Abstract
Purpose
Ewing sarcoma (ES) and desmoplastic small round cell tumors (DSRCTs) are aggressive sarcomas molecularly characterized by EWSR1 gene fusions. As pathognomonic genomic events in these respective tumor types, EWSR1 fusions represent robust potential biomarkers for disease monitoring.
Methods
To investigate the feasibility of identifying EWSR1 fusions in plasma-derived cell-free DNA (cfDNA) from patients with ES and DSRCT, we evaluated two complementary approaches in samples from 17 patients with radiographic evidence of disease. The first approach involved identification of patient-specific genomic EWSR1 fusion breakpoints in formalin-fixed, paraffin-embedded tumor DNA using a broad, hybridization capture-based next-generation sequencing (NGS) panel, followed by design of patient-specific droplet digital polymerase chain reaction (ddPCR) assays for plasma cfDNA interrogation. The second approach used a disease-tailored targeted hybridization capture-based NGS panel applied directly to cfDNA, which included EWSR1 as well as several other genes with potential prognostic use.
Results
EWSR1 fusions were identified in 11 of 11 (100%) ES and five of six (83%) DSRCT cfDNA samples by ddPCR, whereas 10 of 11 (91%) and four of six (67%) were identified by NGS. The ddPCR approach had higher sensitivity, ranging between 0.009% and 0.018%. However, the hybrid capture–based NGS assay identified the precise fusion breakpoints in the majority of cfDNA samples, as well as mutations in TP53 and STAG2, two other recurrent, clinically significant alterations in ES, all without prior knowledge of the tumor genotype.
Conclusion
These results provide a compelling rationale for an integrated approach using both NGS and ddPCR for plasma cfDNA-based biomarker evaluations in prospective cooperative group studies.
INTRODUCTION
Ewing sarcoma (ES) and desmoplastic small round cell tumor (DSRCTs) are aggressive sarcomas with peak incidences in adolescence and young adulthood.1 Both are characterized by fusions involving the EWSR1 gene on chromosome 22q12.2,3 The fusions in ES involve EWSR1 and a member of the ETS family. Most commonly, the partner gene is ETS family member FLI1, with alternative genes such as ERG, ETV1, ETV4, and FEV less commonly observed.4 In DSRCTs, the fusion involves EWSR1 and the Wilms tumor gene WT1. Given their exquisite specificity for ES and DSRCT, respectively, the detection of these fusions in a tumor biopsy has become a standard part of the diagnostic assessment of patients with these sarcomas.
Over the past several decades, numerous trials for ES have been conducted, leading to current therapeutic standards of care that result in long-term cure for approximately 75% of patients who present with localized disease but only approximately 25% in patients who present with metastatic disease.5,6 DSRCT was only recognized as a distinct malignancy 25 years ago, with the characterization of its pathognomonic oncogenic fusion following several years later.3,7 Despite intensive regimens evaluating combinations of chemotherapy, aggressive debulking surgical approaches, radiation therapy, and autologous stem cell rescue, outcomes for DSRCT remain poor, with locoregional recurrences representing the most common type of relapse.8
The establishment of clinically valid prognostic and predictive biomarkers has been challenging in ES and largely unstudied in DSRCT.9 Specifically, numerous studies have been conducted in an effort to establish platforms to identify and follow subclinical levels of disease burden.10-14 The two strategies most commonly used have been reverse transcription polymerase chain reaction (RT-PCR) evaluation for the EWSR1 fusion transcript from cellular RNA extracted from peripheral blood and/or bone marrow, or, for ES, flow cytometric evaluation of peripheral blood and/or bone marrow cells on the basis of a CD99+ gating strategy. These studies have had varying results and, to date, have not satisfied criteria as suitable clinical biomarkers.
More recently, studies in various cancer types have demonstrated the potential use of identifying and following tumor-specific mutations in cell-free DNA (cfDNA) isolated from plasma as a marker for subclinical disease.15-17 A study of various different cancer types by Bettegowda et al18 demonstrated highly variable levels of mutations in baseline cfDNA in patients with different tumor types, suggesting that the use of cfDNA as a clinically relevant signal for subclinical disease will be partly based on the cancer type itself. These studies all rely on the ability to identify tumor-specific aberrations as a marker of disease burden. Tumors such as ES and DSRCTs have low mutational burdens, because it is believed that the pathognomonic EWSR1 fusions are the primary driving oncogenic lesions in these tumor types, with few recurrent secondary alterations.19 Therefore, the identification of tumor-specific EWSR1 fusions in cfDNA as a marker for active disease is an especially attractive option, given that these tumors are defined by these translocations. Moreover, issues such as clonal evolution, which can affect the detection of certain mutations in cfDNA in other tumor types, are less pertinent, because these sarcomas maintain these specific fusions through an individual’s disease course. Importantly, even identical EWSR1 fusion transcripts are the result of genomic breakpoints in the corresponding introns that are unique to each patient.20 Although a genomic DNA-based clinical assay to monitor these translocations in plasma cfDNA should be more robust than RNA-based assays in this context, the heterogeneity of intronic breakpoint regions between tumors from different patients had previously presented a major challenge for the design and routine application of such a DNA-based assay, a hurdle now overcome by the application of more powerful sequencing approaches.
In this study, we directly compared two strategies to identify baseline EWSR1 genomic rearrangements in cfDNA of patients with ES and DSRCT. The first approach uses droplet digital PCR (ddPCR) of cfDNA to detect fusions identified by targeted next-generation sequencing of formalin-fixed paraffin-embedded tumor biopsy material.21 This approach has been successfully used in numerous studies of other tumor types. The second approach uses next-generation sequencing (NGS) via a modified hybridization capture approach to identify EWSR1 fusions as well as potentially prognostic mutations in cfDNA samples. The primary aim of this study was to evaluate each platform, ddPCR and NGS, on paired cfDNA samples from patients with ES and DSRCT with confirmed radiographic evidence of disease.
