Abstract
The gut microbiome plays a crucial role in maintaining human health. Functions performed by gastrointestinal microbes range from regulating metabolism to modulating immune and nervous system development. Scientists have attempted to exploit this importance through the development of engineered probiotics that are capable of producing and delivering small molecule therapeutics within the gut. However, existing synthetic probiotics are simplistic and fail to replicate the complexity and adaptability of native homeostatic mechanisms. In this review, we discuss the ways in which the tools and approaches of synthetic biology have been applied to improve the efficacy of therapeutic probiotics, and the ways in which they might be applied in the future. Simple devices, such as a bistable switches and integrase memory arrays, have been successfully implemented in the mammalian gut, and models for targeted delivery in this environment have also been developed. In the future, it will be necessary to introduce concepts such as logic-gating and biocontainment mechanisms into synthetic probiotics, as well as to expand the collection of relevant biosensors. Ideally, this will bring us closer to a reality in which engineered therapeutic microbes will be able to accurately diagnose and effectively respond to a variety of disease states.
Keywords: biosensors, engineered microbes, microbiome therapeutics, engineered probiotics
1 Introduction: synthetic biology and the gut microbiota
Our gastrointestinal tracts are populated by a diverse ecosystem of commensal microbes commonly known as the gut microbiome or microbiota [1]. In total, the gut microbiome is thought to comprise up to 1014 organisms of over 2,000 distinct bacterial, viral, and eukaryotic species [2–4]. The exact makeup of the gut microbiome is influenced by both environmental (age, diet, rearing environment, method of natal delivery) and genetic (gender, ethnicity, etc.) factors [5–8] – and, as a result, can be highly variable from individual to individual [1,2,9]. The microbiome also changes markedly over the course of an individual's life. It is affected profoundly by the health of its host, and, in turn, shapes its host profoundly, in areas ranging from metabolism to immune development to stress response [1,10–12]. The metabolic capacity of the microbiome – containing over three million genes – is thought to be greater than that of the liver [11]. Gastrointestinal bacteria direct carbohydrate metabolism and energy production, guide immune response, and influence memory [3,13]. Metabolites produced by the microbiome pass through the epithelial lining of the intestines and are taken into the bloodstream and transported throughout the body [11,13]. It is no surprise that gut dysbiosis has been linked to a bevy of disease states, from diabetes [14] to colitis [13,15] to mood disorders [16–18]: the microbiome has an immense effect on every part of human physiology (Fig. 1), in ways that are only now making themselves understood. Indeed, due to its large role in human health, some regard the microbiome as an additional “organ,” no less crucial to human health than the stomach or spleen [19,20].
Figure 1.
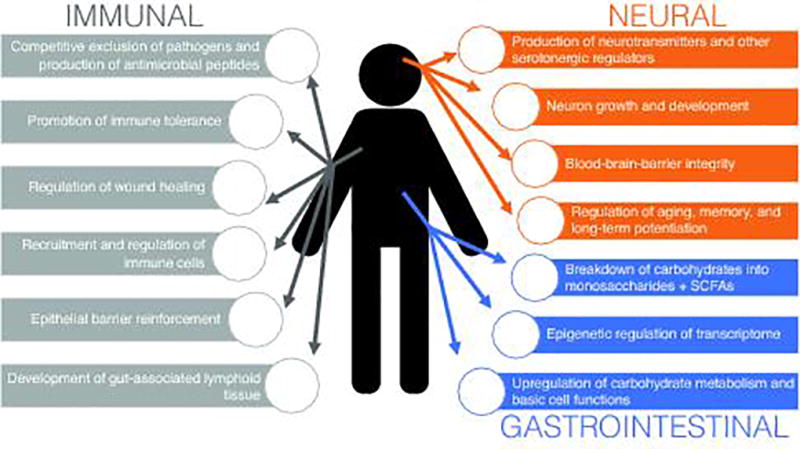
Overview of the ways in which the gut microbiome influences the human body. The microbiota is known to have a profound effect on the gastrointestinal, immune, and neuroendocrine systems.
Because of its interplay with human health, the gut microbiome provides an attractive target for therapeutic applications of synthetic biology. There is a nascent and fast-growing industry dedicated to probiotics, or microbial species that have a beneficial effect on the body, and studies of these species in relation to diseases ranging from inflammatory bowel disease (IBD) [21] to eczema [22,23] to anxiety disorders [24] demonstrate that we can work deep changes on the human body by altering the composition of its gut microbiome. Commensal bacteria generate metabolites, such as short-chain fatty acids or quorum-sensing molecules, that can rewire human metabolism, immune response, and mood [13]. Prebiotics, a related field to probiotics, attempts to identify these relevant compounds and their specific effects. The combination of the two – that is, bacterial species synthetically engineered to produce prebiotic compounds – has been successful in treating obesity, diabetes, and colitis in animal models [25–27]. One particular strain of cytokine-producing Laccococtus lactis has even reached clinical trials [28]. Our challenge now is to expand on that principle. To treat the complex, multifaceted diseases related to gut dysbiosis, we will need engineered systems that mimic the complexity and responsiveness of native microbial species. These synthetic microbes should be able to sense relevant changes in the microenvironment of the gut, such as signs of inflammation or the absence of important signaling molecules, and subsequently respond to correct those imbalances. Ideally, they should work in the way that homeostatic mechanisms in the human body work: not by attempting to overwhelm a pathway at a single point, but by targeting multiple points in the pathway, or multiple pathways simultaneously, to produce an integrated response.
Synthetic biology provides the principles, and is beginning to provide the tools, through which we might create the next generation of probiotics. Researchers have demonstrated that simple devices such as a nitric oxide-sensing switch [29] or an integrase-based memory array [30] can function in the environment of the gut microbiome. Engineered bacteria have also been developed for uses elsewhere in the human body, such as the eradication of infectious disease [31] and cancer diagnostics [32], and these can serve as a basis for similar systems within the gut. As we move into the development of more complex therapeutic systems, we must also consider issues of safety and long-term stability – the risk of horizontal gene transfer, the potential utility of probiotics that colonize the gut in a lasting fashion versus the potential pitfalls of introducing a foreign organism that may be subsequently difficult to remove. This review will address these issues in addition to covering recent advances in the application of synthetic biology techniques to the gut microbiome. We will review the mechanisms by which the gut microbiome influences human health and the ways in which researchers have used engineered probiotics to exploit this connection, before moving on to a discussion of how the synthetic biology approach might be more broadly used in the design of improved therapeutics, and the challenges that may arise in the process.
2 Background: the gut microbiome, pathways, and human health
The microbiome has a metabolic potential that dwarfs that of the human body – the species contained in a typical gut are estimated to encode more than 3.3 million distinct genes, more than 150 times the number found in humans alone [6,19,33]. Further, the necessity of interchange between the GI tract and bloodstream means that many millions of microbes interact intimately with their host, producing bacterial metabolites and modifying those produced by the host [13,34]. Around 10% of the human transcriptome is thought to be regulated by the microbiota [4], primarily through epigenetic means [10]. The vast majority of affected genes are related to functions such as energy production, metabolism, and fat storage [10,13,35–37]. Microbial species typically found in the gut up-regulate carbohydrate metabolism and basic cell-productivity functions such as translation and ribosome biogenesis, while down-regulating genes related to lipid production [38].
The microbiome modifies a wide variety of both natural and man-made compounds [39], but perhaps plays its most influential role in the fermentation of otherwise indigestible fibers and carbohydrates. By breaking these large compounds into monosaccharides, short-chain fatty acids (SCFAs), and other digestible compounds, they provide an additional source of metabolic energy [13,34,40]. In practice, this means that the microbiomes of obese individuals process and store energy more efficiently than those of lean individuals [41,42] – in fact, transplantation of an obese mouse’s microbiome into a lean subject has been sufficient to induce symptoms of obesity [43]. Gnotobiotic animals (those lacking a microbiome) are leaner than their conventionally raised counterparts, despite having a variety of other developmental deficiencies [36]. From this we can conclude that diet influences human health in large part through altering the composition of the microbiome. We know that changes in diet and sanitation associated with a modern Westernized lifestyle have led to a marked decrease in microbiome diversity, a change that is thought to be linked to "Western diseases" such as obesity, diabetes, and allergies[11] [44,45].
