Abstract
Interstitial lung diseases (ILD) involve several abnormal imaging patterns observed in computed tomography (CT) images. Accurate classification of these patterns plays a significant role in precise clinical decision making of the extent and nature of the diseases. Therefore, it is important for developing automated pulmonary computer-aided detection systems. Conventionally, this task relies on experts’ manual identification of regions of interest (ROIs) as a prerequisite to diagnose potential diseases. This protocol is time consuming and inhibits fully automatic assessment. In this paper, we present a new method to classify ILD imaging patterns on CT images. The main difference is that the proposed algorithm uses the entire image as a holistic input. By circumventing the prerequisite of manual input ROIs, our problem set-up is significantly more difficult than previous work but can better address the clinical workflow. Qualitative and quantitative results using a publicly available ILD database demonstrate state-of-the-art classification accuracy under the patch-based classification and shows the potential of predicting the ILD type using holistic image.
Keywords: Interstitial lung disease, convolutional neural network, holistic medical image classification
1. Introduction
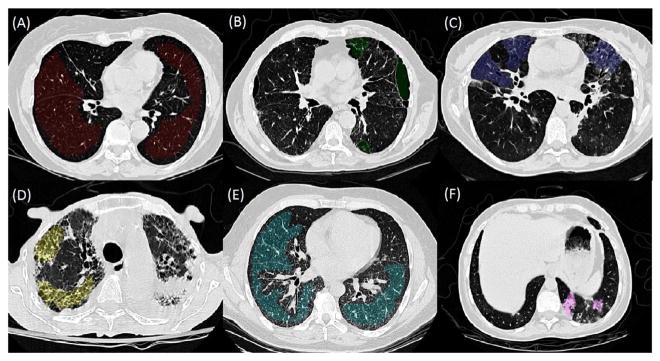
The interstitial lung diseases (ILD) cause progressive scarring of lung tissue, which would eventually affect the patients’ ability to breathe and get enough oxygen into the bloodstream. High-resolution computed tomography (HRCT) is the standard in-vivo radiology imaging tool for visualising normal/abnormal imaging patterns to identify the specific type of ILD (Webb et al. 2014), and to develop appropriate therapy plans. Examples of these lung tissue patterns are shown in Figure 1. Computer-aided detection (CAD)/classification systems are needed for achieving higher recalls on ILD assessment (Bağcı et al. 2012). In particular, the amounts and anatomical positions of abnormal imaging patterns (along with patient history) can help radiologists to optimise their diagnostic decisions, with better quantitative measurements.
Figure 1.
Example images (segment of HRCT axial slices) for each of the six lung tissue types. (A) Normal (NM). (B) Emphysema (EM). (C) Ground Glass (GG). (D) Fibrosis (FB). (E) Micronodules (MN). (F) Consolidation (CD).
There are a vast amount of relevant literature on developing CAD systems of pulmonary diseases, but most of them focus on identifying and quantifying a single pattern such as consolidation or nodules (Bagci et al. 2012). For computer-aided ILD classification, all previous studies have employed a patch-based image representation with the classification results of moderate success (Depeursinge, Van de Ville et al. 2012; Song et al. 2013, 2015; Li et al. 2014). There are two major drawbacks for the image patch-based methods: (1), The image patch sizes or scales in studies (Song et al. 2013, 2015) are relatively small (31 × 31 pixels) where some visual details and spatial context may not be fully captured. The holistic computed tomography (CT) slice holds a lot of details that may be overlooked in the patch-based representation. (2), More importantly, the state-of-the-art methods assume the manual annotation as given. Image patches are consequently sampled within these regions of interest (ROIs). Image patch-based approaches, which depend on the manual ROI inputs, are easier to solve, but unfortunately less clinically desirable. This human demanding process will become infeasible for the large-scale medical image processing and analysis.
In this paper, we propose a new representation/approach to address this limitation. Our method classifies and labels ILD tags for holistic CT slices and can possibly be used to prescreen a large amount of radiology data. Additionally, the prescreened data can be used as feedback to enlarge the training data-set in a loop. This would be the essential component for a successful and practical medical image analysis tool at a truly large scale. Different from Li et al. (2014), Song et al. (2013, 2015), our CNN-based method is formulated as a holistic image recognition task (Russakovsky et al. 2015) that is also considered as a weakly supervised learning problem. Obtaining image tags alone is cost effective and can be obtained very efficiently. On the other hand, our new set-up of using holistic images makes it significantly more challenging than the previous settings (Song et al. 2013, 2015; Li et al. 2014), since the manual ROIs are no longer required. Image patches as classification instances, which are extracted from the annotated ROIs, are well spatially aligned or invariant to their absolute intraslice CT coordinates. On the contrary, in our set-up, only slice-level image labels or tags are needed and no precise contouring of ILD regions are necessary. This weakly supervised learning scheme can scale well with large scale image database. The experimental evaluation on the publicly available data-set demonstrates the state-of-the-art results under the same image patch-based approaches and shows promising results under this new challenging protocol.
