Abstract
MicroRNA (miRNA) in urine has been considered as a potential biomarker for early-stage diagnosis of multiple diseases like urinary system cancer, kidney injury and diabetes, owing to their many demonstrated advantages including long-term stability and noninvasiveness. However, the traditional enrichment and extraction processes of miRNAs from urine are cumbersome and tedious due to the low concentration and multiple carriers of miRNAs. Herein, we present a novel method to collect low concentrations of miRNAs from dilute solutions such as urine and cell culture medium. 10-nm core sized magnetic nanoparticles with carboxylic acid coating can adsorb low-concentration proteins, and form protein corona which makes them easy to aggregate and precipitate for subsequent isolation. In urine and cell culture medium, these nanoparticles can aggregate with proteins, including miRNAs-associated protein Argonaute 2 and microvesicle-related proteins, to form precipitates, so that miRNAs can be easily extracted from pellets by small amount of lysis buffer for subsequent analysis such as real-time PCR. Our method provides a facile way to enrich miRNAs from biofluids without the need of ultracentrifugation and immunoprecipitations, bringing remarkable convenience to miRNAs-based biomarker research.
Keywords: microRNA extraction, magnetic nanoparticles, carboxylic acid functionalized, protein corona, cell-free biofluid
Graphical abstract
1. INTRODUCTION
MicroRNAs (miRNAs) are a group of short non-coding RNA molecules that function in RNA silencing and post-transcriptional regulation of gene expression. They can be found in plants, animals, and some viruses, with well-conserved homology. Different sets of miRNAs are expressed in various cell types and tissues, which are involved in many biological processes including cell proliferation, apoptosis, metabolism and inflammatory. Aberrant miRNA expression profiles are also related to cell dysfunction and disease states1–5. Although most miRNAs are inside cells, some miRNAs, known as extracellular miRNAs, have also been found in the extracellular environment, including various biological fluids, such as serum, urine, breast milk and saliva. These extracellular miRNAs have the potential to become biomarkers for diseases6–8. Analysis of the miRNAs profile in biofluids will provide important insights into the physiological and pathological status of the respective organs. For instance, the decline of miR-145 expression level have been proved to be able to distinguish bladder cancer patients from non-cancer controls in cell-free urine samples5; the elevation of miR-10a and miR-30d was also observed in urine samples from patients with focal segmental glomerulosclerosis9.
As small RNA molecules, biofluidic miRNAs are remarkably stable than mRNAs10–11. This is probably because miRNAs are associated with Argonaute 2 (Ago2)-containing protein complexes or get packaged inside exosomes and microvesicles, and are thus protected from being digested by RNAses12. Although exosomes are a rich source of circulating microRNAs, only approximately 10% of extracellular microRNAs are packaged inside exosomes. In fact, 90% of miRNAs in the circulation is expected to be present in a non-membrane-bound form based on analysis with differential centrifugation and size-exclusion chromatography12–14. However, the ultracentrifugation-based method for biofluidic miRNA isolation is limited to the miRNAs existing in exosomes or microvesicles, leaving the Ago2-bound miRNAs remaining in the supernatant. Co-immunoprecipitation also can be used to collect miRNAs associated proteins, but it is costly and time-consuming. In addition, extraction of miRNAs from water in dilute biofluids like urine or cell culture medium will require the increased usage of chaotropic and precipitant reagents, leading to wastes and cumbersome operations. Therefore, an easy and efficient way to enrich and collect miRNAs from dilute biofluids will be valuable to support miRNAs-based biomarker research.
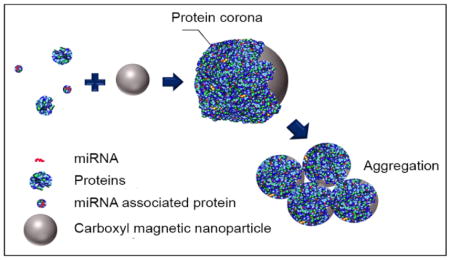
Nanotechnology is holding great promise for diseases diagnosis, targeted delivery of drugs, and biosensors15–17. Recently, it has been demonstrated that proteins bind to the surface of nanoparticles when nanomaterials are introduced to biological fluids, to form clouds of adsorbed proteins known as ‘protein corona’18–19. Although the proteins bind to nanoparticles by noncovalent interactions, their dissociation from nanoparticles might require a combination of detergents, heat, sonication, repeated washings and even acids to chemically dissolve the nanoparticles20. This binding phenomenon could be effectively utilized to capture certain proteins from solutions and biofluids. In this work, we used carboxylic acid functionalized magnetic nanoparticles (MNP) to collect and enrich proteins in urine and cell culture medium. The protein-MNPs complexes contained multiple types of proteins and even small vesicles. And the colloidal stability reduced after protein corona formed, which made the complexes precipitate from solution easily. Moreover, miRNAs were extracted from the pellets by Trizol/chloroform method and analyzed by real-time PCR. The data presented in this study may provide a novel method to extract miRNA from dilute biofluids and cell culture medium, which make the procedure efficient and simple.
2. MATERIAL AND METHOD
2.1 Cell culture
RAW264.7 cells were obtained from Sigma and grown in Dulbecco’s Modified Eagle’s Medium (DMEM) (Cat. 11960-044, Invitrogen) supplemented with 10% heat-inactivated fetal bovine serum (FBS), 4 mM L-Glutamine and penicillin/streptomycin at 37°C in 5% CO2. Cells were split when sub-confluent to 70–80% and dislodged from the flask substrate with a cell scraper. To obtain lipopolysaccharides (LPS, Escherichia coli 0127: B8, Sigma) stimulated cell culture medium, cells were plated into 6-well plates the day before LPS treatment. To obtain cell conditional culture medium, FBS-free DMEM was used to culture cell with or without 10 ng/ml of LPS. Culture medium was collected after incubation with cells for 4, 8 and 16 hours. The freshly collected medium was immediately centrifuged at 2,000 g at 4°C for 10 mins to remove cells and debris and the supernatant was aliquoted and stored at −80°C for subsequent experiments.
2.2 Rat urine sample collection
Adult Sprague Dawley male rats (6–8 weeks, 280–320 g) were used and cared for in the University Laboratory Animal Resources facility of Michigan State University. All animal care procedures and experimental protocols have been approved by the Institutional Animal Care and Use Committee (IACUC) of Michigan State University. For urine collection, a single rat was put into an Allentown cage with a grid frame at the bottom to separate the animal from its excreta that was served with water only. After 3–4 hours, 4 to 8 ml of urine could be collected at the bottom of the cage. The fresh urine samples were immediately centrifuged at 2,000 g at 4°C for 10 mins to remove cells and debris. Supernatants were aliquoted and stored at −80°C for subsequent experiments. The urinary protein concentration of each sample was measured by rat urine protein assay kit (Chondrex, WA).
