Summary
Ancient DNA from Vanuatu and Tonga dating to about 2900-2600 years ago (BP) has revealed that the “First Remote Oceanians” associated with the Lapita archaeological culture were directly descended from the population that, beginning around 5,000 BP, spread Austronesian languages from Taiwan to the Philippines, western Melanesia, and eventually Remote Oceania. Thus, ancestors of the First Remote Oceanians must have passed by the Papuan-ancestry populations they encountered in New Guinea, the Bismarck Archipelago, and the Solomon Islands with minimal admixture [1]. However, all present-day populations in Near and Remote Oceania harbor >25% Papuan ancestry, implying that additional eastward migration must have occurred. We generated genome-wide data for 14 ancient individuals from Efate and Epi Islands in Vanuatu from 2900-150 BP, as well as 185 present-day individuals from 18 islands. We find that people of almost entirely Papuan ancestry arrived in Vanuatu by around 2300 BP, likely reflecting migrations a few hundred years earlier at the end of the Lapita period, when there is also evidence of changes in skeletal morphology and cessation of long-distance trade between Near and Remote Oceania [2, 3]. Papuan ancestry was subsequently diluted through admixture but remains at least 80–90% in most islands. Through a fine-grained analysis of ancestry profiles, we show that the Papuan ancestry in Vanuatu derives from the Bismarck Archipelago rather than the geographically closer Solomon Islands. However, the Papuan ancestry in Polynesia—the most remote Pacific islands—derives from different sources, documenting a third stream of migration from Near to Remote Oceania.
Keywords: Near Oceania, Remote Oceania, Pacific Islanders, Lapita, Migration, Ancient DNA
eTOC Blurb
Lipson, Skoglund et al. analyze ancient DNA from the Pacific island chain of Vanuatu over its entire span of occupation. After humans first arrived around 3,000 years ago, there was a nearly complete replacement of the original inhabitants by 2,300 years ago, and this second wave forms the primary ancestry of people in Vanuatu today.
Results and Discussion
We generated genome-wide data for 14 ancient individuals from central Vanuatu, including 11 newly reported individuals and higher quality data for 3 previously reported individuals [1] (Table 1; Data S1). We identified and selected cochlear bone sections of petrous bones and processed them into powder in dedicated clean rooms at University College Dublin [4]. We shipped the powder to Harvard Medical School, where in a second set of clean rooms we extracted DNA [5, 6] and created individually barcoded Illumina sequencing libraries, some of which we treated with the enzyme Uracil-DNA Glycosylase (UDG) to greatly reduce the characteristic errors associated with ancient DNA [7, 8]. We screened these libraries for evidence of authentic ancient DNA by enriching for DNA overlapping the mitochondrial genome [9], sequencing on an Illumina NextSeq500 instrument, and measuring the rates of cytosine-to-thymine damage in the terminal nucleotide and consistency with the consensus mitochondrial genome (STAR Methods) [10]. For libraries that were promising after screening, we enriched for regions targeting approximately 1.24 million single nucleotide polymorphisms (SNPs) and sequenced the enriched products (STAR Methods). We determined sex by examining the ratio of sequences overlapping the X and Y chromosomes, and for males, we estimated nuclear contamination based on the rate of apparent polymorphism on the X chromosome (present in only one copy in males) (STAR Methods; Data S1). The data for the 14 individuals passing quality control were derived from 46 Illumina libraries (1–8 per individual; Data S1). We assembled direct Accelerator Mass Spectrometry radiocarbon dates for all 14 individuals, including 10 newly reported dates (STAR Methods) (Data S1). Finally, we generated genome-wide SNP genotype data on the Affymetrix Human Origins array for 185 present-day individuals from Vanuatu who gave informed consent for studies of genetic variation (STAR Methods; Data S1).
Table 1.
Details of Ancient Vanuatu Samples Analyzed in this Study
| Sample | Code | Radiocarbon dates (marine-corrected) | Population label | Location | Sex | mtDNA haplogroup | Y haplogroup | SNPs | % Papuan ancestry |
|---|---|---|---|---|---|---|---|---|---|
| I1370 | B17.P3 | 1130-830 calBCE (3083±26 BP, Wk-21026) | Vanuatu_2900BP | Teouma, Efate | F | B4a1a1 | 237405 | −0.2±1.4 | |
| I1369 | B10B.P3 | 1070-800 calBCE (3045±30 BP, Poz-81126) | Vanuatu_2900BP | Teouma, Efate | F | B4a1a1 | 271048 | 3.0±1.4 | |
| I1368 | B30A.P3 | 1040-800 calBCE (2983±32 BP, Wk-22657) | Vanuatu_2900BP | Teouma, Efate | F | B4a1a1 | 185282 | 1.6±1.4 | |
| I5951 | TeoQE | 970-770 calBCE (2955±20 BP, PSUAMS-2411) | Vanuatu_2900BP | Teouma Quarry Edge | M | B4a1a1 | CT | 23107 | 3.9±3.5 |
| I4451 | TAP1 | 410-210 calBCE (2348±32 BP, Wk-20390) | Vanuatu_2300BP | Mele-Taplins, Efate | M | M28a7 | K2b1 | 340152 | 95.8±1.1 |
| I4096 | BURU5B | 580-710 calCE [570-680 calCE (1490±15 BP, PSUAMS-2460), 600-770 calCE (1464±30 BP, Wk-25769)] | Vanuatu_1300BP | Burumba, Epi Island | M | B4a1a1k | K2b1 | 888003 | 92.2±1.2 |
| I3921 | BURU5D | 610-770 calCE [550-670 calCE (1530±20 BP, PSUAMS-1841), 650-780 calCE (1395±15 BP, PSUAMS-2428)] | Vanuatu_1300BP | Burumba, Epi Island | M | P1d1 | K2b1 | 855305 | 91.6±1.1 |
| I5259 | Mang1 | 1320-1620 calCE (559±30 BP, Wk-20030) | Vanuatu_500BP | Mangaliliu, Efate | F | P1f | 799098 | 88.4±1.2 | |
| I4105 | WAMB1 | 1650-1950 calCE (255±20 BP, PSUAMS-1922) | Vanuatu_Epi_150BP | Wambi Bay, Epi Island | M | M28a+204 | O1a2 | 1012081 | 92.6±1.2 |
| I4106 | WAMB2 | 1670-1950 calCE (225±20 BP, PSUAMS-1923) | Vanuatu_Epi_150BP | Wambi Bay, Epi Island | M | B4a1a1a11 | O1a2 | 1020436 | 90.6±1.1 |
| I4450 | SEPU1 | 1520-1950 calCE (305±15 BP, UCIAMS-188793) | Vanuatu_Efate_150BP | Pangpang, Efate | F | P1d2 | 735460 | 90.8±1.2 | |
| I4425 | EF3_2_E | 1680-1950 calCE (200±20 BP, UCIAMS-188795) | Vanuatu_Efate_150BP | Ifira, Efate | F | P2 | 700783 | 73.6±1.4 | |
| I4424 | EF_Pango1 | 1670-1950 calCE (190±15 BP, UCIAMS-188794) | Vanuatu_Efate_150BP | Pango Village, Efate | M | B4a1a1 | M1b | 780469 | 78.6±1.4 |
| I4419 | BB1 | 1690-1930 calCE (135±15 BP, UCIAMS-188792) | Vanuatu_Efate_150BP | Banana Bay, Efate | M | B4a1a1 | K2b1 | 763556 | 86.2±1.2 |
Note: The first three samples listed are previously published individuals [1] but with new libraries now added to increase coverage; the other 11 are newly published individuals. Mitochondrial DNA haplogroups were called after merging data from all libraries. For the mtDNA and Y chromosome columns, underlining indicates typical East Asian (First Remote Oceanian) haplogroups, while lack of underlining indicates typical Australo-Papuan haplogroups (the italicized Y haplogroup CT is unclassified). See also Data S1.
