Abstract
Background:
Oral tumor is a heterogeneous group of tumors, in which it has several different histopathological and molecular features. Recently, genetic and epigenetic alterations are often detected in the development of oral cancer. Gene promoter hypermethylation leads to the silencing of cancer related genes without changes of genes sequence. To clarify the effect of RAS association domain family protein 1a (RASSF1A), retinoic acid receptor beta (RARβ), and E-cadherin (CDH1) promoter hypermethylation on the risk of oral cancer, we performed this meta-analysis.
Methods:
PubMed, Web of Science, Embase, and Chinese National Knowledge Infrastructure (CNKI) databases were retrieved to identify eligible articles. Stata 12.0 software was used to analyze extracted data of the included articles. Odds ratios (ORs) with the corresponding 95% confidence interval (95% CI) were calculated to evaluate the associations of RASSF1A, RARβ, and CDH1 promoter hypermethylation with oral cancer risk.
Results:
Around 23 literatures with 29 studies were included in the final meta-analysis, in which 12 studies were about RASSF1A promoter methylation, 4 studies were about RARβ promoter methylation, and 13 studies were about CDH1 promoter methylation. Overall, the results of this meta-analysis showed that there were significant associations between RASSF1A, RARβ, and CDH1 promoter hypermethylation and oral cancer risk (RASSF1A, OR = 11.8, 95% CI = 6.14–22.66; RARβ, OR = 20.35, 95% CI = 5.64–73.39; CDH1, OR = 13.46, 95% CI = 5.31–34.17). In addition, we found that RASSF1A promoter hypermethylation exerted higher frequency in the tongue tumor than other site tumor in mouth (RASSF1A, tongue tumor vs other site tumor in mouth, unmethylation vs methylation, OR = 0.65, 95%CI = 0.44–0.98).
Conclusion:
RASSF1A, RARβ, and CDH1 promoter hypermethylation might significantly increase the risk of oral cancer.
Keywords: CDH1, oral cancer risk, promoter hypermethylation, RARβ, RASSF1A
1. Introduction
Oral cancer is one of the most frequent tumor of head and neck tumor, in which oral squamous cell carcinoma (OSCC) accounts for approximately 90% of all oral malignancies.[1] As the sixth most common cancer worldwide, OSCC has a high mortality and low cure rate.[2] Although advancements have been made in the prevention and treatment of oral cancer, the 5-year survival rate of OSCC still remained approximately 50%.[3] In addition, invasion, lymph node metastasis, and distant metastasis of tumor cells were often found in the development of mouth cancer. Therefore, early detection was very important to the treatments of oral cancer, and therefore reduced the mortality rates of mouth cancer. It was commonly believed that environmental factors such as drinking, smoking, chewing betel nuts, and ultraviolet radiation modulated multistep process of oral carcinogenesis. Furthermore, some other intrinsic factors such as genetic and epigenetic alternations were also involved in the multistep carcinogenesis of oral cancer.[4] In recent years, DNA hypomethylation, DNA hypermethylation, loss of imprinting, chromosome inactivation, histone acetylation, histone deacetylation, histone methylation, and microRNA changes were referred in the studies of oral cancer.[4] Meanwhile, these studies often focused on gene methylation which regulated the gene expression without DNA sequence changes. And promoter methylation of many tumor suppressor genes were reported in several cancers like breast cancer, thyroid cancer, liver cancer, and nonsmall cell lung cancer.[5–8] In previous studies, promoter aberrant methylation of many candidate genes, including p16, p15, p14, DAPK, p73, APC, WIF1, RUNX3, MGMT, and hMLH1, have been identified in oral cancer.[4,9] It has been reported that the incidence rate of aberrant methylation of these genes in oral cancer tissues were higher than the normal tissues. For example, Don et al[10] have performed a systematic meta-analysis and observed a higher prevalence of methylation of p16, DAPK, and MGMT in OSCC. At the same time, many other case–control or cohort studies have been conducted to explore the potential role of these tumor suppressor genes in the process of oral cancer.[11–13] The results suggested that these genes were often significantly associated with the invasion, metastasis, and differentiation of oral tumor. Thus, these tumor suppressor genes that occurred aberrant methylation might be good biomarkers for the early detection and early treatment of oral cancer.
RASSF1A (RAS association domain family protein 1a), a kind of ras association family (RASSF) proteins, was involved in the Ras/PI3K/AKT signal pathways. RASSF1A played a key role in the cell cycle control, microtubule stabilization, cellular adhesion and motility as well as apoptosis.[14] Moreover, several studies have also found RASSF1A promoter hypermethylation contributed a lot to the gene silencing, and therefore leaded to the tumor occurrence.[15] Based on the previous record, the promoter hypermethylation of RASSF1A was a common phenomenon in many various tumors.[16] The hypermethylation of RASSF1A promoter region was originally detected in lung cancer and breast cancer.[17] Since then, hypermethylation of RASSF1A gene was reported in many different cancers and was described as a good prognostic indicator.[18] Several studies have also been performed to evaluate the relationship between RASSF1A promoter hypermethylation and oral cancer risk. However, the results remained inconsistent. Moreover, the results of other studies indicated that CDH1 and RARβ promoter hypermethylation frequencies were very high in oral cancer patients.[4] Therefore, in order to systematic assess the associations of RASSF1A, CDH1, and RARβ promoter hypermethylation with oral cancer risk, we conducted this meta-analysis.
2. Methods
2.1. Search strategy for included studies
In this study, 2 researchers independently retrieved PubMed, Web of Science, Embase, and CNKI (Chinese National Knowledge Infrastructure) databases and included the relevant articles. The literature research was up to 15 April 2017. The following keywords or medical subject headings (MeSH) words: “oral cancer,” “oral tumor,” “oral carcinoma,” “oral squamous cell carcinoma,” “OSCC,” “Salivary Gland Carcinoma,” “Buccal Carcinoma,” “Salivary Adenoid Cystic Carcinoma,” “RASSF1A,” “CDH1,” “E-cadherin,” “RARβ,” “methylation,” and “hypermethylation” were used to search eligible articles. In addition, references of included articles were reviewed for additional eligible studies. The literature searching was limited to the studies of human disease.
2.2. Study selection criteria
The inclusion criteria were: studies assessing the associations of RASSF1A, RARβ, and CDH1 hypermethylation with oral cancer risk; and case–control or cohort studies that contained data of hypermethylation frequency both in control and case group. If studies did not meet the following criteria, they would be removed: no or incomplete relevant data about RASSF1A, RARβ, and CDH1 methylation data; duplicate data; meta-analysis and review article; and low-quality studies. All relevant articles were evaluated and selected by 2 investigators. If discrepancies occurred in the process of studies selection, the third researcher would help to resolve this problem through discussion with the 2 reviewers. Furthermore, if duplicate data were showed in different studies, the most complete and latest data were selected.
