Abstract
Interactions between galectins and polysaccharides are crucial to many biological processes, and yet these are some of the least understood, usually being limited to studies with small saccharides and short oligosaccharides. The present study is focused on human galectin-3 (Gal-3) interactions with a 60 kDa rhamnogalacturonan RG-I-4 that we use as a model to garner information as to how galectins interact with large polysaccharides, as well as to develop this agent as a therapeutic against human disease. Gal-3 is unique among galectins, because as the only chimera-type, it has a long N-terminal tail (NT) that has long puzzled investigators due to its dynamic, disordered nature and presence of numerous prolines. Here, we use 15N–1H heteronuclear single quantum coherence NMR spectroscopy to demonstrate that multiple sites on RG-I-4 provide epitopes for binding to three sites on 15N-labeled Gal-3, two within its carbohydrate recognition domain (CRD) and one at a novel site within the NT encompassing the first 40 residues that are highly conserved among all species of Gal-3. Moreover, strong binding of RG-I-4 to the Gal-3 NT occurs on a very slow time scale, suggesting that it may be mediated by cis–trans proline isomerization, a well-recognized modulator of many biological activities. The NT binding epitope within RG-I-4 appears to reside primarily in the side chains of the polysaccharide, some of which are galactans. Our results provide new insight into the role of the NT in Gal-3 function.
Keywords: galectin, polysaccharide
Introduction
Galectin-3 (Gal-3) is involved in highly diverse physiological and pathological processes, including mediation of cell adhesion and migration, glycoprotein routing, as well as chemokine and cytokine production (Klyosov et al. 2008; Kaltner and Gabius 2012; Smetana et al. 2013). Like all galectins, Gal-3 has a conserved carbohydrate recognition domain (CRD) that is composed of an 11-stranded, β-sandwich fold that has two faces, the S-face composed of six β-strands to which small saccharides including β-galactosides (Barondes 2008; Nesmelova et al. 2008) and α-galactosides (Miller et al. 2011) bind, and an opposing five β-stranded F-face to which some larger polysaccharides can interact (Nesmelova et al. 2008; Miller, Klyosov, et al. 2009, 2012). However, Gal-3 is unique among galectins, because it is the only chimera-type galectin with a long, proline-rich N-terminal tail (NT) projecting as a dynamic, random coil from its CRD (Nesmelova et al. 2008; Ippel et al. 2016).
Investigators have long been puzzled by the presence of the Gal-3 NT sequence with its numerous prolines (27 of 108 residues in human Gal-3), that is highly conserved among all known species of Gal-3 (Dumic et al. 2006). Although being collagen-like (Mayo 1996), the Gal-3 NT does not form collagen triple helices, and it is not integral to the Gal-3-folded structure because it only transiently interacts with the CRD (Ippel et al. 2016). Moreover, the NT has little effect on carbohydrate binding to the canonical site on the Gal-3 CRD S-face (Dumic et al. 2006; Ippel et al. 2016). Nevertheless, removal of the NT does attenuate or negate Gal-3 functional activity (Dumic et al. 2006). And although the NT is suggested to be involved in Gal-3 pentamer formation (Ahmad et al. 2004), this remains largely unproven. So what is the functional role of the Gal-3 NT with its multiple prolines? By demonstrating a connection to polysaccharide binding, the present study provides one plausible answer to this question.
Information on interactions between lectins like Gal-3 and polysaccharides has been quite limited and is generally viewed through the lens of studies performed with simple, small saccharides and short oligosaccharides (Nesmelova et al. 2008). For some time now, we have been working with a number of large, complex pectin-derived polysaccharides as model systems to garner information as to how galectins interact with carbohydrate-based biomacromolecules (Miller, Klyosov, et al. 2009, 2012) and modify their physicochemical properties (Miller, Klyosov, et al. 2009). Although mostly compositionally unlike their counterparts associated with mammalian cells, some of these polysaccharides do contain numerous galactose residues as possible Gal-3 binding epitopes (Inohara and Raz 1994; Sathisha et al. 2007; Glinsky and Raz 2009). In addition, these model polysaccharides (unlike their mammalian counterparts) can be isolated and fractionated into relatively pure species and in amounts required for most biophysical experiments. In the present study, we provide insight into how Gal-3 interacts with a large, ginseng-derived rhamnogalacturonan (RG-I-4) polysaccharide and the question of the presence and role of the NT in Gal-3. While investigating Gal-3 binding to RG-I-4 (possible structure shown in Figure 1), we discovered that Gal-3 has a novel sugar binding epitope within the NT where strong binding is likely promoted by cis–trans proline isomerization. The present study provides new insight into the possible role of the NT in Gal-3 function and expands glycospace available to this galectin.
Fig. 1.
An illustration showing one possible chemical structure for a fragment of the rhamnogalacturonan I domain RG-I-4 polysaccharide from Ginseng pectin. This figure is slightly modified from a part of Figure 2 in Gao et al. (2013).
Results
Binding of full length Gal-3 to RG-I-4
Figure 2 shows 15N–1H heteronuclear single quantum coherence (HSQC) spectral overlays of 15N-Gal-3 (80 μM) in the absence (black peaks) and presence (red peaks) of RG-I-4 at concentrations of 5 μM (Figure 2A) and 10 μM (Figure 2B). Already at the lower concentration, some resonances are significantly decreased in intensity, an effect that is enhanced at the higher RG-I-4 concentration. Some peaks become so broadened that they fall into the noise, whereas others are only partially reduced in intensity or remain mostly unchanged. Because resonance broadening occurs differentially, the observed increase in line broadening cannot simply be accounted for by a reduction in molecular rotational motion. Therefore, we attribute the increase in resonance broadening primarily to binding of the lectin to the polysaccharide.
Fig. 2.
1H–15N heteronuclear single quantum coherence (HSQC) spectral overlays are shown for 15N-enriched Gal-3 (80 μM) alone (cross-peaks in black) and in the presence (cross-peaks in red) of 5 μM (A) and 10 μM (B) of polysaccharide RG-I-4. Solution conditions are 20 mM potassium phosphate buffer, pH 6.9, 30°C. This figure is available in black and white in print and in color at Glycobiology online.
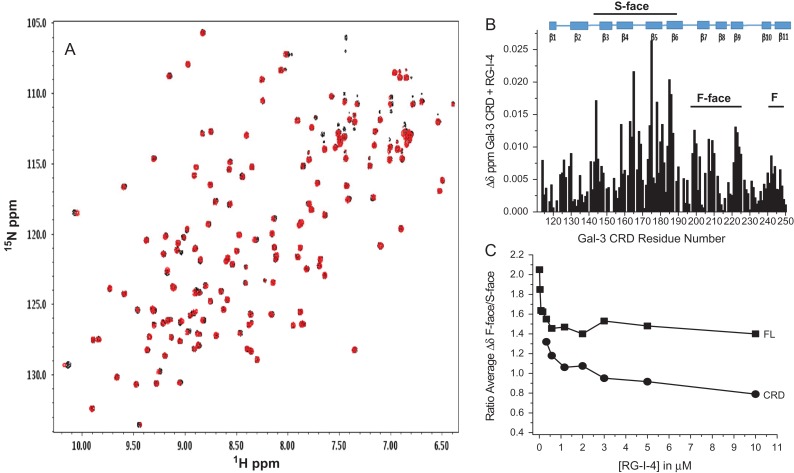
Resonance intensity changes vs. the amino acid sequence of Gal-3 are shown in Figure 3A for an RG-I-4 concentration of 2 μM. At this low polysaccharide concentration, Gal-3 resonances from within its NT (especially the first 20 residues) are the most broadened vis-à-vis those from within the CRD. In addition, resonances from the back side of the CRD β-sandwich (F-face, especially β-strands 7, 8, 9, 11, and at the beginning of β-strand 2 and loop between strands 2 and 3) are more broadened than those within the canonical carbohydrate binding site (S-face). As the titration continues, sites within the S-face eventually become equally broadened (Supplemental Figure S1A,B for 3 μM and 10 μM, respectively). This is perhaps better appreciated in Figure 3B which plots average intensity changes vs. the RG-I-4 concentration. Averaging has been done over all residues, as well as for specific sequences, namely the first 18 residues of the NT, F-face residues 210–225 (β-strands 7, 8, 9), and S-face residues 154–176 (canonical carbohydrate binding site). Once again, note that the Gal-3 NT is affected first at lower RG-I-4 concentrations, and residues within the CRD are affected more at higher polysaccharide concentrations.
