Abstract
Bacterial levansucrases produce β(2,6)-linked levan-type polysaccharides using sucrose or sucrose analogs as donor/acceptor substrates. However, the dominant reaction of Bacillus megaterium levansucrase (Bm-LS) is hydrolysis. Single domain levansucrases from Gram-positive bacteria display a wide substrate-binding pocket with open access to water, challenging engineering for transfructosylation-efficient enzymes. We pursued a shift in reaction specificity by either modifying the water distribution in the active site or the coordination of the catalytic acid/base (E352) and the nucleophile (D95), thus affecting the fructosyl-transfer rate and allowing acceptors other than water to occupy the active site. Two serine (173/422) and two water-binding tyrosine (421/439) residues located in the first shell of the catalytic pocket were modified. Library variants of S173, Y421 and S422, which coordinate the position of D95 and E352, show increased transfructosylation (30–200%) and modified product spectra. Substitutions at position 422 have a higher impact on sucrose affinity, while changes at position 173 and 421 have a strong effect on the overall catalytic rate. As most retaining glycoside hydrolases (GHs) Bm-LS catalyzes hydrolysis and transglycosylation via a double displacement reaction involving two-transition states (TS1 and TS2). Hydrogen bonds of D95 with the side chains of S173 and S422 contribute a total of 2.4 kcal mol−1 to TS1 stabilization, while hydrogen bonds between invariant Y421, E352 and the glucosyl C-2 hydroxyl-group of sucrose contribute 2.15 kcal mol−1 stabilization. Changes at Y439 render predominantly hydrolytic variants synthesizing shorter oligosaccharides.
Keywords: Bacillus megaterium, fructansucrase, hydrolysis partition, levan, levansucrase, transfructosylation
Introduction
Bacterial fructansucrases (FSs) synthesize fructans of varying length by transfer of fructosyl units from sucrose or sucrose analogs to glycoside acceptors (transfructosylation reaction). FSs also catalyze the transfer of fructosyl moieties to water, which results in nonproductive sucrose hydrolysis (hydrolysis reaction) (Ortiz-Soto and Seibel 2014). Bacterial FSs belong to the glycoside hydrolase family 68 (GH68) (Lombard et al. 2014) and according to the linkage of their products they are classified as inulosucrases or levansucrases (producing β(2→1)- or β(2→6)-linked polysaccharides, respectively). GH68 and GH32 enzymes (constituting clan GH-J) share the 5-bladed β-propeller fold of their catalytic domain and some sequence motifs (Meng and Futterer 2003; Lammens et al. 2009; Pijning et al. 2011). Based on the CAZy database (http://www.cazy.org/) (Lombard et al. 2014), members of GH32 include levanases, inulinases, plant fructansucrases and β-fructofuranosidases.
Enzymes from clan GH-J (and in general GHs) catalyze hydrolysis and transfructosylation reactions with inherently different efficiencies and produce characteristic oligosaccharide/polymer spectra (Homann et al. 2007; Ortiz-Soto et al. 2008; Ortiz-Soto and Seibel 2014). Bm-LS catalyzes the formation of glycosidic bonds producing a series of oligosaccharides of variable length; however, this transfructosylating activity is overcome by hydrolysis and the enzyme behaves preferentially as an invertase, hydrolyzing around 90% of the substrate even at high sucrose concentrations (0.5 M) (Homann et al. 2007). Bacillus subtilis and B. megaterium levansucrases share an amino acid sequence identity of 75% (85% similarity) and according to their crystallographic structure display only small structural differences (B. subtilis levansucrase Protein Data Bank (PDB) codes 1oyg/1pt2 and Bm-Ls PDB entry 3om2). The former performs transfructosylation in a more efficient manner at lower substrate concentrations than Bm-LS (Ortiz-Soto et al. 2008). The apo- and sucrose-bound structures of both enzymes show that equivalent residues and water molecules in their active sites are superimposable, with exception of the side chain of R370 (Figure 1) (Meng and Futterer 2003, 2008; Strube et al. 2011). This may indicate that preference for hydrolysis or transfructosylation is either controlled by structural components located not necessarily within the catalytic pocket or that affinity for acceptor molecules other than water may increase as a result of very subtle changes in the central pocket amino acid's side chains. A number of studies focused on identification of structural or molecular determinants regulating the hydrolytic activity in retaining GH have been reported (Ritsema et al. 2006; Schroeven et al. 2008; Lafraya et al. 2011). However, molecular basis of transglycosylation remains elusive, in particular for GH68 enzymes.
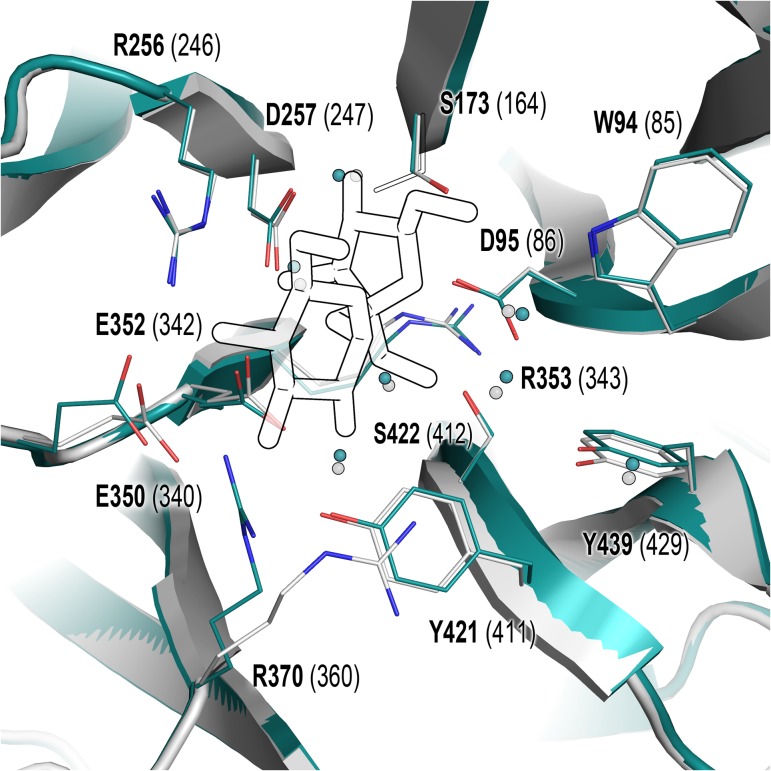
Fig. 1.
Catalytic pocket of B. subtilis and B. megaterium levansucrases in their apo-form. Residues playing different roles in catalysis are displayed as lines. Residue numbers in B. megaterium (cyan, PDB 3om2/3om4) and B. subtilis (white, PDB 1oyg) are depicted in bold font and in brackets, respectively. Sucrose from B. subtilis PDB 1pt2 is shown to define the catalytic pocket. PDB 3om2 belongs to Bm-LS variant D257A, thus residue D257displayed in this figure corresponds to Bm-LS PDB 3om4. Orientation of S173 in 3om2 and 3om4 is shown as full and outlined sticks, respectively. This figure is available in black and white in print and in color at Glycobiology online.
Clan GH-J members are retaining enzymes with a double displacement reaction mechanism involving the formation of a transient fructosyl-enzyme intermediate and their catalytic cycle progresses through two transition states (TS1 and TS2) (Chambert and Gonzy-Treboul 1976; Meng and Futterer 2003; Ozimek et al. 2004). Reported factors contributing to better transglycosylation/hydrolysis partitions in retaining GHs are: (i) a long-lived covalent enzyme-fructose intermediate which could be better disrupted by a saccharide acceptor than by water, (ii) an impaired positioning of the hydrolytic water with respect to the enzyme's covalently linked fructose, and (iii) an increased affinity for saccharide acceptors in the positive (+) subsites. We decided to take a closer look at the inherently consistent coordination network of contacts involving positions 173, 421, 422 and 439 (Bm-Ls numbering) in the catalytic pocket of B. megaterium fructansucrase. We introduced variability in this network via a semi-rational approach based on sequence and structural-alignments of clan GH-J enzymes. Variants yielding more transfer products and altered oligosaccharide spectra were obtained by modifying positions 173, 421 and 422. Changes at residue Y439 rendered predominantly more hydrolytic variants. Although equivalent positions to 173 and 439 were previously mutated in other GHs (Ortiz-Soto et al. 2008; Alvaro-Benito et al. 2010), we perform here for the first time an extensive characterization of mutant libraries of a GH68 family member to analyze the role of these and other noncatalytic amino acids at the −1 and +1 subsites with the focus on transglycosylation/hydrolysis partition.
