Abstract
The Chrysoviridae is a family of small, isometric, non-enveloped viruses (40 nm in diameter) with segmented dsRNA genomes (typically four segments). The genome segments are individually encapsidated and together comprise 11.5–12.8 kbp. The single genus Chrysovirus includes nine species. Chrysoviruses lack an extracellular phase to their life cycle; they are transmitted via intracellular routes within an individual during hyphal growth, in asexual or sexual spores, or between individuals via hyphal anastomosis. There are no known natural vectors for chrysoviruses. This is a summary of the International Committee on Taxonomy of Viruses (ICTV) Report on the taxonomy of the Chrysoviridae, which is available at www.ictv.global/report/chrysoviridae.
Keywords: Chrysoviridae, taxonomy, ICTV report, Penicillium chrysogenum virus, Aspergillus fumigatus chrysovirus, Helminthosporium victoriae virus 145S
Virion
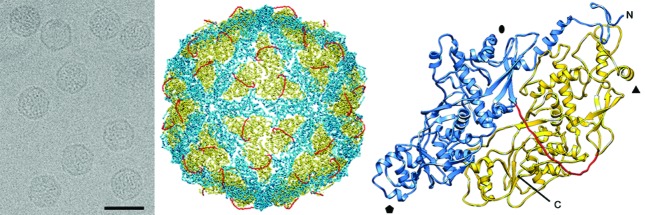
Virions are isometric, non-enveloped and about 40 nm in diameter. The capsid of Penicillium chrysogenum virus comprises 60 copies of a 109 kDa polypeptide arranged on a T=1 icosahedral lattice (Table 1, Fig. 1). The capsid protein is formed by a repeated α-helical domain, indicative of gene duplication despite lack of sequence similarity between the two halves [1]. This domain has a fold that is conserved among dsRNA viruses [2].
Table 1. Characteristics of the family Chrysoviridae.
| Typical member: | Penicillium chrysogenum virus ATCC 9480 (dsRNA1: AF296439; dsRNA2: AF296440; dsRNA3: AF296441; dsRNA4: AF296442), species Penicillium chrysogenum virus, genus Chrysovirus |
|---|---|
| Virion | Isometric, non-enveloped, 40 nm in diameter |
| Genome | A total of 11.5–12.8 kbp of dsRNA in a quadripartite genome with each segment separately encapsidated |
| Replication | Particles containing both dsRNA and ssRNA can be isolated from infected fungal hosts. Virions accumulate in the cytoplasm |
| Translation | From positive-sense transcripts of genomic dsRNAs |
| Host range | Fungi |
| Taxonomy | One genus (Chrysovirus) including nine species |
Fig. 1.
Three-dimensional cryo-EM reconstruction of Penicillium chrysogenum virus virions at a resolution of 4.1 Å. (Left) Cryo-EM image of Penicillium chrysogenum virus (scale bar, 50 nm). (Middle) Atomic model of the Penicillium chrysogenum virus capsid viewed along a twofold axis. (Right) Atomic model of a Penicillium chrysogenum virus CP (top view) showing the N-terminal domain (1–498, blue), the linker segment (499–515, red) and the C-terminal domain (516–982, yellow). Symbols indicate icosahedral symmetry axes.
Genome
The genome consists of four linear, separately encapsidated, dsRNA segments of 2.5–3.6 kbp [3, 4]. The largest segment, dsRNA1, codes for the virion-associated, RNA-dependent RNA polymerase (RdRP; P1) and dsRNA2 codes for the major capsid protein (CP; P2). Both dsRNA 3 and 4 encode proteins of unknown function [5]. Sequences at the 5′- and 3′-UTRs are highly conserved among the four dsRNA segments (Fig. 2). In addition to the absolutely conserved 5′- and 3′-termini, a 40–75 nt region with high sequence identity is present in the 5′-UTR of all four dsRNAs (Box 1, Fig. 2). A second region of strong sequence similarity is present immediately downstream from Box 1 and consists of a stretch of 30–50 nt containing a reiteration of the sequence ‘CAA’. The (CAA)n repeats are similar to the enhancer elements present at the 5′-UTRs of tobamoviruses [6]. The N-terminal region of P3 shares high sequence similarity with the corresponding N-terminal region of RdRP (P7/P-loop domain; possibly a nucleotide triphosphate hydrase domain). P4 is a putative cysteine protease [7].
Fig. 2.
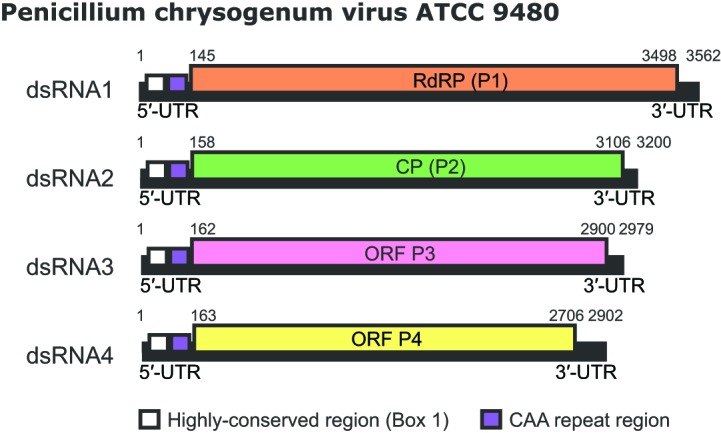
Genome organization of Penicillium chrysogenum virus isolate ATCC 9480 (PcV-ATCC9480). The genome consists of four dsRNA segments, each of which is monocistronic. The RdRP (P1) ORF (nt positions 145–3498 on dsRNA1), the CP (P2) ORF (nt positions 158–3106 on dsRNA2), the P3 ORF (nt positions 162–2900 on dsRNA3) and the P4 ORF (nt positions 163–2706 on dsRNA4) are represented by rectangular boxes.
