Short abstract
In mammalian cells, there are seven members of the sirtuin protein family (SIRT1–7). SIRT1, SIRT6, and SIRT7 catalyze posttranslational modification of proteins in the nucleus, SIRT3, SIRT4, and SIRT5 are in the mitochondria and SIRT2 is in the cytosol. SIRT1 can deacetylate the transcription factor SOX2 and regulate induced pluripotent stem cells (iPSCs) reprogramming through the miR-34a–SIRT1–p53 axis. SIRT2 can regulate the function of pluripotent stem cells through GSK3β. SIRT3 can positively regulate PPAR gamma coactivator 1-alpha (PGC-1α) expression during the differentiation of stem cells. SIRT4 has no direct role in regulating reprogramming but may have the potential to prevent senescence of somatic cells and to facilitate the reprogramming of iPSCs. SIRT5 can deacetylate STAT3, which is an important transcription factor in regulating pluripotency and differentiation of stem cells. SIRT6 can enhance the reprogramming efficiency of iPSCs from aged skin fibroblasts through miR-766 and increase the expression levels of the reprogramming genes including Sox2, Oct4, and Nanog through acetylation of histone H3 lysine 56. SIRT7 plays a regulatory role in the process of mesenchymal-to-epithelial transition (MET), which has been suggested to be a crucial process in the generation of iPSCs from fibroblasts. In this review, we summarize recent findings of the roles of sirtuins in the metabolic reprogramming and differentiation of stem cells and discuss the bidirectional changes in the gene expression and activities of sirtuins in the commitment of differentiation of mesenchymal stem cells (MSCs) and reprogramming of somatic cells to iPSCs, respectively. Thus, understanding the molecular basis of the interplay between different sirtuins and mitochondrial function will provide new insights into the regulation of differentiation of stem cells and iPSCs formation, respectively, and may help design effective stem cell therapies for regenerative medicine.
Impact statement
This is an extensive review of the recent advances in our understanding of the roles of some members of the sirtuins family, such as SIRT1, SIRT2, SIRT3, and SIRT6, in the regulation of intermediary metabolism during stem cell differentiation and in the reprogramming of somatic cells to form induced pluripotent stem cells (iPSCs). This article provides an updated integrated view on the mechanisms by which sirtuins-mediated posttranslational protein modifications regulate mitochondrial biogenesis, bioenergetics, and antioxidant defense in the maintenance and differentiation of stem cells and in iPSCs formation, respectively.
Keywords: Sirtuins, induced pluripotent stem cells, mitochondrial biogenesis, differentiation, metabolic reprogramming, antioxidant defense
The sirtuin family
Metabolic reprogramming is the shift among oxidative phosphorylation (OXPHOS), fatty acid oxidation and glycolysis during stem cell differentiation or reprogramming to stem cells accompanied by changes in the levels of metabolites, redox state, proliferation, and mitochondrial mass. Furthermore, the epigenetic and genetic modifications are also regulated by some metabolites and metabolic reprogramming. Among all epigenetic regulators, the silent information regulator (SIR) protein was originally found to regulate DNA repair and mitosis through its deacetylase activity.1 In yeast, a protein named silent information regulator 2 (Sir2) can regulate life span,2 and this is the first of mammalian sirtuin family proteins. Sir2 domain is highly conserved from bacteria to human in all seven members of this protein family (SIRT1–7).3 Through long time of evolution, the catalytic function-related core domain contains no variant of amino acid residues, while the diversity of its N- and C-terminal regions contributes to its different sub-cellular localization, enzymatic activity, and substrate specificity.4 SIRT1, SIRT6, and SIRT7 are mostly localized in the nucleus but can translocate to the cytoplasm under some conditions.5–7 SIRT2 is mainly present in the cytoplasm but is translocated to the nucleus under special conditions.8,9 Notably, SIRT3, SIRT4, and SIRT5 are mostly present in the mitochondria, but can translocate to the nucleus or cytosol under some exceptional conditions.10–13 These sirtuins not only maintain genome stability and telomere function, but also control the metabolism of glucose and lipids, regulate inflammation, and suppress the development of tumors. In this review, we have summarized and discussed the roles of SIRT1, SIRT2, SIRT3, and SIRT6 in the metabolic reprogramming during the differentiation of stem cells, including mesenchymal stem cells (MSCs), embryonic stem cells (ESCs), and induced pluripotent stem cells (iPSCs). We also provide the current understanding and future perspectives of the potential roles of certain SIRTs in the reprogramming of iPSCs from somatic cells.
SIRT1
SIRT1, the first identified member of sirtuins that regulates various cellular functions through NAD+-dependent protein deacetylase activity. It is not only implicated in longevity, development, tumor suppression, but also in metabolic reprogramming (Table 1). Evidence is mounting to support that the activity of SIRT1 is vital for the maintenance and differentiation of stem cells, especially through metabolic reprogramming. The decrease of the activity of SIRT1 in skeletal muscle stem cells has been demonstrated to be accompanied with a shift from fatty acid β-oxidation to glycolysis by a decrease in the levels of NAD+ and an increase in H4K16ac, one of the substrates of SIRT1-mediated deacetylation.14 Besides, the expression level of SIRT1 in mouse ESCs (mESCs) was found to be higher than that in differentiated cells, which is considered to be required for the survival, differentiation, and speciation of ESCs.15–17 SIRT1 has been postulated to act as a metabolic sensor that directly connects transcriptional output with metabolic function. Ryall et al. showed that epigenetic regulation by SIRT1 plays an integral role in metabolic reprogramming-promoted activation of adult muscle stem cells.14 Deacetylations of peroxisome proliferator-activated receptor γ (PPAR-γ), PPAR γ coactivator 1-alpha (PGC-1α), AMP-activated protein kinase (AMPK), and Forkhead box protein O1 (FoxO1) are important mechanisms underlying SIRT1-mediated regulation of energy metabolism and redox homeostasis, which have recently been linked to the differentiation and speciation processes of MSCs. FoxO1 is one of the Forkhead box O transcription factors involved in stress response, apoptosis, and autophagic regulation. There has been accumulating evidence to indicate that FoxO1 serves as an interface for SIRT1-mediated signaling in the maintenance of stem cell properties and regulation of lineage-specific differentiation of MSCs. Induction of MnSOD and catalase by SIRT1 can increase the capacity of antioxidant defense, which is supported by our findings that antioxidant enzymes are upregulated during osteogenic differentiation of human MSCs (hMSCs).18 SIRT1/FoxO1-mediated signaling cascade may contribute to enhanced antioxidant capacity to scavenge the intracellular reactive oxygen species (ROS) during hMSCs differentiation. Activation of SIRT1 by silencing miR-195 was shown to reverse age-related phenotype and enhance cell proliferation of old MSCs via regulation of telomerase reverse transcriptase (TERT) and FoxO1.19 Conversely, SIRT1 can trigger apoptotic cell death of mESCs in response to an excess amount of ROS through activation of FoxO1.20 In line with its downregulation during adipogenesis, SIRT1 activation by resveratrol has been shown to compromise the expression of adipogenic genes and stimulate apoptosis in bovine intramuscular adipocytes, which is associated with the induction of FoxO1-mediated signaling cascade.21 Repression of SIRT1 transcription by miR-146b can promote adipogenic differentiation of 3T3-L1 through FoxO1 signaling.22
Table 1.
Summary of the sirtuin family proteins that are involved in stem cell differentiation and iPSC formation.
| ActivityReference | Localization Reference | Reprogramming Reference | Pluripotency and stem cell properties Reference | Differentiation Reference | |
|---|---|---|---|---|---|
| SIRT1 | Deacetylation14 | Nucleus (++++) Cytosol (−/+)5–7 |
|
||
| SIRT2 | Deacetylation46,47 | Nucleus (−/+)Cytosol (++++)8,9 | Metabolic shift by acetylation of glycolytic enzymes, such as GAPDH, PGK1, ENO1, PKM, and ALDOA51 | Negative regulation51 | |
| SIRT3 | Deacetylation62 | Nucleus (−/+)Cytosol (−/+)Mitochondria (++++)12,13 | Facilitate the reprogramming of somatic cells from old individuals by downregulation of cellular ROS signaling and aging processes62 | NA | Positive regulation on adipogenesis and osteogenesis by induction of PGC-1α and MnSOD, and FoxO3a signaling60,75 |
| SIRT4 | Deacetylation of ADP-ribosyltransferase82 | Nucleus (−/+)Cytosol (−/+)Mitochondria (++++)11 | Facilitate the reprogramming of somatic cells from old individuals by preventing somatic cell senescence83 | NA | NA |
| SIRT5 | Deacetylation86,87Desuccinylation85,86,123Demalonylation86 | Nucleus (−/+)Cytosol (−/+)Mitochondria (++++)10 | NA | Negative regulation through LIF/JAK/STAT3 axis88,89 | Negative regulation on adipogenesis61 |
| SIRT6 | Deacetylation102 | Nucleus (++++) Cytosol (−/+)5–7 | Facilitate the reprogramming of somatic cells from old individuals by acetylation of Sox2, Oct4, and Nanog100,101 | NA | Positive regulation on adipogenesis and osteogenesis95–97 |
| SIRT7 | Deacetylation | Nucleus (++++) Cytosol (−/+)5–7 | Promoting the MET process111,112 | NA |
GAPDH: glyceraldehyde 3-phosphate dehydrogenase; PGK1: phosphoglycerate kinase 1; ENO1: enolase 1; PKM: phosphoglycerate mutase M; ALDOA: aldolase A; MET: mesenchymal-to-epithelial transition; NA: not available.
Protein localization: −/+: 0–10%, +: 10–25%, ++: 25–50%, +++: 50–75%, ++++: 75–100%.
