Abstract
An important regulatory domain of NF-κB Essential Modulator (NEMO) is a ubiquitin-binding zinc finger, with a tetrahedral CYS3HIS1 zinc-coordinating binding site. Two variations of NEMO’s zinc finger are implicated in various disease states including ectodermal dysplasia and adult-onset glaucoma. To discern structural and dynamical differences between these disease states, we present results of 48-μs of molecular dynamics simulations for three zinc finger systems each in two states, with and without zinc-bound and correspondingly appropriate cysteine thiol/thiolate configurations. The wild-type protein, often studied for its role in cancer, maintains the most rigid and conformationally stable zinc-bound configuration compared with the diseased counterparts. The glaucoma-related protein has persistent loss of secondary structure except within the dominant conformation. Conformational overlap between wild-type and glaucoma isoforms indicate a competitive binding mechanism may be substantial in the malfunctioning configuration, while the alpha-helical disruption of the ectodermal dysplasia suggests a loss of binding selectivity is responsible for aberrant function.
Keywords: zinc fingers, molecular dynamics, NEMO, dysplasia, glaucoma
1. Introduction
Found in most animal cell types, Nuclear Factor kappa-light-chain-enhancer of activated B cells (NF-κB) is an anti-apoptotic protein complex that controls DNA transcription. Its anti-apoptotic nature, found over expressed in cancer, has made it a target to potentiate chemotherapy (Garg & Aggarwal, 2002; Sethi, Sung, & Aggarwal, 2008). One critical regulatory protein of NF-κB, NF-κB Essential Modulator (NEMO) controls activation through various signaling pathways (Huang, Feinberg, Suryanarayanan, & Miyamoto, 2002; McCool & Miyamoto, 2012; Perkins, 2007; Wu, 2006). In particular, the zinc finger of NEMO has been identified as essential for NF-κB activation under certain stress conditions (UV radiation and topoisomerase inhibitors) but is dispensable for rapid, strong inducers of NF-κB, including Tumor Necrosis Factor-α (TNF-α) and lipopolysaccharide (LPS) (Huang et al., 2002). As a means to probe the mechanisms of NF-κB activation via NEMO’s zinc finger, we examine microsecond Molecular Dynamics (MD) simulations of wild type NEMO and two known malfunctioning variants. In previous work, we have shown the importance of long-timescale (microsecond) simulations for zinc fingers as they illuminate rare events unlikely to be seen in short (nanosecond) simulations (Godwin, Gmeiner, & Salsbury, 2015; Godwin, Melvin, Gmeiner, & Salsbury, 2016).
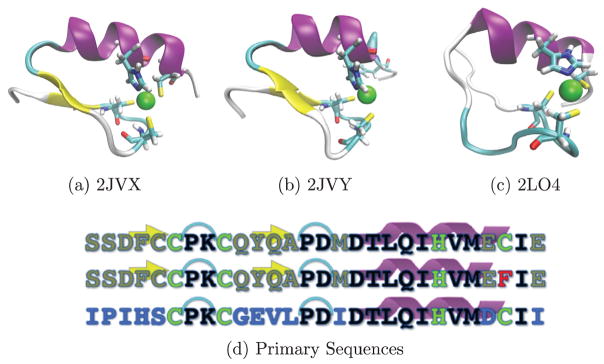
The zinc finger of NEMO belongs to the CYS3HIS1 class of zinc finger with three zinc-binding cysteines and 1 histidine that nominally adopt a ββα–secondary structure motif, as seen in Figure 1, which compares the primary, secondary, and tertiary structures of the data available in the RSCB Protein Data Bank (Berman, Kleywegt, Nakamura, & Markley, 2014). Note that all three variants have an alpha helix, and two of the three (2JVX and 2JVY) have a well-defined beta sheet. Computational modeling of biopolymers, particularly via all-atom molecular dynamics (MD), is emerging as an efficient way to unravel structure-function relationships and generally approach structure-based drug discovery (Durrant & McCammon, 2011; Godwin, Melvin, & Salsbury, 2016; Katsila, Spyroulias, Patrinos, & Matsoukas, 2016; Salsbury, 2010; Vasilyeva et al., 2009). MD provides a wealth of information about a system’s structural, dynamical, and kinematic behavior, in an inexpensive and efficient manner affording sufficient detail for identification of specific mechanisms of action including binding and regulatory function (AbdelHafez et al., 2013; Salsbury et al., 2006). Adept identification of such mechanisms with atomic detail will improve target identification and optimize subsequent drug development (Borhani & Shaw, 2012; Durrant & McCammon, 2011; Sliwoski, Kothiwale, Meiler, & Lowe, 2014). Here, we apply MD to three different zinc finger proteins, probing their conformational and dynamical differences to ascertain relationships from proteins associated with normal functions and with ectodermal dysplasia, and adult-onset glaucoma.
Figure 1.
Crystal structures of the three proteins (a) 2JVX (WT), (b) 2JVY (ED), (c) 2LO4 (AOG).
Notes: Structures are colored by secondary structure with alpha helices in purple, beta-sheets in yellow, while turns are cyan and unstructured regions are white. (d) Sequences of each protein, with (a)–(c) ordered from top to bottom. Color coding differentiates residues that are: common to all three proteins (black), common between 2JVX and 2JVY only (gold), exclusive to 2LO4 (blue), zinc-binding (green), and the helix breaking phenylalanine of 2JVY (red).
In order to understand how these diseases might manifest from malfunctioning zinc fingers, we investigate two zinc fingers associated with known disease states to compare with the functional NEMO wild type (WT) version (PDB ID: 2JVX, Figure 1(a)) via simulation. WT NEMO critically regulates important nuclear translocation processes upstream of activation of the anti-apoptotic NF-κB protein (Miyamoto, 2011). The NEMO WT protein is the zinc-finger domain of NF-κB Essential Modulator (NEMO), a known ubiquitin binder (Cordier et al., 2009). Multiple binding configurations of the zinc finger with ubiquitin have been identified experimentally (Ngadjeua et al., 2013) and preferentially interact with Lys63 linked ubiquitin chains. One of the binding interfaces with ubiquitin exists along the alpha-helical domain of the zinc finger of NEMO. The other proposed site is along the N-terminal region with interactions with the first beta-sheet and the turn before the alpha-helical region where the aspartic acid plays an important role in binding (Ngadjeua et al., 2013). This process of polyubiquitination and associated interactions with NEMO’s zinc finger are important to activation of NF-κB.
The disease-related variant most similar in sequence and structure to the wild type (PDB ID: 2JVY, Figure 1(b)), is associated with anhidrotic ectodermal dysplasia (ED), an uncommon ailment that causes hair, nails, and skin to stiffen (Shifera, 2010). A single point mutation between NEMO WT and the ED mutant (C26F) results in proteins that are 96% sequence identical. In this case, the last zinc-binding cysteine (residue 26) is mutated into a phenylalanine, breaking the α-helix and disrupting tetrahedral zinc coordination - leaving only a trigonal coordinating binding pocket. Hypomorphic genetic mutations have shown that this disease results from disruption of the NF-κB signaling pathway making the zinc-binding region of particular interest (Döffinger et al., 2001). Primary structures of the three different zinc fingers are compared in Figure 1(d).
