Abstract
The cross-talk between epigenetics and miRNA expression plays an important role in human tumorigenesis. Herein, the regulation and role of miR-196b-5p in gastric cancer was investigated. Quantitative real-time RT-PCR (qPCR) demonstrated that miR-196b-5p is significantly overexpressed in human gastric cancer tissues (P<0.01). In addition, it was determined that HOXA10, a homeobox family member and host gene for miR-196b-5p, is overexpressed and positively correlated with miR-196b-5p expression levels (P<0.001). Quantitative pyrosequencing methylation analysis, demonstrated significantly lower levels of DNA methylation at the HOXA10 promoter in gastric cancer, as compared to non-neoplastic gastric mucosa specimens. 5-Aza-2′-deoxycytidine treatment confirmed that demethylation of HOXA10 promoter induces the expression of HOXA10 and miR-196b-5p in gastric cancer cell model systems. Using the Tff1-KO mouse model of gastric neoplasia, hypo-methylation and overexpression of HOXA10 and miR-196b-5p in gastric tumors was observed, as compared to normal gastric mucosa from Tff1-WT mice. Mechanistically, reconstitution of TFF1 in human gastric cancer cells lead to an increased HOXA10 promoter methylation with reduced expression of HOXA10 and miR-196b-5p. Functionally, miR-196b-5p reconstitution promoted human gastric cancer cell proliferation and invasion in vitro. In summary, the current data demonstrates overexpression of miR-196b-5p in gastric cancer and suggests that TFF1 plays an important role in suppressing the expression of miR-196b-5p by mediating DNA methylation of the HOXA10 promoter. Loss of TFF1 expression may promote proliferation and invasion of gastric cancer cells through induction of promoter hypo-methylation and expression of the HOXA10/miR-196b-5p axis.
Keywords: HOX10, miR-196b-5p, methylation, gastric cancer, TFF1
Introduction
Gastric cancer (GC) is the third leading cause of cancer-associated deaths in the world (1,2), responsible for 723,000 deaths, 8.8% of the total cancer-related deaths worldwide (3). Multiple genetic and epigenetic alternations are involved in gastric carcinogenesis, which necessitate the investigation of the underlying molecular mechanisms. Studies of these interactions allow identification of molecular biomarkers that can predict clinical prognoses and improve existing therapeutic strategies in cancer patients (4).
Epigenetic alterations, including DNA methylation of CpG islands and post-translational modifications of histones, are involved in the development of gastric cancer (5–7). DNA methylation is a modification in which a methyl group is added to the cytosine residue at 5Carbon in a CpG dinucleotide (8). Promoter methylation is an important mechanism in regulating gene expression, where hyper-methylation at promoters of tumor suppressor genes is an important tumorigenic step (9,10). On the other hand, hypo-methylation is an independent process where methyl groups are removed, leading to overexpression and activation of genes that can promote tumorigenesis (11). Therefore, it is important to understand the functional significance of the methylation status of gene promoters with respect to the cellular context.
HOXA10 is a member of the homeobox gene family which is well conserved during evolution and participates in a number of biological processes (11). HOXA10 is expressed at a high level in a subset of chronic myelogenous leukemia (CML) advanced phase/blast crisis patients and is involved in the process of self-renewal (12). While constitutively expressed HOXA10 reduces cell invasiveness by regulating P53 expression in human breast cancer (13), its overexpression in ovarian cancer promotes epithelial mesenchymal transition (EMT) (14). Studies in gastric cancer demonstrated that positive HOXA10 expression results in significantly poorer patients’ prognoses than negative expression (4,15). Collectively, these studies indicate that the functional outcome of HOXA10 expression is cell and context dependent.
MicroRNAs (miRNAs) are small noncoding RNAs that inhibit gene expression through post-transcriptional mechanisms (16). The molecular mechanisms regulating miRNA expression include transcriptional regulation, including changes in the host gene expression, methylation of the promoter of the host gene or miRNA, and post-transcriptional mechanisms, including miRNA processing and stability (17). Emerging evidence shows that dysregulation of miRNAs plays a vital role in many biological processes, including proliferation, invasion and apoptosis (18). Hyper-methylation and reduced expression of anti-tumorigenic miRNAs’ promoters are associated with cancer progression in colorectal (19), chronic lymphocytic leukemia (20), cervical (21), breast (22), and gastric cancers (17,23). On the other hand, hypo-methylation of pro-oncogenic miRNAs’ promoters have been reported in hepatocellular carcinoma (24), pancreatic (25), and ERα positive breast cancers (26). In this study, we investigated the expression and molecular relationship between HOXA10 and miR-196b-5p. We also determined the mechanisms that regulate HOXA10 and miR-196b-5p axis and the biological outcome in gastric cancer.
Materials and Methods
Tissue samples
All de-identified gastric tissue samples were obtained following a written informed consent in accordance with the guidelines of the National Institute of Health. The study using de-identified tissue samples was approved by Vanderbilt University Institutional Review Board (IRB# 110076). For DNA, mRNA, and miRNA analysis, 63 de-identified archival human gastric tissue samples (38 non-tumor normal stomach and 25 gastric cancer samples) were used. All adenocarcinomas were classified according to the recent guidelines of the International Union Against Cancer (UICC) TNM classification system.
Cell culture and Reagents
Human gastric cancer cell lines AGS, SNU1, SNU16 and SNU601 purchased from ATCC (American Type Culture Collection). MKN28 and MKN45 cells were obtained from the Riken Cell Bank (Tsukuba, Japan). AGS, SNU1, SNU16 and MKN45 were cultured in F-12 medium (GIBCO, Carlsbad, California, USA) supplemented with 10% fetal bovine serum (FBS, Invitrogen, Life technology) and 1% penicillin/streptomycin (GIBCO, Carlsbad, California, USA). SNU601 and MKN28 were cultured in 1640 medium (GIBCO, Carlsbad, California, USA), supplemented with 10% fetal bovine serum (FBS, Invitrogen, Life technology) and 1% penicillin/streptomycin (GIBCO, Carlsbad, California, USA) at 37°C in an atmosphere containing 5% CO2. All cell lines were ascertained to conform to the original in vitro morphological characteristics and were authenticated by Genetica DNA Laboratories using short tandem repeat (STR) profiling (Genetica DNA Laboratories, Burlington, NC). Mycoplasma was tested and was negative in all cells using the q-PCR method (Thermo Fisher Scientific, USA). De-methylation was induced with 5-Aza-2′ Deoxycytidine (5-Aza) (Selleckchem, Houston, TX, USA) treatment at a concentration of 5μM. Cells were incubated for 72 hours with 5-Aza with replacement of the culture media with fresh media containing 5-Aza every 24 hours.
Expression of TFF1 and miR-196b-5p in cell lines
For reconstitution of TFF1 in cancer cell models, AGS and SNU1 were transfected with the mammalian expression plasmid, pTT5(27) in frame with full coding sequence of human TFF1 gene or PTT5 empty vector with PolyJet (SignaGen, Rockville, USA) for 48 hours (28). For transient overexpression of miR-196b-5p, AGS and SNU1 cells were transfected with 30pmol miR-196b-5p mimic (Applied Biological Materials, ABM, Canada) with LipoJet (SignaGen, Rockville, USA) following the manufacturer’s instructions. The expression of TFF1 and miR-196b-5p was confirmed by quantitative real-time PCR (qRT-PCR).
