Abstract
Although artificial neural networks simulate a variety of human functions, their internal structures are hard to interpret. In the life sciences, extensive knowledge of cell biology provides an opportunity to design visible neural networks (VNNs) which couple the model’s inner workings to those of real systems. Here we develop DCell, a VNN embedded in the hierarchical structure of 2526 subsystems comprising a eukaryotic cell (http://d-cell.ucsd.edu/). Trained on several million genotypes, DCell simulates cellular growth nearly as accurately as laboratory observations. During simulation, genotypes induce patterns of subsystem activities, enabling in-silico investigations of the molecular mechanisms underlying genotype-phenotype associations. These mechanisms can be validated and many are unexpected; some are governed by Boolean logic. Cumulatively, 80% of the importance for growth prediction is captured by 484 subsystems (21%), reflecting the emergence of a complex phenotype. DCell provides a foundation for decoding the genetics of disease, drug resistance, and synthetic life.
Introduction
Deep learning has revolutionized the field of artificial intelligence by enabling machines to perform human activities like seeing, listening and speaking1–6. Such systems are constructed from many-layered, ‘deep’, artificial neural networks (ANNs), inspired by actual neural networks in the brain and how they process patterns. The function of the ANN is created during a training phase, in which the model learns to capture as accurately as possible the correct answer, or output, that should be returned for each example input pattern. In this way, machine vision learns to recognize objects like dogs, people, and faces, and machine players learn to distinguish good from bad moves in games like chess and Go7.
In modern ANN architectures, the connections between neurons as well as their strengths are subject to extensive mathematical optimization, leading to densely entangled network structures that are neither tied to an actual physical system nor based on human reasoning. Consequently, it is typically difficult to grasp how any particular set of neurons relates to system function. For instance, AlphaGo beats top human players7, but examination of its underlying network yields little insight into the rules behind its moves or how these are encoded by neurons. These are so-called ‘black boxes’8, in which the input/output function accurately models an actual system but the internal structure does not (Fig. 1a). Such models, while undoubtedly useful, are insufficient in cases where simulation is needed not only of system function but also of system structure. In particular, many applications in biology and medicine seek to model both functional outcome and the mechanisms leading to that outcome so that these can be understood and manipulated through drugs, genes or environment.
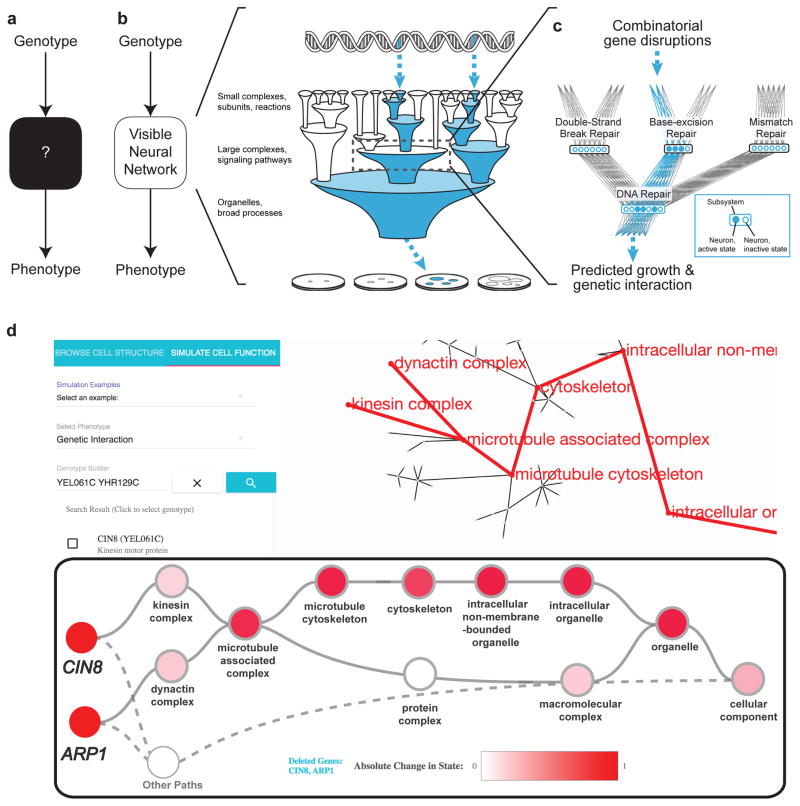
Figure 1. Modeling system structure and function with visible learning.
a, A conventional neural network translates input to output as a black box without knowledge of system structure. b, In a visible neural network, input/output translation is based on prior structural knowledge. In DCell, gene disruption genotypes (top) are translated to cell growth predictions (bottom) through a hierarchy of cell subsystems (middle). c, A neural network is embedded in the prior structure using multiple neurons per subsystem. d, Screen capture of DCell online service, microtubule subsystems.
Here we report DCell, an interpretable or ‘visible’ neural network (VNN) simulating a basic eukaryotic cell. The structure of this model is formulated from extensive prior knowledge of the cell’s hierarchy of subsystems documented for the budding yeast Saccharomyces cerevisiae, drawn from either of two sources: the Gene Ontology (GO), a literature-curated reference database from which we extracted 2526 intracellular components, processes, and functions9; or CliXO, an alternative ontology of similar size inferred from large-scale molecular datasets rather than literature curation10,11. While CliXO and GO overlap in 37% of subsystems, some in CliXO are apparent in large-scale datasets but not yet characterized in literature, whereas some in GO are documented in the literature but difficult to identify in big data. Subsystems in these ontologies are interrelated through hierarchical parent-child relationships of membership or containment. Such hierarchies form a natural bridge from variations in genotype, at the scale of nucleotides and genes, to variations in phenotype, at the scale of cells and organisms12,13.
The function of DCell is learned during a training phase, in which perturbations to genes propagate through the hierarchy to impact parent subsystems that contain them, giving rise to functional changes in protein complexes, biological processes, organelles and, ultimately, a predicted response at the level of cell growth phenotype (Fig. 1b). Previously, we saw that hierarchical groups of genes in an ontology could be used to formulate input features for such phenotypic predictions12,13. However, these features were provided to standard black-box machine learning models which could not be interpreted biologically. Here, we use the biological hierarchy to directly embed the structure of a deep neural network, enabling transparent biological interpretation.
Results
DCell Design
In DCell, the functional state of each subsystem is represented by a bank of neurons (Fig. 1c). Connectivity of these neurons is set to mirror the biological hierarchy, so that they take input only from neurons of child subsystems and send output only to neurons of parent (super)systems, with weights determined during training. The use of multiple neurons (ranging from 20 to 1,075 per system, Online Methods) acknowledges that cellular components can be multifunctional, with distinct states adopting a range of values along multiple dimensions14. The input layer of the hierarchy comprises the genes, while the output layer, or root, is a single neuron representing cell phenotype. By this design, the VNN embedded in GO includes 43,721 neurons while the corresponding model for CliXO includes 22,167 neurons. The depth of both networks is 12 layers, on par with deep neural networks in other fields7.
