Abstract
Objectives:
To determine the applicability of a computer-aided diagnostic system strain elastography system for the classification of breast masses diagnosed by ultrasound and scored using the criteria proposed by the breast imaging and reporting data system ultrasound lexicon and to determine the diagnostic accuracy and interobserver variability.
Methods:
This prospective study was conducted between March 1, 2016, and May 30, 2016. A total of 83 breast masses subjected to percutaneous biopsy were included. Ultrasound elastography images before biopsy were interpreted by 3 radiologists with and without the aid of computer-aided diagnostic system for strain elastography. The parameters evaluated by each radiologist results were sensitivity, specificity, and diagnostic accuracy, with and without computer-aided diagnostic system for strain elastography. Interobserver variability was assessed using a weighted κ test and an intraclass correlation coefficient. The areas under the receiver operating characteristic curves were also calculated.
Results:
The areas under the receiver operating characteristic curve were 0.835, 0.801, and 0.765 for readers 1, 2, and 3, respectively, without computer-aided diagnostic system for strain elastography, and 0.900, 0.926, and 0.868, respectively, with computer-aided diagnostic system for strain elastography. The intraclass correlation coefficient between the 3 readers was 0.6713 without computer-aided diagnostic system for strain elastography and 0.811 with computer-aided diagnostic system for strain elastography.
Conclusion:
The proposed computer-aided diagnostic system for strain elastography system has the potential to improve the diagnostic performance of radiologists in breast examination using ultrasound associated with elastography.
Keywords: ultrasound, elastography, breast tumors, diagnosis and examinations, mass screening
Introduction
Computer-aided diagnosis (CAD) systems have been developed for all types of diagnostic imaging tests. These systems aim to improve the capacity of interpretation of medical imaging by radiologists and help differentiate benign from malignant lesions. They are divided into 2 categories: computer-aided diagnosis system (CADx) and computer-aided detection. The first is designed to diagnose and differentiate benign from malignant lesions, and the second is designed to detect and locate abnormal areas in images.1
Breast cancer is the most common type of cancer in women worldwide and the only cancer with an established screening program that has been adopted on all continents.2 The diagnostic method adopted worldwide for breast cancer screening is mammography, and the benefits of implementing this program are already established. The implementation of mammography has improved the early diagnosis of cancer and decreased morbidity and mortality from the disease.3–5 However, the sensitivity of mammography is poor for dense breasts that are classified into high-density patterns, C (heterogeneously dense) and D (dense), using the breast imaging and reporting data system (BI-RADS) lexicon. Therefore, this technique may fail to detect approximately 10% to 30% of cancers.6,7 For these density patterns, some American states have adopted legislation that recommends the execution of complementary screening tests for cancer using ultrasound (US).8
Therefore, US is not a first-line screening method for breast cancer but a complementary method for patients with breast density patterns C and D.6 Furthermore, US is operator-dependent, and its results are difficult to reproduce and interpret.8,9 Because of its low specificity, additional tools have been developed to improve its diagnostic accuracy. Among these tools, elastography is the most promising to date.10
Ultrasound with elastography has been a promising tool in the diagnosis of breast lesions since its introduction in the fifth edition of the BI-RADS lexicon.11 This method allows the evaluation of the stiffness of the region of interest (ROI) and assumes that malignant lesions are harder. At present, elastography is divided into strain elastography (SE), which involves manual compressions to promote deformity in the tissue evaluated, with the degree of deformity represented by colors, and shear-wave elastography (SWE), in which the device produces vibrational energy to deform the area of interest with US waves. Studies have shown that the 2 methods improve the performance of US for the diagnosis of malignant lesions. Moreover, both approaches allow the quantitative and qualitative analysis of lesions. However, quantitative analysis is performed directly in SWE, whereas the main feature of SE is the qualitative assessment of the lesion. The main limitations of the methods are the lack of standardization, low interobserver agreement, and difficulties in interpreting the results.12
The aim of this study was to evaluate the feasibility of classifying breast masses combining the results from a CADx system for analyzing SE (CADxSE) with the results from B-Mode US and scored using the criteria proposed by the BI-RADS ultrasound lexicon (USdx). For this purpose, the classification of the lesions using USdx was compared to that of USdx combined with CADxSE software (USdx + CADxSE), and the diagnostic accuracy and interobserver variability of the method were determined.
Materials and Methods
The system used to classify the SE images was developed jointly by the University of Pittsburgh, the University of São Paulo at São Carlos, and the Brazilian Institute for Cancer Control (Instituto Brasileiro de Controle do Câncer [IBCC]). Fleury is the software patent holder, registered at the National Institute of Industrial Property (Instituto Nacional da Propriedade Industrial), Brazil, under Patent No. 709.209-1.371-81.
