Abstract
Wheat stem rust, caused by Puccinia graminis f. sp. tritici (Pgt), is a devastating foliar disease. The Ug99 race group has combined virulence to most stem rust (Sr) resistance genes deployed in wheat and is a threat to global wheat production. Here we identified a coiled-coil, nucleotide-binding leucine-rich repeat protein (NLR) completely linked to the Ug99 resistance gene Sr21 from Triticum monococcum. Loss-of-function mutations and transgenic complementation confirmed that this gene is Sr21. Sr21 transcripts were significantly higher at high temperatures, and this was associated with significant upregulation of pathogenesis related (PR) genes and increased levels of resistance at those temperatures. Introgression of Sr21 into hexaploid wheat resulted in lower levels of resistance than in diploid wheat, but transgenic hexaploid wheat lines with high levels of Sr21 expression showed high levels of resistance. Sr21 can be a valuable component of transgenic cassettes or gene pyramids combining multiple resistance genes against Ug99.
Author summary
Wheat stem rust is a devastating disease that is threatening global wheat production. The emergence of new virulent races of this pathogen in Africa, including the Ug99 race group, has prompted global efforts to find effective resistance genes. We report here the identification of stem rust resistance gene Sr21 that is effective against the Ug99 race group. We developed a diagnostic marker to accelerate its deployment in wheat breeding programs and demonstrated that the introduction of two Sr21 copies in transgenic wheat results in high levels of resistance. An unusual characteristic of Sr21 is its increased resistance to stem rust at high temperatures. We show here that this is associated with the ability of Sr21 to coordinate the upregulation of multiple pathogenesis related genes at high temperatures. These genes slow down the growth of the pathogen and result in the characteristic Sr21 intermediate resistance reaction at high temperatures. A better understanding of this temperature dependent resistance mechanism will be useful for controlling the rust pathogens in our changing environments.
Introduction
More than 700 million tons of wheat are produced every year, but further increases are needed to feed a rapidly growing human population. Reducing yield losses caused by wheat pathogens can contribute to this goal. Wheat stem rust, caused by the fungal pathogen Puccinia graminis f. sp. tritici (henceforth Pgt), poses an urgent threat to global wheat production since new virulent races have overcome widely deployed resistance genes.
In 1998, a new Pgt race was detected in Uganda that was virulent to the widely deployed stem rust resistance genes Sr31 and Sr38 [1]. This race, also known as TTKSK using the North American system of nomenclature for Pgt races [2,3], was virulent to roughly 90% of the global wheat cultivars [4]. Since then, the Ug99 race group has spread to more than 13 countries in Africa and the Middle East [4–8] and acquired virulence to additional resistance genes (Sr24, Sr36, Sr9h and SrTmp [3,9–11]), prompting efforts to identify and clone effective resistance genes.
The large and complex nature of the wheat genome has limited the number of cloned Ug99 resistance genes to six, including Sr35 [12], Sr33 [13], Sr50 [14], Sr22 [15], Sr45 [15], and Sr13 [16]. More resistance genes are necessary to diversify the combinations of Pgt resistance genes deployed as gene pyramids or in transgenic cassettes to provide durable resistance [15,17,18].
The stem rust resistance gene Sr21 was discovered in diploid wheat Triticum monococcum (genome Am), which is closely related to T. urartu, the progenitor of the A genome in polyploid wheat [19]. Sr21, which confers a more effective resistance to the Ug99 race group at high temperatures (20–24°C) than at low temperature (16°C) [20], was transferred to hexaploid wheat (T. aestivum) [21]. We previously mapped Sr21 to a 0.19 cM interval on the central region of chromosome arm 2AmL and showed that this region includes a cluster of NBS-LRR resistance genes in the colinear region in the A genome of T. aestivum [20].
Here, we report the identification, validation and characterization of the wheat stem rust resistant gene Sr21, which encodes a coiled-coil nucleotide-binding leucine-rich repeat protein (NLR). Six pathogenesis-related (PR) genes showed increased transcript levels in Sr21-resistant genotypes inoculated with Pgt, only when plants were grown at high temperatures. This result provides a tentative explanation for the more effective Pgt resistance conferred by Sr21 at high temperatures. Finally, we identified five different resistance haplotypes of Sr21 and developed a diagnostic molecular marker to accelerate its deployment in breeding programs.
Results
High-density genetic and physical maps of the Sr21 region
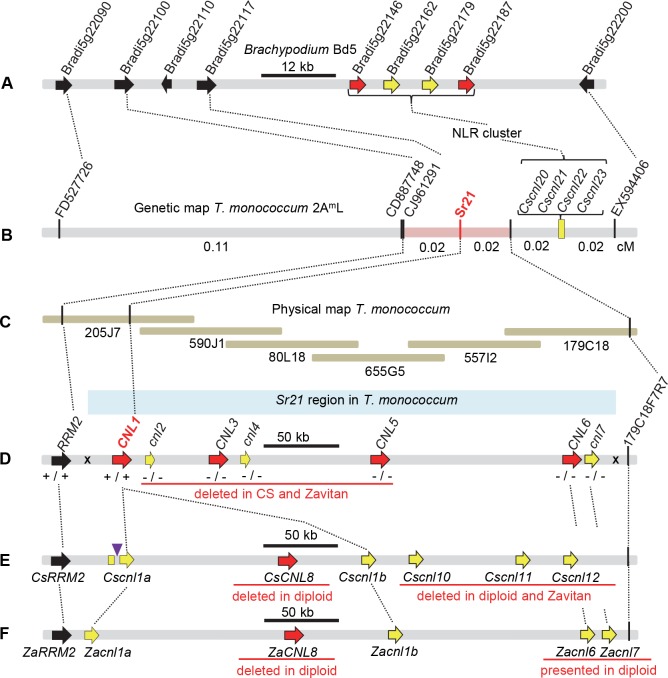
The 0.19 cM region on T. monococcum chromosome arm 2AmL including Sr21 is colinear with an 88-kb region in Brachypodium distachyon chromosome 5 flanked by genes Bradi5g22090 and Bradi5g22200 [20]. In the present study, we identified the wheat orthologs of six B. distachyon genes present in this colinear region (Bradi5g22100, Bradi5g22117, Bradi5g22146, Bradi5g22162, Bradi5g22179 and Bradi5g22187, Fig 1A, S1 Table) and mapped them in ten wheat lines with recombination events between Sr21 flanking markers FD527726 and EX594406 (Fig 1B). Using these new markers, Sr21 was mapped 0.02 cM distal to CJ961291 and 0.04 cM proximal to a group of four linked NLR pseudogenes (Cscnl20, Cscnl21, Cscnl22 and Cscnl23, Fig 1B).
Fig 1. Map-based cloning of Sr21. (A) Brachypodium distachyon chromosome 5 region colinear with the Sr21-candidate region.
Arrows in bracket indicate NLR genes and pseudogenes. (B) High-density genetic map of Sr21 on chromosome arm 2AmL. (C) Physical map of the Sr21 region constructed with overlapping BACs from diploid wheat accession DV92. (D) Diagrammatic representation of the annotated sequence of the Sr21 region (GenBank accession MG582649). NLR genes are represented by red arrows and capital letters (CNL) and pseudogenes by yellow arrows and lower-case letters (cnl). Similar NLR numbers indicate putative orthologous genes. “+” and “-” signs below gene names indicate detection of expression in resistant T. monococcum accessions DV92 (first sign) and G3116 (second sign). (E) Genes in the colinear region of T. aestivum cv. Chinese Spring. (F) Genes in the colinear region of T. turgidum ssp. dicoccoides Zavitan [23]. NLRs with the same number are putative orthologs.
We screened a bacterial artificial chromosome (BAC) library of the resistant T. monococcum accession DV92 [22] using the closest markers, but sequencing of selected BACs showed no connection between proximal and distal groups (S1 Fig). We initiated a chromosome walk from the closest marker (CJ961291, 0.02 cM from the phenotype) in the proximal group (Fig 1C, BAC 205J7). Marker CJ961291 (RRM2) showed a recombination event with gene CNL1, which was completely linked to the phenotype, completing the proximal side of the physical map. After five cycles of library screening, BAC sequencing and marker development, we identified an additional recombination event between the phenotype and marker 179C18F7R7, which closed the distal end of the physical map.
Sequencing the 405-kb candidate region (Fig 1D, blue area, S1 Fig red colored BACs, GenBank accession MG582649) revealed four complete NLR genes (henceforth, CNL1, CNL3, CNL5, and CNL6) and three NLR truncated genes with premature stop codons (henceforth, cnl2, cnl4, and cnl7).
Multiple rearrangements were observed in this NLR cluster in T. aestivum cv. Chinese Spring (419 kb, Fig 1E, IWGSC RefSeq v1.0) and T. turgidum ssp. dicoccoides Zavitan (435 kb, Fig 1F, Zavitan WEWSeq v1.0) [23]. The two polyploid species included the complete gene CsCNL8 (ZaCNL8) that was absent in T. monococcum, whereas the T. monococcum region including cnl2, CNL3, cnl4 and CNL5 was missing in Chinese Spring and Zavitan (Fig 1E and 1F). NLR pseudogenes Cscnl10, Cscnl11 and Cscnl12 identified in CS were not detected in T. monococcum or Zavitan. The CS and Zavitan pseudogenes Cscnl1a (Zacnl1a) and Cscnl1b (Zacnl1b) were similar to T. monococcum CNL1 (~99% identity). Cscnl1a was interrupted by the insertion of a repetitive sequence in CS but not in Zavitan (Fig 1E and 1F).
