SUMMARY
Two or more anions constrained in close proximity within a single pocket are found in a number of natural systems but a less common motif in artificial systems. This review summarizes work on anion receptors capable of stabilizing anionic dimers, trimers, tetramers and clusters in a well-defined fashion. These systems may provide insights into the fundamental chemistry of anion-anion interactions and provide a guide for understanding in greater detail a number of biological and environmental processes, as well as key tenants of relevance to supramolecular chemistry, extraction, transport, crystal engineering, and the like. The primary goal of this review is to provide a general introduction into multi-anion recognition chemistry for the benefit of supramolecular and non-supramolecular chemists alike.
eTOC
This review firstly summarizes artificial receptors capable of recognizing multiple anionic species, namely anionic dimers, trimers, tetramers and clusters, topics that are fundamentally important to biology, environment and material science.
INTRODUCTION
Anion recognition chemistry has emerged as one of the major disciplines within supramolecular chemistry due in part to the rich diversity and structural complexity of anions and their importance in biology, chemistry, energy, resource management and the environment. Although the problem of anion recognition is appreciated as inherently challenging, considerable advances have been made in areas as diverse as anion sensing, transport, extraction and catalysis.1–5 Most anion receptors reported to date were designed to bind a single anion. That is not surprising. Due to Coulomb’s law, and the intuitive appreciation that like charges repel and opposite ones attract (Figure 1A), at first blush it would appear difficult, if not impossible, to constrain two or more anions in close proximity. However, the concordant complexation and recognition of two or more anions is critical to many biological processes. For instance, ion channels, such as chloride anion channels, are characterized by the presence of more than one anion moving through the channels. Fully functioning ion channels are required for full health. Indeed, deficiencies in channel function are linked to a number of disorders involving muscles, kidneys, bones, and the brain. Included among these are myotonia congenita, dystrophia myotonica, cystic fibrosis, osteopetrosis, and epilepsy, which are linked to mutations in the genes encoding for Cl− channels.6 Although a number of synthetic anion channels have been reported in recent years,7 we are still far from being able to apply such artificial channels in clinical practice. Moreover, at this juncture, our understanding of how anions penetrate through the ion channels, in particular how one anion affects another within the same channel and how the putative interactions between different anions affect the ion channel activity, is far from complete. This lack of understanding of how two (or more) anions interact extends to a number of catalytic processes involving biologically relevant anions. For example, the catalytic synthesis of ATP by F1F0-ATP synthase requires the simultaneous binding of two anionic substrates, ADP and phosphate (Pi), within the catalytic sites before formation of ADP-O-P bond can occur8. Currently, we have little idea as to how these two anionic substrates affect one another or how their interactions impact the overall catalytic process. Other transformations involving multiply charged anions, including those associated with the synthesis and hydrolysis of RNA and DNA, also require two or more anion species to be constrained in close proximity within a catalytic domain.9–11 Simple synthetic systems that allow for the co-complexation of multiple anions may provide key insights into how nature recognizes and manipulates co-bound anions.
Figure 1. Schematic representation of Coulomb’s Law and possible approaches to capturing two or more like-charged anions by means of a synthetic receptor.
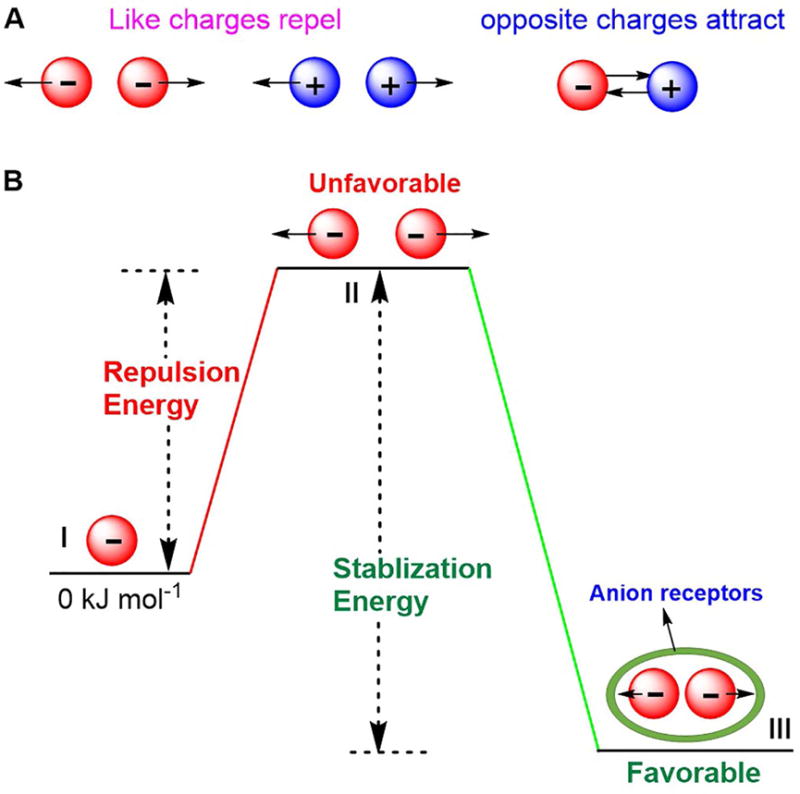
(A) Coulomb’s Law: Repulsion and attraction.
(B) Anions constrained in close proximity stabilized by noncovalent interactions.
All systems, natural and synthetic, wherein anion-anion interactions are established must overcome the like-charge repulsion energies associated with these interactions. While challenging, the inherent Coulombic repulsion between anions can be partially offset by biological or synthetic receptor-derived the stabilization energies (Figure 1B). In the context of supramolecular systems, a variety of noncovalent interactions, including hydrogen bonding, electrostatic interactions, anion-π binding effects, can be used to provide the host-guest stabilization needed to bind concurrently two or more anionic guests. While seemingly straightforward, the design of such receptors is not necessarily simple. The introduction of strong noncovalent interactions, multiple cooperative noncovalent interactions, proper dimensionality of the receptors, as well as reasonable cavity size, are important aspects of the molecular design. In fact, through appropriate design it has proved possible to create synthetic receptors wherein two or more anions are held closer to one another than might be intuitively expected. In this review, we summarize efforts to stabilize anion dimers, trimers, tetramers and clusters within artificial hosts. Receptors that can be used to stabilize two or more anions concurrently are classified as acyclic anion receptors, monocyclic anion receptors, bicyclic anion receptors and tricyclic anion receptors.
ACYCLIC ANION RECEPTORS
Receptors that are capable of providing cleft-like cavities for ion recognition have played a central role in host-guest chemistry. In the early years of anion chemistry, great efforts were devoted to exploring the structure-property relationships between various hosts and guests. In the context of this work, it was found that two or more anions could be entrapped by a single appropriately designed acyclic receptor or by assemblies of separate anion receptors.
In 2009, Arunachalam et al.12 reported an acyclic arene-based tripodal amide receptor 1 designed to recognize and complex the fluoride anion. In this case, a dimeric capsule was formed by the two bowl shaped cavities of 1 in the presence of tetrabutylammonium (TBA) fluoride salt. A hydrated fluoride dimer, namely [F2(H2O)6]2−, was observed in the single crystal X-ray structure (Figure 2B). Apparently, the N–H⋯F (1.95 Å) and N⋯F (2.76 Å) distances are quite short, indicating strong anion binding, while the O–H⋯F distances vary from 1.77 Å to 1.97 Å. The distance between the two fluorides was found to be 3.94 Å. This fluoride dimer was bridged by six water molecules and found to exist with an extensive hydrogen binding network. Presumably, the sum total of these stabilizing interactions serves to overcome the inherent anion-anion repulsion. As opposed to what was seen in the case of the fluoride anion, infinite zipper-like aggregates were found to form when receptor 1 was exposed to the chloride, nitrate and acetate anions, as inferred from single crystal X-ray analyses.
Figure 2. Acyclic arene-based tripodal anion receptors and their dimeric anion complexes.
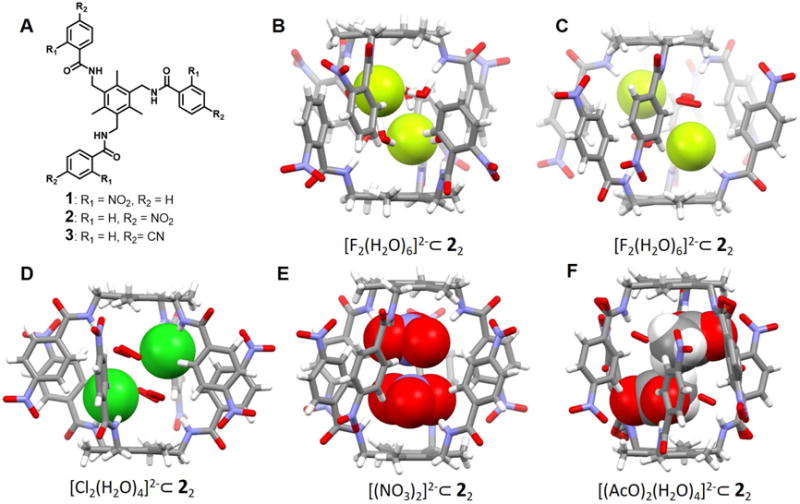
(A) Molecular structures of receptors 1–3.
(B) Single crystal X-ray structure of the complex [F2(H2O)6]2−⊂ 12.
(C) Single crystal X-ray structure of the complex [F2(H2O)6]2−⊂ 22.
(D) Single crystal X-ray structure of the complex Cl2(H2O)4]2−⊂ 22.
(E) Single crystal X-ray structure of the complex [(NO3)2]2−⊂ 22.
(F) Single crystal X-ray structure of the complex [(AcO)2(H2O)4]2−⊂ 22.
All anionic substrates encapsulated within the cavities of the dimeric capsules are shown in space-filling form. Color code: C = gray, N = blue, O = red, F = chartreuse yellow, Cl = green. The counter cations and solvent molecules outside the cavities are not shown.
Receptor 2 is a positional isomer of the triamide 1, wherein the aryl nitro group is in the para, as opposed to ortho, position. In contrast to what was seen in the case of 1, receptor 2 was found to support the formation of staggered dimeric capsules in the solid state upon exposure to the fluoride, chloride, nitrate and acetate anions.13 In the case of the fluoride complex (Figure 2C), six water molecules act as bridges between the fluoride anions and the receptor. The resulting N-H⋯F− and N⋯F− distances were reported to be 1.95 Å and 2.81 Å, respectively, while the O⋯F− and O⋯N distances range from 2.55 Å and 2.90 Å to 2.78 Å and 3.05 Å, respectively. The F−⋯F− distance was found to be 5.16 Å. This larger distance compared to the corresponding structure involving receptor 1, was ascribed to the larger cavity produced via the self-assembly of 2. The chloride dimer complex is characterized by a [Cl2(H2O)4]2− cluster encapsulated within the receptor-derived pocket (Figure 2D). A N⋯Cl− distance of 3.29 Å was observed, while the Cl−⋯Cl− distance was found to be 4.62 Å. The bound chloride anions and associated water molecules are stabilized by multiple hydrogen-bonding interactions. Receptor 2 was also able to capture a nitrate anion dimer and an acetate anion dimer within a capsule-derived binding pocket (Figure 2E and 2F). No co-bound water molecules are observed in the case of the NO3− complex. However, hydrogen-bonding interactions with the -NH and aryl -CH protons of 2 are observed. The N(H)⋯O-NO2− distance range from 2.98 Å to 3.30 Å while the C(H) ⋯O-NO2− distances range between 3.28 Å and 3.45 Å. The two NO3− anions are symmetrically encapsulated inside the capsule in a staggered planar orientation relative to one another. The short nitrate-nitrate contact (N⋯N distance = 3.56 Å) was considered reflective of a strong anion-anion interaction.14–16 Similar structural features were seen in the case of an encapsulated NO3− dimer stabilized within a dimeric capsular assembly produced by the para-cyanophenyl terminated tripodal amide receptor 3, as reported by Arunachalam et al.17 In the case of the acetate complex stabilized by receptor 2, two AcO− anions along with four water molecules are encapsulated inside the dimeric capsule via strong N-H⋯O and C-H⋯O interactions involving the host framework. In addition, hydrogen-bonding interactions involving the encapsulated water molecules are seen.
In 2015, Pandurangan et al.18 reported the design and synthesis of three tripodal receptors, 4–6, based upon an N-methyl-1, 3, 5-benzenetricarboxamide platform bearing three appended aryl urea arms. The complexation and self-assembly of these three receptors with various anions (SO42−, H2PO4−, AcO−, and Cl−) was studied. All three receptors were found to bind the SO42− and H2PO4− ions. Solid-state structural analyses of the complexes formed between receptors 4–6 and the SO42− anion revealed the encapsulation of a single sulfate anion within the capsule. In contrast, a single crystal structure of 4 with H2PO4− revealed the presence of an anionic dimer encapsulated within the cavity of what can be considered as being a pseudo cage (Figure 3). These two dihydrogenphosphate anions were found to be connected to one another via two-fold P-O-H⋯O-P hydrogen bonding interactions, as well as via a bridging water molecule. The closest P⋯P distance is ca. 4.01 Å.
Figure 3. Acyclic tripodal anion receptors and their anionic complexes.
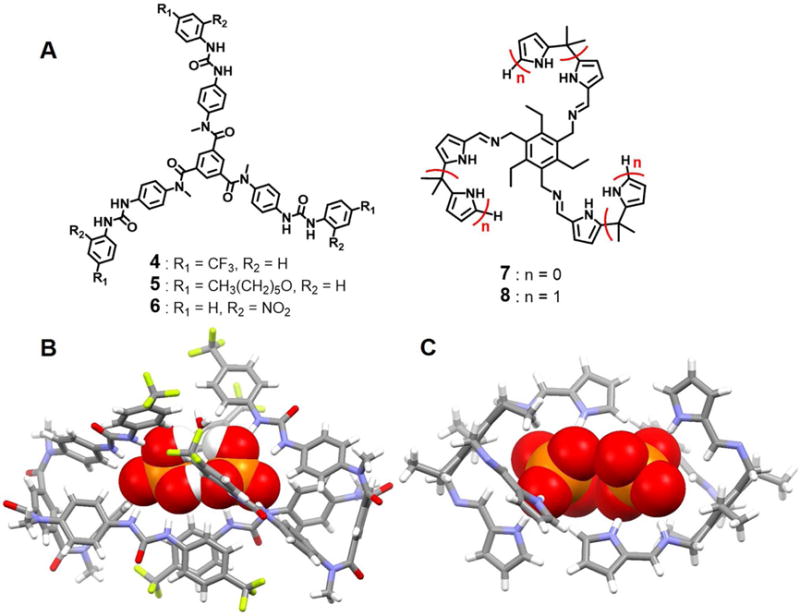
(A) Molecular structures of receptors 4–8.
(B) Single crystal X-ray structure of the complex [(H2PO4)2(H2O)]2− ⊂ 42.
(C) Single crystal X-ray structure of the complex [(H2PO4)2]2− ⊂ 72.
All anions encapsulated in the cavity of the dimeric capsules are shown in space-filling form. Color code: C = gray, N = blue, O = red, and P = yellow. The counter cations and solvent molecules outside the cavities are not shown.
In 2012, Bill et al.19 prepared two new tripodal receptors (7 and 8) that consist of pyrrole- and dipyrromethane-functionalized derivatives of a sterically geared precursor, 1, 3, 5-tris(aminomethyl)-2, 4, 6-triethylbenzene. These two receptors show selective recognition of tetrahedral oxoanions, including HSO4−, H2PO4− and HP2O73− in acetonitrile, as inferred from isothermal titration calorimetry measurements. Large binding constants were seen for H2PO4− in CH3CN in the case of receptors 7 and 8 (Ka = 140 000 and 6 500 000 M−1 respectively). In the solid state, two H2PO4− anions were found to be captured by receptor 7 in a 2 : 2 (host/guest) fashion within a pseudo macrocycle built up from two of the receptor subunits. Close anion-anion contacts (P⋯P distance = ca. 4.20 Å) were inferred from the crystal structure (Figure 3).
In 2009, Arunachalam et al.20 reported a benzimidazole-type receptor 9 which is prone to protonation. Structural analyses revealed that after protonation by HNO3, all three benimidazole substituents are oriented in the same direction and served to form a bowl-like cleft (Figure 4). This same structural analysis revealed the presence of six nitrate anions hydrogen-bonding to the benzimidazolium moieties within what is an overall dimeric capsule-type complex, [H39(NO3)3(H2O)]2, which contains two bridging water molecules. Presumably interactions between the bound nitrate anions and water molecules (O⋯O distances in the range of 2.78–3.36 Å) help overcome the repulsion between the co-bound anions. The pseudo capsule was not stable in the presence of chloride anions. In fact, a non-capsule, trianionic complex, [H39(Cl)3(HCl)(H2O)3nH2O], is formed when HCl is used in lieu of HNO3 in the original protonation step. The formal chloride anion trimer is held together via N-H⋯Cl− and C-H⋯Cl− interactions, as inferred from 1H NMR spectroscopic studies, even though only two of the three benzimidazole arms are oriented on the same side of the central phenyl plane. The pairwise separations between the two chloride anions are 4.79 Å, 5.13 Å and 6.35 Å, respectively.
Figure 4. The arene-based tripodal receptor 9 and its anionic complexes.
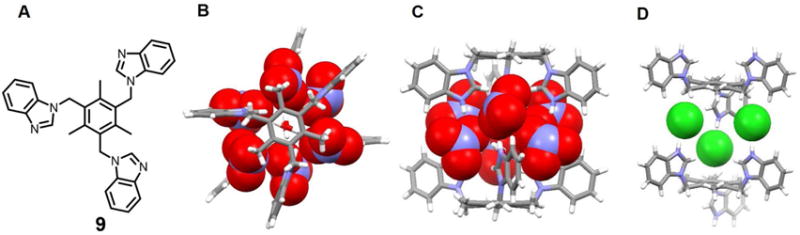
(A) Molecular structure of receptor 9.
(B) Top view of the crystal structure of the complex [(NO3)6(H2O)2]6−⊂ [H39]3+2.
(C) Front view of the crystal structure of the complex [(NO3)6(H2O)2]6−⊂ [H39]3+2.
(D) Single crystal structure of the complex [Cl3]3−⊂ [H39]3+2.
All anions encapsulated in the cavity of the dimeric capsules are shown in space-filling form. Color code: C = gray, N = blue, O = red, and Cl = green. The counter anions and solvent molecules outside the cavities are omitted for clarity.
Basu et al.21 reported a conformationally flexible C3v-symmetric N-bridged tripodal amide receptor 10. It stabilizes halide dimers [X2(H2O)2]2− (X = Cl−/Br−) within what can be viewed as being a self-assemble dimeric capsule (Figure 5). The bound halide dimers are connected to water molecules and presumably stabilized via hydrogen bonding networks. A single crystal X-ray structural analysis of the chloride complex of the protonated form of 10 revealed that all the three arms of the receptor are oriented in one direction so as to create a binding pocket appropriate for chloride anion dimer encapsulation. The [Cl2(H2O)2]2− cluster is completely entrapped by two protonated receptor units and co-stabilized by four N-H⋯Cl−, two N-H⋯OH2, multiple C-H⋯Cl− and C-H⋯OH2 hydrogen bonds. The Cl⋯Cl distance was found to be ca. 4.62 Å. The structure of an essentially isostructural bromide dimer complex (Figure 5C) was also solved. Here, the Br−⋯Br− distance was estimated to be 4.79 Å. In addition, although the host-guest stoichiometry was not mentioned by the authors, 2D-NOESY NMR spectral studies provided evidence for receptor-hydrated halide (Cl− and Br−) interactions in DMSO-d6.
Figure 5. Acyclic N-bridged tripodal anion receptor 10 and its anionic dimer complexes.
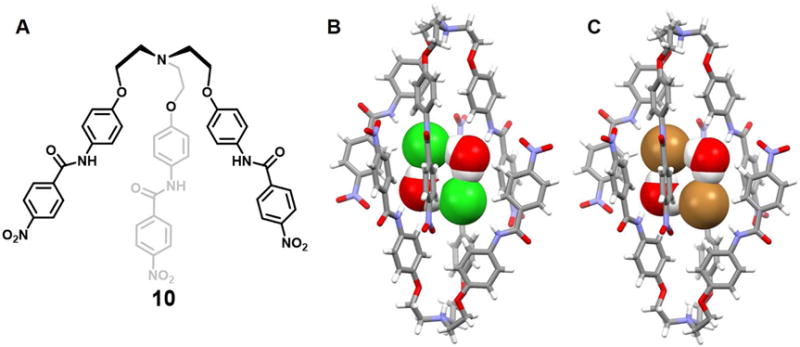
(A) Molecular structure of receptor 10.
(B) Single crystal X-ray structure of the complex [Cl2(H2O)2]2−⊂ [H10]+2.
(C) Single crystal X-ray structure of the complex [Br2(H2O)2]2−⊂ [H10]+2.
All anions and bridging water molecules encapsulated in the cavity of the dimeric capsules are shown in space-filling form. Color code: C = gray, N = blue, O = red, Cl = green and Br = brown. The counter cations and solvent molecules outside the cavities are omitted for clarity.
Jose et al.22 published a tren-based tris(urea) receptor molecule 11 containing a nitrobenzene as a putative chromophoric unit and reported that this system interacts preferentially with the sulfate and phosphate anions. Receptor 11 shows good shape complementarity for these tetrahedral ions and was found to be capable of encapsulating two dianionic sulfate anions, along with three water molecules, within what are formally dimeric capsules formed from two molecules of 11 oriented in a face-to-face fashion (Figure 6C). These water molecules are bridged to the sulfate anions via multiple hydrogen bonding, resulting in an interesting supramolecular architecture - rugby-ball-shaped sulfate-water-sulfate adduct. The length of the dimeric cage and that of sulfate dimer were found to be 15.78 Å and 5.92 Å (S⋯S distance), respectively. It represents a close contact of two repulsive dianionic sulfates, which, in turn, were stabilized by anion receptors via energetically favorable hydrogen binding interactions.
Figure 6. Acyclic tren-based tris(urea) anion receptors and their anionic complexes.
