Abstract
Background and Aims
Ulcerative colitis [UC] is a chronic inflammatory disease of the colon. Colonoscopy remains the gold standard for evaluating disease activity, as clinical symptoms are not sufficiently accurate. The aim of this study is to identify new accurate non-invasive biomarkers based on whole-blood transcriptomics that can predict mucosal lesions and response to treatment in UC patients.
Methods
Whole-blood samples were collected for a total of 152 UC patients at endoscopy. Blood RNA from 25 UC individuals and 20 controls was analysed using microarrays. Genes that correlated with endoscopic activity were validated using real-time polymerase chain reaction in an independent group of 111 UC patients, and a prediction model for mucosal lesions was evaluated. Responsiveness to treatment was assessed in a longitudinal cohort of 16 UC patients who started anti-tumour necrosis factor [TNF] therapy and were followed up for 14 weeks.
Results
Microarray analysis identified 122 genes significantly altered in the blood of endoscopically active UC patients. A significant correlation with the degree of endoscopic activity was observed in several genes, including HP, CD177, GPR84, and S100A12. Using HP as a predictor of endoscopic disease activity, an accuracy of 67.3% was observed, compared with 52.4%, 45.2%, and 30.3% for C-reactive protein, erythrocyte sedimentation rate, and platelet count, respectively. Finally, at 14 weeks of treatment, response to anti-TNF therapy induced alterations in blood HP, CD177, GPR84, and S100A12 transcripts that correlated with changes in endoscopic activity.
Conclusions
Transcriptional changes in UC patients are sensitive to endoscopic improvement and appear to be an effective tool to monitor patients over time.
Keywords: Gene expression, blood biomarkers, ulcerative colitis, anti-TNF-α
1. Introduction
Ulcerative colitis [UC] is a chronic inflammatory disease of the colon. Disease is characterised by continuous mucosal inflammation,1 which in most patients, follows a remitting and relapsing course.2 Disease activity can be determined by assessments of clinical symptoms and endoscopic lesions.3 Nonetheless, the potential discrepancies between clinical symptoms and endoscopic lesions,4 as well as the association of mucosal healing with sustained remission and reduced surgery requirements,2,5 makes mucosal healing a therapeutic objective in UC management.
Although endoscopy is the gold standard for assessing disease activity and severity, it carries a considerable economic cost, discomfort for patients and, in severe cases, risk of procedure complications. Moreover, the need for endoscopic procedures may negatively impact on patient recruitment in clinical trials. Consequently, considerable efforts have been made to identify reliable non-invasive biomarkers that can predict disease activity. Available serological and faecal biomarkers, although widely used in clinical practice and clinical trials, have low diagnostic accuracy for the detection of ongoing inflammation, particularly in cases of mild or moderately active UC.6–8
We and others have previously shown that transcriptional analysis of intestinal biopsies is a useful tool for identifying not only disease biomarkers, but also molecular mechanisms of intestinal inflammation.9–13 Nonetheless, little evidence is available on the usefulness and reliability of transcriptional changes in whole-blood samples in patients suffering from inflammatory bowel disease [IBD].14
Here, we assess the sensitivity of whole-blood transcriptional changes to predict endoscopic lesions in UC. Whole-genome microarray analysis or real-time polymerase chain reaction [RT-PCR] of total blood was used in a total of 152 UC patients undergoing endoscopy to assess disease severity. Our study identifies and validates blood transcriptional biomarkers that can be used in clinical settings as predictors of endoscopic activity. Moreover, the sensitivity of the identified blood biomarkers as surrogate indicators of response to therapy was evaluated using a prospective cohort of patients receiving anti-tumour necrosis factor alpha [TNFα].
2. Materials and Methods
More detailed information is described in the supplementary materials and methods [available as Supplementary data at ECCO-JCC online].
2.1. Patient population and assessment of disease activity
Two cohorts of patients were included in this study. The first cohort came from a cross-sectional multicentre study, and included UC patients who had undergone clinical and endoscopic evaluations. From December 2012 to December 2015, 136 UC patients and 20 non-IBD controls {11 males; median age of 47 years (interquartile range [IQR]: 38–54)} were recruited at the Hospital Clinic Barcelona [Spain], the Hospital Universitari de Bellvitge [Spain], and the Clinical Hospital of the Faculty of Medical Sciences, University of Campinas [Brazil] [Table 1]. Non-IBD controls were those subjects undergoing a screening colonoscopy for colorectal cancer and who had had a normal examination. The second cohort came from an observational prospective study at Hospital Clinic Barcelona [Spain], and included UC patients who had been treated with an anti-TNFα antibody [infliximab, adalimumab or golimumab] and followed up for 14 weeks. All patients underwent clinical and endoscopic evaluations at Weeks 0 and 14. From January 2013 to September 2015, 16 UC patients were included [Table 2]. Patients with concomitant infections were excluded from both cohorts.
Table 1.
Clinical and demographic characteristics of UC patients from cohort 1. Patients with UC are divided into active or remission, based on their endoscopic Mayo scores. Continuous variables are described as median and IQR, and categorical variables as absolute frequencies and percentages; p-values from non-parametric tests are shown.
| Cohort 1: Clinical and demographic characteristics | |||
|---|---|---|---|
| Active UC | Remission UC | p-Value | |
| Total | 103 | 33 | — |
| Microarray [Group 1] | |||
| Total | 17 | 8 | — |
| Gender | 0.39 | ||
| Male | 12 [70.6%] | 4 [50%] | |
| Female | 5 [29.4%] | 4 [50%] | |
| Age [years] | 45 [35–54] | 52 [43–57] | 0.22 |
| Duration of disease [years] | 9 [3–12] | 13 [13–22] | < 0.01 |
| Extent of disease | 0.14 | ||
| Proctitis | 1 [5.9%] | 3 [37.5%] | |
| Left-sided colitis | 12 [70.6%] | 3 [37.5%] | |
| Extensive colitis | 4 [23.5%] | 2 [25%] | |
| Medication | 0.88 | ||
| No treatment | 2 [11.8%] | 2 [25%] | |
| 5-ASA | 10 [58.8%] | 5 [62.5%] | |
| Immunosuppressors | 2 [11.8%] | 1 [12.5%] | |
| Anti-TNF | 2 [11.8%] | 0 [0%] | |
| Steroids | 1 [5.9%] | 0 [0%] | |
| Combined therapy | 0 [0%] | 0 [0%] | |
| CRP [mg/dL] | 1.1 [0.3–1.8] | 0.2 [0.02–0.3] | < 0.01 |
| ESR [mm/h] | 16.0 [7.0–22.8] | 16.0 [12.0–18.3] | 0.94 |
| Platelet count [10^9/L] | 320 [291–381] | 233 [189–386] | 0.14 |
| Global Mayo Score | 7 [5–8] | 0 [0–0] | < 0.01 |
| Endoscopic Mayo Score | — | ||
| Score 0 | 0 [0%] | 8 [100%] | |
| Score 1 | 1 [5.9%] | 0 [0%] | |
| Score 2 | 4 [23.5%] | 0 [0%] | |
| Score 3 | 12 [70.6%] | 0 [0%] | |
| N/L Ratio | 3.3 [2.6–3.5] | 2.6 [2.2–2.9] | 0.20 |
| Real-Time PCR [Group 2] | |||
| Total | 86 | 25 | — |
| Gender | 0.25 | ||
| Male | 53 [61.6%] | 12 [48%] | |
| Female | 33 [38.4%] | 13 [52%] | |
| Age [years] | 45 [36–56] | 50 [37–60] | 0.42 |
| Duration of disease [eayrs] | 9 [4–15] | 15 [11–23] | < 0.01 |
| Extent of disease | 0.38 | ||
| Proctitis | 21 [24.4%] | 3 [12%] | |
| Left-sided colitis | 25 [29.1%] | 9 [36%] | |
| Extensive colitis | 40 [46.5%] | 13 [52%] | |
| Medication | 0.07 | ||
| No treatment | 4 [4.7%] | 4 [16%] | |
| 5-ASA | 32 [37.2%] | 13 [52%] | |
| Immunosuppressors | 18 [20.9%] | 5 [20%] | |
| Anti-TNF | 7 [8.1%] | 2 [8%] | |
| Steroids | 11 [12.8%] | 0 [0%] | |
| Combined therapy | 14 [16.3%] | 1 [4%] | |
| CRP [mg/dL] | 0.84 [0.17–2.24] | 0.12 [0.03–0.5] | < 0.01 |
| ESR [mm/h] | 11.5 [7.0–26.5] | 10.0 [5.5–15.5] | 0.25 |
| Platelet count [10^9/L] | 279 [228–323] | 252 [194–273] | 0.03 |
| Global Mayo Score | 6 [2–8.8] | 0 [0–0] | < 0.01 |
| Endoscopic Mayo Score | — | ||
| Score 0 | 0 [0%] | 25 [100%] | |
| Score 1 | 24 [27.9%] | 0 [0%] | |
| Score 2 | 21 [24.4%] | 0 [0%] | |
| Score 3 | 41 [47.7%] | 0 [0%] | |
| MS | 4 [2–7.8] | 0 [0–0] | — |
| Neutrophils [%] | 64.5 [57.1–68.7] | 61 [54.5–68.2] | 0.27 |
| Neutrophil count [10^9/L] | 3.9 [3.0–5.9] | 3.6 [2.9–4.9] | 0.28 |
| N/L ratio | 2.8 [1.9–3.2] | 2.1 [1.5–3.1] | 0.16 |
UC, ulcerative colitis; IQR, interquartile range; 5-ASA, 5-aminosalicylic acid; TNF, tumour necrosis factor;CRP, C-reactive protein; ESR, erythrocyte sedimentation rate; RT-PCR, real-time polymerase chain reaction; N/L ratio; neutrophil-lymphocyte ratio; MS; Modified Score [sum of endoscopic Mayo score of each segment], only available for patients with complete endoscopy procedure [n = 95].
