Short abstract
FRY like transcription coactivator (Fryl) gene located on chromosome 5 is a paralog of FRY microtubule binding protein (Fry) in vertebrates. It encodes a protein with unknown functions. Fryl gene is conserved in various species ranging from eukaryotes to human. Although there are several reports on functions of Fry gene, functions of Fryl gene remain unclear. A mouse line containing null mutation in Fryl gene by gene trapping was produced in this study for the first time. The survival and growth of Fryl−/− mice were observed. Fryl gene expression levels in mouse tissues were determined and histopathologic analyses were conducted. Most Fryl−/− mice died soon after birth. Rare Fryl−/− survivors showed growth retardation with significantly lower body weight compared to their littermate controls. Although they could breed, more than half of Fryl−/− survivors died of hydronephrosis before age 1. No abnormal histopathologic lesion was apparent in full-term embryo or adult tissues except the kidney. Abnormal lining cell layer detachments from walls of collecting and convoluted tubules in kidneys were apparent in Fryl−/− neonates and full-term embryos. Fryl gene was expressed in renal tubular tissues including the glomeruli and convoluted and collecting tubules. This indicates that defects in tubular systems are associated with Fryl functions and death of Fryl−/− neonates. Fryl protein is required for normal development and functional maintenance of kidney in mice. This is the first report of in vivo Fryl gene functions.
Impact statement
FRY like transcription coactivator (Fryl) gene is conserved in various species ranging from eukaryotes to human. It expresses a protein with unknown function. We generated a Fryl gene mutant mouse line and found that most homozygous mice died soon after their birth. Rare Fryl−/− survivors showed growth retardation with significantly lower body weight compared to their littermate controls. Although they could breed, more than half of Fryl−/− survivors died of hydronephrosis before age 1. Full-term mutant embryos showed abnormal collecting and convoluted tubules in kidneys where Fryl gene was expressed. Collectively, these results indicate that Fryl protein is required for normal development and functional maintenance of kidney in mice. To the best of our knowledge, this is the first report on in vivo Fryl gene functions.
Keywords: Fryl, kidney, nephropathy, mutant, mouse, lethal
Introduction
Two paralogous genes, FRY microtubule binding protein (Fry) and FRY like transcription coactivator (Fryl), have been found in vertebrates including frog, chicken, mouse, and human. However, only one orthologous gene of Fry has been found in yeast, nematode, or fly (named Tao3p in budding yeast, Mor2p in fission yeast and Sax2 in C. elegans).1–5
Diverse functions of the Fry gene have been reported since its first identification from Drosophila in 2001.6–9 This gene is highly conserved during evolution, suggesting that a large selective pressure may have resulted in the conservation of its specific structural and functional characteristics.10 Fry is a protein with a high molecular mass (∼300 kDa). It has five to six conserved regions, including Fry N-terminal domain (FND) consisting of HEAT/Armadillo-like repeats. Additionally, two leucine zipper motifs and coiled-coil motif near the C-terminus have been found in Fry proteins of vertebrates.2,11 Human Fryl and Fry proteins almost have the same structure.10
Most known functions of Fry and its orthologues in invertebrates are associated with nuclear Dbf2-related (NDR) serine/threonine kinases (termed Trc in fly, Sax-1 in nematode, Cbk1p in budding yeast, and Orb6p in fission yeast), wherein Fry functions as an activator or scaffold of these kinases.10 However, in vertebrates, Fry has NDR kinase-unrelated and NDR kinase-related functions.10 Fry can bind to Aurora kinase A and promote the activity of polo-like kinase 1 (Plk1) by binding to Plk1 and facilitating Aurora A-mediated Plk1 phosphorylation during mitosis. In addition, Fry can directly bind to microtubules via multiple microtubule binding sites12 and promote acetylation of microtubules by binding to and suppressing tubulin deacetylase Sirtuin2 (SIRT2).10,13 Xenopus Fry also contributes to axis formation during development by promoting the expression of chordamesodermal genes by regulating the transcription of micro-RNA miR15.14 Collectively, these results suggest that vertebrate Fry and its paralog Fryl have crucial roles in maintaining the integrity of mitotic chromosome alignment, the integrity of spindle pole and spindle bipolarity during early mitosis, and the dynamics of microtubules in mammalian cells.
According to published data on Fryl functions, Fryl gene can be a fusion partner of mixed lineage leukemia (MLL) gene in patients with treatment–related acute lymphoblastic leukemia, suggesting that Fryl may have translational activation properties through its C-terminal leucine zipper motif.1 It has been shown that Fryl is a Notch coactivator in transcriptional activation events subsequent to intracellular domain of Notch 1 recruitment at several Notch-responsive genes.15,16 Notch signaling regulates diverse cell fates throughout embryonic development and in adult tissues.17 Therefore, Fryl may be associated with fate decisions of cells and tissues.
Paralogous proteins Fryl and Fry share 60% sequence identities at amino acid level. They have similar structures in human (74% with conservative substitution).1,10 Fryl may have similar roles as Fry in vertebrates. However, Fryl gene functions remain largely unknown. Therefore, the objective of this study was to determine if Fryl may have distinct and non-redundant roles in mice.
Materials and methods
Generation of Fryl gene mutant mouse
Mutated Fryl gene was introduced into KTPU8 ES cells with pU-21T gene trap vector by electroporation as described previously.18,19 After neomycin selection, ES cell clone with Fryl mutation (clone 8–10) was used to produce Fryl mutant mouse. The exact vector insertion site on the genome was determined by inverse PCR and nucleotide sequencing as described previously.18 PCR primers used for identification of mutant allele were: mutant specific forward primer, 5′-TGTCCTCCAGTCTCCTCCAC-3′; mutant specific reverse primer, 5′-CCAACCTCCAGCATCTTCAT-3′; wild specific forward primer, 5′-TGCGCTTGTTCTACC AACAG-3′; and wild specific reverse primer, 5′-TCGTCTT AGTGGGGGTTGTC-3′. PCR product sizes for wild and mutant alleles were 201 and 247 base pairs, respectively. Mutant ES cells were microinjected into blastocysts from C57BL/6J. These blastocysts were subsequently transferred into pseudo-pregnant mice to obtain chimeric mice. These chimeric male mice were mated with albino C57BL/6J females to establish germ line transmission of mutant allele. Germline transmitted mutant mice were then backcrossed with C57BL/6J for at least six generations before experiments. All animal experiments were conducted with the approval of the ethics committee of the Korea Research Institute of Bioscience and Biotechnology (Approval number: KRIBB-AEC-16027). All animals were bred and maintained in an SPF facility under a 12 h light-dark cycle (lights on at 7:00 and light off at 19:00) at 22 ± 0.5°C and humidity of 55 ± 15%. They were provided free access to food and water.