Patients
This prospective study was approved by the research ethics committee of the IBCC (Protocol No. 012664/2016) and was registered in the Plataforma Brazil (Protocol No. 53543016.2.0000.0072). An informed consent form was signed by all participants. The inclusion criteria consisted of the consecutive evaluation of breast masses in patients subjected to percutaneous biopsy and referred to the breast intervention service at IBCC. Nonmass lesions on US were excluded. In the period between March 2016 and May 2016, 90 consecutive biopsies of breast lesions were conducted in 87 patients. Five patients with 7 lesions were excluded, of which 2 lesions from 1 patient were classified as simple cysts on US, and 4 patients presented 5 nonmass lesions on US in the form of architectural distortion and calcification. The biopsies were performed only in patients without clinical contraindication after the image acquisition and followed the protocol of our service.
Ultrasound and Elastography Examinations
Ultrasound and elastography examinations were performed by the same radiologist with 3 years of experience using elastography. The device used was a Toshiba Aplio 300 (Toshiba, Tokyo, Japan), and the data were analyzed using the commercial software for SE available on the device. Before the biopsy, elastography images were acquired using the technique proposed in previous studies.13 The images that best represented the lesion using conventional US and SE were chosen. The selected images were stored in the picture archiving and communication system (PACS) consecutively without any identification of the nature of the lesions.
Interpretation of the US Images
The US images were interpreted blindly and independently by 3 radiologists, the first with 16 years of experience in breast imaging (reader 1), the second with 8 years of experience (reader 2), and the third with 2 years of experience (reader 3). The lesions were analyzed using the criteria proposed by the BI-RADS USdx, and the parameters evaluated were shape, margins, echotexture, posterior acoustic features, and the relationship to adjacent tissues. These lesions were classified into categories 2, 3, 4 (4a, 4b, and 4c), and 5. The classification was recorded automatically in an Excel spreadsheet (Microsoft, Redmond, Washington) developed specifically for the study. The physician who acquired the images did not participate in image interpretation.
Computer-Aided Diagnosis System for SE Analysis (CADxSE)
The operation of the CADxSE system used in the classification of the lesions with elastography was described in a previous study.14 It consisted of color stratification of the elastography images acquired in the area of interest, which allowed the automatic classification of breast masses considering the percentage of the color spectrum of the hard tissues in the lesion area. The classification involved 4 steps: (1) image load, (2) ROI selection, (3) mass delineation/outlining, and (4) classification (Supplemental Material).
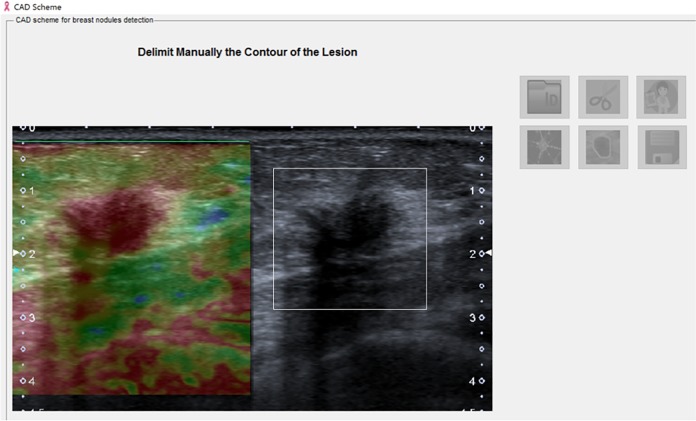
Image load processing: Using the CADxSE software, each reader selected the image of interest from a PACS database. The breast masses were displayed in the split screen, in which one side of the screen showed the US image and the other screen showed the SE image (Figure 1).
Image segmentation: Using a digital pen, the reader selected the area that contained the mass in the US image (Figure 1).
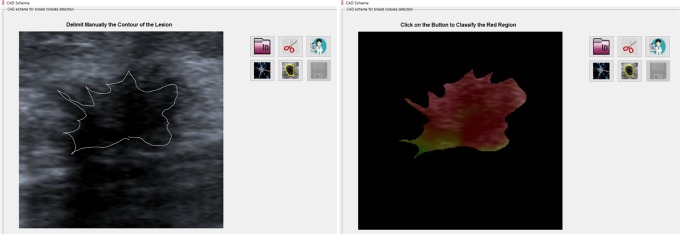
Extraction and selection: After image selection on the screen, the reader outlined the mass using a digital pen following the margins of the lesion (Figure 2).