2.3. Data extraction and methodology quality assessment
Two reviewers independently extracted the data of gene hypermethylation frequency. The following information was extracted from included articles: first author's name, publication year, race, frequency of gene methylation, disease type, detection method of genes methylation, and country of studied population. The methodological quality, including selection of case and control (4 stars), comparability of the groups (2 stars), and ascertainment of exposure (3 stars), were evaluated with the Newcastle–Ottawa scale table. If a study got ≥ 6 stars, the study was considered as high quality and included for this meta-analysis, otherwise it would be removed.
2.4. Statistical methods
STATA software (version 12.0, Stata Corporation, College Station, Texas) was used to conduct all statistical analysis. The associations of RASSF1A, RARβ, and CDH1 methylation with oral cancer risk were evaluated with ORs and 95% CI. All P values were 2-sided in which P < .05 was considered as statistically significant. Heterogeneity among studies were detected by chi-squared test based on Q-statistic test.[19] If P value was < .05 or I2 value was > 50%, which indicated a significant heterogeneity, a random-effects model was used; otherwise, a fixed-effect model would be applied.[20,21] In addition, Z-test was conducted to determine the strength of pooled ORs. In order to assess the publication bias, Egger's test and Begg's test were performed to detect between-study publication bias, in which P < .05 indicated a significant publication bias.[22,23] Moreover, a sensitivity analysis was conducted to further detect the stability of overall ORs by sequential deletion of each study. In the meta-analysis of CDH1 promoter hypermethylation, meta-regression was also performed to explore the source of heterogeneity due to significant heterogeneity.
2.5. Ethics approval
This meta-analysis did not collect clinical sample and applied animals experiment. Moreover, all included studies were approved by relevant Ethics Committee in eligible studies. Therefore, ethical approval was not required.
3. Results
3.1. Characteristics of included studies
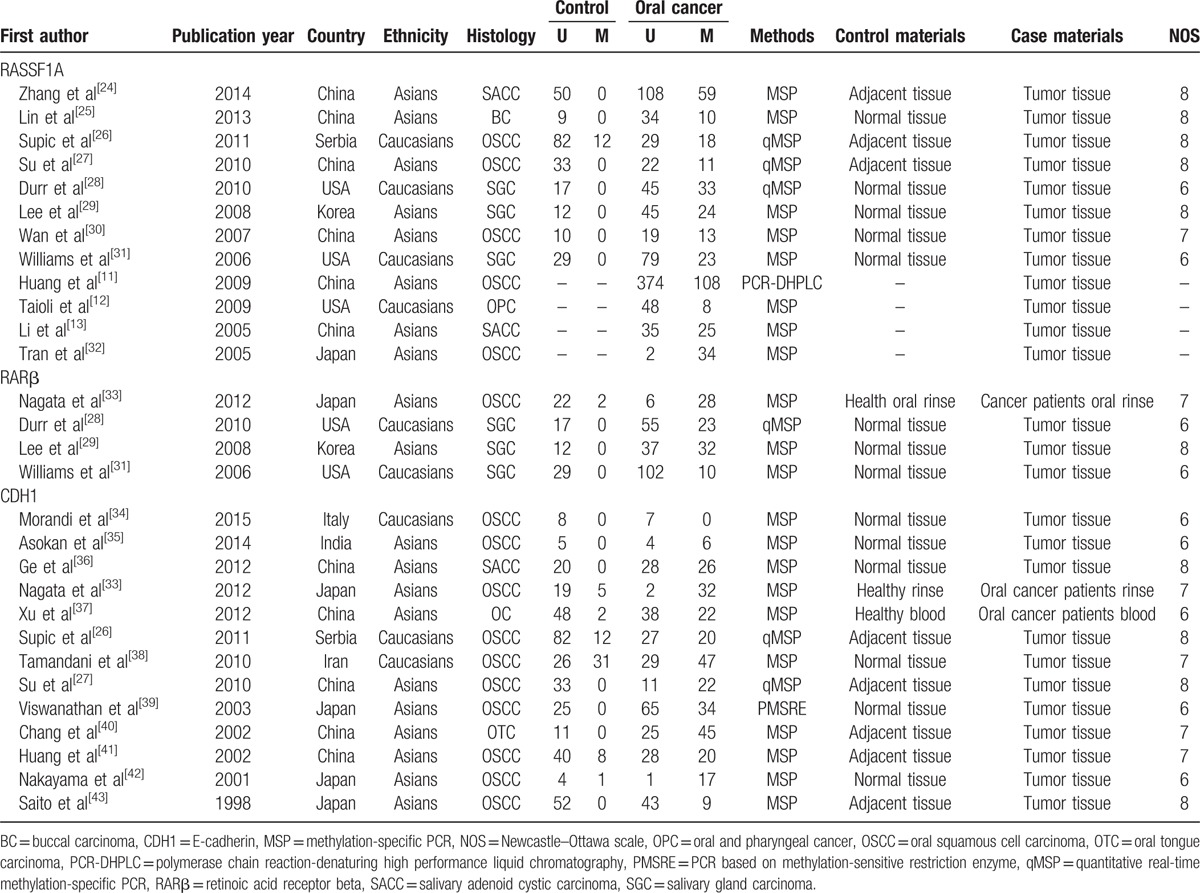
The procedure of literature searching was shown in Figure 1. Around 160 potentially relevant articles were identified in the initial searching. After removing repeat articles, 75 relevant records were reviewed and selected. Through reading titles and abstracts, 55 articles were remained, of which 20 irrelevant literatures were excluded. In addition, the full-text of remained 55 articles were read, in which 32 studies did not contained relevant effective data and were eliminated. Finally, 23 articles with 29 studies were included for the meta-analysis, in which 12 studies with 254 controls and 1238 cases were about RASSF1A, 4 studies with 82 controls and 293 cases were about RARβ, and 13 studies with 432 controls and 608 cases were about CDH1.[24–43] Moreover, all studies obtained a score of ≥ 6, which indicated high methodological quantity of included studies. The characteristics of eligible studies in the present meta-analysis were shown in Table 1.
Figure 1.