Genome-wide clustering analyses
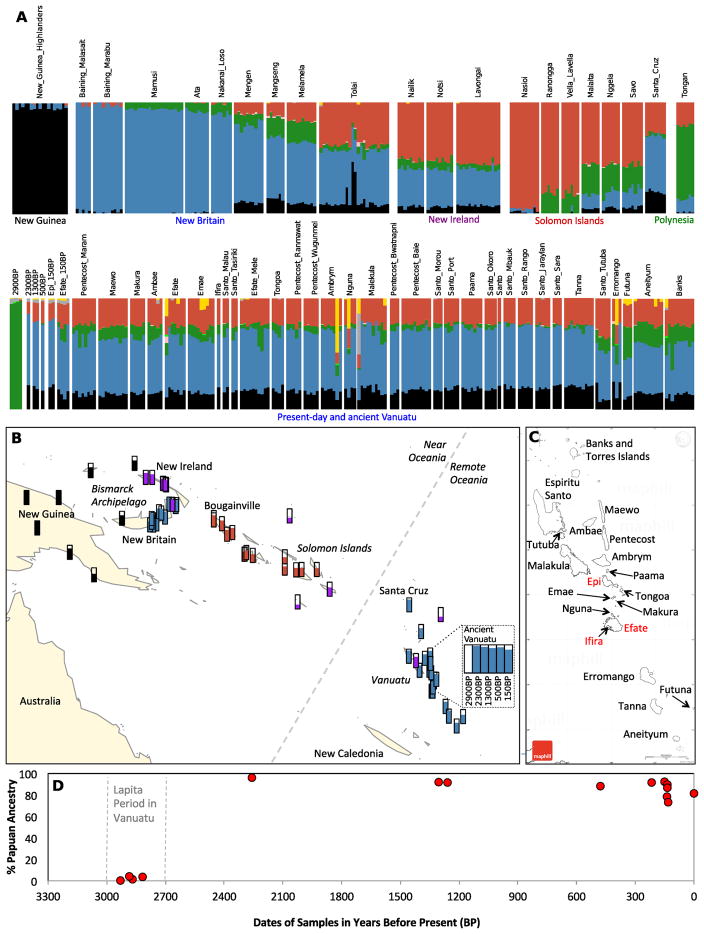
We performed automated clustering analysis with the ADMIXTURE software [11], using a data set consisting of the ancient and present-day Vanuatu samples together with other Oceanian, East Asian, and worldwide populations genotyped on the Human Origins array [1]. At K=8 clusters, four ancestry components were inferred to be widespread in Oceania (Figure 1A; Figure S1). Three correlate (predominantly) to Papuan ancestry, and are maximized in New Guinea (black in the plot), Mamusi and Baining from New Britain (blue), and Nasioi from Bougainville in the Solomon Islands (red). The fourth component (green), correlating to First Remote Oceanian ancestry, is maximized in the ~2900-2600 BP individuals from Vanuatu and Tonga. Other Oceanian populations display variable combinations of these components, forming gradients of ancestry between New Guinea, New Britain and New Ireland in the Bismarck Archipelago, and the Solomon Islands. The great majority of present-day as well as ancient groups from Vanuatu show similar ratios of the three Papuan ancestry components (although their First Remote Oceanian proportions vary), showing that they are consistent with largely deriving their Papuan ancestry from the same source. Among populations in Near Oceania, the most similar to Vanuatu in terms of the Papuan ancestry component ratio (black-to-blue-to-red) are groups from New Britain in the Bismarck Archipelago with a majority of the blue component and smaller contributions of black and red, suggesting that the Papuan ancestry in Vanuatu derives from populations in the Bismarck Archipelago (rather than the geographically closer Solomon Islands). A similar pattern was previously inferred for the Papuan ancestry in Santa Cruz, to the immediate north of Vanuatu [12], a result we replicate here.
Figure 1. Locations and broad-scale genetic structure of analyzed populations.
(A) ADMIXTURE results with K=8 clusters for selected populations (full results in Figure S1). The analysis suggests three primary Papuan components (black maximized in New Guinea, blue in New Britain in the Bismarck Islands, and red maximized in the Solomon Islands) and a component maximized in First Remote Oceanians (green). (B) Population locations with colored clusters assigned based on the ratio of Papuan ancestry components in ADMIXTURE. We loosely adopt the color scheme from ADMIXTURE, with black indicating a New Guinea-like profile, blue a New Britain-like profile, red a Solomon Islands-like profile, and purple a profile mixed between New Britain and Solomon Islands. Level of fill in the bars indicates Papuan ancestry proportion. The map was plotted in R using the ‘maps’ package with data from http://www.naturalearthdata.com/. (C) Close-up of Vanuatu labeling islands for which new data are reported. Islands with ancient samples are indicated in red (for Efate and Ifira, both present-day and ancient individuals). The map was downloaded from http://www.maphill.com/vanuatu/simple-maps/blank-map/no-labels/. (D) Papuan ancestry proportions for ancient samples over time; Efate island is used to represent the present. See also Figure S2 and Data S1.
We also carried out a principal component analysis (Figure S2), which corroborated the findings from ADMIXTURE, with the primary feature being a U-shaped cline from (1) western New Britain in the Bismarck archipelago to (2) eastern New Britain, (3) most of Vanuatu, (4) the atypical Vanuatu island of Tutuba along with the Tolai of New Britain, (5) New Ireland in the Bismarck archipelago, and finally (6) Bougainville in the Solomon Islands. This cline closely correlates to the gradient of decreasing blue and increasing red components in ADMIXTURE (Figure 1A; Figure S1). The position of the Vanuatu samples in the PCA again supports the hypothesis that the inhabitants of the region after the initial Lapita settlement derived ultimately not from populations closely related to those in the closer Solomon Islands but instead from populations related to those from the island of New Britain in the Bismarck Archipelago. We also replicated this result via the statistic f4(Australian, Vanuatu; Solomon Islands, Bismarck Archipelago), which is significantly positive for each choice of populations in the PCA (Z > 2 for all 160 comparisons; median Z > 6; STAR Methods), implying that Vanuatu populations share more alleles with groups from the Bismarck Archipelago than the Solomon Islands.
Papuan and First Remote Oceanian ancestry proportions
It has been shown that the strongest driver of genetic variation in Oceania today is the widespread but highly variable admixture between Papuan and First Remote Oceanian ancestry sources, the former representing original inhabitants of Near Oceania and the latter descendants of an expansion from East and Southeast Asia [1]. From our clustering results, a dramatic turnover is apparent in Vanuatu after around 2900 and before around 2300 years ago, with First Remote Oceanian populations being joined or possibly completely replaced by individuals of (almost) entirely Papuan ancestry. To provide precise estimates of mixture proportions, we used f4-ratio statistics [13], with East Asian reference populations Atayal (aboriginal Taiwanese related to the source population of the Austronesian expansion) and Kankanaey (an Austronesian-speaking population from the Philippines—on the migratory path from Taiwan to Remote Oceania—that has been shown to be descended from the same genetic sources as the First Remote Oceanians) [1] (Figure 1; Table 1; Data S1; STAR Methods). Taking advantage of our increased coverage compared to the first study of Lapita samples, we find that the ~2900 BP Lapita individuals likely had a non-zero proportion of Papuan-related ancestry (2.4 ± 0.9%), although it remains striking that the initial First Remote Oceanian migrants were only minimally admixed. Given the small proportion, we did not have sufficient statistical power to determine whether this Papuan-related ancestry is derived from the region surrounding New Guinea or could perhaps have been acquired elsewhere, such as in the Philippines or eastern Indonesia. Notably, the first post-Lapita sample (2300 BP from the site of Mele-Taplins) had almost entirely Papuan ancestry but with a small amount derived from First Remote Oceanians (4.2 ± 1.1%). The more recent ancient individuals are similar in their proportions to present-day populations: 8–26% First Remote Oceanian ancestry, as compared to a range of 9–38% today (mostly 12–20%, and highest on the island of Futuna, which harbors a “Polynesian Outlier” population, that is, one that speaks a Polynesian language due to east-to-west back-migration from Polynesia [14]). For time points with multiple samples, the individuals’ mixture proportions are statistically indistinguishable, except for 150 BP Efate (point estimates of 9%, 14%, 21%, and 26% First Remote Oceanian). The post-Lapita ancestry turnover is also evident in uniparental markers, as the majority of mtDNA and Y-chromosome haplogroups observed from 2300-150 BP are typical of Papuan populations, albeit with the presence of some East Asian-derived haplogroups in both mtDNA and Y, showing that members of both sexes in both ancestral populations participated in the post-2300 BP Vanuatu admixture process (Table 1).
