Abstract
Members of the family Pleolipoviridae (termed pleolipoviruses) are pseudo-spherical and pleomorphic archaeal viruses. The enveloped virion is a simple membrane vesicle, which encloses different types of DNA genomes of approximately 7–16 kbp (or kilonucleotides). Typically, virions contain a single type of transmembrane (spike) protein at the envelope and a single type of membrane protein, which is embedded in the envelope and located in the internal side of the membrane. All viruses infect extremely halophilic archaea in the class Halobacteria (phylum Euryarchaeota). Pleolipoviruses have a narrow host range and a persistent, non-lytic life cycle. This is a summary of the International Committee on Taxonomy of Viruses (ICTV) Report on the taxonomy of the Pleolipoviridae which is available at www.ictv.global/report/pleolipoviridae.
Keywords: Pleolipoviridae, taxonomy, ICTV, Halorubrum pleomorphic virus 1
Virion
Virions are enveloped pleomorphic membrane vesicles of 40–70 nm diameter (Table 1, Fig. 1a, b) with one or two types of major proteins forming spikes and one or two as internal membrane proteins (Fig. 1c). The spike and internal membrane proteins of Halorubrum pleomorphic virus 1 are VP4 and VP3 respectively. Virions lack a capsid or nucleocapsid.
Table 1. Characteristics of the family Pleolipoviridae.
| Typical member | Halorubrum pleomorphic virus 1 (FJ685651), species Halorubrum virus HRPV1, genus Alphapleolipovirus |
|---|---|
| Virion | Enveloped, pseudo-spherical and pleomorphic virions (diameters 40–70 nm), typically with a single type of spike protein at the envelope and a single type of internal membrane protein embedded in the envelope |
| Genome | Circular ssDNA, circular dsDNA or linear dsDNA, approximately 7–16 kilonucleotides or kbp |
| Replication | Possibly rolling-circle replication for circular molecules; protein-primed replication for linear molecules |
| Translation | Prokaryotic translation using viral mRNA and host ribosomes |
| Host range | Archaea, euryarchaeal Halorubrum, Haloarcula or Halogeometricum strains |
| Taxonomy | Three genera Alphapleolipovirus, Betapleolipovirus and Gammapleolipovirus |
Fig. 1.
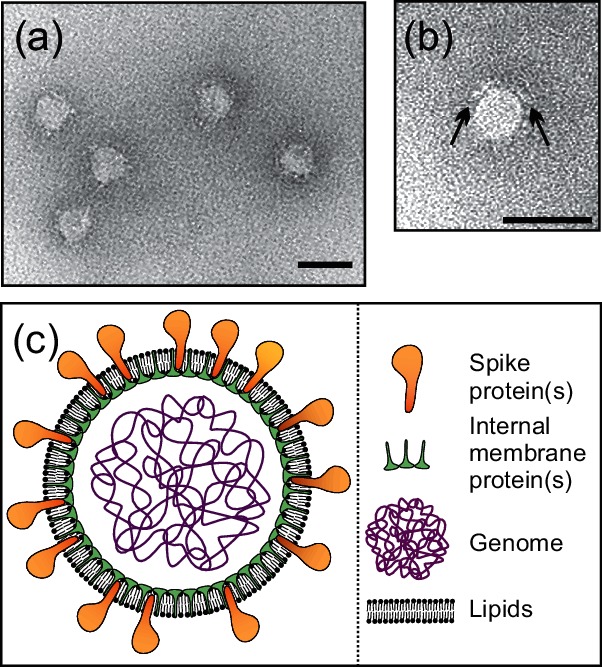
Morphology of pleolipovirus virions. (a) Electron micrograph of negatively stained Halorubrum pleomorphic virus 1 particles. (b) A close-up of one Halorubrum pleomorphic virus 1 virion. The arrows point to the spike structures covering the particle surface. Reproduced with permission from Pietilä et al. [5]. Scale bars in (a) and (b), 50 nm (c) Schematic presentation of the pleolipovirus virion.
Genome
Virus genomes are circular ssDNA of 7.0–10.7 kilonucleotides (Fig. 2), circular dsDNA of 8.1–9.7 kbp, or linear dsDNA of 16 kbp [1]. Members of the genus Alphapleolipovirus have circular ssDNA or dsDNA genomes, members of the genus Betapleolipovirus have circular dsDNA genomes with single-stranded discontinuities, and the only member of the genus Gammapleolipovirus has a linear dsDNA genome. A cluster of five genes/ORFs is conserved among the members of the family (Halorubrum pleomorphic virus 1 genes 3, 4 and 8, ORFs 6 and 7). The cluster includes genes encoding a spike and an internal membrane protein as well as an ORF encoding a putative NTPase. The only member of the genus Gammapleolipovirus is predicted to encode a putative type B DNA-dependent DNA polymerase. The genome ends bear terminal proteins.
Fig. 2.

