Abstract
A novel lanC-like sequence was identified from the dominant human gut bacterium Blautia obeum strain A2-162. This sequence was extended to reveal a putative lantibiotic operon with biosynthetic and transport genes, two sets of regulatory genes, immunity genes, three identical copies of a nisin-like lanA gene with an unusual leader peptide, and a fourth putative lanA gene. Comparison with other nisin clusters showed that the closest relationship was to nisin U. B. obeum A2-162 demonstrated antimicrobial activity against Clostridium perfringens when grown on solid medium in the presence of trypsin. Fusions of predicted nsoA structural sequences with the nisin A leader were expressed in Lactococcus lactis containing the nisin A operon without nisA. Expression of the nisA leader sequence fused to the predicted structural nsoA1 produced a growth defect in L. lactis that was dependent upon the presence of biosynthetic genes, but failed to produce antimicrobial activity. Insertion of the nso cluster into L. lactis MG1614 gave an increased immunity to nisin A, but this was not replicated by the expression of nsoI. Nisin A induction of L. lactis containing the nso cluster and nisRK genes allowed detection of the NsoA1 pre-peptide by Western hybridization. When this heterologous producer was grown with nisin induction on solid medium, antimicrobial activity was demonstrated in the presence of trypsin against C. perfringens, Clostridium difficile and L. lactis. This research adds to evidence that lantibiotic production may be an important trait of gut bacteria and could lead to the development of novel treatments for intestinal diseases.
Keywords: antimicrobial peptide, lantibiotic, nisin, gut pathogens, lachnospiraceae
Introduction
Lantibiotics are small amphiphilic lanthipeptides produced by Gram-positive bacteria and commonly have antimicrobial activity against a wide range of mostly Gram-positive bacteria [1, 2]. While their potential in applications has so far mostly been seen in the areas of preservatives and probiotics [3], they have been of increasing interest since novel therapeutic applications have been discovered [4–6]. Lantibiotics are gene-encoded and the genes involved in their biosynthesis, regulation and immunity are usually clustered. Following synthesis of the precursor peptides on the ribosome, they undergo a series of post-translational modifications, such as serine and threonine dehydration and lanthionine bridge formation, to produce the characteristic lanthionine and methyllanthionine rings that contribute to their stability [7]. Their mode of synthesis makes genetic engineering a powerful tool to create improved peptides and study their biosynthesis [8, 9].
Nisin A is the best characterized example of the type A lantibiotics; nisin is the only bacteriocin to date to be authorized for use as a food preservative, and is also in use for the prevention of bovine mastitis [4]. It is a highly stable peptide with antimicrobial activity against a wide range of Gram-positive organisms, including food-spoilage and food-pathogenic bacteria from genera such as Clostridium, Listeria, Staphylococcus and Bacillus [10]. The nisin A biosynthetic cluster is located within a 70 kb transposon named Tn5307, which has been shown to be transferrable by conjugation [11]. To date, eight natural forms of nisin have been identified from either lactococci (A, Z, F and Q) or streptococci (U, U2, P and H), with nisin H representing the first example from a gastrointestinal tract bacterium [12]. A related nisin homologue has also been identified in the thermophilic bacterium Geobacillus thermodenitrificans [13]. Besides the natural forms of nisin, both random and targeted mutation studies have created libraries of nisin derivatives, with the most notable being a nisin A derivative with an S29G substitution, with enhanced antimicrobial activity against both Gram-positive and Gram-negative pathogens [8]. Hinge region variants such as N20P, M21V [14], K22T [15], N20K and M21K [16] have also led to increases in bioactivity against a range of bacteria.
In addition to the pre-peptide gene nisA, nisin gene clusters typically encode NisB, an enzyme that dehydrates serines and threonines, and NisC, a zinc-dependent metalloprotein that cyclizes dehydrated residues to cysteines, both of which have been shown to be essential for nisin production [17, 18]. In order for these biosynthetic modifications to take place, it is important that the appropriate leader peptide is attached to the bacteriocin precursor. In the case of nisin A, the leader peptide is removed by the protease NisP following transport across the cell membrane by NisT, an ABC transporter that forms a membrane-associated complex with NisB and NisC. Once modified, the mature nisins all contain three dehydrated amino acids and five thioether bridges. NisFEG and NisI control self-immunity, while the two-component sensor histidine kinase system NisRK allows self-induction of the nisA promoter by the mature nisin product [7]. The modified nisin A and U molecules have been shown to have the capacity to act as inducers of their own and each other's promoters, even when attached to their leader peptides [19], while cross-immunity has also been demonstrated between nisins A and Z [20] and A and H [12].
Heterologous expression of lantibiotic clusters and genes, especially when their inducing conditions are not known, has been a powerful aid in the sequencing and characterization of several lantibiotic clusters, such as those of epicidin 280 [21], enterocin A [22] and nukacin ISK-1 [23]. The nisin A biosynthetic cluster has already been expressed successfully in L. lactis and Enterococcus sp. [24]; furthermore, the nisin biosynthetic machinery has been shown to be capable of modifying other non-nisin peptides [25], while the nisin promoter elements have been used extensively for inducible expression of cloned genes [26, 27]. The extreme robustness of the biosynthetic machinery was demonstrated by Majchrzykiewicz et al. [28], who successfully expressed a fully modified and biologically active two-component class II lantibiotic from Streptococcus pneumoniae using the nisin A biosynthetic machinery, while several other studies have demonstrated that the lantibiotic biosynthetic machinery is able to recognize alternative peptides containing the peptide leader sequence of their own lanA gene, and in some cases modify them [29, 30].
The human gut harbours a large number of bacteria, reaching 1012 bacteria per gram of intestinal content, that are diverse in their composition and contain many unknown species; other species, such as Blautia obeum, are recognized as being dominant in the human colon [31]. It is a rich potential source of novel antimicrobials that have evolved to function in the challenging conditions of the gastrointestinal tract, and recent research suggests that bacteriocin production is widespread [32, 33]. Using genome mining of human gut bacteria, new lantibiotic sequences sharing considerable amino acid sequence homology with class AI lantibiotics, and especially nisin U, were discovered from anaerobic bacterium Blautia obeum A2-162. The novel lantibiotic cluster was cloned into L. lactis and evidence of antimicrobial activity and cross-immunity with nisin A was shown.
Methods
Strains, plasmids and growth conditions
The Lactococcus lactis strains and plasmids used in this study are listed in Table 1, while the primers are listed in Table S1 (available in the online Supplementary Material). Ruminococcus obeum A2-162 was previously isolated from human faeces from an adult female consuming a Western-style diet [34–36] and its genome was sequenced as part of the MetaHIT project. R. obeum has subsequently been reclassified as Blautia obeum [37]. B. obeum, C. perfringens NCTC 3110 and C. difficile NCTC 11204 were cultured in pre-reduced brain heart infusion broth (BHI, Oxoid) with complements (50 mg l−1 vitamin K, 5 mg l−1 hemin, 1 mg l−1 resazurin, 0.5 g l−1 l-cysteine) at 37 ˚C in an atmosphere of 5 % CO2, 10 % H2 in N2. L. lactis strains were cultured in M17 medium (Oxoid) supplemented with 5 g l−1 glucose (GM17) at 30 ˚C. E. coli MC1022 was cultured in L medium (Oxoid) at 37 ˚C with shaking. For plasmid selection erythromycin and chloramphenicol were used at 5 µg ml−1 for L. lactis or at 100 and 15 µg ml−1, respectively, for E. coli, and ampicillin was used at 100 µg ml−1.
Table 1. Strains and plasmids used in this study.
| Strains | Relevant characteristics | Reference/source |
|---|---|---|
| L. lactis MG1614 | L. lactis subsp. lactis 712 cured of plasmids and prophage | [71] |
| L. lactis FI5876 | MG1614 with the nisin A biosynthetic cluster | [72] |
| L. lactis FI5876 ΔnisA | Part of nisA deleted (FI7847) | [73] |
| L. lactis FI5876 ΔnisP | nisP deleted (FI8438) | A. Narbad |
| L. lactis FI5876 ΔnisC | nisC deleted (FI8531) | A. Narbad |
| L. lactis FI5876 ΔnisCP | nisC and nisP deleted (FI8532) | A. Narbad |
| L. lactis FI5876 ΔnisB | nisB deleted (FI8620) | [17] |
| L. lactis UKLc10 | nisRK genes integrated on the chromosome | [74] |
| B. obeum A2-162 | Genome mining strain isolated from human GI tract | S. Duncan |
| C. perfringens NCTC 3110 | Indicator strain | National Collection of Type Cultures |
| C. difficile NCTC 11204 | Indicator strain | National Collection of Type Cultures |
| E. coli MC1022 | Shuttle vector cloning strain | [75] |
| Plasmids | ||
| pIL253 | Erythromycin resistance | [76] |
| pJAZZ-OC | Chloramphenicol resistance | (Lucigen Corp, USA) |
| pUK200 | Chloramphenicol resistance | [74] |
| pnisAL-nsoA4 | pUK200 with the nisin leader peptide DNA sequence followed by the nsoA4 DNA sequence under the control of the nisA promoter | This study |
| pnisAL-nsoA1IE | pUK200 with the nisin leader peptide DNA sequence followed by the nsoA1IE DNA sequence under the control of the nisA promoter | This study |
| pnisAL-nsoA1YK | pUK200 with the nisin leader peptide DNA sequence followed by the nsoA1YK DNA sequence under the control of the nisA promoter | This study |
| pnso | nisin O lantibiotic cluster in pIL253 | This study |
| pnsoΔnsoA | nisin O lantibiotic cluster in pIL253 with the nsoA genes deleted | This study |
| pFI2596 | Nisin inducible vector based on pTG262 engineered to contain the nisA promoter and RBS sequences followed by genes encoding the mIL-12 p40 and p35 subunits | [45] |
| pTG262Pn | pFI2596 with mIL-12 removed, empty control vector | This study |
| pTGnsoA1 | pTG262Pn with nsoA1 under the control of the nisA promoter | This study |
| pTGnsoA2 | pTG262Pn with nsoA2 under the control of the nisA promoter | This study |
| pTGnsoA3-nsoA4 | pTG262Pn with nsoA3 and nsoA4 under the control of the nisA promoter | This study |
| pTGnsoI | pTG262Pn with nsoI under the control of the nisA promoter | This study |
| pTGnisI | pTG262Pn with nisI under the control of the nisA promoter | This study |
| pTGnisA | pTG262Pn with nisA under the control of the nisA promoter | This study |
Degenerate oligonucleotide primer design and screening for lanC genes
Genomic DNA from B. obeum A2-162 was extracted using the Qiagen Genomic-tip kit. The degenerate AT-rich primers lanC340 and rlanC460 were designed from the WCYG region (position 294 in SpaC) and the GLIxG region (position 403 in SpaC) of an alignment of the following LanC sequences from the NCBI database: CAA74351 (EciC), AAF99580 (Mut C), CAA48383 (NisC), CAA90026 (PepC) P33115 (SpaC), BAB08164 (SrtC) and P30196 (EpiC), as described previously [38]. The degenerate oligonucleotide primer PCR used GoTaq (Promega) with 10 µM of each degenerate primer, lanC340 and rlanC460. The PCR products were electrophoresed and bands of 200–300 bp were excised and extracted from agarose gels (Qiaex II Gel extraction kit, Qiagen); these were purified using Sureclean (Bioline), ligated into vector pCR2.1 and transformed into chemically competent E. coli TOP10 (TA Cloning Kit, Life Technologies). Positive colonies were identified by colony PCR using GoTaq with universal and reverse primers, and confirmed by sequencing. The sequence was extended using the DNA Walking SpeedUp premix kit (Seegene) and genomic DNA.