METHODS
Patient Population
Between August 2014 and January 2016, patients with a confirmed diagnosis of ES or DSRCT and formalin-fixed paraffin-embedded clinical tumor material available for Memorial Sloan Kettering–Integrated Mutation Profiling of Actionable Cancer Targets (MSK-IMPACT) tumor genotyping were eligible to enroll on this study. Patients with either newly diagnosed or relapsed disease were eligible. All patients consented to tumor genotyping as well as plasma collection on an institutional review board–approved institutional protocol. For each patient, at least one baseline 10-mL blood sample was collected in Streck blood collection tubes (BCTs; Streck, La Vista, NE).
MSK-IMPACT Tumor NGS
Genomic DNA from tumor tissue and patient-matched normal blood were subjected to targeted sequencing using MSK-IMPACT, a custom, deep-coverage targeted sequencing assay approved by the New York State Department of Health as a clinical test.21,22 Tumors were sequenced to an average coverage depth of 766× (range, 405× to 1,251×) in the clinical laboratories of the Memorial Sloan Kettering Cancer Center Molecular Diagnostics Service. Sequence mutations, somatic copy number alterations, and structural rearrangements were called in 341 (version 1) or 410 (version 2) cancer-associated genes using bioinformatics pipelines previously described.21 Gene fusions involving EWSR1 were identified in all patients included in this study using DELLY.23,24
Plasma cfDNA Extraction and Analyses
Whole blood collected in 10-mL cell-free DNA BCTs (Streck) was centrifuged in two steps to separate plasma from cells. In step 1, whole blood was centrifuged at 800 × g for 10 minutes (ambient temperature). Plasma was then separated from RBCs. In step 2, separated plasma was further centrifuged in a high-speed microcentrifuge at 18,000 × g for 10 minutes (ambient temperature). Cell-free plasma was aliquoted and frozen at −80°C until ready to extract. Extraction of cfDNA was performed using a fully automated QIAGEN platform, QIAsymphony SP, and QIAsymphony DSP Virus/Pathogen Midi Kit (catalog #937055; QIAGEN, Valencia, CA). This is a bead-based custom protocol, optimized to work with 3 mL of plasma as starting material. The extraction process includes lysis, binding, wash, and elution steps. The final product is a 60-µL elution of cfDNA, with an average size approximately 170 to 200 bp. Quality and quantity of cfDNA was evaluated with automated electrophoresis using either TapeStation with High Sensitivity D1000 ScreenTape and Reagents (Agilent Technologies, Santa Clara, CA) or Fragment Analyzer with High Sensitivity genomic DNA Analysis Kit (Advanced Analytical, Ankeny, IA).
Breakpoint-Specific ddPCR
For each sample, a fusion-specific assay was designed using Primer3Plus and ordered through BioRad (Hercules, CA). Cycling conditions were tested to ensure optimal annealing/extension temperature as well as optimal separation of positive from empty droplets. All reactions were performed on a QX200 ddPCR system (BioRad). Each sample was evaluated in technical duplicates. PCR reactions contained fusion-specific primers and probes, BioRad validated copy number control primers and probes, and digital PCR Supermix for probes (no 2/-deoxyuridine 5′-triphosphate) and circulating free DNA. Reactions were partitioned into a median of approximately 16,000 droplets per well using the QX200 droplet generator. Emulsified reactions were amplified on a 96-well thermal cycler using cycling conditions identified during the optimization step (95°C 10′; 40 cycles of 94°C 30′′ 58°C 1′, 98°C 10′, 4°C hold). Plates were read and analyzed with the QuantaSoft software to assess the number of droplets positive for mutant DNA, wild-type DNA, both, or neither. The assay threshold sensitivity was set at two mutant droplets.
Targeted Plasma NGS
Cell-free DNA (cfDNA) from Plasma was extracted using the QIAsymphony SP system (QIAGEN). Sequencing libraries were prepared according to the KAPA Hyper protocol (Kapa Biosystems, Wilmington, MA) with the ligation of Illumina sequence adaptors followed by PCR amplification and clean-up. Custom DNA probes targeting all coding exons of STAG2 and TP53 and selected introns of EWSR1 (intron 7 to 13) were synthesized by IDT (Integrated DNA Technologies, Coralville, IA). Precapture libraries were quantified with Qubit (Invitrogen, Carlsbad, CA). An equal amount of each DNA library (200 ng per sample) was pooled for hybridization capture using a customized double-capture protocol modified from the NimbleGen SeqCap Target Enrichment system (Roche Sequencing Solutions, Pleasanton, CA). The first capture was incubated at 55°C for 16 hours and followed by postcapture washes and 16 cycles of PCR amplification. After PCR clean-up, the captured target library was processed by a secondary capture (using the same custom DNA probes) incubated at 65°C for 4 hours and followed by postcapture washes and three to five cycles of PCR amplification. The pooled, cleaned-up libraries containing captured DNA fragments were sequenced on the Illumina HiSeq system with paired end reads (2 × 100 bp).
Analysis was performed using the same bioinformatics pipeline as for MSK-IMPACT tissue sequencing. The same blood DNA samples sequenced with tissue using the MSK-IMPACT platform were used as matched normals for the cfDNA samples captured using the targeted probes. Sequence alignment and mutation calling were performed on reads from all tumor samples, normals, and cfDNA samples simultaneously to ensure consistent realignment around indels and consistent annotation of shared mutations.
The scatterplot for concordance between NGS and ddPCR measurement of fusion molecules was generated using the R package ggplot2. The regression line was calculated using the linear model method.