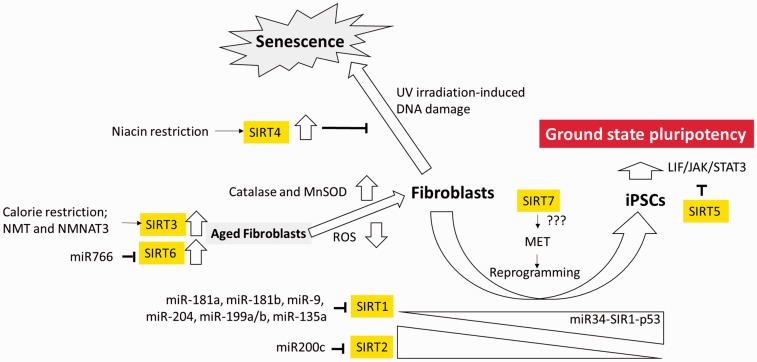
In 2006, Takahashi and Yamanaka reported that mouse somatic cells can be reprogrammed into iPSCs by Oct4, Sox2, Klf4, and c-Myc.23 Oct4 and Sox2 act as the trigger of major endogenous pluripotent genes during reprogramming. The efficiency of reprogramming can be increased by the hypoacetylation of Sox2, and Sox2 can be deacetylated by SIRT1 with the mediation of Oct4. Compared to the wild-type cells, SIRT1 knockout mouse embryonic fibroblasts exhibited a decrease in the efficiency of reprogramming of iPSCs, and SIRT1 overexpression could rescue the defect.24 Furthermore, miR-181a, miR-181b, miR-9, miR-204, miR-199a/b, and miR-135a have been shown to suppress the expression of SIRT1, which provides a new strategy in the regulation of the reprogramming of somatic cells to iPSCs (Figure 1).22,25 Among them, miR-199a negatively regulates the differentiation of iPSCs to endothelial cells through targeting SIRT1.22 Interestingly, Homma et al.26 compared the proliferation, migration, and oxidative stress tolerance among human adult aortic endothelial cells (HAECs), human ESC-derived ECs (ESC-ECs), and human iPSCs-derived ECs (iPSC-ECs). They found that iPSC-ECs and ESC-ECs had higher levels of SIRT1 and were superior to HAECs in the proliferation, migration, and oxidative stress tolerance.26 These findings suggest that SIRT1 plays an important role in regulating the proliferation, migration, and oxidative stress tolerance of ESC-ECs and iPSC-ECs. Jiang et al.27 further demonstrated that overexpression of SIRT1 in iPSC-ECs could overcome early cell senescence to maintain the phenotype and stemness of stem cells. With regard to reprogramming and pluripotency, SIRT1 can facilitate the iPSCs reprogramming through the miR-34a–SIRT1–p53 axis,28 whereas SIRT1, p53, and p38MAPK are detrimental to the survival of Max-null ESCs with different levels of pluripotency.29 Therefore, SIRT1 plays an important role in the regulation of reprogramming and pluripotency of iPSCs (Table 1 and Figure 1).
Figure 1.
Schematic illustration of the regulatory roles of different sirtuins in the reprogramming of human skin fibroblasts to generate induced pluripotent stem cells. Sirtuin1 (SIRT1) has been shown to positively regulate the formation of iPSCs through the miR34–SIR1–p53 axis. Furthermore, many miRNAs, such as miRNA-181a, miRNA181b, miR-9, miR-204, miR-199a/b, and miR135a have been identified to negatively regulate the expression of SIRT1. By contrast, the expression of SIRT2 has been shown to be downregulated during the formation of iPSCs. The expression of SIRT2 can be negatively regulated by miR200c. SIRT3 and SIRT6 have been shown to play important roles to enable senescent skin fibroblasts to be reprogrammed into iPSCs through upregulation of the expression of antioxidant enzymes such as catalase and MnSOD, and thereby leading to a decrease of ROS production. SIRT4 has been shown to negatively regulate the senescence of skin fibroblasts that are exposed to UV irradiation. SIRT5 can negatively regulate the ground state pluripotency of naïve iPSCs through downregulation of the LIF/JAK/STAT3 pathway. The role of SIRT7 in promoting mesenchymal-to-epithelial transition (MET) is yet to be determined since MET is crucial for the successful reprogramming of skin fibroblasts to iPSCs. (A color version of this figure is available in the online journal.)
In addition, SIRT1 can also reduce abnormal DNA methylation in mESCs through interfering with DNA methyltransferase Dnmt3l.15 Evidence from several studies on SIRT1−/− mice suggested that SIRT1 also plays important roles in the defects of embryonic and postnatal development, especially in neurogenesis and spermatogenesis.15 It was found that the delayed differentiation was associated with the downregulation of Oct4, Fgf5, Nanog, Scl, β-globin, and the inactivation of Erk1/2.16 Interestingly, Hayakawa et al.30 showed that a nutrient factor, ManNAc, can induce the expression of Hcrt gene through activation of its epigenetic regulators, including SIRT1, Ogt, and Mgea5, to facilitate the generation of functional neurons from mESCs. As for the deacetylase activity of sirtuins, SIRT1 can deacetylate the cellular retinoid acid (RA) binding protein II (CRABPII) to modulate RA homeostasis and regulate ESCs differentiation. Consequently, downregulation of SIRT1 induces the accumulation of hyperacetylated CRABPII in the nucleus, which in turn enhances RA signaling, myogenic differentiation, and culminates in the developmental defects in mice.17
Recent studies revealed that SIRT1 deficiency induces cell death by the impairment of DNA repair system in human ESCs. Jang et al.31 showed that loss of SIRT1 in human ESCs caused a dramatic reduction of DNA repair proteins and simultaneously induced hyperacetylation of p53, which then triggered an accumulation of DNA damage. Under oxidative stress challenge, SIRT1 was shown to maintain mitochondrial function and facilitate autophagic induction via its inhibitory effect on mechanistic target of rapamycin (mTOR) activity in mESCs.32 As a result of its powerful effect on antioxidant defense, SIRT1 can rescue premature senescence in BM-MSCs when exposed to oxidative stress33 and abate aging-dependent dysfunction of somatic stem cells, an important mechanism linking progressive decline in stemness and differentiation potential through the control of ROS and telomere. It is worth noting that a delicate control of intracellular ROS levels is necessary for the maintenance of cell proliferation, stemness, and differentiation of MSCs. Although our findings showed that differentiated osteoblasts are more resistant to oxidative stress compared with hMSCs, excess production of ROS still hampers osteogenic differentiation of hMSCs.18 Increase in the ROS level was associated with age-related defect in bone formation. Upregulation of SIRT1 in MSCs toward osteo-lineage may facilitate the removal of excess production of ROS accompanied by increased mitochondrial respiration during the differentiation process. Nevertheless, researchers have demonstrated that increase in the ROS levels during adipogenesis and chondrogenesis of MSCs may confer the capacity of differentiation.34,35 However, further study is needed to resolve this contradictory observation. SIRT1 can directly interact with telomeric repeats to attenuate telomere shorting. Palacios et al.36 reported that overexpression of SIRT1 could increase the homologous recombination throughout the whole genome, which links SIRT1 with DNA repair and telomere. SIRT1 can also rescue MSCs from aging-related DNA damage by promoting the expression and telomerase activity of TERT.37
The role of SIRT1 in cell fate decision and differentiation capacity of stem cells is garnering increased attention, and studies in this field have shed light on its epigenetic regulation for differentiation of stem cells. Activation of SIRT1 promotes osteogenic differentiation in both mouse and human MSCs through positively regulating a master transcription factor of osteoblasts, Runx2. However, inhibition of SIRT1 was shown to stimulate the expression of adipogenic genes and increase the number of adipocytes. Abundant evidence indicates that SIRT1 activation favors osteogenesis but interferes with adipogenic and neurogenic differentiations of hMSCs. In the SIRT1 heterozygous mice (SIRT1+/−), a marked reduction in bone formation and enhancement of adipogenesis supports the role of SIRT1 in lineage determination of cell differentiation in vivo.38 MSCs obtained from MSC-specific SIRT1 knockout mice also displayed decreased capacity of osteogenic differentiation, nevertheless, the efficiency of adipogenesis was not significantly changed. Suppression of SIRT1 by miR-132, a repressor of SIRT1, was shown to impair osteogenic differentiation and subsequently lead to diabetic osteoporosis in MC3T3-E1 cells.39 Osteoporosis, characterized by a loss of osteoblasts that results in defective formation and decreased mineral density of bone, is commonly observed in patients with type 2 diabetes. Increase of osteogenic differentiation by activating SIRT1 may be an effective strategy to treat osteoporosis associated with metabolic diseases and aging.
In fact, the SIRT1 expression level is decreased during adipogenic differentiation and activation of SIRT1 was shown to impair adipocytes development. However, the details of the mechanism underlying inhibitory effect of the SIRT1 on adipogenesis of MSCs are still unclear. According to the intendance in the differentiation of MSCs to diverse lineages, SIRT1 represses nuclear receptor PPAR-γ, which contributes to the inhibition of adipogenesis during osteogenic differentiation. This is supported by the finding that resveratrol, a SIRT1 activator, blocks adipogenic differentiation and enhances the expression of osteogenic genes in MSCs. Recent studies indicated that SIRT1 blocks adipogenic differentiation through triggering Wnt/β-catenin signaling, a well-known pathway regulating cell fate determination of MSCs toward osteogenesis,40,41 in C3H10T/2 stem cells,42,43 and hMSCs.44 Deacetylation of β-catenin by SIRT1 promotes its nuclear localization and consequently represses adipogenic gene expressions.43 Zhou et al. also showed that both resveratrol and overexpression of SIRT1 inhibit adipogenesis and enhance myogenic differentiation via Wnt/β-catenin signal cascade in C3H10T/2 cells.45 Metabolic switch to active aerobic metabolism of MSCs is a hallmark during multiple lineage differentiation. A growing body of evidence shows alterations in the bioenergetic function, morphology, and dynamics of mitochondria during stem cell differentiation. However, the details in the regulatory mechanism are still poorly understood. We and other researchers have proved that mitochondrial biogenesis and antioxidant defense capacity are dramatically increased during adipogenic, osteogenic, and myogenic differentiation.