Western blot
Rabbit anti-Fryl polyclonal antibody was purchased from Atlas antibodies (catalog Number HPA031107, Stockholm, Sweden). Ten µg of tissue extract was subjected to 10% SDS-PAGE followed by protein transfer to PVDF membrane (Bio-Rad, Hercules, CA, USA). The membrane was then blocked with 5% skim milk in TBST (50 mM Tris·HCl, pH 7.4, 150 mM NaCl and 0.1% Tween20) and subsequently incubated with primary anti-Fryl antibody (dilution of 1: 500) and anti-GAPDH (catalog number #2118L, Cell Signaling, Danver, MA, USA, dilution of 1: 1,000) diluted with 1% skim milk in TBST. After incubation at 4°C overnight, the membrane was then incubated with secondary antibody (horseradish peroxidase-conjugated anti-rabbit IgG, catalog number #7074S, Cell Signaling, Danvers, MA, USA, 1: 1,000 dilution) at room temperature for 1 h. ECL Super Signal West Dura reagent (BioRad, Hercules, CA, USA) was used for signal detection. These signals were visualized with an Amersham imager 600 (GE Healthcare, Bio-sciences AB, Uppsala, Sweden).
Hematoxylin and eosin stain
Tissues from adult mice or embryos were fixed with 10% neutral buffered formalin (Sigma-Aldrich). Paraffin-embedded tissues were sectioned at 4 µm in thickness using a Microtome (Leica RM2245, Wetzlar, Germany) and subsequently stained with hematoxylin (ScyTek, Logan, UT, USA) and eosin (Sigma-Aldrich).
X-gal stain
X-gal staining was conducted for frozen tissues obtained from nine-weeks-old heterozygous Fryl mice. Fresh tissues were embedded with OCT compound (Tissue-Tek; Sakura Finite, Torrance, CA, USA), frozen, and sectioned at 10 µm in thickness using Cryostat (Leica, Nussloch, Germany). These sectioned tissues were fixed with 0.2% glutaraldehyde (Sigma-Aldrich) in Dulbecco’s Phosphate-Buffered Saline (DPBS; Sigma-Aldrich, St Louis, MO, USA) on ice for 10 min. After washing with DPBS twice, these tissues were rinsed with a solution (2 mM MgCl2, 0.02% igepal, and 0.01% sodium deoxycholate in 0.1 M phosphate buffer, pH 7.5) for 10 min. Tissue slides were then incubated with X-gal staining solution (5 mM K3Fe(CN)6, 5 mM K4Fe(CN)63H2O, 2 mM MgCl2, 0.02% NP40, 0.1% sodium deoxycholate, and 1 mg/ml X-gal in PBS) at 37°C overnight. They were then rinsed with DPBS and fixed in cold 4% paraformaldehyde solution (Affymetrix, Santa Clara, CA, USA) for 10 min. Fixed tissues were washed with DPBS once and distilled water twice (10 min each wash).
Immunohistochemistry for β-galactosidase detection
Neutral formalin-fixed and paraffin-embedded tissues from heterozygous mice were sectioned at 4 µm in thickness using a microtome (Leica RM2245). Rehydrated tissues were autoclaved in an antigen retrieval solution (1×, pH 6.0, DAKO, Carpinteria, CA, USA) at 121°C with high pressure for 30 min and then cooled for 1 h without releasing the pressure. After serial washing with distilled water and PBS, these tissues were incubated with antigen blocking solution (DAKO, Carpinteria, CA, USA) at room temperature for 15 min. They were then incubated with primary antibody against β-galactosidase (rabbit polyclonal, catalog number AHP1292, AbD serotec, Oxford, UK) diluted at 1:700 with antibody diluent solution (DAKO) at 4°C overnight. After rinsing with 1× PBS three times (5 min each), tissues were incubated with polymer-HRP anti-rabbit-labeled secondary antibody (DAKO) at room temperature for 40 min. A chromogen (DAKO) was used to reveal positive signals. Hematoxylin (ScyTek, Logan, UT, USA) was used for counterstaining.
TUNEL assay
Tissues from embryos on embryonic day 18.5 were fixed with formalin and embedded in paraffin. Tissue sections with 4 µm in thickness were rehydrated and used for TUNEL assay using an in situ apoptosis detection kit (Abcam, Cambridge, MA, USA; ab206386), according to the manufacturer’s instructions. In brief, these sections were permeabilized with proteinase K and quenched with 30% H2O2. Sections were then labeled with TdT Labeling Reaction Mixture at 37°C for 1.5 h. After terminating the labeling reaction with Stop Buffer, blocking was performed with blocking buffer. The conjugate was applied to detect the labeled TdT at room temperature for 30 min. After developing with DAB solution, counterstaining was conducted with methyl green to stain the nucleus on these sections. All reagents used were from the kit.
Semi-quantitative RT-PCR analysis
Tissues from C57BL/6J wild-type mice were snap frozen in liquid nitrogen. Total RNA was isolated using TRIzol reagent (Invitrogen, Carlsbad, CA, USA). The RNA concentration was determined with Eppendorf BioPhotometer (Eppendorf AG, Hamburg, Germany). cDNA synthesis was conducted with Superscript IV Reverse-transcriptase System (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions. The cDNA product was amplified by PCR using primers specific for Fryl and Fry genes. Primer sequences were as follows: for Fryl, forward primer 5′-GAACGTGCATATCATTGCGG-3′ and reverse primer 5′-TGATCAAGCTGATGACGCTC-3′; for Fry, forward primer 5′-AGGCCACGAACAAGCAAT AA-3′ and reverse primer 5′-ACTCCGATGACCTCAGCATA-3′. PCR product sizes for Fryl and Fry were 158 and 266 base pairs, respectively. Gapdh (forward primer 5′-AACAGCA ACTCCCACTCTTC-3′ and reverse primer 5′-CCTGTTGC TGTAGCCGTATT-3′) was used as an internal control. PCR was conducted using the following reaction conditions: 94°C for 5 min, followed by 27 cycles of 94°C for 1 min, 55°C for 2 min, and 72°C for 1 min, and a final extension at 72°C for 5 min. RT-PCR products were subjected to 1.5% agarose gel electrophoresis. Integrated density of bands was measured with a BIO-CAPT software (Vilber Lourmat, Marine la Valée, France).