2.3 MNPs and dynamic light scattering measurement
MNPs with carboxylic acid and amine surface coatings were obtained from Sigma with a 10-nm inorganic core surrounded by a monolayer of oleic acid and a monolayer of amphiphilic polymer. MNPs with 20-nm inorganic core and dextran surface coating were synthesized as described previously21. The hydrodynamic radii and zeta potential of each types of MNPs in distilled water or urine were measured by dynamic light scattering (DLS) analyzer Zetasizer (Malvern). 50 μg of each type of MNPs were put into 1 ml of water or rat urine, and measured after brief mixing. The proteins in urine was excessive based on a preliminary test.
2.4 Incubation of MNPs in protein solution
The cell lysate was prepared from RAW264.7 cells treated with NP-40 buffer (150 mM sodium chloride, 1.0% NP-40, 50 mM Tris pH 8.0). Protein concentration was estimated using Bradford Reagent (Cat. B6916, Sigma). To make dilute protein solutions, 10 μg of total cell lysate was added into 1 ml of distilled water. 50 μg of carboxylic acid-coated MNPs (MNPs-COOH), 50 μg of amine-coated MNPs (MNPs-NH2) and 100 μg of dextran-coated MNPs (MNPs-Dextran) were added to the protein solution, respectively. As the size of MNP-dextran is nearly twice the other two types of particles, twice the amount of MNP-dextran was used to ensure the similar surface area for all MNPs in solution. After incubation at room temperature for 10 mins, the MNPs were isolated from the solution by high-speed centrifuge (20,000 g) for 15 mins, before the pellets and the supernatants were collected for subsequent experiments.
2.5 Quantification of urine protein adsorption by MNPs
To determine the amount of urine protein adsorption by MNPs, 40 μg of MNPs with each type of coating was added into a 1.5-ml Eppendorf tube containing 20 μl of rat urine protein standard (4 mg/ml, Chondrex, WA), or 20 μl of phosphate-buffered saline (PBS) as baseline controls. The total volume was adjusted to 100 μl with PBS. The amount of protein well exceeded the amount that MNPs can adsorb which was measured by a preliminary test. All samples were incubated at room temperature for 10 mins followed by centrifugation at 20,000 g for 15 mins. The supernatant was carefully transferred into a 96-well C-bottom microtiter plate. After incubation with 250 μl of 3% sulfosalicylic acid at room temperature for 10 mins, the absorbance was measured at 450 nm using a microplate reader (Tecan Infinite M1000). Urine protein standard curve was made following instructions provided by Chondrex. The net amount of urine protein in the supernatant was corrected by baseline controls and calculated according to the standard curve. The amount of protein adsorbed by MNPs was calculated by subtracting the amount of protein in supernatant from that in total. The assay was performed in replicates.
2.6 Western blot
Urine samples collected from different rat were collected and aliquoted. Rat urine proteins were extracted either by lyophilization or MNPs pull down. To lyophilize the protein, 0.5 ml of rat urine sample frozen at −80°C was put into freeze-drying chamber and lyophilized for 24 hours until all liquids had evaporated. To reconstitute these proteins, 100 μl of distilled water was added and thoroughly vortexed. After boiling with 2× laemmli buffer (Thermal Fisher) for 5 mins, the sample was ready for SDS-PAGE electrophoresis. For MNPs-COOH treated sample, sufficient amount of MNPs-COOH was added into the same aliquoted urine sample according to its adsorption capability and incubated at room temperature for 5 mins. After aggregations formed in solution, pellets can be isolated from solution using low-speed centrifuge (1,000 g). Before SDS-PAGE electrophoresis, the pellets were treated with 30 μl of 2× laemmli buffer and dispersed thoroughly by pipetting up and down. The mixture was boiled for 5 mins before loading. Argonaute 2 (Ago2) antibody (Cat. 2897, C34C6, Cell Signaling Technology) was used for detection, while anti-TSG101 antibody (SAB2702167, Sigma) and anti-CD9 antibody (ab92726, Abcam) were used to identify the presence of exosomes. After the fluorescent image was obtained, the signal intensity was analyzed with Odyssey Fc Imaging System.
2.7 MiRNA extraction, reverse transcription, and real-time PCR
Total miRNA was extracted by the following methods. (1) MNPs-COOH that had been incubated with 0.5 ml of rat urine or 0.5 ml of conditional cell culture medium were pelleted with low-speed centrifuge (1,000 × g) after aggregations were formed. Then the pellets were isolated and treated with 300 μl of Trizol at room temperature for 5 mins. The mixture was dispersed thoroughly by pipetting up and down before 60 μl of chloroform was added. (2) 500 μl of urine not treated with MNPs was directly incubated with 2.5 ml of Trizol for 5 mins at room temperature before 500 μl of chloroform was used. (3) 5 ml of urine was ultra-centrifuged at 150,000 × g for 60 min as described previously22, before the crude exosomes and shed microvesicles were pelleted and incubated with 300 μl of Trizol for 5 mins at room temperature before 60 μl of chloroform was added. Following chloroform treatment in the above-mentioned three methods, the mixture was centrifuged at 12,000 × g at 4°C for 15 mins to separate the phases. The upper, aqueous phase was carefully transferred into a new tube, mixed with 1.5 times the volume of ethanol before passing through the miRNeasy mini spin column (Cat. 217004, QIAGEN). The miRNAs in the solution bound to the silica membrane of the column while other contaminants were washed away. Total miRNAs were then eluted with RNase-free water and quantified by NanoDrop 2000c.
To compare the efficiency of miRNA extraction with or without protein enrichment by MNPs-COOH, miRNA isolation was also performed by using two commercial kits, miRNeasy kit (Qiagen, Cat.217004) and urine microRNA purification kit (Norgen Biotek Cat.29000), following their instructions. The urine samples aliquot was incubated with sufficient MNPs-COOH at room temperature for 5 mins, the aggregated pellets were then separated from the solution by a flat magnet. The total miRNA extracted from these pellets and the original aliquot samples by different kits were quantified.
After quantification, miRNA samples were reverse transcribed into cDNA by TaqMan Advanced miRNA cDNA Synthesis Kit (Thermo Fisher Scientific). Subsequently, individual miRNAs were amplified by PCR using TaqMan® Advanced miRNA Assays for miR-10a (Assay ID, mmu479241_mir), miR-16 (Assay ID, rno481312_mir), mir-30d (Assay ID, rno478606_mir) and miR-19a (Assay ID, mmu479228_mir), with u6 (Assay ID, 001973) for normalization.
