Abstract
In human cells, there are three genes that encode DNA ligase polypeptides with distinct but overlapping functions. Previously small molecule inhibitors of human DNA ligases were identified using a structure-based approach. Three of these inhibitors, L82, a DNA ligase I (LigI)-selective inhibitor, and L67, an inhibitor of LigI and DNA ligases III (LigIII), and L189, an inhibitor of all three human DNA ligases, have related structures that are composed of two 6-member aromatic rings separated by different linkers. Here we have performed a structure-activity analysis to identify determinants of activity and selectivity. The majority of the LigI-selective inhibitors had a pyridazine ring whereas the LigI/III- and LigIII-selective inhibitors did not. In addition, the aromatic rings in LigI-selective inhibitors had either arylhydrazone or acylhydrazone, but not vinyl linkers. Among the LigI-selective inhibitors, L82-G17 exhibited increased activity against and selectivity for LigI compared with L82. Notably. L82-G17 is an uncompetitive inhibitor of the third step of the ligation reaction, phosphodiester bond formation. Cells expressing DNA ligase I were more sensitive to L82-G17 than isogenic LIG1 null cells. Furthermore, cells lacking nuclear LigIIIα, which can substitute for LigI in DNA replication, were also more sensitive to L82-G17 than isogenic parental cells. Together, our results demonstrate that L82-G17 is a LigI-selective inhibitor with utility as a probe of the catalytic activity and cellular functions of LigI and provide a framework for the future design of DNA ligase inhibitors.
INTRODUCTION
DNA ligation is required to generate an intact lagging strand during DNA replication as well as in almost every recombination and DNA repair event. In human cells, this reaction is carried out by the DNA ligases encoded by the three human LIG genes, LIG1, LIG3 and LIG4 (1). Genetic analysis has revealed that there is considerable functional overlap among the DNA ligases encoded by the three LIG genes in nuclear DNA transactions (2–11). A mitochondrial version of DNA ligase IIIα (LigIIIα) is generated by alternative translation initiation (12–15). In addition, alternative splicing of the LIG3 gene in male germ cells results in LigIIIβ, which has a different C-terminal region than LigIIIα (16).
The steady state level of LigI is frequently elevated in cancer cell lines and tumor samples (17,18). This presumably reflects the hyperproliferative state of cancer cells since LigI is the predominant ligase involved in DNA replication (19–21). Unexpectedly, many cancer cell lines exhibit both increased steady state levels of LigIIIα and reduced steady state levels of DNA ligase IV (LigIV), with these reciprocal changes indicative of alterations in the relative contribution of different DNA double-strand break repair pathways between non-malignant and cancer cells (18,22–25). The dysregulation of DNA ligases in cancer cells together with the involvement of these enzymes in the repair of DNA damage caused by agents used in cancer chemotherapy and radiation therapy suggests that DNA ligase inhibitors may have utility as cancer therapeutics.
A set of small molecule LigI inhibitors were identified through an in silico structure-based screen, using the atomic resolution structure of LigI complexed with nicked DNA (18,26). This screen yielded inhibitors that were selective for LigI (L82), inhibited both LigI and LigIII (L67) and inhibited all three human DNA ligases (L189). As expected, subtoxic levels of the DNA ligase inhibitors enhanced the cytotoxicity of DNA damaging agents in cancer cell lines (18). Surprisingly, non-malignant cell lines were not sensitized to DNA damage by the DNA ligase inhibitors under similar conditions, suggesting that there are alterations in genome maintenance pathways between non-malignant and cancer cells (18). Further studies revealed that the repair of DNA double-strand breaks is abnormal in cancer cells with elevated levels of LigIIIα and PARP1 and that these cells are hypersensitive to inhibitors that target LigIIIα and PARP1 (22,23,25).
The DNA ligase inhibitors L82, L67 and L189 are similar in that they are each composed of two 6-member aromatic rings separated by different length linkers (18,26). Here we have examined a series of related compounds in an attempt to identify determinants of activity and selectivity for LigI and LigIIIα. One of the compounds analyzed, L82-G17, is a selective, uncompetitive inhibitor of LigI. Furthermore, the activity of this compound in cell culture assays with genetically-defined cell lines indicates that it inhibits LigI function in cells.
RESULTS
Biochemical activity of L82 derivatives
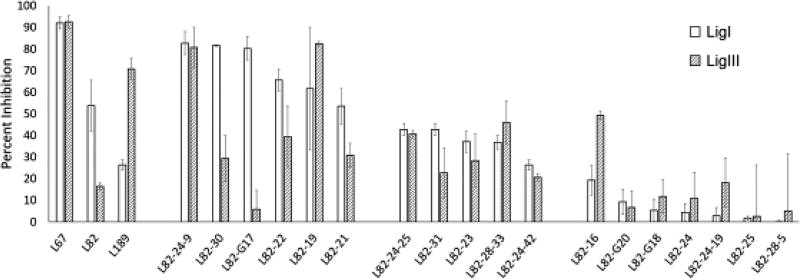
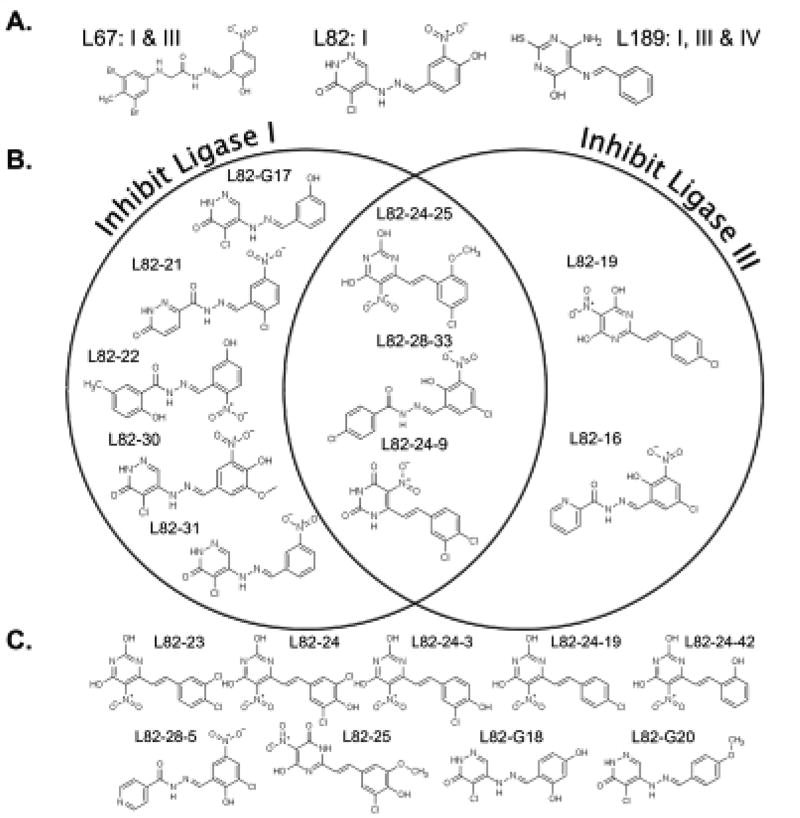
To gain insights into determinants of activity and selectivity of the structurally similar DNA ligase inhibitors, L67, L82 and L189 (18,26), we examined the activity of related derivatives that were either synthesized (L82-GXX) or purchased (L82-XX) on LigI, LigIII and T4 DNA ligase activity. DNA ligase IV/XRCC4 (LigIV/XRCC4) was not included in these assays as the purified enzyme, unlike LigI, LigIII and T4 DNA ligase, acts as a single turnover enzyme (27,28). Any compounds that inhibited T4 DNA ligase, which lacks the DNA binding domain targeted in the structure-based screen (18), were presumed to be non-specific inhibitors and excluded (data not shown). The activities of the remaining compounds were compared with the previously described DNA ligase inhibitors, L82, L67 and L189 at 50 µM (Fig. 1). Among the L82 derivatives, there were 10 compounds that inhibited LigI and/or LigIII by at least 40%. In Figure 2, the structures of L82, L67 and L189 (Fig. 2A) and the active (Fig. 2B) and inactive L82 derivatives (Fig. 2C) are shown with compounds grouped based on their activity against LigI and LigIII. Among the active compounds, 5 compounds preferentially inhibited LigI with two compounds, L82-30 and L82-G17 exhibiting increased selectivity for LigI compared with L82, two compounds preferentially inhibited LigIII and 3 compounds had similar activity against both LigI and LigIII (Fig. 1 and Fig. 2B).
Figure 1. Activity of compounds related to L67, L82 and L189.
The effect of L67, L82, L189 and L82 derivatives (50 µM of each) on DNA joining by LigI (0.6 nM) and LigIII (1.7 nM) was measured in assays with the radiolabeled DNA substrate as described in Materials and Methods. Results are shown graphically with inhibition expressed as a percentage of ligation activity in assays with DMSO alone. The data shown, which is sorted based on activity against LigI, represents the results of at least three independent experiments.
Figure 2. Chemical structures and activity of DNA ligase inhibitors identified by computer aided drug design and their derivatives.
A. Structures and selectivity of three previously described small-molecule inhibitors of human DNA ligases are shown. B. Chemical structures of L82 derivatives that are grouped based on their selectivity for LigI (inhibit LigI more than LigIII by at least 20% and inhibit LigI by at least 40%), selective for LigIII (inhibit LigIII more than LigI by at least 10% and inhibit LigIII by at least 15%)or have similar activity against LigI and LigIII, C. Compounds that have less than 40% activity against both enzymes.
