Abstract
The Global Alliance for Genomics and Health (GA4GH) created the Beacon Project as a means of testing the willingness of data holders to share genetic data in the simplest technical context—a query for the presence of a specified nucleotide at a given position within a chromosome. Each participating site (or “beacon”) is responsible for assuring that genomic data are exposed through the Beacon service only with the permission of the individual to whom the data pertains and in accordance with the GA4GH policy and standards.
While recognizing the inference risks associated with large-scale data aggregation, and the fact that some beacons contain sensitive phenotypic associations that increase privacy risk, the GA4GH adjudged the risk of re-identification based on the binary yes/no allele-presence query responses as acceptable. However, recent work demonstrated that, given a beacon with specific characteristics (including relatively small sample size and an adversary who possesses an individual’s whole genome sequence), the individual’s membership in a beacon can be inferred through repeated queries for variants present in the individual’s genome.
In this paper, we propose three practical strategies for reducing re-identification risks in beacons. The first two strategies manipulate the beacon such that the presence of rare alleles is obscured; the third strategy budgets the number of accesses per user for each individual genome. Using a beacon containing data from the 1000 Genomes Project, we demonstrate that the proposed strategies can effectively reduce re-identification risk in beacon-like datasets.
Keywords: genomic privacy, ga4gh, beacon, re-identification, genomic data sharing
Introduction
The Global Alliance for Genomics and Health (GA4GH)1 conceived the Beacon Project as a means of testing the willingness of international sites to share genomic data in the simplest of all technical contexts: a public web service that any data holder could implement to enable users to submit queries of the form, “Do you have any genomes with nucleotide A at position 100,735 on chromosome 3?,” to which the service would respond with “Yes” or “No.” A site offering this service is called a beacon and is responsible for ensuring that genomic data are exposed through the Beacon service only with the permission of the individual to whom the data pertain and in accordance with the GA4GH ethical framework2 and privacy and security policy.3 Thus, the Beacon service is designed to be technically simple, easy to implement, and privacy protective.
The availability of vast quantities of high-quality genomic and health data is essential to the advancement of biomedical knowledge. Yet, privacy concerns often limit researchers’ ability to access potentially identifiable health data. Indeed, in some cases, privacy laws and regulations actually impede individuals’ ability to make their own data available to researchers.4 This problem is particularly acute in the field of genomics, where the vast majority of variants predicted to be functionally important are extremely rare, occurring in < 0.5% of the population.5 As a result, it is unlikely that any single institution will hold enough data to achieve sufficient statistical power in studying any particular condition. Recognizing the urgent need for federation across organizations, the GA4GH was formed in 2013 to enable responsible sharing of genomic and health-related data by establishing consistent policy and interoperable standards and protocols.
From its inception, the GA4GH has been committed to achieving a responsible and effective balance between data sharing and individual privacy, a challenge that has been extensively explored in the literature.6–9 In 2008, Homer et al.6 showed that statistical techniques can reveal the presence or absence of an individual in a genomic data set, even when the targeted individual’s genome accounts for < 0.1% of the total data. The publication of this paper had a significant impact, prompting several major institutions, including the Wellcome Trust and the US National Institutes of Health, to limit public access to data formerly adjudged to be safely anonymous.10 As this scenario demonstrates, privacy concerns can undermine the ability of researchers to publish and access genomic data.
At the outset, the GA4GH recognized that the Beacon approach can reveal information about the individuals in a data set. However, in performing the risk assessment, the GA4GH recognized several conditions that served to mitigate the risk that any individual would be identified based on Beacon search. First, the Beacon user interface is extremely restrictive, enabling queries only for the presence or absence of the four nucleotides (A, C, T, G) that make up every individual’s genome. Second, the number of individual genomes aggregated in each beacon is very large. Third, for a data seeker to be able to identify an individual through Beacon queries would require as a precondition that the data seeker possess a significant amount of genomic data associated with the targeted individual, such as a variant call format file of the individual’s whole genome sequence. In such a case, a potential adversary would know all variants in the individual’s genome and would have much more efficient means of discovering a disease association than persistent beacon queries. Thus, GA4GH concluded that the risk of a data seeker identifying an individual through Beacon queries was acceptably low, even for the case of a data seeker willing to violate GA4GH’s ethical standards.
