Abstract
Tumor recurrence is a leading cause of death and is thought to arise from a population of residual cells that survive treatment. These residual cancer cells can persist, locally or at distant sites, for years or decades. Therefore, understanding the pathways that regulate residual cancer cell survival may suggest opportunities for targeting these cells to prevent recurrence. Previously it was observed that the pro-apoptotic protein (PAWR/Par-4) negatively regulates residual cell survival and recurrence in mice and humans. However, the mechanistic underpinnings on how Par-4 expression is regulated is unclear. Here, it is demonstrated that Par-4 is transcriptionally upregulated following treatment with multiple drugs targeting the PI3K-Akt-mTOR signaling pathway, and identify the forkhead family of transcription factors as mediators of this upregulation. Mechanistically, Foxo3a directly binds to the Par-4 promoter and activates its transcription following inhibition of the PI3K-Akt pathway. This Foxo-dependent Par-4 upregulation limits the long-term survival of residual cells following treatment with therapeutics that target the PI3K-Akt pathway. Taken together, these results indicate that residual breast cancer tumor cell survival and recurrence requires circumventing Foxo-driven Par-4 upregulation, and suggest that approaches to enforce Par-4 expression may prevent residual cell survival and recurrence.
Keywords: Par-4, Foxo, breast cancer, residual disease, targeted therapy
Introduction
Despite improvements in diagnosis and treatment, breast cancer remains the second-leading cause of cancer-related deaths among women in the United States (1). This is due largely to the recurrence of disease following surgery and adjuvant therapy. Recurrent breast cancer is common, affecting nearly 25% of breast cancer patients, and these recurrent tumors are frequently resistant to drugs used to treat primary breast tumors. Recurrent tumors are thought to arise from a population of residual cells that survive treatment. Consistent with this notion, the extent of residual disease following neoadjuvant therapy is correlated with the risk of developing recurrence (2). In addition, between 30–50% of breast cancer patients have disseminated tumor cells (DTCs) in their bone marrow, and the presence of these cells and their persistence following therapy are strongly correlated with poor prognosis (3–5). Therapies that can eliminate residual tumor cells or prevent their emergence as recurrent breast cancers may prolong the survival of patients with breast cancer. However, the development of such therapies is limited by our poor understanding of the pathways that enable the long-term survival of residual cells following treatment.
We have previously used conditional genetically engineered mouse (GEM) models to identify pathways that mediate the survival and recurrence of residual cells following oncogene inhibition (6). In these models, doxycycline-dependent, mammary gland-specific expression of an oncogene (e.g. Her2, Myc, or Wnt1) drives the formation of invasive mammary adenocarcinomas (7–9). Removal of doxycycline from mice with primary tumors leads to oncogene downregulation and tumor regression. However, a population of residual cells survives oncogene downregulation and persists in a dormant, non-proliferative state (10). Following a variable latency period, these residual cells resume proliferation to form recurrent tumors (6,11).
To identify pathways that regulate the survival of residual cells and their eventual recurrence, we compared gene expression profiles of primary and recurrent tumors from the Her2, Myc, and Wnt1 oncogene models. This analysis revealed that the tumor suppressor protein Par-4 is downregulated in recurrent tumors from all three models (6). Par-4 is a pro-apoptotic protein that induces apoptosis in cancer cells through a variety of mechanisms, primarily through inhibition of the pro-survival pathways NF-κB, Akt, and PKCζ (12). Our functional studies showed that Par-4 is a critical negative regulator of residual cell survival and recurrence. Specifically, cells with low Par-4 expression preferentially survive and persist as residual cells following Her2 down-regulation. Similar results were observed in breast cancer patients treated with neoadjuvant chemotherapy (NAC): low Par-4 expression in primary tumors is associated with increased residual cancer burden following NAC, and residual tumors that remain following NAC have low Par-4 expression (6). These results identify Par-4 as a negative regulator of residual cell survival following therapy.
However, little is known about how Par-4 expression is regulated in response to treatment. Studies in Her2-driven tumors showed that Her2 inhibition leads to acute upregulation of Par-4, thereby limiting the survival of residual tumor cells (6). However, the mechanistic basis of Par-4 upregulation remains unknown. In addition, the relevance of Par-4 in regulating residual tumor cell survival in human cancer cells, and in cells driven by activation of other oncogenic pathways, remains unknown. In the current study we investigate the mechanism and functional significance of Par-4 upregulation following oncogene inhibition in human breast cancer cells. We show that Foxo3a directly binds to the Par-4 promoter and transcriptionally upregulates Par-4 following inhibition of the PI3K-Akt-mTOR pathway. We further show that this Foxo3a-dependent Par-4 expression prevents the long-term survival of residual cells following oncogene inhibition.
Materials and Methods
Cell lines and reagents
Human breast cancer cell lines (BT-474, SKBR3, and MCF-7) and 293T cells were obtained from American Type Culture Collection through the Duke University Cell Culture Facility. BT-474 and MCF-7 cells were maintained in RPMI 1640 medium (Sigma-Aldrich), and SKBR3 and 293T cells were maintained in DMEM (Corning). All media was supplemented with 2mM L-glutamine (ThermoFisher), 100 units/mL of Penicillin-Streptomycin (ThermoFisher), and 10% fetal bovine serum (Corning). All cells were grown in a humidified incubator at 37°C with 5% CO2. Transfections were done using Lipofectamine 2000 (ThermoFisher) per manufacturer’s instructions. Cell viability was measured using CellTiterGlo (Promega) according to the manufacturer’s instructions. Cell lines were tested and found to be negative for mycoplasma contamination by the Duke CCF on the following dates: BT-474 cells, 2/25/14; SKBR3 cells, 10/30/14; MCF-7 cells, 8/28/14. Cells were authenticated by the Duke CCF using Short Tandem Repeat (STR) testing on the following dates: BT-474 cells, 3/11/14; SKBR3 cells, 10/29/14; MCF-7 cells, 9/24/14. All cell lines were used within 10 passages of being thawed.
The following drugs were obtained from Selleck Chemicals: BEZ235, BKM120, RAD001, MK-2206, Lapatinib, Torin1, and PD0325901. 4-Hydroxytamoxifen was purchased from Sigma. All drugs were reconstituted per manufacturer’s instructions and used at concentrations noted in the text.
Western blotting and antibodies
Western blotting was performed as described previously (6). Membranes were probed with the following primary antibodies: total Akt, phospho-Akt S473, Puma, p27 Kip1, mTOR, phospho-S6 S240/244, phospho-HER2/ErbB2 Y1221/1222, Erk1/2, phospho-Erk1/2 T202/Y204, Foxo1, FoxO3a, Foxo4 and TSC1 antibodies were obtained from Cell Signaling and used at 1:1000; FLAG epitope and GAPDH antibodies were obtained from Sigma-Aldrich and used at 1:1000; Par-4 antibody was obtained from Bethyl Laboratories and used at 1:2000; and α-tubulin antibody was purchased from Santa Cruz Biotechnology and used at 1:1000. Secondary antibodies conjugated to IRDye® 800CW were purchased from LI-COR Biosciences and used at 1:5000, and secondary antibodies conjugated to Alexa Fluor® 680 were purchased from ThermoFisher and used at 1:5000. Membranes were scanned using an Odyssey Infrared Imaging System (LI-COR Biosciences).
qRT-PCR
For detection of transcript levels, RNA was extracted and reverse transcribed, and qRT-PCR was performed as described previously (13). Briefly, RNA was extracted using the RNeasy kit (Qiagen) and cDNA was synthesized with the Improm-II Reverse Transcription System (Promega). Expression levels of genes were assessed by real-time qPCR on a CFX384 Touch Real-Time PCR Detection System (Bio-Rad) using the following TaqMan assays: PAWR (Hs01088574_m1), BBC3 (Hs00248075_m1), FOXO3 (Hs00921424_m1), GAPDH (Hs02758991_g1), ACTB (Hs01060665_g1), and TBP (Hs00427620_m1). GAPDH, ACTIN, and TBP were used as control genes.