SIRT2
Cytosolic SIRT2 is involved in the modulation of microtubule dynamics through catalyzing α-tubulin deacetylation. Regulation of cell morphology and mitochondrial distribution by modification of microtubule dynamics is required for neuronal development.46,47 There is no direct evidence to show that SIRT2 can promote reprogramming by deacetylating key reprogramming factors or pluripotency genes. Nonetheless, SIRT2 has shown to directly regulate metabolic transition during somatic reprogramming by controlling the acetylation status of glycolytic enzymes.48 SIRT2 was shown to affect early lineage determination of mESCs via activation of GSK3β,49 which plays a critical role in neurogenic differentiation.50 SIRT2 can regulate caloric restriction-dependent lifespan extension through decreasing the expression level of H4K16Ac during G2/M transition, which indicates that SIRT2 contributes to the alteration of acetylation of histone proteins in cell cycle.8 It is also involved in metabolic shift during reprogramming through acetylation of glycolytic enzymes, such as GAPDH, PGK1, ENO1, PKM, and ALDOA (Table 1).51 In contrast to SIRT1, SIRT2 is downregulated in the iPSCs and is upregulated upon neuronal differentiation.52 Furthermore, overexpression of SIRT2 in human skin fibroblasts reduced iPSC generation by approximately 80%, whereas knockdown of SIRT2 significantly increased the efficiency, which was abrogated by 2-deoxyglucose (2DG), indicating a key role of SIRT2 in metabolic reprogramming during the formation of iPSCs.52 Notably, miR200c has been identified to be induced by Oct4, which in turn downregulates the mRNA and protein expression of SIRT2. This further enhances the efficiency of reprogramming and the pluripotency of iPSCs. Downregulation of SIRT2 by miR-200c upregulates glycolysis by acetylation of glycolytic enzymes GAPDH, PGK1, ENO1, PKM, and ALDOA. Taken together, the miR200c–SIRT2 axis can regulate the reprogramming and pluripotency of iPSCs through metabolic regulation (Figure 1).47
In addition, Jeong and Cho51 recently proved that SIRT2 positively regulates neurogenesis through induction of the ERK1/2 signaling and nuclear cAMP response element-binding protein (CREB), and that the downstream target of SIRT2 can regulate neuronal differentiation and brain neuroplasticity. SIRT2 was found to be highly expressed in the affected tissues of some of the patients with Parkinson's disease (PD) or Huntington's disease (HD) and has thus been implicated in neurodegeneration.53,54 In the neurons with insulin resistance, SIRT2 was shown to negatively regulate insulin-stimulated glucose uptake.54 However, the role of SIRT2 in the progression of neurodegeneration is still controversial. A recent study indicates that SIRT2 protects neuron cells from oxidative stress.54 Overexpression of SIRT2 reduces rotenone-elicited cell death and α-synuclein aggregates in human SH-SY5Y cells, partly effected through the induction of MnSOD. It was suggested that the higher activity of SIRT2 in degenerative brains may be a compensatory effect to cope with the stress. In contrast to the favorable effect on neurogenesis, SIRT2 activity is harmful to adipogenesis and its expression level is downregulated during the initiation of adipogenic differentiation.55 Hyperacetylation of α-tubulin could facilitate mitochondrial transport along the microtubules, which is essential for adipogenic differentiation46 and can initiate cytoskeleton remodeling to facilitate the differentiation and maturation of adipocytes.47 Deacetylation of FoxO1 by SIRT2 was demonstrated to block adipogenic differentiation of MSCs through downregulation of PPAR-γ.49,56 Moreover, it was shown that downregulation of SIRT2 led to hyperacetylation and phosphorylation of FoxO1, which in turn promoted the differentiation of 3T3L1 preadipocytes.57
SIRT3
SIRT3, expressed in tissues with high metabolic activity, is a major sirtuin in mitochondria. It has emerged as a master regulator of oxidative metabolism, redox homeostasis, oxidative response, and longevity. Induction of SIRT3 is required for adipogenic and osteogenic differentiation.58–61 Recently, we demonstrated that the mRNA and protein expression levels of SIRT3 were dramatically increased during the initiation of adipogenic differentiation and maintained at high levels throughout the differentiation process. It was shown that induction of SIRT3 and concurrent changes in the acetylation levels of mitochondrial proteins contribute to the activation of mitochondrial function in adipogenic differentiation of MSCs.62 Recent studies unraveled that the beneficial effect of SIRT3 involves the upregulation of mitochondrial biogenesis, oxidative metabolism, and antioxidant capacity during differentiation of MSCs.58,61 We observed that SIRT3 knockdown caused a decrease in the expression levels of PGC-1α, respiratory enzyme complexes, and antioxidant enzymes in adipogenesis.62 Silencing of SIRT3 led to abnormal differentiation of myoblasts through the decrease in the expression levels of PGC-1α and MnSOD.62 On the other hand, we showed that impaired mitochondrial respiration caused by SIRT3 deficiency induced a metabolic shift to glycolysis. Mitochondrial pyruvate dehydrogenase complex (PDHC) is the rate-limiting enzyme for pyruvate oxidation that produces acetyl-CoA to switch on aerobic metabolism in the mammals. During adipogenic differentiation, activation of PDH63 and downregulation of glycolysis64 are pivotal events for the upregulation of aerobic metabolism. One of our previous studies revealed that a decline of SIRT3 caused hyperacetylation of PDH and consequently attenuated its enzymatic activity in human cells harboring a pathogenic mtDNA mutation.65 It is known that acetylation of PDH-E1 inhibits the PDH activity by triggering phosphoinositide-dependent kinase-1 (PDK1)66-mediated phosphorylation, which promotes glycolysis.67 Deacetylation of PDH by SIRT3 enhances oxidative metabolism through increasing the enzymatic activity of PDH.68 The finding of the upregulation of PDH activity and mitochondrial respiration by overexpression of SIRT3 implies the potential role of SIRT3-mediated deacetylation in modulating the metabolic prolife during adipogenic differentiation.
SIRT3 deficiency has been linked to multiple human diseases and aging-associated syndrome. In a previous study, we found that oxidative stress led to the decline of SIRT3 in human cells harboring pathogenic mtDNA mutations.69 In addition, SIRT3 activity is crucial for brown adipocytes differentiation in vitro.70 Giralt et al.70 demonstrated that increased expression of SIRT3 stimulated CREB phosphorylation, which induced the expressions of PGC-1α, uncoupling protein 1 (UCP1), and an array of mitochondrial biogenesis-related genes in murine brown adipocyte. In contrast to the function of white adipocytes to store energy, brown adipocytes act to increase the energy expenditure and thermogenesis. Many researchers have paid much attention to the development of brown adipocytes from white adipocytes to counteract obesity and insulin resistance.71,72 In miPSCs, PGC-1α overexpression was shown to promote brown-like adipogenic differentiation with the induction of brown adipocyte marker genes such as UCP1 but repressed expression of UCP2, a white adipocyte marker.73 Regarding the potential role of MSCs in the treatment of diabetes,71,72 commanding the adipogenesis of MSCs may be a therapeutic strategy for prevention of obesity-related diseases.
SIRT3−/− cells showed lower ROS levels and decreased expression of antioxidant enzymes such as MnSOD, which supports the notion that SIRT3 plays a role in mediating cellular response to oxidative stress in aging and disease.63 Moreover, the expression level of endogenous SIRT3 was decreased when BM-MSCs were stimulated by a high dose of hydrogen peroxide.63 Decrease in the level of SIRT3 induced by oxidative stress may interfere with the differentiation ability of aged MSCs. Actually, the SIRT3 level was recently found to decrease with replicative senescence of human bone marrow-MSCs (hBM-MSCs) at a later passage.63 Depletion of SIRT3 was found to compromise adipogenesis and osteogenesis. It was shown that exogenous SIRT3 overexpression can ameliorate the differentiation ability and slightly restore aging-related phenotype in senescent hMSCs. A recent study reported that SIRT3 overexpression reduces oxidative stress injuries in neural stem cells (NSCs).74 This neuroprotection of SIRT3 on NSCs involves the increase of mitochondrial membrane potential and attenuation of the ROS levels and apoptotic cell death. Downregulation of SIRT3 can regulate the ROS detoxification by direct deacetylation of FoxO3a and upregulating the expression of MnSOD and catalase, which can attenuate oxidative damage and age-related pathological changes in mice fed with a calorie-restricted diet. SIRT3/FoxO3a signaling can protect mouse BM-MSCs from mitochondrial dysfunction and apoptosis.75 In this study, knockdown of SIRT3 was shown to significantly decrease the protein level of FoxO3a in BM-MSCs. Recently, Lorenowicz and co-workers60 claimed that FoxO3-mediated autophagy is involved in the maintenance of redox homeostasis during osteogenic differentiation of hMSCs. Autophagy is a conserved proteolytic mechanism underlying the degradation of damaged molecules and organelles to maintain intracellular homeostasis. For example, activation of mitophagy, an autophagic degradation of mitochondrion, causes higher mitochondrial turnover whereby maintaining mitochondrial quality during early chondrogenesis.76 Autophagy has been functionally linked to the maintenance of pluripotency and differentiation capacity of stem cells. Lorenowicz’s group used H2O2 stimuli to demonstrate that ROS induced the phosphorylation and activation of FoxO3 by MAPK8/JNK-mediated signaling, by which MSCs could trigger autophagic induction to prevent oxidative stress-elicited damage and cell death.60 The authors showed that FxoO3 knockdown in MSCs elevates the intracellular ROS levels and compromises osteogenic differentiation. Inhibition of the autophagic process was also shown to impair adipogenic differentiation of 3T3-L1 preadipocytes and MSCs. However, the role of autophagy in MSCs and different lineage differentiation is still controversial and its regulation by SIRT3 is worthy of further study. Taken together, the above findings imply that SIRT3 assists stem cells in coping with the external stress, and its induction during differentiation is responsible for mitochondrial activation and meanwhile eliminates the ROS overproduced in the process (Table 1).
It has been shown that the NAD+ levels and NAD+-dependent enzymes are downregulated in the aging process. Decrease of the enzyme activities of sirtuins has been shown to associate with mitochondrial diseases,77 DNA repair defects,78 and deficiency in intermediary metabolism. It has been shown that the efficiency of reprogramming from somatic cells to generate iPSCs declines with age of the donor.79,80 It is thus important to identify regulatory factors that can overcome aging during the reprogramming processes. Interestingly, it was observed that downregulation of p16 signaling could facilitate the cell fate transition and increase SIRT3 protein expression in aging cells to the level of young cells, but no such changes were detected in SIRT1 and SIRT2.81 The iPSCs reprogramming efficiency and protein levels of SIRT3 in mouse iPSCs (miPSCs), generated from tail-tip fibroblasts (TTFs) of old mice, were significantly lower than those of the miPSCs generated from TTFs of young mice. Interestingly, overexpression of nicotinamide nucleotide transhydrogenase (NMT) and nicotinamide mononucleotide adenylyl transferase 3 (NMNAT3) could restore the NAD+ levels and SIRT3 activity in the mitochondria of old TTFs and further enhanced the iPSCs reprogramming efficiency (Figure 1).81 These findings suggest that low NAD+ levels and SIRT3 activity in the mitochondria of somatic cells from aged animal sources are barriers to cell fate transition.
