Abstract
Rapid and continued growth in the generation of glycomic data has revealed the need for enhanced development of basic infrastructure for presenting and interpreting these datasets in a manner that engages the broader biomedical research community. Early in their growth, the genomic and proteomic fields implemented mechanisms for assigning unique gene and protein identifiers that were essential for organizing data presentation and for enhancing bioinformatic approaches to extracting knowledge. Similar unique identifiers are currently absent from glycomic data. In order to facilitate continued growth and expanded accessibility of glycomic data, the authors strongly encourage the glycomics community to coordinate the submission of their glycan structures to the GlyTouCan Repository and to make use of GlyTouCan identifiers in their communications and publications. The authors also deeply encourage journals to recommend a submission workflow in which submitted publications utilize GlyTouCan identifiers as a standard reference for explicitly describing glycan structures cited in manuscripts.
Keywords: database, glycan identifier, GlyTouCan, repository, structure
A bottleneck in glycomics
The inherent complexity of defining glycan structures and the essential importance of understanding how the glycome influences biological function requires the integration of data from multiple disciplines, including cell biology, genetics and molecular biology, as well as structural, analytic and synthetic chemistry. The diverse nature of the experimental approaches underlying these disciplines predicts the need for a common language to communicate glycan structures. In addition, technologic advances across a broad range of analytic approaches, including mass spectrometry, liquid chromatography, capillary electrophoresis and other orthogonal strategies, are generating increasingly expansive glycomic datasets. While this accumulating wealth of diverse datasets presents great opportunities for understanding glycan functions and structural diversity, it also reveals the need for growth in bioinformatic infrastructure to support and enhance data interpretation.
Among “omic” analyses, there is a need for glycomics to catch-up with more established fields like genomics and proteomics. Glycomics currently faces many of the same obstacles that genomics and proteomics resolved successfully in the last two decades. Namely, the adoption of standards for data annotation, data interpretation, data presentation, data archiving and database structure have allowed genomics and proteomics to advance rapidly (MIAME and MIAPE); similar standards for glycomics (MIRAGE) are developing robustly along an accelerating trajectory with the development of MS, glycan array and sample preparation guidelines but are still passing through their early growth stages (Brazmaet al. 2001; Taylor et al. 2007; Kolarich et al. 2013; York et al. 2014; Struwe et al. 2016; Liu et al. 2017).
Major advances in genomics and proteomics were achieved over the last two decades by community acceptance of unique identifiers for genes and proteins. Unique identifiers allow authors to submit and cite unambiguous references to gene sequences, mRNA sequences, translated protein sequences, and explicit protein structures, enhancing the ability of investigators to interrogate published reports and utilize this data to advance their own research. Gene and protein identifiers also facilitated database development by providing interconnectivity and cross-referencing capabilities. Without the development of a similar, broadly accepted infrastructure for submitting and citing glycan structures, the glycomics community will remain handicapped by the need for each individual investigator or groups of investigators to separately generate their own descriptors for publication and communication. While the standardization of graphical representations of glycan structures has enjoyed broad acceptance within the glycomics community (SNFG, symbol nomenclature for glycans), the adoption of SNFG representations for publication does not ensure that glycomics datasets will be accessible or can be interrogated by current or developing database efforts (Varki et al. 2015).
Glycan identifiers, written in a broadly accepted, machine-readable language, will allow authors and investigators to point to explicit glycan structures in their publications and will facilitate the expansion of current knowledge databases through streamlined incorporation of new glycomic discoveries. Full structural characterization of a new or known glycan generally requires multiple analytic approaches, some of which are not amenable to the amounts of material available from many biologic systems. Therefore, unlike genomics or proteomics, unique glycan structure identifiers must be able to incorporate ambiguity to be broadly useful. A unique glycan identifier should register a structure at the level of structural resolution submitted by the experimentalist, who can then use this identifier to reference their work. And, importantly, consumers of this work will be able to use these identifiers to appreciate the level of resolution of the submitted structures in light of the published experimental approaches.