Natural variation in NLR genes linked to Sr21
We sequenced the four NLR genes completely linked to the Sr21 phenotype (CNL1, CNL3, CNL5 and CNL6) from diploid accessions G3116 (Sr21, resistant) and PI 272557 (no-Sr21, susceptible). Using seven different pairs of gene-specific primers, we amplified CNL3 and CNL5 from DV92 but not from G3116 and PI 272557 (similar to CS and Zavitan, Fig 1E and 1F). Since G3116 carries the Sr21 resistant allele, the absence of these two genes suggested that CNL3 and CNL5 were unlikely candidate genes for Sr21. CNL6 was PCR-amplified in all three accessions, but G3116 showed a premature stop codon at position 742 (Q742*), suggesting that this gene was not Sr21. By contrast, CNL1 was PCR-amplified from both resistant accessions (DV92 and G3116) but not from Pgt susceptible accession PI 272557.
Taken together, these results suggested that CNL1 was the best candidate gene for Sr21 among the NLR genes linked to Sr21. This hypothesis was further supported by expression data for these genes in transcriptome databases of DV92 and G3116 [24]. Among the four NLR genes completely linked to Sr21, we only detected transcripts for CNL1 in both DV92 and G3116 databases (Fig 1D).
Validation of CNL1 using EMS mutants
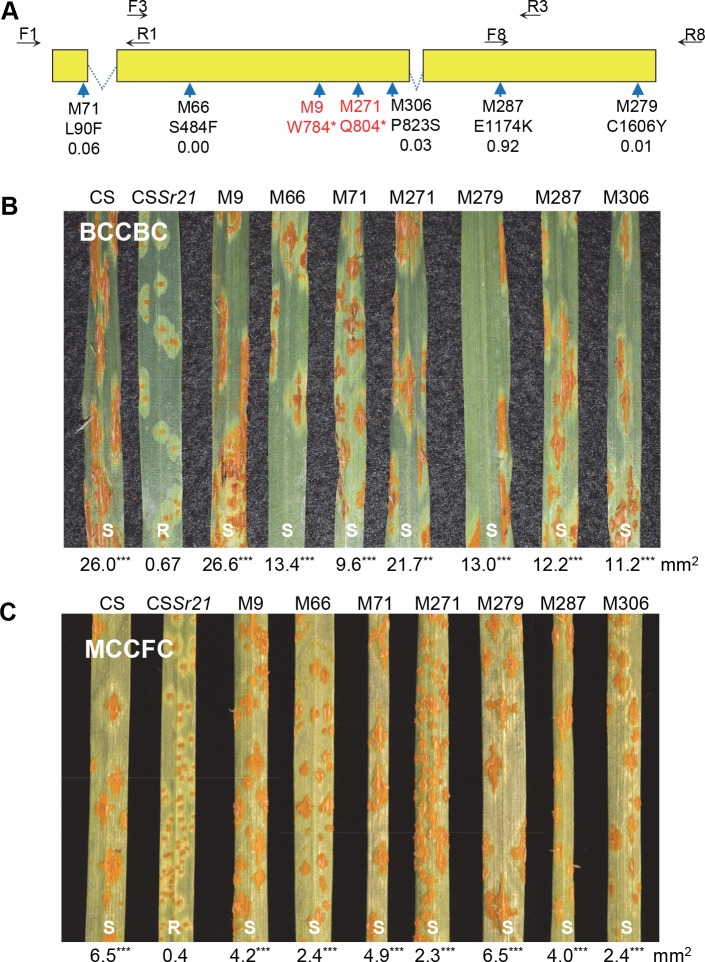
To test if CNL1 was required for Sr21 resistance, we mutagenized the Sr21 introgression lines in Chinese Spring (henceforth CSSr21) with ethyl methane sulfonate (EMS) and generated 1,151 M1 mutant plants. Among the 1,151 M2 mutant families screened with race BCCBC, we identified seven showing susceptible plants (M9, M66, M71, M271, M279, M287 and M306, Fig 2A and 2B). M3 seeds from the susceptible plants were also susceptible to Pgt race MCCFC (Fig 2C).
Fig 2. Validation of the Sr21 candidate gene using EMS mutants.
(A) Structure of gene CNL1 (from start codon to stop codon). Primers used for haplotyping are indicated by black arrows. Dotted lines indicate introns, and yellow rectangles indicate coding exons. The positions of the mutations are indicated by blue arrows. Mutation names in red indicate premature stop codons and in black indicate amino acid changes, which are indicated below the names with the corresponding SIFT scores “Sorting intolerant from tolerant” scores lower than 0.05 are predictive of deleterious missense mutations [50]). (B) Infection types on Chinese Spring (CS), CSSr21, and seven mutants inoculated with Pgt race BCCBC. Numbers below leaves indicate average pustule sizes (n = 3) and superscripts indicate significance of differences with CSSr21 (*** = P< 0.001). (C) Same as B above but with race MCCFC.
Sequencing of CNL1 from the seven susceptible mutants (primers in S1 Table) revealed amino acid changes in five mutants and premature stop codons in two mutants (Fig 2A). Since the probability of a truncation or missense mutation in an annotated gene in a hexaploid wheat line mutagenized with 0.8% EMS is roughly 0.02 [25], the probability of detecting such changes in seven independent mutants by chance is < 1.3×10−12. These results demonstrated that CNL1 is required for Sr21 resistance to Pgt.
Wheat plants transformed with CNL1 were resistant to Ug99
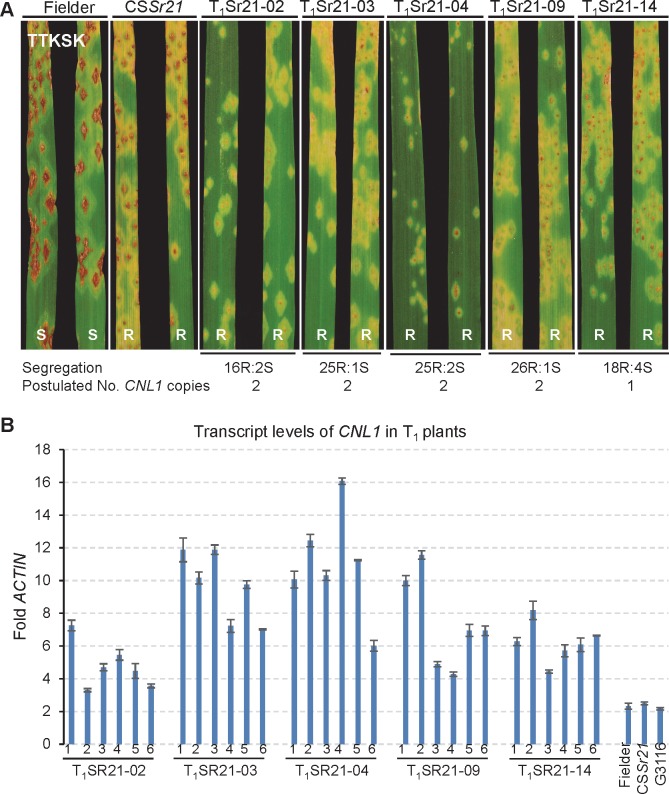
To test if CNL1 was also sufficient to confer resistance to Pgt, we generated thirteen independent transgenic events (T0Sr21) in the susceptible common wheat variety Fielder. Among the 13 T0 plants, we prioritized five that showed higher transcript levels of CNL1 than Fielder, which has a non-functional copy of CNL1 (S2 Fig). The T1 progenies from these five events showed segregation for resistance when challenged with Pgt race TTKSK (isolate 04KEN156/04). Some of the resistant transgenic plants showed even better levels of resistance than the CSSr21 positive control, which carries a Sr21 introgression from T. monococcum (Fig 3A). These results confirmed that CNL1 is sufficient to confer resistance to TTKSK. Taken together, the high-density map, the mutants and the transgenic results confirmed that CNL1 is Sr21.
Fig 3. Sr21 transgenic plants.
(A) Reactions to Pgt race TTKSK (Ug99) in Fielder, CSSr21, and transgenic families T1Sr21-02, T1Sr21-03, T1Sr21-04, T1Sr21-09 and T1Sr21-14. S = Susceptible, R = Resistant. Plants were grown at 25°C during the day and 22°C during the night. The numbers below the figure indicate the observed segregation of resistant or partial resistant versus susceptible lines and the postulated number of independent genes based on χ2 tests (S2 Table). (B) Transcript levels of CNL1 in transgenic T1 families T1SR21-02, T1SR21-03, T1SR21-04, T1SR21-09 and T1SR21-14 (six plants per event). Transcript levels are expressed as fold-ACTIN using the 2-ΔCT method. Fielder has the susceptible CNL1 allele S2, which is expressed at similar levels as the resistant allele in hexaploid CSSr21 and diploid T. monococcum G3116.
CNL1 copy number and expression in transgenic plants
The observed segregation of resistant and susceptible T1 transgenic plants (Fig 3A) suggested that four of these five transgenic lines have more than one independent functional CNL1 insertions. Genotypes from roughly 50 T1 plants from each event showed significant departures from the expected 3:1 segregation ratio (P < 0.001, S2 Table), which suggested the presence of three to six copies of CNL1. This was validated by TaqMan copy number assays [26] (S2 Table). The larger copy number estimates obtained from the genotypic data than from the phenotypic data is likely explained by the presence of non-functional CNL1 insertions and/or by the presence of linked functional copies.
On average, the five selected T1 transgenic events showed 2.6- to 5.7-fold higher CNL1 transcript levels than CSSr21 (S2 Table and Fig 3B), confirming the T0 results (S2 Fig). Copy number based on the CNL1 TaqMan assay was significantly correlated with average transcript levels in the five selected transgenics and CSSr21 (R = 0.83, P = 0.039). A similar correlation (R = 0.81, P = 0.0009) was observed between the T0 expression levels from all 13 transgenic lines and the copy number estimated by the TaqMan assay (S2 Fig). These results suggest that the differences in expression are driven, at least in part, by differences in CNL1 copy number. This was also reflected in the levels of resistance to TTKSK, where a negative correlation was observed between CNL1 copy number and average rust pustule size (R = -0.92, P < 0.0001, S3 Fig). In this experiment, we also compared the CNL1 transcript levels between the Sr21-resistant accessions G3116 (diploid) and CSSr21 (hexaploid) and found no significant differences (Fig 3B).