2.1 Immune homeostasis and development
One point in favor of the above is that “Western diseases” typically have an autoimmune component – for example, obesity can be thought of as “continuous low-grade inflammation [34]” – and commensal microbes are intimately involved in immune development and response. The gut is very large in terms of surface area – if stretched out, it would have approximately the area of a tennis court [1] – and collectively forms the largest area of direct contact between the body and potential foreign invaders [34]. Because nutrients must be taken up through the epithelial lining, the surface of the gut must remain somewhat porous – but at the same time, it must also form enough of a barrier to keep out pathogens. As a result, the gastrointestinal tract itself is commonly thought of as an extension of the immune system [46].
In other words, that commensal microbes shape the environmental conditions of the gut means they also powerfully shape immune response and development [1,2,11]. In fact, the mere presence of commensal species can help protect against disease [13]. Competition for resources itself helps drive out pathogens, and many common gut flora species – Lactobacillus and Bifidobacterium varieties in particular – produce antimicrobial peptides that can stave off infection [1,3]. Commensals also modulate the immune system itself: shoring up epithelial barriers [47], assisting with wound healing [48], recruiting neutrophils and leukocytes [13], and protecting against inflammation. The DNA of commensal bacteria is known to promote immune homeostasis in the gut [13]. In some studies, commensal DNA helps the host respond to potential pathogens – or even stave off cancer [48,49] – by regulating the levels of different types of T-cells [50]. Other commensal DNA motifs have been shown to have a suppressive effect, reducing inflammation [51]. This reflects the complicated and contradictory role of the mucosal immune system, which must defend against invaders while also tolerating the wealth of symbiotic species living there.
In this balancing act, the immune system is aided by members of the gut microbiota, which have a long history of coevolution [7,11] leading to a complex web of interdependence with their host. Metabolites produced by commensal microbes, such as the short-chain fatty acids discussed above, are taken up by the body at high rates [13] and regulate many aspects of homeostasis. Butyrate, for example, suppresses NF-κB signaling through down-regulation of TNF-α [52] and induces mucin synthesis [53], and SCFAs in general are known to interact with neutrophils via the FFAR2 receptor [54]. In addition, commensal species, specifically Bifidobacterium infantis and Clostridium species [55], are known to induce the secretion of anti-inflammatory cytokines such as IL-10. These compounds help trigger the formation of the regulatory T-cells (Treg cells) that modulate and prevent inappropriate immune response. In B. infantis-fed mice, those pathways lead to the elevation of CD4+ T-cell levels [56]. The recruitment of immune cells to the gut by Treg cells also helps maintain the mucosal barrier [13].
With this degree of interdependence, it is no surprise that the gut microbiome is hugely intertwined with, not just immune homeostasis, but immune development [34]. Gnotobiotic mice, which have no microbiota, display striking defects in this department – sharply decreased levels of IgA-producing plasma cells and CD4+ T-cells in the lamina propria (the layer of connective tissue beneath the epithelium) [57], a thinner and abnormally composed mucus layer, and impaired expression of defensins [19]. These abnormalities are widespread and affect the overall structure of the gastrointestinal tract: gut-associated lymphoid tissues (Peyer’s patches, mesenteric lymph nodes) were smaller and comprised fewer cells in gnotobiotic mice compared to mice with an intact microbiota. Even parts of the gut that are not strictly immune-related, such as the shape of microvilli and the size of the cecum [3,15], are affected [13,19]. Accordingly, germ-free mice are more susceptible to bacterial infection [11] and may suffer more severe symptoms of certain diseases [58]. On the other end of things, autoimmune diseases of the gut such as IBD can be thought of as essentially “unrestrained immune-cell activation and pro-inflammatory cytokine production” [11]. These symptoms are widely believed to be triggered by dysbiosis of the gut microbiome. Multiple studies have linked imbalance in levels of Bacteroides to IBD [59], and antibiotics have an palliative effect on animal models [60] of the disease. Certain types of colitis can even be communicated through cross-fostering with affected animals [61]. That some of these defects can be partially ameliorated by transplantation of a conventional microbiota [62] confirms that the influence of these “foreign bodies” can be immense.
2.2 – Nervous system response and development
The gut microbiome is known to have a profound effect on various aspects of the nervous system, including mood, stress, and aging [1,3,12,24,63–65]. We are still uncovering the extent of the interdependence behind what is known as the “gut-brain axis” – but it is apparent that the two systems have a powerful reciprocal effect on one another [66]. For example, physical and psychological stress can disturb the lining of the gut, causing gastrointestinal distress [3], an underlying factor behind many cases of IBD and other gastrointestinal disorders. Similarly, neurological conditions such as autism and mood disorders are associated with irregular digestive function [12]. The vagus nerve is widely thought to be involved in this interplay, since the effects of probiotics on emotional behavior (studied via ingestion of a Lactobacillus strain in mice) are attenuated when it is removed [67]. Through this channel, and perhaps others, signaling molecules from the gut and gut microbiome can be transmitted to the neuroendocrine system. Indeed, many commensal bacterial species produce neurotransmitters (serotonin, dopamine, acetylcholine, among others) and neurotransmitter precursors [12], as well as other regulatory molecules. One notable example are short-chain fatty acids, which in addition to their immunomodulatory effects, can mediate microglial homeostasis [13].
Perhaps because of this production of regulatory molecules, the microbiome has a great effect on the early development of the nervous system, particularly the serotonergic (involving serotonin) system. Germ-free mice exhibit shorter, stubbier, less branched neurons in the hippocampus, as well as lower levels of BDNF (short for brain-derived neurotropic factor), which promotes neuron growth and development, in the cortex and amygdala [3,12]. The hippocampus and amygdala, which mediate social behavior, learning, memory, stress, and mood – and are associated with disorders from autism to anxiety disorders – are both larger in volume, and studies also point to differential regulation of serotonin receptors. A 2015 study by Erny et al. also points to down-regulation of immune-related genes in the brain in germ-free mice, leading, among other things, to the formation of immature microglia [64]. Whether due to these specific defects or as-yet undiscovered ones, gnotobiotic mice display decreased sociability and an increase in stereotyped/repetitive behaviors – an effect that, like many phenotypic differences in germ-free mice, can be partially ameliorated by colonization with conventional microbiota [12]. Finally, the state of immunosenescence that accompanies aging, which can cause a "chronic low-grade inflammatory status in the gut" [68], is also linked to widespread differences in the composition and diversity of the microbiome. These differences, in turn, are likely to influence the progression of healthy aging and the development of neurodegenerative diseases such as Alzheimer's. Blood-brain-barrier integrity, a key component of age-related disease, is at least partially dependent on microbiome composition [69], and researchers have linked the severity of Parkinson’s symptoms to changes in levels of Prevotellaceae and Entereobacteriacae in the gut [12]. Studies have even shown that probiotic treatment can slow the age-related attenuation of long-term potentiation [70].
3 Synthetic probiotic species – engineered for delivery of therapeutic compounds
Since the gut microbiome is important to so many aspects of human health and dysbiosis of the microbiome is closely linked to disease states, it becomes natural for us to attempt to treat these disease states by rebalancing the microbiome through the administration of beneficial microbial species – in other words, through probiotics. Probiotics have shown some success in treating a wide variety of disorders, including virus-induced diarrhea [71], colitis [72], eczema and dermatitis [73], anxiety disorders [18,24], and depression [74]. However, in human clinical trials, results are mixed – the only disease where probiotic treatment consistently shows positive results is C. difficile infection [75] – perhaps because we do not fully understand the physiology of the species we use. Probiotic species are complicated organisms, performing myriad functions, producing complex and sometimes contradictory signals. To create targeted systems that treat specific ailments with precision, we must fine-tune our control of probiotics through bioengineering.
