Abstract
The type and frequency of diagnostic testing was analyzed in a population-based cohort of boys with Duchenne muscular dystrophy or Becker muscular dystrophy. Use of muscle biopsy declined from 66.0% of boys born between January 1982 and September 1987 to 32.6% born between April 1999 and September 2004. DMD mutation was documented for 345 (73.4%) boys. Deletions were more common and point mutations were less common than that has been reported in specialty clinic or laboratory-based cohorts. Deletion of one or more exons was detected in 270 individuals (57.4% of all patients and 78.3% with a DMD mutation). Duplication was identified in 39 individuals (8.3% of all patients and 11.3% with a DMD mutation). Point mutation, small insertion, or small deletion was found in 36 individuals (7.7% of all patients and 10.4% with a DMD mutation). Point mutation analysis was performed in only 37 of 130 (28.5%) individuals with negative deletion and/or duplication testing.
Keywords: epidemiology, genetics, health services research, neuromuscular disease
Duchenne muscular dystrophy (DMD; MIM# 310200) and Becker muscular dystrophy (BMD; MIM# 300376) are allelic disorders caused by mutations of the DMD gene. DMD encodes the dystrophin protein and consists of 79 exons, including an actin-binding domain at the N-terminal, 24 spectrin-like repeats, a cysteine-rich domain, and a C-terminal domain. DMD is the largest gene yet identified. Patients with Duchenne muscular dystrophy have mutations that result in no dystrophin protein production, whereas those with Becker muscular dystrophy produce abnormal dystrophin or a decreased, but not absent, quantity of dystrophin. Becker muscular dystrophy is associated with a milder clinical course than Duchenne muscular dystrophy, but in practice there is an overlap of clinical manifestations among patients with Duchenne muscular dystrophy and Becker muscular dystrophy, so that many clinicians and investigators prefer to use the term “dystrophinopathy” when referring collectively to Duchenne and Becker muscular dystrophy.
The diagnosis of Duchenne and Becker muscular dystrophy rests on the demonstration of either a mutation of DMD or an abnormal quantity or quality of the dystrophin protein on muscle biopsy. Mutation analysis for DMD has undergone progressive refinement since its introduction. The first clinically available testing used multiplex polymerase chain reaction (PCR) and Southern blotting to identify large-scale deletions. In subsequent years, other methods were developed to identify duplications.1,2 Newer methods of analysis have since been introduced into clinical practice, including multiplex amplifiable probe hybridization, multiple ligation probe amplification, and single condition amplification internal primer sequencing, which are designed to detect deletions, duplications, and point mutations.3–6 Using these methods, it is now possible to identify DMD mutations in more than 95% of patients with a clinical diagnosis of Duchenne and Becker muscular dystrophy. Muscle biopsy may also be used for definitive diagnosis of Duchenne and Becker muscular dystrophy. Biopsy samples from skeletal muscle are examined histologically, along with immunochemistry and/or Western blot analysis, for an abnormal quantity or abnormal molecular weight dystrophin protein.7
The Muscular Dystrophy Surveillance Tracking and Research Network is a network of 5 United States geographic regions (Arizona, Colorado, Georgia, Iowa, and western New York) that established the first population-based surveillance system for Duchenne and Becker muscular dystrophy in the United States.8 Muscular Dystrophy Surveillance Tracking and Research Network collects medical information on patients with a possible dystrophinopathy from multiple sources and applies a standard case definition for classification as a definite case, probable case, or noncase. The purpose of this report is to describe the frequency and type of diagnostic testing, changes in testing methods over time, and the types of DMD mutations identified in boys classified as definite or probable Duchenne and Becker muscular dystrophy in Muscular Dystrophy Surveillance Tracking and Research Network.
Methods
The study population includes all residents born or residing in the Muscular Dystrophy Surveillance Tracking and Research Network geographic catchment area between January 1, 1982 and December 31, 2004. The investigations were approved by institutional review boards from each site. Medical records from multiple sources where individuals received medical care were reviewed. These sources include neuromuscular clinics, hospitals and hospital discharge databases, private physicians, service units for children with special health care needs, and birth defect surveillance systems. Using a computerized abstraction instrument, trained medical record abstractors recorded relevant patient information.