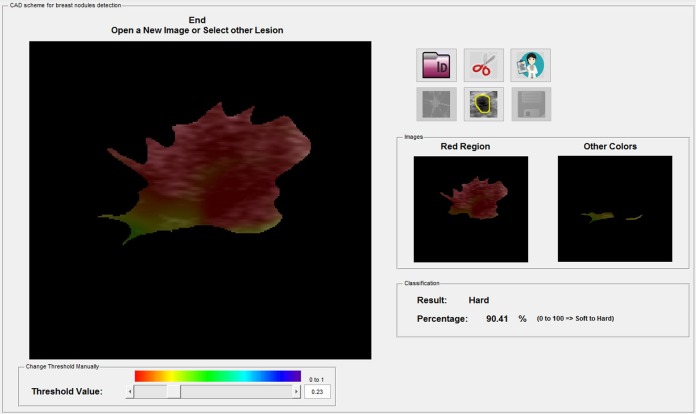
Classification: After image processing, the lesions were classified automatically by the software as soft, intermediate, or hard, as proposed by the fifth edition of the BI-RADS lexicon and in previous studies.15 Two additional images were generated, one showing the color spectrum of the hard tissues and the other showing the color spectrum of the remaining tissue. As proposed in a previous study, soft, intermediate, and hard lesions were considered those that contained less than 50%, 50% to 75% and more than 75% of the hard area inside, respectively14 (Table 1). The software did not allow interference from the readers during this process. The results were automatically transferred to the Excel spreadsheet, where the authors classified the US images. The time taken from image selection to CADxSE final mass classification was on average 25 seconds.
Figure 1.
Computer-aided diagnostic system for strain elastography (CADxSE) system image of the breast mass to be evaluated on the split screen: the left image shows the elastography result, and the right image is in B-mode. The area of interest is selected in B-mode using a digital pen.
Figure 2.
The right image shows the manual segmentation of the margin of the breast mass in B-mode. The left image shows the interposition of elastography with the B-mode image, and the color spectra results are shown within the selected area.
Table 1.
Final Assessment of Breast Masses After the Association of Elastographic Findings (Soft, Intermediate, or Hard) to the BI-RADS.
| Elastography US | Elastography Soft | Elastography Intermediate | Elastography Hard |
|---|---|---|---|
| BI-RADS 3 | BI-RADS 2 | BI-RADS 3 | BI-RADS 4a |
| BI-RADS 4a | BI-RADS 3 | BI-RADS 4a | BI-RADS 4b |
| BI-RADS 4b | BI-RADS 4a | BI-RADS 4b | BI-RADS 4c |
| BI-RADS 4c | BI-RADS 4b | BI-RADS 4c | BI-RADS 5 |
| BI-RADS 5 | BI-RADS 4c | BI-RADS 5 | BI-RADS 5 |
Abbreviation: BI-RADS, breast imaging and reporting data system.
Integration of CADxSE Results With the USdx
The results were integrated as proposed in a previous study (Figure 3) to automatically obtain the BI-RADS classification using USdx + CADxSE. The lesions were classified into categories 2, 3, 4 (4a, 4b, and 4c), and 5.15
Figure 3.
Automatic final analysis by the software (in percentage) and classification of the breast masses as soft, intermediate, or hard. The 2 boxes isolate the color of the hard areas from the color of the other areas.
Statistical Analysis
Statistical analysis was facilitated by including the lesions classified as BI-RADS 4a, 4b, and 4c in the same group (category 4, suspicious findings). The results classified in BI-RADS categories 4 and 5 were considered positive, and the results classified in categories 2 and 3 were considered negative as proposed by BI-RADS. The results of the percutaneous biopsy were used as a reference to determine the nature of the lesions, which were divided into benign and malignant lesions.
The parameters determined were sensitivity, specificity, positive and negative predictive values, and diagnostic accuracy of the final classifications obtained with USdx alone and USdx + CADxSE. The receiver operating characteristic (ROC) curves of the 2 classifications were obtained for each reader, and the diagnostic accuracy was compared using a significance level of 5%.
The performances of the ROC curve of USdx alone and USdx + CADxSE for each reader were compared using a significance level of 5%. The pairwise interobserver agreement of the classifications using USdx alone and US dx + CADxSE was determined using the κ index. The joint agreement between the 3 readers was determined using the intraclass correlation coefficient (ICC). The agreements obtained using the κ index and the ICC were classified as poor (0.0-0.2), small (0.2-0.4), moderate (0.4-0.6), strong (0.6-0.8), and almost perfect (0.8-1.0). All tests were conducted using MedCalc software (MedCalc Software, Ostend, Belgium).
Results
The mean age of the patients submitted to the study was 46.5 years, ranging from 26 to 73 years (SD 8.6, median of 46 years). Positive results for carcinoma were found in 6 (19.3%) patients younger than 40 years, in 11 (35.5%) patients between 40 and 50 years, and in 14 (45.2%) patients older than 50 years.