Plasmids, CRISPR knockout, and lentiviral transduction
To generate cells stably expressing tamoxifen-inducible Foxo3a, we cloned HA-FOXO3a TM-ER (a gift from Michael Greenberg; Addgene #8353), a constitutively active form of Foxo3a with mutations in Akt phosphorylation sites (T32A, S253A, S315A) fused to the estrogen receptor, into the lentiviral vector pCDH-EF1-FHC (a gift from Richard Wood; Addgene #64874). For transient expression of constitutively active Foxo3a we used FOXO3a TM/pcDNA (a gift from William Sellers; Addgene #10709). Myristoylated Akt (pWZL Neo Myr Flag AKT1) was a gift from William Hahn and Jean Zhao (Addgene #20422). Human Par-4 was cloned from cDNA and an N-terminal FLAG tag was appended using the following primers: Forward, 5′-TAACGGATCCATGGACTACAAAGACGATGACGACAAGGCGACCGGTGGCTACCGGAC-3′; Reverse, 5′-TAACGAATTCCTACCTGGTCAGCTGACCCA-3′. Par-4 was cloned into the BamHI and EcoRI sites of pLenti CMV Neo (a gift from Eric Campeau; Addgene #17447). H2B-eGFP (a gift from Geoff Wahl; Addgene #11680) and H2B-mStrawberry (a gift from Robert Benezra; Addgene #20970) were cloned into pLenti CMV Neo. For reporter gene assays, we used luciferase reporter constructs pFHRE-luc (a gift from Michael Greenberg; Addgene #1789), pGL3 control, pGL3 enhancer, and pRL-SV40 (all from Promega).
For CRISPR-mediated knockout, sgRNAs were cloned into lentiCRISPR v2 (a gift from Feng Zhang; Addgene #52961) using previously described protocols (14). The following protospacers were used: non-targeting (NT) sgRNA, GGGCGAGGAGCTGTTCACCG; Par-4 sg#1, GCTGCTGGTCCGGTAGCCAC; Par-4 sg#2, CAGCACCACAGACTTCCTGG.
Lentivirus was generated by transfecting 293T cells with packaging plasmids psPAX2 and pMD2.G (both gifts from Didier Trono; Addgene #12260 and 12259) along with the lentiviral delivery plasmid, as described previously (6). Retrovirus was generated using Amphophoenix packaging cells (National Gene Vector Biorepository), as described previously (6).
Reporter gene assay and chromatin immunoprecipitation
Reporter gene assays were performed essentially as described (15). A BAC clone comprising the Par-4 promoter (RP11-642G2) was purchased from Children’s Hospital Oakland Research Institute. Eight regions of the Par-4 promoter from ~2kb upstream to ~2kb downstream of the transcriptional start site were PCR amplified (see Supplemental Table 1 for primer sequences) and cloned into the multiple cloning site of the pGL3 enhancer plasmid upstream of Firefly luciferase. These constructs were transfected individually into 293T cells along with Renilla luciferase (pRL-SV40) and either empty vector (pcDNA3.1) or constitutively active Foxo3a (FOXO3A AAA/pcDNA3.1). pGL3 control, empty pGL3 enhancer, or FHRE-luc were separately transfected as negative and positive controls. Firefly luciferase values were normalized to Renilla luciferase and are represented as fold-change in luciferase expression in the presence of Foxo3a TM relative to empty vector.
For chromatin immunoprecipitation (ChIP), BT-474 cells were treated with MK-2206 for 24 hours and then fixed with 1% formaldehyde for 10 minutes at room temperature. Crosslinking was quenched with 250 mM glycine for 5 minutes at room temperature, and cells were collected in PBS. Pelleted cells were processed for ChIP and analyzed for Foxo3a occupancy as previously described (16,17). Primers targeting regions #3, 6, and 8 of the Par-4 promoter were used to amplify immunoprecipitated DNA (see Supplemental Table 1 for primer sequences). To calculate Foxo3a occupancy at each region, Ct values were normalized to input DNA and expressed as fold-enrichment relative to control IgG.
Fluorescent competition assay
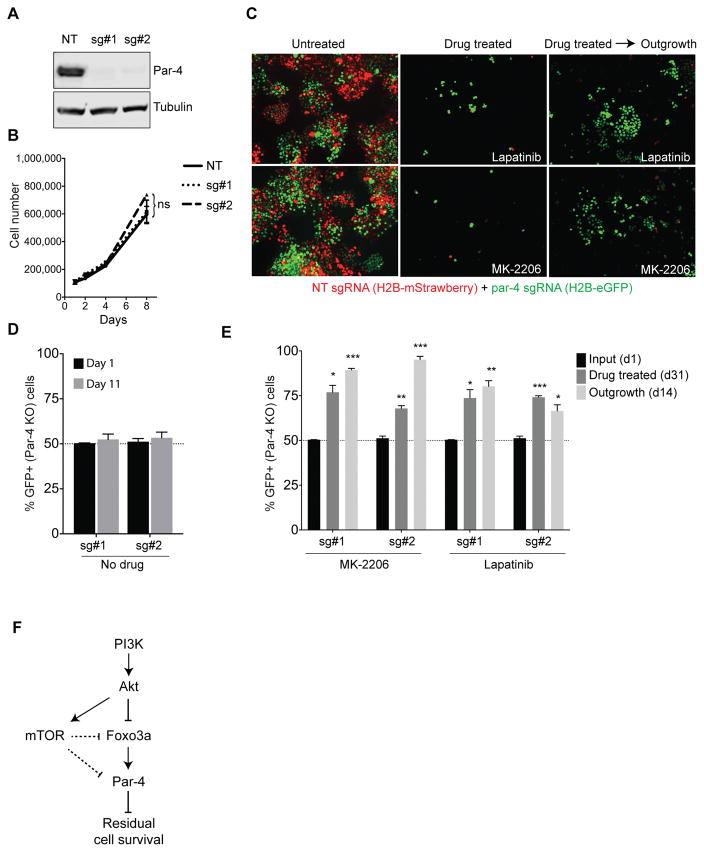
BT-474 cells expressing a non-targeting sgRNA were infected with H2B-mStrawberry and selected in G418. BT-474 cells expressing one of two sgRNAs targeting Par-4 (sg#1 or sg#2) were infected with H2B-eGFP and selected in G418. Control (mStrawberry+) and Par-4 knockout (GFP+) cells were mixed in a 1:1 ratio and plated at 100,000 cells per well in triplicate on a 12-well plate. Fluorescent micrographs were taken at day 1 (input) to confirm equal plating of control and Par-4 knockout cells. For control experiments, cells were left untreated and allowed to grow for 11 days (~5 population doublings) at which point pictures were taken to quantify the ratio of mStrawberry+ to GFP+ cells. For drug treatments, cells were treated with 500 nM Lapatinib or 1 μM MK-2206. Media and drug were replenished every 3 days, and pictures were taken at day 31 to measure the ratio of mStrawberry+ to GFP+ cells in the residual surviving population. At day 31, drug was removed from the media and cells were allowed to grow out for 14 days, and the ratio of mStrawberry+ to GFP+ cells was assessed. Pictures were taken on an EVOS FL Imaging System and analyzed using CellProfiler 2.1.1 software.