3.1 – A survey of engineered probiotic therapies
Thus far, probiotic species engineered to produce therapeutic biomolecules have been used to help fight off infection, reduce inflammation, and treat diet-induced obesity [76]. In a 2011 study by Lagenaur et al., a Lactobacillus jensenii strain modified to express the antiviral protein cyanovirin N successfully decreased simian HIV (SHIV) transmission in macaques by 63% [77]. In the gut itself, researchers have successfully disrupted the virulence patterns of pathogenic species Vibro cholerae, which produces its toxins only at low concentrations, using an altered version of the probiotic E. coli strain Nissle (EcN)[78]. Other approaches, rather than targeting pathogens directly, have attempted to take advantage of the human body's natural protective mechanisms and pathways. N-acylethanolamides (NAEs) are a family of anorexigenic lipids synthesized by the small intestine in response to feeding. Administration of bacteria producing NAE precursors (NAPEs) to a polygenic mouse model of obesity successfully inhibited weight gain, as mice fed NAPE-producing E. coli exhibit a decreased food intake and increased metabolic rate [27]. Elsewhere, commensal species expressing anti-inflammatory compounds such as IL-10 [25], TGF-β1 [79], KGF-2 [80,81], serine protease inhibitors [82] and elafin [83] have been used to treat mouse models of induced colitis (Table 1), and trefoil factor-1 secreting L. lactis can ameliorate the inflammatory effects of chemotherapy and radiation therapy, although studies in this case focused on oral mucositis [84]. As a treatment for IBD, IL-10 L. lactis has even reached phase I clinical trials [28]. There are also indications that engineered probiotics may be able to change cells in a profound and potentially even lasting manner. Another study by Duan et al. attempted to treat a rat model of diabetes through inoculation of an E. coli/Nissle strain expressing GLP-1 [85], a peptide that stimulates production of insulin in intestinal cells in a glucose-dependent manner [86]. This strain successfully managed to “reprogram” a proportion of cells into insulin-producing cells. The cells displayed characteristic markers [79] involved in insulin production and signaling, and the kinetics of insulin secretion in the reprogrammed cells were similar to those in normal (non-diabetic) β cells [85].
Table 1.
A survey of existing studies in which molecular therapeutics are delivered by engineered probiotic species, and their functions in each case.
| Molecular therapeutic |
Producing strain | Purpose |
|---|---|---|
| AI-2 [78] | Escherichia coli strain Nissle 1917 (EcN) | Circumvention of Vibrio cholerae quorum-sensing mechanisms |
| Pyocin S5 [31] | E. coli (EcN) | Toxin targeting Pseudomonas aeruginosa |
| IL-10 [25,88,115,116] | Lactococcus lactis | Anti-inflammatory cytokine |
| TNF-a antibodies [94] | L. lactis | Antagonism of pro-inflammatory cytokines |
| TGF-B1 [82] | L. lactis, Bacteroides ovatus | Anti-inflammatory cytokine |
| KGF-2 [80] | B. ovatus | Growth factor involved in intestinal homeostasis and repair |
| SPLI [82] | L. lactis | Serine protease inhibitor |
| elafin [82,83] | L. lactis, L. casei | Serine protease inhibitor |
| Anti-CTNNB1 miRNA [92] | E. coli | Oncogene silencing |
Notably, previous studies involving intraperitoneal injection of GLP-1 had encountered difficulties due to the short half-life of the active peptide [87]. The use of commensal bacteria as a delivery mechanism was more effective in this regard, demonstrating the potential usefulness of probiotics in applying therapeutics that are difficult to administer by more traditional means [85]. Similarly, tests of IL-10 producing L. lactis in mouse models demonstrate that they are as effective as traditional antibody treatments in ameliorating chemically-induced and genetic (spontaneous, caused by IL-10 −/− phenotype) colitis, at much lower doses than would be necessary using intraperitoneal injection [25]. Subsequent studies by other labs use a cocktail of IL-10- and antioxidant-producing probiotics to prevent colorectal cancer [88], or pair IL-10-secreting bacteria with more traditional antibody therapies to reverse diabetic symptoms [89,90], finding that combination therapies were more effective than any type of individual treatment alone [88,89]. Genetically engineered probiotics might therefore be made even more potent if they are made to express multiple therapeutic compounds with different effects, reprogramming cells by targeting multiple beneficial pathways or overwhelming one.
3.2 – The future of biomolecular therapies
At the moment, probiotic drug delivery is limited to compounds that can be readily synthesized or altered by commensal bacteria species – mostly proteinaceous compounds natively produced by living organisms rather than synthetic drugs [91]. At the same time, however, some studies point to a path through which we might contemplate the integration of this therapeutic approach with other technologies. Work by Xiang et al. establishes a system by which shRNA targeting mammalian genes can be delivered through commensal bacteria. The bacterial chassis used expresses invasin, a protein that allows bacteria to enter mammalian cells, and HlyA, which enables genetic material to pass through vesicles. Using this system, they successfully managed to induce targeted silencing of CTNNB1, a cancer gene, in the intestinal epithelium. This technique can potentially be modified to deliver any number of sequences, enabling combination of siRNA and other non-coding RNA-based technologies with a probiotic delivery system [92]. Elsewhere, Vandenbroucke et al. utilize engineered versions of antibodies known as Nanobodies, which can be produced in vivo by bacteria and yeast and have a number of structural advantages over traditional antibodies [93]. Bivalent versions of these Nanobodies targeting TNF-α (thought to contribute to IBD symptoms) secreted by L. lactis were highly effective in treating both chemically-induced colitis and a genetically induced version (mice lacking IL-10 expression will spontaneously develop enterocolitis) [94]. One important direction for the future will be to continue to expand the biosynthesis capacities of common probiotic species, to improve the versatility of therapies using these species as a drug delivery mechanism. To make full use of probiotics, we will need to move from the production of strictly ribosomally synthesized compounds to peptides that are post-translationally modified, and from there to non-ribosomal compounds, such as, for example, most common antibiotics [91].
4 Devices and pathways – new frontiers in engineered probiotics
The limitations of a system that constitutively produces a single therapeutic compound, as most of the previously discussed studies focus on, are obvious. The gut microbiome is hugely individualized, even between members of the same family, certainly across ethnic and cultural lines [6], and also changes significantly throughout a person's life [68]. Not only that, conditions in the microenvironment of the gut are in constant flux, and homeostasis is a fine balance – an anti-inflammatory cytokine that is helpful in treating IBD under one set of conditions may simply make the body more susceptible to infection under another. There is therefore a powerful need for engineered probiotics that can sense, and subsequently respond to, environmental stimuli, changing behavior based on individualized conditions. Fortunately, these tools are already beginning to be developed by synthetic biologists. Over the past decade, scientists have used natural biological mechanisms as well as electrical circuits as a model for the construction of increasingly complex gene networks: switches, oscillators, counters, and cell-cell communication modules, among others [95]. A few of these devices have even been applied to the gut microbiome: a 2014 study by Kotula et al., for example, successfully implements an aTc-driven bistable switch (Fig. 2A) in E. coli that colonize the guts of mice. Oral feeding of the inducer to host mice switched the bacteria to a lacZ-expressing state, a state which was remembered by the cells even after removal of stimulus, across generations and over the course of days [96]. Similarly, work by Archer et al. demonstrates the viability of a bistable switch (Fig. 2C) in detecting nitric oxide, a crucial marker of inflammation, in a mouse explant model [29]. Both switch systems are irreversible – the nitric oxide switch is recombinase-based – but have potential application as a non-invasive diagnostic that can be detected in fecal matter in addition to the proof of principle they provide.
Figure 2.
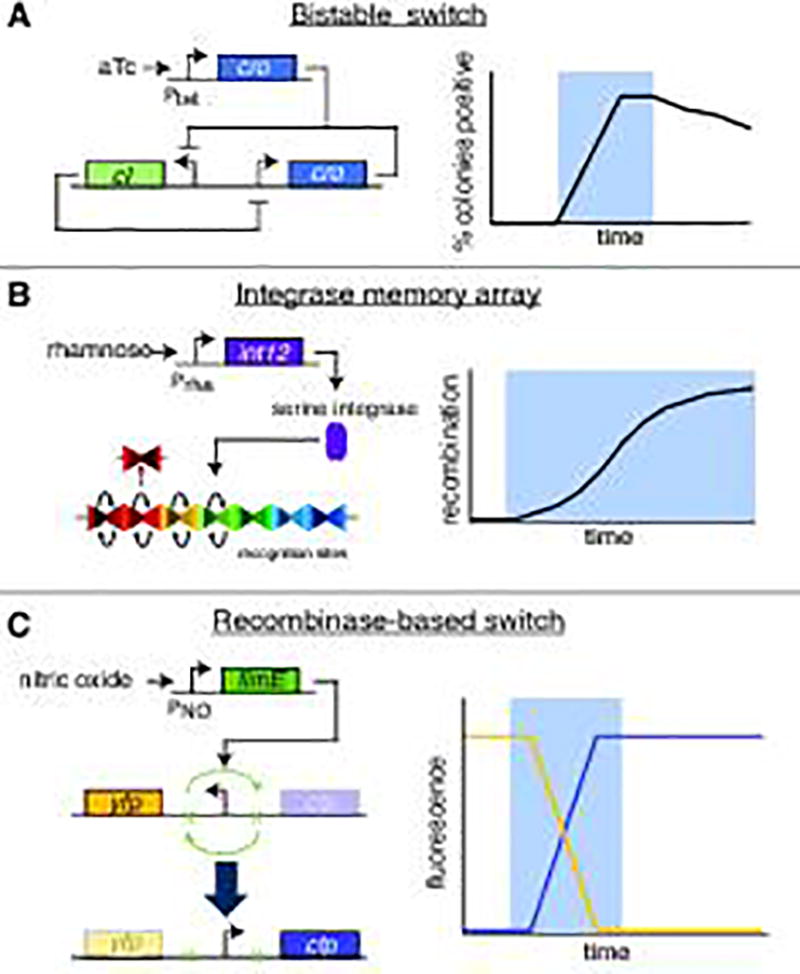
Synthetic biology "devices" applied in the (simulated or otherwise) environment of the mammalian gut, and expected behavior of reporter genes when tested (blue shading signifies addition of inducer). C) was conducted in mouse GI tract explants, while A) and B) were successfully tested in a living mouse model. A) An aTC-triggered variation on the lambda cI/cro bistable switch, in which one of two stable states is maintained over time unless the other state is deliberately triggered. B) A serine integrase-based memory array, consisting of multiple recognition sites which are successively inverted when integrase expression is induced through the presence of rhamnose. C) A nitric oxide-sensing recombinase-based switch which inverts a promoter when activated, switching from production of YFP to CFP.