However, Shringarpure and Bustamante11 describe an attack in which an anonymous adversary, even with knowledge of only a small portion of a target’s genome, can successfully launch a re-identification attack: in a beacon comprising 1000 individuals, for instance, 5000 queries suffice. Such an attack relies on a likelihood ratio test whose power is a function of the responses returned by the beacon, the size of the data set, the allele-frequency spectrum, and the sequencing error rate. Their paper demonstrates that under certain conditions, the anonymous-access model implemented by the Beacon Project does not prevent identification of individuals whose genomes could be exposed through a Beacon interface.
The goal of this paper is to further examine the potential vulnerabilities and risks associated with the Beacon model and to explore ways of mitigating re-identification risks—thus enhancing Beacon privacy protections. Re-identification is the process by which anonymized personal data is matched with its true owner.12 We first analyze the re-identification threat described by Shringarpure and Bustamante and the vulnerability the attack exploited. We then propose an optimized version of the attack that considers an adversary with some background knowledge about the allele frequencies (AFs) in the targeted beacon. We describe three potential strategies for mitigating the risk of re-identification and assess their effectiveness through several experiments with data obtained from the 1000 Genomes Project.13 We conclude the paper by discussing the strengths and weaknesses of the proposed strategies and by providing some recommendations for strengthening Beacon privacy protections.
Materials and Methods
Original re-identification attack
We begin by describing the re-identification attack proposed by Shringarpure and Bustamante.11 In the following, we refer to it as the “SB attack.”
As noted earlier, the setting of the SB attack is similar to that of previous works such as that of Homer et al.9 The attacker is assumed to have access to the variant call format file of a target victim’s genome and queries the beacon at heterozygous positions to determine whether the victim is in the beacon. The SB attack relies on a likelihood-ratio test (LRT) that evaluates the likelihood of the beacon’s responses under two possible hypotheses:
The null hypothesis : The queried victim’s genome is not in the beacon.
The alternative hypothesis : The queried victim’s genome is in the beacon.
The re-identification risk is measured by the power of such a test, i.e., . To make their test as general as possible, Shringarpure and Bustamante assume only that the attacker knows the beacon size , as well as the site frequency spectrum of the beacon population. Formally, the alternate allele frequency of a heterozygous SNP observed in the population is assumed to be distributed as for population parameters . Their LRT further allows for a probability of sequencing errors, resulting in a mismatch between the attacker’s copy of a genome and the copy in the beacon.
Given a set of beacon responses , the log-likelihood of the sequence is
| (1) |
Under , let denote the probability that none of the other genomes in the beacon have an alternate allele at position . Similarly, under , we denote by the probability that none of the genomes in the beacon have an alternate allele at . Then, under the two hypotheses, we have
| (2) |
| (3) |
Shringarpure and Bustamante show that under their assumptions, for any position we have and , where . The log of the LRT is given by
| (4) |
where and are constant for fixed. Thus, (the number of “Yes” responses from the beacon) is a sufficient statistic for the LRT.
“Optimal” attack with real allele frequencies
The SB attack removes direct dependency on AF and sets conservative bounds for the number of queries required for successful re-identification. We consider here a more capable and determined attacker who has access to some background knowledge on AFs and optimizes his attack by querying the rarest alleles in the victim’s genome first. In other words, similarly to best practices in forensics, the attacker makes use of alleles with maximum re-identification power instead of performing random requests. This assumption appears reasonable in practice, as allele frequency information for different ancestries is already publicly available on the Web (e.g., 1000 Genomes Project,14 HapMap Project,15 etc.) and easily accessible even by nonexpert attackers. We show through several experiments (see Results section) that this new attack is significantly more powerful than the original SB attack, even when the attacker has incomplete knowledge of AFs in the beacon.
