Abstract
Although the genus Clavibacter was originally proposed to accommodate all phytopathogenic coryneform bacteria containing B2γ diaminobutyrate in the peptidoglycan, reclassification of all but one species into other genera has resulted in the current monospecific status of the genus. The single species in the genus, Clavibacter michiganensis, has multiple subspecies, which are all highly host-specific plant pathogens. Whole genome analysis based on average nucleotide identity and digital DNA–DNA hybridization as well as multi-locus sequence analysis (MLSA) of seven housekeeping genes support raising each of the C. michiganensis subspecies to species status. On the basis of whole genome and MLSA data, we propose the establishment of two new species and three new combinations: Clavibacter capsici sp. nov., comb. nov. and Clavibacter tessellarius sp. nov., comb. nov., and Clavibacter insidiosus comb. nov., Clavibacter nebraskensis comb. nov. and Clavibacter sepedonicus comb. nov.
Keywords: Clavibacter capsici, Clavibacter michiganensis, Clavibacter insidiosus, Clavibacter nebraskensis, Clavibacter sepedonicus, Clavibacter tessellarius
The genus Clavibacter was originally proposed by Davis et al. [1] to accommodate all phytopathogenic coryneform bacteria containing B2γ diaminobutyrate in the peptidoglycan. This genus originally included six plant pathogenic species: Clavibacter michiganensis, Clavibacter iranicum, Clavibacter rathayi, Clavibacter toxicus, Clavibacter tritici and Clavibacter xyli. Subsequently, the grass-specific pathogens, C. iranicum, C. rathayi, C. toxicus and C. tritici, were reclassified into the genus Rathayibacter on the basis of DNA–DNA hybridization and their unique menaquinone structures [2]. The two subspecies of C. xyli were placed in the genus, Leifsonia [3, 4]. Currently, the genus Clavibacter consists of only one species, C. michiganensis, which is subdivided into seven subspecies of plant pathogenic bacteria with narrow host specificities and two subspecies with close association with tomato and pepper seeds. Five of the subspecies comprise well-known pathogens, namely, C. michiganensissubsp.michiganensis (Cmm; bacterial canker and wilt of tomato), C. michiganensissubsp.sepedonicus (Cms; bacterial ring rot of potato), C. michiganensissubsp.insidiosus (Cmi; wilting and stunting in alfalfa), C. michiganensissubsp.nebraskensis (Cmn; wilt and blight of maize), and C. michiganensissubsp.tessellarius (Cmt; leaf freckles and leaf spots in wheat). More importantly, the first three subspecies are quarantine or regulated pathogens of important agricultural crops in many countries. Recently, C. michiganensissubsp.phaseoli was described as the causal agent of bacterial leaf yellowing on bean [5] and C. michiganensissubsp.capsici (Cmc) as the causal agent of bacterial canker on pepper [6]. Another two subspecies, C. michiganensissubsp.californiensis and C. michiganensissubsp.chilensis were named to include bacterial isolates from tomato and pepper seeds produced in California and Chile, respectively [7]. Among these newly established subspecies, only C. michiganensissubsp.capsici with available genome sequence data (Table 1) was used in this study. The other three recently named subspecies were not included in this study.
Table 1. Bacterial strains and their genome sequences analysed in this study.