Fig. 3.
(A) HSQC resonance broadening of 15N-Gal-3 is shown for the binding of RG-I-4 at 2 μM. Intensity changes are plotted vs. the amino acid sequence of Gal-3, and have been calculated as (1 – Inti/Into), where Inti is the resonance intensity at some concentration of the polysaccharide and Into is the resonance intensity in the absence of polysaccharide. A value of 0 indicates the absence of broadening and a value of 1 indicates that that resonance is no longer observable. (B) Average intensity changes are plotted vs. the concentration of RG-I-4. Averaging has been done over all residues, as well as for specific sequences, namely the first 18 residues of the NT, F-face residues 210–225 (β-strands 7, 8, 9), and S-face residues 154–176 (canonical carbohydrate binding site). (C) Chemical shift changes, Δδ, of resonances in the HSQC spectrum of 15N-Gal-3 are plotted vs. the sequence of Gal-3 for the binding of RG-I-4 at 2 μM. There are 11 β-strands in the Gal-3 CRD β-sandwich; the canonical carbohydrate binding site (S-face of the sandwich) is defined by β-strands 1, 10, 3, 4, 5, and 6, and the opposite F-face of the sandwich is defined by β-strands 11, 2, 7, 8 and 9. The insert to A shows F-face residues involved in RG-I-4 binding; these are highlighted in red (most perturbed in HSQC spectra), orange, and blue (least perturbed in HSQC spectra) on the structure of the Gal-3 CRD (pdb access code: 1A3K (Seetharaman et al. 1998)). The color red indicates chemical shift changes greater than 2 SD above the mean, orange between 1 SD and 2 SD, blue below 1 SD. As present in the crystal structure of this Gal-3 CRD, a molecule of lactose is shown as a stick structure in blue on the S-face. (D) Average chemical shift changes are plotted vs. the concentration of RG-I-4. Chemical shifts were internally referenced to DSS (4,4-dimethyl-4-silapentane-1-sulfonic acid), and chemical shift differences (Δδ) were calculated as [(Δ1H)2 + (0.25Δ15N)2]1/2. Averaging has been done over all residues, as well as for specific sequences, namely the first 18 residues of the NT, F-face residues 210–225 (β-strands 7, 8, 9), and S-face residues 154–176 (canonical carbohydrate binding site). This figure is available in black and white in print and in color at Glycobiology online.
Trends in chemical shift changes (Figure 3C,D) essentially parallel those observed for resonance broadening (Figure 3A,B), with Gal-3 NT and F-face resonances initially being the most perturbed and S-face resonances being more affected at higher concentrations of the polysaccharide (e.g., Figure S1C for Δδ at 10 μM). Because chemical shift changes are small (Figure 3D) and line broadening is considerable (Figure 3B) over the titration, spectral effects can be attributed to net binding interactions that occur in the intermediate exchange regime on the chemical shift time scale (Keeler 2005). In this intermolecular exchange regime, the extent of resonance broadening at a particular 15NH site depends on a combination of factors, including binding affinity and stoichiometry (a.k.a. binding avidity when considered together), exchange between multiple binding sites in the polysaccharide, oligomer exchange, and binding-induced changes in internal motions and conformational exchange dynamics. Therefore, we cannot accurately determine binding affinity or stoichiometry, other than to say that the macroscopic equilibrium dissociation constant, Kd, falls in the ~2 μM to ~100 μM range normally associated with the intermediate exchange regime (Keeler 2005). As measured by SPR and BLI, we previously reported that KD values for RG-I-4 binding to Gal-3 fell in the 13 nM to 69 nM range (Gao et al. 2013; Zhang et al. 2016) which indicate stronger binding, a difference that is likely related to SPR and BLI measuring interactions in the solid phase with immobilized Gal-3 and possible multivalent effects on polysaccharide binding (Zhang et al. 2016).
Even though chemical shift changes are small, they are useful to assess which residues of the lectin have the greatest binding-induced change in chemical environment, especially when analyzed at RG-I-4/Gal-3 molar ratios where most resonances are still observed. The insert to Figure 3A shows the folded structure of the Gal-3 CRD (pdb access code: 1A3K (Seetharaman et al. 1998)) in which residues showing the largest Δδ values (Figure 3C) are highlighted in red, followed by orange and then blue for the RG-I-4 concentration of 2 μM. The Gal-3 CRD is shown oriented on the F-face with β-sheet strands labeled. This again illustrates that the F-face (in particular β-strands 7, 8 and 9) initially binds more strongly to sites within RG-I-4 than does the S-face. However, as the titration progresses with the addition of more RG-I-4, the fraction of residues in the CRD with Δδ values above the average for the S-face increases over that for the F-face (Supplemental Figure S1D). This behavior may seem unusual, but could suggest that initial Gal-3 binding to the polysaccharide changes the distribution and types of binding epitopes available to S- and F-faces, along the lines of Gal-1-induced polysaccharide “decongestion” (Miller, Klyosov, et al. 2009).
Binding of RG-I-4 to truncated Gal-3 CRD
Because spectral changes in full length Gal-3 upon binding RG-I-4 can be complicated by transient interactions between the NT and CRD F-face (Ippel et al. 2016), we tried to clarify S- and F-face binding interactions by performing the RG-I-4 titration with truncated Gal-3 CRD (residues 111–250) that is devoid of the NT. Figure 4A shows the HSQC spectrum of the Gal-3 CRD (80 μM, black peaks) overlaid with that of Gal-3 CRD plus RG-I-4 (20 μM, red peaks). As with full length Gal-3, some CRD peaks are differentially and highly broadened and minimally chemically shifted, again indicating that interactions occur in intermediary exchange on the chemical shift time scale. Figure 4B plots Δδ values vs. the Gal-3 CRD sequence. At this RG-I-4 concentration (20 μM), several resonances arising from S-face residues exhibit greater values than those from the F-face. Nevertheless, changes to F-face residues are generally as significant as those from the S-face, indicating that RG-I-4 interacts with both faces of the CRD.
Fig. 4.
(A) 1H–15N HSQC spectral overlays are shown for 15N-enriched Gal-3 CRD (residues 111–250) alone (80 μM, peaks in black) and in the presence (peaks in red) of 20 μM RG-I-4. Solution conditions are 20 mM potassium phosphate buffer, pH 7, 30°C. (B) Chemical shift changes (Δδ) vs. the residue number for the CRD are shown. (C) The F-face/S-face ratio of chemical shift changes averaged over residues within each face of the CRD in full length (FL) Gal-3 and truncated Gal-3 CRD are plotted. Chemical shifts were internally referenced to DSS (4,4-dimethyl-4-silapentane-1-sulfonic acid), and chemical shift differences (Δδ) were calculated as [(Δ1H)2 + (0.25Δ15N)2]1/2. This figure is available in black and white in print and in color at Glycobiology online.
