Abstract
Novel methods to analyze the tumor microenvironment (TME) are urgently needed to stratify melanoma patients for adjuvant immunotherapy. Tumor infiltrating lymphocyte (TIL) analysis, by conventional pathologic methods, is predictive but is insufficiently precise for clinical application. Quantitative multiplex immunofluorescence (qmIF), allows for evaluation of the TME using multiparameter phenotyping, tissue segmentation, and quantitative spatial analysis (qSA). Given that CD3+CD8+ cytotoxic lymphocytes (CTLs) promote antitumor immunity, whereas CD68+ macrophages impair immunity, we hypothesized that quantification and spatial analysis of macrophages and CTLs would correlate with clinical outcome. We applied qmIF to 104 primary stage II-III melanoma tumors and found that CTLs were closer in proximity to activated (CD68+HLA-DR+) macrophages than non-activated (CD68+HLA-DR−) macrophages (p<0.0001). CTLs were further in proximity from proliferating SOX10+ melanoma cells than non-proliferating ones (p<0.0001). In 64 patients with known cause of death, we found that high CTL and low macrophage density in the stroma (p=0.0038 and p=0.0006, respectively) correlated with disease-specific survival (DSS), but the correlation was less significant for CTL and macrophage density in the tumor (p=0.0147 and p=0.0426, respectively). DSS correlation was strongest for stromal HLA-DR+ CTLs (p=0.0005). CTL distance to HLA-DR− macrophages associated with poor DSS (p=0.0016), whereas distance to Ki67− tumor cells associated inversely with DSS (p=0.0006). A low CTL/macrophage ratio in the stroma conferred a hazard ratio (HR) of 3.719 for death from melanoma and correlated with shortened overall survival (OS) in the complete 104 patient cohort by Cox analysis (p=0.009) and merits further development as a biomarker for clinical application.
Keywords: Melanoma, Biomarker, Novel Assay Technology, Tumor Immunobiology, Immunofluorescence
Introduction
Cutaneous melanoma is an aggressive disease, even when localized, with 40% of patients with stage II-III melanoma developing recurrent disease (1). Current histopathologic methods, including conventional immunohistochemistry (IHC) evaluation of formalin-fixed paraffin-embedded (FFPE) tissues, have been unable to identify precise biomarkers capable of identifying patients at greatest risk for recurrence (2). Advancements in clinically applicable histopathologic methods for evaluating the tumor microenvironment (TME) are essential for patient care and, in particular, selecting patients to receive immunotherapies. In pre-clinical research settings, flow cytometry (FC) offers detailed evaluation of infiltrating immune cells and supplements conventional IHC. However, clinical application of FC is limited because FC can only be performed on fresh tissue. Obtaining fresh tissue is only feasible when a large volume of excess tumor is available, a rare occurrence for primary melanomas, and harvesting fresh tissue requires close coordination between multiple clinicians in an already overburdened health care setting. In contrast, FFPE samples are routinely banked on every patient who has tumor resection and/or biopsy and is a much more practical source of material for immune phenotyping.
In early stage melanoma, traditional tumor, node, and metastasis (TNM) staging does not precisely assess the risk of recurrence of an individual patient. The American Joint Committee on Cancer (AJCC) defines stage II melanoma based on tumor depth >1.01mm with ulceration and >2.01mm with or without ulceration, and stage III disease is defined by locoregional metastasis (1, 3). Stage III disease is further sub-classified based on volume of lymph node metastasis (1, 3). Unfortunately, complete staging is often not performed because patients are reluctant to undergo lymph node dissection, particularly in light of studies demonstrating insignificant impact on clinical outcomes (3, 4). Even for patients who do undergo full clinical staging, a high degree of heterogeneity among patients within the same stage group remains, evidenced by the 50% survival rate at 5 years for stage IIC disease (1). Thus, significant limitations to traditional stage classification in melanoma exist, impeding identification of high-risk patients who might be expected to obtain the greatest benefit from either enhanced radiographic surveillance or adjuvant immunotherapy (IO).
IO, which has changed the clinical landscape of metastatic melanoma, is most effective in lower disease burden, when tumor-mediated immunosuppression is minimal (5). Thus, IO has the potential to be effective in the adjuvant setting. However, although these agents have demonstrated efficacy in melanoma, they also introduce the potential for immune adverse effects (IAE), including colitis, pneumonitis, and myocarditis (6, 7). In the metastatic setting there is a higher tolerance for IAEs. However, the lack of precise predictive tools has contributed to stalling the application of IO to the adjuvant setting due to both the safety concerns and expense of conducting these trials in large populations that include many low-risk patients. Thus, prognostic biomarkers are urgently needed to identify which patients are at greatest risk for recurrence and for stratification of clinical trials (8).
Tumor-infiltrating lymphocytes (TILs), a heterogeneous group of lymphocytes within the TME, are a positive prognosticator in melanoma (9-16). TIL assessment, however, is subject to significant inter-observer variability, thus, limiting clinical utility (11, 12). Detailed analysis of TILs shows that CD8+ cytotoxic T lymphocytes (CTLs) correlate best with survival, but generalizable and reproducible methods to analyze TIL composition have not yet been developed (17). Other cells of importance in the TME are macrophages. Although the role of macrophages has been controversial, discrepancies in reported data is attributable to diverse subgroups of macrophages that either support an effective antitumor response (M1 type) or promote tumor growth (M2 type)(18-21). M2 macrophages are capable of producing enzymes, including indoleamine-pyrrole 2,3-dioxygenase (IDO) and arginase, which inhibit CTL activity through tryptophan and L-arginine depletion (22). M2 macrophages have also been found to express PD-L1,the known inhibitory ligand of the PD-1 receptor found on T-lymphocytes (20). In addition to modulating CTL function, macrophages are implicated in tumor growth, demonstrating capacity to promote neovascularization through production of angiogenic peptides, including VEGF (23). Given that these immunosuppressive mechanisms require either direct contact or close proximity to CTLs, strong biologic rationale exists for pathologic approaches capable of evaluating the spatial relationships between these immune subsets. Although proximity analysis is a limited surrogate for potential cellular interactions, prior clinicopathologic data in melanoma supports the utility of analyzing spatial localization of immune cells within tumors (24). Thus, the immune score takes into account the position of lymphocyte infiltration within the TME. Macrophage infiltration, specifically when present at the invasive front but not within the tumor as a whole, has been shown to correlate with poor survival in stage I/II disease (25). Although CTLs are known to play a protective role and macrophages a deleterious one, these findings have not been applied to clinical practice (26, 27).
In order to address the need for detailed phenotyping of immune infiltrates in FFPE samples, we apply quantitative multiplex immunofluorescence (qmIF) to analyze the TME in primary melanoma tumors (28). QmIF employs a serial IHC approach that includes the application of fluorophores of distinct wavelengths, allowing for staining and visualization of multiple antigens within the same tissue specimen. Subsequently, multispectral, high-resolution images are obtained, which are then analyzed using machine learning algorithms. This approach allows for more sophisticated computational analysis, including automated phenotyping for objective density and spatial assessment of immune cell phenotypes within the TME (28-35). QmIF requires iterative training to develop tissue and cell classification algorithms (Supplementary Fig. S2-5) and can ultimately augment analysis capacities beyond traditional IHC evaluation (31-33, 35).
In addition to enhanced automated quantitation of cell phenotypes over manual review by a pathologist, qmIF is capable of analyzing cells based on tissue segmentation to determine whether cells are located in tumor or stromal compartments. Digital pathology using qmIF offers the ability to perform quantitative spatial analysis (qSA) to evaluate the proximity of individual immune cell subsets to each other, as well as to tumor cells (28). Precise tissue segmentation and localization of immune cells is not evaluable with traditional IHC because the manual nature of analysis is inherently subjective and introduces observer bias. Digital pathology approaches, employing machine-based learning algorithms to evaluate large tissue areas and complex cell phenotypes, have been validated across multiple solid tumor types (28-38). Several studies employ qSA to study tissue segmentation and distance relationships of multiple cell phenotypes (32, 33, 36-39). The importance of tissue segmentation to define precise location of lymphocytes within the TME is established by the Immunoscore method, wherein the leading edge of the tumor is differentiated from the center of the tumor (2). However, location of the leading tumor edge generally requires expert pathology guidance. Thus, Immunoscore has not been applied to primary melanoma, in part, because it is complicated to reproducibly define the leading edge in these small and heterogeneous neoplasms (8, 32-35). Prior IHC work, including Immunoscore, does not provide qSA of the location of immune cells relative to other cells within the tumor.