Formally, the attacker assumes AFs for the SNPs in the victim’s genome. Without loss of generality, we assume the frequencies are already ordered (i.e., ). Then, the attacker will maximize his re-identification power by first querying those SNPs that are least likely to appear in the beacon under , specifically those with the lowest frequency. In this setting, Equations (2) and (3) still hold, but the computation of and is different. Under the alternative hypothesis, we have
Similarly, under , we have .
As the probabilities and now directly depend on the position , we have that the following LRT
| (5) |
We will evaluate the power of this test empirically through experiments in a variety of settings with real data and different levels of adversarial background knowledge. We will estimate the null distribution of the LRT by computing Equation (5) for a number of control individuals known not to be in the beacon. The null hypothesis is rejected if for some threshold . We then let be such that . The power of the test is computed as , where the distribution of given is estimated by querying individuals in the experimental beacon.
Risk mitigation strategies
Based on the “optimal” version of the re-identification attack, we propose three different practical strategies to mitigate the risk. Without loss of generality, we can assume that any defense mechanism that effectively mitigates the “optimal” re-identification attack also effectively mitigates the original SB attack. Our experimental results (see Results section) show the validity of this assumption.
Beacon alteration strategy
The first strategy (S1) relies on the observation that most of the statistical power in the re-identification attack comes from queries targeting unique alleles in the beacon. In particular, S1 alters the beacon by answering a query with “Yes” only if there are at least individuals sharing the queried allele. In other words, is the minimum number of individuals in the beacon sharing the queried allele when returning “Yes.” Current beacons set ; i.e., when there are one or more individuals in the population with the queried allele, the answer will be “Yes.” We assume the value of is made public, hence the attacker will modify the attack to accommodate this change (see Supplementary Appendix A for LRT under S1). Yet, already for , we found that in practice what the attacker can infer is limited (see Results section).
Random flipping strategy
The second strategy (S2) relies on the same observation but instead of altering the beacon response, it introduces noise into the original data. The disadvantage of S1 is that only a subset of variations (e.g., the nonunique SNPs when ) in the beacon population can be queried. In practice, unique alleles that are likely to be the most useful in human genetics research are completely hidden. S2 improves the usability of the beacon over S1 because it hides only a portion of unique alleles, but not all. In other words, a beacon with S2 will add noise by sampling from a binomial distribution with probability only to unique alleles in the database and provide false answers (e.g., “No” instead of “Yes”) to queries targeting these unique alleles. The main goal of S2 is to share as many unique alleles as possible while reducing the likelihood that the information released will be sufficient to re-identify an individual in the database. We assume the value of is public. As for S1, the attacker will adapt the LRT statistic to take it into account (see Supplementary Appendix B for LRT under S2).
Query budget per individual strategy
The third strategy (S3) mitigates the re-identification risk by assigning a budget to every individual in the database; this budget is applied to each authenticated Beacon user. With respect to strategies S1 and S2, S3 leverages two additional assumptions:
Each Beacon user has been identity proofed, holds a single account, is authenticated, and does not collude. If users are allowed to collude, then to be effective, S3 will have a dramatic impact on the utility of the system. This assumption appears reasonable in practice as, in order to collude, a user needs by definition to involve someone else. We assume each user holds a single Beacon account to eliminate the possibility of a single user simulating multiple profiles in collusion, which carries higher risk than either collusion among multiple users or a re-identification attack that can be undertaken at an individual scale. This is because an attack involving multiple accounts all working on behalf of a single attacker does not require exchanging files with other users and could be conducted more quickly than a single-threaded attack.
The attacker has accurate genomic information, which means . This is a worst-case assumption because if we can prevent re-identification under this condition, we can prevent the proposed “optimal” attack, too. Note that in practice, because there are some sequencing errors (i.e., ), the attacker will actually have less power. Hence, this approach is conservative from a re-identification point of view. Moreover, by assuming , we can significantly simplify the analytical treatment of the problem.