For additional insight, Figure 4C plots the F-face/S-face ratio of chemical shift changes averaged over residues within each face for full length (FL) Gal-3 and for truncated Gal-3 CRD. Although the CRD curve is shifted to lower values, overall trends in both instances are essentially the same, with larger F-face/S-face ratios at lower concentrations of RG-I-4. This trend suggests that there are binding epitopes within the polysaccharide that interact more strongly with the F-face than with the S-face, and as the RG-I-4 concentration is increased, the lower affinity S-face binding epitopes display greater avidity due to a greater binding stoichiometry. We interpret higher values for the Gal-3 FL curve to be the effect of RG-I-4 binding on transient interactions between the NT and CRD (Ippel et al. 2016). Of further note, RG-I-4 binding to the F-face primarily affects residues 198–202, 206–210, 222–226, and less so 242–245 and 124–130, whereas transient NT interactions primarily involve residues 211–214, 217–220, and less so for 111–114, 123, 192, 203–205 and 250 (Ippel et al. 2016). Moreover, transient NT-CRD interactions occur in fast exchange reflecting weak binding, whereas interactions with RG-I-4 occur in intermediate exchange reflecting stronger binding.
Binding of RG-I-4 to free NT peptide
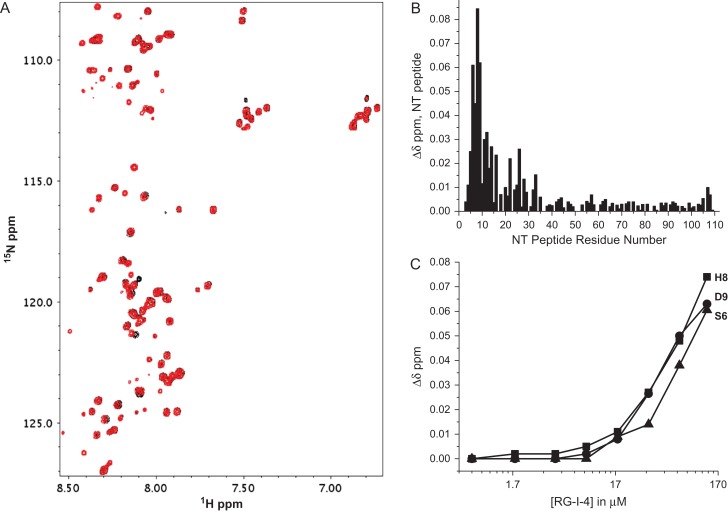
To address the question of whether the NT itself can interact with sites in RG-I-4, we produced 15N-labeled NT peptide residues 1–108 (devoid of the CRD) and acquired HSQC data with RG-I-4. Figure 5A overlays HSQC spectra of 15N-labeled NT peptide (80 μM) in the absence (black peaks) and presence of RG-I-4 (69 μM, red peaks). A number of NT resonances are chemically shifted by the presence of RG-I-4, with the most perturbed ones belonging to residues within the N-terminal part of the NT (Figure 5B) as observed with the NT in full length Gal-3. These data indicate that the Gal-3 NT (independent of the presence of the CRD) binds to one or more sites within the RG-I-4 polysaccharide.
Fig. 5.
(A) 1H–15N HSQC spectral overlays are shown for 15N-enriched Gal-3-NT (residues 1–108) alone (80 μM, cross-peaks in black) and in the presence (cross-peaks in red) of 69 μM RG-I-4 (concentration at the end of the titration). Solution conditions are 20 mM potassium phosphate buffer, pH 7, 30°C. (B) Chemical shift changes (Δδ, difference from the beginning to the end of the titration) vs. the residue number for the N-terminal tail (NT) are shown. (C) Δδ values for three residues (S6, H8 and D9) are plotted vs. the concentration of RG-I-4 during the titration. Chemical shifts were internally referenced to DSS (4,4-dimethyl-4-silapentane-1-sulfonic acid), and chemical shift differences (Δδ) were calculated as [(Δ1H)2 + (0.25Δ15N)2]1/2. This figure is available in black and white in print and in color at Glycobiology online.
However, the story only begins there. The initial part of the titration with RG-I-4 was performed over the period of about 10 hrs (day 1), with the chemical shifts of some of the most perturbed residues plotted in Figure 5C. The following day (day 2), we wanted to continue the titration, but observed that the HSQC spectrum had changed overnight with new NT resonances appearing and original ones displaying attenuated intensities. Because of this, we followed the time course of HSQC spectral changes and discovered that such changes occurred over a number of days. Figure 6 shows expansions from HSQC spectra acquired on days 5, 15 and 29 (red peaks) following addition of RG-I-4 to 15N-labeled NT peptide on day 1 (black peaks). This trend is nicely exemplified with side chain resonances of W22 and W26 Nε and Asn/Gln amides (Figure 6). We previously reported that major and minor peaks observed for W22 and W26 arose from slowly exchanging trans and cis proline isomer states, respectively, most likely associated with P23 (Ippel et al. 2015). The intensities of the new Trp resonances (red peaks) increase from day 5 to day 29. By day 29, all four of these original W22 and W26 Nε resonances have disappeared, and only two new Trp Nε resonances are evident (Figure 6A,C,E), suggesting that NT binding to the polysaccharide stabilizes only one isomer state for each Trp residue. New Asn/Gln side chain amide NH2 resonances also develop over this same time period, as original amide NH2 resonances from free NT disappear (Figure 6B,D,F). By day 29, at least six new pairs of amide NH2 resonances are observed (indicated by dashed lines). In the absence of RG-I-4, only very minor changes to NT resonances are observed by day 35 (Supplemental Figure S2).
Fig. 6.
1H–15N HSQC spectral overlays are shown for 15N-enriched NT peptide (80 μM) alone (peaks in black) and in the presence (peaks in red) of 150 μM RG-I-4 upon standing in solution at 30°C for 5 days (A,B), 15 days (C,D), and 29 days (E,F). Two spectral regions are shown as expansions. Panels A,C,E focus on W22 and W26 HNε resonances, and panels B,D,F focus on Asn and Gln side chain amide 15NH2 resonances. Peaks in black are for data acquired on day 1, and overlaid peaks in red are for the days indicated (5, 15 or 29 days). Dashed lines in B,D,F simply connect amide 15NH2 resonances associated with a single residue. Resonance assignments indicated are for the original resonances as reported by Ippel et al. (2015). Solution conditions are 20 mM potassium phosphate buffer, pH 6.9, 30°C. This figure is available in black and white in print and in color at Glycobiology online.
A number of backbone NH resonances also disappear over time, as new resonances appear (Supplemental Figure S3). The first of these resonances to disappear completely includes those within the S6 to Q20 sequence, most of which were initially perturbed on day 1 of the titration (Figure 5). Resonance assignments shown in Figures 6 and S3 belong to the original NT resonances previously assigned (Ippel et al. 2015). These new NT backbone peaks also do not arise in the absence of the polysaccharide RG-I-4 (Supplemental Figure S4). Although some minor spectral effects are observed by day 35, they are restricted to minimal chemical shift and intensity changes that can be explained most likely by random, cis–trans proline re-arrangements. Therefore, we conclude that binding of the NT peptide to sites within RG-I-4 is causal to time-dependent spectral changes discussed above.