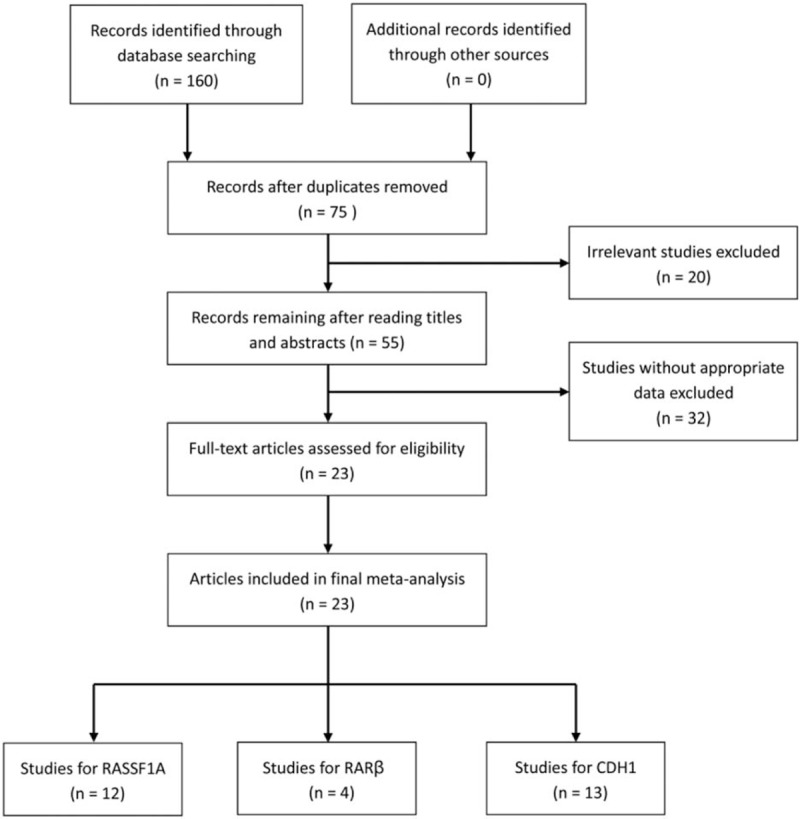
Flowchart of literature selection.
Table 1.
Basic characteristic of the eligible studies.
3.2. RASSF1A, RARβ, and CDH1 promoter hypermethylation and oral cancer risk
The RASSF1A methylation data of 12 case–control studies were pooled together and the ORs were calculated to assess the association between RASSF1A promoter hypermethylation and oral cancer risk. On the basis of the results, the overall pooled ORs clarified that RASSF1A promoter hypermethylation was significantly associated with oral cancer risk (OR = 11.8, 95% CI = 6.14–22.66). According to the results of Q-statistic test, no heterogeneity among studies was found (P = .337, I2 = 12.0%). Then, subgroup analysis based on oral cancer subtype was performed and significant association was detected in OSCC and SGC (salivary gland carcinoma) (OSCC, OR = 6.78, 95% CI = 3.20–14.37; SGC, OR = 18.51, 95% CI = 3.58–95.79). Furthermore, significant associations of RARβ and CDH1 promoter hypermethylation with oral cancer risk were detected in the meta-analysis (RARβ, OR = 20.35, 95% CI = 5.64–73.39; CDH1, OR = 13.46, 95% CI = 5.31–34.17). In the analysis for CDH1 methylation, significant heterogeneity among studies was found (I2 = 73.1%, P = .000). In order to explore the source of heterogeneity, we performed a meta-regression, in which the results indicated that ethnicity might be the mainly source of heterogeneity (P = .028, 95% CI = 0.275–3.976). In the stratified analysis based on ethnicity, CDH1 promoter hypermethylation might significantly increase the oral cancer risk in Asians, but not in Caucasians (Asians, OR = 21.79, 95% CI = 8.66–54.82; Caucasians, OR = 2.57, 95% CI = 0.71–9.31). However, only 3 studies for CDH1 methylation in Caucasians were included to calculate the pooled OR (Table 2, Figs. 2–4).
Table 2.
Subgroup analysis of the association between RASSF1A, RARβ, and CDH1 promoter methylation and oral cancer.
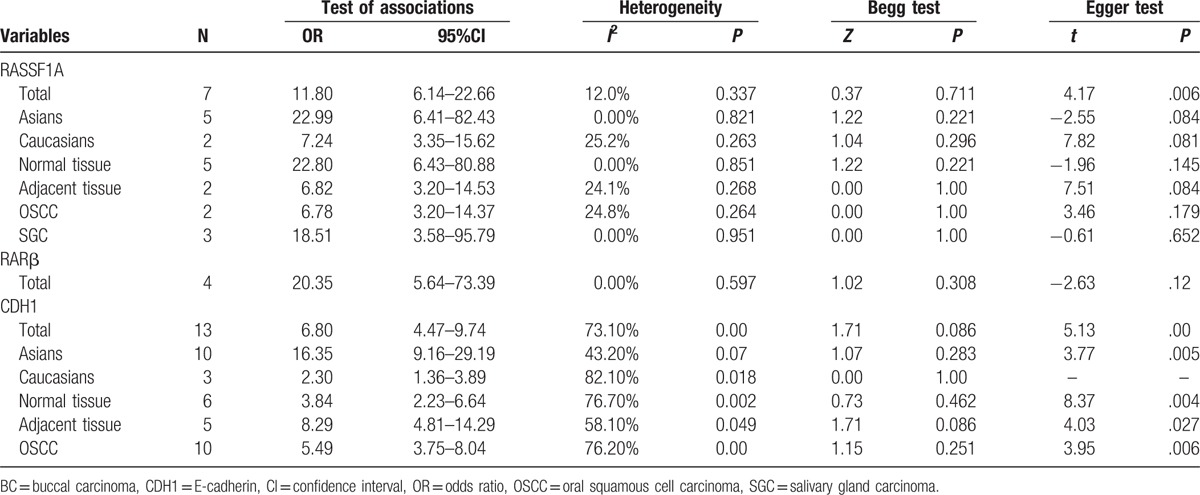
Figure 2.
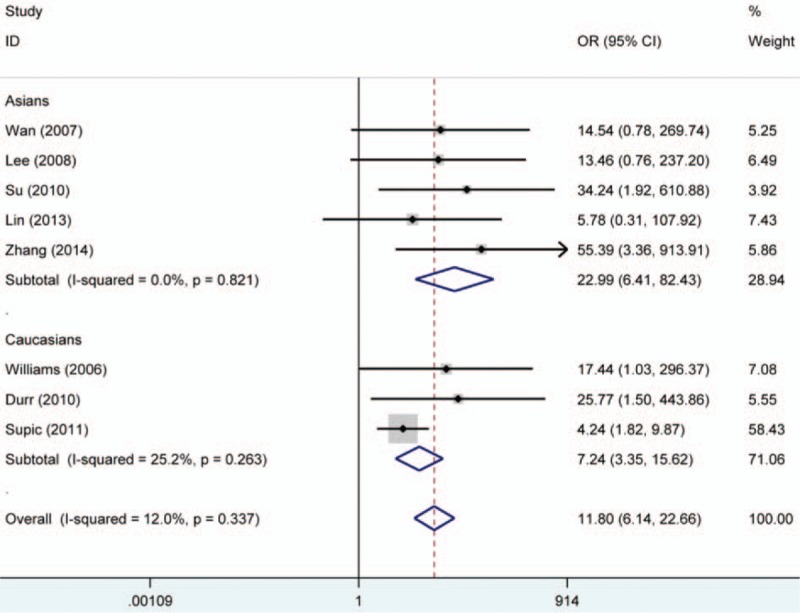
Forest plot on association between RASSF1A promoter hypermethylation and oral cancer risk. The forest plot is drawn to calculate a pooled estimated value of ORs and 95%CIs with stata 12.0 software. For each study, the estimation of ORs and its 95% CI are plotted with a box and a horizontal line which cross the imaginary line. The length of the black line which crosses the imaginary line is proportional to the 95%CIs of included studies. The weight represents the number of elements that give rise to the overall value. According to the chi-squared test based on Q-statistic test, the value of I-squared and P value were used to evaluate the heterogeneity among studies. And no heterogeneity is found in the forest plot. The results indicated that people with RASSF1A promoter hypermethylation were 11.8 times higher risk than those without RASSF1A promoter hypermethylation to suffer from oral cancer. In addition, subgroup analysis based ethnicity was performed. ORs = odds ratios, RASSF1A = RAS association domain family protein 1a.
