Abstract
With the proliferation of heterogeneous health care data in the last three decades, biomedical ontologies and controlled biomedical terminologies play a more and more important role in knowledge representation and management, data integration, natural language processing, as well as decision support for health information systems and biomedical research. Biomedical ontologies and controlled terminologies are intended to assure interoperability. Nevertheless, the quality of biomedical ontologies has hindered their applicability and subsequent adoption in real-world applications. Ontology evaluation is an integral part of ontology development and maintenance. In the biomedicine domain, ontology evaluation is often conducted by third parties as a quality assurance (or auditing) effort that focuses on identifying modeling errors and inconsistencies. In this work, we first organized four categorical schemes of ontology evaluation methods in the existing literature to create an integrated taxonomy. Further, to understand the ontology evaluation practice in the biomedicine domain, we reviewed a sample of 200 ontologies from the National Center for Biomedical Ontology (NCBO) BioPortal—the largest repository for biomedical ontologies—and observed that only 15 of these ontologies have documented evaluation in their corresponding inception papers. We then surveyed the recent quality assurance approaches for biomedical ontologies and their use. We also mapped these quality assurance approaches to the ontology evaluation criteria. It is our anticipation that ontology evaluation and quality assurance approaches will be more widely adopted in the development life cycle of biomedical ontologies.
Keywords: Ontology Evaluation, Biomedical Ontologies, Knowledge Representation, Quality Assurance
Graphical Abstract
1. Introduction
In the big data era, the management and integration of large datasets provide enormous opportunities for the discovery of new knowledge. Ontologies, which organize the domain knowledge in the form of relevant concepts/classes and relationships among them, provide the necessary tools to overcome barriers when integrating data and knowledge from heterogeneous datasets, thereby facilitating knowledge discovery. In the biomedicine domain, biomedical ontologies lay a solid foundation in a variety of healthcare information systems for encoding diagnoses, problem lists [1, 2], laboratory tests in electronic health records [3] as well as in administrative documents such as billing statements [4] and insurance claims. Moreover, with concepts/classes linked by rich taxonomic and lateral relationships, biomedical ontologies play a vital role in knowledge representation and management, data integration, decision support, and natural language processing [5]. Traditionally, ontology development employs a top-down approach, in which ontology engineers and domain experts define ontological elements/axioms through iterative discussions and revisions. Omissions, redundancies, errors, and inconsistencies are inevitable. Thus, ontology evaluation is an integral part of ontology development and maintenance because it can attest that what is being built meets the application requirements [6], increases the availability and reusability of ontologies [7, 8], and lowers the maintenance costs of collaboratively created knowledge bases [7]. Nevertheless, the nomenclature of ontology evaluation, especially in the biomedical domain, causes confusion among researchers. To discuss ontology evaluation more effectively, we first attempt to clarify the different nomenclatures related to ontology evaluation in the following subsections based on our extensive review of the ontology evaluation literature.
1.1. What is ontology evaluation?
Generally, ontology evaluation is a process that determines the quality (and/or correctness) of an ontology in respect to a set of evaluation criteria, depending on what kind of ontologies are being evaluated and for what purpose. A well-known classification of ontology evaluation was proposed by Brank et al. [9], which grouped existing evaluation approaches into four broad categories: 1) approaches that compare the target ontology to a “gold standard” [10]; 2) approaches that use the target ontology in an application and evaluate the application results [11]; 3) those that conduct coverage analysis comparing the target ontology with a source of data (e.g., a collection of documents) about a specific domain [12]; and 4) manual reviews done by human experts that assess how well the target ontology meets a set of predefined criteria, standards, and requirements [13]. A few other variations of ontology evaluation approach classifications were also referred to in recent ontology evaluation publications [12, 14, 15]. Nevertheless, the broad concepts of these classification systems are very similar.
Furthermore, an ontology is a complex structure. A common approach of ontology evaluation is to evaluate different levels/aspects of the ontology separately rather than trying to assess the quality of the ontology as a whole. The ways of how these levels/aspects were defined are slightly different in different ontology evaluation publications.
1.2. Relationships among ontology evaluation, auditing, and quality assurance
Vrandečić [7] defined ontology evaluation as the “task of measuring the quality of an ontology” in order to determine the fitness of the ontology to exchange data, assess the development of the ontology, and to maintain consistency with seven defined criteria. The definition was further enhanced by segmenting the tasks into ontology verification and ontology validation (first introduced by Gómez-Pérez [16] and later adopted by Vrandečić [7]). Verification “deals with building the ontology correctly, that is, ensuring that its definitions implement correctly the requirements.” Validation “refers to whether the meaning of the definitions really models the real world for which the ontology was created.” Essentially, verification examines the intrinsic aspects of the ontology, whereas, validation examines the extrinsic aspects of the ontology. Further details of both of these components and the various aspects to evaluate were discussed in [7]. For brevity, Table 1 summarizes these ideas. Later we will revisit the concepts of validation and verification in the subsequent sections.
Table 1.
Ontology evaluation criteria in Vrandečić [7]
| Criteria | Definition |
|---|---|
| Accuracy | Does the asserted knowledge in the ontology agree with the expert’s knowledge, which is often measured in terms of precision and recall? |
| Completeness | Is the domain of interest appropriately covered (i.e., coverage)? |
| Conciseness | Does the ontology define irrelevant elements with regards to the domain to be covered or redundant representations of the semantics? |
| Consistency | Does the ontology include or allow for contradictions, which is often measured as the number of terms with inconsistent meaning? |
| Computational efficiency | How fast can the tools (e.g., reasoners) work with the ontology? |
| Adaptability | How easy or difficult is it to use an ontology in different contexts? Adaptability often measures coupling (i.e., number of external classes referenced) and cohesion (i.e., modularity of the ontology). |
| Clarity | How effective can the ontology communicate the intended meaning of the defined terms? |
In the biomedicine domain, a number of notable biomedical ontologies were developed as controlled vocabularies/terminologies. The definitions of ontologies and vocabularies/terminologies are interrelated and have some overlaps. Terminologies and vocabularies focus on representing the lexical terms (synonyms) that are used to refer to entities (concepts) with a limited number of relationships (e.g., Lung cancer finding_site Lung structure) among those entities. They are often considered as lightweight ontologies and are most commonly used in biomedicine [17]. Ontologies, on the other hand, provide an explicit representation of meaning defined as a set of terms using a logic-based representation and formal constraints to characterize entities and relationships distinctive to the domain. The formalism of ontologies adds additional reasoning capabilities and expressiveness upon vocabularies/terminologies. They have become more and more popular due to their expressiveness and flexibility. For example, Foundational Model of Anatomy [18] has multilevel granularity of hierarchical relationships with diverse physical relations between anatomical components. Broadly speaking, concepts in a vocabulary/terminology are equivalent to classes in an ontology. Relationships in a vocabulary/terminology are equivalent to the properties in an ontology. In the context of this paper, we use terminologies and ontologies interchangeably.
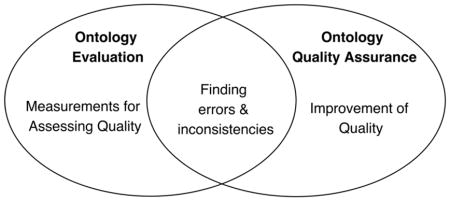
In some literature, evaluation of biomedical ontologies is often referred to as “quality assurance” (QA). When it is conducted by third parties with or without the involvement of the ontology developers, it is also often called “auditing” [19]. Rogers’ framework of biomedical ontology quality assurance included four aspects: philosophical validity, meta-ontological commitment, content correctness, and fitness for purpose [20]. Previously, Zhu et al. conducted a review of auditing methods applied to controlled biomedical terminologies, where they outlined the aspects of ontology auditing of biomedical terminologies and ontologies [19]. We summarized these aspects in Table 2, and mapped each aspect to an ontology evaluation criterion in Vrandečić [7]. Each of the aspects is a close approximation of a criterion in Table 1. Just as ontology evaluation, quality assurance (or auditing) of biomedical ontologies also examines both the intrinsic and the extrinsic aspects of the ontologies. Defined by Zhu et al. [19], the intrinsic aspects are “inherent to ontologies’ content (hierarchy, relationships, lexicon) that can be audited independently from external reference standards. Intrinsic aspects include concept orientation, consistency, soundness, and non-redundancy”. Extrinsic aspects are “contingent on the comprehensive coverage of external user requirements, domain-specific contextual needs, or other external reference standards”. Thus, despite the differences in nomenclature, based on our deduction of the definitions of the criteria, it is reasonable to assume that quality assurance and auditing of the biomedical ontologies is within the scope of ontology evaluation. Nevertheless, in the literature, ontology evaluation and ontology quality assurance do have some noticeable differences in their focus. Ontology evaluation literature often focused on the measurements for assessing the goodness of the ontologies (including the use of competency questions [16]), whereas quality assurance literature focused on identifying modeling errors and inconsistencies and improving the quality of the ontologies. Note that some of the ontology evaluation papers (papers that proposed evaluation methods) did talk about how they can find issues with an ontology. This is logical, as one will have to be able to find issues to come up with a reliable measure. Figure 1 is a Venn diagram showing the differences and commonalities between ontology evaluation and quality assurance in the literature.
Table 2.
Criteria for biomedical ontology auditing and quality assurance adapted from [19].
| Criteria in Zhu et al. (Criteria in Vrandečić) | Definition |
|---|---|
| Concept orientation (Clarity) | Refers to undefined concepts and ambiguous definition of concepts |
| Consistency (Consistency) | Refers to lexical aspects and classification of concepts |
| Non-redundancy (Conciseness) | Refers to redundant classification and concepts |
| Soundness (Accuracy) | Refers to the soundness of classifications and concept descriptions |
| Comprehensive coverage (Completeness) | Refers to the coverage of concepts and related terms, gaps in hierarchal and semantic relationships, and completeness of the definitions of concepts. |
Figure 1.
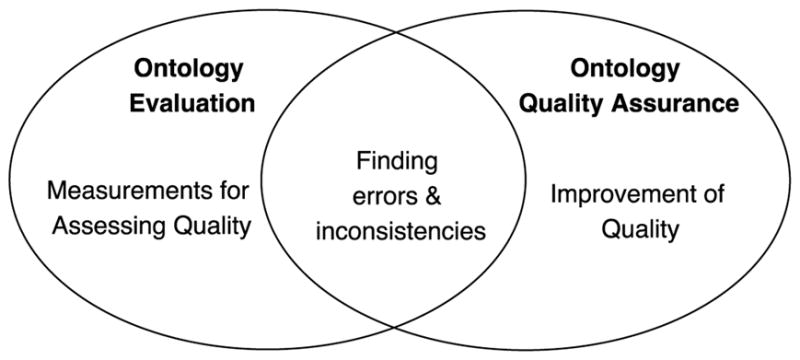
A Venn diagram showing the relationships between ontology evaluation and quality assurance in the literature.