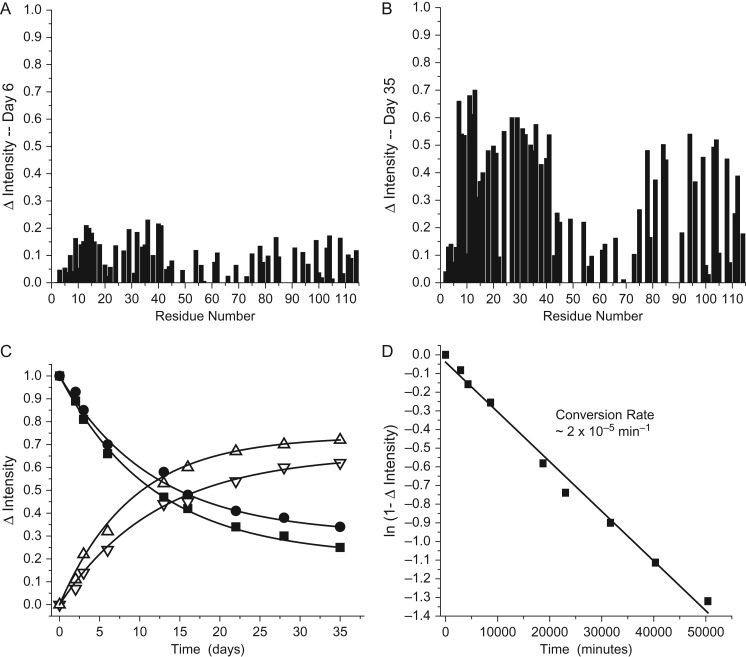
Even though we could not assign new NT resonances due to insufficient signal in 3D heteronuclear NMR experiments (e.g., HNCA and HNCACB), we could assess what occurs when the NT interacts with RG-I-4, because the new resonances should arise from the set of disappearing original resonances whose assignments are known. Figure 7 plots intensity changes of original NT resonances vs. the NT amino acid sequence for time points of day 5 (Figure 7A) and day 29 (Figure 7B) relative to day 1. Although there are some changes that occur within the C-terminal part of the NT (residues ~75–100), the most perturbed resonances belong to residues within the N-terminal part of the NT (residues 1–40). Likewise, seven of the nine Asn/Gln residues in the NT (i.e., N4, N16, N18, Q20, N28, Q29, Q48) perturbed by interactions with RG-I-4 (Figure 6) are also found within the first 40 residues of the NT. Figure 7C plots the change in resonance intensities (ΔIntensity) for two of these NT residues vs. time in days following addition of RG-I-4 to the NT sample. For the original resonances (L11 and G34, solid symbols), the intensity falls smoothly over this time period, while the intensity of new resonances (open symbols) increases proportionally over the same time period. In both instances, 50% change occurs at about day 5, indicating that these events are correlated. Figure 7D plots the logarithmic change in intensity averaged over the first 40 NT residues vs. the incubation time. The straight line fit (regression coefficient of 0.98) indicates the presence of a 2-state process with a conversion rate of 6 × 10−5 min−1.
Fig. 7.
Intensity changes (ΔIntensity) are plotted vs. the amino acid sequence of the NT peptide following 5 days (A) or 29 days (B) of the NT peptide (80 μM) sitting in the presence of 150 μM RG-I-4. ΔIntensity values were calculated as (1 – Inti/Into), where Inti is the resonance intensity at some concentration of the polysaccharide and Into is the resonance intensity in the absence of polysaccharide. A value of 0 indicates the absence of broadening and a value of 1 indicates that that resonance is no longer observable. (C) ΔIntensity is plotted for two original NT residues L11 (solid square) and G34 (solid circle) vs. the incubation time, and ΔIntensity is plotted for two new resonances (assignments presently unknown, open symbols) that arise during the incubation vs. the incubation time. (D) The logarithmic change in resonance intensity averaged over the first 40 NT residues is plotted vs. the incubation time. Data points were fit with a straight line having a regression coefficient of 0.98.
Binding of RG-I-4 to the NT in full length Gal-3
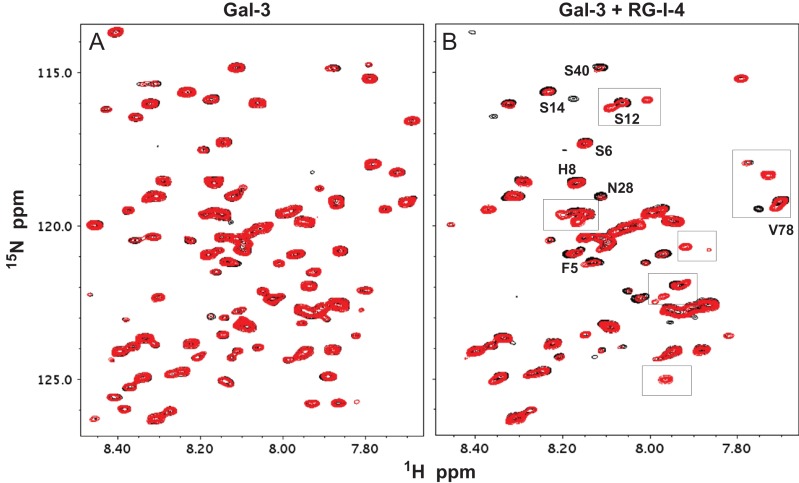
The next question was whether RG-I-4 had the same effect over time on the NT within full length Gal-3. Figure 8 shows HSQC expansions for 15N-labeled Gal-3 sitting in solution in the absence of RG-I-4 for 35 days (Figure 8A,C,E) compared to those for 15N-labeled Gal-3 sitting in the presence of RG-I-4 for 29 days (Figure 8B,D,F). In the absence of the polysaccharide, HSQC spectra remain essentially the same over time. In the presence of RG-I-4, however, the intensities of many NT resonances in full length Gal-3 are decreased significantly by day 29 as new ones appear. These observations essentially parallel those that we observed with free NT peptide. For example, some new resonances found near S12, N28, and V78 (Figure 8D) are observed in both full length Gal-3 NT and NT peptide, and at least three new Asn/Gln side chain amide NH2 resonances (Figure 8B) are evident at similar chemical shift positions in both species. Their similarities are further illustrated in Figures S5A,B,C that show the Asn/Gln side chain NH2 spectral region for Gal-3/RG-I-4 and NT/RG-I-4 on day 1 and day 29. Although there are some spectral differences, this should not be too surprising given that full length Gal-3 consists of both NT and CRD and has three different binding sites for RG-I-4. Moreover, complex formation would be a two-step interaction, with one step being RG-I-4 binding-induced dissociation of the NT from the CRD F-face (Ippel et al. 2016), and the other being NT binding to RG-I-4. Our results with RG-I-4 binding to the truncated Gal-3 CRD (Figure 4) indicate that at least part of the effect with full length Gal-3 is indeed due to RG-I-4 binding-mediated displacement of the weaker, transient interactions between the NT and CRD F-face.
Fig. 8.
(A) 1H–15N HSQC spectral overlays are shown for 15N-enriched full length Gal-3 (80 μM, peaks in black) and after sitting in solution at 30°C for 35 days (peaks in red). (B) 1H–15N HSQC spectral overlays (the same expansion region as in A) are shown for 15N-enriched full length Gal-3 (80 μM, peaks in black) and after sitting in solution with 150 μM RG-I-4 polysaccharide at 30°C for 29 days (peaks in red). This figure is available in black and white in print and in color at Glycobiology online.
As we did in Figure 7 with the NT peptide, Figure 9 plots intensity changes of original NT resonances in full length Gal-3 vs. the amino acid sequence for time points of day 6 (Figure 9A) and day 35 (Figure 9B) relative to day 1. Although the largest effects again occur within the first 40 residues, changes within the C-terminal part of the NT (residues ~75–100) are now relatively greater than with the NT peptide. Moreover, the RG-I-4-induced kinetics of the effect are slower for the NT in full length Gal-3 than with the NT peptide devoid of the CRD. This is shown in Figure 9C which plots the change in Gal-3 NT resonance intensities for two NT residues (L11 and G34, solid symbols) vs. incubation time, along with intensity increases for two new resonances (open symbols). Figure 9D plots the logarithmic change in intensity averaged over the first 40 NT residues vs. the incubation time. The straight line fit (regression coefficient of 0.99) yields a conversion rate of 2 × 10−5 min−1 or three times slower than with the NT peptide.
Fig. 9.
Intensity changes (ΔIntensity) are plotted vs. the amino acid sequence of the NT in full length Gal-3 following 5 days (A) or 29 days (B) of the lectin sitting in the presence of 150 μM RG-I-4. ΔIntensity values were calculated as (1 – Inti/Into), where Inti is the resonance intensity at some concentration of the polysaccharide and Into is the resonance intensity in the absence of polysaccharide. A value of 0 indicates the absence of broadening and a value of 1 indicates that resonance is no longer observable. (C) ΔIntensity is plotted for two original NT residues L11 (solid squares) and G34 (solid circles) vs. the incubation time, and ΔIntensity is plotted for two new resonances (assignments presently unknown) that arise during the incubation vs. the incubation time (open symbols). (D) The logarithmic change in resonance intensity averaged over the first 40 NT residues is plotted vs. the incubation time. Data points were fit with a straight line having a regression coefficient of 0.99.