Table 2.
Clinical and demographic characteristics of UC patients from cohort 2. Patients’ demographics and clinical characteristics at screening [Week 0] and at Week 14. Continuous variables are described as median and IQR, and categorical variables as absolute frequencies and percentages; p-values from paired analysis between Weeks 0 and 14 are shown.
| COHORT 2: Clinical and demographic characteristics | |||
|---|---|---|---|
| Number of cases | 16 | ||
| Gender [male/female] | |||
| Male | 9 [56.3%] | ||
| Female | 7 [43.7%] | ||
| Age [years] | 41 [35–53] | ||
| Duration of disease [years] | 6 [3–18] | ||
| Extent of disease | |||
| Proctitis | 0 [0%] | ||
| Left-sided colitis | 5 [31.3%] | ||
| Extensive colitis | 11 [68.7%] | ||
| Week 0 | Week 14 | p-Value | |
| Medication | < 0.01 | ||
| No treatment | 2 [12.5%] | 0 [0%] | |
| 5-ASA | 1 [6.3%] | 0 [0%] | |
| Immunosuppressors | 10 [62.5%] | 0 [0%] | |
| Anti-TNF | 2 [12.5%]a | 6 [37.5%] | |
| Steroids | 1 [6.3%] | 0 [0%] | |
| Combined therapy | 0 [0%] | 10 [62.5%] | |
| CRP [mg/dL] | 2 [0.4–3.7] | 0.5 [0.04–1.9] | 0.04 |
| ESR [mm/h] | 43 [17.5–66.5] | 32 [15.5–44] | 0.12 |
| Platelet count [10^9/L] | 368 [252–503] | 265 [208–504] | 0.50 |
| Global Mayo Score | 9 [8–9] | 6 [2–9] | < 0.01 |
| Endoscopic Mayo Score | 0.01 | ||
| Score 0 | 0 [0%] | 1 [6.3%] | |
| Score 1 | 0 [0%] | 3 [18.7%] | |
| Score 2 | 3 [18.7%] | 3 [18.7%] | |
| Score 3 | 13 [81.3%] | 9 [56.3%] | |
| MS | 9 [5.8–12] | 7 [2–9.8] | 0.04 |
| Neutrophils [%] | 69.2 [59.6–79.1] | 63.6 [56.7–73.42] | 0.29 |
| Neutrophil count [10^9/L] | 5.8 [3.7–7.5] | 3.2 [2.5–6.2] | 0.09 |
| N/L ratio | 3.7 [2.6–6.2] | 2.8 [2.1–4.4] | 0.02 |
UC, ulcerative colitis; IQR, interquartile range; 5-ASA, 5-aminosalicylic acid; TNF, tumour necrosis factor;CRP, C-reactive protein; ESR, erythrocyte sedimentation rate; N/L ratio; neutrophil-lymphocyte ratio; MS; Modified Score [sum of endoscopic Mayo score of each segment].
aThese two cases were treated with an anti-TNFα drug, but stopped due to no response and started a different one.
Institutional ethics committee approval was required in all centres and written informed consent was obtained for all patients.
As we hypothesised that changes in a systemic biomarker [such as a transcript in blood] may be influenced by the inflammatory burden [whole disease extension and activity at a given time point],15 two different endoscopic scores were reported; the endoscopic Mayo score and the Modified Score [MS, sum of the endoscopic Mayo score for the five colonic segments].16 For those cases with incomplete colonoscopy [n = 16], the MS could not be calculated. Endoscopic remission was defined as an endoscopic Mayo score or MS = 0.
2.2. Blood collection
Whole blood [2.5 ml] for RNA extraction was collected at the time of endoscopy using the PAXgene Blood RNA System [PreAnalytiX GmbH, Switzerland] according to the manufacturer’s instructions. Blood was also collected for standard clinical tests (C-reactive protein [CRP], erythrocyte sedimentation rate [ESR], platelet count, etc.) and to measure haptoglobin concentration [ADVIA Chemistry System, Siemens Healthineers, Germany].
2.3. RNA isolation
Total blood RNA was isolated using the PAXgene Blood RNA Kit [Qiagen, Spain] in a QIAcube instrument [Qiagene, Spain] following the manufacturer’s instructions. Additional DNase Treatment & Removal [Ambion, Life Technologies, USA] was used to safely eliminate DNA contamination from RNA samples. Purity and integrity of the total RNA were assessed with the 2100 Bioanalyzer [Agilent, Germany] and only samples with an RNA integrity number [RIN] > 7.0 were used. RNA was quantified using a NanoDrop spectrophotometer [Nanodrop Technologies, USA].
2.4. Microarray
The derived cDNA from whole blood [Group 1, Table 1], produced by Ovation® PicoSL WTA System V2 [NuGEN, USA], was hybridised to high-density oligonucleotide Affymetrix GeneChip Human Genome U133 Plus 2.0 Arrays [Affymetrix, USA] and raw data were analysed using Bioconductor tools in R [version 3.1.0].17–23
Microarray raw data [.cel files] and processed data have been deposited in the National Center for Biotechnology Information [NCBI] ’s Gene Expression Omnibus24 and are accessible through GEO Series accession number GSE94648 [https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc = GSE94648].
2.5. Quantitative RT-PCR
Microarray transcriptional data was validated using RT-PCR analysis for a selected set of 23 target genes and four endogenous controls [Supplementary Table 1, available as Supplementary data at ECCO-JCC online]. Technical validation was performed in a subset of samples previously evaluated by microarray. For biological validation, an independent group of samples [Group 2, Table 1] was employed.