Figure 4.
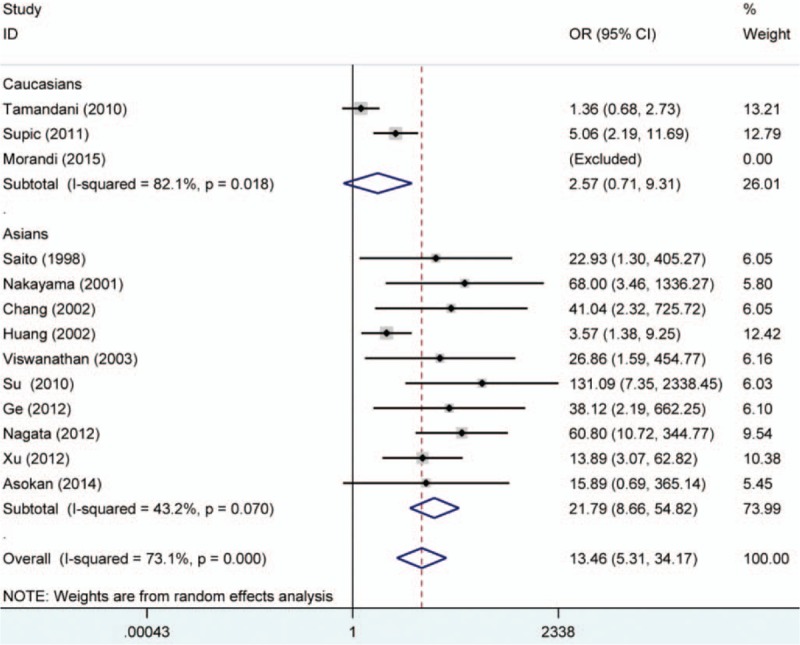
Forest plot on association between CDH1 promoter hypermethylation and oral cancer risk. The forest plot is drawn to calculate a pooled estimated value of ORs and 95%CIs with stata 12.0 software. For each study, the estimation of ORs and its 95% CI are plotted with a box and a horizontal line which cross the imaginary line. The length of the black line which crosses the imaginary line is proportional to the 95%CIs of included studies. The weight represents the number of elements that give rise to the overall value. According to the chi-squared test based on Q-statistic test, the value of I-squared and P value were used to evaluate the heterogeneity among studies. Although heterogeneity is found in the overall analysis, no significant heterogeneity is found in the subgroup-analysis. In the forest plot, the results demonstrated that people with CDH1 promoter hypermethylation were 13.46 times higher risk than those without CDH1 promoter hypermethylation to suffer from oral cancer. CDH1 = E-cadherin, ORs = odds ratios.
Figure 3.
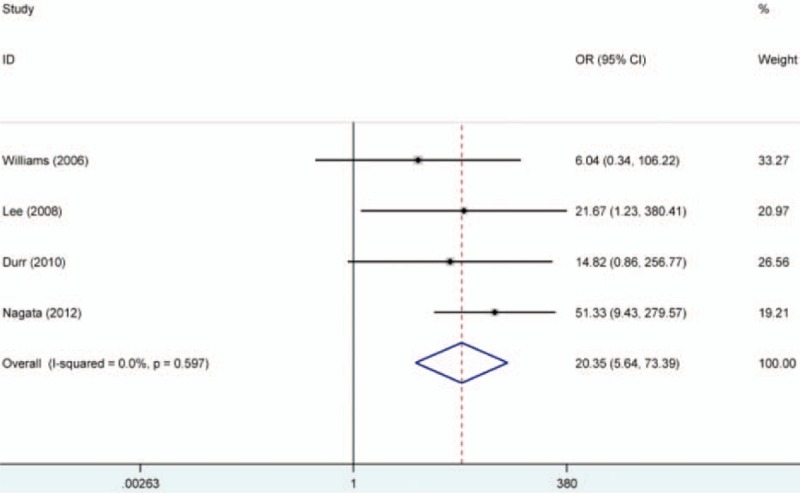
Forest plot on association between RARβ promoter hypermethylation and oral cancer risk. The forest plot is drawn to calculate a pooled estimated value of ORs and 95%CIs with stata 12.0 software. For each study, the estimation of ORs and its 95% CI are plotted with a box and a horizontal line which cross the imaginary line. The length of the black line which crosses the imaginary line is proportional to the 95%CIs of included studies. The weight represents the number of elements that give rise to the overall value. According to the chi-squared test based on Q-statistic test, the value of I-squared and P value were used to evaluate the heterogeneity among studies. And no heterogeneity is found in the analysis. In this forest plot, the results showed that people with RARβ promoter hypermethylation were 20.35 times higher risk than those without RARβ promoter hypermethylation to suffer from oral cancer. ORs = odds ratios, RARβ = retinoic acid receptor beta, RASSF1A = RAS association domain family protein 1a.
3.3. RASSF1A promoter hypermethylation and development of oral cancer
To determine whether promoter methylation of RASSF1A correlates with the development of oral cancer, the statistical analysis of associations of RASSF1A promoter methylation with TNM-stage, tumor-stage, differentiation, and lymph node metastasis of oral cancer were conducted. According to the results, no significant associations were detected. However, there was a remarkably high frequency of RASSF1A promoter hypermethylation in tongue tumor, which suggested that RASSF1A promoter hypermethylation might play an important role in the risk of tongue tumor (tongue tumor vs other site tumor in mouth, OR = 0.65, 95% CI = 0.44–0.98). Furthermore, we discussed the correlation of promoter hypermethylation of RASSF1A with smoking and drinking in oral cancer. Similarly, no significant associations of RASSF1A promoter hypermethylation with smoking and drinking in oral cancer were found. All results were shown in Table 3 (Fig. 5).
Table 3.
Quantitative analyses of RASSF1A promoter hypermethylation and clinicopathological variables of oral cancer.