Quantitative real-time PCR
Total RNA was prepared from cell lines by using the RNeasy Mini Kit (Qiagen, Valencia, California, USA). Total RNA (1ug) was reverse-transcribed to miRNA cDNA using the TaqMan MicroRNA Reverse Transcription kit (Thermo Fisher Scientific, USA) and E. coli Poly A polymerase (New England biolabs, MA, USA). The qRT-PCR was performed using a Bio-Rad CFX Connect Real-time System (Bio-Rad), with the threshold cycle number determined by Bio-Rad CFX manager software V.3.0. mRNA expression was normalized to HPRT (29) and miRNA expression was normalized to miR-16-5p (30,31). The following primers were used for PCR analysis: miR-196b-5p 5′‑TAGGTAGTTTCCTGTTGTTGGG‑′3 (sense) and 5′‑GCGAGCACAGAATTAATACGAC‑′3 (anti-sense), miR-16-5p 5′-TAGCAGCACGTAAATATTGGCG‑′3 (sense), 5′‑GCGAGCACAGAATTAATACGAC‑′3 (anti-sense), HOXA10 5′‑ AGATATTGTCCTAAGTGTCAAGTCCTGA‑′3 (sense), 5′‑GCCATTTCGAGCAGTGGG‑′3(anti-sense), TFF1 5′‑GGTCCTGGTGTCCATGCTG‑′3 (sense), 5′‑ACAGCAGCCCTTATTTGCAC‑′3(anti-sense).
DNA bisulfite treatment and quantitative pyrosequencing analysis
AGS and SNU1 cells were cultured with 5 μM 5-Aza, as described above. DNA was purified by DNeasy Blood & Tissue kits (Qiagen, Valencia, California, USA). The DNA was bisulfite converted using an EZ DNA Methylation Gold Kit (Zymo Research, Orange, California, USA), according to the manufacturer’s protocol. Specific bisulfite PCR and pyrosequencing primers were designed using the PSQ assay design software (Qiagen, Valencia, California, USA) to analyze 138bp of HOXA10 promoter including six CpG dinucleotides sites (-152, -150, -136, -131, -126, -117bp) from -199 to -61, relative to transcription start site (TSS). The following primers were used: forward 5′‑Biotin-GGGTGAGTTTTTTGGTTTATTAATATAGATTAT‑′3, reverse 5′‑ACCAAT CCCCAACCAAAATTTC‑′3, sequencing primer 5′‑CCACCACTCCCAATT‑′3. A 20ng aliquot of modified DNA was amplified by polymerase chain reaction (PCR) of the specific promoter region using the Platinum PCR SuperMix High Fidelity Enzyme Mix (Invitrogen, Carlsbad, CA USA). The PCR products were checked by gel electrophoresis to confirm the size of the product and rule out the formation of primer dimers. The specific PCR products were analyzed using a Biotage PyroMark MD System (Qiagen), according to the manufacturer’s protocol, using PyroMark Gold Reagents (Qiagen, Valencia, California, USA). Based on control normal samples and internal quality controls provided in the software analysis, measurements of DNA with known methylation levels, we used 10% as a cut-off value for identification of DNA hyper-methylation (32).
Transwell cell invasion assay
The invasion assays were performed with Transwell membranes (pore size, 8μm, Corning, Tokyo, Japan) coated with Matrigel™ matrix basement membrane (invasion assay). The AGS and SNU1 (3×105) were suspended in the upper chamber of the Transwell inserts. The medium of the upper chamber was serum-free F-12. F-12 containing 10% FBS was used as a chemoattractant in the lower chamber. After incubation for 24 hours, the cells were washed with PBS twice. Cells in the upper chambers were removed with a cotton swab soaked in PBS. Invading cells were fixed with 100% methanol and stained with 0.05% crystal violet solution and in four randomly selected fields on the underside of the inserts were counted under a light microscope. Data quantification and analysis were performed by ImageJ software (https://imagej.nih.gov/ij/).
Cell proliferation assay
To assess cell proliferation, the Click-iT EdU Assay (Invitrogen, Carlsbad, CA, USA) was performed in AGS and SNU1 following the manufacturer’s protocol. EdU (5-ethynyl-2′-deoxyuridine) is a nucleoside analog of thymidine and is incorporated into DNA during active DNA synthesis. AGS or SNU1 cells were seeded in the 8-chamber plates at a density of 3×104 per well. AGS or SNU1 cells were transfected with miR-196b-5p mimic or control for 48 hours followed by incubation of 1X EdU for 1 hour. The fractions of EdU-positive cells (20 random fields at 20X) were determined using ImageJ software (https://imagej.nih.gov/ij/).
Patients’ survival data analysis
Using Kaplan-Meier survival plots, we analyzed the HOXA10 relationship to overall survival of 876 gastric cancer patients from 3 major cancer research centers including “Berlin dataset” (published in GEO as GSE22377)(33), “Bethesda dataset” (published in GEO as GSE14210)(34), “Melbourne dataset” (published in GEO as GSE51105)(35) and 4 different dataset (GSE15459, GSE29272, GSE62254 and GSE15459)(36) were analyzed using online resource: http://kmplot.com/.
Statistical analyses
All data was expressed as mean ± standard deviation (SD) of three independent experiments. Using the GraphPad Prism (GraphPad Software, San Diego, CA, USA), statistical significance was analyzed by the Student’s t-test, Mann-Whitney test, one way ANOVA and Pearson correlation coefficient. Differences with p values ≤0.05 were considered significantly.
Results
miR-196b-5p overexpression correlates with HOXA10 expression in human gastric cancer
To study the expression level and function of miR-196b-5p in human gastric cancer, the expression of miR-196b-5p in gastric tissue samples was investigated by performing qRT-PCR in 38 human non-tumor normal gastric (NG) tissue samples and 25 human gastric cancer (GC) tissue samples. The data showed that miR-196b-5p was significantly overexpressed in gastric cancer samples as compared to non-tumor normal gastric tissue samples (Figure 1A, p<0.001). When using matched adjacent non-tumor normal and gastric cancer tissues from the same patients, we again found that miR-196b-5p was significantly overexpressed in the gastric cancer tissues (Figure 1B, p<0.01). Furthermore, qRT-PCR of miR-196b-5p expression showed high expression levels in several gastric cancer cell lines, as compared to normal gastric tissues (Figure 1C, p<0.05). Gastric cancer tissue samples and cell lines displayed similar results, as compared to normal tissue samples (Supplementary Figure 1A, p<0.001). In human, miR-196b-5p is located at chromosome band 7p15.2 (Figure 1D) within the HOXA gene cluster between HOXA9 and HOXA10 (37,38). Therefore, we hypothesized that miR-196b-5p shares the same promoter with HOXA10. To examine this possibility, we measured the expression of HOXA10. We found that HOXA10 mRNA was also significantly overexpressed in human gastric cancer tissue samples as compared to non-tumor normal gastric tissue samples (Figure 1E, p<0.001). We also found high expression of HOXA10 in gastric cancer tissue samples and cell lines as compared to normal gastric tissue samples (Supplementary Figure 1B, p<0.05). We also detected a positive correlation between the expression levels of HOXA10 and miR-196b-5p in gastric cancer tissue samples (Figure 1F, p<0.001). Using Kaplan-Meier survival plots for analysis of survival data of 876 gastric cancer patients, we found that patients with high expression levels of HOXA10 had significant poorer prognoses than low HOXA10 expression groups (Figure 1G, p<0.001). Together, these results demonstrated that both miR-196b-5p and HOXA10 were up-regulated in gastric cancer and may play a vital role in human gastric carcinogenesis, patients’ prognosis, and clinical outcome.