Statistical analysis
Data are presented as means and standard deviations. All statistical differences between groups were determined by Student’s t-test using statistical program STATISTICA ver. 8.0 (Tulas, OK, USA). Statistical significance was considered at P < 0.05.
Results
Generation of fryl mutant mice
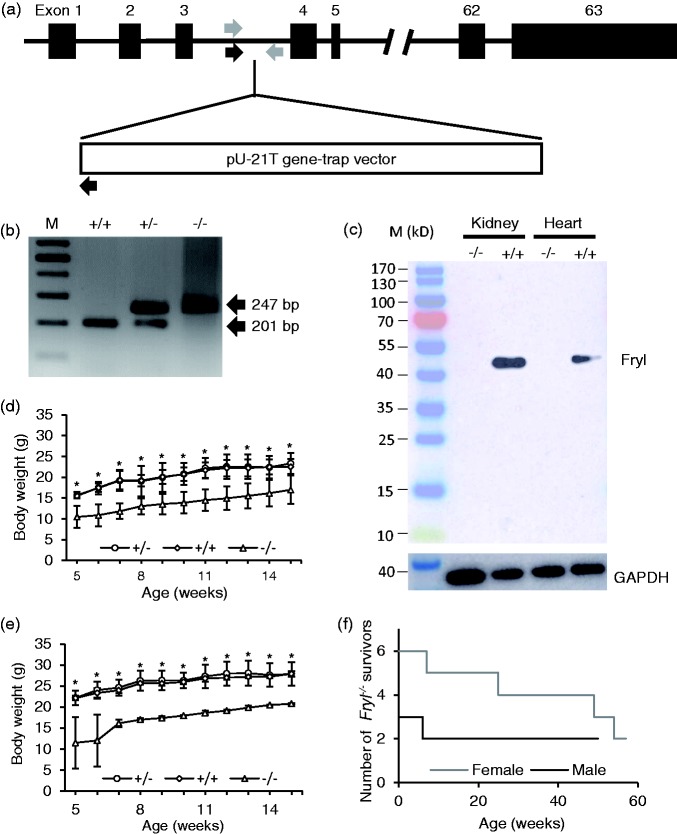
Insertion of pU-21T gene trap vector on intron 3 of Fryl gene was confirmed by inverse PCR and nucleotide sequencing (Figure 1(a)). PCR analysis using selected primer pairs could distinguish between mutant and wild type alleles (Figure 1(b)). Western blot analysis confirmed the absence of Fryl protein expressions in the kidney and heart from Fryl−/− mice, while Fryl protein expression was detected in these organs from Fryl+/+ littermates. There was only one band migrating at around 50 kDa (Figure 1(c)).
Figure 1.
Generation, growth, and survival of Fryl−/− mice. (a) Schematic presentation of truncated Fryl gene by pU-21T gene trap vector. Arrows indicate approximate positions for genotyping primers. Genotyping PCR primer sites for wild type (gray arrows) and mutant alleles (black arrows) were indicated. Exons were shown as black boxes with numbers. (b) Fryl mutant mouse genotyped based on PCR (201 and 247 base pairs for wild and mutant alleles, respectively). M: size marker. (c) Western blot analysis showing no Fryl expression in Fryl−/− mouse. GAPDH was used as internal control. +/+: wild type; +/−: heterozygous; −/−: homozygous. Body weights of Fryl−/− mice were plotted according to gender and genotypes. (d) Body weights of female Fryl+/+ (n = 4), Fryl+/− (n = 4) and Fryl−/− (n = 6) mice were plotted. (e) Body weights of male Fryl+/+ (n = 4), Fryl+/− (n = 4), and Fryl−/− (n = 3) mice were plotted. (f) Among them, one female and one male Fryl−/− mice died at seven and six weeks of age, respectively, as shown in the plot for the number of Fryl−/− survivors. Values are means ± SD. *P < 0.05. (A color version of this figure is available in the online journal.)
Survival and growth of fryl−/− mice
When a total 203 offspring derived from the cross between Fryl+/− parental mice were genotyped at age of two weeks, only nine (six females, three males) pubs (4.4% of the total offspring) were homozygous (Fryl−/−), significantly deviated from Mendelian distribution. This suggests embryonic or neonatal lethality of Fryl−/− mice. However, when embryos from 6 pregnant mice were analyzed on embryonic day 18.5, numbers of homozygous, heterozygous, and wild embryos were 18 (7 females, 11 males), 41 (14 females, 27 males), and 18 (11 females, 7 males), respectively (Table 1), indicating normal Mendelian ratio. All homozygous embryos observed in this study were normal in appearance.
Table 1.
Genotyping results obtained from intercrossing Fryl+/− mutant mice.
| Age | Sex |
Pup numbers (%) |
Total pup numbers | ||
|---|---|---|---|---|---|
| +/+ | +/− | −/− | |||
| E18.5 | Female | 7 (21.9) | 14 (43.8) | 11 (34.4) | 32 |
| Male | 11 (24.4) | 27 (60) | 7 (15.6) | 45 | |
| Total | 18 (23.4) | 41 (53.2) | 18 (23.4) | 77 | |
| Two weeks | Female | 43 (41) | 56 (53.3) | 6 (5.7) | 105 |
| Male | 32 (32.7) | 63 (64.3) | 3 (3.1) | 98 | |
| Total | 75 (36.9) | 119 (58.6) | 9 (4.4) | 203 | |
E18.5: Embryonic day 18.5.
Survived Fryl−/− offspring showed significantly lower body weight gains compared to wild or heterozygous littermates (Figure 1(d) and (e)). Four of six females and one of three males of Fryl−/− mice died at various ages as shown in Figure 1(f).