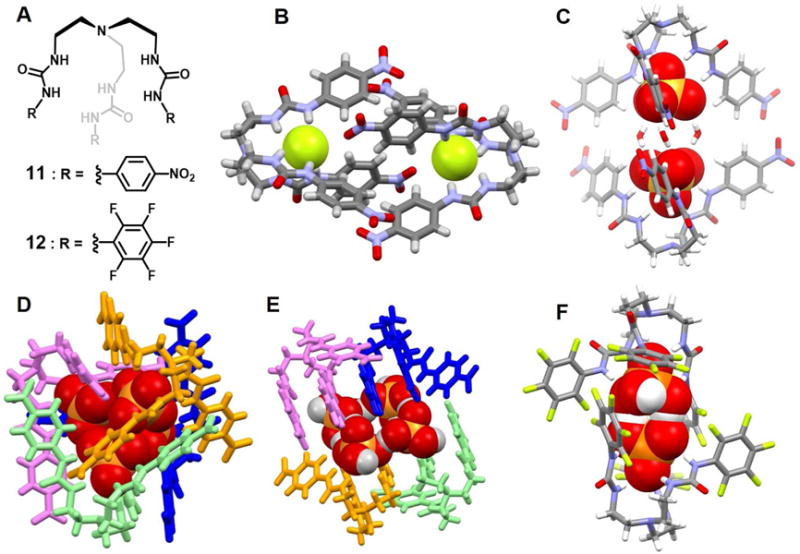
(A) Molecular structures of receptors 11 and 12.
(B) Single crystal X-ray structure of the complex [F2]2−⊂ 112.
(C) Single crystal X-ray structure of the complex [(SO4)2(H2O)3]4−⊂ 112.
(D) Single crystal X-ray structure of the complex [(H2PO4)2(HPO4)2]6−⊂ 114.
(E) Single crystal X-ray structure of the complex [(H2PO4)2(H3PO4)2]2−⊂ 114.
(F) Single crystal X-ray structure of the complex [(H2PO4)2]2−⊂ 122.
All anions encapsulated in the cavities are shown in space-filling form. Color code: C = gray, N = blue, O = red, F = chartreuse yellow and S, P = yellow. For D and E, various colors (pink, blue or aqua) are used to represent different independent ligands. The counter cations and other solvent molecules outside the cavities are omitted for clarity.
In 2015, the same group23 also reported a unique dimeric capsular derived from 11 that contained two encapsulated F− anions (Figure 6B). The F− anions within this 2 : 2 complex are constrained within different parts of the capsule and stabilized via six hydrogen bonds, respectively. The average N-H⋯F− distance is 2.82 Å and the distance between the two fluoride ions is 9.56 Å. Presumed aromatic-CH⋯−O-nitro and nitro-O−⋯π interactions within the cavity also help to stabilize the complex.
In the presence of excess TBAH2PO4, receptor 11 was found to self-assemble into a tetrameric cluster (Figure 6D). This tetrameric cluster contains two pair of H2PO4− • HPO42− pseudo dimers that interact with one another. The distances between the two phosphorus atoms vary between 4.31 and 4.72 Å. The distance between the donor and acceptor were found to range between 2.48 Å and 2.89 Å, a finding that was taken as evidence for a strong O-H⋯O interaction between the H2PO4− and HPO42− anions. Presumably, the tetrameric nature of the ensemble formed from 11 allows the cluster of four phosphate anions to be stabilized through multiple hydrogen bonds (up to 24 are readily observable), as well as providing size and shape complementarity. Average donor-acceptor distances of 2.94 Å (H2PO4− • HPO42−)2 were seen within the anionic cluster.
In the case of H3PO4, the self-assembled form of receptor 11 was found to bind a tetrameric anion-acid (H2PO4− • H3PO4)2 cluster in the form of a sandwich structure stabilized via 24 hydrogen bonds (Figure 6E). In the solid state, a H2PO4− anion is seen to interact with two H3PO4 molecules, which in turn interact with another H2PO4− anion. The net result is a rectangular arrangement. The distances between the phosphorus atoms within the two H2PO4− and H3PO4 pair are 4.09 Å and 4.78 Å, respectively. The O-H⋯O distances corresponding to the separation between the H2PO4− anion and the neutral H3PO4 moiety were found to range from 2.50 to 2.62 Å. Each H3PO4 subunit was found hydrogen bonded to a water molecule and each H2PO4− was found to interact with a DMSO molecule.
Lakshminarayanan et al.24 synthesized a new pentafluorophenyl substituted tripodal urea-based receptor 12. This system was found to form a cage-type capsule in the solid state and to encapsulate a ([H2PO4]−)2 dimer without any bridge molecules. The length of the dimerized capsule is 13.79 Å as inferred from single crystal structural analyses of the 2 : 2 host-guest capsule complex (Figure 6F). In the structure, two C3v symmetric units of 12 are found to encapsulate a dimer of [H2PO4]− via sixteen hydrogen bonding and two anion-π interactions. Within this anion dimer, the closest P⋯P distance is 4.03 Å. Proton NMR spectroscopic titration data, from measurements carried out in DMSO-d6, gave the best fit for an empirical 1 : 1 host-guest stoichiometry. A similar stoichiometric inference was drawn from a Job plot, lending credence to the conclusion that the trapped inorganic phosphate dimer seen in the solid state also exists in DMSO-d6 solution.
Huang et al.25 have recently reported a ferrocenyl-functionalized tripodal hexaurea receptor 13 with a meta-phenylene bridge linking the bis-urea arms. This system was designed to allow anion recognition and electrochemical signaling. Presumably, as the result of its trigonal bipyramidal structure, receptor 13 was found to encapsulate two SO42− anions within the large cavity formed by the three urea-containing arms. The association constants corresponding to the binding of the first and second sulfate anion, were estimated to be log K >4.0 and >2.0, respectively, as obtained from 1H NMR spectroscopic titration experiments carried out in DMSO-d6. It was suggested that the first sulfate ion binds in the “inner” cavity (highlighted in red in Figure 7), whereas the second sulfate ion is complexed within the “outer” cleft (highlighted in blue) produced by the trigonal-bipyrimidal-shaped receptor. Due to the lack of direct structural evidence for a 1 : 2 host/guest stoichiometry in the solid state, the authors carried out molecular modeling studies. On this latter basis, they were able to confirm that the proposed 1 : 2 (host/guest) sulfate anion complex is energetically stable in the case of receptor 13.
Figure 7.
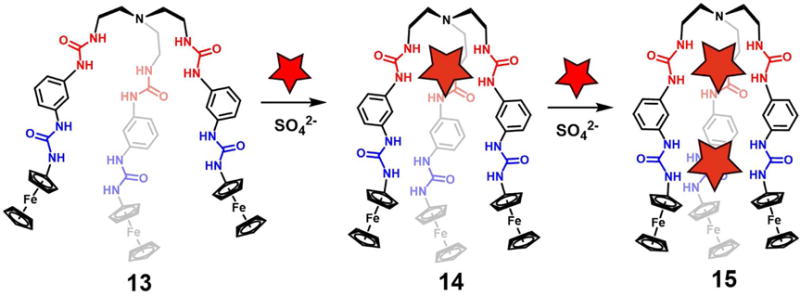
Stepwise encapsulation of sulfate ions by ferrocenyl-functionalized tripodal hexaurea receptor 13
In 2006, Yin et al.26 prepared the tripodal tri(2-pyrrolylmethyl)amine receptor 16. This system is formally C3-symmetric and adopts a cone-like conformation upon anion complexation. A single crystal of the TBAH2PO4 complex was analyzed by X-ray diffraction. The resulting structure revealed an anionic H2PO4− tetramer entrapped within the large cavity produced via the self-assembly of four molecules of 16 (Figure 8B). Each H2PO4− anion was found to be hydrogen-bonded to three pyrrole-NH protons in the solid state. The N-H⋯O distances were found to be 2.84, 2.86 and 2.91 Å, respectively. In addition, each H2PO4− was found to donate and accept two O-H⋯O bonds from the other two bound H2PO4− anions, resulting in formation of a tetrameric anion cluster. The average distance between each pair of phosphorus atoms was 4.25 Å.
Figure 8. Tripodal receptors and their anionic complexes.
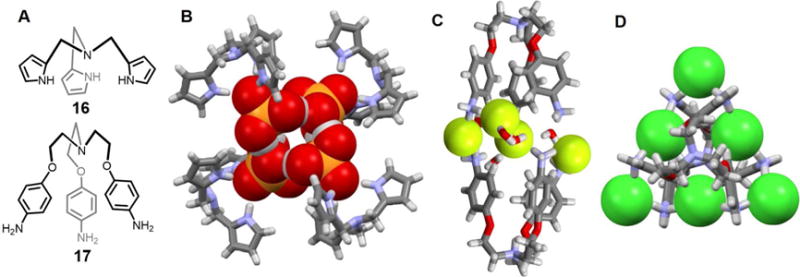
(A) Molecular structures of receptors 16 and 17.
(B) Single crystal structure of the complex [(H2PO4)4]4−⊂ 164.
(C) Single crystal structure of the complex [F4(H2O)5]4−⊂ [H417]4+2.
(D) Single crystal structure of the complex [Cl6]6−⊂ [H417]4+2.
All anions encapsulated in the cavities are shown in space-filling form. Color code: C = gray, N = blue, O = red, F = chartreuse yellow, P = yellow, and Cl = green. Most counter ions and solvent molecules outside the cavities are omitted for clarity.
The polyammonium-based tripodal receptor 17 was reported by Hoque et al.27 Solid-state structural analyses revealed formation of a bimolecular pseudo capsule-like complex containing four fluoride ions and five water molecules (Figure 8C). In the solid state, the capsule appears held together by hydrogen bonding interactions involving the ammonium groups and the bound F− and H2O guests. This results in formation of a [F−-H2O-NH3+] “backbone”, which serves to glue together the two halves of the capsule. The fluoride-fluoride distances range between 4.16 and 4.40 Å. These same researchers also found that receptor 17 stabilizes a similar capsule containing four bound chloride ions, albeit in the absence of co-bound water molecules (Figure 8D). As in the fluoride anion structure, the two cationic tripodal units are held together in the solid state via N-H⋯Cl− hydrogen bonding interactions involving the ammonium groups. The distances between the co-bound chloride anions range from 4.89 Å to 5.05 Å.
Squaramide has been widely employed in the design of new anion receptors. As a general rule squaramide derivatives are characterized by higher anion affinities than their urea counterparts since squaramides are correspondingly stronger hydrogen bond donors. In addition, squaramides typically adopt conformationally rigid square-shaped structures upon anion binding.28–33 The squaramide motif appears in a sulfate-containing complex, reported by Jin et al,34 wherein more than a single anion is co-bound within a receptor-derived ensemble. The system in question involves the tripodal anion receptor 18, which was studied for its ability to recognize various inorganic anions, e.g., Cl−, AcO−, H2PO4− and SO42−. In the presence of a sulfate anion source, receptor 18 was found to form a dimeric pseudo-capsule ensemble wherein two SO42− anions are bound concurrently within the receptor-derived cleft in a 2 : 2 (host/guest) stoichiometry in DMSO-d6 (Figure 9). The three arms of 18 were found to exist in an open relatively non-symmetric conformation. Two of the three squaramide moieties of each tripodal receptor and one squaramide unit of the other tripodal receptor combine to interact with one of the two bound sulfate anions through N-H⋯O hydrogen bonding interactions. Corresponding interactions were found to stabilize the other bound sulfate anion. DOSY NMR spectral analyses, carried out in DMSO-d6, led to the suggestion that a 1 : 1 receptor - sulfate binding stoichiometry dominates in solution rather than a dimeric structure. Both Job plots and HR-ESI-MS experiments proved consistent with the formation of a 1 : 1 SO42− complex in in DMSO-d6 solution.
Figure 9. Acyclic tris-(squaramide) and linear meta-phenylenediamine bis-urea receptors and their dimeric anion complexes.
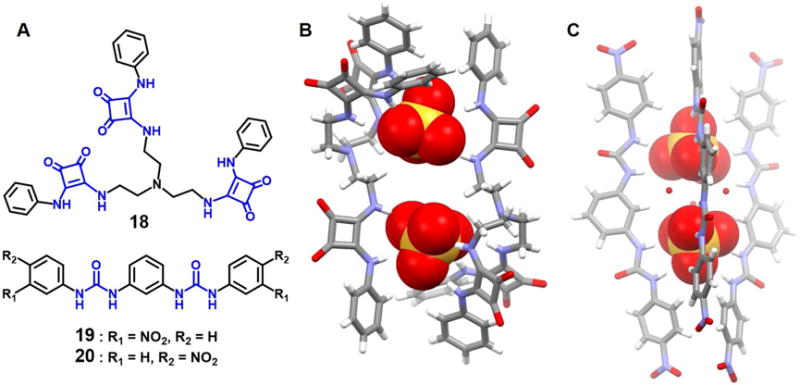
(A) Molecular structures of receptors 18–20.
(B) Single crystal X-ray structure of the complex [(SO4)2]4−⊂ 182.
(C) Single crystal X-ray structure of the complex [(SO4)2]4−⊂ 202.
All anions encapsulated in the cavities are shown in space-filling form. For (B), two disordered SO42− anions were found within the cavity. However, only one set of data is shown for clarity. Color code: C = gray, N = blue, O = red, and S = yellow. The counter cations and solvent molecules outside the cavities are not shown for clarity.
Pores formed via the assembly of two or more functional building units are commonly used to capture specific anions.35–37 Example include a fluoride anion dimer and a sulfate anion dimer stabilized by the isomeric meta-phenylene based bis-urea receptors 19 (meta-nitro) and 20 (para-nitro), respectively, as reported by Manna et al.38 Although both receptors favor a linear conformation, they proved capable of interacting with halide anions as well as planar and tetrahedral oxyanions. In the case of receptor 19, a single crystal X-ray crystallographic analysis revealed that the four urea arms derived from four different molecules of 19 assemble into a barrel-like structure that acts to entrap a fluoride dimer bridged by two water molecules. The F−⋯F− distance was found to be 3.90 Å.
A corresponding solid state structural analysis in the case of complex 20 and bound sulfate anions revealed that three 20 molecules orient in a semicircular fashion along the edges of a barrel-like structure. Relative to the barrel core, the three upper urea subunits combine to encapsulate one sulfate anion while the lower three urea units entrap a sulfate anion; in both cases numerous N-H⋯O hydrogen bonding interactions are seen (Figure 9). Three water molecules function to bridge the two face-to-face oriented sulfate anions resulting in what is oval-shaped, or rugby-ball-like, arrangement with a S⋯S separation of 6.01 Å.
Both the fluoride and sulfate anion complexes of 19 and 20, respectively, were also characterized using 1H NMR spectroscopy in DMSO-d6. Careful examination of the resulting data led to the conclusion that a mixed equilibrium between species with 1 : 1 and 1 : 2 host-guest stoichiometry exists for both complexes in solution.
In 2017, Manna et al.39 also reported the meta-phenylene diamine-based, halomethyl substituted isomeric dipodal bisurea receptors 21 and 22, which were found to bind dianionic carbonate and sulfate in a 4 : 2 (host/guest) mode (Figure 10). Subjecting a mixed DMF/DMSO solution of receptor 21 and excess n-tetrabutylammonium hydroxide (TBAOH) to slow evaporation resulted in the formation of colorless needle-like crystals. A single crystal X-ray diffraction analysis revealed host 21 in the form of an n-tetrabutylammonium carbonate salt, which presumably resulted from the fixation of atmospheric CO2 in the presence of TBAOH. In this complex, two dianionic carbonate anions are entrapped in the form of CO32−−(H2O)2−CO32− clusters with a long, straight tetrameric pillar-like environment produced via the self-assemble of 21. The upper four urea groups within this supramolecular “barrel” entrap one CO32− anion by means of nine N-H⋯O hydrogen bonding interactions and one ortho-aryl C-H⋯O hydrogen bond. In contrast, the four lower urea subunits within the barrel act to trap the other CO32− anion in a reverse triangle fashion via nine N-H⋯O hydrogen bonds and three ortho-aryl C-H⋯O hydrogen bonds. Two water molecules are observed to act as the bridge between the two co-bound anions. The C⋯C distance between the anions trapped within each barrel was 6.22 Å. This relatively short separation lead to the suggestion that the expected repulsive interactions between the two CO32− anions is offset by a total of 31 N-H⋯O and C-H⋯O hydrogen bonding interactions. Interestingly, the analogous 4-bromo-3-methyl difunctionalized isomeric receptor 22 was unable to stabilize a similar structural motif when crystallization studies were carried out in the presence of TBAOH under identical crystallization conditions.
Figure 10. Acyclic bis-urea receptors and their anionic dimer complexes.
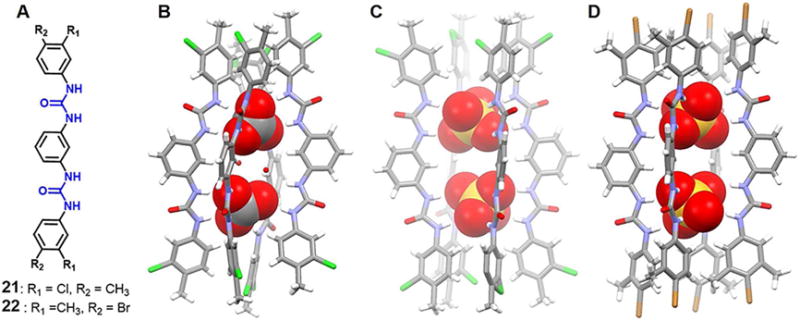
(A) Molecular structures of receptors 21 and 22.
(B) Single crystal X-ray structure of the complex [(CO3)2(H2O)2]4− ⊂ 214.
(C) Single crystal X-ray structure of the complex [(SO4)2]4− ⊂ 214.
(D) Single crystal X-ray structure of the complex [(SO4)2]4− ⊂ 224.
All anions encapsulated within the cavity are shown in space-filling form. Color code: C = gray, N = blue, Cl = green, S = yellow, and Br = brown. The counter cations and solvent molecules outside the cavities are omitted for clarity.
Water-free sulfate dimers were found to be encapsulated in the case of both 21 and 22 when crystallization was carried out in the presence of excess n-TBAHSO4. Structural studies revealed that two pair of symmetry-independent receptor molecules combine to create the edges of a tetrameric linear capsule and entrap fully the two symmetry-equivalent deprotonated hydrogen sulfate anionic guests via numerous hydrogen bonding interactions. The S⋯S distances of adjacent SO42− anions within the barrel-like cavities were found to be 6.05 Å and 6.30 Å for 21 and 22, respectively. The difference in these metric [parameters is attributed to the presence of a stronger electron-withdrawing group (Cl) on 21 compared to 22 (Br).
Manna and coworkers40 also synthesized the two disubstituted bisurea-based receptors 23 and 24 (Figure 11). It was found that the ortho-phenylenediamine based chloromethyl disubstituted receptor 23 can form a trimeric capsular assembly that encapsulates an unusual triangular [Cl3-DMSO]3− cluster. Specifically, in the tris-chelate (Cl)3-DMSO complex, two chloride anions are bound via four N-H⋯Cl− interactions, one TBA-C-H⋯Cl−, and one DMSO-C-H⋯Cl− interaction. The third chloride anion is stabilized via four N-H⋯Cl− interactions. In addition, apparent anion-π interactions, halogen bonding Cl⋯Cl−, as well as several intermolecular C-H⋯Cl− hydrogen bonding also appear to contribute to the stability of this unusual assembly. The average distance between the chloride anions is 6.82 Å. All N-H⋯Cl− distances are less than 2.60 Å, as would be expected for a strongly bound anion complex. Receptor 23 was also found to form a 4 : 2 (host/guest) complex that involves encapsulation of two dianionic carbonate anions via 16 N-H⋯O interactions. In contrast, receptor 24, with two bromo groups on the para-position, forms a 2 : 2 complex. In this latter complex, the two symmetry-identical chloride anions are stabilized via four strong N-H⋯Cl− interactions, two of which are from the urea group on one receptor and the other two are derived from another receptor. The distance between the two co-bound chloride anions is 5.75 Å.
Figure 11. Acyclic bis-urea receptors and their anionic dimer complexes.
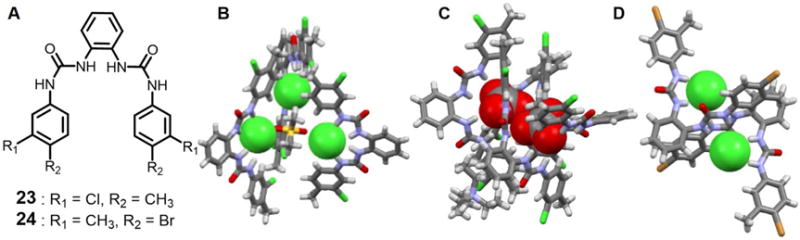
(A) Molecular structures of receptors 23 and 24.
(B) Single crystal X-ray structure of the complex [Cl3 (DMSO)]3− ⊂ 233.
(C) Single crystal X-ray structure of the complex [(CO3)2]4− ⊂ 234.
(D) Single crystal X-ray structure of the complex [Cl2]2− ⊂ 242.
All anions encapsulated within the cavity are shown in space-filling form. Color code: C = gray, N = blue, Cl = green, and Br = brown. The counter cations and solvent molecules outside the cavities are omitted for clarity.
In 2013, Rajbanshi et al.41 reported the co-crystallization of (H2PO4−)n (n = 4, 6) clusters stabilized by multiple hydrogen bonds derived from the polyurea receptors 25 and 26. Single crystal X-ray diffraction analysis revealed that both 25 and 26 were able to capture H2PO4− hexamers, which are sandwiched between two host molecules as shown in Figures 12B and 12C. Four of the six anions exist in the form of a linear chain from which the other two anions branch out. Hydrogen bonds stabilize the dihydrogenphosphate interactions. The intermolecular O⋯O contact distances between dihydrogenphosphate anions are in the range of 2.54–2.68 Å. The four terminal dihydrogenphosphate anions are hydrogen-bonded to two urea moieties present in the host molecules and are characterized by NH⋯O contacts between 1.93–2.34 Å (25) and 1.94–2.30 Å (26). In both cases, 16 urea hydrogen bonds are present in the complex while four more additional water molecules are found hydrogen-bonded to the cluster. Interestingly, co-crystallization of TBA dihydrogenphosphate with 25 also yielded a centrosymmetric H2PO4− tetrameric cluster which displays a linear topology (Figure 12D). Analogous to what was seen in the hexamer complexes, the terminal H2PO4− anions in this latter tetramer cluster are hydrogen-bonded to urea groups with NH⋯O distances between 1.89–2.22 Å. The anions are again connected by hydrogen bonds. The intermolecular O⋯O distances range between 2.57 Å and 2.67 Å.
Figure 12. Acyclic bis-urea receptors and their anionic dimer complexes.