The basic idea is that each time an individual’s genome contributes to a “Yes” answer for a given query (i.e., the individual has the queried allele), her corresponding budget for that Beacon user is reduced by an amount that depends on the frequency of the queried allele. If her budget is less than this amount, her information will not be used to answer that query and the individual will be removed from the dataset, as shown in Algorithm1 in Table 1. In this way, the privacy of the individual will be always preserved at a cost of a slight decrease of utility.
Table 1.
Algorithm describing mitigation strategy S3
| Algorithm1 | |
|---|---|
| Requires: upper bound on test errors | |
| 1. | Set all . |
| 2. | Receive ’th query and check whether it has been asked before. If yes, go to Step 3. If no, go to Step 4. |
| 3. | Return the previous answer, then go to Step 2. |
| 4. | Compute the risk . |
| 5. | Check whether there are any records with the asked variant and . If no, return no and go to Step 2. |
| 6. | For all the individuals with such variant and , reduce their budgets by . Then return yes. |
| 7. | Go back to Step 2 and wait for the next query. |
Let be the set of responses of the beacon; the goal of S3 is to keep track of the power of the attack, which is based on the LRT , to prevent any individual genome from contributing to a query response that can leak identity information with high confidence (see Supplementary Appendix C for formal description of S3).
Experiments with real data
To evaluate the effectiveness of the proposed strategies in reducing risk under the “optimal” attack with real AFs, we designed and ran several experiments on real data with the following setup. We created a beacon composed of 1235 samples of chromosome 10 randomly chosen from the 2504 individuals in phase 3 of the 1000 Genomes Project.13 A total of 31 relatives were removed. The resulting data set consists of individuals with either European, African, admixed American, East Asian, or South Asian ancestries. Among these samples, 100 were selected as the control set. Similarly, from the remaining individuals not in the beacon, 100 were selected as the test set.
The null distribution of the LRT statistic was obtained through the exact-test computation on the 100 individuals in the test set (i.e., not in the beacon). With a false positive rate of , we computed the power () as the proportion of test rejected (i.e., when ) for the control set (i.e., how many individuals in the control set, hence in the beacon, were successfully re-identified).
Results
“Optimal” re-identification attack in single-population Beacon
We evaluated the re-identification power of our attack on a beacon composed of individuals coming from the same ancestry group. From phase 3 of the 1000 Genomes Project, we selected 502 samples of European (EUR) ancestry and randomly picked half of them to set up the beacon. The remaining half was used to compute the EUR population AFs. We considered several scenarios where the attacker has different types of background information.
As expected, results in Figure 1 show that the worst-case scenario is represented by an attacker knowing the exact ancestry of the population in the beacon. With only three SNPs, beacon membership could be re-identified with power and a false positive rate. Yet, because the beacon ancestry information is not always public, a more realistic scenario is to consider an attacker who knows only the AFs of a random population, possibly from a different ancestry than the one of the beacon. Even with the least precise background information (in this case the AFs from East Asian (EAS) ancestry), 36 SNPs are sufficient to re-identify an individual. Figure 2 shows the Kendall rank correlation coefficient16 between the actual AFs in the beacon and the AFs from different ancestry groups. By combining the information in Figures 1 and 2, it is easy to observe that the higher the ordinal association between the beacon AFs and the AFs known by the attacker, the fewer queries needed to re-identify with power and false positive rate (see Supplementary Appendix D for results on the “optimal” attack in a multipopulation beacon).
Figure 1.