The zinc finger domain of optineurin (PDB ID: 2LO4, Figure 1(c)), a NEMO homolog, is a retinal protein associated with adult onset glaucoma (AOG) also thought to mediate apoptosis via the TNF-α signaling pathway, inflammation and vasoconstriction (Rezaie et al., 2002; Zhu, Wu, Zhao, & Ashwell, 2007). Studies have shown the optineurin negatively regulates NF-κB activation induced via the TNFα singaling pathway by competing for ubiquitinated RIP (Zhu et al., 2007). Optineurin activation requires phosphorylation by TBK1 in order to bind ubiquitin. Additionally, optineurin has gained increasing attention as it plays a role in autophagy, amyotrophic lateral sclerosis, and Paget disease in bones (Slowicka, Vereecke, & van Loo, 2016). Both the AOG and the WT NEMO protein adopt comparable alpha-helical secondary structure in X-ray crystal structures and conformations throughout simulation. The two proteins are 57% sequence identical (Table 1). The conserved residues are predominantly the zinc-binding residues and the α-helix while most discrepancies are in the β-sheets as shown in Figure 1.
Table 1.
Summary of the three zinc fingers in this study, 2JVX (NEMO-WT), 2JVY (ED), and 2LO4 (AOG), comparing the number of atoms for the CYNZN/CYS cases, the binding site residues and the sequence identity relative to WT.
| Protein (PDB ID) | Disease | Atoms | Binding site | Sequence identity |
|---|---|---|---|---|
| NEMO-WT (2JVX) | N/A | 423/425 | CCHC | 100% |
| NEMO-C26F (2JVY) | ED | 433/434 | CCH | 96% |
| OPTN-WT (2LO4) | AOG | 433/435 | CCHC | 57% |
2. Methods
2.1. Simulation methods
Using all atom MD, we probe the structure and dynamics of the aforementioned zinc fingers. All simulations were run using ACEMD (Harvey, Giupponi, & Fabritiis, 2009) on Acellera’s MetroCubo GPU workstations with either GeForce GTX 680 or GeForce GTX 780 NVIDIA GPUs. Systems were solvated and ionized to a concentration of 0.15 mol/L NaCl and minimized using ACEMD’s built-in conjugant gradient algorithm. Simulations were then run using periodic boundary conditions and a Particle Mesh Ewald approximation for long-range electrostatics, using the default parameters (Harvey & De Fabritiis, 2009). The simulations used different [box sizes](grid sizes) for simulations due to relaxed limitations requiring cubic water boxes, with 2JVX-[50 × 50 × 50](52 × 52 × 52), 2JVY[48.6 × 41.2× 33.9](52× 44× 38), 2LO4[41.3× 48.0× 38.1](44× 52 × 42). A Langevin thermostat (Feller, Zhang, Pastor, & Brooks, 1995) and Berendsen barostat (Berendsen, Postma, van Gunsteren, DiNola, & Haak, 1984) were used to maintain the temperature to T = 300 K with a damping coefficient of γ = 0.1 and pressure to P = 1.01325 Bar with a relaxation time of 400 fs, respectively. In addition to the speedup offered by parallelization, we also used a hydrogen mass-repartitioning scheme in ACEMD to double the time-step for each calculation, from 2 to 4 fs (Feenstra, Hess, & Berendsen, 1999; Harvey et al., 2009; Hopkins, Le Grand, Walker, & Roitberg, 2015). All bonds to hydrogen atoms were constrained using SHAKE (Ryckaert, Ciccotti, & Berendsen, 1977).
There are two primary forms of the CYS3HIS1 binding site motif that occur under physiological conditions. The first is the zinc-bound configuration with the zinc ion coordinated by thiolate cysteines and the histidine. Proper cysteine deprotonation (referred to throughout as CYN) required a patch (Budiman, Knaggs, Fetrow, & Alexander, 2007; Lee et al., 2004) of the traditional cysteine CHARMM 27 parameters (Foloppe & MacKerell, 2000). The zinc ion in the CYNZN configurations was modeled as a monoatomic spherical particle coordinated with non-bonded interactions and no additional constraints as is standard in the CHARMM force fields. In the second case, the zinc is absent and those same cysteines are instead thiols (CYS). Previous studies have shown that while the CYN, zinc-bound case is structurally stable at physiological ion concentrations; the CYS counterpart is intrinsically disordered (Godwin et al., 2016). Each MD trajectory ran for 1μs, with 8 separate runs for each of the three disease states (2JVX (WT), 2JVY (ED), and 2LO4 (AOG)) and each biological configuration (CYS and CYNZN), for a total of 48μs of data (totaling ~ 348 GB of raw trajectory data).
2.2. Analysis methods
Prior to earnest analysis, we post-process the trajectories by removing the water and sodium and chlorine ions using an interface for VMD’s Catdcd 5.0 plugin (Humphrey, Dalke, & Schulten, 1996; Godwin & Salsbury, 2015), and aligning the conformations to remove rotations and translations introduced by diffusion with VMD’s built-in MultiSeq structure align feature (Roberts, Eargle, Wright & Luthey-Schulten, 2006). After alignment, preliminary RMSD indicates the equilibration phase of the proteins, estimated here to be approximately the first 17 ns (See supplemental Figure 1). Trajectories combined across different proteins used an alpha carbon only basis set. Any combinations of trajectories were realigned to ensure consistency. Data is presented in 80ps intervals unless otherwise specified.
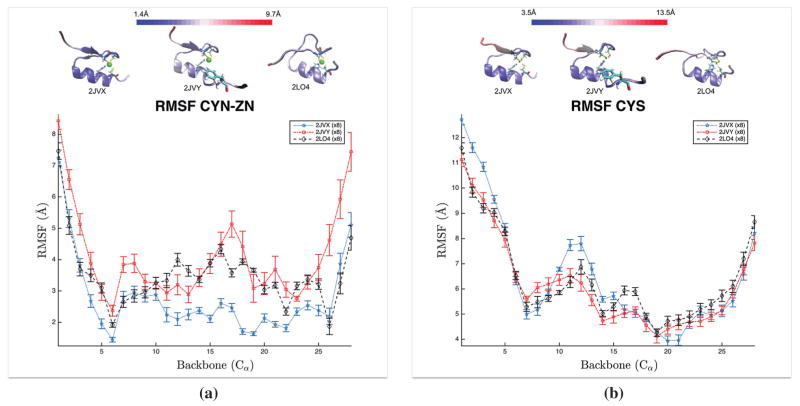
Root Mean Square Fluctuations (RMSF) are spatial variations averaged over time with respect to the mean structure. Error bars in Figure 2 are the standard error across separate trajectories while the data point is the RMSF of all trajectories combined. Calculations employed the measure command built-in to VMD (Humphrey et al., 1996).
Figure 2.
RMSF of Cα of the WT (2JVX), ED (2JVY), and AOG (2LO4) proteins for both binding site configurations – (a) CYNZN and (b) CYS.
Notes: Above the RMSF plots are the corresponding proteins colored by their RMSF value, with blue indicating a low RMSF and red indicating a large RMSF. The error bars for each data point are based on the standard error from the eight separate trajectories.