Figure 1. miR-196b-5p and HOXA10 mRNA are co-overexpressed in gastric cancer tissue samples and cell lines.
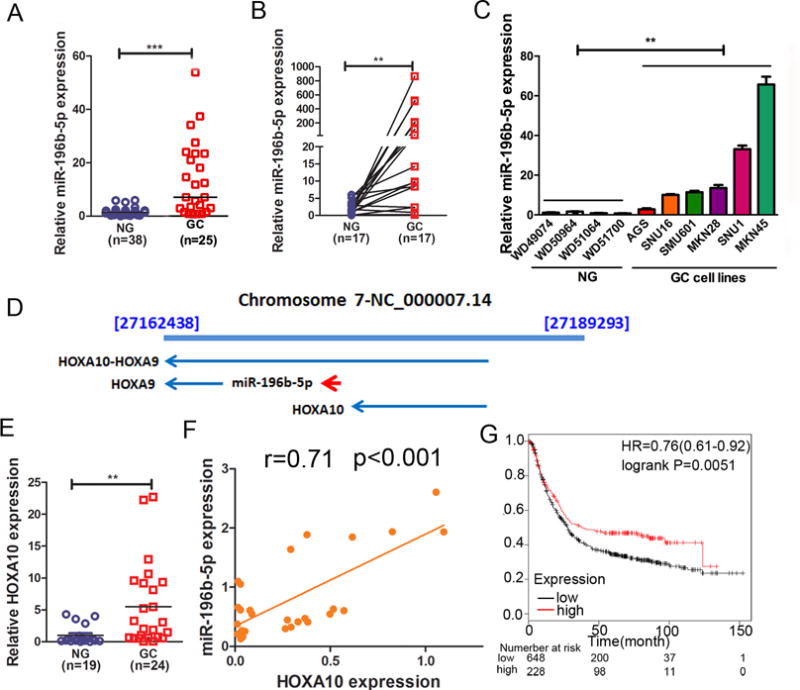
(A) qRT-PCR data showing miR-196b-5p expression level in human gastric cancer (GC, n=25) and non-tumor normal gastric (NG, n=38) tissue samples. The horizontal bar indicates the median, Mann-Whitney test. (B) qRT-PCR results of miR-196b-5p expression in 17 pairs of matched human NG and GC tissue samples. (C) qRT-PCR data of miR-196b-5p expression in 4 human NG samples and 6 human gastric cancer cell lines. (D) miR-196b-5p, HOXA9 and HOXA10 gene location on human chromosome 7. (E) qRT-PCR data of HOXA10 expression in human gastric cancer (GC, n=24) and non-tumor normal gastric (NG, n=19) tissue samples. (F) Pearson correlation coefficient analysis of miR-196b-5p and HOXA10 mRNA expression in human gastric cancer tissue samples (n=27). (G) Kaplan-Meier analysis of HOXA10 related overall survival of 876 human gastric cancer patients. **p<0.01, ***p<0.001.
HOXA10 promoter hypo-methylation mediates HOXA10 overexpression in gastric cancer
Epigenetic mechanisms such as DNA methylation are important mechanisms that control gene expression. Although the overexpression of HOXA10 was previously reported in gastric cancer studies, we investigated if its expression is dependent on the HOXA10 promoter methylation. To this end, we identified CpG sites within the promoter of HOXA10. Our data indicated that there were more than 150 CpG sites within the HOXA10 promoter region -250 to +50 relative to transcript start site (TSS) (Figure 2A). To investigate the effect of DNA methylation on HOXA10 mRNA expression in gastric tissue samples, pyrosequencing was used to quantitatively analyze the DNA methylation levels of six CpG sites in the HOXA10 promoter regions in both human non-tumor normal gastric (n=20) and gastric cancer tissue samples (n=19). Our analysis of the HOXA10 promoter spanned the region from -117 to +33 relative to TSS. Figures 2B and 2C showed representative pyrosequencing profiles of the HOXA10 promoter regions with the percentages of methylation at each of the six CpG sites in non-tumor normal and gastric cancer tissues, respectively. Quantification of the methylation levels in each group showed that there was significant reduction in methylation levels of the HOXA10 promoter in gastric cancer tissues compared to non-tumor normal tissues (Figure 2D, p<0.001). Our data from matched tissue samples of non-tumor normal and gastric cancer tissues also showed significant decrease in the HOXA10 promoter methylation levels in the gastric cancer samples (Figure 2E, p<0.001). The HOXA10 promoter methylation profiles of the six CpG sites of four representative matched samples are shown in Figure 2F. Taken together, these results demonstrated the HOXA10 promoter hypo-methylation in human gastric cancer tissues.
Figure 2. HOXA10 promoter is hypo-methylated in human GC samples.
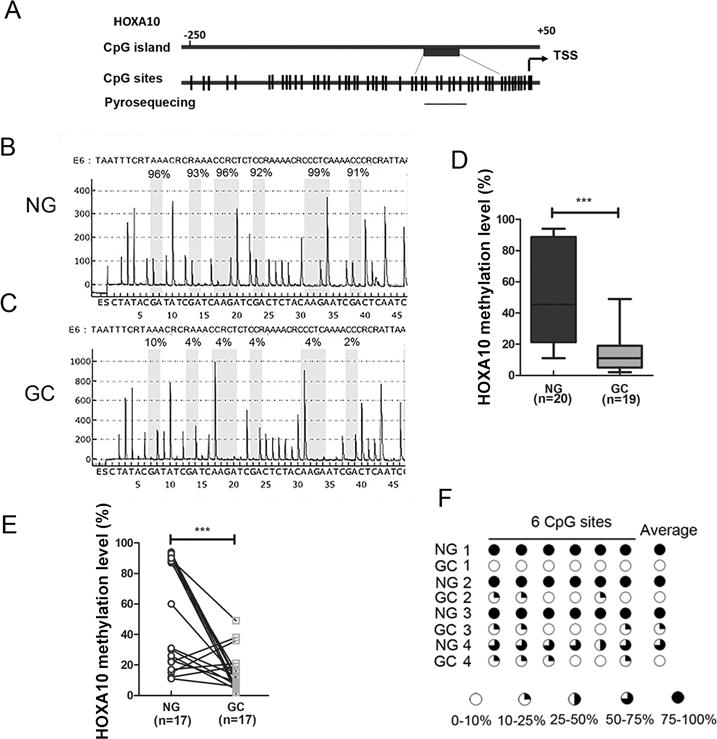
(A) A schematic drawing shows CpG sites in HOXA10 gene promoter and pyrosequencing assay location. Each vertical bar represents a CpG site. TSS, transcription start site. (B) and (C) DNA methylation level of 6 CpG sites in the HOXA10 promoter were quantified by pyrosequencing. Representative pyrosequencing profiles of NG and GC samples are shown. (D) Quantification data of HOXA10 promoter methylation levels in 20 NG and 19 GC samples. The box-and-whisker plots are used to demonstrate the data. The lower and the upper edges of the box mark the 25th and 75th percentile, respectively. The areas between the box and the whisker extend to the 10th and 90th percentile. (E) HOXA10 promoter methylation level in 17 pairs of matched normal and gastric cancer samples. (F) A schematic profile shows HOXA10 promoter methylation level of 6 CpG sites in 4 pairs of matched representative patient samples. ***p<0.001.