| Bacterial strains | Strain no | GenBank accession no | Isolated from | Reference |
|---|---|---|---|---|
| Clavibacter sp. | CF 11 | JROD01000001 | Soil | [22] |
| Clavibacter sp. | LMG 26808 | AZQZ01000000 | unknown | [12] |
| C. michiganensissubsp.insidiosus | LMG 3663T | MZMO00000000 | Alfalfa | This work |
| R1-1 | NZ_CP011043 | Alfalfa | [23] | |
| C. m. subsp. michiganensis | LMG 7333T | MZMP00000000 | Tomato | This work |
| NCPPB 382 | NC_009480 | Tomato | [24] | |
| C. m. subsp. nebraskensis | NCPPB 2581T=LMG 3700T | NC_020891 | Maize | Gartemann unpublished |
| DOAB 397 | LAKL01000001 | Corn | [25] | |
| DOAB 395 | LSOE01000000 | Corn | [21] | |
| C. m. subsp. sepedonicus | ATCC 33113T | NC_010407 | Potato | [26] |
| CFIA-Cs3N | MZMM00000000 | Potato | This work | |
| CFIA-CsR14 | MZMN00000000 | Potato | This work | |
| C. m. subsp. tessellarius | ATCC 33566T | MZMQ00000000 | Wheat | This work |
| C. m. subsp. capsici | PF 008 T | NZ_CP012573 | Pepper | [6] |
| Leifsonia xylisubsp.xyli | 356_LXYL | NZ_JVKI00000000 | Sugarcane | [1] |
| Leifsonia xylisubsp. cynodontis | DSM 46306 | NC_022438 | Bermuda Grass | [1] |
T, Type strain for the subspecies.
To better define the taxonomic position of the subspecies of C. michiganensis, whole-genome sequences of two strains of Cms, six strains of Cmn, two strains of Cmt, and the type strains of Cmm, Cmi, and Cmt were decoded using PacBio single molecule real-time (SMRT) sequencing at Genome Quebec (McGill University and Genome Quebec Innovation Centre, Montreal, Quebec, Canada). The assembled sequences were compared with published sequences of C. michiganensissubsp.michiganensis and subsp. insidiosus, and other clavibacter sequences in GenBank (Table 1). Currently available genome sequences for most type strains of each subspecies of Clavibacter michiganensis were included in this study. The genome sequences generated in this study were deposited in Genbank with accession numbers of MZMQ00000000 (Cmt ATCC 33566), MZMM00000000 (Cms CFIA-Cs3N), MZMN00000000 (Cms CFIA-CsR14), MZMO00000000 (Cmi LMG 3663) and MZMP00000000 (Cmm LMG 7333).
Average nucleotide identity (ANI) values of whole genomes represent the degree of identity/similarity between homologous regions shared by two genomes and has emerged as a powerful genome-based criterion for establishing species identity amongst genetically related micro-organisms [8, 9]. The approach evaluates a large number of genes, including both slow and fast evolving genes, in the calculation and thus minimizes the effect of variable evolutionary rates or horizontal gene transfer events [9]. In this study, ANI was calculated using the JSpecies software [10] with the Nucleotide MUMmer algorithm (NUCmer) and default parameter settings. The degree of pairwise genome-based relatedness was calculated as an ANI value following the blast-based ANI calculation method described by Goris et al. [11]. ANI was calculated based on comparisons between all strains sequenced in this study and those sequenced previously (Table 1).
The ANI values among the subspecies of Clavibacter were generally below the 96 % cutoff value for species delineation suggested by Richter and Rosselló-Móra [10]. ANI values between subspecies were 89.18–95.01 %, whereas ANI values between strains of the same subspecies were >99 % (99.17–99.98 %) (Table 2). Comparative ANI scores of ~90 % for the two strains, CF 11 and LMG 26808, tentatively identified as non-pathogenic isolates of Clavibacter michiganensis [12], were well below the 96 % cutoff for species delineation. The taxonomic status of these strains requires further study.
Table 2. Average nucleotide identity (ANI; lower diagonal) and digital DNA–DNA hybridization (dDDH; upper diagonal) values among Clavibacter michiganensis and related species and subspecies.
Cut-off values for species delineation are 96.0 and 70.0 % for ANI and dDDH, respectively.