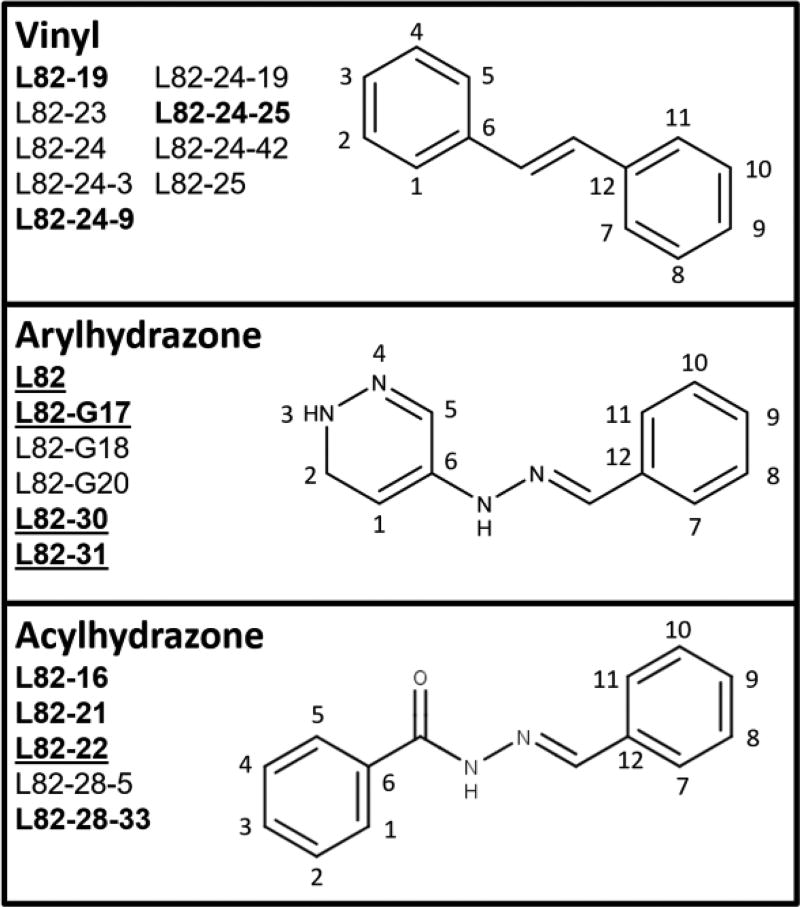
There are three types of linkers between the two aromatic rings of all the DNA ligase inhibitors, vinyl (Fig. 3, upper panel), arylhydrazone (Fig. 3, middle panel), and acylhydrazone (Fig.3, lower panel), linking the rings with 2, 3, and 4 atoms, respectively. None of the LigI-selective inhibitors has a vinyl linker. In addition, all the LigI-selective inhibitors except for L82-22 have a pyridazine ring whereas the LigI/LigIII and LigIII inhibitors do not. The repositioning of a hydroxyl group on the non-pyridazine ring from para to meta, and the removal of a nitro group (Fig. 2), appears to increase the selectivity of L82-G17 for LigI (Fig. 1). Active inhibitors with an arylhydrazone linker have at least one polar group, such as the phenol in L82-G17, at the meta positions (8 and 10, Fig. 3, middle panel). This also occurs in LigI-selective acylhydrazone class inhibitors L82-21 and L82-22, which have either a nitro or a hydroxyl group at their position 10 (Fig. 3, lower panel).
Figure 3. L82 derivatives grouped by their linkers.
All L82 derivatives in this study fall into three structural groups: (A) vinyl-linked, (B) arylhydrazone-linked, (C) and acylhydrazone-linked inhibitors. The members of each group are identified here. Compounds that inhibit either LigI or LigIII are shown in bold text, LigI specific inhibitors are also underlined.
Comparing geometric shape coefficients (29,30), a value that represents a molecule’s potential size based on the connections between atoms, the mean value for LigI-selective inhibitors (7.4) was significantly different (p < 0.05) than that of LigI/III inhibitors (9.5). The mean geometric shape coefficient of LigI-selective inhibitors was also significantly different than that of compounds that do not inhibit either LigI or LigIII. A further point of differentiation between LigI-selective and LigI/LigIII inhibitors is their calculated partition coefficient (LogP). The calculated LogP is significantly lower (p < 0.01) for LigI-selective inhibitors (2.53) than inhibitors of both LigI and LigIII (4.6).
L82-G17 is an uncompetitive inhibitor of LigI
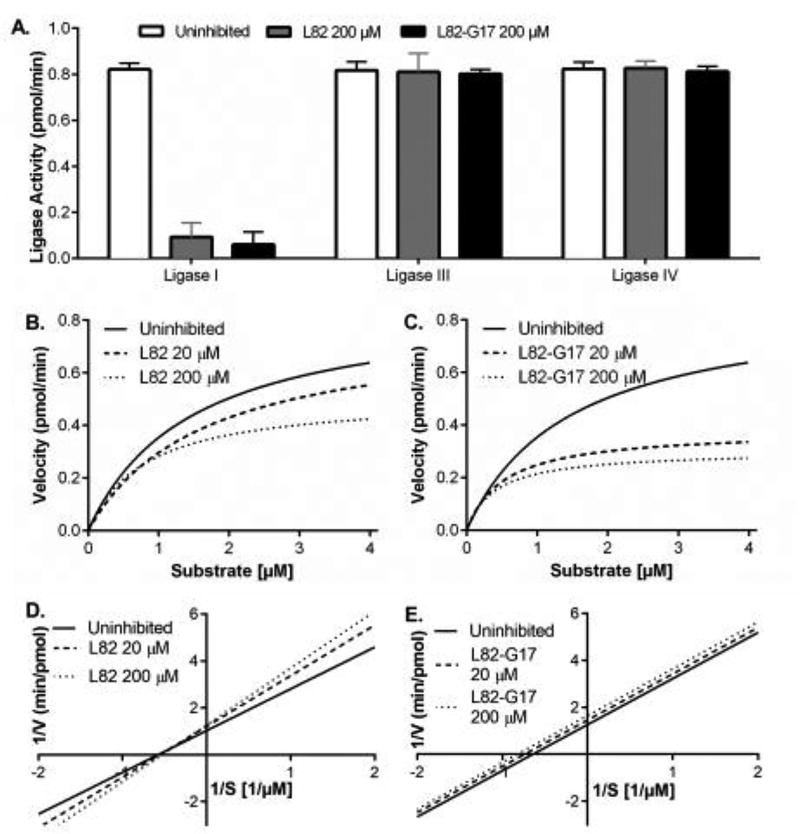
We chose to focus on characterizing L82-G17 because of its higher potency and increased selectivity for LigI. L82-G17 is more related to L82 than L67 with Tanimoto similarity scores of 78% and 32%, respectively, calculated using the Maximum Common Substructure method (31,32). As was observed with L82 (18), L82-G17 did not inhibit LigIV at 200 µM (Fig. 4A). Among the LigI inhibitors identified by computer-aided drug design, L82 was unique in that it appeared to act as an uncompetitive rather than a competitive inhibitor (18). This prompted us to examine the effects of L82 (Fig. 4B) and L82-G17 (Fig. 4C) on the kinetics of ligation by LigI. Under these reaction conditions, the Vmax and Km values for DNA ligase were 0.9 pmol ligations per min and 1.4 µM, respectively. Notably, the addition of L82 increased Km and decreased Vmax, whereas L82-G17 reduced both Km and Vmax. The Lineweaver-Burk plots obtained with L82 (Fig. 4D) indicate that this compound is a mixed inhibitor. In contrast, the Lineweaver-Burk plots obtained with L82-G17 indicate that this compound is an uncompetitive inhibitor (Fig. 4E). It is likely that differences in experimental conditions underlie the discrepancy between the results of the kinetic analyses shown in Figure 4 and the published study (18). Here we used a shorter incubation time (5 min versus 30 min) and higher concentrations of protein and DNA substrate to more accurately measure initial and maximal reaction velocities of LigI.
Figure 4. L82-G17 is a selective uncompetitive inhibitor of LigI.
(A) LigI (500 fmol), LigIII (500 fmol) and LigIV (2 pmol) were pre-incubated with 200 µM of either L82 (gray) or L82-G17 (black) for 30 min at 25°C prior to incubation with the fluorescent nicked DNA substrate (1 pmol) for 5 min at 25°C as described in Materials and Methods. The results of three independent experiments are shown graphically. The effect of L82 (B) and L82-G17 (C) on Michaelis-Menten saturation curves of reaction velocity (pmol/min) versus DNA substrate concentration (µM) derived from at least three independent fluorescence-based ligation assays are shown. Lineweaver-Burk double reciprocal plots of the same data for L82 (D) and L82-G17 (E) calculated using the standard error of mean and showing the extrapolated trendline. Solid line, no inhibitor; long dashes, 20 µM inhibitor; short dashes, 200 µM. Curve fitting was performed with Graphpad PRISM analysis software.