5-Aza-2′ Deoxycytidine (5-Aza) treatment increases HOXA10 and miR-196b-5p expression in human gastric cancer cells
To investigate the effect of HOXA10 promoter methylation on HOXA10 and miR-196b-5p expression levels, SNU1 and AGS (gastric cancer cell lines) were treated with 5-Aza-2′ Deoxycytidine (5-Aza). Analysis of promoter methylation using pyrosequencing and expression using qRT-PCR, following 5-Aza treatment, revealed significant reduction in the HOXA10 promoter methylation and increase in the expression level of HOXA10 and miR-196b-5p in AGS and SNU1 cells (Figure 3). These results suggest that DNA methylation plays an important role in regulating HOXA10 and miR-196b-5p expression levels in gastric cancer cells.
Figure 3. HOXA10 mRNA and miR-196b-5p are induced by DNA demethylation in gastric cancer cells.
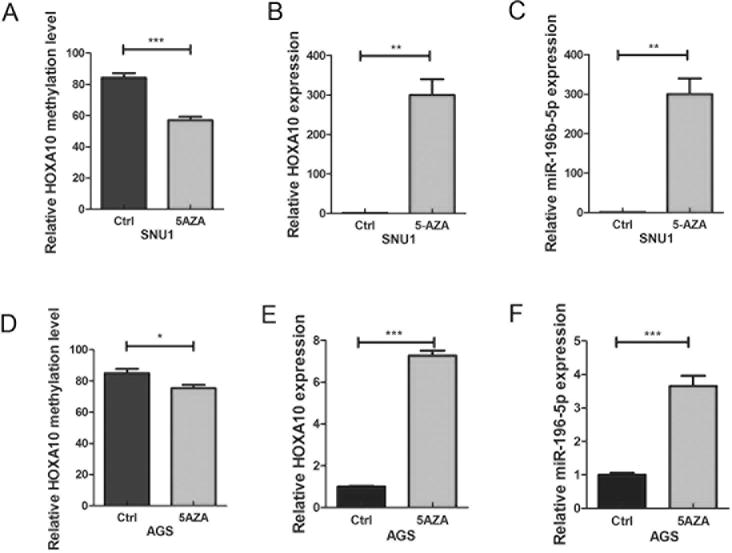
SNU1 and AGS cells that express low-mid range levels of HOXA10 and miR-196b-5p were used for treatment with 5-Aza. (A) Pyrosequencing shows HOXA10 promoter methylation level with and without 5-Aza treatment in SNU1 cells. (B) and (C) qRT-PCR data of HOXA10 mRNA and miR-196b-5p expression with and without 5-Aza treatment in SNU1 cells. (D) Pyrosequencing shows HOXA10 promoter methylation level with and without 5-Aza treatment in AGS. (E) and (F) qRT-PCR of HOXA10 mRNA and miR-196b-5p expression with and without 5-Aza treatment in AGS. *p<0.05, **p<0.01, ***p<0.001.
Reconstitution of TFF1 decreases miR-196b-5p and HOXA10 expression in gastric cancer cells
Our previous studies showed that TFF1 functions as a gastric tumor suppressor playing a vital role in gastric carcinogenesis in vivo (39,40). An earlier study showed that TFF1 reconstitution reduced the expression of miR-504 in gastric cancer cells (28). In this study, we found an increase in HOXA10 mRNA and miR-196b-5p in gastric cancer tissues compared to non-tumor normal gastric tissues. Therefore, we sought to determine if loss of TFF1 expression plays a role in regulating HOXA10 and miR-196b-5p expression in gastric cancer. First, we confirmed TFF1 mRNA expression was significantly decreased in human GC relative to non-tumor normal gastric tissues (Figure 4A, p<0.001). Next, in order to examine miR-196b-5p expression in the absence of TFF1, we compared gastric tissues from Tff1 wild-type and Tff1 knockout mice. Our data demonstrated a significant increase of miR-196b-5p in Tff1 knockout mice gastric cancer tissue samples, as compared with normal gastric tissues (Figure 4B, p<0.05). To investigate whether TFF1 could affect miR-196b-5p and HOXA10 expression in human gastric cancer cells, TFF1 was transiently reconstituted in AGS and SNU1. qRT-PCR was performed to confirm the TFF1 reconstitution in AGS cells (Figure 4C, p<0.01). Our data demonstrated that both HOXA10 and miR-196b-5p expression levels were significantly reduced upon reconstitution of TFF1 in AGS cells (Figure 4D–E, p<0.05). Similar results were obtained in SNU1 cells (Figure 4F–H). Together, these results suggest that TFF1 plays a role in down-regulating HOXA10 and miR-196b-5p expression.
Figure 4. TFF1 reconstitution decreases HOXA10 and miR-196b-5p expression in gastric cancer cells.
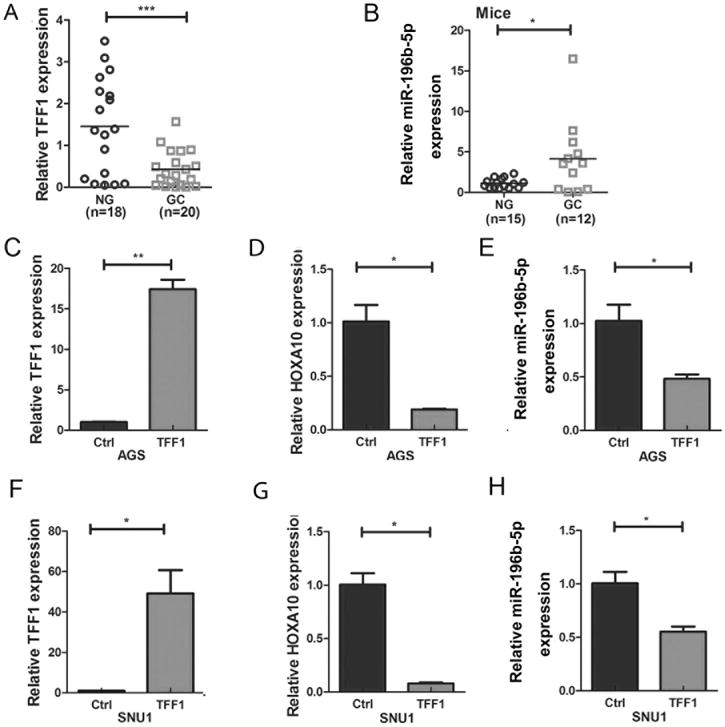
(A) qRT-PCR analysis of TFF1 mRNA expression in human non-tumor normal (NG) and gastric cancer (GC) tissue samples. (B) qRT-PCR analysis of miR-196b-5p expression level in wild type normal (NG) or Tff1 knockout mice gastric cancer (GC) tissue samples. qRT-PCR data of (C) TFF1, (D) HOXA10 and (E) miR-196b-5p in AGS control or TFF1 transiently transfected cells. (F to H) Similar results in SNU1 cells as in C to E. *p<0.05, **p<0.01, ***p<0.001.
Reconstitution of TFF1 expression in gastric cancer cells decreases HOXA10 expression through induction of promoter methylation
To study whether TFF1 regulates the expression of HOXA10 and miR-196b-5p through HOXA10 promoter methylation, quantitative pyrosequencing was performed to determine the methylation levels of the HOXA10 promoter in AGS and SNU1 cells with and without TFF1 transient reconstitution. Pyrosequencing data indicated that TFF1 reconstitution promoted HOXA10 promoter DNA methylation in AGS cells, as compared with control cells (Figure 5A). The HOXA10 promoter methylation profiles of the six CpG sites in AGS cells are shown in Figure 5B. The data from matched CpG sites also showed significant increase in the HOXA10 promoter methylation levels after TFF1 reconstitution (Figure 5C p<0.001). Similar results were obtained in SNU1 cells (Figure 5D–F). These findings suggest a novel role for TFF1 through the downregulation of miR-196b-5p and HOXA10 expression by enhancing promoter methylation.