SIRT4
Among the sirtuin family proteins, SIRT4 is the only member with no deacetylase activity and exhibits catalytic limitation to NAD+-dependent ADP-ribosyl transferase activity.82 SIRT4 is localized in mitochondrial matrix and plays roles in cell metabolism, redox homeostasis, and longevity. Unlike other sirtuins, SIRT4 negatively regulates oxidative metabolism in adult tissues by repressing mitochondrial glutamine metabolism. Upregulation of SIRT4 was observed in senescent spermatogonial stem cells.83 In trophoblast stem cells, SIRT4 activation by mTOR-mediated signaling was found to disturb mitochondrial function and redox homeostasis, which contributed to lysine-specific demethylase 1 deficiency-induced senescence.83 The most common source of somatic cells is skin fibroblasts, which are frequently exposed to UV irradiation and oxidative stress-elicited DNA damage, a known etiology in cell senescence of skin tissues. It was shown that photo-damage could upregulate the mRNA expression of both SIRT1 and SIRT4. Notably, SIRT4 was found to be degraded at the early stage of photo-damage and was then accumulated at the later stage. Upregulation of SIRT2 and SIRT4 by niacin restriction increased DNA damage (Figure 1),84 which implies that SIRT4 may be a promising target to prevent senescence of somatic cells and to facilitate the iPSCs reprogramming (Table 1). However, up to now there has been no direct evidence to support the role of SIRT4 in the reprogramming, pluripotency, and differentiation of iPSCs.
SIRT5
Mitochondrial SIRT5 catalyzes lysine deacylation to remove acetyl, succinyl, malonyl, and glutaryl groups from target proteins and consequently regulates mitochondrial metabolism. SIRT5 is the only sirtuin exhibiting protein desuccinylase and demalonylase activities in mammalian cells. Accumulated evidence suggests that SIRT5 and SIRT3 collaboratively regulate several metabolic pathways, such as β-oxidation of fatty acids and OXPHOS, and they even share the same protein targets. Although SIRT5 was found to be linked to several human diseases including cancer and neurodegenerative disorders, its physiological and pathophysiological functions remain elusive. In addition to regulating the metabolism of lipids,85 SIRT5 was found to be dramatically decreased in the protein level during adipogenic differentiation of hMSCs.61 Moreover, we found that overexpression of SIRT5 could negatively regulate mitochondrial respiration during adipogenic differentiation, which has substantiated the potential role of SIRT5 in stem cell differentiation. It was shown that SIRT5 plays an important role in glycolysis and energy metabolism in cancers.86 Interestingly, Yang et al.87 reported that SIRT5 can deacetylate STAT3, thereby inhibit its function in mitochondrial pyruvate metabolism. Notably, STAT3 is the downstream effector of LIF/JAK signaling, and plays an important role in the maintenance of the ground state pluripotency of iPSCs and is involved in the reprogramming of somatic cells (Figure 1).88,89 These observations suggest that SIRT5 could negatively regulate the pluripotency of iPSCs through down-regulation of the LIF/JAK/STAT3 axis.
SIRT6
SIRT6 has been known as an important regulator of genome stability and is associated with transcription, telomere integrity, genomic repair, and metabolic homeostasis. Its deficiency in mice induced premature aging multiple age-related syndromes, which ultimately led to premature cell death.90 Deletion of SIRT6 in mouse bone marrow cells could reduce osteogenic differentiation and affect the bone mineral density.91 Since SIRT6 has a role in aging, Li and co-workers92 demonstrated a link of the expression of SIRT6 to tooth development by using mouse odontoblasts. They showed that SIRT6 is required for the differentiation of dental MSCs, the formation of the tooth root, tooth eruption, and tooth germs through the regulation of mitochondrial energy metabolism. Impairment of cell proliferation and cell senescence were also observed in hBM-MSCs deficient of SIRT6.93,94 Loss of SIRT6 in hMSCs led to the aberrant redox metabolism and less tolerance to oxidative stress.94 It was found that SIRT6 prevented hMSCs from oxidative stress-induced damage and premature senescence by activating nuclear factor erythroid 2-related factor 2 (Nrf2), a critical redox sensor that modulates the antioxidant responses.94 There has been increasing evidence to suggest that SIRT6 positively regulates osteogenic and adipogenic differentiation of MSCs.95–97 In neurons, oxidative stress results in acute decline in the protein expression level of SIRT6.98 In contrast to the benefit of SIRT6 in adult hippocampal neurogenesis,99 overexpression of SIRT6 was found to induce autophagy via repression of the AKT/ERK signaling, which is responsible for H2O2-induced neuronal cell death.
SIRT6 regulates the balance between pluripotency and differentiation through ten-eleven translocation enzymes (TETs) and 5-hydroxymethylcytosine (5hmC).100 In addition, a combination of SIRT6 and the Yamanaka factors during reprogramming significantly promotes DNA double-strand break (DSB) repair by activating non-homologous end joining (NHEJ) in iPSCs derived from old mice. Therefore, SIRT6 can improve the quality of iPSCs derived from aged cells through the stabilization of their genome.101 Sharma et al.102 demonstrated that SIRT6 can enhance the reprogramming efficiency of iPSCs from aged skin fibroblasts. In addition, miR-766 was identified to regulate SIRT6 and iPSCs reprogramming (Figure 1). Furthermore, SIRT6 was shown to regulate the expression of pluripotent genes, such as Sox2, Oct4, and Nanog, through acetylation of histone H3 lysine 56 (H3K56ac) (Table 1).101
SIRT7
SIRT7 is located in the nucleus and its dysfunction has been linked to the occurrence of cancer and age-related pathologies. Expression of SIRT7 is declined during aging of hematopoietic stem cells (HSCs), its downregulation induces mitochondrial protein folding stress (PFSmt) and contributes to dysfunction of HSCs.103 SIRT7 activation can ameliorate the regenerative capacity of aged HSCs.104 SIRT7 binds to the promoter of NRF1 target genes and thus represses transcription of these genes to impair mitochondrial biogenesis and respiration.105 During osteogenic differentiation, SIRT7 is downregulated and Wnt/β-catenin signaling is activated.106 Conversely, induction of SIRT7 is required for adipogenic differentiation, but the regulation of adipogenesis by SIRT7 has not been fully explored. Loss of SIRT7 impairs the adipogenic differentiation ability of mouse embryonic fibroblasts and 3T3L1 preadipocytes. Besides, SIRT7 knockout mice also exhibited high adiposity.107 Interestingly, the expression level of miR-93, a repressor of SIRT7 that negatively regulates adipogenesis, was found to decrease in ob/ob mice.108 miR-93 has been reported to be associated with the turnover of mature adipocytes and its inhibition promotes fat formation in vivo. These findings suggest a potential role for SIRT7 in promoting obesity. Therefore, it can be expected that SIRT7 is a probable candidate for the treatment of obesity in the future.
It has been reported that SIRT7 also plays an important role in oncogenic transformation and tumor biology.109 It was shown that SIRT7 could regulate metastatic phenotypes in either epithelial or mesenchymal type of cancer cells, and that inactivation of SIRT7 could inhibit metastasis of cancer cells in vivo.110 In addition, SIRT7 plays a regulatory role in the process of mesenchymal-to-epithelial transition (MET) (Table 1), which has been suggested to be a crucial process in the generation of iPSCs from fibroblasts.111,112 However, more studies are required to prove that SIRT7 actually regulates the EMT and MET processes, respectively (Figure 1).
Potential molecular signals to specify the functions of sirtuins
The level of sirtuins and the modulation of their activities are considered to be the key factors in determining stem cell fate. Because of the requirement of NAD+ for the enzyme activity of sirtuins, intracellular ratio of NAD+/NADH is the major factor that controls sirtuin function. Mammalian nicotinamide phosphoribosyltransferase (Nampt) is a rate-limiting enzyme in the process of NAD+ biosynthesis. Its activity is highly related to intracellular levels of NAD+ and SIRT1 activity. Decline of Nampt in senescent MSCs was shown to attenuate the expression and activity of SIRT1. However, Nampt overexpression can restore age-related phenotype in MSCs.113 During osteogenic differentiation, induction of Nampt contributes to elevated NAD+ content and SIRT1 activation.114 Evidence indicates that Nampt promotes osteogenic differentiation and is negatively related to adipogenesis.113,115 A recent study demonstrated that Nampt deficiency by inhibitor or knockdown resulted in a decrease of the NAD+ concentration and SIRT1 activity, and subsequently interfered osteoblastogenesis of BM-MSCs.113 Nampt is also involved in oligodendrocytic lineage determination via modulation of SIRT1 and SIRT2.116 Like SIRT1, SIRT3 has been emerged as a metabolic sensor of the NAD+/NADH level in cells to meet the energy demand. The NAD+ and SIRT3 levels in mitochondria were observed to increase in mice subject to CR or during fasting.117,118 Rui and co-workers observed that Nampt overexpression enhanced mitochondrial function and protected neurons from lethal stress.119 It has been reported that SIRT3 and SIRT4 are involved in the cytoprotection effect of Nampt against genotoxic stress. Actually, AMPK is also involved in the salvage pathway of NAD+ synthesis via regulating Nampt transcription. AMPK activation induced by glucose restriction was shown to increase intracellular ratio of NAD+/NADH and thereby activate SIRT1.120 AMPK activation can enhance SIRT1-mediated deacetylation on its targets such as PGC-1α, FoxO1, and other differentiation-related factors. It has been recently demonstrated that AMPK downstream signaling facilitates osteogenic differentiation and may be associated with the lineage commitment of MSCs61 and reprogramming of iPSCs.121 On the other hand, mitochondrial pools of Nampt and NAD+ are also regulated by PKCɛ activation in an AMPK-dependent manner in rat cortex upon resveratrol treatment or ischemic preconditioning.122 Another research group demonstrated that an increase of mitochondrial Nampt by PKC ε activation could upregulate the expression and desuccinylase activity of SIRT5 in rat neuronal-astrocyte cortical cells, but did not influence SIRT3 activity.123 Moreover, they demonstrated the important role of SIRT5 in PKC ε–Nampt axis-mediated ischemic neuroprotection.123 Interestingly, unlike a downstream target of AMPK pathway, SIRT6 regulates metabolic homeostasis by activating AMPK in skeletal muscle124 and liver.125 Under energy deficient condition, AMPK was induced to directly modulate phosphorylation and subsequent redistribution and degradation of nuclear SIRT7 in cells.126 Collectively, these findings suggest that AMPK and PKC ε represent potential pathways for the regulation of sirtuins during stem cell differentiation.