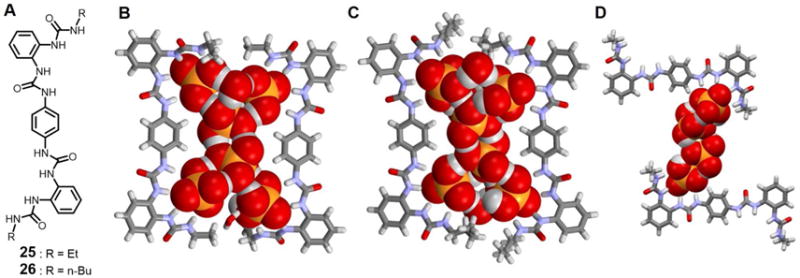
(A) Molecular structures of receptors 25 and 26.
(B) Single crystal X-ray structure of the complex [(H2PO4)6]6− ⊂ 252.
(C) Single crystal X-ray structure of the complex [(H2PO4)6]6− ⊂ 262.
(D) Single crystal X-ray structure of the complex [(H2PO4)4]4− ⊂ 252.
All anions encapsulated within the cavity are shown in space-filling form. Color code: C = gray, N = blue, O = red, and P = yellow. The counter cations and other molecules outside the cavities are omitted for clarity.
Blažek et al.42 prepared two adamantane bisurea derivatives 27 and 28, both of which supported the formation of 4 : 4 complexes with H2PO4− in the solid state (Figure 13). Structural analyses revealed that incorporation of the adamantane unit preorganizes the receptor in a tweezer-like conformation that allows formation of hydrogen bonding networks involving the bound H2PO4− anions. Compared with 27, incorporation of the methylene spacers between the adamantane and the urea moieties (28) appears to increase the stability of the complexes as inferred from UV-vis spectroscopic titration studies carried out in CH3CN (log(K11/M−1) = 5.1 ± 0.2, log(K12/M−2) = 9.1 ± 0.1 for 27 and log(K11/M−1) = 5.8 ± 0.3, log(K12/M−2) = 10.7 ± 0.3 for 28). Analysis of the related complexes 27 and 28 by X-ray crystallography revealed that in both cases, four H2PO4− anions are hydrogen-bonded to each other and co-encapsulated within a large cavity formed via the self-assembly of four receptor molecules. Interestingly, the orientation of complex of 27 does not appear to favor π-π interactions between the two naphthyl subunits, whereas in the complex of 28, the metric parameters were consistent with such an interaction. On the basis of UV-vis spectroscopic studies carried out in CH3CN or DMSO, the authors proposed that both receptors 27 and 28 form 1 : 2 (host/guest) complexes in solution instead of the 1 : 1 (or 4 : 4) complexes seen in the solid state.
Figure 13. Acyclic bisurea-based receptors and their anionic complexes.
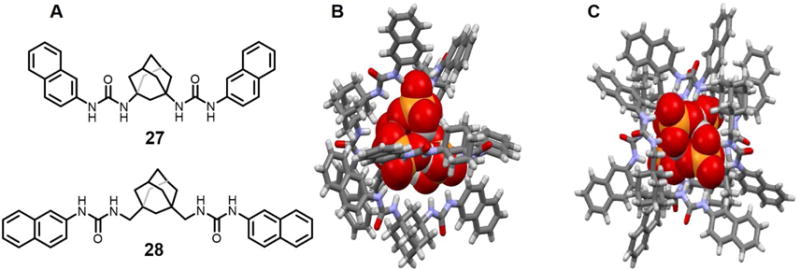
(A) Molecular structures of receptors 27 and 28.
(B) Single crystal X-ray structure of the complex [(H2PO4)4]4− ⊂ 274.
(C) Single crystal X-ray structure of the complex [(H2PO4)4]4− ⊂ 284.
All anions encapsulated within the cavities are shown in space-filling form. Color code: C = gray, N = blue, O = red, and P = yellow. The counter cations and solvent molecules outside the cavities are omitted for clarity.
Over the past couple of decades, urea-based foldamers have been widely investigated due to their potential utility in a number of fields, including molecular recognition, catalysis, and materials science.36, 43–45 Wu et al.46 have developed a series of acyclic ortho-phenylene bridged oligourea receptors 29–32, which were found to interact with a number of anions. Specifically, receptor 29 and 30 were found to recognize the phosphate and sulfate anions.47–48 In the presence of chloride ions, the tri-urea 29 only stabilizes mononuclear chloride anion complex, a finding attributed to the short length of the receptor. However, the longer tetrakis-urea 30, pentakis-urea 31, and hexakis-urea 32, receptors wrap up to produce foldamers, all of which have large enough inner cavities to accommodate chloride anion dimers, as inferred from single crystal X-ray diffraction analyses (Figure 14). Notably, the encapsulated chloride-chloride dimers are hydrogen bonded to multiple urea units arranged along the inner helix with inter-chloride distances of 3.62 Å, 3.83 Å and 4.03 Å for 30, 31 and 32, respectively. Numerous cooperative hydrogen bonds, as well as a strong π-π stacking interaction between the two terminal aryl groups, serve to overcome the electrostatic repulsion associated with the concurrent binding or two anions. Theoretical calculations (Hartree-Fock method) provided support for the suggestion that 30 and 31 form foldamers only in the presence of chloride ions, which act as templates. In contrast, the hexakis(urea) system 32 tends to fold on its own.
Figure 14. Acyclic oligourea-based anion receptors and their anionic dimer complexes.
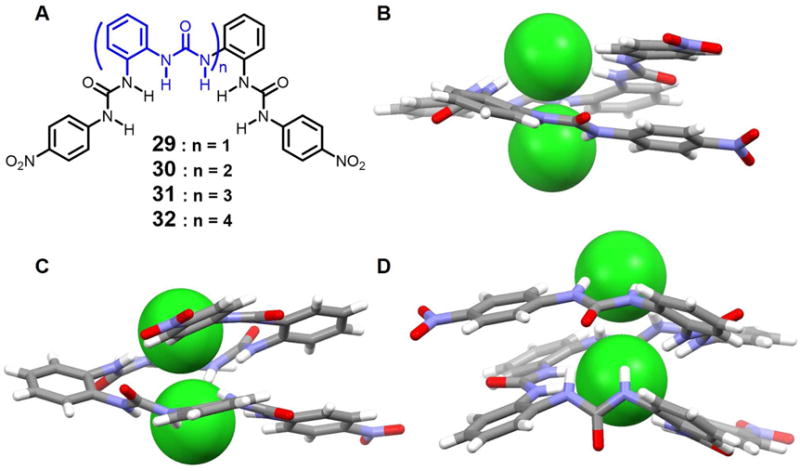
(A) Molecular structures of receptors 29–32.
(B) Single crystal X-ray structure of the complex [Cl2]2− ⊂ 30.
(C) Single crystal X-ray structure of the complex [Cl2]2− ⊂ 31.
(D) Single crystal X-ray structure of the complex [Cl2]2− ⊂ 32.
All anions encapsulated within the cavities are shown in space-filling form. Inter-halide distances: (B) 3.62 Å, (C) 3.83 Å and (D) 4.03 Å. Color code: C = gray, N = blue, O = red, Cl = green. The counter cations and solvent molecules outside the cavities are omitted for clarity.
Chloride anion binding studies, involving 1H NMR spectral titrations carried out in CDCl3, provided support for the conclusion that 30, 31 and 32 form 1 : 2 (host/guest) complexes in solution. In contrast, receptor 29 can stabilize higher order complexes. Two-step changes in the UV-vis spectra were observed upon adding chloride anions to 30–32, supporting the assigned 1 : 2 (host/guest) binding stoichiometry. Similar dinuclear chloride-binding foldamers based on fluorescent oligoureas namely naphthyl- and anthracenyl-decorated 30, 31 and 32, were also reported by the same group.49, Receptor 30 was also found to encapsulate two orthogonally oriented rod-like azide anions within one receptor. This complex is stabilized by N(urea) - H⋯N(azide) hydrogen bonding interactions as inferred from the crystal structure.50 UV-vis spectral titrations performed in DMSO provided further support for the proposed 1 : 2 (host/guest) binding mode in solution.
In 2011, Wang et al.51 described a series of oligo(phenyl-amide-triazole)s (33–35) that can fold into a helical conformations in the presence of halide ions. The short foldamer 33 with one helical turn was found to bind the chloride, bromide and iodide anions in a 1 : 1 mononuclear binding mode. In contrast, the longer foldamers 34 and 35 with two or three helical turns were found to entrap two chloride or bromide anions in a 1 : 2 (H : G) dinuclear fashion, respectively, as inferred from 1H NMR spectroscopic titrations and 1H - 1H NOESY measurements carried out in pyridine-d5 at ambient temperature. An inversion in the sign of the chemical shift displacement is observed during the course of the titrations. Such an observation is consistent with the presence of two (or more) equilibria in the binding processes; that is, oligomer 34 presumably folds first into a helical conformation in the presence of a first chloride ion. Then, the resulting complex is able to bind a second chloride ion (Figure 16A). As expected, the binding isotherms of the amide protons fit well to a 1 : 2 (host/guest) binding model with association constants of 4.9 × 103 · M−1 and 13 ·M−1 being derived for K1 and K2, respectively. This relatively weak second binding event is rationalized in terms of an electrostatic repulsion between the two co-bound chloride anions. Unfortunately, the authors did not report any crystal structures; so, the detailed conformation of the proposed 1 : 2 (host/guest) complexes in the solid state is currently unknown.
Figure 16. Structures of various anionic complexes.
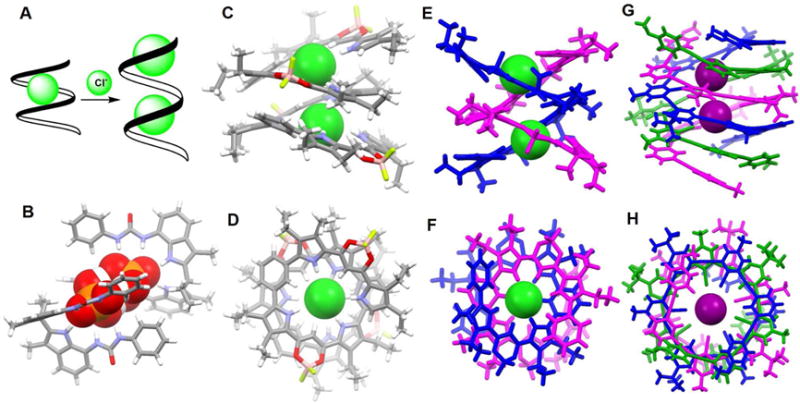
(A) Schematic representation of the complexation of chloride ions by foldamer 34.
(B) Single crystal X-ray structure of the complex [H3(PO4)2]3− ⊂ 362.
(C) Front view of the crystal structure of the complex [Cl2]2− ⊂ 39.
(D) Top view of the crystal structure of the complex [Cl2]2− ⊂ 39.
(E) Front view of the crystal structure of the complex [Cl2]2− ⊂ 382.
(F) Top view of the crystal structure of the complex [Cl2]2− ⊂ 382.
(G) Front view of the crystal structure of the complex [I2]2− ⊂ 403.
(H) Top view of the crystal structure of the complex [I2]2− ⊂ 403.
All anions encapsulated within the cavities are shown in space-filling form. Color code for (B), (C) and (D): C = gray, N = blue, O = red, P = yellow, Cl = green. Various colors (pink, blue or green) are used to represent different independent ligands. The counter cations and other molecules outside the cavities are omitted for clarity.
In 2010, Dydio et al.52 developed acyclic anion receptors based on a neutral diindolylmethane scaffold. Their systems, receptors 36 and 37, were found to bind preferentially tetrahedral oxoanions with association constants of 535 M−1 (36), 265 M−1 (37) and 280 M−1 (36), 90 M−1 (37) for H2PO4− and HSO4−, respectively, in MeOH-d4 at 298 K. As inferred from a single crystal X-ray diffraction analysis of the phosphate complex of 36, an unusual phosphate pseudo-dimer [H3PO4•PO4]3− is trapped within the cavity formed via the self-assembly of two molecules of 36 (Figure 16B). This pseudo-dimer was found even though TBAH2PO4 was used as the anion source to grow the crystals. It appears that upon crystallization, two H2PO4− anions are deprotonated, brought together and bridged by three protons. The P⋯P distance is only 3.68 Å. The “supra-anion” that results is stabilized by interactions with two molecules of 36.
In contrast to the fairly rigid macrocyclic geometries that characterize hemes and cyclodextrins, linear oligomer and polymer structures can exist in the form of spirals and helices that can be considered as mimics for classic protein motifs, such as α-helices and DNA double helices.53–55 Haketa and coworkers56 reported the synthesis of the phenylene-bridged dipyrrolyldiketone boron complexes 38–39, which could be used as π-conjugated acyclic anion receptors. As deduced from single-crystal X-ray analyses of the various anion complexes, receptors 38–39 proved capable of entrapping anions, either Cl− or Br−, in 2 : 2 and 1 : 2 (host/guest) binding modes, respectively. In the case 38 and 39, chloride dimers are encapsulated in the central cavities of the helices formed by the receptors (Figure 16C–16F). Receptor 39 proved long enough to form a single helix and to produce through folding an inner cavity large enough to complex a chloride anion dimer. In contrast, two molecules of 38 are required to create the double helix seen in the solid state structure of the chloride anion dimer complex. As inferred from 1H NMR and UV/Vis spectroscopic studies, along with ESI-TOF MS analyses, the anion-binding mode observed for 38 in CD2Cl2 or CH2Cl2 solution is not fully consistent with that observed in solid state. In the case of 39, a negative cooperative effect (for Cl−, K1 = 1.2× 108 M−1 and K2 = 3.2 × 103 M−1; determined in CH2Cl2 via UV-vis spectroscopy) on the binding was observed; this is consistent a destabilizing electrostatic repulsion between the two anions captured within the helix.
In 2016, Massena et al.57 presented the first triple helicate to encapsulate iodide anion dimers in organic and aqueous media as well as the solid state. In this work, an arylethynyl-based oligomer 40 and its bromo-substituted derivatives were synthesized and used for iodide anion recognition. Single crystal X-ray diffraction analysis of the iodopyridinium salt 40 revealed formation of a triplex that is composed of three intertwined tricationic monameric strands, offset along a common screw axis defined by the two intra-channel iodide anions (Figure 16G–16H). Each iodide is bound tightly by four strong and linear halogen bonds within the helical channel with average halogen-bond C-I⋯I− distance of 3.39 Å. In addition to two iodides captured within the helix (for which the I−⋯I− distance is 5.10 Å), seven other iodide anions appear around the exterior of the helicate. These latter anions, presumably bound through electrostatic interactions, help balance the nine positive charges associated with the cationic strands. The helix demonstrates remarkable stability at high temperatures and in aqueous and organic solvents. This stability is attributed to the presence of eight strong iodine⋯iodide halogen bonds and numerous buried π-surfaces.
In 2010, Arunachalam et al.58 presented a new benzene-based hexasubstituted bistripodal receptor 41 which has two equivalent clefts. Solid state structural analyses revealed that this receptor can adopt two different conformations. In one conformation, the six arms appear alternatively above and below the plane, resulting in a chair-like conformation (ababab). Each tripodal cleft encapsulates two nitrate anions along with a bridging water molecule (Figure 17B). Each nitrate anion interacts with one arm through N-H⋯O contacts, while a water molecule is found bound to the third arm. Another conformation in which three adjacent arms are above the plane and the other three are below (aaabbb) was also observed. In this case, each tripodal cleft holds a single acetate anion. In acetone-d6, 1H NMR spectral studies led to the conclusion that receptor 41 binds only two nitrate anions in solution, instead of the four found bound in the solid state. A Job plot analysis supported a 1 : 2 nitrate binding mode in solution instead of 1 : 4 (host/guest) binding in the solid state. Four AcO− anions were found to bind to one host in DMSO-d6, instead of the two found bound in the solid state. The authors ascribed the differences between the solid state and solution phase binding to conformations that could vary on moving from solution to the solid state.
Figure 17. Arene-based hexapodal amide receptors and their anionic complexes.
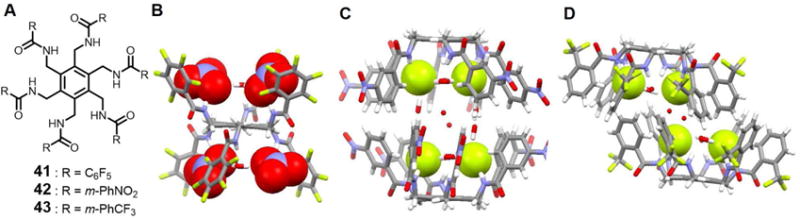
(A) Molecular structures of receptors 41–43.
(B) Single crystal X-ray structure of the complex [(NO3)4(H2O)2]4− ⊂ 41.
(C) Single crystal X-ray structure of the complex [F4(H2O)10]4− ⊂ 422.
(D) Single crystal X-ray structure of the complex [F4(H2O)6]4− ⊂ 432.
All anions encapsulated within the cavity are shown in space-filling form. Color code: C = gray, N = blue, O = red, and F = chartreuse yellow. The counter cations and solvent molecules outside the cavities are omitted for clarity.
The same group used the arene-based hexapodal amide receptor 42 to form a molecular capsule containing a [F4(H2O)10]4− cluster.59 The cluster is stabilized via strong F−-water interactions. Within each half of the capsule, two fluoride ions with short N-H⋯F− contacts and water molecules characterized by strong N-H⋯O hydrogen bonding interactions with the receptor are seen (Figure 17C). The distance between these two fluoride anions is 4.14 Å. Two water molecules are found bound to the anions as inferred from the F−⋯O distances of 2.73, 2.66, 2.72 and 2.63 Å, respectively. All arms of the receptor point in the same direction, allowing for formation of an overall dimeric capsule. Additional structural analyses revealed formation of a 1 : 2 (host/guest) complex with the AcO− anion.
In 2014, Chakraborty et al.60 published a similar hexaamide receptor, 43, which encapsulated a [F4(H2O)6]4− cluster (Figure 17D). In this case, the six arms point in one direction to produce a bowl-like cavity. Two fluoride anions and two water molecules ([F2(H2O)2]2−) are captured within one half of the cavity via N-H⋯F−, N-H⋯O and O-H⋯F− interactions. The two [F2(H2O)2]2− subunits are linked through two bridging water molecules. The net result is formation of a dimeric capsule complex. On the basis of 1H NMR spectral titrations and ITC studies carried out in DMSO-d6 or DMSO, a 1 : 2 binding stoichiometry was inferred for both fluoride and acetate. DFT calculations provided support for the conclusion that the binding energies are sensitive to the conformation of the receptor, as well as the electronegativity of the encapsulated anion.
Over the past 15 years or so, noncovalent contacts between anions and charge-neutral electron-deficient aromatic rings, namely anion-π interactions, have garnered considerable interest as a recognition motif.61 Chifotides, et al.62 exploited anion-π interactions to produce a colorimetric halide sensor (system 44; Figure 18). The presence of conjugated cyano groups, a high molecular polarizability, and a positive quadrupole moment, and a particularly electron-deficient structure was thought to provide multiple sites for stabilizing anion- or lone pair-π contacts. An X-ray structural analysis revealed that 44 is capable of interacting with bromide anions in the solid state. A stacked structure is found wherein three partially occupied bromide ions are distributed over four sites as shown in Figure 18C. In solution (THF or CH3NO2), the formation of 3X− ⊂ 442 (X = Cl−, Br−, I−) complexes was supported by the UV/Vis, 13C, halogen NMR, and ESI-MS data. Strong charge-transfer interactions and anion-π contacts were inferred.
Figure 18. Anionic complexes based on anion-π interactions.
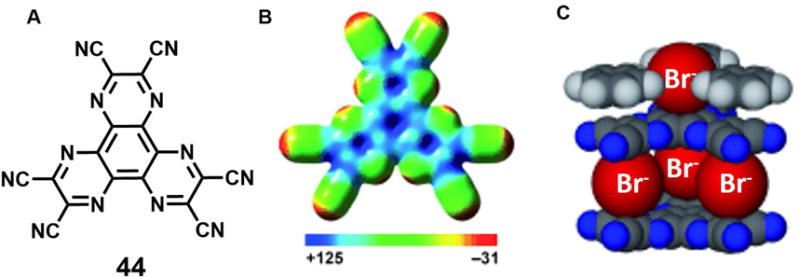
(A) Molecular structure of anion receptor 44.
(B) Electron spin polarization (ESP) map of 44 [kcal mol−1].
(C) Space-filling model of crystal structure of the bromide complex of 44.
Color code: C = black, N = blue, Br = red. The counter cations and other molecules are omitted for clarity.
Taken from Chifotides et al.62 and reproduced with permission. © 2010 Wiley-VCH Verlag GmbH & Co. KGaA.
MONOCYCLIC ANION RECEPTORS
Over the past two decades or so, a number of monocyclic receptors, including azacorands, azaoxacorands, porphyrins, rubyrins, rosarin, and cyclic amide- or urea-based systems have been reported as being capable of recognizing anion dimers and in some instances higher order anion aggregates. This section is designed to provide a review of this work. It is organized by receptor family, rather than in strictly chronological order.
In 2005, Steed and coworkers63 reported a macrocyclic polyaza metacyclophane 45, whose protonated form proved capable of binding two chloride, bromide or iodide anions. On the basis of structural studies, the bound anions were found reside both above and below the mean macrocyclic plane defined by the nitrogen atoms (Figure 19). In the case of the chloride complex, the two chloride anions were found deeply entrapped within the cavity and characterized by a Cl−⋯Cl− distance of 3.51 Å. However, in both the bromide and iodide complexes, the macrocycle 45 much larger inter-halide separations (5.01 Å and 5.30 Å for bromide complex and iodide complex, respectively) are observed. In all cases the binding seen in the solid state was ascribed to multiple receptor NH⋯anion interactions. Unfortunately, no evidence of solution phase binding was reported.
Figure 19. Polyaza metacyclophane anion receptors and its anionic dimer complexes.
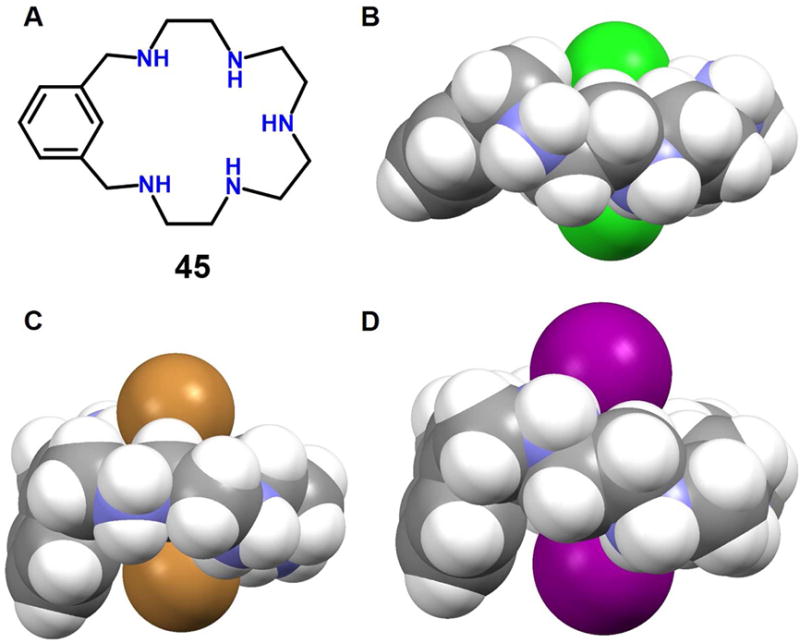
(A) Molecular structure of receptor 45.
