Abstract
Background
Signal-regulatory protein α (SIRPα) is an essential signaling molecule that modulates leukocyte inflammatory responses. However, the regulation of selective SIRPα synthesis and its dynamic changes in leukocytes under inflammatory stimulation remain incompletely understood.
Objective
We sought to identify the microRNAs (miRNAs) that posttranscriptionally regulate SIRPα synthesis and their roles in modulating macrophage inflammatory responses.
Methods
SIRPα was induced in SIRPα-negative promyelocytic cells by retinoic acid or phorbol 12-myristate 13-acetate, and the differential expression of miRNAs was assessed by means of microarray and quantitative RT-PCR assays. The roles of identified miRNAs in controlling SIRPα synthesis in leukocytes and leukocyte inflammatory responses were determined.
Results
We identified SIRPα as a common target gene of miR-17, miR-20a, and miR-106a. During SIRPα induction, levels of these 3 miRNAs were all reduced, and their downregulation by retinoic acid or phorbol 12-myristate 13-acetate occurred through suppression of the c-Myc signaling pathway. All miR-17, miR-20a, and miR-106a specifically bound to the same seed sequence within the SIRPα 3′ untranslated region and correlated inversely with SIRPα protein levels in various cells. In macrophages upregulation of miR-17, miR-20a, and miR-106a by LPS served as the mechanism underlying LPS-induced SIRPα reduction and macrophage activation. Both in vitro and in vivo assays demonstrate that miR-17, miR-20a, and miR-106a regulate macrophage infiltration, phagocytosis, and proinflammatory cytokine secretion through targeting SIRPα.
Conclusion
These findings demonstrate for the first time that miR-17, miR-20a, and miR-106a regulate SIRPα synthesis and SIRPα-mediated macrophage inflammatory responses in a redundant fashion, providing a novel pathway in which a panel of miRNAs can modulate immune polarization through regulation of macrophage activation.
Keywords: Signal-regulatory proteinα, microRNA, macrophage, inflammatory response
As a member of the immunoglobulin superfamily, signal-regulatory protein α (SIRPα) is a receptor-like signaling protein predominantly expressed in myeloid leukocytes, including neutrophils and monocytic lineage cells (monocytes, macrophages, and dendritic cells).1–3 Through its extracellular IgV-like loops, SIRPα binds to its ligand, CD47, a broadly expressed cell-surface protein.4 Following a single transmembrane domain, SIRPα has a long cytoplasmic tail containing 4 tyrosine residues that form 2 immunoreceptor tyrosine-based inhibitory motifs (ITIMs).5 It has been proposed that SIRPα trans-binding to CD47 triggers tyrosine phosphorylation of the SIRPα intracellular ITIMs, which leads to association with the SH2 domain–containing protein tyrosine phosphatases SHP-1 or SHP-2 and initiates negative signaling cascades, resulting in inhibition of leukocyte function.6–11 In macrophages this SIRPα-mediated signaling pathway plays a key role in determining the phagocytic target.12 Specifically, ligation of SIRPα on the macrophage by CD47 on tissue cells elicits phosphorylation of the SIRPα ITIMs and leads to inhibition of phagocytosis, whereas the failure of ITIM-mediated signaling promotes macrophage engulfment of the cell.3,13 By using anti-SIRPα mAbs and soluble CD47 extracellular domains, previous studies have demonstrated that interactions between SIRPα on leukocytes and CD47 on endothelial and epithelial cells also inhibit neutrophil (PMN) and monocyte transmigration.14–18 SIRPα also plays a critical role in controlling alveolar macrophage activity. By binding SIRPα, lung collectins, such as surfactant protein A (SP-A) and surfactant protein D (SP-D), act as surveillance molecules to suppress macrophage phagocytic function and lung inflammation.19,20 Although SIRPα plays an essential role in regulating many aspects of inflammatory responses, the regulation of SIRPα synthesis remains largely un-characterized. At the mRNA level, SIRPα transcripts have been detected in almost every tissue; however, the SIRPα protein is only expressed in myeloid cells and certain neuronal cells.1–3 Furthermore, SIRPα protein levels in macrophages were rapidly reduced when macrophages were stimulated with LPS and thus are inversely correlated with the activation of macrophages.21 However, the mechanism underlying this phenomenon is not clear.
Given the disparity between SIRPα mRNA and protein levels in various human and animal tissues, it is quite likely that a posttranscriptional regulatory mechanism exists. Recently, a class of RNA regulatory genes known as microRNAs (miRNAs) has been discovered and adds a new layer of protein regulation at the posttranscriptional level.22,23 As a novel class of endogenous sequence-specific suppressors of protein translation, miRNAs can block target gene expression. The widespread and important role of these noncoding RNAs, which are approximately 22 nucleotides in length, is highlighted by recent discoveries that they control a wide range of physiologic processes in eukaryotes, including development, differentiation, proliferation, apoptosis, and metabolism.22,23 Thus we propose that miRNAs are involved in posttranscriptional regulation of SIRPα. In the present study we identified SIRPα as a common target of the 3 miRNAs miR-17, miR-20a, and miR-106a and demonstrated that miR-17, miR-20a, and miR-106a regulate macrophage SIRPα synthesis and SIRPα-mediated macrophage inflammatory responses in a redundant fashion. Furthermore, regulation of the levels of miR-17, miR-20a, and miR-106a by various inducers, including retinoic acid (RA), phorbol 12-myristate 13-acetate (TPA), and LPS, and their roles in the modulation of leukocyte maturation and activation have also been studied.
METHODS
Reagents, cells, and antibodies
The human myeloblastic cell lines HL-60 and U937 and the monocyte/macrophage cell line THP-1 were purchased from the American Type Culture Collection (Manassas, Va). All other human cells, including HT-29, 293T, HeLa, HepG2, MDB231, MCF7, SH-SY5Y, and C2C12 cells, were all obtained from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China). HL-60 cells were cultured with Iscove modified Dulbecco medium (Gibco, Carlsbad, Calif) containing 20% FBS, 100 U/mL penicillin, and 100 μg/mL streptomycin. THP-1 and U937 cells were grown in RPMI 1640 (Gibco) containing 10% FBS, 100 U/mL penicillin, 100 μg/mL streptomycin, and 0.05 mmol/L 2-mercaptoethanol. HT-29, 293T, HeLa, HepG2, 231, MCF7, SH-SY5Y, and C2C12 cells were grown in Dulbecco modified Eagle medium (Gibco) containing 10% FBS, 100 U/mL penicillin, and 100 μg/mL streptomycin. The anti-SIRPα (human and mouse) antibodies were obtained from BD Biosciences (San Jose, Calif). The anti–glyceraldehyde-3-phosphate dehydrogenase (GAPDH; human and mouse) antibodies, and the anti–c-Myc antibody were purchased from Santa Cruz Biotechnology (Santa Cruz, Calif). Synthetic RNA molecules, including pre–miR-17, pre–miR-20a, pre–miR-106a, anti–miR-17 antisense oligonucleotides (ASO), anti–miR-20a ASO, and anti–miR-106a ASO and scrambled negative control oligonucleotides (pre-ncRNA and ncRNA), were purchased from Ambion (Carlsbad, Calif).
Isolation of mouse peritoneal and alveolar macrophage
Mouse peritoneal and alveolar macrophages were isolated, as previously described24 with minor modifications. Male C57BL/6J mice (6–8 weeks old, 20–22 g) were purchased from the Model Animal Research Center of Nanjing University (Nanjing, China). Mice were administered 3% sterile thioglycollate intraperitoneally to isolate peritoneal macrophages. After resting for 6 days, mice were killed, and peritoneal lavage was performed with 10 mL of RPMI 1640 (Invitrogen, Carlsbad, Calif). The lavage fluid was centrifuged at 300g for 10 minutes to collect cells. For alveolar macrophage isolation, mice were anesthetized with intraperitoneal pentobarbital and killed by means of exsanguination. Lungs were lavaged through an intratracheal catheter with prewarmed PBS supplemented with 0.6 mmol/L EDTA. A total of 10 mL was used in each mouse in 0.5-mL increments. The lavage fluids were pooled and centrifuged at 300g for 10 minutes to collect cells. Cell preparations were generally greater than 95% enriched for peritoneal or alveolar macrophages. All animal procedures were approved by the Institutional Review Board of Nanjing University.