Hydrogen bonds were treated as a binary consideration, where the bond is present if certain geometrical criteria are met, and absent if not (Gregoret, Rader, Fletterick, & Cohen, 1991; Michaud-Agrawal, Denning, Woolf, & Beckstein, 2011). Namely, we consider intermediate strength hydrogen bonds (Jeffrey & Takagi, 1978), such that if the donor and acceptor pair are within d = 3.2Å of one another and bond angle greater that θ = 120°, the hydrogen bond is ON. Particular bonds highlighted here are those that occur in more than 20% of the time in trajectories for that particular configuration. Defining the hydrogen bond generally in terms of the parameters (d, θ), we have
| (1) |
where x is the donor-acceptor distance and ȡ is the angle between the donor-hydrogen-acceptor triplet.
Alpha carbon clustering employed a Quality Threshold algorithm (QT) based on RMSD between frames via the VMD plugin (Heyer, Kruglyak, & Yooseph, 1999). In QT clustering, the maximum diameter of a cluster along with the maximum number of clusters is required input parameters. The algorithm scans through the trajectory determining all RMSD values between frames and the largest set of frames within the cluster diameter are placed into the first cluster and then removed. With those frames removed, the process is repeated until all frames are clustered or the maximum number of clusters is reached, whichever comes first. All clustering data presented had the following parameters: 100 maximum clusters, 3.5Å RMSD distance cutoff for calculation, and refined trajectories of 160 ps/step.
Correlated motions are the normalized covariance matrix, and quantify how any pair of atoms move together or opposite, throughout a trajectory. Correlations were calculated using internally developed Matlab® code available upon request. The equation for calculating covariance is shown in Equation (2)
| (2) |
where N is the number of frames, r is the xyz vector for atom i or j at frame α. The correlation matrix, Equation (3) is just the normalized covariance such that all values will be between −1 and 1, namely
| (3) |
Principal Component Analysis was then performed on alpha carbon atoms by diagonalizing the correlation matrix and determining the corresponding eigenvalues and eigenvectors, ranking each component’s contribution to the dynamical ensemble, effectively reducing the number of dimensions to analyze while maintaining the largest contributions to the dynamics. Using the eigenvectors as the complete orthonormal basis set we project out and display as a histogram how a trajectory traverses a particular 2D cross-section of the principal component (PC) space. Disease-related trajectories are similarly projected using the wild type principle component eigenvectors of alpha carbon atoms so we can directly compare how these isoforms traverse the wild type PC space. To do this, the xyz coordinates for the system in question were projected onto the basis set by
| (4) |
where k denotes a particular principal component, R⃗ is the vector components of each alpha carbon throughout the trajectory in question and v⃗k is the kth eigenvector and Pc is the resulting projection. Using relative populations from each particular histogram, we construct a free energy surface to show how the particular landscapes change for each protein along the PC reaction coordinate described by Equation (5).
| (5) |
Here, k is Boltzmann’s constant, T is the temperature in Kelvin, P is the density matrix resulting from the PC projections, and Pmax is the maximum non-zero bin set to zero energy.
2.3. Visualization methods
All images were rendered using VMD (Humphrey et al., 1996) with using Tachyon or NVIDIA’s® Gelato® (Stone, 1998). Composite clustering images of representative structures and 1 standard deviation from that center we generated using a python and VMD-based visual statistics package available on FigShare (Melvin & Salsbury, 2015, 2016). Network figures were generated with Cytoscape (Shannon et al., 2003).
3. Results
All three zinc fingers have been identified as ubiquitin binding domains. Ubiquitin biding of zinc fingers is an active area of research in light of the diversity of binding proteins and binding sites (Dikic, Wakatsuki, & Walters, 2009; Suzuki et al., 2016; Toma et al., 2015). Here we show that the ED mutation indeed disrupts the tetrahedral coordination of the ZF, but other hypomorphic phenotypes of ED have been shown to result from different mutations, for example, a D15V mutation. The aspartic acid is conserved in the ubiquitin binders evaluated here. The results herein suggest potential structural and dynamical mechanisms responsible for a change in affinity of ZF ubiquitin binding. Recently published details of zinc finger binding sites with ubiquitin highlight various possible binding arrangements and indicate how the conformational disparity across the zinc fingers herein can interfere with binding process (Ngadjeua et al., 2013).
3.1. The smallest fluctuations belong to the WT zinc-bound configuration
RMSF plots of alpha carbon atoms for all six configurations are shown in Figure 2. The functional, zinc-bound case (CYNZN), Figure 2(a), shows the wild type protein to have the smallest fluctuations from the mean, particularly for residues 11–25, where the two disease-related proteins are on average nearly 2Å larger than that of the wild type. The zinc binding histidine of 2LO4 shows reduced fluctuations compared to that of 2JVY. The phenylalanine of 2JVY has 2Å larger fluctuations than the others, a trend that persists to the C-terminus. Thiol configurations without zinc (CYS) were generally more similar and larger for each alpha carbon than the three variants of the CYNZN configuration. The N-termini ends of CYS fluctuate the most with the largest fluctuations occurring in the wild type protein. The wild type also increases in fluctuations between residues 10–14, the turn between the second beta-sheet and the alpha helix in Figure 2(b). The consistent loss of secondary structure and large fluctuations for all configurations suggests similar intrinsic disorder for all CYS configurations.
3.2. Hydrogen bonds support WT secondary structure
Hydrogen bond motifs of the six different configurations offer details about stabilizing secondary structure interactions. The distribution of hydrogen bond counts for the zinc-bound cases highlight again that the secondary structure of the wild-type protein is the most stable with the largest number of bonds on average, 12.1. The zinc-bound ED protein has an average count of 11 hydrogen bonds. An observant reader will notice that this is slightly less than would be found in the secondary structure elements. We deliberately constrained the hydrogen bond parameters to highlight the strongest bonds, keeping the parameters consistent across the three protein hydrogen bond data-sets. Somewhat surprisingly, the zinc-bound AOG protein has the fewest number of hydrogen bonds of any configurations, including those with protonated cysteines, with just 8.8 hydrogen bonds on average. The loss of β-sheet secondary structure is a primary factor for the small number of optineurin hydrogen bonds. While there are still a relatively large average number of hydrogen bonds in the protonated (CYS) cases the motifs are transient, with the largest sets of unique donor-acceptor pairs. The average hydrogen bond count for the zinc-less configurations for WT, ED, and AOG are 10.2, 9.57, and 9.59, respectively, again showing the similarity between the configurations sans zinc.