Materials and methods
Materials
Unless stated otherwise all chemicals were bought from Merck, Sigma-Aldrich, VWR chemicals or Alfa Aesar.
Generation of Multiple Sequence Alignments MSAs for family 68 and 32
Individual datasets were created for GH family 32 and 68 by extracting all sequences (7544 and 1155, respectively) from UniProt. Identical sequences were clustered and the datasets were further minimized by reducing the redundancy to 95% using BLASTClust (https://toolkit.tuebingen.mpg.de/blastclust) (Alva et al. 2016) to allow Clustal Omega (Sievers et al. 2011) (limit: 4000 sequences) and MUSCLE (Edgar 2004) (limit: 500 sequences) alignments (Supplementary figure S1).
Family GH32. Sequences with less than 400 and more than 700 amino acids and sequences containing unspecified amino acids (“X”) or that were not starting with methionine were removed to further minimize the dataset using BioEdit to yield a dataset with a total of 3651 sequences. A Multiple Sequence Alignment was created with Clustal Omega. The corresponding amino acid positions were identified using the structure similarity tool provided by the PDB-database (http://www.rcsb.org). Amino acid distributions for the positions of interest were analyzed with the positional frequency tool (BioEdit Version 7.0) (Hall 1999).
Family GH68. Sequences with less than 350 amino acids, containing unspecified amino acids (“X”) or not starting with methionine were removed to further minimize the databases using BioEdit. The sequences for Bm-Ls (D5DC07) and Streptococcus oralis Ls (P05655) were added manually to the dataset since they were deleted by BLASTClust. From this dataset (367 sequences), a local BLAST database file was created using BioEdit and Local BLAST 2.0 (Altschul et al. 1997) (internal version in BioEdit 7.0) was performed using Bm-LS sequence (D5DC07) as query. A Multiple Sequence Alignment was created with the MUSCLE webserver and the resulting MSA was sorted by the scoring list from Local BLAST. Amino acid distributions for the positions of interest were analyzed via the positional frequency tool (BioEdit 7.0) (Supplementary data, Table SI).
Creation of a recombination library at positions S173 and S422
The library design is depicted in Table I. The library was generated following the method described in Greiner-Stöffele et al. (2009). In brief, three PCR fragments were generated which contained codon exchanges at position S173, position S422 and a linking fragment, respectively. For each of the desired codon exchange an individual oligonucleotide was ordered and separate PCRs were performed. The resulting PCR fragments (9 for position S173, 10 for position S422 and the linking fragments) were mixed, recombined to the full levansucrase gene and cloned into the expression vector (derivative of pRSF, Novagen). E. coli BL21(DE3) was transformed with the library for expression of the enzyme variants. Inserts from 15 randomly selected clones were sequenced to validate the library quality. No unwanted mutation and an even distribution of the desired codons were obtained. Later on, all biochemically characterized variants were sequenced.
Table I.
Bacillus megaterium levansucrase variants
| Position | Mutation | Theoretical number of clones | Number of screened clones |
|---|---|---|---|
| Library S173/S422 | |||
| S173 | ASTDVGLWF | 90 | 200 |
| S422 | ASHFGQTVLW | ||
| Y421 and Y439 | AVILMFWGNDK | – | – |
Creation of single mutants at residues Y421 and Y439
Single mutant libraries at position 421 and 439 were created with a whole plasmid amplification method using individual oligonucleotides for each desired codon exchange (Table I and Supplementary data, Table SII). All mutants were prepared individually to allow assignment of the sequence with a clone. The resulting plasmids were used to transform E. coli BL21(DE3) and for each mutation three clones were picked and assembled in a 96-well plate. Sequencing was performed on all characterized variants.
Expression of libraries in deep well plates
Single clones from agar plates containing 50 μg/mL kanamycin were used to inoculate 400 μL of ZYM505 medium. The plates were covered with an air permeable foil and grown at 37°C overnight in a TIMIX incubator at 1000 rpm. The final OD600 for each well was around 10–15. The overnight cultures were used to inoculate 1 mL ZYM505 medium with appropriate antibiotic (starting OD600 around 0.05). Plates were shaken at 37°C until an OD600 of 0.6–1.0 was reached. Expression was induced by adding IPTG at a final concentration of 0.5 mM. Cultures were incubated overnight at 20°C. Cells were harvested by centrifugation and resuspended in 100 μL lysis-buffer (50 mM Tris pH 7.0, 2 mM MgCl2, 1× Cellytic B, 0.1 mg/mL lysozyme and 20 U/mL benzonase). Well plates were incubated at 30°C for 45 min at 1000 rpm and centrifuged afterwards at 3000 × g for 30 min. Cleared cell lysates were transferred to 96-well microtiter plates and used directly for activity and product profile screening assays.
Activity screening in microtiter plates
Active variants were identified by following the release of reducing sugars from sucrose using enzyme extracts of the library's variants. Reactions were performed at 37°C by adding 1 μL crude extract to 99 μL sucrose solution (0.5 M sucrose and 50 mM Soerensen-buffer pH 6.6) in 96-well microtiter plates. The reactions were stopped after 10 min by adding 100 μL DNS reagent. Plates were heated at 95°C for 5 min and cooled down for 2 min at 4°C. For quantification, samples were diluted 1:3 with water and the absorbance was measured at 540 nm. Data were processed using a calibration curve of absorbance versus glucose concentration. Active variants showing detectable activity after 10 min incubation with sucrose were incubated with a sucrose solution (same as above) and a sample analyzed after 24 h reaction time on TLC and HPAEC-PAD. Variants showing a product profile and/or transfer/hydrolysis partition different from the wild-type enzyme were further characterized.
Expression and purification of hits
One single colony of freshly transformed E. coli bearing selected plasmids was used to inoculate 10 mL LB-medium containing 50 μg/mL kanamycin. Precultures were incubated overnight at 37°C and used to inoculate 250 mL LB-medium with appropriate antibiotic. Expression of levansucrase was induced at an OD600 of around 0.6 by adding IPTG at a final concentration of 0.5 mM. Cultures were incubated overnight at 20°C. Cells were harvested by centrifugation and resuspended in 7 mL of 20 mM Soerensen-buffer pH 6.0. After sonication, extracts were cleared by centrifugation at 13,000 × g. Cleared lysates were loaded onto a CM-sepharose column and levansucrase eluted with a linear gradient of Soerensen-buffer from 20 mM to 1.0 M. Purification fractions were pooled and concentrated.
Protein determination
An extinction coefficient of 51,014 M−1 cm−1 and a path length of 1 cm were used to calculate concentration of purified enzymes measured at 280 nm.
Determination of kinetic parameters
Catalytic parameters for the global reaction (hydrolysis and transfructosylation) were obtained from reactions performed in 50 mM Soerensen-buffer pH 6.6 containing purified enzyme and varying concentrations of sucrose. Initial rates were determined from the release of reducing sugars over the time course (DNS method) (Miller 1959). KM and kcat values were calculated by fitting the data to the Michaelis–Menten equation using nonlinear curve fitting in OriginPro 9.1 (OriginLab).
ΔΔG# values were calculated for the acceptor/donor substrate (sucrose) for each mutation using the following equation (Fersht et al. 1985; Bissaro et al. 2015):
ΔΔG# = 2.303 RT log (kcat/KM)WT/(kcat/KM)mutant
where R is the universal gas constant and T is the temperature in Kelvin.
HPAEC-PAD analysis
HPAEC-PAD analysis of levansucrase products was performed with a Dionex ICS-5000+ SP system utilizing a CarboPac PA10 column with pulsed amperometric detection. Eluents were 100 mM NaOH (A); 100 mM NaOH, 1 M NaOAc (B). A multistep gradient was programmed as follows: 0–5 min 100% A, 5–30 min 0–50% B, 30–45 min 100% A. Products were analyzed using appropriate standards.
Hydrolysis versus transfructosylation activity
Transfructosylation and hydrolysis were determined from reactions carried out in 0.5 M sucrose solution containing 50 mM Soerensen-buffer pH 6.6 and 2 U/mL levansucrase. Samples were taken after 0, 5, 10, 15, 20, 30, 40, 60, 80, 120, 150, 180, 240, 300 and 1440 min and added to 50 mM NaOH solution to stop the reaction. The amount of glucose, fructose and sucrose was analyzed by HPAEC-PAD. Release of glucose reflects the global activity (hydrolysis and transfructosylation), while fructose only results from hydrolysis. The difference between glucose and fructose concentrations corresponds to transfructosylation. Transfer/hydrolysis ratios were obtained at a sucrose conversion of around 70%.