Figure 5. TFF1 reconstitution induces HOXA10 promoter methylation.
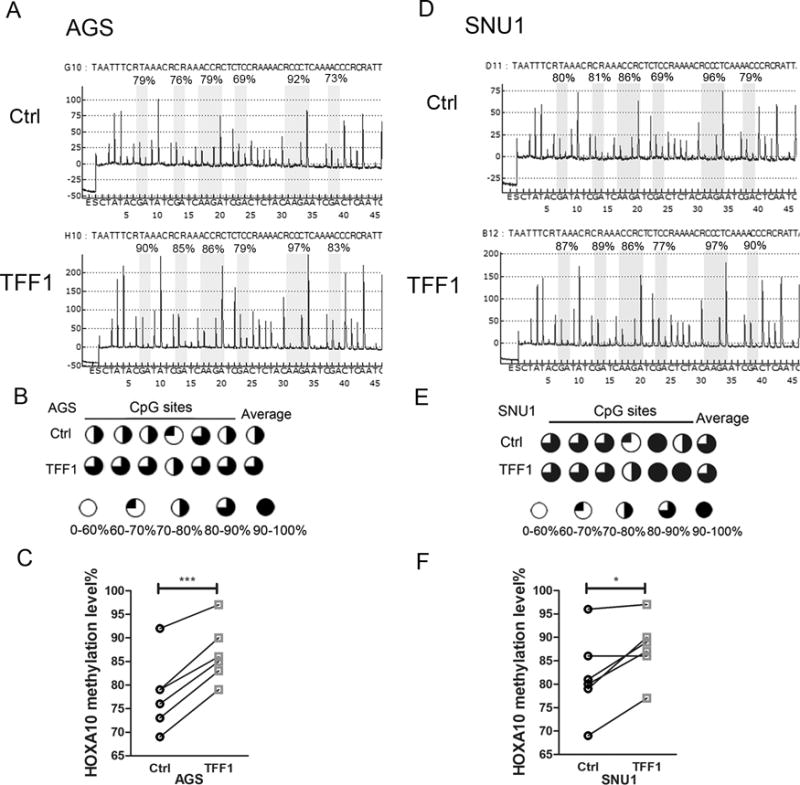
(A) Representative pyrosequencing profile of HOXA10 promoter CpG sites in AGS control (empty vector) or following transient transfection with TFF1. (B) A schematic drawing shows HOXA10 promoter methylation of 6 CpG sites in AGS cells, as in A. (C) Quantification data of B. (D to F) similar results were obtained in SNU1 cells, as in A to C.
Overexpression of miR-196b-5p promotes proliferation and invasion in gastric cancer cells
Recent studies in colorectal cancer demonstrated that miR-196b-5p expression promotes migration and growth of metastases (41). Therefore, to determine the biological outcome of miR-196b-5p overexpression in human gastric cancer, we investigated the influences of miR-196b-5p in gastric cancer cell proliferation and invasion. miR-196b-5p was transiently overexpressed in AGS and SNU1 cells by transfection of miR-196b-5p mimic or empty vector control. The overexpression of miR-196b-5p, following overexpression, was validated using qRT-PCR in the AGS and SNU1 (Figure 6A and B, p<0.01), as compared to their respective control cells. To investigate the function of miR-196b-5p in regulating cancer cell proliferation, EdU assay was used to measure the percent of cells in the DNA synthesis phase of the cell cycle. Figure 6C showed representative photomicrographs of control and miR-196b-5p mimic AGS (upper panels) or SNU1 cells (lower panels) in S phase (green signal). Quantification of the EdU positive cells revealed a significant increase in cellular proliferation, following miR-196b-5p overexpressing AGS (Figure 6D, p<0.05) or SNU1 (Figure 6E, p<0.05) cells, compared to their control cells. Invasive ability was measured by using the Transwell invasion assay with and without transient overexpression of miR-196b-5p in AGS or SUN1 cells. The upregulation of miR-196b-5p was confirmed in both AGS and SNU1 miR-196b-5p overexpressing cells (Figure 6F and G, p<0.001). Figure 6H shows representative photomicrographs of cells which have invaded through the Matrigel matrix to the lower chamber. Quantification of the invading cells revealed a significant increase in invasion, following miR-196b-5p overexpression in AGS (Figure 6I, p<0.05) or SNU1 cells (Figure 6J, p<0.01), as compared to control cells. Taken together, our data suggests that miR-196b-5p promotes cellular proliferation and invasion in gastric cancer.
Figure 6. Reconstitution of miR-196b-5p promotes gastric cancer proliferation and invasion.
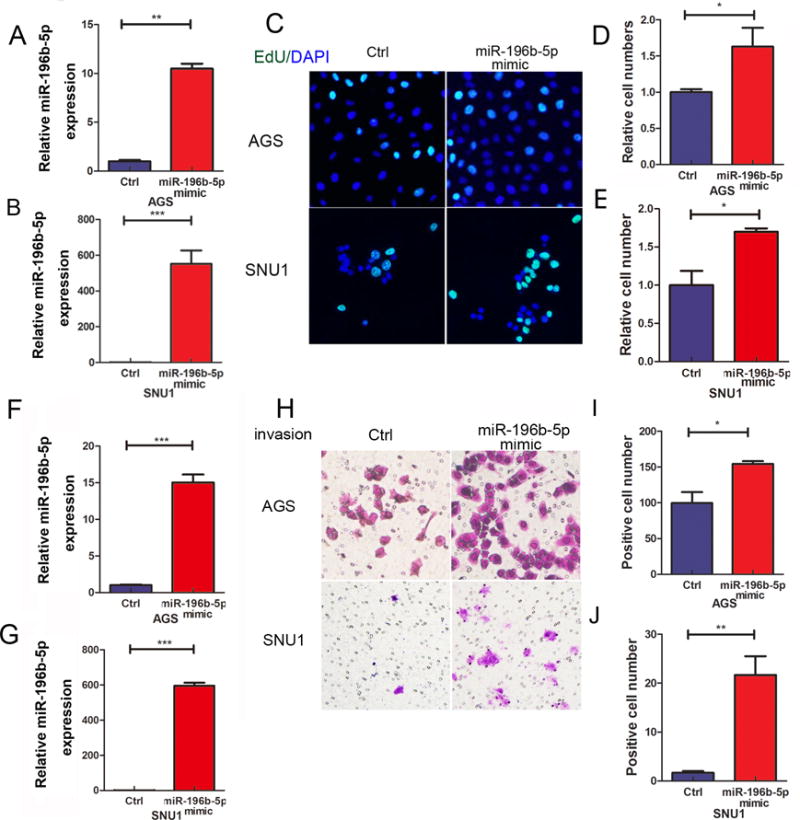
(A) and (B) qRT-PCR data of miR-196b-5p expression levels in AGS and SNU1 cells transfected with control or miR-196b-5p mimic. (C) EdU assay shows proliferating cells in AGS (upper panels) or SUN1 (lower panels) cells as in A and B. Green fluorescence staining indicates proliferating EdU positive cells. Blue staining represents a counterstain with DAPI for cell nucleus. (D) and (E) Quantification data of C. (F) and (G) demonstrate qRT-PCR data of miR-196b-5p expression levels in AGS and SNU1 cells transfected with control or miR-196b-5p mimic. (H) Transwell invasion assay in AGS and SNU1 cells as in F and G. (I) and (J) Quantification data of H. *p<0.05, **p<0.01, ***p<0.001.