Total RNA [500 ng] was transcribed to cDNA and real-time PCR was performed using custom-designed TaqMan Low Density Arrays [TLDA platform, Applied Biosystems, Life Technologies, USA].25
2.6. Statistical analysis
Continuous variables were described as median and IQR, and categorical variables as absolute frequencies. Pearson’s product moment correlation coefficient was applied for continuous variables in order to identify the linear association between paired samples. The Spearman rank correlation coefficient was applied in order to test any associations between continuous and ordinal variables or between continuous variables without assuming a linear relationship. The relationship between categorical variables was assessed by Fisher’s exact test. The Mann-Withney-Wilcoxon and Kruskal-Wallis tests were carried out to analyze the statistical significance of differences between groups [difference between two populations or among more than two populations, respectively]. To correct for multiple testing, the method of Benjamini and Hochberg23 was used. The logistic regression was used to predict endoscopic activity, and the lasso regression analysis method [least absolute shrinkage and selection operator] was performed for variable selection.26 The receiver operating characteristic [ROC] area under the curve [AUC] was then calculated to assess the ability to detect mucosal lesions in our models, and the DeLong’s test was used to determine significant differences between the ROC curves. A p-value ≤ 0.05 was considered statistically significant. All analyses were performed using R statistical environment [V.3.1.0].
3. Results
3.1. Whole-blood transcriptional analysis identifies changes in gene expression associated with the presence of endoscopic mucosal lesions in UC
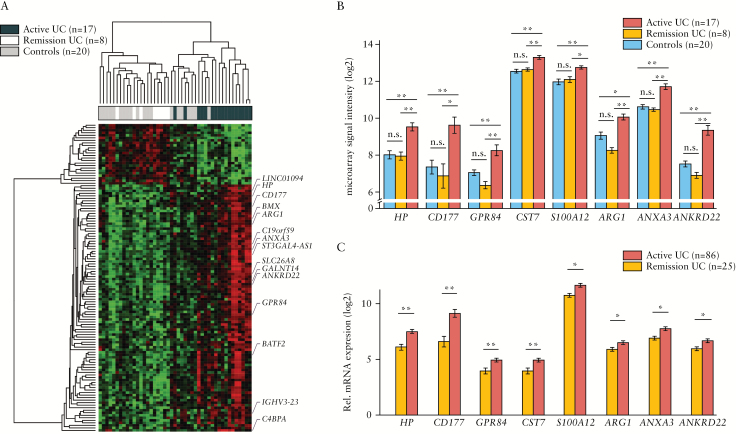
We first carried out whole-blood microarray transcriptional analysis of 25 UC patients with endoscopically active disease [n = 17] or in remission [n = 8] [Table 1, Group 1] and 20 non-IBD controls. This analysis revealed a set of 122 genes [80% upregulated] whose expression significantly changed in the peripheral blood of UC patients with endoscopic lesions compared with those in remission and non-IBD controls [Figure 1A, B; and Supplementary Table 2, available as Supplementary data at ECCO-JCC online]. The changes in blood gene expression associated with disease activity were modest. Only 15 of the identified 122 genes showed a greater than 2-fold change compared with non-IBD individuals and UC patients in endoscopic remission. A sizeable proportion of the identified genes are related to neutrophils: CD177,27 haptoglobin [HP],28 G-protein coupled receptor 84 [GPR84],29 hexokinase 3 [HK3],30,31 arginase 1 [ARG1],32 annexin A3 [ANXA3],33,34 and S100 calcium binding protein A12 [S100A12].35
Figure 1.
Whole-blood transcriptional profile associated with the presence of endoscopic mucosal lesions in UC. [A] Heatmap representation of differentially expressed genes in peripheral blood from UC and non-IBD controls according to microarray analysis [Table 1, Group 1]. A total of 122 differentially expressed genes in active UC patients [endoscopic Mayo score ≥ 1] compared with non-IBD controls and UC patients in remission [endoscopic Mayo score = 0] are represented and the 15 genes with a fold-change above 2 are highlighted. Up-regulated genes are shown in red and down-regulated genes are shown in green. Each row shows one individual gene and each column an experimental sample. Unsupervised hierarchical cluster method using Pearson distance and average linkage method was applied for gene and sample classification. Samples from non-IBD controls [shown in skyblue, n = 20], remission UC patients [shown in gold, n = 8], and active UC patients [shown in indianred, n = 18] are shown. [B] Bar plot representation [mean ± MSE] of microarray data for the eight genes that have been validated by RT-PCR [Table 1, Group 1]. Samples from non-IBD controls [shown in skyblue, n = 20], remission UC patients [shown in gold, n = 8], and active UC patients [shown in indianred, n = 18] are shown. [C] Bar plot representations [mean ± MSE] of the eight genes validated by RT-PCR in peripheral blood from UC patients [Table 1, Group 2]. Samples from remission UC patients [shown in gold, n = 25] and active UC patients [shown in indianred, n = 86] are shown; *P < 0.05, **P < 0.01 by pairwise Wilcoxon test. UC, ulcerative colitis; IBD, inflammatory bowel disease; MSE, mean squared error; RT-PCR, real-time polymerase chain reaction.
In order to validate these findings, we performed RT-PCR for a selected set of 23 genes [Supplementary Tables 1 and 2] in an independent group of UC patients [Table 1, Group 2]. We confirmed eight genes that are significantly altered in endoscopically active [n = 86] compared with non-active [n = 25] UC patients [Figure 1B, C; Supplementary Table 3, available as Supplementary data at ECCO-JCC online].
3.2. Transcriptional changes in blood correlate with the degree of endoscopic disease activity
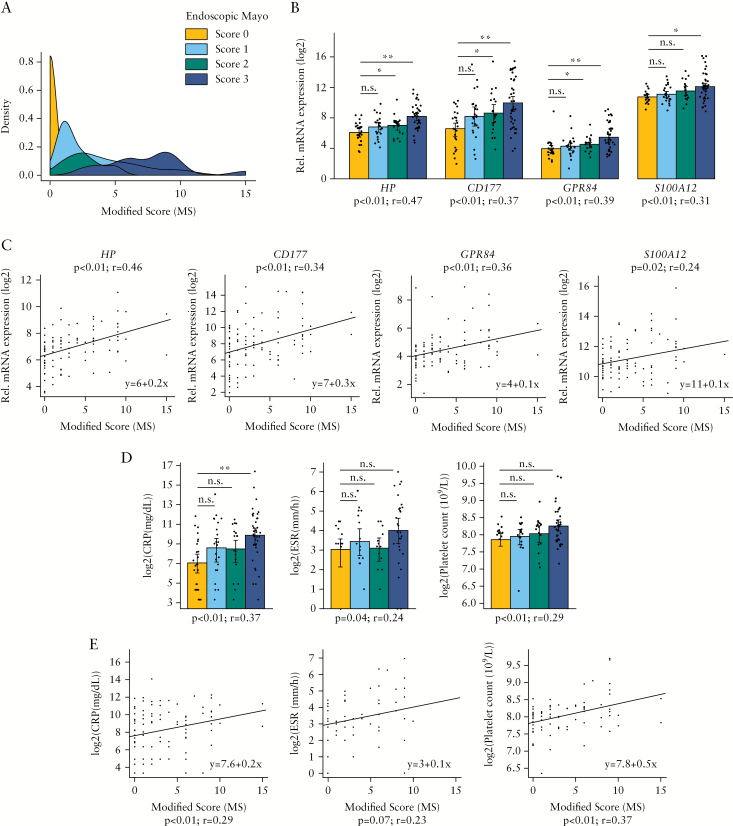
Next, we assessed whether whole-blood transcription could discriminate UC patients based on the degree of endoscopic disease severity [Table 1, Group 2]. We hypothesised that changes in a systemic biomarker [such as a transcript in blood] may be influenced by both severity and extension of the disease [inflammatory burden]. In order to test our hypothesis, two approaches were undertaken. First, we determined the correlation between gene expression and the endoscopic Mayo score; second, we assessed the correlation between gene expression and the MS [Modified Score; as a surrogate measure of inflammatory burden, see Materials and Methods]. The endoscopic Mayo score and the MS were highly correlated [Spearman’s rank correlation test, p < 0.05; rho = 0.9], but MS better reflected the accumulated inflammation along the colon [Figure 2A]. The higher the endoscopic Mayo score, the more flattened was the MS density function due to an increased range of disease extension. Examining the association of gene expression with the endoscopic disease activity, we identified 20 genes that significantly correlated with the endoscopic Mayo score and 18 genes that significantly correlated with MS [Spearman’s rank correlation test, p < 0.05; rho > 0.2] [Table 3]. A total of 16 genes were common, with HP, CD177, and GPR84 being those with the strongest relationship in both cases [Figure 2B, C]. Also significant was the relationship with the inflammation-related molecule S100A12. Moreover, pairwise analysis of the interrogated genes revealed HP, CD177, and GPR84 as the only ones significantly regulated in patients with an endoscopic Mayo score of 2 compared with 0 [Table 3 and Figure 2B]. None of the genes was able to detect changes between patients with an endoscopic Mayo score of 1 compared with 0.