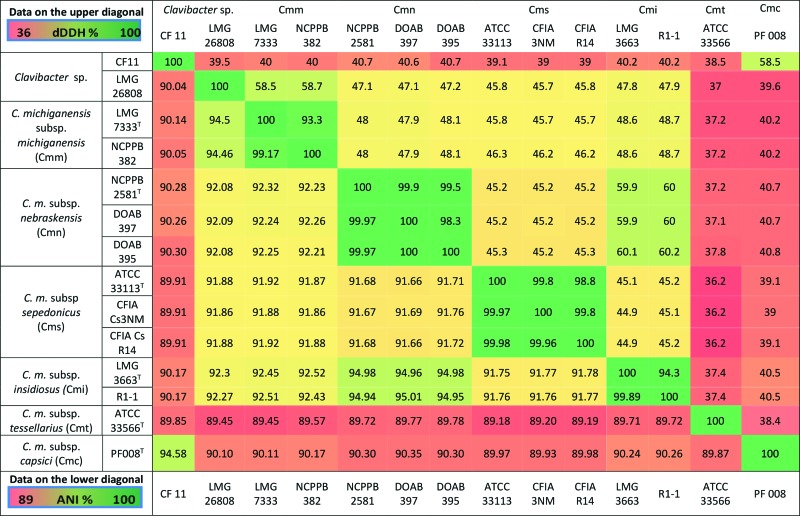 |
While ANI represents core genome homology, genome–genome distance calculation (GGDC) or digital DNA–DNA hybridization (dDDH) [13, 14] measures the genome-to-genome distances between pairs of entirely or partially sequenced genomes. The digital pairwise estimator for the relatedness of genomes serves as an in silico replacement for the wet-lab based DNA–DNA hybridization. In this study dDDH values were calculated using GGDC 2.0 server (http://ggdc.dsmz.de/distcalc2.php) by means of genome-to-genome sequence comparison and pairwise dDDH values were estimated using the GGDC calculator [14]. Consistency with ANI data and dDDH values clearly differentiated the Clavibacter subspecies into distinct clades with high degree of congruency with genomospecies allocation (Table 2). The dDDH values between different subspecies were within the range of 37–60 % (Table 2), below the suggested 70 % cut-off for species delineation [14]. Significantly, but not unexpectedly, evaluations between strains of the same subspecies showed dDDH values of more than 93 % (Table 2).
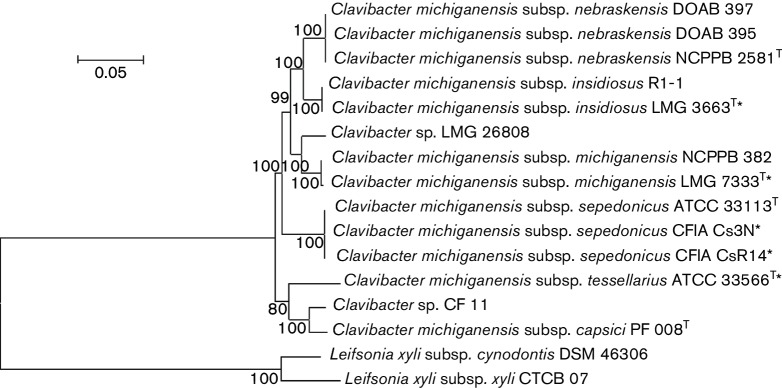
Multi-locus sequence analysis (MLSA) based on concatenated segments of housekeeping genes is used in phylogenetic studies to resolve taxonomic relationships among closely related species [15–17]. MLSA was employed on seven housekeeping genes, acnA, gapA, lcdA, mdh, mtlD, pgi and proA (Fig. 1). Strains within each of the five C. michiganensis subspecies clearly formed five distinct phylogenetic clusters, well-supported by high bootstrap values (Fig. 1). The grouping coincided perfectly with the five apparent genomospecies based on ANI and dDDH values (Table 2). Of the two non-pathogenic strains, LMG 26808 clustered most closely to C. m. subsp. michiganensis but separate from CF11, which formed a unique cluster. In addition, single gene phylogenies confirmed the distinct clustering of the five subspecies studied (Fig. S1, available in the online version of this article).
Fig. 1.