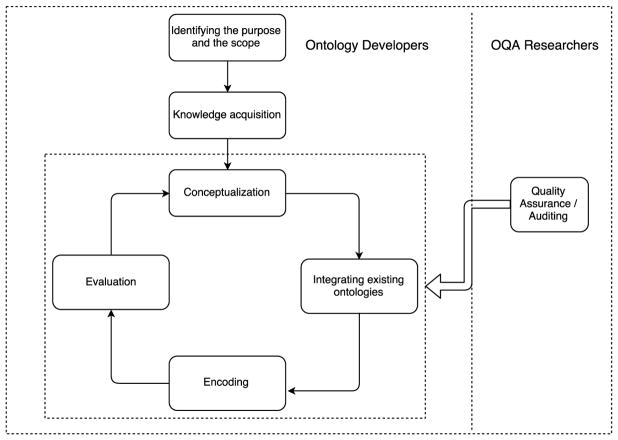
Figure 2 illustrates the relationship between ontology evaluation and quality assurance in the ontology development life cycle. Ontology developers first identify the purpose and the scope of the ontology, then acquire knowledge from different sources and enter the cycle consisting of four steps: 1) conceptualize the knowledge, 2) identify and integrate existing ontologies, 3) encode the concepts, and 4) evaluate the ontology. By reviewing the literature, we found that quality assurance and auditing appear to be mostly conducted by third parties as an additional mechanism to support the maintenance of an ontology. The quality assurance/auditing tasks are thus conducted after the first public release of the ontologies. The auditing results are submitted to the developers to review and then incorporated in the next release of the ontology, at the discretion of the developers.
Figure 2.
Ontology development lifecycle with ontology evaluation and quality assurance.
1.3. Contributions
BioPortal, created by the National Center for Biomedical Ontology (NCBO), is the world’s largest repository and development system of biomedical ontologies [21]. As of January 15, 2018, it contained 686 ontologies encoded in a wide range of formats including the Web Ontology Language (OWL) [22], Resource Description Framework [23], Open Biological and Biomedical Ontologies (OBO) format [24], Protégé frames, and the Rich Release Format of the Unified Medical Language System Metathesaurus [25]. BioPortal provides tools for browsing, searching, annotation, and visualization of its ontologies to support research in the biomedical sciences. Due to its size, variety of quality, coverage, and its liberal submission policy, we used BioPortal as the dataset to assess the evaluation practices of biomedical ontologies.
In this work, we surveyed and discussed current ontology evaluation methods that have been applied in both open domain (i.e., ontologies in the all the domains) and biomedical domain. To assess whether and how ontology evaluation is being practiced in the biomedical ontology development, we reviewed a sample of 200 ontologies in BioPortal and their documented evaluation methods from their corresponding inception papers—the earliest publication that first introduced and described the specific ontology. We summarized the evaluation methods of the sampled ontologies based on the major categories of ontology evaluation methods. Based on our observations, we will discuss the opportunities for ontology developers to adopt the state-of-the-art ontology evaluation (or “quality assurance”) methods to improve the quality of biomedical ontologies. The rest of the paper will be organized as follows: in Section 2, we will review the ontology evaluation types. In Section 3, we will assess the ontology evaluation practices for a sample of 200 ontologies retrieved from BioPortal based on their popularity. In Section 4, we will review the recent quality assurance and auditing methods for biomedical ontologies and their use in various BioPortal ontologies. In Section 5, we will discuss our findings of this study as well as the gaps and opportunities for evaluating and assessing the quality of biomedical ontologies. Section 6 concludes the paper.
2. Review of Ontology Evaluation Types
As mentioned above and further discussed here, Gómez-Pérez categorized the ontology evaluation into ontology verification and ontology validation [16]. According to Gómez-Pérez, ontology verification is “building the ontology correctly, that is, ensuring that its definitions implement correctly the ontology requirements and competency questions, or function correctly in the real world”. It focuses on inherent features of the ontology, such as lexical or syntactical aspects. Ontology validation is “whether the meaning of the ontology definitions really model the real world for which the ontology was created” [16]. In other words, it is concerned with external assessment of the impact of the ontology, basically how effective the ontology preforms or serves its purpose [19]. As Vrandečić described [7], verification “answers if the ontology was built the right way” (i.e., intrinsic [19]) and validation “answers if the right ontology was built” (i.e., extrinsic [19]) [7].
Furthermore, other researchers have expanded and produced their own categorical schemes of ontology evaluation. Brank et al. [9] classified ontology evaluation by approaches: 1) gold standard, 2) application based, 3) data-driven, 4) user-based, and extended by levels: 1) lexical, 2) hierarchical/taxonomy, 3) semantic relations, 4) context, 5) syntactic, and 6) structure, architecture, design. Obrst et al. [26] also mentioned six types of ontology evaluation that would be categorized as ontology validation approaches, according to Vrandečić [7]. These categories include: 1) application-based evaluation, 2) data source comparison, 3) human assessment, 4) NLP-based evaluation, 5) reality benchmarking, and 6) community verification. Tartir [6] introduced a classification of ontology evaluation based on the 1) evolution of the ontology, 2) logic and rules, and 3) metrics. Duque-Ramos [27] classified ontology evaluation by 1) ranking, 2) correctness, and 3) quality.
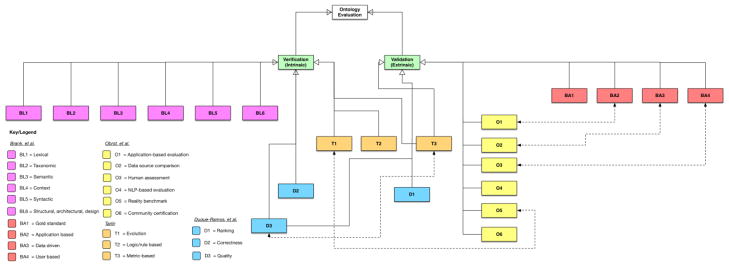
Figure 3 is an integrated taxonomy of ontology evaluation, which is our attempt to organize and explicate the aforementioned heterogeneous categorical schemes of ontology evaluation. In our review of the literature, we noted several authors who introduced their own categorizations of ontology evaluation and mapped them to the classic division of verification and validation. We also noted that some of the classifications were similar. For example, the data-driven evaluation approach was repeated by multiple authors. This was denoted with a dotted line in Figure 3. The BA1–BA4 and BL1–BL6 labels are categorizations proposed by Brank et al. [9]; T1–T3 are from Tartir [6]; D1–D3 are from Duque-Ramos et al. [27]; O1–O6 are derived from Obrst et al. [26]. In the following, we will expound these classification schemes and then discuss the use of them in this study.
Figure 3.
Taxonomy of ontology evaluation
Brank et al. [9]
In their influential paper, they highlighted the importance of categorical evaluation and proposed associated categories across different “levels” (or “aspects” in Vrandečić [7]). One of the evaluation categories was to compare the target ontology with a “gold standard” ontology (BA1). This was rooted in the work of Maedche and Staab [10] where they considered semantic similarity measures at the lexical level and the taxonomic overlap between ontologies at the conceptual level for the comparison of two ontologies (i.e., the target ontology vs. a gold standard ontology). Aside from using a “gold standard”, the authors situated the evaluation of an ontology with the application of the ontology or an application-specific ontology performance evaluation (BA2). In the various ontologies discussed later in Section 3, the application-based evaluation varied depending on the purpose for which the ontology was developed. Brank et al.’s [9] data-driven evaluation (BA3) was based upon the work of Brewster and colleagues [12]. This category (BA3) differs from the “gold standard” evaluation approach, as it compares the ontology with another data source such as a text corpus to assess whether all the concepts in the text are correctly encoded in the ontology. Another category of evaluation approaches defined by Brank et al. involves human derived assessments based on pre-defined criteria or heuristics (BA4). One example of such an approach is the use of competency questions (i.e., requirements) to evaluate an ontology, i.e., whether the ontology meets these predefined goals and answers the competency questions. Much of what is described here so far relates to the early definition of ontology validation (or extrinsic evaluation).
Brank and colleagues expounded their categorization and included levels (i.e., lexical, hierarchical/taxonomy, semantic relations, context, syntactic, and structure/architecture/design) that are related, in our view, to ontology verification. Most of these “levels” defined by Brank et al. are similar to the aspects put forth by Vrandečić [7]. For example, lexical evaluation (BL1) involves the assessment of the vocabulary or nomenclature (e.g., literals, URIs, values) employed by the ontology, which may include assessing the ontology’s adherence to labeling standards or its appropriate use of vocabulary to describe the intended domain. The hierarchical or taxonomic evaluation of an ontology (BL2) concerns the assessment of the hierarchical relationships between concepts (e.g., “IS-A”). Evaluating semantic relationships other than the “IS-A” relationships between concepts is also an important aspect in ontology evaluation (BL3). Context-based evaluation (BL4) accounts for the target ontology’s interactions with other ontologies or whether the ontology has an “impact” when it is integrated into an application. For example, the Burton-Jones et al.’s metric suite [28] contains a categorical metric called “social”, which accounts for how many ontologies within a certain library or sub-library are linked to the target ontology being evaluated. Syntactic-level evaluation (BL5) assesses whether the ontology conforms to the syntactic profiles of formal languages such as Web Ontology Language 2.0 (OWL2) [22] or Resource Description Framework (RDF) [23]. For example, the Burton-Jones et al.’s metric suite has a set of “syntactic” scores, where one of the sub-metrics—”lawfulness”—calculates the “correctness of syntax” [28]. Brank and colleagues also mentioned architectural and structural design evaluation (BL6), which evaluates whether the design of an ontology follows a set of well-defined design principles or heuristics. While Brank and colleagues did not provide any specific examples, one assumed example would be the principles and guidelines for OBO Foundry ontologies [24].
Obrst et al. [26]
Obrst and colleagues proposed a set of ontology evaluation types, which were described by Vrandečić as ontology validation: 1) evaluation with respect to the use of an ontology in an application (O1), 2) evaluation with respect to domain data sources (O2), 3) evaluation by human experts against a set of criteria (O3), 4) evaluation with natural language evaluation techniques (O4), 5) evaluation with the use of reality itself as a benchmark, and 6) accrediting and certifying ontologies that have passed some evaluation criteria (O6), and the notion of an ontology maturity model (O5). The first three evaluation types (O1–O3) defined by Obrst et al. overlap with categories previously discussed by Brank and colleagues [9], namely the application-based (O1 and BA2), data-driven (O2 and BA3), and user assessment (O3 and BA4), respectively. However, the evaluation based on natural language techniques and the use of reality as a benchmark are particularly unique. According to Obrst and colleagues [26], natural language evaluation includes task-based evaluation that specifically uses natural language processing methods such as information extraction and question answering. An example would be using question answering where natural language questions are translated to SPARQL (a recursive acronym for SPARQL Protocol and RDF Query Language) queries to be executed to measure the quality (whether the retrieved triples are appropriate) and quantity (number of correct triples retrieved) of the responses retrieved from the ontologies. The use of reality as a benchmark was inspired by an early study by Ceusters and Smith [29]. Ceusters introduced the notion that an evolving ontology that modifies its “units” (e.g., universals and instances) over time corresponds to a model that is closer to the reality. Obrst et al., thus, introduced the concept of measuring the evolution of an ontology as a way of showing its fidelity to represent reality [26].