Adding an extra step of complexity is 2015 work by Mimee et al., which introduces tools such as sugar-inducible promoters and an integrase-based memory array into a common gut commensal species, Bacteroides thetaiotaomicron. The results show successful induction of three different sugar-dependent constructs via oral feeding: a simple arabinogalactan-inducible reporter construct, an IPTG-dependent implementation of CRISPR-based interference (CRISPRi), and the aforementioned memory array, triggered by administration of rhamnose [30]. Crucially, that all three are sugar- or sugar-mimic-based means that they can potentially be geared to respond to ambient environmental conditions in the gut, not just specific (fed) triggers. Indeed, studies have demonstrated the relevance of lactose and rhamnose in regulating metabolism [97–100]. Moreover, these tools provide the capability for genetic memory and for responsiveness: the inducible luciferase reporter can respond to changing conditions in real-time while the memory array (Fig. 2B) serves as a ‘permanent’ record of gene induction/environmental conditions at points in the past. One can envision the two being combined into a microbial system that can both respond in the near-term to an environmental danger signal with production of a therapeutic, and also record the presence of the signal to aid in later diagnosis.
Of course, the systems discussed above are only tools that, to be useful, need to be combined into an engineered system with a defined function. The next challenge for synthetic biologists will be to apply these tools in the rational design of systems that address problems of gastrointestinal health. Few models for these systems exist, as modulation of the gut microbiome is somewhat understudied as a therapeutic technique. Most existing synthetic biology-based therapeutic systems use mammalian cell vectors [101–103]; however, a few bacterial systems have been developed to target pathogens in the gastrointestinal tract and elsewhere, most notably a toxin delivery system designed to combat Pseudomonas aeruginosa, an opportunistic pathogen that infects the gastrointestinal tract and is resistant to traditional antimicrobials. Saiedi et al. reasoned that bacteriocins might be more effective because, as of yet, there is no evidence that resistance to these ribosomally synthesized peptides can be conferred through horizontal gene transfer [104], and identified pyocin S5, a bacteriocin that displays strong bactericidal activity against P. aeruginosa but not against E. coli. Researchers then utilized the natural quorum-sensing mechanisms of P. aeruginosa to detect the pathogen, coupling these to production of a lysis protein E7, here chosen for its size and modularity. The result was an elegant system that, in co-culture, successfully reduced P. aeruginosa biofilm formation by close to 90%. One additional useful point is that, while the death of the E. coli chassis is a necessary consequence of this mechanism of delivery, it also fits the needs of this system. Since the modified E. coli species is a treatment for an opportunistic infection – in other words, an acute event – there is little utility in having the bacteria persist in the gut environment long-term. (The issue of long-term colonization of the gut by engineered probiotics – both the potential necessity and the potential pitfalls – is something we will discuss further below.) This system has been successfully tested in a C. elegans and murine gut model [105], and this model of targeted delivery, modified thoughtfully according to the function of the molecule being delivered, could be useful in a wide variety of therapeutic applications. [31]
5 Building a system – the biological chassis and biocontainment
We cannot conclude this review without addressing the issue of which bacterial species form the most appropriate chassis for synthetic probiotic systems, and the complicated trade-offs between persistence and safety that this brings to our attention. Existing systems commonly use either L. lactis (or other Lactobacillus species) or E. coli to house their biological machinery, since these species are in common use because of their industrial and laboratory roles. These species, however, are not common inhabitants of the gut – many commonly used Lactobacillus species are not native to the human microbiome at all – and therefore tend to be ‘flushed out’ by better-adapted microbes within a matter of days. Multiple studies have demonstrated that antibiotic treatment is necessary for these strains to colonize the gut in a long-term fashion, which introduces a confounding factor into any such treatment of gut dysbiosis.
Some researchers are beginning to experiment with species that dominate/comprise a larger portion of the native gut microbiome, the most common being B. thetaiotaomicron – unfortunately, however, the sets of parts and devices available in these alternative systems are considerably smaller than those of the traditional probiotic species, which have been in common laboratory use for decades. Moving forward, we may also wish to experiment with genetic engineering techniques to improve the stability of synthetic microbes colonizing the gut. Gibson et al. [106] detail a process that can be used to screen for bacterial genes that enable stable colonization; however, these genes may have unexpected effects on the balance of the gut microbiome. Not only that, stability itself may be thought of as a double-edged sword: there are many risks associated with having one particular species dominate the gut microbiome, particularly one which has been heavily altered for a specific purpose. In practice, this means that a potential synthetic probiotic is unlikely to be approved for clinical use without some kind of failsafe mechanism that can be triggered to initiate lysis or destruction of the engineered bacteria, should that become necessary. Steidler et al., for example, removed a crucial thymidylate synthase gene from the genome of their IL-10 producing L. lactis strain, rendering growth of the species dependent on outside administration of thymidine [107]. A similar approach is seen in the "Deadman" and "Passcode" kill switches designed by Chan et al. [108], in which continuous input of one or more molecular ligands is required to maintain repression of a toxin. The risk is that noise or leaky expression may prematurely activate the self-destruction sequence, an issue that may be ameliorated by increasing the specificity of the destruction circuit, as the use of AND-gating accomplishes in the "Passcode" switch [108]. More mechanisms of this sort could be built on the same principles, utilizing different ligands to enable the orthogonal deactivation of multiple probiotic systems.
6 Future directions: next steps and challenges
A component of the synthetic biology approach is the adoption of a ground-up, parts-based engineering approach to solving biological problems. In this case, we might find clarity in modeling our (desired) biological behavior as a mechanical process or set of decision states. What does an ideal synthetic probiotic (i.e. a bacterial species that detects and treats disease from inside the gut) look like? What behaviors and capabilities would it exhibit? We propose that such a system (Fig. 3) might have some or all of the following properties:
colonize the gut stably for the duration of treatment;
assign (diagnose) a particular disease state based on a series of inputs (presence or absence of symptoms);
based on that assigned disease state, initiates a program of outputs (production of therapeutic genes, alteration of signaling molecules etc.);
is able to adjust output in response to changes in the environment (diet, stress, cessation of symptoms due to treatment)
may have its output adjusted from by the doctor/patient in a non-invasive manner (for example, by feeding a specific compound or drug to the patient); and
when the host reaches a stable, healthy state, can be easily removed from the environment (self-destruction program).
Obviously, significant work must still be done to achieve these goals. The bacterial systems that have been engineered so far for this purpose comprise a very limited set of simple if-A-then-B systems, if that. (The majority of engineered probiotics reviewed above are unable to respond to environmental cues at all.) The set of biosensors (input detectors) that have been verified in vivo is limited; the set of possible products (outputs) is larger but is still bounded by the current capabilities of metabolic engineering. We must continue to expand our capabilities by developing sensors of biologically relevant compounds, improving biosynthesis techniques, and continuing to integrate other technologies into the probiotic model, as has been done with CRISPRi [30] and siRNA [92]. We must also improve mechanisms of delivery and secretion, particularly if we want to use this method to treat distant parts of the body. Lastly, metabolic engineering issues such as metabolic load and yield are equally relevant here – insufficient yield in particular may be the reason IL-10 secreting systems have not progressed beyond phase I clinical trials [109].
Figure 3.
