Abstract
Protein O-fucosyltransferase-1 (POFUT1), which transfers fucose residues to acceptor sites on serine and threonine residues of epidermal growth factor-like repeats of recipient proteins, is essential for Notch signal transduction in mammals. Here, we examine the consequences of POFUT1 loss on the oncogenic signaling associated with certain leukemia-associated mutations of human Notch1, report the structures of human POFUT1 in free and GDP-fucose bound states, and assess the effects of Dowling-Degos mutations on human POFUT1 function. CRISPR-mediated knockout of POFUT1 in U2OS cells suppresses both normal Notch1 signaling, and the ligand-independent signaling associated with leukemogenic mutations of Notch1. Normal and oncogenic signaling are rescued by wild-type POFUT1 but rescue is impaired by an active-site R240A mutation. The overall structure of the human enzyme closely resembles that of the Caenorhabditis elegans protein, with an overall backbone RMSD of 0.93 Å, despite primary sequence identity of only 39% in the mature protein. GDP-fucose binding to the human enzyme induces limited backbone conformational movement, though the side chains of R43 and D244 reorient to make direct contact with the fucose moiety in the complex. The reported Dowling-Degos mutations of POFUT1, except for M262T, fail to rescue Notch1 signaling efficiently in the CRISPR-engineered POFUT1−/− background. Together, these studies identify POFUT1 as a potential target for cancers driven by Notch1 mutations and provide a structural roadmap for its inhibition.
Keywords: Dowling-Degos disease, GDP-fucose, notch signaling, POFUT1, T-ALL, X-ray crystallography
Introduction
Notch signaling is an essential cell–cell communication pathway conserved in all metazoan organisms, influencing numerous cell fate decisions during development and in the mature organism. Tuning the dose and timing of Notch signals is critical for proper developmental progression, and aberrant increases or decreases in signaling are associated with developmental anomalies and cancer. Most notably, mutations of human Notch1 leading to increased ligand-independent signaling are found in more than half of human T cell acute lymphoblastic leukemia/lymphomas (T-ALLs), identifying Notch1 as an important therapeutic target in this disease (Weng et al. 2004).
One important post-translational modification that modulates Notch signal transduction is O-linked glycosylation. The epidermal growth factor (EGF)-like repeats of Notch receptors contain numerous sites with O-linked sugar modifications (Moloney et al. 2000). In particular, the enzyme protein O-fucosyltransferase-1 (POFUT1) is a critical auxiliary regulator of Notch signaling (Stanley and Okajima 2010), as knockout mice lacking POFUT1 exhibit early embryonic lethality resulting from generalized Notch signaling defects (Shi and Stanley 2003).
POFUT1 is an ER-resident protein that catalyzes O-linked fucosylation of an acceptor site on an EGF-like repeat of a recipient protein, using GDP-fucose as the donor substrate. Mutations of human POFUT1 are associated with Dowling-Degos disease (DDD), an autosomal dominant pigmentation disorder. Affected individuals progressively develop pigmented skin lesions which can be found in a variety of sites, including the trunk, face and extremities (Basmanav et al. 2015). DDD can result from nonsense and frameshift mutations, as well as missense mutations, indicating that they are almost certainly loss of function. Though the disease results from haploinsufficiency, it is not clear whether it is due to a Notch signaling defect, or is a consequence of another role for POFUT1 in melanocytes or other epidermal cell types.
In Notch proteins, the consensus fucosylation sites for POFUT1 are serine or threonine residues within Cys-X4−5-Ser/Thr-Cys (where X represents any amino acid) motifs of the Notch EGF-like repeats (Panin et al. 2002). After O-fucosylation by POFUT1, subsequent sugar-chain extension is catalyzed by Fringe glycosyltransferases, which deliver N-acetylglucosamine to the O-linked fucose. Additional elaboration of the O-linked sugar chain can include extension with galactose and sialic acid, generating an O-linked tetrasaccharide when fully elongated (Haltiwanger and Lowe 2004).
Fucosylation of Notch receptor EGF-like domains is required for normal Notch signaling (Stahl et al. 2008). Mutations at fucosylation sites in Notch receptor EGF-like repeat domains, particularly of T466 of EGF12, impair Notch activation, ligand-binding and normal lymphopoiesis in mouse models (Cordle et al. 2008; Ge and Stanley 2008; Ge et al. 2008; Zhang et al. 2009). Genetic knockout or deletion of POFUT1 reduces Notch ligand binding and Notch activation of target gene expression, impairing hematopoiesis and embryonic development. Catalytically compromised OFUT1 mutants support residual Notch function in flies (Okajima et al. 2005), suggesting that there is an enzymatically independent chaperone activity of the Drosophila enzyme facilitating transport of Notch to the cell surface (Okajima et al. 2005). Whether the enzyme-independent chaperone activity of Pofut1 extends to mammals is not clear. Loss of Pofut1 in mice is associated with reduced cell-surface expression of Notch1 in pre-somitic mesoderm (Okamura and Saga 2008), but not in ES cells (Stahl et al. 2008), suggesting that, at least in some cell contexts, Pofut1 does not have a chaperone-like role.
Glycosylation of Notch receptors by POFUT1 and fringe proteins modulates their binding affinities and responsiveness to their various ligands (see Rana and Haltiwanger 2011 for a review). Within the ligand-binding region of human Notch1, at least 21 of the 36 EGF-like repeats are fucosylated (Takeuchi and Haltiwanger 2014). In flies, Fringe tunes ligand-mediated Notch activation by enhancing Notch receptor binding affinity for Delta without having much effect on Serrate binding (Xu et al. 2007). Similarly, Fringe-modified human Notch1 enhances the response to Delta-like1 (DLL1) compared to Jagged1 (Stahl et al. 2008), a conclusion supported by more detailed biochemical studies of ligand-binding by Notch1 fragments. Although the intrinsic affinity of Notch1 for Delta-like4 (DLL4) is approximately an order of magnitude greater than its affinity for DLL1 (Andrawes et al. 2013), sugar modification of T466 in EGF repeat 12 of a ligand-binding fragment of human Notch1 enhances affinity for both DLL1 and Jagged1 ligands (Taylor et al. 2014) so that all three ligands are bound with comparable affinity after both fucosylation and fringe modification have occurred. The structure of a complex between an affinity enhanced N-terminal fragment of DLL4 with EGF-like repeats 11–13 of Notch1 show that the fucose appended to T466 directly contacts three residues from the MNNL domain of DLL4 (Luca et al. 2015). In studies probing the effects of sugar modification of other Notch receptors, the Fringe modification also appears to potentiate activation of human Notch2 by both classes of ligand (Hicks et al. 2000).
How the activity of POFUT1 affects the ligand-independent signaling of Notch receptors with cancer-associated mutations has not yet been investigated. The majority of Notch mutations implicated in T-ALL map to two regions, known as the NRR and PEST domains (Weng et al. 2004). In normal Notch function, the NRR prevents ligand-independent Notch activation by auto-inhibiting proteolysis at the S2 cleavage site (Gordon et al. 2007, 2009a, 2009b; Xu et al. 2015). The increased ligand-independent Notch1 signaling associated with NRR mutations arises from destabilization of the auto-inhibited conformation of the NRR (Malecki et al. 2006). Since these mutations destabilize the structure of the NRR, they also lead to increased intracellular retention of mutated receptors and ER stress, a feature that has been selectively targeted using sarco/ER Ca2+-ATPase (SERCA) inhibition, which creates a potential therapeutic window for mutated receptors preferentially over wild-type receptors (Roti et al. 2013).