(B) Single crystal X-ray structure of the complex [Cl2]2−⊂ [H545]5+.
(C) Single crystal X-ray structure of the complex [Br2]2−⊂ [H545]5+.
(D) Single crystal X-ray structure of the complex [I2]2−⊂ [H545]5+.
All anions encapsulated within the cavity and the protonated 45 are shown in space-filling form. Inter-halide distances: (B) 3.51 Å, (C) 5.01 Å and (D) 5.30 Å. Color code: C = gray, N = blue, Cl = green, Br = brown and I = purple. The counter anions and solvent molecules outside the cavities are omitted for clarity.
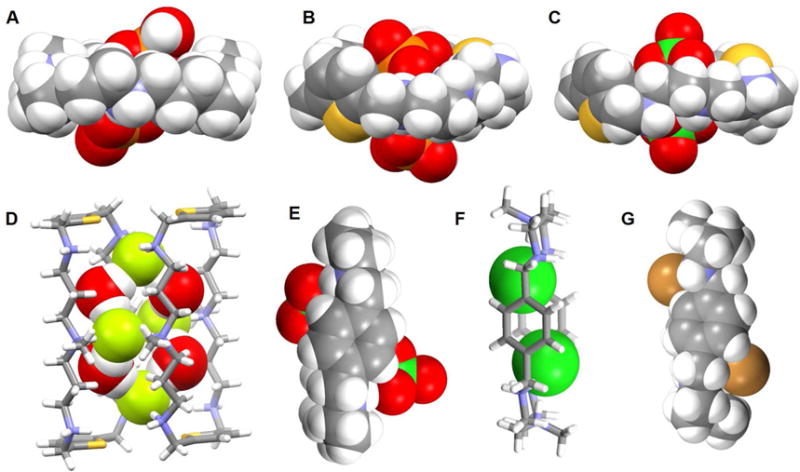
Prior to the contribution of Steed and coworkers, Gerasimchuk et al.64 reported a 26-membered polyammonium macrocycle 46 that contains six readily protonatabled nitrogen centers. A phosphate-containing salt of 46 was isolated and the resulting single crystal X-ray diffraction structure revealed a unique complex containing eight phosphate species, comprising both phosphoric acid and dihydrogenphosphate anions (Figure 21A). A H2PO4− dimer was partially included in the inner cavity of 46, which adopts a relatively flat ellipsoid-like conformation. Each of the three independent nitrogen atoms adopts a different orientation relative to the ring, presumably to enhance the hydrogen bonding interactions with the bound phosphates. Four additional dihydrogenphosphate anions and two neutral phosphoric acid molecules (H3PO4) are found outside the cavity. The ditopic binding of two monohydrogenphosphate anions to one molar equivalent of macrocycle 46 at low pH was confirmed by potentiometric studies.
Figure 21. Crystal structures of polyaza metacyclophane complexes with anionic dimers.
(A) [(H2PO4)2]2− ⊂ [H646]6+ complex shown using a space-filling model.
(B) [(HPO4) · (H2PO4)]3− ⊂ [H647]6+ complex shown using a space-filling model.
(C) [(ClO4)2]2− ⊂ [H647]6+ complex shown using a space-filling model.
(D) [F4(H2O)4]4− ⊂ [H649]6+ complex with the anions and water molecules shown in space-filling form.
(E) [(ClO4)2]2− ⊂ [H450]4+ complex shown using a space-filling model.
(F) [Cl2]2− ⊂ [H651]6+ complex with anions shown using a space-filling model.
(G) [Br2]2− ⊂ [H651]6+ complex shown using a space filling model.
Color code: C = gray, N = blue, O = red, F = chartreuse yellow, Cl = green, Br = brown and P = yellow. The counter anions and solvent molecules outside the cavities are omitted for clarity.
In 2016, a report detailing the study of two thiophene-based azamacrocycles 47 and 48 as phosphate anion receptors was published by Haque et al.65 Binding both in solution, as inferred from 1H and 31P NMR spectroscopic titrations, and in the solid state, as deduced from a single crystal X-ray analysis, was reported. In DMSO-d6, the protonated forms of receptors 47 and 48 bind dihydrogenphosphate anions in a 1 : 2 (host/guest) binding mode with good affinity (log K = 5.2 for 47 and log K = 4.2 for 48). Structural analysis of the phosphate complex of 47 revealed that each macrocycle accommodates two phosphate species, namely one monohydrogenphosphate and one dihydrogenphosphate, which are submerged into the cavity and located slightly above the elliptical plane, respectively (Figure 21B). The complex is stabilized through electrostatic interactions and multiple hydrogen bonds. The phosphate anions in the cavity are connected via P-O-H⋯O-P hydrogen bonds and separated by 4.55 Å (P⋯P distance).
In 2012, Hossain, et al.66 reported a novel [F4(H2O)4]4− cluster entrapped within a dimer of 49. A structural analysis revealed that the bound fluoride anions interact with water molecules to form an anionic [F4(H2O)4]4− tetramer that is presumably stabilized via hydrogen bonding interactions (Figure 21D). The macrocycles are essentially rectangular and each fluoride anion is hydrogen-bonded with two NH groups and two water molecules. The distances between any two fluoride ions bridged by water molecules are 4.50 Å and 4.55 Å. Additionally, the macrocycles also interact with two water molecules and two SiF6− anions through host N-H⋯O bonding interactions. A 1H NMR spectroscopic titration provided evidence for a 1 : 2 (host/guest) binding mode, a conclusion supported by a Job plot analysis.
The perchlorate anion (ClO4−) is characterized by a low charge to radius ratio. As a consequence, this anion is generally found to interact poorly with synthetic hosts.67–69 However, Saeed et al.70 reported two rare examples of perchlorate structures with hexaazamacrocycles 47 and 50 (Figure 21C and 21E). Single crystal X-ray structural analyses revealed that both protonated receptors are capable of hosting two perchlorate species in a 1 : 2 (host/guest) ratio via several hydrogen bonding interactions. However, the structure of the perchlorate complex of 47 differs from that of 50. Specifically, in the structure, receptor 47 is hexaprotonated and the two ClO4− anions are held 4.27 Å apart from one another (Cl⋯Cl distance) in a bipyramidal binding mode.71 In contrast, in the solid state, receptor 50 is only tetraprotonated with the central nitrogen atoms remaining in their neutral forms. A water molecule in the cavity acts as a bridge to connect the two co-bound ClO4− guests. A 1 : 1 binding stoichiometry, which is inconsistent with the solid state findings, was inferred for both systems based on Job plot analyses carried out in D2O at pH = 4.0.72–73
Ahmed and Hossain et al.71, 74–75 recently reported a p xylyl-based azamacrocycle 51 that they studied as a halide anion receptor. Results from solution studies carried out in D2O led to suggestions that receptor 51 preferentially binds smaller halide anions in a 1 : 2 (host/guest) binding mode rather than a 1 : 1 binding mode. Association constants (log K) of 2.82, 2.70, 2.28, and 2.20 for fluoride, chloride, bromide, and iodide, respectively, following this binding strength order: fluoride > chloride > bromide > iodide. X-ray crystallography was used to characterize the solid state structures of the chloride, bromide and iodide anion complexes, respectively, and revealed a 1 : 2 (host/guest) binding stoichiometry for all three anions. In the chloride complex (Figure 21F), all six nitrogen atoms in the macrocycle are fully protonated and the two symmetrical Cl− anions are fully encapsulated within the cavity by what appear to be strong hydrogen bonding interactions. A close Cl−⋯Cl− contact of 4.43 Å was observed. Presumably, the electrostatic repulsion between the two encapsulated Cl− anions is offset by the formation of strong hydrogen bonds with the ammonium groups. In the case of the complexes formed with the relatively larger bromide and iodide anions (Figure 21G) partially encapsulation is observed, with the bromide-bromide and iodide-iodide distances being 7.72 Å and 8.85 Å, respectively, within the two separate complexes. DFT calculations were also performed on [H651]6+ in both the gas phase and in a solvent environment. This led suggests that the receptor binds halides in the order of F− > Cl− > Br− > I− and that a 1 : 2 binding mode is energetically more favorable than a 1 : 1 binding mode. Thus, the calculations proved in full line with the experimental results.
A 36-membered neutral macrocyclic octalactam 52, which contains four 2,6-dicarbamoylpyridine and four ethylene moieties, was reported by Szumna, et al. in 2001.76 Upon cocrystallization of octalactam 52 with TBACl, complex [(Cl)2(52)(H2O)2]2− was obtained. The resulting crystal structure revealed that the macrocycle adopts a symmetrical, fully expanded, stair-like conformation in the solid state. Two Cl− anions along with two bridging H2O molecules are found within the cavity (Figure 22). The distance between the two chloride anions was estimated to be 4.88 Å. The [Cl2(H2O)2]2− guest lies almost within the plane defined by two of the four pyridine rings. The whole system is stabilized by multiple hydrogen bond interactions. The authors suggested that these multiple interactions help overcome the electrostatic repulsion that is inherently associated with constraining two anions in close proximity, as in the cavity of 52.
Figure 22. Cyclic hexaurea-based anion receptor 52 and its chloride dimer complexes.
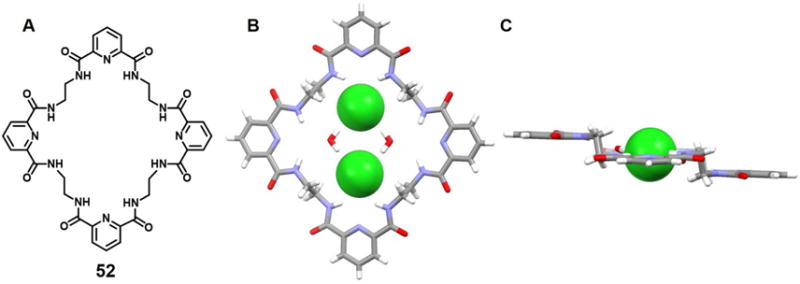
(A) Molecular structure of 52.
(B) Top view of the crystal structure of the complex [Cl2(H2O)2]2−⊂ 52.
(C) Side view of the crystal structure of the complex [Cl2(H2O)2]2−⊂ 52.
All anions encapsulated within the cavity are shown in space-filling form. Color code: C = gray, N = blue, O = red, and Cl = green. The counter cations and solvent molecules outside the cavities are omitted for clarity.
In 2006, Meshcheryakov et al.77 reported an elaborated cyclic hexaurea containing four xanthene and two diphenyl ether units (53). This system was obtained formed via the reaction of two diisocyanate molecules with two diamine molecules in the presence of a chloride anion source. It was proposed that two chloride anions act as templates for the macrocyclization reaction. The crystal structure of the chloride complex of 53 with chloride revealed a “figure-of-eight” shape, which is additionally slightly folded in the middle to leave only a (noncrystallographic) C2 axis perpendicular to the average macrocyclic plane (Figure 23). The net result is that the macrocyclic hexaurea receptor wraps around the two co-bound chloride ions that are separated (Cl⋯Cl) by 6.03 Å. Three adjacent urea units interact with one chloride anion. The observed N⋯Cl distances range between 3.18 Å and 3.34 Å. ESI-MS studies carried out in the negative ionization mode in acetonitrile/THF (3:1 v/v) revealed that, even when no external chloride salts are added, an intense signal for [53+2Cl]2− (m/z 1018.7) was observed. Presumably, traces of chloride, ubiquitous in most laboratory environments, are sufficient to form the 1 : 2 (host/guest) complex observed in the ESI-MS analysis. UV-vis spectroscopic and microcalorimetric titrations in acetonitrile/THF (1 : 3 v/v) at room temperature were consistent with this stoichiometry.
Figure 23. Cyclic hexaurea-based anion receptor 53 and its chloride anion dimer complex.
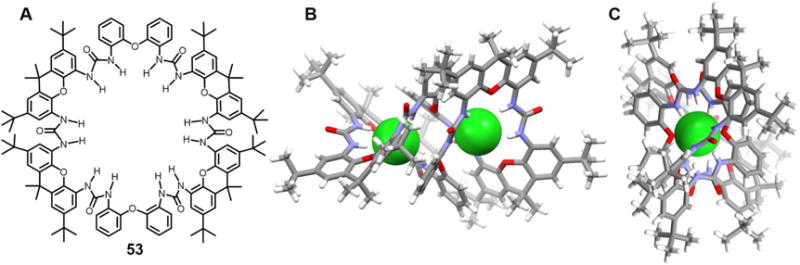
(A) Molecular structure of 53.
(B) Sideview of the crystal structure of the complex [Cl2]2−⊂ 53.
(C) Top view of the crystal structure of the complex [Cl2]2−⊂ 53.
All anions encapsulated within the cavity are shown in space-filling form. Color code: C = gray, N = blue, O = red, and Cl = green. The counter cations and solvent molecules outside the cavities are omitted for clarity.
Since the earliest days of the modern chemical era immense attention has been paid to porphyrins due to their important roles in biological processes such as photosynthesis, respiration, regulation, signal transduction, and so on.78–79 Expanded porphyrins have been investigated for many of these same applications, as well as for their interesting anion recognition properties. These latter features were first reported in 1990 by Ibers, Sessler, and coworkers.80 In that same year, Beckmann et al.81 described the total synthesis of the bisvinylogous coproporphyrin II 54, which was being developed at the time as a potentially improved photosensitizer for use in photodynamic tumor therapy (PDT). A single crystal structure of the di-hydrochloride salt of 54 revealed a macrocycle that was fully protonated and the presence of two chloride anions bound in a crosswise binding mode.71 The distance between the two chloride anions was found to be 3.87 Å. Separately, Sessler et al.82–84 reported the synthesis and structural characterization of alkyl-substituted derivatives of new classes of pentapyrrolic and hexapyrrolic expanded porphyrins, namely rubyrin (55), sapphyrin (56) and rosarin (57). As depicted in Figure 25B–25D, all three receptors (55–57) could readily protonated and capable of binding two Cl− ions with 1 : 2 (macrocycle/anion) stoichiometries. The Cl−⋯Cl− distances were found to be 3.68 Å, 3.59 Å, and 4.18 Å for 55, 56, and 57, respectively. These anion-anion contacts were unusually close for the time (1990s) were thought to reflect stabilization provided by the protonated (and hence cationic) expanded frameworks. However, subtle differences were seen across the series. For instance, protonated rubyrin 55 binds two chloride ions in a bipyramidal binding mode, while 56 hosts two Cl− anions in a crosswise binding fashion. Finally, rosarian 57 was found to encapsulate two Cl− anions within its deep cavity.
Figure 25. Cyclic pyrrole-based anion receptors and their anionic complexes.
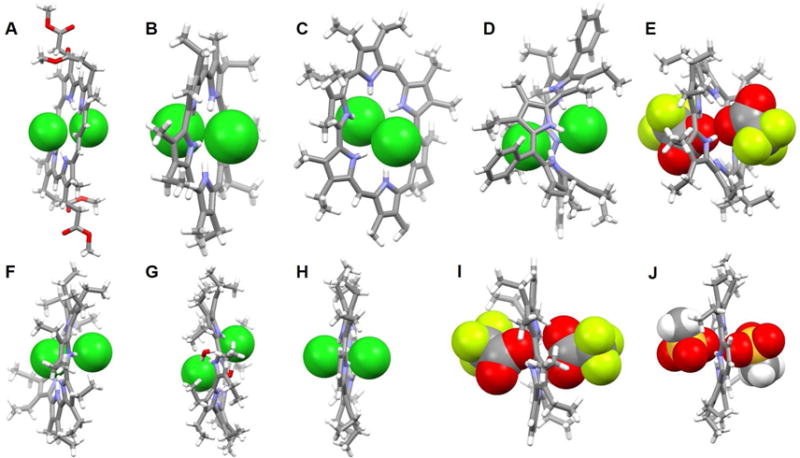
(A) The crystal structure of the complex [Cl2]2−⊂ [H254]2+.
(B) The crystal structure of the complex [Cl2]2−⊂ [H255]2+.
(C) The crystal structure of the complex [Cl2]2−⊂ [H256]2+.
(D) The crystal structure of the complex [Cl2]2−⊂ [H357]3+.
(E) The crystal structure of the complex [(CF3COO)2]2−⊂ [H258]2+.
(F) The crystal structure of the complex [Cl2]2−⊂ [H259]2+ complex.
(G) The crystal structure of the complex [Cl2(H2O)2]2−⊂ [H260]2+.
(H) The crystal structure of the complex [Cl2]2−⊂ [H261]2+.
(I) The crystal structure of the complex [(CF3COO)2]2−⊂ [H261]2+.
(J) The crystal structure of the complex [(CH3SO4)2]2−⊂ [H261]2+.
All anions encapsulated within the cavity are shown in space-filling form. Color code: C = gray, N = blue, O = red, F = chartreuse yellow, S = yellow, and Cl = green. The counter anions and solvent molecules outside the cavities are omitted for clarity.
Sessler and coworkers85,86 reported the synthesis of cyclo[6]pyrrole, cyclo[7]pyrrole and cyclo[8]pyrrole. These meso-free expanded porphyrins were obtained via oxidative coupling of a tetraalkylbipyrrole under FeIII-mediated coupling conditions in the presence of HCl and, in the case of cyclo[8]pyrrole, H2SO4. Structural proof for the binding of anionic dimers by 58, 59 and 60 was obtained from single-X-ray diffraction analyses of their diprotonated forms (Figure 25E–25G). In the case of 58, each receptor in its protonated form binds two CF3COO− ions with an unusual close O⋯O contact of 2.85 Å. The six pyrrolic NH protons of [H258]2+ 2 adopt a 1, 3, 5-alternate conformation, that is, three NHs alternately point toward the anionic oxygen of the CF3COO− anion located above the macrocyclic plane while the other NH protons point toward the lower acetate anion. The fully protonated forms of both 59 and 60 were found to capture two Cl− anions in close proximity (Cl−⋯Cl− distances of 3.75 Å and 4.68 Å were found for 59 and 60, respectively). Two water molecules are found to bridge the two co-bound Cl− anions in the case of the complex stabilized by 60, with the result it is formally a 2Cl−·2H2O complex that is encapsulated within the cavity. In analogy to what was seen in the case of 58, all NH protons of receptors 59 and 60 adopt an alternate conformation and all appear hydrogen bonded to Cl− anions in the solid state.
Recently, Zhang et al.87 reported the synthesis of two hybrid macrocycles, namely the cyclo[m]pyridine[n]pyrroles (m = 2 or 3; n = 4 or 3), that were obtained using a Suzuki cross-coupling strategy. In the case of 61, the addition of acid (e.g., HCl, CF3COOH and CH3SO4H) results in the formation of a diprotonated species that shows antiaromatic character. The counter anions associated with these acids, namely Cl−, CF3COO− and CH3SO4−, were found complexed by 61 in a 1 : 2 (host/guest) stoichiometry. Close anion-anion contacts of 3.72 Å, 3.45 Å and 3.29 Å were seen for Cl−, CF3COO− and CH3SO4−, respectively (Figure 25H–25J). In all cases, compound 61 was found to adopt a slightly ruffled conformation in the solid state, wherein the pyrrole and pyridine rings are tilted off an otherwise planar core. The NH protons adopt a 1, 3, 5-alternate arrangement with three pointing toward the anion bound above the mean macrocyclic plane, while the other three NH protons point to the lower anion.
In 1999, Sessler et al.88 reported the synthesis of [32]octaphyrin(1.0.0.0.1.0.0.0) 62 which was claimed to be the first expanded porphyrin containing a quaterpyrrole subunit. A single-crystal X-ray diffraction analysis of its diprotonated chloride adduct of 62 revealed a macrocycle that deviates substantially from planarity and to which two chloride anions are bound above and below the rim of the macrocyle (Figure 26B). One of the quaterpyrrole units adopts the 1, 2-alternate conformation with two pyrrolic NH protons pointing towards the chloride anion bound above the macrocycle, while the other two NH protons point toward the other Cl− anion. The other quaterpyrrole unit adopts a 1, 3-alternate conformation with one pyrrole NH proton pointing toward the upper chloride anion, one pyrrole NH moiety pointing toward the lower Cl− anion, and the remaining two NH protons interacting with solvent molecules (methanol). The Cl−⋯Cl− distance was found to be 5.31 Å.
Figure 26. Pyrrole-containing macrocycles and their chloride anion complexes.
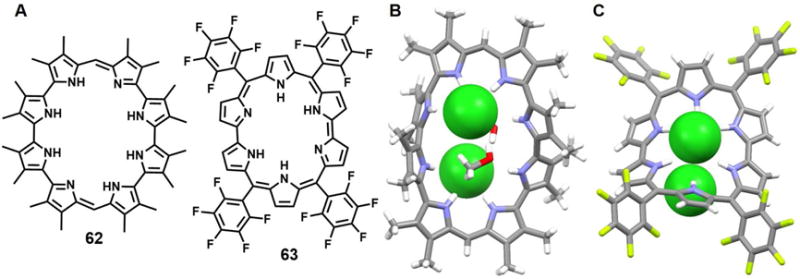
(A) Molecular structures of receptors 62 and 63.
(B) The crystal structure of the complex [(Cl2]2−⊂ [H262]2+.
(C) The crystal structure of the complex [(Cl2]2−⊂ [H263]2+.
All anions encapsulated within the cavities are shown in space-filling form. Color code: C = grey, N = blue, O = red, F = chartreuse yellow and Cl = green. Solvent molecules outside the cavities are omitted for clarity.
In 2005, Shimizu et al.89 published a concise synthesis of the meso-aryl-substituted rubyrin 63 and described the anion-recognition behavior of its diprotonated form. The structure of 63 as its bis-HCl salt was determined by single-crystal X-ray diffraction analysis. This study revealed a dome-like ruffled structure wherein all the pyrrole rings point inward so to support pyrrole NH hydrogen bonding interactions with the two co-bound chloride anions (Figure 26C). One chloride anion was found to lie 1.2 Å above the mean macrocyclic plane and be bound by four bipyrrolic NH protons with NH⋯Cl− distances of 2.34, 2.33, 2.39, and 2.42 Å. The other chloride anion lies slightly off-center and was stabilized by multiple hydrogen-bonding interactions. The Cl−⋯Cl− distance was estimated to be 4.04 Å.