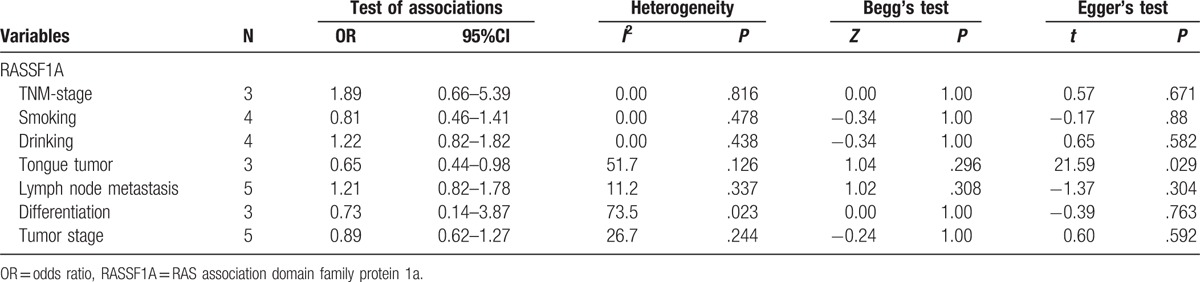
Figure 5.
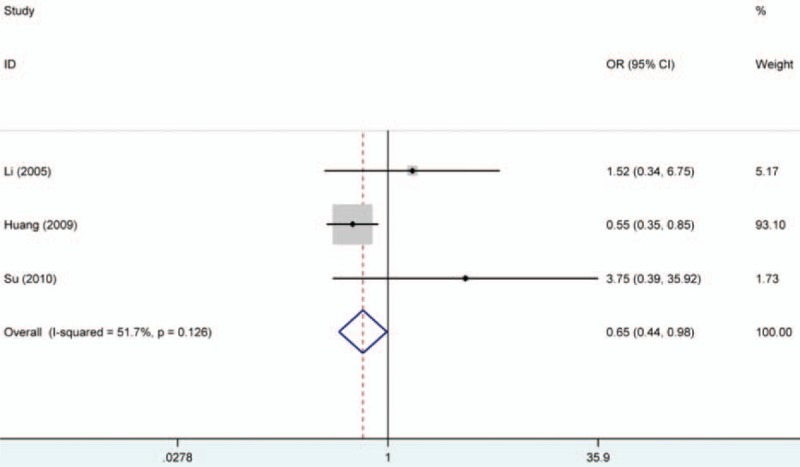
Forest plot on association between RASSF1A promoter hypermethylation and tongue tumor risk. The forest plot is drawn to calculate a pooled estimated value of ORs and 95%CIs with stata 12.0 software. For each study, the estimation of ORs and its 95% CI are plotted with a box and a horizontal line which cross the imaginary line. The length of the black line which crosses the imaginary line is proportional to the 95%CIs of included studies. The weight represents the number of elements that give rise to the overall value. According to the chi-squared test based on Q-statistic test, the value of I-squared and P value were used to evaluate the heterogeneity among studies. And no heterogeneity is found in the analysis. Interestingly, in this forest plot, the results showed that people with RASSF1A promoter hypermethylation had a lower risk than those without RARβ promoter hypermethylation to suffer from tongue cancer. ORs = odds ratios, RASSF1A = RAS association domain family protein 1a.
3.4. Sensitivity analysis and publication bias
Begg's test and Egger's test were all conducted to observe the publication bias in the meta-analysis of RASSF1A, RARβ, and CDH1 hypermethylation. Significant publication bias was found (P < .05) in the analysis of CDH1 aberrant methylation. So the subgroup analysis based on ethnicity was performed. Moreover, the results of funnel plots shown that the most dots were symmetric other than the funnel plot for CDH1 hypermethylation. At the same time, sensitivity analysis was also conducted and the overall pooled ORs did not significantly changed (Figs. 6–8).
Figure 6.
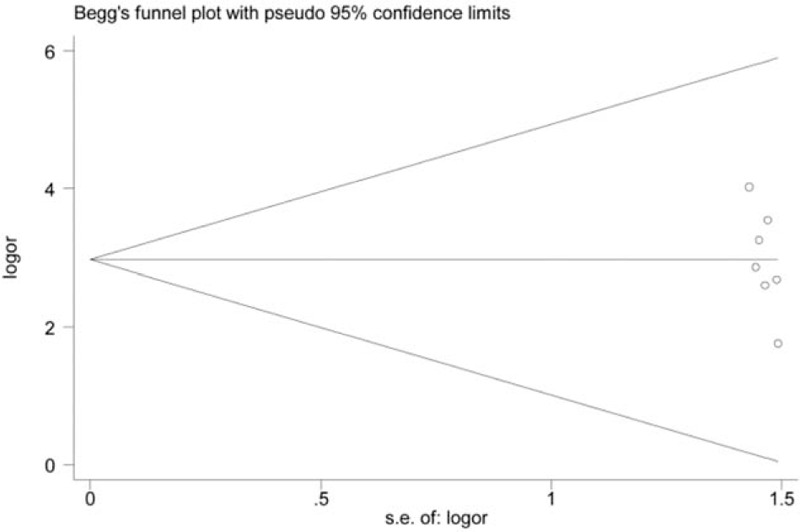
Funnel plot on association between RASSF1A promoter hypermethylation and oral cancer risk (control vs case, unmethylation vs methylation). s.e.of: logOR, standard error of log odds ratio. Log OR, log odds ratio. The funnel plot is used to assess the publication bias among included studies. The horizontal axis represents the effect size of each included study. The area enclosed by the 2 diagonal lines simulates the 95%CIs. If the dot is located out the 95%CIs, publication bias may exist among studies. In addition, these dots that represent included studies should be symmetrical and be distributed on both sides of the angular bisector, which demonstrates no publication bias is found. From the funnel plot of Begg's test, we will acquire a P value. According to the results, the P value of the funnel plot was > 0.05. Thus, no significant publication bias was found in this analysis. Log OR = standard error of log odds ratio, RARβ = retinoic acid receptor beta, ORs = odds ratios, RASSF1A = RAS association domain family protein 1a, s.e.of: log OR = standard error of log odds ratio.
Figure 8.