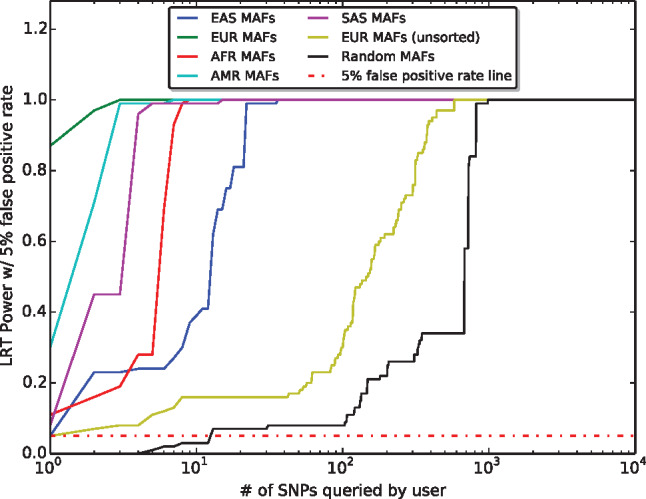
“Optimal” re-identification attack in single-population beacon. Different power rates per number of SNPs queried from an unprotected beacon with a single population (EUR) by an adversary with different types of background knowledge: (green) the attacker knows the allele frequencies (AFs) of a population from the same ancestry (EUR) as the one in the beacon and performs queries following the rare-allele-first logic; (red, cyan, blue, and purple) the attacker knows the AFs of a population from an ancestry different from the one in the beacon and performs queries following the rare-allele-first logic (African [AFR], admixed American [AMR], East Asian [EAS], or South Asian [SAS], respectively); (yellow) the attacker knows the AFs of a distinct population with the same ancestry (EUR) other than the one in the beacon but performs queries in random order; (black) the attacker does not have any information on AFs (i.e., the original attack by Shringarpure and Bustamante11).
Figure 2.

Kendall rank correlation coefficient with respect to true beacon allele frequencies. Kendall rank correlation coefficient between the actual AFs of the single-population beacon of Figure 1 and the AFs of populations with different ancestries. Values closer to represent higher correlation. Color mapping as in Figure 1.
“Optimal” re-identification attack in Beacon with S1
We evaluated the proposed solution S1 by considering an attacker who knows the AFs of the 1000 Genomes Project and the value of threshold parameter . As such, we set up a beacon as described in Materials and Methods section and computed the LRT statistic as described in Supplementary Appendix A. Figure 3 shows that, under such an attack, no individual in the beacon can be re-identified if a “Yes” answer is provided only when the queried allele appears at least times in the database. Yet, the downside of this method is that only a fraction of the alleles that are in the beacon can be shared. For example, in our experimental beacon, only 60% of the alleles are shared by two or more individuals and thus can be shared; the queries to the remaining rare alleles () will receive a “No” answer even though they are actually present in the Beacon database.
Figure 3.

“Optimal” re-identification attack in beacon with S1. Different power rates per number of SNPs randomly queried from a beacon with mitigation S1 by an adversary with knowledge on and on AFs from the 1000 Genomes project: (blue) ; (green) .
“Optimal” re-identification attack in Beacon with S2
To evaluate the effectiveness of S2 against an attack with background knowledge on AFs, we consider an attacker who knows the AFs of the 1000 Genomes Project and the value of the parameter . Figure 4 shows how the statistical power of the attacker decreases when different portions () of unique alleles are hidden. When is set to , the attacker has to query around unique alleles to obtain a strong power of re-identification, compared to queries for re-identification when no random flipping on unique alleles (of strategy) is applied. When ≥ , the re-identification power will not increase above , which will keep the power at an acceptable risk level (i.e., relatively low confidence of re-identification).
Figure 4.