Phagocytosis assays
Phagocytosis of fluorescein isothiocyanate (FITC)–labeled zymosan particles (Sigma-Aldrich, St Louis, Mo) by macrophages was detected in mouse alveolar macrophages and thioglycollate-elicited mouse peritoneal macrophages. Mouse alveolar macrophages were cultured in 12-well plates and transfected with 30 nmol/L miRNA inhibitors (10 nmol/L for each anti-miRNA ASO) or scramble oligonucleotide (ncRNA). Cells were incubated with FITC-zymosan for 30 minutes at 37°C in the presence or absence of 100 ng/mL LPS (Sigma-Aldrich). After 2 washes with cold HBSS, cells were evaluated by gating on FITC-positive cells on a FACSCalibur (BD Biosciences). For thioglycollate-elicited peritoneal macrophages, mice were injected intraperitoneally with 2 mL of 4% thioglycollate solution. On day 3 after injection, mice were administered intraperitoneally a mixture of PEI/30 nmol/L miRNA inhibitors or PEI/ncRNA. After 12 hours, FITC-zymosan was injected intraperitoneally into mice. Macrophages in the peritoneal exudates were obtained 30 minutes later by means of washing the peritoneal cavity with cold PBS and evaluated with the FACSCalibur (BD Biosciences).
Matrigel invasion and Transwell migration assays
Macrophage invasion and migration experiments were performed, as previously described.25 Transwell filters (5-μm pore size; Corning Laboratories, Corning, NY) were left uncoated or coated with 50 μL of 1 mg/mL Matrigel (BD Biosciences). Macrophages isolated from peritoneal exudates (1 × 105) were added to the upper chamber in RPMI 1640 containing 10% FCS, and RPMI 1640 containing 10% FCS and 35 ng/mL colony-stimulating factor-1 was added to the lower chamber. Each assay was performed in triplicate. After 24 hours, cells and Matrigel in the upper chamber were removed, and cells on the bottom of the Transwell filter were fixed and stained with 0.1% crystal violet in 0.1 mol/L borate and 2% ethanol. The images of migrated cells were captured with a photomicroscope (Olympus, Center Valley, Pa). Cell migration was quantified by means of blind counting of migrated cells with 5 fields per chamber.
Cytokine assay and nitrite oxidant detection
Cytokine levels in culture supernatants were determined with commercial ELISA kits for TNF-α and IL-6 (R&D Systems, Minneapolis, Minn), according to the manufacturer’s instructions. Each value represents the mean of triplicate values. For nitric oxide (NO) detection, cells plated in 24-well culture dishes (2 × 105 cells per well) were incubated overnight before stimulation. After the cells were treated with 100 ng/mL LPS for 24 hours, culture supernatants were collected and analyzed with the Griess Reagent kit (Invitrogen). Nitrite concentrations were determined by means of measurement of OD at 570 nm.
Statistical analysis
All images of Western blotting and quantitative RT-PCR are representative of 4 to 6 independent experiments. Real-time PCR was performed in triplicate, and each experiment was repeated at least 3 times. The data shown are presented as means ± SDs of 3 independent experiments; differences are considered statistically significant at a P value of less than .05, as determined by using the Student t test.
RESULTS
Identification of miR-17, miR-20a, and miR-106a as 3 major miRNAs that target SIRPα
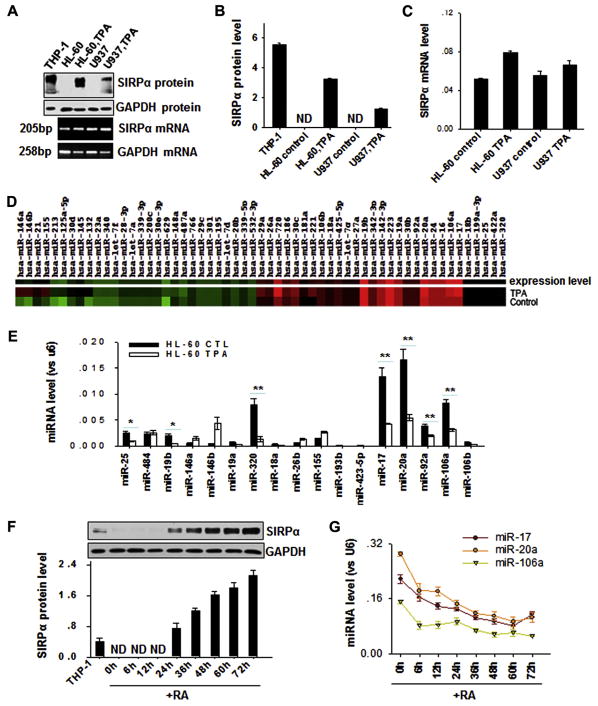
We carefully monitored SIRPα mRNA and protein levels in various tissues and cells by using Western blotting and quantitative RT-PCR assays. As shown in Fig E1 in this article’s Online Repository at www.jacionline.org, all tissues and cells we tested expressed SIRPα mRNA at considerable levels; however, among these tissues and cells, only monocytes, neutrophils, and monocytic THP-1 cells produced SIRPα protein. The results strongly suggest that a posttranscriptional regulatory mechanism exists for SIRPα. To determine whether any miRNAs were involved in posttranscriptional modulation of SIRPα protein production, we monitored the alterations of miRNAs in promyelocytic HL-60 cells using an miRNA microarray after stimulation with TPA. Consistent with the notion that SIRPα is a myeloid differentiation marker, it has been widely reported that treatment of promyelocytic cells, such as HL-60 and U937 cells, with cell differentiation agents, including TPA or RA, can induce the differentiation of promyelocytic HL-60 cells into mature neutrophils or monocytes,26,27 which express high levels of SIRPα. Indeed, as shown in Fig 1, A (upper panel) and B, SIRPα protein production was readily detected in HL-60 and U937 cells after TPA treatment. In contrast, the mRNA levels of SIRPα in HL-60 and U937 cells were not significantly altered by TPA treatment (Fig 1, A [lower panel] C).
FIG. 1.
Downregulation of expression levels of miR-17, miR-20a, and miR-106a in promyelocytic cells during SIRPα protein induction by cell differentiation agents. A–C, Induction of SIRPα protein but not SIRPα mRNA by TPA (30 nmol/L) in promyelocytic HL-60 and U937 cells. Note that the SIRPα protein level (Fig 1, A, upper panel, and B) was significantly increased by TPA, whereas the SIRPα mRNA level (Fig 1, A, lower panel, and C) was not altered. D, Microarray analysis of changes in miRNA expression in TPA-differentiated HL-60 cells. E, TaqMan probe–based quantitative RT-PCR validation of differentially expressed miRNAs. F and G, Inverse correlation between SIRPα protein levels (Fig 1, F) and levels of miR-17/20a/106a (Fig 1, G) in HL-60 cells during RA-induced differentiation process. Data represent means ± SDs of 3 independent experiments performed in triplicate. *P < .05 and **P < .01. ND, Undetectable.
When we compared the expression profile of miRNAs between SIRPα protein–negative HL-60 cells and SIRPα protein–positive TPA-treated HL-60 cells using a low-density human miRNA microarray, we found that a panel of miRNAs, including miR-17, miR-20a, miR-106a, miR-320, and miR-25, were significantly downregulated, whereas miR-146a, miR-146b, and miR-155 were upregulated after TPA-mediated cell differentiation (Fig 1, D). The changes in miRNA expression are detailed in Table E1 in this article’s Online Repository at www.jacionline.org (GEO accession no. GSE29620). We next used a TaqMan probe–based quantitative RT-PCR assay to further validate the expression levels of 17 miR-NAs with significantly altered (fold change > 2) expression levels in the microarray data. As shown in Fig 1, E, which is consistent with the microarray results, miR-17, miR-20a, miR-106a, miR-320, and miR-25 levels were strongly reduced in TPA-treated HL-60 cells compared with those seen in control HL-60 cells. We selected miR-17, miR-20a, miR-106a, and miR-320 as candidates for further study because of their relatively high levels of expression in nontreated promyelocytic HL-60 cells. Other miRNAs with significantly lower levels than those of miR-17, miR-20a, miR-106a, and miR-320, suchas miR-92a and miR-106b, were not further studied, although they were also downregulated by TPA treatment.
The link between the induction of SIRPα protein production and the downregulation of miR-17, miR-20a, and miR-106a was confirmed in RA-treated HL-60 cells (Fig 1, F and G). The results clearly show that SIRPα protein was detected after 24 hours and that its expression level increased in a time-dependent fashion over the course of RA treatment (Fig 1, F). In contrast, miR-17, miR-20a, and miR-106a levels decreased over the course of RA treatment (Fig 1, G). We did not observe the dose-dependent decrease of miR-320 levels induced by RA in HL-60 cells (data not shown).