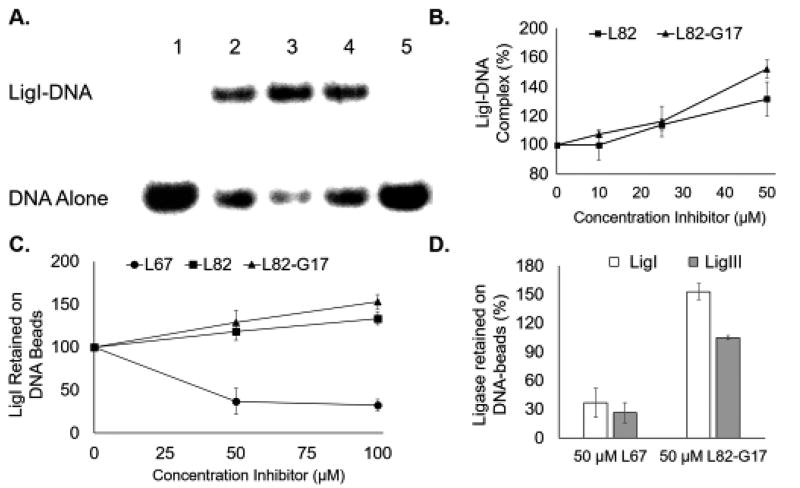
Given the discordant results with L82 in the enzyme kinetic assays, we perfomed an electrophoretic mobility shift assay (EMSA) with a linear duplex containing a single non-ligatable nick that had been used previously to distinguish between uncompetitive and competitive inhibitors of LigI (26). In accord with the published study (26), L82 increased the amount of LigI-DNA complex formed (Fig. 5A, compare lanes 2 and 4, and Fig. 5B) whereas the competitive inhbitor L67 prevented complex formation (Fig. 5A, compare lanes 2 and 5, and B), confirming that L82 is able to act as an uncompetitive inhibitor. As expected, L82-G17 also increased the amount of LigI-DNA complex formed in concentration-dependent manner (Fig. 5A, compare lanes 2 and 3, and Fig. 5B). In pulldown assays with streptavidin beads liganded by a biotinylated linear duplex containing a single non-ligatable nick, both L82 and L82-G17 increased the amount of labeled LigI retained by the beads in a concentration-dependent manner (Fig. 5C) whereas only 14% and 3.6% of LigI was retained by beads liganded by intact double-stranded DNA, and single-stranded DNA, respectively (data not shown). To confirm the selectivity of L82-G17 for LigI, we performed pull down assays with both LigI and LigIII. As expected, the LigI/III competitive inhibitor L67 reduced the binding of both LigI and LigIII to the beads. In contrast, L82-G17 increased the retention of LigI by the beads but had very little effect on LigIII binding (Fig. 5D). Taken together these results demonstrate that L82-G17 is a selective, uncompetitive LigI inhibitor.
Figure 5. L82 and L82-G17 increase LigI binding to non-ligatable nicked DNA binding.
A. A representative gel showing the effect of the inhibitors, L67 (lane 5), L82 (lane 4) and L82-G17 (lane 3), at 100 µM and DMSO alone (lane 2) on DNA-protein complexes formed by LigI with non-ligatable nicked DNA. Lane 1, DNA substrate alone. The position of the DNA substrate (DNA alone) and the protein-DNA complex (LigI-DNA) are shown. B. Results of at least three independent EMSA assays are shown graphically with DNA-protein complex formation calculated as the ratio of bound and unbound DNA and expressed as percentage of the ratio obtained in reactions with LigI alone. L82 (closed squares) and L82-G17 (closed triangles). C. The effect of L82 and L82-G17 on the amount of labeled LigI retained by streptavidin beads liganded by biotinylated nicked DNA. Results of three independent assays are shown graphically and expressed as a percentage LigI retained in assays with DMSO alone. L82 (closed squares), L82-G17 (closed triangles) and L67 (closed circles). D. The effect of L67 and L82-G17 on the amount of labeled LigI (open squares) and LigIII (filled squares) retained by streptavidin beads liganded by biotinylated nicked DNA. Results of three independent assays are shown graphically and expressed as a percentage LigI/LigIII retained compared with assays with DMSO alone.
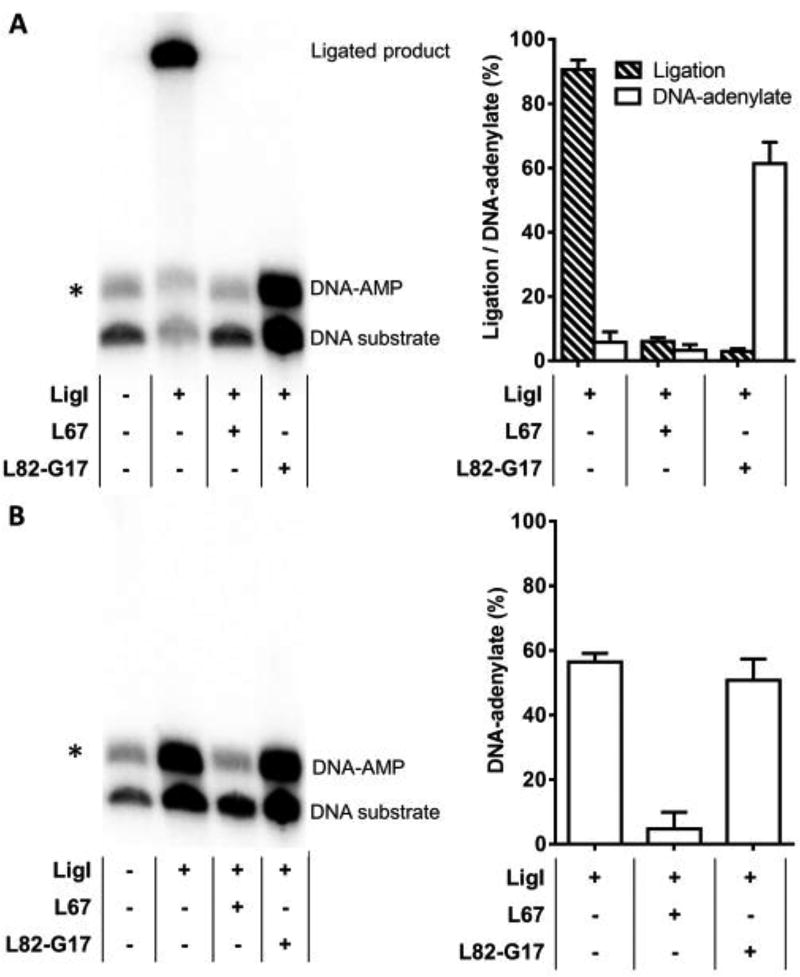
Since L82-G17 stabilizes the LigI-nicked DNA complex, we examined its effect on the reactions, transfer of the adenylate group from LigI to the 5’ phosphate terminus of a DNA nick, and phosphodiester bond formation, that occur within the LigI-nicked DNA complex. In reactions with a ligatable substrate, the competitive inhibitor, L67, inhibited ligation as expected (Fig. 6A). While L82-G17 also inhibited ligation, approximately 50% of the substrate was converted to the DNA-adenylate reaction intermediate whereas no DNA-adenylate was detected in the reactions with L67 (Fig. 6A). To confirm that L82-G17 does not inhibit transfer of the adenylate group to the 5’ phosphate termini at the DNA nick, we performed similar studies with a DNA substrate containing a non-ligatable nick. As expected, the DNA-adenylate intermediate accumulated in reactions with LigI (Fig. 6B). While L67 inhibited DNA-adenylate formation, similar amounts of the DNA-adenylate were detected in the presence of L82-G17 compared with LigI alone (Fig. 6B). Thus, we conclude that L82-G17 is a step 3 inhibitor that stabilizes the complex formed between non-adenylated LigI and the DNA-adenylate.
Figure 6. L82-G17 inhibits step 3 of the ligation reaction, phosphodiester bond formation.
A. LigI incubated with radiolabeled ligatable DNA substrate in the presence or absence of L67 and L82-G17 as described in Materials and Methods. Left panel, a representative gel. Right panel, the results of at least three independent experiments are shown graphically. B. LigI incubated with radiolabeled non-ligatable DNA substrate in the presence or absence of L67 and L82-G17 as described in Materials and Methods. Left panel, a representative gel. Right panel, the results of at least three independent experiments are shown graphically. The positions of the labeled DNA substrate, DNA adenylate and ligated product are indicated. The asterisk indicates a contaminating band from the construction of the radiolabeled oligonucleotide substrates. Ligated product and DNA-adenylate expressed as a percentage of total DNA input after subtraction of the contaminating band.