2.8 Statistical analysis
All data were presented as the mean ± s.e.m. The statistical significance of the differences between mean values for different samples was evaluated by Student’s t-test with a paired, two-tailed distribution. The difference was considered significant when the P value was less than 0.05 (*) or 0.01 (**).
3. RESULTS
3.1 Protein corona formed around MNPs in urine
In biological environments, nanoparticles are enshrouded by a layer of proteins named protein corona. There has been much effort to detect the protein corona and to understand its formation in terms of nanoparticles and protein properties. Although protein corona has been reported when nanoparticles are introduced in serum, it is still not clear whether protein corona can be formed around MNPs in urine or dilute protein solutions. To determine the effect of protein adsorption, three types of MNPs with different surface coating were incubated in distilled water and rat urine, and the hydrodynamic radii and zeta potential of each types of MNPs were analyzed by DLS. The characteristic of these MNPs is listed in Table 1. Compared to MNPs incubated in water, the changes of MNPs’ size and zeta potential in urine indicated that all these MNPs can form protein corona. The increase in MNPs’ hydrodynamic sizes is due to the formation of protein coating layer on their surfaces, making them negatively charged in solution.
Table 1.
Summary of MNPs measured in water and rat urine
| Nanoparticle | Zeta potential (mV) | Hydrodynamic diameter (nm) | ||
|---|---|---|---|---|
| Water | Urine | Water | Urine | |
| MNPs-COOH | −38.1 ± 2.7 | −19.4 ± 3.6 | 44.92 ± 8.3 | 63.80 ± 29.3 |
| MNPs-NH2 | 17.4 ± 3.9 | −10.5 ± 2.9 | 42.20 ± 8.6 | 52.68 ± 7.1 |
| MNPs-dextran | −16.5 ± 4.4 | −13.8 ± 2.5 | 84.37 ± 23.3 | 104.6 ± 29.5 |
We also tested the protein adsorption capability of MNPs in a dilute protein solution prepared by dissolving 10 μg of cell lysate from RAW264.7 in 1 ml of PBS. 50 μg of MNP-NH2 or MNP-COOH, or 100 μg of MNP-dextran was added into dilute protein solution at room temperature for 10 mins, then the pellets of MNPs were isolated by high-speed centrifuge (20,000 g) for 15 mins. The pellets were treated with 30 μL of 2X laemmli buffer, loaded on the SDS-PAGE gel and stained by Coomassie blue after electrophoresis. 20 μl of the dilute protein solutions were used as input. Since the detection limit is around 100 ng for each band with Coomassie brilliant blue R-250 staining, there is no visible band in the lane of input. In contrast, strong bands appeared in the lanes with MNPs-protein pellets collected from protein solutions (Figure 1a). Although all these MNPs could pull down proteins from dilute solutions, MNP-COOH demonstrated the greatest protein adsorption capability.
Figure 1.
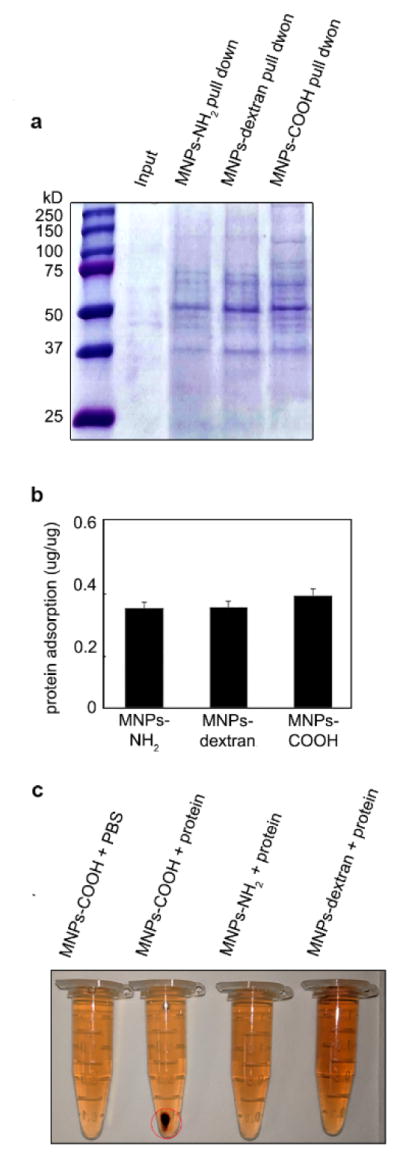
Protein corona formed around MNPs in urine. a, Protein corona forms on MNPs in dilute protein solution. 50 μg of MNPs-NH2, 50 μg of MNPs-COOH or 100 μg of MNPs-dextran was added into 10 μg/ml of dilute protein solution and pelleted by centrifugation. 20 μl of protein solution was used as input. Coomassie brilliant blue staining was used to visualize proteins in pellets on the SDS-PAGE gel with Molecular mass labeled to the left side. b, Quantity of urine protein adsorbed by three types of MNPs. c, Colloidal stability of MNPs in protein solution. Aggregation (red circle) was found in the tube containing MNPs-COOH and protein.
To quantify the protein adsorbed by MNPs in rat urine, rat urine protein assay kit was used to measure the protein amount in the supernatant before and after the protein solution was incubated with the MNPs. The amount of protein adsorbed was calculated by subtraction method and shown in Figure 1b and Table 2. The protein adsorbed on MNPs-COOH is more than that of the other two. However, the protein adsorption capability of these three types of MNPs is not significantly different in this assay, because the protein standard used in this assay contains limited types of urine proteins, such as albumin. Nevertheless, it provides an approximate estimate of protein adsorption of each MNPs.
Table 2.
The quantity of urine proteins adsorbed by MNPs
| NMPs | MNP-NH2 | MNP-dextran | MNP-COOH |
|---|---|---|---|
| Protein adsorption(μg/μg) | 0.31±0.05 | 0.33±0.04 | 0.37±0.05 |
To identify the protein composition in the three MNPs’ corona, LC-MS/MS assay was applied. The proteins detected in protein corona around each type of MNPs were summarized (Table 3). Due to the different functional group on the surface of MNPs, the composition of proteins in each corona varies. Although the size and pI range of these proteins are almost the same, the protein corona from MNP-COOH contained the most types of proteins. The method for LC-MS and detailed data was showed in the supplement material and Table S3.
Table 3.
Summary of the protein character in protein corona around each type of MNPs*
| Protein types | MW | pI | |
|---|---|---|---|
| MNP-COOH | 490 | 7.4–532.1 | 4.34–11.52 |
| MNP-NH2 | 300 | 7.4–571.0 | 4.56–11.52 |
| MNP-Dextran | 293 | 7.4–521.0 | 4.58–11.52 |
The results were filtered with a peptide-level FDR less than 1%. The protein grouping was enabled.