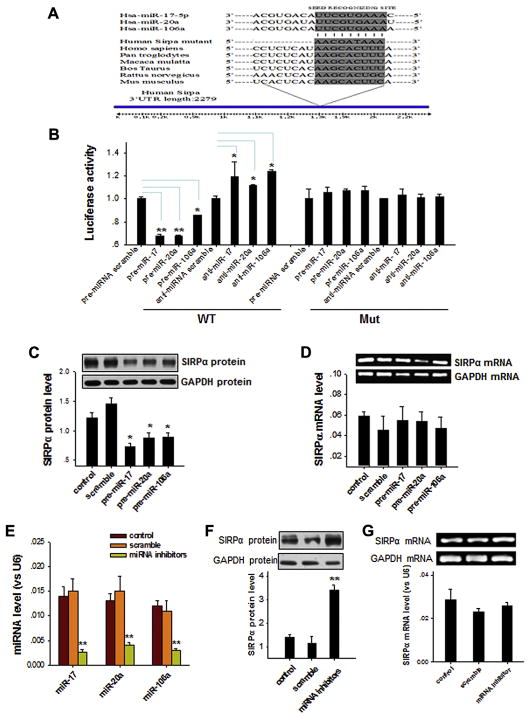
Next, we determined whether the 3 miRNAs miR-17, miR-20a, and miR-106a are involved in SIRPα induction in HL-60 cells during differentiation. As shown in Fig 2, A, bioinformatics analysis with the 3 computer-aided algorithms TargetScan, miRanda, and PicTar predicted that miR-17, miR-20a, and miR-106a can target the same sequence within the 3′ untranslated region (UTR) of SIRPα of various animal species, including human subjects, which suggests that SIRPα might be a common target gene of the 3 miRNAs. To test this possibility, we characterized the binding of the miRNAs to the SIRPα 3′ UTR and the inverse correlation between the levels of miR-17/miR-20a/miR-106a and SIRPα. First, we used a luciferase reporter assay to confirm the specific binding of miR-17, miR-20a, and miR-106a to the 3′ UTR of SIRPα (Fig 2, B). Second, overexpression of miR-17, miR-20a, or miR-106a in THP-1 cells, which greatly increases the levels of each miRNA (see Fig E2 in this article’s Online Repository at www.jacionline.org), significantly decreased the SIRPα level compared with that seen in THP-1 cells transfected with scrambled oligonucleotides (Fig 2, C). In contrast, the SIRPα mRNA level in THP-1 cells was not altered by miR-17, miR-20a, or miR-106a overexpression (Fig 2, D). When miR-17, miR-20a, and miR-106a were depleted from THP-1 cells by means of transfection with a combination of anti–miR-17, anti–miR-20a, and anti–miR-106a ASOs (termed miRNA inhibitors; Fig 2, E), SIRPα protein levels in THP-1 cells were markedly increased (Fig 2, F). However, the SIRPα mRNA level in THP-1 cells was not altered by depleting all miR-17, miR-20a, and miR-106a (Fig 2, G). Taken together, the results suggest that the 3 miRNAs miR-17, miR-20a, and miR-106a, expression levels of which are drastically reduced during promyelocytic cell differentiation, can serve as modulators of SIRPα protein production through targeting the 3′ UTR of SIRPα.
FIG. 2.
Identification of SIRPα as a common target of miR-17, miR-20a, and miR-106a. A, Bioinformatics analysis prediction of the possible target sites of miR-17/20a/106a in the 3′ UTR of SIRPα. B, Validation of binding of miR-17, miR-20a, and miR-106a with the 3′ UTR of SIRPα by using a luciferase reporter assay. Mut, Mutated SIRPα 3′ UTR; WT, wild type SIRPα 3′ UTR. C and D, Levels of SIRPα protein (Fig 2, C) and mRNA (Fig 2, D) in THP-1 cells transfected with pre–miR-17, pre–miR-20a, or pre–miR-106a. E–G, Levels of miRNAs (miR-17, miR-20a, and miR-106a; Fig 2, E), SIRPα protein (Fig 2, F), and SIRPα mRNA (Fig 2, G) in THP-1 after transfection with scrambled or miRNA ASOs. Data represent means ± SDs of 3 independent experiments. *P < .05 and **P < .01.
Modulation of SIRPα protein production in promyelocytic cells by miR-17, miR-20a, and miR-106a
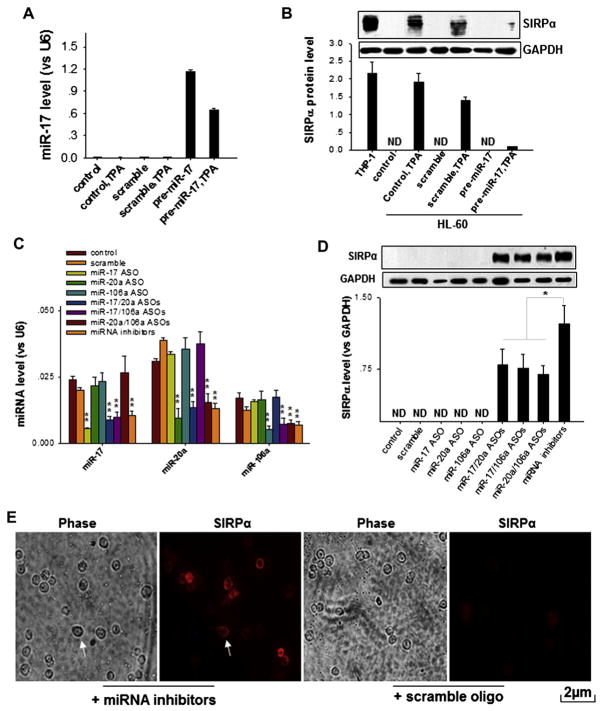
To illustrate the role of miR-17, miR-20, and miR-106a in controlling SIRPα protein expression at the posttranscriptional level, we further characterized SIRPα protein expression in HL-60 cells that had either undergone TPA-induced cell differentiation or had endogenous miRNAs depleted. SIRPα protein–producing THP-1 cells served as a positive control. In agreement with previous results, HL-60 cells treated with TPA produced SIRPα protein. However, when miR-17 was overexpressed in TPA-treated HL-60 cells (Fig 3, A), TPA-induced SIRPα protein production was strongly decreased (Fig 3, B). Scramble oligonucleotides that were used as controls had no effect on SIRPα protein induction by TPA. A similar inhibitory effect on TPA-induced SIRPα protein production was observed by overexpressing miR-20a or miR-106a (see Fig E3 in this article’s Online Repository at www.jacionline.org). In a separate study TPA-treated HL-60 cells were further transfected with miRNA inhibitors to deplete miR-17, miR-20a, and miR-106a. As shown in Fig E4 in this article’s Online Repository at www.jacionline.org, although there was some additional decrease in miRNA levels after treatment with miRNA inhibitors, no significant increase in SIRPα protein levels was observed compared with those seen after TPA treatment alone. We next determined SIRPα protein levels in SIRPα-negative HL-60 cells after knockdown of individual miRNAs by using miRNA ASOs. As shown in Fig 3, C and D, levels of individual miRNAs or multiple miRNAs were successfully reduced by individual miRNA ASOs or a combination of miRNA ASOs. However, although knockdown of single miRNAs of miR-17, miR-20a, or miR-106a showed no effect on SIRPα protein levels, knockdown of multiple miRNAs of miR-17, miR-20a, or miR-106a with a combination of miRNA ASOs significantly induced SIRPα expression in HL-60 cells, even in the absence of TPA stimulation. When miR-17, miR-20a, and miR-106a were depleted altogether from HL-60 cells by means of transfection with miRNA inhibitors, SIRPα protein levels were increased to the highest level. Immunofluorescence labeling showed that the SIRPα protein induced by depleting miR-17, miR-20a, and miR-106a was predominantly located at cell-surface membranes, which is a typical location for SIRPα protein in monocytes (Fig 3, E, arrows). Taken together, the results suggest that miR-17, miR-20a, and miR-106a can directly affect SIRPα protein production and that induction of SIRPα protein by cell differentiation reagents, such as TPA, might also function largely through the downregulation of miR-17, miR-20a, and miR-106a.
FIG. 3.