Dates of admixture
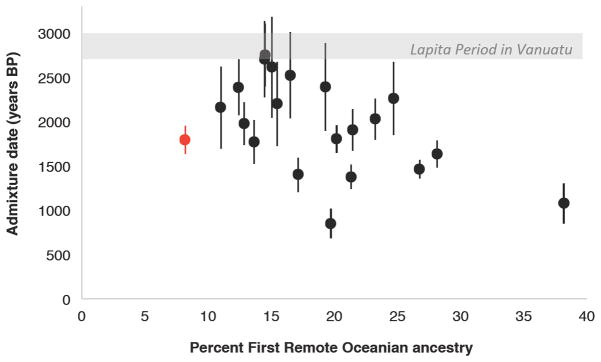
We estimated dates of admixture based on weighted admixture linkage disequilibrium (LD) [15] using ALDER [16], with Ami and New Guinea as references (Figure 2; Data S1). We obtain significant evidence for admixture LD in almost all present-day populations and three ancient population groupings (noting that power is highly sample size-dependent). The date estimates are mostly 40–100 generations before present, or 1,100–2,800 years ago assuming 28 years per generation [17], consistent with admixture having occurred soon after the early settlement of Vanuatu and continuing through time (in cases of multiple pulses of admixture, ALDER produces a single average date). We observe a significant negative correlation between admixture date and First Remote Oceanian ancestry proportion (R2 = 0.33 at p < 0.01 for populations in Figure 2; R2 = 0.21 at p < 0.01 for all present-day populations; STAR Methods), as expected if a subset of populations (e.g., Efate, Emae, Futuna, Makura) received more recent pulses of gene flow from groups with high proportions of First Remote Oceanian ancestry. This scenario is plausible in light of Polynesian (Samoan) cultural influences and language replacement and the establishment of Polynesian Outlier populations on islands such as Ifira and Emae in central Vanuatu and Futuna in southern Vanuatu within the last several hundred years [14, 18].
Figure 2. Ancestry proportions and dates of admixture in Vanuatu.
Black points represent the 20 present-day populations with the most confident admixture date estimates from ALDER (as measured by Z-score for difference from zero), assuming 28 years per generation and showing one standard error in each direction. The red point represents a pair of ancient individuals with a mean calibrated date of 1283 BP (18 ± 6 generations, or 1800 ± 160 years BP). Gray shading indicates the Lapita period in Vanuatu (~3000-2700 BP). See also Data S1.
We also obtain a direct ALDER date of 18 ± 6 generations in the past (500 ± 160 years) for a pair of ancient samples from Vanuatu radiocarbon dated to ~1,300 years ago, coinciding with the typical range of admixture dates in present-day groups (Figure 2). Together with the present-day results, this observation is relevant to the ongoing debate about the timing of admixture between people of East Asian and Papuan ancestry in Remote Oceania. Methods based on wavelet transformations have suggested mixing at a date older than 3,000 BP, prior to the Lapita expansion to Remote Oceania [12, 19], whereas methods based on admixture LD have suggested more recent dates, implying that mixture occurred following later streams of gene flow [20]. It has been argued that the differences may reflect systematic biases of the methods for dates older than a couple of thousand years [12]. Thus, our finding of a definitively post-Lapita date in samples that are closer in time to the admixture provides compelling evidence for the hypothesis of more recent mixture. A plausible scenario is that the initial migration of Papuan populations occurred during the late Lapita period (before ~2700 BP), at which time archaeological evidence such as the transport of New Britain obsidian to Vanuatu documents links between the New Britain region and Remote Oceania (Santa Cruz and Vanuatu) bypassing much of the Solomon Islands [3], a pattern very similar to the population affinities seen in the genetic data. The near-complete population turnover attested to by genetic data may thus correspond to the evidence of transformation at the end of the Lapita period to more localized cultures, initiating a period of hundreds of years when inter-archipelago contacts appear to have nearly ceased [3].
Phylogeny of First Remote Oceanian ancestry
To test whether the First Remote Oceanian ancestry in ancient and present-day groups is more closely related to Lapita samples from Tonga or Vanuatu, we compared the values of the statistics f4(Test, Han; Atayal, Tonga_2600BP) and f4(Test, Han; Atayal, Vanuatu_2900BP) for Oceanian populations as Test (STAR Methods). We found a trend toward greater allele-sharing with Tonga, with significant results in Polynesian and to a lesser degree Polynesian Outlier populations (Data S1). These results show that the First Remote Oceanian ancestry in Polynesians today is derived from a source that was closer to the sampled Lapita-period population from Tonga than to the Vanuatu Lapita population. For post-Lapita populations (ancient and present-day) from Vanuatu, however, we do not have sufficient statistical power to determine which potential First Remote Oceanian source is closer.
Phylogeny of Papuan ancestry
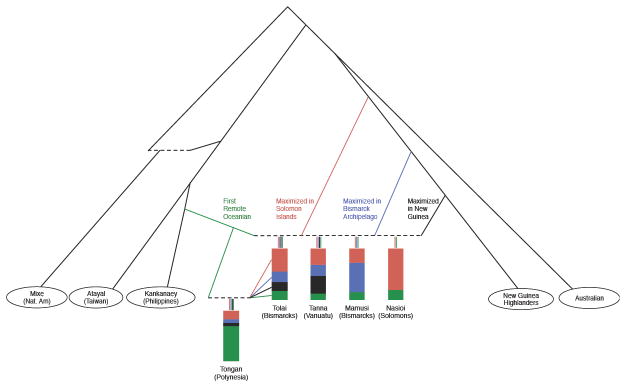
We built admixture graphs to explore in more detail the different streams of Papuan ancestry present in Oceania. We used as reference populations Australia, Kankanaey, Atayal, and Mixe, together with representatives of major poles of Papuan genetic variation inferred from the ADMIXTURE analysis: Vanuatu_Tanna, Mamusi (New Britain), Nasioi (Solomon Islands), New Guinea, and Tolai (New Britain/New Ireland). To avoid overfitting, we adopted a restricted framework in which the ancestry in each population was modeled as a combination of the same set of source lineages, with the exception of the unadmixed New Guinea population. We found that three Papuan source lineages were necessary in order to obtain a good fit for the model—one maximized in Mamusi, one maximized in Nasioi, and one closest to New Guinea—showing that the implied ancestry components from ADMIXTURE (Figure S1) are all well-supported in formal models based on allele-sharing statistics (Figure S3). Additionally, the admixture graph analysis suggests that the blue (Bismarck Archipelago-majority) and red (Solomon Islands-majority) ADMIXTURE components represent admixed ancestry. In particular, both include First Remote Oceanian ancestry (~20% for red and ~5% for blue), and the two are additionally admixed with each other, as we could not fit a Solomon Islands population (e.g., Nasioi) and a Bismarck Archipelago population (e.g., Mamusi or Baining) simultaneously without admixture from one to the other. In our models, we included Solomon Islands-type ancestry in Mamusi (approximately one-third of its total Papuan ancestry), although we were unable to distinguish the direction(s) of gene flow. Vanuatu was confidently inferred to have ancestry from all three Papuan sources (|Z| > 8 for omitting any source).