Development and implementation of a solution
The Complex Carbohydrate Structure Database and CarbBank were early attempts to implement useful identifiers for explicitly describing glycan structures (Doubet et al. 1989; Doubet and Albersheim 1992). By the time funding support for CarbBank ended in the late 1990s, other efforts (CFG, KEGG, JCGGDB, BCSDB) had been initiated around the world, resulting in a proliferation of database identifiers (Hashimoto et al. 2006; Raman et al. 2006; Toukach et al. 2007). GlycomeDB undertook the interconnection and consolidation of multiple databases, potentially providing an opportunity to assign unique identifiers to database entries (Ranzinger et al. 2008). However, the goals of GlycomeDB and other databases are nobly and ambitiously much broader than simply generating unique identifiers. These goals include the capture of as much metadata and analytic structural validation as possible, inevitably requiring significant database curation and expert intervention. The additional responsibility of such oversight is likely to hinder the rapid assignment, archiving and dissemination of unique identifiers for explicit glycan structures, which may be defined at various levels of ambiguity. However, computer algorithms can perform the core function of assigning identifiers to structures with little human intervention, reducing maintenance costs and fostering continuity over the long-term. The durability of the identifiers provided by such a stable core resource is a key requirement for their use as the semantic foundation for mapping, integrating and correlating the data and metadata compiled in more extensive and diverse databases.
Beginning with discussions at the 4th Warren Workshop in Athens, GA in 2012, and continuing through the 5th ACGG-DB (Asian Consortium for Glycobiology and Glycotechnology) meeting in Dalian, China in 2013, the glycomics and glycobioinformatics communities agreed with renewed vigor that unique glycan identifiers were needed in order to enhance data sharing in publications and across database platforms (Aoki-Kinoshita et al. 2013). A consensus was reached that a stand-alone, internationally recognized glycan structure repository should be developed. The function of the repository would be solely to assign unique identifiers to submitted glycan structures and to store minimal metadata, limited only to submitter and submission date/time for each accession number (Aoki-Kinoshita et al. 2016). The simple functions of such a repository would require minimal human management, allowing immediate assignment of accession numbers.
To this end, GlyTouCan was developed and deployed as a website (http://glytoucan.org) in 2015. At the 6th Warren Workshop in Sapporo, Japan in 2016, the assembled glycomics community was presented with the functionalities of the current GlyTouCan release (version 2). Ensuing discussion led to a consensus of support for the broadest possible acceptance of GlyTouCan as the essential resource for obtaining unique identifiers for glycan structures and as the international glycan structure repository of choice.
Based on the pressing need for generating unique glycan structure identifiers that possess appropriate utility for informatics platforms and sufficient stability for journal publications, the authors and the undersigned concurring colleagues strongly endorse GlyTouCan as an accepted, international repository for glycan structures. In order to fully realize the potential of GlyTouCan, the authors encourage the community to submit glycan structures to the repository and use the assigned identifiers in their submitted manuscripts. The authors further recommend that journals endorse the same goal, with the objective of eventually incorporating structural submission to GlyTouCan as an expected part of the manuscript submission workflow for manuscripts that describe the structure or function of glycans.
Enhanced GlyTouCan functionalities are already in place or under development that do or will interconnect structure repository identifiers with glycomic databases such as GlycomeDB (Ranzinger et al. 2008), Carbohydrate Structure Database CSDB (Toukach and Egorova 2016), GlycoEpitope (Okuda et al. 2017), GlycoNAVI (http://glyconavi.org), UniCarb-DB (Hayes et al. 2011), SugarBindDB (Mariethoz et al. 2016) and UniCarbKB (Campbell and Packer 2016); these linked databases will continue to be the appropriate repository for expansive metadata and analytic data that validates structural assignments. The acceptance of GlyTouCan within the glycocommunity allows these databases to link their metadata to specific glycan identifiers (Aoki-Kinoshita et al. 2016). These databases, known as GlyTouCan Partners, can directly register structures into GlyTouCan. Therefore, any associated metadata for a “new” glycan can be submitted to a GlyTouCan Partner, which will subsequently register structures to provide a GlyTouCan link to the associated metadata. After GlyTouCan registration, a submitter will be able to link deposited structures to accepted publications. Registration directly to GlyTouCan can be performed either as a single structure or using batch downloads. Glycan drawing tools derived from familiar resources such as GlycanBuilder facilitate the submission.