Phylogenetic relationship of Clavibacter clades on the basis of multi-locus sequence analysis (MLSA) of concatenated (acnA-gapA-icdA-mdh-mtlD-pgi-proA) sequences of the seven housekeeping genes. The evolutionary distances were computed using the Judes–Cantor method with bootstrap value of 100 (>50 are shown). Phylogenetic analysis was conducted in mega6 [27]. Leifsonia xyli serves as the out group. *, Current work; T, type strain.
Re-classifying C. michiganensis subspecies does not undermine classification based on phenotypic characterization of this group of plant pathogenic bacteria but rather supports their classification as individual species which are easily differentiated by classical bacteriological methods as previously reported [5, 18, 19]. As already noted, each of the C. michiganensis subspecies is highly host-specific and in culture can also be readily differentiated by colony pigmentation on many commonly used growth media and substrate utilization (Table 3). Biochemical and physiological test reactions also differentiate each of the Clavibacter groups (Table 3).
Table 3. Phenotypic characteristics of Clavibacter michiganensis subspecies [5, 6, 18, 19].
| Characteristic | C. michiganensissubsp.michiganensis | C. m. subsp. insidiosus | C. m. subsp. nebraskensis | C. m. subsp. sepedonicus | C. m. subsp. tessellarius | C. m. subsp. capsici |
|---|---|---|---|---|---|---|
| Major host plant | Tomato | Alfalfa | Maize | Potato | Wheat | Pepper |
| Colony pigment | Yellow* | Yellow/blue | Orange/yellow | White | Orange | Orange |
| Colony type | Fluidal | Fluidal | Domed, mucoid | Fluidal | Domed, mucoid | Mucoid |
| Growth on CNS | + | − | + | − | + | n/a |
| Growth on TTC | + | + | − | − | + | + |
| Gelatin liquefaction | + | − | − | − | −† | n/a |
| Levan production | − | − | + | − | + | + |
| Acid from sorbitol | − | − | + | + | + | n/a |
| Acid from mannitol | − | − | − | + | +† | n/a |
| Utilization of melibiose | + | − | + | − | − | + |
| Utilization of trehalose | w | + | + | + | + | + |
| Utilization of fucose | + | − | − | − | − | − |
| Utilization of acetate | + | − | + | + | − | n/a |
| Utilization of glycerol | + | + | + | − | + | n/a |
| Utilization of succinate | + | − | + | + | −† | n/a |
| Hydrolysis of aesculin | + | + | + | + | + | n/a |
| Alkaline phosphatase activity | + | − | + | ± | + | + |
| α-Mannosidase activity | − | + | − | − | − | w |
CNS, Corynebacterium nebraskense semi-selective medium [28]; TTC, 2,3,5 triphenyl tetrazolium chloride medium [29].
*Also various other pigments (e.g. pink, red, orange, white or colourless).
†This work; w, less than 50 % positive results; n/a, not available.
Traditional classification of plant pathogens faces critical challenges in the genome era as sequence data become routinely accessible through next-generation sequencing methods. The growing number of sequenced genomes of plant pathogens provides a rich source of information for new approaches to resolve complex taxonomic questions. In this study, the draft genomes of three type strains of Clavibacter species/subspecies, not previously available, were generated and compared with all publicly available GenBank entries so as to accurately define the taxonomic status of the five subspecies within C. michiganensis. On the basis of the genome data (ANI and dDDH values) and multi-locus phylogenetic analysis presented in this paper and previously reported phenotypic characteristics, we propose that the bacteria presently classified as Clavibacter michiganensis subsp. capsici Oh et al. 2016, Clavibacter michiganensis subsp. nebraskensis (Vidaver and Mandel 1974) Davis et al. 1984, Clavibacter michiganensis subsp. insidiosus (McCulloch 1925) Davis et al. 1984, Clavibacter michiganensis subsp. sepedonicus (Spieckermann and Kotthoff 1914) Davis et al. 1984, and Clavibacter michiganensis subsp. tessellarius (Carlson and Vidaver 1982) Davis et al. 1984 be reclassified as Clavibacter capsici sp. nov., comb. nov., Clavibacter nebraskensis comb. nov., Clavibacter insidiosus comb. nov., Clavibacter sepedonicus comb. nov., and Clavibacter tessellarius sp. nov., comb. nov., respectively. The original type strains of the subspecies become type strains for each of the new species and species descriptions remain the same as for the former descriptions of corresponding subspecies [20].