Non-conjugated pyrrole-containing macrocycles are another class of important anion receptors.90 A number have been found to support complexes containing concurrently more than two anions. For instance, Guchhait et al.91 described the synthesis and structural characterization of several polyanion complexes of the azacalix[2]dipyrrolylmethanes 64 and 65. In analogy to what is seen in the case of calix[4]pyrroles, these receptors have a tendency to undergo a change in conformation upon anion binding. Owing to the presence of tertiary amine nitrogen atoms within the rings, both receptors 64 and 65 can be readily diprotonated. X-ray structural analyses revealed that the protonated form of 64 is able to bind the Cl− and ClO4− anions with a 1 : 2 (host/guest) stoichiometry in the solid state (Figures 27D and 27E). In both complexes, the double protonated macrocycles adopt a 1, 2-alternate conformation such that two pyrrole NH units point towards an anion (Cl− or ClO4−) above the mean macrocycle plane, while the other two pyrrole moieties point toward the anion held below the plane. The host separated anion-anion distances were estimated to be 3.86 Å (Cl−⋯Cl−) and 2.95 Å (Cl-O⋯O-Cl) for the chloride and perchlorate complexes, respectively. In the case of 65, a solid state structural analysis revealed that the receptor was likewise diprotonated and that the macrocycle adopts a 1, 2-alternate conformation in which two adjacent pyrrolic NH groups are pointing towards a BF4− anion held above the effective rim of the macrocycle, while the other two pyrrolic NH groups are pointing towards the other BF4− anion, which is located below the rim (Figure 27F). The host separated B-F⋯F-B contact is about 3.03 Å.
Figure 27. Pyrrole-containing macrocycles and their complexes with various anions.
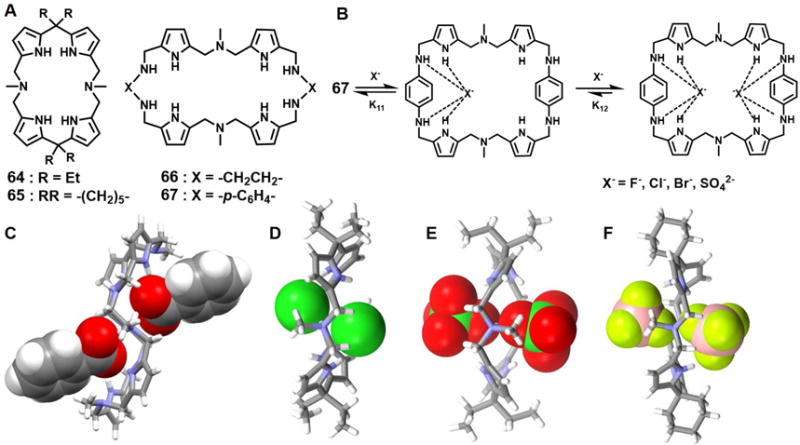
(A) Molecular structures of receptors 64–67.
(B) Schematic representation of the multiple equilibria of the complexes of 67.
(C) The crystal structure of the complex [(C6H5COO)2]2− ⊂ [H266]2+.
(D) The crystal structure of the complex [(Cl2]2− ⊂ [H264]2+.
(E) The crystal structure of the complex [(ClO4)2]2− ⊂ [H264]2+.
(F) The crystal structure of the complex [(BF4)2]2− ⊂ [H265]2+.
All anions encapsulated within the cavities are shown in space-filling form. Color code: C = gray, N = blue, O = red, F = chartreuse yellow, B = peach, and Cl = green.
In 2012 Kumar et al.92 reported a class of large macrocycles 66 and 67 containing N, N-di(pyrrolylmethyl)-N-methylamine moieties that were prepared by exploiting Schiff base-forming condensation reactions. Although both receptors present ostensibly similar pyrrolic and amine NH donors, the observed anion binding is quite different. This difference was ascribed to the different degrees of conformational flexibility provided by the linkers (ethylene vs. phenylene). NMR spectroscopic titration studies, carried out in CDCl3 at room temperature, provide support for the conclusion that 66 binds anions in a 1 : 1 fashion, while 67 bind anions in a sequential 1 : 2 (host/guest) manner thus providing complexes with 1 : 1 as well as 1 : 2 binding stoichiometries (Figure 27B). These receptors showed different affinities for anions, but with the relative order being SO42− > F− > Cl− > Br− > PhCOO− in both cases. A single crystal X-ray diffraction analysis of the salt formed between 66 and PhCOOH revealed a complex wherein only two of the secondary amine nitrogen atoms are protonated and two benzoate anions are bound above and below the rim of the macrocycle (Figure 27C).
In 2013, Sessler et al.93 published a pyrrole-based triazolium-phane (68) that was prepared via the tetraalkylation of a macrocycle originally prepared through “click” chemistry. It was reported that receptor 68 is selective for tetrahedral oxyanions over various test monoanions (e.g., Cl− and Br−) and trigonal planar anions (e.g., NO3− and CH3COO−) in mixed polar organic-aqueous solvent media. In the solid state, 68 was found to bind two H2PO4− or two BF4− anions within its internal cavity as inferred from single crystal X-ray diffraction analyses. These latter analyses revealed P⋯P and B⋯B distances of 4.13 Å and 4.35 Å, respectively (Figure 28). The formation of a dimeric H2PO4− complex was also confirmed by 1H NMR spectroscopic analyses carried out in CD3CN. Receptor 68 contains potential pyrrole N-H, benzene C-H and triazolium C-H donors. However, both experimental and theoretical results support the conclusion that the triazolium (CH)+ -anion interactions are less important in an energetic sense than the neutral aromatic CH-anion interactions, at least in methanol.
Figure 28. The pyrrole-based triazolium-phane and its complexes with anionic dimers.
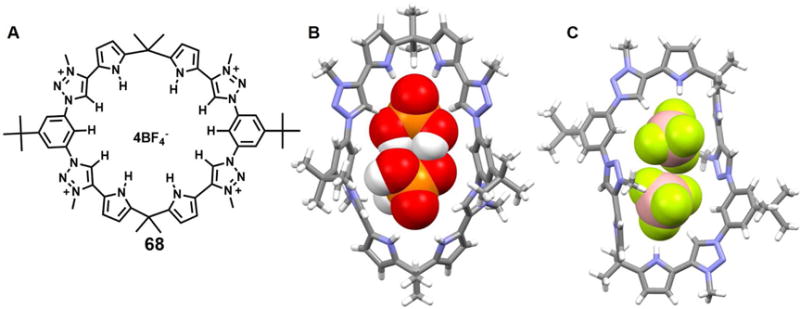
(A) Molecular structure of receptor 68.
(B) The crystal structure of the complex [(H2PO4)2]2−⊂ 682.
(C) The crystal structure of the complex [(BF4)2]2−⊂ 682.
All anions encapsulated within the cavity are shown in space-filling form. Color code: C = gray, N = blue, O = red, F = chartreuse yellow, B = peach and P = yellow. Other counter anions and solvent molecules outside the cavities are omitted for clarity.
In 2016 Fatila et al.94 reported that the cyanostar macrocycle 69 can stabilize a dimer of bisulfate anions bound in a sandwich-like arrangement (Figure 29). As inferred from a single crystal X-ray diffraction structural determination, the complexed bisulfate dimer [HSO4⋯HSO4]2− is characterized by a pair of short OH⋯O hydrogen bonds. The S-O⋯O-S distance between the dimeric guests (HSO4−) in the cavity of the cyanostar receptors is about 2.51 Å, which is shorter than that in the parent salt (2.62 Å).95 Electrospray ionization mass spectrometric (ESI-MS) analyses were carried out in an effort to delineate the possible species in solution. A peak ascribable to a 2 : 2 : 1 ensemble (m/z 2268.2) was seen that could be assigned to a [692(HSO4)2(NBU4)]− species. However, peaks at m/z 1471.2 readily assignable to a triple-decker complex wherein three macrocycles host a pair of bisulfates (i.e.,[693(HSO4)2]2−), are also observed. Taken in concert, such findings support the suggestion that the disulfate dimers stabilized by receptor 69 are stable even in the gas phase.
Figure 29. The cyanostar macrocycle and its complex with bisulfate anion dimers.
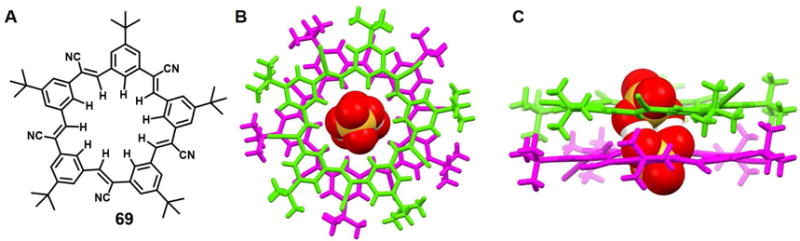
(D) Molecular structure of receptor 69.
(E) Top view of the crystal structure of the complex [(HSO4)2]2−⊂ 692.
(F) Front view of the crystal structure of the complex [(HSO4)2]2−⊂ 692.
All anions encapsulated within the cavity are shown in space-filling form. Color code: O = red, S = yellow. Different colors (pink and green) are used to highlight different independent ligands. The counter cations and solvent molecules outside the cavities are omitted for clarity.
In solution, the 2 : 2 and 3 : 2 (host/guest) dimers were also confirmed by NMR spectroscopic methods. Control studies involving the using a cyanostar that cannot form π-stacked dimers were carried out.96 In the case of the 3 : 2 complex, the authors saw evidence of self-complementary OH⋯O hydrogen bonding in the 1H NMR spectrum: The bisulfate dimer in the 3 : 2 species gives rise to a signature 1H NMR signal at 12.9 ppm, which upon cooling to 218 K appears as two peaks at 13.10 and 13.12 ppm, respectively (Figure 30). In contrast, the bisulfate dimer in the 2 : 2 species gives rise to a 1H NMR peak at 13.75 ppm under these same conditions. Based on the observation of NOE cross-peaks originating from the bisulfate anion and from the cyanostar host at 218 K, the 13.75-ppm peak is correlated with proton Hd at 8.05 ppm and both peaks originate from the 2 : 2 species. In contrast, the 13.2-ppm peak is correlated with a resonance at 8.01 ppm for the 3 : 2 species, further confirming that, in solution, the bisulfate dianion dimer is located inside a pair of cyanostar receptors.
Figure 30. 1H-1H NOESY spectra and 1H NMR spectral signatures.
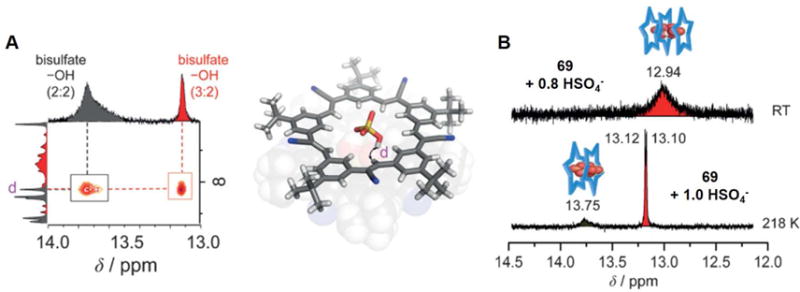
(A) Through-space correlations that were used to determine solution-phase structures; structures with NOE correlations labeled for HSO4− protons proximal to the inner protons of 69 as determined in CD2Cl2 and 1H-1H NOESY spectra with a mixing time of 800 ms for a solution of 69 (10 mm) containing 4 equivalents of NBu4HSO4 (218 K, CD2Cl2).
(B) 1H NMR spectral signatures of the bisulfate dimers stabilized by 69 at RT and 218 K, respectively. Here the blue star represents receptor 69.
Taken from Fatila et al.94 and reproduced with permission. © 2016 Wiley-VCH Verlag GmbH & Co. KGaA.
Further systematic studies of the bisulfate complex of receptor 69 were performed by the same group.97 Here, the authors varied the solvent to tune the formation of various assemblies and took advantage of characteristic NMR signatures to follow the species distribution. They found that in apolar solvents such as CHCl3 it was possible to generate a 2 : 2 : 2 complex composed of π-stacked cyanostars 69 encapsulating the [HSO4⋯HSO4]2− dimer that was capped by tetrabutylammonium cations. In the case of more polar solvents, e.g. CH3CN, three molecules of 69 were observed to form a presumed electropositive pocket within which a 3 : 2 (69 : HSO4−) assembly was stabilized (Figure 31). These two limiting complexes were found to give rise to characteristic 1H NMR spectral signatures. For the 2 : 2 : 2 species, the S-OH⋯O-S protons within the bisulfate dimer were observed at 13.8 ppm at room temperature. These signals shifted to ~ 14 ppm at low temperature (223 K). For the 3 : 2 complex, proton signals seen at 12.9 ppm at room temperature appeared at 13.1 ppm upon cooling to 223 K. Surprisingly, the 2D NOE spectrum of the 3 : 2 species (1 equivalent HSO4−, 233 K) revealed correlations with a residual water peak at 2.5 ppm in the case of both the S-OH⋯O-S signal of the bisulfate dimer and the inner aromatic protons of the cyanostar receptor. This was taken as evidence for a water molecule being bound to the complex in solution. The effect of solvent on the anion recognition process was also studied by the same group.98
Figure 31. Illustrative structures of the complexes and variable concentration 1H NMR spectra.
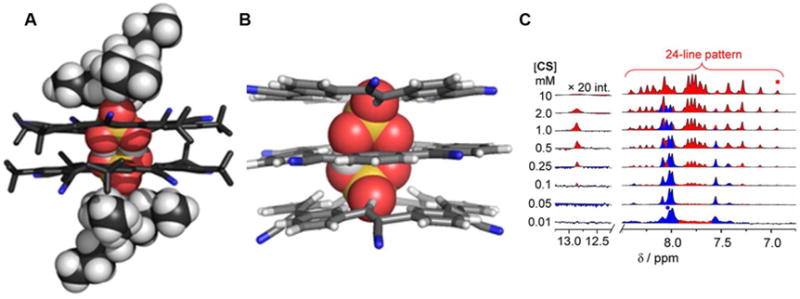
(A) The crystal structure of the 2 : 2 : 2 complex (69 : HSO4− : TBA+).
(B) The energy minimized structure of the 3 : 2 complex (69 : HSO4−).
(C) The variable concentration 1H NMR spectra of 69 (60 : 40 v/v % CD2Cl2/CD3CN) as recorded in the presence of 1 equivalent of NBu4HSO4. Here CS refers to 69.
Taken from Fatila et al.97 and reproduced with permission. © 2017 Wiley-VCH Verlag GmbH & Co. KGaA.
Recently, Mungalpara et al.99–100 reported the cyclic pseudopeptides 70 and 71, containing 1, 4-disubstituted 1, 2, 3-triazole moieties. In the case of 70, conformations are preferred in solution wherein the NH protons and the triazole moieties converge toward the center of the macrocycle. The result is a system that is preorganized for anion binding. Quantitative studies revealed that receptor 70 is capable of binding a H2PO4− dimer in DMSO/water. However, both 1 : 1 and 2 : 1 (host/guest) binding stoichiometries were observed in the presence of the sulfate and hydrogenpyrophosphate anions. A single crystal X-ray diffraction analysis of 70 in the presence of dihydrogenphosphate revealed H2PO4− trimer entrapped within the cavity formed by two molecules 70 in a sandwich-like structure. In the case of 71, four amide NH and four triazole CH groups are available to interact with anions. Receptor 71 proved to bind strongly protonated anions, such as the H2P2O72− and H2PO4− anions, and to a lesser extent the SO42− anion. Structural information provided by X-ray crystallography revealed that both a dihydrogenpyrophosphate dimer and a dihydrogenphosphate tetramer could be captured between molecules of 71 in a 2 : 2 and 2 : 4 (host/guest) stoichiometry for H2P2O72− and H2PO4−, respectively (Figure 32). Although the H2P2O72− complex comprises four individual components and the H2PO4− complex comprises six individual parts, these complexes proved stable enough to persist in solution (2.5 vol% water/DMSO), as evidenced by NMR spectroscopic and ITC titrations. They could also be detected in the gas phase. This stability is attributed to mutually reinforcing interactions between the individual complex components and the shielding of the bound anions from the surrounding solvent by the pseudo sandwich-like cavity created by the receptor.
Figure 32. Cyclic pseudopeptide anion receptors and their complexes with anionic dimers and tetramers.
(A) Molecular structures of receptors 70 and 71.
(B) Top view and side view of the crystal structure of the complex [(H2P2O7)2]4−⊂ 712.
(C) Top view and side view of the crystal structure of the complex [(H2PO4)4]4−⊂ 712.
All anions encapsulated within the cavities are shown in space-filling form. Color code: O = red, P = yellow. Different colors (pink and blue) are used to highlight different independent ligands. The counter cations and other molecules outside the cavities are omitted for clarity.
In 2012, Wang et al.101 reported the electron-deficient phenoxylated oxacalix[2]arene[2]triazine macrocycle 72 that they proposed would function as an anion-π receptor. A [Cl2(H2O)2]2− cluster was found in the cavity created from two molecules of 72. On the basis of a single crystal structural analysis, the resulting rectangular cage structure is stabilized by O-H⋯Cl− hydrogen bonding interactions between water and chloride anions, as well as noncovalent anion-π interactions between the chloride anions and the electron-poor triazine rings and lone pair-π interactions between the water molecules and the other triazine rings (Figure 33). The distance between the chloride anions is 5.33 Å. The two benzene substituents are thought to further stabilize the complex by means of π-π interactions.
Figure 33. Macrocycle 72 designed to act as an anion-π receptor and its chloride anion complex.
(A) The molecular structure of 72.
(B) The crystal structure of the chloride anion complex of 72.
All chloride anions and water molecules encapsulated within the cavity are shown in space-filling form. Color code: C = gray, N = blue, O = red, and Cl = green. Counter cations and solvent outside the cavity are omitted for clarity.
A hierarchical, self-assembled pore that serves to capture and transport chloride anions was reported in 2014 by Miyoshi et al.102 It relies on lithium phthalocyanine (LiPc, 73). LiPc 73 crystallizes in three polymorphic forms: monoclinic α- and β-forms and a tetragonal x-form. It can also exist in the form of a neutral π radical. Electrochemical doping of the x-form of LiPc 73 with Cl− anions yielded single crystals of a neutral π radical LiPc. In this experiment, LiPc crystals on the working electrode were electrochemically oxidized in an acetonitrile solution containing TBACl leading to a crystal-to-crystal transformation and formation of LiPc·Clx (0 < × < 0.5), where the x value was controlled by the electrochemical potential. A series of crystal structures of the LiPc·Clx complexes were successfully solved and the resulting crystal structures revealed a chain of Cl− anions with relatively close Cl−⋯Cl− separation of 3.20 Å within the 1D channels produced by the self-assembly of 73 (Figure 34).
Figure 34. Structure of Li phthalocyanine (LiPc) and the crystal structure of its chloride anion doped complex.
(A) The molecular structure of LiPc 73.
(B) Top view of the crystal structure of the complex LiPc ·Clx (0 < × < 0.5).
(C) Side view of the crystal structure of the complex LiPc ·Clx (0 < × < 0.5).
All chloride anions encapsulated within the channels are shown in space-filling form. Different colors (pink and blue) are used to highlight different independent ligands.
BICYCLIC ANION RECEPTORS
Since the earliest days of anion recognition chemistry, macrobicyclic receptors, containing appropriate binding sites and cavities of suitable size and shape, have been explored for their ability to form stable, selective inclusion complexes with anions; in general, a pronounced cryptate effect on both stability and selectivity is seen relative to the corresponding macromonocyclic control systems.103 A number of macrobicyclic receptors have been found to support the co-complexation of more than one anions. For instance, in 1998 Bowman-James and coworkers.104 described a C3-symmetrical bicyclic polyammonium macrocycle 74 with m-phthalaldehyde spacers, which was designed to bind the trigonal planar nitrate ion. The results of a crystallographic structural analysis revealed to co-encapsulation of two nitrate ions within the bicyclic cavity. More specifically, the complex was shown to be the hexahydronitrate salt of the macrocycle with two molecules of water. Interestingly, two nitrate anions, in the form of a nitrate dimer, were found nestled inside the cavity in an eclipsed conformation (Figure 36A and 36D) with the other four nitrate anions (not shown) associated via predominantly electrostatic and hydrogen bonding interactions outside of the macrocyclic cavity. The two co-bound nitrate anions are almost crystallographically identical and are separated from one another by 3.34 Å (N⋯N distance).The perceived structural stability was attributed to hydrogen bonding contacts between the trigonally oriented macrocyclic amine protons and the bound nitrate anion oxygen atoms. Theoretical gas phase calculations using the CHARMM molecular simulation package provided support for the reasonable assumption that the macrocycle-nitrate attraction in the case of this complex is much stronger than the nitrate-nitrate anion repulsion, thus allowing two anions to be accommodated in close proximity within the macrocycle-derived cavity.
Figure 36. Crystal structures of complexes with anionic dimers.
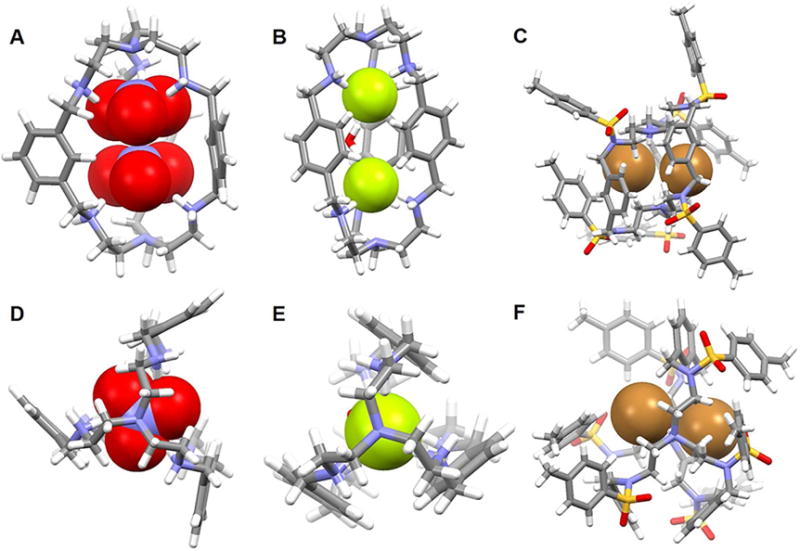
(A) Front view of the crystal structure of the complex [(NO3)2]2− ⊂ [H674]6+.