Statistical Analysis
All experiments were performed a minimum of three independent times. For Western blots, a single experiment is shown that is representative of the results from multiple independent experiments. For gene expression and reporter gene assays, the data are shown as the average from at least three independent experiments plus the standard deviation. The cellular competition assays were performed in triplicate with two independent sgRNAs. P-values were calculated using two-tailed Student’s t-test between continuous variables. All data were graphed and analyzed in GraphPad Prism.
Results
Par-4 is transcriptionally upregulated following inhibition of the Her2-PI3K-Akt-mTOR pathway
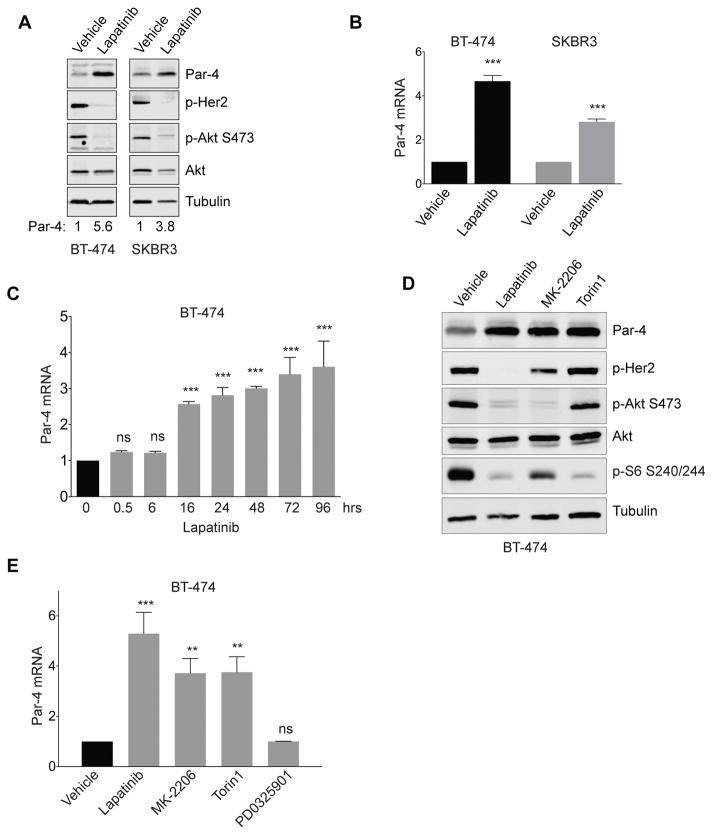
To gain insight into the mechanism of Par-4 upregulation following Her2 inhibition, we used two well-characterized human HER2-amplified breast cancer cell lines, BT-474 and SKBR3. Consistent with our previous findings (6), treatment with Lapatinib, a dual small-molecule Her2/EGFR inhibitor, for two days led to upregulation of Par-4 (Figure 1A). To determine whether this was mediated by changes in transcription of the Par-4 gene, we measured Par-4 mRNA levels by quantitative reverse-transcription PCR (qRT-PCR). We found that Lapatinib treatment for two days led to induction of Par-4 transcript in both BT-474 and SKBR3 cells (Figure 1B). We next performed a time-course of Par-4 mRNA induction in response to Lapatinib in BT-474 cells. We found that Par-4 mRNA increased 16 hours following Lapatinib treatment, and continued to increase up to four days (Figure 1C), suggesting that this effect is relatively direct.
Figure 1.
Drugs targeting the Her2-Akt-mTOR pathway induce Par-4 upregulation in Her2-amplified breast cancer cells. A and B, Treatment of the Her2-amplified cell lines BT-474 and SKBR3 with Lapatinib for 2 days induces Par-4 protein (A) and mRNA (B) upregulation. C, Time-course of Par-4 mRNA upregulation following Lapatinib treatment in BT-474 cells. D and E, Western blot (D) and qPCR (E) analysis showing Par-4 upregulation in BT-474 cells treated with Lapatinib, the Akt inhibitor MK-2206, or the mTOR inhibitor Torin1 for 2 days. Significance was determined by Student’s t-test and data are presented as mean plus standard deviation (SD). **, p<0.01; ***, p<0.001.
Her2 leads to activation of a number of signaling pathways, notably the Ras-Raf-MAPK and PI3K-Akt-mTOR pathways. To dissect the signaling pathway(s) downstream of Her2 that regulate Par-4, we used validated, specific small-molecule inhibitors of each of these pathways. Treatment of BT-474 cells with the allosteric pan-Akt inhibitor MK-2206 for two days induced upregulation of Par-4 protein (Figure 1D) and mRNA (Figure 1E). Similar results were found in SKBR3 cells (Supplemental Figure 1A–B). Similarly, treatment with Torin1, a catalytic mTOR inhibitor that inhibits both mTORC1 and mTORC2, led to upregulation of Par-4 protein and mRNA in BT-474 cells (Figure 1D–E). In contrast, the MEK inhibitor PD0329501 did not affect Par-4 protein or mRNA levels in BT-474 or SKBR3 cells (Figure 1E, Supplemental Figure 1A–B).
To extend these results, we tested the effects on Par-4 expression of other small-molecule inhibitors of this pathway, including the pan-class I PI3K inhibitor BKM120 and the allosteric mTORC1 inhibitor RAD001. Treatment of BT-474 or SKBR3 cells with BKM120 or RAD001 for two days led to upregulation of Par-4 protein and mRNA (Supplemental Figure 1A–D). Taken together, these results indicate that inhibition of the PI3K-Akt-mTOR pathway, but not the Ras-MAPK pathway, is sufficient to induce Par-4 upregulation in Her2-amplified breast cancer cells.