Effects of L82-G17 on replicative DNA synthesis and cell proliferation
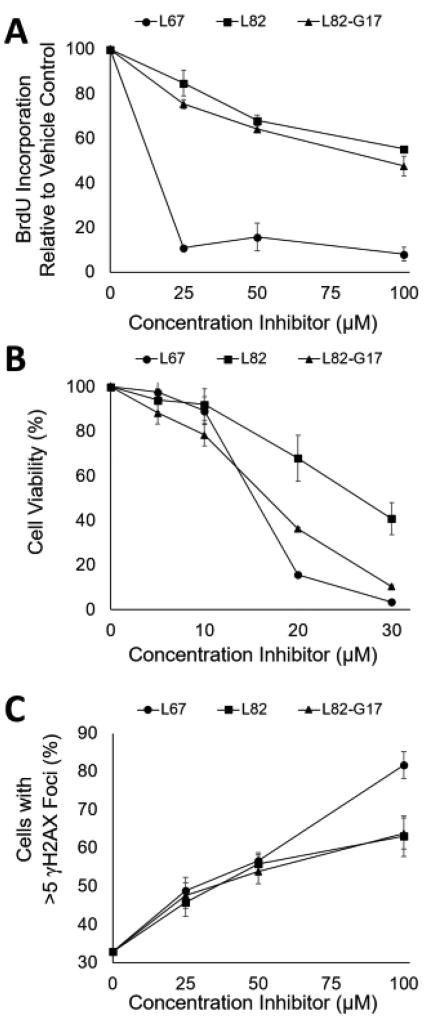
Since LigI is the DNA ligase predominantly responsible for joining Okazaki fragments during replicative DNA synthesis (19,21,33), we determined whether L82-G17 and L82 effected the incorporation of bromodeoxyuridine (BrdU) by asynchronously proliferating HeLa cells. While both L82 and L82-G17 reduced BrdU incorporation, their effects were modest compared with the LigI/III inhbitor L67 (Fig. 7A). This is consistent with studies showing that LigI is not essential for mammalian DNA replication becase of the ability of LigIII to act as a back-up (2,11,33,34). Since the DNA synthesis assay involved a relatively short incubation with the DNA ligase inhibitors, we examined their impact on cell proliferation over a 72 h time period using the MTT assay that quantitates the activity of NAD(P)H-dependent cellular oxidoreductase enzymes, an indicator of metabolic activity that correlates with the number of viable cells. L82-G17 was a more effective inhibitor of proliferation, reducing cell number by about 70% at 20 µM compared with a 30% reduction with 20 µM L82 (Fig. 7B). Similar results were obtained with L82 and L82-G17 using a CYQUANT assay that measures genomic DNA, another indicator of cell number (35) (data not shown).
Figure 7. Effects of ligase inhibitors on DNA synthesis, cell viability and DNA damage.
The effects of L67, (closed circles) L82, (closed squares) or L82-G17 (closed triangles) on BrdU incorporation, cell viability and formation of γH2AX foci were determined as described in Materials and Methods. A. Asynchronous HeLa cells were treated with the ligase inhibitors for 4 hours and were then assayed for BrdU incorporation. Results of three independent assays are shown graphically. B. HeLa cells were incubated with the inhibitors for 5 days prior to the determination of cell viability using the MTT assay. Results of three independent assays are shown graphically. Data points at 20 and 30 µM are significant at p < 0.005. C. HeLa cells were incubated with the ligase inhibitors for 4 hours prior to the detection of γH2AX foci by immunocytochemistry. Cells that contained at least 5 foci were counted as γH2AX positive. At least 100 cells counted per data point. The results of 3 independent assays shown.
Since L82 and L82-G17 had more severe effects on proliferation (Fig. 7B) than replicative DNA synthesis (Fig. 7A), we asked whether these compounds induced DNA damage that, in turn, would activate cell cycle checkpoints. Acute exposure to either L82 or L82-G17 for 4h, resulted in a concentration-dependent increase in formation of γH2AX foci, an indicator of DNA double-strand breaks (DSBs) (Fig. 7C) (36). In accord with the cell proliferation (Fig. 7B) and DNA synthesis (Fig. 7A) assays, L67 was more effective at inducing γH2AX foci than either L82 or L82-G17 (Fig. 7C).
Cells lacking LigI are more resistant to L82 and L82-G17
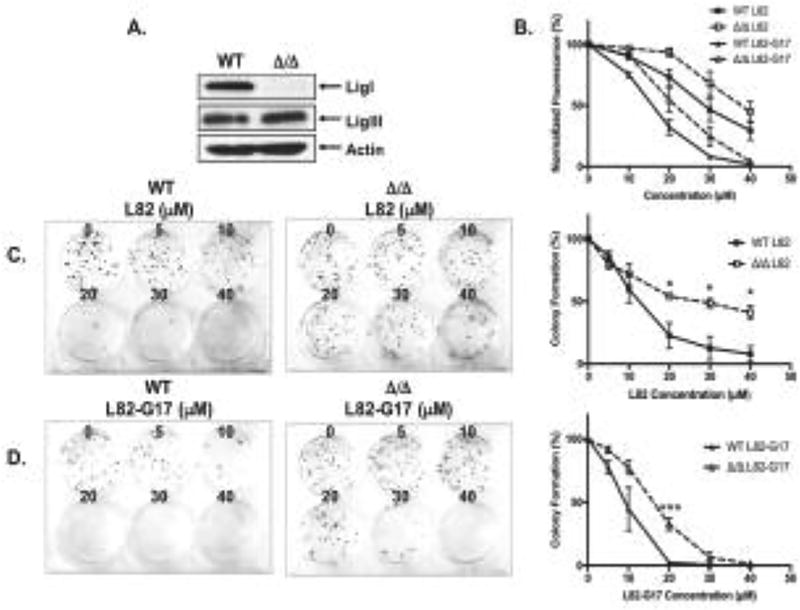
While the effects of L82-G17 and L82 on proliferation, BrdU incorporation and DNA damage are consistent with these inhibitors impacting DNA replication by inhibiting LigI, it is possible that they may be due to off-target effects. This prompted us to compare the effects of L82 and L82-G17 on the parental and LIG1 null derivatives of the mouse B cell line, CH12F3 (Fig. 8A) (2,37). Notably, both L82 and L82-G17 had a greater effect on the proliferation (Fig. 8B) and survival (Fig. 8C and 8D) of the parental CH12F3 cells compared with the LIG1 null derivative, suggesting that the actvity of these inhbitors is dependent, at least in part, on the presence of LigI.
Figure 8. Cells lacking LigI are more resistant to L82 and L82-G17.
A. LigI, LigIII and β-actin proteins were detected in extracts of CH12F3 WT and CH12F3 Δ/Δ cells by immunoblotting. B. Effect of L82 (square) and L82-G17 (triangle) on the proliferation of CH12F3 WT (filled symbols) and CH12F3 Δ/Δ (empty symbols) cells was measured by the CyQUANT assay as described in Materials and Methods. Effect of L82 (C) and L82-G17 (D) on colony formation by CH12F3 WT and CH12F3 Δ/Δ cells. Data shown graphically are the mean ±SEM of three independent experiments and are expressed as a percentage of the values for the untreated cells. * p < 0.05 and *** p < 0.001 using the unpaired two-tailed Student test.
The absence of nuclear LigIIIα increases sensitivity to L82 and L82-G17
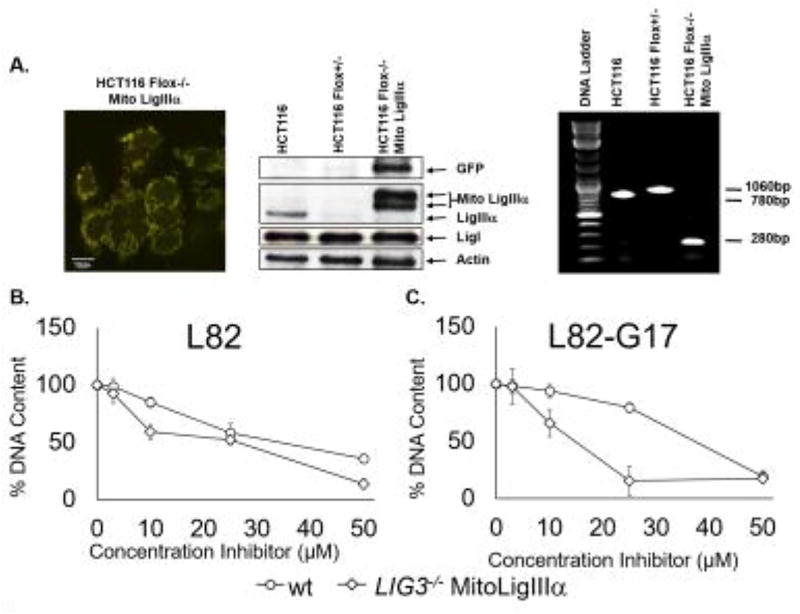
Cells with both nuclear LigI and LigIIIα should be more resistant to a selective LigI inhibitor compared with cells with only nuclear LigI because of the functional redundancy between LigI and LigIIIα in DNA replication (6,11). To confirm this, we examined the effects of L82 and L82-G17 on the proliferation of a derivative of the human colorectal cancer cell line HCT116 that lacks nuclear LigIIIα and its parental cell line (Fig. 9A). The absence of nuclear LigIIIα markedly increased the inhibitory activity of L82-G17 (Fig. 9C) on cell proliferation whereas L82 had a smaller effect (Fig. 9B).Taken together, our results indicate that the activities of L82 and, in particular, L82-G17 in cell culture assays are due, at least in part, to inhibition of LigI.
Figure 9. Cells lacking nuclear LigIIIα are more sensitive to L82 and L82-G17.
A. Fluorescent image of HCT116 Flox−/− expressing YFP-tagged mito LigIIIα. Scale bars, 10 µm (left panel). Immunoblots with extracts of HCT116, HCT116 Flox+/− and HCT116 Flox−/− Mito LigIIIα using antibodies against GFP, LigIII, LigI and β-actin. The positions of YFP-fusion protein (GFP), mito LigIIIα fused to GFP (Mito LigIIIα) and endogenous LigIIIα, LigI and β-actin are indicated (middle panel). Wild type and Floxed LIG3 alleles and the integrated cDNA encoding YFP-tagged Mito LigIIIα were detected in genomic DNA from HCT116, HCT116 Flox+/− and HCT116 Flox−/− Mito Ligase IIIα by PCR (right panel). Proliferation of HCT116 cells (circles) and a derivative lacking nuclear LigIIIα (diamonds) incubated with (B) L82 or (C) L82-G17 for 5 days was measured by CyQUANT as described in Materials and Methods. Results of three independent assays are shown graphically.