Clinical and family history information was used to classify patients into a case definition category. Case definition is determined by a consensus review of neuromuscular specialists from each site. All patients in this report meet the criteria for a probable or definite case. Muscular Dystrophy Surveillance Tracking and Research Network designates a patient as being definite or probable to distinguish between clinically diagnosed patients with Duchenne and Becker muscular dystrophy who have molecular confirmation and those who are clinically diagnosed but who have not had a specific molecular or muscle biopsy abnormality identified. Patients in both these case definition categories have received the clinical diagnosis of Duchenne and Becker muscular dystrophy from a neuromuscular specialist at one of the Muscular Dystrophy Surveillance Tracking and Research Network sites. Definite cases have symptoms referable to a dystrophinopathy and either (1) a documented DMD mutation, (2) a muscle biopsy demonstrating abnormal dystrophin without an alternative explanation, or (3) a creatine kinase level at least 10 times normal, a pedigree compatible with X-linked recessive inheritance, and an affected family member with a DMD mutation or a dystrophin protein abnormality on muscle biopsy. Probable cases have symptoms referable to a dystrophinopathy, a creatine kinase level at least 10 times normal, and either no DMD mutation analysis or muscle biopsy, an inconclusive muscle biopsy, or a negative DMD mutation analysis. A complete description of Muscular Dystrophy Surveillance Tracking and Research Network methods has been published previously. 8 Diagnostic testing information was recorded for all cases. This included muscle biopsy results and results from DMD mutation analyses. When original source documentation of mutation analysis was available, details of the analyses such as the extent of deletions and duplications or the full description of point mutations were recorded.
Results
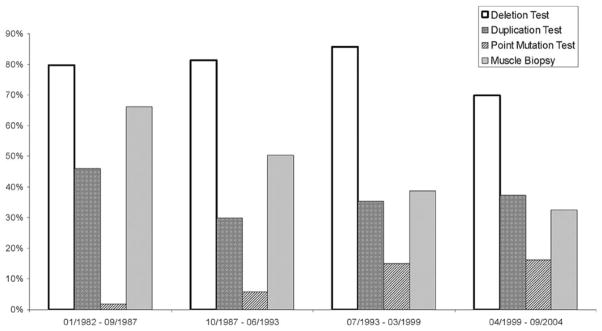
A total of 470 boys with the clinical diagnosis of Duchenne and Becker muscular dystrophy who met the case definition for either definite or probable Duchenne and Becker muscular dystrophy are included in the study population. Diagnostic testing patterns changed over time (Figure 1). The use of muscle biopsy declined from 66.1% of boys born between January 1982 and September 1987 to 32.6% of boys born between April 1999 and September 2004. Point mutation analysis ranged from a low of 1.8% in the January 1982 to September 1987 birth cohort to a high of 16.3% in the April 1999 to September 2004 cohort. No DMD mutation analysis was performed in 41 (8.7%) individuals, and DMD analysis was negative or there was insufficient information to determine conclusively that there was a DMD mutation in an additional 84 (17.9%) individuals (Table 1). A DMD mutation was documented for 345 (73.4%) boys. Deletion of one or more exons was detected in 270 individuals (57.4% of all patients and 78.3% of those with a DMD mutation). Duplication was identified in 39 individuals (8.3% of all patients and 11.3% of those with a DMD mutation). Point mutation, small insertion, or small deletion was found in 36 individuals (7.7% of all patients and 10.4% of those with a DMD mutation; Table 1).
Figure 1.
Percent of definite or probable cases with documented diagnostic test results by date of birth.
Table 1.
Frequency of DMD Mutations in the Muscular Dystrophy Surveillance Tracking and Research Network Cohort
| All Definite and Probable Cases | Cases With Confirmatory Results | |
|---|---|---|
| Negative testing or insufficient records | 84 (17.9%) | – |
| DMD deletiona | 270 (57.4%) | 270 (78.3%) |
| DMD duplicationb | 39 (8.3%) | 39 (11.3%) |
| DMD point mutationc | 36 (7.7%) | 36 (10.4%) |
| No DMD mutation analysis | 41 (8.7%) | – |
| 470 | 345 |
Only 251 of 270 individuals with DMD deletion have mutation details for analysis.
Only 34 of 39 individuals with DMD duplications have mutation details for analysis.