Nephropathy in adult Fryl−/− mice
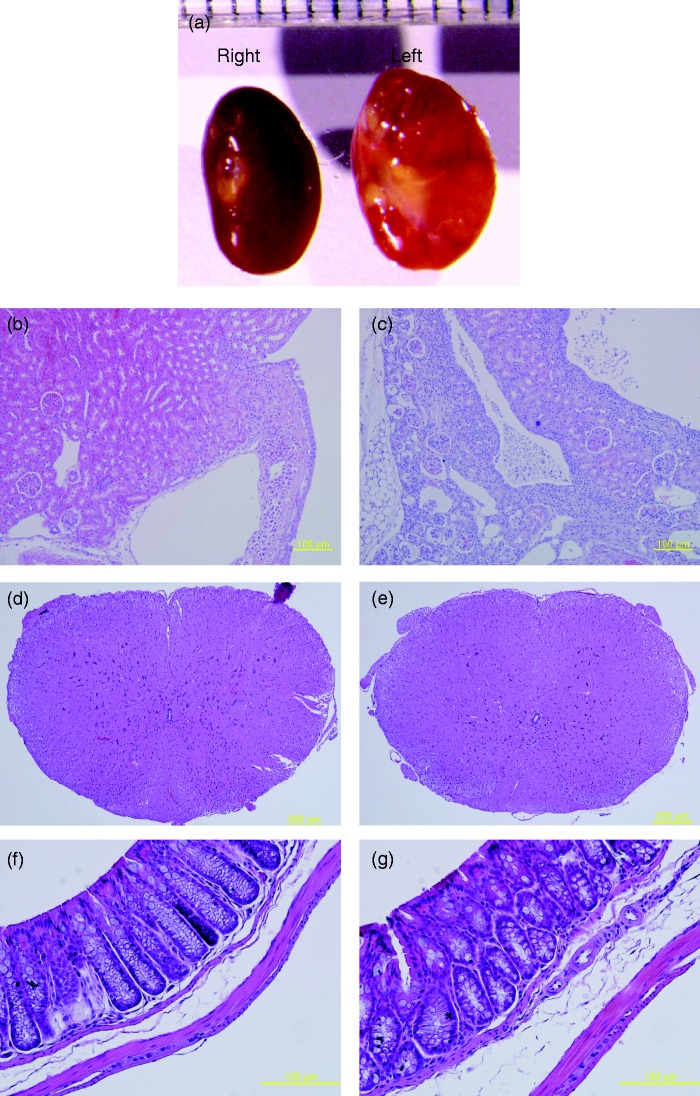
Necropsy for dead Fryl−/− mice revealed pale and bigger kidneys (Figure 2(a)). They were diagnosed as hydronephrosis. In histopathological observation of these hydronephrotic kidneys, chronic progressive nephropathies including fibrosis of the interstitium, mononuclear cell infiltration, and cyst formations within the cortex or medullar as well as pelvic dilation were observed (Figure 2(c)). Kidneys with normal looking appearance also showed chronic nephropathic changes, including cyst formations and eosinophilic infiltrations (Figure 2(b)). Although all four long survivors (two males and two females of Fryl−/− mice) did not show any abnormality in appearance or histopathologic observation including spinal cord and large intestine (Figure 2(d) to (g)), their kidneys also showed weak nephropathic changes such as leukocyte infiltrations and small cyst formation. However, Fryl+/+ or Fryl+/− littermates did not show such changes (data not shown).
Figure 2.
Hydronephrosis observed in Fryl−/− mouse. (a) Representative kidneys with hydronephrosis from one-year-old female mice are shown. (b and c) Histopathologic observation for right kidney (b) and left kidney (c). Normal looking kidneys also showed chronic progressive nephropathy as described in the text. There was no tissue with obvious pathologic changes other than kidney of Fryl−/− mouse. Spinal cord (d and e) and large intestine (f and g) were shown as representative tissues. (d) and (e): Fryl+/+; (f) and (g): Fryl−/−. Magnifications: 100× for (b)–(d); 400× for (f) and (g). (A color version of this figure is available in the online journal.)
Fryl gene expression in adult mouse tissues
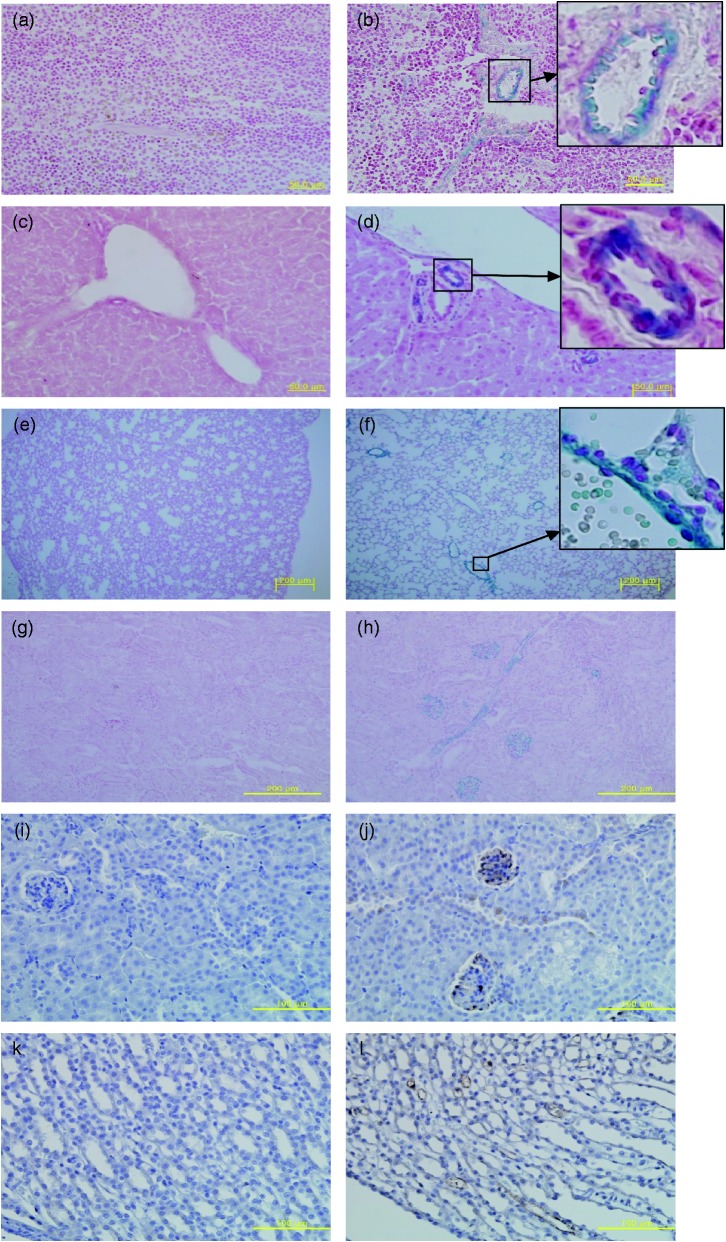
To determine if nephropathies observed in adult Fry−/− mice were associated with Fryl gene expression, in vivo expression of Fryl gene in Fry+/− mice was determined. X-gal staining and immunohistochemistry were conducted for tissues obtained from nine-weeks-old Fryl+/− mice. Based on X-gal staining, Fryl gene expression was clearly observed in tubular structures of several organs such as vascular walls in the spleen (Figure 3(a) and (b)), bile ducts around the portal tract in the liver (Figure 3(c) and (d)), vascular walls in the lung (Figure 3(e) and (f)), and cells constituting glomerulus as well as convoluted tubule in the kidney (Figure 3(g) and (h)). These stained signals were observed mainly on the intimal layer of the tubular walls, although most of the positivity appeared in vessels with thin walls. Fryl expression in glomerular cells, convoluted tubules, and/or collecting ducts in the kidneys from Fry+/− mice was also confirmed by immunohistochemistry staining using antibody against β-galactosidase (Figure 3(i) to (l)). Fryl gene expression was also observed in neuronal tissues such as brain, spinal cord, and retina with weak smear pattern (data not shown).