Discussion
There is emerging interest in the use of DNA repair inhibitors to exploit either cancer cell-specific alterations in genome maintenance pathways or increased dependence upon a specific DNA repair pathway (38). The abnormal expression of DNA ligases in cancer cell lines and samples from cancer patients suggest that DNA ligase inhibitors may have utility as anti-cancer agents either alone or in combination with DNA damaging agents (17,18,22,23,25). Following the determination of the atomic resolution structure of LigI complexed with nicked DNA, small molecule inhibitors with differing activities against three human DNA ligases were identified by computer-aided drug design (18,26). The DNA ligase inhibitors were separated into three groups, LigI-selective, LigI/III-selective and inhibitors of all three human DNA ligases with L82, L67 and L189, respectively, serving as the prototypical compound for each of the groups. Each of these initial compounds contained two aromatic rings but had linkers that differed in length and chemical composition. Here we examined the activity of compounds that are related to L82, L67 and L189 to gain insights into determinants of activity and selectivity. The majority of LigI selective inhibitors had a 3-atom arylhydrazone linker. In addition, the presence of at least one polar group on the right-hand ring appeared to be required for activity. While none of compounds with the arylhydrazone linker had activity against LigIII, indicating the potential utility of this linker in the rational design of LigI-selective inhibitors, the hydrazone linkage is unlikely to be compatible with drug development as this functional group has been shown to be a promiscuous metal scavenger which can lead to toxicity issues (39–41).
Among the compounds related to L82, L67 and L189, we identified one compound, L82-G17 that exhibited increased activity against and increased selectivity for LigI compared with L82. Notably, it has the lowest molecular weight of all arylhydrazone inhibitors with activity against LigI, suggesting that it may represent the minimal requirements for this type of inhibitor. Kinetic analysis revealed that L82-G17 is an uncompetitive inhibitor whereas L82 appears to act by both competitive and uncompetitive mechanisms. As expected, both L82 and L82-G17 enhanced the formation of stable complexes of LigI with nicked DNA. Since L82-G17 inhibits phosphodiester bond formation but not formation of the DNA-adenylate, we conclude that L82-G17 is a step 3 inhibitor that stabilizes the complex formed by non-adenylated LigI with the DNA-adenylate reaction intermediate. Although L82-G17 acts by the same mechanism as topoisomerase (topo) inhibitors, such as camptothecin, and a subset of Poly(ADP-Ribose) Polymerase (PARP) inhibitors that trap topo I-DNA and PARP1-DNA complexes, respectively (42,43), we did not observe an increase in the amount of LigI associated with chromatin in cells exposed to L82-G17 (data not shown). This may be because LigI, unlike PARP1, does not have robust DNA binding activity (42,44–46) and is recruited to sites of DNA replication via an interaction with PCNA (47,48). The increased cytotoxicity of L82-G17 compared with L82 is consistent with the studies showing that PARP inhibitors that trap PARP1-DNA complexes are more cytotoxic than PARP inhibitors that do not (42,45). It is presumed that DNA-protein complexes, even when non-covalent, cause problems because of collisions with either the DNA replication or transcription machinery. In contrast to the trapped DNA-protein complexes formed by topoisomerases and PARPs, the replication machinery is unlikely to encounter trapped LigI-PCNA complexes as these will predominantly formed behind the fork on the lagging strand.
Since the initial identification of DNA ligase inhibitors by a structure-based approach (18,26), there have been several reports describing LigI inhibitors using computer modelling and derivatives of the original DNA ligase inhibitors (49–51). While these studies have shown that the inhibitors have activity against LigI in vitro, their affinity and selectivity appears to be less than that of L82-G17. Furthermore, there is no definitive evidence that these inhibitors target LigI in cells. Here we have shown that LIG1 null cells are more resistant to L82-G17, presumably because there is no formation of trapped LigI-DNA complexes. Furthermore, cells that lack nuclear LigIIIα are more sensitive to L82-G17 as they lack a back-up activity for DNA replication.
The elevated levels of LigI in cancer cells (17,18) and the apparent viability of mammalian cells that lack LigI (2,33,34) suggest that LigI selective inhibitors may preferentially target cancer cells because of their high proliferation rate. Toxicity in normal cells is likely be limited because of the ability of LigIIIα to substitute for LigI in DNA replication and repair (6,11) In the structure-activity studies described here, we have identified and characterized a novel uncompetitive inhibitor of LigI that selectively targets LigI both in in vitro and in cells. Further work to develop improved uncompetitive inhibitors would be facilitated by the determination of atomic resolution structures of inhibitor-bound LigI-DNA complexes (46). Alternatively, the predicted binding site for the LigI inhibitors could be validated using site-directed mutagenesis to generate versions of LigI that retain wild type catalytic activity but are resistant to the inhibitors.
Materials & Methods
Chemicals
L67 (IUPAC: 2-[(3, 5-dibromo-4-methylphenyl)amino]-N'-[(2-hydroxy-5-nitrophenyl)methylidene]acetohydrazide), L82 (IUPAC: 4-chloro-5-{2-[(4-hydroxy-3-nitrophenyl)methylidene]hydrazin-1-yl-2,3-dihydropyridazin-3-one, and L189 (6-amino-5-[(phenylmethylidene)amino]-2-sulfanylpyrimidin-4-ol) were originally purchased from ChemDiv and Specs. Milligram quantities of each of the derivatives reported herein were also purchased from ChemDiv, Specs, or ChemBridge.
Large scale synthesis of L82-G17 (4-chloro-5-{2-[(3-hydroxyphenyl)methylidene]hydrazin-1-yl}-2,3-dihydropyridazin-3-one)
4,5-Dichloropyridazin-3(2H)one (660 mg, 4.0 mmol, 1.0 equiv) was dissolved in methanol (7 mL). Hydrazine monohydrate (380 mg, 7.6 mmol, 1.9 equiv) was dissolved separately in methanol (4 mL). Both solutions were combined and the reaction was heated at reflux for 1.5 hours. Additional methanol (3 mL) was added during heating, as the precipitate of product inhibited stirring. The reaction was cooled and the product was separated from the supernatant through vacuum filtration and dried overnight, yielding 5-chloro-4-hydrazidopyridazin-3(2H)one as a light yellow-brown powder (504 mg, 3.13 mmol, 78%). 1H NMR (DMSO, 400 MHz) δ 12.46 (s, 1H, N-H), 8.07 (s, 1H, aryl), 7.96 (s, 1H, N-H), 4.57 (s, 2H, NH2). This intermediate (100 mg, 0.62 mmol, 1.0 equiv) was dissolved into ethanol (1000 mL) with heat and 3-hydroxybenzaldehyde (91 mg, 0.75 mmol, 1.2 equiv) was added as a solid. The reaction mixture was heated under reflux overnight, then concentrated to 5 mL to precipitate a light brown solid. The product was collected, washed with ethanol, and dried under vacuum, yielding 78 mg, 0.295 mmol, 48%). 1H NMR (DMSO, 400 MHz) δ 12.86 (s, 1H, N-H), 10.74 (s, 1H, N-H), 9.59 (s, 1H, O-H), 8.32 (s, 1H, imine C-H), 8.31 (s, 1H, Ar) 7.23 (t, 1H, Ar), 7.12 (s, 1H, Ar), 7.11 (d, 1H, Ar), 6.80 (d, 1H, Ar).
MarvinSketch v.14.9.8.0 (ChemAxon (http://www.chemaxon.com)) was used for drawing chemical structures. Tanimoto similarity scores were calculated using the Maximum Common Substructure algorithm in online tool ChemMine (32).
Proteins
After expression in E. coli BL21(DE3) cells, N-terminally poly(histidine)-tagged wild type human LigI was purified by HisTrap and HiTrap Q (GE Healthcare Life Sciences) column chromatography as described (52). In addition, a version of LigI with C-terminal poly(histidine), FLAG tags and PKA site was expressed in Rosetta2 cells and then purified by HisTrap HP and HiTrap Q column (GE Healthcare Life Sciences) column chromatography. Purified LigI fractions were pooled, concentrated using a 50 kDa MWCO centrifugal filter (EMD Millipore), and then stored at −80°C. LigIIIβ and the LigIV/XRCC4 complex were purified from E. coli and insect cells, respectively as described previously (53,54).
DNA Ligation Assays
Nick joining was measured using radioactive- and fluorescence-based assays (53,54). The concentrations of oligonucleotides (Integrated DNA Technologies) DNA were measured using absorbance at 260 nm. In radioactive assays, the oligonucleotide (5’ CAAATCTCGAGATCACAGCAACTAGACCA) was 5’ end labeled with polynucleotide kinase and [γ-32P]ATP. After removing excess ATP using a P-30 polyacrylamide spin column (Bio-Rad), the labeled oligonucleotide was annealed with a longer complementary template oligonucleotide (5’ CCATCTTAAGGAGTGCAGTAGAAGTCCAGATCAACGACACTAGAGCTCTAAACGCAAGGCAACTGCATGGACGAACACTCGCGAGTGTTAATG) and another oligonucleotide (5’-GTAATTGTGAGCGCTCACAAGCAGGTACGTCAACGGAACG) that was also complementary to the template oligonucleotide, to generate a linear duplex with a single radiolabeled ligatable nick. DNA ligases and putative inhibitors were pre-incubated for 10 minutes at room temperature prior to the addition of the labeled DNA substrate (500 fmol) and incubation at 25°C for 30 minutes in a final volume of 20 µl containing 25 mM Tris-HCl pH 7.5, 12.5 mM NaCl, 6 mM MgCl2, 0.4 mM ATP, 0.25 mg/mL BSA, 2.5% glycerol, 0.5 mM DTT, and 0.5% DMSO. Reactions were stopped by placing reaction tubes on ice followed by the addition of formamide dye. After heating at 95°C for 5 minutes, labeled oligonucleotides were separated by electrophoresis through a 15% polyacrylamide-urea gel and then quantitated by phosphorimager analysis on a Typhoon FLA 7000, using a 650nm laser (software version 1.2). Images were analyzed using the Fuji Film MultiGauge software (version 3.0).