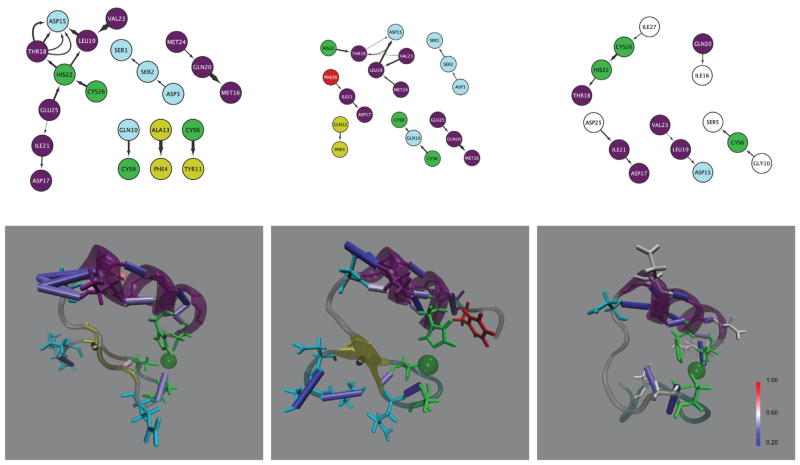
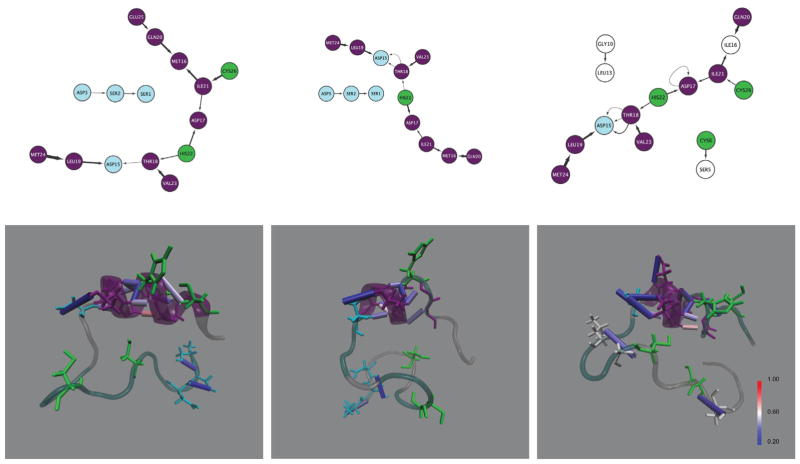
Investigation into particular donor-acceptor pairs that are most common throughout the trajectories provides additional insight into the motifs present in each configuration. Figures 3 and 4 show the hydrogen bond connections in network form, where residues are nodes and hydrogen bonds are edges alongside structural images of hydrogen bonds colors by the occupancy. The zinc-bound wild type protein had five hydrogen bonds that were present more than half of the time, while the others had at most two and the zinc-bound glaucoma protein had no hydrogen bonds persist that much in the simulations. The single hydrogen bond pair present in both active site configurations for the single point mutation are between the same donor-acceptor pair – an amino group of the 20th residue glutamine donates to the carboxyl group of the 16th residue, methionine. This interaction supports the portion of the α-helix opposite the zinc coordinating residues and is present in just over half of both ectordermal dysplasia configurations. This same pair occurs 80% in the wild type zinc-bound case, and in a similar fashion 51% of the time between a glutamine and isoleucine in the zinc-less optineurin configuration. Another hydrogen bond pair that occurs in multiple configurations has the amino group of the 24 residue, methionine (between the zinc-bind histidine and cystine), donating to the carboxyl of the 19th residue, leucine. This pair occurs in the zinc-less (apo) case for both WT (72%) and AOG (65%), and is consistent with an increase in helical radius toward the N-terminus.
Figure 3.
Hydrogen bond networks of the 3 CYNZN configuratoins ((a) 2JVX-CYNZN, (b) 2JVY-CYNZN, (c) 2LO4-CYNZN).
Notes: Edges of the network are directed from donor to acceptor and the width indicates the occupancy – the wider the arrow the more persistent the hydrogen bond. Node labels indicate the amino acid residue and the residue index. Zinc binding nodes are highlighted in green, the 2JVY point-mutation phenylalanine is red, α-helices are purple, β-sheets are yellow turns are light blue, and remaining mutated 2LO4 residues are dark blue.
Figure 4.
Hydrogen bond networks of the three CYNZN configuratoins ((a) 2JVX-CYS, (b) 2JVY-CYS, (c) 2LO4-CYS).
Notes: Edges of the network are directed from donor to acceptor and the width indicates the occupancy – the wider the arrow the more persistent the hydrogen bond. Node labels indicate the amino acid residue and the residue index. Zinc binding nodes are highlighted in green, the 2JVY point-mutation phenylalanine is red, α-helices are purple, β-sheets are yellow turns are light blue, and remaining mutated 2LO4 residues are dark blue.
Additional wild type zinc-bound hydrogen bonds include the zinc binding cysteine donating an amino group to a tyrosine carboxyl (60%) and an amino group from an alanine to a carboxyl phenylalanine (69%), comprising the beta sheet secondary structure. The other two involve the 19th leucine donating to an asparagine (59%) and accepting from a valine (84%), coordinating the α-helix opposite the C-terminus.
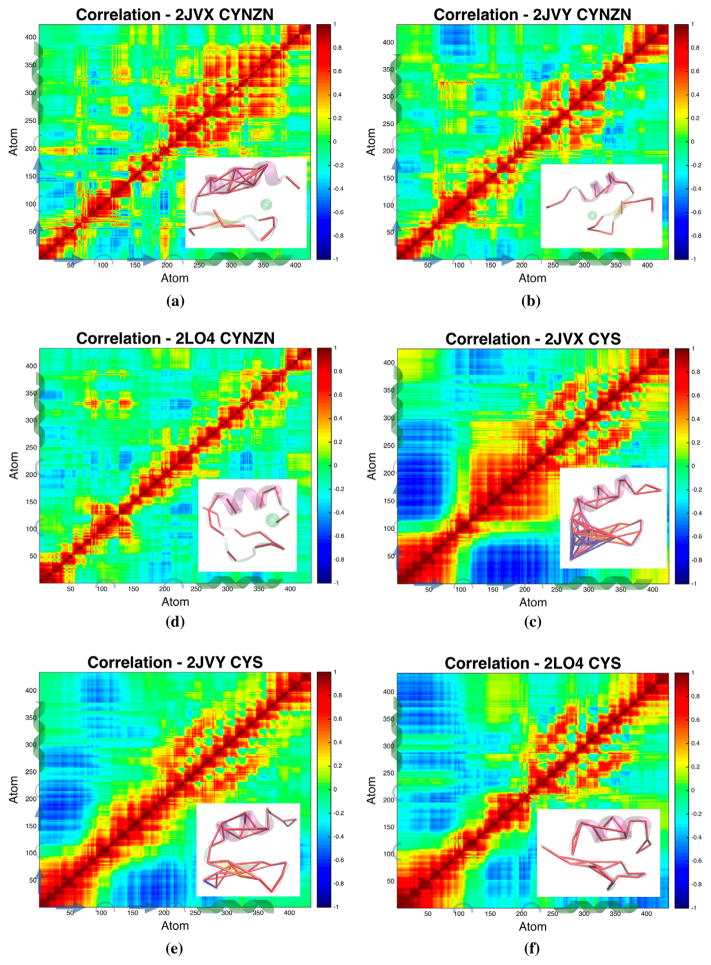
3.3. Atomic fluctuations of secondary structure correlate differently in each configuration
Correlated motions quantifies how much two atoms move together or opposite one another; a value of 1 indicates perfect correlation in the same direction, whereas −1 indicates perfect anti-correlation. Comparing the correlated motions (Table 2) across protein systems we are able to discern which mutated residues most largely influence correlations of the system. Here, Figure 5 shows all CYNZN cases have overall larger correlations than the CYS counterparts, with only small isolated pockets of anti-correlation. Conversely, the CYS configurations have larger regions of anti-correlation that maintain alpha helical signatures across the variants.
Table 2.
Correlation comparison between each set of correlation matrices.
| 2JVX | 2JVY | 2LO4 | ||||
|---|---|---|---|---|---|---|
|
|
|
|||||
| (CYS) | (ZN) | (CYS) | (ZN) | (CYS) | ||
| 2JVX | (ZN) | 0.734 | 0.939 | 0.810 | 0.816 | 0.758 |
| (CYS) | 1 | 0.808 | 0.969 | 0.687 | 0.710 | |
| 2JVY | (ZN) | 1 | 0.860 | 0.882 | 0.757 | |
| (CYS) | 1 | 0.767 | 0.847 | |||
| 2LO4 | (ZN) | 1 | 0.781 | |||
Figure 5.