DNA library construction and lantibiotic cluster sequencing
Genomic DNA from B. obeum A2-162 was used to construct a DNA library (Lucigen Corp, Middleton, WI, USA) using the E. coli vector pJAZZ-OC. For hybridization analysis of the library, a 736 bp DNA probe comprising bases 15 292 to 16 027 of the lantibiotic cluster sequence (accession number KY914474), which includes the C-terminus of the nsoC gene and downstream sequence, was prepared by PCR using the primers 5PrA2162280 and 3PrA2162c, and purified. Hybridization of the probe to filter membranes arrayed with the library was performed by pretreating the membranes for 2 h with gentle shaking in 5× saline sodium citrate buffer (SSC) [39], 0.5 % SDS and 1 mM EDTA (pH 8.0) at 42 °C, scraping them with wet paper towels and rinsing them in 2× SSC, followed by hybridization using the ECL hybridization kit (GE Healthcare). Positive clones identified from the DNA library were cultured and sequenced using primers pJAZZf and nzrevcpJAZZ. The known sequence was extended from the library clones using primer walking and the gaps were filled by PCR until the lantibiotic cluster had been fully sequenced in both directions.
Bioinformatic analysis
Genomic DNA sequences were assembled with the Phred/Phrap program and contigs were assembled in SeqMan (DNASTAR). ORFs were determined by Artemis [40]. Start sites were selected on best match to the consensus ribosome binding site AGGAGG, where present, and to homologous sequences identified using blastp and tblastx searches [41] using the UniProtKB/TrEMBL database. Amino acid alignments were performed using the CLUSTAL W algorithm in Vector NTI (Invitrogen) and edited in Genedoc; the average distance tree was generated using BLOSUM62 from the CLUSTAL WS alignment of the secondary structure prediction of the peptide sequences [42]. Pairwise cluster comparisons were performed using blastn and tblastx from blastall v 2.2.26. The clusters were aligned using mega-cc 7 with the neighbour-joining method (muscle) and a tree was made using RAxML v 8.2.9 with the BS and ML recommended settings. Cluster comparisons were visualized using the tblastx comparison files and RAxML tree in R v 3.3.2 using the genoPlotR package http://genoplotr.r-forge.r-project.org/.
Expression of the nisin O cluster in L. lactis
A 17 438 bp sequence containing the novel lantibiotic cluster was restricted from the identified pJAZZ-OC clone with ClaI and PstI (NEB) and then ligated into vector pIL253 [MspI, PstI restricted and dephosphorylated (Antarctic Phosphatase, NEB)] using Fastlink DNA ligase (Epicentre) to create pnso. The construct was transformed into electrocompetent L. lactis MG1614 using a Gene Pulse Xcell (BioRad, [43]). Plasmid DNA was extracted using the QIAprep miniprep kit (Qiagen) with an additional 15 min at 37 ˚C with 5 mg ml−1 lysozyme and 30 U mutanolysin (Sigma) at the lysis stage, and the insert was confirmed by sequencing with the primers pIL253F and pIL253R.
The region containing the four nsoA genes was deleted from pnso by splice overlap extension PCR [44] using Phusion (Finnzymes). the sequences surrounding the nsoA region were amplified at the 5′ end using the primers splA1 and splA2 and at the 3′ end with the primers splA3 and splA4. These products were spliced and amplified with the primers splA1 and splA4, giving an amplicon of 4818 bp, which was digested with BsaI and StuI and ligated to restricted, dephosphorylated plasmid pnso to produce plasmid pnsoΔnsoA, which was transformed into L. lactis MG1614. Both pnso and pnsoΔnsoA were also transformed into L. lactis UKLc10.
The nsoA genes were cloned separately into a nisin-inducible expression vector. Each nsoA gene was amplified using the primer combinations pTGA13 with a13AleI for gene nsoA1, pTGA23 with a13AleI for gene nsoA2 and pTGA23 with a43AleI for genes nsoA3–nsoA4 to make amplicons nsoA1Ale, nsoA2Ale and nsoA3nsoA4Ale respectively. The region of plasmid pFI2596 [45] containing the nisin promoter PnisA was amplified using the primers pTG262-F with a15pTG to make amplicon nisPa and pTG262-F with a25pTG to make amplicon nisPb. The PnisA and nsoA amplicons were used as DNA templates in splice overlap extension PCR by combining templates nisPa with nsoA1Ale using the primers pTG262-F with a13AleI and nisPb with nsoA2Ale using the primers pTG262-F and a13AleI, and finally nisPb with nsoA3nsoA4Ale using the pTG262-F and a43AleI. The PnisA-IL12 region of pFI2596 was then replaced with the spliced amplicons containing the nsoA genes by SmaI and AleI digestion and ligation to digested, dephosphorylated plasmid pFI2596. The ligation products were transformed into L. lactis MG1614. Clones with PnisA-nsoA1 (pTGnsoA1), PnisA-nsoA2 (pTGnsoA2) and PnisA-nsoA3–nsoA4 (pTGnsoA3–nsoA4) were identified as described previously using the primers p54 and p181, and transformed into electrocompetent MG1614-pnso, MG1614-pnsoΔnsoA, UKLc10-pnso and UKLc10-pnsoΔnsoA. The nisA gene was also subcloned into pTG262 as a positive control; nisA was amplified from L. lactis FI5876 genomic DNA by PCR using the primers NisA-BspHF and NisA-BspHR, restricted with BspHI and ligated into NcoI-restricted pUK200. After transformation into E. coli MC1022 and sequence confirmation, the insert was then excised with SspI and EcoRI and cloned into HindIII-EcoRI-restricted pTG262.
Cloning of hybrid nisAL–nsoA genes into the nisin A biosynthetic system
Hybrid nisAL–nsoA pre-peptides were designed to contain the full NisA leader sequence MSTKDFNLDLVSVSKKDSGASPR (nisAL), followed by the predicted NsoA structural peptides of nsoA1 with possible cleavage sites: nsoA1IE: IEPKYKSKSACTPGCPTGILMTCPLKTATCGCHITGK, nso A1YK: YKSKSACTPGCPTGILMTCPLKTATCGCHITGK or nsoA4: ITSQHSFCTPNCLTGFLCPPKTQLTCTCKLKGQ. The 69 bp nisAL DNA was amplified from L. lactis FI5876 genomic DNA using the primer pr1 combined with each primer 1pr2ie, 1pr2yk or 4pr2 to make nisALIE, nisALYK and nisAL4 amplicons, respectively, and the nsoA1 and nsoA4 structural genes were amplified from plasmid DNA containing the full lantibiotic cluster using the primer 1Pr4 combined with 1Pr3IE, 1Pr3YK or 4Pr3 to make amplicons nsoA1IE, nsoA1YK and nsoA4, respectively. Each hybrid nisAL–nsoA was prepared by splice overlap extension PCR using the template sets nisALIE with nsoA1IE and nisALYK with nsoA1YK with the primers pr1 and lanA14 to make nisAL–nsoA1IE and nisAL–nsoA1YK, respectively, and templates nisAL4 with nsoA4 with the primers pr1 and LanA44 to make nisAL-nsoA4. Purified PCR products were digested with BspHI and XbaI and ligated into NcoI- and XbaI-restricted, dephosphorylated pUK200, and the products were transformed into electrocompetent E. coli MC1022. After sequence confirmation using primers p54 and p181, plasmid DNA from positive clones and the pUK200 vector control was transformed into L. lactis strains FI5876ΔnisA, FI5876ΔnisP, FI5876ΔnisB, FI5876ΔnisC and FI5 876ΔnisCP.
Construction of nsoI and nisI expression vectors
The nsoI gene was amplified from pnso using the primers pTGI3 with iAleIb, and the nisin promoter region of plasmid pFI2596 was amplified with the primers i5pTG and pTG262-F. Splice overlap extension PCR was performed using pTG262-F with iAleIb, the product was ligated into SmaI- and AleI-digested, dephosphorylated pFI2596, and transformed into L. lactis MG1614. Inserts were confirmed by sequencing using the primers pTG262-F and pTG262-R. The nisI gene was amplified from L. lactis FI5876 genomic DNA using the primers spI5 and splI3AleI, and the nisin promoter region of plasmid pFI2596 was prepared using the primers splA1 and pTG262-F. These two amplicons were spliced and amplified using pTG262-F with iAleIb and ligated into vector pFI2596 as described for nsoI.
Measurement of bacterial growth, viability and phenotype
L. lactis FI5876ΔnisA pnisAL–nsoA1YK, L. lactis FI5876ΔnisA pUK200 and L. lactis FI5876ΔnisP pnisAL–nsoA1YK were subcultured from overnight cultures in selective medium and at 2 h were induced with 10 ng ml−1 nisin A. After 18 h growth the cultured cells were prepared for scanning and transmission electron microscopy (SEM and TEM) analysis as described previously [46, 47]. Samples were examined and imaged in a FEI Tecnai G2 20 transmission electron microscope at 200 kV. Bacterial growth was measured using a Labsystems Bioscreen C (Labsystems Oy). Test cultures were subcultured twice from glycerol stocks and induced appropriately overnight for 16 h. Cells were pelleted by centrifugation (10 000 g, 10 min), resuspended in 1 ml PBS, pelleted again and resuspended to an optical density (OD600) of 3.0 in selective medium. Bioscreen plates (honeycomb; Thermo Fisher Scientific) were prepared with 300 µl medium per well, seeded with 1 % of the prepared inoculum in triplicate and grown at 30 ˚C. To measure viability, stationary phase L. lactis strains were washed and resuspended in PBS and then diluted 500-fold in filter-sterilized PBS with 1 µl each of propidium iodide and FM 4-64FX (Life Technologies) before being analysed on a Cytomics FC500 MPL (Beckman Coulter). Flow cytometry data were analysed using Flowjo (Treestar).