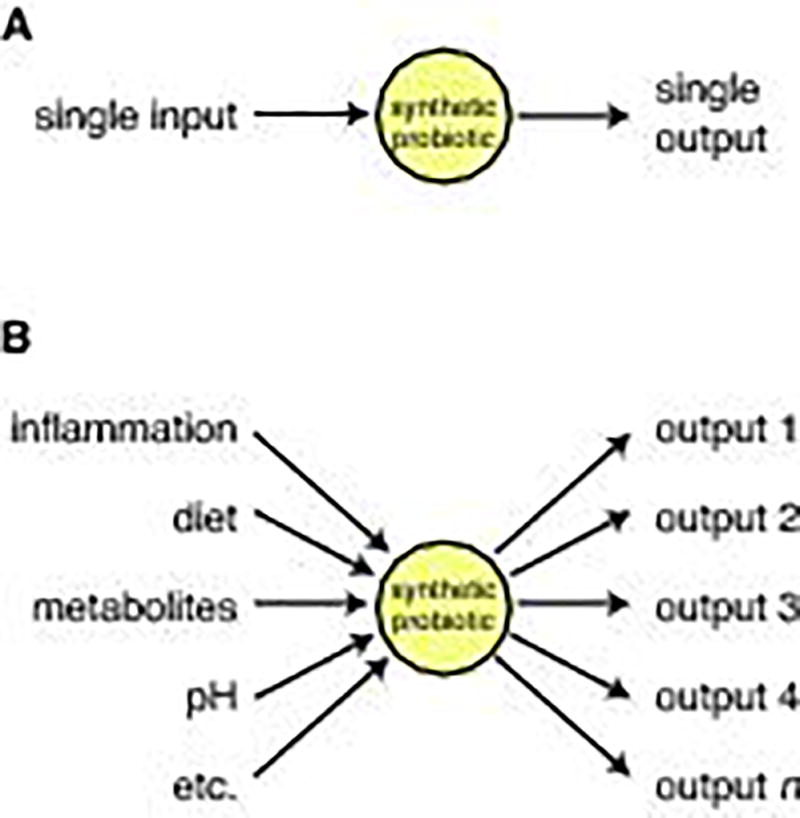
Comparison of existing therapeutic probiotics with proposed future model. a) Existing therapeutic probiotics are either constitutive or respond to a single-molecule input to produce a single-molecule therapeutic. b) Ideally, in the future, we would like engineered probiotics to 1) integrate a variety of different types of information 2) diagnose a disease state based on that information 3) output a therapeutic program of multiple genes if the body is in a disease state.
More than that, however, in order for therapeutic probiotics to be versatile, responsive, and effective, they need to be able to evaluate complex states. In practice, the diagnosis of polygenic, systemic diseases such as diabetes, IBS, and so forth is not based on the presence or absence of a single compound, but on the accumulation of a set of phenotypic symptoms, which may in turn be represented by many different processes on the molecular level. It is in the recognition of complex states that synthetic biology can be most relevant to the development of probiotics, as capturing complexity using biological systems is one of the major preoccupations of the field. The development of AND, OR and NOR gate systems [108,110–113] will enable synthetic organisms to initiate biological programming based on divergent combinations of inputs, as opposed to one singular one. Similarly, attempts to precisely capture analog logic in bioengineered systems will allow us to respond to the extent (severity) and duration (persistence) of symptoms. It will be necessary to move systems such as the chimeric logic gate system described in Shis et al. [111] and the comparators developed by Rubens et al. [114] into an appropriate probiotic chassis and adapt and tune them so that they respond to physiologically relevant changes in the environment.
Similarly, native biological responses to stressors and pathogens are complex and graded, and response states initiated by these synthetic systems must recapitulate that complexity and nuance if they are to be effective. Moving forward, it will be necessary to continue developing systems that are multifaceted in their response to stimuli. Fortunately, this can be accomplished more easily with existing tools such as fusion proteins, common promoters, etc., in combination with the development of a wider library of potential therapeutic biomolecules. The use of synthetic biology devices such as oscillators and counters will also enable us to produce more complex patterns of response, and achieve the spatial and temporal variance of expression that characterizes living organisms. Lastly, many effective therapies involve multiple discrete stages; combination therapy which follows traditional antibody therapy with the administration of immunomodulatory probiotics is significantly more effective in treating type 1 diabetes than either approach alone [89]. Ideally, we would want to recapitulate this in a self-contained probiotic system by including programming for multiple states between sickness and health.
Acknowledgments
Funding for this work was provided by the National Institutes of Health, grant number R01GM117138, and by the Gillson Longenbaugh Foundation.
Abbreviations
- aTc
anhydrotetracycline
- IBD
inflammatory bowel disorder
- SCFA(s)
short-chain fatty acid(s)
Footnotes
Conflicts of interest
The authors declare no financial or commercial conflicts of interest.
References
- 1.Sekirov I, Russell SL, Antunes LCM, Finlay BB, et al. Gut microbiota in health and disease. Physiol Rev. 2010;90:859–904. doi: 10.1152/physrev.00045.2009. [DOI] [PubMed] [Google Scholar]
- 2.Neish AS. Microbes in gastrointestinal health and disease. Gastroenterology. 2009;136:65–80. doi: 10.1053/j.gastro.2008.10.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Grenham S, Clarke G, Cryan JF, Dinan TG. Brain-gut-microbe communication in health and disease. Front Physiol. 2011;2:1–15. doi: 10.3389/fphys.2011.00094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arnold JW, Roach J, Azcarate-Peril MA. Emerging Technologies for Gut Microbiome Research. Trends Microbiol. 2016 doi: 10.1016/j.tim.2016.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chu DM, Ma J, Prince AL, Antony KM, Seferovic MD, Aagaard KM. Maturation of the infant microbiome community structure and function across multiple body sites and in relation to mode of delivery. Nat Med. 2017 doi: 10.1038/nm.4272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huttenhower C, Fah Sathirapongsasuti J, Segata N, Gevers D, et al. Structure, function and diversity of the healthy human microbiome. Nature. 2012;486:207–14. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moeller AH, Li Y, Mpoudi Ngole E, Ahuka-Mundeke S, et al. Rapid changes in the gut microbiome during human evolution. Proc Natl Acad Sci U S A. 2014;111:16431–5. doi: 10.1073/pnas.1419136111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Spor A, Koren O, Ley R. Unravelling the effects of the environment and host genotype on the gut microbiome. Nat Rev Microbiol. 2011;9:279–90. doi: 10.1038/nrmicro2540. [DOI] [PubMed] [Google Scholar]
- 9.Gorlatova M, Sarik J, Grebla G, Cong M, Kymissis I, Zussman G. Movers and shakers: influence of bacteriophages in shaping the mammalian gut microbiota. Gut Microbes. 2014;4:4–16. doi: 10.4161/gmic.22371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Camp JG, Frank CL, Lickwar CR, Guturu H, et al. Microbiota modulate transcription in the intestinal epithelium without remodeling the accessible chromatin landscape. Genome Res. 2014;24:1504–16. doi: 10.1101/gr.165845.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Round J, Mazmanian S. The gut microbiota shapes intestinal immune responses during health and disease. Nat Rev Immunol. 2014;9:313–23. doi: 10.1038/nri2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dinan TG, Cryan JF. Gut Instincts: microbiota as a key regulator of brain development, ageing and neurodegeneration. J Physiol. 2016;2:1–33. doi: 10.1113/JP273106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lin L, Zhang J. Role of intestinal microbiota and metabolites on gut homeostasis and human diseases. BMC Immunol. 2017;18:2. doi: 10.1186/s12865-016-0187-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gülden E, Wong FS, Wen L. The gut microbiota and Type 1 diabetes. Clin Immunol. 2015;159:143–53. doi: 10.1016/j.clim.2015.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gensollen T, Iyer SS, Kasper DL, Blumberg RS. How colonization by microbiota in early life shapes the immune system. Science (80-) 2016;352:539–44. doi: 10.1126/science.aad9378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Logan AC, Katzman M. Major depressive disorder: probiotics may be an adjuvant therapy. Med Hypotheses. 2005;64:533–8. doi: 10.1016/j.mehy.2004.08.019. [DOI] [PubMed] [Google Scholar]
- 17.Logan AC, Jacka FN, Craig JM, Prescott SL. The microbiome and mental health: looking back, moving forward with lessons from allergic diseases. Clin Psychopharmacol Neurosci. 2016;14:131–47. doi: 10.9758/cpn.2016.14.2.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Desbonnet L, Garrett L, Clarke G, Bienenstock J, Dinan TG. The probiotic Bifidobacteria infantis: An assessment of potential antidepressant properties in the rat. J Psychiatr Res. 2008;43:164–74. doi: 10.1016/j.jpsychires.2008.03.009. [DOI] [PubMed] [Google Scholar]
- 19.Sommer F, Bäckhed F. The gut microbiota--masters of host development and physiology. Nat Rev Microbiol. 2013;11:227–38. doi: 10.1038/nrmicro2974. [DOI] [PubMed] [Google Scholar]