RESULTS
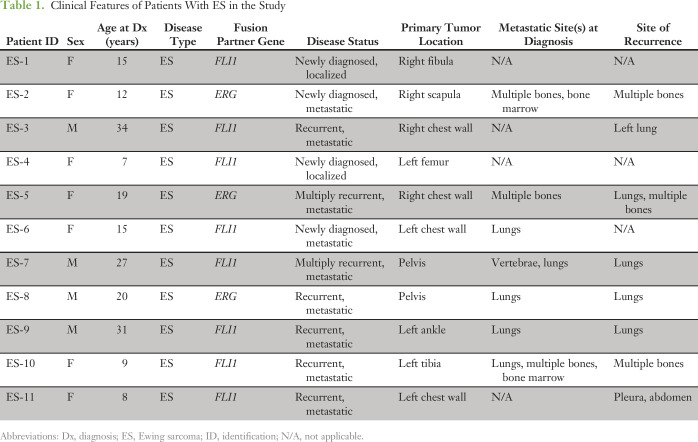
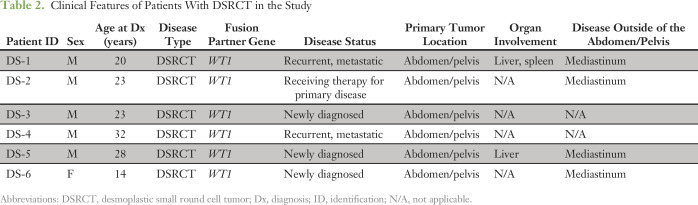
Baseline plasma samples from 17 patients with ES and DSRCT with radiographic evidence of disease were analyzed by both ddPCR and custom capture NGS (Tables 1 and 2). The cohort included 11 patients with ES and six with DSRCT. Four of 11 patients with ES had newly diagnosed disease, with two out of four presenting with localized tumors. The remaining seven patients with ES had recurrent disease at the time of baseline sample analysis. All patients with DSRCT had abdominal/pelvic disease involvement, being the characteristic clinical presentation of this sarcoma, and four out of six patients also had metastatic disease involving the mediastinum.
Table 1.
Clinical Features of Patients With ES in the Study
Table 2.
Clinical Features of Patients With DSRCT in the Study
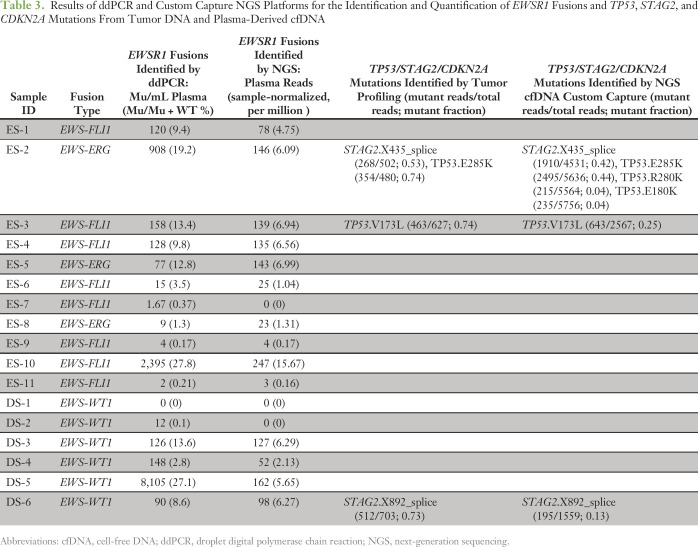
Tumor-specific EWSR1 fusions were successfully identified by MSK-IMPACT in all 17 tumor biopsy–derived DNA samples (Table 3; Data Supplement). Additional aberrations, including previously described recurrent mutations of TP53 and STAG2, were also identified by MSK-IMPACT profiling of biopsy material (Table 3). Concentrations of plasma-derived cfDNA extracted from patient samples ranged from 9.1 to 421.2 ng (mean, 60.8 ng; median, 22.8 ng) per Streck tube (Data Supplement).
Table 3.
Results of ddPCR and Custom Capture NGS Platforms for the Identification and Quantification of EWSR1 Fusions and TP53, STAG2, and CDKN2A Mutations From Tumor DNA and Plasma-Derived cfDNA
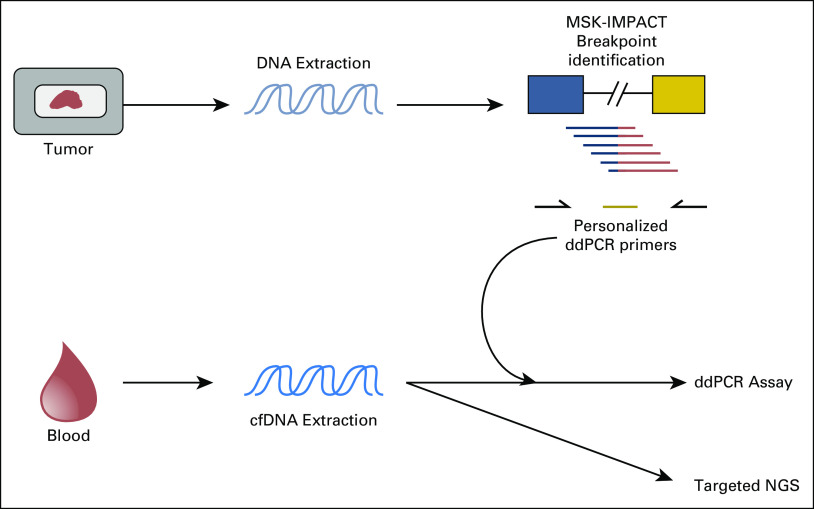
A schema illustrating our approach for comparison of ddPCR and NGS platforms is depicted in Figure 1. For ddPCR-based analysis of baseline plasma samples, primers were designed to amplify the genomic breakpoint region. Serial dilution experiments were performed on eight patient assays (four ES and four DSRCT) to determine the sensitivity of these ddPCR assays (Data Supplement). Fusion detection for all eight assays was stable across the dynamic range. We were able to resolve an average of 13.5 (range, 9 to 17) mutant copies when introduced into 330,000 copies of wild-type background, establishing an average lower limit of fusion detection of 0.013% (range, 0.018% to 0.009%). No false positives were detected in 165,000 wild-type copies.