Preparation and analysis of protein extracts
Pre-warmed medium was inoculated with 1 : 100 v/v overnight culture of L. lactis strains expressing hybrid plasmids. At the mid-exponential phase the cultures were induced with 10 ng ml−1 nisin and incubated from 2 h to overnight. Cells were harvested by centrifugation (4000 g, 40 min, 4 ˚C) and frozen while total protein from filtered (0.45 µm) culture supernatants was precipitated by adding 1 g ml−1 (nisin leader hybrids) or 20 % (nsoA pre-peptides) cold trichloroacetic acid (TCA) and incubating overnight at 4 ˚C. Precipitated proteins were pelleted by centrifugation (13000 g, 30 min, 4 ˚C), washed with ice-cold acetone and resuspended in 0.05 volume 50 mM sodium acetate (pH 5.5). Cells were resuspended in 50 mM sodium acetate (pH 5.5) (MG1614) or 0.2 M Tris HCl (pH 7.4) (UKLc10) and soluble protein extracts produced by bead beating [43]. Proteins were analysed by SDS-PAGE electrophoresis and Western blotting as described previously [43] using 12 % or 4–12 % Bis-Tris NuPAGE gels in MES SDS buffer (Invitrogen) and an antibody (at 1/100 dilution) raised against the nisin A leader peptide or a polyclonal anti-leader peptide antibody raised by Genscript Corp. (NJ, USA) from synthesized N-terminal acetylated NsoA1 leader peptide H2N-AKFDDFDLDVTKTAAQGGC-CONH2 with anti-rabbit IgG-alkaline phosphatase secondary antibody (Sigma).
Antimicrobial assays and optimization
To measure antimicrobial activity using drop tests, strains were cultured from glycerol stocks in the appropriate medium overnight and subcultured twice before the appropriate medium were inoculated. For overlay assays, B. obeum A2-162 was sub-cultured twice in liquid medium before plating or streaking on solid medium or solid medium supplemented with 50 µg ml−1 trypsin. After 1 to 7 d the incubation cultures were killed using chloroform treatment, overlaid with soft medium (0.7 % w/v agar) seeded with indicator bacteria and incubated overnight. For heterologous production, L. lactis strains were sub-cultured with 10 ng ml−1 nisin A, then 5 µl overnight culture spotted on solid agar with 20 mg l−1 NaHCO3 and 10 ng ml−1 nisin, and grown overnight. Bacterial growth was killed by irradiation with UV light for 15 min before overlaying with soft medium containing 1–2 % of an overnight culture of the indicator strain, with or without trypsin (sequencing grade-modified trypsin, Promega, at 1 or 5 µg ml−1, or Tpck-treated trypsin from bovine pancreas, Sigma, at 1, 5, 10 or 15 µg ml−1). Plates were incubated overnight in the conditions preferred by the indicator strain.
To attempt to induce antimicrobial production, B. obeum A2-162 and L. lactis MG1614 and FI5876 were cultured from glycerol stocks in selective medium overnight and then subcultured in a range of test media (BHI pH 5.0, 6.0 and 7.0; BHI with 5 mg l−1 hemin; YCFA medium; PYGS medium; de Man Rogosa Sharp medium; reinforced clostridial medium; Luria broth; lysogeny broth; Rogosa; GM17; M17 supplemented with 5 % of each of lactose, mannitol, cellobiose, mannose, sorbitol, galactose, xylose or inulin, all with and without 50 µg ml−1 trypsin) and inducing agents. The inducing agents included 5 g l−1 yeast extract, a mixture of 2 g l−1 glucose, 1 g l−1 soluble starch and 2 g l−1 cellobiose, a mixture of 2 g l−1 xylose, cellobiose and sorbitol, 2 g l−1 inulin, 2 g l−1 sodium acetate 3-hydrate, 0.31 % volatile fatty acids mix (33 mM acetic acid, 9 mM propionic acid, 1 mM n-valeric acid, 1 mM isovaleric acid and 1 mM isobutyric acid), 50 µg ml−1 trypsin, heat-killed C. perfringens culture, 10 to 1000 ng ml−1 nisin A, and combined trypsin and nisin. Filtered (0.22 µm) stationary-phase culture supernatants from L. lactis strains MG1614-pnso, MG1614-pIL253 or FI5876 with and without nisin (1 : 20 v/v) and L. lactis cell extracts in 50 mM NaOAc or 50 mM NaOAc, 8 M urea buffer and supernatant TCA-extracted proteins were also tested as inducers of activity in B. obeum A2-162, while spent culture of B. obeum was similarly tested as an additive to nisin-induced MG1614-pnso culture. Samples were tested for antimicrobial activity using well diffusion, drop tests and overlay assays [48].
To remove leader peptides, soluble cell extracts (2 µg) and TCA-precipitated culture supernatant extracts from B. obeum A2-162 and L. lactis strains were digested with 0.5 mg ml−1 trypsin for 1 h at 37 ˚C in 50 mM sodium acetate buffer (pH 5.5) [49], or subcultured to medium containing 2.5 µg ml−1 nisin A and 1 : 40 v/v filter-sterilized culture supernatant of Bacillus subtilis. Samples were assayed for antimicrobial activity using well diffusion assays. Additionally, L. lactis MG1614-pnso and its derivative strains were cultured with 100 ng ml−1 of inducing nisin A and cross-streaked with B. subtilis, grown overnight and then overlaid with C. perfringens.
Results
B. obeum A2-162 contains a lantibiotic-like gene cluster
Previous studies have shown that novel lantibiotic genes can be identified by PCR using degenerate primers designed from conserved regions of lantibiotic cluster genes [19, 38]. Here, AT-rich degenerate primers were designed and used to screen a bacterium previously isolated from the human GI tract for lanC-like sequences. We identified a 180 bp DNA sequence from B. obeum A2-162, whose translated product aligned with other LanC proteins. The sequence was extended in both directions and a full lanC-like gene as well as part of a lanT-like gene indicated that the genes belonged to a lantibiotic cluster. A DNA library was prepared using the E. coli linear vector pJAZZ-OC and the surrounding genes were sequenced in both directions by primer walking, identifying a c. 15 kb lantibiotic cluster within a c. 19 kb insert. Comparison to clusters of other nisins and subtilin suggested that the full cluster had been identified. Later publication of the whole draft genome (GenBank FP929054, A. Pajon, K. Turner, J. Parkhill, S. Duncan and H. Flint, unpublished) confirmed this.
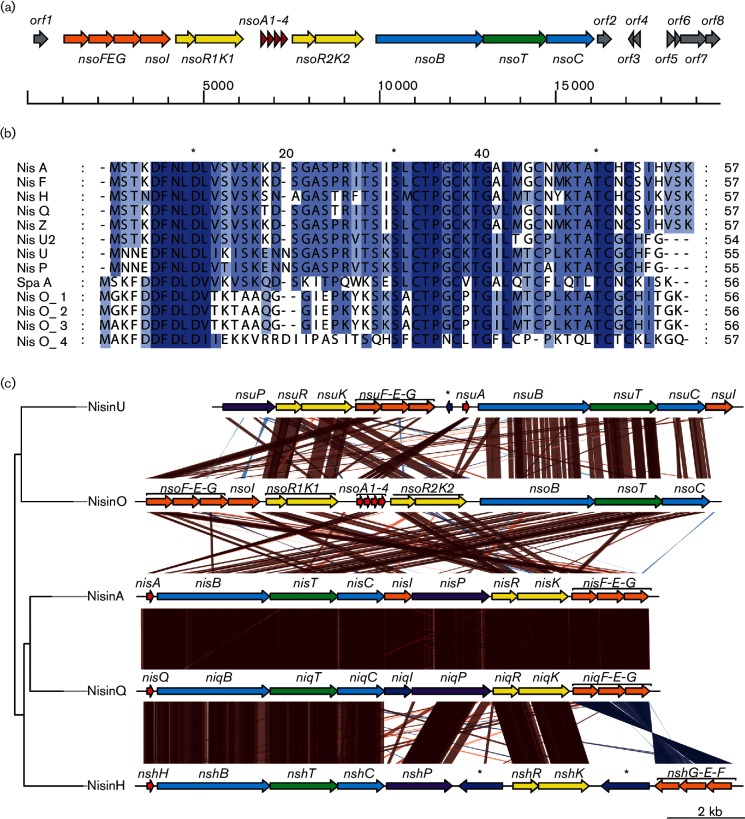
Computational analysis of the cluster identified 15 probable ORFs encoding lantibiotic–associated genes (abbreviated to nso here), whose functions were predicted by blastp analysis (Fig. 1a and Table S2). The genes all had the same orientation and included four nsoA genes, with the first three coding for identical proteins with only one amino acid difference in the leader peptide (Fig. 1b), nsoB and nsoC biosynthetic genes, an nsoT ABC transporter, two sets of nsoRK two-component regulator system genes and nsoI and nsoFEG genes presumed to be involved in immunity. No probable protease genes capable of cleaving the leader sequence were identified; the predicted NsoT protein showed similarity to other NisT-type lantibiotic ABC transporter ATP-binding proteins and did not contain the N-terminal protease domain responsible for leader cleavage in dual-function ABC transporters, which is frequently found in lantibiotic clusters that do not contain a lanP gene. A 506 bp region between the end of nsoK1 and the start of nsoA1 genes displayed no homologies to other known genes commonly found in lantibiotic clusters.
Fig. 1.
(a) Organization of the nisin O lantibiotic cluster and surrounding ORFs from the library clone. (b) Amino acid sequence alignment of the translated nsoA1, nsoA2, nsoA3 and nsoA4 genes to subtilin (SpaA, P10946), nisin Z (CAA79467), nisin U (ABA00878), nisin U2 (ADB43138), nisin F (ABU45463), nisin A (AAA25188), nisin Q (BAG71479), nisin P (BAK30164) and nisin H (AKB95119). Blue, mid-blue, light blue and white correspond to conservation of 100, 80, 60 and <40 %, respectively. (c) Similarity of the nisin O lantibiotic cluster to other lantibiotic clusters from nisin U (DQ146939), nisin A (HM219853), nisin Q (AB362350) and nisin H (KP793707). *, transposase or insertion element sequence.