Tartir [6]
In addition to introducing a metric-based ontology evaluation framework, OntoQA, Tartir also described ontology evaluation in three categories, namely evolution-based (T1), logical/rule-based (T2), and metric-based (T3). Similar to Obrst et al. [26], Tartir shared the idea of measuring the evolution of an ontology as a way of ontology evaluation (T1). However, no clear rationale was given for the evolution-based evaluation, except for the allusion of the growth of the knowledge scope [6]. The concept of logical/rule-based ontology evaluation (T2) is similar to the syntactic level evaluation proposed by Brank and colleagues [9]. The metric-based evaluation (T3) covers all the ontology evaluation approaches that include a quantitative measure of various intrinsic and extrinsic features of an ontology.
Duque-Ramos et al. [27]
Similar to the work by Tartir [6], Duque-Ramos et al. [27] presented their own view of the various perspectives in ontology evaluation: ranking (D1), correctness (D2), and quality (D3). The quality perspective (D3) refers to a holistic quality evaluation of the ontology based on features (similar to Tartir’s metric-based metric (T3) [6]). Correctness (D2) is an intrinsic evaluation that focuses on syntactical adherence, which is similar to a number of aforementioned categories defined by the others (e.g., BL5 and T2), but also incorporates the accuracy of knowledge expressed by the ontology. Ranking (D1) appears to be a unique category defined by Duque-Ramos et al., as it involves any evaluations that have a systematic approach for the selection of an ontology or, as the name implied, ranking the ontology in comparison with others. The classic work of Lozano-Tello and Gómez-Pérez’s OntoMetric [13] is an example of ontology evaluation that would be considered as a ranking-based approach.
3. Review of the Ontology Evaluation Practice for BioPortal’s Ontologies
3.1. Prevalence and documented evidence of evaluation in BioPortal Ontologies
In this paper, to examine the practice of ontology evaluation in biomedicine, we identified a sample of 200 ontologies from NCBO BioPortal based on their popularity in September 2015, and manually reviewed the ontologies with documented evidence of evaluation in peer-reviewed publications. We then analyzed the types of the ontology evaluation approaches employed by these ontologies and reported on the prevalence of the ontology evaluation approaches discussed above in Section 2 [6, 9, 26, 27].
In our previous work, we had collected data for documented evaluation methods of ontologies hosted on the NCBO BioPortal [30, 31]. Based on this previous work, we initially hypothesized that most biomedical ontologies did not conduct any initial evaluations or at least indicate any evidence as such. In this work, we reused this dataset to further explore our hypothesis. The original survey was conducted in September 2015 and comprised of a sample of 200 ontologies (as of September 2015, there were 547 ontologies in total in BioPortal). For each ontology sampled, we searched in PubMed and Google Scholar and selected the earliest paper that introduced the respective ontology. We assessed whether the paper contains a detailed development discussion of the ontology itself. If the paper was chosen as one of the candidates (i.e., as the inception paper of the specific ontology), we took note of whether each one of the evaluation approaches discussed in Section 2 was used [6, 9, 26, 27]. All of the data of our analysis were recorded on a spreadsheet in Appendix I. As shown in Table 3, only 15 of the 200 biomedical ontologies we surveyed from the NCBO BioPortal have documented evaluation in their corresponding inception papers.
Table 3.
The 15 ontologies with documented evaluation and the corresponding publications
| Ontology | Number of Classes | Number of Properties | Release Cycle | Development Paper |
|---|---|---|---|---|
| Bilingual Ontology of Alzheimer’s Disease and Related Diseases (ONTOAD) | 5899 | 182 | Once in 2013 | Drame et al. [32] |
| Ontology for Genetic Susceptibility Factor (OGSF) | 127 | 28 | Once a year between 2013–2015 | Lin et al. [33] |
| Cell Culture Ontology (CCONT) | 19991 | 61 | Twice in 2012; once in 2014 | Ganzinger et al. [34] |
| Genomic Clinical Decision Support Ontology (GENE-CDS) | 2265 | 2 | Once in 2012 | Samwald et al. [35] |
| Parasite Experiment Ontology (PEO) | 143 | 40 | Twice in 2009; once in 2011 | Sahoo et al. [36] |
| Glycomics Ontology (GLYCO) | 230 | 65 | Once in 2012 | Thomas et al. [37] |
| Ontology of Physics for Biology (OPB) | 861 | 66 | Twice in 2015; once in 2017 | Cook et al. [38] |
| Radiation Oncology Ontology (ROO) | 1183 | 211 | Once in 2014; twice in 2015 | Pratt et al. [39] |
| Ontology of Core Data Mining Entities (ONTODM-CORE) | 838 | 91 | Once in 2012; three times in 2016; once in 2017 | Panov et al. [40] |
| Autism Spectrum Disorder Phenotype Ontology (ASDPTO) | 284 | 0 | Once in 2014 | McCray et al. [41] |
| Pharmacogenomic Relationships Ontology (PHARE) | 229 | 83 | Once in 2010; once in 2011 | Coulet et al. [42] |
| DebugIT Core Ontology (DCO-DEBUGIT) | 1029 | 87 | Once in 2013 | Schober et al. [43] |
| Alzheimer’s disease ontology (ADO) | 1565 | 12 | Three times in 2013 | Malhotra et al. [44] |
| Human Physiology Simulation Ontology (HUPSON) | 2920 | 91 | Once in 2014 | Gündel et al. [45] |
| Semantic DICOM Ontology (SEDI) | 1423 | 4606 | Once in 2013; twice in 2014; twice in 2015 | Van Soest et al. [46] |
Within the 200 ontologies from BioPortal that we surveyed, there were 15 OBO Foundry ontologies. Only one ontology (i.e., the Ontology for Genetic Susceptibility Factor) out of the 15 OBO Foundry ontologies had documented evidence of evaluation [33].
3.2. Analysis of the ontology evaluation practice of BioPortal ontologies
We assessed the 15 ontologies with documented evaluation on whether their evaluation methods belong to the classifications defined by the ontology evaluation review papers discussed in Section 2 [6, 9, 26, 27]. The analysis result is given in Table 4. None of the 15 NCBO biomedical ontologies utilized any ranking based evaluation (D1), but close to half of them (53.3%) evaluated the correctness of the information or the formal language (“Correctness” D2). Seven out of the 15 (46.7%) had documented evidence of using some holistic quality evaluation (D3 and T3), mostly structural metrics or metrics that reported on ontological features.
Table 4.
The analysis result of the 15 ontologies with documented evaluation
| Number of Ontologies (n=15) | Completeness | Overall Completeness | |
|---|---|---|---|
| Validation | 9 | 60.00% | 70.00% |
| Verification | 12 | 80.00% | |
| Ranking (D1) | 0 | 0.00% | 31.11% |
| Correctness (D2) | 7 | 46.67% | |
| Quality (D3) | 7 | 46.67% | |
| Lexical (BL1) | 1 | 6.67% | 20.00% |
| Taxonomic (BL2) | 3 | 20.00% | |
| Semantic relationships (BL3) | 2 | 13.33% | |
| Context (BL4) | 3 | 20.00% | |
| Syntactic (BL5) | 1 | 6.67% | |
| Structural, architecture, design (BL6) | 8 | 53.33% | |
| Gold standard (BA1) | 2 | 13.33% | 31.67% |
| Application based (BA2) | 7 | 46.67% | |
| Data driven (BA3) | 5 | 33.33% | |
| User-based (BA4) | 5 | 33.33% | |
| Evolution (T1) | 1 | 6.67% | 28.89% |
| Logic/Rule based (T2) | 5 | 33.33% | |
| Metric-based (T3) | 7 | 46.67% | |
| Application-based evaluation (O1) | 7 | 46.67% | 22.22% |
| Data source comparison (O2) | 5 | 33.33% | |
| Human assessment (O3) | 5 | 33.33% | |
| NLP-based evaluation (O4) | 2 | 13.33% | |
| Reality benchmark (O5) | 0 | 0.00% | |
| Community certification (O6) | 1 | 6.67% |
Brank and colleagues had a more detailed classification of ontology verification and validation [9] in terms of levels. Regarding their verification items, or those that were labeled as “levels”, most of the biomedical ontologies in the sample had evaluations that were of the “structural, architecture, design” type. Some of these included verifying whether their ontologies complied with design principles or checking the integrity of the ontologies using reasoners such as Fact++ [47] and HermiT [48]. “Lexical” or “syntactical” level evaluations were poorly represented, since only one ontology assessed its vocabulary, terms, or syntactic requirements. Regarding Brank and colleagues’ validation items [9], most of the evaluation types employed were assessing the ontology according to an application purpose (46.7%), while others were mostly data-driven-based evaluations (i.e., measuring the ontology against a corpus or database) or user-driven evaluations (e.g., crowdsourcing). Evaluation with a gold standard was limited as only two ontologies employed such an evaluation. Overall the 15 biomedical ontologies covered 20% of Brank and colleagues’ verification items and 31.7% of the validation items.
Some of the categories in Obrst and others’ evaluation classification [26] overlap with Brank et al.’s categories, specifically application-based, data-source comparison, and human assessment. Application-based evaluation was utilized the most according to the Obrst and colleagues’ categorical schemes. However, NLP-based evaluation, reality benchmarking, and community certification were poorly represented. The 15 ontologies covered 22.2% of Obrst et al.’s classification categories.
Duque-Ramos and colleagues delineated ontology evaluation by ranking, correctness, and global quality [27]. Among the 15 ontologies, the only types of assessments performed according to Duque-Ramos’s classification were the latter two, correctness and global quality (46.7%). None of the 15 ontologies applied any ranking-related evaluations.
Lastly, Tartir’s categories [6] include evolution-based, logic/rules-based, and metric-based evaluation. Some ontologies used metric-based evaluation (46.7%); a third utilized logic/rule-based evaluation (33.3%); and one utilized evolution-driven evaluation.
4. Review of Recent Biomedical Ontology Quality Assurance Methods
As mentioned in the Introduction Section, the existing literature on ontology evaluation often focused on the metrics for assessing the goodness of the ontologies, whereas quality assurance focused on identifying modeling errors and inconsistencies and improving the quality of the ontologies. Even though both ontology evaluation and quality assurance cover the identification of modeling errors and inconsistencies based on their definitions, in the field of biomedical informatics, researchers mostly refer to their methods for identifying the errors in the ontology as “quality assurance” methods.