Figure 2.
Association between blood transcripts and serological biomarkers with the degree of endoscopic inflammation. [A] Representations of probability density function of Modified Score [MS] for each endoscopic Mayo score. Density function of UC patients from Group 2 [Table 1] with endoscopic Mayo score equals 0 [n = 25, shown in gold], patients with endoscopic Mayo score = 1 [n = 22; shown in skyblue], patients with endoscopic Mayo score = 2 [n = 19; shown in seagreen], and patients with endoscopic Mayo score = 3 [n = 29; shown in darkblue] are shown. [B] Bar plot representation [mean ± MSE] of HP, CD177, GPR84, and S100A12 gene expression from RT-PCR analysis. Patients are categorised by endoscopic Mayo score; patients with endoscopic Mayo score = 0 [n = 25, shown in gold], patients with endoscopic Mayo score = 1 [n = 24; shown in skyblue], patients with endoscopic Mayo score = 2 [n = 21; shown in seagreen], and patients with endoscopic Mayo score = 3 [n = 41] shown in darkblue] are shown. Additionally, the data point of each observation is represented in the plot; *p < 0.05, **p < 0.01 by pairwise Wilcoxon test. The Spearman’s rho and p-value are also shown. [C] Correlation between MS and gene expression of HP, CD177, GPR84, and S100A12 [RT-PCR data]. The linear regression model and Spearman’s rho and p-value are shown. [D] Bar plot representation [mean ± MSE] of CRP, ESR, and platelet count by endoscopic Mayo score [same observations as panel B]; *p < 0.05, **p < 0.01 by pairwise Wilcoxon test. The Spearman’s rho and p-value are also shown. [E] Correlation between MS and CRP, ESR, and platelet count [same observations as panel C]. The linear regression model and Spearman’s rho and p-value are shown. UC, ulcerative colitis; MSE, mean squared error; RT-PCR, real-time polymerase chain reaction; CRP, C-reactive protein; ESR, erythrocyte sedimentation rate.
Table 3.
List of genes assessed by RT-PCR in UC patients [Table 1, Group 2]. For each target gene, comparisons between inactive [endoscopic Mayo score = 0; n = 25] and active [endoscopic Mayo score ≥ 1; n = 86] UC patients using the Wilcoxon test, the correlation of the transcriptional profile and the endoscopic activity [endoscopic Mayo score and MS] using the Spearman rank correlation coefficient, and the pairwise analysis of endoscopic Mayo score using an adjusted Wilcoxon test for multiple comparisons was carried out. The p-value or Benjamini-Hochberg adjusted p-value [FDR] and the mean fold-change [Fch] are shown for each comparison. The p-value and rho value are shown for the Spearman correlation.
| Wilcoxon | Spearman correlation | Pairwise analysis | Spearman correlation | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Active vs inactive | Endoscopic Mayo score | Score 1 vs 0 | Score 2 vs 0 | Score 3 vs 0 | MS | |||||||
| Gene | p-Value | Fch | P-value | rho | FDR | Fch | FDR | Fch | FDR | Fch | p-Value | rho |
| HP | 0.000 | 2.656 | 0.000 | 0.471 | 0.105 | 1.633 | 0.026 | 1.894 | 0.000 | 4.246 | 0.000 | 0.465 |
| CD177 | 0.001 | 5.825 | 0.000 | 0.370 | 0.120 | 3.000 | 0.043 | 4.091 | 0.001 | 10.294 | 0.001 | 0.338 |
| GPR84 | 0.001 | 1.973 | 0.000 | 0.393 | 0.119 | 1.276 | 0.025 | 1.507 | 0.002 | 2.925 | 0.000 | 0.362 |
| CST7 | 0.005 | 1.518 | 0.001 | 0.321 | 0.199 | 1.249 | 0.192 | 1.280 | 0.007 | 1.842 | 0.002 | 0.319 |
| S100A12 | 0.017 | 1.877 | 0.001 | 0.309 | 0.423 | 1.250 | 0.101 | 1.724 | 0.026 | 2.493 | 0.022 | 0.243 |
| ARG1 | 0.036 | 1.507 | 0.001 | 0.302 | 0.796 | 1.155 | 0.366 | 1.219 | 0.016 | 1.963 | 0.124 | 0.160 |
| ANXA3 | 0.037 | 1.811 | 0.001 | 0.336 | 0.533 | 1.263 | 0.210 | 1.482 | 0.029 | 2.582 | 0.005 | 0.318 |
| ANKRD22 | 0.048 | 1.637 | 0.005 | 0.280 | 0.980 | 1.032 | 0.108 | 1.526 | 0.088 | 2.108 | 0.041 | 0.221 |
| ZDHHC19 | 0.050 | 1.879 | 0.002 | 0.293 | 0.627 | 1.183 | 0.520 | 1.305 | 0.035 | 2.918 | 0.019 | 0.244 |
| HK3 | 0.054 | 1.289 | 0.001 | 0.313 | 0.929 | 1.042 | 0.537 | 1.125 | 0.017 | 1.564 | 0.016 | 0.247 |
| GALNT14 | 0.057 | 1.589 | 0.000 | 0.360 | 0.939 | 1.063 | 0.630 | 1.214 | 0.006 | 2.278 | 0.009 | 0.276 |
| CARD16 | 0.075 | 1.180 | 0.001 | 0.315 | 0.976 | -1.033 | 0.581 | 1.087 | 0.017 | 1.383 | 0.005 | 0.285 |
| CASP5 | 0.081 | 1.263 | 0.050 | 0.187 | 0.440 | 1.205 | 0.440 | 1.194 | 0.440 | 1.338 | 0.033 | 0.220 |
| AIM2 | 0.105 | 1.274 | 0.002 | 0.318 | 0.948 | -1.147 | 0.659 | 1.116 | 0.027 | 1.651 | 0.015 | 0.271 |
| TLR5 | 0.144 | 1.314 | 0.007 | 0.255 | 0.806 | -1.001 | 0.533 | 1.248 | 0.082 | 1.573 | 0.039 | 0.213 |
| MMP9 | 0.147 | 1.448 | 0.008 | 0.252 | 0.759 | 1.053 | 0.421 | 1.182 | 0.116 | 1.927 | 0.042 | 0.209 |
| CA4 | 0.161 | 1.261 | 0.028 | 0.209 | 0.874 | 1.113 | 0.874 | 1.054 | 0.224 | 1.487 | 0.322 | 0.103 |
| C1QB | 0.254 | 1.214 | 0.263 | 0.108 | 0.814 | 1.048 | 0.586 | 1.349 | 0.586 | 1.254 | 0.028 | 0.227 |
| CLEC4D | 0.364 | 1.224 | 0.001 | 0.299 | 0.126 | -1.244 | 0.358 | 1.163 | 0.046 | 1.606 | 0.019 | 0.240 |
| FBXO6 | 0.456 | 1.135 | 0.032 | 0.203 | 0.831 | -1.022 | 0.831 | -1.035 | 0.157 | 1.343 | 0.051 | 0.201 |
| SLC26A8 | 0.465 | 1.168 | 0.011 | 0.241 | 0.327 | -1.236 | 0.948 | 1.050 | 0.095 | 1.530 | 0.035 | 0.216 |
| TDRD9 | 0.659 | 1.238 | 0.030 | 0.206 | 0.623 | -1.064 | 0.329 | -1.081 | 0.088 | 1.690 | 0.268 | 0.115 |
| CD274 | 0.964 | 1.109 | 0.248 | 0.114 | 0.503 | -1.236 | 0.919 | 1.086 | 0.594 | 1.318 | 0.147 | 0.154 |
UC, ulcerative colitis; RT-PCR, real-time polymerase chain reaction; MS, Modified Score.