An interesting fact we noticed in these experiments is that the 10-nm (core-size) carboxyl MNPs, unlike MNPs coated with amine and dextran, were prone to aggregate in protein solutions when their concertation reached a critical value (Figure 1c). This phenomenon can be reproduced when MNPs-COOH was introduced to urine and cell culture medium. When the MNPs-COOH concentration was gradually increased in urine, aggregation formed when the amount of MNPs approached, or slightly exceeded the amount needed to adsorb all the protein in solution. This can be explained by the decreased colloidal stability of carboxyl MNPs with protein corona formed on their surface, or the bridging effects that a macromolecule interacts with more than one NP and therefore leads to the formation of aggregates23–25. This character of MNPs-COOH enables easier separation of MNPs-protein complexes from dilute protein solutions. In our experiments, these pellets could be isolated from solutions simply by a flat magnet or two-minute low speed centrifugation (1,000 × g).
Another favorable feature of carboxylic acid coated MNPs is that they are inert to direct nucleotide binding. This is because the net negative charge in carboxylic acid-functionalized coatings prevent their interactions with negatively-charged DNAs26–27. These properties of carboxylic acid-functionalized MNPs can also be very helpful in miRNA extraction from biofluids. We also tested the nucleotides binding capability of MNPs-COOH and ascertained that no detectable nucleotides can be pulled down directly by these MNPs (supplement Figure S1). Because of their compatibility with subsequent procedures for miRNA extraction, carboxylic acid-functionalized MNPs were chosen for the rest of our experiments.
To test the reproducibility of the protein pulldown by MNPs-COOH, different samples were tested including urine samples from rats and human volunteers with pH value varies from 4.0 to 9.0, and cell culture medium from several cell lines (Raw264.7, HEK293 and HT29). Protein corona could form in each sample and miRNA could be extracted from these MNPs-protein pellets. It should be noted that weak alkali condition (pH 8–9.5) contributed to MNPs-protein complex aggregate in solutions.
3.2 Carboxylic acid-functionalized MNPs enrich proteins in urine
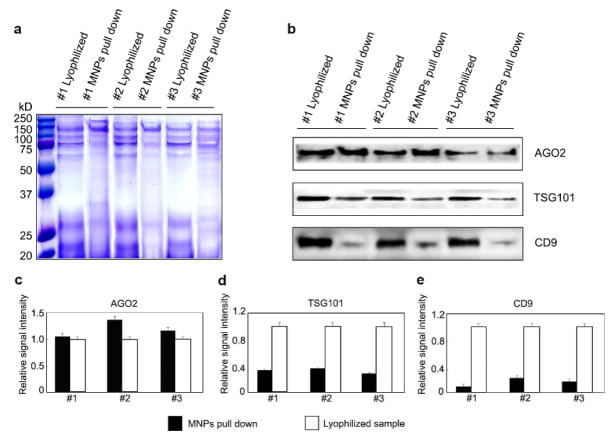
To determine the capability of the MNPs-COOH to pull down proteins from rat urine samples, proteins samples from MNPs-COOH protein pellets and lyophilized powder obtained from the same aliquoted rat urine samples were compared in SDS-PAGE gel. Urine samples from three different rats were collected and aliquoted for analysis. Protein concentration of each sample was measured by rat urinary protein assay kit. 50 μg of lyophilized urine protein was reconstituted. Same aliquoted urine samples containing 50 μg of protein were incubated with sufficient MNPs to obtain MNPs-protein pellets. Coomassie blue staining was performed to visualize protein bands that correspond to the two approaches (Figure 2a). Although protein bands could be detected in both lyophilized samples and MNPs pellets, the band patterns were different between MNPs-protein pellets and the corresponding lyophilized samples, suggesting certain protein-adsorbing preference of MNPs-COOH.
Figure 2.
MNPs-COOH enrich proteins in urine. a, Protein patterns in lyophilized urine samples and MNP-protein pellets. Coomassie brilliant blue staining was used to visualize proteins in pellets on the SDS-PAGE gel. Molecular mass is labelled to the left side. b, Immunoblot analysis of Ago2, TSG101 and CD9 in lyophilized urine samples and MNP-protein pellets. Band signal intensity analysis of Ago2, TSG101 and CD9 were shown in c, d and e. Band signal intensity was obtained by odyssey imaging system. In each group, band signal intensity was normalized by the value of lyophilized sample. AGO2, Argonaute-2; TSG101, Tumor susceptibility gene 101; CD9, CD9 antigen.
Since most of the cell-free miRNAs in urine are protected in Ago2-contained protein complexes or wrapped in microvesicles such as exosomes, we next asked whether these proteins could be found in MNPs-protein pellets. Western blot was performed in three groups of urine samples from three different rats to detect miRNA binding protein Ago2, while anti-TSG101 and anti-CD9 antibody were used to detect exosomes. These proteins were detected in both lyophilized urine samples and MNPs-COOH precipitated pellets (Figure 2b). The quantitative signal intensity showed similar amounts of Ago2 in lyophilized samples and MNPs pellets, indicating Ago2 protein in urine was adsorbed by MNPs completely (Figure 2c). On the other hand, only around 30% of TSG101 and 20% of CD9 could be detected in MNPs pellet compared with lyophilized samples in each group, suggesting that only a portion of exosomes were pulled down by MNPs (Figure 2d, 2e). We also label the exosomes with fluorescent dye and measured the fluorescence intensity of urine samples before and after MNPs treatment. The ratio of exosomes adsorption by MNPs increased with the amount of MNPs added (Table S1). These data indicate that MNPs-COOH can pull down miRNA-associated proteins and exosomes from rat urine, but with better efficiency for Ago2. In fact, exosomes pulling down by MNPs from urine is a complex process, in which the amount of absorbed exosomes increases nonlinearly with the amount of MNPs.
3.3 Urine miRNA extraction from MNPs-protein pellet
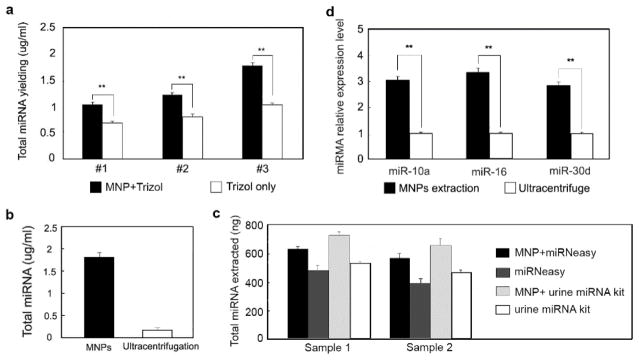
Since miRNA-binding proteins were found in MNPs-protein pellets isolated from rat urine, we further hypothesized that miRNAs could be extracted from these pellets. Trizol and chloroform were used to isolate miRNA from three urine samples collected and aliquoted from different rats. To eliminate the interference of MNPs that had strong absorbance at OD 230–290 nm (Supplement Figure S2), miRNeasy mini spin columns were used to purify miRNA. In comparison, miRNA extraction was also performed by adding Trizol reagent directly into urine samples. The urine protein concentration and the total amount of miRNA were shown in Table 4 and Figure 3a. The extraction yield of total urine miRNA increased by 30–50% when MNPs were used to enrich proteins with reduced reagent use and operating time.