2. Methods
CNN has been successfully exploited in various image classification problems and achieved the state-of-the-art performances in image classification, detection and segmentation challenges such as MNIST, ImageNet, etc. (Krizhevsky et al. 2012; Razavian et al. 2014). The typical image classification approach consists of two steps of feature extraction and classification. However, the most attractive characteristics of the CNN method are that it learns the end-to-end feature extraction and classification simultaneously. CNN also shows promise in medical image analysis applications, such as mitosis detection (Cireşan et al. 2013), lymph node detection (Roth et al. 2014) and knee cartilage segmentation (Prasoon et al. 2013). In previous ILD classification work, hand-crafted local image descriptors (such as LBP, HOG) are used in Depeursinge, Van de Ville et al. (2012), Song et al. (2013, 2015) to capture the image patch appearance.
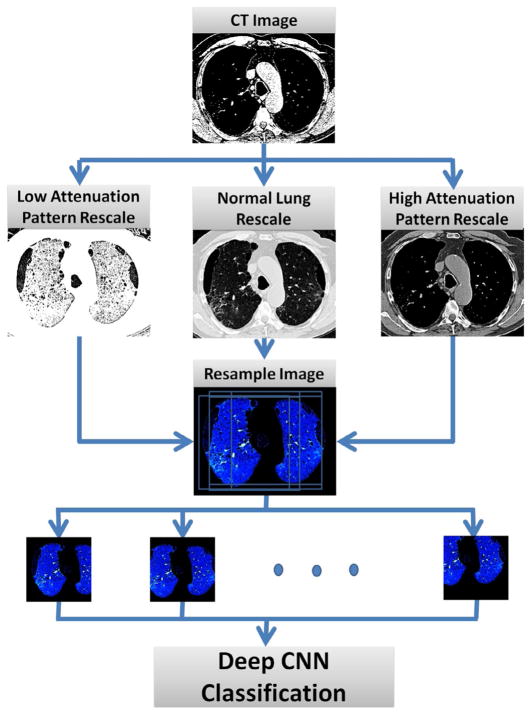
Our proposed framework is illustrated in Figure 2. Three attenuation scales with respect to lung abnormality patterns are captured by rescaling the original CT image in Hounsfield Units to 2-D inputs in training and testing. For this purpose, three different ranges are utilised: one focusing on patterns with lower attenuation, one on patterns with higher attenuation and one for normal lung attenuation. Using three attenuation ranges offers better visibility or visual separation among all six ILD disease categories. Another reason for using the three ranges is to accommodate the CNN architecture that we adapt from ImageNet (Krizhevsky et al. 2012) that uses RGB values of natural images. Finally, for each input 2-D slice, 10 samples (“data augmentation”) are cropped randomly from the original images and resized to 224 × 224 pixels via linear interpolation. This step generates more training data to reduce the overfitting. These inputs, together with their labels, are fed to CNN for training and classification. Each technical component is discussed in details as follows.
Figure 2.
Flowchart of the training framework.
2.1. CNN architecture
The architecture of our CNN is similar to the convolutional neural network proposed by Krizhevsky et al. (2012). CNNs with shallow layers do not have enough discriminative power, while too deep CNNs are computationally expensive to train and easy to be overfitted. Our network contains multiple layers: first five layers are convolutional layers followed by three fully connected (FC) layers and the final softmax classification layer, which is changed from 1000 classes to 6 classes in our application.
It is known from the computer vision community that supervised pre-training on a large auxiliary data-set, followed by the domain-specific fine tuning on a small data-set, is an effective paradigm to boost the performance of CNN models (when the training data are limited Girshick et al. 2014). In our experiments, the training convergence speed is much faster when using pre-trained model than using randomly initialised model. The use of three CT attenuation ranges also accommodates the CNN architecture of three input channels. The output of the last FC layer is formed into a six-way softmax to produce a distribution over the six class labels (with six neurons). We start the training via stochastic gradient descent at a learning rate of 1/10th of the initial pre-training rate (Krizhevsky et al. 2012) expect for the output softmax layer. The adjusted learning rate allows appropriate fine-tuning progresses without ruining the initialisation. The output layer still needs a large learning rate for convergence to the new ILD classification categories.
