreover, DR6 mRNA levels were measured by quantitative real‐time PCR as previously described (Kuhn et al, 2010). Briefly, total RNA was extracted using RNeasy Mini kit (Qiagen) from primary neurons and SCs following the manufacturer's instructions. Concentrations and purities of total RNA were spectrophotometrically assessed at 260 and 280 nm, and total RNA was reverse‐transcribed into cDNA. Real‐time PCR was carried out on a 7500 Fast Real‐Time PCR machine (ABI) with the Power SYBR® Green PCR Master Mix (ABI). DR6 primer sequences were as follows: CAGAACCGGGAGAAATGGATC (F); CTGATAGATGTCCTTCCACTGG (R). The relative expression of DR6 mRNA was normalized to the reference gene (actin).
Primary SC cultures
Sciatic nerves from P3 C57Bl6/J were collected in Leibovitz medium L‐15 (Gibco). Tissue was digested in 1% collagenase (Cultrex) and 2.5% trypsin (Gibco). Single‐cell suspension was plated in poly‐d‐lysine‐coated plates in the culture medium: DMEM‐high glucose plus GlutaMAX (Gibco), 10% FBS, and 1% Pen/strep (Gibco). Proliferating cells were eliminated by incubating primary cultures with 10 μM ARA‐C (Gibco) for 48 h. After the recovery phase in fresh culture medium, cells were lysed for protein and RNA analysis.
Postnatal neuromuscular junction (NMJ) analysis
Immunohistochemistry was performed as described previously (Brill et al, 2011, 2016) using postnatal mice (P9, P11, or P21). The whole thorax was excised as previously described and fixed in 4% paraformaldehyde (PFA) diluted in 0.1 M phosphate buffer (PB; Kerschensteiner et al, 2008; Brill et al, 2013). The triangularis muscle was dissected (Brill et al, 2013) and incubated overnight (4°C) in blocking solution (5% BSA, 0.5% Triton X‐100 in 0.1 M PB) with the following primary antibodies or dyes: anti‐β3‐tubulin conjugated to Alexa 555 (BD Pharmingen); Alexa 647‐conjugated α‐bungarotoxin (Invitrogen); and anti‐Caspr1 (Abcam 34151). Muscles were washed in 0.1 M PB and, in case of the Caspr1 staining, incubated for 1 h at room temperature with an anti‐rabbit secondary antibody coupled to Alexa 647 (Invitrogen). Muscles were washed again in 0.1 M PB and mounted in DAPI Fluoromount‐G (SouthernBiotech) or Vectashield (Vector Laboratories). Z‐stack images were scanned using a confocal microscope (FV1000; Olympus). For figure representation, different channels of confocal image series were combined in pseudo‐color using the “screen” function in Adobe Photoshop and gamma‐adjusted to enhance low‐intensity objects.
The number of SCs is determined by counting the DAPI‐stained nuclei co‐localizing with plp‐GFP (SC marker) signals in association with axons. Terminal SCs were defined by association with the bungarotoxin‐stained postsynaptic sites of the NMJ, and axonal SCs as being associated with the terminal motor axon branch ending on an NMJ. Length of the axonal SCs was measured with the “segmented line” function of Fiji (littermate controlled, n ≥ 5 mice per group, n ≥ 30 NMJ per mouse). Nodal development was determined by the presence of Caspr1‐positive nodes on the terminal branches (littermate controlled, n ≥ 6 mice per group, n ≥ 24 singly innervating branches per mouse).
Semithin and ultrathin sections of sciatic nerves
Semithin and ultrathin sections were conducted as previously described (Quattrini et al, 1996) and obtained from tissues fixed in 2% glutaraldehyde (Sigma‐Aldrich) in 0.12 M phosphate buffer, postfixed in 2% osmium tetroxide (Electron Microscopy Sciences), and embedded in Epon (Sigma‐Aldrich). Semithin sections (1,000 nm thick) were stained with 0.5% toluidine blue and examined by light microscopy (Olympus BX51). Digitized semithin section images were acquired with a digital camera (Leica DFC310 FX) using a 100× objective and were then matched to reconstruct the entire nerve. Ultrathin sections (100–120 nm thick) were stained with uranyl acetate and lead citrate and examined by electron microscopy (Leo 912 Omega).