Akt inhibition is required for Par-4 upregulation following Her2 inhibition
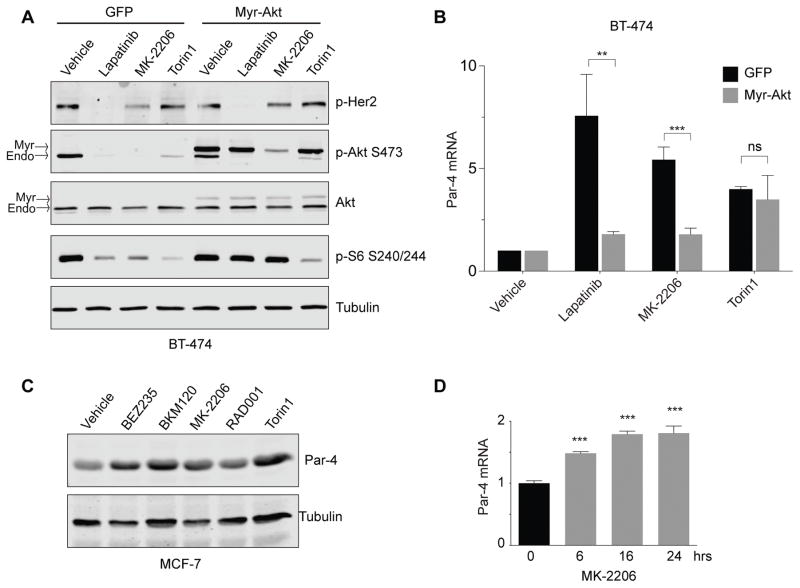
We next wished to address whether inhibition of Akt is necessary for Par-4 upregulation following Her2 inhibition. To do this, we asked whether a constitutively active form of Akt could prevent Par-4 upregulation following Her2 inhibition. We transduced BT-474 cells with retrovirus expressing myristoylated Akt (Myr-Akt) or control GFP virus, and treated cells with Lapatinib, MK-2206, or Torin1. Treatment of control BT-474 cells with Lapatinib for 30 minutes led to reduced phosphorylation of Her2, Akt, and S6 ribosomal protein (Figure 2A). Similarly, MK-2206 treatment caused reductions in phospho-Akt (pAkt) phosphorylation and phospho-S6 (pS6; Figure 2A). Interestingly, Torin1 treatment led to more profound reductions in pS6 than Lapatinib or MK-2206, and induced a partial reduction in pAkt levels (Figure 2A), consistent with previous findings (18). In contrast, expression of Myr-Akt in BT-474 cells partially rescued the decrease in pAkt and pS6 levels induced by Lapatinib and MK-2206, but not by Torin1. These results are consistent with the fact that MK-2206 is an allosteric inhibitor that targets the Pleckstrin homology (PH) domain of Akt and prevents its recruitment to the plasma membrane by phosphatidylinositol–3,4,5-triphosphate (19); Myr-Akt, which induces constitutive membrane localization, has been shown to be resistant to inhibition by MK-2206 (20).
Figure 2.
Inhibition of the Akt pathway is required for Par-4 upregulation following drug treatment. A, BT-474 cells expressing a control vector or myristoylated Akt were treated with the indicated drugs, and the effects on downstream signaling were measured by Western blotting. B, qPCR analysis showing that constitutively active Akt prevents Par-4 upregulation following inhibition of Her2 and Akt but not mTOR. C, The PIK3CA-mutant breast cancer cell line MCF-7 was treated with drugs targeting PI3K (BEZ235 and BKM120), Akt (MK-2206), or mTOR (RAD001 and Torin) for 2 days and Par-4 levels were measured by Western blot. D, Time-course of Par-4 mRNA upregulation following MK-2206 treatment in MCF-7 cells. Data are presented as mean plus SD. **, p<0.01; ***, p<0.001.
Having established that Myr-Akt expression rescues the acute downstream signaling effects of Lapatinib and MK-2206, we next asked whether its expression also prevents Par-4 upregulation. Control BT-474 cells or cells expressing Myr-Akt were treated with Lapatinib, MK-2206, or Torin1 for two days and Par-4 mRNA levels were measured by qRT-PCR. Myr-Akt expression completely blocked Par-4 upregulation following Lapatinib and MK-2206 treatment (Figure 2B). In contrast, Myr-Akt expression did not affect Par-4 upregulation induced by Torin1 treatment (Figure 2B). Together, these data indicate that inhibition of Akt is required for Par-4 upregulation following Lapatinib treatment in Her2-amplified breast cancer cells, and suggest that mTORC1/2 inhibition functions downstream of Akt to induce Par-4 upregulation.
Par-4 upregulation following PI3K-Akt-mTOR inhibition in PIK3CA-mutant breast cancer
We next asked whether the upregulation of Par-4 is a common event following PI3K-Akt-mTOR pathway inhibition in other subtypes of breast cancer. Approximately half of hormone receptor-positive (HR+) breast cancers have mutations in components of the PI3K pathway, including the catalytic subunit PIK3CA, the regulatory subunit PIK3R1, PTEN, and Akt1 (21,22). To determine whether Par-4 upregulation is also observed following inhibition of the PI3K pathway in PI3K-mutant HR+ breast cancer, we used MCF-7 cells, which harbor an E545K activating mutation in PIK3CA and have been used as a preclinical model for this cancer subtype. We treated MCF-7 cells with inhibitors of PI3K, Akt, or mTORC1 for two days and measured Par-4 levels by Western blotting. Inhibition of each protein induced Par-4 upregulation (Figure 2C). A time-course of Par-4 mRNA expression following MK-2206 treatment revealed that Akt inhibition induces acute upregulation of Par-4 (Figure 2D). These results show that Par-4 upregulation is a common response to PI3K-Akt-mTOR inhibition in multiple breast cancer subtypes.
Par-4 is a direct target of Foxo transcription factors
To gain insight into the transcription factor(s) that mediate Par-4 upregulation following Her2-PI3K-Akt pathway inhibition, we first considered transcriptional activators whose activity increases following inhibition of this pathway. We focused on Foxo family transcription factors for several reasons. First, Akt directly phosphorylates Foxo proteins, leading to their cytoplasmic retention and/or degradation; consequently, inhibiting Akt induces Foxo activation and expression of Foxo target genes (23). Second, mTOR inhibition induces Foxo3a upregulation, nuclear accumulation, and the expression of Foxo3a target genes (24). Finally, a recent study in prostate cancer suggested that the natural compound Withaferin-A can induce Par-4 upregulation via activation of Foxo3a (25). We first asked whether Foxo transcription factors are activated concurrent with Par-4 upregulation in these breast cancer cells. Total levels of Foxo1, Foxo3a, and Foxo4 increased following Akt inhibition in BT-474 cells, consistent with the finding that Akt directly phosphorylates these proteins and induces proteasomal degradation (Supplemental Figure 2A). These family members likely have redundant functions (16, 26), and so we focused on a single family member, Foxo3a, for subsequent experiments. We next asked whether Foxo3a can upregulate Par-4 expression in breast cancer cells. Expression of Foxo3a TM, a constitutively active mutant of Foxo3a in which the three serine residues phosphorylated by Akt were mutated to alanine, led to upregulation of Par-4 in both Her2-amplified cell (BT-474 and SKBR3) and PIK3CA-mutant cells (MCF-7; Supplemental Figure 2B–D).