- 20.Martin F-PJ, Sprenger N, Yap IKS, Wang Y, et al. Panorganismal Gut Microbiome−Host Metabolic Crosstalk. J Proteome Res. 2009;8:2090–105. doi: 10.1021/pr801068x. [DOI] [PubMed] [Google Scholar]
- 21.Lee HJ, Choi JK, Ryu HS, Choi CH, et al. Therapeutic Modulation of Gut Microbiota in Functional Bowel Disorders. J Neurogastroenterol Motil. 2017;23:9–19. doi: 10.5056/jnm16124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li D, Wang P, Wang P, Hu X, Chen F. The gut microbiota: A treasure for human health. Biotechnol Adv. 2016;34:1210–24. doi: 10.1016/j.biotechadv.2016.08.003. [DOI] [PubMed] [Google Scholar]
- 23.McFarland LV. Use of probiotics to correct dysbiosis of normal microbiota following disease or disruptive events: a systematic review. BMJ Open. 2014;4:1–18. doi: 10.1136/bmjopen-2014-005047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Savignac HM, Tramullas M, Kiely B, Dinan TG, Cryan JF. Bifidobacteria modulate cognitive processes in an anxious mouse strain. Behav Brain Res. 2015;287:59–72. doi: 10.1016/j.bbr.2015.02.044. [DOI] [PubMed] [Google Scholar]
- 25.Steidler L, Hans W, Schotte L, Neirynck S, et al. Treatment of murine colitis by Lactococcus lactis secreting interleukin-10. Science. 2000;289:1352–5. doi: 10.1126/science.289.5483.1352. [DOI] [PubMed] [Google Scholar]
- 26.Duan F, Curtis KL, March JC. Secretion of insulinotropic proteins by commensal bacteria: Rewiring the gut to treat diabetes. Appl Environ Microbiol. 2008;74:7437–8. doi: 10.1128/AEM.01019-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Davies SS, Chen Z, Guo L, Zhang Y, Stien X, Coulon D. Incorporation of therapeutically modified bacteria into gut microbiota inhibits obesity. Free Radic Biol Med. 2014;124:3381–406. doi: 10.1172/JCI72517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Braat H, Rottiers P, Hommes DW, Huyghebaert N, et al. A Phase I Trial With Transgenic Bacteria Expressing Interleukin-10 in Crohn’s Disease. Clin Gastroenterol Hepatol. 2006;4:754–9. doi: 10.1016/j.cgh.2006.03.028. [DOI] [PubMed] [Google Scholar]
- 29.Archer EJ, Robinson AB, Süel GM. Engineered E. coli that detect and respond to gut inflammation through nitric oxide sensing. ACS Synth Biol. 2012;1:451–7. doi: 10.1021/sb3000595. [DOI] [PubMed] [Google Scholar]
- 30.Mimee M, Tucker AC, Voigt CA, Lu TK. Programming a Human Commensal Bacterium, Bacteroides thetaiotaomicron, to Sense and Respond to Stimuli in the Murine Gut Microbiota. Cell Syst. 2015;1:62–71. doi: 10.1016/j.cels.2015.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Saeidi N, Wong CK, Lo T-M, Nguyen HX, et al. Engineering microbes to sense and eradicate Pseudomonas aeruginosa, a human pathogen. Mol Syst Biol. 2011;7:521. doi: 10.1038/msb.2011.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Danino T, Prindle A, Kwong GA, Skalak M, et al. Programmable probiotics for detection of cancer in urine. Sci Transl Med. 2015;7 doi: 10.1126/scitranslmed.aaa3519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Balzola F, Bernstein C, Ho GT, Lees C. A human gut microbial gene catalogue established by metagenomic sequencing. Inflamm Bowel Dis Monit. 2010;11:28. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shen J, Obin MS, Zhao L. The gut microbiota, obesity and insulin resistance. Mol Aspects Med. 2013;34:39–58. doi: 10.1016/j.mam.2012.11.001. [DOI] [PubMed] [Google Scholar]
- 35.Chuang H-L, Huang Y-T, Chiu C-C, Liao C-D, et al. Metabolomics characterization of energy metabolism reveals glycogen accumulation in gut-microbiota-lacking mice. J Nutr Biochem. 2012;23:752–8. doi: 10.1016/j.jnutbio.2011.03.019. [DOI] [PubMed] [Google Scholar]
- 36.Bäckhed F, Ding H, Wang T, Hooper LV, et al. The gut microbiota as an environmental factor that regulates fat storage. Proc Natl Acad Sci U S A. 2004;101:15718–23. doi: 10.1073/pnas.0407076101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wolf G. Gut microbiota: a factor in energy regulation. Nutr Rev. 2006;64:47–50. doi: 10.1111/j.1753-4887.2006.tb00173.x. [DOI] [PubMed] [Google Scholar]
- 38.Gosalbes MJ, Durbán A, Pignatelli M, Abellan JJ, et al. Metatranscriptomic approach to analyze the functional human gut microbiota. PLoS One. 2011;6 doi: 10.1371/journal.pone.0017447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lu K, Mahbub R, Fox JG. Xenobiotics: Interaction with the intestinal microflora. ILAR J. 2015;56:218–27. doi: 10.1093/ilar/ilv018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tremaroli V, Bäckhed F. Functional interactions between the gut microbiota and host metabolism. Nature. 2012;489:242–9. doi: 10.1038/nature11552. [DOI] [PubMed] [Google Scholar]
- 41.Turnbaugh PJ, Ley RE, Mahowald Ma, Magrini V, Mardis ER, Gordon JI. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444:1027–31. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- 42.Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, et al. A core gut microbiome in obese and lean twins. Nature. 2009;457 doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ellekilde M, Selfjord E, Larsen CS, Jakesevic M, et al. Transfer of gut microbiota from lean and obese mice to antibiotic-treated mice. Sci Rep. 2014;4:5922. doi: 10.1038/srep05922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Richards JL, Yap YA, McLeod KH, Mackay CR, Marino E. Dietary metabolites and the gut microbiota: an alternative approach to control inflammatory and autoimmune diseases. Clin Transl Immunol. 2016;5:e82. doi: 10.1038/cti.2016.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.De Filippo C, Cavalieri D, Di Paola M, Ramazzotti M, et al. Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc Natl Acad Sci. 2010;107:14691–6. doi: 10.1073/pnas.1005963107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nagler-Anderson C. Man the barrier! strategic defences in the intestinal mucosa. Nat Rev Immunol. 2001;1:59–67. doi: 10.1038/35095573. [DOI] [PubMed] [Google Scholar]
- 47.Belkaid Y, Hand TW. Role of the Microbiota in Immunity and Inflammation. Cell. 2014;157:121–41. doi: 10.1016/j.cell.2014.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Erdman SE, Poutahidis T. Gut microbiota modulate host immune cells in cancer development and growth. Free Radic Biol Med. 2016 doi: 10.1016/j.freeradbiomed.2016.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yamamoto M, Matsumoto S, Schwabe R, Jobin C, et al. Gut microbiota and colorectal cancer. Genes Environ. 2016;38:11. doi: 10.1186/s41021-016-0038-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hall JA, Bouladoux N, Sun CM, Wohlfert EA, et al. Commensal DNA Limits Regulatory T Cell Conversion and Is a Natural Adjuvant of Intestinal Immune Responses. Immunity. 2008;29:637–49. doi: 10.1016/j.immuni.2008.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bouladoux N, Hall JA, Grainger JR, dos Santos LM, et al. Regulatory role of suppressive motifs from commensal DNA. Mucosal Immunol. 2012;5:623–34. doi: 10.1038/mi.2012.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Segain JP, Raingeard de la Blétière D, Bourreille A, Leray V, et al. Butyrate inhibits inflammatory responses through NFkappaB inhibition: implications for Crohn’s disease. Gut. 2000;47:397–403. doi: 10.1136/gut.47.3.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Burger-van Paassen N, Vincent A, Puiman PJ, van der Sluis M, et al. The regulation of intestinal mucin MUC2 expression by short-chain fatty acids: implications for epithelial protection. Biochem J. 2009;420 doi: 10.1042/BJ20082222. [DOI] [PubMed] [Google Scholar]