Regarding the fibroglandular background pattern to US of the 83 patients included in the study, 36 patients with 19 (61.3%) positive biopsies showed homogeneous fat, 20 patients with 2 (6.4%) positive biopsies showed homogeneous fibroglandular patterns, and 27 patients had 10 (32.3%) positive biopsies with heterogeneous patterns.
Table 2 shows the results of the final classifications using USdx and USdx + CADxSE for each reader. The sensitivity, specificity, positive and negative predictive values, and diagnostic accuracy of USdx and USdx + CADxSE are shown in Table 2.
Table 2.
Distribution of the Final Classification According to the BI-RADS Lexicon of Breast Masses Using Ultrasound (USdx) and Ultrasound Combined With Elastography (USdx + CADxSE) and According to the Histological Diagnosis (Benign [−] or Malignant [+]) of Each Reader.a
| BI-RADSTM | Reader 1 | Reader 2 | Reader 3 | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| USdx | USdx + CADxSE | USdx | USdx + CADxSE | USdx | + USdx + CADxSE | |||||||||||||
| (−) | (+) | Tt | (−) | (+) | Tt | (−) | (−) | Tt | (−) | (+) | Tt | (−) | (+) | Tt | (−) | (+) | Tt | |
| 2 | 3 | - | 3 | 20 | - | 20 | 2 | 1 | 3 | 19 | - | 19 | - | - | - | 18 | - | 18 |
| 3 | 27 | 1 | 28 | 11 | - | 11 | 29 | 2 | 31 | 14 | - | 14 | 30 | 4 | 34 | 11 | 1 | 12 |
| 4A | 4 | 1 | 5 | 7 | 2 | 9 | 8 | 3 | 11 | 6 | 4 | 10 | 12 | 6 | 18 | 10 | 3 | 13 |
| 4B | 12 | 5 | 17 | 8 | 2 | 10 | 10 | 4 | 14 | 9 | 3 | 12 | 8 | 4 | 12 | 6 | 6 | 12 |
| 4C | 6 | 15 | 21 | 3 | 7 | 10 | 3 | 12 | 15 | 3 | 4 | 7 | 2 | 11 | 13 | 7 | 7 | 14 |
| 5 | - | 9 | 9 | 3 | 20 | 23 | - | 9 | 9 | 1 | 20 | 21 | - | 6 | 6 | - | 14 | 14 |
| Total | 83 | 83 | 83 | 83 | 83 | 83 | ||||||||||||
| Sensitivity | 96.8% | 100% | 90.3% | 100% | 87.1% | 96.8% | ||||||||||||
| Specificity | 57.7% | 59.6% | 59.6% | 63.5% | 57.7% | 55.8% | ||||||||||||
| Positive Predictive Values | 57.7% | 59.6% | 57.1% | 62.0% | 55.1% | 56.6% | ||||||||||||
| Negative Predictive Values | 96.8% | 100% | 91.2% | 100% | 88.2% | 100% | ||||||||||||
| Diagnostic Accuracy | 72.3% | 74.7% | 71.1% | 77.1% | 68.7% | 71.1% | ||||||||||||
Abbreviations: BI-RADS, breast imaging and reporting data system; CADxSE, computer-aided diagnostic system for strain elastography; USdx, ultrasound lexicon.
aThe total number of lesions is represented by Tt. The sensitivity, specificity, positive and negative predictive values, and diagnostic accuracy of each reader for the final classification according to the BI-RADS lexicon for USdx and USdx + CADxSE.
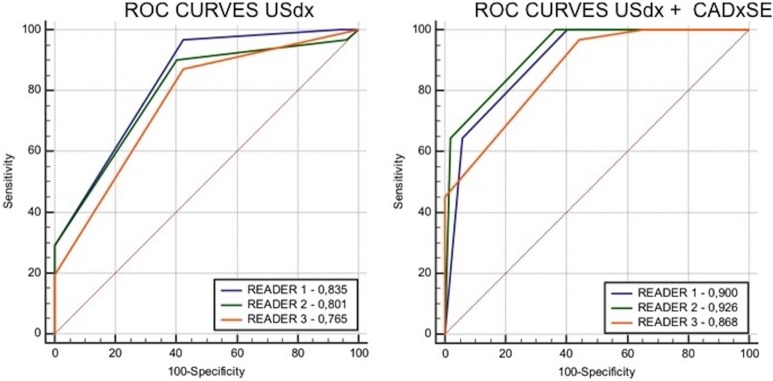
The areas under the ROC curve for USdx and USdx + CADxSE classifications for each reader are shown in Table 3. Comparison of these areas indicated significant (P < .05) improvements in the performance of USdx + CADxSE for all readers.