Conclusions and future perspectives
Throughout the past decade, fast progress in stem cell research has hold great promise for cell therapy and regenerative medicine. Besides epigenetic regulation mechanisms, posttranslational modification of master proteins have been demonstrated to play important roles in the maintenance of pluripotency and differentiation potential of stem cells. Emerging evidence suggests that sirtuin family proteins regulate many protein functions, signaling pathways and cell fates in the differentiation of MSCs and reprogramming of iPSCs from somatic cells. The potential of sirtuins, especially SIRT1 and SIRT3, in regulating metabolic reprogramming and lineage-specific commitment of MSCs and iPSCs have received increasing attention in recent years. In previous studies, we showed that SIRT3 function is important for the upregulation of the mitochondrial biogenesis and respiration, and induction of antioxidant enzymes during adipogenic differentiation of hMSCs.60 However, the underlying mechanisms regulating the expression of sirtuins during these processes are largely unknown. It is important to specifically address the effects of SIRT3 and other sirtuins on oxidative metabolism and further delineate the role of master transcription factors such as FoxO1, FoxO3a, and PGC-1α in the metabolic reprogramming modulated by sirtuins in stem cell differentiation and iPSCs formation. During these processes, the expression levels of different sirtuins and mitochondrial biogenesis-related PGC-1α, NRF-1, NRF2, mtTFA, and respiratory enzyme complexes subunits, and antioxidant enzymes should be examined in an integrated and systemic manner. The potential signaling molecules that govern sirtuins activation, the role of PKCɛ–AMPK–Nampt axis in the regulation of different sirtuins, their crosstalk in mitochondrial protection during differentiation and maintenance of stem cells should be further investigated. Besides, the newly identified role of Sirt5 in catalyzing the succinylation and malonylation of mitochondrial proteins indicates the complexity of lysine acylations and suggests the possible crosstalk between these modifications in mitochondrial metabolism. Since succinylation and acetylation have the same protein targets in mitochondria,85 it is worthy to clarify if different lysine acylation at overlapped or adjacent residues results in an antagonistic or synergistic effect. We assume that Sirt3 could function coordinately with Sirt5 to modulate various metabolic processes during stem cell differentiation, iPSC reprogramming, or in cellular response to environmental stress.
In this article, we have reviewed and summarized the regulatory roles of SIRT1–7 in the reprogramming, maintenance of pluripotency, and differentiation of stem cells. We have also discussed the known miRNAs that target at different sirtuins. It is noteworthy that SIRT1 can be positively regulated by miR34 to promote reprogramming of iPSCs. Furthermore, the roles of miRNA-181a, miRNA181b, miR-9, miR-204, miR-199a/b, and miR135a in the regulation of the expression of SIRT1 have been proved. Besides, SIRT2 has been shown to be downregulated by miR200c during the formation of iPSCs. The miRNAs targeting at SIRT1 and SIRT2 could be applied in promoting stem cell translational medicine to make the reprogramming of iPSCs from somatic cells or stem cell differentiation more efficient and safe. However, the miRNA profile targeting SIRT3–7 remains to be investigated since the potential roles of SIRT3, SIRT4, and SIRT6 in enabling senescent skin fibroblasts to be reprogrammed into iPSCs have remained unclear. SIRT5 is crucial for the ground state pluripotency of naïve iPSCs. The understanding of the functions of sirtuins in stem cell biology are rather limited and are worthy of more investigation. The underlying molecular mechanisms of sirtuin family proteins and related miRNAs in the reprogramming and differentiation of iPSCs and other types of stem cells remain to be investigated. Further studies are warranted to identify potential natural products, chemical compounds, or drugs that specifically target at certain sirtuins to modulate stem cell differentiation or iPSCs formation.
Authors’ contributions
YCH: conception and design, manuscript writing. YTW: conception and design, manuscript writing. CLT: manuscript writing. YHW: conception and design, manuscript writing and revision, administrative support.
Declaration of conflicting interests
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding
The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This review has been prepared on the basis of some of the results obtained from research projects sponsored by grants (MOST104–2314-B-715–003-MY3, MOST104–2627-M-715–002, MOST104–2320-B-715–006-MY2, MOST105–2627-M-715–001, MOST106–2627-M-371–001 and MOST106–2320-B-371–002) from the Ministry of Science and Technology, Taiwan. We acknowledge the support of an intramural research grant from Changhua Christian Hospital, Changhua City, Taiwan.
References
- 1.Haigis MC, Guarente LP. Mammalian sirtuins–emerging roles in physiology, aging, and calorie restriction. Genes Dev 2006; 20:2913. [DOI] [PubMed] [Google Scholar]
- 2.Sinclair DA, Guarente L. Extrachromosomal rDNA circles – a cause of aging in yeast. Cell 1997; 91:1033–42 [DOI] [PubMed] [Google Scholar]
- 3.Frye RA. Phylogenetic classification of prokaryotic and eukaryotic Sir2-like proteins. Biochem Biophys Res Commun 2000; 273:793–8 [DOI] [PubMed] [Google Scholar]
- 4.Sebastian C, Satterstrom FK, Haigis MC, Mostoslavsky R. From sirtuin biology to human diseases: an update. J Biol Chem 2012; 287:42444–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tanno M, Kuno A, Horio Y, Miura T. Emerging beneficial roles of sirtuins in heart failure. Basic Res Cardiol 2012; 107:273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kiran S, Chatterjee N, Singh S, Kaul SC, Wadhwa R, Ramakrishna G. Intracellular distribution of human SIRT7 and mapping of the nuclear/nucleolar localization signal. FEBS J 2013; 280:3451–66 [DOI] [PubMed] [Google Scholar]
- 7.Ford E, Voit R, Liszt G, Magin C, Grummt I, Guarente L. Mammalian Sir2 homolog SIRT7 is an activator of RNA polymerase I transcription. Genes Dev 2006; 20:1075–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vaquero A, Scher MB, Lee DH, Sutton A, Cheng HL, Alt FW, Serrano L, Sternglanz R, Reinberg D. SirT2 is a histone deacetylase with preference for histone H4 Lys 16 during mitosis. Genes Dev 2006; 20:1256–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.North BJ, Marshall BL, Borra MT, Denu JM, Verdin E. The human Sir2 ortholog, SIRT2, is an NAD+-dependent tubulin deacetylase. Mol Cell 2003; 11:437–44 [DOI] [PubMed] [Google Scholar]
- 10.Nakamura Y, Ogura M, Tanaka D, Inagaki N. Localization of mouse mitochondrial SIRT proteins: shift of SIRT3 to nucleus by co-expression with SIRT5. Biochem Biophys Res Commun 2008; 366:174–9 [DOI] [PubMed] [Google Scholar]
- 11.Miyo M, Yamamoto H, Konno M, Colvin H, Nishida N, Koseki J, Kawamoto K, Ogawa H, Hamabe A, Uemura M, Nishimura J, Hata T, Takemasa I, Mizushima T, Doki Y, Mori M, Ishii H. Tumour-suppressive function of SIRT4 in human colorectal cancer. Br J Cancer 2015; 113:492–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Iwahara T, Bonasio R, Narendra V, Reinberg D. SIRT3 functions in the nucleus in the control of stress-related gene expression. Mol Cell Biol 2012; 32:5022–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Scher MB, Vaquero A, Reinberg D. SirT3 is a nuclear NAD+-dependent histone deacetylase that translocates to the mitochondria upon cellular stress. Genes Dev 2007; 21:920–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ryall JG, Dell'Orso S, Derfoul A, Juan A, Zare H, Feng X, Clermont D, Koulnis M, Gutierrez-Cruz G, Fulco M, Sartorelli V. The NAD+-dependent SIRT1 deacetylase translates a metabolic switch into regulatory epigenetics in skeletal muscle stem cells. Cell Stem Cell 2015; 16:171–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Heo J, Lim J, Lee S, Jeong J, Kang H, Kim Y, Kang JW, Yu HY, Jeong EM, Kim K, Kucia M, Waigel SJ, Zacharias W, Chen Y, Kim IG, Ratajczak MZ, Shin DM. Sirt1 regulates DNA methylation and differentiation potential of embryonic stem cells by antagonizing Dnmt3l. Cell Rep 2017; 18:1930–45 [DOI] [PubMed] [Google Scholar]
- 16.Ou X, Chae HD, Wang RH, Shelley WC, Cooper S, Taylor T, Kim YJ, Deng CX, Yoder MC, Broxmeyer HE. SIRT1 deficiency compromises mouse embryonic stem cell hematopoietic differentiation, and embryonic and adult hematopoiesis in the mouse. Blood 2011; 117:440–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tang S, Huang G, Fan W, Chen Y, Ward JM, Xu X, Xu Q, Kang A, McBurney MW, Fargo DC, Hu G, Baumgart-Vogt E, Zhao Y, Li X. SIRT1-mediated deacetylation of CRABPII regulates cellular retinoic acid signaling and modulates embryonic stem cell differentiation. Mol Cell 2014; 55:843–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen CT, Shih YR, Kuo TK, Lee OK, Wei YH. Coordinated changes of mitochondrial biogenesis and antioxidant enzymes during osteogenic differentiation of human mesenchymal stem cells. Stem Cells 2008; 26:960–8 [DOI] [PubMed] [Google Scholar]
- 19.Maiese K. MicroRNAs and SIRT1: a strategy for stem cell renewal and clinical development? J Transl Sci 2015; 1:55–7 [PMC free article] [PubMed] [Google Scholar]
- 20.Chae HD, Broxmeyer HE. SIRT1 deficiency downregulates PTEN/JNK/FOXO1 pathway to block reactive oxygen species-induced apoptosis in mouse embryonic stem cells. Stem Cells Dev 2011; 20:1277–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu X, Zhao H, Jin Q, You W, Cheng H, Liu Y, Song E, Liu G, Tan X, Zhang X, Wan F. Resveratrol induces apoptosis and inhibits adipogenesis by stimulating the SIRT1-AMPKalpha-FOXO1 signalling pathway in bovine intramuscular adipocytes. Mol Cell Biochem 2017;439:213–23 [DOI] [PubMed]
- 22.Li Z, Margariti A, Wu Y, Yang F, Hu J, Zhang L, Chen T. MicroRNA-199a induces differentiation of induced pluripotent stem cells into endothelial cells by targeting sirtuin 1. Mol Med Rep 2015; 12:3711–7 [DOI] [PubMed] [Google Scholar]
- 23.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 2006; 126:663–76 [DOI] [PubMed] [Google Scholar]