Only 32 of 36 individuals with DMD point mutations have mutation details for analysis.
A detailed description of the mutation was available for 317 individuals. Among the 251 deletion cases for which the extent of the deletion was known, 60 (23.9%) had single exon deletions, 110 (43.8%) had deletions of 2 to 5 exons, and 81 (32.3%) had deletions of more than 5 exons (Table 2). The most common deletion observed was deletion of exon 45, present in 13 patients (5.2% of those with deletions and 3.8% of those with a DMD mutation). Deletions of exons 48 to 50 were found in 12 patients (4.8% of those with deletions and 3.5% of those with a DMD mutation), and deletions of exons 45 to 52 were found in 10 patients (4.0% of those with deletions and 2.9% of those with a DMD mutation). In all, 162 (64.5%) were deleted for at least one of the exons within the major hotspot region spanning exons 45 to 53.
Table 2.
Muscular Dystrophy Surveillance Tracking and Research Network Deletion Location, Size, and Frequency
| 1 Exon Deletion | 2 Exon Deletions | 3 Exon Deletions | 4 Exon Deletions | 5 Exon Deletions | >5 Exon Deletions | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
|
|
|
|
||||||||||||
| Exons | # | % | Exons | # | % | Exons | # | % | Exons | # | % | Exons | # | % | Exons | # | % |
| 45 | 13 | 22 | 49–50 | 9 | 31 | 48–50 | 12 | 36 | 48–51 | 5 | 38 | 46–50 | 9 | 25 | 45–52 | 10 | 12 |
| 51 | 9 | 15 | 46–47 | 5 | 17 | 45–47 | 8 | 24 | 3–6 | 2 | 15 | 3–7 | 8 | 22 | 45–50 | 9 | 11 |
| 44 | 7 | 12 | 51–52 | 3 | 10 | 53–55 | 4 | 12 | 8–11 | 2 | 15 | 48–52 | 8 | 22 | 45–54 | 4 | 5 |
| 1 | 7 | 12 | 3–4 | 2 | 7 | 50–52 | 3 | 9 | 45–49 | 4 | 11 | 45–53 | 3 | 4 | |||
| 52 | 5 | 8 | 19–20 | 2 | 7 | 46–48 | 2 | 6 | 12–16 | 2 | 6 | 48–54 | 2 | 2 | |||
| 50 | 4 | 7 | 52–53 | 2 | 7 | ||||||||||||
| 19 | 3 | 5 | 53–54 | 2 | 7 | ||||||||||||
| 21 | 2 | 3 | |||||||||||||||
| Others | 10 | 17 | Others | 4 | 14 | Others | 4 | 12 | Others | 4 | 12 | Others | 4 | 12 | Others | 53 | 65 |
| 60 | 29 | 33 | 13 | 35 | 81 | ||||||||||||
Among the 34 patients with a detailed description of their DMD duplication available, 12 individuals (35.3%) were members of kindreds with multiple affected family members (Table 3). The most common duplication was duplication of exons 10 to 16, seen in 4 patients (11.8% of the 34 patients with duplication information available and 1.2% of all those with a DMD mutation). Most duplications (23 of 34, 67.6%) included at least one exon in the minor hotspot region spanning exons 2 to 20.
Table 3.
Muscular Dystrophy Surveillance Tracking and Research Network Duplications
| Number of Cases | Exon(s) Duplicated | Number of Exons Duplicated |
|---|---|---|
| 4 | 10–16 | 7 |
| 2 | 4–19 | 16 |
| 2 | 8–13 | 6 |
| 2 | 50–55 | 6 |
| 2 | 46–49 | 4 |
| 2 | 5–7 | 3 |
| 2 | 2–3 | 2 |
| 2 | 2 | 1 |
| 1 | 5–44 | 40 |
| 1 | 3–41 | 39 |
| 1 | 13–44 | 32 |
| 1 | 50–62 | 13 |
| 1 | 8–19 | 12 |
| 1 | 2–9 | 8 |
| 1 | 10–17 | 8 |
| 1 | 45–48 | 4 |
| 1 | 61–63 | 3 |
| 1 | 8–9 | 2 |
| 1 | 10–11 | 2 |
| 1 | 20–21 | 2 |
| 1 | 43 | 1 |
| 1 | 53 | 1 |
| 1 | 62 | 1 |
| 1 | 69 | 1 |
Point mutation analysis was performed in only 37 of 130 (28.5%) individuals with negative deletion and duplication testing. Among 32 patients with detailed results of point mutation analysis, 13 (40.6%) had a nonsense mutation that might be amenable to drug treatments that increase ribosomal readthrough of stop codons (Table 4).