The predicted mature protein sequence encoded by nsoA1 was found to be very similar to other NisA proteins, with conservation of the positions of the serine, threonine and cysteine residues; the predicted mature protein sequence of nsoA4 showed a lower similarity but had the majority of serine, threonine and cysteine residues in similar positions to other nisin analogues (Fig. 1b). The spaces separating the nsoA genes were 22, 23 and 19 bp, with the first two gaps exhibiting 95 % identity. blastp searches categorized the three NsoA1-3 pre-peptides as part of the gallidermin/nisin family, with 63 % identity to the nisin U precursor peptide. The N-terminal region of the NsoA1-3 pre-peptides contained an FDLD motif followed by a GG motif and a further PK motif. The most likely leader peptide cleavage sites were therefore presumed to be following either the GG or the PK motif, and the two predicted structural peptides are referred to as NsoA1IE and NsoA1YK, respectively. These exhibited 82 and 90 % sequence identity to nisin U, respectively (Fig. 1b). Only an FDLD leader peptide motif was identified in the N-terminal region of the NsoA4 pre-peptide and an ITS amino acid sequence resembling that of the start of the active nisin A was found in a similar region of the pre-peptide. The predicted NsoA4 structural peptide is one amino acid shorter than nisin A and showed the best similarity to geobacillin I (59 % identity).
blastp analysis of an ORF at the 5′ end of the cluster (orf1) showed sequence similarity to the second half of a transcriptional regulator from B. obeum (Fig. 1a and Table S2); the preceding nucleotide sequence encoded the earlier part of the protein but contained frame shifts. At the 3′ end of the cluster, orf2 showed sequence similarity to hypothetical proteins from several Clostridiales species and to a transposase from an uncultured faecal bacterium (AMP50088, 2e−52). This was followed by sequences that matched short regions of database proteins with the sequence interrupted by frameshifts – orf3 and orf4 had similarity to consecutive regions from a transposase from Blautia wexlerae and to other transposases from a range of other Clostridiales bacteria, while orf5 and orf6 matched consecutive regions of a putative transcriptional regulator. The first seemingly complete protein is orf7, which showed up to 91 % sequence identity with DNA-binding response regulators; orf8 is truncated by the end of the clone and has similarity to ATP-binding proteins/sensory histidine kinases.
The nisin O cluster may have evolved from the nisin U cluster
blastn comparison of the full nisin O cluster to those of the other nisins did not show considerable sequence conservation, but tblastx showed a high similarity between the nisin O cluster and nisins U, A and Q (Fig. 1c). It was interesting to note that the GC percentage of the three highly similar nsoA genes was higher than that of the fourth structural gene and the rest of the cluster (average GC content 31 %) and the producing organism, B. obeum A2-162 (average 41.6 % GC).
Comparison of all nisin and B. obeum A2-162 clusters showed that the clusters of nisins Q and A were highly conserved, while there appeared to be a translocation event between the nisin A and nisin U clusters that was also present in the nisin O cluster. The 3′ end of the nisin U cluster containing nsuA, nsuB, nsuT and nsuC was conserved in the nisin O cluster, while the 5′ end of the cluster containing genes nsuP, nsuRK and nsuFEG showed evidence of a few translocations of genes within the clusters. A blastn search of the region between nsuA and nsuG in the nisin U cluster showed the presence of a ISSmu4-like putative transposase sequence (DQ368682) within the region. However, blastp analysis showed that the Nso translated proteins share the highest sequence identity with proteins from other Clostridiales and Bacillales bacteria, frequently from faecal sources, so any evolution from the nisin U operon in Streptococcus uberis would appear to be ancient.
B. obeum A2-162 exhibits trypsin-induced antimicrobial production on solid medium
A range of growth conditions, culture media and media additives, which included supernatants or cell extracts from spent cultures, were tested for their ability to induce antimicrobial production in B. obeum A2-162. Of these, only those cultures grown for at least 4 days in liquid culture and then plated on solid medium supplemented with 50 µg ml−1 trypsin before overlaying reproducibly showed evidence of antimicrobial production against the indicator strain C. perfringens (Fig. 2); no antimicrobial activity was detectable from culture supernatants by drop tests.
Fig. 2.
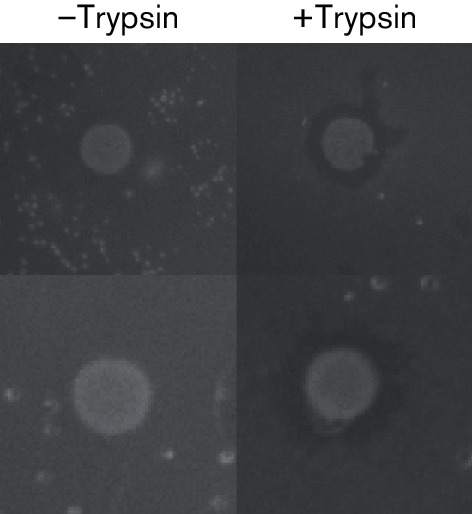
Antimicrobial activity. Overlay assays of B. obeum A2-162 after 6 d (top) or 7 d growth (bottom), grown on solid medium or solid medium supplemented with 50 µg ml−1 trypsin and overlaid with C. perfringens.
Hybrid NisAL–NsoA peptides alter the phenotype of L. lactis when they are expressed in the presence of the nisin modification machinery
To investigate whether the nisin A biosynthetic cluster could modify and produce active NsoA peptides, the predicted leader of each NsoA pre-peptide was replaced with that of nisin A (nisAL) and the hybrid nisAL–nsoA1IE, nisAL–nsoA1YK and nisAL–nsoA4 genes were expressed from vector pUK200 in L. lactis strains FI5876ΔnisA, FI5876ΔnisP, FI5876ΔnisB, FI5876ΔnisC and FI5876ΔnisCP.
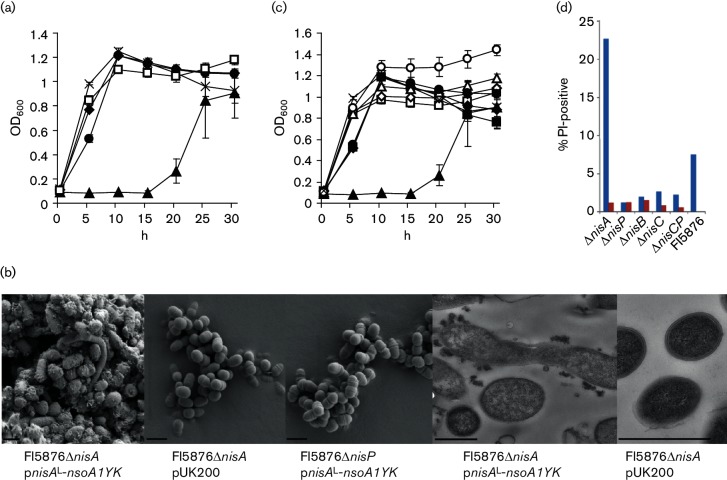
Although plasmids expressing nisAL–nsoA1IE and nisAL–nsoA4 had no effect on growth, strain FI5876ΔnisA pnisAL-nsoA1YK exhibited a longer lag phase and reached lower maximum OD600 values (Fig. 3a). This strain displayed an aggregated phenotype in liquid culture, and TEM and SEM revealed loss of cell shape, extensive aggregation and less defined cell membranes (Fig. 3b), suggesting problems with membrane synthesis or stability. The nisin biosynthetic gene knockout strains FI5876ΔnisB, FI5876ΔnisC and FI5876ΔnisP containing the hybrid plasmid pnisAL–nsoA1YK were not phenotypically different from their empty vector control counterparts and showed similar growth rates, suggesting that the nisin biosynthetic genes were necessary for the slow growth phenotype (Fig. 3c). These results were supported by flow cytometry analysis of stationary-phase cells, showing that FI5876ΔnisA pnisAL-nsoA1YK had an increased percentage of PI-positive cells (Fig. 3d).
Fig. 3.
Effect of hybrid genes on growth and phenotype. (a) Effect of hybrid constructs on the growth of L. lactis. X, FI5876; ●, FI5876ΔnisA pnisAL-nsoA1IE; ▲, FI5876ΔnisA pnisAL-nsoA1YK; ♦, FI5876ΔnisA pnisAL-nsoA4; □, FI5876ΔnisA pUK200. Results are the mean of triplicate measurements ±SD. (b) SEM (left) and TEM (right) analysis of the effect of hybrid construct expression on cell phenotype. Bar, 1 µm. (c) Growth of FI5876ΔnisA (▲), FI5876ΔnisP (●), FI5876ΔnisC (♦) and FI5876ΔnisB (■) containing pnisAL-nsoA1YK (closed symbols) or pUK200 (open symbols). X, FI5876. (d) Viability of stationary-phase L. lactis FI5876 and knockout strains containing pnisAL-nsoA1YK (red) or pUK200 (blue).
Despite the altered growth and phenotype, the strains expressing pnisAL–nsoA hybrids showed no evidence of antimicrobial activity after a number of different antimicrobial detection tests, which included the use of trypsin or filtered culture supernatant as inducers (data not shown). However, using an antibody to the nisin A leader, Western analysis of FI5876ΔnisA containing pnisAL-nsoA1IE, pnisAL-nsoA1YK, pnisAL-nsoA4 or pUK200 identified a band at c. 6 kDa in the FI5876ΔnisA pnisAL-nsoA1YK samples (Fig. 4a). The absence of this band in the other strains suggests that NisAL–NsoA1IE or NisAL–NsoA4 are either not produced or are not modified, causing instability and the rapid degradation of the produced pre-peptides. Examination of nisin biosynthetic gene knockout strains demonstrated that deletions in nisB, nisC or nisCP prevented accumulation of the NisAL–NsoA1YK pre-peptide (Fig. 4b). Western analysis did not detect any cleaved nisin A leader, but it did show the presence of the pre-peptide in TCA-precipitated culture supernatants (Fig. 4a), suggesting that the pre-peptide was either exported or released from damaged or lysed cells during culture.
Fig. 4.
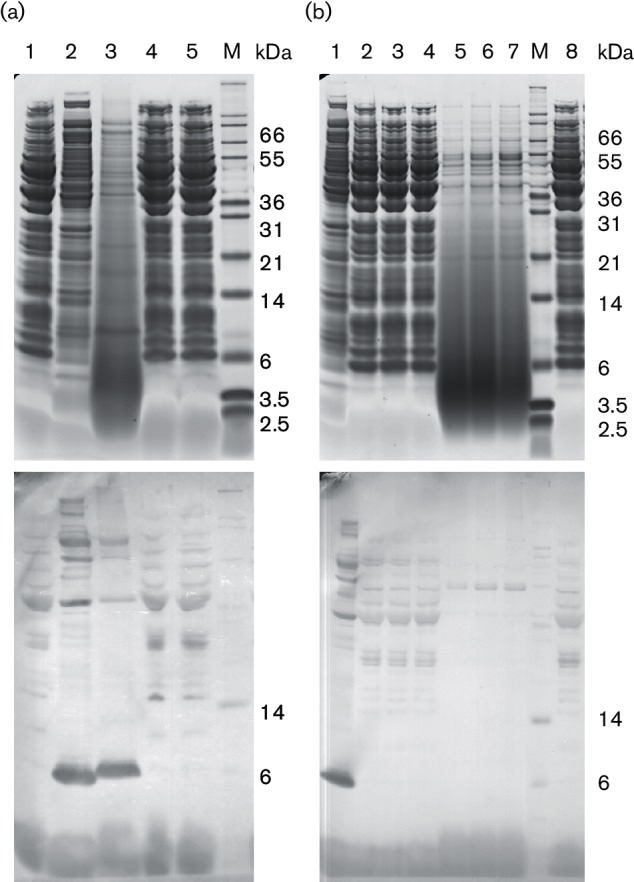
Expression of nisin leader hybrids in L. lactis. SDS-PAGE electrophoresis (top) and Western hybridization (bottom) using the nisin A leader antibody. (a) Extracts from cells (lanes 1, 2, 4 and 5) and TCA-precipitated culture supernatant (lane 3) from FI5876ΔnisA containing pnisAL-nsoA1IE (lane 1), pnisAL-nsoA1YK (lanes 2 and 3), pnisAL-nsoA4 (lane 4) or pUK200 (lane 5). (b) Extracts from cells (lanes 1–4 and 8) and TCA-precipitated culture supernatant (lanes 5–7) from FI5876 biosynthetic gene knockout strains containing either pnisAL-nsoA1YK (lanes 1–7) or pUK200 (lane 8). Lanes 1 and 8, FI5876ΔnisA; lanes 2 and 5, FI5876ΔnisB; lanes 3 and 6, FI5876ΔnisC; lanes 4 and 7, FI5876ΔnisCP. M, marker.