In the fluorescence-based ligation assay (53,54), purified LigI (500 fmol) was incubated in the presence or absence of either L82 or L82-G17 with fluorescent nicked DNA (1 pmol) for 5 min in a final volume of 20 µl containing 60 mM Tris–HCl pH 7.4, 50mM NaCl, 10 mM MgCl2, 5 mM DTT, 1 mM ATP, 50 µg/ml BSA, and 4% DMSO) at 25 °C. Following incubation, reactions were further diluted to 200 µL with a 30-fold molar excess of an unlabeled oligonucleotide (5’-TAGGAGGGCTTTCCTCCTCACGACCGTCAAACGACGGTCA) identical in sequence to the ligated strand in 10 mM Tris–HCl pH 7.4, 50 mM KCl, 1 mM EDTA and 5 mM MgCl2 for 5 min, and then heated to 95 °C for 5 min. After cooling to 4 °C at a rate of 2 °C/min, fluorescence at 519 nm (excitation at 495 nm) was measured immediately using the Synergy H4 microplate reader (BioTek). Data, including Michaelis-Menten and Lineweaver-Burk equations and kinetic values, were calculated and displayed graphically through GraphPad Prism 6.0 software.
DNA binding Assays
For electrophoretic mobility shift assays, a radiolabeled 73 bp linear duplex with a single nick was generated as above except that there was a dideoxycytosine residue at the 3’ terminus of the nick (18,46). Putative inhibitors, 1500 fmol DNA ligase and the 500 fmol nicked substrate were incubated in 50 mM Tris-HCl pH 7.5, 15 mM NaCl, 1 mM DTT, 0.2 mM ATP, 0.005 mg/mL BSA, 5 mM MgCl2, 8.75% glycerol and 0.5% DMSO for 30 min. After electrophoresis through a 5% non-denaturing polyacrylamide gel using 0.5 × TBE pH 8.3 as running buffer, labeled oligonucleotides were detected by phosphorimager and imaged as described above.
For DNA binding assays using magnetic beads, a non-radioactively labeled version of the DNA substrate used in the ligation assay was constructed with non-phosphorylated versions of the same three oligonucleotides except that a biotin molecule was attached to the 5’ end of long template oligo. Purified LigI was 32P-labeled by protein kinase A (New England Biolabs). Labeled LigI (1 pmol) and the biotinylated DNA substrate (1 pmol) bound to streptavidin magnetic beads were mixed in 20 mM HEPES-KOH pH 7.5, 20 mM NaCl, 10 mM MgCl2, 5 µg/mL BSA, and 4% glycerol in a final reaction volume of 40 µL prior to incubation at 25°C for 30 minutes prior to capture of the beads using a magnetic tube rack. The beads were washed 3-times with a 20 mM HEPES-KOH pH 7.5, 100 mM NaCl, and 10 mM MgCl2 before to heating at 95°C for 5 minutes with SDS sample buffer. Eluates from the beads were electrophoresed through a 10% denaturing polyacrylamide gel. Labeled LigI was detected by phosphorimager analysis and imaged as described above.
DNA Adenylation Assay
An oligonucleotide with either a 3’ deoxy-or a 3'dideoxy-nucleotide was 5’ end-labeled and then annealed with two oligonucleotides to generate a radio-labeled 107 bp duplex with a either a ligatable or non-ligatable nick. Purified LigI (1 pmol) was pre-incubated in the presence of either 200 µM L67, L82-G17, or DMSO control for 5 minutes at 25°C prior to the addition of the ligatable or non-ligatable DNA substrate (5 pmol), and incubated for 30 minutes at 20°C in 60 mM Tris–HCl (pH 7.4), 50 mM NaCl, 10 mM MgCl2, 5 mM DTT, 1 mM ATP, 50 µg/ml BSA, and 4% DMSO. Reactions were terminated by transfer to ice and addition of formamide dye. After electrophoresis through a 12.5% polyacrylamide-urea gel, labeled oligonucleotide products were detected and quantitated by phosphorimaging on a Typhoon FLA 7000. The NIH software ImageJ was used for densitometric analysis with data displayed graphically using Graphpad Prism 6.0.
Cell Lines
Human cervical (HeLa) and colorectal (HCT116) cancer cell lines were acquired from the ATCC (Manassas, VA) and grown in the recommended media. The mouse B cell lines CH12F3 Lig1 WT and CH12F3 Lig1 Δ/Δ (2,37) were maintained in RPMI 1640 medium supplemented with 10% fetal bovine serum (FBS), 1% penicillin/streptomycin and 55 µM β-mercaptoethanol. HCT116 cells were maintained in McCoy’s 5A supplemented with 10% FBS and 1% Pen/Strep. HCT116 LIG3Flox+/− cells containing a conditional LIG3 allele and a deletion allele (8) were further engineered to express mitochondrially-targeted human LigIIIα. A full length human LigIIIα cDNA that encodes the mitochondrial leader sequence was mutated so that the internal ATG start codon for nuclear LigIIIα was altered to ATC and then fused in-frame to an EYFP gene in the pCAG-YFP-neo plasmid to generate pCAG-MitoLigIII alpha-YFP-neo. After verification by structure DNA sequencing, this plasmid was transfected into HCT116Flox+/− cells and YFP-positive cells were selected by flow cytometry. To delete the remaining conditional LIG3 allele, cells were infected with an adenovirus type 5 (dE1/E3) virus encoding the Cre recombinase, (Ad-CMV-Cre #1045, Vector Biolabs). After 24 h, cells were washed and then cultured in fresh medium containing 0.5 mg/ml G418. Single cells were isolated in 96-well plates using an SY3200 cell sorter. Extracts of G418-resistant YFP-positive clones were probed by immunoblotting with antibodies to human LigIII (GeneTEX #103172), LigI (55) and GFP (Santa Cruz #8334). The LIG3−/− genotype was confirmed using primers Lig3 Exon 5 F1: 5'-AAA GCA ACC CTC CTG TCT TCT CCT GCA AGT-3' and Lig3 Exon 5 R1: 5'-TGG TAC CAG GGA TAG AGT CAC GGA CAA ACC AA-3'.
Bromodeoxyuridine (BrdU) incorporation
Asynchronous populations of HeLa cells were incubated with the DNA ligase inhibitors for 4 h prior to pulse labeling with BrdU (10 µM) for 45 min. Immunofluorescent staining of cells was performed using a BD BrdU flow Kit (BD Pharmingen) according to the manufacturer’s instructions and then quantitated by flow cytometry using an LSR Fortessa in the UNMCCC Shared Flow Cytometry and High Throughput Screening Resource. Total cellular DNA was stained with 7-AAD prior to analysis by flow cytometry. Data was analyzed using FlowJo (software version 10.1).
Cell Viability, Proliferation and survival assays
Cells were cultured in 96 well plates with either ligase inhibitors, or DMSO alone, for 5 days at 37°C. Cell viability was measured using the MTT assay, in which a tetrazolium dye, 3-(4, 5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide, is metabolized into (E,Z)-5-(4, 5-dimethylthiazol-2-yl)-1,3-diphenylformazan in the mitochondria, resulting in a color change from yellow to purple. After incubation with the MTT reagent (Promega) for one hour at 37°C per the manufacturer’s instructions, absorbance at 570 nm was measured using a PerkinElmer Victor 3V1420 Multilabel Counter. Cell viability is expressed as a percentage of the value obtained with DMSO-treated cells.
To quantitate genomic DNA as a measure of cell proliferation, cells were plated at density of 2000 per well in a 96-well in the presence or absence of L82 and L82-G17 and cultured for 72 h. After washing with 1 × PBS, the CyQUANT NF reagent (CyQUANT NF Cell Proliferation Assay Kit, Invitrogen) was added and incubation continued for 1h at 37°C according to the manufacturer’s instructions. Fluorescence intensities of triplicate samples were measured with a fluorescence microplate reader using excitation at 485 +/− 10nm and fluorescence detection at 530 +/− 15 nm. Cell number is expressed as a percentage of the value obtained with DMSO-treated cells.
Colony forming assays with CH12F3 Lig1 WT and CH12F3 Lig1 Δ/Δ cells were performed in methylcellulose-based media (Cat #HSC001 R&D Systems), which was diluted 1:3 with cell medium (RPMI Medium 1640, 10% FBS, 1% penicillin/streptomycin and 55 µM β-mercaptoethanol) for approximately 30 minutes without disturbance. Cells were counted in order to have 300 cells per well of a 6-well plate. L82 and L82-G17 were added to cell suspensions and vortexed briefly, prior to the addition of 3 mL of methylcellulose-based medium and plating. After incubation for 10 days at 37°C in 5% CO2, colonies were stained overnight with 1 mL of 1 mg/mL iodonitrotetrazolium chloride per well. Colonies were counted using ImageJ Cell Counter.