Table 4.
Muscular Dystrophy Surveillance Tracking and Research Network Point Mutations
| Number of Cases | Point Mutation Effect | Point Mutation | Description |
|---|---|---|---|
| 3 | Frameshift | b c.6390delC | Exon 43 |
| 3 | Nonsense | a c.7031T>A | Exon 48 |
| 2 | Unknown | (IVS58-12 T>G) | Intron 58 |
| 2 | Frameshift | b c.9139insA | Exon 59 |
| 1 | Frameshift | a c.251dupT | Exon 4 |
| 1 | Nonsense | a c.583C>T | Exon 7 |
| 1 | Nonsense | a c.586C>T | Exon 7 |
| 1 | Missense | a c.953C>T | Exon 9 |
| 1 | Nonsense | b c.1301C>T | Exon 10 |
| 1 | Missense | b c.1596G>A | Exon 12 |
| 1 | Nonsense | b c.1823C>T | Exon 14 |
| 1 | Nonsense | b c.1682G>A | Exon 14 |
| 1 | Missense | a c. 3679C>T | Exon 27 |
| 1 | Frameshift | a c.4572delG | Exon 33 |
| 1 | Nonsense | b c. 5461G>T | Exon 39 |
| 1 | Frameshift | b c.5979delAG | Exon 41 |
| 1 | Nonsense | a c.5899C>T | Exon 41 |
| 1 | Missense | a c.9808G>C | Exon 68 |
| 1 | Frameshift | a c.9941insGTAA | Exon 68 |
| 1 | Frameshift | b9897_9898delCA | Exon 68 |
| 1 | Nonsense | a c.9978C>G | Exon 69 |
| 1 | Nonsense | a c.10141C>T | Exon 70 |
| 1 | Nonsense | a c.10171C>T | Exon 70 |
| 1 | Unknown | Exon 5 | |
| 1 | Frameshift | Exon 45 | |
| 1 | Nonsense | Exon 64 |
Verified using the coding DNA reference sequence implemented on January 01, 2003. www.dmd.nl/seqs/murefDMD.html.
Verified using the coding DNA reference sequence based on Koenig et al.2 www.dmd.nl/seqs/murefDMD_old.html.
Discussion
The importance of identifying a DMD mutation in individuals who have neuromuscular symptoms or who are at risk based on a positive family history cannot be overemphasized. Such information is critical for diagnosis, prognosis, recurrence risk, and possibly for selection of future treatments. Identification of a mutation confirms clinical diagnosis, and it is often useful in predicting the likely clinical course of the disease. Although there is some controversy regarding the full validity of what is known as the reading frame rule, it appears that about 90% of individuals with a mutation that disrupts the open reading frame of DMD will have a severe phenotype consistent with Duchenne muscular dystrophy, and a similar percentage of individuals with an in-frame mutation will have a milder clinical course most consistent with Becker muscular dystrophy.9,10
Knowledge of mutation status may also aid in recurrence risk counseling beyond its customary purpose for detection of carrier status in female relatives of affected males. Deletions are more commonly of maternal origin, whereas duplications and point mutations are more typically of grandpaternal origin.11,12 This means that when a duplication or point mutation is identified in a boy with no family history of Duchenne muscular dystrophy or Becker muscular dystrophy, the mother is likely to be a carrier. Furthermore, when a mother is negative for a duplication or point mutation identified in her son, it is unlikely that she has germline mosaicism. Finally, mutation status may in the future be used to direct treatment. In patients with nonsense mutations, drugs that suppress premature termination and selectively induce ribosomal readthrough to produce increased levels of functional protein are being investigated13 and may provide clinical benefit in individuals with this specific class of mutation.