In prior work, to more comprehensively characterize the TME, as well as identify a 53 immune gene mRNA panel, our group applied a NanoString assay, comprised principally of genes associated with a Th1 immune response, which correlates with disease-specific survival (DSS) in two independent patient populations (13). NanoString assays are both quantitative and non-subjective but are more costly than IHC and require homogenization of tissue, precluding evaluation of spatial localization of immune cells. Although the NanoString data did not suggest a prominent role for macrophage lineage cells in tumor progression, this might have been due to the lack of information regarding spatial localization of the macrophages. We hypothesized that further spatial characterization of the macrophages, together with additional phenotypic markers, might clarify the role of macrophages in tumor progression. For example, macrophages at the leading tumor edge mark high-risk patients (40). Markers of activation and antigen presentation, including HLA-DR, aid in the separation of macrophage phenotypes, such that HLA-DR− macrophages are more likely to be inflammatory and HLA-DR+ macrophages are more likely to favor antitumor immunity (41-43).
Thus, to further expand our capacity to assess the TME, we use qmIF to evaluate stage II-III primary melanoma. We included CTLs and macrophages as proposed biomarkers, while adding HLA-DR and Ki67 for further evaluation due to previous studies showing the addition of these markers to be important features of immune cell activation (8, 30, 40, 44). Ina cohort of 104 patients (Table 1), we performed tissue segmentation, cellular phenotyping, automated density, and qSA of immune cell subsets. We find that low CTL/macrophage ratio in the stroma correlated with shortened DSS and overall survival (OS) in stage II-III melanoma.
Table 1. Melanoma Patient Characteristics of the Overall Survival (OS) Cohort.
| Clinical Characteristics | |
| Gender | |
| Male, no. (%) | 75 (72.1) |
| Female, no. (%) | 29 (27.9) |
| Age | |
| Median, no. (range) | 74.5 (22-96) |
| Location of Tumor | |
| Trunk, no. (%) | 61 (58.7) |
| Extremity, no. (%) | 41 (39.4) |
| Unknown, no. (%) | 2 (1.9) |
| Stage | |
| II, no. (%) | 91 (87.5) |
| III, no. (%) | 13 (12.5) |
|
| |
| Pathological Characteristics | |
| Depth (mm) | |
| Median, no. (range) | 2.5 (0.6-26) |
| Ulceration | |
| Absent, no. (%) | 36 (34.6) |
| Present, no. (%) | 65 (62.5) |
| Unknown, no. (%) | 3 (2.9) |
| TILs | |
| Absent, no. (%) | 2 (1.9) |
| Non-Brisk, no. (%) | 59 (56.8) |
| Brisk, no. (%) | 33 (31.7) |
| Unknown, no. (%) | 10 (9.6) |
|
| |
| Outcome Characteristics | |
| Patient Follow Up (months) | |
| Median, no. (range) | 45 (4-173) |
| OS (months) | |
| Alive (at least 2 years), no. (%) | 31 (29.8) |
| Dead, no. (%) | 73 (70.2) |
| DSS (months) | |
| Alive or NED at Death, no. (%) | 42 (40.4) |
| Dead with melanoma, no. (%) | 22 (21.2) |
| Unknown, no. (%) | 40 (38.4) |
(TILs = Tumor Infiltrating Lymphocytes, OS = Overall Survival, DSS = Disease Specific Survival, NED = No Evidence of Disease)
Methods
Patients and Samples
The study was approved by Columbia University Medical Center's (CUMC) Institutional Review Board. A patient database of stage II-III melanoma patients at CUMC was created retrospectively by searching surgical pathology records from 2000-2012 for “melanoma”. Dermatopathology reports were first screened retrospectively for tumor size, finding tumors 1.01– 2.00 mm with ulceration and 2.01 – 10mm. After reviewing 1139 patient records, we identified 765 stage II-III patients. Of these 765 patients, 219 had available survival data, defined as known date of death and/or 24 months documented clinical follow-up. Hematoxylin and eosin (H&E) slides were reviewed with a dermatopathologist, and 183 patients had confirmed tumors, whereas 36 had no melanoma in the residual specimen. Wide local excision and post biopsy specimens were excluded due to concerns for immune infiltrate post biopsy, thus, leaving 142 patients. During the staining process 17 were lost due to tissue destruction, 19 had staining artifact, 4 were too small, and 2 were unable to be analyzed using the image analysis software, leaving a total of 104 analyzable samples (Supplementary Fig. S1 and Supplementary Table S1). 104 patients were analyzed (Table 1), 64 with known recurrence status at the time of last follow-up or death (Supplementary Table S2), and 40 patients with unknown cause of death. This cohort consists of more stage II (n=91) than stage III (n=13) and more males (n=75) than females (n=29). Median depth was 2.5 mm, and 65% of tumors were ulcerated. 59 and 33 patients had non-brisk TILs and brisk TILs, respectively, as determined by dermatopathologist (BH), whereas 2 had absent TILs and 10 were not assessed due to absence of the tumor base (16).
Staining
Full section 5 μm slides of tissue specimens were stained using Opal™ multiplex 6-plex kits, according to the manufacturer's protocol (PerkinElmer), for DAPI, CD3 (clone LN10; Leica (Buffalo Grove, IL); 1:200 dilution), CD8 (clone 4B11; Leica; Ready to use (RTU)), CD68 (clone KP1; Biogenex (Fremont, CA), RTU); SOX10 (clone BC34; Biocare (Pacheco, CA); 1:300), HLA-DR (clone LN-3; Abcam (Cambridge, MA); 1:200 dilution), and Ki67 (clone MIB1; Abcam; RTU). Opal multiplexing is a serial immunohistochemistry method that relies on tyramide signal amplification (TSA)(30, 45), which creates an amplification of signal that then covalently binds to the epitope in a specific manner (27, 46). Primary and secondary antibody complexes are subsequently removed for serial immunofluorescence, while the covalent fluorescent signal remains. Single controls and an unstained slide were stained with each group of slides.
Multispectral Imaging
H&E slides were viewed by a dermatopathologist to determine representative areas for multispectral image capture at 20× magnification using Mantra™ (PerkinElmer Images were analyzed using inform™ software (PerkinElmer) (Fig. 1A-G, Supplementary Fig. S2-5). Five representative areas were chosen by the dermatopathologist: (i) three areas with tumor and up to 50% stroma and (ii) two areas with tumor only (at least 90% tumor). These images were factored equally into the analysis for each patient. Mantra™ captures spectral information from a multiplexed panel of targets using a multispectral camera. For samples of small size, a minimum of two areas meeting the above criteria were required for inclusion. For spectral unmixing, examples of each fluorophore are taken from single stained slides for each antibody, as well as a representative autofluorescence spectrum from an unstained sample. Images from each of these single-stained and unstained slides were used to create a multispectral library in inform™. Intensity of each fluorescent target was extracted from the multispectral data using linear unmixing (47).
Figure 1. Characterization of the tumor immune microenvironment with qmIF, including evaluation of spatial distribution of CTLs relative to tumor cells and CD68+ macrophages.
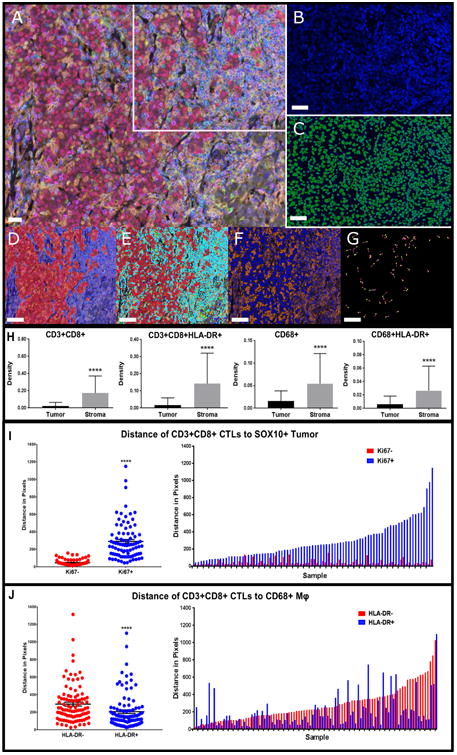
Processing of slides done in inform™ (Perkin Elmer). Steps for analysis using inForm represented using a single image from one patient. A) Multiplex image of a melanoma stained using qmIF. DAPI (nuclei, blue), SOX10 (tumor, red), CD3 (T cells, cyan), CD8 (CTLs, magenta), CD68 (macrophages, green), Ki67 (proliferation marker, yellow), HLA-DR (activation marker, orange). B) Area from the multiplex image (marked by white inset) zoomed in as DAPI only. C) Cell segmentation of zoomed DAPI image. (D-G) are representative analysis steps of the image in (A). D) Tissue segmentation. E) Phenotyping showing base phenotypes:macrophages (green), T cells (cyan), Tumor (red) and Other (blue). F) Scoring with representation of HLA-DR scoring (orange). G) Representative visual example of Nearest Neighbor Analysis to evaluate distance between CD3+CD8+ (pink) and SOX10+Ki67+ (yellow). H) Density of CTLs and CD68+macrophages (n=104). CD3+CD8+ (far left, p<0.0001); CD3+CD8+HLA-DR+ (middle left, p<0.0001); CD68+ (middle right, p<0.0001); CD68+HLA-DR+ (far right, p<0.0001). I) Median distance of CTLs to SOX10+Ki67– (red) or SOX10+Ki67+ (blue) grouped (left, p<0.0001) (n=86). Matched median distance to Ki67− and Ki67+ per patient (right). J) Median distance of CD3+CD8+ to CD68+HLA-DR– (red) or CD68+HLA-DR+ (blue) grouped (left, p<0.0001) (n=97). Matched median distance to HLA-DR− and HLA-DR+ per patient (right). Macrophages: Mφ. Statistical comparison performed using Mann Whitney test. *P≤0.05, **P≤0.01 ***P≤0.001, ****P≤0.0001. A-G images: white bars = 10μm.