Correlated motions of the six configurations.
Notes: Each correlation has at the bottom right a structure pertaining to the dominant conformation of clustering results showing alpha carbons with pairwise correlations given by |Ci j | ≥ 0.6, where red cylinders represent correlation and blue cylinders represent anti-correlation. The top row is the zinc-bound cases (CYNZN) and the bottom row is the protonated, zinc-less cases (CYS). Columns from the left to right are WT, ED, and AOG. That is, (a) WT CYNZN, (b) ED CYNZ, (c) AOG CYNZN, (d) WT CYS, (e) ED CYS, and (f) AOG CYS.
The zinc-bound wild type α-helix correlations are very strong and persists to neighbors that are not hydrogen bonded. Neither disease-related protein shows the strongly correlated alpha helix of the wild type. The mutated phenylalanine of 2JVY clearly disrupts the later part of the normal wild type alpha-helical signature. Even though the alpha-helical residues of the glaucoma-related protein (two are identical to the wild type, the correlated signature is missing. All three CYNZN configurations show correlated pockets between CYS9 and HIS22, both zinc-binding residues.
Correlations of CYS all maintain the alpha helical signature and have pockets of anti-correlation. The anti-correlation of the wild type is between the two beta sheets including the N-terminus. This anti-correlation region is present in AOG, however, a more prominent region of anti-correlation occurs between the two termini.
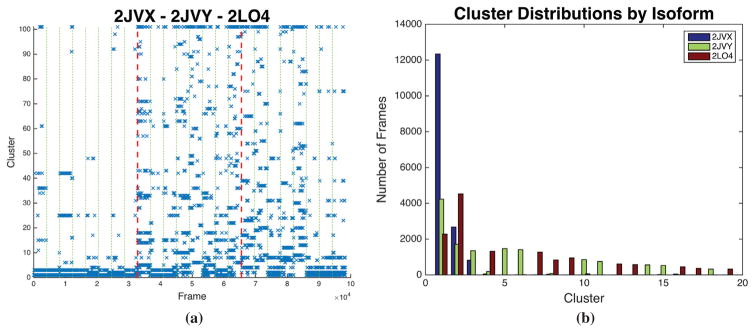
3.4. Wild type is the most conformationally stable configuration
Meaningful partitioning of conformational ensembles provides insight regarding specific geometries, the regularity of those geometries and transitions thereof for a particular system of interest. To do this, we cluster data on an attribute of the trajectory, in this case using RMSD. Figure 6 shows the RMSD based clustering of the three zinc-bound configurations throughout all 24 1-μs trajectories. The WT is the most stable; it samples the fewest number of states and conformations are dominated by the first three clusters, with over 3/4 of its conformations in cluster 1. Comparatively, the ectodermal dysplasia (2JVY) and glaucoma (2LO4) polypeptides have 25.8 and 13.8% in the first cluster, respectively. Cluster 2 is the most populated state for the glaucoma-related polymer (2LO4) at 27.7%, and shows a zinc-displaced conformation with considerable overlap with the wild-type.
Figure 6.
Clustering results show conformational overlap among the disease-related polypeptides and the wild type zinc finger comparing the zinc-bound configurations. (a) A cluster time series plot of cluster versus frame effectively displays the Markov chain. (b) The equilibrium distribution of the clusters for the first 20 clusters. With 100 maximum clusters 3.5Å RMSD cutoff, the first 3 clusters include 61% of all zinc-bound conformations.
Overall, the first three clusters account for 61% of all conformations with 1% of the frames unclustered. Figure 6(b) highlights the result that after clusters 1–3 the remaining clusters are dominated by disease-related conformations with little overlap with WT conformations. The ED mutation (CYS26PHE) shares more conformational overlap with WT (21/24) than the glaucoma mutations (18/24). The phenylalanine disrupts the alpha helix where the otherwise zinc coordinating cysteine would stabilize the structure and this difference dominates the conformational deviations between 2JVX and 2JVY. In general, both disease associated proteins show more conformational disparity than the wild-type properly functional zinc finger.
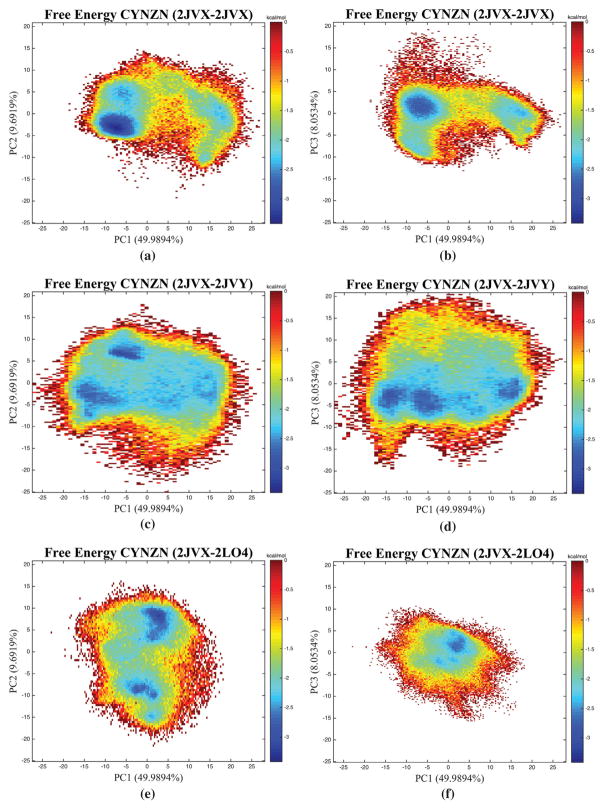
3.5. Principal components isolate configurations
The first three principal components of the wild type, zinc-bound basis set account for 67.7% of the overall variance in protein correlation. The free energy diagrams in Figure 7 shows histograms of the corresponding trajectory projected on the first and second or first and third components in principal component space. Wild type, zinc-bound case (Figure 7(a)) shows a highly populated region of the free energy surface, with outlying regions split along the 1st component. The single point mutation trajectory projected onto the wild type basis set for both components 1–2 and 1–3 show large, shallow surfaces. The circular shape of these surfaces indicate the sampling of the ED mutant (2JVY) trajectory is distributed throughout a large area of the PC space. Conversely, the free energy surfaces of AOG (2LO4) show smaller overall areas, and for PC 1–2 shows two main wells, this time split by the second PC instead of the first.
Figure 7.
Principal Component Projections Histograms of the Zinc-Bound Configurations relative to the WT trajectories. Only showing are the zinc-bound configurations (CYNZN) for (a) WT-WT, [PC1 vs. PC2] (b) WT-WT [PC1 vs. PC3], (c) WT-ED [PC1 vs. PC2], (d) WT-ED [PC1 vs. PC3], (e) WT-AOG [PC1 vs. PC2] (f) WT-AOG [PC1 vs. PC3].