Description of Clavibacter capsici sp. nov., comb. nov.
Clavibacter capsici (cap′si.ci. N.L. neut. gen. n. capsici, referring to Capsicum, the genus name of pepper).
Basonym: Clavibacter michiganensissubsp.capsici Oh et al. 2016.
The species description is unchanged from its description as Clavibacter michiganensissubsp.capsici given by Oh et al. [6].
The type strain is PF008T (=KACC 18448T=LMG 29047T). The type strain was originally isolated from pepper showing bacterial canker disease in Anyang, Republic of Korea.
Description of Clavibacter insidiosus comb. nov.
Clavibacter insidiosus (in.si.di.o′sus. L. masc. adj. insidiosus, deceitful, insidious).
Basonym: Corynebacterium insidiosum (McCulloch 1925) Jensen 1934, Corynebacterium michiganensesubsp.insidiosum (McCulloch 1925) Carlson and Vidaver 1982, Clavibacter michiganensissubspinsidiosus (McCulloch 1925) Davis et al. 1984.
Gram-stain-positive, non-spore forming, aerobic bacterium without flagella. Produces yellowish colonies with blue granules on common laboratory growth media. Grows on TTC but not CNS medium. It does not liquefy gelatin nor produces levan. It does not produce acid from either sorbitol or mannitol. It utilizes glycerol but not acetate or succinate; it hydrolyses aesculin, and has α-mannosidase activity but no alkaline phosphatase activity. It causes bacterial wilt disease of alfalfa (lucerne) (Medicago sativa L.). DNA G+C content of the type strain is 72.7 %. The type strain is LMG 3663T (=ATCC 10253T=NCPPB1109T).
Description of Clavibacter nebraskensis comb. nov.
Clavibacter nebraskensis (ne.bras.ken′sis. N.L. masc. adj. nebraskensis, pertaining to the state of Nebraska, USA).
Basonym: Corynebacterium nebraskense Vidaver and Mandel 1974, Corynebacterium michiganensesubsp.nebraskense (Vidaver and Mandel 1974) Carlson and Vidaver 1982, Clavibacter michiganensissubsp.nebraskensis (Vidaver and Mandel 1974) Davis et al. 1984.
Gram-stain-positive, non-spore forming, aerobic bacterium without flagella. Produces yellow to orange colonies on common laboratory growth media. It grows on CNS but does not grow on TTC medium. It does not liquefy gelatin but it does produce levan. It produces acid from sorbitol but it does not produce acid from mannitol. It utilizes acetate, glycerol and succinate. It hydrolyses aesculin, it has alkaline phosphatase activity, but it does not have α-mannosidase activity. It causes leaf freckles and a wilt disease of maize (Zea mays L.) DNA G+C content of the type strain is 73.0 %. The type strain is NCPPB 2581T (=ATCC 27794T=LMG 3700T).
Description of Clavibacter sepedonicus comb. nov.
Clavibacter sepedonicus (se.pe.do′ni.cus. Gr. n. sepedon rottenness, decay; N.L. masc. adj. sepedonicus, leading to decay).
Basonym: Corynebacterium sepedonicum (Spieckermann and Kotthoff 1914) Skaptason and Burkholder 1942, Corynebacterium michiganensesubsp.sepedonicum (Spieckermann and Kotthoff 1914) Carlson and Vidaver 1982, Clavibacter michiganensissubsp.sepedonicus (Spieckermann and Kotthoff 1914) Davis et al. 1984.