2.2. CT attenuation rescale
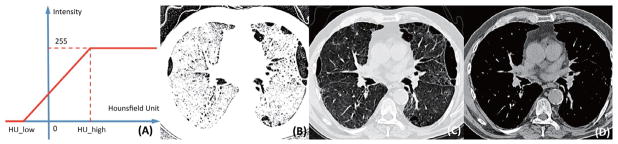
To better capture the abnormal ILD patterns in CT images, we selected three ranges of attenuation and rescaled them to [0, 255] for CNN input. As shown in Figure 3(A), this process is designed to select the attenuation value between HU_low and HU_high so that the value within the range can be highlighted to represent different visual patterns. A linear transformation is applied to rescale the intensities. Specifically, low-attenuation range (Figure 6(B)) is used to capture patterns with lower intensities, such as emphysema; normal range (Figure 3(C)) to represent normal appearance of lung regions; and high attenuation range (Figure 6(D)) for modelling patterns with higher intensities, such as consolidation and nodules. Specific HUs we chose in our experiments are: for low attenuation, HU_ low = −1400 and HU_ high = −950; for normal, HU_ low = −1400 and HU_ high = 200; for high attenuation, HU_ low = −160 and HU_ high = 240.
Figure 3.
(A) CT attenuation range rescale. (B) Low-attenuation range. (C) Normal lung range. (D) High-attenuation range.
2.3. Data augmentation
The most common and effective way to reduce overfitting on image recognition training using CNN is to artificially enlarge or augment the original data-set by label-preserving geometric transformations. We generate new images by randomly jittering and cropping 10 subimages per original CT slice. Although the generated images are interdependent, the scheme would improve the training/testing performance by ~ 5% in classification accuracy. At test time, 10 jittered images are also generated and fed into the trained CNN model for any CT slice. Final per slice prediction is obtained by aggregating (e.g. majority voting, maximal pooling) over the CNN six-class softmax probabilities on 10 jittered images.
3. Experiments and discussions
A publicly available ILD database has been released recently (Depeursinge, Vargas et al. 2012) to improve the detection and classification of a wide range of lung abnormal imaging patterns. This database contains 120 HRCT scans with 512 × 512 pixels per axial slice, where 17 types of lung tissues are annotated on marked regions (i.e. ROIs). Most existing classification methods (Song et al. 2013, 2015; Li et al. 2014) evaluated on the ILD data-set first extract many image patches from ROIs and then only classify patches into five lung tissue classes: normal (NM), emphysema (EM), ground glass (GG), fibrosis (FB) and micronodules (MN). Here, consolidation (CD), as a highly prevalent type of ILD, is also included within our classification scheme. All of the six diseases are prevalent characteristics of ILD and identifying them is critical to determine their ILD types or healthy.
The database contains 2084 ROIs labelled with specific type of ILD disease, out of 120 patients. All patients are randomly split into two subsets at the patient level for training (100 patients) and testing (20 patients). Training/testing data are separated at patient level, i.e. different slices from the same patient will not appear in both training and testing. All images containing the six types of diseases are selected, resulting 1689 images in total for training and testing. Note that previous work (Song et al. 2013, 2015; Li et al. 2014) report performance on patch classification only, rather than performance assessment for the whole image slices or at patient level, which are actually more clinically relevant.
For fair comparisons with previous work, we conduct experiments under two different settings. One is patch-based classification, that is exactly the same as in previous state-of-the-art work (Song et al. 2013, 2015). An overall accuracy of 87.9% is achieved, comparing with 86.1% (Song et al. 2013) accuracy of previous patch methods. The best F-scores are achieved in most classes as shown in Table 1. 31 × 31 patches are extracted from the ROI regions, and then resized to the size of 224 × 224 to accomodate the CNN architecture. Another experiment shows the holistic image classification results. The overall accuracy is 68.6%. Note that our per slice testing accuracy results are not strictly comparable to Song et al. (2013, 2015), Li et al. (2014), reporting classification results only at the image patch level (a significantly less challenging protocol).
Table 1.
F -score of ILD classifications.
| EM | FB | GG | NM | MN | CD | |
|---|---|---|---|---|---|---|
| Song et al. (2013) | 0.753 | 0.841 | 0.782 | 0.840 | 0.857 | – |
| Song et al. (2015) | 0.768 | 0.872 | 0.795 | 0.877 | 0.888 | – |
| Li et al. (2014) | 0.5449 | 0.7624 | 0.7150 | 0.8395 | 0.9096 | – |
| Ours | 1.0000 | 0.8000 | 0.7500 | 0.4000 | 0.5600 | 0.5000 |
| Ours patch setting | 0.8940 | 0.8509 | 0.8159 | 0.8844 | 0.8950 | – |
Note: All comparison are statistically significant (p < 0.05).