Regulation of SIRPα protein expression by miR-17, miR-20a, and miR-106a. A and B, Effect of over-expression of miR-17 on TPA-induced SIRPα protein production in HL-60 cells. Note that forced expression of pre–miR-17 significantly increases miR-17 levels (Fig 3, A) but abolishes SIRPα induction by TPA (Fig 3, B). C and D, Effect of depleting miR-17, miR-20a, and miR-106a on SIRPα protein levels in HL-60 cells. Note that compared with scrambled oligonucleotides, the combined miRNA ASOs, particularly miRNA inhibitors (the combination of 3 miRNA ASOs) in HL-60 cells, strongly decrease levels of the 3 miRNAs (Fig 3, C) and lead to SIRPα induction, even in the absence of differentiation reagents (Fig 3, D). E, Localization of SIRPα induced by depletion of miR-17, miR-20a, and miR-106a in HL-60 cells. Scale bar = 2 α m. Data represent means ± SDs of 3 independent experiments. *P < .05 and **P < .01. ND, Undetectable.
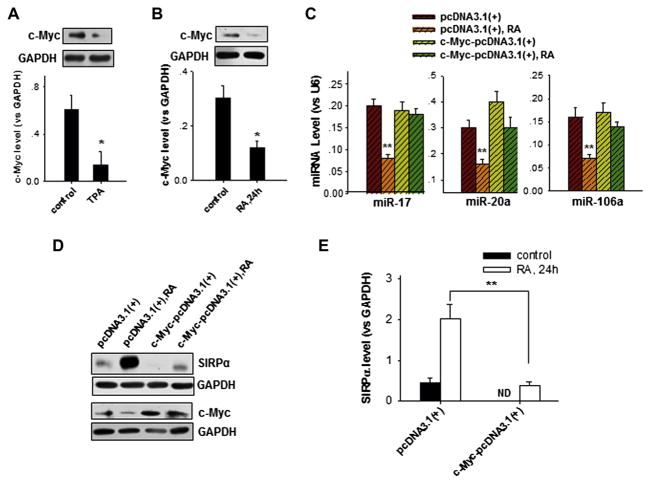
miR-17 and miR-20a are members of a protumorigenic group of miRNAs known as the miR-17-92 cluster. Previous reports have shown that c-Myc is a transcription factor that promotes the expression of the miR-17-92 cluster in tumor cells.28,29 To further identify the mechanism underlying the downregulation of miR-17, miR-20a, and miR-106a caused by cell differentiation reagents in promyelocytic cells, we determined the effect of c-Myc on the modulation of miR-17, miR-20a, and miR-106a expression and SIRPα protein levels in HL-60 cells treated with TPA or RA. As shown in Fig 4, c-Myc levels were significantly reduced in HL-60 cells treated with TPA (Fig 4, A) or RA (Fig 4, B), suggesting that c-Myc might positively regulate the expression of miR-17, miR-20a, and miR-106a. When RA-induced reduction of c-Myc levels in HL-60 cells was reversed by overexpressing c-Myc (see Fig E5 in this article’s Online Repository at www.jacionline.org), downregulation of miR-17, miR-20a, and miR-106a by RA was also reversed (Fig 4, C). Overexpression of c-Myc also significantly blocked RA-induced upregulation of SIRPα protein production (Fig 4, D and E). These results show that the RA-induced downregulation of miR-17, miR-20a, and miR-106a in HL-60 cells likely occurs through c-Myc suppression.
FIG. 4.
Downregulation of miR-17, miR-20a, and miR-106a in HL-60 cells by TPA or RA through suppression of the c-Myc signaling pathway. A and B, Reduction of c-Myc expression in HL-60 cells by TPA (Fig 4, A) and RA (Fig 4, B) treatment. C, Overexpression of c-Myc inhibits RA-mediated downregulation of miR-17, miR-20a, and miR-106a in HL-60 cells. D and E, Overexpression of c-Myc in RA-treated HL-60 cells reverses RA-induced SIRPα protein production. Data represent means ± SDs of 3 independent experiments. *P < .05 and **P < .01. ND, Not determined.
Role of miR-17/20a/106a–mediated regulation of SIRPα in the activation of macrophages by LPS
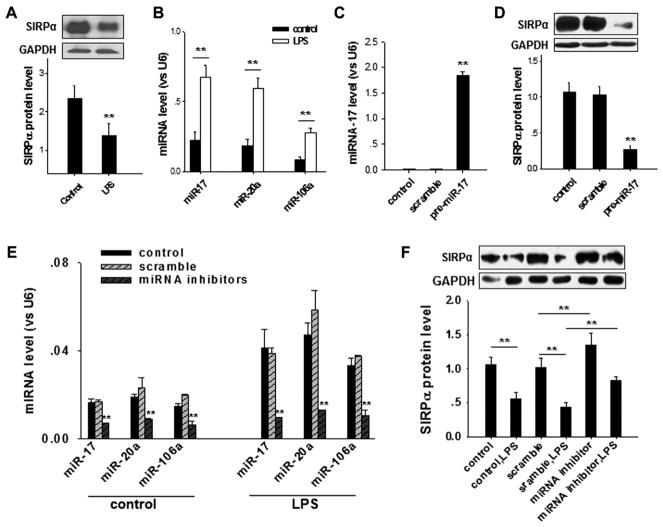
Although the mechanism remains unknown, previous reports have demonstrated that LPS stimulation reduces murine macrophage SIRPα protein levels, which in turn promotes macrophage activation.21 Next, we studied the potential role of miR-17, miR-20a, and miR-106a in the LPS-induced downregulation of SIRPα protein in murine macrophages. In this experiment macrophages were isolated from mouse lungs.20 In agreement with previous findings,21 SIRPα protein levels in mouse alveolar macrophages were rapidly decreased by LPS stimulation (Fig 5, A). However, miR-17, miR-20a, and miR-106a levels in macrophages were strongly increased by LPS in the same timeframe (Fig 5, B). Given that overexpression of any one of these 3 miRNAs, such as miR-17, in murine macrophages could reduce SIRPα protein levels (Fig 5, C), we postulated that increased miR-17, miR-20a, or miR-106a levels in mouse alveolar macrophages under LPS stimulation might cause the LPS-induced SIRPα protein reduction. To test this hypothesis, we performed an experiment to determine whether blockade of LPS-induced upregulation of miR-17, miR-20a, and miR-106a could block LPS-induced SIRPα protein reduction. As shown in Fig 5, E, treatment with the combined miRNA inhibitors significantly depleted miR-17, miR-20a, and miR-106a levels in both control and LPS-stimulated macrophages. More importantly, depletion of miR-17, miR-20a, and miR-106a strongly reversed the LPS-induced SIRPα protein reduction in mouse macrophages (Fig 5, F).
FIG. 5.
Effect of miR-17, miR-20a, and miR-106a on LPS-induced downregulation of SIRPα in murine alveolar macrophages. A and B, LPS stimulation (100 ng/mL) significantly reduces SIRPα protein levels (Fig 5, A) but increases miR-17, miR-20a, and miR-106a levels (Fig 5, B) in macrophages. C and D, Overexpression of miR-17 in macrophages (Fig 5, C) decreases SIRPα protein levels (Fig 5, D). E and F, Effect of depleting miR-17, miR-20a, and miR-106a on LPS-induced SIRPα reduction in mouse alveolar macrophages. Note that transfection with the miRNA inhibitors strongly reverses both LPS-induced miR-17/20a/106a increase (Fig 5, E) and SIRPα reduction (Fig 5, F) in macrophages. Data represent means ± SDs of 3 independent experiments. **P < .01.
Role of miR-17, miR-20a, and miR-106a in regulating macrophage inflammatory responses in vitro and in vivo
Given that miR-17, miR-20a, and miR-106a can target SIRPα and are involved in LPS-induced SIRPα reduction in macrophages, we studied the role of miR-17, miR-20a, and miR-106a in modulating macrophage inflammatory responses.