Here, we investigate the effects of POFUT1 loss on the oncogenic signaling associated with T-ALL mutations of Notch1, report the X-ray structures of human POFUT1 both in its free form and bound to GDP-fucose substrate, and analyze the effects of DDD mutations on POFUT1 activity using a Notch reporter gene assay. Cell-based assays show that normal Notch1 signaling and the autonomous activity associated with T-ALL-associated mutations of Notch1 are both efficiently suppressed upon POFUT1 loss of function. The structure of the human enzyme closely resembles that of the C. elegans enzyme (Lira-Navarrete et al. 2011), with a conserved GDP-fucose binding pocket and an analogous cleft for EGF-repeat binding. Together, these studies clarify the functional effects of Dowling-Degos mutations, identify POFUT1 as a potential target for cancers driven by Notch1 mutations, and provide a structural roadmap for POFUT1 inhibition.
Results and Discussion
Here, we explored whether POFUT1 may be a viable therapeutic target in Notch-dependent T-ALLs with NRR mutations (Weng et al. 2004), and determined the X-ray structure of the human POFUT1 enzyme to provide a roadmap for inhibition. We also evaluated the effects of DDD mutations in a POFUT1-dependent assay for Notch1 signaling, clarifying how each of the four reported missense mutations affect POFUT1 function.
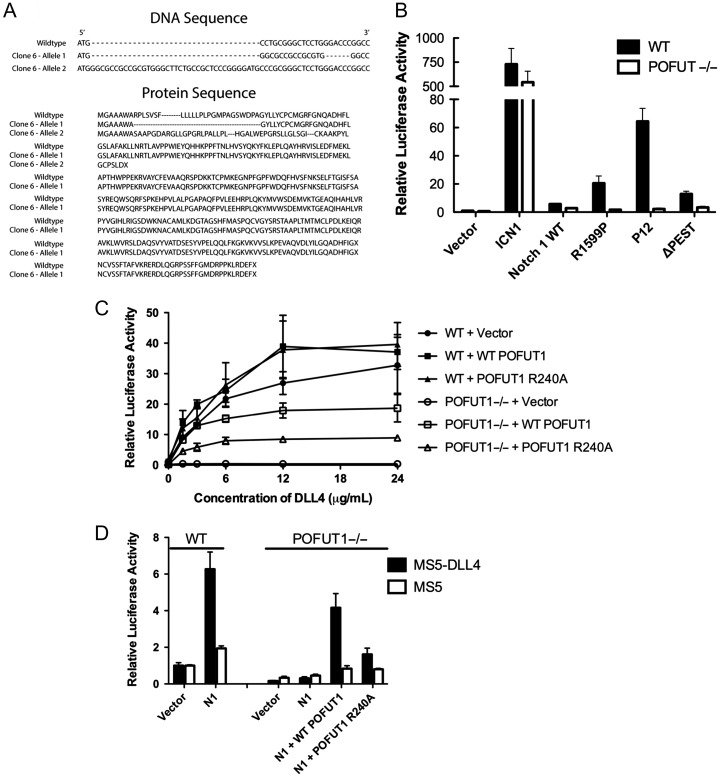
Genetic knock-down of Drosophila OFUT1 traps Notch in the ER, precluding wild-type Notch signaling (Okajima et al. 2005, 2008). We tested whether POFUT1 inactivation silences not only normal Notch1 signaling but also the ligand-independent activity of human Notch1 receptors with leukemia-associated NRR mutations using CRISPR-targeted gene deletion of POFUT1 in U2OS cells. Using a lentiviral vector expressing the Cas9 enzyme and a CRISPR guide sequence targeting exon 1 of the POFUT1 gene (see Methods), we identified a U2OS subclone in which POFUT1 was inactivated with biallelic mutations (Figure 1A). One allele contains a frameshift with an early stop codon, and the other allele bears amino acid substitutions that disrupt the signal sequence.
Fig. 1.
Effect of POFUT1 knockout on Notch signaling in U2OS cells. (A) Sequence results from the PCR product of the CRISPR/Cas9 engineered POFUT1−/− U2OS cell line. One allele generates an early stop codon, and the other allele disrupts the signal sequence. (B) Luciferase reporter assay testing ligand-independent Notch activity in wild-type and POFUT1−/− U2OS cells expressing the indicated Notch1 protein. ICN1 is the intracellular portion of Notch1; R1599P, P12 and ΔPEST are three mutations found in T-ALL. (C) Plated ligand assay testing DLL4-dependent Notch activity in wild-type and POFUT1−/− U2OS cells expressing wild-type Notch1, and empty vector, wild-type POFUT1 or the R240A active-site mutant. The relative luciferase activity of each test condition is plotted as a function of the amount of DLL4 deposited on the surface of the tissue culture well. (D) Co-culture assay testing ligand-dependent Notch activity in wild-type and POFUT1−/− U2OS cells and the ability of POFUT1 to rescue the activity in POFUT1−/− cells. U2OS cells were transfected with wild-type Notch1 along with empty vector, wild-type POFUT1 or the R240A active-site mutant, co-cultured with MS5 stromal cells expressing DLL4, or MS5 cells alone. The relative luciferase activity of each test condition is plotted on the y-axis. Error bars in all panels represent the standard deviation of three biological replicates.
We next carried out reporter gene assays in the isogenic parent and POFUT1 mutated cell lines in order to assess the effect of POFUT1 loss on normal Notch signaling and the ligand-independent signaling associated with T-ALL-associated mutated Notch1 receptors. The data show that signaling by a construct expressing only the intracellular portion of Notch1 is not suppressed by POFUT1 loss, but that ligand-independent signaling associated with class I mutations, such as R1599P, as well as with the class II P12 insertion (Malecki et al. 2006), is suppressed to near basal levels of reporter activity, comparable to that of the wild-type receptor in the absence of ligand (Figure 1B). Normal ligand-induced Notch signal transduction, tested in a plated ligand assay with DLL4 as ligand, is also suppressed by POFUT1 loss, as predicted (Figure 1C).
We also tested whether wild-type POFUT1, or the R240A variant reported to lack GDP-fucose hydrolase activity (Lira-Navarrete et al. 2011), could rescue wild-type Notch1 signaling in the POFUT1 null U2OS cell line, using both a plated ligand assay (Figure 1C) and a co-culture assay with DLL4-expressing cells (Figure 1D). For wild-type Notch1, the rescue seen with wild-type POFUT1 is impaired in the R240A mutant (Figure 1C) in the plated ligand assay, though there is residual detectable Notch1 activity. Forced expression of POFUT1 in a wild-type background also appears to enhance the strength of wild-type Notch1 signaling (Figure 1C) suggesting the possibility that additional POFUT1 even improves the maturation efficiency of the wild-type receptor (when transiently expressed in these cells), and enhances surface delivery. A similar rescue pattern is observed when wild-type and R240A forms of POFUT1 are tested in the co-culture assay: Rescue with wild-type POFUT1 is evident, and the amount of rescue seen is greatly reduced when the reintroduced POFUT1 has the R240A mutation (Figure 1D). Together, these results argue that POFUT1 is required for the ligand-independent signaling associated T-ALL mutations of Notch1, and that the catalytic activity of POFUT1 is necessary for full ligand-dependent signaling. The data also suggest that inhibition of the catalytic activity of POFUT1 may be a rational therapeutic approach to Notch pathway inhibition in cancers where ligand-independent Notch1 signaling is oncogenic.