Genome organization of Halorubrum pleomorphic virus 1. Genes encoding structural proteins are coloured black, ORFs in grey. Rep, replication initiation protein; NTPase, nucleoside triphosphate hydrolase. The position of the first nucleotide (1 nt) is indicated.
Replication
Pleolipoviruses are non-lytic and most likely enter cells by membrane fusion. Either rolling-circle or protein-primed replication may be used; transcription has not been studied. Progeny virions exit host cells continuously retarding host growth with concurrent unselective lipid acquisition implying that virions bud through the cell membrane.
Taxonomy
Alphapleolipovirus
The genus includes the species Haloarcula virus HHPV1, Haloarcula virus HHPV2, Halorubrum virus HRPV1, Halorubrum virus HRPV2 and Halorubrum virus HRPV6. The genomes of alphapleolipoviruses contain 8–15 ORFs, and besides the conserved gene/ORF cluster, an ORF encoding a putative rolling-circle replication protein and one ORF with unknown function [2–4]. Virions contain a single spike and internal membrane protein. Additionally, Halorubrum pleomorphic virus 1 virions contain one minor structural protein predicted to be an NTPase; tomographic reconstruction shows that the glycosylated spikes [5] form irregular arrays on the virion surface [6].
Betapleolipovirus
The genus includes the species Halogeometricum virus HGPV1 and Halorubrum virus HRPV3. The betapleolipovirus genomes contain 12 or 15 ORFs. In addition to the cluster of five genes/ORFs, genomes share two ORFs encoding proteins with unknown function [4]. Virions contain one type of spike protein and one or two types of internal membrane proteins. The spike protein of Halogeometricum pleomorphic virus 1 is lipid-modified [6].
Gammapleolipovirus
Haloarcula virus His2 is the only species of this genus. The member has a genome with 35 ORFs [7] and a virion consisting of one type of internal membrane protein and two types of spike protein as well as one minor structural protein [6]. One of the spike proteins is lipid-modified.
Resources
Full ICTV Online (10th) Report: www.ictv.global/report/pleolipoviridae.
Funding information
Production of this summary, the online chapter and associated resources was funded by a grant from the Academy of Finland (1274748; M. K. P) and by a grant from the Wellcome Trust (WT108418AIA). H. M. O. was supported by University of Helsinki funding for Instruct-FI research infrastructure Virus and Macromolecular Complex Production (ICVIR).
Acknowledgements
Members of the ICTV Report Consortium are Elliot J. Lefkowitz, Andrew J. Davison, Stuart G. Siddell, Peter Simmonds, Sead Sabanadzovic, Donald B. Smith, Richard J. Orton and Andrew M. Kropinski.
Conflicts of interest
The authors declare that there are no conflicts of interest.
Footnotes
Abbreviations: Rep, replication initiation protein; NTPase, nucleoside triphosphate hydrolase.
References
- 1.Pietilä MK, Roine E, Sencilo A, Bamford DH, Oksanen HM. Pleolipoviridae, a newly proposed family comprising archaeal pleomorphic viruses with single-stranded or double-stranded DNA genomes. Arch Virol. 2016;161:249–256. doi: 10.1007/s00705-015-2613-x. [DOI] [PubMed] [Google Scholar]
- 2.Pietilä MK, Roine E, Paulin L, Kalkkinen N, Bamford DH. An ssDNA virus infecting archaea: a new lineage of viruses with a membrane envelope. Mol Microbiol. 2009;72:307–319. doi: 10.1111/j.1365-2958.2009.06642.x. [DOI] [PubMed] [Google Scholar]
- 3.Roine E, Kukkaro P, Paulin L, Laurinavicius S, Domanska A, et al. New, closely related haloarchaeal viral elements with different nucleic acid types. J Virol. 2010;84:3682–3689. doi: 10.1128/JVI.01879-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sencilo A, Paulin L, Kellner S, Helm M, Roine E. Related haloarchaeal pleomorphic viruses contain different genome types. Nucleic Acids Res. 2012;40:5523–5534. doi: 10.1093/nar/gks215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pietilä MK, Laurinavicius S, Sund J, Roine E, Bamford DH. The single-stranded DNA genome of novel archaeal virus Halorubrum pleomorphic virus 1 is enclosed in the envelope decorated with glycoprotein spikes. J Virol. 2010;84:788–798. doi: 10.1128/JVI.01347-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pietilä MK, Atanasova NS, Manole V, Liljeroos L, Butcher SJ, et al. Virion architecture unifies globally distributed pleolipoviruses infecting halophilic archaea. J Virol. 2012;86:5067–5079. doi: 10.1128/JVI.06915-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bath C, Cukalac T, Porter K, Dyall-Smith ML. His1 and His2 are distantly related, spindle-shaped haloviruses belonging to the novel virus group, Salterprovirus. Virology. 2006;350:228–239. doi: 10.1016/j.virol.2006.02.005. [DOI] [PubMed] [Google Scholar]