Training and Performance in Genotype-Phenotype Translation
Given this architecture, we taught DCell to predict phenotypes related to cellular fitness, a model genotype-to-phenotype translation task (Online Methods). Extensive training was made possible by a compendium of yeast growth phenotypes measured for single and double gene deletion genotypes, comprising several million genotype-phenotype training examples15,16. Two related phenotypes were considered: (i) Capacity for growth measured by colony size relative to wild-type cells; (ii) For double gene deletions, genetic interaction score measured as the difference in colony size from that expected from the corresponding single gene deletions. Predicting genetic interaction represents a harder task than predicting absolute growth, as it requires learning of non-linear effects beyond superposition of elemental genotypes. Based on the training examples, the weights of input connections to each neuron were optimized by stochastic gradient descent computed by backpropagation. For execution and inspection of this DCell model, we created an interactive website at http://d-cell.ucsd.edu/ (Fig. 1d).
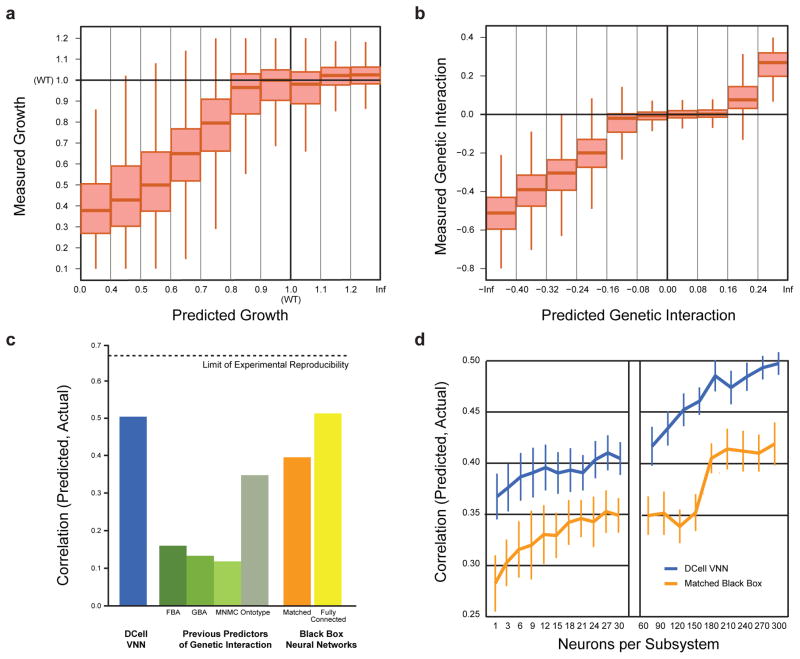
We found that DCell was able to make accurate phenotypic predictions for both growth (Fig. 2a) and genetic interaction (Fig. 2b). It outperformed previous predictors, including those based on metabolic models17 and protein-protein interaction networks18,19, as well as a hierarchical method not related to deep learning (Fig. 2c, Supplementary Fig. 1)13. We also compared performance to black-box ANNs of several types. First, we constructed ANNs with matching structure to DCell but permuting the assignment of genes to subsystems. Predictive performance decreased substantially (Fig. 2c) and was restored only after increasing the number of neurons by an order of magnitude (Fig. 2d). Thus, the biological hierarchy provides significant information not found in randomized versions. Second, we constructed a fully connected ANN with the same number of layers and neurons as DCell but unlimited connectivity between adjacent layers. Despite these extra parameters, performance of this fully connected model was not significantly better (Fig. 2c).
Figure 2. Prediction of cell viability and genetic interaction phenotypes.
a, Measured versus predicted cell viability relative to wild type (WT = 1) on the Costanzo et al.16 dataset. b, Measured versus predicted genetic interaction scores for each double gene disruption genotype; genetic interactions between the disrupted genes can be positive (epistasis), zero (non-interaction), or negative (synthetic sickness or lethality). c, Model performance expressed as the correlation between measured and predicted genetic interaction scores. Performance of DCell (first bar, blue) is compared to previous methods for predicting genetic interactions (second four bars, shades of green): FBA, Flux Balance Analysis17; GBA, Guilt By Association18; MNMC, Multi-Network Multi-Classifier19; Ontotype13. Performance is also shown for matched black-box structures in which gene-to-subsystem mappings are randomly permuted (orange bar, average of 10 randomizations) or for fully-connected neural networks with the same number of layers as DCell (final bar, yellow). Correlations were calculated across gene pairs that meet an interaction significance criterion of p<0.05. d, Predictive performance versus number of neurons per subsystem. Performance measure and two structural hierarchies as in (c).
From Prediction to Mechanistic Interpretation
Unlike standard ANNs, DCell’s simulations were tied to an extensive hierarchy of internal biological subsystems with states that could be queried. This ‘visible’ aspect raised the possibility that DCell could be used for in-silico studies of biological mechanism, of which we focused on four major types:
Explaining a genotype-phenotype association
Prioritizing all important mechanisms in determination of phenotype overall
Characterization of the genetic logic implemented by a process
Discovery of new biological processes and states
Explaining a Genotype-Phenotype Association
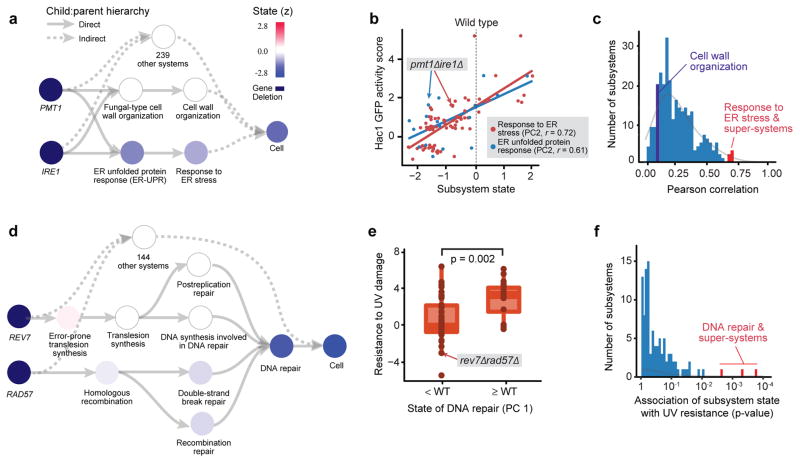
A fundamental goal of genetics is to explain the molecular mechanisms linking changes in genotype to changes in phenotype. To generate such explanations automatically, we used DCell to simulate the impact of a genotypic change, relative to wild type, on the states of all cellular subsystems in the model. Subsystems with significant changes were proposed as candidate explanations in translation of genotype to phenotype, whereas those without state changes – typically the vast majority – were excluded from consideration. For example, to explain the severe growth defect caused by pmt1Δire1Δ, disrupting the genes PMT1 and IRE1, we simulated this genotype with DCell and examined the 243 subsystems incorporating PMT1 or IRE1 at any level of the GO hierarchy (ancestors of one or both genes). These subsystems encompassed functions of PMT1 or IRE1 in the endoplasmic reticulum unfolded protein response (ER-UPR)20,21, cell wall organization and integrity22,23, and many other processes (Fig. 3a). Examining the simulated states of these candidate subsystems (values of their neurons), we found that ER-UPR output was substantially reduced compared to wild type, whereas cell wall organization and other subsystems were relatively unaffected (Fig. 3a).