The NT binding epitope in RG-I-4 resides primarily within its side chains
For insight into the NT binding epitope in RG-I-4, we chemically removed side chains from the polysaccharide as previously reported (Gao et al. 2013). This species, RG-I-4-RG, consists only of the RG-I-4 polysaccharide backbone. When 15N-labeled NT peptide was incubated with RG-I-4-RG for up to 35 days, HSQC spectra remain essentially unchanged. First of all, this indicates that the observed spectral changes do not simply arise from trapping interactions with a relatively large polysaccharide. This view was reinforced by lack of effects from another relatively large polysaccharide, namely a 59 kDa galactomannan [α(1→6)-galactose branched to a β(1→4)-linked mannose backbone], as well as a 5 kDa β(1→4)-linked mannan (the galactomannan backbone) (Miller, Klyosov, et al. 2009; Miller et al. 2016). Overall, our data indicate that the NT binding epitope in RG-I-4 is relatively specific and is located primarily within its side chains, many of which are composed of galactose residues, e.g., galactans.
This proposal was supported by incubating 15N-labeled NT peptide with the pOligo-k galactan (97% galactose, 5.6 kDa) (Gao et al. 2013). In Figure 10, HSQC spectra are overlaid for 15N-labeled NT peptide in the presence of this galactan on day 1 (black peaks) and day 29 (red peaks). After four weeks incubation, new peaks arise as they did in the presence of RG-I-4. Those arising from Asn/Gln amide side chains are connected with dashed lines (Figure 10A), and others arising from NH backbone resonances are boxed (Figure 10B). Some new NT peaks exhibit the same chemical shifts as those observed in the presence of RG-I-4 (see Figures 6 and S3), suggesting that some interacting NT residues are the same. Using the temporal change in intensity averaged over the first 40 NT residues vs. incubation time allows us to derive a conversion rate of about 1 × 10−5 min−1. This indicates that the kinetics of the process with pOligo-k are about six times slower than with RG-I-4. Thus there appears to be something specific with the side chain sugar groups, their structures and/or their structural organization in RG-I-4.
Fig. 10.
1H–15N HSQC spectral overlays are shown for 15N-enriched NT peptide (80 μM) in the presence of pOligo-k galactan (5.6 kDa with 96.9% Gal, 2.8% Ara, 0.3% Glc) (Gao et al. 2013) on day 1 (peaks in black) and on day 29 (peaks in red). Two HSQC expansions are shown: (A) Asn/Gln side chain amide region with some paired peaks connected by dashed lines and (B) NH backbone region where some new peaks are boxed in. Solution conditions are 20 mM potassium phosphate buffer, pH 6.9, 30°C. This figure is available in black and white in print and in color at Glycobiology online.
Discussion
Here, we demonstrate that Gal-3 binds relatively strongly to sites within the 60 kDa rhamnogalacturonan polysaccharide RG-I-4, consistent with our previous results using different techniques (SPR (Gao et al. 2013), BLI (Zhang et al. 2016), fluorescence microscopy (Gao et al. 2013), FLISA (Zhang et al. 2016)). Gal-3 binding to RG-I-4 may not be surprising in itself, because this relatively complex polysaccharide has side chains composed of galactose residues, a well-known galectin binding epitope (Barondes 2008). What is surprising is that RG-I-4 contains multiple binding sites for Gal-3 that interact not only with the lectin's canonical carbohydrate binding site on its CRD S-face, but also at two noncanonical sites, one at the CRD F-face (the opposing β-sheet of the CRD β-sandwich) and a novel one within the NT. Previously, we reported that another large rhamnogalacturonan can bind at both the S- and F-faces of Gal-1 (Miller, Nesmelova, et al. 2009). In addition, we reported that other relatively large polysaccharides (galactomannans) can also bind to the F-face of both Gal-1 (Gal-1) (Miller, Klyosov, et al. 2009, 2012) and Gal-3 (Miller et al. 2016). However, this is the first report that the NT of Gal-3 presents a binding epitope(s) for interaction with a polysaccharide.
The presence of these two noncanonical binding sites (NT and F-face) may explain some previous results that could not be explained when considering interactions to the CRD S-face canonical binding site alone. We reported previously that although the backbone of RG-I-4 binds strongly to Gal-3 and inhibits Gal-3-mediated hemagglutination, binding was independent of the canonical carbohydrate binding site because even a high concentration of lactose did not inhibit Gal-3 binding to the polysaccharide (Gao et al. 2013). The Gal-3 CRD F-face, like the S-face, is composed of many hydrophilic and charged residues: S244, S246, T248 (β-strand 11); R129, L131, T133, L135 (β-strand 2); K199, Q201, L203, E205 (β-strand 7); H208, K210, A212 (β-strand 8); D215 (β-turn), and L218, Q220, N222 (β-strand 9), that could make direct contacts with sugar units from the polysaccharide. And although the F-face does not contain the key galactose-interacting Trp residue found on the S-face, it does contain a number of solvent-exposed, hydrophobic side chains that could make similar contacts. In addition, the well-known concave shape of the canonical binding site, with loops on each side of the β-sheet folding in around a sugar molecule as though it is being grabbed by a hand, is structurally mimicked by the F-face which also has a concave shape (Miller et al. 2016). Therefore, both S- and F-faces have some similar compositional and structural features.
The presence of the Gal-3 NT with its 27 prolines out of 108 residues (human Gal-3) has perplexed researchers for some time. The collagen-like NT does not form collagen triple helices (Mayo et al. 1991), is not integral to the Gal-3-folded structure (Ippel et al. 2016), has questionable involvement in proposed pentamer formation (Ahmad et al. 2004), and has little effect on carbohydrate binding to the CRD canonical site (Dumic et al. 2006; Ippel et al. 2016), yet it is crucial to the functional activity of this unique galectin (Dumic et al. 2006). In the present study, we have provided insight into this conundrum by discovering a polysaccharide binding epitope within the NT, primarily encompassing the first 40 residues that are highly conserved among all known species of Gal-3 (Figure 11). This sequence contains many polar residues (Ser, Asn, Gln), a composition favorable to interactions with polysaccharides. We previously reported that this NT sequence (F5 through N18) forms transient helix structure, followed by less periodic, multiple turn conformations up to about A44 (Ippel et al. 2016). It remains unknown whether this transient NT structure is stabilized (or modified) upon RG-I-4 binding. Furthermore, we find that this NT sequence essentially defines the same RG-I-4 binding epitope in free NT peptide and in the NT within full length Gal-3.
Fig. 11.
The aligned amino acid sequences of full length Gal-3 from six species are shown. For clarity, NT residues are separated from CRD residues as indicated, and totally conserved proline residues within the NT are highlighted in red. This figure is available in black and white in print and in color at Glycobiology online.
The slow spectral changes that occur to resonances from the Gal-3 NT in the presence of RG-I-4 indicate that binding is mediated by slow structural re-arrangements of one or both of the interacting partners. Although we cannot say definitively, the most likely explanation for such slow kinetics is a process involving cis–trans proline isomerization (Keeler 2005). The N-terminal sequence of the NT (residues 1–40) that primarily interacts with the polysaccharide contains six conserved prolines (Figure 11). If only a single proline modulated NT binding, then we would have anticipated reaching the binding end point in a much shorter amount of time. However, the observed time scale is on the order of weeks with an apparent rate constant of about 6 × 10−5 min−1. The rate constant for cis–trans isomerization of a single proline in small linear and cyclic peptides falls in the range of about 1 × 10−1 min−1 to 1 × 10−2 min−1 (Grathwohl and Wüthrich 1981; Stein 1993; Reimer et al. 1998; Fischer 2000). Therefore, the much slower molecular event that we observe could be explained by cis–trans isomerization of multiple prolines. Nevertheless, further investigation is clearly required to prove the hypothesis of a proline isomerization-mediated event.