Table 3.
Area Under the ROC Curve for Each Reader According to the Classification Proposed by the BI-RADS Lexicon for USdx and USdx + CADxSE.a
| Interclass correlation | AUC | SEb | 95% CIc | DBA | SEb | 95% CI | Significanced | |
|---|---|---|---|---|---|---|---|---|
| Reader 1 | USdx | 0.835 | 0.0348 | (0.737-0.907) | 0.0648 | 0.0302 | (0.00557-0.124) | P = .0320 |
| + CADxSE | 0.900 | 0.0291 | (0.814-0.955) | |||||
| Reader 2 | USdx | 0.801 | 0.0458 | (0.698-0880) | 0.125 | 0.0378 | (0.0509-0.199) | P = .0010 |
| + CADxSE | 0.926 | 0.0231 | (0.847-0.972) | |||||
| Reader 3 | USdx | 0.765 | 0.0461 | (0.659-0.851) | 0.103 | 0.0357 | (0.0332-0.173) | P = .0039 |
| + CADxSE | 0.868 | 0.0312 | (0.776-0.932) | |||||
Abbreviations: AUC, area under the curve; BI-RADS, breast imaging and reporting data system; CADxSE, computer-aided diagnostic system for strain elastography; DBA, difference between the curves; ROC, receiver operating characteristic; SE, strain elastography; USdx, ultrasound lexicon; 95% CI, 95% confidence interval.
aThe parameters evaluated were the AUC, SE, 95% CI, DBA, and significance.
bDeLong et al (1988)16.
cBinomial exact.
d P < .05.
Among the 3 readers, there were no significant differences (P < .05) in the areas under the ROC curves between USdx and USdx + CADxSE when evaluated independently (Figure 4).
Figure 4.
Receiver operating characteristic (ROC) curves of the final classification using ultrasound lexicon (USdx) and USdx + CADxSE. The left graphic compares the classification of breast masses using the parameters proposed by the breast imaging and reporting data system (BI-RADS) USdx. The area under the ROC curve is larger for reader 1. The right graphic compares the classification of breast masses with the association of the computer-aided diagnostic system for strain elastography (CADxSE) results with the USdx. An increase in the area under the ROC curve was observed for all readers when compared to USdx alone, especially for reader 2.
The κ interobserver agreement for USdx was strong in the pairwise comparison (Table 4). The κ agreement for USdx + CADxSE was almost perfect between readers 1 and 2 (0.848) and was strong between readers 1 and 3 and readers 2 and 3 (Table 4).
Table 4.
Interobserver Agreement Using the WK and the ICC.a
| Weighted κ (WK)b Test USdx | Weighted κ (WK)b Test USdx + CADxSE | ||||
|---|---|---|---|---|---|
| Reader 2 WK/(95% CI) | Reader 3 WK/(95% CI) | Reader 2 WK/(95% CI) | Reader 3 WK/(95% CI) | ||
| Reader 1 | 0.655 (0.520-0.791) | 0.681 (0.585-0.778) | Reader 1 | 0.848 (0.786-0.910) | 0.799 (0.718-0.880) |
| Reader 2 | 0.672 (0.558-0.786) | Reader 2 | 0.777 (0.688-0.865) | ||
| Interclass Correlation Coefficient USdx(3 readers) | Interclass Correlation Coefficient USdx + CADxSE(3 readers) | ||||
| ICCc | 95% CI | ICCc | 95% CI | ||
| Single measuresd | 0.6713 | (0.5682-0.7604) | Single measuresd | 0.811 | (0.7423-0.8666) |
| Average measurese | 0.8597 | (0.7979-0.9049) | Average measurese | 0.9280 | (0.8963-0.9512) |
Abbreviations: CADxSE, computer-aided diagnostic system for strain elastography; ICC, interclass correlation coefficient; USdx, ultrasound lexicon; WK, weighted κ; 95% CI, 95% confidence interval.
aThe parameters evaluated were WK, 95% CI, and ICC for the final classification using USdx and USdx + CADxSE. Interobserver agreement using the WK and the ICC.
bQuadratic weights.
cThe degree of absolute agreement among measurements.
dEstimates the reliability of single ratings.
eEstimates the reliability of averages of k ratings.
The ICC was strong for the 3 readers together for USdx and almost perfect for USdx + CADxSE (Table 4).
Discussion
Elastography has been used as an additional tool for the classification of breast masses using US since the beginning of this decade.15 It was incorporated in the fifth edition of the BI-RADS lexicon as a complementary tool for the classification of breast masses. However, owing to the lack of standardization of the software used in US devices, the techniques used, and the strategy used for image interpretation, its use in clinical practice is time-consuming.