CNL1 structure and alternative splicing forms
We compared the sequences of the CNL1 transcripts (from the DV92 and G3116 transcriptome database [24]) with the corresponding genomic sequences and determined that the 4,872 bp coding sequence of CNL1 is divided in three exons that encode 1,624 amino acids. The predicted protein includes an N-terminal coiled-coil (CC) domain, a central nucleotide-binding (NB) site, and a leucine-rich repeat (LRR) region, typical of many NLR proteins. A nuclear localization signal (NLS) was predicted in Sr21 between amino acids 141 and 152 (cNLS Mapper http://nls-mapper.iab.keio.ac.jp/cgi-bin/NLS_Mapper_form.cgi).
Comparing different transcripts and genomic sequences, we identified a 210-bp 5’ untranslated region (UTR) and a 2,159-bp 3’ UTR. The 5’ UTR does not include any introns, whereas the 3’ UTR shows 2 to 4 introns depending on the alternative splice forms (S4 Fig). These results were confirmed by 5’ and 3’ rapid amplification of cDNA ends (5’ and 3’ RACE). We identified 10 CNL1 alternative splicing forms, which differ only in their 3’ UTR regions (S4 Fig). We detected five alternative splicing forms (CNL1-1 to CNL1-5) in the transcriptomes of DV92 and G3116 [24] and an additional five (CNL1-6 to CNL1-10) in the 3' RACE reactions.
Since Sr21 resistance is modulated by temperature [20], we characterized the frequency of the different alternative splice forms in 3' RACE reactions from RNAs obtained from G3116 T. monococcum resistant plants grown at 24°C and 16°C. The plants at each temperature were further divided in mock-inoculated and inoculated with Pgt race BCCBC, and RNA samples were extracted six days after inoculation (80 clones per treatment were sequenced). χ2 tests showed no significant differences (P = 0.81) in the frequencies of alternative splice forms in inoculated vs. mock-inoculated plants (averaged across temperatures) but detected significant differences between temperatures (P = 0.002, averaged across inoculation treatments, S3 Table). The CNL1-1, CNL1-5 and CNL1-6 forms were more frequent at 24°C than at 16°C, and the opposite was observed for CNL1-2. We currently do not know the biological significance of these differences.
Effect of temperature and Pgt inoculation on transcript levels of CNL1 and PR genes
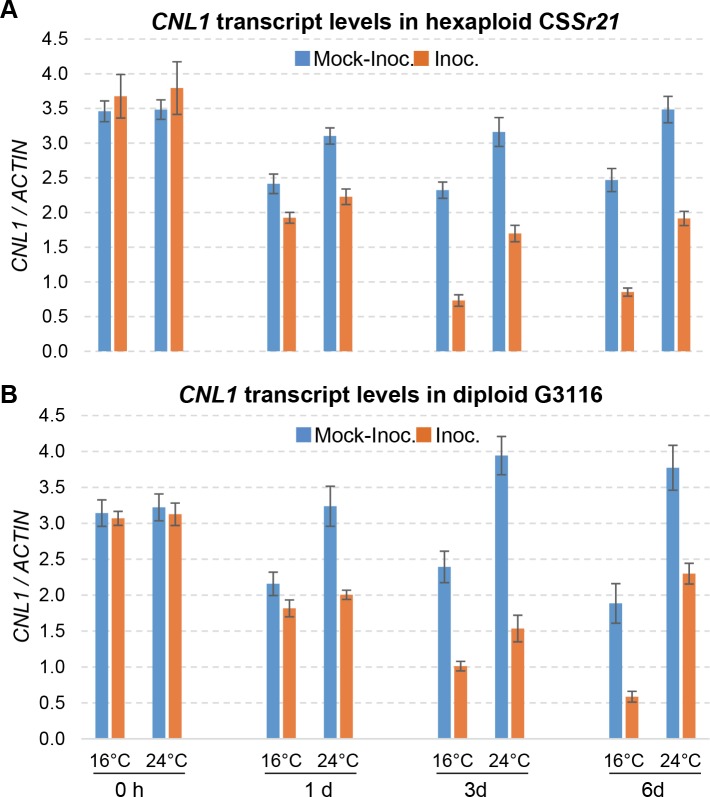
We analyzed CNL1 transcript levels in the four temperature / inoculation combinations described above in resistant accessions G3116 (diploid) and CSSr21 (hexaploid). We took samples immediately after moving the plants inoculated with Pgt from a greenhouse (~20°C) to growth chambers at 16°C and 24°C (time 0 h, Fig 4). As expected, we found no significant differences in CNL1 transcript levels between temperatures or inoculations at the control sampling point (S4A Table). The CNL1 basal transcript levels relative to ACTIN were 14.8% higher in CSSr21 than in G3116 (P = 0.0089, S4A Table).
Fig 4. Effect of temperature and Pgt inoculation (race BCCBC) on CNL1 transcript levels.
(A) Sr21 resistant hexaploid wheat CSSr21. (B) Sr21 resistant diploid wheat G3116. Leaves from both genotypes were collected after inoculation with BCCBC and immediately after moving the plants to chambers at 16°C and 24°C (0 h). The statistical analysis for the 0 h control time point is presented in S4A Table, and the combined analysis for one, three and six days after inoculation in S4B Table. Transcript levels were expressed as fold-ACTIN. Bars are standard errors of the means. Inoc: inoculated with BCCBC, Mock-Inoc: mock inoculated with water.
For the samples collected at 1, 3 and 6 days post inoculation (dpi), we performed a four-way ANOVA for CNL1 transcript levels including genotype, temperature, inoculation treatment and day after inoculation as factors. We found no significant differences between genotypes (diploid G3116 and hexaploid CSSr21), but detected significantly lower transcript levels in plants grown at 16°C than at 24°C (36.5%, P < 0.0001), and in Pgt inoculated plants relative to mock-inoculated plants (45.8%, P < 0.0001) (Fig 4, S4B Table). A smaller effect was detected among days (P = 0.02), but no clear trend was observed.
We used the same RNA samples collected 6 dpi from CSSr21 and G3116 described above and primers described in a previous study [16] to quantify the transcript levels of six pathogenesis-related (PR) genes (PR1, PR2, PR3, PR4, PR5 and PR9 = TaPERO). For all six PR genes, transcript levels were significantly higher (P < 0.0001) in Pgt-inoculated plants than in mock-inoculated plants (S5 Table and S5 Fig). The overall differences in transcript levels between temperatures were not significant for some PR genes (PR1, PR4, PR9) but the interactions between temperature and inoculation were all highly significant in the combined ANOVA (P < 0.001, S5 Table). These interactions are clear in S5 Fig, which shows lower PR transcript levels at 24°C than at 16°C in the mock-inoculated plants but significantly higher levels in the Pgt-inoculated plants at 24°C than at 16°C.
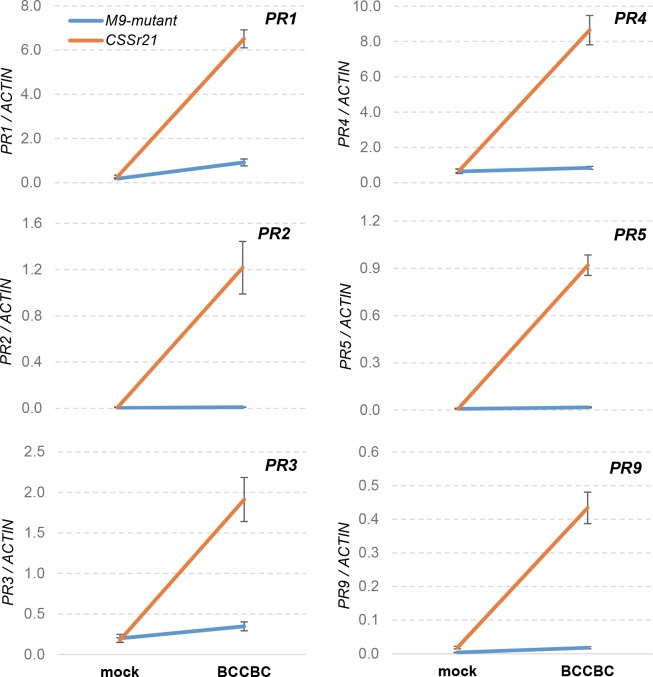
To confirm that Sr21 was required for the coordinated upregulation of the six PR genes at 24°C, we compared the transcript levels of these six genes in the resistant hexaploid line CSSr21 and its derived susceptible sr21-mutant M9 (Fig 5). Plants from both genotypes were inoculated with race BCCBC or were mock inoculated. For all six PR genes, the differences between genotypes, inoculations and the interactions genotype x inoculation were highly significant (P < 0.001) in two-way factorial ANOVAs. In all cases, a strong upregulation of all six PR genes was observed after inoculation with race BCCBC relative to mock inoculation only in the resistant genotype CSSr21 (Fig 5). This result confirmed that the coordinated upregulation of these six PR genes was triggered by Sr21.
Fig 5. Transcript levels of PR genes in the presence and absence of Sr21.
Transcript levels of PR genes (PR1, PR2, PR3, PR4, PR5, and PR9) in inoculated (BCCBC) and mock-inoculated plants with a functional Sr21 resistance gene (CSSr21) and without it (sr21-M9-mutant) grown at 24°C. A strong up-regulation of all PR genes was observed only when both Sr21 and the pathogen were present. Lack of parallelism between lines reflect significant interactions (P < 0.001). Values were calculated using the 2ΔCT method relative to ACTIN as endogenous control. Error bars indicate standard errors of the means. n = 5. Samples were collected six days post inoculation.