Image Analysis
Images were analyzed using inform™ software (PerkinElmer) (Fig. 1A-G). Tissue segmentation was performed by highlighting examples of SOX10+ tumor and SOX10− stroma or non-tumor tissue, allowing the algorithm to ‘learn’ each tissue type and segment image based on gross morphology (Fig. 1D and Supplementary Fig. S2). Supplementary Fig. S2 provides example of tissue segmentation using SOX10 for five images from a single patient. Cellular and subcellular compartments were defined by a counterstain (DAPI) to define the nucleus of each cell (Fig. 1B), with each associated membrane detected via presence of a specific stain (CD3, CD8, or CD68) (Fig. 1C). Cell segmentation was adjusted based on minimum DAPI signal to accurately locate all cells, splitting of cells in order to avoid hypersegmentation and hyposegmentation, and growing and shrinking the nuclei to fit both tumor and immune cells (Fig. 1A-C and Supplementary Fig. S3). Supplementary Fig. S3 shows each of the five images from a single patient with associated DAPI and cell segmentation of the whole 20× image, as well as a zoomed image of the top right corner showing both DAPI and cell segmentation enlarged. Cells were then phenotyped by using the phenotyping step of inform™. 10 – 15 cells for each base variable were selected to train the phenotyping algorithm: tumor (SOX10+, red dots), T cells (CD3+, cyan dots), macrophages (CD68+, green dots), and other (negative for SOX10, CD3, and CD68, blue dots) (Fig. 1E and Supplementary Fig. S4). Supplementary Fig. S4 shows representative phenotyping of five images for a single patient. The last step in inForm was scoring. The images were scored for intensity based on each individual secondary marker for further phenotyping of HLA-DR, CD8, and Ki67 (Fig. 1F and Supplementary Fig. S5). Supplementary Fig. S5 shows scoring in the tumor and stroma for CD8 on five images from a single patient. Finally, data obtained from all representative images was compiled to yield values for each patient. Image data was exported from inform™ version 2.2.1 (PerkinElmer, Hopkinton, MA). The inForm data from all images for each patient were processed in separate software designed in R Studio (version 0.99.896, Boston, MA) (https://github.com/thmshrt/transform_essential). In this software, images were combined and analyzed to concatenate variables (i.e. CD3+CD8+HLA-DR+) and determine density and distance of distinct phenotypes. High and low CTL and macrophage density was defined as above and low a cutoff defined by CART analysis, as well as determined using ROC curves and AUC cutoff.
Statistical Analysis
Analysis was completed with GraphPad Prism Version 7.02 (GraphPad software, La Jolla, CA), and statistical significance was defined as p≤0.05. The need for multiple comparison correction was bypassed by considering unique nearest neighbors in the analysis, which was analyzed using R Studio Version 0.99.896 (CRAN, Boston, MA) and Spotfire software (TIBCO, Palo Alto, CA). Unique nearest neighbor analysis was applied to define spatial relationships between cellular phenotypes. Median distance was calculated across all images for each patient (13).
The effect of the density and distance of immune cell subsets on survival was further analyzed using Classification and Regression Tree (CART) analysis. Kaplan–Meier curves were obtained for DSS and OS in R Version 3.3.1 (CRAN, Boston, MA). P values were calculated using the log-rank (Mantel–Cox) test. Receiver Operating Characteristic (ROC) curves, standard univariate and multivariate Cox proportional hazards analysis, and Pearson correlation matrix testing were performed using R Studio.
Results
CTLs are closer to non-dividing tumor cells and to activated macrophages
We first sought to characterize the TME in primary melanoma using qmIF. Based on published data suggesting that CTLs limit cancer progression, whereas macrophages promote it, we selected CTLs and macrophages as candidate biomarkers (40). We analyzed 104 primary tumors for total number of CTLs and macrophages in both tumor and stroma (Fig. 1H). In order to characterize the level of activation of these cell types, we also included HLA-DR, a marker of cellular activation previously shown to predict response to anti-PD1 therapy, and Ki67, a marker of cell division (11, 40). CTLs and macrophages were evaluated based on expression of HLA-DR (Fig. 1H). All immune cells analyzed were significantly more prevalent in the stroma than in the tumor (Fig. 1H).
We next applied qSA, using nearest neighbor analysis to evaluate relationships between expression of Ki67 and HLA-DR on macrophages and tumor cells and spatial localization of these cells to CTLs. We found that CTLs were spatially closer to tumor cells when they were not proliferating (Ki67−) (p<0.0001; Fig. 1I, left). When evaluating each patient individually, we showed CTLs were closer to non-proliferating tumor cells in nearly all samples having both Ki67+ and Ki67−tumor cells (n=86; Fig. 1I, right). This result is consistent with literature showing that melanomas with higher rates of proliferation have lower frequencies of infiltrating CTLs (18).
We found that HLA-DR is expressed on CD68+ macrophages, CTLs, and, as previously reported, melanoma cells (9). We evaluated the distance of CTLs to SOX10+ tumor and CD68+ macrophages using nearest neighbor analysis. We found that CTLs were closer to CD68+ macrophages when they were HLA-DR+ (p<0.0001; Fig. 1J, left). The median CTL distance to macrophages is shown for individual patients matched for both HLA-DR+ and HLA-DR− cells (n=97; Fig. 1J, right). HLA-DR expression on tumor cells did not significantly impact distance to CTLs (p=0.0846; Supplementary Fig. S6A).
CTL infiltration correlates inversely with risk of death from melanoma
Among the 104 patients included, we performed a subset analysis on 64 patients for whom cause of death was known (Supplementary Table S2) to determine whether density of CTLs in tumor or stroma correlate with DSS. Marked differences in the density of CTLs was observed between patients, as depicted in Fig. 2A and B with tumor specimens demonstrating high and low CTL infiltration, respectively.
Figure 2. Infiltration of CTLs in tumor and stroma and distance of CTLs to non-proliferating tumor cells associates with DSS.