Results and discussion
B. subtilis LS structures (PDB 1pt2 and 1oyg) were used for sucrose bound/unbound state analysis due to the absence of a Bm-LS crystal structure bound to sucrose. However, amino acid numbering used through this document refers to the B. megaterium enzyme. Both levansucrases show the same residues with similar disposition in the catalytic pocket (Figure 1).
Reaction mechanism of retaining GH
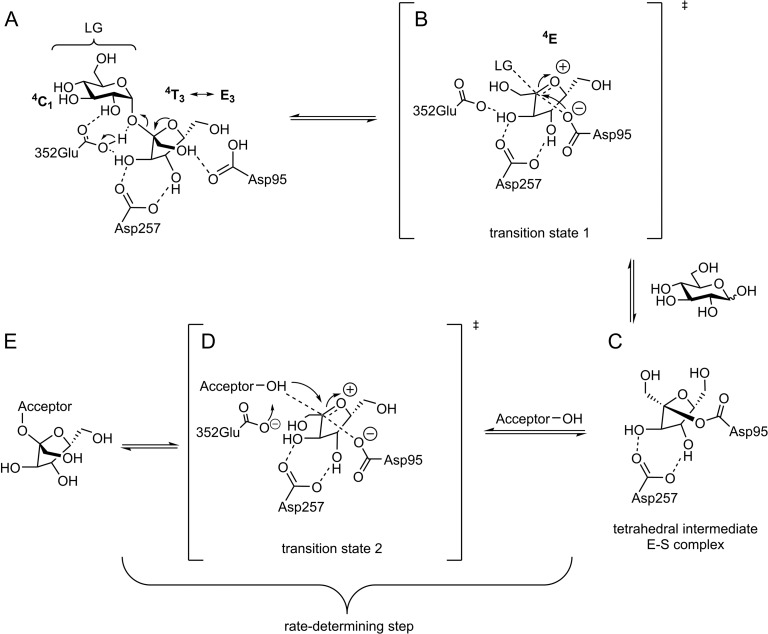
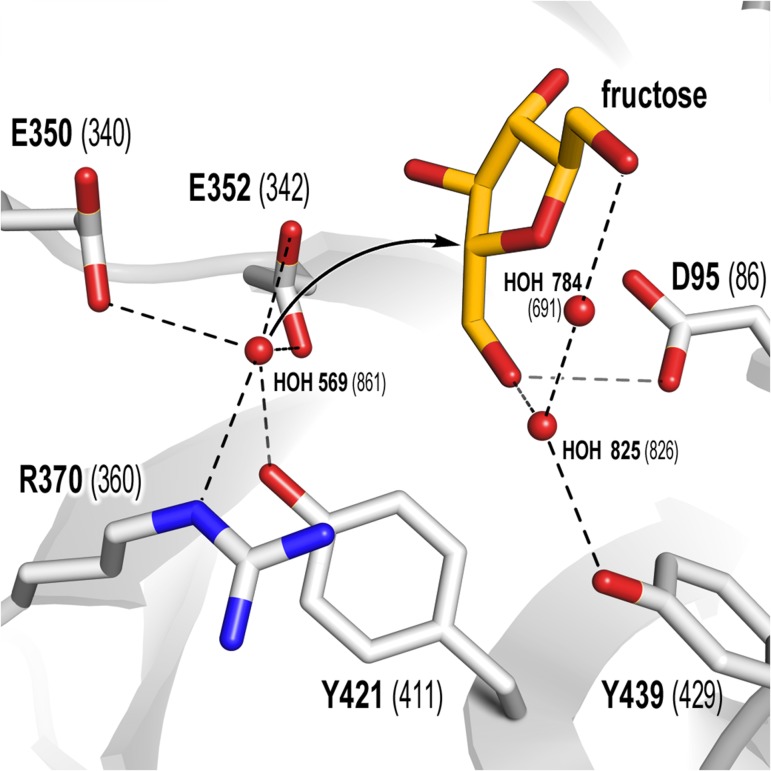
Most retaining GH enzymes catalyze hydrolysis and transglycosylation via a double displacement reaction (Figure 2) involving two-transition states (TS1 and TS2, Figure 2B and D). In GH68 enzymes two aspartate (D95: nucleophile, D257: TS stabilizer) and a glutamate (E352: acid/base catalyst) residues constitute the catalytic triad. Upon sucrose binding D257 forms strong hydrogen bonding interactions with the C-3′- and C-4′ hydroxyls of the fructosyl unit (Figures 2A and 3A) (Chambert and Gonzy-Treboul 1976; Rye and Withers 2000; Meng and Futterer 2003; Ozimek et al. 2004). The latter's furanose ring adopts an envelope shape (Figures 2A and 3A) with a more pronounced amplitude of deformation between 4T3 and E3 (energy minimum conformations in water and most occurring conformations in crystal structures) (Immel and Lichtenthaler 1995; Taha et al. 2013). E352 protonates the glycosidic oxygen of sucrose. The glucopyranoside acts as a leaving group and the fructoside forms an oxocarbenium ion-like transition state (TS1, Figure 2B). Unfortunately, only little is known of the nature of the transition states and the covalent enzyme-substrate complex in fructofuranosidases. The oxocarbenium ion requires a co-planar positioning of C2, C3, C5 and O5 suggesting a 4E or E4 conformation in the transition state (Nelson 1979; Walaszek et al. 1982; Sinnott 1990). The 4E conformation is presumably more favored since it only undergoes a small conformational change starting from 4T3 or E3, all substituents are equatorial and the attack of D95 avoids a 1,3 diaxial repulsion. The nucleophile D95 can now attack the sp2-carbon of the oxonium-ion within the Bürgi-Dunitz criteria (near attack conformation with an angle of 105 ± 5°) (Burgi et al. 1974; Sadiq and Coveney 2015) and form a covalent fructosyl-enzyme intermediate (Figure 2C) by inverting the stereogenic center of C-2 (α-configuration). The retaining mechanism further proceeds through the second transition state (TS2, Figure 2D) and a nucleophilic attack of the acceptor substrate to finally yield the fructosylated product (Meng and Futterer 2003, 2008; Homann et al. 2007). The second transition state is assumed to share very similar features to TS1 (Kempton and Withers 1992; Brás et al. 2010; Bissaro et al. 2015). The properties of TS2 and its interactions with the saccharide/water acceptor are made responsible for the enzyme's hydrolytic versus transfer activity (Bissaro et al. 2015).
Fig. 2.
Possible reaction pathway of GH68 enzymes. (A) Donor (sucrose in this figure) coordinates in the active site of the enzyme; (B) d-glucose is released and a reactive oxocarbenium ion-like transition state (TS1) of the fructosyl-residue is formed and subsequently attacked by Asp95; (C) a covalent fructosyl-enzyme (ES-complex) complex is formed; (D) the acceptor attacks in the second formed transition state (TS2) to (E) form a fructosylated acceptor (Rye and Withers 2000; Bissaro et al. 2015).