Table 4.
Summary of total miRNA extract from urine samples
| Samples number | Protein concentration (μg/ml) | MNPs-COOH applied (μg/ml) | Total miRNA (μg/ml) | |
|---|---|---|---|---|
| MNPs enrichment | Trizol directly | |||
| #1 | 183.1 ± 0.6 | 500 | 1.79 ± 0.04 | 1.04 ± 0.03 |
| #2 | 114.0 ± 0.4 | 310 | 1.05 ± 0.03 | 0.69 ± 0.04 |
| #3 | 128.2 ± 0.5 | 350 | 1.23 ± 0.04 | 0.80 ± 0.06 |
Figure 3.
Urine miRNA extraction from MNPs-protein pellet. a, Total miRNA yield comparison among urine samples with or without MNP-COOH enrichment. In each group, the MNPs treatment can significantly increase total miRNA yield by at least 50%. b, Total miRNA yield comparison between MNPs treated urine sample and the control sample treated by ultracentrifugation. c, Total miRNA yield comparison from two commercial kits with or without protein enrichment by MNPs. d, Real-time PCR was performed using miRNA extracted from pellets obtained from MNPs pull down or ultracentrifugation. MiR-10a, miR-16, miR-30d and U6 were measured using TaqMan advanced miRNA assays. The relative expression of each miRNA was normalized to U6, then compared with the value obtained from ultracentrifuged sample. The relative quantity of miRNA extracted from MNPs pull down pellets is higher than that from ultracentrifugation pellets with significant difference.
We also compared the extraction yield of miRNAs from MNPs pellets and ultracentrifugation pellet of sample #1. Total amount of miRNA extracted from 5 ml of ultracentrifuged urine pellet was 0.8 ± 0.02 μg, which corresponded to 0.16 μg of total miRNA in crude exosome from 1 ml of urine (Figure 3b). This accounted for only 9.0 % of the total miRNA yield from MNPs pellets, which was consistent with the previous report 12–13.
To test the improved miRNA extraction efficiency of MNPs, we carried out miRNA isolation using two commercial kits, miRNeasy kit (Qiagen, Cat.217004) and urine microRNA purification kit (Norgen Biotek Cat.29000), with or without protein enrichment by MNPs. Although both kits could work well in urine miRNA extraction, the additional step of protein enrichment by MNPs could improve miRNA yield by 25–30% (Fig. 3c). This step also saved reagents and time spent in miRNA extraction significantly (Table S2).
Reverse transcription was performed by TaqMan advanced miRNA cDNA synthesis kit on the total miRNAs extracted from sample #1, with MNPs enrichment or ultracentrifugation. Real-time PCR was carried out using TaqMan advanced miRNA assays for miR-10a, miR-16, and miR-30d. These urine miRNAs have been reported to be related to kidney diseases. U6 was chosen to be a reference miRNA as it had been shown to conserve and exist in urine. All these miRNAs got stable amplification. The relative expression level of each miRNA was shown in Figure 3c and Table 5. In addition, the relative expression level of each miRNA extracted from MNPs pellets was about three times higher than that obtained by ultracentrifugation, indicating the majority of miRNAs were in Ago2-bound state. Collectively, these data indicate that MNPs can enrich miRNAs from urine and improve miRNA isolation efficiency.
Table 5.
Relative expression ratio of miRNAs from MNP pellets in comparison with ultracentrifuge
| miRNA | miR-10a | miR-16 | miR-30d |
|---|---|---|---|
| MNP/ultracentrifuge | 3.04±0.04 | 3.34±0.05 | 2.85±0.04 |
3.4 MNPs pull down method can be applied to collect miRNAs from cell culture medium
During the cell culture process, miRNAs could be released to the culture medium which are considered to be related to cell-cell communication, immune response, and other biological processes28. MiRNAs in cell culture medium can be found in stem cells, cancer cell, neuro cell, and in cells from immune system such as macrophages. It can be used to analyze the developmental stage of stem cells and to study the drug effects on cells29. For proof-of-concept demonstration, Raw264.7, a mouse macrophage cell line, was cultured to harvest cell conditioned medium (treated with/out LPS) for miRNA analysis. After Raw264.7 cells grew to 80% confluence, cell culture medium was changed to FBS-free DMEM, with or without LPS. The LPS-free cell culture medium was used as control samples that were collected 8 and 16 hours after the medium was changed. Cell conditional medium was LPS-contain culture medium that was collected at 4, 8, and 16 hours after LPS treatment. Excessive amount of MNP-COOH was added into the control and conditioned cell culture medium, respectively, and aggregation was observed within 5 minutes. To characterize protein adsorption of MNPs in cell culture medium, coomassie brilliant blue staining was applied to SDS-PAGE gel. As shown in Figure 4a, proteins were captured by MNP-COOH. Although the protein patterns from both control and conditional medium were similar, protein intensity was higher in conditional medium, indicating more proteins secreted from Raw264.7 after LPS stimulation which could be adsorbed by MNPs. To identify miRNA-associated proteins in MNPs pellets, western blot was also performed. Ago2, TSG101 and CD9 were detected in MNPs pellets from both control and conditional medium, and the amount of miRNA-associated proteins is higher in LPS stimulated conditional medium than that in control (Figure 4b).
Figure 4.

MiRNA in cell culture medium can be extracted from MNP-protein complex. a, Protein contained in MNP-protein complex was shown on coomassie brilliant blue staining SDS-PAGE gel with molecular mass indicated on the left, indicating the protein band pattern in each group. Protein amount in LPS treated medium is slightly higher than that in control. b, Immunoblot analysis of Ago2, TSG101 and CD9 in MNP-protein pellets from control or LPS treatment medium. c, miR-19a was measured using TaqMan advanced miRNA assays and normalized by U6. The relative expression of miR-19a of all samples were normalized by U6 then compared with control. The expression level of miR-19a increased with prolonged LPS treatment time.