Figure 3. Interpretation of genotype-phenotype associations.
a, Hierarchy of candidate subsystems that can explain the association of the pmt1Δire1Δ genotype with a negative genetic interaction phenotype (synthetic lethality). Subsystem states are represented by their neuron values, reduced to the first two principal components (PCs) which captured >75% of the total variance. The color of the node for each subsystem shows its PC2 expressed as a z-score with zero mean and unit standard deviation. By construction, the wild-type genotype produces z-score = 0 for all subsystems (Online Methods). b, Correspondence between Hac1 GFP activity and the functional states of ER-UPR (blue) or its parent subsystem, Response to ER stress (red). Points represent genotypes, with pmt1Δire1Δ genotype indicated. Subsystem state is the z-score of PC2 as in (a). c, Distribution of correlations between Hac1 GFP activity and the states of every other subsystem containing at least 10 genotypes with measured GFP activity. d, DNA repair is a highly altered subsystem that explains the slow growth phenotype of rev7Δrad57Δ. e, Experimental resistance to UV damage plotted against the simulated state of DNA repair in DCell, separated into two classes: above or below wild-type value. Significance measured by Mann–Whitney U test. f, Distribution of the associations between UV damage resistance and the states of other subsystems containing at least 10 genotypes with measured UV resistance. Panels (a–c) use genetic interaction prediction as the phenotypic readout; (d–f) use growth. All panels implement GO as the structural model.
To validate this simulated decrease, we examined a dataset measuring abundance of Green Fluorescent Protein (GFP) driven by a promoter responsive to Hac1, a key transcriptional activator of ER-UPR, over numerous pairwise gene disruptions24. Hac1 activity was significantly lowered in the pmt1Δire1Δ genotype compared to wild type, consistent with model simulations (Fig. 3b). Moreover, we found that the simulated state of ER-UPR was well correlated with experimental Hac1 activity, not only for this genotype but across all relevant gene disruptions in the dataset (Fig. 3b). To address the concern that Hac1 activity might associate non-specifically with state changes in many diverse subsystems, not just those related to ER, we examined its correlation with the simulated states of every subsystem in DCell. High correlation was observed only for ER-UPR and super-systems (Fig. 3c), demonstrating specific validation. In this way, DCell was used to test among competing mechanistic hypotheses for a genotype-phenotype relationship.
In explaining genotype-phenotype associations, a key requirement is that the state of a subsystem in silico approximate its true state in vivo. To further validate this capability, we examined the subsystem of DNA repair (Fig. 3d) which, like ER-UPR, had been experimentally interrogated over many double gene deletions25. In particular, DNA repair status had been characterized by resistance to ultraviolet radiation (UV), a model DNA damaging agent26. Once again we saw good agreement between model and experiment: the simulated state of DNA repair significantly tracked experimental UV resistance across genotypes (Fig. 3e), in a manner highly specific to this subsystem (Fig. 3f).
Prioritizing all important systems in determination of phenotype overall
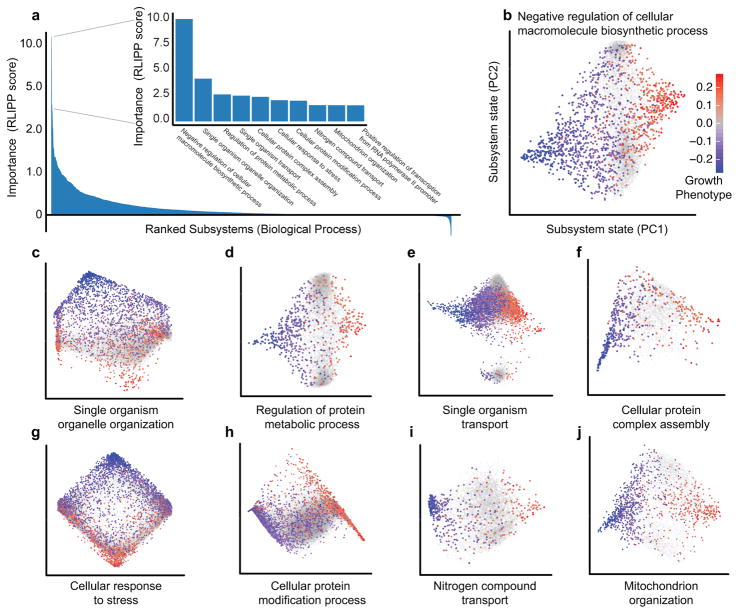
Beyond individual explanations, a critical question was whether a complex phenotype such as growth depends on equal contributions from many subsystems or is dominated by a few. To address this question, we reasoned that the overall importance of a subsystem can be computed quantitatively as the degree to which its state is more predictive of phenotype than the states of its children – a metric we called Relative Local Improvement in Predictive Power (RLIPP, Online Methods). We observed that RLIPP approximately followed a Pareto (power-law) distribution, in which a few subsystems are highly important for model predictions, with a long tail of weakly important systems (Fig. 4a). In particular, 80% of the cumulative importance was captured by 21% of subsystems (the Pareto 80/20 rule27), while >88% of subsystems retained some improvement in phenotypic prediction over their children (RLIPP > 0). The GO subsystem of greatest individual importance was ‘Negative regulation of cellular macromolecule biosynthesis’, which organizes cellular circuits that inhibit biosynthesis and, as evidenced by DCell simulations, can lead to strong increases in growth when disrupted. Other subsystems important for growth related to the proper function of organelles, biomolecular transport, stress response, protein modification, and assembly of complexes (Figs. 4b–j).
Figure 4. Top subsystem states for translation of genotype to growth.
a, Ranking of all cellular subsystems in GO (x-axis) by their importance in determining genetic interactions (RLIPP score, y-axis). Inset: ten highest-scoring subsystems. Positive RLIPP corresponds to increases in predictive power relative to children; negative RLIPP corresponds to decreases in power. b–j, Two-dimensional state maps of informative subsystems (PC2 versus PC1). Supplementary Fig. 2 provides equivalent information for the CliXO hierarchy.