γ‐Secretase cleavage assay
Human full‐length DR6 with Flag tag at the C‐terminus was transiently overexpressed in HEK293E cells. Forty‐eight hours after transfection, cells have been treated either with γ‐secretase inhibitor DAPT (1 μM; Sigma‐Aldrich) or with proteasome inhibitor MG132 (1 μM; Sigma‐Aldrich) for 24 h. Cells were lysed and samples were analyzed on a 12% acrylamide gel followed by Western blotting using an antibody against the Flag tag (#F3165, Flag M2; Sigma‐Aldrich). Similar results were obtained in four independent experiments.
Statistical analysis
For comparison between two groups, unpaired Student's t‐test with two‐tailed distribution was used except for the NMJ analysis (Fig 4; Mann–Whitney test for two samples). For comparison between more than two groups (Fig 5B), one‐way ANOVA followed by Dunnett's multiple comparisons test was used. Fisher's exact test was used on the contingency table with exact numbers (Fig 3E). Data are represented using box plots showing the first and the third quartile together with the median. The whiskers show the maximum and the minimum data point. A value of P < 0.05 was considered significant (*P < 0.05; **P < 0.01; ***P < 0.001).
Author contributions
SFL, AC, H‐EH, TM, and CT designed the experiments. SFL, AC, and H‐EH wrote the manuscript. AC and H‐EH performed the in vitro analysis, MW and MSB the NMJ analysis, and PC and CT the EM study and toluidine stainings. P‐HK, AC, and RF generated the anti‐DR6 antibody (6B6).
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting information
Expanded View Figures PDF
Source Data for Expanded View
Review Process File
Source Data for Figure 1
Acknowledgements
We thank Katrin Moschke for technical support, Paola Podini for technical assistance with electron microscopy and Manuela Budak for animal husbandry. This work was supported by the DFG (SyNergy EXC1010, CIPSM EXC114, FOR2290, SFB870), IWT, the Israel‐Helmholtz Program, the Centers of Excellence in Neurodegeneration Program, and the Breuer Foundation Research Award. T.M.'s work was further supported by the German‐Israeli Foundation (I‐1200‐237.1/2012), the European Research Council under the European Union's Seventh Framework Program (FP/2007‐2013; ERC Grant Agreement no. 616791 with T.M.), and DFG Research grants Mi 694/7‐1 and 8‐1. M.W. was supported by the Graduate School of the Technische Universität München (TUM‐GS) and received a fellowship from TUM School of Medicine. CT's team work was supported by Telethon Foundation (GPP14040) and the Italian Minister of Health (PE‐2013‐02355206).
The EMBO Journal (2018) 37: e97390
References
- Aggarwal BB (2003) Signalling pathways of the TNF superfamily: a double‐edged sword. Nat Rev Immunol 3: 745–756 [DOI] [PubMed] [Google Scholar]
- Brill MS, Lichtman JW, Thompson W, Zuo Y, Misgeld T (2011) Spatial constraints dictate glial territories at murine neuromuscular junctions. J Cell Biol 195: 293–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brill MS, Marinkovic P, Misgeld T (2013) Sequential photo‐bleaching to delineate single Schwann cells at the neuromuscular junction. J Vis Exp 71: e4460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brill MS, Kleele T, Ruschkies L, Wang M, Marahori NA, Reuter MS, Hausrat TJ, Weigand E, Fisher M, Ahles A, Engelhardt S, Bishop DL, Kneussel M, Misgeld T (2016) Branch‐specific microtubule destabilization mediates axon branch loss during neuromuscular synapse elimination. Neuron 92: 845–856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinkmann BG, Agarwal A, Sereda MW, Garratt AN, Muller T, Wende H, Stassart RM, Nawaz S, Humml C, Velanac V, Radyushkin K, Goebbels S, Fischer TM, Franklin RJ, Lai C, Ehrenreich H, Birchmeier C, Schwab MH, Nave KA (2008) Neuregulin‐1/ErbB signaling serves distinct functions in myelination of the peripheral and central nervous system. Neuron 59: 581–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombo A, Repici M, Pesaresi M, Santambrogio S, Forloni G, Borsello T (2007) The TAT‐JNK inhibitor peptide interferes with beta amyloid protein stability. Cell Death Differ 14: 1845–1848 [DOI] [PubMed] [Google Scholar]