NsoA production in the presence of the nisin O biosynthetic machinery in L. lactis
We inserted a 17 438 bp sequence containing the lantibiotic cluster into pIL253 to create plasmid pnso and transformed it into the non-nisin producer L. lactis MG1614. Initial antimicrobial testing of this strain using deferred antagonism tests did not identify any antimicrobial activity. However, a high level of resistance to nisin A was observed in both MG1614-pnso and in a strain where the four nsoA genes had been deleted (MG1614-pnsoΔnsoA) (Fig. 5). The putative nsoI gene and the nisin immunity gene nisI were expressed in MG1614 separately; although pTGnisI increased the immunity of MG1614 to nisin A, pTGnsoI did not (data not shown). Nisin A was detrimental to B. obeum A2-162 at concentrations above 100 ng ml−1, suggesting that the immunity systems conferring resistance to the MG1614-pnso strain were not being expressed.
Fig. 5.
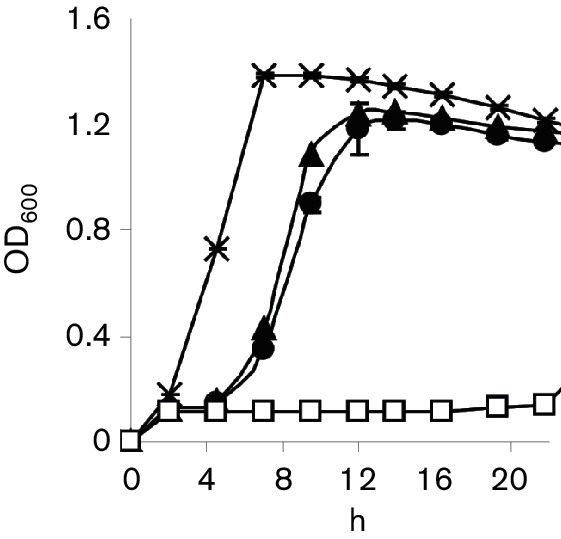
Effect of nisin O genes on immunity to nisin. Growth of L. lactis strains FI5876 (X), and MG1614 with plasmids pIL253 (□), pnso (▲) or pnsoΔnsoA (●) in selective medium supplemented with 1 µg ml−1 nisin A. Results are the mean of triplicate samples±sd
We hypothesized that functional similarities between nisin O and nisin A clusters might allow one of the nsoRK systems to interact with nisin A to induce expression via the nisin A promoter. Genes nsoA1, nsoA2 and nsoA3-nsoA4 were inserted into vector pTG262Pn under the control of the nisin A promoter and co-expressed in MG1614-pnso and MG1614-pnsoΔnsoA with nisin A induction. Western blot analysis with a peptide antibody made to the NsoA1 leader showed hybridization at c. 6 kDa to the MG1614-pnso pTGnsoA3-nsoA4 samples (Fig. 6). There was also faint hybridization to extracts from MG1614-pnso containing pTG262Pn. The 6 kDa band was not detectable in any of the strains expressing just the A1 or A2 sequences, MG1614-pnsoΔnsoA samples, the original producer B. obeum A2-162 or nisin producer FI5876. It was not possible to identify a cleaved leader at c. 2 kDa (cleaving at GG/IE) or c. 2.5 kDa (cleaving at PK) in these cell extracts, possibly due to high background hybridization in this size range or instability of the cleaved leader peptide. None of these cell extracts produced antimicrobial activity. Attempts to cleave the leader peptide and release an active antimicrobial from these strains using treatment of culture supernatants with trypsin, or culture supernatant from B. subtilis, which is known to produce extracellular proteases, or by co-culturing with B. subtilis, failed to produce antimicrobial activity against C. perfringens (data not shown).
Fig. 6.
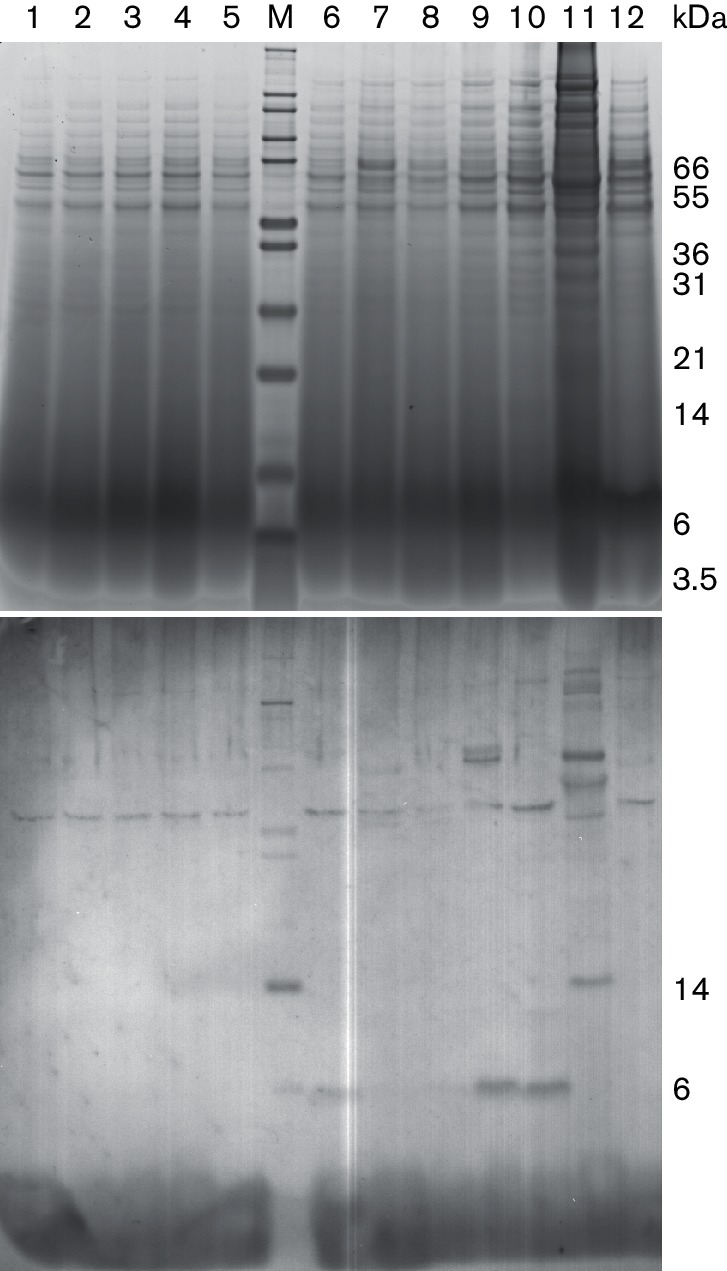
Heterologous expression of the nso cluster. SDS-PAGE analysis (top) and Western hybridization (bottom) using the NsoA1 leader antibody. Comparison of L. lactis TCA-precipitated culture supernatant extracts from MG1614-pnsoΔnsoA (lanes 1 to 5) or MG1614-pnso (lanes 6–10) containing plasmids pTG262Pn (1 and 6), pTGnsoA1 (2 and 7) pTGnsoA2 (3 and 8), pTGnsoA3-nsoA4 (4, 5, 9 and 10) and cell extracts from B. obeum A2-162 (11) and FI5876 (12). M, marker.
Antimicrobial activity after nisin A induction and trypsin treatment
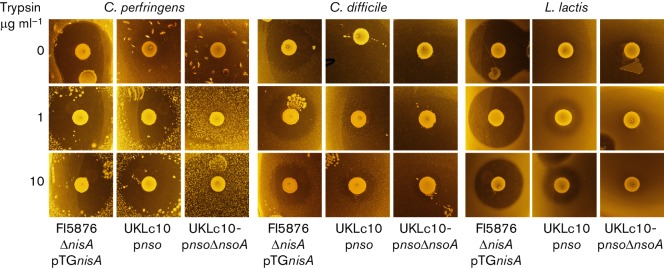
To investigate whether nisin A could act as a heterologous inducer of the nso pre-peptides, pnso or pnsoΔnsoA were expressed in L. lactis UKLc10, which has the nisRK genes integrated in the chromosome, and cultures were induced with 10 ng µl−1 nisin. Western analysis showed improved production of the pre-peptide in both cell extracts and TCA-precipitated supernatants from cultures after nisin induction (Fig. 7). The culture supernatants from these strains did not exhibit antimicrobial activity. However, when strains were grown on solid medium containing nisin A and then overlaid with indicator strains in soft agar containing trypsin, clear activity was seen against C. perfringens, C. difficile and L. lactis (Fig. 8). These zones of inhibition were absent when trypsin was not added to the soft agar, but were evident in the presence of 1, 5, 10 and 15 ng µl−1 trypsin. The activity seen from the positive nisin A control strain FI5876ΔnisA pTGnisA was maintained in the presence of trypsin.
Fig. 7.