To extract miRNAs from cell culture medium, FBS-free DMEM incubated with RAW264.7 for 8 hours was collected, and LPS containing cell culture medium was collected at different time intervals. Excessive amount of MNPs was incubated with control and LPS conditional medium. Aggregated pellets were separated followed by Trizol treatment. MiRNA was purified by miRNeasy mini spin column. After reverse transcription with TaqMan advanced miRNA cDNA synthesis kit, miR-19a and U6 were analyzed by real-time PCR using TaqMan advanced miRNA assays. The miR-19a expression level gradually increased in cell conditional medium after LPS stimulation after 4h to 16h (Figure 4c). After LPS treatment for 16h, the miR-19a in culture medium was 1.8-fold higher than that in control, which was consistent with previous reports, demonstrating the effectiveness of MNPs to extract miRNAs from cell culture medium30.
4. DISCUSSION
MiRNAs have been considered as a stable biomarker that can provide a wealth of information in recent years. Although most miRNAs are kept in cells, there are still plenty of miRNAs found in extracellular environment, such as serum, urine and breast milk. MiRNAs in urine have been exploited as a potential biomarker to diagnose multiple diseases like urinary system cancer, kidney injury and diabetes, and they have already been demonstrated many advantages as stable and noninvasive biomarkers for early stage detection31–34. However, the traditional method to extract miRNA from urine is cumbersome and costly. In our experiment, when Trizol was used directly to extract miRNA from urine, water in urine diluted Trizol, necessitating a larger amount of reagent to maintain the proper chaotropic agent concentration and ionic strength required for miRNA binding to the purification column. In contrast, enrichment of macromolecule from urine by MNPs will reduce agent use and make the procedure as straightforward as miRNA extraction from homogenized tissues.
Protein corona is a phenomenon that proteins in biofluids adsorb to the surface of nanoparticles, forming a cloud of protein layer. Due to its immense complexity, it is hard to predict the composition of protein corona of each type of nanoparticle. The properties of nanoparticle, such as surface chemistry, size, shape and material, can affect the composition of its protein corona35–36. Although protein corona forms because of non-covalent binding, it has enough stability to sustain reasonable levels of detergents, heat, sonication and repeated washings, enabling its practical utilization as a tool to preferentially enrich certain proteins. To the best of our knowledge, no one has utilized “protein corona” to enrich proteins in dilute bio-fluids or cell culture medium and extract miRNAs from it. After comparing several nanoparticles, we found the 10-nm (core size) magnetic nanoparticles, with carboxylic acid functional group coating, have many advantages for miRNA extraction from urine and cell culture medium: this type MNPs adsorb the largest amount of proteins in dilute protein solutions and they tends to form aggregate when covered by protein corona due to reduced colloidal stability. Colloidal stability is related to net surface charge, ionic strength, the composition of colloidal material and the adsorption of macromolecules, etc. DLVO theory combines the effect of van der Waals and double layer force to explain the aggregation of aqueous dispersions. According to DLVO theory, repulsive electrostatic double-layer force will decrease with the reduction of surface potential, and attractive Van der wales force will increase when the distance between two spheres become very close. When the MNP-COOH are introduced into protein solution, their surface potential is reduced by the formation of a protein layer while some large proteins bind to more than one MNPs and reduce the distance between them, causing the MNPs-protein complex to aggregate. The colloidal instability of carboxyl MNP-protein complex is a feature not found in MNPs with the other two types of coating, which can be utilized to enrich proteins from dilute solutions. In our experiments, these aggregated MNPs-protein complex can be easily isolated from solution by a simple flat magnet or low speed centrifuge (1,000×g). Another favorable character of carboxyl-coated MNPs is that they are inert to nucleotides binding. Although it has been reported that nanoparticles are apt to interact with nucleotides due to their high surface to volume ratio, the net negative charge in carboxylic acid-functionalized coatings prevent their interactions with negatively-charged DNAs26, 37–38. This important feature will enable future applications of carboxyl MNPs in miRNA isolation.
When sufficient amount of MNP-COOH was added to urine or cell culture medium, aggregation formed immediately after thorough mixing, and the formation of larger aggregated pellets required longer incubation time. Since protein corona formed rapidly and the protein composition may vary according to incubation time, we compared the amount of miRNA extracted from MNP-protein pellets collected within 5 min, 15 min and 30 min, and found no significant difference.
Nanoparticle surface charge plays an important role in protein interaction. It has been reported that positively charged nanoparticles prefer to adsorb proteins with isoelectric points (pI) <5.5, while the negative surface charge enhances the adsorption of proteins with pI > 5.539. The urine tested by LC-MS showed that most proteins (387 out of 490) had pI > 5.5. According to previous studies, MNP-dextran adsorbed fewer proteins when incubated in serum40, while MNP with negative surface charge adsorbed more serum proteins than the positive or dextran-coated MNP, leading to higher blood circulation time. Our LC-MS results of MNP-COOH, MNP-NH2 and MNP-dextran were consistent with these previous findings. In addition, the Ago2 protein amount is small in urine, and the protein bound on negatively charged MNP is tight even when washed by strong protein elution. This could explain why Ago2 protein was shown in western blot assay but not in our LC-MS results.
In our experiments, MNP-COOH could pulled down proteins by protein corona formation and spontaneous aggregation. When introduced into biofluids, MNPs adsorb proteins to form protein corona. When the amount of MNP was increased, there were not enough proteins to cover all the MNPs. Instead, some proteins bridged different particles and broke colloidal stability. Our results also showed that exosome pulldown by MNP had a nonlinear relationship (Table S1). Higher exosome binding efficiency of MNP was observed after certain amount of MNP input which implied preferential MNP binding. The situation was complex when MNPs bound to exosomes. Although the surface of exosome had plenty of proteins, they couldn’t help the MNP to form a protein layer based on bridge component. That might be the reason why only a portion of exosomes were pulldown. Nevertheless, carboxyl-coated MNPs were proved to adsorb most of Ago2 protein and a portion of exosomes in urine. This pull-down procedure is more convenient and cheaper than traditional protein pull-down assay.
5. CONCLUSION
In this study, we have discovered that carboxyl-coated MNPs, can enrich miRNA associated proteins and exosomes in a manner of protein corona, thus improve the miRNA extraction efficiency from dilute biofluids and cell culture medium. The colloidal instability of carboxyl MNP-protein complex and the negative surface charge of carboxyl MNP also contribute to the significantly improved miRNA extraction efficiency. Moreover, the qPCR results for miRNAs extracted from MNPs-protein pellets were consistent with the previous research. This magnetic nanoparticle provides a novel tool for miRNA extraction in biofluids and will benefit miRNA biomarkers research.
Supplementary Material
Acknowledgments
The authors greatly acknowledge the technical support from Dr. Kristin Parent, Dr. John Dover, Dr. Jian Hu and Dr. Dexin Sui. This research is currently supported by the NIBIB under award number R00EB016753 and the Department of Radiology at Michigan State University. The content of this paper does not necessarily represent official views of the NIH.