- Colombo A, Wang H, Kuhn PH, Page R, Kremmer E, Dempsey PJ, Crawford HC, Lichtenthaler SF (2013) Constitutive alpha‐ and beta‐secretase cleavages of the amyloid precursor protein are partially coupled in neurons, but not in frequently used cell lines. Neurobiol Dis 49: 137–147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeRosa DC, Ryan PJ, Okragly A, Witcher DR, Benschop RJ (2008) Tumor‐derived death receptor 6 modulates dendritic cell development. Cancer Immunol Immunother 57: 777–787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dislich B, Wohlrab F, Bachhuber T, Muller SA, Kuhn PH, Hogl S, Meyer‐Luehmann M, Lichtenthaler SF (2015) Label‐free quantitative proteomics of mouse cerebrospinal fluid detects beta‐site APP cleaving enzyme (BACE1) protease substrates in vivo . Mol Cell Proteomics 14: 2550–2563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleck D, van Bebber F, Colombo A, Galante C, Schwenk BM, Rabe L, Hampel H, Novak B, Kremmer E, Tahirovic S, Edbauer D, Lichtenthaler SF, Schmid B, Willem M, Haass C (2013) Dual cleavage of neuregulin 1 type III by BACE1 and ADAM17 liberates its EGF‐like domain and allows paracrine signaling. J Neurosci 33: 7856–7869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freese C, Garratt AN, Fahrenholz F, Endres K (2009) The effects of alpha‐secretase ADAM10 on the proteolysis of neuregulin‐1. FEBS J 276: 1568–1580 [DOI] [PubMed] [Google Scholar]
- Fujikura D, Ikesue M, Endo T, Chiba S, Higashi H, Uede T (2017) Death receptor 6 contributes to autoimmunity in lupus‐prone mice. Nat Commun 8: 13957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamage KK, Cheng I, Park RE, Karim MS, Edamura K, Hughes C, Spano AJ, Erisir A, Deppmann CD (2017) Death receptor 6 promotes wallerian degeneration in peripheral axons. Curr Biol 27: 890–896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibb DR, El Shikh M, Kang DJ, Rowe WJ, El Sayed R, Cichy J, Yagita H, Tew JG, Dempsey PJ, Crawford HC, Conrad DH (2010) ADAM10 is essential for Notch2‐dependent marginal zone B cell development and CD23 cleavage in vivo . J Exp Med 207: 623–635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu X, Fan Q, Hou H, Yan R (2016) Neurological dysfunctions associated with altered BACE1‐dependent Neuregulin‐1 signaling. J Neurochem 136: 234–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jangouk P, Dehmel T, Meyer Zu Horste G, Ludwig A, Lehmann HC, Kieseier BC (2009) Involvement of ADAM10 in axonal outgrowth and myelination of the peripheral nerve. Glia 57: 1765–1774 [DOI] [PubMed] [Google Scholar]
- Kallop DY, Meilandt WJ, Gogineni A, Easley‐Neal C, Wu T, Jubb AM, Yaylaoglu M, Shamloo M, Tessier‐Lavigne M, Scearce‐Levie K, Weimer RM (2014) A death receptor 6‐amyloid precursor protein pathway regulates synapse density in the mature CNS but does not contribute to Alzheimer's disease‐related pathophysiology in murine models. J Neurosci 34: 6425–6437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerschensteiner M, Reuter MS, Lichtman JW, Misgeld T (2008) Ex vivo imaging of motor axon dynamics in murine triangularis sterni explants. Nat Protoc 3: 1645–1653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn PH, Wang H, Dislich B, Colombo A, Zeitschel U, Ellwart JW, Kremmer E, Rossner S, Lichtenthaler SF (2010) ADAM10 is the physiologically relevant, constitutive alpha‐secretase of the amyloid precursor protein in primary neurons. EMBO J 29: 3020–3032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn PH, Koroniak K, Hogl S, Colombo A, Zeitschel U, Willem M, Volbracht C, Schepers U, Imhof A, Hoffmeister A, Haass C, Rossner S, Brase S, Lichtenthaler SF (2012) Secretome protein enrichment identifies physiological BACE1 protease substrates in neurons. EMBO J 31: 3157–3168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn PH, Voss