Fig 1.
Summary of experimental design. Tumor DNA from clinical formalin-fixed paraffin-embedded biopsy or resection material was extracted from all patients. Profiling of tumor DNA was performed using the Memorial Sloan Kettering–Integrated Mutation Profiling of Actionable Cancer Targets (MSK-IMPACT) next-generation sequencing (NGS) assay to identify tumor-specific EWSR1 genomic breakpoints for downstream droplet digital polymerase chain reaction (ddPCR) assay design, as well as the presence of concurrent TP53, STAG2, and CDKN2A mutations. Consenting patients with radiographically active Ewing sarcoma or desmoplastic small round cell tumors had peripheral blood samples drawn for cell-free DNA (cfDNA) extraction. After primer design and validation of genomic breakpoints identified by MSK-IMPACT, patient-derived cfDNA was evaluated by ddPCR. As a complementary approach, cfDNA samples were also directly profiled using a custom capture–targeted NGS assay designed to identify alterations in EWSR1, TP53, STAG2, and CDKN2A.
We identified tumor-specific fusions as evidence of circulating tumor DNA (ctDNA) in all 11 baseline plasma samples from patients with ES (Table 3). The highest levels were in patients with widespread metastatic disease, as reflected in samples ES-2 and ES-10, which had > 20% of total cfDNA identified as ctDNA. We also identified EWSR1-WT1 fusions in five out of six DSRCT baseline plasma samples.
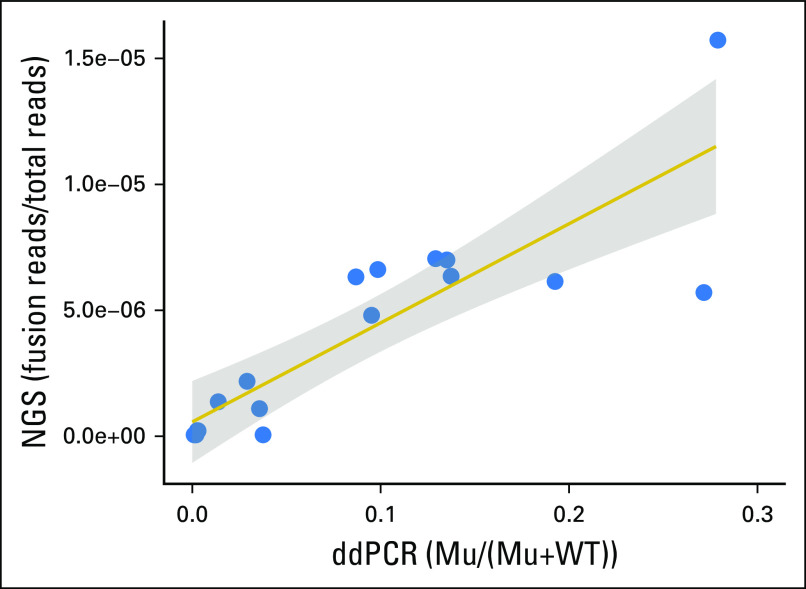
Using our custom capture NGS assay involving EWSR1, TP53, and STAG2, we sequenced parallel cfDNA samples from the cohort of 17 patients to a mean total depth of coverage of 147,549× (mean unique coverage, 3,754× after removing inferred PCR duplicates). More than 92% of sequence reads mapped to the four target genes, owing to the high specificity of our sequential capture methodology. By achieving such deep coverage, we achieved high sensitivity for detecting different EWSR1 fusions in a single universal assay, regardless of the precise location of the genomic breakpoint in EWSR1. Fusions were detected in 14 out of 17 patients (Table 3), including 10 of 11 patients with Ewing sarcoma and four of six patients with DSCRT. In the majority of patients (12 of 17), NGS data revealed the precise location of the DNA breakpoints without prior knowledge from sequencing the tumor tissue (Data Supplement). When we compared the ddPCR and NGS results for each sample, we observed a similar fraction of molecules harboring the rearrangement (Fig 2).
Fig 2.
Concordance between downstream droplet digital polymerase chain reaction (ddPCR) and next-generation sequencing (NGS) for quantifying fusions in plasma cell-free DNA. Scatterplot of fusion molecules in plasma cell-free DNA detected by NGS (reads supporting the fusion divided by total reads sequenced) versus ddPCR (droplets supporting the fusion divided by total droplets with template) for the 17 samples (correlation coefficient, 0.859). The shaded region indicates the area in which the true regression line lies with probability 0.95 (95% confidence region). Mu, mutant droplets; WT, wild-type droplets.
Prior MSK-IMPACT mutation profiling of tumor tissue had previously revealed four mutations in TP53 or STAG2 in these patients, with variant allele fractions ranging from 0.53 to 0.74. All four mutations were independently called in the corresponding cfDNA that we sequenced with our customized NGS panel, with variant allele fractions ranging from 0.13 to 0.44. Furthermore, we identified two additional novel mutations in TP53 in patient ES-2 that had not been seen in the tumor tissue. Altogether, this patient harbored three TP53 mutations in cfDNA, potentially indicative of convergent tumor evolution characterized by multiple independent mutations in the same gene in different subclones (Appendix Fig A1).25
DISCUSSION
ES and DSRCTs are aggressive sarcomas, which lack clinically useful prognostic and predictive biomarkers. Recent investigations into the use of plasma cfDNA in different tumor types have shown considerable promise, but studies in EWSR1 translocation-associated sarcomas have been limited. This is the first report to our knowledge to directly compare two complementary methodologies, ddPCR and hybrid-capture NGS, to evaluate tumor-specific EWSR1 fusions in cfDNA from patients with ES and DSRCT.