Figure 3.
In vivo Fryl gene expression in kidney and other tissues. Frozen mouse tissues were sectioned to 10 µm in thickness and stained with X-gal to detect Fryl gene expression. Nuclear Fast Red was used as counter stain. Tissues from wild (a, c, e, and g) and Fryl+/− (b, d, f, and h) mice were analyzed. Positive staining was found for tubular cells in the spleen (b), liver (d), lung (f), and kidney (h) of Fryl+/−mice. Fryl gene expression in kidneys from nine-week-old wild (i and k) and Fryl+/− (j and l) mice were analyzed by immunohistochemistry staining using anti-β galactosidase antibody. Hematoxylin was used for counterstaining. The glomerulus and collecting tubules showed strong positivity for these staining (j, l). Upper right inlets in b, d, and f show magnified photos of small squares. Magnifications: 400× for (a–d), (i), (j); 100× for e–h; 200× for (k) and (l). (A color version of this figure is available in the online journal.)
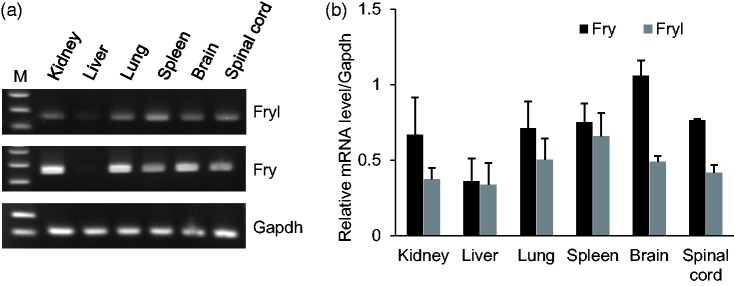
To see the possibility that Fry might compensate the Fryl deficiency in mouse tissues, we have compared the expression levels of Fry and Fryl in various mouse tissues including kidney. Fry expressions were dominant for Fryl in most of the tissues including kidney (Figure 4).
Figure 4.
Fryl and Fry expressions in wild type mouse tissues. (a) Fryl and Fry mRNA expressions in several adult mouse tissues analyzed by RT-PCR. Gapdh was used as a reference gene. (b) Relative mRNA expression levels of Fryl and Fry in mouse tissues depicted in a graph (n = 3).
Nephropathy in embryonic and newborn Fryl−/− mice
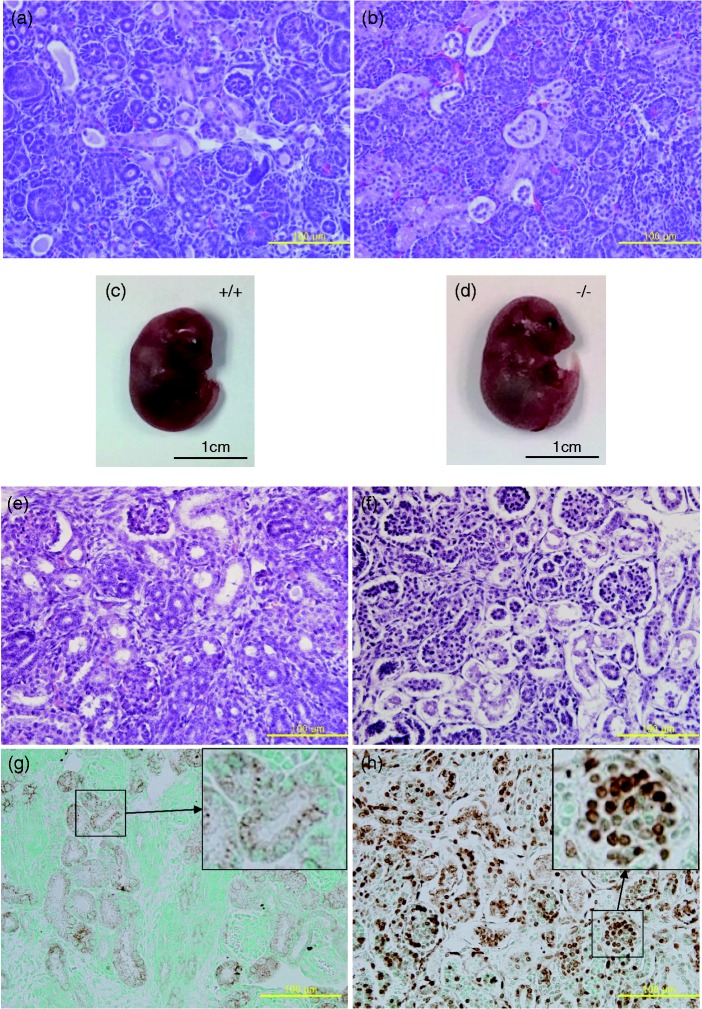
Fryl expression in glomerulus and tubules of the kidney and nephropathies observed in adult Fryl−/− mice led us to scrutinize abnormalities in kidneys from neonates and full-term embryos. Histopathological observation of kidneys from live Fry−/− newborns collected on the day of parturition revealed severe luminal expansion of collecting ducts, detachment of tubular wall lining cell layer from the basal layer, and the presence of materials filling the space between the detached cell layer and the basal layer (Figure 5(a) and (b)). However, these lesions were not found in Fryl+/+ or Fryl+/− littermates.