Furthermore, the association between routinely used blood biomarkers [CRP, ESR, and platelet counts] and endoscopic disease activity was explored [Supplementary Table 3 and Figure 2D, E]. Although significant correlations of the biomarkers with disease severity were observed, pairwise analysis revealed significant changes when comparing mild, moderate, or severe activity with inactive UC only for CRP [elevated exclusively in severe disease]. Additionally, given the neutrophil-related signature we had identified, we explored the potential relationship between neutrophil counts and endoscopic disease activity and found no significant associations [Supplementary Table 3 and Supplementary Figure 1, available as Supplementary data at ECCO-JCC online].
3.3. Haptoglobin [HP] is the best transcript for predicting mucosal lesions in UC patients
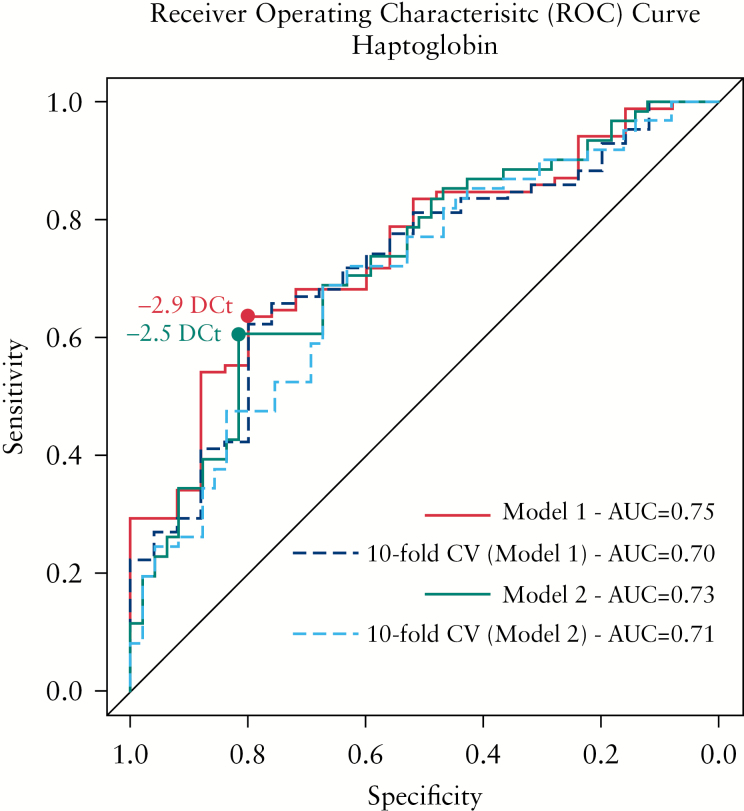
Within the subset of genes whose expression best correlates with endoscopic disease severity, we found HP as the best gene to predict the presence of active mucosal lesions in UC patients. The logistic regression model for endoscopic activity prediction [endoscopic Mayo score: 0 versus ≥ 1] was assessed. The fitted model demonstrated a significant effect of HP [p < 0.001], with an odds ratio [OR] of 1.9 (95% confidence interval [CI]: 1.35–2.78) and an AUC = 0.75 [95% CI: 0.64–0.85]. Defining a cut-off of -2.9 DCt, the sensitivity and specificity were 63.5% and 80.0%, respectively [Table 4 and Figure 3─red line]. To evaluate the reproducibility of the model, a 10-fold cross-validation was done, obtaining an AUC = 0.70 [95% CI: 0.59–0.82] [Figure 3─darkblue dashed line]. Given that no significant differences were found in the transcription of HP in blood between patients with an endoscopic Mayo score of 0 and 1, the power of HP to predict moderate to severe inflammation [endoscopic Mayo score of 2 or 3] compared with no or mild inflammation [endoscopic Mayo score of 0 or 1] was explored. The fitted model similarly revealed a significant effect of HP [p < 0.001], with an odds ratio [OR] of 1.8 [95% CI: 1.36–2.50] and an AUC = 0.73 [95% CI: 0.63–0.82]. Defining a cut-off of -2.5 DCt, the sensitivity and specificity were 60.7% and 81.6%, respectively [Table 4 and Figure 3─green line]. The 10-fold cross-validation showed an AUC = 0.71 [95% CI: 0.61–0.80] [Figure 3─skyblue dashed line]. Both approximations gave similar results, showing HP to have fair discrimination ability in predicting mucosal lesions in UC patients.
Figure 3.
Predictive power of haptoglobin [HP] gene for disease activity in UC patients. The receiver operating characteristic curve [ROC] is shown for different models. Red line shows the ROC curve from Model 1, where the binary response is defined as inactive [endoscopic Mayo score 0] versus active [endoscopic Mayo score 1–3]. The darkblue dashed line shows the 10-fold cross-validation from Model 1. Green line shows the ROC curve from Model 2, where the binary response is defined as inactive [endoscopic Mayo score 0–1] versus active [endoscopic Mayo score 2–3]. The skyblue dashed line shows the 10-fold cross-validation from Model 2. The cut-off points associated with the best specificity and sensitivity are shown [dots] for each Model. The AUC of each ROC curve are shown. UC, ulcerative colitis; AUC, area under the curve.
Table 4.
Diagnostic accuracy of haptoglobin gene expression and serological CRP, ESR, platelet counts, and HP protein for the detection of endoscopically active disease in ulcerative colitis. For each variable, the AUC is given and the sensitivity [Se], specificity [Sp], positive predictive value [PPV], negative predictive value [NPV], and accuracy were calculated and are shown as percentages based on the defined cut-off points. The CRP [normally < 1 mg/dL], ESR [normally < 20 mm/h], and platelet count [normally < 400· 10^9/L] cut-offs were defined according to the normal levels.
| Biomarker | AUC | Cut-off | Se | Sp | PPV | NPV | Accuracy |
|---|---|---|---|---|---|---|---|
| HP transcript [DCt] | 0.75 | -2.9 DCta | 63.5 | 80.0 | 91.5 | 39.2 | 67.3 |
| HP transcript [DCt] | 0.73 | -2.5 DCtb | 60.7 | 81.6 | 80.4 | 62.5 | 70.0 |
| CRP [mg/dL] | 0.72 | 1 mg/dL | 43.2 | 83.3 | 89.7 | 30.3 | 52.4 |
| ESR [mm/h] | 0.60 | 20 mm/h | 34.5 | 86.7 | 90.9 | 25.5 | 45.2 |
| Platelet count [10^9/L] | 0.66 | 400·10^9/L | 12.7 | 100 | 100 | 22.5 | 30.3 |
| HP protein[g/L] | 0.70 | 1.32 g/La | 48.7 | 90.9 | 94.9 | 33.9 | 58.2 |
CRP, C-reactive protein; ESR, erythrocyte sedimentation rate; AUC, area under the curve.
aCut-off point resulted from the regression model with binary response defined as inactive [Endoscopic Mayo score 0] versus active [Endoscopic Mayo score 1–3].
bCut-off point resulted from the regression model with binary response defined as inactive [Endoscopic Mayo score 0–1] versus active [Endoscopic Mayo score 2–3].
Moreover, serological protein levels of HP were measured for the majority of patients [n = 98] from Group 2 [Table 1] and a significant correlation with endoscopic disease activity and a fair predictive accuracy were observed [Table 4; Supplementary Figure 2, available as Supplementary data at ECCO-JCC online].
Although the diagnostic accuracy of HP is superior to that obtained from the widely used clinical biomarkers for IBD activity [CRP, ESR, and platelet count] and serological HP, no significant differences were found between ROC curves [DeLong’s test p-value > 0.05] [Table 4].