Characterization of the genetic logic implemented by a process
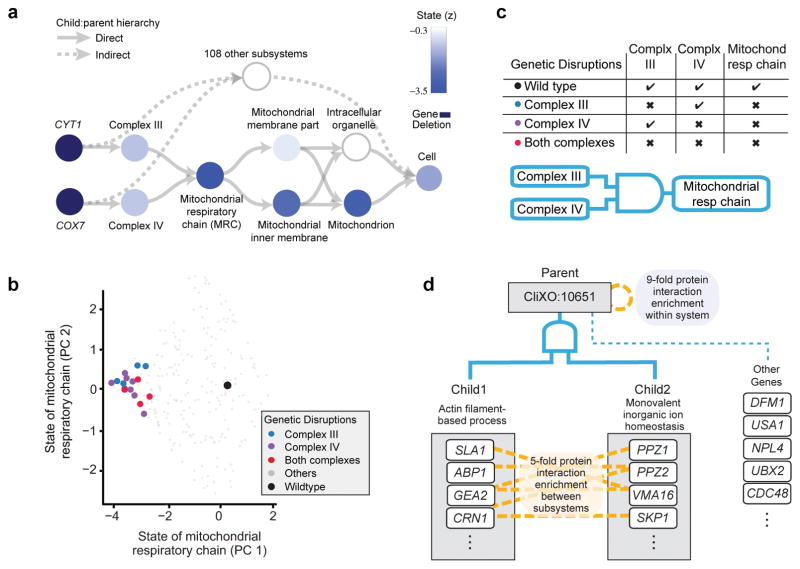
Another type of mechanistic interpretation relates to the mathematical functions by which the neurons representing each subsystem integrate information. We investigated whether these functions could be reduced to simple forms, such as Boolean logic gates, which are easily interpreted (Online Methods). This analysis found 1119 subsystems at least partly governed by Boolean logic (44% of GO, Supplementary Table 1). For instance, the state of Mitochondrial Respiratory Chain (Fig. 5a), while relatively high in wild-type cells, was driven low by disruptions in any of its several enzymatic complexes involved in electron transport, such as complexes III or IV (Fig. 5b). Thus Mitochondrial Respiratory Chain resembles a logical AND gate (Fig. 5c). We also observed many cases of OR, XOR, and (A not B), although the AND configuration arose most frequently. The remaining subsystems did not map clearly to Boolean functions, suggesting machinery that is more complex than an on/off switch.
Figure 5. Analysis of subsystem functional logic.
a, Causal hierarchy of important subsystems involving changes to Mitochondrial Respiratory Chain (MRC), displayed as in Fig. 3a. b, 2D state map of MRC plotted as in Fig. 3b. Genotypes disrupting MRC complex III only, MRC complex IV only, or both complexes are demarcated by colors. c, Truth table relating the state of MRC to the states of its children. The logic resembles an AND gate, pictured. d, Newly identified system (CliXO:10651) encoding a logical AND integrating the states of two well-characterized children. Names of children are cross-annotated from the corresponding enriched GO terms: Actin filament-based process, GO:0030029; Monovalent inorganic ion homeostasis, GO:0030004.
Discovery of new biological processes and states
Finally, since DCell’s hierarchy could be structured from systematic datasets (CliXO) as an alternative to literature (GO), we investigated the extent to which model simulations with CliXO relied on entirely new cellular subsystems not previously appreciated in biology. In total we found 236 subsystems in the CliXO hierarchy that were previously undocumented in GO or elsewhere in literature and had high RLIPP importance scores for genotype-phenotype translation (Supplementary Table 2). One example was CliXO:10651, a previously undocumented process ranking among the top ten systems important for growth prediction. We found that CliXO had inferred this system based on the elevated density of protein-protein interactions observed among its 154 genes (Fig. 5d, 9-fold enrichment, p<10−200). These interactions interconnected two subsystems that were much better understood, relating to actin filaments and ion homeostasis (5-fold enrichment between subsystems, p=0.00029). The simulated state of CliXO:10651 was governed approximately by a Boolean AND of the states of its two subsystems, both being required to maintain wild-type status. These findings were supported by previous reports that homeostasis of ions, such as iron, regulates the level of oxidative stress, which in turn disrupts actin cytoskeletal organization28,29.
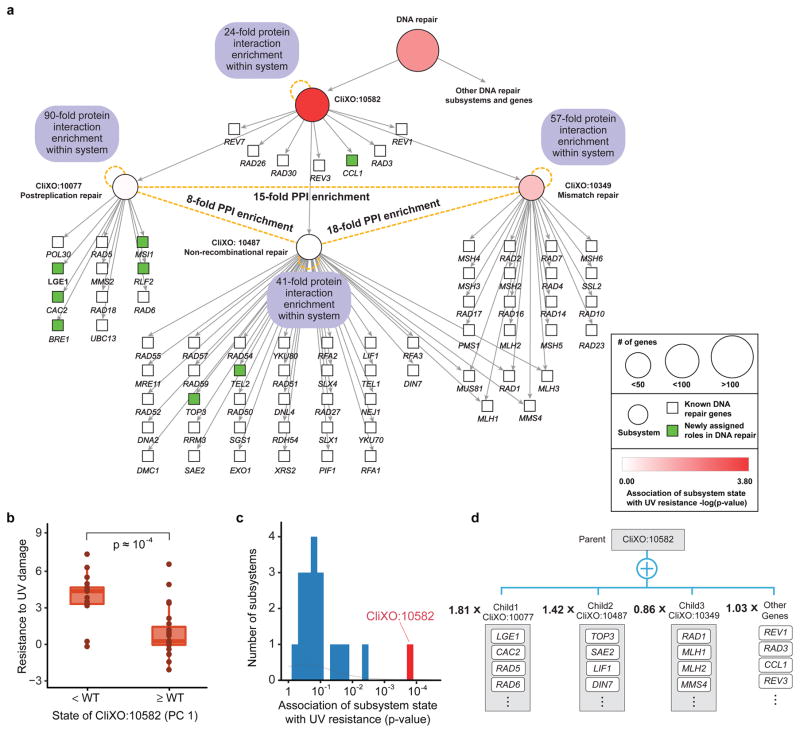
As a second example we considered CliXO:10582, a novel subsystem of 71 genes (Fig. 6a). Although many of these genes had known roles in DNA repair, nothing like this grouping had been previously recognized. Examination of the hierarchical model structure revealed that CliXO:10582 interconnects components of three known DNA repair subsystems, postreplication repair, mismatch repair, and non-recombinational repair, based on a very high density of protein-protein interactions falling among these components (Fig. 6a). Revisiting the experimental data on resistance to UV-induced DNA damage25 (Figs. 3e,f), we saw that the simulated state of CliXO:10582 strongly correlated with experimental UV resistance across genotypes (Fig. 6b). This association was stronger than for any child and, in fact, for any other CliXO subsystem interrogated by the experimental data (Fig. 6c). Mathematically, the state of CliXO:10582 was not well-captured by Boolean logic but by a weighted linear summation of the states of the three child systems, with postreplication repair having the greatest single contribution (Fig. 6d). Thus, DCell had identified a novel organization of subcomponents which specifically coordinate the response to UV damage. For the eight genes in this system not previously known to function in DNA repair (green nodes, Fig. 6a), the evidence summarized by the model – that these gene products physically interact within a larger cluster of known DNA repair factors, and that they functionally manifest with the same UV sensitivity phenotype when disrupted – creates a compelling case for further studies.