Table 2 shows the confusion matrix of the classification results on holistic images. Majority voting-based aggregation from jittered subimages is used. Emphysema is perfectly classified from other diseases. One of the three CT attenuation ranges is specifically designed to emphasise on the patterns with lower attenuation, which boosts the classification performance on emphysema significantly. Healthy images and micronodule patterns are difficult to be separated based on the confusion matrix result. Micronodule patterns are indeed visually challenging to be recognised from one single static CT slice (Bagci et al. 2012). 3D cross-slice image features may be needed. Majority voting performs slightly better (~ 2%) than choosing the highest value from 10 subimage CNN scores per ILD class, and assigning the CT slice into the class corresponding to the maximum of aggregated highest scores. Table 3 shows the confusion matrix of patch-based classification.
Table 2.
Confusion matrix of ILD classification.
| Ground truth | Prediction
|
|||||
|---|---|---|---|---|---|---|
| EM | FB | GG | NM | MN | CD | |
| EM | 1 | 0 | 0 | 0 | 0 | 0 |
| FB | 0 | 0.7111 | 0.0889 | 0.0667 | 0 | 0.1333 |
| GG | 0 | 0 | 0.9375 | 0.0625 | 0 | 0 |
| NM | 0 | 0 | 0 | 0.5 | 0.5 | 0 |
| MN | 0 | 0 | 0 | 0.4615 | 0.5385 | 0 |
| CD | 0 | 0.2 | 0.3333 | 0 | 0 | 0.4667 |
Note: All comparison are statistically significant (p < 0.05).
Table 3.
Confusion matrix of ILD patch classification.
| Ground truth | Prediction
|
||||
|---|---|---|---|---|---|
| EM | FB | GG | NM | MN | |
| EM | 0.9142 | 0.0078 | 0.0237 | 0.0047 | 0.0495 |
| FB | 0.0546 | 0.8270 | 0.0075 | 0.0464 | 0.0646 |
| GG | 0.0558 | 0.0025 | 0.8151 | 0.0930 | 0.0337 |
| NM | 0.0141 | 0.0108 | 0.0494 | 0.8910 | 0.0348 |
| MN | 0.0600 | 0.0070 | 0.0262 | 0.0268 | 0.8799 |
Note: All comparison are statistically significant (p < 0.05).
Our model is implemented in Matlab using MatConvNet package (Vedaldi & Lenc 2015) for the CNN implementation, running on a PC with 3.10 GHz dual processors CPU and 32-GB memory. Training the CNN model consumes about 20–24 h (Figure 4), while classifying a new testing image takes only a few seconds.
Figure 4.
Trained convolutional filters in the first layer.
4. Conclusion and future work
In this paper, we present a new representation and approach for interstitial lung disease classification. Our method with holistic images (i.e. CT slice) as input is significantly different from previous image patch-based algorithms. It addresses a more practical and realistic clinical problem. Our preliminary experimental results have demonstrated the promising feasibility and advantages of the proposed approach.
There are several directions to be explored as future work. The image features learned from the deep convolutional network can be integrated into more sophisticated classification algorithms. There are some cases (~ 5%) with multiple disease tags on the same slice of CT image. Detection with multiple labels at a slice level would be interesting. Understanding the clinical meaning and value of the features learned from the network would also be a direction that we plan to pursue.
Acknowledgments
Funding
This research is supported by Center for Research in Computer Vision (CRCV) of UCF, Center for Infectious Disease Imaging (CIDI), the intramural research program of the National Institute of Allergy and Infectious Diseases (NIAID), the National Institute of Biomedical Imaging and Bioengineering (NIBIB), the Clinical Center (CC), Radiology and Imaging Sciences, Imaging Biomarkers and Computer-Aided Diagnosis Laboratory and Clinical Image Processing Service. Acknowledgment to Nvidia Corp. for donation of K40 GPUs.