3.4. Changes in blood transcriptional biomarkers correlate with response to anti-TNFα therapy
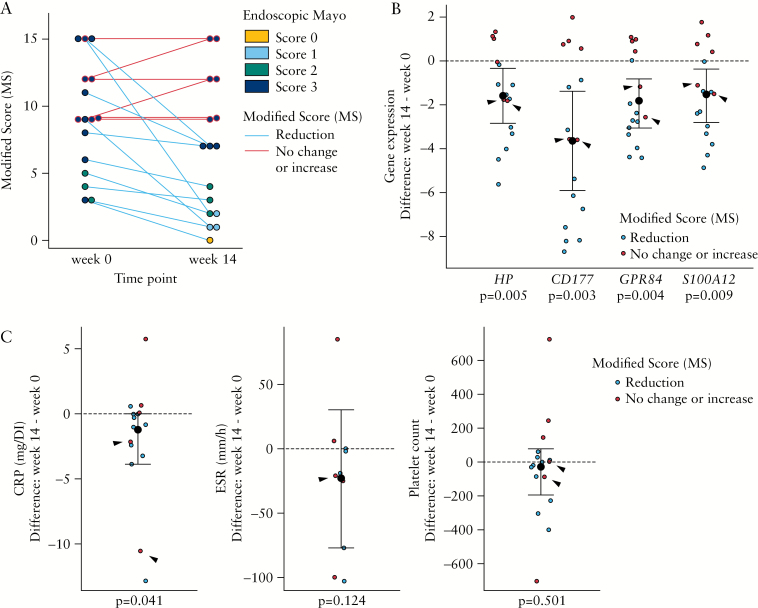
Finally, the ability of the identified transcriptional biomarkers [HP, CD177, GPR84, and S100A12] to evaluate changes in endoscopic activity in response to biological therapy was assessed. A cohort of UC patients with active disease starting anti-TNFα treatment [n = 16] were followed up over 14 weeks. Table 2 reports the clinical and endoscopic parameters at Weeks 0 and 14. Despite the fact that only four patients achieved an endoscopic Mayo score ≤ 1 at Week 14, a decrease in the global severity or extension of the endoscopic lesions was observed in the majority of cases. By reporting the MS, we identify a reduction of MS of at least 1 point in 10 cases, no changes in four, and worsening in two [Figure 4A]. In contrast, only five cases showed an endoscopic response based on the endoscopic Mayo score [reduction in endoscopic Mayo score of at least 1 point] and no changes were detected in the remaining cases. Furthermore, a better concordance is observed between the response based on the Global Mayo score and MS compared with the endoscopic Mayo score [Supplementary Figure 3, available as Supplementary data at ECCO-JCC online]. This highlights the importance of reporting both disease extension and activity to accurately reflect changes in disease activity over time.
Figure 4.
Changes in whole-blood transcriptional profiles are associated with endoscopic progression in UC patients after 14 weeks of anti-TNFα treatment. [A] Distribution of MS in patients with UC before and after anti-TNFα treatment [n = 16, Table 2]. Each dot represents an individual patient at Week 0 or 14. Dot colour corresponds to endoscopic Mayo score and the colour of the connecting line to the endoscopic progression [assessed by MS]. Blue connecting line corresponds to mucosal improvement [n = 10] and red connecting line to no improvement or worsening [n = 6]. [B] Dot plot representation of HP, CD177, GPR84, and S100A12 gene expression changes after 14 weeks of anti-TNFα treatment. Y-axis shows the difference between gene expression at Weeks 14 and 0. [C] Dot plot representation of CRP, ESR, platelet count, and serological HP changes after 14 weeks of anti-TNFα treatment. Y-axis shows the difference between biomarker levels at Weeks 14 and 0. Black dashed line at point 0 denotes no changes in gene expression. Based on MS, individuals with mucosal improvement [shown in blue, n = 10] and those without improvement or worsening [shown in red, n = 6] are shown. Black arrows show two samples without mucosal improvement [based on MS], but decreased gene expression after treatment. Median ± IQR are represented. UC, ulcerative colitis; MS, Modified Score; CRP, C-reactive protein; ESR, erythrocyte sedimentation rate; TNF, tumour necrosis factor; IQR, interquartile range.
Importantly, we found that a decrease in the expression of HP, CD177, GPR84, and S100A12 was associated with a reduction in the MS. As shown in Figure 4B, the changes observed in the transcription of those genes permitted differentiation between those patients who, based on MS, endoscopically improved [blue dots] and those who worsened or did not change [red dots]. Interestingly, in two patients [black arrows], expression of the transcriptional biomarkers decreased at Week 14 despite no improvement in their MS. Although in both patients an improvement in the severity of the disease [a marked decrease in the number of ulcers] was reported, the ulcers persisted in all involved segments and therefore no change in the endoscopic score [endoscopic Mayo score of 3, or MS of 15 and 9] was noted. The behaviour of serological markers as well as serum HP protein before and after treatment was also evaluated and showed no significant association with the mucosal changes with the exception of CRP [Table 2 and Figure 4C].
4. Discussion
The importance of monitoring local inflammatory activity in IBD is becoming increasingly clear. There can, however, be discrepancies between clinical symptoms and the severity of mucosal lesions. In this context, clinical trials relying on clinical scores to assess activity at inclusion or efficacy as an endpoint, may result in high placebo response rates, as well as unreliable un-reproducible results.
Blood transcriptional signatures have been explored in infectious and autoimmune systemic diseases and cancer.36–39 The use of whole-blood transcriptional analysis to detect disease activity nonetheless remains a largely unexplored tool in IBD. Compared with the dramatic transcriptional changes observed in the intestinal mucosa of patients with active inflammation, the degree of transcriptional regulation in blood, even in patients with severe disease, is remarkably low. This is not entirely unexpected, given that circulating cells may only partially reflect changes in the mucosa of UC patients. Nonetheless, using sensitive RT-PCR analysis, we can predict endoscopic mucosal lesions in patients with UC.
Our data reveal the important contribution of neutrophils to the systemic inflammatory response in UC patients. We show that expressions of neutrophil-related genes [including CD177, HP, S100A12, ARG1, and ANXA3], rather than total neutrophil numbers, are surrogate markers of endoscopic activity. Remarkably, an increase in serum and faecal concentrations of S100A12 protein had been previously shown to correlate with disease activity in IBD patients.35,40 In contrast, the increased levels of serum calprotectin [S100A8/A9], described as a new diagnostic and prognostic marker in IBD,41 were not observed here at a transcriptional level. Similarly, the neutrophil gelatinase B-associated lipocalin [LCN2] and the matrix metalloproteinase-9 [MMP9] complex have been proposed as surrogate serum markers for assessing mucosal healing in UC,42 but were not transcriptionally regulated in blood from our patient cohort. Along the same lines, whereas we identified HP transcript as a sensitive predictor of mucosal lesions and therapy response, changes in serum HP are not sensitive to such alterations. HP protein is primarily synthesised by hepatocytes43 but it has also been described as a specific granule protein of neutrophils,28 which could explain the transcript detection at the blood level and the moderate correlation between transcript and protein measurements [r = 0.40]. Altogether, this suggests that although changes in serum proteins due to disease activity can originate in other tissues [eg liver, intestine], transcription consistently reflects changes in the activation and abundance of cells present in the peripheral compartment. Therefore, blood transcription and serum protein concentrations may not necessarily reflect the same biological processes.
Besides secreted or granule-associated proteins, we also identified the cell membrane proteins CD177 and GPR84 as predictors of endoscopic lesions. CD177 is a neutrophil-specific GPI-anchored glycoprotein normally expressed on a subpopulation of neutrophils.44 Although there are controversial results, the preferential recruitment of CD177+ neutrophils towards inflamed tissues has been suggested,45 and the increased protein and transcripts for CD177 have been described in bacterial infections, polycythaemia vera, and ANCA-associated vasculitis as well as in response to G-CSF stimulation.27,46 Interestingly, elevated expression of both CD177 and HP genes in whole blood has been closely associated with ipilimumab-induced colitis in advanced melanoma patients, also suggesting that neutrophils potentially play a broader role in immune-related colonic disorders.47
GPR84, mainly expressed by leukocytes, is a G protein-coupled receptor that has been described as a putative receptor for medium-chain fatty acids [MCFAs] which elicits neutrophil and macrophage chemotaxis.29 The efficacy of inhibiting GPR84 has been studied in a mouse chronic colitis model and in human UC patients [clinical trial: NCT02337608]. Although a diminution of colonic lesions and a reduction in neutrophil infiltrate was observed in mouse,48 results in human showed safety and tolerance but the study did not meet the efficacy endpoints.49 Nevertheless, our data points to the potential use of this gene as a biomarker of disease activity.