Using GlyTouCan
GlyTouCan (Figure 1) has been developed to be as user-friendly as possible. It provides an intuitive portal for searching and depositing structures. Note that glycans with unknown linkages, glycans known only as monosaccharide compositions (e.g. Gal2GlcNAc4Man2), and even glycan compositions with undefined monosaccharides (e.g. Hex4HexNAc4) can all be registered and retrieved.
Fig. 1.

GlyTouCan Logo. GlyTouCan is named by a combination of “Glycan” and “Tou (糖)”, which means “sugar” in Japanese. The word “Can (Jpn. 缶)” of GlyTouCan signifies that it is a container where glycans are accumulated. Therefore, a can is used in the logo to indicate that it is a repository with accumulated glycan data. In addition, a toucan is used as a mascot character since the word “TouCan” is included in GlyTouCan. The GlyTouCan toucan sits, appropriately, on the branch of a Lewis type glycan structure. This figure is available in black and white in print and in color at Glycobiology online.
Searching glycan structures in GlyTouCan
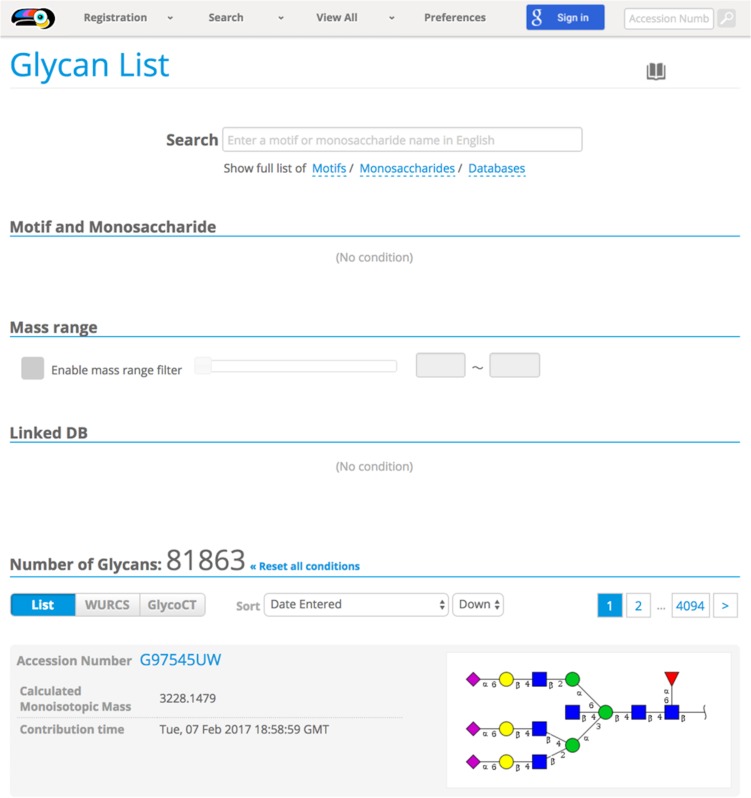
Glycans can be searched by either (1) browsing through the list of registered glycans or (2) specifying a particular glycan (sub)-structure and querying for similar registered structures. The “Glycan List” option under “View All” provides functionality to allow the user to filter down the list of glycans to search. Figure 2 is a snapshot of the full list that is shown initially after choosing the Glycan List option. Here, the list can be filtered by selecting structural components of the glycans that are being searched for, such as Motif (e.g. “Sialyl-Lewis” or “Lactosamine”) or Monosaccharide component. A list of Databases is also available if the target glycan is known to be stored in a particular database. Moreover, a mass range can be specified to filter the Glycan List by mass.
Fig. 2.
Glycan List for browsing/filtering glycans registered in GlyTouCan. This interface is accessible through the pull-down menu under View All on the GlyTouCan home page and provides a rapid, intuitive way to interrogate existing entries. This figure is available in black and white in print and in color at Glycobiology online.