Formation of γH2AX
To detect γH2AX foci by immunocytochemistry, HeLa cells were grown on coverslips as described above in 12-well plates. Each well was seeded with 5000 cells that were allowed to adhere for at least 8 hours prior to incubation with inhibitors or DMSO alone for 4 hours. Cells were then incubated with anti-γH2AX FITC-conjugated antibodies using a kit purchased from BD Pharmingen and 4',6-Diamidino-2-Phenylindole (DAPI) to stain nuclei. After mounting of the coverslips onto slides, cells were imaged using Zeiss AxioObserver microscope in the UNMCCC Fluorescence Microscopy, with a Hamamatsu Flash 4 sCMOS camera and a 63× 1.4 NA objective. Images were viewed and analyzed using SlideBook (version 6.0.4).
Statistical analysis
Data are expressed as mean ± SEM. For comparison of groups, we used the Student two-tailed t test. A level of P < 0.05 was regarded as statistically significant. Conflict of Interest; “Structure-activity relationships among DNA ligase inhibitors; characterization of a selective uncompetitive DNA ligase I inhibitor” by Timothy R.L. Howes, Annahita Sallymr, Rhys Brooks, George E. Greco, Darin E. Jones, Yoshihiro Matsumoto, and Alan E. Tomkinson
Acknowledgments
We thank Dr. Jennifer Gillette for the use of her plate reader and Genevieve Phillips at the UNM Fluorescence Microscopy Shared Resource for her invaluable knowledge and assistance. This work was supported by the University of New Mexico Comprehensive Cancer Center (P30 CA118100) and National Institute of Health Grants R01 GM57479 (to A.E.T.) and P01 CA92584.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
A.E. Tomkinson is a co-inventor on US patents that cover the use of DNA ligase inhibitors as anti-cancer agents. No potential conflicts were disclosed by the other authors
References
- 1.Ellenberger T, Tomkinson AE. Eukaryotic DNA ligases: structural and functional insights. Annual review of biochemistry. 2008;77:313–338. doi: 10.1146/annurev.biochem.77.061306.123941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Han L, Masani S, Hsieh CL, Yu K. DNA ligase I is not essential for Mammalian cell viability. Cell Rep. 2014;7:316–320. doi: 10.1016/j.celrep.2014.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Moser J, Kool H, Giakzidis I, Caldecott K, Mullenders LH, Fousteri MI. Sealing of chromosomal DNA nicks during nucleotide excision repair requires XRCC1 and DNA ligase III alpha in a cell-cycle-specific manner. Molecular cell. 2007;27:311–323. doi: 10.1016/j.molcel.2007.06.014. [DOI] [PubMed] [Google Scholar]
- 4.Frosina G, Fortini P, Rossi O, Carrozzino F, Raspaglio G, Cox LS, Lane DP, Abbondandolo A, Dogliotti E. Two pathways for base excision repair in mammalian cells. The Journal of biological chemistry. 1996;271:9573–9578. doi: 10.1074/jbc.271.16.9573. [DOI] [PubMed] [Google Scholar]
- 5.Simsek D, Brunet E, Wong SY, Katyal S, Gao Y, McKinnon PJ, Lou J, Zhang L, Li J, Rebar EJ, Gregory PD, Holmes MC, Jasin M. DNA ligase III promotes alternative nonhomologous end-joining during chromosomal translocation formation. PLoS Genet. 2011;7:e1002080. doi: 10.1371/journal.pgen.1002080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Arakawa H, Bednar T, Wang M, Paul K, Mladenov E, Bencsik-Theilen AA, Iliakis G. Functional redundancy between DNA ligases I and III in DNA replication in vertebrate cells. Nucleic acids research. 2012;40:2599–2610. doi: 10.1093/nar/gkr1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liang L, Deng L, Nguyen SC, Zhao X, Maulion CD, Shao C, Tischfield JA. Human DNA ligases I and III, but not ligase IV, are required for microhomology-mediated end joining of DNA double-strand breaks. Nucleic acids research. 2008;36:3297–3310. doi: 10.1093/nar/gkn184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Oh S, Harvey A, Zimbric J, Wang Y, Nguyen T, Jackson PJ, Hendrickson EA. DNA ligase III and DNA ligase IV carry out genetically distinct forms of end joining in human somatic cells. DNA repair. 2014;21:97–110. doi: 10.1016/j.dnarep.2014.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang H, Rosidi B, Perrault R, Wang M, Zhang L, Windhofer F, Iliakis G. DNA ligase III as a candidate component of backup pathways of nonhomologous end joining. Cancer research. 2005;65:4020–4030. doi: 10.1158/0008-5472.CAN-04-3055. [DOI] [PubMed] [Google Scholar]
- 10.Audebert M, Salles B, Calsou P. Involvement of poly(ADP-ribose) polymerase-1 and XRCC1/DNA ligase III in an alternative route for DNA double-strand breaks rejoining. The Journal of biological chemistry. 2004;279:55117–55126. doi: 10.1074/jbc.M404524200. [DOI] [PubMed] [Google Scholar]
- 11.Le Chalony C, Hoffschir F, Gauthier LR, Gross J, Biard DS, Boussin FD, Pennaneach V. Partial complementation of a DNA ligase I deficiency by DNA ligase III and its impact on cell survival and telomere stability in mammalian cells. Cell Mol Life Sci. 2012;69:2933–2949. doi: 10.1007/s00018-012-0975-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gao Y, Katyal S, Lee Y, Zhao J, Rehg JE, Russell HR, McKinnon PJ. DNA ligase III is critical for mtDNA integrity but not Xrcc1-mediated nuclear DNA repair. Nature. 2011;471:240–244. doi: 10.1038/nature09773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Simsek D, Furda A, Gao Y, Artus J, Brunet E, Hadjantonakis AK, Van Houten B, Shuman S, McKinnon PJ, Jasin M. Crucial role for DNA ligase III in mitochondria but not in Xrcc1-dependent repair. Nature. 2011;471:245–248. doi: 10.1038/nature09794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lakshmipathy U, Campbell C. The human DNA ligase III gene encodes nuclear and mitochondrial proteins. Molecular and cellular biology. 1999;19:3869–3876. doi: 10.1128/mcb.19.5.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sallmyr A, Matsumoto Y, Roginskaya V, Van Houten B, Tomkinson AE. Inhibiting Mitochondrial DNA Ligase IIIalpha Activates Caspase 1-Dependent Apoptosis in Cancer Cells. Cancer research. 2016;76:5431–5441. doi: 10.1158/0008-5472.CAN-15-3243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mackey ZB, Ramos W, Levin DS, Walter CA, McCarrey JR, Tomkinson AE. An alternative splicing event which occurs in mouse pachytene spermatocytes generates a form of DNA ligase III with distinct biochemical properties that may function in meiotic recombination. Molecular and cellular biology. 1997;17:989–998. doi: 10.1128/mcb.17.2.989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sun D, Urrabaz R, Nguyen M, Marty J, Stringer S, Cruz E, Medina-Gundrum L, Weitman S. Elevated expression of DNA ligase I in human cancers. Clin Cancer Res. 2001;7:4143–4148. [PubMed] [Google Scholar]
- 18.Chen X, Zhong S, Zhu X, Dziegielewska B, Ellenberger T, Wilson GM, MacKerell AD, Jr, Tomkinson AE. Rational design of human DNA ligase inhibitors that target cellular DNA replication and repair. Cancer research. 2008;68:3169–3177. doi: 10.1158/0008-5472.CAN-07-6636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Barnes DE, Tomkinson AE, Lehmann AR, Webster AD, Lindahl T. Mutations in the DNA ligase I gene of an individual with immunodeficiencies and cellular hypersensitivity to DNA-damaging agents. Cell. 1992;69:495–503. doi: 10.1016/0092-8674(92)90450-q. [DOI] [PubMed] [Google Scholar]
- 20.Lasko DD, Tomkinson AE, Lindahl T. Mammalian DNA ligases. Biosynthesis and intracellular localization of DNA ligase I. The Journal of biological chemistry. 1990;265:12618–12622. [PubMed] [Google Scholar]
- 21.Levin DS, McKenna AE, Motycka TA, Matsumoto Y, Tomkinson AE. Interaction between PCNA and DNA ligase I is critical for joining of Okazaki fragments and long-patch base-excision repair. Current biology : CB. 2000;10:919–922. doi: 10.1016/s0960-9822(00)00619-9. [DOI] [PubMed] [Google Scholar]
- 22.Tobin LA, Robert C, Nagaria P, Chumsri S, Twaddell W, Ioffe OB, Greco GE, Brodie AH, Tomkinson AE, Rassool FV. Targeting abnormal DNA repair in therapy-resistant breast cancers. Molecular cancer research : MCR. 2012;10:96–107. doi: 10.1158/1541-7786.MCR-11-0255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tobin LA, Robert C, Rapoport AP, Gojo I, Baer MR, Tomkinson AE, Rassool FV. Targeting abnormal DNA double strand break repair in tyrosine kinase inhibitor-resistant chronic meyloid leukemias. Oncogene. 2013;32:1784–1793. doi: 10.1038/onc.2012.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sallmyr A, Tomkinson AE, Rassool F. Up-regulation of WRN and DNA ligase IIIa in Chromic myeloid leukemia: Consequences for the repair of DNA double strand breaks. Blood. 2008;112:1413–1423. doi: 10.1182/blood-2007-07-104257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Newman EA, Lu F, Bashllari D, Wang L, Opipari AW, Castle VP. Alternative NHEJ Pathway Components Are Therapeutic Targets in High-Risk Neuroblastoma. Molecular cancer research : MCR. 2015;13:470–482. doi: 10.1158/1541-7786.MCR-14-0337. [DOI] [PubMed] [Google Scholar]