Tumor-specific fusions are particularly attractive target substrates for mutation-based biomarker studies of EWSR1 translocation-associated sarcomas. Although tumor cfDNA studies typically target oncogene point mutations, we previously demonstrated the relative scarcity of such recurrent somatic mutations in fusion-associated sarcomas.19 Specifically, in 75 ES and 24 DSRCT samples screened for 275 recurrent point mutations in 29 oncogenes frequently mutated across different cancer types, mutations were identified in only 4% of ES samples, and none of the DSRCT samples. Recent comprehensive whole-exome and whole-genome sequencing studies of ES tumor specimens have validated this finding.26-28 Conversely, EWSR1 fusions are defining molecular features of ES and DSRCTs and have never been reported to undergo modification or clonal evolution through a disease course.
Previous studies using RT-PCR to identify EWSR1 fusion transcripts in peripheral blood or bone marrow samples from patients with ES report successful identification in < 50% of patients.10,11,13 It is perhaps not surprising that the cfDNA-based approach used in our study yielded more sensitive results, because previous studies attempted to identify rare occult tumor cells through the identification of cellular EWSR1 fusion transcripts. Recent studies have established the increased sensitivity of plasma-derived cfDNA assays as compared with circulating tumor cell assays for blood-based identification of disease.18 A similar sensitivity advantage of cfDNA over cell-free RNA is also likely. Furthermore, the RT-PCR methodologies used RNA as a substrate, which undergoes rapid degradation leading to potential false-negative results if samples are not processed immediately. Finally, previous attempts at using RT-PCR used primers for only the most common transcripts seen in ES tumors among the many reported EWSR1-FLI1 fusion types.20 Therefore, a subset of patients whose tumors harbored less common EWSR1-FLI1 fusion types or other EWSR1 fusion partners was uncaptured, reducing the clinical sensitivity of the assay.
In this study, both platforms were highly sensitive at detecting EWSR1 fusions in baseline cfDNA samples, although ddPCR demonstrated increased sensitivity and was able to detect levels ≤ 0.1%. Both approaches also provide potentially significant prognostic genomic information, specifically TP53 and STAG2 mutation status in ES samples. The characterization of TP53 alterations as an adverse prognostic finding was proposed on the basis of several retrospective studies29-32 but was not confirmed in a recent prospective study.33 However, another recent large-scale genomic profiling study of ES identified tumors with co-mutation of STAG2 and TP53 as defining a particularly high-risk subset of patients.28 It is therefore important that future biomarker studies continue to collect these mutational data in an effort to establish more definitively their clinical use.
A significant advantage of the NGS approach is that it can be performed directly on cfDNA without a priori knowledge of tumor-specific mutations. This is a particularly attractive aspect of this methodology, because reliable acquisition of baseline tumor tissue has proven difficult in North American cooperative group ES banking studies.34 In this study, we demonstrated the ability to identify fusion breakpoint sequences in the majority of plasma cfDNA samples, along with potentially prognostic alterations in TP53 and STAG2. Although NGS was successful at identifying fusions in the majority of baseline cfDNA samples, there were several examples in which ddPCR was able to identify a low level of fusion positivity that was not identified by NGS. This finding is not unexpected, because the sensitivity of our ddPCR assays on the basis of serial dilution experiments was approximately 0.01%. A similar NGS-based approach was recently described in patients with advanced lung cancer, with 88% and 100% sensitivity and specificity, respectively, for detecting previously characterized mutations present at 0.1% allele frequency or higher.35 The higher sensitivity of a ddPCR approach is potentially clinically important for identifying early evidence of relapse in follow-up samples during and after therapy.
Our study highlights the advantages of both platforms. Targeted NGS has the unique advantage of obviating the need for parallel tumor tissue profiling for breakpoint identification. Furthermore, NGS provides the ability to capture the heterogeneity of a patient’s disease more completely than ddPCR, as reflected in the patient cfDNA sample (ES-2) with multiple TP53 mutations. Conversely, ddPCR provides significantly higher sensitivity, which may be a critical feature for the early detection of relapse. The patient cohort in our study represents the population seen at a sarcoma referral center, which is enriched for high-risk and relapsed disease. Therefore, it will be important to study a more typical population in forthcoming studies. Nonetheless, these results provide important data for the establishment of an optimal strategy for future cooperative group prospective biomarker studies of EWSR1-defined sarcomas. Given these findings, we propose a hybrid approach for future clinical trials (Fig 3). Our data suggest that the majority of patients will have identifiable disease at baseline using our targeted NGS approach. This will also allow for identification of the precise fusion breakpoint as well as additional TP53 and STAG2 mutational status on the majority of patients, without the need to profile tumor material. Identification of the fusion breakpoint sequence will then allow for primer design for subsequent ddPCR evaluation of subsequent follow-up samples, providing the enhanced sensitivity for detection of minimal residual disease.
Fig 3.
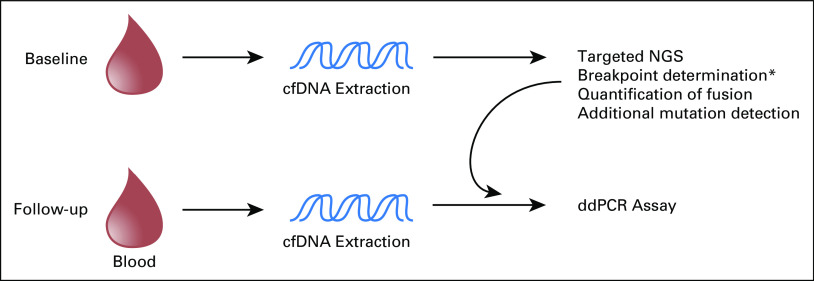
Proposed schema for prospective EWSR1 fusion-based cell-free DNA (cfDNA) studies. The depicted proposed schema for future prospective studies evaluating the use of cfDNA as a marker for subclinical disease in Ewing sarcoma or desmoplastic small round cell tumors incorporates the advantages of each methodology (next-generation sequencing [NGS] and downstream droplet digital polymerase chain reaction [ddPCR]). Breakpoint determination, as well as quantification of EWSR1 fusion, and identification of additional mutation data are identifiable on the majority of baseline plasma cfDNA samples using a targeted custom-capture NGS approach. This approach obviates the need to collect tumor biopsy material for breakpoint identification from the majority of patients. Subsequent follow-up samples can be followed using ddPCR though primer design against breakpoints identified by the baseline NGS evaluation of cfDNA samples. This approach offers higher sensitivities compared with NGS, potentially improving the ability to detect subclinical disease. (*) For cases where NGS fails to identify genomic fusion sequences, tissue samples will need to be collected for breakpoint determination through tumor profiling.