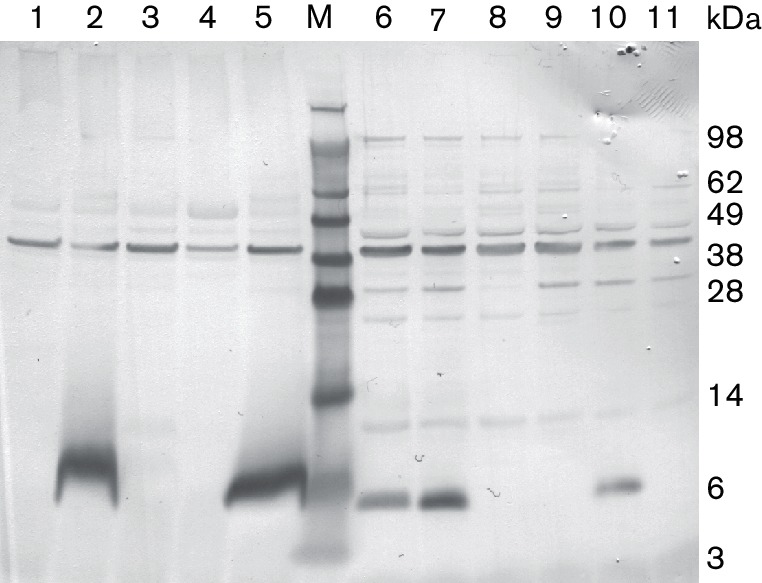
Western hybridization using the NsoA1 leader antibody to detect pre-peptide production in UKLc10. Comparison of L. lactis TCA-precipitated culture supernatant extracts (lanes 1–5) or cell extracts (lanes 6–11) from UKLc10 (lanes 1, 2, 4–8, 10 and 11) or MG1614 (lanes 3 and 9) containing plasmids pnsoAΔA (lanes 1 and 11), pnsoA (lanes 2, 6 and 10), pnsoAΔA, pTGnsoA3-nsoA4 (lanes 3 and 9), pIL253 (lanes 4 and 8) and pnsoA pTGnsoA3-nsoA4 (lanes 5 and 7). Samples were induced with nisin for 3 h, except for lane 6 (2 h). M, marker.
Fig. 8.
Antimicrobial activity in the presence of trypsin. Overlay assays of L. lactis strains grown on solid agar with NaHCO3 and 10 ng ml−1 nisin, and then overlaid with soft agar with or without trypsin and the indicator strain.
Discussion
In this work a novel type A lantibiotic cluster with a unique gene arrangement was discovered in the genome of B. obeum A2-162 and heterologous production of the structural peptides in L. lactis was investigated using either the native or the nisin A biosynthetic machinery. According to Sahl et al. [50], lantibiotic natural variants can be defined as having only a few amino acid substitutions, essentially the same ring pattern, and cross-immunity between producing strains. The novel cluster contained a triplicate structural peptide that showed close sequence similarity to other nisins and conservation of the predicted ring positions, while the lantibiotic cluster provided immunity to exogenous nisin A. Consequently, the predicted lantibiotic was regarded as a member of the nisin group and named nisin O. However, it was interesting to note that the native producer B. obeum showed sensitivity to nisin A, suggesting that the immunity system may require induction. O’Connor et al. [12] also found that the native nisin U-producing strain S. uberis was inhibited by supernatant from a nisin A producer.
The nisin O cluster is unusual in that it is the first nisin cluster to have more than one copy of a nisin-like structural gene, two sets of lanRK genes and no identifiable protease. Differences in nisin cluster gene arrangements have been described before [12, 19, 51] and have been proposed to be a consequence of horizontal gene transfer, but up to now only nisin H has been found to be different in its gene content, with the absence of a detectable nisI [12]. At the pre-peptide level, the nsoA genes deviated from the conserved leader peptide and cleavage sequences found in other nisA genes. Class I lantibiotic leader peptides share conserved F(N/D)LD boxes and C-terminal PQ or PR amino acid sequences, while class II lantibiotic leader peptides contain the motif ELXXBXG (B=V,L or I) and usually end in a GG motif [52]; only the F(N/D)LD box was present in all the NsoA leader peptides.
This is also the first report of a nisin-like cluster in the genus Blautia. B. obeum-like organisms can make up a significant percentage of the faecal microbiome [37]. Increased levels of Blautia in the human gut have been associated with a reduced risk of death from graft-versus-host disease [53], as well as good cognition and reduced inflammation [54], while decreased levels have been associated with the occurrence of type I diabetes in children [55] and increased risk of colorectal cancer [56]. The amount of influence and the mechanisms that lie behind the associations of intestinal Blautia with these conditions, and whether lantibiotic production is important to their ecology, are currently unknown.
The nisins discovered to date are produced by L. lactis (A, Z, F and Q), Streptococcus uberis (U), Streptococcus agalactiae (U2), Streptococcus gallolyticus and Streptococcus suis (P), and, more recently, another gut-derived strain, Streptococcus hyointestinales (H) [12]. An in silico study of the genomes of gut bacteria from the Human Microbiome Project identified lantibiotic-associated genes from a range of genera, including R. obeum A2-162 [33]. Other Clostridiales have been shown to produce the bacteriocins albusin B, a type III bacteriocin [57], and the lantibiotics ruminococcin A and ruminococcin C [58, 59], but these were not found to have any sequence similarities to the nisin O cluster. The discrepancy between the GC content of the structural gene region, the remaining cluster and the producer organism, and the presence of transposase-like sequences at the 3′ end of the cluster could signify that some ORFs have been acquired by horizontal gene transfer. The high gene and intergenic sequence similarity between the nsoA1 genes suggests that the triplication occurred by consecutive duplication events. This is not unprecedented – ruminococcin A, also found in the gut and induced by trypsin, contains three rumA genes in its cluster that code for the same peptide [60]. Two-component lantibiotics that contain two active structural genes are not uncommon [61]. McAuliffe et al. [62] observed that in most cases the sequence of two pre-peptides in two-component lantibiotics is c. 25 % conserved, while many contain different enzymes for the post-translational modification of each peptide. It is not known whether nsoA4 encodes a functional lantibiotic peptide that is active on its own or in combination with nsoA123 peptides – further work in heterologous systems or the original host is required to determine its contribution.
Several lantibiotics have been produced successfully using the nisin A biosynthetic machinery [28, 63]. Slow growth, an altered phenotype and reduced viability effects in nisin leader hybrid-expressing strains suggest that the NsoA1YK peptide can be stably expressed, but this is detrimental to L. lactis in the presence of the nisin biosynthetic machinery. These effects, combined with the visualization of hybrid pre-peptides, suggest that the YK site is the correct start of the mature NsoA1. However, despite extensive experimentation using extracts from B. obeum A2-162 and hybrid NisAL–NsoA-producing strains, we did not identify any inducing agents able to produce antimicrobial activity in liquid culture. This suggests that the pre-peptides are produced but not cleaved to the active product. As with subtilin [64, 65] and mutacin I [66], an extracellular protease encoded elsewhere in the genome might be necessary for cleavage of the NsoA leader peptides to activate the B. obeum A2-162 lantibiotics, and under the culture conditions used this protease was either not expressed from the native strain or was not effective. The differences in the NsoA leader peptides and the starts of the active peptides compared to other nisin analogues support the hypothesis that processing uses a different type of protease. Experiments using trypsin, filter-sterilized B. subtilis spent culture supernatants or co-culturing with B. subtilis strains before overlaying with C. perfringens did not show reliable evidence of antimicrobial activity. However, antimicrobial activity against C. perfringens was observed when B. obeum A2-16 was cultured on solid medium with trypsin. Lantibiotic regulation by trypsin has been seen before with ruminococcin A, a response that suggests adaption to its environment in the gut [67]. Given the presence of two nsoRK systems and the low antimicrobial production it could be that a further inducing factor is involved in regulation in the native host. This factor and/or the antimicrobial itself may be expressed in low quantities by B. obeum A2-162 and could be concentrated around the culture in solid medium, but would be too dilute in liquid medium, explaining our inability to detect antimicrobial activity from culture supernatants. The yield of nisin H in culture supernatants from gut bacterium S. hyointestinalis was also found to be low compared to that of nisin A [12]. Production of mutacin I, the Bifidobacterium longum DJO10A lantibiotic and the two component haloduracin from Bacillus halodurans was also only seen on solid media [66, 68, 69], and it has been proposed that the dense colonization necessary for mutacin I production is reminiscent of a biofilm condition [66]. A putative lantibiotic cluster in Streptococcus pneumoniae has recently been shown to be controlled by quorum sensing, with expression being induced at high cell densities and depending on the carbon source [70]. Alternatively, the mechanism of trypsin may rely on pre-peptide cleavage rather than induction; in vitro biosynthesis of nisin using just nisABC successfully produced active nisin after treatment with trypsin [49]. Trypsin is known to cleave after arginine or lysine residues and there is a lysine immediately before the proposed NsoA1,2,3 YK peptides, so trypsin activity could be generating the mature peptide in the absence of a suitable host protease, as appeared to be the case where trypsin was included in the soft agar of overlay assays of the nso cluster in L. lactis. In either case, trypsin could be a useful tool to identify novel lantibiotic activity from gut bacteria.
The nso cluster was able to confer immunity to nisin A in L. lactis MG1614-pnso, and the use of nisin A to induce coexpression of nsoA genes allowed the visualization of bands that hybridized to the NsoA1 leader antibody. As nisin variants have been shown to induce the production of alternative nisin genes [19], we investigated whether nisin A was able to induce nso gene expression using a strain with the NisRK two-component regulatory system integrated into the chromosome. This increased production to levels high enough to identify antimicrobial activity, as long as trypsin was present in the overlaying agar, presumably to release the active peptide from the leader sequence. Given that the full peptide is expected to be only a small fraction of the peptides generated by trypsin digestion, the resultant activity is impressive and suggests that further understanding and production of this lantibiotic could provide a novel weapon against clostridial pathogens. Future production of mature peptides may allow us to test whether, like nisin A, the system is self-regulating and can be induced in the original host strain to produce the native modified peptide.
In this work screening of gut bacterial isolates for lantibiotic biosynthetic genes revealed a novel lantibiotic cluster from B. obeum with four structural peptides and an unusual leader peptide sequence. Cross-immunity of the nisin O cluster to nisin A was demonstrated and heterologous expression of the novel cluster with the structural peptides on a nisin A inducible system showed evidence of antimicrobial activity against the pathogens C. perfringens and C. difficile in the presence of trypsin. Further work on the regulation of this novel cluster and its spectrum of antimicrobial activity will expand our understanding of the evolution of type I lantibiotics and may lead to the development of novel antimicrobials to target gut pathogens.
Funding information
The Institute of Food Research is funded by the BBSRC (strategic core grant IFR/08/1, Institute Strategic Programme grant BB/J004529/1); S. H. D. and H. J. F. acknowledge support from the Scottish Government Food Land and People programme. D. H. and C. G. F. received BBSRC PhD studentship grants.
Acknowledgements
We are grateful to Kathryn Cross for the EM analysis and Neil Rigby for helpful advice.
Conflicts of interest
The authors declare that there are no conflicts of interest.
Supplementary Data
Footnotes
Abbreviations: NCTC, National Collection of Type Cultures; OD, optical density; SEM, scanning electron microscopy; SSC, saline sodium citrate; TCA, trichloroacetic acid; TEM, transmission electron microscopy.