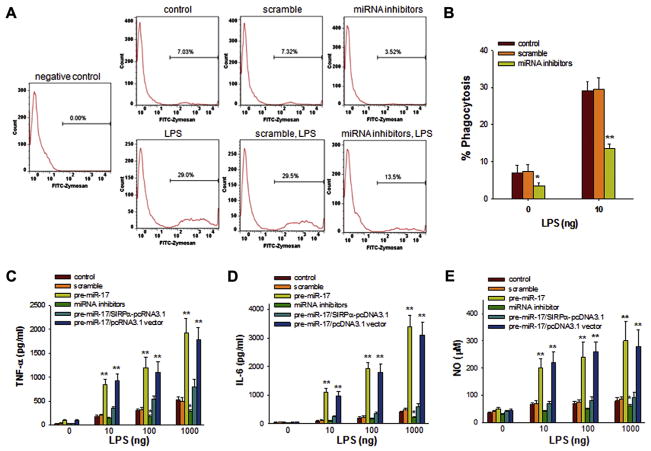
First, phagocytic activity and inflammatory cytokine secretion were assessed in macrophages isolated from mouse lung tissue. Because SIRPα serves as an inhibitory modulator of macrophage phagocytosis,1,2,21,30,31 we investigated whether depletion of miR-17, miR-20a, and miR-106a in mouse macrophages, which causes an increase in SIRPα protein levels, could suppress their phagocytic activity. Macrophages were incubated with fluorescently labeled zymosan particles for 30 minutes at 37°C in the presence or absence of LPS to stimulate phagocytosis. After washing off free zymosan particles, macrophages were assayed by means of flow cytometry, and phagocytosis was presented as the ratio of fluorescently labeled macrophages to total cells. As shown by the flow cytometric results in Fig 6, A and B, when miR-17, miR-20a, and miR-106a levels were decreased in macrophages by means of transfection with miRNA inhibitors, the uptake of fluorescently labeled zymosan particles by mouse lung macrophages was significantly inhibited.
FIG. 6.
Role of miR-17, miR-20a, and miR-106a in modulating inflammatory responses of mouse alveolar macrophages. A and B, Phagocytic activity of macrophages detected by means of flow cytometry. Macrophages were transfected with or without scrambled oligonucleotides or combined miRNA inhibitors. Cells were then incubated with fluorescently labeled zymosan particles at 37°C in the presence or absence of 100 ng/mL LPS. C–E, secretion of TNF-α (Fig 6, C), IL-6 (Fig 6, D), and NO (Fig 6, E) by macrophages. Data represent means ± SDs of 3 independent experiments. *P < .05 and **P < .01.
Second, we investigated whether SIRPα reduction by miR-17, miR-20a, and miR-106a directly controls cytokine production in mouse alveolar macrophages on LPS treatment. Macrophages overexpressed with or without miR-17, control oligonucleotides, or the miRNA inhibitors were stimulated with various concentrations of LPS for 12 hours to measure the production of cytokines or for 24 hours to detect NO in the conditioned medium. In response to LPS stimulation, miR-17–overexpressing macrophages produced substantially more TNF-α, IL-6, and NO than macrophages transfected with the miRNA inhibitors or scrambled oligonucleotides (Fig 6, C–E). As expected, because of their higher level of SIRPα protein, macrophages transfected with the miRNA inhibitors produced the least TNF-α, IL-6, and NO. Interestingly, when maintaining SIRPα levels in miR-17–overexpressing macrophages through transfection of macrophages with SIRPα-pcDNA3.1, the effect of miR-17 on TNF-α, IL-6, and NO production is blocked (Fig 6, C–E). This result supports that miR-17 modulates macrophage inflammatory responses through targeting SIRPα.
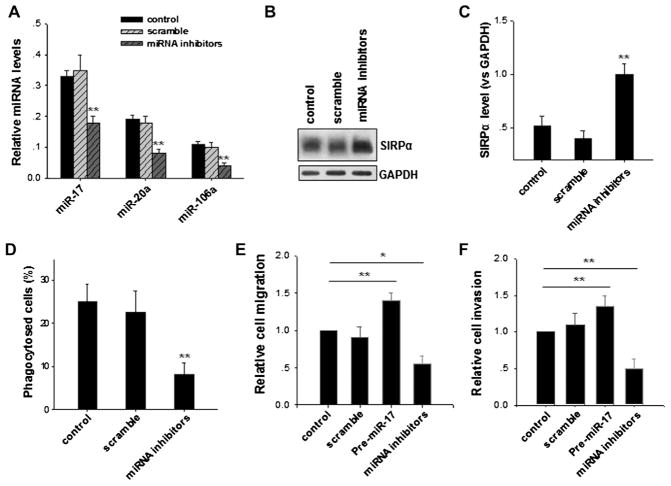
To study the role of miR-17/miR-20a/miR-106a–targeting SIRPα in regulating macrophage inflammatory responses under pathophysiologic conditions, we measured the effect of depletion of miR-17, miR-20a, and miR-106a on the phagocytic activity of thioglycollate-elicited peritoneal macrophages from C57BL/6 mice. In this experiment C57BL/6 mice were injected intraperitoneally with 2 mL of 4% thioglycollate, and subsequently, mice were injected intraperitoneally with a mixture of galactosylated low-molecular-weight chitosan (PEI)/miRNA inhibitors or PEI/scramble oligonucleotides once per day.32 Three days later, fluorescently labeled zymosan particles were injected intraperitoneally into mice. After 3 hours, cells in the peritoneal exudates were isolated by washing the peritoneal cavity with ice-cold HBSS. Collected cells were directly assayed for miRNA expression, SIRPα levels, and zymosan particle uptake. As shown in Fig 7, miR-17, miR-20a, and miR-106a levels in thioglycollate-elicited peritoneal macrophages were significantly reduced by means of PEI delivery of miRNA inhibitors (Fig 7, A). In agreement with this, SIRPα protein levels in peritoneal macrophages treated with PEI/miRNA inhibitors were increased compared with those in normal mice or mice treated with PEI/scramble oligonucleotide (Fig 7, B and C). More importantly, flow cytometric data clearly demonstrated that peritoneal macrophages treated with PEI/miRNA inhibitors had much lower phagocytic activity than control macrophages or macrophages treated with PEI/scrambled oligonucleotide (Fig 7, D). Consistent with this observation, peritoneal macrophages in which miR-17, miR-20a, and miR-106a were depleted also showed less CSF-1–induced Matrigel invasion (Fig 7, E) and migration across Transwell filters (Fig 7, F) than control macrophages or macrophages treated with PEI/scramble oligonucleotide. Together, the results suggest that upregulation of SIRPα protein in mouse macrophages by depleting miR-17, miR-20a, and miR-106a significantly suppresses inflammatory responses, including phagocytosis and migration.
FIG. 7.
Modulation of mouse macrophage inflammatory responses by depletion of miR-17, miR-20a, and miR-106a in thioglycollate-elicited peritoneal macrophages. A, Specific downregulation of miR-17, miR-20a, and miR-106a in peritoneal macrophages by PEI/miRNA inhibitors. B and C, Increased SIRPα protein levels in peritoneal macrophages by means of depletion of miR-17, miR-20a, and miR-106a. D, Reduced phagocytic activity of peritoneal macrophages with miR-17, miR-20a, and miR-106a depletion. E and F, Decreased Matrigel invasion (Fig 7, E) and migration across Transwell filters (Fig 7, F) of peritoneal macrophages on depletion of miR-17, miR-20a, and miR-106a. Data represent means ± SDs of 3 independent experiments. *P < .05 and **P < .01.
DISCUSSION
In the present study, through miRNA microarray screening of downregulated miRNAs during SIRPα protein induction in promyelocytic HL-60 cells and gain-of-function and loss-of-function assays in various leukocytes, we present the first evidence that the miRNAs miR-17, miR-20a, and miR-106a modulate SIRPα protein synthesis at the posttranscriptional level. Our results further suggest that miR-17, miR-20a, and miR-106a effectively regulate macrophage inflammatory responses through modulating leukocyte SIRPα synthesis.