In biomedicine, ontologies are designed to represent both the explicit and implied concepts (classes) used in a particular biomedical discipline, and the relationships between those concepts [49]. As such, the evaluation of the OWL-based ontologies usually examines both the explicitly defined concepts and their relationships, as well as the implicit information that can be derived by a reasoner with a computer algorithm implemented in a description logic engine. Due to the complex nature of the biomedicine domain, biomedical ontologies are often large and complex, with many relationships among concepts. Therefore, inconsistencies and modeling errors, especially missing concepts, missing/redundant semantic relationships and missing synonyms, are inevitable and hard to detect with limited manual evaluation and quality assurance (QA) resources. Based on the open domain ontology evaluation criteria, sophisticated QA methods that were designed specifically for biomedical ontologies have been shown to be effective and efficient in detecting errors and inconsistencies, providing concrete and actionable ways to improve their quality. Nevertheless, these QA methods were mostly developed by the academic researchers outside of the ontology development teams (i.e., third parties) as an auditing effort to provide an additional layer for ontology evaluation. Therefore, they are not likely to be used in the initial development of the ontologies.
In 2009, Zhu et al. presented a methodology review paper on the auditing of controlled biomedical terminologies [19]. In this section, we will focus on the biomedical ontology quality assurance (OQA) methods that were published between 2009 and 2017. Recent OQA methods can be broadly categorized into structural [50–55], lexical [56–60], semantic [61–65], abstraction-network-based [66–76], big-data-based [77–79], crowd-sourcing-based [80–82], cross-validation [83–86], hybrid (mix of structural, lexical, and description-logic-based approach) [87–89], corpus-based [90, 91], and miscellaneous approaches [1, 92, 93]. These methods have been shown to be effective in detecting erroneous classifications, missing concepts, missing or redundant semantic relationships, missing synonyms, and modeling inconsistencies in an automated or semi-automated fashion. Most of the OQA methods reviewed here can be categorized as intrinsic evaluation methods (ontology verification). As most of the aforementioned OQA methods aim to meet multiple criteria for the ontology evaluation, we categorized them based on the approaches and mapped them to the evaluation criteria in Table 5. Representative examples for each of the categories listed above are given below.
Table 5.
Categorization of the ontology quality assurance methods based on ontology evaluation criteria
| Criteria | Structure -based |
Lexical- based |
Semantic -based |
Abstraction -Network- based |
Big- data- based |
Crowd- sourcing- based |
Cross- validation |
Hybrid | Corpus -based |
Miscell aneous |
|---|---|---|---|---|---|---|---|---|---|---|
| Concept orientation (Clarity) | [50] [53] [54] [55] | [56–60] | [61–65] | [66–76] | [80–82] | [83] [84] [85] [86] | [89] | [1] [92] [93] | ||
| Consistency (Consistency) | [50–55] | [56–60] | [61–65] | [66–76] | [77–79] | [80–82] | [83] [84] [85] [86] | [87] [88] [89] | [1] [92] [93] | |
| Non-redundancy (Conciseness) | [51] [52] [54] [55] | [61] [63] | [66–76] | [77–79] | [83] [84] [85] [86] | [87] [88] [89] | [1] [92] [93] | |||
| Soundness (Accuracy) | [50–55] | [56–60] | [61–65] | [66–76] | [77–79] | [80–82] | [83] [84] [85] [86] | [87] [88] [89] | [1] [92] [93] | |
| Comprehensi ve coverage (Completenes s) | [51] [52] [55] | [64, 65] | [66–76] | [89] | [90] [91] | [92] [93] |
Structure-based approaches
Structure-based OQA methods aim to detect missing and redundant relationships. Existing methods often focus on the Foundational Model of Anatomy (FMA) ontology due to its complex inner structures of concept names and relationship types. Gu et al. [51] investigated the transitive structural relationships and categorized them into five major categories of possibly incorrect relationships: circular, mutually exclusive, redundant, inconsistent, and missed entries in FMA. They examined four types of relationships including “subclass”, “part_of”, “branch_of”, and “tributary_of” to determine whether the relationship assignments are in accord with the principles as declared by the FMA for the representation of these anatomical entities. For example, in the category of “missing relationships”, they found that Articular circumference of head of radius is “part_of” Surface of proximal epiphysis of radius. However, the children Articular circumference of head of right radius and Articular circumference of head of left radius do not have a “part_of” relationship to the concepts Surface of proximal epiphysis of right radius and Surface of proximal epiphysis of left radius, respectively. They suggested that the “part_of” relationships should be added. Zhang et al. [50] proposed a method called Motif Checking to study the effect of multi-relation type interactions for detecting logical inconsistencies and anomalies after representing FMA as an RDF graph and motifs as SPARQL queries. The two-node motif involves the cases where 1) A is-a B and also A is a part-of B at the same time, 2) A is-a B, and A and B involve antonyms in their class names; and 3) A is a part-of B, and A and B involve antonyms in their class names. Luo et al. [53] leveraged the taxonomy and partonomy information for disambiguating terms in FMA. Mougin et al. [52] investigated concepts in the Unified Medical Language System (UMLS) associated through multiple relationships, which extended Gu et al.’s work in identifying redundant relationships in the UMLS [54]. Later, Mougin [55] used a similar approach to find missing or redundant relationships in Gene Ontology.
Lexical-based approaches
Luo et al. [57] devised an automated method to audit symmetric concepts by leveraging bi-similarity and linguistic structure in the concept names. They identified concepts with symmetric modifiers such as “left” and “right” and enumerated all possible structural types according to their subsumption hierarchy. This approach was extended by Agrawal et al. [56] who algorithmically identified lexical similarity sets of SNOMED CT concepts to detect the inconsistencies in their formal definitions and semantic relationships. They defined a similarity set as a collection of concepts whose fully specified names have lexical similarity. Based on this definition, they formulated and tested five hypotheses about the concepts in thse lexical similarity sets such as “Similarity sets whose concepts exhibit different number of parents are more likely to harbor inconsistencies than randomly selected similarity sets”. Rector [58] analyzed the common qualifiers of the SNOMED CT concepts and the accuracy of the definitions of pre-coordinated SNOMED concepts as a proxy for the expected accuracy of the post-coordinated expressions. Quesada-Martínez et al. [59] proposed a notion of lexical regularities as a group of consecutive tokens that appears in several labels of an ontology to audit SNOMED CT concepts to find missing relations. Bodenreider [60] represented logical definitions according to the lexical features of concept names with OWL and then inferred hierarchical relations among the concepts using the ELK reasoner. This method is effective in identifying missing hierarchical relations in SNOMED CT.
Semantic-based approaches
The Unified Medical Language System (UMLS) is an important terminological resource in biomedicine. The Metathesaurus of the 2017AB release of the UMLS contains approximately 3.64 million concepts and 13.9 million unique concept names from 201 source vocabularies (e.g., SNOMED CT, FMA, RxNORM, ICD). Each of the 3.64 million concepts in the UMLS is assigned one or more of the 127 semantic types in the UMLS Semantic Network. The Refined Semantic Network (RSN) for the UMLS was devised to complement the UMLS Semantic Network [94]. The RSN partitions the UMLS Metathesaurus into disjoint groups of concepts. Each such group is semantically uniform. In subsequent work, He et al. [63] performed a longitudinal study on the RSN for the last 17 releases of the UMLS (10 years) with the goal of reducing the size of the RSN. This goal was achieved by correcting inconsistencies and errors in the semantic type assignments in the UMLS, which additionally helped identify and correct ambiguities, inconsistencies, and errors in source terminologies widely used in the realm of public health. The audit was focused on Intersection Semantic Types (ISTs, simultaneous assignments of multiple semantic types to a concept) with a few concepts assigned to them. Many errors were found in the extents of ISTs with few concepts such as “Experimental Model of Disease ∩ Neoplastic Process”. Geller et al. [61] identified a set of inclusion and exclusion instructions in the UMLS Semantic Network documentation and derived corresponding rule-categories from the UMLS concept content. They designed an algorithm adviseEditor based on these rule-categories. The algorithm specifies rules for a UMLS editor how to proceed when considering a tuple (pair, triple, quadruple, quintuple) of semantic types to be assigned to a concept. They further designed and developed a Web-based adviseEditor system, a computational platform to inform UMLS editors on the permissible or prohibited semantic type assignments to UMLS concepts [61].
Mougin et al. [62] performed an audit of the concept categorization in the UMLS by analyzing the association of a concept with multiple Semantic Groups, each of which contains only a few semantic types as a surrogate for polysemy. They created semantically homogeneous clusters for these concepts and found that concepts exhibit limited semantic compatibility with their parent and child concepts. Jiang et al. [64] audited the semantic completeness of SNOMED CT concepts by formalizing normal forms of SNOMED CT expressions using formal concept analysis. Recently, Zhu et al. [65] developed a scalable framework that leverages the Spark platform to evaluate the semantic completeness of SNOMED CT using formal concept analysis.
Abstraction-network-based approaches
Biomedical ontologies are often large and complex. Abstraction networks overlay an ontology’s underlying network structure at a higher level of abstraction [67]. They can summarize the structure and content of the ontologies to support their quality assurance [67]. Previously, area and partial-area taxonomies, which are derived from a partition of an ontology hierarchy based on the relationships of its concepts, were shown to effectively highlight areas with modeling errors and inconsistencies in large biomedical ontologies such as SNOMED CT [66] and National Cancer Institute Thesaurus (NCIt) [75]. This technique has also been proved to effectively support the OQA for various OWL-based application ontologies such as Ontology for Clinical Research (OCRe) [69], Sleep Domain Ontology (SDO) [70], Ontology for Drug Discovery Investigations (DDI) [68], Gene Ontology [74], and National Drug File – Reference Terminology (NDFRT) [76].
The development of auditing techniques that are applicable to one ontology at a time is labor-intensive. To support the development of QA techniques for hundreds of domain ontologies in BioPortal, He et al. [95] introduced a family-based QA framework that uses uniform abstraction network derivation techniques and accompanying QA methodologies, applicable to whole families of structurally similar ontologies. Based on this framework, Ochs et al. [71] developed a structural meta-ontology for classifying ontologies into structurally similar families, enabling the derivation of uniform QA methods for the whole families. Tribal abstraction networks were proposed to highlight modeling errors in SNOMED CT hierarchies without attribute relationships [96]. Further, an algorithmic technique called diff partial-area taxonomy derivation has recently been developed to summarize and visualize the structural changes during the evolution of biomedical ontologies [72].
Semi-automated techniques that concentrate on auditing selected sets of similar concepts, identified with the help of abstraction networks, are expected to have high QA yield [66]. Halper et al. [67] reviewed a variety of abstraction networks that can visually summarize the structure and content of certain ontologies to support their QA. Although introduced in the context of well-established controlled vocabularies such as SNOMED CT, the abstraction networks such as diff partial-area taxonomies [72] and partial-area taxonomies [66], can be applied to other OWL ontologies as long as they have hierarchical and attribute relationships. For ontologies without attribute relationships, tribal abstraction networks can be used for summarization and quality assurance [96]. Traditionally, these auditing methods for biomedical ontologies have been adopted by ontology developers on an ad hoc manner, which can be observed from the collaborative publications by the OQA researchers and the ontology developers. Examples of such efforts for ontologies in BioPortal include SNOMED CT [97], Gene Ontology [74], NCIt [75], OCRe [69], SDO [70], Cancer Chemoprevention Ontology (CanCo) [95], and DDI [68].