As addressed in a recent publication by the Children’s Oncology Group Ewing Sarcoma Biology Committee, there are significant challenges facing biomarker development in rare cancer types, and certain characteristics are imperative for the successful development of predictive markers in this clinical context.9 Our proposed approach is especially compelling given these criteria. First, our approach should capture almost 100% of patients with ES and DSRCT, because the platforms are designed to capture any EWSR1 fusion type, including those with rare variant EWSR1 translocations. As a substrate, cfDNA is advantageous over bone marrow aspirates, given the lack of need for an invasive procedure. This is especially important in a disease that largely occurs in a pediatric population, where sedation is generally required for even minor procedures. Furthermore, we demonstrated the feasibility and efficacy of using specific cfDNA BCTs (Streck BCTs) for collection of patient plasma samples. Cell-free specific BCTs contain stabilizing reagents, which have been shown to minimize both ctDNA degradation and sample contamination with blood cell–derived genomic DNA.36,37 This is particularly important for potential biomarkers in rare diseases, where cooperative group studies require long-distance sample deliveries, necessitating strategies to eliminate the issue of substrate degradation. Finally, the technical platforms are particularly appropriate in terms of reproducibility and cost effectiveness. ddPCR has been established as an attractive platform for circulating nucleic acids on the basis of the ease of quantitation of mutant fragments as compared with real-time PCR.38 Furthermore, broad, hybrid-capture NGS platforms are becoming increasingly used for clinical use, as exemplified by the MSK-IMPACT sequencing platform, with well over 10,000 patient tumor samples studied to date,39 and even greater numbers studied by similar NGS-based assays at commercial reference laboratories.
Our primary aim in this study was to establish a robust methodology suitable for the unique and stringent requirements for biomarker development in prospective studies of rare diseases. Given our promising results with baseline samples from patients with ES and DSRCT, we believe that the proposed hybrid approach incorporating complementary NGS custom capture and ddPCR platforms provides an attractive methodological platform for a prospective biomarker study in patients with EWSR1-driven cancers.
ACKNOWLEDGMENT
We thank Chintan Patel, Jose Sosa, and Liliana Villafania from the Memorial Sloan Kettering Cancer Center Department of Laboratory Medicine for performing plasma cfDNA extraction in this study.
Appendix
Fig A1.
Integrative genomics viewer screenshot of ES-2 bam file showing TP53 R280K (right; frequency, 0.04; identified in plasma) and E285K (left; frequency, 0.44; identified in both plasma and tissue) mutations. The screenshot illustrates the fact that the two mutations are mutually exclusive; that is, two distinct sets of reads carry these two mutations, supporting the possibility of convergent evolution of distinct subclones sampled in cfDNA.
Footnotes
Supported by use of the Integrated Genomics Operation Core, which was funded by the National Cancer Institute Cancer Center Support Grant (Grant No. P30 CA008748), Cycle for Survival, and the Marie-Josée and Henry R. Kravis Center for Molecular Oncology.
M.F.B. and M.L. contributed equally to this work.
AUTHOR CONTRIBUTIONS
Conception and design: Neerav N. Shukla, Heather Magnan, Jiabin Tang, Paul A. Meyers, Marc Ladanyi
Administrative support: Neerav N. Shukla
Financial support: Neerav N. Shukla
Provision of study material or patients: Neerav N. Shukla, Paul A. Meyers, Agnes Viale
Collection and assembly of data: Neerav N. Shukla, Juber A. Patel, Heather Magnan, Ahmet Zehir, Daoqi You, Jiabin Tang, Fanli Meng, Aliaksandra Samoila, Emily K. Slotkin, Srikanth R. Ambati, Alexander J. Chou, Leonard H. Wexler, Ellinor I. Peerschke, Michael F. Berger
Data analysis and interpretation: Neerav N. Shukla, Juber A. Patel, Heather Magnan, Ahmet Zehir, Daoqi You, Aliaksandra Samoila, Paul A. Meyers, Agnes Viale, Michael F. Berger, Marc Ladanyi
Manuscript writing: All authors
Final approval of manuscript: All authors
Accountable for all aspects of the work: All authors
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to www.asco.org/rwc or po.ascopubs.org/site/ifc.