M, Haug‐Kroper M, Schroder B, Schepers U, Brase S, Haass C, Lichtenthaler SF, Fluhrer R (2015) Secretome analysis identifies novel signal Peptide peptidase‐like 3 (sppl3) substrates and reveals a role of sppl3 in multiple Golgi glycosylation pathways. Mol Cell Proteomics 14: 1584–1598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn PH, Colombo AV, Schusser B, Dreymueller D, Wetzel S, Schepers U, Herber J, Ludwig A, Kremmer E, Montag D, Muller U, Schweizer M, Saftig P, Brase S, Lichtenthaler SF (2016) Systematic substrate identification indicates a central role for the metalloprotease ADAM10 in axon targeting and synapse function. eLife 5: e12748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lastun VL, Grieve AG, Freeman M (2016) Substrates and physiological functions of secretase rhomboid proteases. Semin Cell Dev Biol 60: 10–18 [DOI] [PubMed] [Google Scholar]
- Laurent SA, Hoffmann FS, Kuhn PH, Cheng Q, Chu Y, Schmidt‐Supprian M, Hauck SM, Schuh E, Krumbholz M, Rubsamen H, Wanngren J, Khademi M, Olsson T, Alexander T, Hiepe F, Pfister HW, Weber F, Jenne D, Wekerle H, Hohlfeld R et al (2015) gamma‐Secretase directly sheds the survival receptor BCMA from plasma cells. Nat Commun 6: 7333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichtenthaler SF, Haass C, Steiner H (2011) Regulated intramembrane proteolysis–lessons from amyloid precursor protein processing. J Neurochem 117: 779–796 [DOI] [PubMed] [Google Scholar]
- Luo X, Prior M, He W, Hu X, Tang X, Shen W, Yadav S, Kiryu‐Seo S, Miller R, Trapp BD, Yan R (2011) Cleavage of neuregulin‐1 by BACE1 or ADAM10 protein produces differential effects on myelination. J Biol Chem 286: 23967–23974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo X, He W, Hu X, Yan R (2014) Reversible overexpression of bace1‐cleaved neuregulin‐1 N‐terminal fragment induces schizophrenia‐like phenotypes in mice. Biol Psychiat 76: 120–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallon BS, Shick HE, Kidd GJ, Macklin WB (2002) Proteolipid promoter activity distinguishes two populations of NG2‐positive cells throughout neonatal cortical development. J Neurosci 22: 876–885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maretzky T, Schulte M, Ludwig A, Rose‐John S, Blobel C, Hartmann D, Altevogt P, Saftig P, Reiss K (2005) L1 is sequentially processed by two differently activated metalloproteases and presenilin/gamma‐secretase and regulates neural cell adhesion, cell migration, and neurite outgrowth. Mol Cell Biol 25: 9040–9053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marik SA, Olsen O, Tessier‐Lavigne M, Gilbert CD (2013) Death receptor 6 regulates adult experience‐dependent cortical plasticity. J Neurosci 33: 14998–15003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIlwain DR, Lang PA, Maretzky T, Hamada K, Ohishi K, Maney SK, Berger T, Murthy A, Duncan G, Xu HC, Lang KS, Haussinger D, Wakeham A, Itie‐Youten A, Khokha R, Ohashi PS, Blobel CP, Mak TW (2012) iRhom2 regulation of TACE controls TNF‐mediated protection against Listeria and responses to LPS. Science 335: 229–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer zu Horste G, Derksen A, Stassart R, Szepanowski F, Thanos M, Stettner M, Boettcher C, Lehmann HC, Hartung HP, Kieseier BC (2015) Neuronal ADAM10 promotes outgrowth of small‐caliber myelinated axons in the peripheral nervous system. J Neuropathol Exp Neurol 74: 1077–1085 [DOI] [PubMed] [Google Scholar]
- Mi S, Lee X, Hu Y, Ji B, Shao Z, Yang W, Huang G, Walus L, Rhodes K, Gong BJ, Miller RH, Pepinsky RB (2011) Death receptor 6 negatively regulates oligodendrocyte survival, maturation and myelination. Nat Med 17: 816–821 [DOI] [PubMed] [Google Scholar]
- Nathanson JL, Yanagawa Y, Obata K, Callaway EM (2009) Preferential labeling of inhibitory and excitatory cortical neurons by endogenous tropism of adeno‐associated virus and lentivirus vectors. Neuroscience 161: 441–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naus S, Richter M, Wildeboer D, Moss M, Schachner M, Bartsch JW (2004) Ectodomain shedding of the neural recognition molecule CHL1 by the metalloprotease‐disintegrin ADAM8 promotes neurite outgrowth and suppresses neuronal cell death. J Biol Chem 279: 16083–16090 [DOI] [PubMed] [Google Scholar]