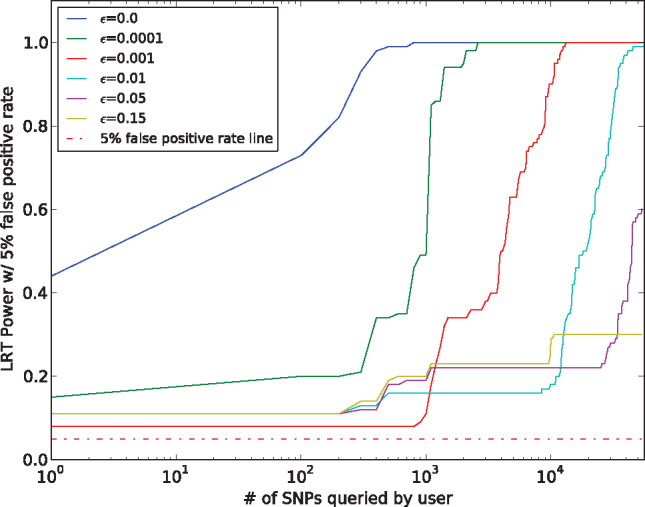
“Optimal” re-identification attack in beacon with S2. Different power rates per number of SNPs queried (with rare-first logic) from a beacon with mitigation S2 by an adversary with knowledge on and on AFs from the 1000 Genomes project. Different colors for different values of .
Budget evaluation in Beacon with S3
We evaluated strategy S3 with the same experimental setting as for S1 and S2. By default, we set p = .05, which means the statistical power of attack cannot exceed 0.95. Differently from experiments performed on solutions S1 and S2, which show an increase in re-identification risk given certain levels of utility of the beacon, we evaluate the efficacy of S3 by computing the decrease of utility across queries for a certain level of re-identification risk.
To this purpose, we emulate the query behavior of a typical honest beacon user by generating queries based on the distribution of query frequency per allele frequency extracted from ExAC browser17 logs over a period of 12 weeks (data on beacon query frequencies were not available at the time of this work.) During this time frame, a total of 1 345 291 queries were asked on 934 680 variants present in ExAC. Table 2 shows the proportion of queries per range of AFs.
Table 2.
Proportions of queries (over a period of 12 weeks) for each range of allele frequency
| Allele frequency | < 0.001 | 0.001–0.01 | 0.01–0.05 | 0.05–0.5 | > 0.5 |
|---|---|---|---|---|---|
| Queries in ExAC | 0.853 | 0.076 | 0.023 | 0.033 | 0.014 |
Figure 5 shows how the number of individuals with enough budget decreases with respect to the number of queries answered by the beacon. Note that the beacon’s utility is completely preserved for the first 2000 queries.
Figure 5.

Budget evaluation in beacon with S3. Behaviors of individual budgets per number of SNPs queried according to the typical user’s query profile obtained from ExAC log data. The cyan curve represents the number of individuals with enough budget to answer “Yes” to queries targeting alleles with AF = 0.001. Red, green, and blue curves correspond to 0.002, 0.005, and 0.01, respectively.
Discussion
In this paper, we have analyzed in detail the beacon re-identification attack originally proposed by Shringarpure and Bustamante and a new and “optimal” version of it by considering a smarter adversary who makes use of public information on AFs. We evaluated the power of our new attack through several experiments on real data by considering different conditions of adversarial background knowledge. Our results show that our attack always outperforms the original SB attack. As one might expect, we have observed that the power of an adversary’s re-identification attack is directly related to the completeness and accuracy of the adversary’s knowledge of the AF of the targeted Beacon. As already analyzed by Shringarpure and Bustamante, the underlying LRT test can be extremely harmful when a beacon is linked to sensitive phenotypes. Yet it is important to emphasize that, although our attack further reinforces SB’s concern, the re-identification risk is relative to each beacon. These attacks fundamentally rely on the assumption that the attacker already has access to the genome of the victim.
Despite such a strong assumption, several research efforts in genomic privacy have studied the problem of re-identification of membership in genetic databases and have shown that this is extremely hard to prevent and sometimes even impossible.18
Based on the “optimal” re-identification attack, we have proposed three different strategies aimed at effectively thwarting beacon membership re-identification. Because the accuracy of the beacon re-identification attack depends on the power and false positive rate of the LRT test, the probability that a test behaves correctly (rejecting the null hypothesis when it is false and failing to reject when it is true) is given by Power * (Probability of alternative hypothesis) + (1 − False positive rate) * (Probability of null hypothesis). From the perspective of a beacon administrator, the attacker’s test should be incorrect most of the time; i.e., power should be low and/or the false positive rate should be high.