Gram-stain-positive, non-spore forming, aerobic bacterium without flagella. Produces white mucoid colonies at an optimum growth temperature of 20–23 °C. It does not grow on CNS or TTC media. It does not liquefy gelatin nor produces levan. It differs from the other Clavibacter species in producing acid from both sorbitol and mannitol. It utilizes acetate and succinate but not glycerol; it hydrolyses aesculin; alkaline phosphatase activity is weak, and α-mannosidase activity is lacking. It causes bacterial ring rot disease of potato (Solanum tuberosum L). DNA G+C content of the type strain is 72.4 %. The type strain is ATCC 33113T (=LMG 2889T=NCPPB 2137T).
Description of Clavibacter tessellarius sp. nov. comb. nov.
Clavibacter tessellarius (tes.sel.la′ri.us. L. masc. n. tessellarius a mosaic stone maker).
Basonym: Clavibactermichiganensesubsp.tessellarius, Clavibacter michiganensissubsp.tessellarius (Carlson and Vidaver 1982) Davis et al. 1984.
The species description is unchanged from its description as Clavibacter michiganensissubsp.tessellarius given by Carlson and Vidaver, 1982 [20].
The type strain is ATCC 33566T (=NCPPB 3664T=LMG 7294T).
This new taxonomy not only resolves the long-standing problem of having only a single species within the well-established genus, Clavibacter, but it also provides a practical solution for plant pathologists and policy makers dealing with quarantine and regulated plant pathogens. C. michiganensis, C. sepedonicus and C. insidiosus are quarantine or regulated pathogens of important agricultural crops in many countries, while C. capsici is a newly described plant pathogen for which the range of distribution and risk to agriculture need to be assessed. The revised classification, and accordingly a simpler nomenclature, uncomplicates regulatory documents and more accurately reflects biological reality.
While this manuscript was under review, one of the co-authors [21] of this manuscript carried out an independent investigation titled ‘Comparative genomics of Clavibacter michiganensis subspecies, pathogens of important agricultural crops’. It is quoted here ‘the study also assessed the taxonomic position of the subspecies based on 16S rRNA and genome-based DNA homology and concludes that there is ample evidence to elevate some of the subspecies to species-level’.
Funding information
This study was partially funded by the Canadian Safety and Security Program (CRTI 09-462RD) and Genome Research and Development Initiatives (GRDI) funding.
Acknowledgements
The technical assistance of Jingbai Nie is gratefully acknowledged.
Conflicts of interest
The authors declare that there are no conflicts of interest.
Supplementary Data
Footnotes
Abbreviations: ANI, average nucleotide identity; Cmc, C. michiganensis subsp. capsici; Cmi, C. michiganensis subsp. insidiosus; Cmm, C. michiganensis subsp. michiganensis; Cmn, C. michiganensis subsp. nebraskensis; Cms, C. michiganensis subsp. sepedonicus; Cmt, C. michiganensis subsp. tessellarius; dDDH, digital DNA-DNA hybridization; MLSA, multi-locus sequence analysis; SMRT, single molecule real-time.
One supplementary figure is available with the online version of this article.