Assuming that cis–trans proline isomerization is causal to what we observe, the following model can be proposed as shown in equation (1):
 |
(1) |
where NTU is some “nonbinding” distribution of cis-tran isomer states of the multiple prolines within the NT, and NTB is the correct “binding” distribution of those isomer states. In this model, the NTB population is very small compared to that of NTU. However, once NTB is formed, it rapidly binds to its epitope(s) within the polysaccharide “RG” to form a strong complex, i.e., RG/NTB. Although the actual binding epitope within RG-I-4 remains unknown, it appears to reside primarily with its side chains, because slow binding kinetic effects are not observed with the RG backbone stripped of its side chains, yet are observed with a poly-galactose polysaccharide (p-oligo K), albeit to a much lower/slower extent. Furthermore, NTB binding to epitope(s) in RG should be relatively strong, i.e., RG/NTB dissociation is minimal, because by the end of the process all original resonances (primarily residues 1–40) completely disappear and are replaced by new resonances whose chemical shifts are quite different from the original ones. In this regard, formation of the NTB state is the rate limiting step, thus explaining the apparent first order kinetics observed with either free NT or NT in Gal-3 (Figures 7 and 9, respectively).
Proline isomerization is known to induce large conformational shifts in proteins and modulate their intra- and intermolecular interactions (Andreotti 2006). Because of this, it has been reported to mediate a number of biological processes, e.gs. cell signaling (Zhou et al. 2000; Brazin et al. 2002; Wulf et al. 2005; Min et al. 2005; Mayfield et al. 2015), neurodegeneration (Pastorino et al. 2006); transcription (Werner-Allen et al. 2011); channel gating (Lummis et al. 2005), ligand binding (Breheny et al. 2003; Santiveri et al. 2004; Pletneva et al. 2006), amyloidogenesis (Eakin et al. 2006), enzyme activity (Grochulski et al. 1994; Bartholomew et al. 2005; Alag et al. 2013; Bottoni et al. 2016; Quistgaard et al. 2016Shaw 2007; Belvitch et al. 2014), phage infection (Eckert et al. 2005), auto-inhibition of the Crk adaptor protein (Sarkar et al. 2007), and protein folding (Osvath and Gruebele 2003; Glover et al. 2015). Even in light of these studies, the biological relevance of NT binding to this pectin-derived polysaccharide RG-I-4 is unclear. However, our observation that the polysaccharide binding site lies within the N-terminal region of NT whose amino acid sequence is highly conserved among Gal-3 sequences from different species does lend credence to its relevance. In any event, the slow time scale of Gal-3 NT structural conversion suggests that any biological effect would have to be correlated to long term exposure of Gal-3 as could occur with some pathological disorders like Gal-3-induced fibrosis (Li et al. 2014) or atherosclerosis (Pugliese et al. 2015).
In summary, our results indicate that Gal-3 binds relatively strongly to a rhamnogalacturonan polysaccharide. Binding interactions occur at three epitopes in Gal-3, two within its CRD (S- and F-faces) and a novel one within its NT. Given the very slow kinetic changes observed, we propose here that binding of the NT to RG-I-4 is mediated by cis–trans isomerization of conserved, multiple prolines. This work expands our understanding of how Gal-3 interacts with polysaccharides, provides a reasonable explanation for the presence of the NT in Gal-3, and sets the stage for further investigations into the binding of galectins to other complex polysaccharides.
Materials and methods
Preparation of full length and the NT of galectin-3
15N-labeled full length galectin-3 (15N-Gal-3) and 15N-labeled truncated galectin-3 CRD (15N-Gal-3 CRD) were prepared according to Gao et al. (2012). Briefly, E. coli BL21 (DE3) cells were transformed with the expression plasmid for full length Gal-3 that was constructed by inserting the cDNA for Gal-3 (residues 1 to 250) or Gal-3 CRD (residues 111–250) into the vector pET-22b(+) between the NdeI and BamHI cut sites. Bacteria were grown in minimal medium with 15NH4Cl as the nitrogen source, and expression was induced by incubating with 0.2 mM IPTG for 16 h at 25°C. The protein was extracted and purified using a lactose-Sepharose CL-6B column. Purity was assessed by SDS-PAGE.
The 15N-labeled N-terminal tail (15N-NT) of galectin-3 was prepared by thrombin digestion of 15N-labeled His-tagged NT (15N-His-NT). Briefly, E. coli BL21 (DE3) cells were transformed with the expression plasmid for His-NT that was constructed by inserting the cDNA for this sequence (residues 1 to 108) into the pET28a vector between BamHI and EcoRI cut sites. Bacteria were grown in minimal medium with 15NH4Cl as the nitrogen source, with 15N-His-NT expression being induced by using 0.2 mM IPTG at 25°C for 16 h. The cell lysate was then incubated with Ni-NTA Agrose beads, and after extensive washing, the beads with bound 15N His-NT were digested by using 2 U/mg thrombin. 15N-NT was released into the buffer and recovered after dialysis. Purity was assessed by SDS-PAGE.
Polysaccharide preparation
Dried water-soluble ginseng polysaccharide (WGP) from ginseng roots was a gift from Hongjiu Company (Jilin, China). WGP was fractionated according to the procedure established in our lab (Zhang et al. 2009). In brief, WGP was applied to DEAE–Cellulose, eluted with distilled water to give a neutral fraction (WGPN) and then with 0.5 M NaCl to give an acid fraction (WGPA). WGPA was then hydrolyzed by Endo-PG, and the fractionation of the hydrolysates was performed by a combination of anion-exchange and gel-permeation chromatography, as described in our previous publication (Yu et al. 2010).
Total carbohydrate contents were determined by using the phenol–sulfuric acid method (Dubois et al. 1956). The standard was the mixture of monosaccharides that constituted the polysaccharide to be tested. Uronic acid contents were determined by the m-hydroxydiphenyl method (Blumenkrantz and Asboe-Hansen 1973) using galacturonic acid as standard. Gel-permeation and anion-exchange chromatographies were monitored by assaying the total sugar and uronic acid contents.
Sugar composition analysis was performed as described by Zhang et al. (2009). Each of polysaccharide samples (2 mg) was hydrolyzed first with anhydrous methanol containing 1 M HCl at 80°C for 16 h and then with 2 M TFA at 120°C for 1 h. Released monosaccharides were derived by 1-phenyl-3-methyl-5-pyrazolone (PMP), and derivatives were analyzed by HPLC. Molecular weights were estimated by gel-permeation chromatography on a TSK-gel G-3000PWXL column (7.8–300 mm, TOSOH, Japan) coupled to a Shimadzu HPLC system as described by Zhang et al. (2009). The column was precalibrated by standard dextrans (50, 25, 12, 5 and 1 kDa) using linear regression.
These procedures provided us with ginseng-derived rhamnogalacturonan (RG-I-4), with weight averaged molecular weight of 60 kDa and sugar composition of 21.8% Rha, 33.8% GalA, 19.5% Gal and 9.2% Ara.
NMR spectroscopy
Uniformly 15N-labeled Gal-3 was dissolved at a concentration of 80 μM in 20 mM potassium phosphate buffer at pH 7.0, made up using a 95% H2O/ 5% D2O mixture. 1H–15N HSQC NMR experiments were used to investigate binding of polysaccharides. 1H and 15N resonance assignments for recombinant Gal-3 were previously reported by Ippel et al. (2015).
NMR experiments were carried out at 30°C on a Bruker Avance 700 MHz spectrometer equipped with a H/C/N triple-resonance probe and x/y/z triple-axis pulse field gradient unit. A gradient sensitivity-enhanced version of two-dimensional 1H–15N HSQC was applied with 256 (t1) × 2048 (t2) complex data points in nitrogen and proton dimensions, respectively. Raw data were converted and processed by using NMRPipe (Delaglio et al. 1995) and were analyzed by using NMRview (Johnson and Blevins 1994).