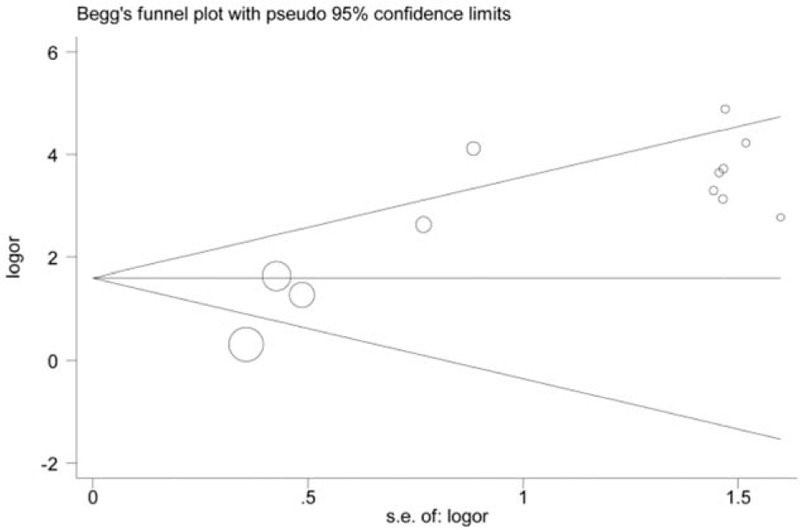
Funnel plot on association between CDH1 promoter hypermethylation and oral cancer risk (control vs case, unmethylation vs methylation). s.e.of: logOR, standard error of log OR. The funnel plot is applied to evaluate the publication bias among included studies. The horizontal axis represents the effect size of each included study. The area enclosed by the 2 diagonal lines simulates the 95%CIs. If the dot is located out the 95%CIs, publication bias may exist among studies. In addition, these dots that represent included studies should be symmetrical and be distributed on both sides of the angular bisector, which demonstrates no publication bias is found. From the funnel plot of Begg's test, we will acquire a P value. From the funnel plot, the P value was < 0.05. Therefore, publication bias may exist among these studies involving the analysis of association between CDH1 promoter hypermethylation and oral cancer risk. CDH1 = E-cadherin, log OR = log odds ratio, s.e.of: log OR = standard error of log odds ratio.
Figure 7.
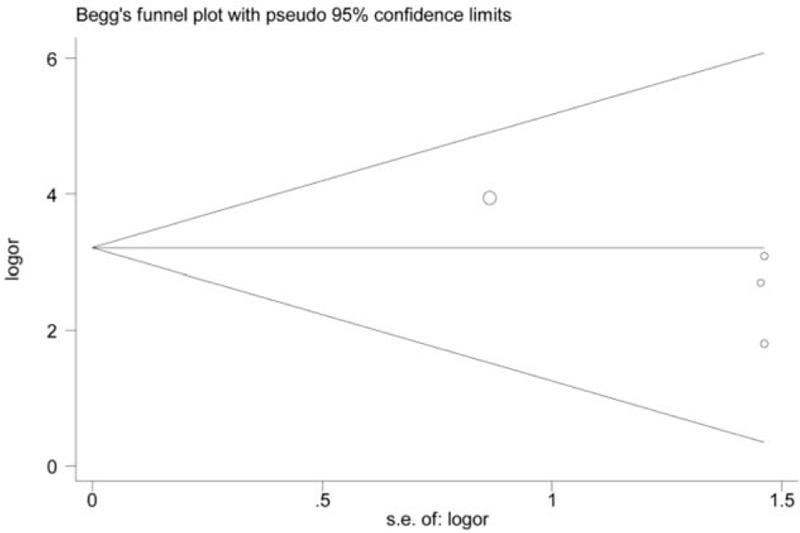
Funnel plot on association between RARβ promoter hypermethylation and oral cancer risk (control vs case, unmethylation vs methylation). s.e.of: logOR, standard error of log OR. The funnel plot is applied to evaluate the publication bias among included studies. The horizontal axis represents the effect size of each included study. The area enclosed by the 2 diagonal lines simulates the 95%CIs. If the dot is located out the 95%CIs, publication bias may exist among studies. In addition, these dots that represent included studies should be symmetrical and be distributed on both sides of the angular bisector, which demonstrates no publication bias is found. From the funnel plot of Begg's test, we will acquire a P value. From the results, the P value of the funnel plot was > 0.05. Therefore, no significant publication bias was found in this analysis. Log OR = standard error of log odds ratio, RARβ = retinoic acid receptor beta, s.e.of: log OR = standard error of log odds ratio.
4. Discussion
DNA mutations and gene epigenetic inactivation affected the function of many genes, which they were commonly detected in tumor suppressor genes. These changes affected the normal growth control of cells in which cell cycle was disturbed and cell abnormal proliferation was driven. Although several changes of chromosome and genes were described in OSCC, the fundamental cause was the inactivation of tumor suppressor genes.[44] For example, one of the most important tumor suppressor genes was p16, as we all known, it regulated the cell growth just like p53.[45] Much of the work of genetic and epigenetic changes in oral cancer have been done, however, complex interactions between the gene products and signal pathways were very complicated.[46] MDM (murine double minute), class of oncogenes, had a multiple role in tumor inhibition, in which MDM2 (murine double minute 2) inhibited p53 if p53 mutations were absent. Moreover, the tumor cell cycle was regulated by p16 and p14 which stabilized p53 protein by MDM2.[45] Although a lot of studies about tumor suppressor genes in OSCC were conducted, there might be some different patterns of gene silencing among oral cancer patients. Therefore, more tumor suppressor genes might be studied to discuss the potential role in the development of oral cancer.
According to the results of literature search, this was the first meta-analysis to evaluate the associations of RASSF1A, RARβ, and CDH1 promoter hypermethylation with oral cancer risk. In the present study, 12 studies were combined to assess the association between RASSF1A promoter hypermethylation and oral cancer risk, in which the results indicated a significant association. In the included studies, these results were consistent with the results of Su et al, Zhang et al, Williams et al, and Durr et al.[24,27,28,31] At the same time, no significant heterogeneity among studies was detected in which P value was > .05 and I2 value was < 50%. So although oral cancer included many subtypes, theses studies could be put together to calculate the pooled ORs and assess the association between levels of genes methylation and oral cancer risk. Meanwhile, subgroup analysis was still conducted to obtain more accurate results in different races. From the results, we found that RASSF1A promoter hypermethylation was significantly associated with oral cancer risk in Asians and Caucasians. However, the number of studies was very small after subgroup analysis based on oral cancer subtype was performed. In addition, the significant correlation between RASSF1A promoter hypermethylation and oral cancer risk was observed both in normal tissue and adjacent tissue of control group. But the results should be carefully taken into consideration due to the small sample size. Furthermore, this study demonstrated that RARβ and CDH1 promoter hypermethylation were significantly associated with the oral cancer risk. In previous studies, Williams et al and Durr et al believed that there were no association between RARβ methylation and oral cancer risk, however, according to the results of the meta-analysis, the frequency of RARβ promoter hypermethylation in oral cancer group was higher than control group.[28,29,31,33] In addition, we discussed the relationship between RASSF1A promoter hypermethylation and clinicopathological features in oral cancer patients. To achieved accurate statistic data, we conducted heterogeneity analysis for all clinicopathological variables of oral cancer and environmental factors such as: smoking and drinking. Significant interstudy heterogeneity was only detected among studies for differentiation of oral cancer (I2 = 73.5%, P = .023), and random effects model was therefore applied to calculate the pooled ORs and 95% CI. Based on the ORs and 95% CI derived from the present meta-analysis, a significantly increased risk of tongue cancer with RASSF1A promoter hypermethylation was found. However, no significant associations were found between RASSF1A promoter hypermethylation and tumor stage, lymph node metastasis, differentiation, and TNM-stage of oral cancer. Although there was no clear evidence of significant associations between RASSF1A promoter hypermethylation and clinicopathologic features of oral cancer, we still needed to explore these associations due to the small sample size. Furthermore, more larger scale, multicenter, and more reasonable study should be performed to confirm the predictive value of RASSF1A promoter hypermethylation in the development of oral cancer.