Discussion
The aim of this study was to investigate the aberrant overexpression and oncogenic functions of HOXA10 and miR-196b-5p in gastric cancer. HOXA10 deregulation has been controversial showing overexpression in some malignancies, whereas reduced expression was noted in others. Overexpression of miR-196b-5p was reported in endometrial cancer (42), ovarian cancer (43), and pancreatic cancer (44). Conversely, its reduced expression was noted in breast cancer (45,46). These discordant findings suggest that HOXA10 plays different roles in human tumorigenesis in cellular and context-dependent manners. Our data showed concordant overexpression of HOXA10 and miR-196b-5p in mouse and human gastric cancers. We also found that promoter methylation of HOXA10, host gene of miR-196b-5p, plays a role in regulating miR-196b-5p expression. Of note, we demonstrate, for the first time, that TFF1, a frequently silenced tumor suppressor gene in gastric carcinogenesis, plays an important role in regulating methylation of HOXA10 promoter and suppressing expression of HOXA10 and miR-196b-5p.
The results demonstrated a positive correlation between HOXA10 mRNA and miR-196b-5p expression levels in human gastric cancer tissue samples. miR-196b-5p overexpression has been reported in gastrointestinal cancer types, such as pancreatic (47), colorectal (41), and gastric (48). However, the molecular relationship between miR-196b-5p and HOXA10 is not clear. A growing body of evidence suggests that epigenetic alterations are involved in the initiation and progression of gastric cancer (5,7,49). DNA methylation in human cancers provide insightful understanding of the aberrant methylation and subsequent gene silencing that disturb signal pathways and contribute to the development of cancers (8,50). For example, methylation of miR-342-3p and its host gene EVL correlated with low expression of miR-342-3p and EVL in multiple myeloma (51). Similarly, epigenetic regulation of miR-486-5p and its host gene ANK1 was reported in non-small cell lung cancer (52). In the present study, our findings not only demonstrate a positive correlation between the expression of miR-196b-5p and HOXA10, but also suggest a common regulatory molecular mechanism that involves promoter methylation of HOXA10 in gastric cancer. We found that methylation levels of HOXA10 are lower in gastric cancer tissues, as compared to normal tissue samples, suggesting that DNA hypo-methylation as a potential mechanism that induce overexpression of HOXA10 and miR-196b-5p in gastric cancer. This conclusion is supported by results using a 5-Aza demethylation agent which reduced methylation of HOXA10 with a significant increase in the expression of HOXA10 and miR-196b-5p expression.
We next investigated possible molecular mechanisms that promote HOXA10 promoter methylation and miR-196b-5p aberrant expression in gastric cancer. Our findings that HOXA10 and miR-196b-5p are overexpressed in the Tff1-KO mouse model of gastric cancer raised a question of whether this is an outcome of a tumorigenic cascade or if TFF1 can play a role in their regulation. TFF1 is a gastric mucosa specific protein that plays an essential role in protecting the gastric mucosa integrity (53,54). Loss of TFF1 promotes gastric carcinogenesis in a gastric cancer mouse model (39,55). Several studies have shown that TFF1 is an anti-tumorigenic protein that has tumor suppressive functions. For example, TFF1 regulates gastric cancer cell apoptosis, proliferation, and cell viability by activating p53 (56) and suppressing β-catenin (40), miR-504 (28), and NF-κB (39). Our results provide an important additional insight where TFF1 reconstitution in gastric cancer cells enhanced the HOXA10 promoter methylation with downregulation of both HOXA10 and miR-196b-5p. This observation adds to the spectrum of TFF1 tumor suppressor functions in gastric mucosa, given the pro-proliferative and invasive functions of HOXA10 and miR-196b-5p. We acknowledge that detailed investigations are needed to elucidate the mechanisms by which TFF1 alters methylation of HOXA10. Uncovering these epigenetic mechanisms in future studies is likely to reveal several genes that are epigenetically regulated by TFF1.
The association between HOXA10 expression and clinical outcome has not been consistent. While some studies suggested that HOXA10 overexpression is associated with a better prognosis (4), Lim and colleagues’ (15) findings indicate that HOXA10 functions as an oncogene with poor clinical outcome. Our analysis of several large public data sets suggests that the HOXA10 high expression levels denote a poor clinical outcome. Our findings that miR-196b-5p, overexpressed and co-regulated by the HOXA10 promoter, promoting cellular proliferation and invasion, are consistent with a pro-tumorigenic role for HOXA10 that is associated with poor prognosis.
In summary, we report, for the first time, that TFF1 can play a role in epigenetic regulation of HOXA10 and miR-196b-5p expression levels. Aberrant hypo-methylation and overexpression of HOXA10 and miR-196-5p promote gastric cancer cell proliferation and invasion. Further studies are needed to determine the mechanisms by which TFF1 promotes DNA methylation and its impact on shaping the epigenome in gastric carcinogenesis.
Supplementary Material
Implications.
This study indicates that loss of TFF1 promotes the aberrant overexpression of HOXA10 and miR-196b-5p by demethylation of the HOXA10 promoter, which provides a new perspective of TFF1/HOXA10/miR-196b-5p functions in human gastric cancer.
Acknowledgments
This study was supported by the U.S. National Institutes of Health (R01CA93999 and R01CA177372), Research Career Scientist award (1IK6BX003787), and a merit award (I01BX001179) from the U.S. Department of Veterans affairs (W. El-Rifai). The contents of this work are solely the responsibility of the authors and do not necessarily represent the official views of the Department of Veterans Affairs, National Institutes of Health, or University of Miami.
Footnotes
The authors declare no conflict of interest.