Figure 6. Analysis of a new DNA repair subsystem.
a, Hierarchical organization of DNA repair including subsystems newly identified by CliXO. b, Experimental resistance to UV damage plotted against the state of CliXO:10582, separated into two classes: above or below wild-type value. Significance measured by Mann–Whitney U test. c, Distribution of associations between UV resistance and the states of all CliXO subsystems with at least 10 genotypes with measured UV resistance. d, Approximate mathematical function implemented by CliXO:10582 neurons.
Discussion
A direct route to interpretable neural networks is to encode not only function but form. Here, we have explored such visible learning in the context of cell biology, by incorporating an unprecedented collection of knowledge10,11,30 and data15,16,31 to simultaneously simulate cell hierarchical structure and function. DCell captured nearly all phenotypic variation in cellular growth, a classic complex phenotype, including much of the less-understood non-additive portion due to genetic interactions (Figs. 2a–c). Armed with this explanatory power, the model simulated the intermediate functional states of thousands of cellular subsystems. Knowledge of these states enabled in-silico studies of molecular mechanism, including dissection of subsystems important to growth phenotype, identification of new subsystems, and reduction of subsystem functions, where possible, to Boolean logic (Figs. 3–5).
Methodologically, our approach works towards a synthesis of statistical genetics and systems biology. State-of-the-art methods in statistical genetics32,33 are based on linear regression of phenotype against the independent effects of genetic polymorphisms, without modeling the underlying molecular mechanisms that give rise to nonlinearity and genetic interaction. Separately, studies in systems biology capture molecular mechanisms using mathematical models18,34,35, but such models typically do not have the breadth for large-scale genetic dissection of phenotype. DCell bridges these two avenues: Its neural network encodes a complex nonlinear regression, an extension of statistical genetics, in which the additional complexity is enabled by a hierarchical mechanistic model, an extension of systems biology. In contrast to other mechanistic models that have attempted large-scale genotype-phenotype prediction13,36, the framework of hierarchical neural networks is very general and expressive, such that a large class of biological structures and functions can be represented. For example, our earlier approach13 used hierarchical knowledge of subsystems to create new features based on the number of gene disruptions in a subsystem, but these features were predetermined before modeling and thus nothing was learned about the real functions encoded by subsystems.
It is also instructive to view DCell in context of previous research in interpretable machine learning, in which the notion of interpretability has been defined in different ways37. One direction has been to perform a post-hoc examination of an ANN that has already been trained, by inspecting neurons and rationalizing their decisions. A model trained to identify images of dogs might, upon later inspection, be seen to have neurons capturing interpretable properties like “tail” or “furry”38–40. A limitation of post-hoc interpretation is that it is disconnected from training, leaving no guarantees as to what level of human understanding can be achieved41. Therefore, in attention-based neural networks42,43, a separate module preselects key “interpretable” features for input to a black-box model. For example, in a model predicting emotional attitude of a blog author (positive or negative, angry or calm), the key interpretable feature might come from a key phrase preselected from text (LOL, I’m so upset). While DCell has some similarity to these attention-based approaches, its deep hierarchical structure captures many different clusters of features at multiple scales, pushing interpretation from the model input to internal features representing biological subsystems.
In several case studies, involving genotypes impacting ER-UPR and DNA repair subsystems, the subsystem states learned by DCell could be directly confirmed by molecular measurements. Notably, no information about subsystem states was provided during model training. These states emerged from translating genotypes (model inputs) to growth phenotypes (model outputs) under the structural constraints of the subsystem hierarchy; together, the input/output data and hierarchical structure were sufficient to guide subsystem neurons to learn a biologically correct function. In future, one might directly supervise a VNN to learn potentially multiple subsystem states and/or complex phenotypes, in which case training data could be provided at any level: genotype, phenotype, or points in between.
In some applications of machine learning, predictive performance is all that matters. Indeed, in these cases it is often possible to build a large number of alternative models that, while different in structure, all make excellent near-optimal functional predictions. In biology, however, prediction is not enough. The key additional question is which of the many excellent predictive models is the one actually used by the living system, as optimized not by computation but by evolution. DCell provides proof-of-concept of a system that, while optimizing functional prediction, respects biological structure. Such models are of immediate interest in genome-wide association studies of human disease44, in which different patient genotypes can influence disease outcomes by complex mechanisms hidden from black-box statistical approaches. Once trained on sufficient data, these models have application in personalized therapy by analyzing a patient’s genotype in combination with potential points of intervention targeted by drugs. We also see compelling uses in design of synthetic organisms, in which candidate genotypes can be efficiently evaluated in silico prior to validation in vivo. Finally, beyond the architecture of the cell, biological systems at other scales may benefit from this type of constrained learning, including modeling of neural connections in the brain.
Online Methods
Preparation of Ontologies
We guided the deep neural network structure using a biological ontology, consisting of terms representing cellular subsystems, child-parent relations representing containment of one term by another, and gene-to-term annotations. The first ontology considered was the Gene Ontology (GO), in which all three branches of GO (biological process, cellular component, and molecular function) were joined under a single root. We used the following criteria to filter (remove) terms from GO:
Terms with the evidence code “inferred by genetic interaction” (IGI), to avoid potential circularity in predicting genetic interactions in the genotype-phenotype samples.
Terms containing fewer than six yeast genes disrupted in the available genotypes (with “containment” defined as all genes annotated to that term or its descendants).
Terms that are redundant with respect to their children terms in the ontology.
When a term was removed, all children were connected directly to all parent terms to maintain the hierarchical structure. The remaining 2526 terms were used to define the hierarchy of DCell subsystems.