4. Discussion
While the precise mechanisms of how NEMO’s zinc finger regulates the NF-κB signaling pathway are not known, the importance of ubiquitin binding is widely accepted (Cordier, Vinolo, Véron, Delepierre, & Agou, 2008, Cordier et al., 2009; Ngadjeua et al., 2013; Tang, Wang, Xiong, & Guan, 2003). Various mutations disrupt the nominal function of NEMO to regulate the NF-κB pathway. One such mutation not directly studied here, K392R (a known ubiquitin-binding site of the zinc finger), disrupts Lys63 linked polyubiquitination and, combined with deficiency of cylindromatosis, is lethal in early embryogenesis in mice (Zhao et al., 2015). Details of the simulation results highlight the dynamical and structural differences across these proteins.
In previous work, we proposed that the correlation signature, combined with the large fluctuations and loss of secondary structure was coordinated as a mechanism of zinc detection and binding (Godwin et al., 2016). We see here, that the CYS configurations of all three zinc-fingers exhibit similar characteristics with larger contrast in the zinc-bound configurations. Zinc-bound configurations show pockets of long range correlation for WT and both mutations, with WT showing the most pockets of long range correlation. The α-helix region of WT shows a strong, long range correlation signature, particularly near the C-terminus. While there are slight perturbations in the results 2JVY from WT they are more pronounced for 2LO4 in both protonation states (thiol and thiolate cysteines).
4.1. Ectodermal dysplasia (2JVY) vs. wild type (2JVX)
The single point mutation of the zinc-binding cysteine has a significant effect on the structure and dynamics of the protein. Increased fluctuations between residues 15–22 (atoms 203–329), as shown in Figure 2, show that the C-terminal mutation disrupts the stability of the entire α-helix. However, the correlation in Figure 5 maintains the strong α-helical signature of the WT zinc bound case in this region of the comparable ED (2JVY) mutant. Naturally, there is deviation in correlation around the single-point mutation; the phenyalanine creates an anti-correlation pocket that appears in ED between atoms Met24-MET16 (indexes 350-225) is correlated in WT simulations. Along the first two principal components, a dominant well exists for WT located near (−6, −3) and less populated wells occur near (−5, 5) and (17,0) in Figure 7(a). In the ED single-point mutation case the dominant well is now located near (−5, 6) and the main well from the WT configuration is less populated, suggesting an inversion of the first two most populated states from WT to ED in Figure 7(b). Additionally, the basin for ED is shallower and larger than that of WT offering the mutation greater access to the PC space along the first two components.
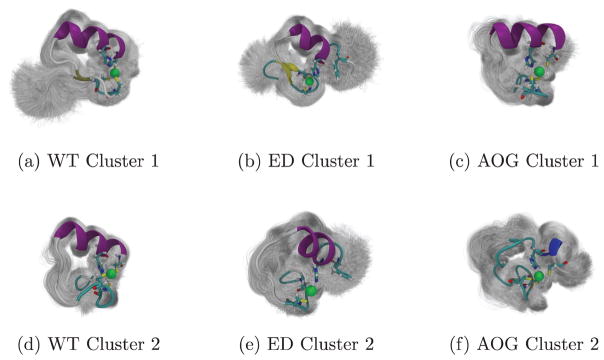
Hydrogen bond analysis evinced candidates responsible for the change in the RMSF and correlation figures in the alpha helical region between WT and ED. In particular, GLU25 is donated to the zinc-binding histidine, effectively pinching the α-helix nearest the C-terminus to a π-helical configuration. Clustering analysis of these simulation suites indicate that the tetrahedral coordination morphs into a trigonal coordination altering the dominant conformations between the two as shown Figure 8. When clustering the two configurations separately, using the same parameters as before (3.5Å RMSD cutoff for 100 clusters), shows a displaced zinc by 4.9Å exposing more of the zinc than when coordinated as a tetrahedral with all 3 cysteines.
Figure 8.
Isolated clustering conformations of thiolate zinc-bound conformations for the first (a) 2JVX (b) 2JVY (c) 2LO4, and the second (d) 2JVX (e) 2JVY (f) 2LO4 clusters showing one standard deviation of each cluster with its representative colored by secondary structure.
4.2. Phenylalanine mutation reduces binding selectivity
The phenylalanine mutation clearly disrupts the structure and dynamics, particularly near the N-terminus (β-sheet region) of the protein. The disruption to the alpha-helix caused by the mutation shortens the secondary structure element by a half-turn in the most populated clusters in Figure 8. Additionally, the displacement of the zinc ion between the dominant configurations resulting from the transition from the tetrahedral coordination to trigonal coordination is likely to disrupt binding selectivity. While the details of the NEMO-Ubiquitin protein-protein interface are an active area of research (Cordier et al., 2009; Ngadjeua et al., 2013; Toma et al., 2015), we hypothesize that the disruption to conformations and correlations of the zinc bound ED mutation result in decreased ubiquitin binding selectivity.
4.3. Adult onset glaucoma (2LO4) vs. wild type (2JVX)
Alteration of the beta-sheet region of the zinc fingers disrupts the secondary structure motifs compared to the wild type configuration as shown in the hydrogen bond and clustering results. This disruption of secondary structure leads to a much larger sampling of conformational space. When present, the zinc maintains a modicum of structure with the tetrahedral coordination throughout the trajectories, but the loss of alpha helix and beta sheets leaves loop-like structure along the second beta sheet to the alpha helix pinched by the zinc. If it does stay in a stable conformation it is more likely to do so in a conformation consistent with cluster 2 (see Figure 6). Interestingly, on rare occasion the WT protein samples this conformation, a kinetic trap, and does not readily return to the dominant conformation.
4.4. Conformational overlap motivates competitive binding hypothesis
In addition to extracting unique structures, clustering analysis highlights the conformational overlap between the disease-related proteins and the wild type in Figure 8. The second cluster of combined clustering, shown in Figure 2(b), is the most populated AOG conformation (27.7% of AOG conformations) and second most WT conformation (16.3% of WT conformations). The conformation is characterized by and extension of the second turn, between the β-sheet and the α-helix, and a perpendicular reorientation of the two β-sheets. When looking at the RMSD clustering of each configuration separately, Figure 8, we see that cluster 2 of the WT (2JVX) is very similar to that of cluster 1 of AOG (2LO4). The median of each are compared in the supplemental Figure 5. The second cluster of the WT is a rare conformational kinetic trap as indicated by Markov analysis of the clustering results (supplemental Figures 2–4) and conformations are 80% likely to stay in that cluster after entering. The average RMSD of these two clusters combined is 2.8Å, smaller than the cluster cutoff used in the QT analysis. This conformational overlap indicates that the two proteins likely have competitive binding in cells where they are both present, potentially leading to the AOG phenotype.
5. Conclusion
Detailed atomic motions revealed by MD highlight specific physical interactions of proteins and provide insights to mechanisms of malfunction that are difficult to ascertain experimentally. Here, we show the results of 16 μs of MD simulations for 3 different ββα-motif zinc fingers, one wild-type and two mutations associated with disease states. Simulation results for each of the configurations highlight unique conformational and dynamics, that are likely to play a role in aberrant function.
In particular, the ED mutation showed clear deviation from the wild type structure and dynamics, particularly in the region of the CYS26PHE mutation as well as shifting the zinc-coordination motif that will likely disrupt nominal ubiquitin binding. Specifically, the single point mutation in the ED protein is predicted to cause a decrease in binding selectivity of the ubiquitin-binding domain of NEMO resulting in down regulation of NF-κB activation. Restoration of normal function might involve docking a small ligand capable of strong interaction with the phenylalanine to make the C-terminal domain more rigid.