- 24.Mu WL, Wang YJ, Xu P, Hao DL, Liu XZ, Wang TT, Chen F, Chen HZ, Lv X, Liu DP. Sox2 deacetylation by Sirt1 is involved in mouse somatic reprogramming. Stem Cells 2015; 33:2135–47 [DOI] [PubMed] [Google Scholar]
- 25.Saunders LR, Sharma AD, Tawney J, Nakagawa M, Okita K, Yamanaka S, Willenbring H, Verdin E. miRNAs regulate SIRT1 expression during mouse embryonic stem cell differentiation and in adult mouse tissues. Aging 2010; 2:415–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Homma K, Sone M, Taura D, Yamahara K, Suzuki Y, Takahashi K, Sonoyama T, Inuzuka M, Fukunaga Y, Tamura N, Itoh H, Yamanaka S, Nakao K. Sirt1 plays an important role in mediating greater functionality of human ES/iPS-derived vascular endothelial cells. Atherosclerosis 2010; 212:42–7 [DOI] [PubMed] [Google Scholar]
- 27.Jiang B, Jen M, Perrin L, Wertheim JA, Ameer GA. SIRT1 overexpression maintains cell phenotype and function of endothelial cells derived from induced pluripotent stem cells. Stem Cells Dev 2015; 24:2740–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee YL, Peng Q, Fong SW, Chen AC, Lee KF, Ng EH, Nagy A, Yeung WS. Sirtuin 1 facilitates generation of induced pluripotent stem cells from mouse embryonic fibroblasts through the miR-34a and p53 pathways. PLoS One 2012; 7:e45633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hishida T, Nozaki Y, Nakachi Y, Mizuno Y, Iseki H, Katano M, Kamon M, Hirasaki M, Nishimoto M, Okazaki Y, Okuda A. Sirt1, p53, and p38(MAPK) are crucial regulators of detrimental phenotypes of embryonic stem cells with Max expression ablation. Stem Cells 2012; 30:1634–44 [DOI] [PubMed] [Google Scholar]
- 30.Hayakawa K, Hirosawa M, Tabei Y, Arai D, Tanaka S, Murakami N, Yagi S, Shiota K. Epigenetic switching by the metabolism-sensing factors in the generation of orexin neurons from mouse embryonic stem cells. J Biol Chem 2013; 288:17099–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jang J, Huh YJ, Cho HJ, Lee B, Park J, Hwang DY, Kim DW. SIRT1 enhances the survival of human embryonic stem cells by promoting DNA repair. Stem Cell Rep 2017; 9:629–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ou X, Lee MR, Huang X, Messina-Graham S, Broxmeyer HE. SIRT1 positively regulates autophagy and mitochondria function in embryonic stem cells under oxidative stress. Stem Cells 2014; 32:1183–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhou L, Chen X, Liu T, Gong Y, Chen S, Pan G, Cui W, Luo ZP, Pei M, Yang H, He F. Melatonin reverses H2O2-induced premature senescence in mesenchymal stem cells via the SIRT1-dependent pathway. J Pineal Res 2015; 59:190–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim KS, Choi HW, Yoon HE, Kim IY. Reactive oxygen species generated by NADPH oxidase 2 and 4 are required for chondrogenic differentiation. J Biol Chem 2010; 285:40294–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang W, Zhang Y, Lu W, Liu K. Mitochondrial reactive oxygen species regulate adipocyte differentiation of mesenchymal stem cells in hematopoietic stress induced by arabinosylcytosine. PLoS One 2015; 10:e0120629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Palacios JA, Herranz DD, Bonis ML, Velasco S, Serrano M, Blasco MA. SIRT1 contributes to telomere maintenance and augments global homologous recombination. J Biol Chem 2010; 191:1299–313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen H, Liu X, Zhu W, Chen H, Hu X, Jiang Z, Xu Y, Wang L, Zhou Y, Chen P, Zhang N, Hu D, Zhang L, Wang Y, Xu Q, Wu R, Yu H, Wang J. SIRT1 ameliorates age-related senescence of mesenchymal stem cells via modulating telomere shelterin. Front Aging Neurosci 2014; 6:103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Perez-Campo FM, Riancho JA. Epigenetic mechanisms regulating mesenchymal stem cell differentiation. Curr Genomics 2015; 16:368–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gong K, Qu B, Wang C, Zhou J, Liao D, Zheng W, Pan X. Peroxisome proliferator-activated receptor alpha facilitates osteogenic differentiation in MC3T3-E1 Cells via the sirtuin 1-dependent signaling pathway. Mol Cells 2017; 40:393–400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Prestwich TC, Macdougald OA. Wnt/beta-catenin signaling in adipogenesis and metabolism. Curr Opin Cell Biol 2007; 19:612–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li HX, Luo X, Liu RX, Yang YJ, Yang GS. Roles of Wnt/beta-catenin signaling in adipogenic differentiation potential of adipose-derived mesenchymal stem cells. Mol Cell Endocrinol 2008; 291:116–24 [DOI] [PubMed] [Google Scholar]
- 42.Zhou Y, Song T, Peng J, Zhou Z, Wei H, Zhou R, Jiang S, Peng J. SIRT1 suppresses adipogenesis by activating Wnt/beta-catenin signaling in vivo and in vitro. Oncotarget 2016; 7:77707–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yuan Z, Li Q, Luo S, Liu Z, Luo D, Zhang B, Zhang D, Rao P, Xiao J. PPAR gamma Wnt signaling in adipogenic and osteogenic differentiation of mesenchymal stem cells. Curr Stem Cell Res Ther 2016; 11:216–25 [DOI] [PubMed] [Google Scholar]
- 44.Feng G, Zheng K, Song D, Xu K, Huang D, Zhang Y, Cao P, Shen S, Zhang J, Feng X, Zhang D. SIRT1 was involved in TNF-alpha-promoted osteogenic differentiation of human DPSCs through Wnt/beta-catenin signal. In Vitro Cell Dev Biol Anim 2016; 52:1001–11 [DOI] [PubMed] [Google Scholar]
- 45.Zhou Y, Zhou Z, Zhang W, Hu X, Wei H, Peng J, Jiang S. SIRT1 inhibits adipogenesis and promotes myogenic differentiation in C3H10T1/2 pluripotent cells by regulating Wnt signaling. Cell Biosci 2015; 5:61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Inoue T, Hiratsuka M, Osaki M, Yamada H, Kishimoto I, Yamaguchi S, Nakano S, Katoh M, Ito H, Oshimura M. SIRT2 a tubulin deacetylase, acts to block the entry to chromosome condensation in response to mitotic stress. Oncogene 2007; 26:945–57 [DOI] [PubMed] [Google Scholar]
- 47.Liu TM, Shyh-Chang N. SIRT2 and glycolytic enzyme acetylation in pluripotent stem cells. Nat Cell Biol 2017; 19:412–4 [DOI] [PubMed] [Google Scholar]
- 48.Yang W, Guo X, Thein S, Xu F, Sugii S, Baas PW, Radda GK, Han W. Regulation of adipogenesis by cytoskeleton remodelling is facilitated by acetyltransferase MEC-17-dependent acetylation of alpha-tubulin. Biochem J 2013; 449:605–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Si X, Chen W, Guo X, Chen L, Wang G, Xu Y, Kang J. Activation of GSK3beta by Sirt2 is required for early lineage commitment of mouse embryonic stem cell. PLoS One 2013; 8:e76699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yoo DY, Kim DW, Kim MJ, Choi JH, Jung HY, Nam SM, Kim JW, Yoon YS, Choi SY, Hwang IK. Sodium butyrate, a histone deacetylase Inhibitor, ameliorates SIRT2-induced memory impairment, reduction of cell proliferation, and neuroblast differentiation in the dentate gyrus. Neurol Res 2015; 37:69–76 [DOI] [PubMed] [Google Scholar]
- 51.Jeong SG, Cho GW. The tubulin deacetylase sirtuin-2 regulates neuronal differentiation through the ERK/CREB signaling pathway. Biochem Biophys Res Commun 2017; 482:182–7 [DOI] [PubMed] [Google Scholar]
- 52.Cha Y, Han MJ, Cha HJ, Zoldan J, Burkart A, Jung JH, Jang Y, Kim CH, Jeong HC, Kim BG, Langer R, Kahn CR, Guarente L, Kim KS. Metabolic control of primed human pluripotent stem cell fate and function by the miR-200c-SIRT2 axis. Nat Cell Biol 2017; 19:445–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Eo SH, Choi SY, Kim SJ. PEP-1-SIRT2-induced matrix metalloproteinase-1 and -13 modulates type II collagen expression via ERK signaling in rabbit articular chondrocytes. Exp Cell Res 2016; 348:201–8 [DOI] [PubMed] [Google Scholar]
- 54.Arora A, Dey CS. SIRT2 regulates insulin sensitivity in insulin resistant neuronal cells. Biochem Biophys Res Commun 2016; 474:747–52 [DOI] [PubMed] [Google Scholar]
- 55.Nahhas F, Dryden SC, Abrams J, Tainsky MA. Mutations in SIRT2 deacetylase which regulate enzymatic activity but not its interaction with HDAC6 and tubulin. Mol Cell Biochem 2007; 303:221–30 [DOI] [PubMed] [Google Scholar]
- 56.Wang F, Tong Q. SIRT2 suppresses adipocyte differentiation by deacetylating FOXO1 and enhancing FOXO1's repressive interaction with PPARgamma. Mol Biol Cell 2009; 20:801–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jing E, Gesta S, Kahn CR. SIRT2 regulates adipocyte differentiation through FoxO1 acetylation/deacetylation. Cell Metab 2007; 6:105–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ding Y, Yang H, Wang Y, Chen J, Ji Z, Sun H. Sirtuin 3 is required for osteogenic differentiation through maintenance of PGC-1α-SOD2-mediated regulation of mitochondrial function. Int J Biol Sci 2017; 13:254–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Huh JE, Shin JH, Jang ES, Park SJ, Park DR, Ko R, Seo DH, Kim HS, Lee SH, Choi Y, Kim HS, Lee SY. Sirtuin 3 (SIRT3) maintains bone homeostasis by regulating AMPK-PGC-1beta axis in mice. Sci Rep 2016; 6:22511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gomez-Puerto MC, Verhagen LP, Braat AK, Lam EW, Coffer PJ, Lorenowicz MJ. Activation of autophagy by FOXO3 regulates redox homeostasis during osteogenic differentiation. Autophagy 2016; 12:1804–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hsu YC, Wu YT, Yu TH, Wei YH. Mitochondria in mesenchymal stem cell biology and cell therapy: from cellular differentiation to mitochondrial transfer. Semin Cell Dev Biol 2016; 52:119–31 [DOI] [PubMed] [Google Scholar]
- 62.Abdel Khalek W, Cortade F, Ollendorff V, Lapasset L, Tintignac L, Chabi B, Wrutniak-Cabello C. SIRT3, a mitochondrial NAD+-dependent deacetylase, is involved in the regulation of myoblast differentiation. PLoS One.2014;9:e114388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Denu RA . SIRT3 enhances mesenchymal stem cell longevity and differentiation. Oxid Med Cell Longev 2017; 2017:5841716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhang Y, Marsboom G, Toth PT, Rehman J. Mitochondrial respiration regulates adipogenic differentiation of human mesenchymal stem cells. PLoS One 2013; 8:e77077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pan CW. Effect of mitochondrial sirtuins-mediated posttranslational modification of pyruvate dehydrogenase on oxidative metabolism in human cells harboring mitochondrial DNA with A8344G mutation. Master’s thesis, Institute of Biochemistry and Molecular Biology, National Yang-Ming University, Taipei, 2015.