The three proposed strategies all address the mitigation problem by controlling the power or the false positive rate. The first (S1) and second (S2) strategies reduce the power to nearly zero when the LRT must have a small false positive rate, whereas in the third solution (S3), the test always has 100% power but a high false positive rate. In particular, S1 and S2 directly alter the beacon to reduce the inference power of the attacker, whereas S3 introduces a new idea of personal budget that decreases when the genome of the individual is used to positively answer a query.
Results of our experiments have shown that all proposed mitigation strategies have advantages and disadvantages, as summarized in Table 3. S1 effectively mitigates the attack by keeping the power of the LRT to 0.2 if all unique alleles are flipped. Yet it generates a significant loss in utility of the beacon because the majority of the queries of a typical beacon user usually target rare alleles. We define the utility of a beacon as the proportion of true answers it can provide. S2 can be considered a more sophisticated version of S1 because it flips only a portion of unique alleles, affording more fine-grained control over the utility vs privacy trade-off. The attack inference power can be confined to a secure level by flipping only of unique alleles (which means a drop in utility of against of S1). Note that the utility of a beacon adopting either S1 or S2 is fixed a priori and does not change along with the power of the attack.
Table 3.
Summary of advantages and disadvantages of the 3 proposed mitigation strategies
| Risk mitigation strategy | Disadvantages | Advantages |
|---|---|---|
| S1: Beacon alteration | Eliminates possibility of querying for unique alleles highly likely to be most useful in genetic research | Protects privacy of individuals possessing variants most likely to be targeted by attackers |
| S2: Random flipping | Decreases rate of true answers returned from querying unique alleles likely to be useful in genetic research | Permits some unique alleles to be discoverable and to fine-tune the privacy–utility trade-off |
| S3: Query budget per individual | Requires the assumption of Beacon user being nonanonymous and holding no more than one Beacon account; may require complicated accounting scheme | Enables all alleles to be discoverable until budget is exceeded |
Finally, results of experiments on S3 show that, given a certain assurance level (p = .05), the beacon utility is completely preserved for the first 2000 queries. Yet S3 relies on the assumption that the beacon system is not anonymous and has a controlled level of access with user authentication and identity proofing. Based on data collected from the ExAC browser logs, a budget of 2000 queries per beacon user seems a reasonable compromise between re-identification risk and utility.
Preventing inference attacks on large databases is widely known to be one of the most daunting of database security challenges.19 This fact has been a major consideration in the development of GA4GH’s framework for responsible sharing of genomic and health-related data, privacy and security policy, and security infrastructure. Effective risk management must leverage policy, technology, and community governance to address re-identification risks. Effective risk management is fundamental to facilitating and promoting data sharing across the GA4GH global community. We emphasize that security and privacy are components of risk management. Technical risk-management strategies such as those proposed in this paper are practical and can be adapted according to the context of each beacon. Therefore, they represent a valuable set of options for assessing and mitigating risk within the GA4GH community.
Supplementary Material
ACKNOWLEDGMENTS
The authors would like to thank the GA4GH for its continuous support and Konrad Karczewski and Marc Duby from the Broad Institute for their valuable feedback and for having provided the data on query access patterns for the ExAC browser. Dixie Baker and Paul Flicek are co-chairs of the Security Working Group of the GA4GH and David Lloyd is a member of the GA4GH Secretariat.
Funding
This work was supported by National Institute of Health/ National Heart, Lung, and Blood Institute grant number U54HL108460; National Institute of Health/National Human Genome Research Institute grant numbers R01HG007078, K99HG008175, and R00HG008175; and National Institute of Health/National Library of Medicine grant numbers T15LM011271, R00LM011392, and R21LM012060. Wellcome Trust grant number WT201535/Z/16/Z to PF and the European Molecular Biology Laboratory to PF and DL
Competing Interests
LO-M is editor-in-chief of JAMIA. The other co-authors have no competing interests to declare.