The second popular search option is the “Graphic Input” option under the “Search” menu, which allows the user to draw their target glycan, using GlycanBuilder (Tsuchiya et al. 2017), and subsequently use it as a query. The resulting list of matching similar glycans will be shown, and if the query is already registered, its accession number will be displayed.
Depositing glycan structures to GlyTouCan
It is possible to deposit a glycan structure directly to GlyTouCan by signing in to a Google account. There is no need to enter or remember a new password as long as the user has a Google account (other types of accounts will be supported in the future). After signing in, a “Registration” menu will be displayed, via which glycan structure(s) can be added using Graphic Input (similar to Graphic Search via GlycanBuilder), Text Input using GlycoCT{condensed} or WURCS format, or File Upload. Every submitted structure will first be compared with the existing GlyTouCan registrations to ensure that duplicate deposits are not generated. A confirmation screen is shown (1) to list existing GlyTouCan IDs for those that are already registered and (2) to display images of the new structure(s) that will be registered.
Future perspectives
As has been true for all database efforts over the past 30 years, those that are heavily used and that prove to be most useful (e.g. UniProt, GenBank and PDB) are likely to achieve stable funding and long-term support. It will ultimately be in the hands of the glycomics community to demonstrate that GlyTouCan is essential infrastructure worth the continued investment of financial resources.
Existing proteomic databases (e.g. Uniprot and PDB) offer minimal characterization of glycoproteins and currently indicate only the positions of predicted or experimentally validated N- and O-linked glycosylation sites or GPI-anchor attachment sites. As unique GlyTouCan identifiers are integrated with existing proteomic resources, future queries will allow elucidation of the functional significance of specific glycan features across or within protein families to permit advances in the biomedical application of glycomics.
Moreover, with the inclusion of glycan identifiers in other omics databases, the authors and Supporting Investigators anticipate that glycomic data will achieve ever higher visibility and enhanced appreciation within the life science research community.
Supporting Investigators as of May, 2017 (in alphabetical order of last name)
Friedrich Altmann, University of Natural Resources and Life Sciences, Vienna, Austria
Antony Bacic, University of Melbourne, Australia
Christopher B. Barnett, University of Cape Town, South Africa
Júlia Costa, Laboratory of Glycobiology, ITQB NOVA, Portal
Vivien J. Coulson-Thomas, University of Houston, USA
Tamara L. Doering, Washington University School of Medicine, USA
Nathan Edwards, Georgetown University, USA
Michiko Ehara, Asahi University, Japan
Tamao Endo, Tokyo Metropolitan Institute of Gerontology, Tokyo, Japan
Ten Feizi, Imperial College London, UK
Martin Frank, Biognos AB, Sweden
Morihisa Fujita, Jiangnan University, China
Koichi Fukase, Osaka University, Japan
Yuzuru Ikehara, AIST and Chiba University, Japan
Makoto Ito, Kyushu University, Japan
Yukishige Ito, RIKEN, Japan
Kenji Kadomatsu, Nagoya University Graduate School of Medicine, Japan
Osamu Kanie, Tokai University, Japan
Takane Katayama, Kyoto University, Japan
Toshisuke Kawasaki, Ritsumeikan University, Japan
Hiroto Kawashima, Chiba University, Japan
Carsten Kettner, Beilstein Institut, Germany
Kshitij Khatri, Boston University, USA
Yoshinobu Kimura, Okayama University, Japan
Hiroshi Kitagawa, Kobe Pharmaceutical University, Japan
Shinobu Kitazume, RIKEN, Japan
Yuriy A. Knirel, N.D. Zelinsky Institute of Organic Chemistry, Russian Academy of Sciences, Moscow, Russia
Kyoko Kojima-Aikawa, Ochanomizu University, Japan