Large-scale investigations of the distribution of DMD mutations in Duchenne and Becker muscular dystrophy suggest that single or multiple exon deletions account for about 60% of mutations, exon duplications represent about 10%, and point mutations, microinsertions, and microdeletions account for the remaining 30%.14–16 The largest database of known DMD mutations is the Leiden Duchenne muscular dystrophy mutation database, which includes information on more than 4700 individuals, including published cases and those reported to their web-accessible database.9 In the Leiden database, intragenic deletions of one or more exons make up about 72% of cases, duplications about 7%, and small deletions, insertions or point mutations approximately 20%. In the Muscular Dystrophy Surveillance Tracking and Research Network cohort, we found deletions in 57.4% of the entire population of individuals with Duchenne and Becker muscular dystrophy (Table 1). When only DMD mutation–positive individuals were included, however, deletions were seen in 78.3%, which is higher than reported in the Leiden database and much higher than most other case series. Among the 251 individuals for whom a detailed description of the deletion was available, 64.3% included at least 1 exon from the major hotspot region spanning exons 45 to 53. Whole exon deletions are detectable by multiplex PCR analysis, which is a relatively low-cost and high-throughput diagnostic assay. In countries where more time-consuming, high-cost, and high-technology analyses may not be available, multiplex PCR could therefore be expected to identify conclusively a mutation in about two thirds of affected individuals. The most commonly encountered deletion, a single exon deletion of exon 45 (Table 2), is also the most commonly reported deletion in the Leiden muscular dystrophy database.9
Among the 34 patients with an identified DMD duplication, 1 duplication spanning exons 10 to 16 was seen in 4 individuals, and 7 duplications were seen in 2 patients each (Table 3). In keeping with the understanding that duplications tend to arise in spermatogenesis are often of grandpaternal origin and are therefore often familial, all but 2 of these recurrent duplications were present in multiple members of the same kindred. The single exon duplication of exon 2 is particularly common and has been found in 6% of all patients with duplications in the Leiden Duchenne muscular dystrophy database.9 In contrast with DMD deletions, 23 of 34 (67.6%) duplications in the Muscular Dystrophy Surveillance Tracking and Research Network cohort were found in the minor hotspot region that spans exons 2 to 20. Previous reports have pointed to this region as the most common site for duplications, and more in-depth investigations have suggested that the mechanism for these duplications does not involve unequal crossing over of sister chromatids.17
As shown in Figure 1, point mutation analysis was undertaken in 37 of 130 individuals with negative deletion and duplication testing. Of the 37 individuals, 36 (97%) were found to have a point mutation, microinsertion, or microdeletion, which compares favorably with the approximately 90% detection rate reported in 2 large case series.4,14 These 36 individuals comprised 7.7% of all cases and 10.4% of those with DMD mutations, which is considerably lower than the estimated 30% that is reported in previous investigations.4,5 Of the 36 individuals, 32 had additional information available regarding the point mutation, and 13 (40.6%) of these had a nonsense mutation that might be amenable to treatments that can induce ribosomal readthrough and produce increased amounts of functional protein. As therapies become available for patients with specific types of mutations, there will likely be renewed interest in a comprehensive approach toward genotyping all patients with unknown DMD mutations.18
The high percentage of deletion-positive cases in the Muscular Dystrophy Surveillance Tracking and Research Network cohort appears to be related to the relatively low percentage of point mutations tested for and detected. For many individuals in the Muscular Dystrophy Surveillance Tracking and Research Network, point mutation analysis has never been performed. As noted previously and depicted in Figure 1, among the 130 patients who have not had a deletion or duplication identified, point mutation testing has been completed in only 37 (28.5%). Ideally, a full complement of deletion, duplication, and point mutation testing would be available in all patients, but the patients we report represent a diverse clinical cohort, for whom extensive testing may not be practical or desirable. The availability of point mutation testing on a clinical rather than research basis is a relatively new phenomenon, which may explain its low use in this cohort. Other reasons why point mutation testing has not been performed are unknown, but may include lack of insurance to pay for diagnostic testing, insurers’ denial of authorization for such testing in patients with a clinical diagnosis of Duchenne muscular dystrophy or Becker muscular dystrophy, lack of perceived benefit by patients, families, and health care providers, or lack of familiarity with this type of test by treating physicians. However, the high frequency of deletions reported herein may reflect real differences between the Muscular Dystrophy Surveillance Tracking and Research Network cohort and the populations studied in previous investigations, because our data are population-based and not derived from a specialty clinic population or diagnostic laboratory sample. The rigorous case definition standards of Muscular Dystrophy Surveillance Tracking and Research Network are likely to eliminate patients who have another neuromuscular disorder or who may be included in other mutation frequency investigations.