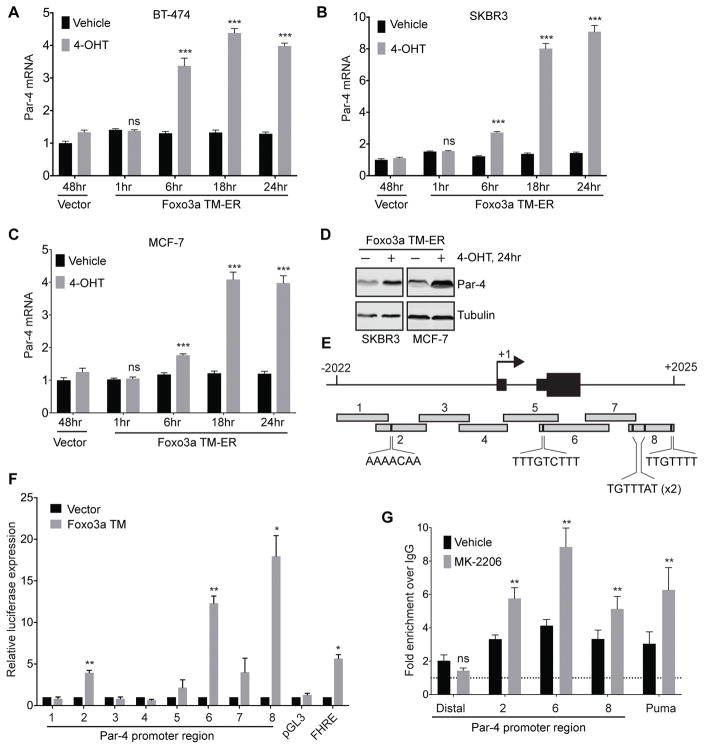
To gain insight into whether Par-4 is a direct transcriptional target of Foxo3a, we next assessed the timing of Par-4 mRNA upregulation following Foxo3a activation. To do this, we generated stable MCF-7, SKBR3, and BT-474 cells in which constitutively active Foxo3a TM is fused to the ligand-binding domain of the estrogen receptor (ER). Under basal conditions, this Foxo3a TM-ER construct is sequestered in the cytoplasm and inactive. Upon treatment with 4-OH Tamoxifen (TAM), Foxo3a TM-ER translocates to the nucleus to drive expression of target genes. Treatment of cells expressing Foxo3A TM-ER with 4-OH TAM led to time-dependent increases in Par-4 mRNA levels, beginning as soon as 6 hours following treatment (Figure 3A–C). This timing paralleled the upregulation of Puma, a known direct Foxo3a target (Supplemental Figure 2E–G). This translated to an increase in Par-4 protein levels (Figure 3D). Importantly, 4-OH TAM treatment of cells expressing an empty vector had no effect on Par-4 levels (Figure 3A–C), indicating that Par-4 upregulation in these cells was mediated through Foxo3a, and not through inhibition of endogenous estrogen receptor signaling.
Figure 3.
Par-4 is a direct target of Foxo3a. A – C, Time-course of Par-4 mRNA upregulation following 4-OH Tamoxifen treatment in BT-474 (A), SKBR3 (B), or MCF-7 (C) cells stably expressing Foxo3a TM-ER fusion. D, Western analysis showing Par-4 upregulation 24 hours after 4-OHT treatment. E, Schematic of the Par-4 promoter surrounding the transcriptional start site. Eight regions (~500bp each) were cloned upstream of luciferase for use in reporter gene experiments. Putative Foxo3a binding sites in region 2, 6, and 8 are shown. F, 293T cells were transfected with luciferase constructs containing each promoter region together with empty vector or constitutively active Foxo3a TM, and luciferase expression was measured 24 hours later. pGL3: empty vector (negative control); FHRE: forkhead response element (positive control) G, Chromatin immunoprecipitation (ChIP) analysis of Foxo3a occupancy at indicated regions of the Par-4 promoter in BT-474 cells treated with vehicle or MK-2206 for 24 hours. A distal region (-10 kb) of the Par-4 promoter was used as a negative control, and the Puma promoter was used as a positive control. Data are expressed as fold-enrichment over IgG IP. Significance was determined by Student’s t-test and data are presented as mean plus SD. **, p<0.01; ***, p<0.001.
We next wished to identify the region of the Par-4 promoter that mediates Foxo3a-dependent transcription. We focused on a ~4-kb region surrounding the transcriptional start site (TSS) that, based upon ENCODE data, is likely to contain regulatory elements (data not shown). We cloned eight 500-bp fragments of this region upstream of Firefly luciferase (Figure 3E). These constructs were then transfected individually into 293T cells together with Renilla luciferase and constitutively active Foxo3A TM, and luciferase expression was measured 24 hours later (Figure 3F). We identified three regions of the Par-4 promoter, Regions 2, 6, and 8, that drove Foxo3a-dependent transcription (Figure 3F). The induction of luciferase expression by these regions in response to Foxo3A was comparable to or greater than that conferred by the positive control construct, Forkhead response element (FHRE; Figure 3F). Importantly, each of these regions contains a putative Foxo3a binding site (Figure 3E).
Finally, we used chromatin immunoprecipitation (ChIP) to determine whether Foxo3a binds directly to these regions of the Par-4 promoter. BT-474 cells were treated with MK-2206 to inhibit Akt and activate Foxo3a, and the binding of endogenous Foxo3a to the Par-4 promoter was measured by ChIP followed by qPCR. MK-2206 treatment led to a 2-fold increase in Foxo3a occupancy at these regions, similar to what was observed at the Puma promoter (Figure 3G). Foxo3a was not present at a distal region of the Par-4 promoter ~10 kb upstream of the TSS. Taken together, these results indicate that Foxo3a directly binds to the Par-4 promoter and activates Par-4 transcription following inhibition of the Akt pathway.
mTOR pathway inhibition induces upregulation of Foxo3a and Par-4
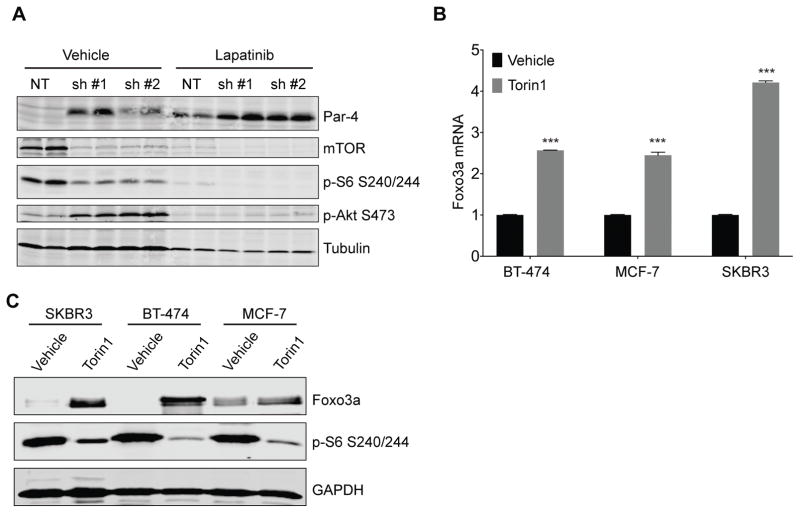
Foxo3a and mTORC1 are generally considered to be parallel pathways downstream of Akt (23), and so it was not clear whether Foxo3a mediates Par-4 upregulation in response to mTORC1 inhibition. However, as described above, mTOR inhibition has recently been shown to induce Foxo3a upregulation, nuclear accumulation, and the expression of Foxo3a target genes (24), suggesting that Foxo3a may mediate Par-4 upregulation downstream of mTORC1 inhibition. To address this, we explored the mechanism of Par-4 upregulation following treatment with drugs targeting the mTOR pathway. We first confirmed that these drugs were acting through on-target effects by performing genetic knockdown of mTOR. BT-474 cells were transduced with lentivirus expressing a control NT shRNA or one of two shRNAs targeting mTOR. Cells were selected in puromycin for 4 days, and then treated with vehicle or Lapatinib for 3 days. mTOR knockdown led to a decrease in phosphorylation of its downstream target S6, and an increase in Akt S473 phosphorylation (Figure 4A), consistent with previous reports (27), indicating that mTOR knockdown effectively suppressed downstream signaling. mTOR knockdown also led to an increase in Par-4 levels, confirming the pharmacological results obtained with Torin1 and RAD001 (Figure 4A). Lapatinib treatment further augmented the increase in Par-4 expression observed in mTOR knockdown cells, and this was concomitant with a reduction in Akt activation (Figure 4A). Taken together, these results indicate that inhibition of Akt and mTORC1 can independently induce Par-4 upregulation, and suggest that these proteins cooperate in mediating Par-4 upregulation in response to inhibition of the Her2-PI3K-Akt pathway.