Daniel Kolarich, Griffith University, Australia
Matthew R. Kudelka, Emory University, USA
Todd L. Lowary, Canadian Glycomics Network Scientific Director and University of Alberta, Canada
Thomas Luetteke, ITech Progress GmbH, Germany
Shino Manabe, RIKEN, Japan
David Matten, University of Cape Town, South Africa
Raja Mazumder, George Washington University, USA
Eiji Miyoshi, Osaka University, Japan
Antonio Molinaro, University of Napoli Federico II, Italy
Yasu S. Morita, University of Massachusetts Amherst, USA
Toni M. Mueller, University of Alabama at Birmingham, USA
Shunji Natsuka, Niigata University, Japan
Shoko Nishihara, Soka University, Japan
Sriram Neelamegham, State University of New York, USA
Tetsuya Okajima, Nagoya University School of Medicine, Japan
Shujiro Okuda, Niigata University, Japan
Noorjahan Panjwani, Tufts University School of Medicine, USA
Dayoung Park, University of California, Davis, USA
Serge Perez, France
Salomé S. Pinho, University of Porto and Institute for Research and Innovation in Health, Portugal
Melody Porterfield, University of Georgia, USA
Alka Rao, CSIR-Institute of Microbial Technology, Chandigarh, India
Celso A. Reis, University of Porto, Portugal
Sylvie Ricard-Blum, University of Lyon 1, France
Rafael Ricci de Azevedo, University of Sao Paulo, Brazil
Nancy Schwartz, University of Chicago, USA
Siro Simizu, Keio University, Japan
Avadhesha Surolia, Indian Institute of Science, Bangalore, India
Naoyuki Taniguchi, RIKEN, Japan
Carlo Unverzagt, University of Bayreuth, Germany
Ajit Varki, University of California, San Diego, USA
Masahiro Wakao, Kagoshima University, Japan
Christopher M. West, University of Georgia, USA
Robert J. Woods, University of Georgia, USA
Ajit Varki, University of California, San Diego, USA
Yoshiki Yamaguchi, RIKEN, Japan
Kazuo Yamamoto, The University of Tokyo, Japan
Heng Yin, Dalian Institute of Chemical Physics, Chinese Academy of Sciences, China
Joseph Zaia, Boston University, USA
Funding
Integrated Database Project sponsored by the Japan Science and Technology Agency (JST) and the National Bioscience Database Center of Japan to GlyTouCan. Contributions of The National Center for Biomedical Glycomics (Grant P41GM103490) and the National Center for Functional Glycomics (Grant P41GM103694) were supported by The National Institute of General Medical Sciences, a part of the United States National Institutes of Health.
Conflict of interest statement
None declared.
Abbreviation
SNFG, Symbol Nomenclature for Glycans.
References
- Aoki-Kinoshita K, Agravat S, Aoki NP, Arpinar S, Cummings RD, Fujita A, Fujita N, Hart GM, Haslam SM, Kawasaki T et al. . 2016. GlyTouCan 1.0—The international glycan structure repository. Nucleic Acids Res. 44:D1237–D1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoki-Kinoshita KF, Sawaki H, An HJ, Campbell MP, Cao Q, Cummings R, Hsu DK, Kato M, Kawasaki T, Khoo K-H et al. . 2013. The fifth ACGG-DB meeting report: Towards an International Glycan Structure Repositorty. Glycobiology. 23:1422–1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brazma A, Hingamp P, Quackenbush J, Sherlock G, Spellman P, Stoeckert C, Aach J, Ansorge W, Ball CA, Causton HC et al. . 2001. Minimum information about a microarray experiment (MIAME)-toward standards for microarray data. Nat Genet. 29:365–371. [DOI] [PubMed] [Google Scholar]
- Campbell MP, Packer NH. 2016. UniCarbKB: New database features for integrating glycan structure abundance, compositional glycoproteomics data, and disease associations. Biochim Biophys Acta. 1860(8):1669–1675. [DOI] [PubMed] [Google Scholar]
- Doubet S, Albersheim P. 1992. CarbBank. Glycobiology. 2:505. [DOI] [PubMed] [Google Scholar]
- Doubet S, Bock K, Smith D, Darvill A, Albersheim P. 1989. The complex carbohydrate structure database. Trends Biochem Sci. 14:475–477. [DOI] [PubMed] [Google Scholar]