Figure 5.
Kidneys of Fryl−/− newborns and embryos. Kidney sections from wild (a) and Fryl−/− (b) live neonates were stained with hematoxylin and eosin. Detachment of tubular wall cell layer and materials filling the space were observed in live Fryl−/− neonates. Wild (c) and Fryl−/− (d) embryos aged 18.5 days showed normal appearance. However, almost all cells in kidneys from Fryl−/− embryos (f) had picnotic nuclei apart from their wild littermates (e). Cells with picnotic nuclei were apoptotic cells as demonstrated by TUNEL assay for wild (g) and Fryl−/− (h) embryos. Upper right inlets in G and H show magnified photos of small squares. Counterstain; methyl green. Magnification: 400× for (a), (b), (e), (f), (g), and (h). (A color version of this figure is available in the online journal.)
We also conducted histopathological observation for kidneys from embryos at embryonic day 18.5. All embryos showed normal looking appearance irrespective of their genotypes (Figure 5(c) and (d)). However, kidneys from Fryl−/− embryos showed much more severe histopathological lesions (Figure 5(f)) compared to those from live Fryl−/− neonates on the day of parturition (Figure 5(b)). Most cells constituting the renal parenchyma of Fryl−/− embryos had pycnotic nucleus. Detachment of tubular wall lining cell layer from the basal layer was also observed in their embryonic kidneys. These lesions were not observed in kidneys from Fryl+/− or Fryl+/+ embryos (Figure 5(e)). The space in the kidney from Fryl−/− embryos was almost empty. This was different from that from live Fryl−/− neonates (Figure 5(b) and (f)).
To determine if cells with pycnotic nucleus were apoptotic cells, we conducted TUNEL staining with sections from embryonic kidneys. Almost all cells in the kidney from Fryl−/− embryos showed TUNEL positivity with nuclear staining pattern (Figure 5(h)). Although some tubular cells in the kidney from Fryl+/+ embryos showed TUNEL positivity, most of these positive signals were cytoplasmic (Figure 5(g)).
Discussion
In this study, we produced Fryl gene mutant mice with gene trap vector inserted on intron 3, thus eliminating the transcription of downstream 60 exons. Fryl protein in vertebrates possesses conserved FND domain near its N-terminal region.10 The vector insertion site is located in front of the FND domain. This mutation is expected to produce a null mutation in the Fryl gene. This was supported by results of Western blot showing undetectable Fryl expression in mutant mouse tissues (Figure 1(c)).
In this study, we found only one Fryl band in Western blot. The molecular weight was approximately 50 kDa which was consistent with the information from the antibody manufacturer. Recently, Rippe et al.15 reported a major Fryl band with molecular weight just above 150 kDa in mice. The difference of the molecular weight may be the reflection of difference of the epitops recognized by antibodies.
At present, there are only a few reports indicating that Fryl is associated with transcriptional activator. Hayette et al.1 found that the C-terminal region of Fryl gene could be a fusion partner with MLL in human leukemia cells by translocation. Such fusion protein is a potential transcriptional activator through the leucine zipper motif in the fused C-terminal domain of Fryl.
Although biological functions of Fryl protein remain mostly unknown, its paralog Fry in vertebrate has been reported to have various cellular functions. Because human Fry and Fryl share specific structural characteristics,10 Fryl may have similar functions as Fry. It has been reported that Fry proteins in most invertebrate model organisms can interact with NDR serine/threonine kinases and these model organisms with mutant Fry gene show phenotypes similar to those of NDR mutants.10,20 However, vertebrate Fry protein is known to have NDR kinase-associated and non-associated roles.10
FRY and FRYL in human share 60% identities at amino acid level (74% with conservative substitution).1 In this study, Fryl−/− mice showed significantly reduced survival in the perinatal period, indicating that Fryl may have functions that do not overlap with Fry in mice. On the other hand, a few Fryl−/− neonates survived for several months even though they showed clear growth retardation and nephropathies. Although the mechanism involved in their survival is currently unclear, Fry may have compensated for defects incurred by Fryl deficiency during the survival period of Fryl−/− mice. Fry expression was superior to Fryl expression in various mouse tissues including kidney. It may be possible that Fry compensated the Fryl deficiency in Fryl−/− survivors. Fryl protein expression level in the kidney was stronger than that in the heart and other tissues including liver, brain, spinal code (data not shown). This may indicate that the kidney was the major organ affected by Fryl deficiency. Further detailed studies on the function of Fryl could enable us clarify the possibility.
Although Fryl expression was observed in vascular or tubular structures of various tissues including lung, spleen, liver, and kidney, kidney was the only organ in which clear cellular or structural changes were detected, suggesting specific roles of Fryl in renal development. If Fryl has indispensable roles in the development of kidney, Fryl deficient newborn mice may not eliminate harmful metabolites that can be effectively removed from fetus through fetal–placental exchange system before parturition. This may be the cause of neonatal death of Fryl deficient mice after birth. This may also explain the growth retardation, death, and chronic hydronephrosis observed in Fryl−/− survivors, although we have no direct evidence for defects of kidney functions in Fryl−/− mice.