We next asked if we could add Polynesians (Tongan) as a mixture of a component related to one of the other Oceanian populations along with additional First Remote Oceanian ancestry. Such a model was successful only in one configuration, with Tongan as a mixture of a population related to the Tolai of New Britain plus additional First Remote Oceanian ancestry (all f-statistics fit to within 2.0 standard errors of their observed values except for one residual, f4(Kankanaey, Tongan; Australian, Vanuatu_Tanna), at Z = 2.7; Figure 3; Figure S3). Our choice to include Tolai in the model was guided by the ADMIXTURE analysis, in which the Papuan ancestry profile in Polynesians appears to match that in Tolai (and Tutuba, from near Santo island in Vanuatu) more closely than other populations. The Tolai are known to be descended from relatively recent mixture between groups from New Ireland and New Britain (resulting from displacement caused by the eruption of the Rabaul caldera ~1400 BP [18]), so their ancestors cannot represent the true source population of the Papuan ancestry in Polynesians. However, the similarity of Tolai Papuan ancestry to Polynesians suggests that the Papuan component in Polynesians could similarly be from a mixture of multiple Near Oceanian sources. Given that Tolai are genetically intermediate between populations from New Britain and New Ireland (the latter with higher Solomon Islands-related ancestry), Polynesians could plausibly have acquired New Britain-related ancestry from Vanuatu or Santa Cruz, along with ancestry more closely related to that in New Ireland or the Solomon Islands via a distinct stream of migration.
Figure 3. Inferred admixture graph model with diverse present-day Oceanian populations.
Dotted lines denote admixture events. For five populations, the proportions of four fitted ancestry sources maximized in First Remote Oceanians (green), Solomon Islands (red), Bismarck Archipelago (blue) and New Guinea (black) are shown. Papuan ancestry is inferred to be highly simlar in the Tolai and in the Tongan pouplation, allowing Tongan to be fit as a mixture of a group with ancestry similar to Tolai and additional ancestry from First Remote Oceanians. Colors are chosen to be correlated to the components inferred from ADMIXTURE (Figure S1), but the ADMIXTURE components represent combinations of the sources given here, and hence the ratios differ between the methods. Full model parameters for the admixture graph are shown in Figure S3.
As suggested by similar mixtures of components in ADMIXTURE, the ancient Vanuatu individuals are broadly consistent with descent from the same common ancestral population as present-day groups from Vanuatu. In the admixture graphs, we could fit most of the ancient sample groups as sister populations to Vanuatu_Tanna, albeit with different proportions of First Remote Oceanian ancestry. The one exception was the 150 BP grouping of individuals from Efate (with ~18% First Remote Oceanian ancestry), which showed significant un-modeled allele sharing with Tongan (maximum residual Z = 3.7, after accounting for excess First Remote Oceanian ancestry). Some present-day Vanuatu populations, such as Efate, Makura, and Polynesian Outliers, show a similar pattern when tested in the model, likely reflecting migration of Polynesians to Vanuatu in the last thousand years or less.
Conclusion
By analyzing a time transect of central Vanuatu from initial settlement through the present, combined with dense geographical sampling of present-day populations from 18 islands in Vanuatu and dozens of populations outside Vanuatu, we document a series of dramatic genetic shifts associated with consistently high human mobility through a total of at least four distinct streams of migration and admixture. First, the initial human migration to central Vanuatu involved First Remote Oceanians associated with the Lapita culture. Second, by ~2300 BP, these groups were almost completely displaced by Papuan-ancestry populations originally from the Bismarck Archipelago, who remain the source for most of the ancestry of people in Vanuatu today. Third, in Polynesia, we find evidence for a different Papuan ancestry type that reflects a distinct migration. Finally, these streams of ancestry reconnected in parts of Vanuatu, influenced by back-migration from Polynesia. These results highlight the importance of multiple episodes of migration and mixture in shaping the human diversity of Oceania.
STAR Methods
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, David Reich (reich@genetics.med.harvard.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Archaeological Context on Ancient Individuals with New Genome-Wide Data
We newly report data from 14 ancient skeletons. For 3 of these skeletons we are reporting new ancient DNA data increasing the quality of the dataset beyond the data reported from the same samples in a previous study [1]. For 11 samples the data are entirely new:
Teouma, Efate Island (~2900 BP) – Lapita Culture (n=4 samples)
The Teouma Lapita culture cemetery and settlement site is discussed in detail in the Supplementary Information of Skoglund et al. 2016 and references [1]. The additional sample I5951 was displaced during quarrying activities before controlled archaeological excavations began at the site in 2004. Given its radiocarbon date it is highly likely to have been from a disturbed burial context of Lapita age and can be legitimately considered with the other Lapita-age skeletons from the site.
-
I5951 (TeoQE), Vanuatu_2900BP
Newly reported sample
Genetic Sex: Male
Radiocarbon Date: 970-770 calBCE (2955±20 BP, PSUAMS-2411, marine corrected)
-
I1370 (B17.P3), Vanuatu_2900BP
Previously reported in [1]; here we report higher coverage data
Genetic Sex: Female
Radiocarbon Date: 1130-830 calBCE (3083±26 BP, Wk-21026, marine corrected)
-
I1369 (B10B.P3), Vanuatu_2900BP
Previously reported in [1]; here we report higher coverage data
Genetic Sex: Female
Radiocarbon Date: 1070-800 calBCE (3045±30 BP, Poz-81126, marine corrected)
-
I1368 (TB30A.P3), Vanuatu_2900BP
Previously reported in [1]; here we report higher coverage data
Genetic Sex: Female
Radiocarbon Date: 1040-800 calBCE (2983±32 BP, Wk-22657, marine corrected)
Mele-Taplins, Efate Island (~2300 BP) (n=1 sample)
The Mele-Taplins site is described by Valentin and colleagues [21]. The skeleton comes from a subsurface grave in a rockshelter (Taplins 1) at the base of a cliff, excavated by Graeme Ward of The Australian National University in 1973-4 and curated at Otago University, Dunedin, New Zealand. Other burials from the Taplins 2 shelter were of broadly similar age.
-
I4451 (TAP1), Vanuatu_2300BP
Newly reported sample
Genetic Sex: Male
Radiocarbon Date: 410-210 calBCE (2348±32 BP, Wk-20390, marine corrected)
Burumba, Epi Island (~1300 BP) (n=2 samples)
The Burumba site is described by Valentin and colleagues [21] and excavated in 2006 by Frederique Valentin and Jacques Bole. The graves of nine adults were excavated from an open site at Kalala Plantation 200m from the current beach, dug into sterile sand. Burial 5 was an assemblage of cranial remains of five individuals placed on a pile of coral slabs and blocks.
-
I3921 (BURU5D), Vanuatu_1300BP
Newly reported sample
Genetic Sex: Male
Radiocarbon Date: 610-770 calCE [550-670 calCE (1530±20 BP, PSUAMS-1841), 650-780 calCE (1395±15 BP, PSUAMS-2428), marine corrected]
-
I4096 (BURU5B), Vanuatu_1300BP
Newly reported sample
Genetic Sex: Male
Radiocarbon Date: 580-710 calCE [570-680 calCE (1490±15 BP, PSUAMS-2460), 600-770 calCE (1464±30 BP, Wk-25769), marine corrected]
Mangaliliu, Efate Island (~500 BP) (n=1 sample)
The burial was excavated from a test pit in Mangaliliu village by Richard Shing in 2002 and described in detail by Valentin and colleagues [22]. The originally reported age of the burial was reassessed after direct dating of the skeleton [20].