Flow cytometric analysis of surface Notch1 was analyzed in various POFUT1 backgrounds to test whether loss of POFUT1 disables signaling by preventing delivery of Notch1 to the cell surface. For these studies, we created a FLAG-Notch1-eGFP allele, which has an N-terminal FLAG tag in the extracellular part of the protein and a C-terminal eGFP fusion intracellularly. This form of Notch1 allowed us to monitor GFP fluorescence as a measure of total protein, and anti-FLAG-APC fluorescent staining as a measure of Notch1 at the cell surface. The flow data show that transfected Notch1 is readily delivered to the cell surface in wild-type U2OS cells, but remains intracellularly trapped in the CRISPR-engineered U2OS cells lacking POFUT1 (Supplementary data, Figure S1). Co-transfection of wild-type POFUT1 restores Notch1 surface delivery, whereas surface delivery is greatly reduced in cells co-transfected with the R240A mutant of POFUT1 (Supplementary data, Figure S1A). Whether the partial activity of R240A results from residual catalytic activity of this mutant or a chaperone-like activity, as previously proposed, remains unclear. Western blotting shows that the abundance of POFUT1 protein in the rescue studies with wild-type enzyme is equal to or exceeds the amount present in wild-type U2OS cells (Supplementary data, Figure S1B). Western blotting was also used to probe the extent to which Notch1 is proteolytically processed at site S1 from its immature (intracellular) precursor form into the mature cell-surface heterodimer (Blaumueller et al. 1997; Sanchez-Irizarry et al. 2004) as a function of POFUT1 status (Supplementary data, Figure S1B). Consistent with the flow cytometry studies, Notch1 matures readily in unmodified U2OS cells, and fails to mature in the CRISPR-engineered U2OS cells lacking POFUT1. Addition of wild-type POFUT1 into the null background restores proteolytic processing, as anticipated from the flow results.
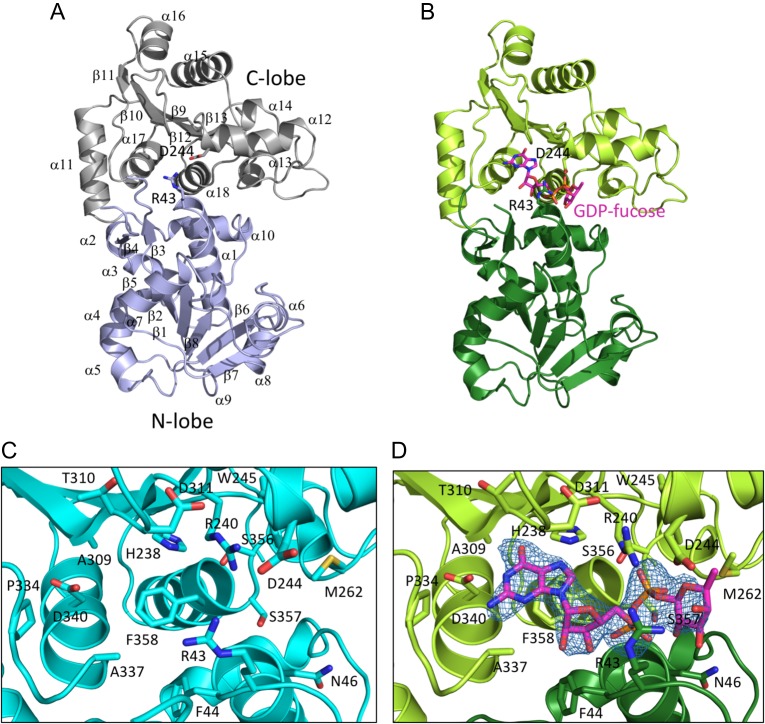
To facilitate efforts to target POFUT1 in T-ALL and other cancers, we determined X-ray structures of human POFUT1 in its free and GDP-fucose bound states to 2.1 Å and 2.4 Å resolution, respectively, by molecular replacement using the structure of the C. elegans protein as a search model (Table I). Comparison of the substrate complex with the unliganded enzyme shows that limited global movement takes place upon substrate binding (Figure 2A and B and Supplementary data, Figure S2A). The nucleotide binding pocket is preorganized, with little movement of the residues that contact the adenine ring of the substrate, and the C-terminal lobe makes a hinge movement of less that 1 Å upon GDP-fucose binding. Upon complexation, the side chain of R43 approaches within hydrogen-bonding distance of the fucose ring, and D244 also moves to accommodate the bound ligand (Figure 2C and D).
Table I.
Crystallographic data collection and refinement
| Apo | GDP-fucose | |
|---|---|---|
| Data collection | ||
| Space group | P 1 21 1 | P 1 21 1 |
| Cell dimensions | ||
| a, b, c (Å) | 50.20, 132.24, 72.06 | 50.23, 133.92, 70.01 |
| α, β, γ (°) | 90, 106.44, 90 | 90, 104.13, 90 |
| Wavelength (Å) | 0.9792 | 0.9792 |
| Resolution (Å) | 50–2.09 (2.20–2.09) | 50–2.41 (2.46–2.41) |
| Rmeas | 15.1 (136.9) | 19.5 (184.7) |
| CC(1/2) | 99.2 (36.5) | 98.8 (44.1) |
| Completeness (%) | 98.2 (97.6) | 98.0 (92.1) |
| I/σ(I) | 7.42 (0.87) | 5.28 (0.97) |
| Redundancy | 2.7 (2.6) | 4.5 (4.0) |
| Refinement | ||
| Resolution (Å) | 46.09–2.09 | 47.67–2.41 |
| No. reflections | 52548(5126) | 33915(3207) |
| Rwork/Rfree | 0.182/0.228 | 0.240/0.292 |
| No. atoms | ||
| Protein | 5635 | 5534 |
| Water | 635 | 72 |
| Ligand | 76 | |
| B-factors | ||
| Protein | 32.39 | 67.45 |
| Water | 35.68 | 54.16 |
| Ligand | 57.53 | |
| RMS deviations | ||
| Bond lengths (Å) | 0.004 | 0.004 |
| Bond angles (°) | 0.710 | 0.865 |
| Ramachandran (%) | ||
| Favored | 97.29 | 96.09 |
| Allowed | 2.29 | 3.77 |
| Outlier | 0.43 | 0.14 |
| Rotamer outliers (%) | 2.17 | 0.69 |
| Clashscore | 2.68 | 8.36 |
Fig. 2.