Figure 4.
Inhibition of the mTOR pathway induces upregulation of Par-4 and Foxo3a. A, BT-474 cells were transduced with lentivirus expressing a control shRNA or one of two shRNAs targeting mTOR. Four days later, cells were treated with vehicle or Lapatinib for 3 days and levels of Par-4 and components of the mTOR pathway were measured by Western blotting. B and C, BT-474, MCF-7, and SKBR3 cells were treated with Torin1 for 2 days and levels of Foxo3a were measured by qRT-PCR (B) or Western blot (C). Significance was determined by Student’s t-test and data are presented as mean plus SD. **, p<0.01; ***, p<0.001.
As described above, mTOR inhibition has recently been shown to induce Foxo3a upregulation (24). We therefore asked whether Foxo3a expression increases following mTOR inhibition in BT-474, MCF-7, and SKBR3 cells. Treatment of each cell line with Torin1 for 2 days led to a 2- to 4-fold increase in Foxo3a transcript and protein levels (Figure 4B and C). This suggests are consistent with Foxo3a mediating Par-4 upregulation following mTOR pathway inhibition.
Par-4 does not regulate the acute survival of cells following Her2-PI3K-Akt pathway inhibition
We previously showed that Par-4 is a negative regulator of residual cell survival in both mice and humans (6). In mouse mammary tumors, Par-4 knockdown increases the number of residual cells that survive following Her2 down-regulation. In breast cancer patients, tumors with low Par-4 expression have more extensive residual cancer burden following neoadjuvant therapy, and neoadjuvant therapy selects for residual cancer cells with low Par-4 expression (6). These results led us to hypothesize that Par-4 may regulate residual cell survival following inhibition of the Her2-PI3K-Akt pathway in human breast cancer cells.
We addressed the role of Par-4 in regulating cell survival in response to inhibition of the PI3K-Akt-mTOR pathway by determining whether Par-4 expression is required for cell death in response to inhibition of this pathway. We used CRISPR-Cas9 to knock out Par-4 in BT-474 cells. Expression of either of two independent sgRNAs targeting Par-4 led to complete reduction in protein levels (Figure 5A). Control and Par-4 knockout cells grew at equal rates (Figure 5B), consistent with our previous findings in mouse tumors that Par-4 knockdown does not affect the growth or survival of untreated primary tumor cells (6). We next treated control or Par-4 knockout cells with drugs targeting the Her2-PI3K-Akt pathway and measured viability after three days. Lapatinib, MK-2206, BKM120, BEZ235, and Torin1 all produced dose-dependent decreases in viability in BT-474 cells (Supplemental Figure 3A–E). However, control and Par-4 knockout cells responded to all drugs similarly, and the IC50 of these drugs was identical in control and Par-4 knockout cells (Supplemental Figure 3A–E). Similar results were obtained in PIK3CA-mutant MCF-7 cells (data not shown). Consistent with this, Par-4 knockout did not affect the extent of apoptosis induced by Lapatinib treatment for three days (Supplemental Figure 3F). These results indicate that Par-4 knockout has no effect on cell viability in response to PI3K-Akt-mTOR pathway inhibition at early time-points.
Figure 5.
Par-4 expression prevents the long-term survival of cells following inhibition of the PI3K-Akt pathway. A, Western blot showing Par-4 expression in BT-474 cells expressing a non-targeting sgRNA or one of two sgRNAs targeting Par-4. B, Growth curves for control and Par-4 knockout BT-474 cells. C, Fluorescent micrographs from a cellular competition assay. Control cells labeled with H2B-mStrawberry were mixed in a 1:1 ratio with Par-4 knockout cells labeled with H2B-eGFP. Cells were left untreated or treated with Lapatinib or MK-2206 for 31 days, at which point the drug was removed and surviving cells were allowed to grow out for 2 weeks. D, Quantification of the percentage of Par-4 knockout (GFP+) cells in untreated cells at day 1 and after 5 population doublings (d11). E, Quantification of the percentage of Par-4 knockout (GFP+) cells in the residual population that survives following treatment with Lapatinib or MK-2206 (d31), and after removal of drug to allow the outgrowth of residual cells (d14). F, Model for Par-4 regulation following therapies targeting the PI3K-Akt-mTOR pathway. Data are presented as the mean of three biological replicates plus SD. *, p<0.05; **, p<0.01; ***, p<0.001.
Par-4 regulates the long-term survival of residual cells following Her2-PI3K-Akt pathway inhibition
To determine the effect of Par-4 knockout on the long-term survival of BT-474 cells after PI3K-Akt pathway inhibition, we performed a cellular competition experiment. Control cells were labeled with H2B-mStrawberry, and Par-4 knockout cells were labeled with H2B-eGFP. Cells were mixed in a 1:1 ratio, and the ratio of GFP+ to mStrawberry+ cells was measured at various time-points. In the absence of drug treatment, the proportion of control and Par-4 knockout cells remained constant (Figure 5C–D), again indicating that Par-4 knockout has no effect on tumor cell survival in the absence of drug treatment. In contrast, long-term treatment with Lapatinib or MK-2206 led to a dramatic selection for Par-4 knockout cells; strikingly, the residual cells that survived after 31 days of drug treatment were predominantly GFP+ Par-4 knockout cells (Figure 5C and E). To test whether these residual cells were viable and competent to re-initiate proliferation in the absence of drug, we removed the drugs and allowed residual cells to grow out. Two weeks after drug removal, residual cells had resumed proliferation, and the vast majority of cells in the cultures were GFP+ Par-4 knockout cells (Figure 5C and E). Taken together, these results show that Par-4 expression prevents the long-term survival of residual cells following Her2 or Akt inhibition.
Discussion
We have elucidated the mechanism of Par-4 regulation in response to therapies targeting the Her2-PI3K-Akt pathway and the functional significance of Par-4 in regulating the long-term survival of residual cells in breast cancer (Figure 5F). We found that Par-4 is upregulated in response to inhibition of PI3K, Akt, and mTOR, but not the Ras-MAPK pathway. We identified Foxo proteins as the transcription factors that mediate these effects. Foxo3a directly binds to the Par-4 promoter following Akt inhibition and activates Par-4 transcription, and expression of a constitutively active form of Foxo3a lacking the Akt phosphorylation sites is sufficient to induce Par-4 expression. Interestingly, we also found that mTOR inhibition alone is sufficient to induce Par-4 upregulation. While the mechanism by which mTOR inhibition induces Par-4 upregulation remains to be elucidated, we found that mTOR inhibition leads to an increase in Foxo3a levels. This is consistent with previous findings suggesting that mTORC1 inhibition leads to increased expression and nuclear accumulation of Foxo3a (24), and suggests that Foxo3a could mediate Par-4 upregulation in response to drugs targeting the mTOR pathway, as well. It is important to note that while our functional experiments focused on Foxo3a, the Foxo family members Foxo1 and Foxo4 are also activated following Akt inhibition, and are likely to serve redundant functions in Par-4 upregulation. Taken together, our results show that Par-4 upregulation is a common response to treatment with drugs targeting the PI3K-Akt-mTOR pathway.