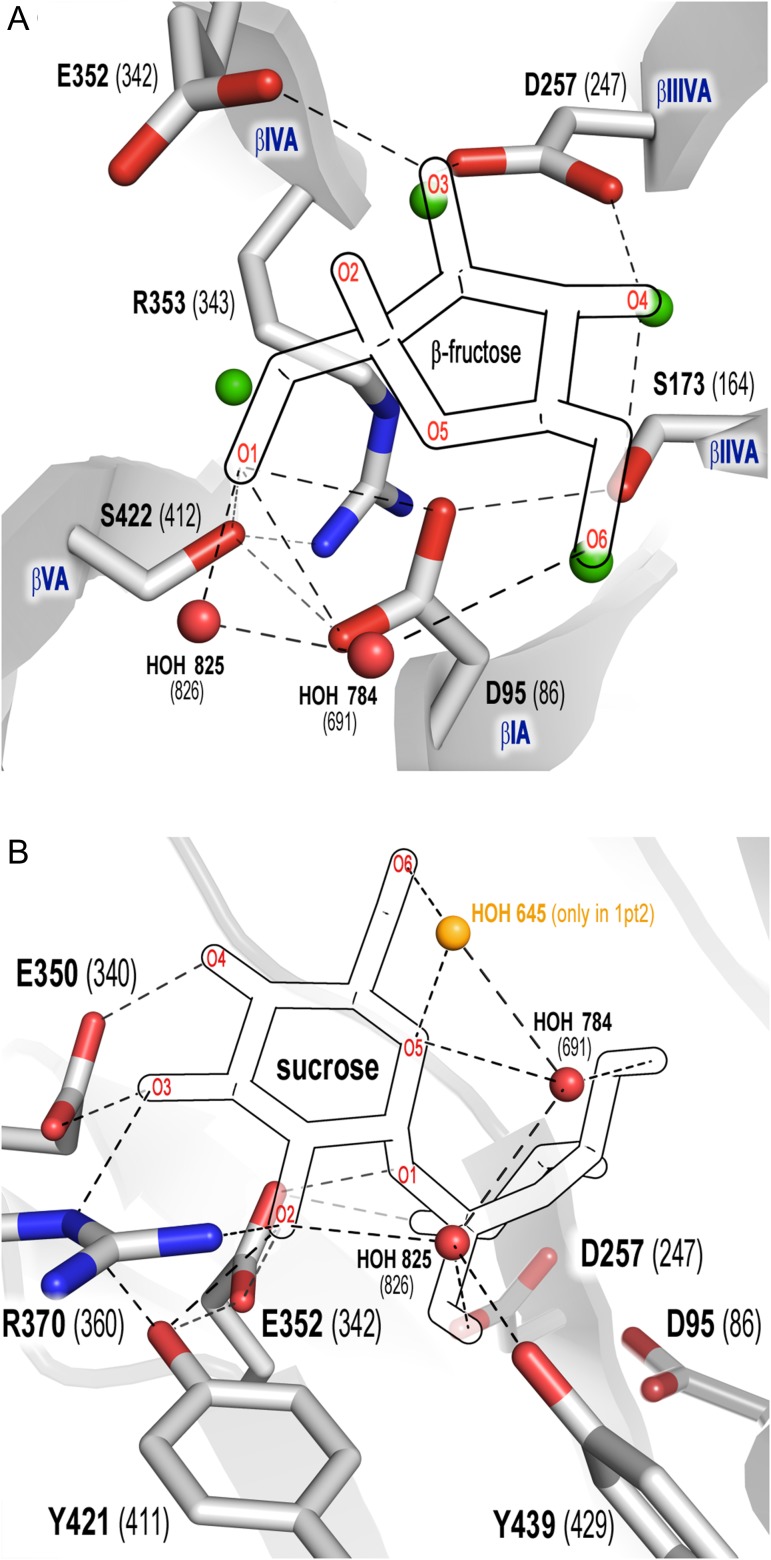
Fig. 3.
Network of contacts within the −1 (A) and +1 (B) subsites of B. subtilis levansucrase (1oyg, numbering in brackets). Fructose (A), glucose (B) and structural water molecules were superimposed from 1pt2. Dashed lines: contacts within 2.5–3.5 Å. β-sheets I–VA are labeled in blue font. (A) Contacts of fructose with the catalytic amino acids, S173, S422 and R353; (B) network of contacts of R353 within the active site connecting the β-sheets. This figure is available in black and white in print and in color at Glycobiology online.
Selection of targets for mutagenesis and MSA analysis
Four amino acids (S173, S422, Y421 and Y439) were selected for mutagenesis. As S173 and S422 (subsite −1) are in direct contact to the nucleophile D95 in B. megaterium and B. subtilis enzymes (Figure 3A) a library including both positions was created to allow compensatory mutations (Goldsmith and Tawfik 2013). S173 forms additional H-bonds to the 4-hydroxyl-group (out of plane in 4E, E4 or 4T3 conformation, Figure 3A) of the fructosyl-moiety, while S422 coordinates the 1-hydroxyl-group and R353 (invariant residue in GH family 68) of the 349DEIER353 motif (Supplementary data, Figure S2) (Meng and Futterer 2003). These noncatalytic residues are (semi-) conserved in clan GH-J (Supplementary data, Table SI), but may play a crucial role in transglycosylation/hydrolysis partition based on evidence that modification of interactions at negative subsites of invertases and other GHs favors transglycosylation via destabilization of the water-mediated deglycosylation step (Feng et al. 2005; Bissaro et al. 2015). Y421 is located between the −1 and +1 subsites and forms polar contacts with E352 (general acid/base). Residues Y421 and Y439 (which has only direct contacts with water) could potentially play a crucial role in acceptor/structural water binding (Figure 3B).
To narrow down the number of possible combinations in library S173/S422, the amino acid distribution at both positions in the primary structure alignment of clan GH-J enzymes was analyzed. Serine is the predominant residue at position 173 within GH68 and GH32 families (67 and 80%, respectively) with alanine being the sole substitute in GH68 and threonine in GH32 (Supplementary data, Table SI). Serine 422 is highly conserved in the GH68 family. From 367 aligned sequences of experimentally characterized and putative fructansucrases, 92% contain serine at this position while the rest show an alanine instead. In turn, an alanine residue is highly conserved (94% out of 3951 sequences) in the GH32 family.
For GH32 the most occurring pairs at 173/422 positions are Ser/Ala (76%) and Thr/Ala (16%). While the pair Ser/Ser (59%) – followed by Ala/Ser (33%) and Ser/Ala (8%) – is the predominant pair within GH68 family, it is absent from GH32 family (Supplementary data, Table SI). In addition to the amino acids mentioned above, other substitutions were allowed to cover more bulky, charged and aromatic side chains allowing compensatory effects (Goldsmith and Tawfik 2013) (Table I).
Y421 is 100 and 97% conserved in GH68 and GH32, respectively. At position 439 only tyrosine (38%) and phenylalanine (61%) are present within GH68 family while tryptophan (96%) is conserved within family GH32.
Analysis of structured water within the active site
Clan GH-J enzymes exhibit a 5-bladed β-propeller architecture numbered I–V with the active site formed in the central cleft. Each β-sheet is formed of four antiparallel β-strands labeled as A-D which are connected by variable loops, displaying a characteristic “W” topology (Paoli 2001; Meng and Futterer 2003; Pijning et al. 2011; Strube et al. 2011). The catalytic triad consisting of D95, D257 and E352 is situated at β-sheets IA, IIIA and IVA, respectively. S422 is located in β-sheet VA and forms an additional bond with Nε of the invariant R353. The latter residue forms – via its guanidinium group and main chain – a network of contacts involving waters, D257, S96, G174 and S422. Such interactions connect all five β-sheets that model the active site of both B. subtilis and B. megaterium enzymes (Supplementary data, Figure S2).
Structured water molecules in the apo- and bound X-ray structures of B. subtilis (1oyg and 1pt2 at 1.5 and 2.1 Å resolution, respectively) and in the apo-structure of Bm-Ls (3om2, 1.9 Å resolution) show the same conservation pattern in the active site (Strube et al. 2011) (Supplementary data, Figure S3). Both serine side chains (173/422 in B. megaterium and 164/412 in B. subtilis) bind water molecules at similar positions in the apo-structures. Since well-defined water molecules can be observed starting from 2.7 Å resolution (Wlodawer et al. 2008), it is reasonable to gain knowledge on the effect of structural water in B. megaterium enzyme (in the bound state) by inferences using B. subtilis LS structure (PDB 1pt2).
Effect of variability at positions S171/S422 on kinetics
Library variants showing detectable activity during screening (10 min incubation with sucrose) and showing a different product spectrum and/or transfer/hydrolysis ratio than the wild-type enzyme (determined by TLC and HPAEC-PAD) were selected and further analyzed. These mutants contain combinations of Ala, Ser, Gly or Thr at positions 173 and 422. Since side chains of S173 and S422 participate in hydrogen bonds to nucleophile D95 and sucrose (Figure 3A) (Meng and Futterer 2003; Strube et al. 2011), bulky amino acids or residues with large charged side chains have a detrimental effect on activity and sucrose binding and can not be rendered active through compensatory effects.