Structure overview. (A) Ribbon diagram of the unliganded enzyme, showing overall architecture of the enzyme. The N-terminal lobe is colored slate blue, and the C-terminal lobe is gray. Secondary structure elements are indicated; R43 and D244 are shown as sticks. (B) Ribbon diagram of the enzyme complexed with GDP-fucose. The N-terminal lobe is dark green, the C-terminal lobe is light green, and the GDP-fucose is in CPK colors, with carbon atoms colored magenta. R43 and D244 are shown as sticks. (C) Zoomed in view of the active site of the unliganded enzyme. Side chains of residues in the active site are in CPK colors, with carbon atoms in cyan. (D) Zoomed in view of the active site of the enzyme with GDP-fucose bound. Side chains of residues in the active site are rendered as sticks in CPK colors, with carbon atoms in green. The bound GDP-fucose is rendered as sticks in CPK colors, with carbon atoms in magenta. An Fo–Fc difference map for the GDP-fucose ligand is shown in blue mesh, contoured at 3σ. This figure is available in black and white in print and in color at Glycobiology online.
The overall structure of the human enzyme closely resembles that of the C. elegans protein (Lira-Navarrete et al. 2011), with an overall backbone RMSD of 0.93 Å over 355 residues when the two GDP-fucose complexed structures are superimposed (Supplementary data, Figure S2B), despite primary sequence identity of only 39% in the mature protein (Supplementary data, Figure S3). The most substantial structural divergence between the two proteins takes place in loops connecting secondary structural elements, most notably in the loop C-terminal to the α12 helix that lines the GDP-fucose binding pocket.
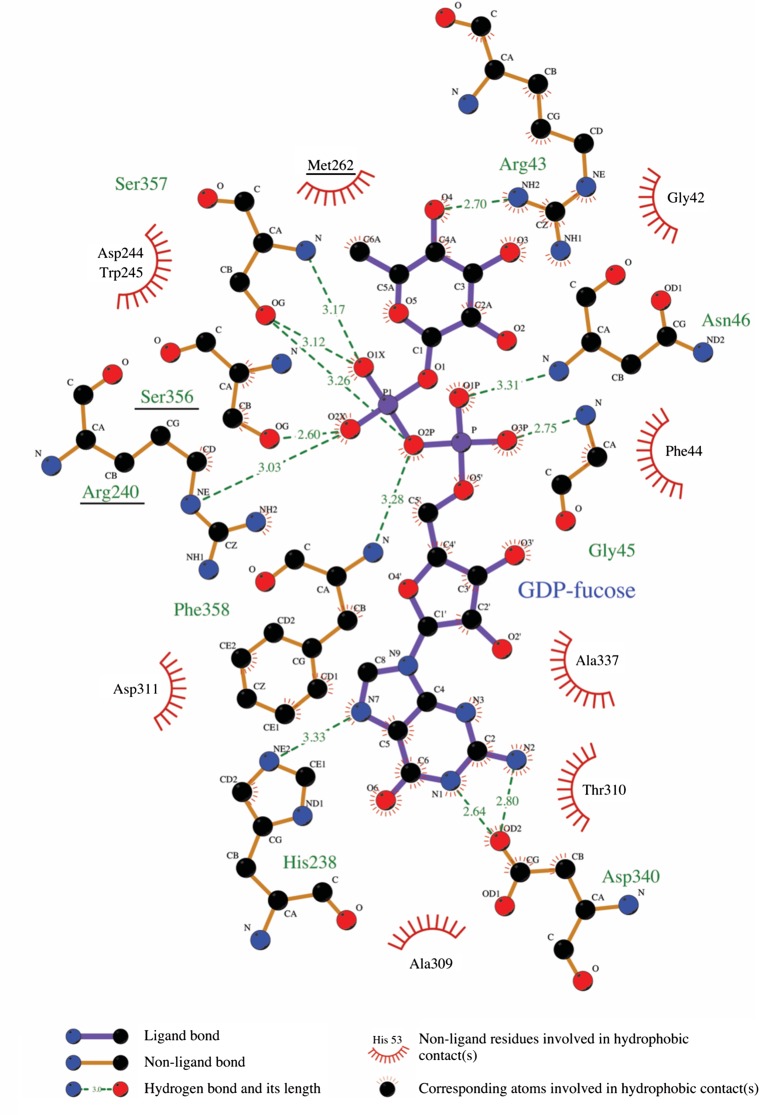
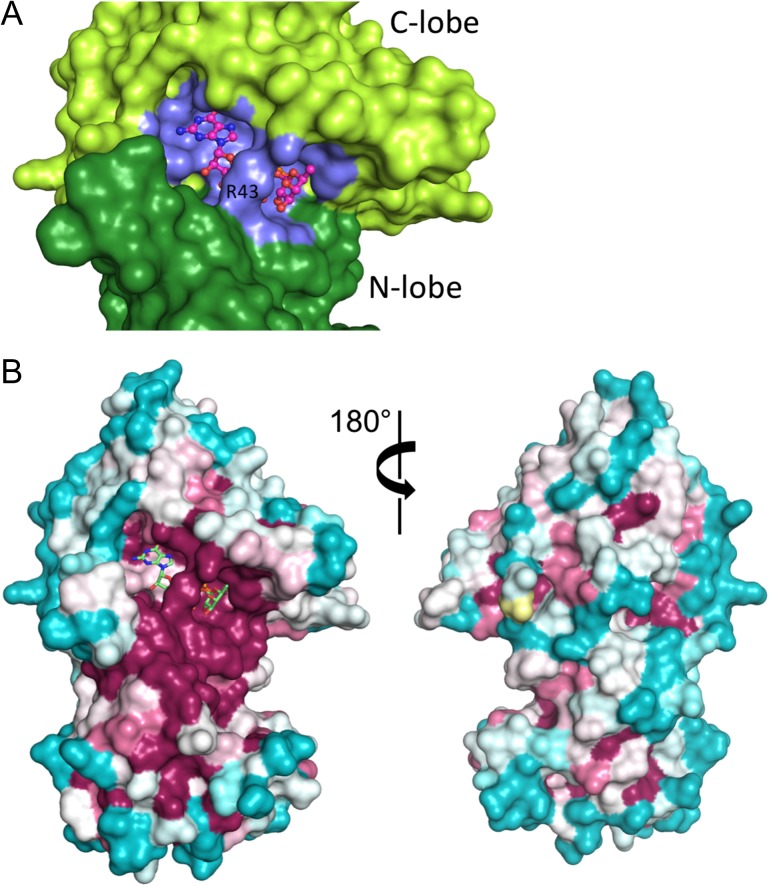
As with the worm enzyme, the GDP-fucose substrate binds in a cleft between the two lobes of the enzyme, with most of the contact interface derived from the C-terminal lobe (Figure 2C and D). Residues of the C-terminal lobe that approach within hydrogen-bonding distance of the guanine ring include H238 and D340 (Figure 3). The phosphate oxygens of the GDP-fucose are positioned by hydrogen bonds with backbone amides from G45 and D46 of the N-lobe, S356, S357 and F358 of the C-lobe, and with the side chains of R240, S356 and S357; the side chain of R43 reorients in the GDP-fucose complex to hydrogen bond with O4 position of the fucose residue (Figure 3). Not surprisingly, mapping of sequence conservation onto the protein surface shows that the most highly conserved region corresponds to the nucleotide binding pocket and the surrounding cleft (Figure 4A and B), presumed to be the pocket for binding of EGF-like protein substrates (Lira-Navarrete et al. 2011).
Fig. 3.
Schematic illustration of contacts between POFUT1 and GDP-fucose. Interaction diagram generated using the program Ligplot+ (Laskowski and Swindells 2011). GDP-fucose is situated in the center of the diagram, with interacting residues from POFUT1 around the periphery, and interactions schematized according to the key at the base of the figure. Residues mutated in DDD are underlined. This figure is available in black and white in print and in color at Glycobiology online.