To complement the GO structure, we also constructed a data-driven gene ontology using the method of Clique Extracted Ontologies (CliXO) as previously described11. Briefly, data on gene pairs were sourced from YeastNet v331, which lists 68 experimental studies of 8 data types, excluding genetic interactions to avoid circularity similar to criterion 1 above. All features were integrated to create a single gene-gene similarity network following a previously described procedure11, in which each gene-gene pair is assigned a weighted similarity based on a combination of the YeastNet data. This network was subsequently analyzed with the CliXO algorithm, which identifies nested cliques as the threshold gene-gene similarity becomes progressively less stringent. This process yields a hierarchy (directed acyclic graph) of parent-child relations among cliques at different similarity thresholds.
DCell Architecture and Training Algorithm
DCell trains a deep neural network to predict phenotype from genotype, with architecture that exactly mirrors the hierarchical structure of an ontology of cellular subsystems. Each cellular subsystem is represented by a group of hidden variables (neurons) in the neural network, and each parent-child relation is represented by a set of edges that fully connect these groups of hidden variables. The depth of this architecture (12 layers) presents two challenges for training: 1) There is no guarantee that each subsystem will learn new patterns instead of copying those of its child subsystems; 2) Gradients tend to vanish lower in the hierarchy. To tackle these challenges, we borrow ideas from two previous systems, GoogLeNet45 and Deeply-Supervised Net46, which improve the transparency and discriminative power of hidden variables and reduce the effect of vanishing gradients.
We denote our input training dataset as D = {(X1,y1),(X2,y2),…,(XN,yN)}, where N is the number of samples. For each sample i, Xi ∈ RM denotes the genotype, represented as a binary vector of states on M genes (1 = disrupted; 0 = wild type), and yi ∈ R denotes the observed phenotype, which can be either relative growth rate or genetic interaction value. The multi-dimensional state of each subsystem t, denoted by the output vector , is defined by a nonlinear function of the states of all of its child subsystems and annotated genes, concatenated in the input vector :
| (1) |
is a linear transformation of defined as . Let denote the length of , representing the number of values in the state of t and determined by:
| (2) |
Intuitively, larger subsystems have larger state vectors to capture potentially more complex biological responses. Similarly, let denote the length of . In Eqn. (1), W(t) is a weight matrix with dimensions and b(t) is a column vector with size . W(t) and b(t) provide the parameters to be learned for subsystem t. Tanh is the nonlinear transforming hyperbolic tangent function. BatchNorm47 is a normalizing function that reduces the impact of internal covariate shift caused by different scales of weights in W(t). Batch normalization can be viewed as a type of regularization of model weights and reduces the need for the traditional dropout step in deep learning. We perform the training process by minimizing the objective function:
| (3) |
Here, Loss is the squared error loss function, and r is the root of the hierarchy. Note that we compare yi with not only the root’s output, , but also the outputs of all other subsystems, . Linear in (3) denotes linear functions transforming multi-dimensional vector into a scalar. In this way, every subsystem is optimized to serve its parents as features and to predict the phenotype itself, as used previously by GoogLeNet45; the parameter α (=0.3) balances these two contributions. λ is a l2 norm regularization factor determined by four-fold cross validation. To train the DCell model, we initialize all weights uniformly at random between −0.001 and 0.001. We optimize the objective function using ADAM48, a popular stochastic gradient descent algorithm, with mini-batch size of 15,000. Gradients with respect to model parameters are computed by standard back-propagation49. Note that while other hyperparameters might influence the overall predictive performance, they are unrelated to our focus on biological interpretation as long as the same settings are applied to both DCell and the black-box models we use as controls (Fig. 2d). We implemented DCell using the Torch7 library (https://github.com/torch/torch7) on Tesla K20 GPUs.
Training Genotype-Phenotype Data
Several forms of the model were employed in this study, trained on either Costanzo et al. 2010 (~3 million training examples)16 or a more recently published update in 2016 (~8 million training examples)15. The first model was used for all results and figures in the main text to enable comparisons against previous approaches to predict genetic interactions. The latter model with updated data is provided at d-cell.ucsd.edu.
Alternative Genotype-Phenotype Translation Methods
We compared DCell to three state-of-the-art non-hierarchical approaches for predicting genetic interactions: flux balance analysis (FBA)35, multi-network multi-classifier (MNMC)19, and guilt-by-association (GBA)18. FBA uses a model of metabolism to assess the impact on cell growth of gene deletions in metabolic pathways. MNMC is an ensemble supervised learning system that uses many different datasets as features to predict genetic interactions. GBA predicts the genetic interaction score of pairwise gene deletions based on the phenotypes of their network neighbors. We also compared against our previous prediction method (Ontotype)13 which applies prior knowledge from a hierarchy like GO or CliXO but does not use deep learning nor simulate the internal states of subsystems. Ontotype counts the number of genes knocked in every GO term and uses these counts as features in a random forest regression.
Relative Local Improvement in Predictive Power (RLIPP)
The RLIPP score was used to quantify and compare the importance of DCell’s internal subsystems in prediction of phenotype. To calculate the RLIPP score of a subsystem, we compared two different linear models for phenotypic prediction. In the first model, the subsystem’s neurons were used as features in a l2-norm penalized linear regression (Supplementary Fig. 3a). In the second model, the neurons of the subsystem’s children were used as the features instead. Each model was trained separately, with the optimal hyper-parameter associated with the l2-norm penalty determined in five-fold cross validation. The performance of each of these two models was calculated as the Spearman correlation between the predicted and measured phenotype, here taken as genetic interaction scores (Supplementary Fig. 3b,c). The RLIPP score was defined as the performance of the parent model relative to that of the children (Supplementary Fig. 3d). A positive RLIPP score indicates that the state of the parent subsystem is more predictive of phenotype than the states of its children. This situation can occur when the parent learns complex (nonlinear) patterns from the children, as opposed to merely copying or adding their values. The intuition behind the RLIPP score is similar to a related ‘linear probe’ technique developed in a previous study to characterize the utility of each layer of a deep neural network50.
Identification of subsystems that mimic Boolean logic gates
As one means to interpret the mechanisms by which DCell translates genotype to phenotype, we evaluated each subsystem for the extent to which it approximates Boolean logic. In particular, we considered all trios of subsystems, each consisting of a parent subsystem and two of its children, and tested whether their binary states (S,C1,C2) were well-approximated by non-trivial Boolean logic (Supplementary Table 1). For each genotype, the binary state of each child subsystem was defined as either ‘Wild Type’ (True) or ‘Disrupted’ (False), by comparing PC1 to the wild-type state. The binary state of each parent subsystem was defined as either ‘≤Wild Type’ (True) or ‘>Wild Type’ (False), by comparing PC1 to the wild-type state. For each combinatorial state (C1,C2) of two child subsystems, the parent state S implied by DCell was determined based on the majority parent states of genotypes annotated to (C1,C2). For instance, suppose that for all the genotypes that induce (C1=True, C2=False) in the two children, DCell transforms 80% to parent state S=True and 20% to state S=False. We conclude the underlying logic for the parent subsystem to translate the signal from children subsystems is (True, False)→True. By checking the parent states for all four possible (C1,C2) combinations, we can decide whether this trio of subsystems exhibits Boolean logic (Supplementary Table 1). A trio belongs to none of the logic functions if >50% of all the genotypes or <4 genotypes are annotated to any (C1,C2) combinatorial state, or none of the annotated genotypes yield significant genetic interactions (|ε|<=0.08). For those subsystems exhibiting Boolean logic, we excluded ‘trivial’ functions in which the parent is always True, always False, or follows one of the children without dependence on the other.