- 66.Zhang S, Hulver MW, McMillan RP, Cline MA, Gilbert ER. The pivotal role of pyruvate dehydrogenase kinases in metabolic flexibility. Nutr Metab 2014; 11:10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bigrigg JK, Heigenhauser GJ, Inglis JG, LeBlanc PJ, Peters SJ. Carbohydrate refeeding after a high-fat diet rapidly reverses the adaptive increase in human skeletal muscle PDH kinase activity. Am J Physiol Regul Integr Comp Physiol 2009; 297:R885–91 [DOI] [PubMed] [Google Scholar]
- 68.Jing E, O'Neill BT, Rardin MJ, Kleinridders A, Ilkeyeva OR, Ussar S, Bain JR, Lee KY, Verdin EM, Newgard CB, Gibson BW, Kahn CR. Sirt3 regulates metabolic flexibility of skeletal muscle through reversible enzymatic deacetylation. Diabetes 2013; 62:3404–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wu YT, Lee HC, Liao CC, Wei YH. Regulation of mitochondrial FoF1ATPase activity by Sirt3-catalyzed deacetylation and its deficiency in human cells harboring 4977 bp deletion of mitochondrial DNA. Biochim Biophys Acta 2013; 1832:216–27 [DOI] [PubMed] [Google Scholar]
- 70.Giralt A, Hondares E, Villena JA, Ribas F, Díaz-Delfín J, Giralt M, Iglesias R, Villarroya F. Peroxisome proliferator-activated receptor-gamma coactivator-1alpha controls transcription of the Sirt3 gene, an essential component of the thermogenic brown adipocyte phenotype. J Biol Chem 2011; 286:16958–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Poher AL, Altirriba J, Veyrat-Durebex C, Rohner-Jeanrenaud F. Brown adipose tissue activity as a target for the treatment of obesity/insulin resistance. Front Physiol 2015; 6:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Roman S, Agil A, Peran M, Alvaro-Galue E, Ruiz-Ojeda FJ, Fernández-Vázquez G, Marchal JA. Brown adipose tissue and novel therapeutic approaches to treat metabolic disorders. Transl Res 2015; 165:464–79 [DOI] [PubMed] [Google Scholar]
- 73.Huang PI, Chou YC, Chang YL, Chien Y, Chen KH, Song WS, Peng CH, Chang CH, Lee SD, Lu KH, Chen YJ, Kuo CH, Hsu CC, Lee HC, Yung MC. Enhanced differentiation of three-gene-reprogrammed induced pluripotent stem cells into adipocytes via adenoviral-mediated PGC-1alpha overexpression. Int J Mol Sci 2011; 12:7554–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Jiang DQ, Wang Y, Li MX, Ma YJ, Wang Y. SIRT3 in neural stem cells attenuates microglia activation-induced oxidative stress injury through mitochondrial pathway. Front Cell Neurosci 2017; 11:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wang S, Zhang C, Niyazi S, Zheng L, Li J, Zhang W, Xu M, Rong R, Yang C, Zhu TA. novel cytoprotective peptide protects mesenchymal stem cells against mitochondrial dysfunction and apoptosis induced by starvation via Nrf2/Sirt3/FoxO3a pathway. J Transl Med 2017; 15:33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Marycz K, Kornicka K, Grzesiak J, Smieszek A, Szlapka J. Macroautophagy and selective mitophagy ameliorate chondrogenic differentiation potential in adipose stem cells of equine metabolic syndrome: new findings in the field of progenitor cells differentiation. Oxid Med Cell Longev 2016; 2016:3718468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Gomes AP, Price NL, Ling AJ, Moslehi JJ, Montgomery MK, Rajman L, White JP, Teodoro JS, Wrann CD, Hubbard BP, Mercken EM, Palmeira CM, de Cabo R, Rolo AP, Turner N, Bell EL, Sinclair DA. Declining NAD+ induces a pseudohypoxic state disrupting nuclear-mitochondrial communication during aging. Cell 2013; 155:1624–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Fang EF, Scheibye-Knudsen M, Brace LE, Kassahun H, SenGupta T, Nilsen H, Mitchell JR, Croteau DL, Bohr VA. Defective mitophagy in XPA via PARP-1 hyperactivation and NAD+/SIRT1 reduction. Cell 2014; 157:882–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wang B, Miyagoe-Suzuki Y, Yada E, Ito N, Nishiyama T, Nakamura M, Ono Y, Motohashi N, Segawa M, Masuda S, Takeda S. Reprogramming efficiency and quality of induced pluripotent stem cells (iPSCs) generated from muscle-derived fibroblasts of mdx mice at different ages. PLoS Curr 2011; 3:RRN1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lapasset L, Milhavet O, Prieur A, Besnard E, Babled A, Aït-Hamou N, Leschik J, Pellestor F, Ramirez JM, Vos Lehmann DJ, Lemaitre SJM. Rejuvenating senescent and centenarian human cells by reprogramming through the pluripotent state. Genes Dev 2011; 25:2248–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Son MJ, Kwon Y, Son T, Cho YS. Restoration of mitochondrial NAD+ levels delays stem cell senescence and facilitates reprogramming of aged somatic cells. Stem Cells 2016; 34:2840–51 [DOI] [PubMed] [Google Scholar]
- 82.Haigis MC, Mostoslavsky R, Haigis KM, Fahie K, Christodoulou DC, Murphy AJ, Valenzuela DM, Yancopoulos GD, Karow M, Blander G, Wolberger C, Prolla TA, Weindruch R, Alt FW, Guarente L. SIRT4 inhibits glutamate dehydrogenase and opposes the effects of calorie restriction in pancreatic beta cells. Cell 2006; 126:941–54 [DOI] [PubMed] [Google Scholar]
- 83.Castex J, Willmann D, Kanouni T, Arrigoni L, Li Y, Friedrich M, Schleicher M, Wöhrle S, Pearson M, Kraut N, Méret M, Manke T, Metzger E, Schüle R, Günther T. Inactivation of Lsd1 triggers senescence in trophoblast stem cells by induction of Sirt4. Cell Death Dis 2017; 8:e2631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Benavente CA, Schnell SA, Jacobson EL. Effects of niacin restriction on sirtuin and PARP responses to photodamage in human skin. PLoS One 2012; 7:e42276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Park J, Chen Y, Tishkoff DX, Peng C, Tan M, Dai L, Xie Z, Zhang Y, Zwaans BM, Skinner ME, Lombard DB, Zhao Y. SIRT5-M lysine desuccinylation impacts diverse metabolic pathways. Mol Cell 2013; 50:919–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Bringman-Rodenbarger LR, Guo AH, Lyssiotis CA, Lombard DB. Emerging roles for SIRT5 in metabolism and cancer. Antioxid Redox Signal. Epub ahead of print 26 October 2017. doi: 10.1089/ars.2017.7264 [DOI] [PMC free article] [PubMed]
- 87.Yang J, van Oosten AL, Theunissen TW, Guo G, Silva JC, Smith A. Stat3 activation is limiting for reprogramming to ground state pluripotency. Cell Stem Cell 2010; 7:319–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Tang Y, Luo Y, Jiang Z, Ma Y, Lin CJ, Kim C, Carter MG, Amano T, Park J, Kish S, Tian XC. Jak/Stat3 signaling promotes somatic cell reprogramming by epigenetic regulation. Stem Cells 2012; 30:2645–56 [DOI] [PubMed] [Google Scholar]
- 89.Tang Y, Tian XC. JAK-STAT3 and somatic cell reprogramming. Jakstat 2013; 2:e24935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Lombard DB, Schwer B, Alt FW, Mostoslavsky R. SIRT6 in DNA repair, metabolism and ageing. J Intern Med 2008; 263:128–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Zhang DM, Cui DX, Xu RS, Zhou YC, Zheng LW, Liu P, Zhou XD. Phenotypic research on senile osteoporosis caused by SIRT6 deficiency. Int J Oral Sci 2016; 8:84–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Liao X, Feng B, Zhang D, Liu P, Zhou X, Li R, Ye L. The Sirt6 gene: does it play a role in tooth development?. PLoS One 2017; 12:e0174255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Zhai XY, Yan P, Zhang J, Song HF, Yin WJ, Gong H, Li H, Wu J, Xie J, Li RK. Knockdown of SIRT6 enables human bone marrow mesenchymal stem cell senescence. Rejuvenation Res. Epub ahead of print 14 March 2016. DOI: 10.1089/rej.2015.1770 [DOI] [PubMed]
- 94.Pan H, Guan D, Liu X, Li J, Wang Wu J, Zhou J, Zhang W, Ren R, Zhang W, Li Y, Yang J, Hao Y, Yuan T, Yuan G, Wang H, Ju Z, Mao Z, Li J, Qu J, Tang F, Liu GH. SIRT6 safeguards human mesenchymal stem cells from oxidative stress by coactivating NRF2. Cell Res 2016; 26:190–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Sun H, Wu Y, Fu D, Liu Y, Huang C. SIRT6 regulates osteogenic differentiation of rat bone marrow mesenchymal stem cells partially via suppressing the nuclear factor-kappaB signaling pathway. Stem Cells 2014; 32:1943–55 [DOI] [PubMed] [Google Scholar]