- Hashimoto K, Goto S, Kawano S, Aoki-Kinoshita K, Ueda N, Hamajima M, Kawasaki T, Kanehisa M. 2006. KEGG as a glycome informatics resource. Glycobiology. 16(5):63R–70R. [DOI] [PubMed] [Google Scholar]
- Hayes CA, Karlsson NG, Struwe WB, Lisacek F, Rudd PM, Packer NH, Campbell MP. 2011. UniCarb-DB: A database resource for glycomic discovery. Bioinformatics. 27(9):1343–1344. [DOI] [PubMed] [Google Scholar]
- Kolarich D, Rapp E, Struwe WB, Haslam SM, Zaia J, McBride R, Agravat S, Campbell MP, Kato M, Ranzinger R et al. . 2013. The minimum information required for a glycomics experiment (MIRAGE) project: Improving the standards for reporting mass-spectrometry-based glycoanalytic data. Mol Cell Proteomics. 12(4):991–995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, McBride R, Stoll M, Palma AS, Silva L, Agravat S, Aoki-Kinoshita KF, Campbell MP, Costello CE, Dell A et al. . 2017. The minimum information required for a glycomics experiment (MIRAGE) project: Improving the standards for reporting glycan microarray-based data. Glycobiology. 27(4):280–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariethoz J, Khatib K, Alocci D, Campbell MP, Karlsson NG, Packer NH, Mullen EH, Lisacek F. 2016. SugarBindDB, a resource of glycan-mediated host–pathogen interactions. Nucleic Acids Res. 44(D1):D1243–D1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuda S, Nakao H, Kawasaki T. 2017. GlycoEpitope In: Aoki-Kinoshita KF, editor. A Practical Guide to Using Glycomics Databases. Tokyo, Japan: Springer Japan; p. 227–245. [Google Scholar]
- Raman R, Venkataraman M, Ramakrishnan S, Lang W, Raguram S, Sasisekharan R. 2006. Advancing glycomics: Implementation strategies at the Consortium for Functional Glycomics. Glycobioloby. 16(5):82R–90R. [DOI] [PubMed] [Google Scholar]
- Ranzinger R, Herget S, Wetter T, von der Lieth CW. 2008. GlycomeDB—Integration of open-access carbohydrate structure databases. BMC Bioinformatics. 9:384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struwe WB, Agravat S, Aoki-Kinoshita KF, Campbell MP, Costello CE, Dell A, Feizi T, Haslam SM, Karlsson NG, Khoo KH et al. . 2016. The minimum information required for a glycomics experiment (MIRAGE) project: Sample preparation guidelines for reliable reporting of glycomics datasets. Glycobiology. 26(9):907–910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor CF, Paton NW, Lilley KS, Binz PA, Julian RK Jr., Jones AR, Zhu W, Apweiler R, Aebersold R, Deutsch EW et al. . 2007. The minimum information about a proteomics experiment (MIAPE). Nat Biotechnol. 25:887–893. [DOI] [PubMed] [Google Scholar]
- Toukach P, Egorova K. 2016. Carbohydrate structure database merged from bacterial, archaeal, plant and fungal parts. Nucleic Acids Res. 44(D1):D1229–D1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toukach P, Joshi H, Ranzinger R, Knirel Y, von der Lieth CW. 2007. Sharing of worldwide distributed carbohydrate-related digital resources: Online connection of the Bacterial Carbohydrate Structure DataBase and GLYCOSCIENCES.de. Nucleic Acids Res. 35(Database issue):D280–D286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchiya S, Aoki NP, Shinmachi D, Matsubara M, Yamada I, Aoki-Kinoshita KF, Narimatsu H. 2017. Implementation of GlycanBuilder to draw a wide variety of ambiguous glycans. Carbohydr Res. 445:104–116. [DOI] [PubMed] [Google Scholar]
- Varki A, Cummings RD, Aebi M, Packer NH, Seeberger PH, Esko JD, Stanley P, Hart G, Darvill A, Kinoshita T et al. . 2015. Symbol nomenclature for graphical representations of glycans. Glycobiology. 25:1323–1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- York WS, Agravat S, Aoki-Kinoshita KF, McBride R, Campbell MP, Costello CE, Dell A, Feizi T, Haslam SM, Karlsson N et al. . 2014. MIRAGE: The minimum information required for a glycomics experiment. Glycobiology. 24:402–406. [DOI] [PMC free article] [PubMed] [Google Scholar]