-
I5259 (burial 1, Mang1), Vanuatu_500BP, Mangaliliu (Efate Island)
Newly reported sample
Genetic Sex: Female
Radiocarbon Date: 1320-1620 calCE (559±30 BP, Wk-20030, marine corrected)
Pangpang, Efate Island (~150 BP) (n=1 sample)
This burial, in a flexed position, was excavated by Richard Shing and Iarawai Phillip during archaeological impact assessment related to the Efate Ring Road construction between the villages of Pangpang and Forari. The body was adorned with ornaments composed of numerous tiny Conus shell and shark vertebrae beads and a large pearl shell pendant. This range of ornaments has been recorded in burial contexts of the last 400 years, prior to and during the initial phases of European contact (unpublished field notes, Vanuatu National Museum).
-
I4450 (SEPU1, Sepulture 1), Vanuatu_Efate_150BP
Newly reported sample
Genetic Sex: Female
Radiocarbon Date: 1520-1950 calCE (305±15 BP, UCIAMS-188793, marine corrected)
Wam Bay, Epi Island (~150 BP) (n=2 samples)
The site appears to have been a largely Mission period cemetery of the late 19th to early 20th centuries. Three burials were exposed in proximity to a combustion feature associated with the making of lime-plaster for construction, a European introduced practice. The site was excavated by Frederique Valentin and Matthew Spriggs in 2006 (unpublished field notes, Vanuatu National Museum).
-
I4105 (WAMB1), Vanuatu_Epi_150BP
Newly reported sample
Genetic Sex: Male
Radiocarbon Date: 1650-1950 calCE (255±20 BP, PSUAMS-1922, marine corrected)
-
I4106 (WAMB2), Vanuatu_Epi_150BP
Newly reported sample
Genetic Sex: Male
Radiocarbon Date: 1670-1950 calCE (225±20 BP, PSUAMS-1923, marine corrected)
Ifira, Efate Island (~150 BP) (n=1 sample)
This tightly flexed burial from a feature containing skeletal remains of two individuals was excavated by Mary Elizabeth and Richard Shutler, Jr, in June 1964 on the small island of Ifira in Vila Harbor, Port Vila, during a test pit survey of the island. It is briefly mentioned in Shutler and Shutler [23]. Unpublished field notes relating to the excavation are held in the files of the Vanuatu National Museum. Ifira is notable as one of the Vanuatu Polynesian Outlier islands and this burial would date to the period of Polynesian cultural influence.
-
I4425 (EF3_2_E, Pit 2; Loc E), Vanuatu_Efate_150BP
Newly reported sample
Genetic Sex: Female
Radiocarbon Date: 1680-1950 calCE (200±20 BP, UCIAMS-188795, marine corrected)
Pango Village, Efate Island (~150 BP) (n=1 sample)
This is one of two individuals excavated by Mary Elizabeth and Richard Shutler, Jr. on the Pango Peninsula opposite the small island of Ifira in Vila Harbour, Port Vila. Unpublished field notes relating to the burial are held in the files of the Vanuatu National Museum, but the notes provide limited detail.
-
I4424 (EF_Pango1), Vanuatu_Efate_150BP
Newly reported sample
Genetic Sex: Male
Radiocarbon Date: 1670-1950 calCE (190±15 BP, UCIAMS-188794, marine corrected)
Banana Bay, Efate Island (~150 BP) (n=1 sample)
The burial was excavated by Richard Shing and Iarawai Phillip during archaeological impact assessment related to the Efate Ring Road construction in the Banana Bay area, southeast Efate. The body, lying on the back, was adorned with ornaments including numerous tiny Conus shell beads and a few European glass beads (unpublished field notes, Vanuatu National Museum).
-
I4419 (BB1, Burial 1), Vanuatu_Efate_150BP, Banana Bay (Efate Island)
Newly reported sample
Genetic Sex: Male
Radiocarbon Date: 1690-1930 calCE (135±15 BP, UCIAMS-188792, marine corrected)
Genotyping Data from Present-Day Vanuatu
We genotyped 185 present-day individuals from 32 populations from Vanuatu spanning 18 islands. All individuals gave informed verbal consent for studies of population history and human health, especially as they may shed light on anemia, consistent with the standards prevailing at the time the data were collected. Samples of whole blood were collected as part of a range of research projects undertaken from the late 1970s in collaborations between multiple sites and institutions in Vanuatu and the University of Oxford investigating population differences at the genetic level. In accordance with participant consent, DNA was extracted, anonymized, and stored in batches analyzable only by geographic location of participant origin. Use of the samples for genome-wide analyses including studies of population history was reviewed by the Oxford Tropical Research Ethics Community at the University of Oxford and formally approved in a letter dated July 2, 2014 (OXTREC Reference: 537-14). The use of the samples for genetic analysis was also approved by the Vanuatu Cultural Centre in a formal letter dated May 30, 2017.
METHOD DETAILS
Ancient DNA laboratory work
In dedicated clean rooms at University College Dublin, we used a dental sandblaster to separate cochlear sections from petrous bones. We milled these samples into fine powder, and shipped them to Harvard Medical School.
In dedicated clean rooms at Harvard Medical School, we extracted DNA following a previously published protocol [5], with two modifications. First, we replaced the combination of a funnel and a MinElute column with Roche columns [6]. Second, we eluted two times in 45μl, obtaining 90μl of extract for each sample (Data S1).
We prepared libraries from the extracts using a double-stranded protocol, affixing 7-base-pair sequences to either end to allow multiplexing of the libraries and to prevent contamination from affecting the samples after barcodes were added. We prepared some of the libraries in the presence of the enzyme UDG to remove characteristic damage associated with ancient DNA (Data S1) [7].
We enriched the libraries in solution for sequences overlapping the mitochondrial genome [9] as well as for 3000 nuclear positions, and sequenced on an Illumina NextSeq500 instrument for 2×76cycles + 2×7 cycles after adding a pair of unique 7-base-pair indices. For libraries that were promising after screening, we enriched for sequences overlapping approximately 1.24 million SNPs on the nuclear genome [10, 24–26]. We added two unique 7-base-pair indices to each enriched library and sequenced a multiplexed pool of samples with an Illumina NextSeq500 instrument for 2×76cycles + 2×7cycles. We iteratively sequenced more from each sample until the number of new SNPs covered per additional sequences generated was less than about 1 in 100.
For samples for which we wished to obtain more coverage, we prepared additional libraries from existing extract or new extract, leading to a total of up to 8 libraries for some samples. We pooled data from all libraries for further analysis. We also prepared versions of the sample data using only UDG-treated libraries. We use the suffix “_all” to refer to the versions of each sample with all libraries in Data S1 and Figure S2. We use the “_all” versions for our primary analyses, but also perform some analyses on the entirely UDG-treated versions to assess if there is evidence that any results are influenced by ancient DNA artifacts (all appear to be robust).
Bioinformatic processing
We demultiplexed reads from the NextSeq500 lanes into individual libraries based on the sequences of their two indices and two barcodes. To assign a read pair to a library we required no more than one mismatch to the total of four expected 7 base pair sequences. We merged sequences using SeqPrep (github.com/jstjohn/SeqPrep), requiring at least 15 base pairs of overlap. At positions of overlap, we used the allele and quality score of the read of higher quality.
We aligned merged sequences to the mitochondrial RSRS genome [27] (for mitochondrial DNA analyses) and to the hg19 reference (for whole genome analyses) using the command “samse” from BWA with default parameters (version 0.6.1) [28]. For non-UDG treated libraries, which are expected to have higher mismatch rates compared to the reference genome, we used more relaxed alignment parameters, “-n 0.01 -o 2 -l 16500”. This setting disables seeding, allowing for less conservative alignments and helping to align damaged sequences. We chose one allele at random per site (“pseudo-haploid” genotypes) from aligned sequences to use in analyses.