The GenBank accession number for the nucleotide sequence of the nisin O cluster is KY914474.
Two supplementary tables are available with the online Supplementary Material.
Edited by: M. Holden and J. Stulke
References
- 1.Arnison PG, Bibb MJ, Bierbaum G, Bowers AA, Bugni TS, et al. Ribosomally synthesized and post-translationally modified peptide natural products: overview and recommendations for a universal nomenclature. Nat Prod Rep. 2013;30:108–160. doi: 10.1039/C2NP20085F. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cotter PD, Ross RP, Hill C. Bacteriocins – a viable alternative to antibiotics? Nat Rev Microbiol. 2013;11:95–105. doi: 10.1038/nrmicro2937. [DOI] [PubMed] [Google Scholar]
- 3.Hegarty JW, Guinane CM, Ross RP, Hill C, Cotter PD. Bacteriocin production: a relatively unharnessed probiotic trait? F1000Res. 2016;5:2587. doi: 10.12688/f1000research.9615.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dischinger J, Basi Chipalu S, Bierbaum G. Lantibiotics: promising candidates for future applications in health care. Int J Med Microbiol. 2014;304:51–62. doi: 10.1016/j.ijmm.2013.09.003. [DOI] [PubMed] [Google Scholar]
- 5.Hammami R, Fernandez B, Lacroix C, Fliss I. Anti-infective properties of bacteriocins: an update. Cell Mol Life Sci. 2013;70:1–21. doi: 10.1007/s00018-012-1202-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Malik DK, Bhatia D, Nimbriya A, Kumar S. Lactic acid bacteria and bacteriocin: a review. J Pharm Res. 2012;5:2510–2513. [Google Scholar]
- 7.Chatterjee C, Paul M, Xie L, van der Donk WA. Biosynthesis and mode of action of lantibiotics. Chem Rev. 2005;105:633–684. doi: 10.1021/cr030105v. [DOI] [PubMed] [Google Scholar]
- 8.Field D, Begley M, O'Connor PM, Daly KM, Hugenholtz F, et al. Bioengineered nisin A derivatives with enhanced activity against both gram positive and gram negative pathogens. PLoS One. 2012;7:e46884. doi: 10.1371/journal.pone.0046884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Montalbán-López M, Van Heel AJ, Kuipers OP. Employing the promiscuity of lantibiotic biosynthetic machineries to produce novel antimicrobials. FEMS Microbiol Rev. 2017;41:5–18. doi: 10.1093/femsre/fuw034. [DOI] [PubMed] [Google Scholar]
- 10.Shin JM, Gwak JW, Kamarajan P, Fenno JC, Rickard AH, et al. Biomedical applications of nisin. J Appl Microbiol. 2016;120:1449–1465. doi: 10.1111/jam.13033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Horn N, Swindell S, Dodd H, Gasson M. Nisin biosynthesis genes are encoded by a novel conjugative transposon. Mol Gen Genet. 1991;228:129–135. doi: 10.1007/BF00282457. [DOI] [PubMed] [Google Scholar]
- 12.O'Connor PM, O'Shea EF, Guinane CM, O'Sullivan O, Cotter PD, et al. Nisin H is a new Nisin variant produced by the gut-derived strain Streptococcus hyointestinalis DPC6484. Appl Environ Microbiol. 2015;81:3953–3960. doi: 10.1128/AEM.00212-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Garg N, Tang W, Goto Y, Nair SK, Van der Donk WA. Lantibiotics from Geobacillus thermodenitrificans. Proc Natl Acad Sci USA. 2012;109:5241–5246. doi: 10.1073/pnas.1116815109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Field D, Connor PM, Cotter PD, Hill C, Ross RP. The generation of nisin variants with enhanced activity against specific gram-positive pathogens. Mol Microbiol. 2008;69:218–230. doi: 10.1111/j.1365-2958.2008.06279.x. [DOI] [PubMed] [Google Scholar]
- 15.Field D, Quigley L, O'Connor PM, Rea MC, Daly K, et al. Studies with bioengineered Nisin peptides highlight the broad-spectrum potency of Nisin V. Microb Biotechnol. 2010;3:473–486. doi: 10.1111/j.1751-7915.2010.00184.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yuan J, Zhang ZZ, Chen XZ, Yang W, Huan LD. Site-directed mutagenesis of the hinge region of nisinZ and properties of nisinZ mutants. Appl Microbiol Biotechnol. 2004;64:806–815. doi: 10.1007/s00253-004-1599-1. [DOI] [PubMed] [Google Scholar]
- 17.Karakas Sen A, Narbad A, Horn N, Dodd HM, Parr AJ, et al. Post-translational modification of nisin. The involvement of NisB in the dehydration process. Eur J Biochem. 1999;261:524–532. doi: 10.1046/j.1432-1327.1999.00303.x. [DOI] [PubMed] [Google Scholar]
- 18.Ra R, Beerthuyzen MM, de Vos WM, Saris PE, Kuipers OP. Effects of gene disruptions in the nisin gene cluster of Lactococcus lactis on nisin production and producer immunity. Microbiology. 1999;145:1227–1233. doi: 10.1099/13500872-145-5-1227. [DOI] [PubMed] [Google Scholar]
- 19.Wirawan RE, Klesse NA, Jack RW, Tagg JR. Molecular and genetic characterization of a novel Nisin variant produced by Streptococcus uberis. Appl Environ Microbiol. 2006;72:1148–1156. doi: 10.1128/AEM.72.2.1148-1156.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.De Vos WM, Mulders JW, Siezen RJ, Hugenholtz J, Kuipers OP. Properties of nisin Z and distribution of its gene, nisZ, in Lactococcus lactis. Appl Environ Microbiol. 1993;59:213–218. doi: 10.1128/aem.59.1.213-218.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Heidrich C, Pag U, Josten M, Metzger J, Jack RW, et al. Isolation, characterization, and heterologous expression of the novel lantibiotic epicidin 280 and analysis of its biosynthetic gene cluster. Appl Environ Microbiol. 1998;64:3140–3146. doi: 10.1128/aem.64.9.3140-3146.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.O'Keeffe T, Hill C, Ross RP. Characterization and heterologous expression of the genes encoding enterocin a production, immunity, and regulation in Enterococcus faecium DPC1146. Appl Environ Microbiol. 1999;65:1506–1515. doi: 10.1128/aem.65.4.1506-1515.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Aso Y, Nagao J, Koga H, Okuda K, Kanemasa Y, et al. Heterologous expression and functional analysis of the gene cluster for the biosynthesis of and immunity to the lantibiotic, nukacin ISK-1. J Biosci Bioeng. 2004;98:429–436. doi: 10.1016/S1389-1723(05)00308-7. [DOI] [PubMed] [Google Scholar]
- 24.Li H, O'Sullivan DJ. Heterologous expression of the Lactococcus lactis bacteriocin, nisin, in a dairy Enterococcus strain. Appl Environ Microbiol. 2002;68:3392–3400. doi: 10.1128/AEM.68.7.3392-3400.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rink R, Kuipers A, de Boef E, Leenhouts KJ, Driessen AJ, et al. Lantibiotic structures as guidelines for the design of peptides that can be modified by lantibiotic enzymes. Biochemistry. 2005;44:8873. doi: 10.1021/bi050081h. [DOI] [PubMed] [Google Scholar]
- 26.Arora T, Wegmann U, Bobhate A, Lee YS, Greiner TU, et al. Microbially produced glucagon-like peptide 1 improves glucose tolerance in mice. Mol Metab. 2016;5:725–730. doi: 10.1016/j.molmet.2016.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mierau I, Kleerebezem M. 10 years of the nisin-controlled gene expression system (NICE) in Lactococcus lactis. Appl Microbiol Biotechnol. 2005;68:705–717. doi: 10.1007/s00253-005-0107-6. [DOI] [PubMed] [Google Scholar]
- 28.Majchrzykiewicz JA, Lubelski J, Moll GN, Kuipers A, Bijlsma JJ, et al. Production of a class II two-component lantibiotic of Streptococcus pneumoniae using the class I nisin synthetic machinery and leader sequence. Antimicrob Agents Chemother. 2010;54:1498–1505. doi: 10.1128/AAC.00883-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kluskens LD, Kuipers A, Rink R, De Boef E, Fekken S, et al. Post-translational modification of therapeutic peptides by NisB, the dehydratase of the lantibiotic nisin. Biochemistry. 2005;44:12827–12834. doi: 10.1021/bi050805p. [DOI] [PubMed] [Google Scholar]
- 30.Rink R, Kluskens LD, Kuipers A, Driessen AJ, Kuipers OP, et al. NisC, the cyclase of the lantibiotic Nisin, can catalyze cyclization of designed nonlantibiotic peptides. Biochemistry. 2007;46:13179–13189. doi: 10.1021/bi700106z. [DOI] [PubMed] [Google Scholar]
- 31.Flint HJ, Scott KP, Duncan SH, Louis P, Forano E. Microbial degradation of complex carbohydrates in the gut. Gut Microbes. 2012;3:289–306. doi: 10.4161/gmic.19897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Birri DJ, Brede DA, Tessema GT, Nes IF. Bacteriocin production, antibiotic susceptibility and prevalence of haemolytic and gelatinase activity in faecal lactic acid bacteria isolated from healthy ethiopian infants. Microb Ecol. 2013;65:504–516. doi: 10.1007/s00248-012-0134-7. [DOI] [PubMed] [Google Scholar]
- 33.Walsh CJ, Guinane CM, Hill C, Ross RP, O'Toole PW, et al. In silico identification of bacteriocin gene clusters in the gastrointestinal tract, based on the human microbiome project's reference genome database. BMC Microbiol. 2015;15:183. doi: 10.1186/s12866-015-0515-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Barcenilla A. Diversity of the Butyrate-Producing Microflora of the Human Gut. 1999. Aberdeen, UK, Robert Gordon University, PhD Thesis. [Google Scholar]
- 35.Dabek M, Mccrae SI, Stevens VJ, Duncan SH, Louis P. Distribution of beta-glucosidase and β-glucuronidase activity and of β-glucuronidase gene gus in human colonic bacteria. FEMS Microbiol Ecol. 2008;66:487–495. doi: 10.1111/j.1574-6941.2008.00520.x. [DOI] [PubMed] [Google Scholar]
- 36.Flint HJ, Duncan SH, Scott KP, Louis P. Interactions and competition within the microbial community of the human colon: links between diet and health. Environ Microbiol. 2007;9:1101–1111. doi: 10.1111/j.1462-2920.2007.01281.x. [DOI] [PubMed] [Google Scholar]
- 37.Lawson PA, Finegold SM. Reclassification of Ruminococcus obeum as Blautia obeum comb. nov. Int J Syst Evol Microbiol. 2015;65:789–793. doi: 10.1099/ijs.0.000015. [DOI] [PubMed] [Google Scholar]
- 38.Mayer M, Dodd H, Narbad A, Gasson M. Identifying lantibiotic gene clusters and novel lantibiotic genes. 2006. PCT/GB2006/001429.