Neerav N. Shukla
No relationship to disclose
Juber A. Patel
No relationship to disclose
Heather Magnan
No relationship to disclose
Ahmet Zehir
No relationship to disclose
Daoqi You
No relationship to disclose
Jiabin Tang
No relationship to disclose
Fanli Meng
No relationship to disclose
Aliaksandra Samoila
No relationship to disclose
Emily K. Slotkin
No relationship to disclose
Srikanth R. Ambati
No relationship to disclose
Alexander J. Chou
No relationship to disclose
Leonard H. Wexler
Consulting or Advisory Role: AstraZeneca
Paul A. Meyers
Stock and Other Ownership Interests: Amgen, Bayer, EI du Pont, Henry Schein, Jazz Pharmaceuticals, MEDNAX, Novartis, Procter & Gamble, Sigma-Aldrich
Honoraria: France Foundation (I)
Consulting or Advisory Role: Boehringer Ingelheim (I)
Speakers' Bureau: France Foundation (I)
Travel, Accommodations, Expenses: Takeda, InterMune (I)
Ellinor I. Peerschke
Honoraria: Roche Diagnostics, Sysmex, Becton Dickinson
Consulting or Advisory Role: Roche Diagnostics, Becton Dickinson, Sight Diagnostics
Research Funding: Sysmex, Abbott Diagnostics, Siemens Healthcare Diagnostics
Patents, Royalties, Other Intellectual Property: 60.11, 74.4.2 monoclonal antibodies licensed for sale and distribution by Stony Brook University; US patent No 8-883-153, method for preventing and treating angioedema (I)
Travel, Accommodations, Expenses: Sysmex
Agnes Viale
No relationship to disclose
Michael F. Berger
Consulting or Advisory Role: Cancer Genetics, Sequenom
Marc Ladanyi
Honoraria: Merck (I)
Consulting or Advisory Role: National Comprehensive Cancer Network/Boehringer Ingelheim Afatinib Targeted Therapy Advisory Committee, National Comprehensive Cancer Network/AstraZeneca Tagrisso Request for Proposals Advisory Committee
Research Funding: Loxo Oncology (Inst)
REFERENCES
- 1. Fletcher C, Bridge J, Hogendoorn P, et al (eds): World Health Organization Classification of Tumours: Pathology and Genetics of Tumours of Soft Tissue and Bone (ed 4). Lyon, France, IARC Press, 2013. [Google Scholar]
- 2.Delattre O, Zucman J, Plougastel B, et al. Gene fusion with an ETS DNA-binding domain caused by chromosome translocation in human tumours. Nature. 1992;359:162–165. doi: 10.1038/359162a0. [DOI] [PubMed] [Google Scholar]
- 3.Ladanyi M, Gerald W. Fusion of the EWS and WT1 genes in the desmoplastic small round cell tumor. Cancer Res. 1994;54:2837–2840. [PubMed] [Google Scholar]
- 4.Lessnick SL, Ladanyi M. Molecular pathogenesis of Ewing sarcoma: New therapeutic and transcriptional targets. Annu Rev Pathol. 2012;7:145–159. doi: 10.1146/annurev-pathol-011110-130237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. doi: 10.1200/JCO.2011.41.5703. Womer RB, West DC, Krailo MD, et al: Randomized controlled trial of interval-compressed chemotherapy for the treatment of localized Ewing sarcoma: A report from the Children’s Oncology Group. J Clin Oncol 30:4148-4154, 2012 [Erratum: J Clin Oncol 33:814, 2015] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kolb EA, Kushner BH, Gorlick R, et al. Long-term event-free survival after intensive chemotherapy for Ewing’s family of tumors in children and young adults. J Clin Oncol. 2003;21:3423–3430. doi: 10.1200/JCO.2003.10.033. [DOI] [PubMed] [Google Scholar]
- 7.Gerald WL, Miller HK, Battifora H, et al. Intra-abdominal desmoplastic small round-cell tumor. Report of 19 cases of a distinctive type of high-grade polyphenotypic malignancy affecting young individuals. Am J Surg Pathol. 1991;15:499–513. [PubMed] [Google Scholar]
- 8.Desai NB, Stein NF, LaQuaglia MP, et al. Reduced toxicity with intensity modulated radiation therapy (IMRT) for desmoplastic small round cell tumor (DSRCT): An update on the whole abdominopelvic radiation therapy (WAP-RT) experience. Int J Radiat Oncol Biol Phys. 2013;85:e67–e72. doi: 10.1016/j.ijrobp.2012.09.005. [DOI] [PubMed] [Google Scholar]
- 9.Shukla N, Schiffman J, Reed D, et al. Biomarkers in Ewing sarcoma: The promise and challenge of personalized medicine. A report from the Children’s Oncology Group. Front Oncol. 2013;3:141. doi: 10.3389/fonc.2013.00141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zoubek A, Ladenstein R, Windhager R, et al. Predictive potential of testing for bone marrow involvement in Ewing tumor patients by RT-PCR: A preliminary evaluation. Int J Cancer. 1998;79:56–60. doi: 10.1002/(sici)1097-0215(19980220)79:1<56::aid-ijc11>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- 11.Schleiermacher G, Peter M, Oberlin O, et al. Increased risk of systemic relapses associated with bone marrow micrometastasis and circulating tumor cells in localized Ewing tumor. J Clin Oncol. 2003;21:85–91. doi: 10.1200/JCO.2003.03.006. [DOI] [PubMed] [Google Scholar]
- 12.Dubois SG, Epling CL, Teague J, et al. Flow cytometric detection of Ewing sarcoma cells in peripheral blood and bone marrow. Pediatr Blood Cancer. 2010;54:13–18. doi: 10.1002/pbc.22245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Avigad S, Cohen IJ, Zilberstein J, et al. The predictive potential of molecular detection in the nonmetastatic Ewing family of tumors. Cancer. 2004;100:1053–1058. doi: 10.1002/cncr.20059. [DOI] [PubMed] [Google Scholar]
- 14.Ash S, Luria D, Cohen IJ, et al. Excellent prognosis in a subset of patients with Ewing sarcoma identified at diagnosis by CD56 using flow cytometry. Clin Cancer Res. 2011;17:2900–2907. doi: 10.1158/1078-0432.CCR-10-3069. [DOI] [PubMed] [Google Scholar]