Par-4 upregulation in response to PI3K-Akt-mTOR pathway inhibition was observed in both Her2-amplified breast cancers and breast cancers with activating PI3K mutations, which together constitute nearly half of breast cancers. Drugs targeting Her2 – including the small molecule Lapatinib and the monoclonal antibodies trastuzumab and pertuzumab – are mainstays in the clinical management of Her2-positive breast cancer, and drugs targeting PI3K are in clinical development for PIK3CA-mutant cancers. Thus Par-4 upregulation in response to drugs targeting Her2 and PI3K is relevant in a significant fraction of breast cancer patients.
Several Foxo3a target genes have been implicated in the response and resistance of tumor cells to targeted therapies. The cell cycle inhibitor p27 and the pro-apoptotic proteins Bim and Puma are direct targets of Foxo3a that are upregulated following oncogene inhibition (26,28–30). In an elegant study, Bean et al. dissected the contribution of Bim and Puma upregulation to cell death and tumor regression following Her2 inhibition in breast cancer. They found that both proteins are required for maximal apoptosis at early time-points – between one and five days – following Her2 inhibition (26). Furthermore, knockout of each protein impaired tumor regression measured two days following Her2 down-regulation in vivo (26). These results establish Puma and Bim as critical mediators of the acute apoptotic response at short time-points following Her2 inhibition.
The results described here represent a significant advance by identifying Foxo-driven Par-4 expression as a critical pathway regulating the long-term survival of cells following targeted therapies. In contrast to Puma and Bim, Par-4 knockout does not affect the survival of cells at short time-points following inhibition of the Her2-PI3K-Akt pathway. This is consistent with the notion that acute cell death following inhibition of these pathways is mediated through the intrinsic mitochondrial pathway that is regulated by Bcl-2 family members. Bim and Puma both directly bind to and inhibit anti-apoptotic Bcl-2 family members, tilting the balance of mitochondrial apoptotic proteins to favor apoptosis (31,32). In contrast, Par-4 does not directly interact with Bcl-2 family members, can induce apoptosis even in the presence of high expression of Bcl-2 and Bcl-xL, and is not thought to directly influence the mitochondrial apoptotic pathway (33,34). Consistent with this, we found that Par-4 knockout has no effect on Caspase 3/7 activity measured three days following Lapatinib treatment (Supplemental Figure 3F).
In contrast, cells lacking Par-4 exhibited a profound competitive advantage at longer time-points following Her2 down-regulation, and at one month post-treatment Par-4 knockout cells predominate in these cultures. This is consistent with our previous findings in mice, in which Par-4 knockdown improves the survival of residual tumor cells that persist one month following Her2 down-regulation. Importantly, these Par-4 knockout cells have improved survival even though Puma expression in intact, suggesting that expression of Par-4, and not Puma, is a key determinant of cell death at late time-points. These results suggest the intriguing possibility that the pathways regulating the acute vs. long-term survival of cells following targeted therapy may be distinct, with Puma and Bim regulating cell death at early time points and Par-4 regulating cell death at later times. This represents to our knowledge the first evidence that the long-term survival of residual cells may be controlled by distinct pathways. It is possible that targeting pathways required for the long-term survival of cells may be effective in eliminating residual tumor cells and preventing recurrence.
Consistent with the notion that Par-4 upregulation limits the survival and recurrence of residual cells, Par-4 is silenced in recurrent tumors arising in three independent conditional GEM models and downregulated in residual cancers following NAC in breast cancer patients (6). This suggests that primary tumor cells in which Par-4 is silenced are able to circumvent Foxo3a-dependent Par-4 upregulation and thereby preferentially survive targeted therapy. By elucidating the mechanism of Par-4 silencing, it may be possible to reverse this silencing to prevent residual cell survival and recurrence. There may also be other mechanisms by which tumor cells are able to overcome Foxo3a-driven Par-4 expression. Understanding in more detail how these tumor suppressors act together to limit the long-term survival of residual cells may offer additional opportunities for therapeutic intervention. Finally, a number of approaches are being developed to enforce Par-4 expression (35) and these may be effective in eliminating residual cancer cells.
Supplementary Material
Acknowledgments
We thank members of the Alvarez lab for helpful discussions and suggestions on the manuscript. This work was funded by the National Cancer Institute (R01 CA208042 to J.V. Alvarez and F31CA220957 to A. Walens) and by startup funds from the Duke Cancer Institute, the Duke University School of Medicine and the Whitehead Foundation (J.V. Alvarez).
Footnotes
The authors declare that they have no conflicts of interest relevant to this work.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66:7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 2.Symmans WF, Peintinger F, Hatzis C, Rajan R, Kuerer H, Valero V, et al. Measurement of residual breast cancer burden to predict survival after neoadjuvant chemotherapy. J Clin Oncol. 2007;25:4414–22. doi: 10.1200/JCO.2007.10.6823. [DOI] [PubMed] [Google Scholar]
- 3.Bednarz-Knoll N, Alix-Panabieres C, Pantel K. Clinical relevance and biology of circulating tumor cells. Breast Cancer Res. 2011;13:228. doi: 10.1186/bcr2940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pantel K, Muller V, Auer M, Nusser N, Harbeck N, Braun S. Detection and clinical implications of early systemic tumor cell dissemination in breast cancer. Clin Cancer Res. 2003;9:6326–34. [PubMed] [Google Scholar]
- 5.Janni W, Vogl FD, Wiedswang G, Synnestvedt M, Fehm T, Juckstock J, et al. Persistence of disseminated tumor cells in the bone marrow of breast cancer patients predicts increased risk for relapse--a European pooled analysis. Clin Cancer Res. 2011;17:2967–76. doi: 10.1158/1078-0432.CCR-10-2515. [DOI] [PubMed] [Google Scholar]
- 6.Alvarez JV, Pan TC, Ruth J, Feng Y, Zhou A, Pant D, et al. Par-4 downregulation promotes breast cancer recurrence by preventing multinucleation following targeted therapy. Cancer Cell. 2013;24:30–44. doi: 10.1016/j.ccr.2013.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gunther EJ, Moody SE, Belka GK, Hahn KT, Innocent N, Dugan KD, et al. Impact of p53 loss on reversal and recurrence of conditional Wnt- induced tumorigenesis. Genes Dev. 2003;17:488–501. doi: 10.1101/gad.1051603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Moody SE, Sarkisian CJ, Hahn KT, Gunther EJ, Pickup S, Dugan KD, et al. Conditional activation of Neu in the mammary epithelium of transgenic mice results in reversible pulmonary metastasis. Cancer Cell. 2002;2:451–61. doi: 10.1016/s1535-6108(02)00212-x. [DOI] [PubMed] [Google Scholar]