The use of muscle biopsy declined considerably over time, from a high of 66.1% in individuals born between January 1982 and September 1987 to 32.6% of those born between April 1999 and September 2004 (Figure 1). In addition to the high operative, hospital, and laboratory cost of open muscle biopsy, complications of bleeding, infection, and patient distress make it a much less attractive alternative to DMD mutation analysis, which requires only venipuncture. Although it has been presumed that mutation analysis would largely replace muscle biopsy as a primary diagnostic tool, this report provides clear evidence of this change. It is likely, however, that the muscle biopsy will continue to play a role in differentiating Duchenne and Becker muscular dystrophy from other neuromuscular diseases in the DMD mutation–negative individual. In rarer circumstances, muscle biopsy may also be used to investigate for immunohistological and immunohistochemical features that may distinguish between the milder course of Becker muscular dystrophy and the more severe course of Duchenne muscular dystrophy.19
In conclusion, we report on the first population-based sample of boys with Duchenne and Becker muscular dystrophy in the United States. Although comprehensive mutation analysis is increasingly used for all patients with possible Duchenne and Becker muscular dystrophy, results from Muscular Dystrophy Surveillance Tracking and Research Network suggest that muscle biopsy use will continue to decrease and will play an increasingly smaller role as a primary diagnostic test. The distribution of mutations in this population-based cohort differed from previous reports of specialty clinic or laboratory-based samples. Deletions were encountered more frequently, and point mutations were identified with a much lower frequency, presumably because point mutation testing was often not performed in individuals who were DMD deletion and/or duplication negative. The reasons for this low use are unknown, but future advances in DMD mutation analysis and Duchenne and Becker muscular dystrophy treatment may yield more clinically useful information and increase the number of patients who undergo this testing.
Acknowledgments
This research was supported by CDC Cooperative Agreement 5U01DD000187. We acknowledge the efforts of the following personnel without whom this work would not have been feasible: Rebeca Arias, April Breen, April Bryant, Shawnell Damon, Patricia Ennis, Juliana Evans, Carrie Fall, Katy Fox, Xiomara Harris, Laura Hunter, Brad McDowell, Shelley Reed, Patti Stevens, Cynthia Vogel, and Christina Westfield for record abstraction; Susan Apkon, Marques Harvey, Bobby Lyles, Zöe Powis, and Carrie Stephan for abstract review; Valerie Cwik and John Sladky for clinical review; and Florence Foo, Deborah Fox, Bobby Lyles, and Russ Roberge for data management. Statistical analysis was conducted by Jennifer Andrews, MBA, University of Arizona, and Katherine James, MSPH, Colorado Department of Health.
Footnotes
The authors have no conflicts of interest to disclose with regard to this article.
References
- 1.Diagnosis of Duchenne and Becker muscular dystrophies by polymerase chain reaction. A multicenter study. JAMA. 1992;267:2609–2615. doi: 10.1001/jama.1992.03480190051030. [DOI] [PubMed] [Google Scholar]
- 2.Koenig M, Hoffman EP, Bertelson CJ, Monaco AP, Feener C, Kunkel LM. Complete cloning of the Duchenne muscular dystrophy (DMD) cDNA and preliminary genomic organization of the DMD gene in normal and affected individuals. Cell. 1987;50:509–517. doi: 10.1016/0092-8674(87)90504-6. [DOI] [PubMed] [Google Scholar]