Taken together, we provide insights on new and promising minimally invasive biomarkers capable of predicting endoscopic mucosal lesions in patients with UC. We realise that the lack of faecal calprotectin [FC] represents the major drawback of our study, as FC has demonstrated to be the best biomarker of endoscopic activity.15 Despite considerable efforts to collect this biomarker in our study cohorts, there remained a large number of missing values, rendering it impossible to include this datum in the analysis and restricting our study to the use of other available biomarkers with poor correlation with endoscopic activity, like CRP, ESR, and platelet counts. The difficulties in FC sampling were also pointed out in the first clinical study that used FC as a primary endpoint.50 Nonetheless, the results obtained by Sandborn et al.51 regarding the ability of FC to predict endoscopic remission are remarkably similar to the prediction power of HP in our study [Supplementary File 1, available as Supplementary data at ECCO-JCC online]. Whereas additional studies should be done, as the total number of patients with endoscopically inactive disease in our cohort is limited, we would like to stress that with potentially comparable predictive values, whole-blood biomarkers can offer practical advantages in clinical practice compared with faecal biomarkers.
Another limitation, as distinct inflammatory mechanisms have been suggested between early and late inflammatory bowel diseases,52–54 could be the difference in disease duration between our active and inactive patients. However, no significant correlation between disease duration and expression of the selected transcriptional biomarkers was found.
Importantly, our data reveal the sensitivity of the identified blood biomarkers to mirror changes in local inflammatory activity in UC patients following induction therapy. Although our results are promising, they are derived from a relatively small number of patients and restricted to anti-TNFα therapy. It will be necessary to validate the usefulness of the identified biomarkers in predicting response to drugs with mechanisms of action that differ from those of TNFα blockade. Interestingly, whole-blood CD177, HP, and GPR84 transcripts were found at higher levels in corticosteroid-resistant children with severe UC compared with corticosteroid-responsive children,14 suggesting these are also sensitive to the effect of corticosteroids.
In summary, our study has the merit of assessing in a multicentric cohort the usefulness of transcriptional blood analysis to predict endoscopic disease activity. By starting with a whole-genome approach, we were able to identify transcripts with the potential to change based on the presence of endoscopic lesions. Importantly, we validated a subset of genes in a large group of patients with a wide range of endoscopic activity and disease extent. More importantly, the transcriptional blood biomarkers were tested for responsiveness in a prospective cohort and proved to be sensitive to changes in endoscopic disease activity.
Funding
This work was supported by Boehringer Ingelheim Pharmaceuticals and the Leona and Harry Helmsley Trust.
Conflict of Interest
SV is an employee of Boehringer Ingelheim Pharmaceuticals. The authors declare that they have no competing interests.
Author Contributions
AS and JP designed the study; MCM, IO, IJA, ER, AR, RFL, MLSA, LR, and JG recruited patients; LR, IO, IJA, ER, AR, RFL, MLSA, and CSRC assessed clinical disease activity; RFL, NP, MCM, and ME performed the experiments; NP and JJL carried out bioinformatic and biostatistic analysis; SV participated in the discussion; and AS, NP, and JP wrote the manuscript.
Supplementary Data
Supplementary data are available at ECCO-JCC online.
Supplementary Material
Acknowledgments
We gratefully acknowledge all patients for their selfless participation. We also thank Joe Moore for editorial assistance.
References
- 1. Ordás I, Eckmann L, Talamini M, Baumgart DC, Sandborn WJ. Ulcerative colitis. Lancet 2012;380:1606–19. [DOI] [PubMed] [Google Scholar]
- 2. Solberg IC, Lygren I, Jahnsen J et al. ; IBSEN Study Group. Clinical course during the first 10 years of ulcerative colitis: results from a population-based inception cohort [IBSEN Study]. Scand J Gastroenterol 2009;44:431–40. [DOI] [PubMed] [Google Scholar]
- 3. Schroeder KW, Tremaine WJ, Ilstrup DM. Coated oral 5-aminosalicylic acid therapy for mildly to moderately active ulcerative colitis. A randomized study. N Engl J Med 1987;317:1625–9. [DOI] [PubMed] [Google Scholar]
- 4. Rosenberg L, Lawlor GO, Zenlea T et al. . Predictors of endoscopic inflammation in patients with ulcerative colitis in clinical remission. Inflamm Bowel Dis 2013;19:779–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Colombel JF, Rutgeerts P, Reinisch W et al. . Early mucosal healing with infliximab is associated with improved long-term clinical outcomes in ulcerative colitis. Gastroenterology 2011;141:1194––201.. [DOI] [PubMed] [Google Scholar]
- 6. Stidham RW, Higgins PD. Value of mucosal assessment and biomarkers in inflammatory bowel disease. Expert Rev Gastroenterol Hepatol 2010;4:285–91. [DOI] [PubMed] [Google Scholar]
- 7. D’Haens G, Ferrante M, Vermeire S et al. . Fecal calprotectin is a surrogate marker for endoscopic lesions in inflammatory bowel disease. Inflamm Bowel Dis 2012;18:2218–24. [DOI] [PubMed] [Google Scholar]
- 8. Lemmens B, Arijs I, Van Assche G et al. . Correlation between the endoscopic and histologic score in assessing the activity of ulcerative colitis. Inflamm Bowel Dis 2013;19:1194–201. [DOI] [PubMed] [Google Scholar]
- 9. Boland BS, Boyle DL, Sandborn WJ et al. . Validation of gene expression biomarker analysis for biopsy-based clinical trials in Crohn’s disease. Inflamm Bowel Dis 2015;21:323–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Leal RF, Planell N, Kajekar R et al. . Identification of inflammatory mediators in patients with Crohn’s disease unresponsive to anti-TNFα therapy. Gut 2015;64:233–42. [DOI] [PubMed] [Google Scholar]
- 11. Planell N, Lozano JJ, Mora-Buch R et al. . Transcriptional analysis of the intestinal mucosa of patients with ulcerative colitis in remission reveals lasting epithelial cell alterations. Gut 2013;62:967–76. [DOI] [PubMed] [Google Scholar]
- 12. Román J, Planell N, Lozano JJ et al. . Evaluation of responsive gene expression as a sensitive and specific biomarker in patients with ulcerative colitis. Inflamm Bowel Dis 2013;19:221–9. [DOI] [PubMed] [Google Scholar]
- 13. Wu F, Dassopoulos T, Cope L et al. . Genome-wide gene expression differences in Crohn’s disease and ulcerative colitis from endoscopic pinch biopsies: insights into distinctive pathogenesis. Inflamm Bowel Dis 2007;13:807–21. [DOI] [PubMed] [Google Scholar]
- 14. Kabakchiev B, Turner D, Hyams J et al. . Gene expression changes associated with resistance to intravenous corticosteroid therapy in children with severe ulcerative colitis. PLoS One 2010;5:e13085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Peyrin-Biroulet L, Panes J, Sandborn WJ et al. . Defining disease severity in inflammatory bowel diseases: Current and future directions. Clin Gastroenterol Hepatol 2016;14:348–54.e17. [DOI] [PubMed] [Google Scholar]
- 16. Lobatón T, Bessissow T, De Hertogh G et al. . The Modified Mayo Endoscopic Score [MMES]: a new index for the assessment of extension and severity of endoscopic activity in ulcerative colitis patients. J Crohns Colitis 2015;9:846–52. [DOI] [PubMed] [Google Scholar]
- 17. Gentleman RC, Carey VJ, Bates DM et al. . Bioconductor: open software development for computational biology and bioinformatics. Genome Biol 2004;5:R80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. R Development Core Team. R: A Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing, 2014. [Google Scholar]