Analyzed variants showed lower activity than the wild-type enzyme (Table II), with modification of position 173 having a much larger effect on the enzyme's catalysis (kcat) than modifications at position 422. While the effect of the mutation on sucrose binding is low, single variant S173A/S422S has a kcat 25-times lower than variant S173S/S422A, which corroborates the essential role the contact between S173 and D95 has on GH68 enzymes’ catalysis (Homann et al. 2007; Ortiz-Soto et al. 2008; Alvaro-Benito et al. 2010). This result is supported by the kcat value observed for double mutant S173A/S422A (2.8 s−1), which is only slightly lower than that of S173A/S422S (3.8 s−1). Exchange of S173 by threonine is sterically permitted and may still maintain bonding contacts with D95 (based on the sucrose-bound structure of B. subtilis PDB 1pt2). This is in agreement with the catalytic behavior observed for mutant S173T/S422S, which recovers partially the wild-type performance. Introduction of two methyl-groups in the double mutant S173T/S422T decreases kcat by 6-fold comparing to the wild-type enzyme due to additional steric hindrance. Mutation S164K (173 in Bm-Ls) in the levansucrase from B. subtilis renders the enzyme inactive, while mutation S164A results in a 25-fold reduction in activity and increased affinity compared to the wild-type enzyme (Ortiz-Soto et al. 2008). Analog mutation (S111T) in invertase/β-fructofuranosidase from Schwanniomyces occidentalis, an enzyme with transfructosylating activity that synthesizes 6-kestose and 1-kestose, does not affect its catalytic efficiency, but increases both KM and kcat by 3-fold without modifying its transferase activity (Alvaro-Benito et al. 2010).
Table II.
Catalytic parameters of Bm-LS variants from library S173/S422
| Enzyme | KM (mM) | kcat (s−1) | kcat/KM (s−1 mM−1) | ΔΔG# (kcal mol−1) |
|---|---|---|---|---|
| S173S/S422S (WT) | 10.8 ± 1.5 | 197.9 ± 3.3 | 18.33 | 0 |
| S173S/S422A | 19.9 ± 2.4 | 97.9 ± 4 | 4.91 | 0.8 |
| S173A/S422S | 3.9 ± 0.3 | 3.8 ± 0.1 | 0.97 | 1.8 |
| S173A/S422A | 7.9 ± 0.9 | 2.8 ± 0.1 | 0.35 | 2.4 |
| S173A/S422G | 23.7 ± 2.9 | 5.9 ± 0.1 | 0.24 | – |
| S173A/S422T | 43.1 ± 3.7 | 17.8 ± 0.4 | 0.41 | – |
| S173G/S422S | 27.7 ± 5.5 | 8.7 ± 0.4 | 0.31 | 2.48 |
| S173G/S422G | 50.1 ± 7.2 | 28.3 ± 1.3 | 0.56 | 2.14 |
| S173G/S422A | 52.6 ± 5.3 | 45.8 ± 1.6 | 0.87 | – |
| S173T/S422S | 41.8 ± 7.0 | 107.9 ± 4.2 | 2.58 | – |
| S173T/S422T | 82.2 ± 9.4 | 30.3 ± 1.8 | 0.36 | – |
Substitutions at position S422 have a higher impact on KM than modifications of position S173, which could be explained by the indirect modification of the position of its neighbor residue Y421, which participates in a contact with the C-2 hydroxyl-group of glucose in the sucrose-bound state (Figure 3B). While KM of variant S173A/S422S was improved (Table II), mutant S173S/S422A showed a 2-fold increase. It is then somewhat expected that KM of the double variant S173A/S422A (7.9) was similar to the value of the wild-type enzyme (10.8). Exchange by threonine at any of the two positions affects sucrose binding due to the additional bulky methyl group. The absence of side chains affects sucrose binding in every analyzed combination. For instance, KM of variant S173G/S422S increases by 2.5-fold and around 4.6-fold in the double mutant S173G/S422G.
Effect of suppressing interactions between the nucleophile and S173/S422 on the ground- and the first transition state (TS1, Figure 2B) was investigated. Values of ΔΔG# were calculated based upon changes in the kcat/KM value (for sucrose) induced by mutations relative to the value of the wild-type enzyme. kcat/KM describes the first glycosylation step in most retaining β-glycosidases, while kcat describes the second deglycosylation step and approaches k3 (Figure 2D, E) only in cases when the donor substrates bear a good leaving group (pKa < 8), thus becoming the limiting step (Zechel and Withers 2001; Bissaro et al. 2015; Strazzulli et al. 2017). Hydrogen bonds of D95 with S173 and S422 contribute a total of 2.4 kcal mol−1 to TS1 stabilization. S173A mutation results in 18-fold decrease in activity, which corresponds to a decrease of 1.8 kcal mol−1 in TS1 stabilization by disruption of the interaction of D95 carboxylate with the side chain hydroxyl-group of S173. The sum of energy contributed by single mutations is 2.6 kcal mol−1, in accordance with the results obtained for the double mutant (Table II). The overall decrease in transition-state stabilization caused by the absence of side chain at S173 is 2.5 kcal mol−1, while the lack of side chain at both positions (S173 and S422) results in a decrease of 2.2 kcal mol−1 TS1 stabilization, highlighting again the importance of the former position in catalysis (Table II).
Four water molecules occupy the active site in absence of sucrose (Figure 3A, green) and are replaced by side chain contacts with the C-1’, C-3’, C-4’ and C-6’ hydroxyl-group of the fructose unit in the sucrose-bound state. A possible explanation for the improved KM of mutant S173A/S422S is the inability of the alanine side chain to establish a hydrogen bond to water in the unbound state so that there is a loss in entropy upon sucrose binding due to the absence of water to be displaced by the C-4’ hydroxyl-group of the fructose moiety. Although the contact with fructose is also lost, the C-4’ hydroxyl-group can still be stabilized by hydrogen bonding to D257. S422 is also bound to a water molecule in the apo-form, however, substitution of this reside by alanine or other residues not only disrupts the contact with fructose C-1’ hydroxyl, but also with Nε of the invariant R353. As mentioned above, this residue is located at the bottom of the active site cleft and establishes contacts with the β-sheets containing the catalytic amino acids (Supplementary figure S2). Thus, any modification of its side chain conformation may alter the position of D95 and E352 and possibly the substrate binding.
In general, KM for sucrose decreases with substitution in the order A > S > G > T in position S173. In turn, kcat increases when S173 is exchanged by S > T > G > A. This comparison is not entirely possible for position S422 as the equivalent single mutants were not found within the active variants of the library. However, from the analyzed variants at position 422, KM decreases in the order S > A and kcat increases in the same order (Table II). Interestingly, combinations of Ala and Gly do not occur in clan GH-J (Supplementary data, Table SI), highlighting the necessity of at least one hydrogen bond enabling/providing side chain at these positions for optimal catalysis.
Effect of mutagenesis on kinetics of Y421 and Y439 variants
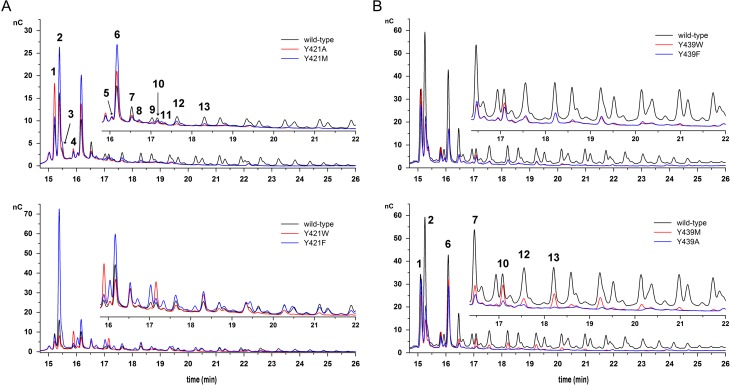
We evaluated the impact of a modified coordination of the second catalytic amino acid (the general acid/base E352) on activity. The side chain of residue Y421 forms a strong hydrogen bond to the carboxylate of E352 (Figure 3B) and substitution by alanine decreases kcat/KM by 560-fold (Table III). Here we modified this position by introducing other aromatic residues and allowing again residues with different functionalities (Table I). Only exchange by aromatic amino acids and methionine produces (slightly) active variants, retaining as little as 6.5–9.5% of the wild-type kcat (Table III). KM of all active variants, except that of the phenylalanine mutant, increases drastically, and none of the variants is able to contact neither E352 side chain nor sucrose (Figure 3B). The contribution to stabilization of TS1 caused by the tyrosine OH-group is of 2.15 kcal mol−1. Substitution by alanine results in a loss of 3.96 kcal mol−1 stabilization.
Table III.