Effect of temperature and Sr21 on Pgt growth in diploid and hexaploid wheat
Sr21 showed higher levels of resistance to BCCBC when plants were grown at 24°C than when grown at 16°C (Figs 6A, 6B and S6). Similar differences were observed before for Sr21 resistance response to TTKSK at 20°C and 16°C [20]. These observations were confirmed in a three-way ANOVA for average pustule size 14 days post inoculation (dpi) (S6 Table). This analysis showed highly significant (P < 0.0001) effects for ploidy level and genotype (presence or absence of Sr21), and a very strong interaction between temperature and genotype (P < 0.0001). Plants having the Sr21 resistance gene showed smaller sporulation areas at high temperatures, whereas those without Sr21 showed significantly larger sporulation areas at high temperatures (Fig 6A and 6B). These opposite effects masked the main effect of temperature for race BCCBC (S6 Table).
Fig 6. Pgt race BCCBC growth at 16°C and 24°C in different genotypes.
(A, C, E) Diploid wheat. Sr21 resistant genotype G3116 (orange) and susceptible genotype PI 272557 (blue). (B, D, E) Hexaploid wheat. Sr21 resistant genotype CSSr21 (R) (orange) and susceptible genotype CS (S) (blue). (A, B) Interaction graphs for average pustule size at 14 dpi n = 6). (C, D) Interaction graphs for Pgt DNA relative to wheat DNA at 5 dpi (n = 6). (E, F) Interaction graphs for average size of individual fungal infection areas estimated by fluorescence microscopy at 5 dpi (n = 6). Lack of parallelism between lines in all graphs reflects significant interactions (P<0.0001, S6 Table).
We also quantified the differences in BCCBC growth at five dpi by measuring the ratio of Pgt DNA relative to wheat DNA (Fig 6C and 6D) and average infection areas using microscopy and a fluorescent dye that stains the pathogen (Figs 6E, 6F and S7). Both methods showed highly significant (P < 0.0001) differences between genotypes and between temperatures. As with the sporulation area, we also detected highly significant interactions between genotype and temperature for both methods (P < 0.0001, S6 Table), which reflected the larger differences between genotypes at high than at low temperature (Fig 6C–6F). Average areas for sporulation at 14 dpi (TTKSK and BCCBC) and for pathogen growth (BCCBC determined by fluorescence) were larger in the Sr21-resistant hexaploid than in the resistant diploid plants, which agrees with a previous report [20].
Fluorescent images of Pgt growth in diploid and hexaploid wheat plants lacking Sr21 at high temperature show a diffuse network of hyphae at the borders of the infected areas, which seems to be expanding without host resistance. By contrast, in the presence of Sr21, the infected areas are smaller and their borders are denser (S7 Fig), which suggests that the expansion of the hyphae is facing opposition from the host. At the macroscopic level, a chlorotic halo was observed around the Pgt pustules in the Sr21 resistant reactions, which in some cases resulted in cell death. However, Pgt resistance conferred by Sr21 is mostly associated with a delay in the progression of the disease (partial resistance) rather than with a rapid hypersensitive reaction.
Haplotype analysis of CNL1
We evaluated 114 T. monococcum and T. urartu accessions with Pgt races MCCFC and BCCBC and confirmed the presence of Sr21 in 44 of them and its absence in 70. We sequenced the complete CNL1 gene from these accessions using three pairs of primers (S1 Table and Fig 2A) and identified four susceptible haplotypes (S1 to S4) and five resistant haplotypes (R1 to R5, S8 Table and S8 Fig). The presence of Sr21 in the resistant diploid wheat accessions is based on inoculation with Pgt races TRTTF, TTKSK, TTTTF, QFCSC, and MCCFC (S7 Table).
Of the 44 resistant accessions, 28 were classified as haplotype R1 (MG582649, equal to DV92), six as R2 (MG601519), one as R3 (MG601520), six as R4 (MG601521) and three as R5 (MG601522, S8 Table). The R1 haplotype differed from the other four by one (R2), three (R3), two (R4) and three (R5) amino acid changes, respectively (S8 Fig).
Of the 30 T. monococcum susceptible accessions, 25 showed identical sequences and were classified as haplotype S1 (MG601523, 90.2% identical at the cDNA level to CNL1 haplotype R1). S1 is likely a non-functional gene since it carries a frame shift mutation. Surprisingly, polymorphisms between R1 and S1 were concentrated in the CC-NBS domains (87.2% identity) whereas the LRR was more similar to CNL1 (99.2% identity), which suggests a recombination or conversion event. A phylogenetic analysis including the DNA coding regions of the different NLR genes in the CNL1 region, and the closest paralogues in B. distachyon and H. vulgare showed that the CNL1 S1 haplotype is more closely related to cnl7 than to the cluster including the different CNL1 haplotypes (S9 Fig).
All 40 T. urartu accessions showed haplotype S2 (MG601524), which was similar to R1 (99.2% identical). This haplotype also includes three accessions classified as T. monococcum subsp. aegilopoides in the NSGC. However, analysis of 12 additional genes showed T. urartu haplotypes in these three accessions, suggesting that they are misclassified (we counted them as T. monococcum in the numbers reported above). The S2 haplotype from T. urartu, and the related haplotypes found in wild tetraploid Zavitan and hexaploid cultivars Chinese Spring and Fielder corresponds to the pseudogene Cscnl1b from Fig 1 and shares a 2-bp deletion at positions 3,255 and 3,256 (cDNA coordinates). When this shift mutation is manually corrected, these accessions share 20 amino acid polymorphisms compared to the R1 haplotype (S8 Fig). The cultivated tetraploid Kronos shares 14 of the 20 amino acid polymorphisms found in Chinese Spring, Fielder and Zavitan. Kronos does not have the 2-bp frame shift mutation, but has two different 1-bp deletions at positions 928 (a missing A) and 3,667 (a missing T). The susceptible haplotypes S3 (MG601525) and S4 (MG601526) were very similar to R1 (> 99.9% identical) and included a single accession each (S8 Table). Three accessions from the Balkans, classified as S1 based on their CNL1 sequence (PI 355538, PI 362610 and PI 377668), showed a resistance reaction when inoculated with race MCCFC but were susceptible to BCCBC. Since Sr21 is resistant to both BCCBC and MCCFC, this result suggested that these three accessions carry a Pgt resistance gene different from Sr21.
To develop a diagnostic marker for the Sr21 resistant haplotypes introgressed into polyploid wheat, we designed primers based on the C1228W diagnostic polymorphism and the T1500R and H1501S polymorphisms that separate Sr21 resistant and susceptible haplotypes. PCR amplification with primers Sr21TRYF5R5 (S1 Table) at an annealing temperature of 56°C generates a 951-bp fragment when the T. monococcum haplotypes are present and no amplification in T. urartu, tetraploid or hexaploid wheat (S10 Fig). Treatment of the amplified PCR products with restriction enzyme NsiI generated two bands of 836 and 115 bp for the T. monococcum accessions carrying the Sr21 susceptible haplotypes and a single 951-bp band for the accessions carrying the resistant haplotypes (S10 Fig).
Discussion
The T. monococcum genomic region encompassing the Sr21 resistance gene includes four NLR genes and three pseudogenes (Fig 1). The clustering of NLR genes facilitates the generation of novel variants through recombination and conversion events, increasing genetic variability in resistance genes [27]. Ectopic recombination events can generate deletions and duplications, and these were frequent in the Sr21 region. We detected several rearrangements between T. monococcum, hexaploid wheat Chinese Spring and wild tetraploid wheat Zavitan [23] (S1 Fig) that diverged less than one million years ago [28]. A T. monococcum region of more than 150-kb including cnl2, CNL3, cnl4 and CNL5 was not detected in any of the polyploid species. Similarly, a large region in Chinese Spring between CsCNL8 and Cscnl12 was deleted in T. monococcum and part of it in Zavitan. Finally, a region including CNL6 and cnl7 was detected in diploid and tetraploid wheat but was absent in hexaploid wheat. These large differences suggest that this NLR cluster has experienced rapid evolutionary changes. The presence of four related NLR genes and pseudogenes in the colinear region of Brachypodium distachyon chromosome 5 (Figs 1 and S9) suggests that this NLR cluster has a long evolutionary history that extends beyond the divergence between the Triticum and Brachypodium lineages more than 30 million years ago [29].
When we characterized Sr21 expression, we detected multiple alternative splicing forms of CNL1, which differed in the intron structure of the 3’ UTR region (S4 Fig). Alternative splicing forms have been identified in several CC-NBS-LRR genes, including Pi-ta in rice [30], Mla in barley [31], and Lr10 [32] in wheat. Complex UTR regions with multiple introns have been also described for other wheat NLR genes involved in resistance to Pgt including Sr35 [12] and Sr13 [16]. In Sr21, we observed differences in the frequencies of the main alternative splicing forms with temperature, but the role of these differences is currently unknown.
Results presented here and in a previous study [20] indicate that Sr21 is less effective when present in a hexaploid background than in a diploid background. Since the hexaploid Sr21 gene was introgressed directly from T. monococcum [21], the encoded proteins are expected to be identical. In addition, we found no significant differences between diploid and hexaploid wheat in Sr21 transcript levels. Therefore, differences downstream of Sr21 transcription are likely responsible for the reduced resistance conferred by Sr21 in hexaploid than in diploid wheat. Reduced stability of the Sr21 protein in hexaploid wheat or reduced compatibility between the T. monococcum Sr21 protein and some of its downstream T. aestivum protein interactors are possible explanations. Both hypotheses can explain the stronger upregulation of downstream PR genes observed in diploid than in hexaploid Sr21-resistant wheat accessions (1.5 fold for PR2 to 14.6 fold for PR9, S5 Table). In both species, the coordinated upregulation of PR genes was observed only in the presence of the pathogen and the resistance gene, suggesting that the Sr21 protein needs to be activated through interactions with a Pgt effector or a wheat protein modified by Pgt.