Footnotes
AUTHOR CONTRIBUTIONS
S.X. designed this research and undertook experimentation and data analysis. S.H. contributed to the synthesis of nanoparticles and DLS experiment. X.Z. assisted in urine samples collection. D.C. and L.S. performed LC-MS. S.X, X.H and C.Q wrote the manuscript.
References
- 1.Wendler F, Favicchio R, Simon T, Alifrangis C, Stebbing J, Giamas G. Extracellular vesicles swarm the cancer microenvironment: from tumor–stroma communication to drug intervention. Oncogene. 2017;36(7):877–884. doi: 10.1038/onc.2016.253. [DOI] [PubMed] [Google Scholar]
- 2.Ma H, Morey R, O’Neil RC, He Y, Daughtry B, Schultz MD, Hariharan M, Nery JR, Castanon R, Sabatini K. Abnormalities in human pluripotent cells due to reprogramming mechanisms. Nature. 2014;511(7508):177. doi: 10.1038/nature13551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pua HH, Steiner DF, Patel S, Gonzalez JR, Ortiz-Carpena JF, Kageyama R, Chiou N-T, Gallman A, de Kouchkovsky D, Jeker LT. MicroRNAs 24 and 27 suppress allergic inflammation and target a network of regulators of T helper 2 cell-associated cytokine production. Immunity. 2016;44(4):821–832. doi: 10.1016/j.immuni.2016.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tran ND, Kissner M, Subramanyam D, Parchem RJ, Laird DJ, Blelloch RH. A miR-372/let-7 Axis Regulates Human Germ Versus Somatic Cell Fates. Stem Cells. 2016;34(7):1985–1991. doi: 10.1002/stem.2378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yun SJ, Jeong P, Kim W-T, Kim TH, Lee Y-S, Song PH, Choi Y-H, Kim IY, Moon S-K, Kim W-J. Cell-free microRNAs in urine as diagnostic and prognostic biomarkers of bladder cancer. International journal of oncology. 2012;41(5):1871–1878. doi: 10.3892/ijo.2012.1622. [DOI] [PubMed] [Google Scholar]
- 6.Williams Z, Ben-Dov IZ, Elias R, Mihailovic A, Brown M, Rosenwaks Z, Tuschl T. Comprehensive profiling of circulating microRNA via small RNA sequencing of cDNA libraries reveals biomarker potential and limitations. Proceedings of the National Academy of Sciences. 2013;110(11):4255–4260. doi: 10.1073/pnas.1214046110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Klingenberg M, Matsuda A, Diederichs S, Patel T. Non-coding RNA in hepatocellular carcinoma: Mechanisms, biomarkers and therapeutic targets. Journal of Hepatology. 2017 doi: 10.1016/j.jhep.2017.04.009. [DOI] [PubMed] [Google Scholar]
- 8.McManus DD, Freedman JE. MicroRNAs in platelet function and cardiovascular disease. Nat Rev Cardiol. 2015;12(12):711–717. doi: 10.1038/nrcardio.2015.101. [DOI] [PubMed] [Google Scholar]
- 9.Wang N, Zhou Y, Jiang L, Li D, Yang J, Zhang C-Y, Zen K. Urinary microRNA-10a and microRNA-30d serve as novel, sensitive and specific biomarkers for kidney injury. PloS one. 2012;7(12):e51140. doi: 10.1371/journal.pone.0051140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mall C, Rocke DM, Durbin-Johnson B, Weiss RH. Stability of miRNA in human urine supports its biomarker potential. Biomarkers in medicine. 2013;7(4):623–631. doi: 10.2217/bmm.13.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova-Agadjanyan EL, Peterson A, Noteboom J, O’Briant KC, Allen A. Circulating microRNAs as stable blood-based markers for cancer detection. Proceedings of the National Academy of Sciences. 2008;105(30):10513–10518. doi: 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Arroyo JD, Chevillet JR, Kroh EM, Ruf IK, Pritchard CC, Gibson DF, Mitchell PS, Bennett CF, Pogosova-Agadjanyan EL, Stirewalt DL, Tait JF, Tewari M. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proceedings of the National Academy of Sciences. 2011;108(12):5003–5008. doi: 10.1073/pnas.1019055108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhu H, Fan G-C. Extracellular/circulating microRNAs and their potential role in cardiovascular disease. American journal of cardiovascular disease. 2011;1(2):138. [PMC free article] [PubMed] [Google Scholar]
- 14.Turchinovich A, Weiz L, Langheinz A, Burwinkel B. Characterization of extracellular circulating microRNA. Nucleic acids research. 2011;39(16):7223–7233. doi: 10.1093/nar/gkr254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang X, Liu Y, Yung B, Xiong Y, Chen X. Nanotechnology-Enhanced No-Wash Biosensors for In Vitro Diagnostics of Cancer. ACS nano. 2017 doi: 10.1021/acsnano.7b02618. [DOI] [PubMed] [Google Scholar]
- 16.Wei H, Wang E. Nanomaterials with enzyme-like characteristics (nanozymes): next-generation artificial enzymes. Chemical Society Reviews. 2013;42(14):6060–6093. doi: 10.1039/c3cs35486e. [DOI] [PubMed] [Google Scholar]
- 17.Pushpavanam K, Narayanan E, Chang J, Sapareto S, Rege K. A Colorimetric Plasmonic Nanosensor for Dosimetry of Therapeutic Levels of Ionizing Radiation. ACS Nano. 2015;9(12):11540–50. doi: 10.1021/acsnano.5b05113. [DOI] [PubMed] [Google Scholar]
- 18.Hamad-Schifferli K. How can we exploit the protein corona? Nanomedicine. 2013;8(1):1–3. doi: 10.2217/Nnm.12.179. [DOI] [PubMed] [Google Scholar]
- 19.Tenzer S, Docter D, Kuharev J, Musyanovych A, Fetz V, Hecht R, Schlenk F, Fischer D, Kiouptsi K, Reinhardt C. Rapid formation of plasma protein corona critically affects nanoparticle pathophysiology. Nature nanotechnology. 2013;8(10):772–781. doi: 10.1038/nnano.2013.181. [DOI] [PubMed] [Google Scholar]
- 20.Walkey CD, Chan WC. Understanding and controlling the interaction of nanomaterials with proteins in a physiological environment. Chemical Society Reviews. 2012;41(7):2780–2799. doi: 10.1039/c1cs15233e. [DOI] [PubMed] [Google Scholar]