Although how Fryl affected apoptosis observed in kidneys of Fryl−/− embryos is currently unknown, the mechanism may be associated with NDR kinase pathway. Fryl paralog Fry can interact with NDR kinases.12 NDR1 is upregulated in human progressive ductal carcinoma.21 NDR kinases are central players within the conserved Hippo signal pathway.22,23 Functionally, NDR kinase pathway is involved in cell proliferation, apoptosis, and to lesser extent, cell migration.22,23 Thus, it is possible that Fryl deficiency can induce a defect in NDR kinase activity as in Fry deficiency, thus affecting cell proliferation and leading to apoptotic fate of ductal cells in kidneys of Fryl−/− embryos. On the other hand, other studies have suggested that NDR1 and NDR2 can mediate apoptosis in association with tumor suppressor of MST kinases.24,25 In addition, Fry can interact with NDR kinase and act as a scaffold in the assembly.26 Thus, altered NDR kinase activities in Fryl−/− mice may have association with apoptosis found in the developing kidney of this study.
Apparent detachment of tubular lining cell layer from the basal layer was observed in kidneys of Fryl−/− mice. Although Fryl function related to this has not been reported, Fry in vertebrates has NDR kinase-unrelated functions besides NDR kinase-related functions.10 Fry can bind directly to microtubules via multiple microtubule binding sites12 and promote acetylation of microtubules by binding to and suppressing tubulin deacetylase Sirtuin 2 (Sirt2).10,13 Sirt1 and Sirt3 are associated with protection against renal disease. Such protective effects of Sirtuins are associated with regulation of acetylation status of microtubules.27–29 In addition, phosphorylated YAP in Hippo pathway is associated with cytoplasmic retention in cell–cell tight junction and adhesion junction.22,30 YAP also regulates genes that control cell signaling and cell structure during the development of kidney, thereby affecting kidney development.23,30 YAP is also closely related to genes that promote cell proliferation and inhibition of cell death.23,30 These results support the hypothesis that Fryl, similar to Fry, may disturb the acetylation status of microtubules through Sirtuins and/or Hippo pathway in the kidney, thus leading to tubular wall cell-layer detachment as observed in the kidney of Fryl−/−mouse in this study.
During the preparation of this manuscript, Yatim et al.16 have reported that Fryl functions as a transcriptional coactivator for Notch in human T cell acute lymphoblastic leukemia (T-ALL) cell lines. Fryl acts as a Notch coactivator. It plays a role in RNA polymerase II assembly subsequent to recruitment of intracellular domain of Notch1 at several Notch-responsive genes. Nuclear responses downstream of Notch activation are tightly regulated and diverse.31 Notch signaling can be arbiter of survival versus death, proliferation versus growth arrest, or differentiation versus stemness depending on the cellular context.32 Therefore, apoptosis observed in the kidney of Fryl−/− embryos may have association with disturbance of Notch signaling. Most recently, Fryl is also shown to be associated with hypertension. Gucy1a3 and Gucy1b3 genes as subunits of soluble guanylyl cyclase (sGC) are Notch-target genes that require Fryl as transcriptional coactivator. sGC is the major nitric oxide (NO) receptor in the vascular wall.33 Whether Fryl deficiency may have association with hypertension due to reduced sGC expression and subsequent NO-mediated vasodilatation merits further study. Whether hypertension may have association with renal abnormalities observed in this study also merits further study.
In summary, we demonstrated that Fryl deficiency was associated with defective kidney development and function in mice, resulting in lethality to most neonates. Rare Fryl−/− survivors also suffered from chronic nephropathy. Therefore, Fryl may be a valuable therapeutic target to develop treatment for nephropathies in human.
Authors’ contributions
All authors participated in the design, interpretation of the studies and analysis of the data and review of the manuscript: YSB, EKK, KL, and WKY conducted the experiments and wrote the manuscript, KA, KY, YSW and HCK reviewed the manuscript, KCC and KHN designed and interpreted the data and review the manuscript.
Declaration of Conflicting Interests
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding
This study was supported by a grant from the KRIBB Research Initiative Program and the Korea Mouse Phenotyping Project (2013M3A9D5072559) of the Korean Ministry of Science, ICT and Future Planning through the National Research Foundation.
References
- 1.Hayette S, Cornillet-Lefebvre P, Tigaud I, Struski S, Forissier S, Berchet A, Doll D, Gillot L, Brahim W, Delabesse E, Magaud JP, Rimokh R. AF4p12, a human homologue to the furry gene of Drosophila, as a novel MLL fusion partner. Cancer Res 2005; 65:6521–5 [DOI] [PubMed] [Google Scholar]
- 2.Goto T, Fukui A, Shibuya H, Keller R, Asashima M. Xenopus furry contributes to release of microRNA gene silencing. Proc Natl Acad Sci U S A 2010; 107:19344–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.He B, Adler PN. The genetic control of arista lateral morphogenesis in Drosophila. Dev Genes Evol 2002; 212:218–29 [DOI] [PubMed] [Google Scholar]
- 4.Hirata D, Kishimoto N, Suda M, Sogabe Y, Nakagawa S, Yoshida Y, Sakai K, Mizunuma M, Miyakawa T, Ishiguro J, Toda T. Fission yeast Mor2/Cps12, a protein similar to Drosophila Furry, is essential for cell morphogenesis and its mutation induces Wee1-dependent G(2) delay. EMBO J 2002; 21:4863–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kanai M, Kume K, Miyahara K, Sakai K, Nakamura K, Leonhard K, Wiley DJ, Verde F, Toda T, Hirata D. Fission yeast MO25 protein is localized at SPB and septum and is essential for cell morphogenesis. EMBO J 2005; 24:3012–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cong J, Geng W, He B, Liu J, Charlton J, Adler PN. The furry gene of Drosophila is important for maintaining the integrity of cellular extensions during morphogenesis. Development 2001; 128:2793–802 [DOI] [PubMed] [Google Scholar]