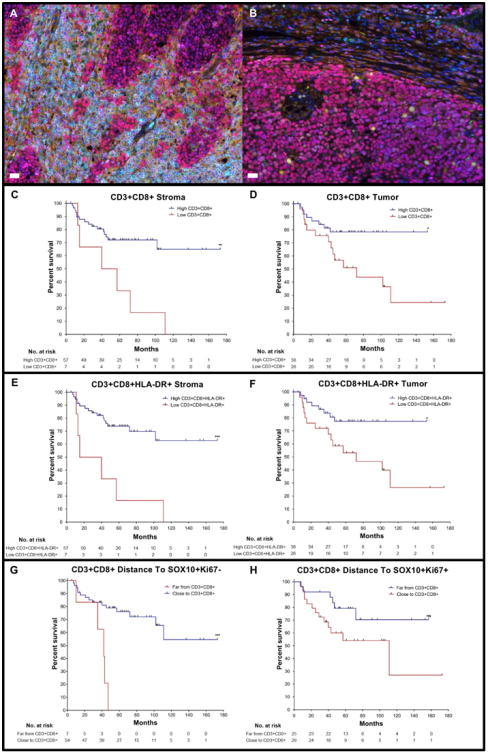
Melanoma slides were stained for qmIF with DAPI (blue), SOX10 (red), CD3 (cyan), CD8 (magenta), CD68 (green), Ki67 (yellow), and HLA-DR (orange). Multiplex images of melanoma showing A) high and B) low infiltration of CTLs in tumor. Kaplan Meier (KM) curves were created using classification and regression tree (CART) analysis for each variable shown in Figures C-H. C) High (n=57) and low (n=7) density of CD3+CD8+ cells in the stroma (p=0.0038). D) High (n=38) and low (n=26)CD3+CD8+ cells in the tumor (p=0.0147). E) High (n=57) and low (n=7) density of CD3+CD8+ HLA-DR+ cells in the stroma (p=0.0005). F) High (n=38) and low (n=26) density ofCD3+CD8+HLA-DR+ cells in the tumor (p=0.0167). G) Far (n=7) and Close (n=54) sistance of CD3+CD8+ cells to SOX10+Ki67− tumor cells (p=0.0006). H) Far (n=25) and Close (n=29) distance from CD3+CD8+ cells to proliferating (SOX10+Ki67+) tumor cells (p=0.0618).Statistical comparison performed using Log-rank (Mantel-Cox) test. ns: not significant (P>0.05), *P≤0.05, **P≤0.01 ***P≤0.001, ****P≤0.0001. White bars in A and B = 10μm.
We next evaluated density of CTLs in the stroma and tumor using CART analysis to determine whether density could stratify patients in terms of DSS. Specifically, using CART cutoffs, we created KM curves to assess the effect of CTL density on DSS in both tumor and stroma. We found that a high density of CTLs in the stroma (p=0.0038) and tumor (p=0.0147) correlated with DSS (Fig. 2C and D), consistent with prior reports that CTLs conferred favorable prognosis (10-12, 17, 43). We next evaluated whether subsets of CTLs might predict prognosis more accurately. The density of CTLs expressing HLA-DR (activated T cells) correlated significantly with DSS in the stroma (p=0.0005; Fig. 2E) but was less significantly correlated in tumor (p=0.0167; Fig. 2F). Ki67-expressing CTLs, however, did not correlate with DSS in the stroma (p=0.1737; Supplementary Fig. S6B) or tumor (p=0.2161;Supplementary Fig. S6C). Thus, assessment of an activation marker appeared to enhance correlation with DSS, whereas including a proliferation marker reduced the correlation between CTL density and DSS.
We further analyzed CTLs by evaluating their distance to proliferating (SOX10+Ki67+) and non-proliferating (SOX10+Ki67−) tumor cells. As stated above (Fig. 1I), CTLs were preferentially located closer to SOX10+Ki67− cells. We found that close distance of CTLs to non-proliferating tumor cells (Ki67−) stratified DSS (p=0.0006; Fig. 2G). However, the distance of CTLs to proliferating tumor cells (Ki67+) did not significantly correlate with DSS (p=0.0618; Fig. 2H). The distance of CTLs to tumor cells, with or without HLA-DR expression, also did not effect DSS (p=0.8436 and p=0.0899, respectively; Supplementary Fig. S6D and E).
HLA-DR− macrophage density correlates with high-risk of death from melanoma
Because we determined that CTLs, particularly in the stroma, correlated with a favorable prognosis, we next evaluated density of CD68+ macrophages in each patient with complete clinical follow-up. Fig. 3A shows a representative multiplex image from a melanoma tumor with CD68+ macrophage infiltrate, and Fig. 3B shows the same image with only CD68 and DAPI staining to demonstrate how these cells were visualized. Using CART cutoffs, we created KM curves evaluating the effect of CD68+ macrophage density on DSS in both the tumor and stroma. In contrast to CTLs, high density of macrophages in the stroma (p=0.0006) and tumor (p=0.0426) associated with poor DSS (Fig. 3C and D). Evaluation of macrophages with and without HLA-DR, we found that high density of HLA-DR− macrophages in stroma (p=0.0013, Fig. 3E) and tumor (p=0.0163, Supplementary Fig. S6F) indicated poor DSS. However, density of HLA-DR+ macrophages in the stroma (p=0.0637) and tumor (p=0.1027) had no significant impact on survival (Fig. 3F and Supplementary Fig. S6G). Thus, CD68+ macrophages in the stroma, but not in the tumor, correlated with DSS, and this correlation was substantially driven by the HLA-DR−macrophage subset.
Figure 3. High infiltration of CD68+macrophages in the tumor and stroma and close distance of CTLs to HLA-DR− macrophages associates with poor DSS.
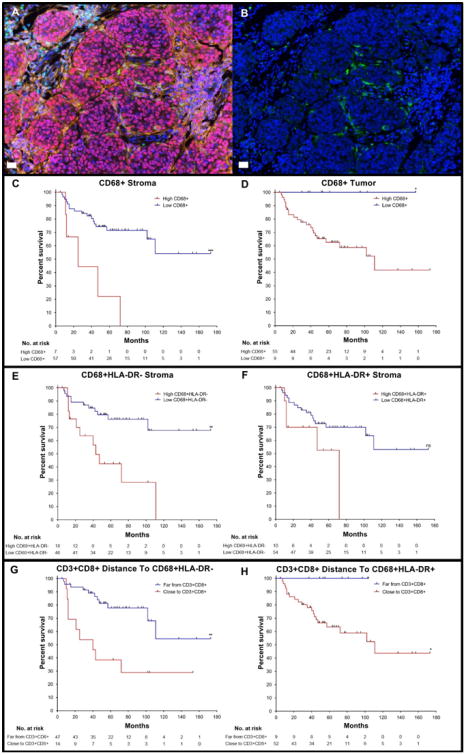
A) Multiplex image of a melanoma slide stained using qmIF for DAPI (blue), SOX10 (red), CD3 (cyan), CD8 (magenta), CD68 (green), Ki67 (yellow), and HLA-DR (orange). B) Multiplex image of melanoma showing DAPI (blue) and CD68 (green) for macrophages. KM curves were created using CART analysis for each variable shown in Figures C-H. C) High (n=7) and low(n=57) density of CD68+ macrophages in the stroma (p=0.0006). D) High (n=55) and low (n=9) density of CD68+macrophages in the tumor (p=0.0426). E) High (n=18) and low (n=46) density of CD68+HLA-DR− macrophages in the stroma (p=0.0013). F) High (n=10) and low (n=54) density of CD68+HLA-DR+ macrophages in stroma (p=0.0637). G) Far (n=47) and Close (n=14) distance of CTLs to HLA-DR− macrophages (p=0.0016). H) Far (n=9) and Close (n=52) distance of CTLs to HLA-DR+macrophages (p=0.0388). Statistical comparison performed using Log-rank (Mantel-Cox) test. ns: not significant (P>0.05), *P≤0.05, **P≤0.01 ***P≤0.001, ****P≤0.0001). White bars in A and B = 10μm.
Given that macrophage density associated with poor survival, we next tested whether distance of CTLs to macrophages with or without HLA-DR correlated with survival. We found that close distance of CTLs to CD68+ HLA-DR− macrophages segregated with shorter DSS (p=0.0016, Fig. 3G). Close distance of CTLs to CD68+ HLA-DR+ macrophages also indicated poor prognosis but was less robust (p =0.0388, Fig. 3H). Thus, HLA-DR– macrophages in the strom a conferred a poor prognosis, and the distance between HLA-DR− macrophages and CTLs also associated with survival, with a shorter distance indicating poor outcome.
Low CTL/macrophage is associated with inferior survial in stage II-III melanoma
Because we determined that CTLs associated with prolonged survival, whereas macrophages in the stroma associated with shortened survival, we sought to integrate these cell types into one biomarker for clinical application. We first tested whether the distance between the two most impactful populations in terms of survival, CTLs and HLA-DR− macrophages, segregated with clinical outcome. Fig. 4A shows a representative multiplex image of a melanoma tumor containing CD68+HLA-DR− macrophages and a selected region containing CD68+ HLA-DR− macrophages, designated by white arrows (Fig. 4A, inset). We further analyzed and compared distance of CTLs to HLA-DR− macrophages using a ROC curve to find the area under the curve (AUC) for death from melanoma over the time of follow-up. We found that the distance of CTLs to HLA-DR− macrophages was significant via ROC (AUC = 0.682, p=0.011, cutoff = 165.54 pixels; Fig. 4B, left). Using the AUC cutoff (165.54) to create a binary analysis of the 61 patients with distance values, excluding 3 patients with no CTLs, to evaluate DSS and Cox proportional hazards (Supplementary Table S3), we found that close distance of CTLs to HLA-DR− macrophages correlated with shorter survival (p=0.0077; Fig. 4B, right).