Supplementary Material
Supplementary data
Funding
Funding for NMR instrumentation at the University of Minnesota was provided by the Office of the Vice President for Research, the Medical School, the College of Biological Sciences, NIH, NSF and the Minnesota Medical Foundation.
Conflict of interest statement
None declared.
Abbreviations
CRD, carbohydrate recognition domain; Gal-3, galectin-3; Gal-1, galectin-1; NT, N-terminal tail of Gal-3; NMR, nuclear magnetic resonance; HSQC, heteronuclear single quantum coherence; SPR, surface plasmon resonance; BLI, biolayer interferometry; FLISA, fluorescence-linked immunosorbent assay.
References
- Ahmad N, Gabius H-J, André S, Kaltner H, Sabesan S, Roy R, Liu B, Macaluso F, Brewer CF. 2004. Galectin-3 precipitates as a pentamer with synthetic multivalent carbohydrates and forms heterogeneous cross-linked complexes. J Biol Chem. 279:10841–10847. [DOI] [PubMed] [Google Scholar]
- Alag R, Balakrishna AM, Rajan S, Qureshi IA, Shin J, Lescar J, Grüber G, Yoon HS. 2013. Structural insights into substrate binding by PvFKBP35, a peptidylprolyl cis-trans isomerase from the human malarial parasite Plasmodium vivax. Eukaryot Cell. 12:627–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreotti AH. 2006. Opening the pore hinges on proline. Nat Chem Biol. 2:13–14. [DOI] [PubMed] [Google Scholar]
- Barondes SH. 2008. Stumbling on galectins. 2008 In: Klyosov AA, Witczak ZJ, Platt D, editors. Galectins. Hoboken, NJ: Wiley; p. 1–8. [Google Scholar]
- Bartholomew PJ, Jones CW, Benware A, Chernoff J, La Flamme SE. 2005. Refulation of thecatalytic activity of PTP1B: Roles for cell adhesion, tyrosine residue 66, and proline residues 309 and 310. Exp Cell Res. 311:294–306. [DOI] [PubMed] [Google Scholar]
- Belvitch P, Adyshev D, Elangovan VR, Brown ME, Naureckas C, Rizzo AN, Siegler JH, Garcia JG, Dudek SM. 2014. Proline-rich region of non-muscle myosin light chain kinase modulates kinase activity and endothelial cytoskeletal dynamics. Microvasc Res. 95:94–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blumenkrantz N, Asboe-Hansen G. 1973. New method for quantitative determination of uronic acids. Analytical Biochem. 54:484–489. [DOI] [PubMed] [Google Scholar]
- Bottoni C, Perilli M, Marcoccia F, Piccirilli A, Pellegrini C, Colapietro M, Sabatini A, Celenza G, Kerff F, Amicosante G et al. 2016. Kinetic studies on CphA mutants reveal the role of the P158-P172 loop in activity vs carbapenems. Antimicrob Agents Chemother. 660:3123–3126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brazin KN, Mallis RJ, Fulton DB, Andreotti AH. 2002. Regulation of the tyrosine kinase Itk by the peptidylprolyl isomerase cyclophilin A. Proc Natl Acad Sci USA. 99:1899–1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breheny PJ, Laederach A, Fulton DB, Andreotti AH. 2003. Ligand specificity modulated by prolyl imide bond Cis/Trans isomerization in the Itk SH2 domain: A quantitative NMR study. J Am Chem Soc. 125:15706–15707. [DOI] [PubMed] [Google Scholar]
- Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. 1995. NMRPipe: A multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 6:277–293. [DOI] [PubMed] [Google Scholar]
- Dubois M, Gilles KA, Hamilton JK, Rebers PA, Smith F. 1956. Colorimetric method for determination of sugars and related substances. Analytical Chem. 28:350–356. [Google Scholar]
- Dumic J, Dabelic S, Flögel M. 2006. Galectin-3: An open-ended story. Biochim Biophys Acta. 1760:616–635. [DOI] [PubMed] [Google Scholar]
- Eakin CM, Berman AJ, Miranker AD. 2006. A native to amyloidogenic transition regulated by a back bone trigger. Nat Struct Mol Biol. 13:202–208. [DOI] [PubMed] [Google Scholar]
- Eckert B, Martin A, Balbach J, Schmid FX. 2005. Prolyl isomerization as a molecular timer in phage infection. Nat Str Mol Biol. 7:619–623. [DOI] [PubMed] [Google Scholar]
- Fischer G. 2000. Chemical aspects of peptide bond isomerization. Chem Soc Rev. 29:119–127. [Google Scholar]
- Gao X, Liu D, Fan Y, Li X, Xue H, Ma Y, Zhou Y, Tai G. 2012. The two endocytic pathways mediated by the carbohydrate recognition domain and regulated by the collagen-like domain of galectin-3 in vascular endothelial cells. PLoS One. 7:e52430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X, Zhi Y, Sun L, Peng X, Zhang T, Xue H, Tai G, Zhou Y. 2013. The inhibitory effects of a rhamnogalacturonan I (RG-I) domain from ginseng pectin on galectin-3 and its structure-activity relationship. J Biol Chem. 288:33953–33965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glinsky VV, Raz A. 2009. Modified citrus pectin anti-metastatic properties: One bullet, multiple targets. Carbohyd Res. 344:1788–1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glover MS, Bellinger EP, Radivojac P, Clemmer DE. 2015. Penultimate proline in neuropeptides. Anal Chem. 87:8466–8472. [DOI] [PubMed] [Google Scholar]
- Grathwohl C, Wüthrich K. 1981. NMR studies of the rates of proline cis-trans isomerization in Oligopeptides. Biopolymers. 20:2623–2633. [Google Scholar]
- Grochulski P, Li Y, Schrag JD, Cygler M. 1994. Two conformational states of Candida rugose lipase. Protein Sci. 3:82–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inohara H, Raz A. 1994. Effects of natural complex carbohydrate (citrus pectin) on murine melanoma cell properties related to galectin-3 functions. Glycoconj J. 11:527–532. [DOI] [PubMed] [Google Scholar]
- Ippel H, Berbis MA, Suylen D, André S, Hackeng T, Cañada FJ, Weber C, Gabius H-J, Jiménez-Barbero J, Mayo KH. 2015. 1H, 13C, and 15N backbone and side-chain chemical shift assignments for the 29 kDa human chimera-type galectin-3, an adhesion/growth-regulatory lectin. Biol NMR Assign. 9:59–63. [DOI] [PubMed] [Google Scholar]
- Ippel H, Miller MC, Vértesy S, Zheng Y, Cañada FJ, Suylen D, Umemoto K, Romano C, Hackeng T, Tai G et al. 2016. Intra- and intermolecular interactions of human galectin-3: Assessment by full-assignment-based NMR. Glycobiol. 26:888–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson BA, Blevins RA. 1994. NMR View: A computer program for the visualization and analysis of NMR data. J Biomol NMR. 4:603–614. [DOI] [PubMed] [Google Scholar]
- Kaltner H, Gabius H-J. 2012. A toolbox of lectins for translating the sugar code: The galectin network in phylogenesis and tumors. Histol Histopathol. 27:397–416. [DOI] [PubMed] [Google Scholar]
- Keeler J. 2005. Understanding NMR Spectroscopy. New York: Wiley & Sons. [Google Scholar]
- Klyosov AA, Witczak ZJ, Platt D, editors. 2008. Galectins. Hoboken, NJ: Oxford University Press. [Google Scholar]
- Li LC, Li J, Gao J. 2014. Functions of galectin-3 and its role in fibrotic diseases. J Pharmacol Exp Ther. 351:336–343. [DOI] [PubMed] [Google Scholar]
- Lummis SC, Beene DL, Lee LW, Lester HA, Broadhurst RW, Dougherty DA. 2005. Cis-trans isomerization at a proline opens the pore of a neurotransmitter-gated ion channel. Nature. 438:248–252. [DOI] [PubMed] [Google Scholar]