- 19. Gautier L, Cope L, Bolstad BM, Irizarry RA. affy–analysis of Affymetrix GeneChip data at the probe level. Bioinformatics 2004;20:307–15. [DOI] [PubMed] [Google Scholar]
- 20. Dai M, Wang P, Boyd AD et al. . Evolving gene/transcript definitions significantly alter the interpretation of GeneChip data. Nucleic Acids Res 2005;33:e175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Johnson WE, Li C, Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics 2007;8:118–27. [DOI] [PubMed] [Google Scholar]
- 22. Ritchie ME, Phipson B, Wu D et al. . limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 2015;43:e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Benjamini Y, Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J R Stat Soc B 1995;57:289–300. [Google Scholar]
- 24. Edgar R, Domrachev M, Lash AE. Gene expression omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res 2002;30:207–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Vandesompele J, De Preter K, Pattyn F et al. . Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 2002;3:RESEARCH0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Efron B, Hastie T, Johnstone I, Tibshirani R. Least angle regression. Ann Stat 2004;32:407–99. [Google Scholar]
- 27. Hu N, Mora-Jensen H, Theilgaard-Mönch K et al. . Differential expression of granulopoiesis related genes in neutrophil subsets distinguished by membrane expression of CD177. PLoS One 2014;9:e99671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Theilgaard-Mönch K, Jacobsen LC, Nielsen MJ et al. . Haptoglobin is synthesized during granulocyte differentiation, stored in specific granules, and released by neutrophils in response to activation. Blood 2006;108:353–61. [DOI] [PubMed] [Google Scholar]
- 29. Suzuki M, Takaishi S, Nagasaki M et al. . Medium-chain fatty acid-sensing receptor, GPR84, is a proinflammatory receptor. J Biol Chem 2013;288:10684–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Rijksen G, Staal GE, Beks PJ, Streefkerk M, Akkerman JW. Compartmentation of hexokinase in human blood cells. Characterization of soluble and particulate enzymes. Biochim Biophys Acta 1982;719:431–7. [DOI] [PubMed] [Google Scholar]
- 31. Federzoni EA, Valk PJ, Torbett BE et al. . PU.1 is linking the glycolytic enzyme HK3 in neutrophil differentiation and survival of APL cells. Blood 2012;119:4963–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Munder M, Mollinedo F, Calafat J et al. . Arginase I is constitutively expressed in human granulocytes and participates in fungicidal activity. Blood 2005;105:2549–56. [DOI] [PubMed] [Google Scholar]
- 33. Le Cabec V, Maridonneau-Parini I. Annexin 3 is associated with cytoplasmic granules in neutrophils and monocytes and translocates to the plasma membrane in activated cells. Biochem J 1994;303[Pt 2]:481–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Mirsaeidi M, Gidfar S, Vu A, Schraufnagel D. Annexins family: insights into their functions and potential role in pathogenesis of sarcoidosis. J Transl Med 2016;14:89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Foell D, Kucharzik T, Kraft M et al. . Neutrophil derived human S100A12 [EN-RAGE] is strongly expressed during chronic active inflammatory bowel disease. Gut 2003;52:847–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lill M, Kõks S, Soomets U et al. . Peripheral blood RNA gene expression profiling in patients with bacterial meningitis. Front Neurosci 2013;7:33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hu X, Yu J, Crosby SD, Storch GA. Gene expression profiles in febrile children with defined viral and bacterial infection. Proc Natl Acad Sci U S A 2013;110:12792–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kam SH, Singh A, He JQ et al. . Peripheral blood gene expression changes during allergen inhalation challenge in atopic asthmatic individuals. J Asthma 2012;49:219–26. [DOI] [PubMed] [Google Scholar]
- 39. Galamb O, Sipos F, Solymosi N et al. . Diagnostic mRNA expression patterns of inflamed, benign, and malignant colorectal biopsy specimen and their correlation with peripheral blood results. Cancer Epidemiol Biomarkers Prev 2008;17:2835–45. [DOI] [PubMed] [Google Scholar]
- 40. Kaiser T, Langhorst J, Wittkowski H et al. . Faecal S100A12 as a non-invasive marker distinguishing inflammatory bowel disease from irritable bowel syndrome. Gut 2007;56:1706–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kalla R, Kennedy NA, Ventham NT et al. . Serum calprotectin: a novel diagnostic and prognostic marker in inflammatory bowel diseases. Am J Gastroenterol 2016;111:1796–805. [DOI] [PubMed] [Google Scholar]
- 42. de Bruyn M, Arijs I, Wollants WJ et al. . Neutrophil gelatinase B-associated lipocalin and matrix metalloproteinase-9 complex as a surrogate serum marker of mucosal healing in ulcerative colitis. Inflamm Bowel Dis 2014;20:1198–207. [DOI] [PubMed] [Google Scholar]
- 43. Quaye IK. Haptoglobin, inflammation and disease. Trans R Soc Trop Med Hyg 2008;102:735–42. [DOI] [PubMed] [Google Scholar]
- 44. Stroncek D. Neutrophil-specific antigen HNA-2a [NB1, CD177]: serology, biochemistry, and molecular biology. Vox Sang 2002;83[Suppl 1]:359–61. [DOI] [PubMed] [Google Scholar]
- 45. Wang L, Ge S, Agustian A, Hiss M, Haller H, von Vietinghoff S. Surface receptor CD177/NB1 does not confer a recruitment advantage to neutrophilic granulocytes during human peritonitis. Eur J Haematol 2013;90:436–7. [DOI] [PubMed] [Google Scholar]
- 46. Göhring K, Wolff J, Doppl W et al. . Neutrophil CD177 [NB1 gp, HNA-2a] expression is increased in severe bacterial infections and polycythaemia vera. Br J Haematol 2004;126:252–4. [DOI] [PubMed] [Google Scholar]
- 47. Shahabi V, Berman D, Chasalow SD et al. . Gene expression profiling of whole blood in ipilimumab-treated patients for identification of potential biomarkers of immune-related gastrointestinal adverse events. J Transl Med 2013;11:75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Dupont S, Arijs I, Blanque R et al. . Gpr84 inhibition as a novel therapeutic approach in IBD: Mechanistic and translational studies. In: 10th Congress of the European Crohn’s and Colitis Organisation [ECCO]; February 1821, 2015, Barcelona, Spain. [Google Scholar]
- 49. Vanhoutte F, Dupont S, Van Kaem T et al. . Human safety, pharmacokinetics and pharmacodynamics of the gpr84 antagonist glpg1205, a potential new approach to treat IBD. J Crohns Colitisournal of Crohns & Colitis 2015;9[Suppl 1]:5387. [Google Scholar]
- 50. Reinisch W, Panés J, Khurana S et al. . Anrukinzumab, an anti-interleukin 13 monoclonal antibody, in active UC: efficacy and safety from a phase IIa randomised multicentre study. Gut 2015;64:894–900. [DOI] [PubMed] [Google Scholar]
- 51. Sandborn WJ, Panés J, Zhang H, Yu D, Niezychowski W, Su C. Correlation between concentrations of fecal calprotectin and outcomes of patients with ulcerative colitis in a phase 2 trial. Gastroenterology 2016;150:96–102. [DOI] [PubMed] [Google Scholar]
- 52. Kugathasan S, Saubermann LJ, Smith L et al. . Mucosal T-cell immunoregulation varies in early and late inflammatory bowel disease. Gut 2007;56:1696–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Desreumaux P, Brandt E, Gambiez L et al. . Distinct cytokine patterns in early and chronic ileal lesions of Crohn’s disease. Gastroenterology 1997;113:118–26. [DOI] [PubMed] [Google Scholar]
- 54. Veny M, Esteller M, Ricart E, Piqué JM, Panés J, Salas A. Late Crohn’s disease patients present an increase in peripheral Th17 cells and cytokine production compared with early patients. Aliment Pharmacol Ther 2010;31:561–72. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.