Fig. 4.
GDP-fucose binding and conservation analysis. (A) Surface representation showing GDP-fucose bound at the cleft between N- and C-terminal lobes (dark and light green, respectively). Residues at the contact interface with GDP-fucose are colored slate blue. GDP-fucose is in CPK colors, with carbon atoms colored magenta. (B) Conservation analysis. The structure of the enzyme is shown in surface representation, colored on a sliding scale from maroon (most conserved) to teal (least conserved) using the program ConSurf (Ashkenazy et al. 2016). The GDP-fucose is shown in CPK colors, with the carbon atoms colored green. This figure is available in black and white in print and in color at Glycobiology online.
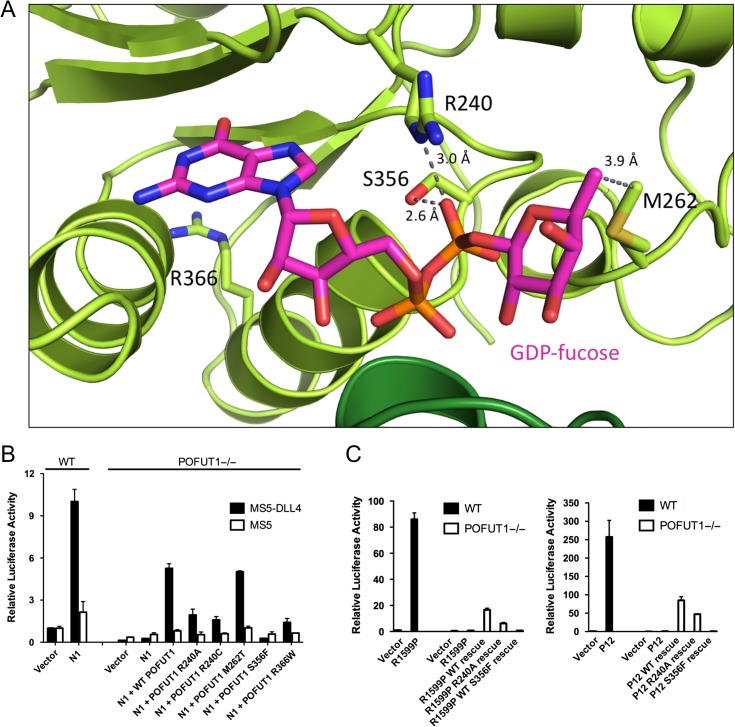
There are four known DDD missense mutations: R240C, M262T, S356F and R366W (Basmanav et al. 2015), each of which resides in the C-terminal lobe (Figure 5A). To test the prediction that these mutations result in POFUT1 loss of function, we evaluated the ability of each mutated enzyme to rescue Notch1 signal transduction using a co-culture assay, comparing the effect of each mutant enzyme to that of wild-type POFUT1 (Figure 5B). A robust Notch1 signaling response is induced in parental U2OS cells upon co-culture with MS5-DLL4 cells, but there is no detectable signaling response in our CRISPR-engineered POFUT1 null U2OS cells, and co-transfection of wild-type POFUT1 substantially rescues the Notch1 signal in the POFUT1 null background (Figures 1D, 5B). The Dowling-Degos mutants exhibit varying effects in the rescue assay, ranging from complete loss of activity in the case of S356F to rescue indistinguishable from the wild-type enzyme for M262T (Figure 5B). Western blotting suggests that the variation in rescue activity cannot simply be attributed to differences in the amount of protein produced, as the level of protein for each mutant tested in the rescue experiments is equal to or exceeds the endogenous amount present in unmodified U2OS cells (Supplementary data, Figure S4). It is true, however, that there is less total S356F protein in comparison to the other mutants, leaving open the possibility that a reduced steady-state level of this protein also contributes to the complete loss of observable activity. The complete loss of function from S356F is likely attributable to the importance of the side chain hydroxyl of S356 as an H-bond donor to the β-phosphoryl oxygen of GDP-fucose (Figure 5A); an S356F mutation will likely sterically interfere with GDP-fucose binding (and even if GDP-fucose could bind, the S356F mutation would eliminate an important means of stabilizing the increasing negative charge on a β-phosphoryl oxygen of the GDP-fucose in the transition state for fucose transfer). R240 forms a salt-bridge with a β-phosphoryl oxygen of the GDP-fucose substrate, both contributing to GDP-fucose binding (Lira-Navarrete et al. 2011), and in position to stabilize the increasing negative charge on the leaving group upon fucosyl transfer (Figure 5A). The R240C mutation will eliminate or change the character of this interaction, likely reducing affinity for GDP-fucose and attenuating stabilization of the additional negative charge developing on the phosphate group in the transition state, akin to the published loss-of-function effect of the R240A mutation of the worm enzyme (Lira-Navarrete et al. 2011). It is notable that both R240A and R240C appear to result in partial loss of function, as there is residual activity of both these mutations in all of the Notch1 signaling assays, unlike the total loss of function observed with S356F. R366W changes the shape, charge and steric bulk of a buried residue in the C-terminal lobe, suggesting that the primary effect of this mutation is likely to be structural disruption of the protein (Figure 5A). Lastly, although M262 comes into van der Waals contact with the methyl group of the fucose ring (Figure 5A), the M262T substitution appears to be well tolerated, and any loss of function that may be associated with this mutation is too subtle to be detected with our Notch1 signaling rescue assay.
Fig. 5.
Sites of DDD mutations and effects on Notch1 signaling. (A) Ribbon diagram of the active site, with residues mutated in DDD labeled and rendered as sticks in CPK colors, with the carbon atoms colored green. The GDP-fucose is shown as sticks in CPK colors, with the carbon atoms colored magenta. Dashed lines, with distances shown in Å, indicate hydrogen-bonding and non-polar interactions of mutated residues. (B) Effects of Dowling-Degos mutations on Notch1 signaling. Parent U2OS cells or POFUT1 null U2OS cells were transfected with Notch1 and the indicated POFUT1 constructs, and co-cultured with MS5 cells expressing DLL4, or MS5 cells alone. The Notch1 signaling activity was determined using a luciferase reporter assay (see Methods). Error bars represent the standard deviation of two (MS5 cells) or three (MS5-DLL4 cells) biological replicates. (C) Luciferase reporter assay testing rescue of ligand-independent oncogenic Notch1 activity in POFUT1 null U2OS cells transfected with various forms of POFUT1. Cells were transfected with the R1599P (left panel) or P12 (right panel) T-ALL mutant form of Notch1 along with empty vector, wild-type POFUT1, the R240A active-site mutant or the S356F Dowling-Degos mutant. Error bars in all panels represent the standard deviation of three biological replicates. This figure is available in black and white in print and in color at Glycobiology online.