Big data approaches
To facilitate automated large-scale QA, Zhang et al. [79] developed a lattice-based structural method for auditing large biomedical ontologies such as SNOMED CT, which was implemented through automated SPARQL queries. The lattice is a structure in an ontology in which two concepts do not share more than one minimal common ancestor. They developed a method called Lattice-based Structural Auditing (LaSA), which exhaustively checks concept pairs for conformation to the requirement of being a part of a lattice. Its performance was later enhanced by using the MapReduce pipeline with a 30-node Hadoop local cloud [77]. They systematically extracted non-lattice fragments in 8 SNOMED CT versions from 2009 to 2014, with an average total computing time of less than 3 hours per version. The lattice-based structural auditing principle, which focuses on the order structure induced by the hierarchical relationships, provides an error-agnostic method for auditing ontologies [78, 79].
Crowdsourcing-based approaches
Mortensen et al. [80–82] leveraged the “wisdom of the crowd” to find erroneous hierarchical relationships in SNOMED CT. They asked the crowd workers simple true/false questions, such as “Diabetes Mellitus is a kind of Disorder of abdomen” and meanwhile listed the definitions of these two concepts. They showed that the crowd can identify errors in SNOMED CT that match experts’ findings with a reasonably good accuracy between 61.3% to 70.8% for different qualification types (i.e., biology, medicine, oncology, none). The crowd may be particularly useful in situations where an expert is unavailable, budget is limited, or an ontology is too large for manual error checking.
Cross-validation approaches
Rector et al. [83] examined the hierarchies for SNOMED CT concepts in the CORE Problem List Subset published by the UMLS for their appropriateness in the description logic modeling and classification process. Gu et al. [84] employed a cross-validation approach to audit the semantic types of the UMLS concepts in the same semantic group based on SNOMED CT’s hierarchy and semantic tags but with inconsistent semantic type assignments. They first examined the set of UMLS semantic types assigned to concepts in each hierarchy of SNOMED CT. They then partitioned the SNOMED CT hierarchies into concept groups such that each subset contains all the concepts in a hierarchy with the same combination of UMLS semantic type assignments. Then, a domain expert reviewed small subsets, which have a higher likelihood of containing errors. The reviewed groups exhibited statistically significantly more errors than the controlled samples. Wei et al. [85] created semantic uniformity groups of SNOMED CT concepts based on each concept’s properties and hierarchical information to identify UMLS semantic type assignment errors. Cui et al. [86] developed a cross-ontology analysis method to detect inconsistencies and possible errors in hierarchical relations across multiple ontologies using the UMLS as a proxy.
Hybrid approaches
Agrawal et al. [87] proposed an algorithm that combines the lexical and structural indicators to identify inconsistent modeling in SNOMED CT. Wei et al. [88] showed that the area and partial-area taxonomies (abstraction networks) and the description logic reasoning can complement each other in identifying errors in SNOMED CT. Recently, Cui et al. [89] exploited the lexical patterns of the non-lattice subgraphs mined using the LaSA method to indicate missing hierarchical relations or concepts. This method has been proved effective through a manual review of a random sample of small subgraphs in SNOMED CT.
Corpus-based approaches
Yao et al. [90] assessed the conceptual coverage and parsimony of four commonly used medical ontologies (i.e., ICD-9-CM, CCPSS, SNOMED CT, and MeSH) by applying biomedical named-entity recognition (using MetaMap1) and standard information retrieval measures on a text corpus of medical documents. Park et al. [91] used diabetes-related text corpora from the Tumblr blogs and the Yahoo! Answers social questions and answers (social Q&A) forum to assess the conceptual coverage of SNOMED CT and the Consumer Health Vocabulary.
Miscellaneous approaches
The issues with synonyms and the coverage of conceptual content in biomedical ontologies have also been investigated by researchers. For example, the high percentage of errors in well-curated terminologies such as SNOMED CT is notable. Agrawal et al. [1] performed a concept analysis by comparing Problem List concepts and general SNOMED CT concepts. The error rate for the widely used concepts of the CORE Problem List, a subset of SNOMED CT, is lower, but still high [93]. He et al. [92] used simulated clinical scenarios involving various term-based searches of concepts mapped into UMLS concepts to assess whether SNOMED’s concept descriptors provide sufficient differentiation to enable possible concept selection between similar terms.
We further categorized these OQA approaches based on the ontology evaluation criteria defined in [19]. The definition of each criterion can be seen in Table 1. For the criterion “Non-redundancy (Conciseness)”, we focused on the approaches to addressing the issues of redundant concepts and redundant relationships. For the criterion “Comprehensive Coverage (Completeness)”, we focused on the approaches to addressing the issues of missing concepts and missing relationships. As shown in Table 5, recent OQA methods and studies mostly focus on the concept orientation, consistency, non-redundancy, and the accuracy but not coverage. Some recent work by He et al. [98–100], even though not based on OQA methods, attempted to use a topological-pattern-based approach that leverages the hierarchical structure and the native term mapping of the UMLS to improve the conceptual coverage of a biomedical ontology integrated into the UMLS.
5. Discussion
5.1. Findings of this study
Biomedical ontologies and controlled vocabularies play an important role in various health information systems and biomedical research. However, even for SNOMED CT, the world’s most comprehensive clinical reference terminology with a large development team, its quality is often questioned [101–103]. Ceusters [103] applied evolutional terminology auditing over 18 versions of SNOMED CT and found that at “the level of the concepts minimal improvements are obtained”. Elhanan et al.’s 2010 survey among SNOMED CT users found that 92% of users use the Clinical Finding hierarchy and they requested improvement of its quality [102].
For the smaller application ontologies in BioPortal, their quality is even harder to be assured. The result of our survey showed that only 15 out of 200 sampled ontologies from the NCBO BioPortal have documented evidence of evaluation in their corresponding inception papers. We did not account for later published research that might have had some form of evaluation. Nevertheless, our point is that ontology evaluation should have been started during the development phase. Only one out of the 15 ontologies was an OBO Foundry ontology. In January 2018, there exist 686 ontologies in BioPortal. As stated before, one of the driving forces for ontology evaluation is to assure researchers, developers, and other users that the ontology is of “good” quality [104]. The NCBO BioPortal is a multi-million dollar national initiative and yet if only a handful of ontologies have been assessed for quality, it raises a concern about the quality of these ontologies provided to the research community. We believe that highlighting this concern stresses the need for further research in quality evaluation of biomedical ontologies.
Using our sample of 15 ontologies, we presented a taxonomic representation of various previous authors’ perspectives on the types of ontology evaluation, and mapped the techniques found in each of the ontology’s inception paper to the specific categories. Some of the categories were similar, e.g., the metrics in Tartir’s [6] (T1–T3) and Duque-Ramos et al. [27] (D1–D3), or the application-based approaches in Brank et al. [9] (BL1–BL6, BA1–BA4) and Obrst et al. [26] (O1–O6). With validation and verification methods, biomedical ontology evaluations appear to be concerned with intrinsic-level evaluation of the various types discussed in Section 2. We assumed that assessing whether the ontology was “built the right way” (verification) may be more convenient to test, versus whether the “right ontology was built” (validation). Also, the evaluation of six out of the 15 ontologies covered both validation and verification, which may indicate attempts of comprehensive evaluation. For the remaining ontologies, while they did have either validation-based or verification-based evaluation, the evaluation may not have been thorough enough to provide assurances of their “good” quality.
Aside from the dichotomy of validation and verification, there were four categorical schemes of ontology evaluation presented in the literature (i.e., Brank et al. [9], Obrst et al. [26], Tartir [6], and Duque-Ramos [27]). We measured the completeness, which is a percentage value of how well each category of ontology evaluation is covered by the evaluation practice of ontologies in BioPortal. Going strictly by analyzing which scheme provided the most “completeness” for ontologies, Duque-Ramos et al.’s [27] and Brank et al.’s [9] approaches (D1, D2, D3 and BA1, BA2, BA3, BA4 from Table 4, respectively) appear to provide a comprehensive description of evaluation types (31.1% and 31.7%, respectively). We refrain from making any assumption that a particular scheme is better suited. Nonetheless, it does highlight the deficiency in types of evaluation that are not considered in practice, such as lexical and syntactic level evaluations. Reality benchmarking from Obrst et al. [26], though an interesting approach, has not been made use of by any ontologies. The same can be said about community certification. Also worth mentioning, some techniques are similar in concepts (e.g., reporting metrics and competency questions), but based on our review of the literature, ontology evaluation techniques varied and there do not appear to be any standard or universally agreed upon approaches.
Overall, investigating and introducing evaluation techniques to help ontology developers make use of these criteria is a worthy endeavor. Past researchers have noted the need to automate the evaluation [28], and this direction can lead to a greater adoption of ontology evaluation. For example, Aruna and colleagues have surveyed various software tools that implement ontology evaluation, e.g., OntoAnalyser, OntoGenerator, OntoClean plugin, ONE-T, and S-OntoEval [105].
After surveying the recent OQA methods for biomedical ontologies published in the last seven years, we drew the same conclusion as Zhu et al. [19] that most of the OQA methods focused on content correctness, i.e., identifying modeling errors and inconsistencies of the components (e.g., classes, attributes, relations, and axioms) of the ontologies. The goals are mostly to improve the concept clarity, modeling consistency, non-redundancy, and soundness of biomedical ontologies. The fitness for purpose, on the other hand, is assumed to be automatically assured by the good quality of content. In addition, most of the recent methods make use of intrinsic ontology-specific knowledge as well as extrinsic domain knowledge of the human experts. The purely extrinsic resources are used mostly to assess the conceptual coverage of the ontologies. Compared with the review paper of Zhu et al. [19] that reviewed ontology-specific OQA methods, we found that quite a few recent OQA methods are designed to be applicable to multiple ontologies that exhibit similar characteristics (e.g., with object properties, with multiple parents). This addressed the need in OQA that is partly associated with the popularity of OWL-based application ontologies and the infrastructure such as Protégé and BioPortal to facilitate the development and reuse of ontologies. Similar to the finding in Zhu et al. [19], most of the OQA methods are largely automated, as many ontologies are large and complex. In contrast, some open domain ontology evaluation methods can be conducted manually.
5.2. Gaps in Ontology Quality Assurance Methods and Future Opportunities
While reviewing both open domain ontology evaluation and biomedical ontology quality assurance methods, we observed a few important gaps in OQA, including 1) the lack of systematic approaches to evaluate the OQA methods, 2) the lack of gold standards for ontology evaluation in the biomedicine domain, 3) the lack of collaborations between ontology evaluation and OQA communities, and 4) the lack of tools for ontology evaluation and OQA:
Lack of systematic approaches to evaluate the OQA methods
We observed that the evaluation of the OQA methods often involves domain experts to manually review the samples of algorithmically identified errors and inconsistencies, or subsets of concepts/relationships with a high likelihood of errors. In many OQA methods, domain experts were given blind samples of case and control to show the effectiveness of the OQA methods in generating a sample with more modeling errors and inconsistencies than a random sample. However, it may not always be feasible to make the samples completely blind. For example, a recent paper by He et al. [106] investigated the assignments of top-level semantic types to UMLS concepts, where the case sample included UMLS concepts assigned to one of the 10 top-level semantic types, whereas the control sample included UMLS concepts assigned to non-top-level semantic types. It is not feasible to make the samples blind because the auditors had to know the semantic type to which a concept is assigned. For such a case, the evaluation of the OQA methods may be biased. Systematic approaches to evaluate the OQA methods are thus needed to reduce the bias in the manual evaluation.