As an important signaling molecule, SIRPα modulates many aspects of leukocyte inflammatory responses, including activation, chemotaxis, and phagocytosis. Although the molecular basis is unclear, many have noted the disparity between SIRPα mRNA and protein synthesis in various tissues and cells. As shown in Fig E1 and Fig 1, most tissues and cells tested expressed SIRPα mRNA, but only leukocytes produced SIRPα protein. Our finding that miR-17, miR-20a, and miR-106a promote the downregulation of SIRPα explains this disparity. In agreement with the notion of miRNA-based posttranscriptional regulation of SIRPα, SIRPα-negative cells generally express high levels of miR-17, miR-20a and miR-106a (see Fig E6 in this article’s Online Repository at www.jacionline.org), whereas downregulation of cellular miR-17, miR-20a, and miR-106a levels through miRNA inhibitors or TPA-or RA-induced cell differentiation induced SIRPα protein synthesis. Identification of the role of miR-17, miR-20a, and miR-106a in SIRPα protein regulation also provides a novel mechanism for the rapidly decreasing or increasing SIRPα protein levels in leukocytes during inflammatory stimulation. Instead of de novo synthesis or blockade of SIRPα mRNA, leukocytes can effectively control SIRPα protein levels through altering levels of miR-17, miR-20a, or miR-106a. For example, when macrophages were stimulated with LPS, miR-17, miR-20a, and miR-106a levels were all rapidly increased, and any one of these miRNAs could decrease leukocyte SIRPα protein levels. At this stage, it remains unknown whether other proinflammatory or anti-inflammatory factors can also alter the expression of miR-17, miR-20a, and miR-106a. Interestingly, we also observed that levels of several miRNAs, particularly miR-146a, miR-146b, and miR-155, are significantly increased in HL-60 cells on TPA treatment (see Table E1). Because the increased expression of miR-146a, miR-146b, and miR-155 generally promotes macrophage or immune cell activation or polarization, our results might indicate that TPA-induced promyelocytic HL-60 cell differentiation is a proinflammatory process and relevant to macrophage polarization.
miR-17 and miR-20a are members of the miR-17-92 cluster (encoding miR-17, 18a, 19a/b, 20a, and 92a). Sequence analysis showed that miR-92a contains the same seed sequence targeting SIRPα as the 3 miRNAs studied here; however, its role in modulating SIRPα expression was not characterized because of its relatively low expression level in promyelocytic HL-60 cells. miR-17, miR-20a, and miR-106a are a protumorigenic group of miRNAs. They are highly expressed in tumor cells, and overexpression of these miRNAs promotes cell proliferation and inhibits the differentiation of lung epithelial progenitor cells.33,34 These findings are in agreement with our finding that these miRNAs are strongly reduced when promyelocytic cells differentiate into mature monocytes and neutrophils (Fig 2). Many target genes have been identified for miR-17, miR-20a, and miR-106a in their modulation of tumorigenesis; these molecules include signal transducer and activator of transcription 3,35 suppressor of cytokine signaling 1,36 and hypoxia-inducible factor 1α,37 which have been shown to play an essential role in modulating macrophage functions. However, these 3 molecules are generally rapidly activated or increased by LPS stimulation,38–40 which is not in agreement with the inhibitory role of miR-17, miR-20a, and miR-106a. This might suggest that miR-17, miR-20a, and miR-106a are not directly involved in regulating levels of signal transducer and activator of transcription 3, suppressor of cytokine signaling 1, and hypoxia-inducible factor 1α under the conditions under which our study was performed. Identifying SIRPα as a common target gene of miR-17, miR-20a, and miR-106a extends our understanding of the role these miRNAs in controlling cell proliferation and differentiation. Because SIRPα-derived signals provide a critical negative regulation for cell growth and survival,2,3,21,41 downregulation of SIRPα protein by miR-17, miR-20a, and miR-106a would promote tumor-like proliferation and survival, whereas induction of SIRPα protein by depleting cellular miR-17, miR-20a, and miR-106a would trigger a SIRPα-mediated proapoptotic signal.
In the present study we demonstrate that, in addition to the differentiation of promyelocytic cells, miR-17, miR-20a, and miR-106a also play a critical role in dynamically modulating leukocyte inflammatory responses. Through targeting SIRPα, miR-17, miR-20a, and miR-106a actually serve as activators of macrophage inflammatory reactions. In macrophages isolated from mouse lung tissues, forced expression of any one of miR-17, miR-20a, and miR-106a strongly enhanced cell phagocytic function and secretion of various inflammatory cytokines, whereas depletion of all 3 miRNAs suppressed macrophage phagocytosis and cytokine secretion (Fig 6). Furthermore, LPS-induced SIRPα reduction and consequent macrophage activation were associated with upregulation of miR-17, miR-20a, and miR-106a and could be completely blocked by depleting these 3 miRNAs. The inhibitory effect of the combined miRNA inhibitors on macrophage phagocytic function was also confirmed by using thioglycollate-elicited mouse peritoneal macrophages (Fig 7).
In summary, we have shown that SIRPα is posttranscriptionally modulated by miR-17, miR-20a, and miR-106a, and through targeting SIRPα, these 3 miRNAs modulate macrophage phagocytic function and secretion of various inflammatory cytokines. Identification of SIRPα targeting by miR-17, miR-20a, and miR-106a and its associated role in modulating leukocyte inflammatory responses provides a novel therapeutic target or strategy for anti-inflammatory treatment.
METHODS
Plasmid construction and luciferase reporter assay
A luciferase reporter assay was performed to test the binding of miR-17/20a/106a to the target gene SIRPα. A 1954-bp segment of the human SIRPα 3′ UTR containing a presumed miR-17/20a/106a complementary site (seed sequence, GCACTTT) was amplified by means of PCR with human genomic DNA as a template. The PCR product was inserted into the pMIR-REPORT plasmid (Applied Biosystems), and insertion was confirmed by means of sequencing. To test the binding specificity, we mutated the seed sequence from GCACTTT to CGTGAAA. For the luciferase reporter assays, 1 μg of firefly luciferase reporter plasmid, 1 μg of β-galactosidase expression vector (Applied Biosystems), and 30 nmol/L of each pre-miRNA, anti-miRNA ASO, or scramble control RNA were transfected into 293T cells cultured in 24-well plates with Lipofectamine 2000 (Invitrogen), according to the manufacturer’s instructions. The β-galactosidase vector was used as a transfection control. One day after transfection, the cells were assayed with a luciferase assay kit (Promega, Madison, Wis).
Construction of eukaryotic expression vector plasmid and transfection
The full-length human c-Myc gene sequence was amplified by means of RT-PCR with total RNA template extracted from HL-60. The primers were designed as follows: sense, 5′-CCGGAATTCATTCTGCCCATTTGG GGACACTT-3′; antisense, 5′-CCGCTCGAGTTTTCCTTACGCACAAGA GTTCC-3′. After purification with a QIAquick gel extraction kit (Qiagen, Hilden, Germany), the fragment was identified by means of dual digestion of the restriction endonuclease EcoRI and XhoI and confirmed further by using DNA sequencing. The eukaryotic expression vector pcDNA3.1 (+) was digested by EcoRI and XhoI. The digested fragment was purified and then ligated with T4 ligase at 16°C overnight, followed by transformation into Escherichia coli. The HL-60 cells were seeded on 6-well plates overnight and transfected the following day with Lipofectamine 2000 (Invitrogen), according to the manufacturer’s instructions. Briefly, the cells were transfected with c-Myc–pcDNA3.1 (+) or pcDNA3.1 (+) as a negative control, respectively, and harvested 48 hours after transfection for further analysis.
RNA isolation and quantitative RT-PCR of miRNAs or mRNAs
Total RNA was extracted from cells or tissues with TRIzol Reagent (Invitrogen), according to the manufacturer’s instructions. Quantitative RT-PCR was performed with TaqMan miRNA probes (Applied Biosystems). Briefly, total RNA was reverse transcribed to cDNA with AMV reverse transcriptase (Takara) and a stem-loop RT primer (Applied Biosystems) or RT primer. Real-time PCR was performed with aTaqMan PCR kit and an Applied Biosystems 7900 Sequence Detection System (Applied Biosystems). All reactions, including no-template controls, were run in triplicate. After the reaction, the cycle threshold values were determined by using fixed threshold settings. Expression of miRNA in cells or tissues was normalized against U6 small nuclear RNA, and mRNA expression in cells or tissues was normalized against GAPDH. Sequences of primers used are as follows: SIRPα sense, 5′-AGCACTAAGCAACATCTCGCTGTG GACG-3′; SIRPα antisense, 5′-CAAACTGTTAAACCTCAGACTTCACAA GACCC-3′; mouse GAPDH sense, 5′-GGTGAAGGTCGGTGTGAACG-3′; mouse GAPDH antisense, 5′-CTCGCTCCTGGAAGATGGTG-3′; human GAPDH sense, 5′-AGAAGGCTGGGGCTC ATTTG-3′; and human GAPDH antisense, 5′-AGGGGCCATCCACAGTCTTC-3′.