Our rationale for investigating POFUT1 as a therapeutic target in Notch-driven leukemias derived from the prior observation that knock-down of Drosophila OFUT1 led to trapping of Notch in the ER, precluding Notch signaling (Okajima et al. 2005, 2008). This effect resembles the increased ER stress imposed by SERCA inhibition (Roti et al. 2013). Indeed, CRISPR/Cas9-mediated deletion of POFUT1 silenced the ligand-independent signaling associated with Notch1 receptors bearing oncogenic NRR mutations, as well as signals transduced by wild-type Notch1 receptors, and blocked delivery of wild-type Notch1 to the cell surface. We also tested the R240A and S356F mutants of POFUT1 in a rescue assay for the ligand-independent signaling activity of the P12 and R1599P oncogenic Notch1 mutants: The silencing of the signal seen in the POFUT1-null background is rescued by wild-type POFUT1, but rescue of the ligand-independent signaling is greatly attenuated by the R240A mutation, and completely abrogated by the S356F form of POFUT1 (Figure 5C), paralleling the findings seen with ligand-dependent signaling in the co-culture assay. The difference between S356F and the R240A mutant in the rescue studies suggests that either (i) there is residual fucosyltransferase activity in the R240A protein, or that (ii) the S356F is disruptive enough to not only render the enzyme catalytically inactive but also interfere either with EGF-repeat binding or POFUT1 protein maturation, two possibilities that can be examined in future studies. The reduced activity of both mutants in the wild-type Notch1 rescue assay also indicates that any chaperone-like activity of human POFUT1 that may exist in this particular assay context is insufficient to restore Notch1 signaling activity fully. Although it remains to be seen whether there is an effective therapeutic window (in which normal Notch signaling can be spared while ligand-independent signaling by mutated receptors is suppressed), POFUT1 inhibition appears to warrant further investigation as a therapeutic avenue in T-ALL or in other diseases with aberrant Notch signaling.
Materials and Methods
Materials and DNA constructs
MIT's CRISPR guide design server (crispr.mit.edu) was used to generate four guides targeting exon 1 and 2 of the POFUT1 gene. DNA encoding guide sequences were purchased (IDT Technologies) and annealed in 10 mM Tris, 50 mM NaCl, pH 8. The annealed, double-stranded oligonucleotides were inserted into a pSpCas9 WT-2A-GFP vector using a BsmBI restriction site compatible with overhangs designed into the annealed duplex. For expression of protein used in the crystallographic studies, human POFUT1 (residues 24–384, containing the N160A mutation to eliminate one of the glycosylation sites) was cloned into a pSecTag2 expression vector, so that the encoded protein contained an additional glycine followed by a hexahistidine tag at the C-terminus. For Notch1 signaling rescue experiments, the hexahistidine tag was inserted at the N-terminus of POFUT1, between the pSecTag2-encoded signal sequence and POFUT1 residue 24, without the N160A mutation. Plasmids encoding the Notch1 T-ALL mutations investigated here have been previously described (Malecki et al. 2006). The FLAG-Notch1-GFP cDNA was assembled in a pcDNA5/FRT/TO backbone, with the FLAG sequence inserted between the signal peptide and the first EGF repeat; the sequence encoding eGFP was installed at the Notch1 C-terminus after a six amino acid linker. GDP-fucose was purchased from Carbosynth and used without further purification.
Cell line generation
To generate POFUT1 knockout cells, U2OS cells were transfected with a modified pSpCas9 WT-2A-GFP plasmid designed for POFUT1 disruption using Lipofectamine 2000 (Invitrogen). Cells expressing GFP were sorted using flow cytometry 48–72 h after transfection. Then, 72 h after this initial sorting step used to identify transfectants, cells were sorted into 96-well plates containing DMEM with 10% fetal bovine serum (FBS) and 1% Penicillin/Streptomycin (Pen/Strep). After 2–3 weeks of clonal expansion, genomic DNA was extracted from individual clones. For each clone, the targeted POFUT1 locus was amplified by PCR. POFUT1 PCR fragments were TOPO cloned (Invitrogen) into pcDNA5 for sequencing. The single cell isolate used for subsequent studies had the mutations shown in Figure 1A.
Protein expression
293T cells were transfected with a modified pSecTag2 plasmid designed to secrete mature POFUT1 (residues 24–384, containing the N160A mutation) and stabilized using hygromycin. Cells were grown in 15 cm dishes in DMEM containing 10% FBS, 1% Pen/Strep and 100 µg/mL hygromycin. Cells from 15 cm dishes were transferred to suspension culture (106 cells/mL) in Freestyle media (Invitrogen) containing 1% FBS and 1% Pen/Strep and allowed to grow and secrete protein for 5 days. On day 5, the media was harvested and the cells were placed in new Freestyle media for an additional 5 days, or as long as viability remained above 75% as assessed by Trypan Blue staining. Conditioned media was centrifuged at low speed (20 min at 1000 g) to remove any remaining cells or large cell debris. A high-speed spin was then performed (30 min at 5000 g) to remove finer cellular debris. Conditioned media was then filtered through a 0.2 µm filter and the pH was adjusted to 7.4 using HEPES buffer (1 M, pH 7.4) containing 20 mM imidazole. The recovered conditioned media was then passed over Ni-NTA resin at room temperature. The resin was washed with HEPES-buffered saline (20 mM HEPES, 150 mM NaCl, pH 7.4; HBS) containing 20 mM imidazole. Bound POFUT1 was eluted in HBS containing 250 mM imidazole. The eluate was concentrated and applied onto on an S200 Superdex column. Fractions containing POFUT1, which eluted as a single sharp peak around 16 mL, were concentrated and stored at 4°C. An enzyme concentration of at least 10 mg/mL was used for crystallization trials. Protein yields typically ranged from 1 to 2 mg/L in Freestyle media.
Ligand-independent luciferase assays
On day 1, parental or POFUT1 knockout U2OS cells in six-well plates were transfected with a Notch1 responsive TP1-firefly luciferase reporter (Zimber-Strobl et al. 1994) and a control Renilla luciferase reporter using Lipofectamine 2000 (Invitrogen). Cells were also co-transfected with empty pcDNA3 vector or pcDNA3-encoding a Notch1 variant to examine the effect of POFUT1 loss on Notch receptor activity. The final amounts of plasmid per well were 750 ng Notch1 variant or empty pcDNA3 vector, 980 ng TP1-luciferase and 20 ng pRL-TK internal control Renilla luciferase plasmid. In add-back experiments, 250 ng empty pcDNA3 vector or pSecTag2-encoding N-terminally hexahistidine-tagged wild-type or mutant POFUT1 plasmid was additionally co-transfected. Transfected cells were incubated at 37°C. On day 3, media was aspirated and cells were washed with PBS. Luciferase reporter activity in whole-cell lysates was then assessed using a Dual Luciferase Assay Kit (Promega) and a luminometer (Turner Systems). Measurements were normalized to the empty vector-transfected wild-type cell control, which was assigned a value of 1. All conditions in each experiment were performed in triplicate.
Plated ligand-dependent luciferase assays
Plated ligand assays analyzing wild-type Notch1 signaling were performed as described previously with minor adaptations (Andrawes et al. 2013). On day 1, 80 μL of PBS containing 0–24 μg/mL DLL4 (R&D Systems) and 5 μg/mL RetroNectin (Takara) were added to each well of a non-tissue culture-treated 96-well plate and incubated overnight at 4°C. In addition, parental or POFUT1 knockout U2OS cells in six-well plates were transfected with plasmids encoding various Notch1 and POFUT1 constructs or empty pcDNA3 vector using Lipofectamine 2000 (Invitrogen) and incubated overnight at 37°C. The final amounts of plasmid per well were 750 ng Notch1 WT or pcDNA3 empty vector, 250 ng POFUT1 WT or R240A or empty vector, 980 ng TP1-luciferase and 20 ng pRL-TK internal control Renilla luciferase plasmid. On day 2, the PBS solution was removed from the wells of the 96-well plate, 100 μL of transfected U2OS cells resuspended in 7.5 mL media was added to each well, and the cells were incubated overnight at 37°C. On day 3, cells were processed and relative luciferase activity was measured and normalized as above. All conditions in each experiment were performed in triplicate.