Mitochondrial DNA haplogroup determination
We determined mitochondrial DNA haplogroups for each library separately as well as for pools of libraries for each sample using Haplogrep2, which provides a ranking score measuring the reliability of the haplogroup assignments [29] (Data S1). The procedure used here is designed to extract the maximally informative data from the sample during haplogroup assignment by building a mitochondrial consensus sequence in multiple ways and then using the rank score to select the most confident call. First, we restricted to sequences with characteristic ancient DNA damage in their terminal nucleotides (a procedure that removes potentially contaminating sequences at the cost of greatly reduced coverage). To restrict to damaged sequences, we used the PMDtools software [30], requiring a minimum score of pmdscore=3, and then trimmed the sequences by 5 base pairs on either side to remove nucleotides likely to be deaminated before calling a haplogroup with Haplogrep2. As a second approach, we trimmed sequences by 0–7 base pairs on either side to eliminate characteristic ancient DNA damage and fed these sequences to Haplogrep2 without damage restriction (hence retaining more data), calling a haplogroup at each trim level.
Direct Accelerator Mass Spectrometry (AMS) Radiocarbon Dates
We prepared 10 new bone samples for AMS radiocarbon dating at the Human Paleoecology and Isotope Geochemistry Laboratory at the Pennsylvania State University. After preparation, we AMS dated the samples either at the W.M. Keck Carbon Cycle Accelerator Mass Spectrometry Laboratory at the University of California, Irvine (lab code: UCIAMS) or at the Accelerator Mass Spectrometer Laboratory at the Pennsylvania State University (lab code: PSUAMS). We co-analyzed our 10 newly generated dates with 6 previously published dates generated by other laboratories: five by the Radiocarbon Dating Laboratory at the University of Waikato (lab code: Wk) and one by the Poznan Radiocarbon Laboratory (lab code: Poz) [1, 21, 31].
Bone samples for the newly and previously reported direct AMS 14C dates were manually cleaned and demineralized in weak HCl and soaked in an alkali bath (NaOH) at room temperature to remove contaminating soil humates. Samples were then rinsed to neutrality in Nanopure H2O and gelatinized in HCL [32]. The resulting gelatin was lyophilized and weighed to determine percent yield as a measure of collagen preservation (% crude gelatin yield). Collagen was either directly AMS 14C dated (Wk-20390, Wk-20030) or further purified using ultrafiltration prior to analysis. At PSUAMS we hydrolyzed two bone samples with low collagen yields and purified the resulting amino acids using XAD chromotography [33].
We use stable carbon and nitrogen isotopic analysis of bone collagen (or amino acids) as an additional quality control measure. For all samples, we examined the %C, %N and C:N ratios. C:N ratios for well-preserved samples fall between 2.9 and 3.6 [34], and all our samples met this criterion. We also used stable carbon and nitrogen isotope measurements to determine the marine reservoir correction for each individual as described below. The detailed bone preparation and quality control methods we used for the newly reported dates from PSUAMS and UCIAMS are reported elsewhere [33, 35].
To calibrate the dates, we began with an adjustment for the marine reservoir effect, applying a correction (ΔR) of 40±44 BP based on marine shell measurements to adjust for local oceanic variation in 14C levels around Vanuatu as previously described by Petchey and colleagues [31]. We then corrected for mass-dependent fractionation with measured 13C values [36] and calibrated in OxCal 4.3 [37] via a mixture of the IntCal13 Southern Hemisphere and Marine13 calibration curves [38] using the Mix_Curves function in OxCal according to the marine component of each individual’s diet (estimated by δ13C values). This method uses δ13C endpoints of -21 and -12‰, the former representing a highly terrestrial diet and the latter indicating a diet rich in marine foods, with the percentage of marine dietary contribution estimated via linear interpolation [31]. In two cases, individuals were sampled twice for radiocarbon dating and stable isotope analysis: I3921 (PSUAMS-1841, PSUAMS-2428) and I4096 (PSUAMS-2460, Wk-25769). In these instances, we combined radiocarbon dates using the R Combine function in OxCal 4.3 and estimated the marine contribution according to the mean of two δ13C measurements.
QUANTIFICATION AND STATISTICAL ANALYSIS
Analysis dataset
All analyses are based on 593,124 SNPs on chromosomes 1–22 genotyped on the Affymetrix Human Origins array [39], with the newly reported data from ancient and present-day Vanuatu individuals merged with published Human Origins data [1]. Based on the ADMIXTURE results, we excluded 26 present-day individuals from analyses as outliers: 14 from Vanuatu (2 Efate, 3 Emae, 2 Malekula, 1 Ambrym, 1 Nguna, 1 Erromango, 3 Aneityum, and 1 Banks) with evidence of European admixture, 4 from Samoa also with evidence of European admixture, and 7 Tolai and 1 Tutuba with evidence of recent admixture or otherwise non-representative ancestry profiles.
Clustering analyses
We performed ADMIXTURE [11] clustering analysis using default parameters, with the cluster components (K) ranging from K=2 to K=8. We carried out principal component analysis (PCA) using the “lsqproject” and “autoshrink” options in smartpca [40, 41], computing axes using the present-day populations and projecting ancient samples. For PCA, we restricted to populations within a narrow range of Papuan ancestry proportions in order to minimize the variance due to Papuan versus First Remote Oceanian ancestry and capture components related to variation in Papuan ancestry sources. The specific range (~80% Papuan ancestry) was chosen as the highest possible that included groups from all of the major Oceanian island chains and genetic clusters. We did not project the Vanuatu_2900BP individuals because of their near-zero Papuan ancestry.
For the Papuan ancestry clusters defined in Figure 1B, we manually assigned populations based on their majority Papuan component in ADMIXTURE (out of red, blue, and black). For borderline populations with large red and blue components and small black components, we created a mixed cluster containing Kuot_Kabil, Kuot_Lamalaua, Lavongai, Madak, Nailik, Notsi, and Tigak (New Ireland); Ontong Java, Rennell and Bellona, and Tikopia (Polynesian Outliers); Makira (Solomon Islands); Tolai (New Britain); and Tutuba (Vanuatu). We assigned two borderline populations with large black components to the black cluster (Kove and Mussau).
Allele-sharing statistics
We computed allele-sharing statistics (f-statistics) in ADMIXTOOLS [39], with standard errors obtained by block jackknife. To replicate the findings from ADMIXTURE and PCA and test for differential allele sharing of populations from Vanuatu with groups from the Solomon Islands versus the Bismarck Archipelago, we computed the statistic f4(Australian, Vanuatu; Solomon Islands, Bismarck Archipelago). We used the same populations as in the PCA (10 from Vanuatu, 8 from the Bismarck Archipelago, and 2 from the Solomon Islands; Figure S2) to avoid confounding from differential proportions of First Remote Oceanian ancestry.
We computed Papuan ancestry proportions using the f4-ratio statistic f4(Atayal, Australian; Kankanaey, Test)/f4(Atayal, Australian; Kankanaey, New_Guinea_Highlander). The form of this statistic assumes a topology of (Atayal, (Kankanaey, First Remote Oceanian)) for the First Remote Oceanian ancestry in the Test population.
We measured differential affinity to Lapita samples from Vanuatu and Tonga by computing the difference between the statistics f4(Test, Han; Atayal, Tonga_2600BP) and f4(Test, Han; Atayal, Vanuatu_2900BP), using the qp4diff program (“allsnps” mode). In expectation, this difference is equal to the single statistic f4(Test, Han; Vanuatu_2900BP, Tonga_2600BP), but our formulation allows us to increase power by utilizing the union rather than the intersection of the SNPs covered by the relatively low-coverage Vanuatu_2900BP and Tonga_2600BP samples.
Dates of admixture
We estimated dates of admixture using ALDER [14]. As reference populations we used published Human Origins data for Ami (aboriginal Taiwanese; n = 10 individuals) and New Guinea Highlanders (n = 19). In computing the correlation between ancestry proportions and dates of admixture, we performed a weighted linear regression of present-day Vanuatu population groups with ALDER date estimates (treated independently; either all 31 such groups or only the 20 with most confident estimates, as shown in Figure 2), weighting by the Z-score for the difference of the date estimate from zero [42].