Catalytic parameters of Bm-LS variants at positions 421 and 439
| Enzyme | KM (mM) | kcat (s−1) | kcat/KM(s−1 mM−1) | ΔΔG# (kcal mol−1) |
|---|---|---|---|---|
| Wild-type | 10.8 ± 1.5 | 197.9 ± 4 | 18.32 | – |
| Y421A | 421.0 ± 42.3 | 13.8 ± 1.3 | 3.27 × 10−2 | 3.96 |
| Y421M | 214.0 ± 14.5 | 13.0 ± 0.9 | 6.07 × 10−2 | |
| Y421F | 31.7 ± 2.8 | 17.5 ± 1.6 | 0.55 | 2.15 |
| Y421W | 106.1 ± 15.7 | 18.9 ± 2.8 | 0.18 | |
| Y439A | 621.7 ± 66.2 | 5.4 ± 0.6 | 0.86 × 10−2 | 4.71 |
| Y439M | 441.9 ± 38.6 | 61.0 ± 1.9 | 0.14 | |
| Y439F | 92.9 ± 19.6 | 183.6 ± 7.6 | 1.98 | 1.36 |
| Y439W | 368.2 ± 33.3 | 167.4 ± 4.5 | 0.45 |
Bm-LS displays a wide-open catalytic pocket with ample water accessibility and Y439 is part of a network which seems to coordinate the water supply to the catalytic center. Besides its binding to two structural waters, Y439 does not maintain any direct contact to other side chains in its vicinity. While kcat is not substantially affected upon modification by tryptophan or phenylalanine, KM increases for both, dramatically for the later. Exchange by methionine also results in a low catalytic efficiency. Substitution of tyrosine by alanine raises the KM and lowers kcat, with the variant retaining only 0.5% of the wild-type kcat/KM value (Table III). The hydroxyl-group of Y439 contributes 1.36 kcal mol−1 to TS1 stabilization, while substitution by alanine results in a loss of 4.71 kcal mol−1 stabilization (Table III).
Modulation of transfructosylation/hydrolysis partition through site directed mutagenesis
The efficiency of the Bm-LS variants for transfructosylation was studied in reactions employing sucrose as donor/acceptor. Due to the difference in specific activities shown by some variants relative to the wild-type, comparison was performed using the same enzymatic activities. Only variants from the library S173/S422 and mutant Y421F showed higher transfructosylation than wild-type Bm-LS (Table IV).
Table IV.
Hydrolysis and transfructosylation values for Bm-LS on sucrose
| Enzyme | Transfructosylation (%) | Hydrolysis (%) |
|---|---|---|
| Library S173/S422 | ||
| WT | 11.0 | 89.0 |
| S173S/S422A | 15.6 | 84.4 |
| S173A/S422S | 32.3 | 67.7 |
| S173A/S422A | 25.2 | 74.8 |
| S173A/S422G | 18.6 | 81.4 |
| S173A/S422T | 25.8 | 74.2 |
| S173G/S422S | 16.8 | 83.2 |
| S173G/S422G | 20.6 | 79.4 |
| S173G/S422A | 14.7 | 85.3 |
| S173T/S422S | 9.4 | 90.6 |
| S173T/S422T | 20.3 | 79.7 |
| Variants at other positions | ||
| Y421A | 8.1 | 91.9 |
| Y421M | 12.1 | 87.9 |
| Y421F | 23.3 | 76.7 |
| Y421W | 9.3 | 90.7 |
| Y439A | 8.1 | 91.9 |
| Y439M | 9.8 | 90.2 |
| Y439F | 8.7 | 91.3 |
| Y439W | 8.3 | 91.7 |
All variants with an alanine in position 173 yielded higher amounts of transfructosylation products (Figure 4). Single mutant S173A/S422S showed the best performance with hydrolysis reduced by around 3-fold compared to the wild-type enzyme. Equivalent mutation in B. subtilis levansucrase increased transfructosylation by only 18% (Ortiz-Soto et al. 2008).
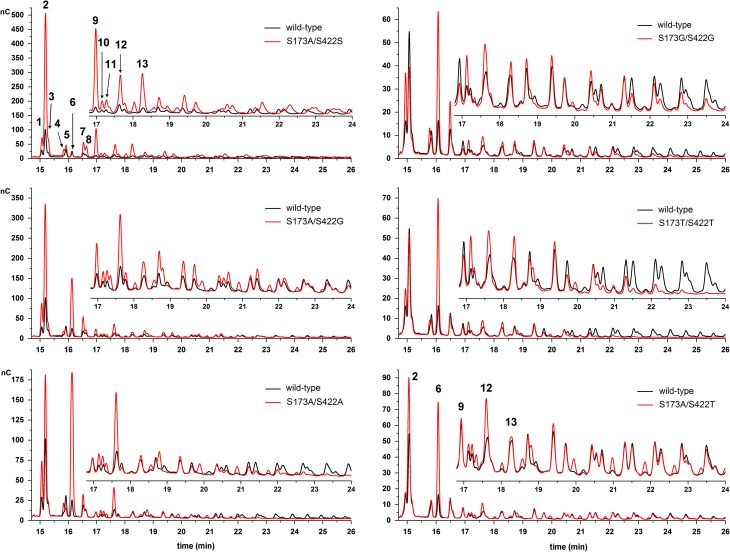
Fig. 4.
HPAEC-PAD product spectra of variants from the S173/S422 library. Products were obtained from reactions containing 0.5M sucrose and 2 U/mL of enzymatic activity in 50 mM Soerensen buffer pH 6.6. Samples were evaluated at sucrose conversion of around 70%. (1) 1-kestose, (2) blastose, (3) disaccharide, (4) 6-kestose, (5) disaccharide, (6) neokestose, (7) tetrasaccharide Glu-(Fru)3, (8) disaccharide, (9) tetrasaccharide, (10) 6-nystose, (11) tetrasaccharide, (12) pentasaccharide and (13) hexasaccharide. Samples from the left and right panels belong to different sample batches. Signal intensities may vary between measurements, thus, wild-type Bm-LS was measured with every sample batch to allow comparison. This figure is available in black and white in print and in color at Glycobiology online.
Variants S173G/S422G and S173T/S422T also produce significantly more oligosaccharides; 2-times the amount of the wild-type LS.
Rare disaccharide blastose (β-d-fructofuranosyl(2→6)-d-glucopyranose) is the most abundant oligosaccharide produced by wild-type Bm-LS. All S173/S422 variants produce the trisaccharide neokestose (β-d-fructofuranosyl(2→6)-α-d-glucopyranosyl(1→2)-β-d-fructofuranoside), the blastose precursor (Homann et al. 2007), more efficiently. Variant S173A/S422S yields overall more oligosaccharides than wild-type Bm-LS of both low and high molecular weight, while double mutants S173G/S422G, S173A/S422A and S173T/S422T produce only small oligosaccharides.
Variant S173G/S422S produces the same oligosaccharide pattern as the wild-type enzyme and the product spectrum of variant S173S/S422A is similar to that of S173A/S422G. This supports the assumption that only modifications at position S422 decrease the affinity for large acceptor oligosaccharides.
Disruption of the hydrogen bonding network among the nucleophile (D26), W23 and N25 in a vacuolar invertase (a bonding network characteristic of GH32 plant invertases) shifts transfer from water to sucrose favoring the synthesis of 1-kestose over sucrose hydrolysis (Schroeven et al. 2008), an effect that was also observed for other invertases (Ritsema et al. 2006).
Not unexpectedly, substitution of Y421 by phenylalanine resulted in enhance transfructosylation (Table IV and Figure 5A). In the catalytic process, departing glucose leaves the +1 subsite vacant, in a step preceding water or carbohydrate acceptors binding (Figure 6). Y421F increases the hydrophobicity of the +1 subsite, probably diminishing water availability for deglycosylation or hindering the productive orientation of the water's lone electron pair, as previously observed for variant Y198F of an inverting GHs turned an efficient glycosynthase (Hidaka et al. 2010). Changes at a structural homolog position of E350 (D239) in A. thaliana cell wall invertase (also at +1 subsite) by hydrophobic amino acids almost abolishes sucrose hydrolysis, while the variants retained activity towards 1-kestose (Le Roy et al. 2007).
Fig. 5.