- 26.Zhong S, Chen X, Zhu X, Dziegielewska B, Bachman KE, Ellenberger T, Ballin JD, Wilson GM, Tomkinson AE, MacKerell AD., Jr Identification and validation of human DNA ligase inhibitors using computer-aided drug design. Journal of medicinal chemistry. 2008;51:4553–4562. doi: 10.1021/jm8001668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Riballo E, Woodbine L, Stiff T, Walker SA, Goodarzi AA, Jeggo PA. XLF-Cernunnos promotes DNA ligase IV-XRCC4 re-adenylation following ligation. Nucleic acids research. 2009;37:482–492. doi: 10.1093/nar/gkn957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen X, Tomkinson AE. Yeast Nej1 is a key participant in the initial end binding and final ligation steps of nonhomologous end joining. The Journal of biological chemistry. 2011;286:4931–4940. doi: 10.1074/jbc.M110.195024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Petitjean M. Application of the radius-diameter diagram to the classification of topological and geometrical shapes of chemical compounds. J. Chem. Inf. Comput. Sci. 1992;32:331–337. [Google Scholar]
- 30.C WY. PaDEL-descriptor: an open source software to calculate molecular descriptors and fingerprints. J. Comput. Chem. 2011;32:1466–1474. doi: 10.1002/jcc.21707. [DOI] [PubMed] [Google Scholar]
- 31.Bajusz D, Racz A, Heberger K. Why is Tanimoto index an appropriate choice for fingerprint-based similarity calculations? J Cheminform. 2015;7:20. doi: 10.1186/s13321-015-0069-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Backman TW, Cao Y, Girke T. ChemMine tools: an online service for analyzing and clustering small molecules. Nucleic acids research. 2011;39:W486–491. doi: 10.1093/nar/gkr320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bentley DJ, Harrison C, Ketchen AM, Redhead NJ, Samuel K, Waterfall M, Ansell JD, Melton DW. DNA ligase I null mouse cells show normal DNA repair activity but altered DNA replication and reduced genome stability. Journal of cell science. 2002;115:1551–1561. doi: 10.1242/jcs.115.7.1551. [DOI] [PubMed] [Google Scholar]
- 34.Bentley D, Selfridge J, Millar JK, Samuel K, Hole N, Ansell JD, Melton DW. DNA ligase I is required for fetal liver erythropoiesis but is not essential for mammalian cell viability. Nature genetics. 1996;13:489–491. doi: 10.1038/ng0896-489. [DOI] [PubMed] [Google Scholar]
- 35.Jones LJ, Gray M, Yue ST, Haugland RP, Singer VL. Sensitive determination of cell number using the CyQUANT cell proliferation assay. J Immunol Methods. 2001;254:85–98. doi: 10.1016/s0022-1759(01)00404-5. [DOI] [PubMed] [Google Scholar]
- 36.Bonner WM, Redon CE, Dickey JS, Nakamura AJ, Sedelnikova OA, Solier S, Pommier Y. GammaH2AX and cancer. Nat Rev Cancer. 2008;8:957–967. doi: 10.1038/nrc2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nakamura M, Kondo S, Sugai M, Nazarea M, Imamura S, Honjo T. High frequency class switching of an IgM+ B lymphoma clone CH12F3 to IgA+ cells. Int Immunol. 1996;8:193–201. doi: 10.1093/intimm/8.2.193. [DOI] [PubMed] [Google Scholar]
- 38.Jackson SP, Helleday T. DNA REPAIR. Drugging DNA repair. Science. 2016;352:1178–1179. doi: 10.1126/science.aab0958. [DOI] [PubMed] [Google Scholar]
- 39.Baell JB, Holloway GA. New substructure filters for removal of pan assay interference compounds (PAINS) from screening libraries and for their exclusion in bioassays. Journal of medicinal chemistry. 2010;53:2719–2740. doi: 10.1021/jm901137j. [DOI] [PubMed] [Google Scholar]
- 40.Charkoudian LK, Pham DM, Franz KJ. A pro-chelator triggered by hydrogen peroxide inhibits iron-promoted hydroxyl radical formation. J Am Chem Soc. 2006;128:12424–12425. doi: 10.1021/ja064806w. [DOI] [PubMed] [Google Scholar]
- 41.Peng X, Tang X, Qin W, Dou W, Guo Y, Zheng J, Liu W, Wang D. Aroylhydrazone derivative as fluorescent sensor for highly selective recognition of Zn2+ ions: syntheses, characterization, crystal structures and spectroscopic properties. Dalton Trans. 2011;40:5271–5277. doi: 10.1039/c0dt01590c. [DOI] [PubMed] [Google Scholar]
- 42.Pommier Y, O'Connor MJ, de Bono J. Laying a trap to kill cancer cells: PARP inhibitors and their mechanisms of action. Sci Transl Med. 2016;8:362ps317. doi: 10.1126/scitranslmed.aaf9246. [DOI] [PubMed] [Google Scholar]
- 43.Staker BL, Hjerrild K, Feese MD, Behnke CA, Burgin AB, Jr, Stewart L. The mechanism of topoisomerase I poisoning by a camptothecin analog. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:15387–15392. doi: 10.1073/pnas.242259599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Langelier MF, Planck JL, Roy S, Pascal JM. Structural basis for DNA damage-dependent poly(ADP-ribosyl)ation by human PARP-1. Science. 2012;336:728–732. doi: 10.1126/science.1216338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Murai J, Huang SY, Das BB, Renaud A, Zhang Y, Doroshow JH, Ji J, Takeda S, Pommier Y. Trapping of PARP1 and PARP2 by Clinical PARP Inhibitors. Cancer research. 2012;72:5588–5599. doi: 10.1158/0008-5472.CAN-12-2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pascal JM, O'Brien PJ, Tomkinson AE, Ellenberger T. Human DNA ligase I completely encircles and partially unwinds nicked DNA. Nature. 2004;432:473–478. doi: 10.1038/nature03082. [DOI] [PubMed] [Google Scholar]
- 47.Cardoso MC, Joseph C, Rahn HP, Reusch R, Nadal-Ginard B, Leonhardt H. Mapping and use of a sequence that targets DNA ligase I to sites of DNA replication in vivo. The Journal of cell biology. 1997;139:579–587. doi: 10.1083/jcb.139.3.579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Montecucco A, Rossi R, Levin DS, Gary R, Park MS, Motycka TA, Ciarrocchi G, Villa A, Biamonti G, Tomkinson AE. DNA ligase I is recruited to sites of DNA replication by an interaction with proliferating cell nuclear antigen: identification of a common targeting mechanism for the assembly of replication factories. The EMBO journal. 1998;17:3786–3795. doi: 10.1093/emboj/17.13.3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Krishna S, Singh DK, Meena S, Datta D, Siddiqi MI, Banerjee D. Pharmacophore-based screening and identification of novel human ligase I inhibitors with potential anticancer activity. J Chem Inf Model. 2014;54:781–792. doi: 10.1021/ci5000032. [DOI] [PubMed] [Google Scholar]
- 50.Pandey M, Kumar S, Goldsmith G, Srivastava M, Elango S, Shameem M, Bannerjee D, Choudhary B, Karki SS, Raghavan SC. Identification and characterization of novel ligase I inhibitors. Mol Carcinog. 2017;56:550–566. doi: 10.1002/mc.22516. [DOI] [PubMed] [Google Scholar]
- 51.Shameem M, Kumar R, Krishna S, Kumar C, Siddiqi MI, Kundu B, Banerjee D. Synthetic modified pyrrolo[1,4] benzodiazepine molecules demonstrate selective anticancer activity by targeting the human ligase 1 enzyme: An in silico and in vitro mechanistic study. Chem Biol Interact. 2015;237:115–124. doi: 10.1016/j.cbi.2015.05.024. [DOI] [PubMed] [Google Scholar]
- 52.Peng Z, Liao Z, Matsumoto Y, Yang A, Tomkinson AE. Human DNA Ligase I Interacts with and Is Targeted for Degradation by the DCAF7 Specificity Factor of the Cul4-DDB1 Ubiquitin Ligase Complex. The Journal of biological chemistry. 2016;291:21893–21902. doi: 10.1074/jbc.M116.746198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chen X, Ballin JD, Della-Maria J, Tsai MS, White EJ, Tomkinson AE, Wilson GM. Distinct kinetics of human DNA ligases I, IIIalpha, IIIbeta, and IV reveal direct DNA sensing ability and differential physiological functions in DNA repair. DNA repair. 2009;8:961–968. doi: 10.1016/j.dnarep.2009.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chen X, Pascal J, Vijayakumar S, Wilson GM, Ellenberger T, Tomkinson AE. Human DNA ligases I, III, and IV-purification and new specific assays for these enzymes. Methods in enzymology. 2006;409:39–52. doi: 10.1016/S0076-6879(05)09003-8. [DOI] [PubMed] [Google Scholar]
- 55.Peng Z, Liao Z, Dziegielewska B, Matsumoto Y, Thomas S, Wan Y, Yang A, Tomkinson AE. Phosphorylation of serine 51 regulates the interaction of human DNA ligase I with replication factor C and its participation in DNA replication and repair. The Journal of biological chemistry. 2012;287:36711–36719. doi: 10.1074/jbc.M112.383570. [DOI] [PMC free article] [PubMed] [Google Scholar]