(B) Front view of the crystal structure of the complex [F2(H2O)]2− ⊂ [H675]6+.
(C) Front view of the crystal structure of the complex [Br2]2− ⊂ [H276]2+.
(D) Top view of the crystal structure of the complex [(NO3)2]2− ⊂ [H674]6+.
(E) Top view of the crystal structure of the complex [F2(H2O)]2− ⊂ [H675]6+.
(F) Top view of the crystal structure of the complex [Br2]2− ⊂ [H276]2+.
All anions encapsulated within the cavity are shown in space-filling form. Color code: C = gray, O = red, N = blue, F = chartreuse yellow, S = yellow, and Br = brown. The counter anions and solvent molecules outside the cavities are omitted for clarity.
Several years later the Bowman-James group105 reported PEAcryp 75 that can be considered as being a slightly enlarged version of 74. Using the p-xylyl bridged receptor 75, this team succeeded in capturing two discrete fluoride ions within an azacryptand environment, instead of the far more common bifluoride anion (F-H⋯F−).106 In this case, structural analyses revealed that receptor 75 was hexaprotonated and that the two fluoride ions reside almost within the center of the cavity. One water molecule points out from the central core and may contribute to the cryptand adopting an elongated ellipsoidal shape (Figure 36B and 36E). The two fluoride ions in the cavity are almost equidistant from their respective bridgehead amines and found at F−⋯N distances of 2.99 Å and 3.00 Å, respectively. Each fluoride anion is coordinated within a very distorted tetrahedral environment and stabilized through apparent hydrogen-bonding interactions involving three protonated secondary amines of a tren subunit, as well the bridging water molecule seen in the structure. The resulting F−⋯F− distance is 4.74 Å.
With an early notable exception,107 the tosyl group is viewed as a useful synthetic protecting group with the context of azacryptand anion recognition chemistry, rather than a functionality that abets anion recognition. However, in 2002 Morehouse et al.108 reported a tosylated azacryptand 76, whose protonated form acts a ditopic anion receptor. The crystal structure of the complex [Br2]2− ⊂ [H276]2+ produced from this tosylated system revealed the presence of two bromide anion guests encapsulated concurrently within the cryptand cavity. Each of the bromide anions lies off-center between two of the cryptand arms and is hydrogen bonded with one of the endo-oriented amine protons (Figure 36C and 36F). Compared with the neutral 76, the bromide complex is somewhat elongated. This likely reflects electrostatic repulsion between the two endo-oriented protons, as well as between the relatively large bromide anions. The Br−⋯Br− separation in [Br2]2− ⊂ [H276]2+ was found to be ca. 4.05 Å.
In 2010, Saeed et al.109 reported a thiophene-based octaazacryptand 77 that contains six secondary amines, as well as two bridgedhead nitrogen atoms. Structural analysis of the nitrate salt of 77 provided support for the notion that the receptor was fully protonated and that three nitrate anions are co-complexed within the internal cavity (Figure 37A). Both protons on the central bridgehead nitrogen atoms point toward the cavity and help to stabilize an overall trigonal bipyramidal dianion substructure. Almost all the secondary nitrogen protons are directed out from the cavity and are involved in coordinating external nitrates counter anions. Each of the three internal nitrate anions is bridged through one oxygen with two endo-oriented protons on the bridgehead nitrogen atoms. The three bridging nitrate oxygens are separated by 2.49, 2.60, and 3.00 Å, while the remaining six oxygen atoms in the co-bound nitrate anions lie between the arms of the macrocycle to minimize anion-anion repulsion. Support for the energetically favorable formation of a tris-nitrate anion complex came from density functional theory (DFT) calculations carried out using the hybrid density functional B3LYP with a 6-311G** basis set.
Figure 37. Crystal structures of complexes with anions.
(A) Crystal structure of the complex [(NO3)3]3− ⊂ [H877]8+.
(B) Crystal structure of the complex [(NO3)2]2− ⊂ [H478]4+.
(C) Crystal structure of the complex [(HSO4)2]2− ⊂ [H578]5+.
(D) Crystal structure of the complex [Cl2(H2O)]2− ⊂ [H279]2+.
All anions encapsulated within the cavities are shown in space-filling form. Color code: C = grey, O = red, N = blue, S = yellow, and Cl = green. The counter anions and solvent molecules outside the cavities are omitted for clarity.
Studies of the binding of nitrate and bisulfate anions to the bifunctional and laterally asymmetric azacryptand 78 were reported by Das et al.110 Crystallization of 78 with either nitrate or bisulfate was accomplished by dissolving 78 in water or in aqueous methanol in the presence of the appropriate acid, followed by slow evaporation at room temperature. Single crystal X-ray diffraction studies revealed that two nitrate or bisulfate anions may be readily sandwiched between two arms of a single cryptand (Figure 37B and 37C). Receptor 78 proved to be tetra- and penta-protonated in the case of the nitrate and bisulfate complexes, respectively. Two of the bridges making up the protonated cryptand fold in one direction so as to trap one of the nitrate or bisulfate anions, while the other nitrate or bisulfate is seen to reside between the other two arms of the cryptand. Based on the metric parameters, both anionic dimers appear stabilized by multiple hydrogen bonding interactions.
Kang et al.111 designed receptor 79 as part of an effort to explore the effect of topicity and charge on the anion recognition features of amidocryptands. A single crystal structure of the dichloride complex of 79 revealed that the receptor folds with two of the diamide loops directed in one direction, while the third points in the opposite direction. This results in a face-to-face orientation of the pyridine-containing spacers. The two bridgehead amines are protonated and hydrogen-bonded to each of the two co-bound chloride anions and presumably assists in the recognition process through charge complementarity (Figure 37D). The two entrapped chloride anions are bridged by a “cascading” water molecule. The authors noted that this cascade effect is reminiscent of what is observed in the case of transition metal cations complexed with neutral azacryptands, where the bridging species is an anion. The Cl−⋯Cl− separation was ca. 4.95 Å.
In 2008, Arunachalam et al.112 reported a C3-symmetric drum-shaped homoditopic hexaamino bicyclic cyclophane 80 with spatially substituted phenyl caps and an m-xylyl spacer along with its chloride and iodide anion complexes. Single-crystal X-ray analyses served to confirm that the hexaprotonated state of 80 is capable of encapsulating readily only a single iodide anion within its cavity. However, in the case of the corresponding chloride complex, two water molecules are stabilized within the inner cavity while a chloride anion (three total) is captured in each of the three side pockets. Extensive hydrogen bonding interactions, involving the water molecules, chloride anions and receptor framework, are observed (Figure 38B).
Figure 38. Bicyclic anion receptors and their crystal structures of complexes with anions.
(A) Molecular structures of receptors 80 and 81.
(B) Top view and front view of the crystal structure of the complex [Cl3]3− ⊂ [H680]6+.
(C) Crystal structure of the complex [(NO3)2]2− ⊂ [H680]6+.
(D) Crystal structure of the complex [(ClO4)3]3− ⊂ [H680]6+.
(E) Crystal structure of the complex [Cl2(H2O)]2− ⊂ 81.
All anions encapsulated within the cavity are shown in space-filling form. Color code: C = grey, O = red, N = blue and Cl = green. The counter anions or cations and solvent molecules outside the cavities are omitted for clarity.
Recently, Chakraborty et al.113 reported that 80 could be also used to bind other anions, as well as π-electron rich neutral guests. A single crystal X-ray structural analysis of the nitrate complex revealed that, as expected, all six nitrogen atoms of 80 are protonated but also that it encapsulates two nitrate anions as well as six water molecules (Figure 38C). One of the nitrates is nestled within the cavity while the other one resides in one of the clefts of the receptor; both guests are presumably stabilized via numerous N-H⋯O and O-H⋯O hydrogen bonding interactions. One oxygen atom of the nitrate anion bound within the cleft is oriented above, and points towards, the central nitrogen atom of the other nitrate anion (i.e., the one bound within the cavity). This structural motif presages what was recently seen in the case of certain solid state nitrate-nitrate interactions.114 When receptor 80 was subject to protonation with HClO4, a hexaprotonated cyclophane cage is produced. Two units of this hexaprotonated species, [H680]6+, combine to encapsulate three perchlorate ions both within the cavity and in the cleft-like pockets that are produced from the protonated receptor (Figure 38D). Two of the perchlorate anions are entrapped within the inner receptor cavities while the third perchlorate is held in a cleft-like pocket. The result is a perchlorate anion trimer that does not contain any anion-bridging water molecules.
As early as 1997, Bisson et al.115 noted that the amide-linked C3-symmetric bicyclic cyclophane 81 is capable of recognizing anions exclusively through neutral hydrogen bonding interactions. X-ray quality crystals of the chloride complex were obtained from a CH3Cl/CH3CN solution in the presence of excess TBACl. The resulting crystal structure revealed that the bicyclic cyclophane provides six potential amide hydrogen bond donors that converge in towards the center of the binding pocket. A 2 : 1 chloride-to-host binding stoichiometry was found with each Cl− anion hydrogen bonded to the amide groups of one acyl pyridine (Figure 38E). A water molecule occupies the remaining space of the cavity and is hydrogen bonded to the two chloride anions as well as to an acyl pyridine unit. The Cl−⋯Cl− separation was found to be about 5.42 Å. However, in contrast to what was seen in the solid state, the stoichiometry of the complexes in solution was determined to be 1 : 1 on the basis of a Job plot.
The Sessler group is credited with publishing the first cryptand-like calixpyrrole containing both pyrrole subunits and carbon bridgeheads in 2001.116 A decade later, Guchhait et al.117 exploited the Mannich reaction to create the azacryptand 82, which contains dipyrrolylmethane subunits and nitrogen atom bridgeheads. As inferred from variable-temperature 1H NMR spectral analyses, the neutral form of 82 shows a dynamic equilibrium between a staggered conformation as exhibited in the solid state and an eclipsed conformation that appears to dominate in solution. A single crystal X-ray structure of the fluoride ion complex of 82 revealed that a fluoride ion was accommodated in its cavity and that all the pyrrolic NHs are involved in hydrogen bonding interactions. This results in a distorted trigonal prismatic geometry around the F− center. The neutral form of 82 was found to bind the nitrate anion (as TBANO3 in acetone-d6), albeit extremely weakly (~26 M−1 by 1H NMR). However, once diprotonated, receptor 82 was found to host two nitrate anions. These later co-bound guests are oriented asymmetrically and were found bound to two of the three available clefts. The bound anion substrates were presumed stabilized via a combination of both hydrogen bonds and electrostatic interactions involving the protonated bridgehead nitrogen atoms (Figure 39B). One water molecule was encapsulated within the center of the cavity, bridging the two co-bound nitrate anions via hydrogen bonds.
Figure 39. Bicyclic anion receptors and crystal structures of selected anion complexes.
(A) Molecular structures of receptors 82 and 83.
(B) The crystal structure of the complex [(NO3)2]2− ⊂ [H282]2+.
(C) The crystal structure of the complex [(Cl2]2− ⊂ [H883]8+.
All anions encapsulated within the cavity are shown in space-filling form. Color code: C = grey, O = red, N = blue and Cl = green. The counter anions and solvent molecules outside the cavities are omitted for clarity.
Dynamic covalent chemistry (DCC) has been used to construct organic cages with three dimensional cavities. Many of these contain functional groups and have been widely applied in molecular recognition, gas sorption, separation, and sensing applications.118–122 In some cases, more than one anion is bound within a cage produced via DCC. One example comes from Jana et al.123 who reported the synthesis and structural characterization of macrobicycles and macrotricycles obtained via Schiff base condensations. The solid state structure of the chloride anion complex of the macrobicycle 83 in the protonated form revealed that the eight amine nitrogen atoms of the neutral bicycle molecule including the two bridgehead nitrogen atoms are fully protonated. Two chloride anions are encapsulated inside the cavity of the macrobicycle (Figure 39C). A short Cl−⋯Cl− separation of 3.69 Å, which falls within the sum of the van der Waals radii of the Cl atoms (3.40–3.80 Å), is seen.124 Presumably, the expected strong repulsion between these two chloride anions is offset by a combination of electrostatic interactions and hydrogen bonding interactions.
In 2012, Guchhait et al.125 reported the tripyrrolylmethane-based triazacryptand 84 containing bridgehead carbon atoms. This cryptand as well as several acyclic molecules were obtained via a Mannich reaction involving a formylated tripyrrolylmethane, a secondary amine hydrochloride, and formaldehyde. In the case of 84, size-selective anion binding behavior was observed. Specifically, both experimental and computational studies revealed that the neutral form of 84 has a very high affinity for the F− anion with good selectivity among the halide ions. As revealed by the crystal structures of the chloride and bromide complexes, the triprotonated form of 84 is capable of binding three Cl− or Br− anions within the three cleft-like pockets of the triply positively charged triazacryptand in the solid state (Figure 40B and 40C). Each halide anion is almost symmetrically bound by two pyrrolic NH and one N+H hydrogen bonds. The pairwise chloride-chloride distances between the co-bound chloride anions are 4.20, 4.22 and 4.31 Å, respectively, while the corresponding distances in the case of the trisbromide anion complex are 4.76, 4.77 and 4.86 Å, respectively.
Figure 40. Bicyclic anion receptors and their crystal structures of complexes with anions.
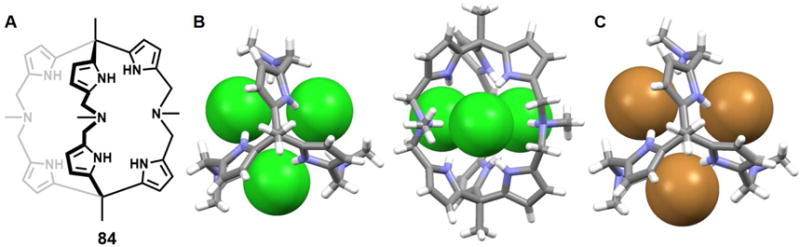
(A) Molecular structure of receptor 84.
(B) Top view and front view of the crystal structure of the complex [(Cl3]3− ⊂ [H384]3+.
(C) The crystal structure of the complex [(Br3]3− ⊂ [H384]3+.
All anions encapsulated within the cavity are shown in space-filling form. Color code: C = grey, O = red, N = blue, Cl = green, and Br = brown. The counter anions and solvent molecules outside the cavities are omitted for clarity.
Setsune et al.126 used three dipyrrylpyridine chains to construct the cryptand-like porphyrinoid 85. Bicycle 85 is defined not just by an inner cavity but also by three crevices that could prove suitable for guest binding. In the case of dichloroacetic acid (DCA), X-ray crystallography of the resulting complex revealed that all three pyridine nitrogen atoms in 85 were protonated and that three dichloroacetate counter anions are each bound to one of the three crevices defined by the tricationic receptor (Figure 41). The resulting complex adopts a pseudo-D3 symmetric conformation. A binding isotherm for the binding of Cl2CHCO2H as inferred from a UV-vis spectroscopic titration was characterized by a sigmoidal response characteristic of positive cooperativity. Presumably, this reflects interactions between the three binding sites. Cooperativity was evaluated using the Hill equation and the resulting coefficient (n = 2.7 ±0.2) and derived association constant (log K = 13.6 ± 1.8) supported the assumption of homotropic positive allosteric binding. These effects were rationalized by the authors in terms of intramolecular interactions that are enhanced upon protonation.
Figure 41. Cryptand-like receptor 85 and crystal structures of its dichloroacetate anion complex.
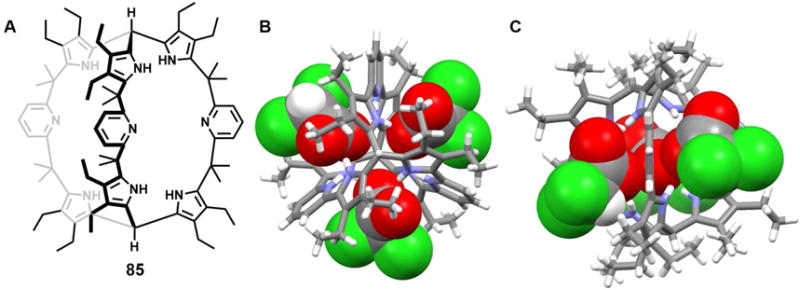
(A) Molecular structure of receptor 85.
(B) Top view of the crystal structure of the complex [(CHCl2COO)3]3− ⊂ [H385]3+.
(C) Side view of the crystal structure of the complex [(CHCl2COO)3]3− ⊂ [H385]3+.
All anions encapsulated within the cavity are shown in space-filling form. Color code: C = grey, N = blue, O = red and Cl = green. Solvent molecules outside the cavities are omitted for clarity.
TRICYCLIC ANION RECEPTORS
In a quest to expand the dimensionality of anion receptors, in 2008, the Bowman-James group127 synthesized and characterized the multidimensional and multitopic cyclophane-capped anion receptor 86. This system contains four diamide-monoamine chains and provides one large inner cavity and four side binding pockets in its tetraprotonated form. In the solid state, [H486]4+ proved capable of entrapping various phosphate anions, including H2PO4−, H2P2O72− and H3P3O102− as inferred from X-ray crystallographic analyses. These complexes, obtained by treating the neutral form of 86 with the corresponding acid, were characterized by the presence of two H2PO4−, two H2P2O72−, and two H3P3O102− anions, respectively, trapped within the pocket-like cavities of the tetraprotonated receptor (Figure 42). The apical phenyl rings stack at distances of 4.17, 3.94, and 3.87 Å, in [(H2PO4)2]2− ⊂ [H486]4+, [(H2P2O7)2]4− ⊂ [H486]4+, and [(H3P3O10)2]4− ⊂ [H486]4+, respectively, and adopt eclipsed conformations. In [(H2P3O7)2]4− ⊂ [H486]4+, one PO3H− subunit is nestled in one of the side pockets and stabilized via four hydrogen bonds arising from the amide hydrogen atoms and the protonated amines, each of which provides two NH donors. The other PO3H− subunit is not directly hydrogen bonded to the macrocycle. In contrast, in [(H3P3O10)2]4− ⊂ [H486]4+, the triphosphate is bound to the receptor via five hydrogen bonds involving both the amides and protonated amines as NH donors.
Figure 42. Tricyclic receptor 86 and solid state structures of selected anion complexes.
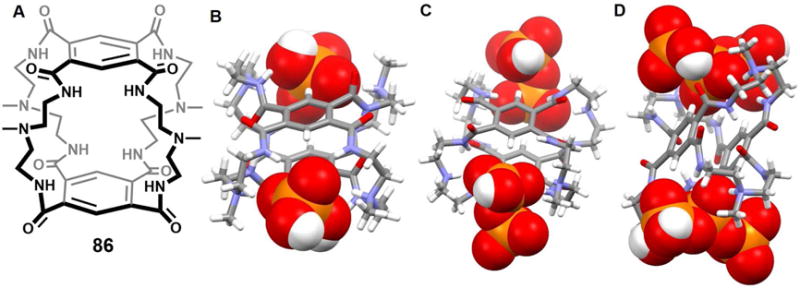
(A) Molecular structure of receptor 86.
(B) The crystal structure of the complex [(H2PO4)2]2− ⊂ [H486]4+.
(C) The crystal structure of the complex [(H2P2O7)2]4− ⊂ [H486]4+.
(D) The crystal structure of the complex [(H3P3O10)2]4− ⊂ [H486]4+.
All anions encapsulated in the cavity are shown in space-filling form. Color code: C = gray, N = blue, O = red and P = yellow. Solvent molecules outside the cavities are omitted for clarity.
Two years later, the same group128 designed and synthesized a tricyclic host 87 for anions. This system consists of two tetraamide monocycles linked by two ethylene chains. Structural and binding studies revealed that the receptor is selective for linear anions, especially for the simple triatomic FHF− and N3− ions. In the presence of excess H2SO4, the four bridgehead amines present in 87 become protonated. The resulting tetraprotonated form of the receptor was found to exist in the form of the bis-sulfate complex [SO4]24− ⊂ [H 87]4+ (Figure 43). A crystallographic analysis revealed that the macrocycle folds to adopt a conformation with approximate C2h symmetry. The pseudo-C2 axis bisects the two ethylene linkers and the pseudo-mirror plane passes through the four pyridine nitrogen atoms. Two SO42− anions along with two H2O molecules are entrapped by two of the larger side pockets of the protonated receptor. Each of the co-bound SO42− oxygen atoms is stabilized by hydrogen bonding interactions involving either the amide hydrogen atoms, protonated amines, or water molecules.
Figure 43. Tricyclic receptor 87 and single crystal structure of its sulfate anion complex.
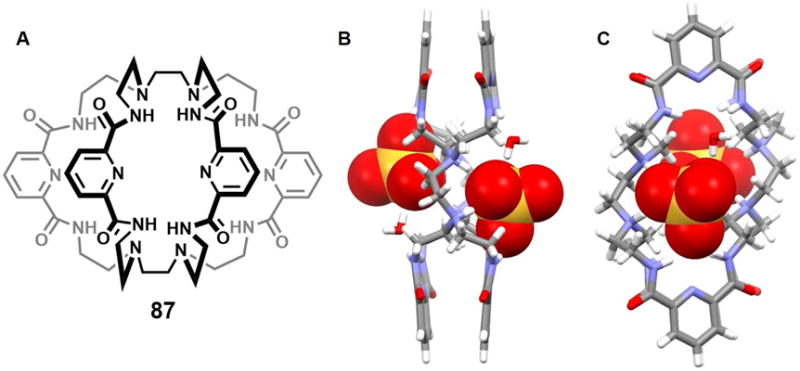
(A) Molecular structure of receptor 87.
(B) Top view of the crystal structure of the complex [(SO4)2]4− ⊂ [H487]4+.
(C) Side view of the crystal structure of the complex [(SO4)2]4− ⊂ [H487]4+.
All anions encapsulated in the cavity are shown in space-filling form. Color code: C = grey, N = blue, O = red and S = yellow. Solvent molecules outside the cavities are omitted for clarity.