- 96.Zhang P, Liu Y, Wang Y, Zhang M, Lv L, Zhang X, Zhou Y. SIRT6 promotes osteogenic differentiation of mesenchymal stem cells through BMP signaling. Sci Rep 2017; 7:10229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Chen Q, Hao W, Xiao C, Wang R, Xu X, Lu H, Chen W, Deng CX. SIRT6 is essential for adipocyte differentiation by regulating mitotic clonal expansion. Cell Rep 2017; 18:3155–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Shao J, Yang X, Liu T, Zhang T, Xie QR, Xia W. Autophagy induction by SIRT6 is involved in oxidative stress-induced neuronal damage. Protein Cell 2016; 7:281–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Okun E, Marton D, Cohen D, Griffioen K, Kanfi Y, Illouz T, Madar R, Cohen HY. Sirt6 alters adult hippocampal neurogenesis. PLoS One 2017; 12:e0179681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Chen W, Liu N, Zhang H, Qiao J, Jia W, Zhu S, Mao Z, Kang J. Sirt6 promotes DNA end joining in iPSCs derived from old mice. Cell Rep 2017; 18:2880–92 [DOI] [PubMed] [Google Scholar]
- 101.Etchegaray JP, Chavez L, Huang Y, Ross KN, Choi J, Martinez-Pastor B, Walsh RM, Sommer CA, Lienhard M, Gladden A, Kugel S, Silberman DM, Ramaswamy S, Mostoslavsky G, Hochedlinger K, Goren A, Rao A, Mostoslavsky R. The histone deacetylase SIRT6 controls embryonic stem cell fate via TET-mediated production of 5-hydroxymethylcytosine. Nat Cell Biol 2015; 17:545–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Sharma A, Diecke S, Zhang WY, Lan F, He C, Mordwinkin NM, Chua KF, Wu JC. The role of SIRT6 protein in aging and reprogramming of human induced pluripotent stem cells. J Biol Chem 2013; 288:18439–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Mohrin M, Shin J, Liu Y, Brown K, Luo H, Xi Y, Haynes CM, Chen D. Stem cell aging. A mitochondrial UPR-mediated metabolic checkpoint regulates hematopoietic stem cell aging. Science 2015; 347:1374–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Haynes CM, Fiorese CJ, Lin YF. Evaluating and responding to mitochondrial dysfunction: the mitochondrial unfolded-protein response and beyond. Trends Cell Biol 2013; 23:311–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Barber MF, Michishita-Kioi E, Xi Y, Tasselli L, Kioi M, Moqtaderi Z, Tennen RI, Paredes S, Young NL, Chen K, Struhl K, Garcia BA, Gozani O, Li W, Chua KF. SIRT7 links H3K18 deacetylation to maintenance of oncogenic transformation. Nature 2012; 487:114–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Chen EEM, Zhang W, Ye CCY, Gao X, Jiang LLJ, Zhao TTF, Pan ZZJ, Xue DDT. Knockdown of SIRT7 enhances the osteogenic differentiation of human bone marrow mesenchymal stem cells partly via activation of the Wnt/beta-catenin signaling pathway. Cell Death Dis 2017; 8:e3042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Kurylowicz A. In search of new therapeutic targets in obesity treatment: sirtuins. Int J Mol Sci 2016; 17:E572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Shin J, He M, Liu Y, Paredes S, Villanova L, Brown K, Qiu X, Nabavi N, Mohrin M, Wojnoonski K, Li P, Cheng HL, Murphy AJ, Valenzuela DM, Luo H, Kapahi P, Krauss R, Mostoslavsky R, Yancopoulos GD, Alt FW, Chua KF, Chen D. SIRT7 represses Myc activity to suppress ER stress and prevent fatty liver disease. Cell Rep 2013; 5:654–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Paredes S, Villanova L, Chua KF. Molecular pathways: emerging roles of mammalian Sirtuin SIRT7 in cancer. Clin Cancer Res 2014; 20:1741–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Malik S, Villanova L, Tanaka S, Aonuma M, Roy N, Berber E, Pollack JR, Michishita-Kioi E, Chua KF. SIRT7 inactivation reverses metastatic phenotypes in epithelial and mesenchymal tumors. Sci Rep 2015; 5:9841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Li R, Liang J, Ni S, Zhou T, Qing X, Li H, He W, Chen J, Li F, Zhuang Q, Qin B, Xu J, Li W, Yang J, Gan Y, Qin D, Feng S, Song H, Yang D, Zhang B, Zeng L, Lai L, Esteban MA, Pei D. A mesenchymal-to-epithelial transition initiates and is required for the nuclear reprogramming of mouse fibroblasts. Cell Stem Cell 2010; 7:51–63 [DOI] [PubMed] [Google Scholar]
- 112.Saito K, Fukuda N, Shinohara K, Masuhiro Y, Hanazawa S, Matsuda H, Fujiwara K, Ueno T, Soma M. Modulation of the EMT/MET process by pyrrole-imidazole polyamide targeting human transforming growth factor-beta1. Int J Biochem Cell Biol 2015; 66:112. [DOI] [PubMed] [Google Scholar]
- 113.Ma C, Pi C, Yang Y, Lin L, Shi Y, Li Y, Li Y, He X. Nampt expression decreases age-related senescence in rat bone marrow mesenchymal stem cells by targeting Sirt1. PLoS One 2017; 12:e0170930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Li Y, He J, He X, Li Y, Lindgren U. Nampt expression increases during osteogenic differentiation of multi- and omnipotent progenitors. Biochem Biophys Res Commun 2013; 434:117–23 [DOI] [PubMed] [Google Scholar]
- 115.He X, He J, Shi Y, Pi C, Yang Y, Sun Y, Ma C, Lin L, Zhang L, Li Y, Li Y. Nicotinamide phosphoribosyltransferase (Nampt) may serve as the marker for osteoblast differentiation of bone marrow-derived mesenchymal stem cells. Exp Cell Res 2017; 352:45–52 [DOI] [PubMed] [Google Scholar]
- 116.Stein LR, Imai S. Specific ablation of Nampt in adult neural stem cells recapitulates their functional defects during aging. EMBO J 2014; 33:1321–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Pham TX, Lee J. Dietary regulation of histone acetylases and deacetylases for the prevention of metabolic diseases. Nutrients 2012; 4:1868–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Haigis MC, Sinclair DA. Mammalian sirtuins: biological insights and disease relevance. Annu Rev Pathol 2010; 5:253–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Wang P, Xu TY, Guan YF, Tian WW, Viollet B, Rui YC, Zhai QW, Su DF, Miao CY. Nicotinamide phosphoribosyl transferase protects against ischemic stroke through SIRT1-dependent adenosine monophosphate-activated kinase pathway. Ann Neurol 2011; 69:360–74 [DOI] [PubMed] [Google Scholar]
- 120.Fulco M, Cen Y, Zhao P, Hoffman EP, McBurney MW, Sauve AA, Sartorelli V. Glucose restriction inhibits skeletal myoblast differentiation by activating SIRT1 through AMPK-mediated regulation of Nampt. Dev Cell 2008; 14:661–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Vazquez-Martin A, Vellon L, Quirós PM, Cufí S, Ruiz de Galarreta E, Oliveras-Ferraros C, Martin AG, Martin-Castillo B, López-Otín C, Menendez JA. Activation of AMP-activated protein kinase (AMPK) provides a metabolic barrier to reprogramming somatic cells into stem cells. Cell Cycle 2012; 11:974–89 [DOI] [PubMed] [Google Scholar]
- 122.Morris-Blanco KC, Cohan CH, Neumann JT, Sick TJ, Perez-Pinzon MA. Protein kinase C epsilon regulates mitochondrial pools of Nampt and NAD+ following resveratrol and ischemic preconditioning in the rat cortex. J Cereb Blood Flow Metab 2014; 34:1024–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Morris-Blanco KC, Dave KR, Saul I, Koronowski KB, Stradecki HM, Perez-Pinzon MA. Protein kinase C epsilon promotes cerebral ischemic tolerance via modulation of mitochondrial sirt5. Sci Rep 2016; 6:29790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Cui X, Yao L, Yang X, Gao Y, Fang F, Zhang J, Wang Q, Chang Y. SIRT6 regulates metabolic homeostasis in skeletal muscle through activation of AMPK. Am J Physiol Endocrinol Metab 2017; 313:E493–E505 [DOI] [PubMed] [Google Scholar]
- 125.Elhanati S, Kanfi Y, Varvak A, Roichman A, Carmel-Gross I, Barth S, Gibor G, Cohen HY. Multiple regulatory layers of SREBP1/2 by SIRT6. Cell Rep 2013; 4:905–12 [DOI] [PubMed] [Google Scholar]
- 126.Sun L, Fan G, Shan P, Qiu X, Dong S, Liao L, Yu C, Wang T, Gu X, Li Q, Song X, Cao L, Li X, Cui Y, Zhang S, Wang C. Regulation of energy homeostasis by the ubiquitin-independent REGgamma proteasome. Nat Commun 2016; 7:12497. [DOI] [PMC free article] [PubMed] [Google Scholar]