Transfection of cells with ncRNA, pre-miRNA, or miRNA inhibitors
Cells were seeded on 6-well plates overnight and transfected the following day with Lipofectamine 2000 (Invitrogen), according to the manufacturer’s instructions. For the overexpression of miRNA, 30 nmol/L pre-miRNA was used. For the inhibition of 3 miRNAs altogether, a total of 30 nmol/L miRNA inhibitors (10 nmol/L of each anti-miRNA ASO) were used. Scrambled oligonucleotides (30 nmol/L) were used as controls for pre-miRNA or miRNA inhibitors, respectively. Cells were harvested 24 hours after transfection for real-time PCR analysis and Western blotting.
miRNA profiling with TaqMan low-density arrays
RNA was extracted from samples by using the mirVana miRNA Isolation Kit (Applied Biosystems). For each group of samples (control and stimulated), 1 μg of total RNA was used for multiplex reverse transcription reactions with miRNA-specific RT primers (Megaple RT Primers, Human Pool Set v3.0, Applied Biosystems). Subsequently, expression of miRNAs was tested by using quantitative PCR with the TaqMan Low Density Array Human MicroRNA Panel 1.0 (Applied Biosystems) run on an Applied Biosystems 7900HT instrument with a detection limit set to a cycle threshold of 33, according to the manufacturer’s instructions. The data presented are normalized to U6 and identified as a suitable endogenous control.
Immunofluorescence labeling
HL-60 cells transfected with a total of 30 nmol/L miRNA inhibitors (10 nmol/L each) or scrambled oligonucleotides were harvested and washed twice in PBS. Cells were fixed and permeabilized with cold alcohol (−20°C) for 30 minutes and then blocked for 30 minutes with 3% BSA. Cells were stained with rabbit anti-human SIRPα antibodies for 1 hour, followed by detection with Alexa Fluor–conjugated mouse anti-rabbit secondary antibodies. Samples were mounted with antifade solution for observation under laser scanning confocal microscopy (FV1000, Olympus).
Western blot analysis
Samples were lysed in lysis buffer (50 mmol/LTris-HCl [pH 7.4], 150 mmol/L NaCl, 1% NP-40, and 0.1% SDS) at 4°C and centrifuged at 12,000g (4°C for 10 minutes). The supernatant fraction was collected, and the protein concentration was determined by using the BCA assay (Pierce, Rockford, Ill). Aliquots of proteins (20–40 μg) were separated on 10% SDS-PAGE and transferred to polyvinylidene difluoride membranes. The membranes were blocked with 5% non-fat milk and then incubated overnight at 4°C with primary antibodies diluted in blocking solution. After 3 × 15-minute washes, the blots were incubated with the appropriate horseradish peroxidase–conjugated secondary antibody and detected with an enhanced chemiluminescence reagent (Cell Signaling). The autoradiographic intensity of each protein band was quantified by using BandScan software (Informer Technologies, Inc) and normalized against GAPDH.
Cytokine assay and nitrite oxidant detection
Cytokine levels in culture supernatants were determined with commercial ELISA kits for TNF-α and IL-6 (R&D Systems), according to the manufacturer’s instructions. Each value represents the mean of triplicate values. For NO detection, cells plated in 24-well culture dishes (2 × 105 cells per well) were incubated overnight before stimulation. After the cells were treated with 100 ng/mL LPS for 24 hours, culture supernatants were collected and analyzed with the Griess Reagent kit. Nitrite concentrations were determined based on the measurement of OD at 570 nm.
Supplementary Material
Key messages.
SIRPα is a common target gene of miR-17, miR-20a, and miR-106a.
miR-17, miR-20a, and miR-106a regulate SIRPα production and SIRPα-mediated macrophage inflammatory responses.
Inflammatory stimuli, such as LPS, activate macrophages by increasing miR-17, miR-20a, and miR-106a levels, leading to reduction of SIRPα.
Abbreviations used
- ASO
Antisense oligonucleotide
- FITC
Fluorescein isothiocyanate
- GAPDH
Glyceraldehyde-3-phosphate dehydrogenase
- ITIM
Immunoreceptor tyrosine-based inhibitory motif
- miRNA
Micro RNA
- ncRNA
Normal control RNA
- NO
Nitric oxide
- PEI
Galactosylated low-molecular-weight chitosan
- pre-ncRNA
Pre-normal control RNA
- RA
Retinoic acid
- SIRPα
Signal-regulatory protein α
- SP-A
Surfactant protein A
- SP-D
Surfactant protein D
- TPA
Phorbol 12-myristate 13-acetate
- UTR
Untranslated region
Footnotes
Disclosure of potential conflict of interest: The authors declare that they have no relevant conflicts of interest.
References
- 1.Barclay AN, Brown MH. The SIRP family of receptors and immune regulation. Nat Rev Immunol. 2006;6:457–64. doi: 10.1038/nri1859. [DOI] [PubMed] [Google Scholar]
- 2.van Beek EM, Cochrane F, Barclay AN, van den Berg TK. Signal regulatory proteins in the immune system. J Immunol. 2005;175:7781–7. doi: 10.4049/jimmunol.175.12.7781. [DOI] [PubMed] [Google Scholar]
- 3.Matozaki T, Murata Y, Okazawa H, Ohnishi H. Functions and molecular mechanisms of the CD47-SIRPalpha signalling pathway. Trends Cell Biol. 2009;19:72–80. doi: 10.1016/j.tcb.2008.12.001. [DOI] [PubMed] [Google Scholar]
- 4.Liu Y, Tong Q, Zhou Y, Lee HW, Yang JJ, Buhring HJ, et al. Functional elements on SIRPalpha IgV domain mediate cell surface binding to CD47. J Mol Biol. 2007;365:680–93. doi: 10.1016/j.jmb.2006.09.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kharitonenkov A, Chen Z, Sures I, Wang H, Schilling J, Ullrich A. A family of proteins that inhibit signalling through tyrosine kinase receptors. Nature. 1997;386:181–6. doi: 10.1038/386181a0. [DOI] [PubMed] [Google Scholar]
- 6.Fujioka Y, Matozaki T, Noguchi T, Iwamatsu A, Yamao T, Takahashi N, et al. A novel membrane glycoprotein, SHPS-1, that binds the SH2-domain-containing protein tyrosine phosphatase SHP-2 in response to mitogens and cell adhesion. Mol Cell Biol. 1996;16:6887–99. doi: 10.1128/mcb.16.12.6887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Takada T, Matozaki T, Takeda H, Fukunaga K, Noguchi T, Fujioka Y, et al. Roles of the complex formation of SHPS-1 with SHP-2 in insulin-stimulated mitogen-activated protein kinase activation. J Biol Chem. 1998;273:9234–42. doi: 10.1074/jbc.273.15.9234. [DOI] [PubMed] [Google Scholar]
- 8.Kontaridis MI, Liu X, Zhang L, Bennett AM. SHP-2 complex formation with the SHP-2 substrate-1 during C2C12 myogenesis. J Cell Sci. 2001;114:2187–98. doi: 10.1242/jcs.114.11.2187. [DOI] [PubMed] [Google Scholar]
- 9.Oh ES, Gu H, Saxton TM, Timms JF, Hausdorff S, Frevert EU, et al. Regulation of early events in integrin signaling by protein tyrosine phosphatase SHP-2. Mol Cell Biol. 1999;19:3205–15. doi: 10.1128/mcb.19.4.3205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ochi F, Matozaki T, Noguchi T, Fujioka Y, Yamao T, Takada T, et al. Epidermal growth factor stimulates the tyrosine phosphorylation of SHPS-1 and association of SHPS-1 with SHP-2, a SH2 domain-containing protein tyrosine phosphatase. Biochem Biophys Res Commun. 1997;239:483–7. doi: 10.1006/bbrc.1997.7489. [DOI] [PubMed] [Google Scholar]
- 11.Veillette A, Thibaudeau E, Latour S. High expression of inhibitory receptor SHPS-1 and its association with protein-tyrosine phosphatase SHP-1 in macrophages. J Biol Chem. 1998;273:22719–28. doi: 10.1074/jbc.273.35.22719. [DOI] [PubMed] [Google Scholar]
- 12.Ishikawa-Sekigami T, Kaneko Y, Okazawa H, Tomizawa T, Okajo J, Saito Y, et al. SHPS-1 promotes the survival of circulating erythrocytes through inhibition of phagocytosis by splenic macrophages. Blood. 2006;107:341–8. doi: 10.1182/blood-2005-05-1896. [DOI] [PubMed] [Google Scholar]