Admixture graph fitting
We constructed admixture graphs using the qpGraph utility in ADMIXTOOLS [41]. The position of Mixe (a Native American population from present-day Mexico) as an outgroup relative to the other populations (in an unrooted sense) means that its eastern and western Eurasian ancestry components can be collapsed into a single lineage with no change in the model. Similarly, we can omit explicit inclusion of Denisovan admixture because of the symmetry of such ancestry in the right-hand clade of the model (as displayed in Figure S3).
DATASET AND SOFTWARE AVAILABILITY
Raw sequences from the 14 individuals are available from the European Nucleotide Archive at accession number PRJEB24938. Genotype files are available at https://reich.hms.harvard.edu/datasets. To access data for the newly genotyped present-day individuals from Vanuatu, researchers should send a signed letter to D.R. containing the following text: “(a) I will not distribute the data outside my collaboration; (b) I will not post the data publicly; (c) I will make no attempt to connect the genetic data to personal identifiers for the samples; (d) I will use the data only for studies of population history; (e) I will not use the data for any selection studies; (f) I will not use the data for medical or disease-related analyses; (g) I will not use the data for commercial purposes.”
Supplementary Material
Data S1. Supplementary data: (a) Data on ancient samples, (b) Data on next generation sequencing libraries, (c) Data on radiocarbon dates, (d) Data on newly reported present-day individuals, (e) Summary of information on newly-reported present-day populations, (f) Data on mixture proportions and dates of admixture, and (f) Data on allele sharing rates with Lapita-period samples from Vanuatu and Tonga. Related to Table 1, Figure 1, and Figure 2.
Highlights.
The population of Vanuatu in the Pacific was largely replaced 2900-2300 years ago
This second wave of migrants came from New Britain, east of New Guinea
A third wave spread different ancestry to the far-flung islands of Polynesia
Acknowledgments
We are grateful to Fiona Petchey and Tomasz Goslar for sharing unpublished information on previously reported radiocarbon dates generated at the University of Waikato and the Poznan Accelerator Mass Spectrometry laboratories. The Teouma research was supported by the Australian Research Council (Discovery Grants DP0880789 and DP1101014 15, M.S. and S.B.), the National Geographic Society (M.S. and S.B.), the Australia-Pacific Science Foundation (M.S. and S.B.), the Royal Society of New Zealand Marsden Fund (UOO0917, H.B. and S.B.), and a University of Otago Research Grant (H.B.). We are grateful to the late Richard Shutler Jr. for access to his original field notes, and to David Burley for contributing further Shutler archives to the Vanuatu Cultural Centre which aided in interpretation. Ralph Regenvanu, former Director of the Vanuatu Cultural Centre, gave ethical guidance on the use of the present-day samples for this project. A.J.M. was supported by a Wellcome Trust Clinical Research Training Fellowship grant reference 106289/Z/14/Z. We thank Professors John Clegg, David Weatherall, Donald Bowden and their colleagues for their work establishing the Oceanic sample collection at the University of Oxford in the U.K, with support from the Wellcome Trust and Medical Research Council. F.V. was supported by CNRS-UMR 7041. P.S. was supported by the Swedish Research Council (VR grant 2014-453). Accelerator Mass Spectrometry radiocarbon dating work at Pennsylvania State University (D.J.K) was supported by the NSF Archaeometry program (BCS-1460369). D.R. was supported by NIH grant GM100233, by NSF HOMINID grant BCS-1032255, and by an Allen Discovery Center of the Paul Allen Foundation, and is a Howard Hughes Medical Institute investigator.
Footnotes
Author Contributions
R.P. and D.R. supervised the study. M.S., F.V., S.B., R.S., H.B., I.P., G.W., and R.P. provided ancient samples and assembled archaeological and anthropological information. N.R., N.B., O.C., M.F., M.M., J.O., K.Si., and K.St. performed ancient DNA laboratory work. T.K.H. and D.J.K. carried out and analyzed radiocarbon dating data. K.A., A.H., K.M., S.J.O., T.P., K.R., T.N.W., and A.J.M. provided data from present-day populations. M.L., P.S., S.M., and D.R. analyzed genetic data. M.L., P.S., M.S., and D.R. wrote the manuscript.
Declaration of Interests: The authors declare no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Skoglund P, Posth C, Sirak K, Spriggs M, Valentin F, Bedford S, Clark GR, Reepmeyer C, Petchey F, Fernandes D, et al. Genomic insights into the peopling of the Southwest Pacific. Nature. 2016;538:510–513. doi: 10.1038/nature19844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Valentin F, Detroit F, Spriggs MJ, Bedford S. Early Lapita skeletons from Vanuatu show Polynesian craniofacial shape: Implications for Remote Oceanic settlement and Lapita origins. Proceedings of the National Academy of Sciences of the United States of America. 2016;113:292–297. doi: 10.1073/pnas.1516186113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Valentin F, Herrscher E, Bedford S, Spriggs M, Buckley H. Evidence for Social and Cultural Change in Central Vanuatu Between 3000 and 2000 BP: Comparing Funerary and Dietary Patterns of the First and Later Generations at Teouma, Efate. The Journal of Island and Coastal Archaeology. 2014;9:381–399. [Google Scholar]
- 4.Pinhasi R, Fernandes D, Sirak K, Novak M, Connell S, Alpaslan-Roodenberg S, Gerritsen F, Moiseyev V, Gromov A, Raczky P, et al. Optimal Ancient DNA Yields from the Inner Ear Part of the Human Petrous Bone. PloS One. 2015;10:e0129102. doi: 10.1371/journal.pone.0129102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dabney J, Knapp M, Glocke I, Gansauge MT, Weihmann A, Nickel B, Valdiosera C, Garcia N, Pääbo S, Arsuaga JL, et al. Complete mitochondrial genome sequence of a Middle Pleistocene cave bear reconstructed from ultrashort DNA fragments. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:15758–15763. doi: 10.1073/pnas.1314445110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Korlevic P, Gerber T, Gansauge MT, Hajdinjak M, Nagel S, Aximu-Petri A, Meyer M. Reducing microbial and human contamination in DNA extractions from ancient bones and teeth. BioTechniques. 2015;59:87–93. doi: 10.2144/000114320. [DOI] [PubMed] [Google Scholar]
- 7.Rohland N, Harney E, Mallick S, Nordenfelt S, Reich D. Partial uracil- DNA-glycosylase treatment for screening of ancient DNA. Philosophical Transactions of the Royal Society of London Series B, Biological Sciences. 2015;370:20130624. doi: 10.1098/rstb.2013.0624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Briggs AW, Stenzel U, Meyer M, Krause J, Kircher M, Pääbo S. Removal of deaminated cytosines and detection of in vivo methylation in ancient DNA. Nucleic Acids Research. 2010;38:e87. doi: 10.1093/nar/gkp1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Maricic T, Whitten M, Pääbo S. Multiplexed DNA sequence capture of mitochondrial genomes using PCR products. PloS One. 2010;5:e14004. doi: 10.1371/journal.pone.0014004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fu Q, Meyer M, Gao X, Stenzel U, Burbano HA, Kelso J, Pääbo S. DNA analysis of an early modern human from Tianyuan Cave, China. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:2223–2227. doi: 10.1073/pnas.1221359110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Alexander DH, Novembre J, Lange K. Fast model-based estimation of ancestry in unrelated individuals. Genome Research. 2009;19:1655–1664. doi: 10.1101/gr.094052.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pugach I, Duggan AT, Merriwether DA, Friedlaender FR, Friedlaender JS, Stoneking M. The gateway from Near into Remote Oceania: New insights from genome-wide data. Molecular Biology and Evolution. 2018 doi: 10.1093/molbev/msx333. Advance online publication January 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Reich D, Thangaraj K, Patterson N, Price AL, Singh L. Reconstructing Indian population history. Nature. 2009;461:489–494. doi: 10.1038/nature08365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Garanger J. Archéologie des Nouvelles-Hébrides. Paris: ORSTOM; 1972. [Google Scholar]