Footnotes
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- Bağcı U, Bray M, Caban J, Yao J, Mollura DJ. Computer-assisted detection of infectious lung diseases: a review. CMIG. 2012;36:72–84. doi: 10.1016/j.compmedimag.2011.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagci U, Yao J, Wu A, Caban J, Palmore T, Suffredini A, Aras O, Mollura D. Automatic detection and quantification of tree-in-bud (TIB) opacities from CT scans. TBME. 2012;59:1620–1632. doi: 10.1109/TBME.2012.2190984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cireşan DC, Giusti A, Gambardella LM, Schmidhuber J. Mitosis detection in breast cancer histology images with deep neural networks. In: Mori K, Sakuma I, Sato Y, Barillot C, Navab N, editors. Medical Image Computing and Computer-Assisted Intervention -- MICCAI; 2013 16th International Conference Proceeding Part II; Nagoya, Japan. September 22–26, 2013; Berlin, Heidelberg: Berlin Heidelberg: Springer ; 2013. pp. 411–418. [DOI] [PubMed] [Google Scholar]
- Depeursinge A, Van de Ville D, Platon A, Geissbuhler A, Poletti P-A, Muller H. Near-affine-invariant texture learning for lung tissue analysis using isotropic wavelet frames. IEEE Trans Inf Technol Biomed. 2012;16:665–675. doi: 10.1109/TITB.2012.2198829. [DOI] [PubMed] [Google Scholar]
- Depeursinge A, Vargas A, Platon A, Geissbuhler A, Poletti P-A, Müller H. Building a reference multimedia database for interstitial lung diseases. CMIG. 2012;36:227–238. doi: 10.1016/j.compmedimag.2011.07.003. [DOI] [PubMed] [Google Scholar]
- Girshick R, Donahue J, Darrell T, Malik J. CVPR. Columbus, OH: IEEE; 2014. Rich feature hierarchies for accurate object detection and semantic segmentation; pp. 580–587. [Google Scholar]
- Krizhevsky A, Sutskever I, Hinton GE. Imagenet classification with deep convolutional neural networks. NIPS. 2012:1097–1105. [Google Scholar]
- Li Q, Cai W, Wang X, Zhou Y, Feng DD, Chen M. Medical image classification with convolutional neural network. Paper presented at 2014 13th International Conference on Control Automation Robotics & Vision ( In ICCAR); Singapore. 2014. [Google Scholar]
- Prasoon A, Petersen K, Igel C, Lauze F, Dam E, Nielsen M. Deep feature learning for knee cartilage segmentation using a triplanar convolutional neural network. In: Mori K, Sakuma I, Sato Y, Barillot C, Navab N, editors. Medical Image Computing and Computer-Assisted Intervention – MICCAI; 2013 16th International Conference Proceedings, Part II; Nagoya, Japan. September 22–26, 2013; Berlin, Heidelberg: Springer Berlin Heidelberg; 2013. pp. 246–253. [DOI] [PubMed] [Google Scholar]
- Razavian AS, Azizpour H, Sullivan J, Carlsson S. CNN features off-the-shelf: an astounding baseline for recognition. In Computer Vision and Pattern Recognition Workshops (CVPRW); IEEE; 2014. pp. 512–519. [Google Scholar]
- Roth HR, Lu L, Seff A, Cherry KM, Hoffman J, Wang S, Liu J, Turkbey E, Summers RM. A new 2.5 D representation for lymph node detection using random sets of deep convolutional neural network observations. In: Golland P, Hata N, Barillot C, Hornegger J, Howe R, editors. Medical Image Computing and Computer-Assisted Intervention -- MICCAI; 2014 17th International Conference Proceedings, Part I; Boston, MA, USA. September 14–18, 2014; Cham: Springer International Publishing; 2014. pp. 520–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russakovsky O, Deng J, Su H, Krause J, Satheesh S, Ma S, Huang Z, Karpathy A, Khosla A, Bernstein M, et al. Imagenet large scale visual recognition challenge. Int J Comput Vision. 2005;115:211–252. doi: 10.1007/s11263-015-0816-y. [DOI] [Google Scholar]
- Song Y, Cai W, Huang H, Zhou Y, Feng D, Wang Y, Fulham M, Chen M. Large margin local estimate with applications to medical image classification. TMI. 2015;34:1362–1377. doi: 10.1109/TMI.2015.2393954. [DOI] [PubMed] [Google Scholar]
- Song Y, Cai W, Zhou Y, Feng DD. Feature-based image patch approximation for lung tissue classification. TMI. 2013;32:797–808. doi: 10.1109/TMI.2013.2241448. [DOI] [PubMed] [Google Scholar]
- Vedaldi A, Lenc K. Matconvnet-convolutional neural networks for matlab. Proceedings of the 23rd ACM International Conference on Multimedia, MM ’15; Brisbane, Australia. 2015; New York, NY: ACMSA; 2014. pp. 689–692. [DOI] [Google Scholar]
- Webb WR, Muller NL, Naidich DP. High-resolution CT of the lung. Philadelphia, PA: Lippincott Williams & Wilkins; 2014. [Google Scholar]