The stronger resistance response observed at 24°C suggests that temperature modulates some of the involved processes. Elevated growth temperatures have been reported to affect plant resistance to diseases by reducing steady-state levels of resistance protein at high temperatures [33], affecting temperature-sensing NB-LRR proteins [34], or affecting salicylic acid (SA) regulation [35,36]. A similar inhibition of the resistance response by high temperatures has been reported for wheat Pgt resistance genes Sr6, Sr10, Sr15, and Sr17. By contrast, Sr13- [16] and Sr21-mediated Pgt resistances are more effective at higher temperatures. Both genes also show a coordinated upregulation of the same PR genes at high temperatures, which suggests that they may share some common mechanisms.
Although the mechanisms by which Sr21 and Sr13 [16] coordinate the upregulation of PR genes are currently unknown, information from other species suggests that the NPR1 pathway or WRKY transcription factors may be involved. Previous studies in Arabidopsis have shown that NPR1 interactions with TGA transcription factors play an important role in the regulation of several PR genes [37,38]. This was also observed in barley, in which overexpression of a conserved protein from the stripe rust pathogen that competes with TGA transcription factors for the binding with NPR1, reduced the induction of several PR genes in a leaf region adjacent to a bacterial infection [39]. WRKY transcription factors are also interesting candidates because the promoter of several PR genes contain W-box elements recognized by these proteins [40,41]. In addition, NLR proteins from barley (MLA) and rice (Pb1) have been shown to interact with WRKY transcription factors to regulate their defense responses [42,43]. It would be interesting to determine if Sr21 or Sr13 can interact with NPR1, WRKY or other transcription factors to coordinate the upregulation of wheat PR genes.
The results presented here provide useful information for the utilization of Sr21 in agriculture. Since Sr21 is susceptible to some Pgt races, it needs to be deployed in combination with other Pgt resistance genes or in transgenic cassettes including multiple resistance genes. Given the better Ug99 resistance levels observed in transgenic plants carrying more than one active copy of Sr21, it might be valuable to include at least two copies of Sr21 in the transgenic cassettes. It might be also advisable to avoid combining Sr21 and Sr13, since both genes seem to operate by a similar mechanism involving the activation of multiple PR genes at high temperature. Even if Sr21 and Sr13 recognize different effectors, the pathogen could bypass both resistance genes simultaneously by attacking a single target if the two genes share a common downstream signaling pathway. Combining genes that operate by different mechanisms may reduce the probability that a single change in the pathogen can defeat multiple pyramided genes [44].
Sr21 confers only partial resistance to Ug99, but this might be useful in programs that aim to combine multiple partial resistance genes and avoid major all-stage resistance genes, a strategy that has been proposed to increase the durability of wheat resistance to rusts [45]. The deployment of Sr21 in commercial tetraploid or hexaploid commercial varieties can be accelerated by the diagnostic marker developed in this study (S10 Fig).
Materials and methods
Segregating populations and stem rust assays
A total of 7,168 recombinant gametes from two segregating populations were used to construct a high-resolution genetic map of Sr21. These populations included 734 F2 plants from population PI 272557 × DV92 and 2,850 from population PI 272557 × G3116. Plants with informative recombination events were challenged with races MCCFC (isolate 59KS19) and TTKSK (isolate 04KEN156/04) at the USDA-ARS Cereal Disease Laboratory and with race BCCBC (isolate 09CA115-2) at the University of California, Davis (UCD). Assays of response to race TTKSK were performed at 25°C during the day and 22°C during the night with a 16 h photoperiod (Fig 3) and those for BCCBC at 16°C (low temperature) and 24°C (high temperature). Procedures for inoculation and statistical analyses of infection types were reported previously [20].
BAC library screening and sequence annotation
A Bacterial Artificial Chromosome (BAC) library from the resistant parent DV92 [22] was used to generate the physical map by chromosome walking. DNAs from the selected BACs were extracted using QIAGEN Large-Construct Kit. BACs were fingerprinted using restriction enzyme HindIII, were sequenced using a combination of Illumina Hi Seq2500 at the Beijing Genomic Institute (Sacramento, CA, USA) and WideSeq at Purdue Genomics Core Facility (https://www.purdue.edu/hla/sites/genomics/wideseq-2/), and were assembled using Galaxy [46,47]. We identified and annotated the repetitive elements in the Sr21 region using the Triticeae Repeat Sequence Database (http://wheat.pw.usda.gov/ITMI/Repeats/blastrepeats3.html) and the genes using BLASTN / BLASTX searches in GenBank (http://www.ncbi.nlm.nih.gov/). These three websites were last accessed February 26, 2018.
EMS mutants screening
Mutant lines were generated by treating 10,000 seeds from the hexaploid wheat line CSSr21 (Sr21 resistance haplotype R1) with 0.8% ethyl methane sulphonate (EMS). Seeds from 1,151 independent M1 mutants were harvested, and 25 M2 seeds per family were planted and inoculated with Pgt race BCCBC (isolate 09CA115-2) at UCD. Twenty-five M3 seeds from susceptible M2 plants were retested with race MCCFC (isolate 59KS19) at the USDA-ARS Cereal Disease Laboratory.
5' and 3' race
Rapid amplification of cDNA ends (RACE) was performed using total RNA extracted from leaves of resistant parent DV92. Both 5' RACE and 3' RACE were performed using the FirstChoice RLM-RACE Kit (Invitrogen) following the manufacturer’s instruction. The PCR products from Nested PCR amplifications were cloned using the TA cloning kit (Invitrogen).
Wheat transformation and CNL1 copy number assays
A 10,463-bp genomic DNA fragment including Sr21 was amplified from DV92 BAC clone 205J7 using Phusion High-Fidelity DNA Polymerase (New England BioLabs Inc.). This fragment, including the complete Sr21 (5,208 bp) coding region and introns, 2,772 bp upstream from the start codon, and 2,483 bp downstream from the stop codon, was cloned into the binary vector pLC41Hm. The resulting plasmid pLC41HmSr21 was transformed into Agrobacterium strain EHA105, and was transformed into the Ug99-susceptible wheat variety Fielder (CNL1 haplotype S2) at the UC Davis transformation facility (http://ucdptf.ucdavis.edu/). Primers HptmikiF/R developed from the hygromycin resistance gene and primers S21CNL1F5R5 and Sr21TRYF5R5 developed from CNL1 were used to confirm the presence of transgene (S1 Table). A TaqMan Copy Number Assay was used to estimate the number of copies inserted in every transgenic event as described before [16].
Alternative splicing
Sequences from the 3' RACE described above and from the transcriptome databases of DV92 and G3116 [24] revealed the presence of several alternative splicing forms at the 3’UTR region of Sr21. To determine the number of alternative splicing variants, we first extracted total RNAs from G3116 T. monococcum plants grown at 16°C and 24°C six days after inoculation. We used these RNAs for 3’RACE reactions and cloned the PCR products using the TA cloning kit (Invitrogen). Eighty colonies from every 3' RACE reaction were PCR-amplified and sequenced using the Sanger method.
qRT-PCR analysis
Total RNA was extracted from leaves of Sr21-resistant diploid T. monococcum ssp. aegilopoides accession G3116 and hexaploid CSSr21 using Spectrum Plant Total RNA Kit (Sigma-Aldrich). First strand cDNA was synthesized from 1 µg of total RNA using a High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems). Samples were collected from plants grown under the four treatments resulting from the combination of two temperatures (16°C or 24°C) and two inoculation treatments (Pgt race BCCBC or mock inoculation). Samples were collected immediately after inoculation (0 h) and 1, 3 and 6 days post inoculation (dpi). We quantified Sr21 expression using primers Sr21qRTF1R1 (S1 Table). The same cDNA samples were used to quantify the transcript levels of six pathogenesis-related (PR) genes (PR1, PR2, PR3, PR4, PR5 and PR9 = TaPERO) using primers described before [16].
We performed the qRT-PCR reactions on an ABI 7500 Fast Real-Time PCR System (Applied Biosystems) using Fast SYBR GREEN Master Mix. Transcript levels were expressed as fold-ACTIN levels (the number of molecules in the target / the number of ACTIN molecules) using the 2ΔCT method as described before [48]. We calculated the significance of the differences in expression levels using factorial ANOVAs and the SAS program version 9.4.
Haplotype analysis of CNL1
From a previous study [49], we selected 44 T. monococcum accessions (28 cultivated and 16 wild type) resistant to races MCCFC and TTKSK and susceptible to races TTTTF, TRTTF and QFCSC, which were previously postulated to carry only Sr21 (S7 Table). We also selected 30 accessions of T. monococcum and 40 from T. urartu that were susceptible to all five races (S7 Table). Seeds were obtained from the U.S. Department of Agriculture National Small Grains Collection (NSGC).
All T. monococcum and T. urartu accessions were re-evaluated with races MCCFC (isolate 59KS19) and BCCBC (isolate 09CA115-2) (at 24°C) for this study. We also confirmed the absence of Sr35 with the diagnostic marker developed from the cloned Sr35 gene [12]. Seeds from three T. monococcum accessions showed heterogeneous resistance reactions to MCCFC (PI 427971, PI 119422 and PI 225164) and were listed twice in S8 Table (as–S and–R) resulting in a total of 74 T. monococcum accessions. The sequenced genomes of diploid wheat T. urartu (http://plants.ensembl.org/Triticum_urartu/Info/Index), tetraploid wheat Zavitan (https://wheat.pw.usda.gov/GG3/wildemmer), and hexaploid wheat Chinese Spring RefSeq v1 (https://urgi.versailles.inra.fr/blast_iwgsc/blast.php) were used to detect the corresponding CNL1 alleles.