HPAEC-PAD product spectra of Y421 (A) and Y439 (B) variants. Products were obtained from reactions containing 0.5 M sucrose and 2 U/mL of enzymatic activity in 50 mM Soerensen buffer pH 6.6. Samples were evaluated at sucrose conversion of around 70%. (1) 1-kestose, (2) blastose, (3) disaccharide, (4) 6-kestose, (5) disaccharide, (6) neokestose, (7) tetrasaccharide Glu-(Fru)3, (8) disaccharide, (9) tetrasaccharide, (10) 6-nystose, (11) tetrasaccharide, (12) pentasaccharide and (13) hexasaccharide. Signal intensities may vary between measurements, thus, wild-type Bm-LS was measured with every sample batch to allow comparison, except for samples in Figure 5B. This figure is available in black and white in print and in color at Glycobiology online.
Fig. 6.
Putative trajectory of hydrolytic water onto the ES complex. Waters 825 and 784 are conserved in 3om2 (Bm-Ls), 1oyg (white, apo-structure) and 1pt2 (orange, sucrose-bound structure), while water 569 is only present in the apo-structures. Fructose is superimposed from 1pt2. Contacts within 3.5 Å are shown as dashed lines. This figure is available in black and white in print and in color at Glycobiology online.
Variants at position 439 synthesize smaller products than the wild-type LS (Figure 5B) and keep a transfructosylation/hydrolysis balance similar to that of the wild-type enzyme. Exchange by phenylalanine was better tolerated than other amino acids from the kinetic point of view (Table III); yet, little changes were observed regarding the oligosaccharide distribution compared to other variants at the same position. Although the strategy of manipulating water accessibility to the active site appears to be straightforward (Teze et al. 2013) and has proven successful for some GHs, i.e., an agarase (AgaD) from Zobellia galactanivorans (David et al. 2017), observation of water accessible active sites of hydrolases and transferases suggest that restricting water supply does not guarantee lesser hydrolytic activities (Leemhuis and Dijkhuizen 2003). The presence of hydrophobic residues near the catalytic acid/base (+1 subsite), however, has proven beneficial for better transfer/hydrolysis partitions in α-amylase enzyme families, in accordance with the results obtained in this work for variant Y421F. Further strategies may target putative channels supplying water in the direction of E352.
Conclusions
Although disruption of contacts between D95, S173 and S422 proved to be detrimental for activity, our results indicate that modification of the nucleophile-coordinating network is beneficial for the transferase activity of the levansucrase from B. megaterium, in agreement with results published for other GHs enzymes (Ritsema et al. 2006; Schroeven et al. 2008; Bissaro et al. 2015). Slowing down the second step in the reaction mechanism may result in a long-lived enzyme-fructose covalent complex and allow saccharide acceptors to compete in the acceptor binding site of the enzyme to become deprotonated by the general acid/base instead of water (Teze et al. 2013).
The role of tyrosine residues in the catalytic cleft as water-binding determinants has long been established for various GHs. Here we showed that only the modification of the invariant tyrosine coordinating the general acid/base position (Y421) by phenylalanine augments transfructosylation. Improved transfer may be the result of either a hindered water positioning or a reduced water availability at the +1 subsite (Figure 6).
Levansucrase from B. subtilis displays similar global kinetics as the B. megaterium enzyme; yet it synthesizes fructans (levan) in a more efficient fashion without compromising activity. This is also the case of the levansucrase and inulosucrase from Lactobacillus reuteri 121 (Ozimek et al. 2006). Gram negative levansucrases from Pseudomonas syringae pv. tomato and Gluconacetobacter diazotrophicus also show high transfructosylation activities, as they transfer more than 70% of converted sucrose to produce levan/fructooligosaccharides (Tambara et al. 1999; Visnapuu et al. 2011). While Y421 is fully conserved in GH68, there is not a correlation between serine/alanine found at either 173 or 422 positions with a preferred hydrolytic or transferase activity. However, Gram-negative fructansucrases contain additional structural elements (they often share as little as 15% similarity with their Gram-positive counterparts), which could be responsible for their improved performance, maybe promoting an optimal binding of oligosaccharides acceptors and/or restricting water accessibility. A remarkable difference between B. subtilis and B. megaterium levansucrases – sharing high similarity at structural and amino acid level – is the molecular weight of their products. Under similar reaction conditions the former produces almost exclusively high molecular weight levan (a bimodal distribution of 8.3 and 3500 kDa, respectively) (Ortiz-Soto et al. 2008), while the latter only synthesizes oligosaccharides, with accumulation of blastose at the end of the reaction (Homann et al. 2007; Strube et al. 2011). A higher affinity for saccharide acceptors seems to be a plausible explanation for a better transfer in the catalytically efficient enzyme from B. subtilis. The concept of fine-tune molecular adjustments being responsible for transglycosylation in GHs rather than larger structural changes was previously suggested based on the close evolutionary relationship between transglycosylases (GH enzymes that mainly catalyze transfer) and predominantly hydrolytic GHs (Leemhuis and Dijkhuizen 2003; Bissaro et al. 2015).
Here, we demonstrated that better transglycosylation/hydrolysis partitions in B. megaterium levansucrase may be achieved by inducing a long-lived covalent enzyme-fructose intermediate (−1 subsite), as well as by modifying the optimal positioning of the catalytic water (at the +1 subsite) regarding the covalent enzyme-fructose complex.
Supplementary Material
Supplementary data
Funding
Financial support from the 7th Framework Programme for Research and Technological Development (project “SuSy”: Sucrose synthase as effective mediator of glycosylation) is acknowledged.
Conflict of interest statement
None declared.
Abbreviations
Bm-LS, Bacillus megaterium levansucrase; DP, degree of polymerization; FS, fructansucrase; GH, glycoside hydrolase; HPAEC-PAD, high-performance anion exchange chromatography with pulsed amperometric detection; IPTG, isopropyl β-D-1-thiogalactopyranoside; IS, inulosucrase; LS, levansucrase; MSA, multiple sequence alignment; PDB, Protein Data Bank.
References
- Altschul SF, Madden TL, Schaffer AA, Zhang JH, Zhang Z, Miller W, Lipman DJ. 1997. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 25:3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alva V, Nam SZ, Soding J, Lupas AN. 2016. The MPI bioinformatics toolkit as an integrative platform for advanced protein sequence and structure analysis. Nucleic Acids Res. 44:W410–W415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvaro-Benito M, de Abreu M, Portillo F, Sanz-Aparicio J, Fernandez-Lobato M. 2010. New Insights into the fructosyltransferase activity of Schwanniomyces occidentalis beta-fructofuranosidase, emerging from nonconventional codon usage and directed mutation. Appl Environ Microb. 76:7491–7499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bissaro B, Monsan P, Fauré R, O'Donohue Michael J. 2015. Glycosynthesis in a waterworld: New insight into the molecular basis of transglycosylation in retaining glycoside hydrolases. Biochem J. 467:17–35. [DOI] [PubMed] [Google Scholar]
- Brás NF, Ramos MJ, Fernandes PA. 2010. DFT studies on the β-glycosidase catalytic mechanism: The deglycosylation step. J Mol Struct: THEOCHEM. 946:125–133. [Google Scholar]
- Burgi HB, Dunitz JD, Lehn JM, Wipff G. 1974. Stereochemistry of reaction paths at carbonyl centers. Tetrahedron. 30:1563–1572. [Google Scholar]
- Chambert R, Gonzy-Treboul G. 1976. Levansucrase of Bacillus subtilis. Eur J Biochem. 71:493–508. [DOI] [PubMed] [Google Scholar]
- David B, Irague R, Jouanneau D, Daligault F, Czjzek M, Sanejouand Y-H, Tellier C. 2017. Internal water dynamics control the transglycosylation/hydrolysis balance in the agarase (AgaD) of Zobellia galactanivorans. ACS Catalysis. 7:3357–3367. [Google Scholar]
- Edgar RC. 2004. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32:1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng HY, Drone J, Hoffmann L, Tran V, Tellier C, Rabiller C, Dion M. 2005. Converting a beta-glycosidase into a beta-transglycosidase by directed evolution. J Biol Chem. 280:37088–37097. [DOI] [PubMed] [Google Scholar]
- Fersht AR, Shi JP, Knilljones J, Lowe DM, Wilkinson AJ, Blow DM, Brick P, Carter P, Waye MMY, Winter G. 1985. Hydrogen-bonding and biological specificity analyzed by protein engineering. Nature. 314:235–238. [DOI] [PubMed] [Google Scholar]
- Goldsmith M, Tawfik DS. 2013. Enzyme engineering by targeted libraries. Methods Enzymol. 523:257–283. [DOI] [PubMed] [Google Scholar]
- Greiner-Stöffele T, Feller C, Struhalla M. 2009. Process for generating a variant library of dna sequences. EP2297321B1: C-Lecta Gmbh.