References
- 1.Davis MJ, Gillaspie AG, Vidaver AK, Harris RW. Clavibacter: a new genus containing some phytopathogenic coryneform bacteria, including Clavibacter xyli subsp. xyli sp. nov., subsp. nov. and Clavibacter xyli subsp. cynodontis subsp. nov., pathogens that cause ratoon stunting disease of sugarcane and bermudagrass stunting disease. Int J Syst Bacteriol. 1984;34:107–117. doi: 10.1099/00207713-34-2-107. [DOI] [Google Scholar]
- 2.Zgurskaya HI, Evtushenko LI, Akimov VN, Kalakoutskii LV. Rathayibacter gen. nov., Including the species Rathayibacter rathayi comb. nov., Rathayibacter tritici comb. nov., Rathayibacter iranicus comb. nov., and six strains from annual grasses. Int J Syst Bacteriol. 1993;43:143–149. doi: 10.1099/00207713-43-1-143. [DOI] [Google Scholar]
- 3.Evtushenko LI, Dorofeeva LV, Subbotin SA, Cole JR, Tiedje JM. Leifsonia poae gen. nov., sp. nov., isolated from nematode galls on Poa annua, and reclassification of 'Corynebacterium aquaticum' Leifson 1962 as Leifsonia aquatica (ex Leifson 1962) gen. nov., nom. rev., comb. nov. and Clavibacter xyli Davis et al. 1984 with two subspecies as Leifsonia xyli (Davis et al. 1984) gen. nov., comb. nov. Int J Syst Evol Microbiol. 2000;50:371–380. doi: 10.1099/00207713-50-1-371. [DOI] [PubMed] [Google Scholar]
- 4.Suzuki KI, Suzuki M, Sasaki J, Park YH, Komagata KK. Leifsonia gen. nov., a genus for 2,4-diaminobutyric acid-containing actinomycetes to accommodate "Corynebacterium aquaticum" Leifson 1962 and Clavibacter xyli subsp. cynodontis Davis et al. 1984. J Gen Appl Microbiol. 1999;45:253–262. doi: 10.2323/jgam.45.253. [DOI] [PubMed] [Google Scholar]
- 5.González AJ, Trapiello E. Clavibacter michiganensis subsp. phaseoli subsp. nov., pathogenic in bean. Int J Syst Evol Microbiol. 2014;64:1752–1755. doi: 10.1099/ijs.0.058099-0. [DOI] [PubMed] [Google Scholar]
- 6.Oh EJ, Bae C, Lee HB, Hwang IS, Lee HI, et al. Clavibacter michiganensis subsp. capsici subsp. nov., causing bacterial canker disease in pepper. Int J Syst Evol Microbiol. 2016;66:4065–4070. doi: 10.1099/ijsem.0.001311. [DOI] [PubMed] [Google Scholar]
- 7.Yasuhara-Bell J, Alvarez AM. Seed-associated subspecies of the genus Clavibacter are clearly distinguishable from Clavibacter michiganensis subsp. michiganensis. Int J Syst Evol Microbiol. 2015;65:811–826. doi: 10.1099/ijs.0.000022. [DOI] [PubMed] [Google Scholar]
- 8.Kim M, Oh HS, Park SC, Chun J. Towards a taxonomic coherence between average nucleotide identity and 16S rRNA gene sequence similarity for species demarcation of prokaryotes. Int J Syst Evol Microbiol. 2014;64:346–351. doi: 10.1099/ijs.0.059774-0. [DOI] [PubMed] [Google Scholar]
- 9.Konstantinidis KT, Tiedje JM. Genomic insights that advance the species definition for prokaryotes. Proc Natl Acad Sci USA. 2005;102:2567–2572. doi: 10.1073/pnas.0409727102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Richter M, Rosselló-Móra R. Shifting the genomic gold standard for the prokaryotic species definition. Proc Natl Acad Sci USA. 2009;106:19126–19131. doi: 10.1073/pnas.0906412106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goris J, Konstantinidis KT, Klappenbach JA, Coenye T, Vandamme P, et al. DNA–DNA hybridization values and their relationship to whole-genome sequence similarities. Int J Syst Evol Microbiol. 2007;57:81–91. doi: 10.1099/ijs.0.64483-0. [DOI] [PubMed] [Google Scholar]