- 9.Boxer RB, Jang JW, Sintasath L, Chodosh LA. Lack of sustained regression of c-MYC-induced mammary adenocarcinomas following brief or prolonged MYC inactivation. Cancer Cell. 2004;6:577–86. doi: 10.1016/j.ccr.2004.10.013. [DOI] [PubMed] [Google Scholar]
- 10.Feng Y, Pan TC, Pant DK, Chakrabarti KR, Alvarez JV, Ruth JR, et al. SPSB1 promotes breast cancer recurrence by potentiating c-MET signaling. Cancer Discov. 2014;4:790–803. doi: 10.1158/2159-8290.CD-13-0548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moody SE, Perez D, Pan TC, Sarkisian CJ, Portocarrero CP, Sterner CJ, et al. The transcriptional repressor Snail promotes mammary tumor recurrence. Cancer Cell. 2005;8:197–209. doi: 10.1016/j.ccr.2005.07.009. [DOI] [PubMed] [Google Scholar]
- 12.Hebbar N, Wang C, Rangnekar VM. Mechanisms of apoptosis by the tumor suppressor Par-4. J Cell Physiol. 2012;227:3715–21. doi: 10.1002/jcp.24098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Alvarez JV, Belka GK, Pan TC, Chen CC, Blankemeyer E, Alavi A, et al. Oncogene pathway activation in mammary tumors dictates FDG-PET uptake. Cancer Res. 2014;74:7583–98. doi: 10.1158/0008-5472.CAN-14-1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sanjana NE, Shalem O, Zhang F. Improved vectors and genome-wide libraries for CRISPR screening. Nat Methods. 2014;11:783–4. doi: 10.1038/nmeth.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Alvarez JV, Greulich H, Sellers WR, Meyerson M, Frank DA. Signal transducer and activator of transcription 3 is required for the oncogenic effects of non-small-cell lung cancer-associated mutations of the epidermal growth factor receptor. Cancer Res. 2006;66:3162–8. doi: 10.1158/0008-5472.CAN-05-3757. [DOI] [PubMed] [Google Scholar]
- 16.Chandarlapaty S, Sawai A, Scaltriti M, Rodrik-Outmezguine V, Grbovic-Huezo O, Serra V, et al. AKT inhibition relieves feedback suppression of receptor tyrosine kinase expression and activity. Cancer Cell. 2011;19:58–71. doi: 10.1016/j.ccr.2010.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Norris JD, Chang CY, Wittmann BM, Kunder RS, Cui H, Fan D, et al. The homeodomain protein HOXB13 regulates the cellular response to androgens. Mol Cell. 2009;36:405–16. doi: 10.1016/j.molcel.2009.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Serra V, Scaltriti M, Prudkin L, Eichhorn PJ, Ibrahim YH, Chandarlapaty S, et al. PI3K inhibition results in enhanced HER signaling and acquired ERK dependency in HER2-overexpressing breast cancer. Oncogene. 2011;30:2547–57. doi: 10.1038/onc.2010.626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hirai H, Sootome H, Nakatsuru Y, Miyama K, Taguchi S, Tsujioka K, et al. MK-2206, an allosteric Akt inhibitor, enhances antitumor efficacy by standard chemotherapeutic agents or molecular targeted drugs in vitro and in vivo. Mol Cancer Ther. 2010;9:1956–67. doi: 10.1158/1535-7163.MCT-09-1012. [DOI] [PubMed] [Google Scholar]
- 20.Sefton EC, Qiang W, Serna V, Kurita T, Wei JJ, Chakravarti D, et al. MK-2206, an AKT inhibitor, promotes caspase-independent cell death and inhibits leiomyoma growth. Endocrinology. 2013;154:4046–57. doi: 10.1210/en.2013-1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cancer Genome Atlas N. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thorpe LM, Yuzugullu H, Zhao JJ. PI3K in cancer: divergent roles of isoforms, modes of activation and therapeutic targeting. Nat Rev Cancer. 2015;15:7–24. doi: 10.1038/nrc3860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Manning BD, Cantley LC. AKT/PKB signaling: navigating downstream. Cell. 2007;129:1261–74. doi: 10.1016/j.cell.2007.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mori S, Nada S, Kimura H, Tajima S, Takahashi Y, Kitamura A, et al. The mTOR pathway controls cell proliferation by regulating the FoxO3a transcription factor via SGK1 kinase. PLoS One. 2014;9:e88891. doi: 10.1371/journal.pone.0088891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Das TP, Suman S, Alatassi H, Ankem MK, Damodaran C. Inhibition of AKT promotes FOXO3a-dependent apoptosis in prostate cancer. Cell Death Dis. 2016;7:e2111. doi: 10.1038/cddis.2015.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bean GR, Ganesan YT, Dong Y, Takeda S, Liu H, Chan PM, et al. PUMA and BIM are required for oncogene inactivation-induced apoptosis. Sci Signal. 2013;6:ra20. doi: 10.1126/scisignal.2003483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.O’Reilly KE, Rojo F, She QB, Solit D, Mills GB, Smith D, et al. mTOR inhibition induces upstream receptor tyrosine kinase signaling and activates Akt. Cancer Res. 2006;66:1500–8. doi: 10.1158/0008-5472.CAN-05-2925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.You H, Pellegrini M, Tsuchihara K, Yamamoto K, Hacker G, Erlacher M, et al. FOXO3a-dependent regulation of Puma in response to cytokine/growth factor withdrawal. J Exp Med. 2006;203:1657–63. doi: 10.1084/jem.20060353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Essafi A, Fernandez de Mattos S, Hassen YA, Soeiro I, Mufti GJ, Thomas NS, et al. Direct transcriptional regulation of Bim by FoxO3a mediates STI571-induced apoptosis in Bcr-Abl-expressing cells. Oncogene. 2005;24:2317–29. doi: 10.1038/sj.onc.1208421. [DOI] [PubMed] [Google Scholar]
- 30.Tang L, Wang Y, Strom A, Gustafsson JA, Guan X. Lapatinib induces p27(Kip1)-dependent G(1) arrest through both transcriptional and post-translational mechanisms. Cell Cycle. 2013;12:2665–74. doi: 10.4161/cc.25728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Adams JM, Cory S. The Bcl-2 apoptotic switch in cancer development and therapy. Oncogene. 2007;26:1324–37. doi: 10.1038/sj.onc.1210220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Faber AC, Ebi H, Costa C, Engelman JA. Apoptosis in targeted therapy responses: the role of BIM. Adv Pharmacol. 2012;65:519–42. doi: 10.1016/B978-0-12-397927-8.00016-6. [DOI] [PubMed] [Google Scholar]
- 33.El-Guendy N, Rangnekar VM. Apoptosis by Par-4 in cancer and neurodegenerative diseases. Exp Cell Res. 2003;283:51–66. doi: 10.1016/s0014-4827(02)00016-2. [DOI] [PubMed] [Google Scholar]
- 34.Hebbar N, Shrestha-Bhattarai T, Rangnekar VM. Cancer-selective apoptosis by tumor suppressor par-4. Adv Exp Med Biol. 2014;818:155–66. doi: 10.1007/978-1-4471-6458-6_7. [DOI] [PubMed] [Google Scholar]
- 35.Hebbar N, Burikhanov R, Shukla N, Qiu S, Zhao Y, Elenitoba-Johnson KSJ, et al. A Naturally Generated Decoy of the Prostate Apoptosis Response-4 Protein Overcomes Therapy Resistance in Tumors. Cancer Res. 2017 doi: 10.1158/0008-5472.CAN-16-1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.