Contributors
Conception and design: JLR, FT, ZJ, DB, YZ, KC, SW, XJ, HT, XFW, and JPH. Algorithm implementation: JLR, FT, ZJ, and DB. Data analysis and interpretation, writing of manuscript, final approval of manuscript: JLR, FT, ZJ, DB, YZ, KC, DL, HS, DB, PF, SS, CB, SW, XJ, OML, HT, XFW, and JPH. JLR, FT, ZJ, DB share first co-authorship.
SUPPLEMENTARY MATERIAL
Supplementary material is available at Journal of the American Medical Informatics Association online.
References
- 1. The Global Alliance for Genomics and Health. A federated ecosystem for sharing genomic, clinical data. Science 2016;3526291:1278–80. [DOI] [PubMed] [Google Scholar]
- 2. Framework for Responsible Sharing of Genomic and Health-related Data. 2014.[Online]. https://genomicsandhealth.org/about-the-global-alliance/key-documents/framework-responsible-sharing-genomic-and-health- related-data. Accessed May 25, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. GA4GH privacy and security policy . 2015. . [Online]. https://genomicsandhealth.org/work-products-demonstration-projects/privacy-and-security-policy. Accessed May 25, 2016. [Google Scholar]
- 4. Terry SF, Shelton R, Biggers G, Baker D, Edwards K. The haystack is made of needles. Genetic Testing Mol Biomarkers 2013;173:175–7. [DOI] [PubMed] [Google Scholar]
- 5. Tennessen JA, Bigham AW, O’Connor TD et al. Evolution and functional impact of rare coding variation from deep sequencing of human exomes. Science 2012;3376090:64–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Homer N, Szelinger S, Redman M et al. Resolving individuals contributing trace amounts of DNA to highly complex mixtures using high-density SNP genotyping microarrays. PLoS Genet 2008;48: e1000167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sankararaman S, Obozinski G, Jordan MI, Halperin E. Genomic privacy and limits of individual detection in a pool. Nat Genetics 2009;419:965–7. [DOI] [PubMed] [Google Scholar]
- 8. El Emam K, Jonker E, Arbuckle L, Malin B. A systematic review of re-identification attacks on health data. PLoS One 2011;612:e28071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gymrek M, McGuire AL, Golan D, Halperin E, Erlich Y. Identifying personal genomes by surname inference. Science 2013;3396117: 321–4. [DOI] [PubMed] [Google Scholar]
- 10. Greenbaum D, Sboner A, Mu XJ, Gerstein M. Genomics and privacy: implications of the new reality of closed data for the field. PLoS Comput Biol. 2011;712:e1002278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Shringarpure SS, Bustamante CD. Privacy risks from genomic data-sharing beacons. Am J Hum Genet 2015;975:631–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. EPIC, https://epic.org/privacy/reidentification/. Accessed May 25, 2016. [Google Scholar]
- 13. The 1000 Genomes Project Consortium. An integrated map of genetic variation from 1,092 human genomes. Nature 2012;4917422:56–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. The 1000 Genomes Project Consortium. A global reference for human genetic variation. Nature 2015;5267571:68–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gibbs RA, Belmont JW, Hardenbol P et al. The international HapMap project. Nature 2003;4266968:789–96. [DOI] [PubMed] [Google Scholar]
- 16. Kendall MG. Rank Correlation Methods, New York: Hafner Publishing Co. 1955, 196 pp. [Google Scholar]
- 17. Lek M, Karczewski KJ, Minikel E V., et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature 2016;536:285–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Erlich Y, Narayanan A. Routes for breaching and protecting genetic privacy. Nat Rev Genet 2014;156:409–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Adam NR, Worthmann JC. Security-control methods for statistical databases: a comparative study. ACM Computing Surveys (CSUR). 1989;214:515–56. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