- 13.Ikeda H, Okazawa H, Ohnishi H, Murata Y, Oldenborg PA, Matozaki T. Mutational analysis of the mechanism of negative regulation by SRC homology 2 domain-containing protein tyrosine phosphatase substrate-1 of phagocytosis in macrophages. J Immunol. 2006;177:3123–32. doi: 10.4049/jimmunol.177.5.3123. [DOI] [PubMed] [Google Scholar]
- 14.Liu Y, Buhring HJ, Zen K, Burst SL, Schnell FJ, Williams IR, et al. Signal regulatory protein (SIRPalpha), a cellular ligand for CD47, regulates neutrophil transmigration. J Biol Chem. 2002;277:10028–36. doi: 10.1074/jbc.M109720200. [DOI] [PubMed] [Google Scholar]
- 15.Cooper D, Lindberg FP, Gamble JR, Brown EJ, Vadas MA. Transendothelial migration of neutrophils involves integrin-associated protein (CD47) Proc Natl Acad Sci U S A. 1995;92:3978–82. doi: 10.1073/pnas.92.9.3978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.de Vries HE, Hendriks JJ, Honing H, De Lavalette CR, van der Pol SM, Hooijberg E, et al. Signal-regulatory protein alpha-CD47 interactions are required for the transmigration of monocytes across cerebral endothelium. J Immunol. 2002;168:5832–9. doi: 10.4049/jimmunol.168.11.5832. [DOI] [PubMed] [Google Scholar]
- 17.Lindberg FP, Bullard DC, Caver TE, Gresham HD, Beaudet AL, Brown EJ. Decreased resistance to bacterial infection and granulocyte defects in IAP-deficient mice. Science. 1996;274:795–8. doi: 10.1126/science.274.5288.795. [DOI] [PubMed] [Google Scholar]
- 18.Rosseau S, Selhorst J, Wiechmann K, Leissner K, Maus U, Mayer K, et al. Monocyte migration through the alveolar epithelial barrier: adhesion molecule mechanisms and impact of chemokines. J Immunol. 2000;164:427–35. doi: 10.4049/jimmunol.164.1.427. [DOI] [PubMed] [Google Scholar]
- 19.Gardai SJ, Xiao YQ, Dickinson M, Nick JA, Voelker DR, Greene KE, et al. By binding SIRPalpha or calreticulin/CD91, lung collectins act as dual function surveillance molecules to suppress or enhance inflammation. Cell. 2003;115:13–23. doi: 10.1016/s0092-8674(03)00758-x. [DOI] [PubMed] [Google Scholar]
- 20.Janssen WJ, McPhillips KA, Dickinson MG, Linderman DJ, Morimoto K, Xiao YQ, et al. Surfactant proteins A and D suppress alveolar macrophage phagocytosis via interaction with SIRP alpha. Am J Respir Crit Care Med. 2008;178:158–67. doi: 10.1164/rccm.200711-1661OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kong XN, Yan HX, Chen L, Dong LW, Yang W, Liu Q, et al. LPS-induced down-regulation of signal regulatory protein α contributes to innate immune activation in macrophages. J Exp Med. 2007;204:2719–31. doi: 10.1084/jem.20062611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–97. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 23.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–5. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 24.Steele C, Marrero L, Swain S, Harmsen AG, Zheng M, Brown GD, et al. Alveolar macrophage-mediated killing of Pneumocystis carinii f. sp. muris involves molecular recognition by the Dectin-1 beta-glucan receptor. J Exp Med. 2003;198:1677–88. doi: 10.1084/jem.20030932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wheeler AP, Wells CM, Smith SD, Vega FM, Henderson RB, Tybulewicz VL, et al. Rac1 and Rac2 regulate macrophage morphology but are not essential for migration. J Cell Sci. 2006;119:2749–57. doi: 10.1242/jcs.03024. [DOI] [PubMed] [Google Scholar]
- 26.Oliveira NL, Kalf GF. Induced differentiation of HL-60 promyelocytic leukemia cells to monocyte/macrophages is inhibited by hydroquinone, a hematotoxic metabolite of benzene. Blood. 1992;79:627–33. [PubMed] [Google Scholar]
- 27.Sokoloski JA, Sartorelli AC, Rosen CA, Narayanan R. Antisense oligonucleotides to the p65 subunit of NF-kappa B block CD11b expression and alter adhesion properties of differentiated HL-60 granulocytes. Blood. 1993;82:625–32. [PubMed] [Google Scholar]
- 28.Chang TC, Yu D, Lee YS, Wentzel EA, Arking DE, West KM, et al. Widespread microRNA repression by Myc contributes to tumorigenesis. Nat Genet. 2008;40:43–50. doi: 10.1038/ng.2007.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.O’Donnell KA, Wentzel EA, Zeller KI, Dang CV, Mendell JT. c-Myc-regulated microRNAs modulate E2F1 expression. Nature. 2005;435:839–43. doi: 10.1038/nature03677. [DOI] [PubMed] [Google Scholar]
- 30.Liu DQ, Li LM, Guo YL, Bai R, Wang C, Bian Z, et al. Signal regulatory protein alpha negatively regulates beta2 integrin-mediated monocyte adhesion, transendothelial migration and phagocytosis. PLoS One. 2008;3:e3291. doi: 10.1371/journal.pone.0003291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu SQ, Alkema PK, Tieche C, Tefft BJ, Liu DZ, Li YC, et al. Negative regulation of monocyte adhesion to arterial elastic laminae by signal regulatory protein alpha and Src homology 2 domain-containing protein-tyrosine phosphatase-1. J Biol Chem. 2005;280:39294–301. doi: 10.1074/jbc.M503866200. [DOI] [PubMed] [Google Scholar]
- 32.Zuo L, Huang Z, Dong L, Xu L, Zhu Y, Zeng K, et al. Targeting delivery of anti-TNFalpha oligonucleotide into activated colonic macrophages protects against experimental colitis. Gut. 2010;59:470–9. doi: 10.1136/gut.2009.184556. [DOI] [PubMed] [Google Scholar]
- 33.Lu Y, Thomson JM, Wong HY, Hammond SM, Hogan BL. Transgenic over-expression of the microRNA miR-17-92 cluster promotes proliferation and inhibits differentiation of lung epithelial progenitor cells. Dev Biol. 2007;310:442–53. doi: 10.1016/j.ydbio.2007.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Matozaki T, Murata Y, Saito Y, Okazawa H, Ohnishi H. Protein tyrosine phosphatase SHP-2: a proto-oncogene product that promotes Ras activation. Cancer Sci. 2009;100:1786–93. doi: 10.1111/j.1349-7006.2009.01257.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Foshay KM, Gallicano GI. miR-17 family miRNAs are expressed during early mammalian development and regulate stem cell differentiation. Dev Biol. 2009;326:431–43. doi: 10.1016/j.ydbio.2008.11.016. [DOI] [PubMed] [Google Scholar]
- 36.Pichiorri F, Suh SS, Ladetto M, Kuehl M, Palumbo T, Drandi D, et al. MicroRNAs regulate critical genes associated with multiple myeloma pathogenesis. Proc Natl Acad Sci U S A. 2008;105:12885–90. doi: 10.1073/pnas.0806202105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Taguchi A, Yanagisawa K, Tanaka M, Cao K, Matsuyama Y, Goto H, et al. Identification of hypoxia-inducible factor-1 alpha as a novel target for miR-17-92 mi-croRNA cluster. Cancer Res. 2008;68:5540–5. doi: 10.1158/0008-5472.CAN-07-6460. [DOI] [PubMed] [Google Scholar]
- 38.Bode JG, Ehlting C, Haussinger D. The macrophage response towards LPS and its control through the p38(MAPK)-STAT3 axis. Cell Signal. 2012;24:1185–94. doi: 10.1016/j.cellsig.2012.01.018. [DOI] [PubMed] [Google Scholar]
- 39.Kinjyo I, Hanada T, Inagaki-Ohara K, Mori H, Aki D, Ohishi M, et al. SOCS1/JAB is a negative regulator of LPS-induced macrophage activation. Immunity. 2002;17:583–91. doi: 10.1016/s1074-7613(02)00446-6. [DOI] [PubMed] [Google Scholar]
- 40.Kim YS, Shin SI, Kang KL, Chung JH, Herr Y, Bae WJ, et al. Nicotine and lipopolysaccharide stimulate the production of MMPs and prostaglandin E(2) by hypoxia-inducible factor-1α up-regulation in human periodontal ligament cells. J Periodontal Res. 2012;47:719–28. doi: 10.1111/j.1600-0765.2012.01487.x. [DOI] [PubMed] [Google Scholar]
- 41.Stofega MR, Argetsinger LS, Wang H, Ullrich A, Carter-Su C. Negative regulation of growth hormone receptor/JAK2 signaling by signal regulatory protein alpha. J Biol Chem. 2000;275:28222–9. doi: 10.1074/jbc.M004238200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.