Co-culture ligand-dependent luciferase assays
Co-culture ligand assays analyzing wild-type Notch1 signaling were performed as described previously with minor adaptations (Andrawes et al. 2013). On day 1, parental or POFUT1 knockout U2OS cells in six-well plates were transfected as above and incubated overnight at 37°C. In addition, 12,500 murine control (MS5) or ligand-expressing (MS5-DLL4) were plated in each well of a 96-well plate and incubated overnight at 37°C. On day 2, the media was removed from the wells of the 96-well plate, 100 μL of transfected U2OS cells resuspended in 7.5 mL media was added to each well, and the cells were co-cultured overnight at 37°C. On day 3, cells were processed and relative luciferase activity was measured and normalized as above. All conditions in each experiment were performed in duplicate (MS5 cells) or triplicate (MS5-DLL4 cells).
Flow cytometry
On day 1, parental or POFUT1 knockout U2OS cells in six-well plates were transfected with 1.75 µg FLAG-Notch1-GFP, 250 ng pSecTag2-POFUT1 (wild-type or R240A) and 500 ng empty pcDNA3 vector using Lipofectamine 2000 (Invitrogen) and incubated at 37°C. On day 3, cells were briefly trypsinized, resuspended in media and washed twice in 1 mL cold PBS + 0.1% BSA. Cells were counted, and 100,000 to 200,000 cells were incubated with rotation in 100 μL cold PBS + 0.1% BSA containing 10 μg/mL APC-conjugated anti-FLAG M2 antibody (Perkin-Elmer) in the dark for 30 min at 4°C. Cells were washed twice in 1 mL cold PBS + 0.1% BSA and resuspended in 100 μL for flow cytometry analysis on an Accuri C6 flow cytometer (BD Biosciences).
S1 cleavage assays
On day 1, parental or POFUT1 knockout U2OS cells in six-well plates were transfected as above, and incubated at 37°C. On day 3, cells were briefly trypsinized, resuspended in media, and washed in cold PBS before freezing. Cell pellets were resuspended in 1× Laemmli buffer in PBS, then sonicated briefly on setting three of a tip sonicator (Sonic Dismembrator Model F550, Fisher Scientific). Equal volumes were loaded on 4–20% SDS-PAGE gels (Bio-Rad), transferred to nitrocellulose membranes, and probed with antibodies recognizing the anti-Notch1 transcriptional activation domain (Wang et al. 2011), POFUT1 (Abcam, ab154051) and GAPDH (14C10, Cell Signaling Technology) at 1:2000, 1:1000 and 1:5000, respectively.
Crystallization and data collection
POFUT1 crystals were obtained at 4°C by vapor diffusion using the hanging drop method. Crystallization occurred with 16 mg/mL protein in 16.5% v/v PEG3350, 0.05% w/v octyl-β-D-glucoside, 0.5% v/v glycerol and 100 mM sodium citrate pH 5.0. For cryopreservation, crystals were transferred to a solution containing 25% PEG3350, 0.05% w/v octyl-β-D-glucoside, 20% v/v glycerol, and 100 mM sodium citrate, pH 5.0, and were then plunged into liquid nitrogen. The ligand-bound structure was obtained by soaking crystals for 15 min in cryopreservation solution containing 2 mM GDP-Fucose immediately prior to freezing. Data were obtained at the Advanced Photon Source, beam-line 23-ID-D for the apo structure and 24-ID-C for the ligand-bound structure (NE-CAT).
Structure determination
Diffraction images were indexed and integrated using XDS (Kabsch 2010). Initial phases were produced by molecular replacement in Phenix (Adams et al. 2010) using PDB 3ZY4 as a search model. A starting model for human POFUT1 was generated using Phenix auto-build, with subsequent manual building and review in Coot (Emsley et al. 2010). Structure refinement was performed in Phenix (Adams et al. 2010).
Coordinates have been deposited into the protein data bank. The PDB accession code for the free POFUT1 structure is 5UX6, and the code for POFUT1 complexed with GDP-fucose is 5UXH.
Supplementary Material
Acknowledgements
We thank Kelly Arnett for helpful discussions and Roberta Pascolutti for assistance with flow cytometry experiments. This work is based upon research conducted at the Northeastern Collaborative Access Team beamlines, which are funded by the National Institute of General Medical Sciences from the National Institutes of Health (P41 GM103403). The Pilatus 6 M detector on 24-ID-C beam line is funded by a NIH-ORIP HEI grant (S10 RR029205). This research used resources of the Advanced Photon Source, a U.S. Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory under Contract No. DE-AC02-06CH11357.
Supplementary data
Funding
National Institutes of Health [CA092433 to S.C.B.], and a Leukemia and Lymphoma Society Postdoctoral Fellowship (to M.L.).