- 15.Moorjani P, Patterson N, Hirschhorn JN, Keinan A, Hao L, Atzmon G, Burns E, Ostrer H, Price AL, Reich D. The history of African gene flow into Southern Europeans, Levantines, and Jews. PLoS Genetics. 2011;7:e1001373. doi: 10.1371/journal.pgen.1001373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Loh PR, Lipson M, Patterson N, Moorjani P, Pickrell JK, Reich D, Berger B. Inferring admixture histories of human populations using linkage disequilibrium. Genetics. 2013;193:1233–1254. doi: 10.1534/genetics.112.147330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moorjani P, Sankararaman S, Fu Q, Przeworski M, Patterson N, Reich D. A genetic method for dating ancient genomes provides a direct estimate of human generation interval in the last 45,000 years. Proceedings of the National Academy of Sciences of the United States of America. 2016;113:5652–5657. doi: 10.1073/pnas.1514696113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Spriggs M. The Island Melanesians. Oxford: Blackwell; 1997. [Google Scholar]
- 19.Xu S, Pugach I, Stoneking M, Kayser M, Jin L Consortium HPAS. Genetic dating indicates that the Asian-Papuan admixture through Eastern Indonesia corresponds to the Austronesian expansion. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:4574–4579. doi: 10.1073/pnas.1118892109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lipson M, Loh PR, Patterson N, Moorjani P, Ko YC, Stoneking M, Berger B, Reich D. Reconstructing Austronesian population history in Island Southeast Asia. Nature Communications. 2014;5:4689. doi: 10.1038/ncomms5689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Valentin F, Spriggs M, Bedford S, Buckley H. Vanuatu mortuary practices over three millennia: Lapita to the early contact period. Journal of Pacific Archaeology. 2011;2:49–65. [Google Scholar]
- 22.Valentin F, Shing R, Spriggs M. Des restes humains datés du début de la période de Mangaasi (2400-1800 BP) découverts à Mangaliliu (Efate, Vanuatu) Comptes Rendus Palévol. 2005;4:420–427. [Google Scholar]
- 23.Shutler ME, Shutler R. A preliminary report of archaeological explorations in the Southern New Hebrides. Asian Perspectives. 1966;9:157–166. [Google Scholar]
- 24.Fu Q, Hajdinjak M, Moldovan OT, Constantin S, Mallick S, Skoglund P, Patterson N, Rohland N, Lazaridis I, Nickel B, et al. An early modern human from Romania with a recent Neanderthal ancestor. Nature. 2015;524:216–219. doi: 10.1038/nature14558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Haak W, Lazaridis I, Patterson N, Rohland N, Mallick S, Llamas B, Brandt G, Nordenfelt S, Harney E, Stewardson K, et al. Massive migration from the steppe was a source for Indo-European languages in Europe. Nature. 2015;522:207–211. doi: 10.1038/nature14317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mathieson I, Lazaridis I, Rohland N, Mallick S, Patterson N, Roodenberg SA, Harney E, Stewardson K, Fernandes D, Novak M, et al. Genome-wide patterns of selection in 230 ancient Eurasians. Nature. 2015;528:499–503. doi: 10.1038/nature16152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Behar DM, van Oven M, Rosset S, Metspalu M, Loogvali EL, Silva NM, Kivisild T, Torroni A, Villems R. A “Copernican” reassessment of the human mitochondrial DNA tree from its root. American journal of human genetics. 2012;90:675–684. doi: 10.1016/j.ajhg.2012.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li H, Durbin R. Fast and accurate long-read alignment with Burrows- Wheeler transform. Bioinformatics. 2010;26:589–595. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Weissensteiner H, Pacher D, Kloss-Brandstatter A, Forer L, Specht G, Bandelt HJ, Kronenberg F, Salas A, Schonherr S. HaploGrep 2: mitochondrial haplogroup classification in the era of high-throughput sequencing. Nucleic Acids Research. 2016;44:W58–63. doi: 10.1093/nar/gkw233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Skoglund P, Northoff BH, Shunkov MV, Derevianko AP, Pääbo S, Krause J, Jakobsson M. Separating endogenous ancient DNA from modern day contamination in a Siberian Neandertal. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:2229–2234. doi: 10.1073/pnas.1318934111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Petchey F, Spriggs M, Bedford S, Valentin F, Buckley H. Radiocarbon dating of burials from the Teouma Lapita cemetery, Efate, Vanuatu. J Archaeol Sci. 2014;50:227–242. [Google Scholar]
- 32.Longin R. New method of collagen extraction for radiocarbon dating. Nature. 1971;230:241–242. doi: 10.1038/230241a0. [DOI] [PubMed] [Google Scholar]
- 33.Lohse JC, Madsen DB, Culleton BJ, Kennett DJ. Isotope paleoecology of episodic mid-to-late Holocene bison population expansions in the Southern Plains, U.S.A. Quaternary Science Reviews. 2014;102:14–26. [Google Scholar]
- 34.Van Klinken GJ. Bone collagen quality indicators for palaeodietary and radiocarbon measurements. J Archaeol Sci. 1999;26:687–695. [Google Scholar]
- 35.Kennett DJ, Plog S, George RJ, Culleton BJ, Watson AS, Skoglund P, Rohland N, Mallick S, Stewardson K, Kistler L, et al. Archaeogenomic evidence reveals prehistoric matrilineal dynasty. Nature Communications. 2017;8:14115. doi: 10.1038/ncomms14115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stuiver M, Polach HA. Reporting of C-14 Data-Discussion. Radiocarbon. 1977;19:355–363. [Google Scholar]
- 37.Bronk Ramsay C. Bayesian analysis of radiocarbon dates. Radiocarbon. 2009;51:337–360. [Google Scholar]
- 38.Reimer PJ, Bard E, Bayliss A, Beck JW, Blackwell PG, Ramsey CB, Buck CE, Cheng H, Edwards RL, Friedrich M, et al. Intcal13 and Marine13 Radiocarbon Age Calibration Curves 0–50,000 Years cal BP. Radiocarbon. 2013;55:1869–1887. [Google Scholar]
- 39.Patterson N, Moorjani P, Luo Y, Mallick S, Rohland N, Zhan Y, Genschoreck T, Webster T, Reich D. Ancient admixture in human history. Genetics. 2012;192:1065–1093. doi: 10.1534/genetics.112.145037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Patterson N, Price AL, Reich D. Population structure and eigenanalysis. PLoS Genetics. 2006;2:e190. doi: 10.1371/journal.pgen.0020190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Galinsky KJ, Bhatia G, Loh PR, Georgiev S, Mukherjee S, Patterson NJ, Price AL. Fast Principal-Component Analysis Reveals Convergent Evolution of ADH1B in Europe and East Asia. American Journal of Human Genetics. 2016;98:456–472. doi: 10.1016/j.ajhg.2015.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lipson M, Szécsényi-Nagy A, Mallick S, Pósa A, Stégmár B, Keerl V, Rohland N, Stewardson K, Ferry M, Michel M, et al. Parallel palaeogenomic transects reveal complex genetic history of early European farmers. Nature. 2017;551:368–372. doi: 10.1038/nature24476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jostins L, Xu Y, McCarthy S, Ayub Q, Durbin R, Barrett J, Tyler-Smith C. YFitter: Maximum likelihood assignment of Y chromosome haplogroups from low-coverage sequence data. 2014 arXiv preprint arXiv:1407.7988. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data S1. Supplementary data: (a) Data on ancient samples, (b) Data on next generation sequencing libraries, (c) Data on radiocarbon dates, (d) Data on newly reported present-day individuals, (e) Summary of information on newly-reported present-day populations, (f) Data on mixture proportions and dates of admixture, and (f) Data on allele sharing rates with Lapita-period samples from Vanuatu and Tonga. Related to Table 1, Figure 1, and Figure 2.