Lack of gold standards for ontology evaluation and OQA in the biomedicine domain
Due to the lack of gold standard ontologies in biomedicine, “gold standard” ontology evaluation (BA1 of Brank et al. [9]) was rarely seen. In fact, it is almost impossible to create a gold standard biomedical ontology. As such, the performance of extant OQA methods is often evaluated by domain experts with respect to precision but not recall. Nevertheless, even without a “gold standard”, one can leverage cumulative changes of high-quality ontologies such as SNOMED CT as a partial and surrogate standard to evaluate the performance of OQA methods for the ontology itself [107].
Lack of collaborations between ontology evaluation and OQA communities
We also observed that some approaches used in open domain ontology evaluation are omitted by the OQA research community. For example, application-specific ontology performance evaluation (BA1 of Brank et al. [9] and O1 of Obrst et al. [26]), and evaluation with competency questions and heuristics (BA4 of Brank et al. [9] and O5 of Obrst et al. [26]) are mostly omitted in the OQA literature. We hope to see more novel and effective OQA methods as well as closer collaboration of these two seemingly isolated communities in the future.
Lack of tools for ontology evaluation and OQA
Brank and colleagues have suggested in their seminal study, that automated tools are needed to ensure that high quality ontologies are developed [9]. Aruna and colleagues surveyed apparently rarely-used ontology evaluation tools [105] which included a tool that implemented Guarino and Welty’s OntoClean evaluation system. To our knowledge, there are still few known dedicated ontology evaluation tools that can help ontology engineers design and improve their ontologies. Future research is needed to evaluate the ontology evaluation and OQA tools.
Based on a recently proposed family-based QA framework [95] and a structural meta-ontology for ontology QA [71], the Ontology Abstraction Framework (OAF), a unified software framework for deriving, visualizing, and exploring abstraction networks for OWL and OBO-based ontologies [108], was recently released as both a standalone software tool as well as a plugin for Protégé 5, the most often used tool for ontology development in the world. With the better integration of the QA tool with the ontology development tool (i.e., Protégé), it is recommended that more and more ontology developers will use OAF to improve the quality of their ontologies, which will further improve their utility in knowledge management, knowledge integration, data analysis, and decision support. As Protégé is the most popular ontology development tool, more OQA plugins should be developed for Protégé to help ontology designers better integrate OQA into the ontology development life cycle.
5.3. Limitations
A few limitations should be noted for this study. Our initial survey was based on a sample of 200 ontologies from the NCBO BioPortal, which contains 686 ontologies in total (29%). This may be a limitation as there may be more ontologies that have early documented evidence of ontology evaluation but are not in our sample. We also restricted selection criteria to evaluation that took place at the time of development. We assumed that if ontologies are to be released to the community that it would be appropriate, for the original developers, to formally evaluate it. We believe it is good practice, similar to software development practices, to test and validate one’s ontology before deploying it for public consumption. However, evaluation, in the form of quality assurance or auditing, may have occurred after the original developers released their ontology artifact. So this was not accounted for in the survey.
Another direction would have been to focus on the OBO Foundry. While the sample did include some OBO Foundry ontologies, we only accounted for 15 of them. The NCBO BioPortal was chosen in this study because it includes a relatively big number of biomedical ontologies. On the same note, this survey was limited to ontologies that were biomedical ontologies. The survey may be more indicative of the ontologies from the biomedical domain than say the legal domain ontologies or cross-domain ontologies.
Another limitation to note is the nomenclature of OQA and ontology evaluation. We conducted an extensive literature review to understand the difference between ontology evaluation and quality assurance. It appears that even though they have much overlap with respect to methodologies, aspects, and tools, they do have some differences with respect to 1) timing (during the initial development vs. after the first public release), 2) major objective (measuring the quality of the ontology vs. identifying and correcting modeling errors and inconsistencies), and 3) the responsible party (original ontology developers vs. external QA researchers).
6. Conclusions
Ontology evaluation is an integral part of ontology development and maintenance. In this work, we organized four categorical schemes of ontology evaluation methods in the existing literature to create an integrated taxonomy of ontology evaluation. Further, to understand the ontology evaluation practice in the biomedicine domain, we reviewed a sample of 200 ontologies in NCBO BioPortal and observed that only 15 of these ontologies have documented evaluation in their corresponding inception papers. We then surveyed the recent quality assurance approaches for biomedical ontologies and their use. It is our expectation that ontology evaluation and quality assurance approaches will be more widely adopted in the development life cycle of the biomedical ontologies.
Highlights.
Ontology evaluation is an integral part of ontology development and maintenance.
We assessed the ontology evaluation practice of a sample of 200 BioPortal ontologies.
We reviewed recent ontology quality assurance and auditing techniques.
We identified the gaps between ontology evaluation and quality assurance.
Acknowledgments
This research was partially supported by the National Library of Medicine of the National Institutes of Health under Award Number R01 LM011829, the National Institute of Allergy and Infectious Diseases of the National Institutes of Health under Award Number R01 AI130460, the National Institutes of Health BD2K program under Award Number U01 HG009454, the National Cancer Institute of the National Institutes of Health under Award Number U24 CA194215, and the Cancer Prevention Research Institute of Texas (CPRIT) Training Grant #RP160015. The work was also partially supported by National Center for Advancing Translational Sciences under the Clinical and Translational Science Award UL1TR001427 (PI: Nelson & Shenkman). The content is solely the responsibility of the authors and does not represent the official view of the funding agencies.
Footnotes
MetaMap, https://metamap.nlm.nih.gov/
Declarations of interest: none
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Agrawal A, He Z, Perl Y, Wei D, Halper M, Elhanan G, Chen Y. The readiness of SNOMED problem list concepts for meaningful use of electronic health records. Artif Intell Med. 2013;58(2):73–80. doi: 10.1016/j.artmed.2013.03.008. [DOI] [PubMed] [Google Scholar]
- 2.Matney SA, Warren JJ, Evans JL, Kim TY, Coenen A, Auld VA. Development of the nursing problem list subset of SNOMED CT(R) J Biomed Inform. 2012;45(4):683–8. doi: 10.1016/j.jbi.2011.12.003. [DOI] [PubMed] [Google Scholar]
- 3.Rector A, Qamar R, Marley T. Binding ontologies and coding systems to electronic health records and messages. Applied Ontology. 2009;4(1):51–69. [Google Scholar]
- 4.Finnegan R. ICD-9-CM coding for physician billing. J Am Med Rec Assoc. 1989;60(2):22–3. [PubMed] [Google Scholar]
- 5.Bodenreider O. Biomedical ontologies in action: role in knowledge management, data integration and decision support. Yearb Med Inform. 2008:67–79. [PMC free article] [PubMed] [Google Scholar]
- 6.Tartir S. Ontological Evaluation and Validation. 2010. [Google Scholar]
- 7.Vrandečić D. Ontology Evaluation. In: Staab S, Studer R, editors. Handbook on Ontologies. Springer Berlin Heidelberg; Berlin, Heidelberg: 2009. pp. 293–313. [Google Scholar]
- 8.Kamdar MR, Tudorache T, Musen MA. A systematic analysis of term reuse and term overlap across biomedical ontologies. Semantic Web. 2016 doi: 10.3233/sw-160238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brank J, Grobelnik M, Mladenic D. A survey of ontology evaluation techniques. Proceedings of the Conference on Data Mining and Data Warehouses (SiKDD 2005); 2005. [Google Scholar]
- 10.Maedche A, Staab S. Measuring similarity between ontologies. Proceedings of the 13th International Conference on Knowledge Engineering and Knowledge Management. Ontologies and the Semantic Web; 2002. [Google Scholar]
- 11.Porzel R, Malaka R. A task-based approach for ontology evaluation. ECAI 2004 Workshop Ontology Learning and Population; 2004. [Google Scholar]
- 12.Brewster C, Alani H, Dasmahapatra S, Wilks Y. Data driven ontology evaluation. International Conference on Language Resources and Evaluation (LREC 2004); 2004; Lisbon, Portugal. [Google Scholar]
- 13.Lozano-Tello A, Gómez-Pérez A. Ontometric: A method to choose the appropriate ontology. J Datab Mgmt. 2004;15(2):1–18. [Google Scholar]
- 14.Hlomani H, Stacey AD. Contributing evidence to data- driven ontology evaluation: Workflow ontologies perspective. Proceedings of the 5th International Conference on Knowledge Engineering and Ontology Development; 2013; Vilamoura, Portugal. [Google Scholar]
- 15.Ouyang L, Zou B, Qu M, Zhang C. A method of ontology evaluation based on coverage, cohesion and coupling. The Eighth International Conference on Fuzzy Systems and Knowledge Discovery (FSKD); 2011; Shanghai, China. [Google Scholar]
- 16.Gómez-Pérez A. Ontology Evaluation. In: Staab S, Studer R, editors. Handbook on Ontologies. Springer; Berlin, Heidelberg: 2004. pp. 251–274. [Google Scholar]
- 17.Gómez-Pérez A, Fernández-López M, Corcho O. In: Ontological engineering: with examples from the areas of knowledge management, e-commerce and the SemanticWeb. Gomez-Perez A, Fernandez-Lopez M, Corcho O, editors. London: Springer; [Google Scholar]
- 18.Cook DL, Mejino JL, Rosse C. The foundational model of anatomy: a template for the symbolic representation of multi-scale physiological functions. Conf Proc IEEE Eng Med Biol Soc. 2004;7:5415–8. doi: 10.1109/IEMBS.2004.1404513. [DOI] [PubMed] [Google Scholar]
- 19.Zhu X, Fan JW, Baorto DM, Weng C, Cimino JJ. A review of auditing methods applied to the content of controlled biomedical terminologies. J Biomed Inform. 2009;42(3):413–25. doi: 10.1016/j.jbi.2009.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rogers JE. Quality assurance of medical ontologies. Methods of Information in Medicine. 2006;45:267–274. [PubMed] [Google Scholar]
- 21.Salvadores M, Alexander PR, Musen MA, Noy NF. BioPortal as a Dataset of Linked Biomedical Ontologies and Terminologies in RDF. Semant Web. 2013;4(3):277–284. [PMC free article] [PubMed] [Google Scholar]
- 22.OWL Web Ontology Language Overview. 2017 Apr 6; Available from: http://www.w3.org/TR/owl-features.