- 3.Armour JA, Sismani C, Patsalis PC, Cross G. Measurement of locus copy number by hybridisation with amplifiable probes. Nucleic Acids Res. 2000;28:605–609. doi: 10.1093/nar/28.2.605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Flanigan KM, von Niederhausern A, Dunn DM, Alder J, Mendell JR, Weiss RB. Rapid direct sequence analysis of the dystrophin gene. Am J Hum Genet. 2003;72:931–939. doi: 10.1086/374176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Buzin CH, Feng J, Yan J, et al. Mutation rates in the dystrophin gene: a hotspot of mutation at a CpG dinucleotide. Hum Mutat. 2005;25:177–188. doi: 10.1002/humu.20132. [DOI] [PubMed] [Google Scholar]
- 6.Gatta V, Scarciolla O, Gaspari AR, et al. Identification of deletions and duplications of the DMD gene in affected males and carrier females by multiple ligation probe amplification (MLPA) Hum Genet. 2005;117:92–98. doi: 10.1007/s00439-005-1270-7. [DOI] [PubMed] [Google Scholar]
- 7.Hoffman EP, Fischbeck KH, Brown RH, et al. Characterization of dystrophin in muscle-biopsy specimens from patients with Duchenne’s or Becker’s muscular dystrophy. N Engl J Med. 1988;318:1363–1368. doi: 10.1056/NEJM198805263182104. [DOI] [PubMed] [Google Scholar]
- 8.Miller LA, Romitti PA, Cunniff C, et al. The muscular Dystrophy Surveillance Tracking and Research Network (MD STAR-net): surveillance methodology. Birth Defects Res A Clin Mol Teratol. 2006;76:793–797. doi: 10.1002/bdra.20279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Aartsma-Rus A, Van Deutekom JC, Fokkema IF, Van Ommen GJ, Den Dunnen JT. Entries in the Leiden Duchenne muscular dystrophy mutation database: an overview of mutation types and paradoxical cases that confirm the reading-frame rule. Muscle Nerve. 2006;34:135–144. doi: 10.1002/mus.20586. [DOI] [PubMed] [Google Scholar]
- 10.Monaco AP, Bertelson CJ, Liechti-Gallati S, Moser H, Kunkel LM. An explanation for the phenotypic differences between patients bearing partial deletions of the DMD locus. Genomics. 1988;2:90–95. doi: 10.1016/0888-7543(88)90113-9. [DOI] [PubMed] [Google Scholar]
- 11.Grimm T, Meng G, Liechti-Gallati S, Bettecken T, Müller CR, Müller B. On the origin of deletions and point mutations in Duchenne muscular dystrophy: most deletions arise in oogenesis and most point mutations result from events in spermatogenesis. J Med Genet. 1994;31:183–186. doi: 10.1136/jmg.31.3.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hu XY, Ray PN, Murphy EG, Thompson MW, Worton RG. Duplicational mutation at the Duchenne muscular dystrophy locus: its frequency, distribution, origin, and phenotypegenotype correlation. Am J Hum Genet. 1990;46:682–695. [PMC free article] [PubMed] [Google Scholar]
- 13.Welch EM, Barton ER, Zhuo J, et al. PTC124 targets genetic disorders caused by nonsensemutations. Nature. 2007;447:87–91. doi: 10.1038/nature05756. [DOI] [PubMed] [Google Scholar]
- 14.Yan J, Feng J, Buzin CH, et al. Three-tiered noninvasive diagnosis in 96% of patients with Duchenne muscular dystrophy (DMD) Hum Mutat. 2004;23:203–204. doi: 10.1002/humu.10307. [DOI] [PubMed] [Google Scholar]
- 15.Muntoni F, Torelli S, Ferlini A. Dystrophin and mutations: one gene, several proteins, multiple phenotypes. Lancet Neurol. 2003;2:731–740. doi: 10.1016/s1474-4422(03)00585-4. [DOI] [PubMed] [Google Scholar]
- 16.White S, Kalf M, Liu Q, et al. Comprehensive detection of genomic duplications and deletions in the DMD gene, by use of multiplex amplifiable probe hybridization. Am J Hum Genet. 2002;71:365–374. doi: 10.1086/341942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.White SJ, Aartsma-Rus A, Flanigan KM, et al. Duplications in the DMD gene. Hum Mutat. 2006;27:938–945. doi: 10.1002/humu.20367. [DOI] [PubMed] [Google Scholar]
- 18.Aurino S, Nigro V. Readthrough strategies for stop codons in Duchenne muscular dystrophy. Acta Myol. 2006;25:5–12. [PubMed] [Google Scholar]
- 19.Tuffery-Giraud S, Saquet C, Chambert S, et al. The role of muscle biopsy in analysis of the dystrophin gene in Duchenne muscular dystrophy: experience of a national referral centre. Neuromuscul Disord. 2004;14:650–658. doi: 10.1016/j.nmd.2004.05.002. [DOI] [PubMed] [Google Scholar]