Figure 4. Close distance of CD68+HLA-DR− macrophages to CTLs associates with poor prognosis in stage II-III melanoma.
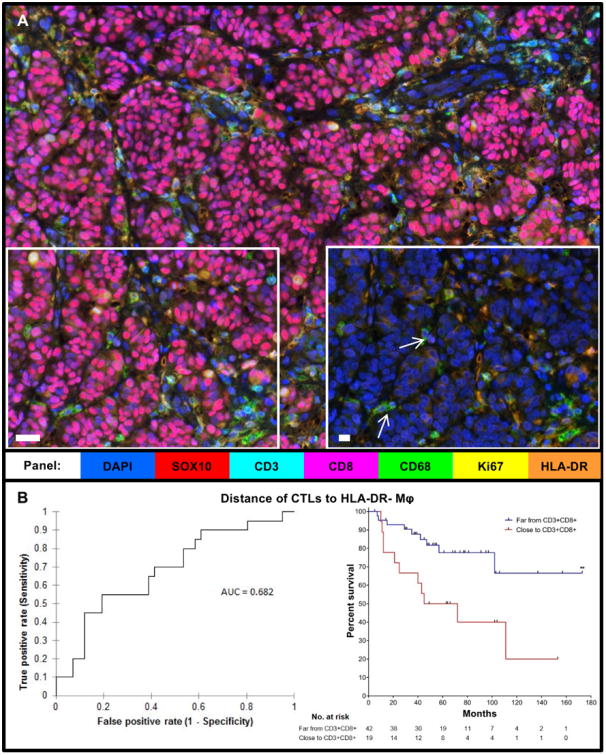
A) Multiplex image of melanoma with selected region (left) shown again, including only HLA-DR, DAPI, and CD68 stains (right).White arrows: HLA-DR− macrophages. B) Receiver Operating Characteristic (ROC) Curve for distance of CTLs to HLA-DR− macrophages (Mφ) (n=61, AUC = 0.682, p=0.011) and KM curve using the AUC cutoff (p=0.0077), Far (n=42), Close (n-19).Statistical comparison performed using Log-rank (Mantel-Cox) test. **P≤0.01. White bars in A and inset = 10μm.
Consistent with our hypothesis that the density of CTLs and macrophages both have a significant impact on survival, we sought to analyze the ratio of these cells in each patient to create a more clinically impactful biomarker that would also allow us to include patients for whom there are no CTLs detected in the sample. We found that a low CTL/macrophage ratio was significant for death from melanoma using ROC (AUC = 0.724, p=0.026, cut off = 2.557; Fig. 5A). We then used the AUC cutoff to create a binary analysis of all 64 patients with DSS to analyze, using a KM curve. We found that a low CTL/macrophage ratio predicted poor prognosis (p=0.0033; Fig. 5B). Univariable Cox analysis showed that a low CTL/macrophage ratio indicated poor prognosis (p = 0.006, Hazards Ratio (HR) = 3.719, 95% Confidence Interval (CI) = 1.451 - 9.533; Fig. 5D). In this population of 64 patients, among other clinical parameters including stage, gender, age, location and depth, only stage was found to be significant (Fig. 5D). Multivariable Cox regression was most significant when stage, ulceration, and CTL/macrophage ratio were included (p=0.002; Fig. 5D). Further analysis found a that low CTL/macrophage ratio also correlated with poor DSS at the two-year and five-year time points. specifically (Supplementary Fig. S7).
Figure 5. Establishing CTL/macrophage ratio in stroma as a biomarker for stage II-III melanoma.
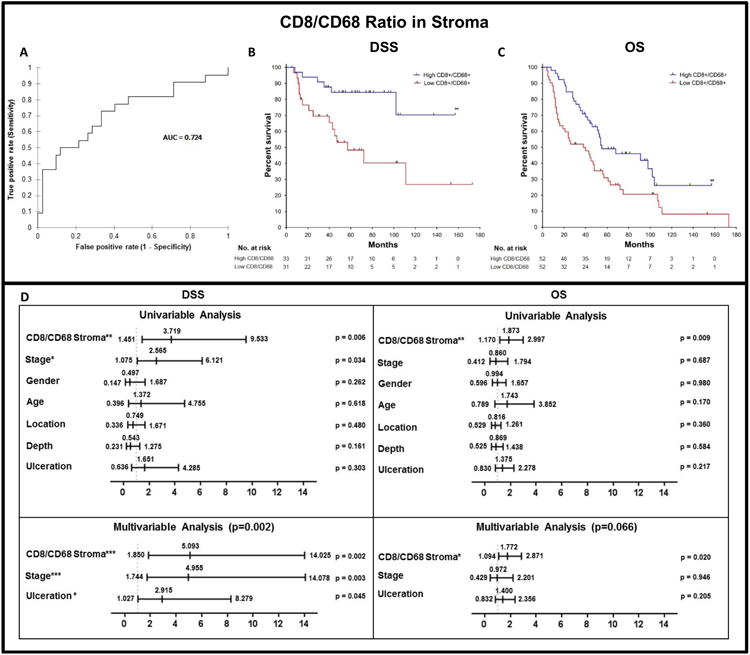
CTL/macrophage ratio in the stroma using ROC curve and Cox Proportional Hazards Model. A) ROC curve for the CTL/macrophage ratio in the stroma (n=64, AUC = 0.724, p=0.026, cut off = 2.557). B) DSS KM curve using AUC cutoff (p=0.0033 in 64 patients, High (n=33), Low (n=31)). C) Overall survival (OS) KM curve using the AUC cutoff (n=104, p=0.0076, High (n=52), Low (n=52)). D) Univariable and multivariable Cox analysis of DSS (n=64) and OS (n=104) patients. Statistical comparison for DSS and OS performed using Log-rank (Mantel-Cox) test. Values in bold are significant at P≤0.05.*P≤0.05, **P≤0.01 ***P≤0.001, ****P≤0.0001.
To further evaluate the robustness of the CTL/macrophage ratio in predicting prognosis, we analyzed all 104 patients for OS (note: cause of death was not known for all the patients). Using the AUC cutoff (2.557) to create a binary of all patients to evaluate using a KM curve, we found that a low CTL/macrophage ratio predicted poor OS (p=0.0076; Fig. 5C). Univariable Cox analysis showed that a low CTL/macrophage ratio indicated poor prognosis (p=0.009, HR=1.873, 95% CI = 1.170-2.997),whereas all other variables were not found to be significant (Fig. 5D). Contrary to DSS, multivariable analysis in the OS cohort of 104 patients showed that ulceration and stage did not enhance the robustness of the model (p=0.066; Fig. 5D). We found that the CTL/macrophage ratio in the stroma correlated with the presence of TILs, using Pearson Correlation Matrix (Table 2). No significant correlation between the densities of CTLs and macrophages was found, suggesting that they play independent roles in tumor progression (p=0.225, Table 2, Supplementary Table S4).
Table 2. Pearson Correlation Matrix.
Correlation of CTL/macrophage ratio with TILs (p=0.044), CD8+ stroma (p<0.0001), and CD68+ stroma (p<0.0001). Correlation with depth, ulceration, age, and stage also shown. n=58; 6 patients removed due to unknown status for ulceration or TILs).