- 54.Maslowski KM, Vieira AT, Ng A, Kranich J, et al. Regulation of inflammatory responses by gut microbiota and chemoattractant receptor GPR43. Nature. 2009;461:1282–6. doi: 10.1038/nature08530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Atarashi K, Tanoue T, Oshima K, Suda W, et al. T reg induction by a rationally selected mixture of Clostridia strains from the human microbiota. Nature. 2013;500 doi: 10.1038/nature12331. [DOI] [PubMed] [Google Scholar]
- 56.O’Mahony C, Scully P, O’Mahony D, Murphy S, et al. Commensal-Induced Regulatory T Cells Mediate Protection against Pathogen-Stimulated NF-κB Activation. PLoS Pathog. 2008;4:e1000112. doi: 10.1371/journal.ppat.1000112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Macpherson AJ, Harris NL. Interactions between commensal intestinal bacteria and the immune system. Nat Rev Immunol. 2004;4:478–85. doi: 10.1038/nri1373. [DOI] [PubMed] [Google Scholar]
- 58.Nardi RM, Silva ME, Vieira EC, Bambirra EA, Nicoli JR. Intragastric infection of germfree and conventional mice with Salmonella typhimurium. Brazilian J Med Biol Res = Rev Bras Pesqui Medicas E Biol. 1989;22:1389–92. [PubMed] [Google Scholar]
- 59.Zhou Y, Zhi F. Lower Level of Bacteroides in the Gut Microbiota Is Associated with Inflammatory Bowel Disease: A Meta-Analysis. Biomed Res Int. 2016;2016:1–9. doi: 10.1155/2016/5828959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Peloquin JM, Nguyen DD. The microbiota and inflammatory bowel disease: Insights from animal models. Anaerobe. 2013;24:102–6. doi: 10.1016/j.anaerobe.2013.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Garrett WS, Lord GM, Punit S, Lugo-Villarino G, et al. Communicable Ulcerative Colitis Induced by T-bet Deficiency in the Innate Immune System. Cell. 2007;131:33–45. doi: 10.1016/j.cell.2007.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mazmanian SK, Liu CH, Tzianabos AO, Kasper DL. An Immunomodulatory Molecule of Symbiotic Bacteria Directs Maturation of the Host Immune System. Cell. 2005;122:107–18. doi: 10.1016/j.cell.2005.05.007. [DOI] [PubMed] [Google Scholar]
- 63.Rea K, Dinan TG, Cryan JF. The microbiome: A key regulator of stress and neuroinflammation. Neurobiol Stress. 2016;4:23–33. doi: 10.1016/j.ynstr.2016.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Erny D, Hrabě de Angelis AL, Jaitin D, Wieghofer P, et al. Host microbiota constantly control maturation and function of microglia in the CNS. Nat Neurosci. 2015;18:965–77. doi: 10.1038/nn.4030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Navarro F, Liu Y, Rhoads JM. Can probiotics benefit children with autism spectrum disorders? World J Gastroenterol J Gastroenterol. 2016;22:10093–102. doi: 10.3748/wjg.v22.i46.10093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Aidy S, El, Dinan TG, Cryan JF. Immune modulation of the brain-gut-microbe axis. Front Microbiol. 2014;5 doi: 10.3389/fmicb.2014.00146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bravo JA, Forsythe P, Chew MV, Escaravage E, et al. Ingestion of Lactobacillus strain regulates emotional behavior and central GABA receptor expression in a mouse via the vagus nerve. Proc Natl Acad Sci U S A. 2011;108:16050–5. doi: 10.1073/pnas.1102999108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kumar M, Babaei P, Ji B, Nielsen J. Human gut microbiota and healthy aging: Recent developments and future prospective. Nutr Heal Aging. 2016;4:3–16. doi: 10.3233/NHA-150002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Braniste V, Al-Asmakh M, Kowal C, Anuar F, et al. The gut microbiota influences blood-brain barrier permeability in mice. Sci Transl Med. 2014;6:263ra158. doi: 10.1126/scitranslmed.3009759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lynch MA. Long-Term Potentiation and Memory. Physiol Rev. 2004;84 doi: 10.1152/physrev.00014.2003. [DOI] [PubMed] [Google Scholar]
- 71.Aoki-Yoshida A, Saito S, Fukiya S, Aoki R, et al. Lactobacillus rhamnosus GG increases Toll-like receptor 3 gene expression in murine small intestine ex vivo and in vivo. Benef Microbes. 2016;7:421–9. doi: 10.3920/BM2015.0169. [DOI] [PubMed] [Google Scholar]
- 72.Westendorf AM, Gunzer F, Deppenmeier S, Tapadar D, et al. Intestinal immunity of Escherichia coli Nissle 1917: a safe carrier for therapeutic molecules. FEMS Immunol Med Microbiol. 2005;43:373–84. doi: 10.1016/j.femsim.2004.10.023. [DOI] [PubMed] [Google Scholar]
- 73.Martín R, Miquel S, Ulmer J, Kechaou N, Langella P, Bermúdez-Humarán LG. Role of commensal and probiotic bacteria in human health: a focus on inflammatory bowel disease. Microb Cell Fact. 2013;12:71. doi: 10.1186/1475-2859-12-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wallace CJK, Milev R. The effects of probiotics on depressive symptoms in humans: a systematic review. Ann Gen Psychiatry. 2017;16:14. doi: 10.1186/s12991-017-0138-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Spinler JK, Ross CL, Savidge TC. Probiotics as adjunctive therapy for preventing Clostridium difficile infection – What are we waiting for? Anaerobe. 2016;41:51–7. doi: 10.1016/j.anaerobe.2016.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Mimee M, Citorik RJ, Lu TK. Microbiome therapeutics - Advances and challenges. Adv Drug Deliv Rev. 2016 doi: 10.1016/j.addr.2016.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lagenaur LA, Sanders-Beer BE, Brichacek B, Pal R, et al. Prevention of vaginal SHIV transmission in macaques by a live recombinant Lactobacillus. Mucosal Immunol. 2011;4:648–57. doi: 10.1038/mi.2011.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Duan F, March JC. Engineered bacterial communication prevents Vibrio cholerae virulence in an infant mouse model. Proc Natl Acad Sci U S A. 2010;107:11260–4. doi: 10.1073/pnas.1001294107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hamady ZZR, Scott N, Farrar MD, Wadhwa M, et al. Treatment of colitis with a commensal gut bacterium engineered to secrete human TGF-β1 under the control of dietary xylan. Inflamm Bowel Dis. 2011;17:1925–35. doi: 10.1002/ibd.21565. [DOI] [PubMed] [Google Scholar]
- 80.Hamady ZZR, Scott N, Farrar MD, Lodge JPA, et al. Xylan-regulated delivery of human keratinocyte growth factor-2 to the inflamed colon by the human anaerobic commensal bacterium Bacteroides ovatus. Gut. 2010;59:461–9. doi: 10.1136/gut.2008.176131. [DOI] [PubMed] [Google Scholar]
- 81.Hamady Z. Novel xylan-controlled delivery of therapeutic proteins to inflamed colon by the human anaerobic commensal bacterium. Ann R Coll Surg Engl. 2013;95:235–40. doi: 10.1308/003588413X13511609958217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Bermúdez-Humarán LG, Motta J-P, Aubry C, Kharrat P, et al. Serine protease inhibitors protect better than IL-10 and TGF-β anti-inflammatory cytokines against mouse colitis when delivered by recombinant lactococci. Microb Cell Fact. 2015;14:26. doi: 10.1186/s12934-015-0198-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Motta J-P, Bermúdez-Humarán LG, Deraison C, Martin L, et al. Food-Grade Bacteria Expressing Elafin Protect Against Inflammation and Restore Colon Homeostasis. Sci Transl Med. 2012;4 doi: 10.1126/scitranslmed.3004212. [DOI] [PubMed] [Google Scholar]
- 84.Caluwaerts S, Vandenbroucke K, Steidler L, Neirynck S, et al. AG013, a mouth rinse formulation of Lactococcus lactis secreting human Trefoil Factor 1, provides a safe and efficacious therapeutic tool for treating oral mucositis. Oral Oncol. 2010;46:564–70. doi: 10.1016/j.oraloncology.2010.04.008. [DOI] [PubMed] [Google Scholar]