- 39.Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Plainview, NY: Cold Spring Harbor lab Press; 1989. [Google Scholar]
- 40.Rutherford K, Parkhill J, Crook J, Horsnell T, Rice P, et al. Artemis: sequence visualization and annotation. Bioinformatics. 2000;16:944–945. doi: 10.1093/bioinformatics/16.10.944. [DOI] [PubMed] [Google Scholar]
- 41.Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Larkin MA, Blackshields G, Brown NP, Chenna R, Mcgettigan PA, et al. CLUSTAL W and CLUSTAL X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 43.Mayer MJ, Narbad A, Gasson MJ. Molecular characterization of a Clostridium difficile bacteriophage and its cloned biologically active endolysin. J Bacteriol. 2008;190:6734–6740. doi: 10.1128/JB.00686-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Horton RM, Hunt HD, Ho SN, Pullen JK, Pease LR. Engineering hybrid genes without the use of restriction enzymes: gene splicing by overlap extension. Gene. 1989;77:61–68. doi: 10.1016/0378-1119(89)90359-4. [DOI] [PubMed] [Google Scholar]
- 45.Fernandez A, Horn N, Wegmann U, Nicoletti C, Gasson MJ, et al. Enhanced secretion of biologically active murine interleukin-12 by Lactococcus lactis. Appl Environ Microbiol. 2009;75:869–871. doi: 10.1128/AEM.01728-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dertli E, Colquhoun IJ, Gunning AP, Bongaerts RJ, Le Gall G, et al. Structure and biosynthesis of two exopolysaccharides produced by Lactobacillus johnsonii FI9785. J Biol Chem. 2013;288:31938–31951. doi: 10.1074/jbc.M113.507418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pitino I, Randazzo CL, Cross KL, Parker ML, Bisignano C, et al. Survival of Lactobacillus rhamnosus strains inoculated in cheese matrix during simulated human digestion. Food Microbiol. 2012;31:57–63. doi: 10.1016/j.fm.2012.02.013. [DOI] [PubMed] [Google Scholar]
- 48.Ryan MP, Rea MC, Hill C, Ross RP. An application in cheddar cheese manufacture for a strain of Lactococcus lactis producing a novel broad-spectrum bacteriocin, lacticin 3147. Appl Environ Microbiol. 1996;62:612–619. doi: 10.1128/aem.62.2.612-619.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cheng F, Takala TM, Saris PE. Nisin biosynthesis in vitro. J Mol Microbiol Biotechnol. 2007;13:248–254. doi: 10.1159/000104754. [DOI] [PubMed] [Google Scholar]
- 50.Sahl HG, Jack RW, Bierbaum G. Biosynthesis and biological activities of lantibiotics with unique post-translational modifications. Eur J Biochem. 1995;230:827–853. doi: 10.1111/j.1432-1033.1995.tb20627.x. [DOI] [PubMed] [Google Scholar]
- 51.Richards VP, Lang P, Bitar PD, Lefébure T, Schukken YH, et al. Comparative genomics and the role of lateral gene transfer in the evolution of bovine adapted Streptococcus agalactiae. Infect Genet Evol. 2011;11:1263–1275. doi: 10.1016/j.meegid.2011.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Plat A, Kluskens LD, Kuipers A, Rink R, Moll GN. Requirements of the engineered leader peptide of nisin for inducing modification, export, and cleavage. Appl Environ Microbiol. 2011;77:604–611. doi: 10.1128/AEM.01503-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jenq RR, Taur Y, Devlin SM, Ponce DM, Goldberg JD, et al. Intestinal Blautia is associated with reduced death from graft-versus-host disease. Biol Blood Marrow Transplant. 2015;21:1373–1383. doi: 10.1016/j.bbmt.2015.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bajaj JS, Hylemon PB, Ridlon JM, Heuman DM, Daita K, et al. Colonic mucosal microbiome differs from stool microbiome in cirrhosis and hepatic encephalopathy and is linked to cognition and inflammation. Am J Physiol Gastrointest Liver Physiol. 2012;303:G675–G685. doi: 10.1152/ajpgi.00152.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Murri M, Leiva I, Gomez-Zumaquero JM, Tinahones FJ, Cardona F, et al. Gut microbiota in children with type 1 diabetes differs from that in healthy children: a case-control study. BMC Med. 2013;11:46. doi: 10.1186/1741-7015-11-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chen W, Liu F, Ling Z, Tong X, Xiang C. Human intestinal lumen and mucosa-associated microbiota in patients with colorectal cancer. PLoS One. 2012;7:e39743. doi: 10.1371/journal.pone.0039743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chen J, Stevenson DM, Weimer PJ. Albusin B, a bacteriocin from the ruminal bacterium Ruminococcus albus 7 that inhibits growth of Ruminococcus flavefaciens. Appl Environ Microbiol. 2004;70:3167–3170. doi: 10.1128/AEM.70.5.3167-3170.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Crost EH, Ajandouz EH, Villard C, Geraert PA, Puigserver A, et al. Ruminococcin C, a new anti-Clostridium perfringens bacteriocin produced in the gut by the commensal bacterium Ruminococcus gnavus E1. Biochimie. 2011;93:1487–1494. doi: 10.1016/j.biochi.2011.05.001. [DOI] [PubMed] [Google Scholar]
- 59.Dabard J, Bridonneau C, Phillipe C, Anglade P, Molle D, et al. Ruminococcin A, a new lantibiotic produced by a Ruminococcus gnavus strain isolated from human feces. Appl Environ Microbiol. 2001;67:4111–4118. doi: 10.1128/AEM.67.9.4111-4118.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Marcille F, Gomez A, Joubert P, Ladiré M, Veau G, et al. Distribution of genes encoding the trypsin-dependent lantibiotic Ruminococcin A among bacteria isolated from human fecal microbiota. Appl Environ Microbiol. 2002;68:3424–3431. doi: 10.1128/AEM.68.7.3424-3431.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lawton EM, Ross RP, Hill C, Cotter PD. Two-peptide lantibiotics: a medical perspective. Mini Rev Med Chem. 2007;7:1236–1247. doi: 10.2174/138955707782795638. [DOI] [PubMed] [Google Scholar]
- 62.Mcauliffe O, Hill C, Ross RP. Identification and overexpression of ltnl, a novel gene which confers immunity to the two-component lantibiotic lacticin 3147. Microbiology. 2000;146:129–138. doi: 10.1099/00221287-146-1-129. [DOI] [PubMed] [Google Scholar]
- 63.Piper C, Hill C, Cotter PD, Ross RP. Bioengineering of a Nisin A-producing Lactococcus lactis to create isogenic strains producing the natural variants Nisin F, Q and Z. Microb Biotechnol. 2011;4:375–382. doi: 10.1111/j.1751-7915.2010.00207.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Corvey C, Stein T, Düsterhus S, Karas M, Entian KD. Activation of subtilin precursors by Bacillus subtilis extracellular serine proteases subtilisin (AprE), WprA, and Vpr. Biochem Biophys Res Commun. 2003;304:48–54. doi: 10.1016/S0006-291X(03)00529-1. [DOI] [PubMed] [Google Scholar]
- 65.Stein T, Entian KD. Maturation of the lantibiotic subtilin: matrix-assisted laser desorption/ionization time-of-flight mass spectrometry to monitor precursors and their proteolytic processing in crude bacterial cultures. Rapid Commun Mass Spectrom. 2002;16:103–110. doi: 10.1002/rcm.552. [DOI] [PubMed] [Google Scholar]
- 66.Qi F, Chen P, Caufield PW. The group I strain of Streptococcus mutans, UA140, produces both the lantibiotic mutacin I and a nonlantibiotic bacteriocin, mutacin IV. Appl Environ Microbiol. 2001;67:15–21. doi: 10.1128/AEM.67.1.15-21.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gomez A, Ladiré M, Marcille F, Fons M. Trypsin mediates growth phase-dependent transcriptional regulation of genes involved in biosynthesis of Ruminococcin A, a lantibiotic produced by a Ruminococcus gnavus strain from a human intestinal Microbiota. J Bacteriol. 2002;184:18–28. doi: 10.1128/JB.184.1.18-28.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lee JH, Li X, O'Sullivan DJ. Transcription analysis of a lantibiotic gene cluster from Bifidobacterium longum DJO10A. Appl Environ Microbiol. 2011;77:5879–5887. doi: 10.1128/AEM.00571-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mcclerren AL, Cooper LE, Quan C, Thomas PM, Kelleher NL, et al. Discovery and in vitro biosynthesis of haloduracin, a two-component lantibiotic. Proc Natl Acad Sci USA. 2006;103:17243–17248. doi: 10.1073/pnas.0606088103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hoover SE, Perez AJ, Tsui HC, Sinha D, Smiley DL, et al. A new quorum-sensing system (TprA/PhrA) for Streptococcus pneumoniae D39 that regulates a lantibiotic biosynthesis gene cluster. Mol Microbiol. 2015;97:229–243. doi: 10.1111/mmi.13029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gasson MJ. Transfer of sucrose fermenting ability, nisin resistance and nisin production into Streptococcus lactis 712. FEMS Microbiol Lett. 1984;21:7–10. doi: 10.1111/j.1574-6968.1984.tb00176.x. [DOI] [Google Scholar]
- 72.Dodd HM, Horn N, Gasson MJ. Analysis of the genetic determinant for production of the peptide antibiotic nisin. J Gen Microbiol. 1990;136:555–556. doi: 10.1099/00221287-136-3-555. [DOI] [PubMed] [Google Scholar]
- 73.Dodd HM, Horn N, Giffard CJ, Gasson MJ. A gene replacement strategy for engineering nisin. Microbiology. 1996;142:47–55. doi: 10.1099/13500872-142-1-47. [DOI] [PubMed] [Google Scholar]
- 74.Wegmann U, Klein JR, Drumm I, Kuipers OP, Henrich B. Introduction of peptidase genes from Lactobacillus delbrueckii subsp. lactis into Lactococcus lactis and controlled expression. Appl Environ Microbiol. 1999;65:4729–4733. doi: 10.1128/aem.65.11.4729-4733.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Casadaban MJ, Cohen SN. Analysis of gene control signals by DNA fusion and cloning in Escherichia coli. J Mol Biol. 1980;138:179–207. doi: 10.1016/0022-2836(80)90283-1. [DOI] [PubMed] [Google Scholar]
- 76.Simon D, Chopin A. Construction of a vector plasmid family and its use for molecular cloning in Streptococcus lactis. Biochimie. 1988;70:559–566. doi: 10.1016/0300-9084(88)90093-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.