- 23.Resource Description Framework. 2017 Apr 6; Available from: http://www.w3.org/RDF/
- 24.The OBO Foundry Principles. 2017 Apr 6; Available from: http://www.obofoundry.org/principles/fp-000-summary.html.
- 25.Bodenreider O. The Unified Medical Language System (UMLS): integrating biomedical terminology. Nucleic Acids Res. 2004;32(Database issue):D267–70. doi: 10.1093/nar/gkh061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Obrst L, Ceusters W, Mani I, Ray S, Smith B. The Evaluation of Ontologies. Semantic Web. 2007:139–158. [Google Scholar]
- 27.Duque-Ramos A, Fernández-Breis JT, Stevens R, Aussenac-Gilles N. OQuaRE: A square-based approach for evaluating the quality of ontologies. Journal of Research and Practice in Information Technology. 2011;43:159–176. [Google Scholar]
- 28.Burton-Jones A, Storey CV, Sugumaran V, Ahluwalia P. A semiotic metrics suite for assessing the quality of ontologies. Data & Knowledge Engineering. 2005;55(1):84–102. [Google Scholar]
- 29.Ceusters W, Smith B. A realism-based approach to the evolution of biomedical ontologies. AMIA … Annual Symposium proceedings/AMIA Symposium. AMIA Symposium; 2006. pp. 121–125. [PMC free article] [PubMed] [Google Scholar]
- 30.Amith M, Tao C. A Web Application Towards Semiotic-based Evaluation of Biomedical Ontologies. 2015 [Google Scholar]
- 31.Amith M, Tao C. Modulated Evaluation Metrics for Drug-Based Ontologies. Journal of biomedical semantics. 2017;8(1):17. doi: 10.1186/s13326-017-0124-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Drame K, Diallo G, Delva F, Dartigues JF, Mouillet E, Salamon R, Mougin F. Reuse of termino-ontological resources and text corpora for building a multilingual domain ontology: an application to Alzheimer’s disease. J Biomed Inform. 2014;48:171–82. doi: 10.1016/j.jbi.2013.12.013. [DOI] [PubMed] [Google Scholar]
- 33.Lin Y, He Y. The ontology of genetic susceptibility factors (OGSF) and its application in modeling genetic susceptibility to vaccine adverse events. Journal of biomedical semantics. 2014;5:19. doi: 10.1186/2041-1480-5-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ganzinger M, He S, Breuhahn K, Knaup P. On the Ontology Based Representation of Cell Lines. PLoS ONE. 2012:7. doi: 10.1371/journal.pone.0048584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Samwald M, Miñarro Giménez JA, Boyce RD, Freimuth RR, Adlassnig K-p, Dumontier M. Pharmacogenomic knowledge representation, reasoning and genome-based clinical decision support based on OWL 2 DL ontologies. BMC Medical Informatics and Decision Making. 2015;15:12. doi: 10.1186/s12911-015-0130-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sahoo SS, Weatherly DB, Mutharaju R, Anantharam P, Sheth A, Tarleton RL. Ontology-driven provenance management in eScience: An application in parasite research. Lecture Notes in Computer Science (including subseries Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics) 2009:992–1009. [Google Scholar]
- 37.Thomas CJ, Sheth AP, York WS. Modular ontology design using canonical building blocks in the biochemistry domain. Frontiers in Artificial Intelligence and Applications. 2006:115–127. [Google Scholar]
- 38.Cook DL, Mejino JLV, Neal ML, Gennari JH. Bridging Biological Ontologies and Biosimulation: The Ontology of Physics for Biology. AMIA Annual Symposium Proceedings. 2008;2008:136–140. [PMC free article] [PubMed] [Google Scholar]
- 39.Pratt J, Pandian V, Morrison E, Miller AA. Developing a tool for crowd-sourced verification of a radiation oncology ontology: a summer project. 2014 [Google Scholar]
- 40.Panov P, Soldatova L, Džeroski S. Ontology of core data mining entities. Data Mining and Knowledge Discovery. 2014;28:1222–1265. [Google Scholar]
- 41.McCray AT, Trevvett P, Frost HR. Modeling the autism spectrum disorder phenotype. Neuroinformatics. 2014;12:291–305. doi: 10.1007/s12021-013-9211-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Coulet A, Garten Y, Dumontier M, Altman RB, Musen Ma, Shah NH. Integration and publication of heterogeneous text-mined relationships on the Semantic Web. Journal of biomedical semantics. 2011;2(Suppl 2):S10. doi: 10.1186/2041-1480-2-S2-S10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schober D, Boeker M, Bullenkamp J, Huszka C, Depraetere K, Teodoro D, Nadah N, Choquet R, Daniel C, Schulz S. The DebugIT core ontology: Semantic integration of antibiotics resistance patterns. Studies in Health Technology and Informatics. 2010;160:1060–1064. [PubMed] [Google Scholar]
- 44.Malhotra A, Younesi E, Gündel M, Müller B, Heneka MT, Hofmann-Apitius M. ADO: A disease ontology representing the domain knowledge specific to Alzheimer’s disease. Alzheimer’s and Dementia. 2014;10:238–246. doi: 10.1016/j.jalz.2013.02.009. [DOI] [PubMed] [Google Scholar]
- 45.Gündel M, Younesi E, Malhotra A, Wang J, Li H, Zhang B, de Bono B, Mevissen H-T, Hofmann-Apitius M. HuPSON: the human physiology simulation ontology. Journal of biomedical semantics. 2013;4:35. doi: 10.1186/2041-1480-4-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Van Soest J, Lustberg T, Grittner D, Marshall MS, Persoon L, Nijsten B, Feltens P, Dekker A. Towards a semantic PACS: Using Semantic Web technology to represent imaging data. Studies in Health Technology and Informatics. 2014:166–170. [PMC free article] [PubMed] [Google Scholar]
- 47.Tsarkov D, Horrocks I. FaCT++ description logic reasoner: system description. Proceedings of the Third international joint conference on Automated Reasoning; 2006; Seattle, WA. [Google Scholar]
- 48.Shearer R, Motik B, Horrocks I. HermiT: a highly-efficient OWL reasoner. Proc 5th International Workshop on OWL: Experiences and Directions (OWLED); 2008. [Google Scholar]
- 49.Gruber TR. Toward Principles for the Design of Ontologies Used for Knowledge Sharing. International Journal of Human-Computer Studies. 1993;43:907–28. [Google Scholar]
- 50.Zhang GQ, Luo L, Ogbuji C, Joslyn C, Mejino J, Sahoo SS. An analysis of multi-type relational interactions in FMA using graph motifs with disjointness constraints. AMIA Annu Symp Proc. 2012;2012:1060–9. [PMC free article] [PubMed] [Google Scholar]
- 51.Gu HH, Wei D, Mejino JL, Jr, Elhanan G. Relationship auditing of the FMA ontology. J Biomed Inform. 2009;42(3):550–7. doi: 10.1016/j.jbi.2009.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mougin F, Grabar N. Auditing the multiply-related concepts within the UMLS. J Am Med Inform Assoc. 2014;21(e2):e185–93. doi: 10.1136/amiajnl-2013-002227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Luo L, Xu R, Zhang GQ. Dissecting the Ambiguity of FMA Concept Names Using Taxonomy and Partonomy Structural Information. AMIA Jt Summits Transl Sci Proc. 2013;2013:157–61. [PMC free article] [PubMed] [Google Scholar]
- 54.Gu H, Elhanan G, Halper M, He Z. Questionable Relationship Triples in the UMLS. Proceedings of 2012 IEEE-EMBS International Conference on Biomedical and Health Informatics; 2012; Shenzhen, China. [Google Scholar]
- 55.Mougin F. Identifying redundant and missing relations in the gene ontology. Stud Health Technol Inform. 2015;210:195–9. [PubMed] [Google Scholar]
- 56.Agrawal A, Elhanan G. Contrasting lexical similarity and formal definitions in SNOMED CT: consistency and implications. J Biomed Inform. 2014;47:192–8. doi: 10.1016/j.jbi.2013.11.003. [DOI] [PubMed] [Google Scholar]
- 57.Luo L, Mejino JL, Jr, Zhang GQ. An analysis of FMA using structural self-bisimilarity. J Biomed Inform. 2013;46(3):497–505. doi: 10.1016/j.jbi.2013.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rector A, Iannone L. Lexically suggest, logically define: quality assurance of the use of qualifiers and expected results of post-coordination in SNOMED CT. J Biomed Inform. 2012;45(2):199–209. doi: 10.1016/j.jbi.2011.10.002. [DOI] [PubMed] [Google Scholar]
- 59.Quesada-Martinez M, Fernandez-Breis JT, Karlsson D. Suggesting Missing Relations in Biomedical Ontologies Based on Lexical Regularities. Stud Health Technol Inform. 2016;228:384–8. [PubMed] [Google Scholar]
- 60.Bodenreider O. Identifying Missing Hierarchical Relations in SNOMED CT from Logical Definitions Based on the Lexical Features of Concept Names. ICBO/BioCreative. 2016;2016 [PMC free article] [PubMed] [Google Scholar]
- 61.Geller J, He Z, Perl Y, Morrey CP, Xu J. Rule-based support system for multiple UMLS semantic type assignments. J Biomed Inform. 2013;46(1):97–110. doi: 10.1016/j.jbi.2012.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mougin F, Bodenreider O, Burgun A. Analyzing polysemous concepts from a clinical perspective: application to auditing concept categorization in the UMLS. J Biomed Inform. 2009;42(3):440–51. doi: 10.1016/j.jbi.2009.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.He Z, Morrey CP, Perl Y, Elhanan G, Chen L, Chen Y, Geller J. Sculpting the UMLS Refined Semantic Network. Online J Public Health Inform. 2014;6(2):e181. doi: 10.5210/ojphi.v6i2.5412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jiang G, Chute CG. Auditing the semantic completeness of SNOMED CT using formal concept analysis. J Am Med Inform Assoc. 2009;16(1):89–102. doi: 10.1197/jamia.M2541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhu W, Cui L, Zhang GQ. Spark-MCA: Large-scale, Exhaustive Formal Concept Analysis for Evaluating the Semantic Completeness of SNOMED CT. AMIA Annu Symp Proc. 2017;2017:1914–23. [PMC free article] [PubMed] [Google Scholar]
- 66.Wang Y, Halper M, Wei D, Gu H, Perl Y, Xu J, Elhanan G, Chen Y, Spackman KA, Case JT, Hripcsak G. Auditing complex concepts of SNOMED using a refined hierarchical abstraction network. J Biomed Inform. 2012;45(1):1–14. doi: 10.1016/j.jbi.2011.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Halper M, Gu H, Perl Y, Ochs C. Abstraction networks for terminologies: Supporting management of “big knowledge”. Artif Intell Med. 2015;64(1):1–16. doi: 10.1016/j.artmed.2015.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.He Z, Ochs C, Soldatova L, Perl Y, Arabandi S, Geller J. Auditing Redundant Import in Reuse of a Top Level Ontology for the Drug Discovery Investigations Ontology. International Workshop on Vaccine and Drug Ontology Studies; 2013; Montreal, QC, Canada. [Google Scholar]