In 2017, He et al.129 described a large capsule-like biscalix[4]pyrrole 88, which represents a rare example of a multidimentional neutral metal-free macrocycle that can capture more than one anion. Receptor 88 was synthesized by condensing a bis-dipyrromethane containing a pyridinyldiamido units with acetone in the presence of BF3 and provided a different approach to creating bis-calixpyrroles than that used previously by the groups of Ballester130–126 and Lee.131 Both the Ballester and Lee systems were found capable of forming bis-anion complexes. However, the available evidence leads to the conclusion that the co-bound anions do not make direct contact within the relatively large receptor cavity. In contrast, and as evidenced by X-ray crystal structures, the neutral form of receptor 88 was found capable of encapsulating either two H2PO4− anions, two SO42− anions, or two H2P2O72− anions within the large inner cavity (Figure 44). In the case of the bis-H2PO4− and bis-SO42− anion complexes, one or two water molecules are seen to bridge the co-bound anions creating interesting anion-water-anion clusters. Hydrogen bond interactions involving the pyrrolic and amide NH protons, as well as those provided by water molecules, serve to stabilize the complexes. Solution phase studies carried out using 1H NMR (in a mixture of CD2Cl2/CD3OD (9/1, v/v) and UV-vis spectroscopies (in 1, 2-dichloroethane (DCE)), as well as ITC analyses (in DCE), revealed a 1 : 2 (host/guest) stoichiometry for all three anionic substrates, thus matching what was seen in the solid state. The bis-anion recognition chemistry of 88 was also supported by DFT calculations.
Figure 44. Bis-calix[4]pyrrole 88 and single crystal structures of its anion complexes.
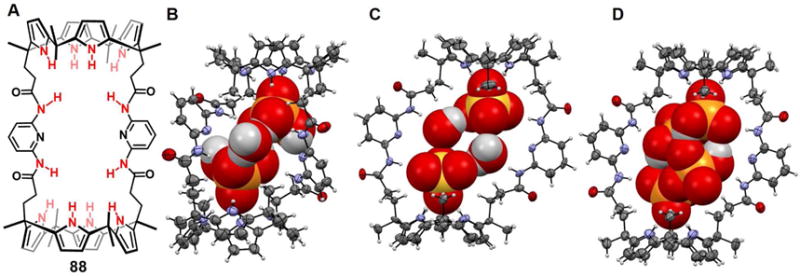
(A) Molecular structure of receptor 88.
(B) The crystal structure of the complex [(H2PO4)2 (H2O)]2− ⊂ 88.
(C) The crystal structure of the complex [(SO4)2 (H2O)2]4− ⊂ 88.
(D) The crystal structure of the complex [(H2P2O7)2]4− ⊂ 88.
All anions and water molecules encapsulated in the cavity are shown in space-filling form. Color code: C = gray, N = blue, O = red, P, S = yellow. Counter cations and solvent molecules outside the cavities are omitted for clarity.
Taken from He et al.129 and reproduced with permission. © 2017 American Chemical Society.
CONCLUSIONS AND FUTURE PERSPECTIVES
Since the pioneering work on anion recognition by Simmons, et al.132 in the late 1960s, the field has blossomed. Anion recognition has been widely applied to anion sensors, anion-responsive materials, anion-driven organocatalysis, extraction and separations, as well as through lipid anion transport.1, 133 To date, much of the focus have been placed on the recognition of a single anionic substrate. However, anion-anion interactions are prevalent in nature. Anion-anion interactions and anion-anion pairs are also commonly seen in the crystal engineering, where weak interactions may make significant contributions to the crystal packing.16 As detailed in this review, the specific recognition of like-charged anionic species within synthetic receptors is a subfield that is starting to come into its own. It is likely that the systems reported to date and reviewed here, as well as new ones still to be conceived, will contribute to advances across a number of applications areas, including many that extend beyond efforts to model natural polyanion receptors and engineer crystals. However, at present our ability to control the specific binding and orientation of multiple anions at the molecular levels remains a tremendous challenge. This is particularly true for systems that constrain more than two anions in close proximity. However, we predict that the future will see this challenge met with increasingly levels of sophistication. This, in turn, should translate into important contributions to a variety of fields where supramolecular chemistry is already playing a dominant role, such as environmental remediation, biological modeling, materials development, and medicinal chemistry. It is hoped that this review helps sow the seeds of this future growth.
Figure 15.
Molecular structures of receptors 33–40
Figure 20.
Molecular structures of azamacrocycles 46–51.
Figure 24.
Molecular structures of receptors 54–61.
Figure 35.
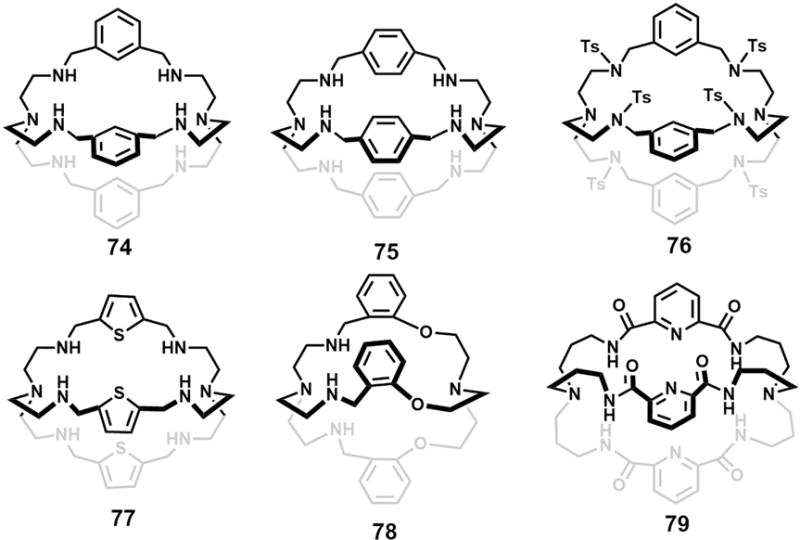
Molecular structures of macrobicyclic receptors 74–79.
THE BIGGER PICTURE.
To date, intensive interest has been focused on designing and synthesizing functional anionic receptors for a single anion. A limited number of acyclic, monocyclic, bicyclic and tricyclic anionic receptors have shown promise for recognizing of anionic dimers, trimers, tetramers and clusters, wherein more than one negatively charged species is constrained in close proximity. The ability to manipulate and bind two or more anions within a single receptor environment is crucial to understanding many biological processes and environmental effects. It may also allow inter alia for the fabrication of high quality ionic crystals and the construction of improved extraction systems. To our knowledge, this is the first review to focus on artificial receptors capable of recognizing multiple anionic species.
Acknowledgments
The work in Austin was supported by the National Institutes of Health (Grant No. GM103790 to J.L.S.) and the Robert Welch Foundation (F-0018 to J.L.S.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
SUPPLEMENTARY INFORMATION
All crystal structures used in the figures in this review were generated using data downloaded from the Cambridge Crystallographic Data Centre (CCDC) unless stated otherwise. CCDC numbers: 733118, 732741, 732740, 732742, 732739 for Figure 2; 1046452, 833333 for Figure 3; 711822, 711820 for Figure 4; 922581, 922582 for Figure 5; 1009054, 623880, 1009053, 1009052, 655560 for Figure 6; 275964, 961079, 961080 for Figure 8; 889595, 1450231 for Figure 9; 1536469, 1536470, 1536473 for Figure 10; 1484815, 1484817, 1484818 for Figure 11; 944527, 944528, 944529 for Figure 12; 889131, 889133 for Figure 13; 823697, 823698, 823700 for Figure 14; 778928, 740497, 1476727 for Figure 16; 735310, 812289, 996874 for Figure 17; 272040, 272039, 272038 for Figure 19; 144341, 1035333, 739057, 860049, 739058, 753654, 742965 for Figure 21; 164973 for Figure 22; 286528 for Figure 23; 1196690, 1198530, 1190223, 1190538, 213326, 213327, 784483, 878910, 878909, 878911 for Figure 25; 138745, 258965 for Figure 26; 879576, 1455741, 1455743, 1455740 for Figure 27; 849578, 849576 for Figure 28; 1486436 for Figure 29; 1555955, 1555956 for Figure 32; 897875 for Figure 33; 979680 for Figure 34; 1151397, 172118, 228605 for Figure 36; 713455, 727166, 727164, 288776 for Figure 37; 714050, 890512, 890513, 100347 for Figure 38; 844342, 1002123 for Figure 39; 912749, 912750 for Figure 40; 673262 for Figure 41; 702035, 702036, 702037 for Figure 42; 796749 for Figure 43; 1535973, 1535974, 1535975 for Figure 44.
AUTHOR CONTRIBUTIONS
Conceptualization, Q.H., and J.L.S; Writing - Original Draft, Q.H., and P.T.; Writing - Review & Editing, Q.H., P.T., and J.L.S.
REFERENCES AND NOTES
- 1.Busschaert N, Caltagirone C, Van Rossom W, Gale PA. Applications of supramolecular anion recognition. Chem Rev. 2015;115:8038–8155. doi: 10.1021/acs.chemrev.5b00099. [DOI] [PubMed] [Google Scholar]
- 2.Evans NH, Beer PD. Advances in anion supramolecular chemistry: from recognition to chemical applications. Angew Chem Int Ed. 2014;53:11716–11754. doi: 10.1002/anie.201309937. [DOI] [PubMed] [Google Scholar]
- 3.Gale PA, Busschaert N, Haynes CJE, Karagiannidis LE, Kirby IL. Anion receptor chemistry: highlights from 2011 and 2012. Chem Soc Rev. 2014;43:205–241. doi: 10.1039/c3cs60316d. [DOI] [PubMed] [Google Scholar]
- 4.Kang SO, Llinares JM, Day VW, Bowman-James K. Cryptand-like anion receptors. Chem Soc Rev. 2010;39:3980–4003. doi: 10.1039/c0cs00083c. [DOI] [PubMed] [Google Scholar]
- 5.Wenzel M, Hiscock JR, Gale PA. Anion receptor chemistry: highlights from 2010. Chem Soc Rev. 2012;41:480–520. doi: 10.1039/c1cs15257b. [DOI] [PubMed] [Google Scholar]
- 6.Gupta SP, Kaur PK. Chloride ion channels: structure, functions, and blockers. In: Gupta SP, editor. ion channels and their inhibitors. Springer Berlin Heidelberg; Berlin, Heidelberg: 2011. pp. 309–339. [Google Scholar]
- 7.Gokel GW, Negin S. Synthetic ion channels: from pores to biological applications. Acc Chem Res. 2013;46:2824–2833. doi: 10.1021/ar400026x. [DOI] [PubMed] [Google Scholar]
- 8.Senior AE, Nadanaciva S, Weber J. The molecular mechanism of ATP synthesis by F1F0-ATP synthase. BBA-Bioenergetics. 2002;1553:188–211. doi: 10.1016/s0005-2728(02)00185-8. [DOI] [PubMed] [Google Scholar]
- 9.Doublie S, Tabor S, Long AM, Richardson CC, Ellenberger T. Crystal structure of a bacteriophage T7 DNA replication complex at 2.2 angstrom resolution. Nature. 1998;391:251–258. doi: 10.1038/34593. [DOI] [PubMed] [Google Scholar]
- 10.Kiefer JR, Mao C, Braman JC, Beese LS. Visualizing DNA replication in a catalytically active Bacillus DNA polymerase crystal. Nature. 1998;391:304–307. doi: 10.1038/34693. [DOI] [PubMed] [Google Scholar]
- 11.Cramer P, Bushnell DA, Fu JH, Gnatt AL, Maier-Davis B, Thompson NE, Burgess RR, Edwards AM, David PR, Kornberg RD. Architecture of RNA polymerase II and implications for the transcription mechanism. Science. 2000;288:640–649. doi: 10.1126/science.288.5466.640. [DOI] [PubMed] [Google Scholar]
- 12.Arunachalam M, Ghosh P. Recognition and complexation of hydrated fluoride anion: F2(H2O)62− templated formation of a dimeric capsule of a tripodal amide. Chem Commun. 2009:5389–5391. doi: 10.1039/b910014h. [DOI] [PubMed] [Google Scholar]
- 13.Arunachalam M, Ghosh P. A versatile tripodal amide receptor for the encapsulation of anions or hydrated anions via formation of dimeric capsules. Inorg Chem. 2010;49:943–951. doi: 10.1021/ic901644h. [DOI] [PubMed] [Google Scholar]
- 14.Rocchicciolideltcheff C, Fournier M, Franck R, Thouvenot R. Vibrational investigations of polyoxometalates.2. evidence for anion anion interactions in molybdenum(VI) and tungsten(VI) compounds related to the Keggin structure. Inorg Chem. 1983;22:207–216. [Google Scholar]
- 15.Chesman ASR, Hodgson JL, Izgorodina EI, Urbatsch A, Turner DR, Deacon GB, Batten SR. Anion-anion interactions in the crystal packing of functionalized methanide anions: an experimental and computational study. Cryst Growth Des. 2014;14:1922–1932. [Google Scholar]
- 16.Nelyubina YV, Antipin MY, Lyssenko KA. Anion - anion interactions: their nature, energy and role in crystal formation. Russ Chem Rev. 2010;79:167–187. [Google Scholar]
- 17.Arunachalam M, Ghosh P. Recognition of nitrates in a discrete dimeric capsular assembly of a triamide half-capsule. Indian J Chem A. 2011;50:1343–1349. [Google Scholar]
- 18.Pandurangan K, Kitchen JA, Blasco S, Boyle EM, Fitzpatrick B, Feeney M, Kruger PE, Gunnlaugsson T. Unexpected self-sorting self-assembly formation of a [4:4] sulfate: ligand cage from a preorganized tripodal urea ligand. Angew Chem Int Ed. 2015;54:4566–4570. doi: 10.1002/anie.201411857. [DOI] [PubMed] [Google Scholar]
- 19.Bill NL, Kim DS, Kim SK, Park JS, Lynch VM, Young NJ, Hay BP, Yang YJ, Anslyn EV, Sessler JL. Oxoanion recognition by benzene-based tripodal pyrrolic receptors. Supramol Chem. 2012;24:72–76. [Google Scholar]
- 20.Arunachalam M, Ghosh P. Formation of a nitrate zipped dimeric capsule and un-zipping by chloride doping. Chemical Communications. 2009:3184–3186. doi: 10.1039/b823018h. [DOI] [PubMed] [Google Scholar]
- 21.Basu A, Das G. Encapsulation of a discrete cyclic halide water tetramer [X−2(H2O)2]2−, X = Cl−/Br− within a dimeric capsular assembly of a tripodal amide receptor. Chem Commun. 2013;49:3997–3999. doi: 10.1039/c3cc40955d. [DOI] [PubMed] [Google Scholar]
- 22.Jose DA, Kumar DK, Ganguly B, Das A. Rugby-ball-shaped sulfate-water-sulfate adduct encapsulated in a neutral molecular receptor capsule. Inorg Chem. 2007;46:5817–5819. doi: 10.1021/ic062466+. [DOI] [PubMed] [Google Scholar]
- 23.Chutia R, Dey SK, Das G. Self-assembly of a tris(urea) receptor as tetrahedral cage for the encapsulation of a discrete tetrameric mixed phosphate cluster (H2PO4−·HPO42−)2. Cryst Growth Des. 2015;15:4993–5001. [Google Scholar]
- 24.Lakshminarayanan PS, Ravikumar I, Suresh E, Ghosh P. Trapped inorganic phosphate dimer. Chem Commun. 2007:5214–5216. doi: 10.1039/b713365k. [DOI] [PubMed] [Google Scholar]
- 25.Huang XJ, Wu B, Jia CD, Hay BP, Li MR, Yang XJ. Stepwise encapsulation of sulfate ions by ferrocenyl-functionalized tripodal hexaurea receptors. Chem -Eur J. 2013;19:9034–9041. doi: 10.1002/chem.201300275. [DOI] [PubMed] [Google Scholar]
- 26.Yin Z, Zhang Y, He J, Cheng JP. A new tripodal anion receptor with selective binding for H2PO4− and F− ions. Tetrahedron. 2006;62:765–770. [Google Scholar]
- 27.Hoque MN, Das G. Hydrated anion glued capsular and non-capsular assembly of a tripodal host: Solid state recognition of bromide-water [Br5(H2O)6]5− and iodide-water [I2-(H2O)4]2− clusters in cationic tripodal receptor. CrystEngComm. 2014;16:4447–4458. [Google Scholar]
- 28.Quinonero D, Frontera A, Suner GA, Morey J, Costa A, Ballester P, Deya PM. Squaramide as a binding unit in molecular recognition. Chem Phys Lett. 2000;326:247–254. [Google Scholar]
- 29.Arosio D, Fontanella M, Baldini L, Mauri L, Bernardi A, Casnati A, Sansone F, Ungaro R. A synthetic divalent cholera toxin glycocalix[4]arene ligand having higher affinity than natural GM1 oligosaccharide. J Am Chem Soc. 2005;127:3660–3661. doi: 10.1021/ja0444029. [DOI] [PubMed] [Google Scholar]
- 30.Yum JH, Walter P, Huber S, Rentsch D, Geiger T, Nuesch F, De Angelis F, Gratzel M, Nazeeruddin MK. Efficient far red sensitization of nanocrystalline TiO2 films by an unsymmetrical squaraine dye. J Am Chem Soc. 2007;129:10320–10321. doi: 10.1021/ja0731470. [DOI] [PubMed] [Google Scholar]
- 31.Malerich JP, Hagihara K, Rawal VH. Chiral squaramide derivatives are excellent hydrogen bond donor catalysts. J Am Chem Soc. 2008;130:14416–14417. doi: 10.1021/ja805693p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Amendola V, Bergamaschi G, Boiocchi M, Fabbrizzi L, Milani M. The squaramide versus urea contest for anion recognition. Chem -Eur J. 2010;16:4368–4380. doi: 10.1002/chem.200903190. [DOI] [PubMed] [Google Scholar]
- 33.Rostami A, Wei CJ, Guerin G, Taylor MS. Anion detection by a fluorescent poly(squaramide): self-assembly of anion-binding sites by polymer aggregation. Angew Chem Int Ed. 2011;50:2059–2062. doi: 10.1002/anie.201006884. [DOI] [PubMed] [Google Scholar]
- 34.Jin C, Zhang M, Wu L, Guan YF, Pan Y, Jiang JL, Lin C, Wang LY. Squaramide-based tripodal receptors for selective recognition of sulfate anion. Chem Commun. 2013;49:2025–2027. doi: 10.1039/c3cc00196b. [DOI] [PubMed] [Google Scholar]
- 35.Barboiu M. Artificial water channels - incipient innovative developments. Chem Commun. 2016;52:5657–5665. doi: 10.1039/c6cc01724j. [DOI] [PubMed] [Google Scholar]
- 36.Yashima E, Ousaka N, Taura D, Shimomura K, Ikai T, Maeda K. Supramolecular helical systems: helical assemblies of small molecules, foldamers, and polymers with chiral amplification and their functions. Chem Rev. 2016;116:13752–13990. doi: 10.1021/acs.chemrev.6b00354. [DOI] [PubMed] [Google Scholar]
- 37.Chen BL, Xiang SC, Qian GD. Metal-organic frameworks with functional pores for recognition of small molecules. Acc Chem Res. 2010;43:1115–1124. doi: 10.1021/ar100023y. [DOI] [PubMed] [Google Scholar]
- 38.Manna U, Chutia R, Das G. Entrapment of cyclic fluoride-water and sulfate-water-sulfate cluster within the self-assembled structure of linear meta-phenylenediamine based bis-urea receptors: positional isomeric effect. Cryst Growth Des. 2016;16:2893–2903. [Google Scholar]
- 39.Utsab Manna SK, Soham Samanta, Gopal Das. Fixation of atmospheric CO2 as novel carbonate-(water)2-carbonate cluster and entrapment of double sulfate within a linear tetrameric barrel of a neutral bis-urea scaffold. Dalton Trans. 2017;46:10374–10386. doi: 10.1039/c7dt01697b. [DOI] [PubMed] [Google Scholar]
- 40.Manna U, Nayak B, Das G. Dual guest [(chloride)3-DMSO] encapsulated cation-sealed neutral trimeric capsular assembly: meta-substituent directed halide and oxyanion binding discrepancy of isomeric neutral disubstituted bis-urea receptors. Cryst Growth Des. 2016;16:7163–7174. [Google Scholar]
- 41.Rajbanshi A, Wan S, Custelcean R. Dihydrogen phosphate clusters: trapping H2PO4− tetramers and hexamers in urea-functionalized molecular crystals. Cryst Growth Des. 2013;13:2233–2237. [Google Scholar]
- 42.Blazek V, Molcanov K, Mlinaric-Majerski K, Kojic-Prodic B, Basaric N. Adamantane bisurea derivatives: anion binding in the solution and in the solid state. Tetrahedron. 2013;69:517–526. [Google Scholar]
- 43.Gellman SH. Foldamers: a manifesto. Acc Chem Res. 1998;31:173–180. [Google Scholar]
- 44.Hill DJ, Mio MJ, Prince RB, Hughes TS, Moore JS. A field guide to foldamers. Chem Rev. 2001;101:3893–4011. doi: 10.1021/cr990120t. [DOI] [PubMed] [Google Scholar]
- 45.Yoo SH, Lee HS. Foldectures: 3D molecular architectures from self-assembly of peptide foldamers. Acc Chem Res. 2017;50:832–841. doi: 10.1021/acs.accounts.6b00545. [DOI] [PubMed] [Google Scholar]
- 46.Wu B, Jia CD, Wang XL, Li SG, Huang XJ, Yang XJ. Chloride coordination by oligoureas: from mononuclear crescents to dinuclear foldamers. Org Lett. 2012;14:684–687. doi: 10.1021/ol2031153. [DOI] [PubMed] [Google Scholar]
- 47.Li SG, Jia CD, Wu B, Luo Q, Huang XJ, Yang ZW, Li QS, Yang XJ. A triple anion helicate assembled from a bis(biurea) ligand and phosphate ions. Angew Chem Int Ed. 2011;50:5720–5723. doi: 10.1002/anie.201180593. [DOI] [PubMed] [Google Scholar]
- 48.Jia CD, Wu BA, Li SG, Yang ZW, Zhao QL, Liang JJ, Li QS, Yang XJ. A fully complementary, high-affinity receptor for phosphate and sulfate based on an acyclic tris(urea) scaffold. Chem Commun. 2010;46:5376–5378. doi: 10.1039/c0cc00937g. [DOI] [PubMed] [Google Scholar]
- 49.Yang PJ, Wang JM, Jia CD, Yang XJ, Wu BA. Dinuclear chloride-binding foldamers based on fluorescent oligoureas. Eur J Org Chem. 2013:3446–3454. [Google Scholar]
- 50.Wang XL, Jia CD, Huang XJ, Wu B. Azide anion encapsulation in a tetraurea receptor. Inorg Chem Commun. 2011;14:1508–1510. [Google Scholar]