DCell server construction
The DCell server (http://d-cell.ucsd.edu/) comprises several interconnected components working in unison to collect user input, run simulations, and transcode results to the web interface. On the backend, the DCell neural network model runs on the Torch library on a dedicated multi-GPU machine. On the front end, the web interface is built on cytoscape.js51 and an in-house D352 graph visualizer to display a subgraph of the hierarchy, and React53 for agile DOM (Document Object Model) editing54. To respond to user input, including searching and viewing details of model subsystems, a low-latency proxy service translates between plain text fetched from the front end and binary data used by the backend. An Elasticsearch cluster55 caches and indexes data for fast lookup and predictions. All web services run on a Kubernetes-based cloud infrastructure (http://kubernetes.io/) that auto-scales to heavy workloads. The result of these efforts is to allow easy visualization and interactivity of the model.
Life Sciences Reporting Summary
Further information on experimental design is available in the Life Sciences Reporting Summary.
Availability
Server, software implementation and the dataset used in this work are available at http://d-cell.ucsd.edu/
Supplementary Material
Acknowledgments
We gratefully acknowledge support for this work provided by grants from the National Institutes of Health to TI (TR002026, GM103504, HG009979). We also wish to thank Dr. Terry Sejnowski and Dr. Michael Kramer for very helpful comments during development of this work.
Footnotes
Author contributions
J.M., M.K.Y, S.F., R.S. and T.I. designed the study and developed the conceptual ideas. J.M. implemented the main algorithm. M.K.Y collected all the input sources. J.M. and S.F. implemented all other computational methods and conducted analysis. J.M., M.K.Y., S.F. and T.I. wrote the manuscript with suggestions from the other authors. J.M., M.K.Y, S.F., K.O., E.S. and B.D. designed and developed the server.
Competing financial interests
Trey Ideker is co-founder of Data4Cure, Inc. and has an equity interest. Trey Ideker has an equity interest in Ideaya BioSciences, Inc. The terms of this arrangement have been reviewed and approved by the University of California, San Diego in accordance with its conflict of interest policies.
References
- 1.Farabet C, Couprie C, Najman L, Lecun Y. Learning hierarchical features for scene labeling. IEEE Trans Pattern Anal Mach Intell. 2013;35:1915–1929. doi: 10.1109/TPAMI.2012.231. [DOI] [PubMed] [Google Scholar]
- 2.Mikolov T, Deoras A, Povey D, Burget L, Černocký J. Strategies for training large scale neural network language models. in. 2011 IEEE Workshop on Automatic Speech Recognition Understanding; 2011. pp. 196–201. [Google Scholar]
- 3.Hinton G, et al. Deep Neural Networks for Acoustic Modeling in Speech Recognition: The Shared Views of Four Research Groups. IEEE Signal Process Mag. 2012;29:82–97. [Google Scholar]
- 4.Sainath TN, Mohamed Ar, Kingsbury B, Ramabhadran B. Deep convolutional neural networks for LVCSR. 2013 IEEE International Conference on Acoustics, Speech and Signal Processing; 2013. pp. 8614–8618. [Google Scholar]
- 5.Collobert R, et al. Natural Language Processing (Almost) from Scratch. J Mach Learn Res. 2011;12:2493–2537. [Google Scholar]
- 6.LeCun Y, Bengio Y, Hinton G. Deep learning. Nature. 2015;521:436–444. doi: 10.1038/nature14539. [DOI] [PubMed] [Google Scholar]
- 7.Silver D, et al. Mastering the game of Go with deep neural networks and tree search. Nature. 2016;529:484–489. doi: 10.1038/nature16961. [DOI] [PubMed] [Google Scholar]
- 8.Brosin HW. An Introduction to Cybernetics. Br J Psychiatry. 1958;104:590–592. [Google Scholar]
- 9.The Gene Ontology Consortium. Expansion of the Gene Ontology knowledgebase and resources. Nucleic Acids Res. 2016 doi: 10.1093/nar/gkw1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dutkowski J, et al. A gene ontology inferred from molecular networks. Nat Biotechnol. 2013;31:38–45. doi: 10.1038/nbt.2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kramer M, Dutkowski J, Yu M, Bafna V, Ideker T. Inferring gene ontologies from pairwise similarity data. Bioinformatics. 2014;30:i34–42. doi: 10.1093/bioinformatics/btu282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carvunis AR, Ideker T. Siri of the cell: what biology could learn from the iPhone. Cell. 2014;157:534–538. doi: 10.1016/j.cell.2014.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yu MK, et al. Translation of Genotype to Phenotype by a Hierarchy of Cell Subsystems. Cell Syst. 2016;2:77–88. doi: 10.1016/j.cels.2016.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Copley SD. Moonlighting is mainstream: paradigm adjustment required. Bioessays. 2012;34:578–588. doi: 10.1002/bies.201100191. [DOI] [PubMed] [Google Scholar]
- 15.Costanzo M, et al. A global genetic interaction network maps a wiring diagram of cellular function. Science. 2016;353 doi: 10.1126/science.aaf1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Costanzo M, et al. The genetic landscape of a cell. Science. 2010;327:425–431. doi: 10.1126/science.1180823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Szappanos B, et al. An integrated approach to characterize genetic interaction networks in yeast metabolism. Nat Genet. 2011;43:656–662. doi: 10.1038/ng.846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee I, et al. Predicting genetic modifier loci using functional gene networks. Genome Res. 2010;20:1143–1153. doi: 10.1101/gr.102749.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pandey G, et al. An integrative multi-network and multi-classifier approach to predict genetic interactions. PLoS Comput Biol. 2010;6 doi: 10.1371/journal.pcbi.1000928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xu C, Wang S, Thibault G, Ng DTW. Futile protein folding cycles in the ER are terminated by the unfolded protein O-mannosylation pathway. Science. 2013;340:978–981. doi: 10.1126/science.1234055. [DOI] [PubMed] [Google Scholar]
- 21.Free SJ. Fungal Cell Wall Organization and Biosynthesis. Advances in Genetics. 2013:33–82. doi: 10.1016/B978-0-12-407677-8.00002-6. [DOI] [PubMed] [Google Scholar]