- 69.Ochs C, Agrawal A, Perl Y, Halper M, Tu SW, Carini S, Sim I, Noy N, Musen M, Geller J. Deriving an abstraction network to support quality assurance in OCRe. AMIA Annu Symp Proc. 2012;2012:681–9. [PMC free article] [PubMed] [Google Scholar]
- 70.Ochs C, He Z, Perl Y, Arabandi S, Halper M, Geller J. Choosing the Granularity of Abstraction Networks for Orientation and Quality Assurance of the Sleep Domain Ontology. The 4th International Conference on Biomedical Ontology; 2013; Montreal, QC, Canada. [Google Scholar]
- 71.Ochs C, He Z, Zheng L, Geller J, Perl Y, Hripcsak G, Musen MA. Utilizing a structural meta-ontology for family-based quality assurance of the BioPortal ontologies. J Biomed Inform. 2016;61:63–76. doi: 10.1016/j.jbi.2016.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ochs C, Perl Y, Geller J, Haendel M, Brush M, Arabandi S, Tu S. Summarizing and visualizing structural changes during the evolution of biomedical ontologies using a Diff Abstraction Network. J Biomed Inform. 2015;56:127–44. doi: 10.1016/j.jbi.2015.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Perl Y, Ochs C, de Coronado S, Thomas N. Visualizing the “Big Picture” of Change in NCIt’s Biological Processes. International Conference on Biomedical Ontology. CEUR-ws.org; 2016. [Google Scholar]
- 74.Ochs C, Perl Y, Halper M, Geller J, Lomax J. Quality assurance of the gene ontology using abstraction networks. J Bioinform Comput Biol. 2016;14(3):1642001. doi: 10.1142/S0219720016420014. [DOI] [PubMed] [Google Scholar]
- 75.Min H, Zheng L, Perl Y, Halper M, De Coronado S, Ochs C. Relating Complexity and Error Rates of Ontology Concepts. More Complex NCIt Concepts Have More Errors. Methods Inf Med. 2017 doi: 10.3414/ME16-01-0085. [DOI] [PubMed] [Google Scholar]
- 76.Zheng L, Yumak H, Chen L, Ochs C, Geller J, Kapusnik-Uner J, Perl Y. Quality assurance of chemical ingredient classification for the National Drug File - Reference Terminology. J Biomed Inform. 2017;73:30–42. doi: 10.1016/j.jbi.2017.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhang GQ, Zhu W, Sun M, Tao S, Bodenreider O, Cui L. MaPLE: A MapReduce Pipeline for Lattice-based Evaluation and Its Application to SNOMED CT. Proc IEEE Int Conf Big Data. 2014;2014:754–759. doi: 10.1109/BigData.2014.7004301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Cui L, Tao S, Zhang GQ. Biomedical Ontology Quality Assurance Using a Big Data Approach. ACM Transactions on Knowledge Discovery from Data. 2016;10(4):41. [Google Scholar]
- 79.Zhang GQ, Bodenreider O. Large-scale, Exhaustive Lattice-based Structural Auditing of SNOMED CT. AMIA Annu Symp Proc. 2010;2010:922–6. [PMC free article] [PubMed] [Google Scholar]
- 80.Mortensen JM, Minty EP, Januszyk M, Sweeney TE, Rector AL, Noy NF, Musen MA. Using the wisdom of the crowds to find critical errors in biomedical ontologies: a study of SNOMED CT. J Am Med Inform Assoc. 2015;22(3):640–8. doi: 10.1136/amiajnl-2014-002901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Mortensen JM, Telis N, Hughey JJ, Fan-Minogue H, Van Auken K, Dumontier M, Musen MA. Is the crowd better as an assistant or a replacement in ontology engineering? An exploration through the lens of the Gene Ontology. J Biomed Inform. 2016;60:199–209. doi: 10.1016/j.jbi.2016.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Mortensen JM, Musen MA, Noy NF. Crowdsourcing the verification of relationships in biomedical ontologies. AMIA Annu Symp Proc. 2013;2013:1020–9. [PMC free article] [PubMed] [Google Scholar]
- 83.Rector AL, Brandt S, Schneider T. Getting the foot out of the pelvis: modeling problems affecting use of SNOMED CT hierarchies in practical applications. J Am Med Inform Assoc. 2011;18(4):432–40. doi: 10.1136/amiajnl-2010-000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Gu H, Chen Y, He Z, Halper M, Chen L. Quality Assurance of UMLS Semantic Type Assignments Using SNOMED CT Hierarchies. Methods Inf Med. 2016;55(2):158–65. doi: 10.3414/ME14-01-0104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wei D, Halper M, Elhanan G. Using SNOMED semantic concept groupings to enhance semantic-type assignment consistency in the UMLS. Proceedings of the 2nd ACM SIGHIT International Health Informatics Symposium; 2012; Washington D.C. [Google Scholar]
- 86.Cui L. COHeRE: Cross-Ontology Hierarchical Relation Examination for Ontology Quality Assurance. AMIA Annu Symp Proc. 2015;2015:456–65. [PMC free article] [PubMed] [Google Scholar]
- 87.Agrawal A, Perl Y, Ochs C, Elhanan G. Algorithmic detection of inconsistent modeling among SNOMED CT concepts by combining lexical and structural indicators. 2015 IEEE International Conference on Bioinformatics and Biomedicine (BIBM); 2015; Washington, DC. [Google Scholar]
- 88.Wei D, Bodenreider O. Using the abstraction network in complement to description logics for quality assurance in biomedical terminologies - a case study in SNOMED CT. Stud Health Technol Inform. 2010;160(Pt 2):1070–4. [PMC free article] [PubMed] [Google Scholar]
- 89.Cui L, Zhu W, Tao S, Case JT, Bodenreider O, Zhang GQ. Mining non-lattice subgraphs for detecting missing hierarchical relations and concepts in SNOMED CT. J Am Med Inform Assoc. 2017;24(4):788–798. doi: 10.1093/jamia/ocw175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Yao L, Divoli A, Mayzus I, Evans JA, Rzhetsky A. Benchmarking ontologies: bigger or better? PLoS Comput Biol. 2011;7(1):e1001055. doi: 10.1371/journal.pcbi.1001055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Park MS, He Z, Chen Z, Oh S, Bian J. Consumers’ Use of UMLS Concepts on Social Media: Diabetes-Related Textual Data Analysis in Blog and Social Q&A Sites. JMIR Med Inform. 2016;4(4):e41. doi: 10.2196/medinform.5748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.He Z, Halper M, Perl Y, Elhanan G. Clinical Clarity versus Terminological Order -The Readiness of SNOMED CT Concept Descriptors for Primary Care. MIXHS 12 (2012) 2012;2012:1–6. doi: 10.1145/2389672.2389674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Fung KW, Xu J. An exploration of the properties of the CORE problem list subset and how it facilitates the implementation of SNOMED CT. J Am Med Inform Assoc. 2015;22(3):649–58. doi: 10.1093/jamia/ocu022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Gu H, Perl Y, Geller J, Halper M, Liu LM, Cimino JJ. Representing the UMLS as an object-oriented database: modeling issues and advantages. J Am Med Inform Assoc. 2000;7(1):66–80. doi: 10.1136/jamia.2000.0070066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.He Z, Ochs C, Agrawal A, Perl Y, Zeginis D, Tarabanis K, Elhanan G, Halper M, Noy N, Geller J. A family-based framework for supporting quality assurance of biomedical ontologies in BioPortal. AMIA Annu Symp Proc. 2013;2013:581–90. [PMC free article] [PubMed] [Google Scholar]
- 96.Ochs C, Geller J, Perl Y, Chen Y, Agrawal A, Case JT, Hripcsak G. A tribal abstraction network for SNOMED CT target hierarchies without attribute relationships. J Am Med Inform Assoc. 2015;22(3):628–39. doi: 10.1136/amiajnl-2014-003173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Ochs C, Geller J, Perl Y, Chen Y, Xu J, Min H, Case JT, Wei Z. Scalable quality assurance for large SNOMED CT hierarchies using subject-based subtaxonomies. J Am Med Inform Assoc. 2015;22(3):507–18. doi: 10.1136/amiajnl-2014-003151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.He Z, Geller J, Elhanan G. Categorizing the Relationships between Structurally Congruent Concepts from Pairs of Terminologies for Semantic Harmonization. AMIA Jt Summits Transl Sci Proc. 2014;2014:48–53. [PMC free article] [PubMed] [Google Scholar]
- 99.He Z, Geller J, Chen Y. A comparative analysis of the density of the SNOMED CT conceptual content for semantic harmonization. Artif Intell Med. 2015;64(1):29–40. doi: 10.1016/j.artmed.2015.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.He Z, Chen Y, de Coronado S, Piskorski K, Geller J. Topological-Pattern-Based Recommendation of UMLS Concepts for National Cancer Institute Thesaurus. AMIA Annu Symp Proc. 2016;2016:618–627. [PMC free article] [PubMed] [Google Scholar]
- 101.Wang Y, Halper M, Min H, Perl Y, Chen Y, Spackman KA. Structural methodologies for auditing SNOMED. J Biomed Inform. 2007;40(5):561–81. doi: 10.1016/j.jbi.2006.12.003. [DOI] [PubMed] [Google Scholar]
- 102.Elhanan G, Perl Y, Geller J. A survey of SNOMED CT direct users, 2010: impressions and preferences regarding content and quality. J Am Med Inform Assoc. 2011;18(Suppl 1):i36–44. doi: 10.1136/amiajnl-2011-000341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Ceusters W. Applying Evolutionary Terminology Auditing to SNOMED CT. AMIA Annu Symp Proc. 2010;2010:96–100. [PMC free article] [PubMed] [Google Scholar]
- 104.Gómez-Pérez A. Some ideas and examples to evaluate ontologies. 11th Conference of Artificial Intelligence for Applications; 1995. p. 299. [Google Scholar]
- 105.Aruna T, Saranya K, Bhandari C. A survey on ontology evaluation tools. Proceedings of 2011 International Conference on Process Automation, Control and Computing, PACC 2011; 2011. [Google Scholar]
- 106.He Z, Perl Y, Elhanan G, Chen Y, Geller J, Bian J. Auditing the Assignments of Top-Level Semantic Types in the UMLS Semantic Network to UMLS Concepts. Proceedings of 2017 IEEE International Conference on Bioinformatics and Biomedicine; 2017; Kansas City, MO: IEEE; pp. 1262–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Zhang GQ, Huang Y, Cui L. Can SNOMED CT Changes Be Used as a Surrogate Standard for Evaluating the Performance of Its Auditing Methods? AMIA Annu Symp Proc. 2017;2017:1886–95. [PMC free article] [PubMed] [Google Scholar]
- 108.Ochs C, Geller J, Perl Y, Musen MA. A unified software framework for deriving, visualizing, and exploring abstraction networks for ontologies. J Biomed Inform. 2016;62:90–105. doi: 10.1016/j.jbi.2016.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]