Almost all US companies offer devices with built-in SE software. Its availability makes SE the most commonly used method for the classification of breast lesions compared to SWE. In a recent study, Barr and Zhang17 showed that the diagnostic accuracies of SE and SWE were similar. The qualitative analysis of SE images involves the evaluation of the distribution and frequency of colors within the lesion and allows the classification of these lesions as soft, intermediate, or hard, as proposed by the BI-RADS lexicon. The interobserver agreement is a weakness in the SE method because this method is operator-dependent and the results are interpretative.
Therefore, CAD systems have the potential to be used as a complement in imaging diagnosis.18-20 Moreover, these systems are even more important for breast lesions since mammography is the only imaging method tested and well established for breast cancer screening. Patients aged 40 to 70 years should be subjected to an annual mammography. The main advantage of mammography is the high negative predictive value, that is, negative values indicate a small probability of breast cancer. However, the disadvantages include false-positive results that require healthy women without breast cancer to undergo invasive procedures. Consequently, the greater the numbers of patients who adhere to screening programs, the greater the number of diagnoses of lesions via mammography and of US screening for patients with dense breasts (BI-RADS lexicon categories C and D). To date, the false-positive results in US are the main limitation for the introduction of US into screening programs.
Breast mass classification is a medical act for diagnostic purpose, in the author’s opinion. The authors believe that the delineation of the lesion has to be performed manually by the physician and not by the computer. This has a medical-legal implication where the physician has to have direct and active participation in the classification of the masses, rather than a passive behavior when it was determined by a computer software. In the present model, CADxSE serves as an excellent auxiliary tool for breast mass classification.
To facilitate the classification technique proposed by the CADxSE system and to minimize the factors that could influence the results, the area of interest was segmented using B-mode images (better sharpness) instead of the SE images. B-mode images are routinely used by radiologists for their evaluations. Furthermore, from the manual segmentation of the image until the final classification, radiologists cannot interfere the results. The result of the classification is expressed in a box that shows: the percentage of hard area in the lesion; the elastographic classification (soft, intermediate, or hard); and 2 boxes that make up the lesion, one with the colors corresponding to the hard areas and the other with the colors of the other areas. This format allows a third party to validate the results and to assess whether the segmentation chosen for evaluation was adequate.
The results herein indicated that the use of USdx + CADxSE improved the diagnostic performance of the 3 readers involved in the study compared with the USdx alone. In addition, there were no statistically significant changes in the areas under the ROC curves for the classification of the images obtained with USdx and USdx + CADxSE between the 3 readers when analyzed independently, demonstrating that their performances were similar in both methods.
The evaluation of the interobserver agreement using pairwise comparisons for the USdx + CADxSE classification indicated that the best agreement was found between the 2 specialists with greater experience (almost perfect agreement) and that the worst was observed between those with less experience (strong agreement). This result was expected because imaging experience enhances the reviewer’s ability to interpret the images. Therefore, classification using USdx + CADxSE improved interobserver agreement compared with classification using USdx.
Similar results were obtained when the ICC was evaluated between the 3 readers; this correlation increased from 0.671 (strong agreement) for USdx to 0.811 (almost perfect agreement) for USdx + CADxSE.
It is of note that the comparison of the distribution of classifications made by the authors for the lesions classified as 3 using USdx and USdx + CADxSE indicated a tendency of reduction in the number of lesions classified as category 3 with migration to category 2. For reader 1, the number of lesions decreased from 28 to 11 (reduction of 60.7%), for reader 2, it decreased from 31 to 14 (reduction of 54.8%), and for reader 3, it decreased from 34 to 12 (reduction of 64.7%). In addition, even with the downgrading of the final category of BI-RADS, there was no interference in the sensitivity obtained by the 3 readers. This result occurred because only 1 category can be downgraded from positive to negative, going from category 4a to category 3. Category 3 corresponds to lesions with typical benign characteristics but that need to be controlled because they are diagnosed most often in the first screening examination of the patient. Category 4a consists of lesions with benign characteristics but that changed between 2 screening examinations. Because most benign biopsied lesions correspond to fibroadenomas and fibrocystic changes and these lesions often belong to categories 3 and 4a, downgrades are usually made for these types of lesions.
The adoption of this classification system helps to prevent the change in a true-positive result using USdx into a false-negative result using USdx + CADxSE. For example, masses classified as category 4b using USdx can never be reclassified as category 3, even if the CADxSE is soft (a false negative). This procedure prevents the false-negative results in mucinous and papillary carcinomas for example.