- 85.Duan FF, Liu JH, March JC. Engineered commensal bacteria reprogram intestinal cells into glucose-responsive insulin-secreting cells for the treatment of diabetes. Diabetes. 2015;64:1794–803. doi: 10.2337/db14-0635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Duan F, March JC. Interrupting Vibrio cholerae infection of human epithelial cells with engineered commensal bacterial signaling. Biotechnol Bioeng. 2008;101:128–34. doi: 10.1002/bit.21897. [DOI] [PubMed] [Google Scholar]
- 87.Suzuki A, Nakauchi H, Taniguchi H. Glucagon-like peptide 1 (1–37) converts intestinal epithelial cells into insulin-producing cells. Proc Natl Acad Sci. 2003;100:5034–9. doi: 10.1073/pnas.0936260100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.del Carmen S, de Moreno de LeBlanc A, Levit R, Azevedo V, et al. Anti-cancer effect of lactic acid bacteria expressing antioxidant enzymes or IL-10 in a colorectal cancer mouse model. Int Immunopharmacol. 2017;42:122–9. doi: 10.1016/j.intimp.2016.11.017. [DOI] [PubMed] [Google Scholar]
- 89.Takiishi T, Korf H, Van Belle TL, Robert S, et al. Reversal of autoimmune diabetes by restoration of antigen-specific tolerance using genetically modified Lactococcus lactis in mice. J Clin Invest. 2012;122:1717–25. doi: 10.1172/JCI60530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Robert S, Gysemans C, Takiishi T, Korf H, et al. Oral Delivery of Glutamic Acid Decarboxylase (GAD)-65 and IL10 by Lactococcus lactis Reverses Diabetes in Recent-Onset NOD Mice. Diabetes. 2014;63 doi: 10.2337/db13-1236. [DOI] [PubMed] [Google Scholar]
- 91.Claesen J, Fischbach MA. Synthetic microbes as drug delivery systems. ACS Synth Biol. 2015;4:358–64. doi: 10.1021/sb500258b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Xiang S, Fruehauf J, Li CJ. Short hairpin RNA-expressing bacteria elicit RNA interference in mammals. Nat Biotechnol. 2006;24:697–702. doi: 10.1038/nbt1211. [DOI] [PubMed] [Google Scholar]
- 93.Els Conrath K, Lauwereys M, Wyns L, Muyldermans S. Camel single-domain antibodies as modular building units in bispecific and bivalent antibody constructs. J Biol Chem. 2001;276:7346–50. doi: 10.1074/jbc.M007734200. [DOI] [PubMed] [Google Scholar]
- 94.Vandenbroucke K, de Haard H, Beirnaert E, Dreier T, et al. Orally administered L. lactis secreting an anti-TNF Nanobody demonstrate efficacy in chronic colitis. Mucosal Immunol. 2010;3:49–56. doi: 10.1038/mi.2009.116. [DOI] [PubMed] [Google Scholar]
- 95.Ruder WC, Lu T, Collins JJ. Synthetic biology moving into the clinic. Science. 2011;333:1248–52. doi: 10.1126/science.1206843. [DOI] [PubMed] [Google Scholar]
- 96.Kotula JW, Kerns SJ, Shaket LA, Siraj L, et al. Programmable bacteria detect and record an environmental signal in the mammalian gut. Proc Natl Acad Sci U S A. 2014;111:4838–43. doi: 10.1073/pnas.1321321111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Bovo F, Lenzi RM, Yamassaki FT, Messias-Reason IJ, et al. Modulating Effects of Arabinogalactans from Plant Gum Exudates on Human Complement System. Scand J Immunol. 2016;83:314–20. doi: 10.1111/sji.12427. [DOI] [PubMed] [Google Scholar]
- 98.Darzi J, Frost GS, Swann JR, Costabile A, Robertson MD. L-rhamnose as a source of colonic propionate inhibits insulin secretion but does not influence measures of appetite or food intake. 2016 doi: 10.1016/j.appet.2015.12.011. [DOI] [PubMed] [Google Scholar]
- 99.Conterno L, Fava F, Viola R, Tuohy KM. Obesity and the gut microbiota: Does up-regulating colonic fermentation protect against obesity and metabolic disease? Genes Nutr. 2011;6:241–60. doi: 10.1007/s12263-011-0230-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Ritchie ML, Romanuk TN, McFarland L, Sazawal S, et al. A Meta-Analysis of Probiotic Efficacy for Gastrointestinal Diseases. PLoS One. 2012;7:e34938. doi: 10.1371/journal.pone.0034938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Ye H, Charpin-El Hamri G, Zwicky K, Christen M, Folcher M, Fussenegger M. Pharmaceutically controlled designer circuit for the treatment of the metabolic syndrome. Proc Natl Acad Sci U S A. 2013;110:141–6. doi: 10.1073/pnas.1216801110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Xue S, Yin J, Shao J, Yu Y, et al. A Synthetic-Biology-Inspired Therapeutic Strategy for Targeting and Treating Hepatogenous Diabetes. Mol Ther. 2017;25:443–55. doi: 10.1016/j.ymthe.2016.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Bai P, Ye H, Xie M, Saxena P, et al. A synthetic biology-based device prevents liver injury in mice. J Hepatol. 2016;65:84–94. doi: 10.1016/j.jhep.2016.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Williams SR, Gebhart D, Martin DW, Scholl D. Retargeting R-type pyocins to generate novel bactericidal protein complexes. Appl Environ Microbiol. 2008;74:3868–76. doi: 10.1128/AEM.00141-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Hwang IY, Koh E, Wong A, March JC, et al. Engineered probiotic Escherichia coli can eliminate and prevent Pseudomonas aeruginosa gut infection in animal models. Nat Commun. 2017;8:15028. doi: 10.1038/ncomms15028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Gibson MK, Pesesky MW, Dantas G. The Yin and Yang of Bacterial Resilience in the Human Gut Microbiota. J Mol Biol. 2014;426:3866–76. doi: 10.1016/j.jmb.2014.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Steidler L, Neirynck S, Huyghebaert N, Snoeck V, et al. Biological containment of genetically modified Lactococcus lactis for intestinal delivery of human interleukin 10. Nat Biotechnol. 2003;21:785–9. doi: 10.1038/nbt840. [DOI] [PubMed] [Google Scholar]
- 108.Chan CTY, Lee JW, Cameron DE, Bashor CJ, Collins JJ. “Deadman” and “Passcode” microbial kill switches for bacterial containment. Nat Chem Biol. 2015;12:82–6. doi: 10.1038/nchembio.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Asadullah K, Sterry W, Volk HD. Interleukin-10 Therapy—Review of a New Approach. Pharmacol Rev. 2003;55 doi: 10.1124/pr.55.2.4. [DOI] [PubMed] [Google Scholar]
- 110.Shis DL, Bennett MR. Library of synthetic transcriptional AND gates built with split T7 RNA polymerase mutants. Proc Natl Acad Sci. 2013;110:5028–33. doi: 10.1073/pnas.1220157110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Shis DL, Hussain F, Meinhardt S, Swint-Kruse L, Bennett MR. Modular, Multi-Input Transcriptional Logic Gating with Orthogonal LacI/GalR Family Chimeras. ACS Synth Biol. 2014;3:645–51. doi: 10.1021/sb500262f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Wang L, Qian K, Huang Y, Jin N, et al. SynBioLGDB: a resource for experimentally validated logic gates in synthetic biology. Sci Rep. 2015;5:8090. doi: 10.1038/srep08090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Tamsir A, Tabor JJ, Voigt CA. Robust multicellular computing using genetically encoded NOR gates and chemical “wires”. Nature. 2011;469:212–5. doi: 10.1038/nature09565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Rubens JR, Selvaggio G, Lu TK. Synthetic mixed-signal computation in living cells. Nat Commun. 2016;7:11658. doi: 10.1038/ncomms11658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Del Carmen S, Rosique RM, Saraiva T, Zurita-Turk M, et al. Protective effects of lactococci strains delivering either IL-10 protein or cDNA in a TNBS-induced chronic colitis model. J Clin Gastroenterol. 2014;48:S12–7. doi: 10.1097/MCG.0000000000000235. [DOI] [PubMed] [Google Scholar]
- 116.Robert S, Gysemans C, Takiishi T, Korf H, et al. Oral delivery of glutamic acid decarboxylase (GAD)-65 and IL10 by lactococcus lactis reverses diabetes in recent-onset NOD mice. Diabetes. 2014;63:2876–87. doi: 10.2337/db13-1236. [DOI] [PubMed] [Google Scholar]