Supporting information
(A) Proximal chromosome walk group: 22 BACs from the region proximal to Sr21. All BACs were selected from the DV92 BAC library and fingerprinted with restriction enzyme HindIII [22]. Markers CJ961291 and 179C18F7R7 delimit a 405-kb candidate region for Sr21 (blue shaded square). (B) Distal chromosome walk group: 17 T. monococcum BACs from the region distal to Sr21. In both panels, BACs in the minimum tilling path that were sequenced are indicated in red. Colored ovals represent markers used for chromosome walking, whereas colored arrows represent genes identified in the BAC sequences.
(PDF)
Transcript levels of CNL1 in 13 transgenic T0 plants based on three technical replicates from a single plant. Numbers below rectangles indicate the copy number of transgenes based on the TaqMan copy number assay. Transcript levels are expressed as fold-ACTIN using the 2ΔCT method. Fielder has a non-functional copy of CNL1.
(PDF)
(A) Six T1 plants from family T1Sr21-03 inoculated with Pgt race TTKSK (Ug99). (B) Six T1 plants from family T1Sr21-04 inoculated with TTKSK. The first row of numbers below the figure indicates the average pustule size estimated using the image analysis software ASSESS v.2.0 (n = 3). The second row of numbers is the estimated number of CNL1 copies in the transgenic plants based on a TaqMan copy number assay relative to CSSr21. Pustule size was inversely correlated with the estimated number of CNL1 copies (R = -0.92, P < 0.0001).
(PDF)
Yellow rectangles indicate coding regions and dotted blue lines introns. Gray rectangles represent 3’ and 5’ untranslated regions (UTR). The transcriptome databases of DV92 and G3116 and sequencing of multiple clones from 3' RACE reactions revealed ten alternative splice variants of CNL1 3’UTR (CNL1-1 to CNL1-10).
(PDF)
Transcript levels of Pathogenesis Related genes PR1, PR2, PR3, PR4, PR5, and PR9 (TaPERO). Diploid T. monococcum (G3116) n = 4 and hexaploid T. aestivum (CSSr21) n = 3. Values were calculated using the 2ΔCT method relative to ACTIN endogenous control (scales are comparable across genotypes). Error bars indicate standard errors of the means. Lack of parallelism between lines in all graphs indicate significant interactions (P < 0.0001, S5 Table).
(PDF)
(A-B) Inoculation with Pgt race BCCBC at 16°C (A) and 24°C (B). The hexaploid susceptible line is Chinese Spring (CS, “-”) and the resistant line is CSSr21 (“Sr21”). The diploid susceptible line is T. monococcum PI 272557 (“-”) and the resistant line is G3116 (“Sr21”). Similar differences were observed before for Sr21 resistance to TTKSK at 16°C and 20°C [20].
(PDF)
(A-D) Race BCCBC growth at 16°C. (A-B) Hexaploid wheat, (C-D) diploid wheat. (E-H) Race BCCBC growth at 24°C. (E-F) Hexaploid wheat, (G-H) diploid wheat. CSSr21 and G3116 are the hexaploid and diploid resistant lines carrying Sr21, respectively. CS and PI 272557 are the hexaploid and diploid susceptible lines, respectively. Pictures were taken five days post inoculation (dpi). Infected leaves were cleared with KOH and stained with WGA-FITC. Interaction graphs are presented in Fig 6E and 6F, and statistical analyses in S6 Table. Scale bars represent 500 μm.
(PDF)
T. monococcum Ug99 resistant (R) and susceptible (S) accessions. Four bottom lines are CNL1 closest homologs from T. urartu and A-genome of polyploid wheat. Red highlight indicates alleles present in a single haplotype. The first part of haplotype S1 is too divergent and is not presented here.
(PDF)
Neighbor-Joining tree analysis including CNL1 haplotypes, linked CNL3, CNL5 and cnl7 genes (blue squares), and the closest predicted genes from T. urartu (T.u.), T. dicoccoides (Tdic), Triticum aestivum (Traes), Hordeum vulgare (HORVU), and Brachypodium distachyon (Bradi). Coding DNA sequences were aligned with muscle as implemented in Mega 7, and phylogenetic trees were then generated using the pair-wise deletion method (bootstrap confidence values based on 1000 iterations).
(PDF)
The undigested 951-bp PCR products (black arrow) are present in genotypes DV92 (R1), G3116 (R3), PI 306540 (R2), CSSr21 (R1), PI 427971-R (R4) and PI 427796 (R5) carrying the different Sr21 resistant haplotypes. A digested band of 836-bp (yellow arrowhead, 115 bp band out of the gel) was detected in T. monococcum genotypes PI 427971-S and PI 538540 that carry the Sr21 susceptible haplotypes S3 or S4. No amplification product was detected with these primers for susceptible genotypes of T. monococcum PI 272557 (S1), T. urartu PI 428227 (S2), PI 428183 (S2), and the related susceptible haplotypes present in Kronos, Fielder and Chinese Spring (CS) (S8 Fig and S8 Table).
(PDF)
Primers for high density genetic map, screening of the T. monococcum BAC library, haplotype analysis, mutant screening, marker-assisted selection (MAS), expression analysis, 3’ and 5’ RACE, cloning and screening for transgenic studies, and copy number assays.
(PDF)
Number of insertions in each transgenic event based on T1 plants and a TaqMan copy number assay of the transgenic plants relative to CSSr21 (1 copy).
(PDF)
Alternative splicing forms were identified from multiple 3' RACE reactions (total RNAs of G3116 from 24°C / Inoculated / 6 d, 24°C / Mock-inoculated / 6 d, 16°C / Inoculated / 6 d and 16°C / Mock-inoculated / 6 d). Eighty colonies (using TA-cloning) from every 3' RACE reaction were PCR amplified and sequenced using the Sanger method. Eighty clones were analyzed for each of the four conditions. For the χ2 analysis, the five less frequent alternative splice forms were merged (CNL1-3, CNL1-4, CNL1-7, CNL1-8, CNL1-9, and CNL1-10).
(PDF)
Transcript levels at two temperatures (16°C and 24°C), two Pgt treatments (inoculated and mock-inoculated), and four collection time points after inoculation (0 hour, 1 dpi, 3 dpi and 6 dpi). (A) Three-way ANOVA at time 0 immediately after inoculation and transfer from greenhouse at 20°C to the chambers at 16°C and 24°C. (B) Four-way ANOVA at 1, 3 and 6 dpi.
(PDF)
P values of transcript levels of PR genes in CSSr21 and G3116 plants 6 dpi with Pgt race BCCBC or mock-inoculated at high (24°C) and low (16°C) temperatures. CSSr21 and G3116 indicate P values for the individual two-way ANOVAS. The last row indicates the ratio between the diploid and hexaploid average expression of the PR gene in the Pgt inoculated plants at 24°C.
(PDF)
Sporulation size was determined at 14 dpi, whereas Pgt/host DNA ratio and area of fungal growth detected by fluorescence were determined at 5 dpi. Experiments with BCCBC were performed at 16°C and 24°C and those for TTKSK were performed at 16°C and 20°C and were reported before [20]. Six replications were used per treatment combination, with the exception of sporulation size for race TTKSK where only three replications were used. Normality of residuals of the transformed data was confirmed using Shapiro-Wilk test.
(PDF)
Infection types in diploid accessions (Triticum monococcum subsp. aegilopoides, Triticum monococcum subsp. monococcum and Triticum urartu) to five Pgt races TRTTF, TTKSK, TTTTF, QFCSC, and MCCFC. Infection types shown here were based on the previous study [49].
(PDF)
CNL1 haplotypes among a collection of 74 T. monococcum and 40 T. urartu accessions.
(PDF)
Data Availability
The sequences reported in this study are available in the GenBank database as accession numbers MG582649 and MG601519-MG601526. Data supporting the findings of this study are within the manuscript or the supplementary file.