- 22.Walter P, Ron D. The Unfolded Protein Response: From Stress Pathway to Homeostatic Regulation. Science. 2011;334:1081–1086. doi: 10.1126/science.1209038. [DOI] [PubMed] [Google Scholar]
- 23.Scrimale T, Didone L, de Mesy Bentley KL, Krysan DJ. The unfolded protein response is induced by the cell wall integrity mitogen-activated protein kinase signaling cascade and is required for cell wall integrity in Saccharomyces cerevisiae. Mol Biol Cell. 2009;20:164–175. doi: 10.1091/mbc.E08-08-0809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jonikas MC, et al. Comprehensive characterization of genes required for protein folding in the endoplasmic reticulum. Science. 2009;323:1693–1697. doi: 10.1126/science.1167983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Srivas R, et al. A UV-induced genetic network links the RSC complex to nucleotide excision repair and shows dose-dependent rewiring. Cell Rep. 2013;5:1714–1724. doi: 10.1016/j.celrep.2013.11.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cadet J, Sage E, Douki T. Ultraviolet radiation-mediated damage to cellular DNA. Mutat Res. 2005;571:3–17. doi: 10.1016/j.mrfmmm.2004.09.012. [DOI] [PubMed] [Google Scholar]
- 27.Pareto V, Page AN. Translation of Manuale di economia politica (‘Manual of political economy’) AM Kelley. 1971 [Google Scholar]
- 28.Farrugia G, Balzan R. Oxidative Stress and Programmed Cell Death in Yeast. Front Oncol. 2012;2 doi: 10.3389/fonc.2012.00064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pujol-Carrion N, de la Torre-Ruiz MA. Glutaredoxins Grx4 and Grx3 of Saccharomyces cerevisiae play a role in actin dynamics through their Trx domains, which contributes to oxidative stress resistance. Appl Environ Microbiol. 2010;76:7826–7835. doi: 10.1128/AEM.01755-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gene Ontology Consortium. Gene Ontology Consortium: going forward. Nucleic Acids Res. 2015;43:D1049–56. doi: 10.1093/nar/gku1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim H, et al. YeastNet v3: a public database of data-specific and integrated functional gene networks for Saccharomyces cerevisiae. Nucleic Acids Res. 2014;42:D731–6. doi: 10.1093/nar/gkt981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang J, et al. Genetic variance estimation with imputed variants finds negligible missing heritability for human height and body mass index. Nat Genet. 2015;47:1114–1120. doi: 10.1038/ng.3390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang J, Zaitlen NA, Goddard ME, Visscher PM, Price AL. Advantages and pitfalls in the application of mixed-model association methods. Nat Genet. 2014;46:100–106. doi: 10.1038/ng.2876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen WW, Niepel M, Sorger PK. Classic and contemporary approaches to modeling biochemical reactions. Genes Dev. 2010;24:1861–1875. doi: 10.1101/gad.1945410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Szappanos B, Kovács K, Szamecz B, Honti F. An integrated approach to characterize genetic interaction networks in yeast metabolism. Nature. 2011 doi: 10.1038/ng.846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Karr JR, et al. A whole-cell computational model predicts phenotype from genotype. Cell. 2012;150:389–401. doi: 10.1016/j.cell.2012.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lipton ZC. The mythos of model interpretability. 2017 Preprint at https://arxiv.org/abs/1606.03490.
- 38.Mahendran A, Vedaldi A. Understanding deep image representations by inverting them. Proceedings of the IEEE conference on computer vision and pattern recognition; 2015. pp. 5188–5196. [Google Scholar]
- 39.Vondrick C, Khosla A, Malisiewicz T, Torralba A. Hoggles: Visualizing object detection features. Proceedings of the IEEE International Conference on Computer Vision; 2013. pp. 1–8. [Google Scholar]
- 40.Weinzaepfel P, Jégou H, Pérez P. Reconstructing an image from its local descriptors. CVPR 2011. 2011:337–344. [Google Scholar]
- 41.Chakraborty S, et al. Interpretability of deep learning models: a survey of results. 2017 DAIS.
- 42.Bahdanau D, Cho K, Bengio Y. Neural machine translation by jointly learning to align and translate. 2016 Preprint at https://arxiv.org/abs/1409.0473.
- 43.Lei T, Barzilay R, Jaakkola T. Rationalizing neural predictions. 2016 Preprint at https://arxiv.org/abs/1606.04155.
- 44.Visscher PM, Brown MA, McCarthy MI, Yang J. Five years of GWAS discovery. Am J Hum Genet. 2012;90:7–24. doi: 10.1016/j.ajhg.2011.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Szegedy C, et al. Going deeper with convolutions. 2015 IEEE Conference on Computer Vision and Pattern Recognition (CVPR); IEEE; pp. 1–9. [Google Scholar]
- 46.Lee CY, Xie S, Gallagher PW, Zhang Z, Tu Z. Deeply-Supervised Nets. AISTATS. 2015;2:5. [Google Scholar]
- 47.Ioffe S, Szegedy C. Batch Normalization: Accelerating Deep Network Training by Reducing Internal Covariate Shift. 2015 Preprint at https://arxiv.org/abs/1502.03167.
- 48.Kingma DP, Ba J. Adam: A Method for Stochastic Optimization. 2017 Preprint at https://arxiv.org/abs/1412.6980.
- 49.Rumelhart DE, Hinton GE, Williams RJ. Learning representations by back-propagating errors. Cognitive modeling. 1988;5:1. [Google Scholar]
- 50.Alain G, Bengio Y. Understanding intermediate layers using linear classifier probes. 2016 Preprint at https://arxiv.org/abs/1610.01644.
- 51.Franz M, et al. Cytoscape.js: a graph theory library for visualization and analysis. Bioinformatics. 2016;32:309–311. doi: 10.1093/bioinformatics/btv557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bostock M, Ogievetsky V, Heer J. D3: Data-Driven Documents. IEEE Trans Vis Comput Graph. 2011;17:2301–2309. doi: 10.1109/TVCG.2011.185. [DOI] [PubMed] [Google Scholar]
- 53.Stefanov S. React: Up & Running: Building Web Applications. O’Reilly Media, Inc; 2016. [Google Scholar]
- 54.Wood L, Nicol G, Robie J, Champion M, Byrne S. Document Object Model (DOM) level 3 core specification. 2004. [Google Scholar]
- 55.Gormley C, Tong Z. Elasticsearch: The Definitive Guide: A Distributed Real-Time Search and Analytics Engine. O’Reilly Media, Inc; 2015. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.