- Mayfield JE, Fan S, Wei S, Zhang M, Li B, Ellington AD, Etzkorn FA, Zhang YJ. 2015. Chemical tools to decipher regulation of phosphatases by proline isomerization on eukaryotic RNA polymerase II. ACS Chem Biol. 10:2405–2414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayo KH. 1996. NMR and X-ray studies of collagen model peptides. Biopolymers. 40:359–370. [DOI] [PubMed] [Google Scholar]
- Mayo KH, Parra-Diaz D, McCarthy JB, Chelberg M. 1991. Cell adhesion promoting peptide GVKGDKGNPGWPGAP from the collagen type IV triple helix: cis/trans proline-induced multiple 1H NMR conformational and evidence for a KG/PG multiple turn repeat motif in the all-trans proline state. Biochem. 30:8251–8267. [DOI] [PubMed] [Google Scholar]
- Miller MC, Ribeiro JP, Roldos V, Martín-Santamaría S, Cañada FJ, Nesmelova IA, André S, Pang M, Klyosov AA, Baum LG, et al. 2011. Structural aspects of binding of α-linked digalactosides to human galectin-1. Glycobiology. 21:1627–1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller MC, Klyosov AA, Mayo KH. 2009. The α-galactomannan Davanat binds galectin-1 at a site different from the conventional galectin carbohydrate binding domain. Glycobiol. 19:1034–1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller MC, Nesmelova IV, Platt D, Klyosov AA, Mayo KH. 2009. Carbohydrate binding domain on galectin-1 is more extensive for a complex glycan than for simple saccharides: Implications for galectin-glycan interactions at the cell surface. Biochem J. 421:211–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller MC, Klyosov AA, Platt D, Mayo KH. 2009. Using pulse field gradient NMR diffusion measurements to define molecular weight distributions in glycan preparations. Carbohydr Res. 344:1205–1212. [DOI] [PubMed] [Google Scholar]
- Miller MC, Klyosov AA, Mayo KH. 2012. Structural features for α-galactomannan binding to galectin-1. Glycobiology. 22:543–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller MC, Ippel H, Suylen D, Klyosov AA, Traber PG, Hackeng T, Mayo KH. 2016. Binding of polysaccharides to human galectin-3 at a non-canonical site in its carbohydrate recognition domain. Glycobiol. 26:88–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min L, Fulton DB, Andreotti AH. 2005. A case study of proline isomerization in cell signaling. Front Biosci. 10:385–397. [DOI] [PubMed] [Google Scholar]
- Nesmelova IV, Dings RPM, Mayo KH. 2008. Chapter 2–Understanding galectin structure-function relationships to design effective antagonists In: Klyosov AA, editor. Galectins. New York: Oxford University Press, p. 11–26. [Google Scholar]
- Osvath S, Gruebele M. 2003. Proline can have opposite effects on fast and slow protein folding phases. Biophys J. 85:1215–1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastorino L, Sun A, Lu PJ, Zhou XZ, Balastik M, Finn G, Wulf G, Lim J, Li SH, Li X et al. 2006. The prolyl isomerase Pin1 regulates amyloid precursor protein processing and amyloid-beta production. Nature. 440:528–534. [DOI] [PubMed] [Google Scholar]
- Pletneva EV, Sundd M, Fulton DB, Andreotti AH. 2006. Molecular details of Itk activation by prolyl isomerization and phospho ligand binding: the NMR structure of the Itk SH2 domain bound to a phosphopeptide. J Mol Biol. 357:550–561. [DOI] [PubMed] [Google Scholar]
- Pugliese G, Iacobini C, Blasetti Fantauzzi C, Menini S. 2015. The dark and bright side of atherosclerosis calcification. Atherosclerosis. 238:220–230. [DOI] [PubMed] [Google Scholar]
- Quistgaard EM, Weininger U, Ural-Blimke Y, Modig K, Nordlund P, Akke M, Löw C. 2016. Molecular insights into substrate recognition and catalytic mechanism of the chaperone and FKBP peptidyl-prolyl isomerase SlyD. BMC Biol. 14:82–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reimer U, Scherer G, Drewello M, Kruber S, Schutkowski M, Fischer G. 1998. Side-chain effects on peptidyl-prolyl cis/trans isomerization. J Mol Biol. 279:449–460. [DOI] [PubMed] [Google Scholar]
- Santiveri CM, Perez-Canadillas JM, Vadivelu MK, Allen MD, Rutherford TJ, Watkins NA, Bycroft M. 2004. NMR structure of the alpha-hemoglobin stabilizing protein: Insights into conformational heterogeneity and binding. J Biol Chem. 279:34963–34970. [DOI] [PubMed] [Google Scholar]
- Sarkar P, Reichman C, Saleh T, Birge RB, Kalodimos CG. 2007. Proline cis-trans isomerization controls auto-inhibition of a signaling protein. Mol Cell. 25:413–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sathisha UV, Jayaram S, Harish Nayaka MA, Dharmesh SM. 2007. Inhibition of galectin-3 mediated cellular interactions by pectic polysaccharides from dietary sources. Glycoconj J. 24:497–507. [DOI] [PubMed] [Google Scholar]
- Seetharaman J, Kanigsberg A, Slaaby R, Leffler H, Barondes SH, Rini JM. 1998. X-ray crystal structure of the human galectin-3 carbohydrate recognition domain at 2.1-A resolution. J Biol Chem. 273:13047–13052. [DOI] [PubMed] [Google Scholar]
- Shaw PE. 2007. Peptidyl-prolyl cis/trans isomerases and transcription: Is there a twist in the tail? EMBO Rep. 8:40–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smetana K, André S, Kaltner H, Kopitz J, Gabius H-J. 2013. Context-dependent multifunctionality of galectin-1: A challenge for defining the lectin as therapeutic target. Expert Opin Ther Targets. 17:379–392. [DOI] [PubMed] [Google Scholar]
- Stein RL. 1993. Mechanism of enzymatic and nonenzymatic prolyl cis–trans isomerization. Adv Protein Chem. 44:1–24. [DOI] [PubMed] [Google Scholar]
- Werner-Allen JW, Lee CJ, Liu P, Nicely NI, Wang S, Greenleaf AL, Zhou P. 2011. Cis-Proline mediated Ser(P)5 dephosphorylation by the RNA poltmerase II C-terminal domain phosphatase Ssu72. J Biol Chem. 286:5717–5726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wulf G, Finn G, Suizu F, Lu KP. 2005. Phosphorylation-specific prolyl isomerization: Is there an underlying theme? Nat Cell Biol. 7:435–441. [DOI] [PubMed] [Google Scholar]
- Yu L, Zhang X, Li S, Liu X, Sun L, Liu H, Iteku J, Zhou Y, Tai G. 2010. Rhamnogalacturonan I domains from ginseng pectin. Carbohyd Polym. 79:811–817. [Google Scholar]
- Zhang X, Yu L, Bi H, Li X, Ni W, Han H, Li N, Wang B, Zhou Y, Tai G. 2009. Total fractionation and characterization of the water-soluble polysaccharides isolated from Panax ginseng C.A. Meyer. Carbohyd Polym. 77:544–552. [Google Scholar]
- Zhang T, Zheng Y, Zhao D, Yan J, Sun C, Zhou Y, Tai G. 2016. Multiple approaches to assess pectin binding to galectin-3. Int J Biol Macromol. 91:994–1001. [DOI] [PubMed] [Google Scholar]
- Zhou XZ, Kops O, Werner A, Lu PJ, Shen M, Stoller G, Kullertz G, Stark M, Fischer G, Lu KP. 2000. Pin1-dependent prolyl isomerization regulates dephosphorylation of Cdc25C and tau proteins. Mol Cell. 6:873–883. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.