- Hall TA. 1999. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl Acids Symp Ser. 41:95–98. [Google Scholar]
- Hidaka M, Fushinobu S, Honda Y, Wakagi T, Shoun H, Kitaoka M. 2010. Structural explanation for the acquisition of glycosynthase activity. J Biochem. 147:237–244. [DOI] [PubMed] [Google Scholar]
- Homann A, Biedendieck R, Gotze S, Jahn D, Seibel J. 2007. Insights into polymer versus oligosaccharide synthesis: Mutagenesis and mechanistic studies of a novel levansucrase from Bacillus megaterium. Biochem J. 407:189–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Immel S, Lichtenthaler FW. 1995. Molecular modeling of saccharides 0.7. The conformation of sucrose in water - a molecular-dynamics approach. Liebigs Ann. 11:1925–1937. [Google Scholar]
- Kempton JB, Withers SG. 1992. Mechanism of Agrobacterium beta-glucosidase: Kinetic studies. Biochemistry. 31:9961–9969. [DOI] [PubMed] [Google Scholar]
- Lafraya A, Sanz-Aparicio J, Polaina J, Marin-Navarro J. 2011. Fructo-oligosaccharide synthesis by mutant versions of Saccharomyces cerevisiae invertase. Appl Environ Microb. 77:6148–6157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lammens W, Le Roy K, Schroeven L, Van Laere A, Rabijns A, Van den Ende W. 2009. Structural insights into glycoside hydrolase family 32 and 68 enzymes: Functional implications. J Exp Bot. 60:727–740. [DOI] [PubMed] [Google Scholar]
- Le Roy K, Lammens W, Verhaest M, De Coninck B, Rabijns A, Van Laere A, Van den Ende W. 2007. Unraveling the difference between invertases and fructan exohydrolases: A single amino acid (Asp-239) substitution transforms Arabidopsis cell wall invertase1 into a fructan 1-exohydrolase. Plant Physiol. 145:616–625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leemhuis H, Dijkhuizen L. 2003. Engineering of hydrolysis reaction specificity in the transglycosylase cyclodextrin glycosyltransferase. Biocatal Biotransfor. 21:261–270. [Google Scholar]
- Lombard V, Golaconda Ramulu H, Drula E, Coutinho PM, Henrissat B. 2014. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 42:D490–D495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng G, Futterer K. 2003. Structural framework of fructosyl transfer in Bacillus subtilis levansucrase. Nat Struct Biol. 10:935–941. [DOI] [PubMed] [Google Scholar]
- Meng G, Futterer K. 2008. Donor substrate recognition in the raffinose-bound E342A mutant of fructosyltransferase Bacillus subtilis levansucrase. BMC Struct Biol. 8:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller GL. 1959. Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal Chem. 31:426–428. [Google Scholar]
- Nelson CR. 1979. Conformation of 2,3,4-Tri-O-acetyl-D-xylono-1,5-lactone. Carbohyd Res. 68:55–60. [Google Scholar]
- Ortiz-Soto ME, Rivera M, Rudino-Pinera E, Olvera C, Lopez-Munguia A. 2008. Selected mutations in Bacillus subtilis levansucrase semi-conserved regions affecting its biochemical properties. Protein Eng Des Sel. 21:589–595. [DOI] [PubMed] [Google Scholar]
- Ortiz-Soto ME, Seibel J. 2014. Biotechnological synthesis and transformation of valuable sugars in the food and pharmaceutical industry. Curr Org Chem. 18:964–986. [Google Scholar]
- Ozimek LK, Kralj S, van der Maarel MJ, Dijkhuizen L. 2006. The levansucrase and inulosucrase enzymes of Lactobacillus reuteri 121 catalyse processive and non-processive transglycosylation reactions. Microbiology. 152:1187–1196. [DOI] [PubMed] [Google Scholar]
- Ozimek LK, van Hijum SAFT, van Koningsveld GA, van der Maarel MJEC, van Geel-Schutten GH, Dijkhuizen L. 2004. Site-directed mutagenesis study of the three catalytic residues of the fructosyltransferases of Lactobacillus reuteri 121. FEBS Lett. 560:131–133. [DOI] [PubMed] [Google Scholar]
- Paoli M. 2001. Protein folds propelled by diversity. Prog Biophys Mol Biol. 76:103–130. [DOI] [PubMed] [Google Scholar]
- Pijning T, Anwar MA, Boger M, Dobruchowska JM, Leemhuis H, Kralj S, Dijkhuizen L, Dijkstra BW. 2011. Crystal structure of inulosucrase from Lactobacillus: Insights into the substrate specificity and product specificity of GH68 fructansucrases. J Mol Biol. 412:80–93. [DOI] [PubMed] [Google Scholar]
- Ritsema T, Hernandez L, Verhaar A, Altenbach D, Boller T, Wiemken A, Smeekens S. 2006. Developing fructan-synthesizing capability in a plant invertase via mutations in the sucrose-binding box. Plant J. 48:228–237. [DOI] [PubMed] [Google Scholar]
- Rye CS, Withers SG. 2000. Glycosidase mechanisms. Curr Opin Chem Biol. 4:573–580. [DOI] [PubMed] [Google Scholar]
- Sadiq SK, Coveney PV. 2015. Computing the role of near attack conformations in an enzyme-catalyzed nucleophilic bimolecular reaction. J Chem Theory Comput. 11:316–324. [DOI] [PubMed] [Google Scholar]
- Schroeven L, Lammens W, Van Laere A, Van den Ende W. 2008. Transforming wheat vacuolar invertase into a high affinity sucrose:sucrose 1-fructosyltransferase. New Phytol. 180:822–831. [DOI] [PubMed] [Google Scholar]
- Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li WZ, Lopez R, McWilliam H, Remmert M, Soding J et al. . 2011. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol. 7:539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinnott ML. 1990. Catalytic mechanisms of enzymatic glycosyl transfer. Chem Rev. 90:1171–1202. [Google Scholar]
- Strazzulli A, Cobucci-Ponzano B, Carrillo S, Bedini E, Corsaro MM, Pocsfalvi G, Withers SG, Rossi M, Moracci M. 2017. Introducing transgalactosylation activity into a family 42 β-galactosidase. Glycobiology. 27:425–437. [DOI] [PubMed] [Google Scholar]
- Strube CP, Homann A, Gamer M, Jahn D, Seibel J, Heinz DW. 2011. Polysaccharide synthesis of the levansucrase SacB from Bacillus megaterium is controlled by distinct surface motifs. J Biol Chem. 286:17593–17600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taha HA, Richards MR, Lowary TL. 2013. Conformational analysis of furanoside-containing mono- and oligosaccharides. Chem Rev. 113:1851–1876. [DOI] [PubMed] [Google Scholar]
- Tambara Y, Hormaza JV, Perez C, Leon A, Arrieta J, Hernandez L. 1999. Structural analysis and optimised production of fructo-oligosaccharides by levansucrase from Acetobacter diazotrophicus SRT4. Biotechnol Lett. 21:117–121. [Google Scholar]
- Teze D, Hendrickx J, Dion M, Tellier C, Woods VL, Tran V, Sanejouand YH. 2013. Conserved water molecules in family 1 glycosidases: A DXMS and molecular dynamics study. Biochemistry. 52:5900–5910. [DOI] [PubMed] [Google Scholar]
- Visnapuu T, Mardo K, Mosoarca C, Zamfir AD, Vigants A, Alamae T. 2011. Levansucrases from Pseudomonas syringae pv. tomato and P. chlororaphis subsp. aurantiaca: Substrate specificity, polymerizing properties and usage of different acceptors for fructosylation. J Biotechnol. 155:338–349. [DOI] [PubMed] [Google Scholar]
- Walaszek Z, Horton D, Ekiel I. 1982. Conformational studies on aldonolactones by NMR-spectroscopy – conformations of D-glucono-1,5-lactone and D-mannono-1,5-lactone in solution. Carbohyd Res. 106:193–201. [Google Scholar]
- Wlodawer A, Minor W, Dauter Z, Jaskolski M. 2008. Protein crystallography for non-crystallographers, or how to get the best (but not more) from published macromolecular structures. FEBS J. 275:1–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zechel DL, Withers SG. 2001. Dissection of nucleophilic and acid-base catalysis in glycosidases. Curr Opin Chem Biol. 5:643–649. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.