- 15.Roschewski M, Dunleavy K, Pittaluga S, et al. Circulating tumour DNA and CT monitoring in patients with untreated diffuse large B-cell lymphoma: a correlative biomarker study. Lancet Oncol. 2015;16:541–549. doi: 10.1016/S1470-2045(15)70106-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hyman DM, Diamond EL, Vibat CR, et al. Prospective blinded study of BRAFV600E mutation detection in cell-free DNA of patients with systemic histiocytic disorders. Cancer Discov. 2015;5:64–71. doi: 10.1158/2159-8290.CD-14-0742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dawson SJ, Tsui DW, Murtaza M, et al. Analysis of circulating tumor DNA to monitor metastatic breast cancer. N Engl J Med. 2013;368:1199–1209. doi: 10.1056/NEJMoa1213261. [DOI] [PubMed] [Google Scholar]
- 18.Bettegowda C, Sausen M, Leary RJ, et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci Transl Med. 2014;6:224ra24. doi: 10.1126/scitranslmed.3007094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shukla N, Ameur N, Yilmaz I, et al. Oncogene mutation profiling of pediatric solid tumors reveals significant subsets of embryonal rhabdomyosarcoma and neuroblastoma with mutated genes in growth signaling pathways. Clin Cancer Res. 2012;18:748–757. doi: 10.1158/1078-0432.CCR-11-2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Plougastel B, Zucman J, Peter M, et al. Genomic structure of the EWS gene and its relationship to EWSR1, a site of tumor-associated chromosome translocation. Genomics. 1993;18:609–615. doi: 10.1016/s0888-7543(05)80363-5. [DOI] [PubMed] [Google Scholar]
- 21.Cheng DT, Mitchell TN, Zehir A, et al. Memorial Sloan Kettering-Integrated Mutation Profiling of Actionable Cancer Targets (MSK-IMPACT): A hybridization capture-based next-generation sequencing clinical assay for solid tumor molecular oncology. J Mol Diagn. 2015;17:251–264. doi: 10.1016/j.jmoldx.2014.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Shah R: IMPACT-pipeline: First pre-release to run MSK-IMPACT framework. [DOI]
- 23. Shah R: IMPACT-SV: A Perl-based framework to call, annotate structural variants using delly and dRanger. [DOI]
- 24.Rausch T, Zichner T, Schlattl A, et al. DELLY: Structural variant discovery by integrated paired-end and split-read analysis. Bioinformatics. 2012;28:i333–i339. doi: 10.1093/bioinformatics/bts378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gerlinger M, Rowan AJ, Horswell S, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med. 2012;366:883–892. doi: 10.1056/NEJMoa1113205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. doi: 10.1371/journal.pgen.1004475. Brohl AS, Solomon DA, Chang W, et al: The genomic landscape of the Ewing sarcoma family of tumors reveals recurrent STAG2 mutation. PLoS Genet 10:e1004475, 2014 [Erratum: PLoS Genet 10:e1004629, 2014] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Crompton BD, Stewart C, Taylor-Weiner A, et al. The genomic landscape of pediatric Ewing sarcoma. Cancer Discov. 2014;4:1326–1341. doi: 10.1158/2159-8290.CD-13-1037. [DOI] [PubMed] [Google Scholar]
- 28.Tirode F, Surdez D, Ma X, et al. Genomic landscape of Ewing sarcoma defines an aggressive subtype with co-association of STAG2 and TP53 mutations. Cancer Discov. 2014;4:1342–1353. doi: 10.1158/2159-8290.CD-14-0622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.de Alava E, Antonescu CR, Panizo A, et al. Prognostic impact of P53 status in Ewing sarcoma. Cancer. 2000;89:783–792. [PubMed] [Google Scholar]
- 30.Huang HY, Illei PB, Zhao Z, et al. Ewing sarcomas with p53 mutation or p16/p14ARF homozygous deletion: A highly lethal subset associated with poor chemoresponse. J Clin Oncol. 2005;23:548–558. doi: 10.1200/JCO.2005.02.081. [DOI] [PubMed] [Google Scholar]
- 31.Tsuchiya T, Sekine K, Hinohara S, et al. Analysis of the p16INK4, p14ARF, p15, TP53, and MDM2 genes and their prognostic implications in osteosarcoma and Ewing sarcoma. Cancer Genet Cytogenet. 2000;120:91–98. doi: 10.1016/s0165-4608(99)00255-1. [DOI] [PubMed] [Google Scholar]
- 32.Wei G, Antonescu CR, de Alava E, et al. Prognostic impact of INK4A deletion in Ewing sarcoma. Cancer. 2000;89:793–799. doi: 10.1002/1097-0142(20000815)89:4<793::aid-cncr11>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- 33.Lerman DM, Monument MJ, McIlvaine E, et al. Tumoral TP53 and/or CDKN2A alterations are not reliable prognostic biomarkers in patients with localized Ewing sarcoma: A report from the Children’s Oncology Group. Pediatr Blood Cancer. 2015;62:759–765. doi: 10.1002/pbc.25340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Borinstein SC, Beeler N, Block JJ, et al. A decade in banking Ewing sarcoma: A report from the Children’s Oncology Group. Front Oncol. 2013;3:57. doi: 10.3389/fonc.2013.00057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Paweletz CP, Sacher AG, Raymond CK, et al. Bias-corrected targeted next-generation sequencing for rapid, multiplexed detection of actionable alterations in cell-free DNA from advanced lung cancer patients. Clin Cancer Res. 2016;22:915–922. doi: 10.1158/1078-0432.CCR-15-1627-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kang Q, Henry NL, Paoletti C, et al. Comparative analysis of circulating tumor DNA stability in K3EDTA, Streck, and CellSave blood collection tubes. Clin Biochem. 2016;49:1354–1360. doi: 10.1016/j.clinbiochem.2016.03.012. [DOI] [PubMed] [Google Scholar]
- 37.Denis MG, Knol AC, Théoleyre S, et al. Efficient detection of BRAF mutation in plasma of patients after long-term storage of blood in cell-free DNA blood collection tubes. Clin Chem. 2015;61:886–888. doi: 10.1373/clinchem.2015.238352. [DOI] [PubMed] [Google Scholar]
- 38.Hudecova I. Digital PCR analysis of circulating nucleic acids. Clin Biochem. 2015;48:948–956. doi: 10.1016/j.clinbiochem.2015.03.015. [DOI] [PubMed] [Google Scholar]
- 39.Zehir A, Benayed R, Shah RH, et al. Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat Med. doi: 10.1038/nm.4333. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]