- Nikolaev A, McLaughlin T, O'Leary DD, Tessier‐Lavigne M (2009) APP binds DR6 to trigger axon pruning and neuron death via distinct caspases. Nature 457: 981–989 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Peschon JJ, Slack JL, Reddy P, Stocking KL, Sunnarborg SW, Lee DC, Russell WE, Castner BJ, Johnson RS, Fitzner JN, Boyce RW, Nelson N, Kozlosky CJ, Wolfson MF, Rauch CT, Cerretti DP, Paxton RJ, March CJ, Black RA (1998) An essential role for ectodomain shedding in mammalian development. Science 282: 1281–1284 [DOI] [PubMed] [Google Scholar]
- Prox J, Bernreuther C, Altmeppen H, Grendel J, Glatzel M, D'Hooge R, Stroobants S, Ahmed T, Balschun D, Willem M, Lammich S, Isbrandt D, Schweizer M, Horre K, De Strooper B, Saftig P (2013) Postnatal disruption of the disintegrin/metalloproteinase ADAM10 in brain causes epileptic seizures, learning deficits, altered spine morphology, and defective synaptic functions. J Neurosci 33: 12915–12928, 12928a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quattrini A, Previtali S, Feltri ML, Canal N, Nemni R, Wrabetz L (1996) Beta 4 integrin and other Schwann cell markers in axonal neuropathy. Glia 17: 294–306 [DOI] [PubMed] [Google Scholar]
- Reiss K, Saftig P (2009) The “A Disintegrin And Metalloprotease” (ADAM) family of sheddases: physiological and cellular functions. Semin Cell Dev Biol 20: 126–137 [DOI] [PubMed] [Google Scholar]
- Saftig P, Lichtenthaler SF (2015) The alpha secretase ADAM10: a metalloprotease with multiple functions in the brain. Prog Neurobiol 135: 1–20 [DOI] [PubMed] [Google Scholar]
- Schmidt CS, Liu J, Zhang T, Song HY, Sandusky G, Mintze K, Benschop RJ, Glasebrook A, Yang DD, Na S (2003) Enhanced B cell expansion, survival, and humoral responses by targeting death receptor 6. J Exp Med 197: 51–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strilic B, Yang L, Albarran‐Juarez J, Wachsmuth L, Han K, Muller UC, Pasparakis M, Offermanns S (2016) Tumour‐cell‐induced endothelial cell necroptosis via death receptor 6 promotes metastasis. Nature 536: 215–218 [DOI] [PubMed] [Google Scholar]
- Tam EM, Morrison CJ, Wu YI, Stack MS, Overall CM (2004) Membrane protease proteomics: isotope‐coded affinity tag MS identification of undescribed MT1‐matrix metalloproteinase substrates. Proc Natl Acad Sci USA 101: 6917–6922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassar R, Kuhn PH, Haass C, Kennedy ME, Rajendran L, Wong PC, Lichtenthaler SF (2014) Function, therapeutic potential and cell biology of BACE proteases: current status and future prospects. J Neurochem 130: 4–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Yang L, Wang Z, Zheng H (2007) Amyolid precursor protein mediates presynaptic localization and activity of the high‐affinity choline transporter. Proc Natl Acad Sci USA 104: 14140–14145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weyer SW, Zagrebelsky M, Herrmann U, Hick M, Ganss L, Gobbert J, Gruber M, Altmann C, Korte M, Deller T, Muller UC (2014) Comparative analysis of single and combined APP/APLP knockouts reveals reduced spine density in APP‐KO mice that is prevented by APPsalpha expression. Acta Neuropathol Commun 2: 36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J (2005) Preparation, culture, and immortalization of mouse embryonic fibroblasts. Curr Protoc Mol Biol Chapter 28: Unit 28.1 [DOI] [PubMed] [Google Scholar]
- Zhao H, Yan M, Wang H, Erickson S, Grewal IS, Dixit VM (2001) Impaired c‐Jun amino terminal kinase activity and T cell differentiation in death receptor 6‐deficient mice. J Exp Med 194: 1441–1448 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Expanded View Figures PDF
Source Data for Expanded View
Review Process File
Source Data for Figure 1