- 7.Emoto K, He Y, Ye B, Grueber WB, Adler PN, Jan LY, Jan YN. Control of dendritic branching and tiling by the Tricornered-kinase/Furry signaling pathway in Drosophila sensory neurons. Cell 2004; 119:245–56 [DOI] [PubMed] [Google Scholar]
- 8.Fang X, Adler PN. Regulation of cell shape, wing hair initiation and the actin cytoskeleton by Trc/Fry and Wts/Mats complexes. Dev Biol 2010; 341:360–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fang X, Lu Q, Emoto K, Adler PN. The Drosophila Fry protein interacts with Trc and is highly mobile in vivo. BMC Dev Biol 2010; 10:40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nagai T, Mizuno K. Multifaceted roles of Furry proteins in invertebrates and vertebrates. J Biochem 2014; 155:137–46 [DOI] [PubMed] [Google Scholar]
- 11.Gallegos ME, Bargmann CI. Mechanosensory neurite termination and tiling depend on SAX-2 and the SAX-1 kinase. Neuron 2004; 44:239–49 [DOI] [PubMed] [Google Scholar]
- 12.Chiba S, Ikeda M, Katsunuma K, Ohashi K, Mizuno K. MST2- and Furry-mediated activation of NDR1 kinase is critical for precise alignment of mitotic chromosomes. Curr Biol 2009; 19:675–81 [DOI] [PubMed] [Google Scholar]
- 13.Nagai T, Ikeda M, Chiba S, Kanno S, Mizuno K. Furry promotes acetylation of microtubules in the mitotic spindle by inhibition of SIRT2 tubulin deacetylase. J Cell Sci 2013; 126:4369–80 [DOI] [PubMed] [Google Scholar]
- 14.Du LL, Novick P. Pag1p, a novel protein associated with protein kinase Cbk1p, is required for cell morphogenesis and proliferation in Saccharomyces cerevisiae. Mol Biol Cell 2002; 13:503–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rippe C, Zhu B, Krawczyk KK, Bavel EV, Albinsson S, Sjolund J, Bakker E, Sward K. Hypertension reduces soluble guanylyl cyclase expression in the mouse aorta via the Notch signaling pathway. Sci Rep 2017; 7:1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yatim A, Benne C, Sobhian B, Laurent-Chabalier S, Deas O, Judde JG, Lelievre JD, Levy Y, Benkirane M. NOTCH1 nuclear interactome reveals key regulators of its transcriptional activity and oncogenic function. Mol Cell 2012; 48:445–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Artavanis-Tsakonas S, Rand MD, Lake RJ. Notch signaling: cell fate control and signal integration in development. Science 1999; 284:770–6 [DOI] [PubMed] [Google Scholar]
- 18.Nakahara M, Tateyama H, Araki M, Nakagata N, Yamamura K, Araki K. Gene-trap mutagenesis using Mol/MSM-1 embryonic stem cells from MSM/Ms mice. Mammal Genome 2013; 24:228–39 [DOI] [PubMed] [Google Scholar]
- 19.Taniwaki T, Haruna K, Nakamura H, Sekimoto T, Oike Y, Imaizumi T, Saito F, Muta M, Soejima Y, Utoh A, Nakagata N, Araki M, Yamamura K, Araki K. Characterization of an exchangeable gene trap using pU-17 carrying a stop codon-beta geo cassette. Dev Growth Differ 2005; 47:163–72 [DOI] [PubMed] [Google Scholar]
- 20.Cornils H, Stegert MR, Hergovich A, Hynx D, Schmitz D, Dirnhofer S, Hemmings BA. Ablation of the kinase NDR1 predisposes mice to the development of T cell lymphoma. Sci Signal 2010; 3:ra47. [DOI] [PubMed] [Google Scholar]
- 21.Millward TA, Heizmann CW, Schafer BW, Hemmings BA. Calcium regulation of Ndr protein kinase mediated by S100 calcium-binding proteins. EMBO J 1998; 17:5913–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hergovich A, Stegert MR, Schmitz D, Hemmings BA. NDR kinases regulate essential cell processes from yeast to humans. Nat Rev Mol Cell Biol 2006; 7:253–64 [DOI] [PubMed] [Google Scholar]
- 23.Pan D. The hippo signaling pathway in development and cancer. Dev Cell 2010; 19:491–505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hergovich A, Kohler RS, Schmitz D, Vichalkovski A, Cornils H, Hemmings BA. The MST1 and hMOB1 tumor suppressors control human centrosome duplication by regulating NDR kinase phosphorylation. Curr Biol 2009; 19:1692–702 [DOI] [PubMed] [Google Scholar]
- 25.Vichalkovski A, Gresko E, Cornils H, Hergovich A, Schmitz D, Hemmings BANDR. kinase is activated by RASSF1A/MST1 in response to Fas receptor stimulation and promotes apoptosis. Curr Biol 2008; 18:1889–95 [DOI] [PubMed] [Google Scholar]
- 26.Emoto K. The growing role of the Hippo–NDR kinase signalling in neuronal development and disease. J Biochem 2011; 150:133–41 [DOI] [PubMed] [Google Scholar]
- 27.North BJ, Marshall BL, Borra MT, Denu JM, Verdin E. The human Sir2 ortholog, SIRT2, is an NAD+-dependent tubulin deacetylase. Mol Cell 2003; 11:437–44 [DOI] [PubMed] [Google Scholar]
- 28.He W, Wang Y, Zhang MZ, You L, Davis LS, Fan H, Yang HC, Fogo AB, Zent R, Harris RC, Breyer MD, Hao CM. Sirt1 activation protects the mouse renal medulla from oxidative injury. J Clin Invest 2010; 120:1056–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kitada M, Kume S, Takeda-Watanabe A, Kanasaki K, Koya D. Sirtuins and renal diseases: relationship with aging and diabetic nephropathy. Clin Sci 2013; 124:153–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reginensi A, Scott RP, Gregorieff A, Bagherie-Lachidan M, Chung C, Lim DS, Pawson T, Wrana J, McNeill H. Yap- and Cdc42-dependent nephrogenesis and morphogenesis during mouse kidney development. PLoS Genet 2013; 9:e1003380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Andersson ER, Sandberg R, Lendahl U. Notch signaling: simplicity in design, versatility in function. Development 2011; 138:3593–612 [DOI] [PubMed] [Google Scholar]
- 32.South AP, Cho RJ, Aster JC. The double-edged sword of Notch signaling in cancer. Sem Cell Develop Biol 2012; 23:458–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ruetten H, Zabel U, Linz W, Schmidt HH. Downregulation of soluble guanylyl cyclase in young and aging spontaneously hypertensive rats. Circ Res 1999; 85:534–41 [DOI] [PubMed] [Google Scholar]