| Variables | Details | Age | Stage | Location | Depth | Ulceration | TILs | CD68+ Stroma | CD8/ CD68 Stroma | CD8+ Stroma |
|---|---|---|---|---|---|---|---|---|---|---|
| Age | ≥55 vs <55 | 1 | -0.333 | 0.026 | -0.006 | -0.071 | -0.008 | -0.038 | -0.066 | -0.051 |
| Stage | III vs II | -0.333 | 1 | -0.127 | -0.139 | -0.215 | -0.115 | -0.089 | -0.238 | -0.031 |
| Location | Extremity vs Trunk | 0.026 | -0.127 | 1 | -0.006 | -0.051 | 0.171 | -0.039 | -0.002 | 0.083 |
| Depth | ≥2mm vs <2mm | -0.006 | -0.139 | -0.006 | 1 | -0.269 | 0.054 | -0.079 | -0.060 | -0.224 |
| Ulceration | Positive vs Negative | -0.071 | -0.215 | -0.051 | -0.269 | 1 | 0.005 | 0.026 | 0.044 | 0.145 |
| TILs | Absent, Non-brisk, Brisk | -0.008 | -0.115 | 0.171 | 0.054 | 0.005 | 1 | -0.072 | -0.265 | -0.504 |
| CD68+ Stroma | High vs Low | -0.038 | -0.089 | -0.039 | -0.079 | 0.026 | -0.072 | 1 | 0.584 | 0.162 |
| CD8/ CD68 Stroma | Low vs High | -0.066 | -0.238 | -0.002 | -0.060 | 0.044 | -0.265 | 0.584 | 1 | 0.564 |
| CD8+ Stroma | Low vs High | -0.051 | -0.031 | 0.083 | -0.224 | 0.145 | -0.504 | 0.162 | 0.564 | 1 |
Discussion
Technological advances using machine learning allow for precise and reproducible quantitation of immune populations within the TME using qmIF (30, 41, 48-50). The Opal staining method combined with qmIF imaging and analysis, unlike other multiplex methods, allows for evaluation using a single slide rather than layering of stains using multiple slides (43). Multiplex staining of a single slide using qmIF provides high-resolution of each individual cell, thereby, allowing for tissue segmentation, multiparameter phenotyping of cells, and precise qSA, all of which was not previously possible. Although pathology expertise is required for the initial training algorithm, automation of subsequent image analysis minimizes subjectivity of interpretation. Because qmIF offers the advantage of using a single slide for processing and automation for analysis, this method will be more clinically applicable, especially for predictive biomarkers, and may also be useful diagnostically. QmIF has been successfully performed in multiple tumor types, including melanoma, breast cancer, lung cancer, and Hodgkin's lymphoma (41, 48-50). Specifically, this method was successfully employed using 6-plex qmIF to evaluate 17 patients with melanoma, finding that the ratio of CTLs to PD-L1 or FOXP3 is capable of predicting TIL generation following adoptive T-cell therapy (51). For our study, we proposed a biomarker developed using this technique that incorporates both density assessment, including precise tissue segmentation and multiparameter phenotyping, and qSA in early stage melanoma patients (13, 52).
We achieved precise tissue segmentation with automated differentiation of tumor from stroma through qmIF analysis of the TME. This analysis built on prior IHC literature, demonstrating that macrophages in the peri-tumor stroma were more significant to clinical outcome than those within the tumor proper. This builds on prior research showing that macrophages at the “leading edge” of the tumor are prognostic, but our automated method for tissue segmentation allows for potential rapid clinical application across institutions (13, 24). Because the qmIF method allows for quantification of cells with multiple concatenated markers, as well as automated assessment of morphology, we were able to accurately define the CTL/macrophage ratio specifically within the tumor-associated stroma, allowing for a precise and quantifiable biomarker. The potential for qmIF to allow for qSA exists, by evaluating the minimum distance between different cell phenotypes using nearest neighbor analysis. Using qSA, we can analyze distance to and from different immune cells, as well as immune cell distance to individual tumor cells.
In terms of prognosis, also consistent with prior IHC literature, we found that high density of CTLs, particularly in the stroma, was a favorable indicator (13). Specifically, we found that CTLs in the stroma were significantly more prognostic than overall CD8 density with both ROC and univariable Cox analysis. The fact that CTL density within each compartment of the tissue was more significant than total density without tissue segmentation may indicate that there is a biologic difference between tumor and stroma, as identified by our algorithm, such that each is best evaluated independently.
The CTL density in the tumor was also favorable but less significant. This may have been due to the increased CTL density in the stroma in comparison to the tumor, impacting statistical analysis, but may also indicate that the biology of the stroma is more critical for progression. Among CTLs, the activated HLA-DR+ subset was prognostic, whereas dividing, Ki67+ CTLs had less effects, suggesting that activation status of CTLs impacts clinical outcome. Using qSA, our findings showed that CTLs were spatially closer to non-dividing tumor cells (Ki67−) and activated macrophages (HLA-DR+), consistent with established findings that melanomas with a high proliferation rate are less infiltrated (13). CTLs may inhibit melanoma differentiation, or rapidly growing tumor may exclude CTLs. Close spatial proximity of CTLs to non-proliferating tumor (Ki67−) segregated with good DSS, perhaps because proximity allowed for cytotoxic antitumor activity.
Although spatial proximity is a limited surrogate for interaction between cells, our finding that close proximity of CTLs to HLA-DR− macrophages was associated with poor survival is consistent with the hypothesis that interactions between CTLs and macrophages promote inflammation and impair antitumor immunity. Further functional studies are needed to delineate the role spatial proximity plays in facilitating cellular communication within the TME. In the evaluation of macrophage density segmentation, our findings showed that high density of CD68+ macrophages, particularly in the stroma, strongly conferred an unfavorable prognosis in stage II-III melanoma and was driven by HLA-DR− macrophages, as HLA-DR+ macrophages did not significantly affect prognosis. No direct correlation between CTL density and macrophage density was found, implying two distinct biologic processes. Macrophages, thus, may cause local tissue destruction, increasing tumor growth via a mechanism not directly related to CTLs or antitumor immunity (53-55).
Our proposed biomarker, the CTL/macrophage ratio, is consistent with our original hypothesis that CTLs play a favorable role, whereas macrophages play a deleterious one. Distance between CTLs and HLA-DR− macrophages was also prognostic, raising the possibility that interactions between these two subsets may inhibit immune surveillance by the CTLs. Specifically, the stromal CTL/macrophage ratio demonstrated superior prognostic value over independent CTL and macrophage density, with the highest AUC on ROC analysis. Consistent with prior literature showing that macrophage density within the leading edge, but not the center of the tumor, is prognostic, we found no correlation by Cox between macrophage density within the tumor and clinical outcome. Thus, focusing on the stroma avoids including additional macrophage data that may dilute the predictive power of the biomarker.
One limitation to our study was that IHC analysis was subject to an inherent sampling bias due to heterogeneity. In order to address tissue heterogeneity, we sampled five representative areas determined by a trained dermatopathologist who was blinded to the clinical data. Notably, pathologists have been able to reproducibly evaluate TIL infiltration based on representative areas of the tumor suggesting that there is limited variability in assessment between pathologists (12, 55). Although pathology expertise is required for the initial training algorithm, automation of subsequent image analysis minimizes subjectivity of interpretation. In the future, it may be possible to apply additional machine learning algorithms to eliminate the role of the pathologist and, thus, any possible subjectivity in image selection. A second limitation of our study was that all 104 patients came from a single center. We also were not able to analyze 42 patients due to poor tissue quality, suggesting that further technical refinement is needed, particularly in specimens stored for longer than 10 years and for very small tumors. Validation in additional populations is needed for clinical application.
In this work, we present a quantitative analysis combining assessment of multiparameter immune phenotypes, tissue segmentation, and qSA in primary melanoma using qmIF. Using this method, we also identifed a biomarker for potential clinical application, demonstrating the value of this approach. Our findings were consistent with known biology of primary melanoma tumors and provide a more quantitative and accurate biomarker in the population studied than standard TIL analysis, which demonstrated only borderline significance. An accurate prognostic clinical biomarker in early stage melanoma is needed to select patients in the development of therapeutic adjuvant protocols. This is of particular importance given that melanoma may recur years after diagnosis, further complicating the development of predictive biomarkers in an ever-changing immunotherapy landscape. QmIF as a platform has significant, and untapped, potential for defining novel biomarkers. The CTL/macrophage ratio in the stroma of primary melanoma tumors merits further development as a biomarker for clinical application.
Supplementary Material
Acknowledgments
This publication was supported by the National Center for Advancing Translational Sciences, National Institutes of Health through Grant Number 1TL1TR001875-01 (Gartrell, Chung PI) and National Cancer Institute Cancer Clinical Investigator Team Leadership Award, supplement to Herbert Irving Comprehensive Cancer Center Support through Grant Number P30 CA013696-41 (Saenger, Emerson PI). The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH. This project also received funding from Swim Across America (Gartrell) and research supported by the 2014 LANDON FOUNDATION-AACR INNOVATOR AWARD FOR CANCER IMMUNOLOGY RESEARCH, Grant Number 14-20-27-SAEN.
Financial Support: This publication was supported by the National Center for Advancing Translational Sciences, National Institutes of Health through Grant Number 1TL1TR001875-01 (Gartrell, Chung PI) and National Cancer Institute Cancer Clinical Investigator Team Leadership Award, supplement to Herbert Irving Comprehensive Cancer Center Support through Grant Number P30 CA013696-41 (Saenger, Emerson PI). The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH. This project also received funding from Swim Across America (Gartrell) and also from research supported by Landon Foundation-AACR Innovator Award for Cancer Immunology Research (Saenger).
Footnotes
Conflicts of Interest: The authors declare no potential conflicts of interest.