- 12.Załuga J, Stragier P, Baeyen S, Haegeman A, van Vaerenbergh J, et al. Comparative genome analysis of pathogenic and non-pathogenic Clavibacter strains reveals adaptations to their lifestyle. BMC Genomics. 2014;15:392. doi: 10.1186/1471-2164-15-392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Auch AF, von Jan M, Klenk HP, Göker M. Digital DNA–DNA hybridization for microbial species delineation by means of genome-to-genome sequence comparison. Stand Genomic Sci. 2010;2:117–134. doi: 10.4056/sigs.531120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Meier-Kolthoff JP, Auch AF, Klenk HP, Göker M. Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinformatics. 2013;14:60. doi: 10.1186/1471-2105-14-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mulet M, Lalucat J, García-Valdés E. DNA sequence-based analysis of the Pseudomonas species. Environ Microbiol. 2010;12:1513–1530. doi: 10.1111/j.1462-2920.2010.02181.x. [DOI] [PubMed] [Google Scholar]
- 16.González AJ, Cleenwerck I, de Vos P, Fernández-Sanz AM. Pseudomonas asturiensis sp. nov., isolated from soybean and weeds. Syst Appl Microbiol. 2013;36:320–324. doi: 10.1016/j.syapm.2013.04.004. [DOI] [PubMed] [Google Scholar]
- 17.Tambong JT, Xu R, Bromfield ESP. Pseudomonas canadensis sp. nov., a biological control agent isolated from a field plot under long-term mineral fertilization. Int J Syst Evol Microbiol. 2017;67:889–895. doi: 10.1099/ijsem.0.001698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Davis MJ, Vidaver AK. Coryneform plant pathogens. In: Schaad NW, Jones JB, Chun W, editors. Laboratory Guide for Identification of Plant Pathogenic Bacteria. 3rd ed. St Paul, MN: APS Press; 2001. pp. 218–234. (editors) [Google Scholar]
- 19.Eichenlaub R, Gartemann K-H, Burger A. Clavibacter michiganensis, a group of Gram-positive phytopathogenic bacteria. In: Gnanamanickam SS, editor. Plant-Associated Bacteria. Dordrecht: Springer; 2006. pp. 385–422. (editor) [Google Scholar]
- 20.Carlson RR, Vidaver AK. Taxonomy of Corynebacterium plant pathogens, including a new pathogen of wheat, based on polyacrylamide gel electrophoresis of cellular proteins. Int J Syst Bacteriol. 1982;32:315–326. doi: 10.1099/00207713-32-3-315. [DOI] [Google Scholar]
- 21.Tambong JT. Comparative genomics of Clavibacter michiganensis subspecies, pathogens of important agricultural crops. PLoS One. 2017;12:e0172295. doi: 10.1371/journal.pone.0172295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.du Y, Yuan B, Zeng Y, Meng J, Li H, et al. Draft genome sequence of the cellulolytic bacterium Clavibacter sp. CF11, a strain producing cold-active cellulase. Genome Announc. 2015;3:e01304-14. doi: 10.1128/genomeA.01304-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lu Y, Samac DA, Glazebrook J, Ishimaru CA. Complete genome sequence of Clavibacter michiganensis subsp. insidiosus R1-1 using pacbio single-molecule real-time technology. Genome Announc. 2015;3:e00396-15. doi: 10.1128/genomeA.00396-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gartemann KH, Abt B, Bekel T, Burger A, Engemann J, et al. The genome sequence of the tomato-pathogenic actinomycete Clavibacter michiganensis subsp. michiganensis NCPPB382 reveals a large island involved in pathogenicity. J Bacteriol. 2008;190:2138–2149. doi: 10.1128/JB.01595-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tambong JT, Xu R, Adam Z, Cott M, Rose K, et al. Draft genome sequence of Clavibacter michiganensis subsp. nebraskensis Strain DOAB 397, isolated from an infected field corn plant in Manitoba, Canada. Genome Announc. 2015;3:e00768-15. doi: 10.1128/genomeA.00768-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bentley SD, Corton C, Brown SE, Barron A, Clark L, et al. Genome of the actinomycete plant pathogen Clavibacter michiganensis subsp. sepedonicus suggests recent niche adaptation. J Bacteriol. 2008;190:2150–2160. doi: 10.1128/JB.01598-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gross DC. A selective medium for isolation of Corynebacterium nebraskense from soil and plant parts. Phytopathology. 1979;69:82–87. doi: 10.1094/Phyto-69-82. [DOI] [Google Scholar]
- 29.Kelman A. The relationship of pathogenicity in Pseudomonas solanacearum to colony appearance on tetrazolium medium. Phytopathol. 1954;44:693–695. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.