The AOG mutation, on the other hand, has a consistent loss of the β-sheet secondary structure, while maintaining a strong α-helical structure. The most stable AOG conformation overlaps with a rare WT kinetic trap conformation, highlighting the possibility of competitive binding between these two proteins. In this case, it is unclear whether the rare WT conformation binds with the Optineurin binding partner, or vice-versa, particularly in light of the conformational diversity of the AOG mutation. These observations and predictions will inspire hopefully further experimental and computational research.
Supplementary Material
Acknowledgments
Funding
This work was partially supported by National Institutes of Health [grant number R01CA129373] to FRS. The authors wish to acknowledge the support of the Wake Forest Baptist Comprehensive Cancer Center Crystallography & Computational Biosciences Shared Resource, supported by the National Cancer Institutes Cancer Center Support [grant award number P30CA012197]. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Cancer Institute. RCG acknowledges SCB training National Institute of General Medical Sciences [grant number T32GM095440] for partially supporting this research. FRS also acknowledges a Reynolds Research leave from Wake Forest University. Some computations were performed on the Wake Forest University DEAC Cluster, a centrally managed resource with support provided in part by the University.
Footnotes
Communicated by Ramaswamy H. Sarma
Disclosure statement
No potential conflict of interest was reported by the authors.
The supplementary material for this paper is available online at https://doi.10.1080/07391102.2017.1363662.
References
- AbdelHafez EMN, Diamanduros A, Negureanu L, Luy Y, Bean JH, Zielke K, … King SB. Computational and synthetic studies towards improving rescinnamine as an inducer of MSH2-dependent apoptosis in cancer treatment. Molecular Cancer Biology. 2013;1(1):1–13. [PMC free article] [PubMed] [Google Scholar]
- Berendsen HJC, Postma JPM, van Gunsteren WF, DiNola A, Haak JR. Molecular dynamics with coupling to an external bath. The Journal of Chemical Physics. 1984;81:3684–3690. [Google Scholar]
- Berman HM, Kleywegt GJ, Nakamura H, Markley JL. The Protein Data Bank archive as an open data resource. Journal of Computer-Aided Molecular Design. 2014;28:1009–14. doi: 10.1007/s10822-014-9770-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borhani DW, Shaw DE. The future of molecular dynamics simulations in drug discovery. Journal of Computer-Aided Molecular Design. 2012;26:15–26. doi: 10.1007/s10822-011-9517-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budiman ME, Knaggs MH, Fetrow JS, Alexander RW. Using molecular dynamics to map interaction networks in an aminoacyl-tRNA synthetase. Proteins: Structure, Function, and Bioinformatics. 2007;68:670–689. doi: 10.1002/prot.21426. [DOI] [PubMed] [Google Scholar]
- Cordier F, Grubisha O, Traincard F, Véron M, Delepierre M, Agou F, … Agou F. The zinc finger of NEMO is a functional ubiquitin-binding domain. Journal of Biological Chemistry. 2009;284:2902–2907. doi: 10.1074/jbc.M806655200. [DOI] [PubMed] [Google Scholar]
- Cordier F, Vinolo E, Véron M, Delepierre M, Agou F. Solution structure of NEMO zinc finger and impact of an anhidrotic ectodermal dysplasia with immunodeficiency-related point mutation. Journal of Molecular Biology. 2008;377:1419–1432. doi: 10.1016/j.jmb.2008.01.048. [DOI] [PubMed] [Google Scholar]
- Dikic I, Wakatsuki S, Walters KJ. Ubiquitin-binding domains – From structures to functions. Nature Reviews Molecular Cell Biology. 2009;10:659–671. doi: 10.1038/nrm2767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Döffinger R, Smahi A, Bessia C, Geissmann F, Feinberg J, Durandy A, … Casanova JL. X-linked anhidrotic ectodermal dysplasia with immunodeficiency is caused by impaired NF-kappaB signaling. Nature Genetics. 2001;27:277–285. doi: 10.1038/85837. [DOI] [PubMed] [Google Scholar]
- Durrant JD, McCammon JA. Molecular dynamics simulations and drug discovery. BMC Biology. 2011;9:71. doi: 10.1186/1741-7007-9-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feenstra KA, Hess B, Berendsen HJC. Improving efficiency of large time-scale molecular dynamics simulations of hydrogen-rich systems. Journal of Computational Chemistry. 1999;20:786–798. doi: 10.1002/(SICI)1096-987X(199906)20:8<786::AID-JCC5>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- Feller SE, Zhang Y, Pastor RW, Brooks BR. Constant Pressure Molecular Dynamics Simulation: The Langevin Piston Method. The Journal of Chemical Physics. 1995;103:4613–4631. [Google Scholar]
- Foloppe N, MacKerell ADJ. All-atom empirical force field for nucleic acids: 1. parameter optimization based on small molecule and condensed phase macromolecular target data. Journal of Computational Chemistry. 2000;21:86–104. [Google Scholar]
- Garg A, Aggarwal BB. Nuclear transcription factor-kappaB as a target for cancer drug development. Leukemia. 2002;16:1053–1068. doi: 10.1038/sj.leu.2402482. [DOI] [PubMed] [Google Scholar]
- Godwin R, Salsbury FR. Catdcd Interface Code. 2015 Retrieved from https://figshare.com/articles/Catdcd_Interface/1613888/1.