References
- 1.Balch CM, Gershenwald JE, Soong SJ, Thompson JF, Atkins MB, Byrd DR, et al. Final version of 2009 AJCC melanoma staging and classification. J Clin Oncol. 2009;27(36):6199–206. doi: 10.1200/JCO.2009.23.4799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Galon J, Fox BA, Bifulco CB, Masucci G, Rau T, Botti G, et al. Immunoscore and Immunoprofiling in cancer: an update from the melanoma and immunotherapy bridge 2015. Journal of translational medicine. 2016;14:273. doi: 10.1186/s12967-016-1029-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Balch CM, Gershenwald JE, Soong SJ, Thompson JF, Ding S, Byrd DR, et al. Multivariate analysis of prognostic factors among 2,313 patients with stage III melanoma: comparison of nodal micrometastases versus macrometastases. J Clin Oncol. 2010;28(14):2452–9. doi: 10.1200/JCO.2009.27.1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Faries MB, Thompson JF, Cochran AJ, Andtbacka RH, Mozzillo N, Zager JS, et al. Completion Dissection or Observation for Sentinel-Node Metastasis in Melanoma. The New England journal of medicine. 2017;376(23):2211–22. doi: 10.1056/NEJMoa1613210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gulley JL, Arlen PM, Madan RA, Tsang KY, Pazdur MP, Skarupa L, et al. Immunologic and prognostic factors associated with overall survival employing a poxviral-based PSA vaccine in metastatic castrate-resistant prostate cancer. Cancer immunology, immunotherapy : CII. 2010;59(5):663–74. doi: 10.1007/s00262-009-0782-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eggermont AM, Chiarion-Sileni V, Grob JJ, Dummer R, Wolchok JD, Schmidt H, et al. Prolonged Survival in Stage III Melanoma with Ipilimumab Adjuvant Therapy. The New England journal of medicine. 2016;375(19):1845–55. doi: 10.1056/NEJMoa1611299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Johnson DB, Balko JM, Compton ML, Chalkias S, Gorham J, Xu Y, et al. Fulminant Myocarditis with Combination Immune Checkpoint Blockade. The New England journal of medicine. 2016;375(18):1749–55. doi: 10.1056/NEJMoa1609214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sivendran S, Chang R, Pham L, Phelps RG, Harcharik ST, Hall LD, et al. Dissection of immune gene networks in primary melanoma tumors critical for antitumor surveillance of patients with stage II-III resectable disease. The Journal of investigative dermatology. 2014;134(8):2202–11. doi: 10.1038/jid.2014.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee N, Zakka LR, Mihm MC, Jr, Schatton T. Tumour-infiltrating lymphocytes in melanoma prognosis and cancer immunotherapy. Pathology. 2016;48(2):177–87. doi: 10.1016/j.pathol.2015.12.006. [DOI] [PubMed] [Google Scholar]
- 10.Clemente CG, Mihm MC, Jr, Bufalino R, Zurrida S, Collini P, Cascinelli N. Prognostic value of tumor infiltrating lymphocytes in the vertical growth phase of primary cutaneous melanoma. Cancer. 1996;77(7):1303–10. doi: 10.1002/(SICI)1097-0142(19960401)77:7<1303::AID-CNCR12>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 11.Azimi F, Scolyer RA, Rumcheva P, Moncrieff M, Murali R, McCarthy SW, et al. Tumor-infiltrating lymphocyte grade is an independent predictor of sentinel lymph node status and survival in patients with cutaneous melanoma. J Clin Oncol. 2012;30(21):2678–83. doi: 10.1200/JCO.2011.37.8539. [DOI] [PubMed] [Google Scholar]
- 12.Busam KJ, Antonescu CR, Marghoob AA, Nehal KS, Sachs DL, Shia J, et al. Histologic classification of tumor-infiltrating lymphocytes in primary cutaneous malignant melanoma. A study of interobserver agreement. Am J Clin Pathol. 2001;115(6):856–60. doi: 10.1309/G6EK-Y6EH-0LGY-6D6P. [DOI] [PubMed] [Google Scholar]
- 13.Gajewski TF, Schreiber H, Fu YX. Innate and adaptive immune cells in the tumor microenvironment. Nat Immunol. 2013;14(10):1014–22. doi: 10.1038/ni.2703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ladanyi A. Prognostic value of tumor-infiltrating immune cells in melanoma. Magyar onkologia. 2013;57(2):85–95. [PubMed] [Google Scholar]
- 15.Mihm MC, Jr, Mule JJ. Reflections on the Histopathology of Tumor-Infiltrating Lymphocytes in Melanoma and the Host Immune Response. Cancer immunology research. 2015;3(8):827–35. doi: 10.1158/2326-6066.CIR-15-0143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Elder DE, Guerry Dt, VanHorn M, Hurwitz S, Zehngebot L, Goldman LI, et al. The role of lymph node dissection for clinical stage I malignant melanoma of intermediate thickness (1.51-3.99 mm) Cancer. 1985;56(2):413–8. doi: 10.1002/1097-0142(19850715)56:2<413::aid-cncr2820560234>3.0.co;2-t. [DOI] [PubMed] [Google Scholar]
- 17.Erdag G, Schaefer JT, Smolkin ME, Deacon DH, Shea SM, Dengel LT, et al. Immunotype and immunohistologic characteristics of tumor-infiltrating immune cells are associated with clinical outcome in metastatic melanoma. Cancer research. 2012;72(5):1070–80. doi: 10.1158/0008-5472.CAN-11-3218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Komohara Y, Jinushi M, Takeya M. Clinical significance of macrophage heterogeneity in human malignant tumors. Cancer science. 2014;105(1):1–8. doi: 10.1111/cas.12314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ostuni R, Kratochvill F, Murray PJ, Natoli G. Macrophages and cancer: from mechanisms to therapeutic implications. Trends in immunology. 2015;36(4):229–39. doi: 10.1016/j.it.2015.02.004. [DOI] [PubMed] [Google Scholar]
- 20.Schupp J, Krebs FK, Zimmer N, Trzeciak E, Schuppan D, Tuettenberg A. Targeting myeloid cells in the tumor sustaining microenvironment. Cellular immunology. 2017 doi: 10.1016/j.cellimm.2017.10.013. [DOI] [PubMed] [Google Scholar]
- 21.Mills CD, Kincaid K, Alt JM, Heilman MJ, Hill AM. M-1/M-2 macrophages and the Th1/Th2 paradigm. Journal of immunology. 2000;164(12):6166–73. doi: 10.4049/jimmunol.164.12.6166. [DOI] [PubMed] [Google Scholar]
- 22.van Baren N, Van den Eynde BJ. Tumoral Immune Resistance Mediated by Enzymes That Degrade Tryptophan. Cancer immunology research. 2015;3(9):978–85. doi: 10.1158/2326-6066.CIR-15-0095. [DOI] [PubMed] [Google Scholar]
- 23.Corliss BA, Azimi MS, Munson JM, Peirce SM, Murfee WL. Macrophages: An Inflammatory Link Between Angiogenesis and Lymphangiogenesis. Microcirculation. 2016;23(2):95–121. doi: 10.1111/micc.12259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jensen TO, Schmidt H, Moller HJ, Hoyer M, Maniecki MB, Sjoegren P, et al. Macrophage markers in serum and tumor have prognostic impact in American Joint Committee on Cancer stage I/II melanoma. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2009;27(20):3330–7. doi: 10.1200/JCO.2008.19.9919. [DOI] [PubMed] [Google Scholar]
- 25.Tsujikawa T, Kumar S, Borkar RN, Azimi V, Thibault G, Chang YH, et al. Quantitative Multiplex Immunohistochemistry Reveals Myeloid-Inflamed Tumor-Immune Complexity Associated with Poor Prognosis. Cell reports. 2017;19(1):203–17. doi: 10.1016/j.celrep.2017.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Garcia-Martinez E, Gil GL, Benito AC, Gonzalez-Billalabeitia E, Conesa MA, Garcia Garcia T, et al. Tumor-infiltrating immune cell profiles and their change after neoadjuvant chemotherapy predict response and prognosis of breast cancer. Breast cancer research : BCR. 2014;16(6):488. doi: 10.1186/s13058-014-0488-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mansfield JR. Multispectral imaging: a review of its technical aspects and applications in anatomic pathology. Veterinary pathology. 2014;51(1):185–210. doi: 10.1177/0300985813506918. [DOI] [PubMed] [Google Scholar]