Previous studies validate the reproducibility of CADxSE systems for elastography, which present better diagnostic accuracy when compared to visual analysis.14,21,22 In 2014, Zhan et al presented a study using a CAD system for SE.22 The system proposed differs from ours in 3 main points: (1) In our system, a 3-score classification based on the quantitative evaluation of the rigid strain within the delimited area is used, while the anterior one is based on 5-score visual classification including the periphery of the mass using as reference the classification proposed by Itho et al 23; (2) the current system uses the gray-scale image as a standard image to delimit the lesion manually, so the radiologist is the one who determines the area to be studied, without the interference of the CAD system; (3) the cutoff point used by this study for malignancy was 75% of hard strain within the mass, while Zhan et al adopted 80% as the cutoff point.
Our previous studies demonstrated a better diagnostic accuracy when we used the cutoff point of 75% when compared with the threshold of 80%. The area under the curve for each cutoff point was respectively 0.837 and 0.832.24 Another point that differentiates our work from Zhan et al’s is the incorporation of USdx to the CADxSE.
This study has some limitations. To test the proposed CADxSE in isolation as an additional tool for the classification of breast lesions, it was decided that only 1 radiologist would acquire the images to be evaluated and would not participate in image interpretation because he already knew the results. This strategy was adopted because it is believed to reduce the interference of image acquisition between the radiologists. Another limitation was the small sample size with a predominance of negative results. Because we opted for the inclusion of consecutive breast biopsies in a given period, the results reflect what was found in the clinical practice of our service. It is important to emphasize that this study did not aim to evaluate the examination technique to obtain the elastographic images already discussed in other studies, but the applicability of the CADxSE system. Another limitation of the CADxSE is that its results are closely dependent on the quality of the image obtained by the SE examination. However, most manufactures provide visual data to determine whether the SE image is optima for clinical usage.
A multicenter study with a larger sample should be performed to validate the application of the CADxSE in the clinical practice. We also believe that the results obtained by the CADxSE may be associated in the future with the USdx classification using artificial intelligence, such as deep learning, where it would reduce the interference of the interobserver interpretation.
The use of computer-assisted systems such as CADxSE will be increasingly present in clinical diagnostic practice to improve radiologist performance, standardize the interpretation of tests, and facilitate data sharing with physicians in other specialties. Our results demonstrate that the proposed CADxSE system increases the diagnostic accuracy of examinations using USdx combined with elastography and improves the interobserver agreement for the classification of breast masses.
Supplementary Material
Abbreviations
- BI-RADS
breast imaging and reporting data system
- CADx
computer-aided diagnostic system
- CADxSE
computer-aided diagnostic system for strain elastography
- ICC
intraclass correlation coefficient
- PACS
picture archiving and communication system
- ROC
receiver operating characteristics
- ROI
region of interest
- SE
strain elastography
- SWE
shear-wave elastography
- US
ultrasound
- USdx
final category assessment adopting the criteria proposed by the BI-RADS ultrasound lexicon
- US + CADxSE
final category assessment adopting the criteria proposed by the BI-RADS ultrasound lexicon combined to results from the computer-aided diagnostic system for strain elastography
- WK
weighted κ test
- 95% CI
95% confidence interval.
Footnotes
Authors’ Note: The authors guarantee that the article has been submitted only to Technology in Cancer Research & Treatment.
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This study was approved by the research ethics committee of the IBCC (Protocol No. 012664/2016) and was registered in the Plataforma Brazil (Protocol No. 53543016.2.0000.0072).
Supplemental Material: Supplementary material for this article is available online.