References
- 1.Zhu S, Soutto M, Chen Z, Peng D, Romero-Gallo J, Krishna US, et al. Helicobacter pylori-induced cell death is counteracted by NF-kappaB-mediated transcription of DARPP-32. Gut. 2017;66(5):761–2. doi: 10.1136/gutjnl-2016-312141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136(5):E359–86. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 3.Siegel R, Ma J, Zou Z, Jemal A. Cancer statistics, 2014. CA: a cancer journal for clinicians. 2014;64(1):9–29. doi: 10.3322/caac.21208. [DOI] [PubMed] [Google Scholar]
- 4.Sentani K, Oue N, Naito Y, Sakamoto N, Anami K, Oo HZ, et al. Upregulation of HOXA10 in gastric cancer with the intestinal mucin phenotype: reduction during tumor progression and favorable prognosis. Carcinogenesis. 2012;33(5):1081–8. doi: 10.1093/carcin/bgs121. [DOI] [PubMed] [Google Scholar]
- 5.Calcagno DQ, Gigek CO, Chen ES, Burbano RR, de Smith MA. DNA and histone methylation in gastric carcinogenesis. World journal of gastroenterology. 2013;19(8):1182–92. doi: 10.3748/wjg.v19.i8.1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ziogas D, Roukos D. Epigenetics in gastric cancer: challenges for clinical implications. Annals of surgical oncology. 2009;16(7):2077–8. doi: 10.1245/s10434-009-0472-y. [DOI] [PubMed] [Google Scholar]
- 7.Qu Y, Dang S, Hou P. Gene methylation in gastric cancer. Clinica chimica acta; international journal of clinical chemistry. 2013;424:53–65. doi: 10.1016/j.cca.2013.05.002. [DOI] [PubMed] [Google Scholar]
- 8.Pan Y, Liu G, Zhou F, Su B, Li Y. DNA methylation profiles in cancer diagnosis and therapeutics. Clinical and experimental medicine. 2017 doi: 10.1007/s10238-017-0467-0. [DOI] [PubMed] [Google Scholar]
- 9.Singal R, Wang SZ, Sargent T, Zhu SZ, Ginder GD. Methylation of promoter proximal-transcribed sequences of an embryonic globin gene inhibits transcription in primary erythroid cells and promotes formation of a cell type-specific methyl cytosine binding complex. The Journal of biological chemistry. 2002;277(3):1897–905. doi: 10.1074/jbc.M105580200. [DOI] [PubMed] [Google Scholar]
- 10.Jones PA. The DNA methylation paradox. Trends in genetics: TIG. 1999;15(1):34–7. doi: 10.1016/s0168-9525(98)01636-9. [DOI] [PubMed] [Google Scholar]
- 11.Cheng W, Jiang Y, Liu C, Shen O, Tang W, Wang X. Identification of aberrant promoter hypomethylation of HOXA10 in ovarian cancer. Journal of cancer research and clinical oncology. 2010;136(8):1221–7. doi: 10.1007/s00432-010-0772-4. [DOI] [PubMed] [Google Scholar]
- 12.Oakley K, Han Y, Vishwakarma BA, Chu S, Bhatia R, Gudmundsson KO, et al. Setbp1 promotes the self-renewal of murine myeloid progenitors via activation of Hoxa9 and Hoxa10. Blood. 2012;119(25):6099–108. doi: 10.1182/blood-2011-10-388710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Park SM, Choi EY, Bae M, Choi JK, Kim YJ. A long-range interactive DNA methylation marker panel for the promoters of HOXA9 and HOXA10 predicts survival in breast cancer patients. Clinical epigenetics. 2017;9:73. doi: 10.1186/s13148-017-0373-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang HY, Li JH, Li G, Wang SR. Activation of ARK5/miR-1181/HOXA10 axis promotes epithelial-mesenchymal transition in ovarian cancer. Oncology reports. 2015;34(3):1193–202. doi: 10.3892/or.2015.4113. [DOI] [PubMed] [Google Scholar]
- 15.Lim JY, Yoon SO, Seol SY, Hong SW, Kim JW, Choi SH, et al. Overexpression of miR-196b and HOXA10 characterize a poor-prognosis gastric cancer subtype. World J Gastroenterol. 2013;19(41):7078–88. doi: 10.3748/wjg.v19.i41.7078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nature reviews Cancer. 2006;6(11):857–66. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 17.Gulyaeva LF, Kushlinskiy NE. Regulatory mechanisms of microRNA expression. Journal of translational medicine. 2016;14(1):143. doi: 10.1186/s12967-016-0893-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–97. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 19.Qin J, Ke J, Xu J, Wang F, Zhou Y, Jiang Y, et al. Downregulation of microRNA-132 by DNA hypermethylation is associated with cell invasion in colorectal cancer. OncoTargets and therapy. 2015;8:3639–48. doi: 10.2147/OTT.S91560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Deneberg S, Kanduri M, Ali D, Bengtzen S, Karimi M, Qu Y, et al. microRNA-34b/c on chromosome 11q23 is aberrantly methylated in chronic lymphocytic leukemia. Epigenetics. 2014;9(6):910–7. doi: 10.4161/epi.28603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jimenez-Wences H, Martinez-Carrillo DN, Peralta-Zaragoza O, Campos-Viguri GE, Hernandez-Sotelo D, Jimenez-Lopez MA, et al. Methylation and expression of miRNAs in precancerous lesions and cervical cancer with HPV16 infection. Oncology reports. 2016;35(4):2297–305. doi: 10.3892/or.2016.4583. [DOI] [PubMed] [Google Scholar]
- 22.Augoff K, McCue B, Plow EF, Sossey-Alaoui K. miR-31 and its host gene lncRNA LOC554202 are regulated by promoter hypermethylation in triple-negative breast cancer. Molecular cancer. 2012;11:5. doi: 10.1186/1476-4598-11-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yin H, Song P, Su R, Yang G, Dong L, Luo M, et al. DNA Methylation mediated down-regulating of MicroRNA-33b and its role in gastric cancer. Scientific reports. 2016;6:18824. doi: 10.1038/srep18824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.He Y, Cui Y, Wang W, Gu J, Guo S, Ma K, et al. Hypomethylation of the hsa-miR-191 locus causes high expression of hsa-mir-191 and promotes the epithelial-to-mesenchymal transition in hepatocellular carcinoma. Neoplasia. 2011;13(9):841–53. doi: 10.1593/neo.11698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li A, Omura N, Hong SM, Vincent A, Walter K, Griffith M, et al. Pancreatic cancers epigenetically silence SIP1 and hypomethylate and overexpress miR-200a/200b in association with elevated circulating miR-200a and miR-200b levels. Cancer research. 2010;70(13):5226–37. doi: 10.1158/0008-5472.CAN-09-4227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.de Souza Rocha Simonini P, Breiling A, Gupta N, Malekpour M, Youns M, Omranipour R, et al. Epigenetically deregulated microRNA-375 is involved in a positive feedback loop with estrogen receptor alpha in breast cancer cells. Cancer research. 2010;70(22):9175–84. doi: 10.1158/0008-5472.CAN-10-1318. [DOI] [PubMed] [Google Scholar]
- 27.Shi C, Shin YO, Hanson J, Cass B, Loewen MC, Durocher Y. Purification and characterization of a recombinant G-protein-coupled receptor, Saccharomyces cerevisiae Ste2p, transiently expressed in HEK293 EBNA1 cells. Biochemistry. 2005;44(48):15705–14. doi: 10.1021/bi051292p. [DOI] [PubMed] [Google Scholar]
- 28.Soutto M, Chen Z, Saleh MA, Katsha A, Zhu S, Zaika A, et al. TFF1 activates p53 through down-regulation of miR-504 in gastric cancer. Oncotarget. 2014;5(14):5663–73. doi: 10.18632/oncotarget.2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.El-Rifai W, Moskaluk CA, Abdrabbo MK, Harper J, Yoshida C, Riggins GJ, et al. Gastric cancers overexpress S100A calcium-binding proteins. Cancer research. 2002;62(23):6823–6. [PubMed] [Google Scholar]