References
- Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. 2010. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 66:213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrawes MB, Xu X, Liu H, Ficarro SB, Marto JA, Aster JC, Blacklow SC. 2013. Intrinsic selectivity of Notch 1 for Delta-like 4 over Delta-like 1. J Biol Chem. 288:25477–25489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashkenazy H, Abadi S, Martz E, Chay O, Mayrose I, Pupko T, Ben-Tal N. 2016. ConSurf 2016: an improved methodology to estimate and visualize evolutionary conservation in macromolecules. Nucleic Acids Res. 44:W344–W350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basmanav FB, Fritz G, Lestringant GG, Pachat D, Hoffjan S, Fischer J, Wehner M, Wolf S, Thiele H, Altmuller J, et al. 2015. Pathogenicity of POFUT1 in Dowling-Degos disease: additional mutations and clinical overlap with reticulate acropigmentation of kitamura. J Invest Dermatol. 135:615–618. [DOI] [PubMed] [Google Scholar]
- Blaumueller CM, Qi H, Zagouras P, Artavanis-Tsakonas S. 1997. Intracellular cleavage of Notch leads to a heterodimeric receptor on the plasma membrane. Cell. 90:281–291. [DOI] [PubMed] [Google Scholar]
- Cordle J, Redfieldz C, Stacey M, van der Merwe PA, Willis AC, Champion BR, Hambleton S, Handford PA. 2008. Localization of the delta-like-1-binding site in human Notch-1 and its modulation by calcium affinity. J Biol Chem. 283:11785–11793. [DOI] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, Cowtan K. 2010. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 66:486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge C, Liu T, Hou X, Stanley P. 2008. In vivo consequences of deleting EGF repeats 8-12 including the ligand binding domain of mouse Notch1. BMC Dev Biol. 8:48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge C, Stanley P. 2008. The O-fucose glycan in the ligand-binding domain of Notch1 regulates embryogenesis and T cell development. Proc Natl Acad Sci USA. 105:1539–1544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon WR, Roy M, Vardar-Ulu D, Garfinkel M, Mansour MR, Aster JC, Blacklow SC. 2009. a. Structure of the Notch1-negative regulatory region: implications for normal activation and pathogenic signaling in T-ALL. Blood. 113:4381–4390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon WR, Vardar-Ulu D, Histen G, Sanchez-Irizarry C, Aster JC, Blacklow SC. 2007. Structural basis for autoinhibition of Notch. Nat Struct Mol Biol. 14:295–300. [DOI] [PubMed] [Google Scholar]
- Gordon WR, Vardar-Ulu D, L'heureux6 S, Ashworth T, Malecki MJ, Sanchez-Irizarry C, Mcarthur DG, Histen G, Mitchell JL, Aster JC, et al. 2009. b. Effects of S1 cleavage on the structure, surface export, and signaling activity of human Notch1 and Notch2. PLoS One. 4:e6613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haltiwanger RS, Lowe JB. 2004. Role of glycosylation in development. Annu Rev Biochem. 73:491–537. [DOI] [PubMed] [Google Scholar]
- Hicks C, Johnston SH, diSibio G, Collazo A, Vogt TF, Weinmaster G. 2000. Fringe differentially modulates Jagged1 and Delta1 signalling through Notch1 and Notch2. Nat Cell Biol. 2:515–520. [DOI] [PubMed] [Google Scholar]
- Kabsch W. 2010. Xds. Acta Crystallogr D Biol Crystallogr. 66:125–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskowski RA, Swindells MB. 2011. LigPlot+: multiple ligand-protein interaction diagrams for drug discovery. J Chem Inf Model. 51:2778–2786. [DOI] [PubMed] [Google Scholar]
- Lira-Navarrete E, Valero-Gonzalez J, Villanueva R, Martinez-Julvez M, Tejero T, Merino P, Panjikar S, Hurtado-Guerrero R. 2011. Structural insights into the mechanism of protein O-fucosylation. PLoS One. 6:e25365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luca VC, Jude KM, Pierce NW, Nachury MV, Fischer S, Garcia KC. 2015. Structural biology. Structural basis for Notch1 engagement of Delta-like 4. Science. 347:847–853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malecki MJ, Sanchez-Irizarry C, Mitchell JL, Histen G, Xu ML, Aster JC, Blacklow SC. 2006. Leukemia-associated mutations within the NOTCH1 heterodimerization domain fall into at least two distinct mechanistic classes. Mol Cell Biol. 26:4642–4651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moloney DJ, Shair LH, Lu FM, Xia J, Locke R, Matta KL, Haltiwanger RS. 2000. Mammalian Notch1 is modified with two unusual forms of O-linked glycosylation found on epidermal growth factor-like modules. J Biol Chem. 275:9604–9611. [DOI] [PubMed] [Google Scholar]
- Okajima T, Reddy B, Matsuda T, Irvine KD. 2008. Contributions of chaperone and glycosyltransferase activities of O-fucosyltransferase 1 to Notch signaling. BMC Biol. 6:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okajima T, Xu A, Lei L, Irvine KD. 2005. Chaperone activity of protein O-fucosyltransferase 1 promotes notch receptor folding. Science. 307:1599–1603. [DOI] [PubMed] [Google Scholar]
- Okamura Y, Saga Y. 2008. Pofut1 is required for the proper localization of the Notch receptor during mouse development. Mech Dev. 125:663–673. [DOI] [PubMed] [Google Scholar]
- Panin VM, Shao L, Lei L, Moloney DJ, Irvine KD, Haltiwanger RS. 2002. Notch ligands are substrates for protein O-fucosyltransferase-1 and Fringe. J Biol Chem. 277:29945–29952. [DOI] [PubMed] [Google Scholar]
- Rana NA, Haltiwanger RS. 2011. Fringe benefits: functional and structural impacts of O-glycosylation on the extracellular domain of Notch receptors. Curr Opin Struct Biol. 21:583–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roti G, Carlton A, Ross KN, Markstein M, Pajcini K, Su AH, Perrimon N, Pear WS, Kung AL, Blacklow SC, et al. 2013. Complementary genomic screens identify SERCA as a therapeutic target in NOTCH1 mutated cancer. Cancer Cell. 23:390–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Irizarry C, Carpenter AC, Weng AP, Pear WS, Aster JC, Blacklow SC. 2004. Notch subunit heterodimerization and prevention of ligand-independent proteolytic activation depend, respectively, on a novel domain and the LNR repeats. Mol Cell Biol. 24:9265–9273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi S, Stanley P. 2003. Protein O-fucosyltransferase 1 is an essential component of Notch signaling pathways. Proc Natl Acad Sci USA. 100:5234–5239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahl M, Uemura K, Ge C, Shi S, Tashima Y, Stanley P. 2008. Roles of Pofut1 and O-fucose in mammalian Notch signaling. J Biol Chem. 283:13638–13651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanley P, Okajima T. 2010. Roles of glycosylation in Notch signaling. Curr Top Dev Biol. 92:131–164. [DOI] [PubMed] [Google Scholar]
- Takeuchi H, Haltiwanger RS. 2014. Significance of glycosylation in Notch signaling. Biochem Biophys Res Commun. 453:235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor P, Takeuchi H, Sheppard D, Chillakuri C, Lea SM, Haltiwanger RS, Handford PA. 2014. Fringe-mediated extension of O-linked fucose in the ligand-binding region of Notch1 increases binding to mammalian Notch ligands. Proc Natl Acad Sci USA. 111:7290–7295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Zou J, Zhao B, Johannsen E, Ashworth T, Wong H, Pear WS, Schug J, Blacklow SC, Arnett KL, et al. 2011. Genome-wide analysis reveals conserved and divergent features of Notch1/RBPJ binding in human and murine T-lymphoblastic leukemia cells. Proc Natl Acad Sci USA. 108:14908–14913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng AP, Ferrando AA, Lee W, Morris J.P.t., Silverman LB, Sanchez-Irizarry C, Blacklow SC, Look AT, Aster JC. 2004. Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science. 306:269–271. [DOI] [PubMed] [Google Scholar]
- Xu A, Haines N, Dlugosz M, Rana NA, Takeuchi H, Haltiwanger RS, Irvine KD. 2007. In vitro reconstitution of the modulation of Drosophila Notch-ligand binding by Fringe. J Biol Chem. 282:35153–35162. [DOI] [PubMed] [Google Scholar]
- Xu X, Choi SH, Hu T, Tiyanont K, Habets R, Groot AJ, Vooijs M, Aster JC, Chopra R, Fryer C, et al. 2015. Insights into autoregulation of Notch3 from structural and functional studies of its negative regulatory region. Structure. 23:1227–1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Argaw AT, Gurfein BT, Zameer A, Snyder BJ, Ge C, Lu QR, Rowitch DH, Raine CS, Brosnan CF, et al. 2009. Notch1 signaling plays a role in regulating precursor differentiation during CNS remyelination. Proc Natl Acad Sci USA. 106:19162–19167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimber-Strobl U, Strobl LJ, Meitinger C, Hinrichs R, Sakai T, Furukawa T, Honjo T, Bornkamm GW. 1994. Epstein-Barr virus nuclear antigen 2 exerts its transactivating function through interaction with recombination signal binding protein RBP-J kappa, the homologue of Drosophila Suppressor of Hairless. EMBO J. 13:4973–4982. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.