- Godwin RC, Gmeiner WH, Salsbury FRJ. Importance of long-time simulations for rare event sampling in zinc finger proteins. Journal of Biomolecular Structure and Dynamics. 2015;1102:1–20. doi: 10.1080/07391102.2015.1015168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godwin RC, Melvin R, Salsbury FR. Computer-aided drug discovery. Vol. 34. New York, NY: Springer; 2016. [Google Scholar]
- Godwin RC, Melvin RL, Gmeiner WH, Salsbury FR., Jr Binding site configurations probe structure and dynamics of the zinc-finger NEMO. Biochemistry. 2016;56:623–633. doi: 10.1021/acs.biochem.6b00755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregoret LM, Rader SD, Fletterick RJ, Cohen FE. Hydrogen bonds involving sulfur atoms in proteins. Proteins. 1991;9:99–107. doi: 10.1002/prot.340090204. [DOI] [PubMed] [Google Scholar]
- Harvey MJ, De Fabritiis G. An implementation of the smooth particle mesh ewald method on GPU hardware. Journal of Chemical Theory and Computation. 2009;5:2371–2377. doi: 10.1021/ct900275y. [DOI] [PubMed] [Google Scholar]
- Harvey MJ, Giupponi G, Fabritiis GD. ACEMD: Accelerating biomolecular dynamics in the microsecond time scale. Journal of Chemical Theory and Computation. 2009;5:1632–1639. doi: 10.1021/ct9000685. [DOI] [PubMed] [Google Scholar]
- Heyer LJL, Kruglyak S, Yooseph S. Exploring expression data: Identification and analysis of coexpressed genes. Genome Research. 1999;9:1106–1115. doi: 10.1101/gr.9.11.1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopkins CW, Le Grand S, Walker RC, Roitberg AE. Long-time-step molecular dynamics through hydrogen mass repartitioning. Journal of Chemical Theory and Computation. 2015;11:1864–1874. doi: 10.1021/ct5010406. [DOI] [PubMed] [Google Scholar]
- Huang TT, Feinberg SL, Suryanarayanan S, Miyamoto S. The zinc finger domain of NEMO is selectively required for NF-kappa B activation by UV radiation and topoisomerase inhibitors. Molecular and Cellular Biology. 2002;22:5813–5825. doi: 10.1128/MCB.22.16.5813-5825.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphrey W, Dalke A, Schulten K. VMD: Visual molecular dynamics. Journal of Molecular Graphics. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- Jeffrey GA, Takagi S. Hydrogen-bond structure in carbohydrate crystals. Accounts of Chemical Research. 1978;109:264–270. [Google Scholar]
- Katsila T, Spyroulias GA, Patrinos GP, Matsoukas MT. Computational approaches in target identification and drug discovery. Computational and Structural Biotechnology Journal. 2016;14:177–184. doi: 10.1016/j.csbj.2016.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Park CJ, Shin JS, Ikegami T, Akutsu H, Choi BS. NMR structure of the DNA decamer duplex containing double T.G mismatches of cis-syn cyclobutane pyrimidine dimer: Implications for DNA damage recognition by the XPC-hHR23B complex. Nucleic Acids Research. 2004;32:2474–2481. doi: 10.1093/nar/gkh568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCool KW, Miyamoto S. DNA damage-dependent NF-κB activation: NEMO turns nuclear signaling inside out. Immunological Reviews. 2012;246(1):311–326. doi: 10.1111/j.1600-065X.2012.01101.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melvin R, Salsbury F. Visual Statistics Code. 2015 doi: 10.6084/m9.figshare.1601897.v1. Retrieved from . [DOI]
- Melvin RL, Salsbury FR. Visualizing ensembles in structural biology. Journal of Molecular Graphics and Modelling. 2016;67:44–53. doi: 10.1016/j.jmgm.2016.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaud-Agrawal N, Denning EJ, Woolf TB, Beckstein O. MD Analysis: A tool kit for the analysis of molecular dynamics simulations. Journal of Computational Chemistry. 2011;32:2319–2327. doi: 10.1002/jcc.21787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamoto S. Nuclear initiated NF-κB signaling: NEMO and ATM take center stage. Cell Research. 2011;21:116–130. doi: 10.1038/cr.2010.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngadjeua F, Chiaravalli J, Traincard F, Raynal B, Fontan E, Agou F. Two-sided ubiquitin binding of NF-κB essential modulator (NEMO) zinc finger unveiled by a mutation associated with anhidrotic ectodermal dysplasia with immunodeficiency syndrome. Journal of Biological Chemistry. 2013;288:33722–33737. doi: 10.1074/jbc.M113.483305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins ND. Integrating cell-signalling pathways with NF-kappaB and IKK function. Nature Reviews Molecular Cell Biology. 2007;8:49–62. doi: 10.1038/nrm2083. [DOI] [PubMed] [Google Scholar]
- Rezaie T, Child A, Hitchings R, Brice G, Miller L, Coca-Prados M, … Sarfarazi M. Adult-onset primary open-angle glaucoma caused by mutations in optineurin. Science (New York, NY ) 2002;295:1077–1079. doi: 10.1126/science.1066901. [DOI] [PubMed] [Google Scholar]
- Roberts E, Eargle J, Wright D, Luthey-Schulten Z. MultiSeq: Unifying sequence and structure data for evolutionary analysis. BMC Bioinformatics. 2006;7:382. doi: 10.1186/1471-2105-7-382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryckaert J, Ciccotti G, Berendsen H. Numerical integration of the cartesian equations of motion of a system with constraints: Molecular dynamics of n-alkanes. Journal of Computational Physics. 1977;341:327–341. [Google Scholar]
- Salsbury FR, Clodfelter JE, Gentry MB, Hollis T, Scarpinato KD, Jr, Clodfelter JE, … Scarpinato KD. The molecular mechanism of DNA damage recognition by MutS homologs and its consequences for cell death response. Nucleic Acids Research. 2006;34:2173–85. doi: 10.1093/nar/gkl238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salsbury FR., Jr Molecular dynamics simulations of protein dynamics and their relevance to drug discovery. Current Opinion in Pharmacology. 2010;10:738–44. doi: 10.1016/j.coph.2010.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sethi G, Sung B, Aggarwal BB. Nuclear factor-kappaB activation: From bench to bedside. Experimental Biology and Medicine (Maywood) 2008;233:21–31. doi: 10.3181/0707-MR-196. [DOI] [PubMed] [Google Scholar]
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, … Ideker T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Research. 2003;13:2498–504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shifera AS. The zinc finger domain of IKKγ (NEMO) protein in health and disease. Journal of Cellular and Molecular Medicine. 2010;14:2404–2414. doi: 10.1111/j.1582-4934.2010.01054.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sliwoski G, Kothiwale S, Meiler J, Lowe EW. Computational methods in drug discovery. Pharmacological Reviews. 2014;66:334–95. doi: 10.1124/pr.112.007336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slowicka K, Vereecke L, van Loo G. Cellular functions of optineurin in health and disease. Trends in Immunology. 2016;37:621–633. doi: 10.1016/j.it.2016.07.002. [DOI] [PubMed] [Google Scholar]
- Stone JE. Masters thesis. University of Missouri; Rolla, MO: 1998. An efficient library for parallel ray tracing and animation. [Google Scholar]
- Suzuki N, Rohaim A, Kato R, Dikic I, Wakatsuki S, Kawasaki M. A novel mode of ubiquitin recognition by the ubiquitin-binding zinc finger domain of WRNIP1. FEBS Journal. 2016;283:2004–2017. doi: 10.1111/febs.13734. [DOI] [PubMed] [Google Scholar]
- Tang ED, Wang CY, Xiong Y, Guan KL. A role for NF-κB essential modifier/IκB kinase-γ (NEMO/IKKγ) ubiquitination in the activation of the IκB kinase complex by tumor necrosis factor. Journal of Biological Chemistry. 2003;278:37297–37305. doi: 10.1074/jbc.M303389200. [DOI] [PubMed] [Google Scholar]
- Toma A, Takahashi TS, Sato Y, Yamagata A, Goto-Ito S, Nakada S, … Fukai S. Structural basis for ubiquitin recognition by ubiquitin-binding zinc finger of FAAP20. PLOS ONE. 2015;10:e0120887. doi: 10.1371/journal.pone.0120887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasilyeva A, Clodfelter JE, Rector B, Hollis T, Scarpinato KD, Salsbury FR. Small molecule induction of MSH2-dependent cell death suggests a vital role of mismatch repair proteins in cell death. DNA Repair. 2009;8:103–113. doi: 10.1016/j.dnarep.2008.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu ZH. Molecular linkage between the kinase ATM and NF-kB signaling in response to genotoxic stimuli. Science. 2006;311:1141–1146. doi: 10.1126/science.1121513. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Ma CA, Wu L, Iwai K, Ashwell JD, Oltz EM, … Jain A. CYLD and the NEMO zinc finger regulate tumor necrosis factor signaling and early embryogenesis. Journal of Biological Chemistry. 2015;290:22076–22084. doi: 10.1074/jbc.M115.658096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu G, Wu CJ, Zhao Y, Ashwell JD. Optineurin negatively regulates TNFα-induced NF-κB activation by competing with NEMO for ubiquitinated RIP. Current Biology. 2007;17:1438–1443. doi: 10.1016/j.cub.2007.07.041. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.