Additionally, significant heterogeneity among studies was detected in the meta-analysis for CDH1 promoter hypermethylation. Thus, the subgroup analysis based on ethnicity and meta-regression was carried out. The effects of publication year, country, disease type, control type, detection method of methylation, and ethnicity on the association between RASSF1A promoter methylation and oral cancer risk were evaluated by meta-regression. The results showed these factors were not the mainly cause of significant heterogeneity other than ethnicity in which P value was < .05. The same results were found in the stratified analysis based on race which no significant heterogeneity was detected in the subgroup analysis. Finally, the pooled ORs of CDH1 and RARβ were stable on the basis of the results of sensitivity analysis. However, significant publication bias for the meta-analysis of RASSF1A promoter hypermethylation was detected according to the results of Egger's test (P = .006 < .05), while sensitivity analysis results of RASSF1A promoter hypermethylation indicated that the ORs were significantly changed after the study of Supic et al[26] was removed. So the study of Supic et al was eliminated and the heterogeneity and publication bias were significantly reduced.
Notably, although many studies were included to explore these associations, several limitations should be noted in this meta-analysis: the studied population only included Asians and Caucasians; all eligible studies in this meta-analysis were retrospective, some biases might exist in the selection of samples; the cut-off value or evaluation criteria of genes methylation detection was unclear which might bring heterogeneity among different studies; the clinical information of oral patients was too small to get more accurate results; few negative and unpublished studies were included and a tendency for positive results might increase the publication bias.
5. Conclusion
In summary, our meta-analysis demonstrated that PASSF1A, RARβ, and CDH1 promoter hypermethylation were significantly associated with the oral cancer risk. Considering limitations in this meta-analysis, additional multicenter validation studies were still needed to evaluate the associations between RASSF1A, RARβ, and CDH1 promoter hypermethylation and oral cancer risk in the future.
Footnotes
Abbreviations: 95% CI = 95% confidence interval, CDH1 = E-cadherin, CNKI = Chinese National Knowledge Infrastructure, log OR = log odds ratio, MDM = murine double minute, MDM2 = murine double minute 2, NOS = Newcastle–Ottawa scale, ORs = odds ratios, OSCC = oral squamous cell carcinoma, RARβ = retinoic acid receptor beta, RASSF = RAS association family, RASSF1A = RAS association domain family protein 1a, s.e.of: log OR = standard error of log odds ratio, SGC = salivary gland carcinoma.
GW and HW contributed equally to this work.
The authors have no conflicts of interest to disclose.
References
- [1].Stewart BW, Greim H, Shuker D, et al. Defence of IARC monographs. Lancet 2003;361:1300. [DOI] [PubMed] [Google Scholar]
- [2].Stransky N, Egloff AM, Tward AD, et al. The mutational landscape of head and neck squamous cell carcinoma. Science 2011;333:1157–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Kantola S, Parikka M, Jokinen K, et al. Prognostic factors in tongue cancer—relative importance of demographic, clinical and histopathological factors. Br J Cancer 2000;83:614–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Mascolo M, Siano M, Ilardi G, et al. Epigenetic disregulation in oral cancer. Int J Mol Sci 2012;13:2331–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Luo S, Chen J, Mo X. The association of PTEN hypermethylation and breast cancer: a meta-analysis. Onco Targets Ther 2016;9:5643–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Shou F, Xu F, Li G, et al. RASSF1A promoter methylation is associated with increased risk of thyroid cancer: a meta-analysis. Onco Targets Ther 2017;10:247–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Yang X, Yang L, Dai W, et al. Role of p14ARF and p15INK4B promoter methylation in patients with lung cancer: a systematic meta-analysis. Onco Targets Ther 2016;9:6977–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Zhang X, He H, Zhang X, et al. RUNX3 promoter methylation is associated with hepatocellular carcinoma risk: a meta-analysis. Cancer Invest 2015;33:121–5. [DOI] [PubMed] [Google Scholar]
- [9].Shao C, Sun W, Tan M, et al. Integrated, genome-wide screening for hypomethylated oncogenes in salivary gland adenoid cystic carcinoma. Clin Cancer Res 2011;17:4320–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Don KR, Ramani P, Ramshankar V, et al. Promoter hypermethylation patterns of P16, DAPK and MGMT in oral squamous cell carcinoma: a systematic review and meta-analysis. Indian J Dent Res 2014;25:797–805. [DOI] [PubMed] [Google Scholar]
- [11].Huang KH, Huang SF, Chen IH, et al. Methylation of RASSF1A, RASSF2A, and HIN-1 is associated with poor outcome after radiotherapy, but not surgery, in oral squamous cell carcinoma. Clin Cancer Res 2009;15:4174–80.1. [DOI] [PubMed] [Google Scholar]
- [12].Taioli E, Ragin C, Wang XH, et al. Recurrence in oral and pharyngeal cancer is associated with quantitative MGMT promoter methylation. BMC Cancer 2009;9:354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Li J, El-Naggar A, Mao L. Promoter methylation of p16INK4a, RASSF1A, and DAPK is frequent in salivary adenoid cystic carcinoma. Cancer 2005;104:771–6. [DOI] [PubMed] [Google Scholar]
- [14].Donninger H, Vos MD, Clark GJ. The RASSF1A tumor suppressor. J Cell Sci 2007;120:3163–72. [DOI] [PubMed] [Google Scholar]
- [15].Huang KH, Huang SF, Chen IH, et al. Methylation of RASSF1A, RASSF2A, and HIN-1 is associated with poor outcome after radiotherapy, but not surgery, in oral squamous cell carcinoma. Clin Cancer Res 2009;15:4174–80. [DOI] [PubMed] [Google Scholar]
- [16].Grawenda AM, O’Neill E. Clinical utility of RASSF1A methylation in human malignancies. Br J Cancer 2015;113:372–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Dammann R, Li C, Yoon JH, et al. Epigenetic inactivation of a RAS association domain family protein from the lung tumour suppressor locus 3p21.3. Nat Genet 2000;25:315–9. [DOI] [PubMed] [Google Scholar]