Funding Statement
Work at JD laboratory was supported by the Howard Hughes Medical Institute and by the Agriculture and Food Research Initiative Competitive Grant 2017-67007-25939 (WheatCAP) from the USDA National Institute of Food and Agriculture (NIFA). Work at MNR laboratory was supported by USDA-ARS appropriated project 5062-21220-021-00-D, the USDA-ARS National Plant Disease Recovery System, and NRI Competitive Grant 2017-67007-25939 from the USDA National Institute of Food and Agriculture (NIFA). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Pretorius ZA, Singh RP, Wagoire WW, Payne TS. Detection of virulence to wheat stem rust resistance gene Sr31 in Puccinia graminis f. sp. tritici in Uganda. Plant Dis. 2000; 84: 203. [DOI] [PubMed] [Google Scholar]
- 2.Jin Y, Singh RP, Ward RW, Wanyera R, Kinyua M, et al. Characterization of seedling infection types and adult plant infection responses of monogenic Sr gene lines to race TTKS of Puccinia graminis f. sp tritici. Plant Dis. 2007; 91: 1096–9. [DOI] [PubMed] [Google Scholar]
- 3.Jin Y, Szabo LJ, Pretorius ZA, Singh RP, Ward R, et al. Detection of virulence to resistance gene Sr24 within race TTKS of Puccinia graminis f. sp tritici. Plant Dis. 2008; 92: 923–6. [DOI] [PubMed] [Google Scholar]
- 4.Singh RP, Hodson DP, Huerta-Espino J, Jin Y, Bhavani S, et al. The emergence of Ug99 races of the stem rust fungus is a threat to world wheat production. Annu Rev Phytopathol. 2011; 49: 465–81. doi: 10.1146/annurev-phyto-072910-095423 [DOI] [PubMed] [Google Scholar]
- 5.Nazari K, Mafi M, Yahyaoui A, Singh RP, Park RF. Detection of wheat stem rust (Puccinia graminis f. sp. tritici) race TTKSK (Ug99) in Iran. Plant Dis. 2009; 93: 317. [DOI] [PubMed] [Google Scholar]
- 6.Newcomb M, Rouse OMN, Rouse MN, Szabo LJ, Johnson J, et al. Kenyan Isolates of Puccinia graminis f. sp tritici from 2008 to 2014: Virulence to SrTmp in the Ug99 race group and implications for breeding programs. Phytopathology. 2016; 106: 729–36. doi: 10.1094/PHYTO-12-15-0337-R [DOI] [PubMed] [Google Scholar]
- 7.Fetch T, Zegeye T, Park R, Hodson D, Wanyera R. Detection of wheat stem rust races TTHSK and PTKTK in the Ug99 race group in Kenya in 2014. Plant Dis. 2016; 100: 1495. [Google Scholar]
- 8.Pretorius Z, Bender C, Visser B, Terefe T. First report of a Puccinia graminis f. sp. tritici race virulent to the Sr24 and Sr31 wheat stem rust resistance genes in South Africa. Plant Dis. 2010; 94: 784. [DOI] [PubMed] [Google Scholar]
- 9.Jin Y, Szabo LJ, Rouse MN, Fetch T, Pretorius ZA, et al. Detection of virulence to resistance gene Sr36 within the TTKS race lineage of Puccinia graminis f. sp tritici. Plant Dis. 2009; 93: 367–70. [DOI] [PubMed] [Google Scholar]
- 10.Rouse MN, Nirmala J, Jin Y, Chao SM, Fetch TG, et al. Characterization of Sr9h, a wheat stem rust resistance allele effective to Ug99. Theor Appl Genet. 2014; 127: 1681–8. doi: 10.1007/s00122-014-2330-y [DOI] [PubMed] [Google Scholar]
- 11.Patpour M, Hovmøller MS, Shahin AA, Newcomb M, Olivera P, et al. First report of the Ug99 race group of wheat stem rust, Puccinia graminis f. sp. tritici, in Egypt in 2014. Plant Dis. 2016; 100: 863. [Google Scholar]
- 12.Saintenac C, Zhang W, Salcedo A, Rousse M, Trick H, et al. Identification of wheat gene Sr35 that confers resistance to Ug99 stem rust race group. Science. 2013; 341: 783–6. doi: 10.1126/science.1239022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Periyannan S, Moore J, Ayliffe M, Bansal U, Wang X, et al. The gene Sr33, an ortholog of barley Mla genes, encodes resistance to wheat stem rust race Ug99 Science. 2013; 341: 786–9. doi: 10.1126/science.1239028 [DOI] [PubMed] [Google Scholar]
- 14.Mago R, Zhang P, Vautrin S, Šimková H, Bansal U, et al. The wheat Sr50 gene reveals rich diversity at a cereal disease resistance locus. Nat Plants. 2015; 1: 15186 doi: 10.1038/nplants.2015.186 [DOI] [PubMed] [Google Scholar]
- 15.Steuernagel B, Periyannan SK, Hernandez-Pinzon I, Witek K, Rouse MN, et al. Rapid cloning of disease-resistance genes in plants using mutagenesis and sequence capture. Nat Biotechnol. 2016; 34: 652–5. doi: 10.1038/nbt.3543 [DOI] [PubMed] [Google Scholar]
- 16.Zhang W, Chen S, Abate Z, Nirmala J, Rouse MN, et al. Identification and characterization of Sr13, a tetraploid wheat gene that confers resistance to the Ug99 stem rust race group. Proc Natl Acad Sci USA. 2017; 114: E9483–E9492. doi: 10.1073/pnas.1706277114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Singh RP, Hodson DP, Jin Y, Lagudah ES, Ayliffe MA, et al. Emergence and spread of new races of wheat stem rust fungus: continued threat to food security and prospects of genetic control. Phytopathology. 2015; 105: 872–84. doi: 10.1094/PHYTO-01-15-0030-FI [DOI] [PubMed] [Google Scholar]
- 18.McDonald BA, Linde C. Pathogen population genetics, evolutionary potential, and durable resistance. Annu Rev Phytopathol. 2002; 40: 349–79. doi: 10.1146/annurev.phyto.40.120501.101443 [DOI] [PubMed] [Google Scholar]
- 19.Dvorak J, McGuire PE, Cassidy B. Apparent sources of the A genomes of wheats inferred from the polymorphism in abundance and restriction fragment length of repeated nucleotide sequences. Genome. 1988; 30: 680–9. [Google Scholar]
- 20.Chen SS, Rouse MN, Zhang WJ, Jin Y, Akhunov E, et al. Fine mapping and characterization of Sr21, a temperature-sensitive diploid wheat resistance gene effective against the Puccinia graminis f. sp tritici Ug99 race group. Theor Appl Genet. 2015; 128: 645–56. doi: 10.1007/s00122-015-2460-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.The T. Chromosome location of genes conditioning stem rust resistance transferred from diploid to hexaploid wheat. Nature New Biol. 1973; 241: 256 [DOI] [PubMed] [Google Scholar]
- 22.Lijavetzky D, Muzzi G, Wicker T, Keller B, Wing R, et al. Construction and characterization of a bacterial artificial chromosome (BAC) library for the A genome of wheat. Genome. 1999; 42: 1176–82. [PubMed] [Google Scholar]
- 23.Avni R, Nave M, Barad O, Baruch K, Twardziok SO, et al. Wild emmer genome architecture and diversity elucidate wheat evolution and domestication. Science. 2017; 357: 93–7. doi: 10.1126/science.aan0032 [DOI] [PubMed] [Google Scholar]
- 24.Fox SE, Geniza M, Hanumappa M, Naithani S, Sullivan C. De novo transcriptome assembly and analyses of gene expression during photomorphogenesis in diploid wheat Triticum monococcum. PLOS One. 2014; 9: e96855 doi: 10.1371/journal.pone.0096855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Uauy C, Wulff BB, Dubcovsky J. Combining traditional mutagenesis with new high-throughput sequencing and genome editing to reveal hidden variation in polyploid wheat. Annu Rev Genet. 2017; 51: 435–54. doi: 10.1146/annurev-genet-120116-024533 [DOI] [PubMed] [Google Scholar]
- 26.Diaz A, Zikhali M, Turner AS, Isaac P, Laurie DA. Copy number variation affecting the Photoperiod-B1 and Vernalization-A1 genes is associated with altered flowering time in wheat (Triticum aestivum). PLOS One. 2012; 7: e33234 doi: 10.1371/journal.pone.0033234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jacob F, Vernaldi S, Maekawa T. Evolution and conservation of plant NLR functions. Front Immunol. 2013; 4: 297 doi: 10.3389/fimmu.2013.00297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dubcovsky J, Dvorak J. Genome plasticity a key factor in the success of polyploid wheat under domestication. Science. 2007; 316: 1862–6. doi: 10.1126/science.1143986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bossolini E, Wicker T, Knobel PA, Keller B. Comparison of orthologous loci from small grass genomes Brachypodium and rice: implications for wheat genomics and grass genome annotation. Plant J. 2007; 49: 704–17. doi: 10.1111/j.1365-313X.2006.02991.x [DOI] [PubMed] [Google Scholar]
- 30.Costanzo S, Jia Y. Alternatively spliced transcripts of Pi-ta blast resistance gene in Oryza sativa. Plant Sci. 2009; 177: 468–78. [Google Scholar]
- 31.Halterman DA, Wei F, Wise RP. Powdery mildew-induced Mla mRNAs are alternatively spliced and contain multiple upstream open reading frames. Plant Physiol. 2003; 131: 558–67. doi: 10.1104/pp.014407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sela H, Spiridon LN, Petrescu AJ, Akerman M, Mandel‐Gutfreund Y, et al. Ancient diversity of splicing motifs and protein surfaces in the wild emmer wheat (Triticum dicoccoides) LR10 coiled coil (CC) and leucine‐rich repeat (LRR) domains. Mol Plant Pathol. 2012; 13: 276–87. doi: 10.1111/j.1364-3703.2011.00744.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bieri S, Mauch S, Shen QH, Peart J, Devoto A, et al. RAR1 positively controls steady state levels of barley MLA resistance proteins and enables sufficient MLA6 accumulation for effective resistance. Plant Cell. 2004; 16: 3480–95. doi: 10.1105/tpc.104.026682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhu Y, Qian WQ, Hua J. Temperature modulates plant defense responses through NB-LRR proteins. PLOS Pathog. 2010; 6: e1000844 doi: 10.1371/journal.ppat.1000844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang Y, Bao ZL, Zhu Y, Hua J. Analysis of temperature modulation of plant defense against biotrophic microbes. Mol Plant Microbe In. 2009; 22: 498–506. [DOI] [PubMed] [Google Scholar]
- 36.Yang S, Hua J. A haplotype-specific Resistance gene regulated by BONZAI1 mediates temperature-dependent growth control in Arabidopsis. Plant Cell. 2004; 16: 1060–71. doi: 10.1105/tpc.020479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Després C, DeLong C, Glaze S, Liu E, Fobert PR. The Arabidopsis NPR1/NIM1 protein enhances the DNA binding activity of a subgroup of the TGA family of bZIP transcription factors. Plant Cell. 2000; 12: 279–90. [PMC free article] [PubMed] [Google Scholar]
- 38.Kinkema M, Fan WH, Dong XN. Nuclear localization of NPR1 is required for activation of PR gene expression. Plant Cell. 2000; 12: 2339–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang X, Yang B, Li K, Kang Z, Cantu D, et al. A conserved Puccinia striiformis protein interacts with wheat NPR1 and reduces induction of Pathogenesis-Related genes in response to pathogens. Mol Plant Microbe In. 2016; 29: 977–89. [DOI] [PubMed] [Google Scholar]
- 40.Kalde M, Barth M, Somssich IE, Lippok B. Members of the Arabidopsis WRKY group III transcription factors are part of different plant defense signaling pathways. Mol Plant Microbe In. 2003; 16: 295–305. [DOI] [PubMed] [Google Scholar]
- 41.Turck F, Zhou A, Somssich IE. Stimulus-dependent, promoter-specific binding of transcription factor WRKY1 to its native promoter and the defense-related gene PcPR1-1 in parsley. Plant Cell. 2004; 16: 2573–85. doi: 10.1105/tpc.104.024810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Inoue H, Hayashi N, Matsushita A, Liu XQ, Nakayama A, et al. Blast resistance of CC-NB-LRR protein Pb1 is mediated by WRKY45 through protein-protein interaction. Proc Natl Acad Sci USA. 2013; 110: 9577–82. doi: 10.1073/pnas.1222155110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chang C, Yu DS, Jiao J, Jing SJ, Schulze-Lefert P, et al. Barley MLA immune receptors directly interfere with antagonistically acting transcription factors to initiate disease resistance signaling. Plant Cell. 2013; 25: 1158–73. doi: 10.1105/tpc.113.109942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lowe I, Cantu D, Dubcovsky J. Durable resistance to the wheat rusts: integrating systems biology and traditional phenotype-based research methods to guide the deployment of resistance genes. Euphytica. 2011; 179: 69–79. doi: 10.1007/s10681-010-0311-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Singh R, Huerta-Espino J, Rajaram S. Achieving near-immunity to leaf and stripe rusts in wheat by combining slow rusting resistance genes. Acta Phytopathol Hun. 2000; 35: 133–9. [Google Scholar]
- 46.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012; 19: 455–77. doi: 10.1089/cmb.2012.0021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Afgan E, Baker D, Van den Beek M, Blankenberg D, Bouvier D, et al. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2016 update. Nucleic Acids Res. 2016; 44: W3–W10. doi: 10.1093/nar/gkw343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pearce S, Vanzetti LS, Dubcovsky J. Exogenous gibberellins induce wheat spike development under short days only in the presence of VERNALIZATION 1. Plant Physiol. 2013; 163: 1433–45. doi: 10.1104/pp.113.225854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rouse M, Jin Y. Stem rust resistance in A-genome diploid relatives of wheat. Plant Dis. 2011; 95: 941–4. [DOI] [PubMed] [Google Scholar]
- 50.Ng PC, Henikoff S. SIFT: Predicting amino acid changes that affect protein function. Nucleic Acids Res. 2003; 31: 3812–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) Proximal chromosome walk group: 22 BACs from the region proximal to Sr21. All BACs were selected from the DV92 BAC library and fingerprinted with restriction enzyme HindIII [22]. Markers CJ961291 and 179C18F7R7 delimit a 405-kb candidate region for Sr21 (blue shaded square). (B) Distal chromosome walk group: 17 T. monococcum BACs from the region distal to Sr21. In both panels, BACs in the minimum tilling path that were sequenced are indicated in red. Colored ovals represent markers used for chromosome walking, whereas colored arrows represent genes identified in the BAC sequences.