Replication
Replication has not been characterized in detail. Particles containing a single molecule of dsRNA, as well as particles containing both dsRNA and ssRNA, can be isolated from an infected fungal host [3]. Virions accumulate in the cytoplasm.
Taxonomy
The family Chrysoviridae includes a single genus with nine species, whose members infect ascomycetous or basidiomycetous fungi.
Species demarcation criteria include nucleotide and deduced amino acid sequence data (≤70 % and ≤53 % aa sequence identity in the RdRP and CP, respectively). Chrysoviruses cause latent persistent infections in their fungal hosts. Unclassified, chrysovirus-related viruses with 3-segmented dsRNA genomes infect plants with no apparent damage [8]. Some chrysovirus-related viruses with five dsRNA genomic segments, however, cause deleterious effects in their fungal hosts [9]. blast searches using a Penicillium chrysogenum virus RdRP amino acid sequence show high sequence identity (37.6–70.2 %) to the RdRPs of members of the genus Chrysovirus and to related, unclassified viruses. Phylogenetic analysis based on the complete deduced amino acid sequences of RdRPs of members of the family Chrysoviridae, and of related, unclassified viruses with 3–5 dsRNA segments, leads to the identification of two large distinct clusters: cluster I corresponds to members of the genus Chrysovirus and related, unclassified viruses with three genome segments. Cluster II comprises related, unclassified viruses with four or five genome segments.
Resources
Full ICTV Online (10th) Report: www.ictv.global/report/chrysoviridae.
Funding information
Production of this summary, the online chapter and associated resources was funded by a grant from the Wellcome Trust (WT108418AIA).
Acknowledgements
Members of the ICTV Report Consortium are Elliot J. Lefkowitz, Andrew J. Davison, Stuart G. Siddell, Sead Sabanadzovic, Donald B. Smith, Richard J. Orton and Peter Simmonds.
Conflicts of interest
The authors declare that there are no conflicts of interest.
Footnotes
Abbreviations: CP, capsid protein; RdRP, RNA-dependent RNA polymerase.
References
- 1.Castón JR, Luque D, Gómez-Blanco J, Ghabrial SA. Chrysovirus structure: repeated helical core as evidence of gene duplication. Adv Virus Res. 2013;86:87–108. doi: 10.1016/B978-0-12-394315-6.00004-0. [DOI] [PubMed] [Google Scholar]
- 2.Luque D, Gómez-Blanco J, Garriga D, Brilot AF, González JM, et al. Cryo-EM near-atomic structure of a dsRNA fungal virus shows ancient structural motifs preserved in the dsRNA viral lineage. Proc Natl Acad Sci USA. 2014;111:7641–7646. doi: 10.1073/pnas.1404330111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Buck KW, Girvan RF. Comparison of the biophysical and biochemical properties of Penicillium cyaneo-fulvum virus and Penicillium chrysogenum virus. J Gen Virol. 1977;34:145–154. doi: 10.1099/0022-1317-34-1-145. [DOI] [PubMed] [Google Scholar]
- 4.Jiang D, Ghabrial SA. Molecular characterization of Penicillium chrysogenum virus: reconsideration of the taxonomy of the genus Chrysovirus. J Gen Virol. 2004;85:2111–2121. doi: 10.1099/vir.0.79842-0. [DOI] [PubMed] [Google Scholar]
- 5.Ghabrial SA. Chrysoviruses. In: Mahy BWJ, Van Regenmortel MHV, editors. Encyclopedia of Virology. 3rd ed. Oxford: Elsevier; 2008. pp. 503–513. (editors) [Google Scholar]
- 6.Gallie DR, Walbot V. Identification of the motifs within the tobacco mosaic virus 5'-leader responsible for enhancing translation. Nucleic Acids Res. 1992;20:4631–4638. doi: 10.1093/nar/20.17.4631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Covelli L, Coutts RH, di Serio F, Citir A, Açikgöz S, et al. Cherry chlorotic rusty spot and Amasya cherry diseases are associated with a complex pattern of mycoviral-like double-stranded RNAs. I. Characterization of a new species in the genus Chrysovirus. J Gen Virol. 2004;85:3389–3397. doi: 10.1099/vir.0.80181-0. [DOI] [PubMed] [Google Scholar]
- 8.Li L, Liu J, Xu A, Wang T, Chen J, et al. Molecular characterization of a trisegmented chrysovirus isolated from the radish Raphanus sativus. Virus Res. 2013;176:169–178. doi: 10.1016/j.virusres.2013.06.004. [DOI] [PubMed] [Google Scholar]
- 9.Urayama S, Sakoda H, Takai R, Katoh Y, Minh Le T, et al. A dsRNA mycovirus, Magnaporthe oryzae chrysovirus 1-B, suppresses vegetative growth and development of the rice blast fungus. Virology. 2014;448:265–273. doi: 10.1016/j.virol.2013.10.022. [DOI] [PubMed] [Google Scholar]