- [18].Meng RW, Li YC, Chen X, et al. Aberrant methylation of RASSF1A closely associated with HNSCC, a meta-analysis. Sci Rep 2016;6:20756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Higgins JP, Thompson SG, Deeks JJ, et al. Measuring inconsistency in meta-analyses. BMJ 2003;327:557–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials 1986;7:177–88. [DOI] [PubMed] [Google Scholar]
- [21].Mantel N, Haenszel W. Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst 1959;22:719–48. [PubMed] [Google Scholar]
- [22].Song F, Gilbody S. Bias in meta-analysis detected by a simple, graphical test. Increase in studies of publication bias coincided with increasing use of meta-analysis. BMJ 1998;316:471. [PMC free article] [PubMed] [Google Scholar]
- [23].Peters JL, Sutton AJ, Jones DR, et al. Comparison of two methods to detect publication bias in meta-analysis. JAMA 2006;295:676–80. [DOI] [PubMed] [Google Scholar]
- [24].Zhang CY, Zhao YX, Xia RH, et al. RASSF1A promoter hypermethylation is a strong biomarker of poor survival in patients with salivary adenoid cystic carcinoma in a Chinese population. PLoS One 2014;9:e110159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Lin HY, Huang TT, Lee MS, et al. Unexpected close surgical margin in resected buccal cancer: very close margin and DAPK promoter hypermethylation predict poor clinical outcomes. Oral Oncol 2013;49:336–44. [DOI] [PubMed] [Google Scholar]
- [26].Supic G, Kozomara R, Jovic N, et al. Prognostic significance of tumor-related genes hypermethylation detected in cancer-free surgical margins of oral squamous cell carcinomas. Oral Oncol 2011;47:702–8. [DOI] [PubMed] [Google Scholar]
- [27].Su PF, Huang WL, Wu HT, et al. p16(INK4A) promoter hypermethylation is associated with invasiveness and prognosis of oral squamous cell carcinoma in an age-dependent manner. Oral Oncol 2010;46:734–9. [DOI] [PubMed] [Google Scholar]
- [28].Durr ML, Mydlarz WK, Shao C, et al. Quantitative methylation profiles for multiple tumor suppressor gene promoters in salivary gland tumors. PLoS One 2010;5:e10828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Lee ES, Issa JP, Roberts DB, et al. Quantitative promoter hypermethylation analysis of cancer-related genes in salivary gland carcinomas: comparison with methylation-specific PCR technique and clinical significance. Clin Cancer Res 2008;14:2664–72. [DOI] [PubMed] [Google Scholar]
- [30].Wan Y, Gao WY, Chen YX, et al. Methylation and expression of RASSF1A in oral premalignant lesions and squamous cell carcinomas. J Modern Stomatol 2007;21:30–3. [Google Scholar]
- [31].Williams MD, Chakravarti N, Kies MS, et al. Implications of methylation patterns of cancer genes in salivary gland tumors. Clin Cancer Res 2006;12:7353–8. [DOI] [PubMed] [Google Scholar]
- [32].Tran TN, Liu Y, Takagi M, et al. Frequent promoter hypermethylation of RASSF1A and p16INK4a and infrequent allelic loss other than 9p21 in betel-associated oral carcinoma in a Vietnamese non-smoking/non-drinking female population. J Oral Pathol Med 2005;34:150–6. [DOI] [PubMed] [Google Scholar]
- [33].Nagata S, Hamada T, Yamada N, et al. Aberrant DNA methylation of tumor-related genes in oral rinse: a noninvasive method for detection of oral squamous cell carcinoma. Cancer 2012;118:4298–308. [DOI] [PubMed] [Google Scholar]
- [34].Morandi L, Gissi D, Tarsitano A, et al. DNA methylation analysis by bisulfite next-generation sequencing for early detection of oral squamous cell carcinoma and high-grade squamous intraepithelial lesion from oral brushing. J Craniomaxillofac Surg 2015;43:1494–500. [DOI] [PubMed] [Google Scholar]
- [35].Asokan GS, Jeelani S, Gnanasundaram N. Promoter hypermethylation profile of tumour suppressor genes in oral leukoplakia and oral squamous cell carcinoma. J Clin Diagn Res 2014;8:ZC09–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Ge MH, Ling ZQ, Tan Z, et al. Expression and significance of E-cadherin in adenoid cystic carcinoma of salivary glands. Natl Med J China 2012;92:106–9. [PubMed] [Google Scholar]
- [37].Xu C, Zhao J, Loo WT, et al. Correlation of epigenetic change and identification of risk factors for oral submucous fibrosis. Int J Biol Markers 2012;27:e314–21. [DOI] [PubMed] [Google Scholar]
- [38].Kordi-Tamandani DM, Moazeni-Roodi AK, Rigi-Ladiz MA, et al. Promoter hypermethylation and expression profile of MGMT and CDH1 genes in oral cavity cancer. Arch Oral Biol 2010;55:809–14. [DOI] [PubMed] [Google Scholar]
- [39].Viswanathan M, Tsuchida N, Shanmugam G. Promoter hypermethylation profile of tumor-associated genes p16, p15, hMLH1, MGMT and E-cadherin in oral squamous cell carcinoma. Int J Cancer 2003;105:41–6. [DOI] [PubMed] [Google Scholar]
- [40].Chang HW, Chow V, Lam KY, et al. Loss of E-cadherin expression resulting from promoter hypermethylation in oral tongue carcinoma and its prognostic significance. Cancer 2002;94:386–92. [DOI] [PubMed] [Google Scholar]
- [41].Yeh KT, Shih MC, Lin TH, et al. The correlation between CpG methylation on promoter and protein expression of E-cadherin in oral squamous cell carcinoma. Anticancer Res 2002;22:3971–5. [PubMed] [Google Scholar]
- [42].Nakayama S, Sasaki A, Mese H, et al. The E-cadherin gene is silenced by CpG methylation in human oral squamous cell carcinomas. Int J Cancer 2001;93:667–73. [DOI] [PubMed] [Google Scholar]
- [43].Saito Y, Takazawa H, Uzawa K, et al. Reduced expression of E-cadherin in oral squamous cell carcinoma: relationship with DNA methylation of 5’ CpG island. Int J Oncol 1998;12:293–8. [DOI] [PubMed] [Google Scholar]
- [44].Roepman P, Wessels LF, Kettelarij N, et al. An expression profile for diagnosis of lymph node metastases from primary head and neck squamous cell carcinomas. Nat Genet 2005;37:182–6. [DOI] [PubMed] [Google Scholar]
- [45].Scully C, Bagan J. Oral squamous cell carcinoma overview. Oral Oncol 2009;45:301–8. [DOI] [PubMed] [Google Scholar]
- [46].Ha PK, Chang SS, Glazer CA, et al. Molecular techniques and genetic alterations in head and neck cancer. Oral Oncol 2009;45:335–9. [DOI] [PMC free article] [PubMed] [Google Scholar]