- 28.Ali HR, Dariush A, Provenzano E, Bardwell H, Abraham JE, Iddawela M, et al. Computational pathology of pre-treatment biopsies identifies lymphocyte density as a predictor of response to neoadjuvant chemotherapy in breast cancer. Breast cancer research : BCR. 2016;18(1):21. doi: 10.1186/s13058-016-0682-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chang AY, Bhattacharya N, Mu J, Setiadi AF, Carcamo-Cavazos V, Lee GH, et al. Spatial organization of dendritic cells within tumor draining lymph nodes impacts clinical outcome in breast cancer patients. Journal of translational medicine. 2013;11:242. doi: 10.1186/1479-5876-11-242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stack EC, Wang C, Roman KA, Hoyt CC. Multiplexed immunohistochemistry, imaging, and quantitation: a review, with an assessment of Tyramide signal amplification, multispectral imaging and multiplex analysis. Methods. 2014;70(1):46–58. doi: 10.1016/j.ymeth.2014.08.016. [DOI] [PubMed] [Google Scholar]
- 31.Peck AR, Girondo MA, Liu C, Kovatich AJ, Hooke JA, Shriver CD, et al. Validation of tumor protein marker quantification by two independent automated immunofluorescence image analysis platforms. Modern pathology : an official journal of the United States and Canadian Academy of Pathology, Inc. 2016;29(10):1143–54. doi: 10.1038/modpathol.2016.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Carstens JL, Correa de Sampaio P, Yang D, Barua S, Wang H, Rao A, et al. Spatial computation of intratumoral T cells correlates with survival of patients with pancreatic cancer. Nature communications. 2017;8:15095. doi: 10.1038/ncomms15095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Feng Z, Bethmann D, Kappler M, Ballesteros-Merino C, Eckert A, Bell RB, et al. Multiparametric immune profiling in HPV- oral squamous cell cancer. JCI insight. 2017;2(14) doi: 10.1172/jci.insight.93652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kawakami F, Sircar K, Rodriguez-Canales J, Fellman BM, Urbauer DL, Tamboli P, et al. Programmed cell death ligand 1 and tumor-infiltrating lymphocyte status in patients with renal cell carcinoma and sarcomatoid dedifferentiation. Cancer. 2017 doi: 10.1002/cncr.30937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Carey CD, Gusenleitner D, Lipschitz M, Roemer MGM, Stack EC, Gjini E, et al. Topological analysis reveals a PD-L1 associated microenvironmental niche for Reed-Sternberg cells in Hodgkin lymphoma. Blood. 2017 doi: 10.1182/blood-2017-03-770719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yeong J, Thike AA, Lim JC, Lee B, Li H, Wong SC, et al. Higher densities of Foxp3+ regulatory T cells are associated with better prognosis in triple-negative breast cancer. Breast cancer research and treatment. 2017;163(1):21–35. doi: 10.1007/s10549-017-4161-4. [DOI] [PubMed] [Google Scholar]
- 37.Nawas S, H A, Koelbe K, Yuan Y. Beyond immune density: critical role of spatial. Modern Pathology. 2015:766–77. doi: 10.1038/modpathol.2015.37. [DOI] [PubMed] [Google Scholar]
- 38.Algars A, Irjala H, Vaittinen S, Huhtinen H, Sundstrom J, Salmi M, et al. Type and location of tumor-infiltrating macrophages and lymphatic vessels predict survival of colorectal cancer patients. Int J Cancer. 2012;131(4):864–73. doi: 10.1002/ijc.26457. [DOI] [PubMed] [Google Scholar]
- 39.Galon J, Bindea G, Mlecnik B, Angell H, Lagorce C, Todosi AM, et al. Intratumoral immune microenvironment and survival: the immunoscore. Med Sci (Paris) 2014;30(4):439–44. doi: 10.1051/medsci/20143004020. [DOI] [PubMed] [Google Scholar]
- 40.Johnson DB, Estrada MV, Salgado R, Sanchez V, Doxie DB, Opalenik SR, et al. Melanoma-specific MHC-II expression represents a tumour-autonomous phenotype and predicts response to anti-PD-1/PD-L1 therapy. Nature communications. 2016;7:10582. doi: 10.1038/ncomms10582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Barbieri G, Rimini E, Costa MA. Effects of human leukocyte antigen (HLA)-DR engagement on melanoma cells. International journal of oncology. 2011;38(6):1589–95. doi: 10.3892/ijo.2011.988. [DOI] [PubMed] [Google Scholar]
- 42.Ma J, Liu L, Che G, Yu N, Dai F, You Z. The M1 form of tumor-associated macrophages in non-small cell lung cancer is positively associated with survival time. BMC cancer. 2010;10:112. doi: 10.1186/1471-2407-10-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Colloby PS, West KP, Fletcher A. Is poor prognosis really related to HLA-DR expression by malignant melanoma cells? Histopathology. 1992;20(5):411–6. doi: 10.1111/j.1365-2559.1992.tb01011.x. [DOI] [PubMed] [Google Scholar]
- 44.Tumeh PC, Harview CL, Yearley JH, Shintaku IP, Taylor EJ, Robert L, et al. PD-1 blockade induces responses by inhibiting adaptive immune resistance. Nature. 2014;515(7528):568–71. doi: 10.1038/nature13954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bobrow MN, Harris TD, Shaughnessy KJ, Litt GJ. Catalyzed reporter deposition, a novel method of signal amplification. Application to immunoassays. Journal of immunological methods. 1989;125(1-2):279–85. doi: 10.1016/0022-1759(89)90104-x. [DOI] [PubMed] [Google Scholar]
- 46.Bobrow MN, Shaughnessy KJ, Litt GJ. Catalyzed reporter deposition, a novel method of signal amplification. II. Application to membrane immunoassays. Journal of immunological methods. 1991;137(1):103–12. doi: 10.1016/0022-1759(91)90399-z. [DOI] [PubMed] [Google Scholar]
- 47.Bainer R, Frankenberger C, Rabe D, An G, Gilad Y, Rosner MR. Gene expression in local stroma reflects breast tumor states and predicts patient outcome. Scientific reports. 2016;6:39240. doi: 10.1038/srep39240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rubin JT, Elwood LJ, Rosenberg SA, Lotze MT. Immunohistochemical correlates of response to recombinant interleukin-2-based immunotherapy in humans. Cancer research. 1989;49(24 Pt 1):7086–92. [PubMed] [Google Scholar]
- 49.Guidolin D, Ruggieri S, Annese T, Tortorella C, Marzullo A, Ribatti D. Spatial distribution of mast cells around vessels and glands in human gastric carcinoma. Clinical and experimental medicine. 2017 doi: 10.1007/s10238-017-0452-7. [DOI] [PubMed] [Google Scholar]
- 50.Feng Z, Puri S, Moudgil T, Wood W, Hoyt CC, Wang C, et al. Multispectral imaging of formalin-fixed tissue predicts ability to generate tumor-infiltrating lymphocytes from melanoma. Journal for immunotherapy of cancer. 2015;3:47. doi: 10.1186/s40425-015-0091-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mitchell RT, Camacho-Moll ME, Macdonald J, Anderson RA, Kelnar CJ, O'Donnell M, et al. Intratubular germ cell neoplasia of the human testis: heterogeneous protein expression and relation to invasive potential. Modern pathology : an official journal of the United States and Canadian Academy of Pathology, Inc. 2014 doi: 10.1038/modpathol.2013.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vasaturo A, Halilovic A, Bol KF, Verweij DI, Blokx WA, Punt CJ, et al. T cell landscape in a primary melanoma predicts the survival of patients with metastatic disease after their treatment with dendritic cell vaccines. Cancer research. 2016 doi: 10.1158/0008-5472.CAN-15-3211. [DOI] [PubMed] [Google Scholar]
- 53.Jadus MR, Irwin MC, Irwin MR, Horansky RD, Sekhon S, Pepper KA, et al. Macrophages can recognize and kill tumor cells bearing the membrane isoform of macrophage colony-stimulating factor. Blood. 1996;87(12):5232–41. [PubMed] [Google Scholar]
- 54.Keller R, Keist R, Wechsler A, Leist TP, van der Meide PH. Mechanisms of macrophage-mediated tumor cell killing: a comparative analysis of the roles of reactive nitrogen intermediates and tumor necrosis factor. International journal of cancer. 1990;46(4):682–6. doi: 10.1002/ijc.2910460422. [DOI] [PubMed] [Google Scholar]
- 55.Piris A, Mihm MC., Jr Progress in melanoma histopathology and diagnosis. Hematology/oncology clinics of North America. 2009;23(3):467–80. viii. doi: 10.1016/j.hoc.2009.03.012. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