(PDF)
Transcript levels of CNL1 in 13 transgenic T0 plants based on three technical replicates from a single plant. Numbers below rectangles indicate the copy number of transgenes based on the TaqMan copy number assay. Transcript levels are expressed as fold-ACTIN using the 2ΔCT method. Fielder has a non-functional copy of CNL1.
(PDF)
(A) Six T1 plants from family T1Sr21-03 inoculated with Pgt race TTKSK (Ug99). (B) Six T1 plants from family T1Sr21-04 inoculated with TTKSK. The first row of numbers below the figure indicates the average pustule size estimated using the image analysis software ASSESS v.2.0 (n = 3). The second row of numbers is the estimated number of CNL1 copies in the transgenic plants based on a TaqMan copy number assay relative to CSSr21. Pustule size was inversely correlated with the estimated number of CNL1 copies (R = -0.92, P < 0.0001).
(PDF)
Yellow rectangles indicate coding regions and dotted blue lines introns. Gray rectangles represent 3’ and 5’ untranslated regions (UTR). The transcriptome databases of DV92 and G3116 and sequencing of multiple clones from 3' RACE reactions revealed ten alternative splice variants of CNL1 3’UTR (CNL1-1 to CNL1-10).
(PDF)
Transcript levels of Pathogenesis Related genes PR1, PR2, PR3, PR4, PR5, and PR9 (TaPERO). Diploid T. monococcum (G3116) n = 4 and hexaploid T. aestivum (CSSr21) n = 3. Values were calculated using the 2ΔCT method relative to ACTIN endogenous control (scales are comparable across genotypes). Error bars indicate standard errors of the means. Lack of parallelism between lines in all graphs indicate significant interactions (P < 0.0001, S5 Table).
(PDF)
(A-B) Inoculation with Pgt race BCCBC at 16°C (A) and 24°C (B). The hexaploid susceptible line is Chinese Spring (CS, “-”) and the resistant line is CSSr21 (“Sr21”). The diploid susceptible line is T. monococcum PI 272557 (“-”) and the resistant line is G3116 (“Sr21”). Similar differences were observed before for Sr21 resistance to TTKSK at 16°C and 20°C [20].
(PDF)
(A-D) Race BCCBC growth at 16°C. (A-B) Hexaploid wheat, (C-D) diploid wheat. (E-H) Race BCCBC growth at 24°C. (E-F) Hexaploid wheat, (G-H) diploid wheat. CSSr21 and G3116 are the hexaploid and diploid resistant lines carrying Sr21, respectively. CS and PI 272557 are the hexaploid and diploid susceptible lines, respectively. Pictures were taken five days post inoculation (dpi). Infected leaves were cleared with KOH and stained with WGA-FITC. Interaction graphs are presented in Fig 6E and 6F, and statistical analyses in S6 Table. Scale bars represent 500 μm.
(PDF)
T. monococcum Ug99 resistant (R) and susceptible (S) accessions. Four bottom lines are CNL1 closest homologs from T. urartu and A-genome of polyploid wheat. Red highlight indicates alleles present in a single haplotype. The first part of haplotype S1 is too divergent and is not presented here.
(PDF)
Neighbor-Joining tree analysis including CNL1 haplotypes, linked CNL3, CNL5 and cnl7 genes (blue squares), and the closest predicted genes from T. urartu (T.u.), T. dicoccoides (Tdic), Triticum aestivum (Traes), Hordeum vulgare (HORVU), and Brachypodium distachyon (Bradi). Coding DNA sequences were aligned with muscle as implemented in Mega 7, and phylogenetic trees were then generated using the pair-wise deletion method (bootstrap confidence values based on 1000 iterations).
(PDF)
The undigested 951-bp PCR products (black arrow) are present in genotypes DV92 (R1), G3116 (R3), PI 306540 (R2), CSSr21 (R1), PI 427971-R (R4) and PI 427796 (R5) carrying the different Sr21 resistant haplotypes. A digested band of 836-bp (yellow arrowhead, 115 bp band out of the gel) was detected in T. monococcum genotypes PI 427971-S and PI 538540 that carry the Sr21 susceptible haplotypes S3 or S4. No amplification product was detected with these primers for susceptible genotypes of T. monococcum PI 272557 (S1), T. urartu PI 428227 (S2), PI 428183 (S2), and the related susceptible haplotypes present in Kronos, Fielder and Chinese Spring (CS) (S8 Fig and S8 Table).
(PDF)
Primers for high density genetic map, screening of the T. monococcum BAC library, haplotype analysis, mutant screening, marker-assisted selection (MAS), expression analysis, 3’ and 5’ RACE, cloning and screening for transgenic studies, and copy number assays.
(PDF)
Number of insertions in each transgenic event based on T1 plants and a TaqMan copy number assay of the transgenic plants relative to CSSr21 (1 copy).
(PDF)
Alternative splicing forms were identified from multiple 3' RACE reactions (total RNAs of G3116 from 24°C / Inoculated / 6 d, 24°C / Mock-inoculated / 6 d, 16°C / Inoculated / 6 d and 16°C / Mock-inoculated / 6 d). Eighty colonies (using TA-cloning) from every 3' RACE reaction were PCR amplified and sequenced using the Sanger method. Eighty clones were analyzed for each of the four conditions. For the χ2 analysis, the five less frequent alternative splice forms were merged (CNL1-3, CNL1-4, CNL1-7, CNL1-8, CNL1-9, and CNL1-10).
(PDF)
Transcript levels at two temperatures (16°C and 24°C), two Pgt treatments (inoculated and mock-inoculated), and four collection time points after inoculation (0 hour, 1 dpi, 3 dpi and 6 dpi). (A) Three-way ANOVA at time 0 immediately after inoculation and transfer from greenhouse at 20°C to the chambers at 16°C and 24°C. (B) Four-way ANOVA at 1, 3 and 6 dpi.
(PDF)
P values of transcript levels of PR genes in CSSr21 and G3116 plants 6 dpi with Pgt race BCCBC or mock-inoculated at high (24°C) and low (16°C) temperatures. CSSr21 and G3116 indicate P values for the individual two-way ANOVAS. The last row indicates the ratio between the diploid and hexaploid average expression of the PR gene in the Pgt inoculated plants at 24°C.
(PDF)
Sporulation size was determined at 14 dpi, whereas Pgt/host DNA ratio and area of fungal growth detected by fluorescence were determined at 5 dpi. Experiments with BCCBC were performed at 16°C and 24°C and those for TTKSK were performed at 16°C and 20°C and were reported before [20]. Six replications were used per treatment combination, with the exception of sporulation size for race TTKSK where only three replications were used. Normality of residuals of the transformed data was confirmed using Shapiro-Wilk test.
(PDF)
Infection types in diploid accessions (Triticum monococcum subsp. aegilopoides, Triticum monococcum subsp. monococcum and Triticum urartu) to five Pgt races TRTTF, TTKSK, TTTTF, QFCSC, and MCCFC. Infection types shown here were based on the previous study [49].
(PDF)
CNL1 haplotypes among a collection of 74 T. monococcum and 40 T. urartu accessions.
(PDF)
Data Availability Statement
The sequences reported in this study are available in the GenBank database as accession numbers MG582649 and MG601519-MG601526. Data supporting the findings of this study are within the manuscript or the supplementary file.