References
- 1. Jalalian A, Mashohor SB, Mahmud HR, Saripan MI, Ramli AR, Karasfi B. Computer-aided detection/diagnosis of breast cancer in mammography and ultrasound: a review. Clin Imaging. 2013;37(3):420–426. [DOI] [PubMed] [Google Scholar]
- 2. Deandrea S, Molina-Barceló A, Uluturk A, et al. Presence, characteristics and equity of access to breast cancer screening programmes in 27 European countries in 2010 and 2014. Results from an international survey. Prev Med. 2016;91:250–263. [DOI] [PubMed] [Google Scholar]
- 3. Tabàr L, Fagerberg G, Duffy SW, Day NE, Gad A, Gröntoft O. Update of the Swedish two-county program of mammographic screening for breast cancer. Radiol Clin North Am. 1992;30(1):187–210. [PubMed] [Google Scholar]
- 4. Lynge E, Olsen AH, Fracheboud J, Patnick J. Reporting of performance indicators of mammography screening in Europe. Eur J Cancer Prev. 2003;12(3):213–222. [DOI] [PubMed] [Google Scholar]
- 5. Klabunde CN, Ballard-Barbash R; for the International Breast Cancer Screening Network. Evaluating population-based screening mammography programs internationally. Semin Breast Dis. 2007;10(2):102–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Sanders LM, King AB, Goodman KS. Impact of the New Jersey breast density law on imaging and intervention volumes and breast cancer diagnosis. J Am Coll Radiol. 2016;13(10):1189–1194. [DOI] [PubMed] [Google Scholar]
- 7. Bahl M, Baker JA, Bhargavan-Chatfield M, Brandt EK, Ghate SV. Impact of breast density notification legislation on radiologists’ practices of reporting breast density: a multi-state study. Radiology. 2016;280(3):701–706. [DOI] [PubMed] [Google Scholar]
- 8. Freer PE, Slanetz PJ, Haas JS, et al. Breast cancer screening in the era of density notification legislation: summary of 2014 Massachusetts experience and suggestion of an evidence-based management algorithm by multi-disciplinary expert panel. Breast Cancer Res Treat. 2015;153(2):455–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cho KR, Seo BK, Woo OH, et al. Breast cancer detection in a screening population: comparison of digital mammography, computer-aided detection applied to digital mammography and breast ultrasound. Breast Cancer. 2016;19(3):316–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hooley RJ, Scoutt LM, Philpotts LE. Breast ultrasonography: state of the art. Radiology. 2013;268(3):642–659. [DOI] [PubMed] [Google Scholar]
- 11. ACR. ACR Breast Imaging Reporting and Data System, Breast Imaging Atlas. 5th ed Reston, VA: American College of Radiology; 2003. [Google Scholar]
- 12. Duma MM, Chiorean AR, Chiorean M, et al. Breast diagnosis: concordance analysis between the BI-RADS classification and Tsukuba Sonoelastography score. Clujul Med. 2014;87(4):250–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Fleury Ede F, Fleury JC, Piato S, Roveda D., Jr New elastographic classification of breast lesions during and after compression. Diagn Interv Radiol. 2009;15(2):96–103. [PubMed] [Google Scholar]
- 14. Marcomini KD, Fleury EFC, Schiabel H, Nishikawa RM. Proposal of semi-automatic classification of breast lesions for strain sonoelastography using a dedicated CAD system In: Tingberg A, et al. eds. IWDM 2016, LNCS 9699. Cham, Switzerland: Springer International Publishing; 2016:454–460. [Google Scholar]
- 15. Fleury EF. The importance of breast elastography added to the BI-RADS® (5th edition) lexicon classification. Rev Assoc Med Bras. 2015;61(4):313–316. [DOI] [PubMed] [Google Scholar]
- 16. DeLong ER, DeLong DM, Clarke-Pearson DL. Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach. Biometrics. 1988;44(3):837–845. [PubMed] [Google Scholar]
- 17. Barr RG, Zhang Z. Shear-wave elastography of the breast: value of a quality measure and comparison with strain elastography. Radiology. 2015;275(1):45–53. [DOI] [PubMed] [Google Scholar]
- 18. Chen CM, Chou YH, Han KC, et al. Breast lesions on sonograms: computer-aided diagnosis with nearly setting-independent features and artificial neural networks. Radiology. 2003;226(2):504–514. [DOI] [PubMed] [Google Scholar]
- 19. Wu WJ, Moon WK. Ultrasound breast tumor image computer-aided diagnosis with texture and morphological features. Acad Radiol. 2008;15(7):873–880. [DOI] [PubMed] [Google Scholar]
- 20. Jeffers AM, Sieh W, Lipson JA, et al. Breast cancer risk and mammographic density assessed with semiautomated and fully automated methods and BI-RADS. Radiology. 2017;282(2):348–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Xiao Y, Zeng J, Niu L, et al. Computer-aided diagnosis based on quantitative elastographic features with supersonic shear wave imaging. Ultrasound Med Biol. 2014;40(2):275–286. [DOI] [PubMed] [Google Scholar]
- 22. Zhang X, Xiao Y, Zeng J, et al. Computer-assisted assessment of ultrasound real-time elastography: initial experience in 145 breast lesions. Eur J Radiol. 2014;83(1):e1–e7. [DOI] [PubMed] [Google Scholar]
- 23. Itho A, Ueno E, Tohno E, et al. Breast disease: clinical application of US elastography for diagnosis. Radiology. 2006;239(2):341–350. [DOI] [PubMed] [Google Scholar]
- 24. Marcomini KD, Fleury EFC, Oliveira VM, Carneiro AOA, Schiabel H, Nishikawa RM. Agreement between a computer-assisted tool and radiologist to classify lesions in breast elastography images. Proc SPIE. 2017;10134:101342T. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.