- 30.Chang KH, Mestdagh P, Vandesompele J, Kerin MJ, Miller N. MicroRNA expression profiling to identify and validate reference genes for relative quantification in colorectal cancer. BMC cancer. 2010;10:173. doi: 10.1186/1471-2407-10-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rinnerthaler G, Hackl H, Gampenrieder SP, Hamacher F, Hufnagl C, Hauser-Kronberger C, et al. miR-16-5p Is a Stably-Expressed Housekeeping MicroRNA in Breast Cancer Tissues from Primary Tumors and from Metastatic Sites. International journal of molecular sciences. 2016;17(2) doi: 10.3390/ijms17020156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Peng DF, Razvi M, Chen H, Washington K, Roessner A, Schneider-Stock R, et al. DNA hypermethylation regulates the expression of members of the Mu-class glutathione S-transferases and glutathione peroxidases in Barrett’s adenocarcinoma. Gut. 2009;58(1):5–15. doi: 10.1136/gut.2007.146290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Forster S, Gretschel S, Jons T, Yashiro M, Kemmner W. THBS4, a novel stromal molecule of diffuse-type gastric adenocarcinomas, identified by transcriptome-wide expression profiling. Modern pathology: an official journal of the United States and Canadian Academy of Pathology, Inc. 2011;24(10):1390–403. doi: 10.1038/modpathol.2011.99. [DOI] [PubMed] [Google Scholar]
- 34.Kim HK, Choi IJ, Kim CG, Kim HS, Oshima A, Yamada Y, et al. Three-gene predictor of clinical outcome for gastric cancer patients treated with chemotherapy. The pharmacogenomics journal. 2012;12(2):119–27. doi: 10.1038/tpj.2010.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Busuttil RA, George J, Tothill RW, Ioculano K, Kowalczyk A, Mitchell C, et al. A signature predicting poor prognosis in gastric and ovarian cancer represents a coordinated macrophage and stromal response. Clinical cancer research: an official journal of the American Association for Cancer Research. 2014;20(10):2761–72. doi: 10.1158/1078-0432.CCR-13-3049. [DOI] [PubMed] [Google Scholar]
- 36.Szasz AM, Lanczky A, Nagy A, Forster S, Hark K, Green JE, et al. Cross-validation of survival associated biomarkers in gastric cancer using transcriptomic data of 1,065 patients. Oncotarget. 2016;7(31):49322–33. doi: 10.18632/oncotarget.10337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tsai KW, Hu LY, Wu CW, Li SC, Lai CH, Kao HW, et al. Epigenetic regulation of miR-196b expression in gastric cancer. Genes, chromosomes & cancer. 2010;49(11):969–80. doi: 10.1002/gcc.20804. [DOI] [PubMed] [Google Scholar]
- 38.Popovic R, Riesbeck LE, Velu CS, Chaubey A, Zhang J, Achille NJ, et al. Regulation of mir-196b by MLL and its overexpression by MLL fusions contributes to immortalization. Blood. 2009;113(14):3314–22. doi: 10.1182/blood-2008-04-154310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Soutto M, Belkhiri A, Piazuelo MB, Schneider BG, Peng D, Jiang A, et al. Loss of TFF1 is associated with activation of NF-kappaB-mediated inflammation and gastric neoplasia in mice and humans. The Journal of clinical investigation. 2011;121(5):1753–67. doi: 10.1172/JCI43922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Soutto M, Romero-Gallo J, Krishna U, Piazuelo MB, Washington MK, Belkhiri A, et al. Loss of TFF1 promotes Helicobacter pylori-induced beta-catenin activation and gastric tumorigenesis. Oncotarget. 2015;6(20):17911–22. doi: 10.18632/oncotarget.3772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stiegelbauer V, Vychytilova-Faltejskova P, Karbiener M, Pehserl AM, Reicher A, Resel M, et al. miR-196b-5p Regulates Colorectal Cancer Cell Migration and Metastases through Interaction with HOXB7 and GALNT5. Clinical cancer research: an official journal of the American Association for Cancer Research. 2017 doi: 10.1158/1078-0432.CCR-17-0023. [DOI] [PubMed] [Google Scholar]
- 42.Zhang L, Wan Y, Jiang Y, Ma J, Liu J, Tang W, et al. Upregulation HOXA10 homeobox gene in endometrial cancer: role in cell cycle regulation. Medical oncology. 2014;31(7):52. doi: 10.1007/s12032-014-0052-2. [DOI] [PubMed] [Google Scholar]
- 43.Jiang Y, Chu Y, Tang W, Wan Y, Zhang L, Cheng W. Transcription factor WT1 and promoter CpG hypomethylation coactivate HOXA10 expression in ovarian cancer. Current pharmaceutical design. 2014;20(11):1647–54. doi: 10.2174/13816128113199990545. [DOI] [PubMed] [Google Scholar]
- 44.Cui XP, Qin CK, Zhang ZH, Su ZX, Liu X, Wang SK, et al. HOXA10 promotes cell invasion and MMP-3 expression via TGFbeta2-mediated activation of the p38 MAPK pathway in pancreatic cancer cells. Digestive diseases and sciences. 2014;59(7):1442–51. doi: 10.1007/s10620-014-3033-6. [DOI] [PubMed] [Google Scholar]
- 45.Chen Y, Zhang J, Wang H, Zhao J, Xu C, Du Y, et al. miRNA-135a promotes breast cancer cell migration and invasion by targeting HOXA10. BMC cancer. 2012;12:111. doi: 10.1186/1471-2407-12-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mustafa M, Lee JY, Kim MH. CTCF negatively regulates HOXA10 expression in breast cancer cells. Biochemical and biophysical research communications. 2015;467(4):828–34. doi: 10.1016/j.bbrc.2015.10.058. [DOI] [PubMed] [Google Scholar]
- 47.Skrha P, Horinek A, Pazourkova E, Hajer J, Fric P, Skrha J, et al. Serum microRNA-196 and microRNA-200 in pancreatic ductal adenocarcinoma of patients with diabetes mellitus. Pancreatology: official journal of the International Association of Pancreatology. 2016;16(5):839–43. doi: 10.1016/j.pan.2016.05.005. [DOI] [PubMed] [Google Scholar]
- 48.Lee SW, Park KC, Kim JG, Moon SJ, Kang SB, Lee DS, et al. Dysregulation of MicroRNA-196b-5p and MicroRNA-375 in Gastric Cancer. Journal of gastric cancer. 2016;16(4):221–9. doi: 10.5230/jgc.2016.16.4.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ushijima T, Asada K. Aberrant DNA methylation in contrast with mutations. Cancer science. 2010;101(2):300–5. doi: 10.1111/j.1349-7006.2009.01434.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jin Z, Cheng Y, Gu W, Zheng Y, Sato F, Mori Y, et al. A multicenter, double-blinded validation study of methylation biomarkers for progression prediction in Barrett’s esophagus. Cancer research. 2009;69(10):4112–5. doi: 10.1158/0008-5472.CAN-09-0028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li Z, Wong KY, Chan GC, Chng WJ, Chim CS. Epigenetic silencing of EVL/miR-342 in multiple myeloma. Transl Res. 2017 doi: 10.1016/j.trsl.2017.11.005. [DOI] [PubMed] [Google Scholar]
- 52.Tessema M, Yingling CM, Picchi MA, Wu G, Ryba T, Lin Y, et al. ANK1 Methylation regulates expression of MicroRNA-486-5p and discriminates lung tumors by histology and smoking status. Cancer letters. 2017;410:191–200. doi: 10.1016/j.canlet.2017.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hoffmann W, Jagla W. Cell type specific expression of secretory TFF peptides: colocalization with mucins and synthesis in the brain. International review of cytology. 2002;213:147–81. doi: 10.1016/s0074-7696(02)13014-2. [DOI] [PubMed] [Google Scholar]
- 54.Hoffmann W, Jagla W, Wiede A. Molecular medicine of TFF-peptides: from gut to brain. Histology and histopathology. 2001;16(1):319–34. doi: 10.14670/HH-16.319. [DOI] [PubMed] [Google Scholar]
- 55.Katsha A, Soutto M, Sehdev V, Peng D, Washington MK, Piazuelo MB, et al. Aurora kinase A promotes inflammation and tumorigenesis in mice and human gastric neoplasia. Gastroenterology. 2013;145(6):1312–22. e1–8. doi: 10.1053/j.gastro.2013.08.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Busch M, Grosse-Kreul J, Wirtz JJ, Beier M, Stephan H, Royer-Pokora B, et al. Reduction of the tumorigenic potential of human retinoblastoma cell lines by TFF1 overexpression involves p53/caspase signaling and miR-18a regulation. International journal of cancer. 2017;141(3):549–60. doi: 10.1002/ijc.30768. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