- 21.Kamat M, El-Boubbou K, Zhu DC, Lansdell T, Lu X, Li W, Huang X. Hyaluronic acid immobilized magnetic nanoparticles for active targeting and imaging of macrophages. Bioconjugate Chem. 2010;21(11):2128–2135. doi: 10.1021/bc100354m. [DOI] [PubMed] [Google Scholar]
- 22.Greening DW, Xu R, Ji H, Tauro BJ, Simpson RJ. A Protocol for Exosome Isolation and Characterization: Evaluation of Ultracentrifugation, Density-Gradient Separation, and Immunoaffinity Capture Methods. In: Posch A, editor. Proteomic Profiling: Methods and Protocols. Springer New York; New York, NY: 2015. pp. 179–209. [DOI] [PubMed] [Google Scholar]
- 23.Gambinossi F, Mylon SE, Ferri JK. Aggregation kinetics and colloidal stability of functionalized nanoparticles. Advances in colloid and interface science. 2015;222:332–349. doi: 10.1016/j.cis.2014.07.015. [DOI] [PubMed] [Google Scholar]
- 24.Moore TL, Rodriguez-Lorenzo L, Hirsch V, Balog S, Urban D, Jud C, Rothen-Rutishauser B, Lattuada M, Petri-Fink A. Nanoparticle colloidal stability in cell culture media and impact on cellular interactions. Chemical Society Reviews. 2015;44(17):6287–6305. doi: 10.1039/c4cs00487f. [DOI] [PubMed] [Google Scholar]
- 25.Biggs S, Habgood M, Jameson GJ. Aggregate structures formed via a bridging flocculation mechanism. Chemical Engineering Journal. 2000;80(1):13–22. doi: 10.1016/S1383-5866(00)00072-1. [DOI] [Google Scholar]
- 26.Goodman CM, Chari NS, Han G, Hong R, Ghosh P, Rotello VM. DNA-binding by Functionalized Gold Nanoparticles: Mechanism and Structural Requirements. Chemical biology & drug design. 2006;67(4):297–304. doi: 10.1111/j.1747-0285.2006.00372.x. [DOI] [PubMed] [Google Scholar]
- 27.Li K, Zhao X, Hammer KB, Du S, Chen Y. Nanoparticles inhibit DNA replication by binding to DNA: modeling and experimental validation. ACS nano. 2013;7(11):9664–9674. doi: 10.1021/nn402472k. [DOI] [PubMed] [Google Scholar]
- 28.Turchinovich A, Weiz L, Burwinkel B. Extracellular miRNAs: the mystery of their origin and function. Trends in biochemical sciences. 2012;37(11):460–465. doi: 10.1016/j.tibs.2012.08.003. [DOI] [PubMed] [Google Scholar]
- 29.Zhang Y, Feng GH, Xu K, Wang L, Cui P, Li Y, Wang C, Teng F, Hao J, Wan HF. A non-invasive method to determine the pluripotent status of stem cells by culture medium microRNA expression detection. Sci Rep-Uk. 2016;6:22380. doi: 10.1038/Srep22380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ortega FJ, Moreno M, Mercader JM, Moreno-Navarrete JM, Fuentes-Batllevell N, Sabater M, Ricart W, Fernández-Real JM. Inflammation triggers specific microRNA profiles in human adipocytes and macrophages and in their supernatants. Clinical epigenetics. 2015;7(1):49. doi: 10.1186/s13148-015-0083-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Snowdon J, Boag S, Feilotter H, Izard J, Siemens DR. A pilot study of urinary microRNA as a biomarker for urothelial cancer. Canadian Urological Association Journal. 2013;7(1–2):28. doi: 10.5489/cuaj.11115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fredsøe J, Rasmussen AK, Thomsen AR, Mouritzen P, Høyer S, Borre M, Ørntoft TF, Sørensen KD. Diagnostic and Prognostic MicroRNA Biomarkers for Prostate Cancer in Cell-free Urine. European Urology Focus. 2017 doi: 10.1016/j.euf.2017.02.018. [DOI] [PubMed] [Google Scholar]
- 33.Debernardi S, Massat NJ, Radon TP, Sangaralingam A, Banissi A, Ennis DP, Dowe T, Chelala C, Pereira SP, Kocher HM. Noninvasive urinary miRNA biomarkers for early detection of pancreatic adenocarcinoma. American journal of cancer research. 2015;5(11):3455. [PMC free article] [PubMed] [Google Scholar]
- 34.Cardenas-Gonzalez M, Srivastava A, Pavkovic M, Bijol V, Rennke HG, Stillman IE, Zhang X, Parikh S, Rovin BH, Afkarian M. Identification, Confirmation, and Replication of Novel Urinary MicroRNA Biomarkers in Lupus Nephritis and Diabetic Nephropathy. Clinical Chemistry. 2017 doi: 10.1373/clinchem.2017.274175. clinchem. 2017.274175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lundqvist M, Stigler J, Elia G, Lynch I, Cedervall T, Dawson KA. Nanoparticle size and surface properties determine the protein corona with possible implications for biological impacts. Proceedings of the National Academy of Sciences. 2008;105(38):14265–14270. doi: 10.1073/pnas.0805135105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tenzer S, Docter D, Rosfa S, Wlodarski A, Kuharev, Rekik A, Knauer SK, Bantz C, Nawroth T, Bier C. Nanoparticle size is a critical physicochemical determinant of the human blood plasma corona: a comprehensive quantitative proteomic analysis. ACS nano. 2011;5(9):7155–7167. doi: 10.1021/nn201950e. [DOI] [PubMed] [Google Scholar]
- 37.You C-C, Chompoosor A, Rotello VM. The biomacromolecule-nanoparticle interface. Nano Today. 2007;2(3):34–43. doi: 10.1016/S1748-0132(07)70085-3. [DOI] [Google Scholar]
- 38.Nel AE, Mädler L, Velegol D, Xia T, Hoek EM, Somasundaran P, Klaessig F, Castranova V, Thompson M. Understanding biophysicochemical interactions at the nano-bio interface. Nat Mater. 2009;8(7):543. doi: 10.1038/nmat2442. [DOI] [PubMed] [Google Scholar]
- 39.Aggarwal P, Hall JB, McLeland CB, Dobrovolskaia MA, McNeil SE. Nanoparticle interaction with plasma proteins as it relates to particle biodistribution, biocompatibility and therapeutic efficacy. Adv Drug Deliver Rev. 2009;61(6):428–437. doi: 10.1016/j.addr.2009.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sakulkhu U, Mahmoudi M, Maurizi L, Salaklang J, Hofmann H. Protein Corona Composition of Superparamagnetic Iron Oxide Nanoparticles with Various Physico-Chemical Properties and Coatings. Sci Rep-Uk. 2014:4. doi: 10.1038/Srep05020. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.