- 51.Wang Y, Xiang JF, Jiang H. Halide-guided oligo(aryl-triazole-amide)s foldamers: receptors for multiple halide ions. Chem -Eur J. 2011;17:613–619. doi: 10.1002/chem.201001560. [DOI] [PubMed] [Google Scholar]
- 52.Dydio P, Zielinski T, Jurczak J. 7,7′-Diureido-2, 2′-diindolylmethanes: anion receptors effective in a highly competitive solvent, methanol. Org Lett. 2010;12:1076–1078. doi: 10.1021/ol1000395. [DOI] [PubMed] [Google Scholar]
- 53.Yashima E, Maeda K, Furusho Y. Single- and double-stranded helical polymers: Synthesis, structures, and functions. Acc Chem Res. 2008;41:1166–1180. doi: 10.1021/ar800091w. [DOI] [PubMed] [Google Scholar]
- 54.Haldar D, Schmuck C. Metal-free double helices from abiotic backbones. Chem Soc Rev. 2009;38:363–371. doi: 10.1039/b803553a. [DOI] [PubMed] [Google Scholar]
- 55.Juwarker H, Suk JM, Jeong KS. Foldamers with helical cavities for binding complementary guests. Chem Soc Rev. 2009;38:3316–3325. doi: 10.1039/b909034g. [DOI] [PubMed] [Google Scholar]
- 56.Haketa Y, Maeda H. From helix to macrocycle: anion-driven conformation control of pi-conjugated acyclic oligopyrroles. Chem -Eur J. 2011;17:1485–1492. doi: 10.1002/chem.201002748. [DOI] [PubMed] [Google Scholar]
- 57.Massena CJ, Wageling NB, Decato DA, Rodriguez EM, Rose AM, Berryman OB. A halogen-bond-induced triple helicate encapsulates iodide. Angew Chem Int Ed. 2016;55:12398–12402. doi: 10.1002/anie.201605440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Arunachalam M, Ghosh P. Bistripodand amide host for compartmental recognition of multiple oxyanions. Org Lett. 2010;12:328–331. doi: 10.1021/ol9026649. [DOI] [PubMed] [Google Scholar]
- 59.Arunachalam M, Ghosh P. Encapsulation of [F4(H2O)10]4− in a dimeric assembly of an unidirectional arene based hexapodal amide receptor. Chem Commun. 2011;47:6269–6271. doi: 10.1039/c1cc10742a. [DOI] [PubMed] [Google Scholar]
- 60.Chakraborty S, Dutta R, Wong BM, Ghosh P. Anion directed conformational diversities of an arene based hexa-amide receptor and recognition of the [F4(H2O)6]4− cluster. RSC Adv. 2014;4:62689–62693. [Google Scholar]
- 61.Giese M, Albrecht M, Rissanen K. Anion-pi interactions with fluoroarenes. Chem Rev. 2015;115:8867–8895. doi: 10.1021/acs.chemrev.5b00156. [DOI] [PubMed] [Google Scholar]
- 62.Chifotides HT, Schottel BL, Dunbar KR. The pi-accepting arene HAT(CN)6 as a halide receptor through charge transfer: multisite anion interactions and self-assembly in solution and the solid state. Angew Chem Int Ed. 2010;49:7202–7207. doi: 10.1002/anie.201001755. [DOI] [PubMed] [Google Scholar]
- 63.Ilioudis CA, Steed JW. Polyaza metacyclophanes as ditopic anion receptors. Org Biomol Chem. 2005;3:2935–2945. doi: 10.1039/b506828b. [DOI] [PubMed] [Google Scholar]
- 64.Gerasimchuk OA, Mason S, Llinares JM, Song MP, Alcock NW, Bowman-James K. Binding of phosphate with a simple hexaaza polyammonium macrocycle. Inorg Chem. 2000;39:1371–1375. doi: 10.1021/ic9911116. [DOI] [PubMed] [Google Scholar]
- 65.Hague SA, Berkley RS, Fronczek FR, Hossain MA. Solution and structural binding studies of phosphate with thiophene-based azamacrocycles. Inorg Chem Commun. 2016;70:121–124. doi: 10.1016/j.inoche.2016.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hossain MA, Saeed MA, Pramanik A, Wong BM, Haque SA, Powell DR. A self-assembled fluoride-water cyclic cluster of [F(H2O)]44− in a Molecular Box. J Am Chem Soc. 2012;134:11892–11895. doi: 10.1021/ja3043855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Arranz P, Bencini A, Bianchi A, Diaz P, Garcia-Espana E, Giorgi C, Luis SV, Querol M, Valtancoli B. Thermodynamics of sulfate anion binding by macrocyclic polyammonium receptors. J Chem Soc, Perkin Trans. 2001;2:1765–1770. [Google Scholar]
- 68.Hynes MJ, Beatrice MB, McKee V, Town RM, Nelson J. Protonated azacryptate hosts for nitrate and perchlorate. J Chem Soc Dalton. 2000:2853–2859. [Google Scholar]
- 69.Morgan G, Mckee V, Nelson J. Caged Anions - Perchlorate and Perfluoroanion Cryptates. J Chem Soc Chem Comm. 1995:1649–1652. [Google Scholar]
- 70.Saeed MA, Thompson JJ, Fronczek FR, Hossain MA. Ditopic binding of perchlorate anion to hexaazamacrocyclic hosts. Crystengcomm. 2010;12:674–676. doi: 10.1039/b918152k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hossain MA, Saeed MA, Fronczek FR, Wong BM, Dey KR, Mendy JS, Gibson D. Charge-assisted encapsulation of two chlorides by a hexaprotonated azamacrocycle. Cryst Growth Des. 2010;10:1478–1481. doi: 10.1021/cg100110f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Warden AC, Warren M, Hearn MTW, Spiccia L. Binding of inorganic oxoanions to macrocyclic ligands: interactions of sulfate and dithionate with protonated forms of [18]aneN(6) New J Chem. 2004;28:1301–1308. [Google Scholar]
- 73.Hossain A, Morehouse P, Powell D, Bowman-James K. Tritopic (cascade) and ditopic complexes of halides with an azacryptand. Inorg Chem. 2005;44:2143–2149. doi: 10.1021/ic048937e. [DOI] [PubMed] [Google Scholar]
- 74.Ahmed L, Rhaman MM, Mendy JS, Wang J, Fronczek FR, Powell DR, Leszczynski J, Hossain MA. Experimental and theoretical studies on halide binding with a p-xylyl-based azamacrocycle. J Phys Chem A. 2015;119:383–394. doi: 10.1021/jp511040p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Gibson D, Dey KR, Fronczek FR, Hossain MA. A new hexaaminomacrocycle for ditopic binding of bromide. Tetrahedron Lett. 2009;50:6537–6539. doi: 10.1016/j.tetlet.2009.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Szumna A, Jurczak J. Unusual encapsulation of two anions in the cavity of neutral macrocyclic octalactam - preliminary communication. Helv Chim Acta. 2001;84:3760–3765. [Google Scholar]
- 77.Meshcheryakov D, Bohmer V, Bolte M, Hubscher-Bruder W, Arnaud-Neu F, Herschbach H, Van Dorsselaer A, Thondorf I, Mogelin W. Two chloride ions as a template in the formation of a cyclic hexaurea. Angew Chem Int Ed. 2006;45:1648–1652. doi: 10.1002/anie.200503766. [DOI] [PubMed] [Google Scholar]
- 78.Setsune J. 2,2′-Bipyrrole-Based Porphyrinoids. Chem Rev. 2017;117:3044–3101. doi: 10.1021/acs.chemrev.6b00430. [DOI] [PubMed] [Google Scholar]
- 79.Osuka A, Saito S. Expanded porphyrins and aromaticity. Chem Commun. 2011;47:4330–4339. doi: 10.1039/c1cc10534e. [DOI] [PubMed] [Google Scholar]
- 80.Sessler JL, Cyr MJ, Lynch V, Mcghee E, Ibers JA. Synthetic and structural studies of sapphyrin, a 22-pi-electron pentapyrrolic expanded porphyrin. J Am Chem Soc. 1990;112:2810–2813. [Google Scholar]
- 81.Beckmann S, Wessel T, Franck B, Honle W, Borrmann H, Vonschnering HG. Novel Porphyrinoids.11. Coproporphyrin-II for photodynamic therapy. Angew Chem Int Ed. 1990;29:1395–1397. [Google Scholar]
- 82.Sessler JL, Morishima T, Lynch V. Rubyrin - a new hexapyrrolic expanded porphyrin. Angew Chem Int Ed. 1991;30:977–980. [Google Scholar]
- 83.Shionoya M, Furuta H, Lynch V, Harriman A, Sessler JL. Diprotonated sapphyrin - a fluoride selective halide anion receptor. J Am Chem Soc. 1992;114:5714–5722. [Google Scholar]
- 84.Sessler JL, Weghorn SJ, Morishima T, Rosingana M, Lynch V, Lee V. Rosarin - a new, easily prepared hexapyrrolic expanded porphyrin. J Am Chem Soc. 1992;114:8306–8307. [Google Scholar]
- 85.Seidel D, Lynch V, Sessler JL. Cyclo[8]pyrrole: a simple-to-make expanded porphyrin with no meso bridges. Angew Chem Int Ed. 2002;41:1422–1425. doi: 10.1002/1521-3773(20020415)41:8<1422::aid-anie1422>3.0.co;2-o. [DOI] [PubMed] [Google Scholar]
- 86.Kohler T, Seidel D, Lynch V, Arp FO, Ou ZP, Kadish KM, Sessler JL. Formation and properties of cyclo[6]pyrrole and cyclo[7]pyrrole. J Am Chem Soc. 2003;125:6872–6873. doi: 10.1021/ja035089y. [DOI] [PubMed] [Google Scholar]
- 87.Zhang Z, Lim JM, Ishida M, Roznyatovskiy VV, Lynch VM, Gong HY, Yang XP, Kim D, Sesser JL. Cyclo[m]pyridine[n]pyrroles: hybrid macrocycles that display expanded pi-conjugation upon protonation. J Am Chem Soc. 2012;134:4076–4079. doi: 10.1021/ja211985k. [DOI] [PubMed] [Google Scholar]
- 88.Sessler JL, Seidel D, Lynch V. Synthesis of [28]heptaphyrin(1.0.0.1.0.0.0) and [32]octaphyrin(1.0.0.0.1.0.0.0) via a directed oxidative ring closure: the first expanded porphyrins containing a quaterpyrrole subunit. J Am Chem Soc. 1999;121:11257–11258. [Google Scholar]
- 89.Shimizu S, Taniguchi R, Osuka A. Meso-aryl-substituted [26]hexaphyrin(1.1.0.1.1.0) and [38]nonaphyrin(1.1.0.1.1.0.1.1.0) from oxidative coupling of a tripyrrane. Angew Chem Int Ed. 2005;44:2225–2229. doi: 10.1002/anie.200463054. [DOI] [PubMed] [Google Scholar]
- 90.Vargas-Zúñiga GI, Sessler JL. Pyrrole N-H anion complexes. Coord Chem Rev. 2017;345:281–296. doi: 10.1016/j.ccr.2017.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Guchhait T, Mani G, Schulzke C. Synthesis and structural characterization of anion complexes with azacalix[2]dipyrrolylmethane: effect of anion charge on the conformation of the macrocycle. Dalton Trans. 2016;45:11781–90. doi: 10.1039/c6dt01396a. [DOI] [PubMed] [Google Scholar]
- 92.Kumar R, Guchhait T, Mani G. Synthesis and X-ray structures of novel macrocycles and macrobicycles containing N,N-di(pyrrolylmethyl)-N-methylamine moiety: preliminary anion binding study. Inorg Chem. 2012;51:9029–38. doi: 10.1021/ic3011822. [DOI] [PubMed] [Google Scholar]
- 93.Cai JJ, Hay BP, Young NJ, Yang XP, Sessler JL. A pyrrole-based triazolium-phane with NH and cationic CH donor groups as a receptor for tetrahedral oxyanions that functions in polar media. Chem Sci. 2013;4:1560–1567. [Google Scholar]
- 94.Fatila EM, Twum EB, Sengupta A, Pink M, Karty JA, Raghavachari K, Flood AH. Anions stabilize each other inside macrocyclic hosts. Angew Chem Int Ed. 2016;55:14057–14062. doi: 10.1002/anie.201608118. [DOI] [PubMed] [Google Scholar]
- 95.Light ME, G PA, Hursthouse MB. Anion-anion dimerization in tetrabutylammonium hydrogensulfate. Acta Cryst. 2001;E57:o705–o706. [Google Scholar]
- 96.Qiao B, Liu Y, Lee SM, Pink M, Flood AH. A high-yield synthesis and acid-base response of phosphate-templated [3] rotaxanes. Chem Commun. 2016;52:13675–13678. doi: 10.1039/c6cc08113d. [DOI] [PubMed] [Google Scholar]
- 97.Fatila EM, Twum EB, Karty JA, Flood AH. Ion pairing and co-facial stacking drive high-fidelity bisulfate assembly with cyanostar macrocyclic hosts. Chem -Eur J. 2017;23:10652–10662. doi: 10.1002/chem.201701763. [DOI] [PubMed] [Google Scholar]
- 98.Liu Y, Sengupta A, Raghavachari K, Flood AH. Anion binding in solution: beyond the electrostatic regime. Chem. 2017;3:411–427. doi: 10.1002/chem.201802657. [DOI] [PubMed] [Google Scholar]
- 99.Mungalpara D, Kelm H, Valkonen A, Rissanen K, Keller S, Kubik S. Oxoanion binding to a cyclic pseudopeptide containing 1, 4-disubstituted 1, 2, 3-triazole moieties. Org Biomol Chem. 2017;15:102–113. doi: 10.1039/c6ob02172g. [DOI] [PubMed] [Google Scholar]
- 100.Disha Mungalpara AV, Kari Rissanenb, Stefan Kubik. Efficient stabilisation of a dihydrogenphosphate tetramer and a dihydrogenpyrophosphate dimer by a cyclic pseudopeptide containing 1, 4-disubstituted 1, 2, 3-triazole moieties. Chem Sci. 2017;8:6005–6013. doi: 10.1039/c7sc02700a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Wang DX, Fa SX, Liu Y, Hou BY, Wang MX. Anion-directed assembly of a rectangular supramolecular cage in the solid state with electron-deficient phenoxylated oxacalix[2]arene[2]triazine. Chem Commun. 2012;48:11458–11460. doi: 10.1039/c2cc36465d. [DOI] [PubMed] [Google Scholar]
- 102.Miyoshi Y, Yoshikawa H, Awaga K. Crystal-to-crystal transformation in solid-state electrochemical doping of Cl− ions to the nanoporous neutral radical lithium phthalocyanine: revelation of electron-electron correlations in the 1D half-filled system. Crystengcomm. 2014;16:9266–9272. [Google Scholar]
- 103.Lehn JM. Cryptates - macropolycyclic inclusion complexes. Pure Appl Chem. 1977;49:857–870. [Google Scholar]
- 104.Mason S, Clifford T, Seib L, Kuczera K, Bowman-James K. Unusual encapsulation of two nitrates in a single bicyclic cage. J Am Chem Soc. 1998;120:8899–8900. [Google Scholar]
- 105.Hossain MA, Llinares JM, Mason S, Morehouse P, Powell D, Bowman-James K. Parallels in cation and anion coordination: a new class of cascade complexes. Angew Chem Int Ed. 2002;41:2335–2338. doi: 10.1002/1521-3773(20020703)41:13<2335::AID-ANIE2335>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 106.Dietrich B, Guilhem J, Lehn JM, Pascard C, Sonveaux E. Molecular recognition in anion coordination chemistry - structure, binding constants and receptor-substrate complementarity of a series of anion cryptates of a macrobicyclic receptor molecule. Helv Chim Acta. 1984;67:91–104. [Google Scholar]
- 107.Dietrich B, Fyles TM, Hosseini MW, Lehn JM, Kaye KC. Proton coupled membrane-transport of anions mediated by cryptate carriers. J Chem Soc Chem Comm. 1988:691–692. [Google Scholar]
- 108.Morehouse P, Hossain MA, Llinares JM, Powell D, Bowman-James K. A ditopic azacryptate proton cage. Inorg Chem. 2003;42:8131–8133. doi: 10.1021/ic034972u. [DOI] [PubMed] [Google Scholar]
- 109.Saeed MA, Fronczek FR, Huang MJ, Hossain MA. Unusual bridging of three nitrates with two bridgehead protons in an octaprotonated azacryptand. Chem Commun. 2010;46:404–406. doi: 10.1039/b923013k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Das MC, Ghosh SK, Bharadwaj PK. Diversity of binding of sulfate and nitrate anions with laterally asymmetric aza cryptands. Crystengcomm. 2010;12:413–419. [Google Scholar]
- 111.Kang SO, Powell D, Bowman-James K. Anion binding motifs: topicity and charge in amidocryptands. J Am Chem Soc. 2005;127:13478–13479. doi: 10.1021/ja054332l. [DOI] [PubMed] [Google Scholar]
- 112.Arunachalam M, Ravikumar I, Ghosh P. A new hexaaza bicyclic cyclophane with dual binding sites. J Org Chem. 2008;73:9144–9147. doi: 10.1021/jo801762b. [DOI] [PubMed] [Google Scholar]
- 113.Chakraborty S, Arunachalam M, Bose P, Ghosh P. Binding studies on an arene-capped bicyclic cyclophane with pi-rich neutral guests and anions. Cryst Growth Des. 2013;13:3208–3215. [Google Scholar]
- 114.Bauza A, Frontera A, Mooibroek TJ. NO3− anions can act as Lewis acid in the solid state. Nat Commun. 2017;8 doi: 10.1038/ncomms14522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Bisson AP, Lynch VM, Monahan MKC, Anslyn EV. Recognition of anions through NH-pi hydrogen bonds in a bicyclic cyclophane-selectivity for nitrate. Angew Chem Int Ed. 1997;36:2340–2342. [Google Scholar]
- 116.Bucher C, Zimmerman RS, Lynch V, Sessler JL. First cryptand-like calixpyrrole: synthesis, X-ray structure, and anion binding properties of a bicyclic[3,3,3]nonapyrrole. J Am Chem Soc. 2001;123:9716–9717. doi: 10.1021/ja016629z. [DOI] [PubMed] [Google Scholar]
- 117.Guchhait T, Mani G. Dipyrrolylmethane-based macrobicyclic azacryptand: Synthesis, X-ray structures, conformational and anion binding properties. J Org Chem. 2011;76:10114–10121. doi: 10.1021/jo201969f. [DOI] [PubMed] [Google Scholar]
- 118.Voloshin Y, Belaya I, Krämer R. the encapsulation phenomenon: synthesis, reactivity and applications of caged ions and molecules. Springer International Publishing; Cham: 2016. Encapsulation by covalent capsules; pp. 9–138. [Google Scholar]
- 119.Setsune JI, Watanabe K. Cryptand-like porphyrinoid assembled with three dipyrrylpyridine chains: Synthesis, structure, and homotropic positive allosteric binding of carboxylic acids. J Am Chem Soc. 2008;130:2404–2405. doi: 10.1021/ja710424n. [DOI] [PubMed] [Google Scholar]
- 120.Tozawa T, Jones JTA, Swamy SI, Jiang S, Adams DJ, Shakespeare S, Clowes R, Bradshaw D, Hasell T, Chong SY, Tang C, Thompson S, Parker J, Trewin A, Bacsa J, Slawin AMZ, Steiner A, Cooper AI. Porous organic cages. Nat Mater. 2009;8:973–978. doi: 10.1038/nmat2545. [DOI] [PubMed] [Google Scholar]
- 121.Mitra T, Jelfs KE, Schmidtmann M, Ahmed A, Chong SY, Adams DJ, Cooper AI. Molecular shape sorting using molecular organic cages. Nat Chem. 2013;5:276–281. doi: 10.1038/nchem.1550. [DOI] [PubMed] [Google Scholar]
- 122.Brutschy M, Schneider MW, Mastalerz M, Waldvogel SR. Porous organic cage compounds as highly potent affinity materials for sensing by quartz crystal microbalances. Adv Mater. 2012;24:6049–6050. doi: 10.1002/adma.201202786. [DOI] [PubMed] [Google Scholar]
- 123.Jana D, Das S, Mani G. Self assembled macrobicycle and tricycle cages containing pyrrole rings by dynamic covalent chemistry method. J Incl Phenom Macro. 2015;82:461–470. [Google Scholar]
- 124.Bondi A. Van Der Waals volumes + radii. J Phys Chem. 1964;68:441–451. [Google Scholar]
- 125.Guchhait T, Mani G, Schulzke C, Anoop A. A tripyrrolylmethane-based macrobicyclic triazacryptand: X-ray structure, size-selective anion binding, and fluoride-ion-mediated proton-deuterium exchange studies. Inorg Chem. 2012;51:11635–11644. doi: 10.1021/ic301546v. [DOI] [PubMed] [Google Scholar]
- 126.Setsune J, Watanabe K. Cryptand-like porphyrinoid assembled with three dipyrrylpyridine chains: synthesis, structure, and homotropic positive allosteric binding of carboxylic acids. J Am Chem Soc. 2008;130:2404–2405. doi: 10.1021/ja710424n. [DOI] [PubMed] [Google Scholar]
- 127.Kang SO, Day VW, Bowman-James K. Cyclophane capsule motifs with side pockets. Org Lett. 2008;10:2677–2680. doi: 10.1021/ol800762j. [DOI] [PubMed] [Google Scholar]
- 128.Kang SO, Day VW, Bowman-James K. Tricyclic host for linear anions. Inorg Chem. 2010;49:8629–8636. doi: 10.1021/ic1013297. [DOI] [PubMed] [Google Scholar]
- 129.He Q, Kelliher M, Bahring S, Lynch VM, Sessler JL. A bis-calix[4]pyrrole enzyme mimic that constrains two oxoanions in close proximity. J Am Chem Soc. 2017;139:7140–7143. doi: 10.1021/jacs.7b02329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Valderrey V, Escudero-Adan EC, Ballester P. Polyatomic anion assistance in the assembly of [2]pseudorotaxanes. J Am Chem Soc. 2012;134:10733–10736. doi: 10.1021/ja301900s. [DOI] [PubMed] [Google Scholar]
- 131.Saha I, Lee JH, Hwang H, Kim TS, Lee CH. Remarkably selective, non-linear allosteric regulation of anion binding by a tetracationic calix[4] pyrrole homodimer. Chem Commun. 2015;51:5679–5682. doi: 10.1039/c5cc00487j. [DOI] [PubMed] [Google Scholar]
- 132.Park CH, Simmons HE. Macrobicyclic amines .III. Encapsulation of halide ions by in,in-1,(k+2)-diazabicyclo[k.l.m]alkane-ammonium ions. J Am Chem Soc. 1968;90:2431–2432. [Google Scholar]
- 133.Gale PA, Howe ENW, Wu X. Anion receptor chemistry. Chem. 2016;1:351–422. [Google Scholar]


